Abstract
Spectrin, a large cytoskeletal protein, consists of a heterodimeric structure comprising α and β subunits. Here, we have studied the mechanics of spectrin filament as a major constituent of dendrites and dendritic spines. Given the intricate biological details and compact biological construction of spectrin, we've developed a constitutive model of spectrin that describes its continuous deformation over three distinct stages and it’s progressive failure mechanisms. Our model closely predicts both the force at which uncoiling begins and the ultimate force at which spectrin fails, measuring approximately 93 ~ 100 pN. Remarkably, our predicted failure force closely matches the findings from AFM experiments focused on the uncoiling of spectrin repeats, which reported a force of 90 pN. Our theoretical model proposes a plausible pathway for the potential failure of dendrites and the intricate connection between strain and strain rate. These findings deepen our understanding of how spectrin can contribute to traumatic brain injury risk analysis.
Subject terms: Computational biophysics, Molecular biophysics
Introduction
To evaluate the comprehensive mechanical injury threshold1 for the brain, it is needed to understand the injury mechanisms of the brain at the cellular level2. Neuron cells are the unique cellular-level building blocks of brain tissue. The neuronal cytoskeleton is mainly comprised of axons, Dendrites, and the Cell Body. Recent studies on the mechanical properties of neuron cells are primarily focused on the mechanics of axons, stemming from the relatively simple structural form of axon3 and the availability of imaging and structural data of its sub-cellular component. For example, a major constituent of axons is microtubules (MT) and this element has been studied extensively and repeatedly4–6 Understanding MT properties7 subsequently paves the way for modeling axons for mechanical damage evaluation and neuronal signal transmission8–11. However, to determine the comprehensive injury threshold for neuron cells, a model should include the mechanical behavior of dendrites and soma. Of these, dendrites are projections of neurons that are specialized to receive signals from other neurons12. They resemble a tree-like structure that forms projections and thus receives stimuli from other neurons. Recent studies using 3D electron microscopy (3DEM) suggest that while enhancing synaptic connectivity might not be the primary function of dendrites, their precise structural role remains to be conclusively determined12,13.
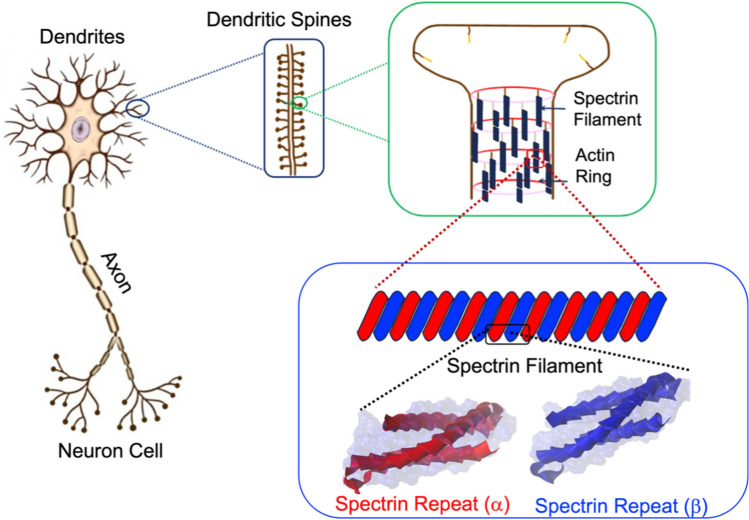
The tree-like structure and branching of dendrites can vary to a large extent and it has been reported that dendrites differ widely in dimensions, distributions, and intracellular composition. Such structural and compositional variations make the complete scale modeling of dendrites challenging. Yet, efforts have been made to define dendrite’s structural features over multiple length scales. Figure 1 outlines the multiscale structural breakdown of dendrites14,15. Dendrites contain dendritic spines which are very small protrusions raised from dendritic branches16. Internally, dendritic spines contain periodic actin-spectrin lattice structures, actin patches, longitudinal actin fibers13, spectrin filaments, and other intracellular constituents. Such microstructural arrangement of dendrites is significantly different from axons. The actin-spectrin lattice structure is very dominant in dendrites and dendritic spines 17 and acts as key load-bearing components in the circumference of dendrites and dendritic spines. A few studies represent some variability about the different spectrin configurations in dendrites18,19. In this, we attempted to develop our model using a common form of spectrin. Although spectrin present in dendrites have slightly different conformations in different areas, as a biopolymer the key structural aspect of spectrin remains unchanged when it is considered for a full-scale modeling of dendrites along with other sub-cellular components. So, the mechanical property of spectrin we have described in our model will be fitted for dendrites at a sub-cellular level as a single component. As such, to predict the injury risk of dendrites we need to understand the mechanical properties of spectrin, most importantly its deformation mechanisms and failure force threshold.
Figure 1.
Schematic representation of concentrated area in this study which is sub cellular level of dendrites in neuron cell.
Although previous models of axons had explored the growth and properties of actin-spectrin-based cytoskeleton membrane 20–22, those studies didn’t report the constitutive and failure properties of spectrin filaments. However, understanding the detailed mechanical behavior of spectrin is essential for the development of constitutive properties of dendrites. A very recent study of the axonal plasma membrane skeleton has outlined a computational approach for the actin-spectrin ring, but the study mostly focused on the axonal membrane rather than investigating the individual properties of spectrin filament23. Nevertheless, it is well-understood that spectrin’s structural stability is directly related to neural functions24. For instance, recent studies on lab-grown mice with αII spectrin-deficient peripheral sensory neurons suggest the absence of spectrin in neurons can lead to rapid neuronal degeneration 25. Another study reveals that a lack of spectrin in neurons may lead to dysfunction of the nervous system26. In vitro studies on functionally knockdown spectrin show that it has a direct impact on the modulus of elasticity of axons 27. It can be inferred from these studies that spectrin filaments play critical roles in the overall function and mechanical stability of dendrites and axons. Structurally, spectrin is an antiparallel dimer with an -chain and -chain lying side by side. These arrangements collectively give spectrin a long, spring-like filament structure. At the same time, locally it remains in the extensive foldable form like other biopolymers28–33. There are biological studies to explain the detailed structural formation of different types of spectrin filaments. Almost all these studies are mainly focused on the biological properties and functionalities of Spectrin34–41.
Here, our key point of interest is to determine the mechanical properties of spectrin, specifically its deformation process and the failure force. With very limited literature data42 on the structure–property relations of spectrin, we have computationally constructed the structure of spectrin from its building blocks, namely from the available structural data of -chain and -chain repeats. Based on the imaging data and other studies, we then constructed the compact biological form of spectrin filaments. Next, we developed an equivalent spring model for spectrin and obtained the non-linear molecular physics-informed constitutive relations for spectrin. We proposed a progressive deformation process for spectrin and estimated the failure force assuming failure is driven by LJ-type molecular force. We compared our findings with the available experimental data.
Materials and methods
Spectrin molecular structure
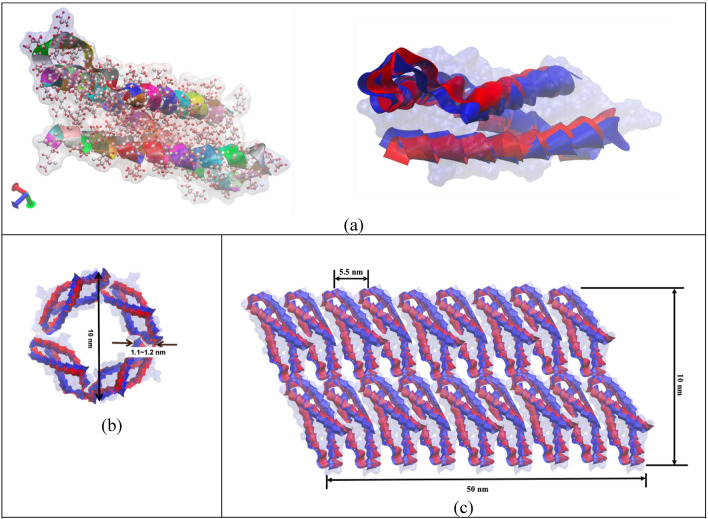
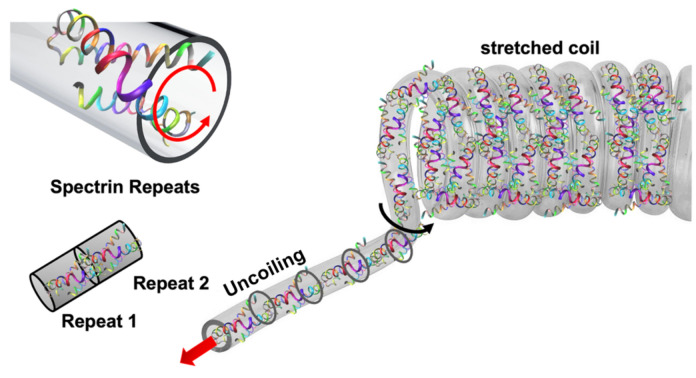
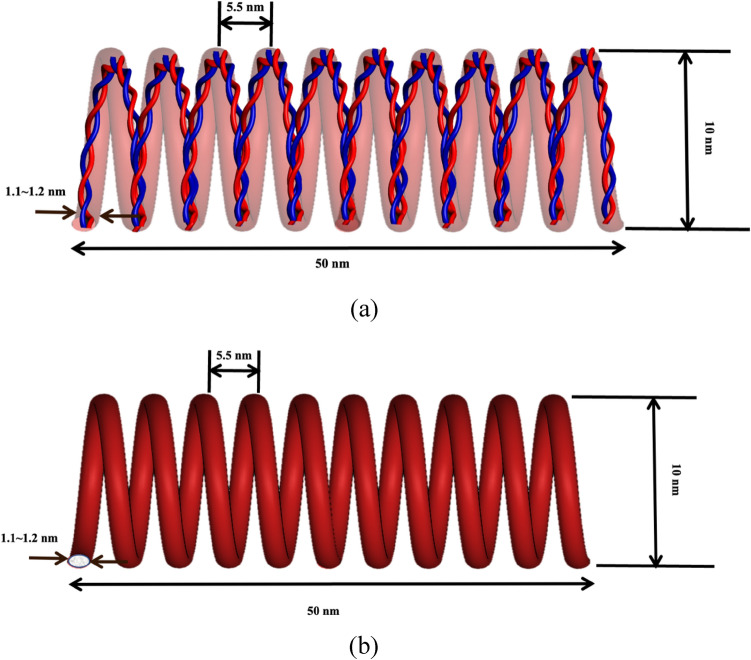
As a heterotetramer biopolymer, spectrin filament comprises of and spectrin. One spectrin is connected with one spectrin and this forms the one strand of spectrin. Similarly, another strand is parallelly formed, and these two strands then connected with each other as a twisted rope. Together, they form a spring like configuration. Structurally, each spectrin has 20 repeat units while each repeat unit is made with 106 amino acids. Each spectrin has 17 repeat units, and each repeat unit is similarly made with 106 amino acids. In one full strand of spectrin, there are about 37 repeats in total43. A spectrin filament is formed when two such strands are twisted together and then arranged to a helical spring configuration. Although there are variations of amino acids in different types of spectrin but for simplicity we have modeled our system with the most observed amino acid sequences in spectrin43 and also, we have conceptualized the idea of compact spectrin spring described in an earlier study44. As described earlier, in compact biological form, spectrin filament takes the form of a helical spring45,46that can be characterized by a pitch p = 5.5 nm, mean diameter D = 10 nm, wire diameter d ~ 1.1–1.2 nm and total free length L = 50 nm. One full turn of this spring covers mutually twisted 4 repeats of Spectrin and 4 repeats of Spectrin. Based on this spring geometry, the most common biological state of spectrin comprises of ~ 9 complete turns. The contour length of each turn is about 20 nm. So, in the maximum stretched state, the spectrin filament could be extended up to 180 ~ 190 nm47Fig. 47c shows the compact biological form representation of the spectrin filament, which is constructed from the spectrin repeat building block (Fig. 2a). Here, the right image in Fig. 2a shows the repeat in simple ribbon form, where red and blue ribbons represent the and repeats. The left image in Fig, 2a shows the molecular detail spectrin, which is obtained from RCSB Protein Data Bank (The PDB file ID is “1aj3”). This visualization has been done with the openly available molecular visualization tool VMD48. Figure 2b shows the schematic representation of one single turn of the spectrin spring formed by longitudinally connecting four repeats46. Note that each repeat contains both and spectrin molecules that are wrapped around each other in spiral form.
Figure 2.
(a) Long-chain connected amino acids form a ribbon representation of a single repeat of spectrin (left), single alpha (a) and beta (b) repeats are shown in ribbon form (b) schematic representation of one turn of the Spectrin helical coil with 4 spectrin repeats. (c) truncated schematic for the compact biological representation of single spectrin filament in helical coil form.
Progressive tensile deformation of spectrin
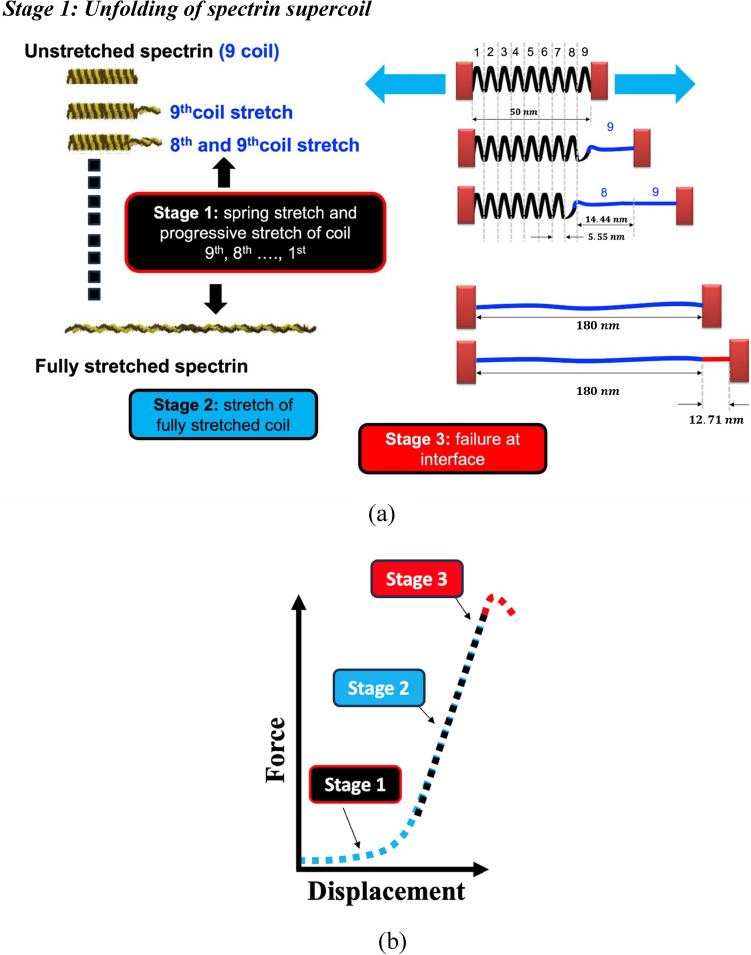
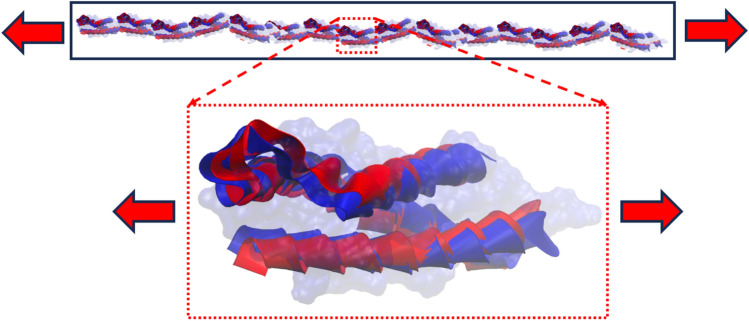
As outlined earlier, the compact biological form of spectrin filament is like a helical spring with ~ 9 full turns (also referred to as supercoil). Although there are and spectrin filaments that are innately twisted with each other, we are considering this as a single coil for the simplicity of our analysis. Now each strand of the supercoil has about 4 repeats (we are considering the repeats of spectrin), while the whole compact spring model has a total of 37 repeats. We hypothesize that upon the application of tensile force on the spectrin filament, each turn/supercoil starts to unfold one at a time from the loading end. When all 9 coils are uncoiled, then the entire uncoiled coil experiences the applied tensile force. Note that each coil is constructed by longitudinally connecting 37 repeats. At this stage, both the joints between repeats and the internal structure of the repeats respond to the applied force. Further application of force will then lead to spectrin failure, either cohesively or at the repeat-repeat interface. The proposed deformation mechanism is outlined in Fig. 3.
Figure 3.
(a) proposed progressive deformation process of spectrin filament that can be broken into three stages (b) during stage 1, each supercoil uncoils, one at a time. When all 9 supercoils are uncoiled, then the fully uncoiled spectrin filament is stretched (stage 2). This deformation is governed by the local structural response of a single repeat. Finally, failure of spectrin is observed in stage 3.
Stage 1: unfolding of spectrin supercoil
As outlined in Figs. 3 and 4, the first stage of spectrin deformation is governed by the uncoiling of spectrin filament, one at a time. After that the stretching of fully uncoiled coil will take place. Finally, the failure will be occurred at the interface connections of repeats in spectrin filament. It can be realized that the unfolding of spectrin is governed by torsional deformation of the spectrin coil.
Figure 4.
Schematic representation of biological form of Spectrin.
Helical spring model for spectrin
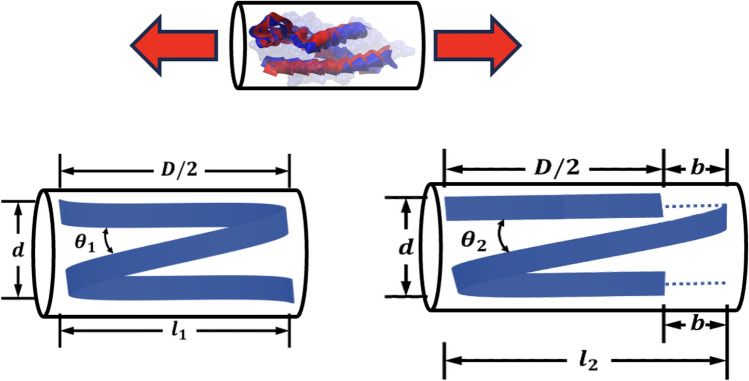
As outlined in the previous sections, spectrin filament can be considered as a spring when it is in the compact biological form. To obtain force–displacement relation for this spring, we need to replace the twisted 2-strand coil into one single coil. Figure 5 describes how the equivalent spring model is obtained. Critical to this equivalent representation is determining average coil diameter. We studied the molecular topology of spectrin coil in several regions and estimated the average coil diameter, d ~ 1.1–1.2 nm. This Details are provided in the supplementary information (Appendix A).
Figure 5.
(a) Schematic showing how the alpha (a) and beta (b) repeats are arranged to form the twisted coil in the spectrin filament spring. The average cross-sectional area coverage of this twisted pair is estimated and the corresponding spring coil diameter d ~ 1.1 − 1.2 nm is obtained. (b) Equivalent spring model with matching coil diameter, mean diameter, pitch and free length.
Considering the spring with mean dia D and coil dia d (coil area A) is subjected to a compressive force , we can take the force balance on the coil cross section using standard spring theory49.
Now, equilibrium forces at cut section anywhere in the body of the spring indicate direct shear and torsion. This implies maximum shear stress comprises of torsional shear and direct shear, as depicted in in Fig. 6.
| 1 |
Figure 6.
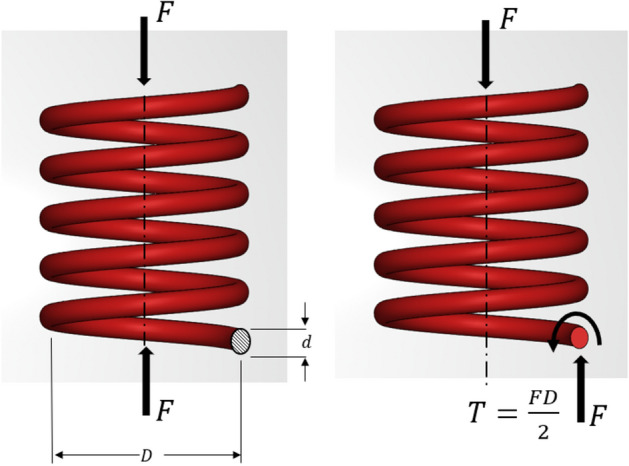
Helical spring subjected to force F.
Here, maximum developed shear stress, .
Torque developed due to applied force ,
Now, to find out the deflection of the spring, we can use Castigliano’s method to relate force and deflection. Accordingly, total strain energy, will be.
| 2 |
Here, refers to the shear modulus or modulus of rigidity.
Substituting the terms to simplify the Eq. (2),
For deflection, , we obtain.
| 3 |
If , from Eq. (3) we can simplify,
| 4 |
Here K is the effective spring constant of the helical spring. This spring constant will be applicable to uncoil each single coil of spectrin spring. The spring constant depends on the number of coils (i.e., N = 1, here always one single coil needs to be considered to get the uncoiling force), the mean diameter of the spring (D = 10 nm), and the torsional shear modulus G of the coil. As this value is not readily available, we estimated this parameter by taking the energy balance from torsional strain energy and molecular level torsional energy.
As we have got from the helical spring model in Eq. (4). Incorporating the G with this K we will get the unfolding force for each coil in the helical spring of Spectrin.
| 5 |
As such, the deformation process will be also dominated by the modulus of rigidity (G) of the supercoil, which, based on our estimation G is ~ 1.127 GPa. We will use this G value to estimate the uncoiling force during the first stage of stretching. Details for the calculation of G are provided in the supplementary information (Appendix B).
Stage 2: Tensile stretch of uncoiled coil
Once all 9 coils are uncoiled and the spectrin is fully stretched, then, as shown in Fig. 7, tensile deformation is governed by the modulus of elasticity (E) of spectrin.
Figure 7.
Force and deformation of single repeat during tension.
After the coils are being uncoiled, then each repeat of spectrin will face a fully stretching force due to tension. Similar to the previous method, here the strain energy due to stretching of the Z-shaped single repeat structure will be equated with the molecular level bond energy and angle energy experienced by spectrin repeat during tensile deformation. The parameters and will be used here respectively for bond and angle energy calculation.
Now the molecular level energy due to stretch is contributed by the bond energy and bend energy.
| 6 |
Now, from continuum mechanics standpoint, the axial strain energy can be estimated as
Now, , from the above derivation.
Now, putting all these values in Eq. (6)
| 7 |
We can define this as , The corresponding value can be estimated as:
| 8 |
We have seen that there is almost no variation of E value in this stage with respect to extension of Z shaped structure, which is obvious. So, we will have a using this E and this will be applicable for all 37 repeats.
Now, once the spectrin filament is fully uncoiled, then all 37 repeats line up in series. Like any other long chain biopolymer, the internal structure of the repeats is built on strong covalently bonded primary bonds and relatively weaker secondary and tertiary bonds. As outlined in Fig. 8, an applied tensile force on an uncoiled spectrin is felt by the 37 repeats and the 36 joints. It is known that the repeats are comprised of amino acids so we can fairly assume that these repeats are connected primarily by carbon–nitrogen bonds. The interactions between the repeats can be modeled with Morse-type50 or Lennard–Jones (LJ)51 type potential functions. Accordingly, we have estimated the average interface LJ energy and Morse Energy of the repeat-repeat interfaces. Then the corresponding interfacial spring constant (note that in our study could be obtained from either Morse or LJ potential functions) of each interface has been estimated. The spring constant for the single repeat is also estimated and referred as . Detail method on how the values for the parameters have been obtained for are discussed in the supplementary document (Appendix C and Appendix D). Detail method and value for are discussed in the supplementary document (Appendix E).To find the effective spring constant for the uncoiled spectrin, we consider that 37 repeats (each with spring constant ) and 36 interfaces (each having spring constant are connected in series. The corresponding will be:
| 9 |
Figure 8.
Stretching the completely uncoiled spectrin coil is controlled by the axial modulus of the spectrin coil.
Stage 3: bond separation and tensile failure of spectrin
As outlined in the Supplementary file (Appendix E), the fully uncoiled spectrin can be stretched up to nm. Further stretch will lead to repeat-repeat separation failure.
The failure force for the interfaces can then be estimated as:
| 10 |
Here maximum stretching is allowed after the coils of spring of spectrin is fully stretched.
We argue that is the maximum total extension of the uncoiled spectrin repeat-repeat junction. For each repeat-repeat junction, the possible extension is the distance between the Van der Waals/Morse bond separation length and equilibrium carbon–nitrogen (C-N) bond length.
Results and discussions
As outlined in "Materials and Methods" section, spectrin goes through 2 stages of tensile deformation before it separates at the repeat-repat interfaces. Here, based on the proposed models, we estimate the value of the parameters necessary for the development of the constitutive relation for spectrin deformation.
Initial deformation and uncoiling force for coils of helical spring model
For Eq. (4), . To get the uncoiling force of a single coil while subjected to stretching, we need to accumulate these in Eq. (4), from where we will get the K for a single repeat uncoiling.
Finally accumulating this in Eq. (5), we will get the uncoiling force a single coil. For Eq. (5), as outlined earlier,
Putting all these values in Eq. (4), we have got the .
As each of the coils is the same in the helical spring form of the spectrin filament, it is assumed that every coil will need this same force to uncoil. After a coil is fully uncoiled, it comes to rest, and then the next coil starts to uncoil, as schematically shown in Fig. 3.
Stretching of fully uncoiled spectrin and the failure force
As outlined earlier, the tensile deformation of the fully uncoiled spectrin is dependent upon the parameter and . To get the value of , we need the value of as outlined in Eq. (8). Putting and values from Supplementary File (Appendix D) in Eq. (7) we will obtain the value for the
The variation of E with stretch of single repeat is shown in Fig. 9. It is observed that E doesn’t vary significantly with the stretch of single repeat. As such, we can take a single value of as presented in Fig. 9. From here we are considering,
Figure 9.
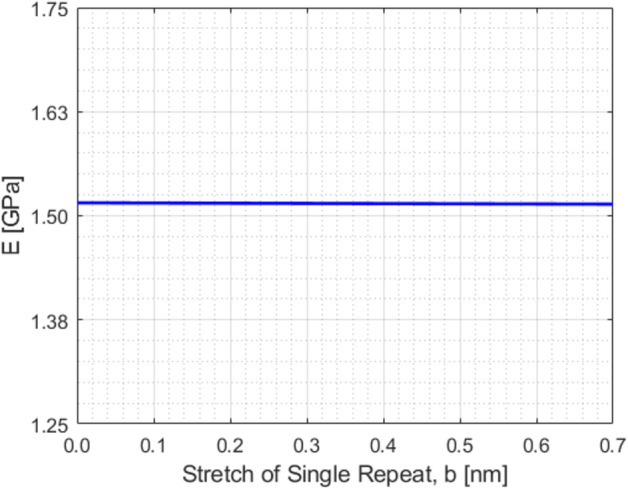
E value estimation.
Putting all the values in Eq. (8) we get, .
Now, we need to calculate the , which can be obtained through the Lennard Jones/Morse potential profile for the Carbon–Nitrogen bond. The details of this calculation about is in the supplementary file (Appendix E). Note that = if the LJ potential is used. Similarly, = if the Morse potential is used. The corresponding value will be also changed. Also, the corresponding total force has been denoted as and consecutively for using LJ-type potential and Morse-type potential.
Now, Using the LJ potential parameters in Eq. (9), we get,
Now, putting the values of and in Eq. (10), we get
So, the total failure force will be .
Using the and instead of and in Eqs. (9) described above, we get,
It should be observed that both approaches predict nearly the same total failure force.
Although LJ potentials are primarily used for describing non-bonded interactions, such potential is also employed to model covalent bonds in some complex molecular systems51. It is slightly less precise for these systems because it does not account for the environmental influences on bonding, but the depth of LJ potential is strongly related to the covalent bond strength within complex molecular systems. It is found from our results that the LJ potential doesn’t bring any significant difference in estimating the failure force values compared to the Morse potential. This is because stage 3 of our model is to approximate a critical separation length for complexly connected spectrin repeats.
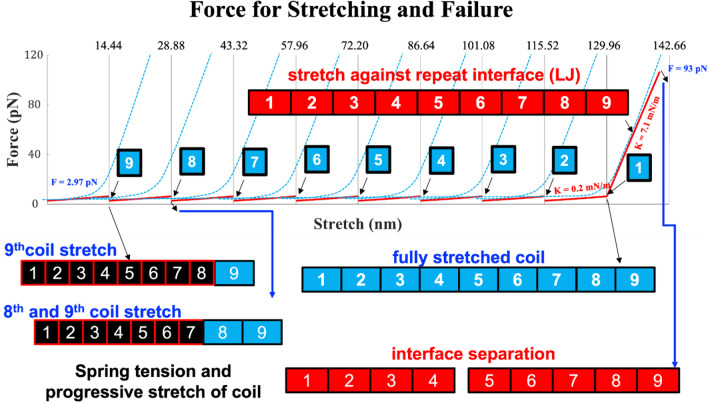
This study has introduced a comprehensive theoretical model to estimate the failure force of spectrin filament. These calculations are based on a three-stage deformation and damage mechanism of spectrin filament that includes the progressive unfolding of the compact biological form of spectrin, stretching of the fully uncoiled filament, and then estimating the failure force of spectrin. This force vs stretching data is plotted and shown in Fig. 10. Through our model, we have estimated the probable failure force of spectrin filament to be . Interestingly, a previous AFM experimental study on spectrin filament reported a potential value for spectrin uncoiling and failure about at 90 pN52.
Figure 10.
Force vs Stretching combined representation.
It is encouraging that our model is closely aligned with the reported experimental results, as shown in Fig. 11. However, it must be considered that the predicted failure force of spectrin, both by our model and by the experiments, may deviate from the actual value due to its reliance on the specific types of amino acids comprising spectrin repeats and the potential complexity of these amino acid chains. In addition, both theory and experiments require making certain assumptions. Nevertheless, despite these limitations, our approach offers a simplified and more accessible method for further advancing the study. Previous studies, such as42, have attempted to determine the modulus of elasticity of spectrin and actin through molecular dynamics (MD) simulations. However, these studies faced significant challenges in accurately modeling the spectrin structure and the associated time-intensive and computationally demanding processes. Furthermore, these studies primarily focused on the constitutive behavior of spectrin filament, ignoring properties like failure force. In contrast to the extensive research on actin filament, where studies have reported on its strain and strain rate responses, research on spectrin filament remains remarkably limited, particularly concerning its failure force. In a nutshell, the previous findings focused on the actin-binding domain, which was mostly a primary-level study to address the simple mechanical property (modulus of elasticity) based on some predicted protein structure through molecular dynamics simulation. No unfolding and failure forces could be extracted from the work. However, these properties are essential if someone wants to develop a multi-scale model to determine injury risks of dendrites where failure properties at the sub cellular level (i.e. spectrin properties and the like) are incorporated in the full-scale finite element modeling of dendrites. In our present theoretical model, we have attempted to embody a very comprehensive and extensive mechanics-based model that will not only predict the uncoiling and failure force of spectrin but also explain the rationale behind the mechanism by keeping the utmost alignment with the biological details of spectrin. So, it can be stated that the previous findings have motivated us to take a path for a more complicated and novel theoretical approach to achieve our desired parameters by avoiding the complex, time-consuming simulation works. As spectrin is a major component of dendrites, understanding the failure force range of spectrin could eventually help predict dendritic failure.
Figure 11.
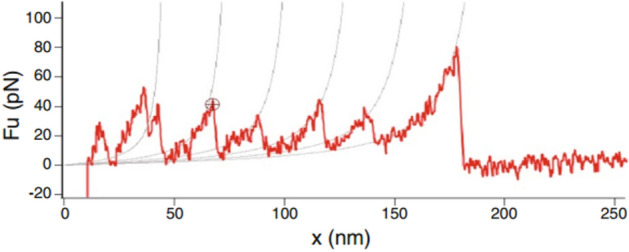
AFM experiment of Spectrin repeats unfolding52.
Conclusion
This study has introduced a new approach to theoretically analyze the mechanical characteristics of a primary component found in neuron cell dendrites. Considering molecular-level interactions, this study has laid the groundwork for predicting the failure force of Spectrin. While previous studies have mainly focused on axons, understanding, and predicting neuron cell injuries requires holistic determination of the injury thresholds for dendrites and other components as well. In a broader context, our work is paving the way for predicting dendritic failures by examining its crucial structural element, the spectrin. Additionally, this approach has the potential to predict the mechanical properties of structurally similar biopolymers, eliminating the need for extensive simulations and complex experimental efforts generally associated with such tasks. Although our proposed work is based on a few assumptions that are currently speculative, the methodology offers an opportunity for further research studies to validate them through experimental or simulation-based approaches.
Supplementary Information
Acknowledgements
This study is supported by the PANTHER program (URL: https://www.panther.engr.wisc.edu/; director: Dr. Christian Franck, University of Wisconsin Madison) through the U.S. Office of Naval Research (ONR) award number N00014-21-1-2855(0000001556) award—Program Director Dr. Timothy Bentley.
Author contributions
Md Nahian Bin Hossain contributed to the hypothesis, methodology, results, and analysis. Dr. Ashfaq Adnan contributed to concept generation, validation, writing review and editing, and funding possession.
Data availability
We affirm all data analyzed during this study are included in this main article and its supplementary information file. For further details on the datasets analyzed during the current study, the corresponding author (Dr. Ashfaq Adnan—aadnan@uta.edu) can be contacted.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-67500-0.
References
- 1.Zhang L, Yang KH, King AI. A proposed injury threshold for mild traumatic brain injury. J. Biomech. Eng. 2004;126(2):226–236. doi: 10.1115/1.1691446. [DOI] [PubMed] [Google Scholar]
- 2.Levitan IB, Kaczmarek LK. The Neuron. Oxford: Oxford University Press; 2015. [Google Scholar]
- 3.Bagnard D, editor. Axon Growth and Guidance. New York: Springer; 2007. [DOI] [PubMed] [Google Scholar]
- 4.Baas PW, Rao AN, Matamoros AJ, Leo L. Stability properties of neuronal microtubules. Cytoskeleton. 2016;73(9):442–460. doi: 10.1002/cm.21286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holzbaur ELF, Scherer SS. Microtubules, axonal transport, and neuropathy. N. Engl. J. Med. 2011;365(24):2330–2332. doi: 10.1056/NEJMcibr1112481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wagner O, Zinke J, Dancker P, Grill W, Bereiter-Hahn J. Viscoelastic properties of f-actin, microtubules, f-actin/α-actinin, and f-actin/hexokinase determined in microliter volumes with a novel nondestructive method. Biophys. J. 1999;76(5):2784–2796. doi: 10.1016/S0006-3495(99)77432-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soheilypour M, Peyro M, Peter SJ, Mofrad MRK. Buckling behavior of individual and bundled microtubules. Biophys. J. 2015;108(7):1718–1726. doi: 10.1016/j.bpj.2015.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Rooij R, Kuhl E, Miller KE. Modeling the axon as an active partner with the growth cone in axonal elongation. Biophys. J. 2018;115(9):1783–1795. doi: 10.1016/j.bpj.2018.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rattay F. Analysis of models for external stimulation of axons. IEEE Trans. Biomed. Eng. 1986;BME-33(10):974–977. doi: 10.1109/TBME.1986.325670. [DOI] [PubMed] [Google Scholar]
- 10.Bernal R, Pullarkat PA, Melo F. Mechanical properties of axons. Phys. Rev. Lett. 2007;99(1):018301. doi: 10.1103/PhysRevLett.99.018301. [DOI] [PubMed] [Google Scholar]
- 11.Hasan F, Al Mahmud K, Khan MdI, Adnan A. Viscoelastic damage evaluation of the axon. Front. Bioeng. Biotechnol. 2022 doi: 10.3389/fbioe.2022.904818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stuart G, Spruston N, Häusser M, editors. Dendrites. Oxford: Oxford University Press; 2007. [Google Scholar]
- 13.Konietzny A, Bär J, Mikhaylova M. Dendritic actin cytoskeleton: structure, functions, and regulations. Front. Cell Neurosci. 2017 doi: 10.3389/fncel.2017.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dharani K, Physiology of the neuron. In The Biology of Thought, 31–52 (Elsevier, Amsterdam). 10.1016/B978-0-12-800900-0.00002-6 (2015).
- 15.Li B-Z, Sumera A, Booker SA, McCullagh EA. Current best practices for analysis of dendritic spine morphology and number in neurodevelopmental disorder research. ACS Chem. Neurosci. 2023;14(9):1561–1572. doi: 10.1021/acschemneuro.3c00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pchitskaya E, Bezprozvanny I. Dendritic spines shape analysis—classification or clusterization? Perspective. Front. Synaptic Neurosci. 2020 doi: 10.3389/fnsyn.2020.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lorenzo DN, Edwards RJ, Slavutsky AL. Spectrins: molecular organizers and targets of neurological disorders. Nat. Rev. Neurosci. 2023;24(4):195–212. doi: 10.1038/s41583-022-00674-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sidenstein SC, D’Este E, Böhm MJ, Danzl JG, Belov VN, Hell SW. Multicolour multilevel STED nanoscopy of actin/spectrin organization at synapses. Sci. Rep. 2016;6(1):26725. doi: 10.1038/srep26725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D’Este E, Kamin D, Göttfert F, El-Hady A, Hell SW. STED nanoscopy reveals the ubiquity of subcortical cytoskeleton periodicity in living neurons. Cell Rep. 2015;10(8):1246–1251. doi: 10.1016/j.celrep.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 20.Xu K, Zhong G, Zhuang X. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science (1979) 2013;339(6118):452–456. doi: 10.1126/science.1232251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhong G, et al. Developmental mechanism of the periodic membrane skeleton in axons. Elife. 2014 doi: 10.7554/eLife.04581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Abiraman K, Li H, Pierce DM, Tzingounis AV, Lykotrafitis G. Modeling of the axon membrane skeleton structure and implications for its mechanical properties. PLoS Comput. Biol. 2017;13(2):e1005407. doi: 10.1371/journal.pcbi.1005407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chai Z, Gu S, Lykotrafitis G. Dynamics of the axon plasma membrane skeleton. Soft. Matter. 2023;19(14):2514–2528. doi: 10.1039/D2SM01602H. [DOI] [PubMed] [Google Scholar]
- 24.Das A, Dubreuil RR. Spectrin: organization and function in neurons. Encycl. Neurosci. 2009 doi: 10.1016/B978-008045046-9.00731-2. [DOI] [Google Scholar]
- 25.Liu C-H, Rasband MN. Axonal spectrins: nanoscale organization, functional domains and spectrinopathies. Front. Cell Neurosci. 2019 doi: 10.3389/fncel.2019.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu C-H, et al. β spectrin-dependent and domain specific mechanisms for Na+ channel clustering. Elife. 2020 doi: 10.7554/eLife.56629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dubey S, et al. The axonal actin-spectrin lattice acts as a tension buffering shock absorber. Elife. 2020 doi: 10.7554/eLife.51772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bennett V, Davis J, Fowler WE. Brain spectrin, a membrane-associated protein related in structure and function to erythrocyte spectrin. Nature. 1982;299(5879):126–131. doi: 10.1038/299126a0. [DOI] [PubMed] [Google Scholar]
- 29.Holley MC, Ashmore JF. Spectrin, actin and the structure of the cortical lattice in mammalian cochlear outer hair cells. J. Cell Sci. 1990;96(2):283–291. doi: 10.1242/jcs.96.2.283. [DOI] [PubMed] [Google Scholar]
- 30.Liem RKH. Cytoskeletal integrators: the spectrin superfamily. Cold Spring Harb. Perspect. Biol. 2016;8(10):a018259. doi: 10.1101/cshperspect.a018259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gardner K, Bennett V. Modulation of spectrin–actin assembly by erythrocyte adducin. Nature. 1987;328(6128):359–362. doi: 10.1038/328359a0. [DOI] [PubMed] [Google Scholar]
- 32.Liu SC, Derick LH, Palek J. Visualization of the hexagonal lattice in the erythrocyte membrane skeleton. J. Cell Biol. 1987;104(3):527–536. doi: 10.1083/jcb.104.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ford C. Principles of medical biochemistry (3rd edn) Ann. Clin. Biochem. Int. J. Lab. Med. 2012;49(6):613–613. doi: 10.1258/acb.2012.201216. [DOI] [Google Scholar]
- 34.Black JD, Koury ST, Bankert RB, Repasky EA. Heterogeneity in lymphocyte spectrin distribution: ultrastructural identification of a new spectrin-rich cytoplasmic structure. J. Cell Biol. 1988;106(1):97–109. doi: 10.1083/jcb.106.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stokke BT, Mikkelsen A, Elgsaeter A. Spectrin, human erythrocyte shapes, and mechanochemical properties. Biophys. J. 1986;49(1):319–327. doi: 10.1016/S0006-3495(86)83644-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scott KA, Batey S, Hooton KA, Clarke J. The folding of spectrin domains i: wild-type domains have the same stability but very different kinetic properties. J. Mol. Biol. 2004;344(1):195–205. doi: 10.1016/j.jmb.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 37.Ralston GB. Physical-chemical studies of spectrin. J. Supramol. Struct. 1978;8(3):361–373. doi: 10.1002/jss.400080313. [DOI] [PubMed] [Google Scholar]
- 38.Calvert R, Bennett P, Gratzer W. Properties and structural role of the subunits of human spectrin. Eur. J. Biochem. 2005;107(2):355–361. doi: 10.1111/j.1432-1033.1980.tb06036.x. [DOI] [PubMed] [Google Scholar]
- 39.Burns NR, Ohanian V, Gratzer WB. Properties of brain spectrin (fodrin) FEBS Lett. 1983;153(1):165–168. doi: 10.1016/0014-5793(83)80140-9. [DOI] [PubMed] [Google Scholar]
- 40.Djinovic-Carugo K, Gautel M, Ylänne J, Young P. The spectrin repeat: a structural platform for cytoskeletal protein assemblies. FEBS Lett. 2002;513(1):119–123. doi: 10.1016/S0014-5793(01)03304-X. [DOI] [PubMed] [Google Scholar]
- 41.Kirkpatrick FH. Spectrin: current understanding of its physical, biochemical, and functional properties. Life Sci. 1976;19(1):1–17. doi: 10.1016/0024-3205(76)90368-4. [DOI] [PubMed] [Google Scholar]
- 42.Khan MI, Ferdous SF, Adnan A. Mechanical behavior of actin and spectrin subjected to high strain rate: a molecular dynamics simulation study. Comput. Struct. Biotechnol. J. 2021;19:1738–1749. doi: 10.1016/j.csbj.2021.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lusitani DM, et al. The first human alpha-spectrin structural domain begins with serine. J. Biol. Chem. 1994;269(42):25955–25958. doi: 10.1016/S0021-9258(18)47141-4. [DOI] [PubMed] [Google Scholar]
- 44.McGough AM, Josephs R. On the structure of erythrocyte spectrin in partially expanded membrane skeletons. Proc. Natl. Acad. Sci. 1990;87(13):5208–5212. doi: 10.1073/pnas.87.13.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, Lykotrafitis G. Erythrocyte membrane model with explicit description of the lipid bilayer and the spectrin network. Biophys J. 2014;107(3):642–653. doi: 10.1016/j.bpj.2014.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown JW, Bullitt E, Sriswasdi S, Harper S, Speicher DW, McKnight CJ. The physiological molecular shape of spectrin: a compact supercoil resembling a Chinese finger trap. PLoS Comput. Biol. 2015;11(6):e1004302. doi: 10.1371/journal.pcbi.1004302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nans A, Mohandas N, Stokes DL. Native ultrastructure of the red cell cytoskeleton by cryo-electron tomography. Biophys. J. 2011;101(10):2341–2350. doi: 10.1016/j.bpj.2011.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 49.Keith Nisbett J, Budynas RG. Shigley’s Mechanical Engineering Design. 10. New York: McGraw-Hill; 2015. [Google Scholar]
- 50.Plastino A, Plastino AR. Inference approach to ground states of quantum systems. Handb. Stat. 2022 doi: 10.1016/bs.host.2022.07.002. [DOI] [Google Scholar]
- 51.Adams JB. Bonding energy models. Encycl. Mater. Sci. Technol. 2001 doi: 10.1016/B0-08-043152-6/00146-7. [DOI] [Google Scholar]
- 52.Renn JP, et al. Mechanical unfolding of spectrin reveals a super-exponential dependence of unfolding rate on force. Sci. Rep. 2019;9(1):11101. doi: 10.1038/s41598-019-46525-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We affirm all data analyzed during this study are included in this main article and its supplementary information file. For further details on the datasets analyzed during the current study, the corresponding author (Dr. Ashfaq Adnan—aadnan@uta.edu) can be contacted.