Abstract
Replication of the genome of human papillomaviruses (HPV) is initiated by the recruitment of the viral E1 helicase to the origin of DNA replication by the viral E2 protein, which binds specifically to the origin. We determined, for HPV type 11 (HPV-11), that the C-terminal 296 amino acids of E1 are sufficient for interaction with the transactivation domain of E2 in the yeast two-hybrid system and in vitro. This region of E1 encompasses the ATP-binding domain. Here we have examined the role of this ATP-binding domain, and of ATP, on E2-dependent binding of E1 to the origin. Several amino acid substitutions in the phosphate-binding loop (P loop), which is implicated in binding the triphosphate moiety of ATP, abolished E2 binding, indicating that the structural integrity of this domain is essential for the interaction. The structural constraints imposed on the E1 P loop may differ between HPV-11 and bovine papillomavirus type 1 (BPV-1), since the P479S substitution that inactivates BPV-1 E1 is tolerated in the HPV-11 enzyme. Other substitutions in the E1 P loop, or in two other conserved motifs of the ATP-binding domain, were tolerated, indicating that ATP binding is not essential for interaction with E2. Nevertheless, ATP-Mg stimulated the E2-dependent binding of E1 to the origin in vitro. This stimulation was maximal at the physiological temperature (37°C) and did not require ATP hydrolysis. In contrast, ATP-Mg did not stimulate the E2-dependent binding to the origin of an E1 protein containing only the C-terminal domain (353 to 649) or that of mutant E1 proteins with alterations in the DNA-binding domain. These results are discussed in light of a model in which the E1 ATP-binding domain is required for formation of the E2-binding surface and can, upon the binding of ATP, facilitate and/or stabilize the interaction of E1 with the origin.
The E1 and E2 proteins of papillomaviruses are the only two viral proteins that are required, in addition to the host cell machinery, to replicate the viral genome (reviewed in references 37 and 55). E1 is a helicase that unwinds the viral origin of DNA replication, as well as the DNA template, ahead of the replication fork (18, 28, 50, 63). E1 and E2 initiate DNA replication by binding to the viral origin of replication. Although E1, by itself, can bind to the origin in vitro (49–51, 56, 58, 60, 62), its specificity for the origin is low. The binding specificity of E1 towards the origin is increased by the viral E2 protein, which binds specifically and with high affinity to sites located in the origin (reviewed in reference 37). E1 and E2 form a complex and, as a result, bind to the origin in a cooperative and highly sequence-specific manner (5, 18, 31, 33, 39, 46, 48, 49, 62).
Domains of E1 and E2 that participate in protein-DNA and protein-protein interactions have been identified. A DNA-binding domain was identified between residues 121 and 311 of human papillomavirus type 1 (HPV-1) E1 (27). Similarly, residues located C-terminal to amino acid 162 of bovine papillomavirus type 1 (BPV-1) E1 (corresponding to amino acids 168 and 206 in HPV-1 and HPV-11, respectively) were found to be essential for DNA binding (45, 56). For E2, the C-terminal portion of the protein was shown to contain a DNA-binding–dimerization domain (reviewed in reference 37). Approximately 200 amino acids at the N terminus of E2 are required for interaction with E1 and for cooperative binding to the origin (2, 3, 61, 64) (see below). This N-terminal domain of E2 also functions as a transcription activation domain required for expression of the viral early genes (reviewed in reference 37). Although encoded by the same domain, the replication and transcription functions of E2 can be uncoupled by mutations (1, 8, 9, 12, 15, 21, 43). Recently, a 15-amino-acid peptide derived from HPV-16 E2 was shown to inhibit the interaction between E1 and E2 (25).
Studies aimed at identifying domains of E1 that interact with E2 have led to different results, perhaps due to differences in approaches or to the use of different E1 proteins from different types of papillomaviruses. Both N- and C-terminal fragments of E1 have been implicated in E2 binding. Furthermore, temperature-sensitive effects on the interaction of E1 with E2 have been noted (see below). For example, a C-terminal fragment of HPV-33 E1, encompassing amino acids 312 to 644 was sufficient for interaction with E2 in vitro at 22 and 4°C, whereas a minimal E2-binding domain located between residues 312 and 450 was active only at 22°C (40). Recently, amino acids 432 to 649 of HPV-16 E1 were shown to be sufficient for E2 binding in vitro at all temperatures tested (4, 20, and 37°C) (36). This is in contrast to the observation by Yasugi et al. (64) that amino acids 420 to 439 of HPV-16 could bind to E2 only at low temperature (4°C) whereas a longer fragment, comprising residues 144 to 649, was required for interaction at the physiological temperature. For HPV-11 E1, amino acids 346 to 649 were shown recently to be sufficient for binding to E2 in solution but not for forming a stable complex with E2 at the origin (53). In this last study, the effect of temperature on the interaction of E1 with E2 was not examined. For BPV-1, an E2-binding region was identified between amino acids 200 and 605 of E1, but a slightly larger domain (165 to 605) was required for cooperative binding with E2 at the origin (45). Point mutations and deletions in the BPV-1 E1 C-terminal domain (31), as well as the study of HPV-11–HPV-16 chimeric E1 proteins (66), also indicated a requirement for this region in interaction with E2.
An N-terminal region of BPV-1 E1 was identified that bound E2 in a cold-sensitive manner in vitro (2, 27, 56). In one study, this domain of E1 was found to interact with the N-terminal 91 amino acids of BPV-1 E2 (2). In subsequent studies, the N-terminal E2-binding domain was mapped to a region overlapping the E1 DNA-binding domain (10, 27) and was found to interact with the DNA-binding domain of E2 rather than with its transactivation domain (3, 10). This interaction between the DNA-binding domains of BPV-1 E1 and E2 is essential for DNA replication only in the context of an origin like that of BPV-1 and unlike those of HPVs, in which the binding sites for E1 and E2 are adjacent (3). A single study reported an interaction between the N terminus of E1 and E2 from HPV (type 31b), although this interaction was not sufficient for complex formation at the origin (16).
Binding of E1 to the origin probably proceeds in an ordered manner, beginning with the formation of an E1-E2-ori ternary complex (33, 47). In a process that is still poorly understood, additional E1 molecules are subsequently recruited to the origin and E1 oligomers are formed (10, 33, 46–50). The role of ATP and magnesium in the binding of E1 to the origin remains controversial. For BPV-1 E1 and E2 proteins, one study indicated that Mg and ATP could stimulate the formation and stability of the E1-E2-ori complex (31), whereas another reported an effect only on the half-life of the complex (48). Lusky et al. (33) found that in the presence of ATP, E2 is not stably retained within the complex. This led to the hypothesis that the dissociation of E2 from the origin could result from ATP-driven conformational changes in E1, such as those associated with the formation of E1 oligomers (33).
The ATP-binding domain of E1 is located in the C-terminal domain of the protein. E1 belongs to superfamily 3 of NTP-binding proteins, which comprises several viral proteins with demonstrated or postulated helicase activity, including the large T antigens of simian virus 40 (SV40) and polyomaviruses (19, 20). Proteins in superfamily 3 share three conserved motifs, termed A, B, and C. Motifs A and B correspond to the classical Walker A and B boxes, which are diagnostic of the mononucleotide-binding fold (59). Structural analysis of several proteins containing these two motifs has supported the notion that they all share a common topology (65). Motif A, of the consensus sequence GXXXXGK(T/S), forms a loop (phosphate-binding loop, or P loop) that binds the triphosphate tail of the nucleotide. Motif B is characterized by a doublet of negatively charged residues, aspartate or glutamate, located at the C terminus of a beta strand in the vicinity of motif A. This motif participates in the coordination of the Mg2+ ion associated with the nucleotide. In addition, proteins of superfamily 3 also contain motif C, whose function is unknown and which is characterized by an invariant Asn residue that is often preceded by a doublet of Ser and/or Thr residues (20). Genetic analysis of the poliovirus 2C protein, which belongs to superfamily 3, suggested that motifs A, B, and C are in close proximity in the three-dimensional structure of this enzyme (57). Motifs A to C of the ATP-binding domain are distinct but related to another set of conserved regions in E1, termed A to D, which were identified as regions that are highly conserved between papillomavirus E1 and the large T antigens of SV40 and other polyomaviruses (11). These sequences are related in that motifs A to C of the ATP-binding domain are contained within conserved regions B and C of E1 (see Fig. 4A).
FIG. 4.
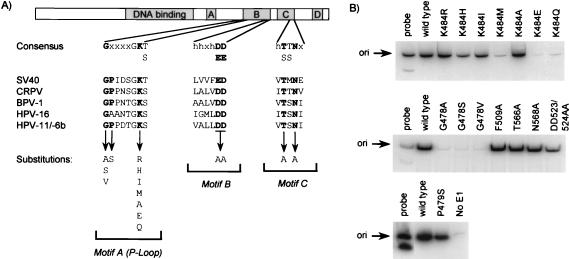
Effects of substitutions in the ATP-binding domain of E1 on the E2-dependent binding of E1 to the origin. (A) Diagram of the E1 helicase. The shaded boxes indicate the positions of conserved regions A, B, C, and D and of the DNA-binding domain. The locations of conserved motifs A, B, and C that are present in members of superfamily 3 of the NTP-binding proteins (20) are indicated. The consensus amino acid sequence of each motif is indicated, as well as a sequence alignment between SV40 T antigen and the E1 proteins of cottontail rabbit papillomavirus, BPV-1, HPV-16, HPV-11, and HPV-6b. Residues that were mutated are shown in boldface. (B) E2-dependent binding of mutant E1 proteins to the origin (ori). DNA-protein complexes were assembled with E2 and the indicated E1 protein made by in vitro translation. The immunoprecipitation of complexes was performed with the rabbit K72 polyclonal antibody. Detection of bound DNA was done as described in the legend to Fig. 2A.
In this study we show that the region of E1 that binds to E2 encompasses the ATP-binding domain. Mutagenesis of highly conserved residues of the ATP-binding domain indicated that its structural integrity is essential for formation of the E2-binding surface and that ATP is not essential for interaction with E2. However, ATP-Mg stimulates the E2-dependent binding of E1 to the origin at a temperature-sensitive step that involves interaction of E1 with DNA and which is distinct from the interaction of E1 with E2. These results indicate that the ATP-binding domain of E1 plays an important structural role in the recruitment of E1 to the origin.
MATERIALS AND METHODS
Yeast two-hybrid system.
Pairs of plasmids expressing a GAL4 DNA-binding domain fusion and a GAL4 activation domain fusion were used to cotransform Saccharomyces cerevisiae Y153 (MATa leu2-3,112 ura3-52 trp1-901 his3-Δ200 ade2-101 gal4Δ gal80Δ URA3::GAL-lacZ LYS::GAL-HIS3) (14) to tryptophan and leucine prototrophy essentially as described previously (17). Typically, each pair of plasmids was introduced into yeast twice, using independent plasmid isolates. For each transformation, β-galactosidase activity was measured in duplicate. Therefore, the value assigned to each combination of plasmids is the average of four independent β-galactosidase activity measurements, which typically varied by no more than 20%. To measure β-galactosidase activity, the transformed yeast cells were pregrown in liquid synthetic defined medium lacking leucine and tryptophan and then used to inoculate yeast extract-peptone-dextrose cultures. These cultures were grown at 30°C until they reached an optical density at 600 nm (OD600) of approximately 0.6. The cells were then harvested, washed, and permeabilized by two cycles of freezing and thawing. β-Galactosidase activity was then measured spectrophotometrically (at 578 nm) with the substrate chlorophenyl red-β-d-galactopyranoside (Boehringer Mannheim) as described in the Clontech Matchmaker Library Protocol. Enzymatic activity was calculated by the following equation: Miller units = (1,000 × OD578)/(elapsed minutes × 1.5 ml of culture × OD600).
Plasmid construction. (i) In-vitro transcription-translation.
The plasmids used for synthesis of HPV-11 E1 and E2 in vitro were derived either from pCR3 or pCR3.1 (Invitrogen, Carlsbad, Calif.) or from pTM1 (obtained from Bernard Moss, National Institutes of Health). In these plasmids, the encoded protein can be expressed in vitro from the phage T7 promoter. When used in a coupled transcription-translation system (TNT coupled reticulocyte lysate system; Promega), plasmids derived from pTM1 directed the synthesis of higher levels of proteins, presumably because this plasmid encodes the encephalomyocarditis virus internal ribosome entry site), which stimulates translation (data not shown). To construct pCR3-E1 and pCR3-E2, the complete HPV-11 E1 and E2 open reading frames (ORFs) were amplified separately by PCR with the following pairs of oligonucleotides: E1, 5′-CAAGGATGGCGGACGATTCA-3′ and 5′-TCTTCATAAAGTTCTAACAAC-3′, and E2, 5′-GAAGATGGAAGCAATAGCCAA-3′ and 5′-ATGGTTACAATAAATGTAATGAC-3′ (the ATG and stop codons of E1 and E2 are underlined). The DNA template used for PCR was baculovirus construct Ac11E1 or Ac11E2 (obtained from R. Rose, University of Rochester). The E1 and E2 PCR products were each cloned under the control of the cytomegalovirus immediate-early promoter in plasmid pCR3, using the TA cloning kit (Invitrogen), to generate pCR3-E1 and pCR3-E2. To construct plasmid pCR3.1 FLAG-E1, a NcoI-XhoI fragment encoding the FLAG-E1 ORF was cut from pTM1-FLAG-E1 (see description below), the DNA ends were filled in with the Klenow fragment of DNA polymerase I, and the resulting blunt fragment was cloned into the EcoRV site of plasmid pCR3.1 (Invitrogen). Plasmid pCR3-FLAG-E1, which expresses E1 (amino acids 2 to 649) fused at its N terminus to the FLAG epitope (Met Asp Tyr Lys Asp Asp Asp Asp Lys; Kodak) was constructed by PCR amplification of the E1 ORF with the following two oligonucleotides: 5′-CCCATGGACTACAAGGACGACGATGACAAGGCGGACGATTCAGGTACAGAAAAT-3′ and 5′-GGGATCCTTATTATAAAGTTCTAACAACTGATCCTGGCAC-3′ (the portion encoding the FLAG epitope is underlined). The resulting PCR product was cloned into plasmid pCR3 (Invitrogen) with the TA cloning kit. To construct plasmid pTM1-E1, the E1 ORF was amplified by PCR with the following two oligonucleotides: 5′-GTACGATCCCATGGCGGACGATTCAGGTACAGAAAAT-3′ and 5′-GTACGATGGGATCCTTATTATAAAGTTCTAACAACTGATCCTGGCAC-3′. The resulting PCR product was digested with NcoI and BamHI (encoded by the two oligonucleotides) and inserted between the NcoI and BamHI sites of plasmid pTM1. Plasmid pTM1-FLAG-E1, which expresses E1 (amino acids 2 to 649) fused at its N terminus to the FLAG epitope, was constructed by PCR amplification of the E1 ORF with the following two oligonucleotides: 5′-CCCATGGACTACAAGGACGACGATGACAAGGCGGACGATTCAGGTACAGAAAAT-3′ and 5′-GGGATCCTTATTATAAAGTTCTAACAACTGATCCTGGCAC-3′ (the portion encoding the FLAG epitope is underlined). The resulting PCR product was digested with NcoI and BamHI (encoded by the two oligonucleotides) and inserted between the NcoI and BamHI sites of plasmid pTM1. Plasmids to express truncated E1 proteins in vitro were constructed by amplification of the desired portion of the E1 ORF with specific primers bearing an NcoI site (forward primer) and a BamHI site (reverse primer). The PCR products were digested with NcoI and BamHI and inserted between the NcoI and BamHI sites of pTM1. The sequences of the different E1 forward and reverse primers will be provided upon request. Plasmid pTM1-FLAG-E1 (2 to 507) was constructed by digestion of pTM1-FLAG-E1 with PstI (which cuts at codon 507 of E1 and in the polylinker of pTM1 downstream of the E1 stop codon) and religation.
(ii) Yeast two-hybrid system.
Unless described otherwise, plasmids to express truncated E1 proteins in the yeast two-hybrid system were constructed by amplification of the desired portion of the E1 ORF with specific primers bearing an NcoI site (forward primer) and a BamHI site (reverse primer). The PCR products were digested with NcoI and BamHI and inserted between the NcoI and BamHI sites of pAS1 (GAL4 DNA-binding domain) and pACT2 (GAL4 activation domain) (14). Two hybrid plasmids encoding the complete E1 protein (amino acids 1 to 649) were constructed in a similar way with the exception that the forward primer contained a BamHI site instead of an NcoI site. Details of the construction of these plasmids will be provided upon request.
(iii) Truncated HPV-11 origins.
Plasmids encoding truncated origins (nucleotides 7902 to 61, 7902 to 53, and 7902 to 34, respectively) were generated by PCR amplification with the forward primer, 5′-CCCAGATCTTACCCACACCCTACATATTTCC-3′ (nucleotide 7902 underlined), and one of the following reverse primers: 5′-CCCGGATCCACCGTTTTCGGTTGAACCG-3′ (nucleotide 61 underlined), 5′-CCCGGATCCCGGTTGAACCGTTTTCGG-3′ (nucleotide 53 underlined), or 5′-CCCGGATCCCCTCCTCTTTTTTAAACTAAG-3′ (nucleotide 34 underlined). The template for PCR was plasmid pN9 (30), which was obtained from D. McCance (University of Rochester) and contains the complete origin of replication of HPV-11 (nucleotides 7884 to 61) cloned into pBluescriptII SK(+) (Stratagene). The PCR products were digested with BamHI and BglIII and were cloned into the BamHI site of plasmid pBluescriptII SK(+). Similar plasmids, but carrying truncated origins 7884 to 53 and 7884 to 34, were constructed in a similar way but using a different forward primer: 5′-CTGCAGCCCGGGGGATC-3′. The resulting PCR products were digested with BamHI and cloned into the BamHI site of plasmid pBluescriptII SK(+). For all of these origin-containing plasmids, the inserted truncated origin is in the same orientation as that of the complete origin in plasmid pN9.
(iv) Transient HPV replication.
The following plasmids, which were used in transient HPV DNA replication assays to express E1 and E2 in transfected cells, were all derived from pCR3: pCR3-E1, pCR3-FLAG-E1 (wild-type and mutant E1), and pCR3-E2. The HPV-11 origin-containing plasmid was pN9. These plasmids are described above.
Site-directed mutagenesis.
Site-directed mutagenesis was performed with the QuickChange site-Directed Mutagenesis kit (Stratagene) according to the instructions supplied by the manufacturer.
Expression and purification of GST fusion proteins.
The plasmid used to express the transactivation domain (TAD; amino acids 1 to 209) of HPV-11 E2 as a fusion with glutathione S-transferase (GST) was constructed as follows. First, the following two oligonucleotides were used to amplify by PCR the region of the E2 ORF encoding the TAD: 5′-GGATCCGACGACGATGACAAGATGGAAGCAATAGCCAAG-3′ (the initiator ATG of the E2 ORF is underlined, and a sequence encoding a synthetic enterokinase cleavage site is double underlined) and 5′-GGATCCTTATCAAGCAATGGATACTTCTCGTAC-3′ (the stop codon is underlined). The resulting PCR fragment was digested with BamHI and cloned into the BamHI site of pGEX-2T (Pharmacia). GST and GST-E2 proteins were expressed in Escherichia coli BL21(DE3)/pLysS. Bacterial cultures were grown to an OD650 of approximately 1.4 in Circle Grow medium (Bio 101), and protein expression was induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 6 h at 20°C. The bacterial cells were then collected by centrifugation and frozen. The cells (approximately 40 g) were lysed by thawing in 5 volumes of binding buffer (50 mM HEPES [pH 7.5], 100 mM NaCl, 2 mM dithiothreitol [DTT], 0.2 mM EDTA) containing protease inhibitors and sonicated. The bacterial extract was then clarified by centrifugation at 16,000 rpm for 30 min in a JA-17 rotor (Beckman). The supernatant was then incubated with 16 ml of glutathione resin (Pharmacia) for 1 h at room temperature. Subsequent steps were carried out at 4°C. The glutathione beads were washed sequentially with 400 ml of binding buffer and 200 ml of buffer containing 1 M NaCl. Bound GST or GST-E2 protein was eluted with 5 bed volumes of 10 mM glutathione and then dialyzed against 50 mM Na phosphate (pH 7.5)–100 mM NaCl. The GST-E2 protein was then concentrated to approximately 50 mg/ml and applied to a gel filtration column (Superdex 200; Pharmacia) in buffer containing 50 mM Na phosphate (pH 7.5)–100 mM NaCl. The peak fractions were collected, pooled, and stored at −80°C.
GST pull-down assay.
Purified GST and GST-E2 TAD were immobilized on gluthathione beads (Amersham Pharmacia Biotech) at a concentration of 1 mg/ml. The beads were then washed with 20 volumes of buffer A (20 mM Tris [pH 7.6], 1 M NaCl, 4 mM MgCl2, 2 mM DTT, 0.5% Nonidet P-40). The beads were then equilibrated with 20 volumes of binding buffer (20 mM Tris [pH 7.6], 50 mM NaCl, 4 mM MgCl2, 2 mM DTT, 0.5% Nonidet P-40) containing 5 mg of bovine serum albumin (BSA)/ml followed by 20 volumes of the same binding buffer containing 1 mg of BSA/ml. For each binding reaction, 20 μl of 35S-labeled in vitro-translated E1 protein (or luciferase as a control), generated with the TNT coupled reticulocyte lysate system (Promega), were diluted with 180 μl of binding buffer containing 1 mg of BSA/ml and then incubated with 40 μl of GST or GST-E2 beads at room temperature for 1 h. The beads were then washed three times with 20 volumes of binding buffer. The bound proteins were eluted first with 120 μl of high-salt buffer (20 mM Tris [pH 7.6], 500 mM NaCl, 1 mM DTT) followed by a second elution with 120 μl of high-salt buffer containing 1% sodium dodecyl sulfate (SDS). Aliquots of the input protein and of the high-salt and SDS eluates were then analyzed by SDS-polyacrylamide gel electrophoresis and autoradiography. Quantification was done with a PhosphorImager (Molecular Dynamics).
E2-dependent binding of E1 to the origin.
Binding of E1 and E2 to the HPV origin was examined by using a DNA-protein coimmunoprecipitation assay similar to the one described for BPV E1 and E2 (45). The DNA probe used in this assay was generated in three steps. First, plasmid pN9 was digested with XmaI. Second, the ends of this linearized plasmid were labeled with [32P]dCTP and the Klenow fragment of DNA polymerase I. Third, the labeled DNA was subjected to a second digestion with PvuII. This resulted in the generation of three DNA fragments, two of which are labeled. One of the two labeled fragments contains the HPV origin (370 bp), whereas the other (186 bp) does not and serves as a control for the specificity of binding. The TNT coupled reticulocyte lysate system was used to produce the E1 and E2 proteins by coupled transcription-translation in vitro. The lysate was programmed with 4 μg of the appropriate plasmid per 50 μl of TNT reticulocyte lysate according to the protocol supplied by the manufacturer. By Western blotting of different amounts of E1-containing lysates and of purified E1 as standards, we estimated that approximately 5 to 20 ng of E1 per μl of transcription-translation reaction mixture was synthesized under the conditions recommended by the manufacturer and when the reaction was programmed with pTM1-E1 (data not shown). Smaller amounts (approximately threefold) were produced when the lysate was programmed with pCR3-E1 (data not shown). Similar analysis done with E2 indicated that approximately 10 ng of E2 per μl of reticulocyte lysate was synthesized when programmed with pCR3-E2 (data not shown). When needed, the proteins were radiolabeled by incorporation of [35S]methionine. Binding reactions were performed by mixing 50 μl of lysate containing E1 with 15 μl of lysate containing E2 and with 200 ng of a radiolabeled DNA probe. The mixture was diluted twofold with 2× binding buffer (40 mM Tris-HCl [pH 7.6], 300 mM NaCl, 30 mM MgCl2, 2 mM EDTA, 2 mM DTT) and incubated at room temperature for 1 h. Unless otherwise indicated, ATP was also added to the binding reaction at a final concentration of 5 mM. For binding reactions performed in the absence of E2, 15 μl of lysate programmed with plasmid pCR3 was used instead of the E2-containing lysate. The binding reactions were allowed to proceed at the indicated temperatures for 90 min. When indicated, ATP (or a related nucleotide) and MgCl2 were added to the binding reactions at final concentrations of 5 and 3 mM, respectively. DNA-protein complexes were immunoprecipitated either with the anti-FLAG M2 monoclonal antibody (Eastman Kodak) when using FLAG-tagged E1 or with the K72 polyclonal antibody, which was raised in rabbits against a peptide derived from the C-terminal 15 amino acids of HPV-11 E1. Before use in immunoprecipitation, the antibodies were prebound to either protein G Sepharose beads (when using anti-FLAG) or protein A Sepharose beads (for K72). The immunoprecipitation of protein-DNA complexes was carried out for 1 h at the binding reaction temperature. The complexes were washed three times with 200 μl (or 100 μl) of wash buffer (50 mM Tris [pH 7.6], 150 mM NaCl, 0.1% Triton X-100). The DNA present in these complexes was extracted with phenol-chloroform and precipitated with ethanol in the presence of carrier yeast tRNA. The precipitated radiolabeled DNA fragments were resolved on a 5% polyacrylamide Tris-borate-EDTA gel and visualized by autoradiography.
Transient HPV DNA replication assay.
CHO-K1 cells (obtained from the American Type Culture Collection) were grown to 40 to 60% confluence in 35-mm-diameter tissue culture dishes in Ham F12 medium containing 10% fetal bovine serum and gentamicin sulfate. The cells were transfected with 250 ng of pCR3-E1 (or pCR3-FLAG-E1 mutant), 25 ng of pCR3-E2, and 250 ng of pN9 plasmids by using Lipofectamine (Gibco BRL). The presence of the FLAG epitope at the N terminus of E1 does not affect its ability to support transient HPV DNA replication (data not shown). The cells were harvested 72 h posttransfection, and total DNA was isolated with the QIAmp blood kit (Qiagen). Replicated pN9 plasmid DNA was detected by PCR amplification of an origin-containing fragment with DpnI-digested total DNA as a template and the following pair of primers: 5′-CTGCAACCGGTTTCGGTTACCCACACCCT-3′ (corresponding to nucleotides 7885 to 7913 of the HPV-11 genome) and 5′-CGTTCCACTGAGCGTAGACCCCGTAGAA-3′ (corresponding to nucleotides 1848 to 1820 of pSK+). As a control, a fragment of the pCR3-E1 plasmid was amplified in the same PCR with the following pair of primers, which hybridize within the E1 ORF: 5′-GCTTTGGGCTGTCATTTG-3′ and 5′-TGTCAGGTGGCCCTACAA-3′ (corresponding to nucleotides 1475 to 1492 and 2275 to 2258, respectively, of the HPV-11 genome). The PCR conditions consisted of an initial denaturation step at 95°C for 1 min, followed by 20 rounds of denaturation at 94°C for 30 s, annealing at 51°C for 1 min, and extension at 72°C for 1 min 30 s, ending with a final extension at 72°C for 3 min. The PCR products were made radioactive by the addition of [α-33P]dCTP to the PCRs and were visualized by agarose-gel electrophoresis and autoradiography.
RESULTS
Interaction between HPV-11 E1 and the TAD of E2 in yeast.
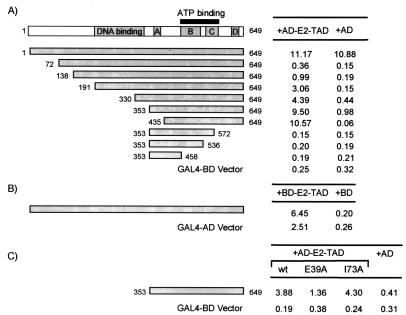
Previous studies implicated several regions of E1 in binding to E2 (see the introduction). To confirm and extend these findings to E1 of HPV-11, we used the yeast two-hybrid system (14) to confirm that it can interact with the TAD of E2 and to map a region of E1 involved in this interaction. In these experiments, the entire E1 molecule (amino acids 1 to 649) or portions of it were fused to the GAL4 DNA-binding domain and were tested for interaction with the E2 TAD (amino acids 1 to 209) fused to the GAL4 activation domain (Fig. 1A). Interactions between fusion proteins were detected by measuring the amount of β-galactosidase activity expressed from a GAL1-LacZ reporter gene. When tested in yeast, the entire E1 molecule fused to the GAL4-binding domain activated transcription of GAL1-LacZ by itself (Fig. 1A) and therefore could not be used to detect an interaction with E2. To overcome this problem, additional fusion proteins were constructed in which the first 71 or more amino acids of E1 were missing. These shorter proteins did not activate transcription on their own, perhaps due to the loss of a highly negatively charged region located between amino acids 34 and 42 of E1 which could potentially act as a transcriptional activation domain in yeast. Fusion proteins encompassing the C-terminal half of E1 interacted specifically with the E2 TAD, albeit weakly in the case of longer proteins containing portions of the E1 N terminus. The smallest region of E1 that interacted with the E2 TAD was composed of amino acids 435 to 649. Attempts to shorten this region by deleting sequences at its N or C terminus resulted in fusion proteins that did not interact with the E2 TAD (Fig. 1 and data not shown). These results suggested that the entire C-terminal region of E1 is required for binding to E2. The interaction between E1 and the E2 TAD was also tested with a different configuration of “bait and prey” by fusing E1 to the GAL4 activation domain and the E2 TAD to the GAL4-binding domain. In this way, a specific interaction between the entire E1 molecule and E2 was detected despite the fact that the GAL4-E2 TAD activates transcription weakly on its own (Fig. 1B).
FIG. 1.
Interaction of E1 with the TAD of E2 in the yeast two-hybrid system. (A) Diagram of the E1 protein (top). The shaded boxes labeled A, B, C, and D represent regions of E1 that have high sequence similarity with large T antigens of SV40 and polyomaviruses (11). A region that is essential for DNA binding (see the introduction) is also shown as a shaded box. The solid box indicates the position of the ATP-binding domain of E1. β-Galactosidase activity detected in yeast cells expressing either E1 or a fragment of E1 fused to the GAL4 DNA-binding domain (BD) or the GAL4 BD alone and also expressing either the E2 TAD (amino acids 1 to 209 of E2) fused to the GAL4 activation domain (AD) or the GAL4 AD alone is shown below. The portions of E1 fused to the GAL4-BD are diagrammed as shaded boxes. The amino acids at the N and C terminus of each E1 fragment are indicated. (B) β-Galactosidase activity detected in yeast cells expressing either the E2 TAD fused to the GAL4 BD or the GAL4 BD alone and also expressing either E1 fused to the GAL4 AD or the GAL4 AD alone. (C) Effect of substitutions in the TAD of E2 on interaction with E1. β-Galactosidase activity detected in yeast cells expressing either E1 (amino acids 353 to 649) fused to the GAL4 BD or the GAL4 BD only and also expressing either the wild-type (WT) E2 TAD or a mutant E2 TAD (E39A or I73A) fused to the GAL4 AD or the GAL4 AD alone. The numbers on the right indicate the amounts of β-galactosidase activity (in Miller units) measured in S. cerevisiae for each combination of plasmids.
The interaction between the E1 C terminus and the E2 TAD appears to be biologically relevant, since it was reduced approximately threefold by the E39A substitution in E2 (Fig. 1C), which affects E1 binding in vitro and HPV DNA replication in vivo (12, 43). In contrast, and as expected, the interaction of E1 with E2 in yeast was not affected by the I73A substitution (Fig. 1C), which affects primarily the transactivation function of E2 (43). Both mutant E2 fusion proteins were expressed in yeast at levels similar to that of the wild-type fusion (data not shown).
Cooperative binding of E1 and E2 to the HPV origin in vitro.
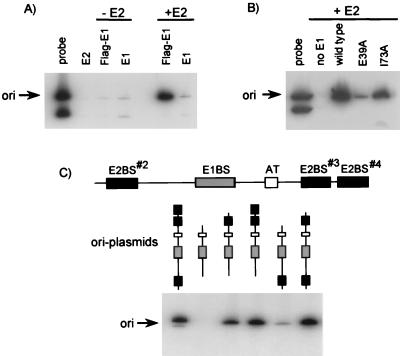
The results presented above indicated that the C-terminal domain of HPV-11 E1 is sufficient for interaction with E2. To address the question of whether this domain is also sufficient for interaction with E2 in vitro, we established an immunoprecipitation assay to monitor the binding of E1 and E2 to the HPV origin. This assay is similar in principle to those described previously for the binding of BPV E1 and E2 (45) and SV40 large T antigen (38) to their respective origins. In these experiments, HPV-11 E1 protein, which was tagged at its N terminus with a FLAG epitope, and HPV-11 E2 were both synthesized in vitro in a rabbit reticulocyte lysate and incubated with a mixture of two radiolabeled DNA fragments, only one of which contained the HPV origin. E1-containing complexes were immunoprecipitated with an anti-FLAG monoclonal antibody, and the coprecipitated DNA was visualized by polyacrylamide gel electrophoresis and autoradiography. Binding of E1 to the origin was detected only in the presence of E2 (Fig. 2A), presumably because of the large excess of competitor DNA used in these binding reactions (plasmid DNA used to program the reticulocyte lysate serves as competitor DNA in these reactions). By analogy to what has been described for the binding of BPV-1 E1 and E2 to their cognate origin (see the introduction and references therein), it is likely that different types of HPV E1-containing complexes are formed in this assay. These complexes may differ in the oligomerization status of E1 and in whether E2 has remained in the complex (see Discussion). As a control for the specificity of the antibody, we showed that replacing FLAG-E1 with an untagged E1 protein did not result in the precipitation of origin-containing DNA (Fig. 2A).
FIG. 2.
E2-dependent binding of E1 to the origin (ori). (A) Origin DNA-protein complexes were formed with E2 alone (E2) or with E1 that was either tagged at its N terminus with a FLAG epitope (Flag-E1) or untagged (E1). Complexes were formed either in the absence (−E2) or presence (+E2) of E2 and were immunoprecipitated with the M2 anti-FLAG monoclonal antibody (Eastman Kodak). Fifty percent of the coprecipitated DNA, along with 0.5% of the amount of probe used in the binding reaction, was visualized by electrophoresis and autoradiography. The arrow indicates the fragment of the probe that contains the origin. (B) Effect of the E39A and I73A substitutions in E2 on formation of the E1-E2-ori complex. Protein-DNA complexes were formed with E2 alone (no E1) or with a combination of E1 plus one of three E2 proteins: wild type, E39A mutant, or I73A mutant. The immunoprecipitation of complexes and detection of bound DNA were done as described for panel A. (C) Effect of deleting one, two, or all three E2-binding sites in the origin. A diagram of the HPV-11 origin is shown that highlights the three E2-binding sites (E2BS; solid boxes), the AT-rich region (AT; open box), and the palindromic E1-binding site (E1BS; shaded box). Complexes were formed with in vitro-translated E1 and E2 proteins and the indicated origin. Each probe was composed of two fragments (not shown), of which only the ori-containing fragment was coprecipitated. Similar amounts of each probe were used in these binding reactions (data not shown). The immunoprecipitation of complexes and detection of bound DNA were done as described for panel A.
In this assay, the E2-dependent binding of E1 to the origin appears to be biologically relevant, since it was affected by the E39A substitution in E2, which affects E1 binding, but was not affected by the I73A substitution, which affects the transcription function of E2 (Fig. 2B). Furthermore, we showed that the ability of mutant HPV-11 origins to support the cooperative binding of E1 and E2 in vitro correlated well with their ability to support transient HPV DNA replication in cells (Fig. 2C). This was done with a set of truncated HPV-11 origins that lack one, two, or all three E2-binding sites and which have been characterized previously for their ability to support transient HPV DNA replication (30). As an example of the excellent correlation between the in vitro and in vivo results, we found that a minimum of two E2-binding sites was required for optimal complex formation in vitro, similar to what is needed for optimal DNA replication in cells (30, 54).
The C terminus of E1 is sufficient for interaction with E2 in vitro.
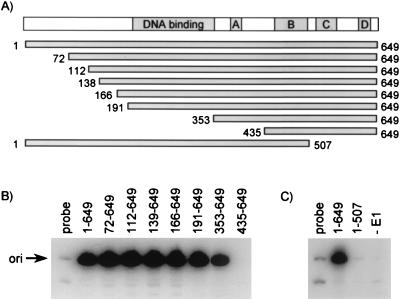
A series of truncated E1 proteins (diagrammed in Fig. 3A) was then used to map the smallest domain of E1 that can be recruited to the origin by E2. The smallest domain of E1 that could be recruited to the origin was composed of amino acids 353 to 649 (Fig. 3B). Further truncation of this domain from the N terminus resulted in loss of activity (Fig. 3B). Similarly, truncated E1 proteins lacking the C-terminal 142 amino acids (E1 [1 to 507] [Fig. 3C]), or lacking 305 or 322 residues at the C terminus (data not shown) were inactive. These results indicated that the C terminus of E1 is necessary and sufficient for interaction with E2 at the origin. However, because E1 (353 to 649) was approximately fivefold less active than wild-type E1 in this assay (for quantification, see Fig. 6C), we surmise that sequences at the N terminus of E1 (between residues 191 and 353) are important for maximal binding of E1 to the origin. These sequences, which encompass the E1 DNA-binding domain (see the introduction), could play a direct role in the recruitment of E1 to the origin and/or in the stability of the assembled protein-DNA complex (see below).
FIG. 3.
Mapping of the minimal domain of E1 that can be recruited to the origin by E2. (A) Diagram of the wild-type E1 protein indicating the positions of conserved regions A, B, C, and D and of the DNA-binding domain. Truncated mutant E1 proteins are diagrammed, and the amino acids at the N and C terminus of each truncated protein are indicated. (B and C) E2-dependent origin-binding assay with truncated E1 proteins. DNA-protein complexes were assembled with E2 and the indicated E1 protein made by in vitro translation. The immunoprecipitation of the complexes was performed with the rabbit K72 polyclonal antibody raised against the C-terminal 15 amino acids of HPV-11 E1 (B) or with the M2 anti-FLAG monoclonal antibody (C). The proteins shown in panel C were tagged at their N termini with the FLAG epitope. Detection of bound DNA was done as described in the legend to Fig. 2A. −E1, no E1 present.
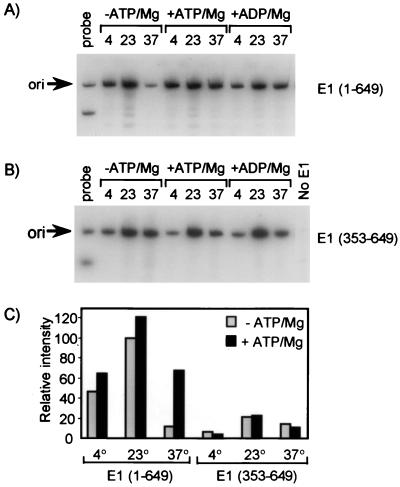
FIG. 6.
Effect of temperature and of ATP on the E2-dependent recruitment of wild-type E1 or E1 (353 to 649) to the origin (ori). DNA-protein complexes were assembled with E2 and with either wild-type E1 (A) or E1 (353 to 649) (B) made by in vitro translation. Due to differences in their levels of expression in reticulocyte lysate in vitro, approximately twice as much E1 (353 to 649) protein was present in the assay shown in panel B than wild-type E1 (1 to 649) in the assay shown in panel A. The immunoprecipitation of complexes was done with the K72 polyclonal antibody. Binding reactions and immunoprecipitations were carried out at the indicated temperature (4, 23, or 37°C) and in the presence (+) or absence (−) of ATP (or ADP) and magnesium at concentrations of 5 and 3 mM, respectively. Detection of bound DNA was done as described in the legend to Fig. 2A. The autoradiograms in panels A and B were obtained after exposure of the gels for 3 and 15 h, respectively. (C) Graph generated by quantification of the data in panels A and B with a PhosphorImager. For each panel, the intensity of each ori band was normalized to the amount of ori probe loaded in lane 1. The amount of probe loaded in lane 1 was the same for both gels. The intensity of the ori band detected with wild-type E1 at 23°C in the absence of ATP-Mg was set arbitrarily at 100%.
Effect of amino acid substitutions in the E1 ATP-binding domain on the E2-dependent binding of E1 to the origin.
The results presented above indicated that the C-terminal domain of E1 is sufficient for interaction with E2 in yeast and in vitro. This region of E1 encompasses the ATP-binding domain, which contains three amino acid motifs, termed A, B, and C, that are highly conserved in superfamily 3 of NTP-binding proteins (see the introduction) (Fig. 4A). By analogy with other proteins, the A and B motifs of E1 are thought to be directly involved in the binding of ATP as a magnesium chelate (see the introduction). To test if the ATP-binding domain was involved in interaction with E2, 15 mutant E1 proteins (amino acids 1 to 649) were constructed that bear replacements of highly conserved amino acid residues in motifs A, B, and C. In addition, phenylalanine 509 of E1, which is not part of motif A, B, or C, was also mutated, since it is conserved in superfamily 3, with most members carrying a phenylalanine or a tryptophan at this position (20). These mutant proteins were then tested in the E2-dependent E1 origin-binding assay described above. Substitutions in motifs B and C, as well as of Phe509, had little effect on formation of the E2-dependent binding of E1 to the origin, indicating that these conserved residues are not essential for binding to E2 (Fig. 4B). In contrast, certain substitutions in motif A, the P loop, dramatically reduced the E2-dependent binding of E1 to the origin (K484M, K484E, K484Q, G478A, G478S, and G478V). These results indicated that the structural integrity of the P loop is essential for E2 binding. Not all amino acid substitutions in motif A were deleterious. Most of the tolerated substitutions are predicted to affect ATP binding on the basis of their effects in other ATP-binding enzymes or in other E1 proteins from other papillomavirus types or because they dramatically reduce the ATPase activity of HPV-11 E1 (41). Therefore, these results suggested that the binding of ATP by E1 is not essential for the E2-dependent binding of E1 to the origin. This is important, since ATP, together with other nucleotides, was present in the E1- and E2-containing lysates used in this origin-binding assay.
Effect of the P479S, K484Q, and K484E substitutions in E1 on interaction with E2 in yeast and in vitro.
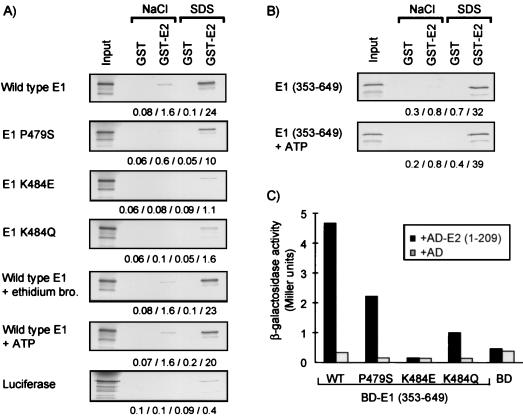
The results presented above indicated that some substitutions in the E1 P loop affect the E2-dependent binding of E1 to the origin. In principle, these substitutions could affect the binding of E1 to E2 or they could act indirectly, for example, by affecting the binding of E1 to DNA. Our finding that the C terminus of E1 is sufficient for E1 to form a complex with E2 at the origin (see above), albeit not optimally, made the former possibility more likely. To determine if these P-loop substitutions affect the interaction of E1 with E2, we tested the effect of three substitutions in E1, P479S, K484E, and K484Q, on binding to the TAD of E2 (amino acids 1 to 209) by using a GST pull-down assay. Specifically, we tested the binding of 35S-labeled E1 (wild type and mutant) to GST-E2 and GST. E1 proteins bound to GST or GST-E2 were eluted sequentially with 0.5 mM NaCl and then with 1% SDS. Aliquots of the input proteins and of the salt and SDS eluates were analyzed by gel electrophoresis and autoradiography, and quantification was performed with a PhosphorImager (Molecular Dynamics). Using this protocol, we detected binding of wild-type E1 specifically to GST-E2 and not to GST (Fig. 5A). The majority of the E1 protein bound to E2 was not eluted with high salt but only with SDS, indicating that the interaction between E1 and E2 is mostly nonionic in nature. A similar observation was made recently for HPV-16 E1 and E2 (36). Under the conditions of our assay, approximately 24% of the input E1 protein could be eluted from GST-E2 with SDS. Under the same conditions, only 0.1% of E1, approximately, was eluted from GST. As a specificity control, we showed that an irrelevant protein, luciferase, did not bind to GST-E2 (only 0.4% of the input protein was eluted from GST-E2 with SDS) (Fig. 5A). The fact that the binding of E1 to E2 is salt resistant is a strong indication that this interaction is not mediated by nucleic acids (either DNA or RNA present in the E1-containing reticulocyte lysate). To further rule out any involvement of nucleic acids, we demonstrated that the binding of E1 to GST-E2 was not affected by ethidium bromide (20 μg/ml) (Fig. 5A). In addition, we showed that the C-terminal domain of E1 (amino acids 353 to 649) could interact with GST-E2 as efficiently as wild-type E1. Since both the E1 and E2 proteins used in this last experiment lack their respective DNA-binding domains, this result provided further evidence that the interaction between E1 and E2 detected in our assay was not mediated by nucleic acids.
FIG. 5.
Effect of amino acid substitutions in the ATP-binding domain of E1 on binding to E2 in vitro and in the yeast two-hybrid system. (A) GST pull-down assays demonstrating the specific binding of wild-type E1 to the TAD of E2 and the effect of three substitutions in the ATP-binding domain of E1 (P479S, K484E, and K484Q) on this interaction. Also shown are the effects of ATP (5 mM) and ethidium bromide (bro.) (20 μg/ml) on this interaction. For each binding reaction, 1/40 of the input protein and 1/10 of the NaCl and SDS eluates were analyzed on a gel. For each lane, the amount of eluted E1 protein was quantified with a PhosphorImager. The percentages of the input protein eluted from the GST and GST-E2 proteins with NaCl and SDS, respectively, are written below each gel. (B) Binding of the C-terminal domain of E1 (353 to 649) to the TAD of E2 and the effect of ATP (5 mM) on this interaction. The gels were loaded and analyzed as described for panel A. (C) β-Galactosidase activity detected in yeast cells expressing either wild-type (WT) E1 (amino acids 353 to 649) fused to the GAL4-binding domain (BD) or one of three mutant derivatives (P479S, K484E, or K484Q) and also expressing either the E2 TAD fused to the GAL4 activation domain (AD) or the GAL4 AD alone.
We then tested the binding of the three mutant E1 proteins to GST-E2 and GST. Both E1 K484E and E1 K484Q were severely defective; only 1.1 and 1.6% of the input protein, respectively, bound to GST-E2 (SDS eluates). In contrast, the P479S mutant could still bind to E2, albeit with reduced efficiency. Binding of the P479S mutant was reduced approximately twofold relative to wild-type E1 (approximately 10% of the input P479S mutant protein could be eluted from GST-E2 with SDS) (Fig. 5A).
Finally, we determined that the interaction of E1 (wild type or 353 to 649) with GST-E2 was not dramatically altered by the addition of 5 mM ATP to the binding reactions. The amounts of input protein bound to GST-E2 (SDS eluates) in the presence or absence of supplemented ATP were 20 and 24% for wild-type E1 (Fig. 5A) and 39 and 32% for E1 (353 to 649) (Fig. 5B).
To substantiate the results obtained in vitro, we also tested the effect of the three P-loop substitutions on E2 binding in vivo in the yeast two-hybrid system. This assay measures the interaction of the E1 C-terminal domain (353 to 649) with the E2 TAD. As can be seen in Fig. 5C, the K484E substitution abolished the interaction with the E2 TAD. In this assay, the K484Q substitution was less detrimental than the K484E substitution. The P479S substitution still allowed substantial interaction with the E2 TAD. Qualitatively, these results were in good agreement with those obtained in vitro and confirmed that the integrity of the E1 P loop is essential for formation of the E2-binding surface.
Effect of temperature and ATP-magnesium on the E2-dependent binding of E1 to the origin.
It was reported previously that the interaction between E1 and E2 from either HPV-16 or HPV-33 is temperature sensitive (see the introduction). To test if the interaction between HPV-11 E1 and E2 is temperature sensitive, we carried out the E2-dependent E1 origin-binding assay at three temperatures (4, 23, and 37°C). In these experiments, we used both the entire E1 protein and a shorter fragment (amino acids 353 to 649) that contains the E2-binding domain. We also determined the effect of supplementing the binding reactions with ATP and magnesium at concentrations of 5 and 3 mM, respectively. The E2-dependent binding of both wild-type E1 and E1 (353 to 649) to the origin was reduced at 4 and 37°C compared to that at 23°C (Fig. 6). These results indicated that the interaction between the C terminus of E1 and E2 is temperature sensitive. The most dramatic effect of temperature was observed at 37°C, in the absence of supplemented ATP-Mg. Under these conditions, the E2-dependent binding of wild-type E1 to the origin was consistently reduced more than fivefold and was similar to that of E1 (353 to 649). Supplementing the binding reactions with ATP-Mg could reverse this effect. In contrast, ATP-Mg had little effect on the binding of E1 (353 to 649) to the origin at all temperatures tested. Therefore, the negative effect of high temperature (37°C) on the E2-dependent binding of wild-type E1 to the origin, and its reversal by ATP-Mg, must occur at a step other than the interaction of the C-terminal domain of E1 with E2. We found that, similarly to ATP-Mg, ADP-Mg could overcome the inhibition by high temperature, indicating that nucleotide binding, but not its hydrolysis, is required (Fig. 6A). These results suggested that the E2-dependent binding of E1 to the origin involves at least two steps: binding of the C terminus of E1 to E2 and a second, temperature-sensitive step, which can be stimulated by ATP-Mg. These results are consistent with a model in which ATP-Mg is not required for E1 to bind to E2 but promotes another step in the binding of E1 to the origin, such as its ability to bind to DNA and/or oligomerize at the origin (see Discussion).
Effect of amino acid substitutions in the E1 DNA-binding domain.
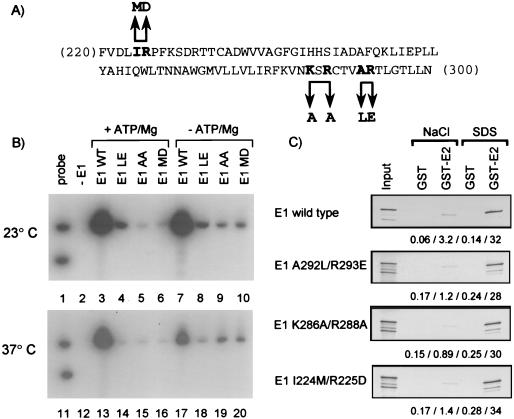
The results presented above indicated that the N-terminal 352 amino acids of E1, although not essential for binding to E2, contribute to the binding of E1 to the origin. Furthermore, these sequences are required for ATP-Mg to stimulate the binding of E1 to the origin at high temperature. Because the N terminus of E1 encompasses the E1 DNA-binding domain, we tested directly whether this domain was required for maximal binding to the origin and for ATP-Mg stimulation. This was accomplished by introducing three specific amino acid substitutions in the E1 DNA-binding domain and determining their effect on the E2-dependent binding of E1 to the origin. The three substitutions that were created are shown in Fig. 7A. Each substitution changes two amino acids. Isoleucine 224 and arginine 225 were replaced by methionine and aspartic acid. Lysine 286 and arginine 288 were both replaced by alanine. Alanine 292 and arginine 293 were replaced by leucine and glutamic acid. For BPV-1 E1, mutagenesis of the residues corresponding to Arg 225 and Arg 293 showed that they were required for binding to the origin (56). Similarly, a double substitution in BPV-1 E1 corresponding to K286A-R288A abolished E1 DNA binding (56). For two of the HPV-11 E1 mutants (K286A-R288A and A292L-R293E), we verified that they were defective in binding to the HPV-11 origin (7). We also demonstrated, using a GST pull-down assay, that all three mutant E1 proteins are capable of binding E2 to the same extent as the wild-type protein (Fig. 7C). For wild-type E1 and E1 mutants I224M-R225D, K286A-R288A, and A292L-R293E, the amounts of input protein that bound to GST-E2 (SDS eluate) were 32, 34, 30, and 28%, respectively (Fig. 7C). When tested in the E2-dependent E1 origin-binding assay, all three mutants were found to be severely defective, in both the absence and presence of ATP-Mg and at both temperatures tested. Importantly, the binding of these E1 mutants to the origin was not stimulated by ATP-Mg; in fact, ATP-Mg exacerbated their defects at high temperature (Fig. 7C, compare lanes 14, 15, and 16 with lanes 18, 19, and 20). These results indicated that the E1 DNA-binding domain is required for maximal E2-dependent binding of E1 to the origin and for its stimulation by ATP-Mg at high temperature.
FIG. 7.
Effect of amino acid substitutions in the E1 DNA-binding domain on the E2-dependent binding of E1 to the origin. (A) Amino acid sequence of a portion of the HPV-11 E1 DNA-binding domain between residues 220 and 300. The locations of three double-amino-acid substitutions are indicated (double arrows), as well as the resulting amino acid changes (boldface). (B) E2-dependent binding of mutant E1 proteins to the origin. DNA-protein complexes were assembled with E2 and the indicated E1 protein made by in vitro translation. The complexes were assembled at the indicated temperatures and in the presence (+) or absence (−) of ATP-Mg. The immunoprecipitation of the complexes was done with the K72 polyclonal antibody, and the detection of bound DNA was done as described in the legend to Fig. 2A. WT, wild type; LE, A292L-R293E; AA, K286A-R288A; MD, I224M-R225D. (C) GST pull-down assays demonstrating similar binding of wild-type E1 and of the three mutant E1 proteins to the TAD of E2. The percentages of the input protein eluted from the GST and GST-E2 proteins with NaCl and SDS are written below each gel. The gels were loaded and analyzed as described in the legend to Fig. 5.
The P479S E1 mutant protein can support transient HPV DNA replication.
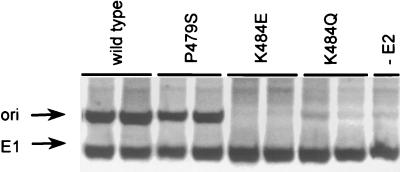
During the course of this work, we tested some of the mutant E1 proteins for the ability to support transient HPV DNA replication in cells. This was accomplished by using an assay that measures transient replication of an HPV origin-containing plasmid in CHO cells, following cotransfection with E1 and E2 expression vectors (see Materials and Methods). In this assay, the amount of newly replicated DNA was measured by quantitative PCR amplification of a portion of the origin-containing plasmid from DpnI-digested total cellular DNA. Because the region of the origin-containing plasmid that served as a template for amplification contains several DpnI sites, digestion with DpnI (which cleaves unreplicated or methylated DNA) ensured that only replicated DNA was amplified. To control for transfection efficiency and variations in the recovery of genomic DNA, the amount of E1 expression vector contained in total DNA was also determined. This was accomplished in the same PCR but with a different pair of primers that amplify a fragment devoid of DpnI sites. As can be seen in Fig. 8, the K484E and K484Q substitutions greatly reduced HPV DNA replication. This result was expected, since these substitutions affect the recruitment of E1 to the origin in vitro (see above) in addition to changing an amino acid, K484, that is essential for the ATPase and helicase activities of E1. In contrast, the P479S E1 protein could support HPV DNA replication, a result which supported our observation that this mutant protein is capable of binding with E2 to the origin in vitro. However, this result was not anticipated, given that this substitution abrogates the enzymatic activities of E1 from BPV-1 and HPV-6b (22, 23, 34, 52, 63) as well as the binding of BPV-1 E1 to E2 (31). Clearly, our results indicate that the P479S E1 protein is enzymatically active (Fig. 8) and capable of binding to E2 (Fig. 4B) and suggest that structural constraints imposed on the HPV-11 E1 P loop differ from those that impinge on the E1 P loops of BPV-1 and HPV-6b (see Discussion).
FIG. 8.
Effect of substitutions in the ATP-binding domain of E1 on transient HPV DNA replication. The amount of replicated origin-containing plasmid (ori), or of an internal control plasmid (E1), was detected by quantitative PCR analysis. PCR amplification was performed on genomic DNA isolated from cells transfected with a plasmid expressing E1 (−E2) or transfected with a combination of plasmids expressing E2 and wild-type E1 or one of three mutant E1 proteins (P479S, K484E, or K484Q). The PCR products were visualized by electrophoresis and autoradiography.
DISCUSSION
Recruitment of E1 by E2 to the HPV origin is an essential step in the initiation of viral DNA replication. In this study we determined that the C-terminal domain of E1 (amino acids 353 to 649) is sufficient for interaction with the TAD of E2 (amino acids 1 to 209) and for recruitment of E1 to the origin by E2. This relatively large domain of E1 could not be truncated without loss of E2 binding, indicating that the entire domain is likely to be required for formation of an E2-binding surface. The interaction between the C terminus of E1 and E2 appears biologically relevant, since it was affected by the E39A substitution in E2, which affects E1 binding in vitro and transient HPV DNA replication in transfected cells (12, 43). In general, our results agree with those obtained with HPV-16 E1 (36, 64) and those of a recent study with E1 and E2 proteins from HPV-11 (53). Similar to what Sun et al. (53) observed for HPV-11 (53), we found that amino acids 353 to 649 of E1 are sufficient for interaction with E2, but unlike those authors, we found that such a fragment is capable of forming a complex with E2 at the origin, albeit not as efficiently as the entire E1 protein. Our results also differ slightly from those of Masterson et al. (36) in that we could not detect an interaction between amino acids 435 to 649 of E1 and E2 in vitro but only in the yeast two-hybrid system. In vitro, in the presence or absence of ATP-Mg and under a wide range of temperatures, amino acids 353 to 649 of E1 were required for E2 to form a complex with E1 at the origin (Fig. 8). In vitro, residues 353 to 435 of E1 could be required to stabilize the conformation of the 435-to-649 domain. In the yeast two-hybrid system, the conformation of the 435-to-649 region may be stabilized artificially as a result of fusion to the GAL4 activation or DNA-binding domain. The notion that the 435-to-649 domain may be conformationally unstable is also supported by results obtained with HPV-16 E1 and E2. In this case residues 420 to 649 of E1 could bind E2 only at low temperature, and a larger fragment (144 to 649) was needed for binding at the physiological temperature (64). It is also formally possible that residues 353 to 434 of E1 stabilize the binding of E1 to E2 in vitro by making additional contacts with E2. A single study, using HPV-33 E1, indicated that this region could bind E2 (40). However, our studies and those of others (36, 53, 64), both with yeast and in vitro, failed to detect an interaction between this region and E2, indicating that this additional interaction either is too weak or does not exist with E1 from HPV-11 or HPV-16. An interaction between the DNA-binding domains of E1 and E2 has been described for BPV-1 (10, 27) that has not been detected between the cognate proteins of HPV-11 or HPV-16 (references 36, 53, and 64 and this study). This interaction appears to be required for DNA replication, in addition to the interaction between the TAD of E2 and the C-terminal domain of E1, only in the context of an origin like that of BPV-1 and unlike that of HPV-11, in which the E1- and E2-binding sites are in close proximity. In the case of the HPV-11 origin, Berg and Stenlund (3) have shown that an interaction between HPV-11 E2 (TAD) and the C-terminal domain of E1 (BPV E1 was used in this study) is sufficient for cooperative binding to the HPV-11 origin in vitro and for transient HPV DNA replication in vivo. Therefore, even if an interaction between the DNA-binding domains of E1 and E2 from HPV-11 were to exist, it may not be required for transient replication of the HPV-11 genome.
In this study, we analyzed further the role of the ATP-binding domain of E1, and the role of ATP-Mg, in the E2-dependent binding of HPV-11 E1 to the origin. The following observations were made. (i) ATP-Mg is not required for the interaction of E1 with E2. (ii) Mutational analysis of the E1 ATP-binding domain indicated that the structural integrity of the P loop is essential for binding to E2. (iii) The effect of the P479S substitution suggests that the structural constraints imposed on the P loop of HPV-11 differ from those imposed on the BPV-1 E1 P loop. (iv) Although the C terminus of E1 (353 to 649) is sufficient for interaction with E2 and recruitment to the origin, the E1 DNA-binding domain is essential for optimal recruitment of E1 to the origin. (v) ATP-Mg can stimulate the E2-dependent binding of wild-type E1, but not of E1 (353 to 649), to the origin at the physiological temperature (37°C). (vi) This stimulation by ATP-Mg requires an intact E1 DNA-binding domain, in addition to the ATP-binding domain. These observations are discussed below.
Our mutational analysis of the E1 ATP-binding domain revealed that highly conserved residues in motifs B and C are not essential for formation of a complex with E2 at the origin. Certain amino acid substitutions in motif A, the P loop, were also tolerated. Most of these substitutions are predicted to affect the binding of ATP. In support of this notion, we found that among four mutant E1 proteins (K484I, K484R, K484A, and K484H) that were tested for ATPase activity, all were severely defective (41). Together, these results suggest that ATP binding is not essential for E1 to bind to E2. In separate experiments, we also used purified recombinant E1 and E2 proteins to show that their interaction does not require ATP-Mg (data not shown).
Despite the fact that ATP is not required for E1 and E2 to interact, we found that certain substitutions in the P loop, like those affecting Gly478, abolish binding to E2. The negative effect of the conservative replacement of Gly478 by alanine, which is often found at this position in the P loops of other proteins, suggests that glycine may be the only residue tolerated at this position. An absolute requirement for glycine was observed in the P loop of the RecA protein of E. coli, a protein involved in DNA recombination (26). As noted by Konola et al. (26), this was an expected result, since the crystal structure of RecA bound to ADP revealed that unusual torsion angles occur in this region of the protein, which can be accommodated only by glycine. Our findings are therefore suggestive that the P loop of E1 is under similar constraints. The analogy between E1 and RecA might extend to other residues of the P loop, since all three helicases whose structures have been solved (Bacillus stearothermophilus PcrA, E. coli Rep, and hepatitis C virus RNA helicase) have nucleotide binding domains similar to that of RecA (reviewed in references 4 and 35).
Binding of E1 to E2 was also affected by substitutions that changed lysine 484 of the E1 P loop. Not all substitutions at position 484 had the same effect on E2 binding. The smaller residues Ala, Ile, and His were tolerated. In contrast, residues with longer side chains, such as Met, Gln, and Glu, were deleterious to E2 binding, with the exception of arginine, which most closely resembles lysine. These deleterious substitutions could affect E2 binding in several ways. One possibility is that they affect the folding of E1. In general, little is known about the role, if any, of P loops in folding of ATP-binding domains. Our results, however, clearly indicate that lysine 484 of E1 is not essential for folding, since it can be changed to a number of residues without loss of E2 binding. Based on the observation that only residues with longer side chains are not tolerated, we favor the alternative possibility that these substitutions distort the correct geometry of the ATP-binding site and of the E2-binding surface. This is particularly likely as the P loop is probably involved in tertiary, intramolecular interactions with the rest of the protein that are required to organize the overall structure of the E1 C-terminal domain. Indeed, analysis of the structures of some of the best-characterized P-loop-containing proteins has provided evidence that the helical conformation of the GKS/T motif of the P loop is maintained by several intramolecular interactions with the rest of the protein (13). Together, the results of our mutational analyses of lysine 484 and glycine 479 suggest that the E1 P loop is under strict structural constraints and plays a key role in formation of the E2-binding surface. These findings also indicate that we must exert caution in interpreting the effect of substitutions in the P loop of E1, and perhaps also in that of other NTP-binding proteins, and not assume that these substitutions only affect NTP binding and hydrolysis. Finally, the notion that the precise geometry of the E1 ATP-binding domain is critical for function is also supported by the recent finding that chimeric E1 proteins, in which the junction between HPV-11 and HPV-16 sequences was within the ATPase domain, were unable to support HPV DNA replication (66).
The structural constraints imposed on the HPV-11 E1 P loop may differ slightly from those imposed on the P loop of BPV-1 E1. This is suggested by the observation that one P-loop substitution, P479S, inactivates BPV-1 E1 but is tolerated in HPV-11 E1. For BPV E1, this substitution reduces ATP binding (34, 52), helicase activity (63), binding to E2 (31), and replication of the genome (34, 52) and increases slightly the binding of E1 to the origin in vitro (56). For HPV-6b E1, the enzyme most closely related to HPV-11 E1, the P479S substitution reduces ATP binding as well as ATPase and helicase activities (22, 23). Our results indicated that the P479S substitution reduces only slightly the ability of HPV-11 E1 to complex with E2 in yeast and in vitro and to support DNA replication in transiently transfected cells. The latter result clearly indicates that the P479S substitution in HPV-11 E1 does not completely abolish the enzymatic activities of this protein in vivo. It may be that the P479S substitution has some effect on the ATPase and helicase activities of HPV-11 E1 but that this reduction in enzymatic activity is tolerated for HPV DNA replication. In support of this notion is the observation that E1 from HPV-16 has naturally lower ATPase and helicase activities due to the presence in its P loop of two alanine residues instead of prolines (corresponding to residues 479 and 480 in HPV-11 E1 [Fig. 4A]) for most HPV types (42).
In this study, we found that ATP-Mg can stimulate the E2-dependent binding of E1 to the origin and that this stimulation can be abolished by substitutions in the E1 DNA-binding domain. These results suggested that upon binding ATP-Mg, the E1 C-terminal domain might affect the binding of E1 to DNA, directly or indirectly. Binding of ATP by E1 could modulate the contacts between the DNA-binding domain and the origin. Alternatively, or in addition, ATP may stabilize the interaction of E1 with the origin by promoting its oligomerization. In this respect, it is noteworthy that the binding and oligomerization of E1 to the origin, in the absence of E2, is also temperature sensitive and stimulated by ATP-Mg (7). Moreover, we have found recently that the C-terminal domain of E1 (amino acids 353 to 649) is sufficient for oligomerization and that mutations in conserved residues of the ATP-binding domain affect oligomerization (7). Stimulation of origin binding by ATP-Mg is likely to occur through conformational changes in E1 that would be brought about upon ATP binding and relayed to the E1 DNA-binding domain and/or the oligomerization domain. This is not an unlikely proposal, since conformational changes are at the heart of the mechanism by which helicases unwind DNA and/or translocate along it in response to binding of ATP or ADP or single-stranded DNA (reference 24; reviewed in references 4, 29, and 35). The fact that the E2-dependent binding of E1 to the origin is temperature sensitive is consistent with the occurrence of conformational changes. In addition to our results, two lines of evidence suggest that changes in the conformation of the C-terminal domain can affect the ability of E1 to bind DNA. First, mutations in the ATP-binding domain (in the P loop) of BPV-1 E1 slightly increase the affinity of the protein for the origin (56). Second, ATP-Mg stimulates binding of BPV-1 E1 to DNA (32, 33, 49, 50) and changes its footprint at the origin (18). Binding of E2 to the C-terminal domain of E1 may also affect DNA binding. This is suggested by the observation that in the absence of E2-binding sites, E2 suppresses the binding of E1 to the origin (32) and to nonspecific DNA (6). Together with our results, these findings support the notion that conformational changes in the E1 C-terminal domain, brought about upon binding ATP-Mg, can affect the interaction of E1 with DNA. For BPV-1 E1 and E2, it has been shown that ATP-Mg regulates the conversion of the E1-E2-ori complex to an E1-ori complex in which E2 has been displaced from the origin (33, 44). Our results suggest that binding of ATP-Mg by E1 is not sufficient to break the interaction between E1 and E2. This conclusion is based on the observation that E1 (353 to 649), which cannot bind to DNA, could be tethered to the origin simply via an interaction with E2, regardless of whether the binding reactions were supplemented with ATP-Mg or not (Fig. 6). Similarly, we have found (in GST pull-down assays) that in the absence of origin DNA, ATP has little effect on the binding of E1 (wild type or 353 to 649) to the TAD of E2 (Fig. 5). Together, these results rule out the possibility that ATP-Mg is sufficient to break the interaction between E1 and E2 in solution. From these results and others, we favor the hypothesis that ATP-Mg affects the interaction between E1 and E2 primarily in an indirect way, for example, by promoting the binding or oligomerization of wild-type E1 at the origin. This hypothesis is supported by our observation that ATP-Mg stimulation of the E2-dependent binding of E1 to the origin requires an intact E1 DNA-binding domain. It is also supported by our observation (7) and those of others (32, 49) that ATP-Mg can promote the binding or oligomerization of E1 at the origin in the absence of E2.
Keeping in mind the role of the ATP-binding domain of E1 in the formation of the E2-binding surface and in modulating the interaction of E1 with DNA, the following model for the recruitment of E1 to the origin can be proposed. An initial step would be the interaction of the E2 TAD with the C terminus of E1 that would result in recruitment of E1 to the origin via a combination of two mechanisms: suppression of the nonspecific DNA-binding activity of E1 and direct binding of E2 to the origin. In the case of BPV-1, an additional interaction between the DNA-binding domains of E1 and E2 may also play a role in this initial loading of E1 molecules on the origin (3). However, the latter interaction appears to be essential only in the context of certain origins, such as that of BPV-1 and unlike that of HPV-11, in which the E1- and E2-binding sites are closely juxtaposed (3). Since a minimum of two E2-binding sites is required for optimal replication of the HPV-11 (or -18) genome (30, 54), and for optimal recruitment of E1 to the origin (Fig. 2), we imagine that two E1 molecules can be recruited to the origin by interaction with two separate E2 dimers. This would bring into close proximity two E1 monomers and might serve as a mechanism to initiate the assembly of an E1 oligomer. Because E1, in the absence of E2, cannot bind stably to the origin as a monomer but rather assembles as an oligomer in a highly cooperative manner (10, 33, 46–50), we assume that conformational changes brought about by binding to the origin also favor E1 oligomerization. These conformational changes, which would be favored by ATP-Mg and disfavored by high temperature, may be required to stabilize the interaction of E1 with DNA and/or to promote the interaction between E1 monomers. Interaction between E1 monomers at the origin may disrupt their interaction with E2, as previously suggested by Lusky et al. (33), and allow for additional E1 molecules to be recruited to the complex. In this context, ATP-Mg may favor the dissociation of E1 and E2, at least in part, by promoting the binding or oligomerization of E1 at the origin. In this model, the two E2-binding sites in the origin would serve as loading docks for two E2 dimers to recruit E1 molecules until a complete oligomer is assembled. This model highlights what we believe to be two key structural roles played by the E1 ATP-binding domain. One role is in the formation of the E2-binding surface. The other, which requires binding of ATP-Mg, is in facilitating conformational changes in E1 which occur upon binding to the origin.
REFERENCES
- 1.Abroi A, Kurg R, Ustav M. Transcriptional and replicational activation functions in the bovine papillomavirus type 1 E2 protein are encoded by different structural determinants. J Virol. 1996;70:6169–6179. doi: 10.1128/jvi.70.9.6169-6179.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benson J D, Howley P M. Amino-terminal domains of the bovine papillomavirus type 1 E1 and E2 participate in complex formation. J Virol. 1995;69:4364–4372. doi: 10.1128/jvi.69.7.4364-4372.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg M, Stenlund A. Functional interactions between papillomavirus E1 and E2 proteins. J Virol. 1997;71:3853–3863. doi: 10.1128/jvi.71.5.3853-3863.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bird L E, Subramanya H S, Wigley D B. Helicases: a unifying structural theme? Curr Opin Struct Biol. 1998;8:14–19. doi: 10.1016/s0959-440x(98)80004-3. [DOI] [PubMed] [Google Scholar]
- 5.Blitz I L, Laimins L A. The 68-kilodalton E1 protein of bovine papillomavirus is a DNA binding phosphoprotein which associates with the E2 transcriptional activator in vitro. J Virol. 1991;65:649–656. doi: 10.1128/jvi.65.2.649-656.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonne-Andrea C, Tillier F, McShan G D, Wilson V G, Clertant P. Bovine papillomavirus type 1 DNA replication: the transcriptional activator E2 acts as a specificity factor. J Virol. 1997;71:6805–6815. doi: 10.1128/jvi.71.9.6805-6815.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brault, K., S. Titolo, A. Pelletier, and J. Archambault. Unpublished data.
- 8.Breiding D E, Grossel M J, Androphy E J. Genetic analysis of the bovine papillomavirus E2 transcriptional activation domain. Virology. 1996;221:34–43. doi: 10.1006/viro.1996.0350. [DOI] [PubMed] [Google Scholar]
- 9.Brokaw J L, Blanco M, McBride A A. Amino acids critical for the functions of the BPV-1 E2 transactivator. J Virol. 1996;70:23–29. doi: 10.1128/jvi.70.1.23-29.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen G, Stenlund A. Characterization of the DNA-binding domain of the bovine papillomavirus replication initiator E1. J Virol. 1998;72:2567–2576. doi: 10.1128/jvi.72.4.2567-2576.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clertant P, Seif I. A common function for polyoma virus large-T and papillomavirus E1 proteins? Nature. 1984;311:276–279. doi: 10.1038/311276a0. [DOI] [PubMed] [Google Scholar]
- 12.Cooper C S, Upmeyer S N, Winokur P L. Identification of single amino acids in the human papillomavirus 11 E2 protein critical for the transactivation and replication functions. Virology. 1998;241:312–322. doi: 10.1006/viro.1997.8941. [DOI] [PubMed] [Google Scholar]
- 13.Cronet P, Bellsolell L, Sander C, Coll M, Serrano L. Investigating the structural determinants of the p21-like triphosphate and Mg2+ binding site. J Mol Biol. 1995;249:654–664. doi: 10.1006/jmbi.1995.0326. [DOI] [PubMed] [Google Scholar]
- 14.Durfee T, Becherer K, Chen P-L, Yeh S-H, Yang Y, Kilburn A E, Lee W-H, Elledge S. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 15.Ferguson M K, Botchan M R. Genetic analysis of the activation domain of bovine papillomavirus protein E2: its role in transcription and replication. J Virol. 1996;70:4193–4199. doi: 10.1128/jvi.70.7.4193-4199.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frattini M G, Laimins L A. The role of the E1 and E2 proteins in the replication of human papillomavirus type 31b. Virology. 1994;204:799–804. doi: 10.1006/viro.1994.1596. [DOI] [PubMed] [Google Scholar]
- 17.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gillette T G, Lusky M, Borowiec J A. Induction of structural changes in the bovine papillomavirus type 1 origin of replication by the viral E1 and E2 proteins. Proc Natl Acad Sci USA. 1994;91:8846–8850. doi: 10.1073/pnas.91.19.8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorbalenya A E, Koonin E V. Helicases: amino acid sequence comparisons and structure-function relationships. Curr Opin Struct Biol. 1993;3:419–429. [Google Scholar]
- 20.Gorbalenya A E, Koonin E V, Wolf Y I. A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett. 1990;262:145–148. doi: 10.1016/0014-5793(90)80175-i. [DOI] [PubMed] [Google Scholar]
- 21.Grossel M J, Sverdrup F, Breiding D E, Androphy E J. Transcriptional activation function is not required for stimulation of DNA replication by BPV-1 E2. J Virol. 1996;70:7264–7269. doi: 10.1128/jvi.70.10.7264-7269.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hughes F J, Romanos M A. E1 protein of human papillomavirus is a DNA helicase/ATPase. Nucleic Acids Res. 1993;21:5817–5823. doi: 10.1093/nar/21.25.5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenkins O, Earnshaw D, Sarginson G, Vecchio A D, Tsai J, Kallender H, Amegadzie B, Browne M. Characterization of the helicase and ATPase activity of human papillomavirus type 6b protein. J Gen Virol. 1996;77:1805–1809. doi: 10.1099/0022-1317-77-8-1805. [DOI] [PubMed] [Google Scholar]
- 24.Jezewska M J, Bujalowski W. Global conformational transitions in Escherichia coli primary replicative helicase DnaB protein induced by ATP, ADP, and single-stranded DNA binding. J Biol Chem. 1996;271:4261–4265. doi: 10.1074/jbc.271.8.4261. [DOI] [PubMed] [Google Scholar]
- 25.Kasukawa H, Howley P M, Benson J D. A fifteen-amino-acid peptide inhibits human papillomavirus E1-E2 interaction and human papillomavirus DNA replication in vitro. J Virol. 1998;72:8166–8173. doi: 10.1128/jvi.72.10.8166-8173.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konola J T, Logan K M, Knight K L. Functional characterization of residues in the P-loop motif of the RecA protein ATP binding site. J Mol Biol. 1994;237:20–34. doi: 10.1006/jmbi.1994.1206. [DOI] [PubMed] [Google Scholar]
- 27.Leng X, Ludes-Meyer J H, Wilson V G. Isolation of an amino-terminal region of bovine papillomavirus type 1 E1 protein that retains origin binding and E2 interaction capacity. J Virol. 1997;71:848–852. doi: 10.1128/jvi.71.1.848-852.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J-S, Kuo S-R, Broker T R, Chow L T. The functions of human papillomavirus type 11 E1, E2, and E2C proteins in cell-free DNA replication. J Biol Chem. 1995;270:27283–27291. doi: 10.1074/jbc.270.45.27283. [DOI] [PubMed] [Google Scholar]
- 29.Lohman T M, Bjornson K P. Mechanisms of helicase-catalyzed DNA unwinding. Annu Rev Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 30.Lu J Z-J, Sun Y-N, Rose R C, Bonnez W, McCance D J. Two E2 binding sites (E2BS) alone or one E2BS plus an A/T-rich region are minimal requirements for the replication of the human papillomavirus type 11 origin. J Virol. 1993;67:7131–7139. doi: 10.1128/jvi.67.12.7131-7139.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lusky M, Fontane E. Formation of the complex of bovine papillomavirus E1 and E2 proteins is modulated by E2 phosphorylation and depends upon sequence within the carboxy terminus of E1. Proc Natl Acad Sci USA. 1991;88:6363–6367. doi: 10.1073/pnas.88.14.6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lusky M, Hurwitz J, Seo Y-S. Cooperative assembly of the bovine papilloma virus E1 and E2 proteins on the replication origin requires an intact E2 binding site. J Biol Chem. 1993;268:15795–15803. [PubMed] [Google Scholar]
- 33.Lusky M, Hurwitz J, Seo Y-S. The bovine papillomavirus E2 protein modulates the assembly of but is not stably maintained in a replication-competent multimeric E1-replication origin complex. Proc Natl Acad Sci USA. 1994;91:8895–8899. doi: 10.1073/pnas.91.19.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MacPherson P, Thorner L, Parker L M, Botchan M. The bovine papilloma virus E1 protein has ATPase activity essential to viral DNA replication and efficient transformation in cells. Virology. 1994;204:403–408. doi: 10.1006/viro.1994.1544. [DOI] [PubMed] [Google Scholar]
- 35.Marians, K. J. Helicase structures: a new twist on DNA unwinding. Structure 5:1129–1134. [DOI] [PubMed]
- 36.Masterson P J, Stanley M A, Lewis A P, Romanos M A. A C-terminal helicase domain of the human papillomavirus E1 protein binds E2 and the DNA polymerase alpha-primase p68 subunit. J Virol. 1998;72:7407–7419. doi: 10.1128/jvi.72.9.7407-7419.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McBride A, Myers G. The E2 proteins. In: Myers G, Baker C, Münger K, Sverdrup F, McBride A, Bernard H-U, editors. Human papillomaviruses 1997. Los Alamos, N.Mex: Los Alamos National Laboratory; 1997. pp. 37–53. [Google Scholar]
- 38.McKay R D G. Binding of a simian virus T antigen-related protein to DNA. J Mol Biol. 1981;145:471–478. doi: 10.1016/0022-2836(81)90540-4. [DOI] [PubMed] [Google Scholar]
- 39.Mohr I J, Clark R, Sun S, Androphy E J, MacPherson P, Botchan M R. Targeting the E1 replication protein to the papillomavirus origin of replication by complex formation with the E2 transactivator. Science. 1990;250:1694–1699. doi: 10.1126/science.2176744. [DOI] [PubMed] [Google Scholar]
- 40.Müller F, Sapp M. Domains of the E1 protein of human papillomavirus type 33 involved in binding to the E2 protein. Virology. 1996;219:247–256. doi: 10.1006/viro.1996.0242. [DOI] [PubMed] [Google Scholar]
- 41.Pelletier, A., and P. White. Unpublished data.
- 42.Raj K, Stanley M A. The ATP-binding and ATP-ase activities of human papillomavirus type 16 E1 are significantly weakened by the absence of prolines in its ATP-binding domain. J Gen Virol. 1995;76:2949–2956. doi: 10.1099/0022-1317-76-12-2949. [DOI] [PubMed] [Google Scholar]
- 43.Sakai H, Yasugi T, Benson J D, Dowhanick J J, Howley P M. Targeted mutagenesis of the human papillomavirus type 16 E2 transactivation domain reveals separable transcriptional activation and DNA replication functions. J Virol. 1996;70:1602–1611. doi: 10.1128/jvi.70.3.1602-1611.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanders C M, Stenlund A. Recruitment and loading of the E1 initiator protein: an ATP-dependent process catalysed by a transcription factor. EMBO J. 1998;17:7044–7055. doi: 10.1093/emboj/17.23.7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarafi T R, McBride A A. Domains of the BPV-1 E1 replication protein required for origin-specific DNA binding and interaction with the E2 transactivator. Virology. 1995;211:385–396. doi: 10.1006/viro.1995.1421. [DOI] [PubMed] [Google Scholar]
- 46.Sedman J, Stenlund A. Co-operative interaction between the initiator E1 and the transcriptional activator E2 is required for replicator specific DNA replication of bovine papillomavirus in vivo and in vitro. EMBO J. 1995;14:6218–6228. doi: 10.1002/j.1460-2075.1995.tb00312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sedman J, Stenlund A. The initiator protein E1 binds to the bovine papillomavirus origin as a trimeric ring-like structure. EMBO J. 1996;15:5085–5092. [PMC free article] [PubMed] [Google Scholar]
- 48.Sedman T, Sedman J, Stenlund A. Binding of the E1 and E2 proteins to the origin of replication of bovine papillomavirus. J Virol. 1997;71:2887–2896. doi: 10.1128/jvi.71.4.2887-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seo Y-S, Müller F, Lusky M, Gibbs E, Kim H-Y, Phillips B, Hurwitz J. Bovine papilloma virus (BPV)-encoded E2 protein enhances binding of E1 to the BPV replication origin. Proc Natl Acad Sci USA. 1993;90:2865–2869. doi: 10.1073/pnas.90.7.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seo Y-S, Müller F, Lusky M, Hurwitz J. Bovine papilloma virus (BPV)-encoded E1 protein contains multiple activities required for BPV DNA replication. Proc Natl Acad Sci USA. 1993;90:702–706. doi: 10.1073/pnas.90.2.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spalholz B A, McBride A A, Sarafi T, Quintro J. Binding of bovine papillomavirus to the origin is not sufficient for DNA replication. Virology. 1993;193:201–212. doi: 10.1006/viro.1993.1116. [DOI] [PubMed] [Google Scholar]
- 52.Sun S, Thorner L, Lentz M, MacPherson P, Botchan M. Identification of a 68-kilodalton nuclear ATP-binding phosphoprotein encoded by bovine papillomavirus type 1. J Virol. 1990;64:5093–5105. doi: 10.1128/jvi.64.10.5093-5105.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun Y, Han H, McCance D J. Active domains of human papillomavirus type 11 E1 protein for origin replication. J Gen Virol. 1998;79:1651–1658. doi: 10.1099/0022-1317-79-7-1651. [DOI] [PubMed] [Google Scholar]
- 54.Sverdrup F, Khan S A. Two E2 binding sites alone are sufficient to function as the minimal origin of replication of human papillomavirus type 18 DNA. J Virol. 1995;69:1319–1323. doi: 10.1128/jvi.69.2.1319-1323.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sverdrup F, Myers G. The E1 proteins. In: Myers G, Baker C, Münger K, Sverdrup F, McBride A, Bernard H-U, editors. Human papillomaviruses 1997. Los Alamos, N.Mex: Los Alamos National Laboratory; 1997. pp. 37–53. [Google Scholar]
- 56.Thorner L K, Lim D A, Botchan M R. DNA-binding domain of bovine papillomavirus type 1 E1 helicase: structural and functional aspects. J Virol. 1993;67:6000–6014. doi: 10.1128/jvi.67.10.6000-6014.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tolskaya E A, Romanova L I, Kolesnikova M S, Gmyl A P, Gorbalenya A E, Agol V I. Genetic studies on the poliovirus 2C protein, an NTPase. A plausible mechanism of guanidine effect on the 2C function and evidence for the importance of 2C oligomerization. J Mol Biol. 1994;236:1310–1323. doi: 10.1016/0022-2836(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 58.Ustav M, Ustav E, Szymanski P, Stenlund A. Identification of the origin of replication of bovine papillomavirus and characterization of the viral origin recognition factor E1. EMBO J. 1991;10:4321–4329. doi: 10.1002/j.1460-2075.1991.tb05010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilson V G, Ludes-Meyers J. A bovine papillomavirus E1-related protein binds specifically to bovine papillomavirus DNA. J Virol. 1991;69:395–402. doi: 10.1128/jvi.65.10.5314-5322.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winokur P, McBride A A. Separation of the transcriptional activation and replication functions of the bovine papillomavirus-1 E2 protein. EMBO J. 1992;11:4111–4118. doi: 10.1002/j.1460-2075.1992.tb05504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang L, Li R, Mohr I L, Clark R, Botchan M. Activation of BPV-1 replication in vitro by the transcription factor E2. Nature (London) 1991;353:628–632. doi: 10.1038/353628a0. [DOI] [PubMed] [Google Scholar]
- 63.Yang L, Mohr I, Fouts E, Lim D A, Nohaile M, Botchan M. The E1 protein of bovine papilloma virus 1 is an ATP-dependent DNA helicase. Proc Natl Acad Sci USA. 1993;90:5086–5090. doi: 10.1073/pnas.90.11.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yasugi T, Benson J D, Sakai H, Vidal M, Howley P M. Mapping and characterization of the interaction domains of human papillomavirus type 16 E1 and E2 proteins. J Virol. 1997;71:891–899. doi: 10.1128/jvi.71.2.891-899.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoshida M, Amano T. A common topology of proteins catalyzing ATP-triggered reactions. FEBS Lett. 1995;359:1–5. doi: 10.1016/0014-5793(94)01438-7. [DOI] [PubMed] [Google Scholar]
- 66.Zou N, Liu J-S, Kuo S-R, Broker T R, Chow L T. The carboxyl-terminal region of the human papillomavirus type 16 E1 protein determines E2 protein specificity during DNA replication. J Virol. 1998;72:3436–3441. doi: 10.1128/jvi.72.4.3436-3441.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]