Abstract
We previously reported on the in vivo adaptation of an infectious molecular simian/human immunodeficiency virus (SHIV) clone, SHIVSF33, into a pathogenic biologic viral variant, designated SHIVSF33A. In the present study, we show that SHIVSF33A is resistant to neutralization by human immunodeficiency virus (HIV) and SHIV antisera. Multiple amino acid substitutions accumulated over time throughout the env gene of SHIVSF33A; some of them coincided with the acquisition of the neutralization resistance of the virus. Of interest are changes that resulted in the removal, repositioning, and addition of potential glycosylation sites within the V1, V2, and V3 regions of envelope gp120. To determine whether potential glycosylation changes within these principal neutralization domains of HIV type 1 formed the basis for the resistance to serum neutralization of SHIVSF33A, mutant viruses were generated on the backbone of parental SHIVSF33 and tested for their neutralization sensitivity. The mutations generated did not alter the in vitro replication kinetics or cytopathicity of the mutant viruses in T-cell lines. However, the removal of a potential glycosylation site in the V1 domain or the creation of such a site in the V3 domain did allow the virus to escape serum neutralization antibodies that recognized parental SHIVSF33. The combination of the V1 and V3 mutations conferred an additive effect on neutralization resistance over that of the single mutations. Taken together, these data suggest that (i) SHIV variants with changes in the Env SU can be selected in vivo primarily by virtue of their ability to escape neutralizing antibody recognition and (ii) carbohydrates play an important role in conferring neutralization escape, possibly by altering the structure of envelope gp120 or by shielding principal neutralization sites.
Viral diversity is a hallmark of human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) infections. The ability of these viruses to continually evolve in the host may contribute to their ability to persist in an individual despite an active specific immune response against them. Accordingly, characterizing virus variants that evolve during the course of infection and establishing the basis for their selection within the host should provide insight into viral persistence and hence pathogenesis and assist in the design of therapeutic approaches.
Phenotypic and immunologic variants have been reported to emerge over the course of both HIV and SIV infections (for reviews, see references 9, 25, 34, and 38). Indeed, variants resistant to neutralization by autologous sera can be detected in vivo and can also be generated by prolonged culturing in the presence of neutralizing antibodies in vitro (1, 2, 8, 14, 23, 30, 33, 36, 42, 45, 56). The majority of neutralizing antibodies present in sera from individuals infected with HIV type 1 (HIV-1) or immunized with recombinant HIV-1 proteins or in experimentally infected animals are directed either to the V3 loop of envelope gp120 or to epitopes overlapping the CD4-binding site of gp120 (10, 15). For SIV, the V1 and V4 domains appear to contain the principal neutralizing determinants (9, 46, 47). It is generally accepted that anti-V3 loop antibodies are type or sequence specific, whereas anti-CD4-binding-site antibodies are broadly cross-neutralizing (11, 41, 54, 55). Neutralization resistance can be acquired either directly by a point mutation within the antibody-binding site that reduces or abrogates the binding of the antibody or indirectly by a point mutation elsewhere in the envelope gene that alters the conformation of the antibody-binding site (4, 30, 33, 42, 53, 59). Resistance can also be conferred by epitope masking. In this regard, N-glycans have been shown to play a critical role in the shielding of neutralizing epitopes of both HIV-1 and SIV (3, 14, 20, 47, 49). Furthermore, carbohydrate side chains have been reported to modulate immune responses (5, 6, 44) and to play a role in maintaining the proper expression and function of envelope gp120 (17, 21, 27, 31, 37, 40, 60).
Although a temporal relationship between sequence changes in the extracellular envelope glycoprotein and neutralization sensitivity has been demonstrated for viruses that evolve during the natural course of SIV infection (8, 14, 39, 47), similar studies have not been reported for HIV-1. Toward this end, we examined temporal changes in the sequence and immunological properties of the HIV-1 env gene in viruses that evolve during the course of simian/human immunodeficiency virus (SHIV) infection of macaques. SHIVs are chimeric viruses constructed between molecular clones of SIVmac and various strains of HIV-1 (38). These chimeras contain an HIV-1 DNA fragment carrying the tat, rev, vpu, and env genes cloned into the genome of the proviral form of pathogenic SIVmac239 (26, 29, 48, 50). We previously infected four juvenile macaques with SHIVSF33 (29). One of these four macaques (Mnu25814) exhibited an increase in virus load at about 16 months after infection (Table 1) (28, 29) concomitant with a decline in the level of CD4+ T cells and the development of simian AIDS. Virus recovered from this animal in the symptomatic stage (i.e., 104 weeks postinfection), designated SHIVSF33A, caused fatal immunodeficiency in juvenile and infant rhesus macaques. In vitro, the SHIVSF33A biologic isolate displayed growth and cytopathicity properties that differed from those of the parental SHIVSF33 molecular clone (28).
TABLE 1.
Viral load over time in Mnu25814a
| Wk postinfection | Cell-associated virus load (TCID50) per 106 PBMC |
|---|---|
| 2 | 10,000 |
| 4 | 470 |
| 8 | 100 |
| 32 | 10 |
| 52 | 1 |
| 72 | 47 |
| 91 | 4,700 |
| 104 | 100 |
See text for details.
In the present study, we show that in contrast to the parental SHIVSF33 clone, SHIVSF33A is resistant to neutralization by HIV antisera and autologous SHIV antisera. The evolution of SHIVSF33 into SHIVSF33A in the infected host therefore provides a system to assess the temporal relationship between specific sequence changes in the HIV-1 envelope that are selected for over time and the establishment of neutralization resistance in vivo. We find that sequence changes that modulate potential N-linked glycosylation of the HIV-1 envelope are selected for within the infected host and play an important role in conferring escape from immune recognition.
MATERIALS AND METHODS
Cells and virus.
RhPBMC (rhesus peripheral blood mononuclear cells [PBMC]) were obtained from healthy rhesus macaques free of simian type D retroviruses, SIV, and simian T-lymphotropic virus by Ficoll gradient centrifugation (lymphocyte separation medium; BioWhittaker, Walkersville, Md.). Purified cells were stimulated with 5 μg of staphylococcal entertoxin B (Sigma Biochemicals) per ml for 72 h and propagated in RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum, l-glutamine, penicillin, streptomycin, and 10 U of recombinant interleukin 2 (Hoffmann-La Roche) per ml. CEMX174 cells, a human hybrid T-B cell line provided by J. Hoxie (University of Pennsylvania, Philadelphia), were maintained in RPMI 1640 medium supplemented with 10% and antibiotics. Cell-associated virus load was determined by coculturing 106 PBMC (and serial 1:10 dilutions thereof) from Mnu25814 with 2.5 × 105 CEMX174 cells per well, with four wells per dilution. Titers were calculated by determining the numbers of infected PBMC per 106 total PBMC. SHIV variants were recovered over time from infected animal Mnu25814 by cocultivation of PBMC with CEMX174 cells (29). Stocks of cell-free SHIV (SHIVSF33, SHIVSF33A, and glycosylation mutant viruses) were prepared by passage in CEMX174 cells. Culture supernatants were collected at 7 to 10 days postinfection, passed through a 0.45-μm-pore-size filter, and frozen in 1-ml aliquots. The 50% tissue culture infective doses (TCID50) of these viruses in CEMX174 cells were determined as described previously (32).
In vitro viral infections.
For in vitro infection studies, 2 × 106 RhPBMC and 105 CEMX174 cells were infected with 100 TCID50 of each virus for 3 h at 37°C. The viral inocula were removed, and cells were washed twice in Hanks’ buffered saline solution (HBSS) and maintained in culture media as described above. At various times postinfection, p27 antigen production in culture supernatants was determined by the RETRO-TEK SIV type 1 p27gag antigen enzyme-linked immunosorbent assay (Cellular Products Inc.) according to the manufacturer’s instructions.
PCR and sequencing of viral DNA.
Viral DNA sequences containing the HIV-1 env gene were amplified from Mnu25814 PBMC by a nested PCR with ED3 and ED14 as first-round primers and ED5 and ED12 as second-round primers as described previously (22). The amplified products were cloned into the TA vector (Invitrogen, Carlsbad, Calif.), and the env clone sequences were determined with [α-33P]ATP and the AmpliCycle sequencing kit (Perkin-Elmer) according to the manufacturer’s instructions. The consensus sequence for PCR products was obtained by direct sequencing of PCR products.
Neutralization assay.
Neutralization experiments were performed with CEMX174 cells in 96-well plates as previously described (16, 52). Briefly, serum samples from HIV-infected individuals and SHIVSF33-infected macaque Mnu25814 were heat inactivated (56°C, 30 min). A 50-μl serial dilution of each serum sample was incubated in triplicate wells with an equal volume of each virus (100 TCID50) for 1 hour at room temperature. Subsequently, 2 × 104 cells in a 100-μl volume of medium were added to the virus-serum mixtures, and incubation was continued at 37°C for an additional 3 h. At the end of this incubation period, the cells were washed three times with HBSS and resuspended in 200 μl of culture medium. Control cultures received virus incubated with preimmune sera or in the absence of antisera. Culture supernatants were assayed for p27 antigen production at 7 days postinfection. A neutralization curve was generated by plotting the percent neutralization versus the serum dilution. The dilution of antiserum that resulted in 90% inhibition (IC90) was then extrapolated from this curve.
Generation of glycosylation mutants.
Site-directed mutagenesis to alter the potential glycosylation sites in the V1, V2, and V3 domains of the HIV-1 env gene in the 3′ genomic fragment of SHIVSF33 was performed with a Quick-Change mutagenesis kit according to the manufacturer’s instructions (Stratagene, San Diego, Calif.). The presence of the mutation was confirmed by DNA sequencing. Mutant viruses were recovered by cotransfection of the SphI-linearized mutagenized 3′ SHIVSF33 proviral DNA together with the 5′ SHIVSF33 proviral DNA into 293T cells as described previously (29), followed by cocultivation with CEMX174 cells. In most cases, two independent clones of each mutated envelope were obtained and characterized to ensure that spontaneous mutations distant from the desired mutation were not responsible for the observed phenotype.
RESULTS
SHIVSF33A is resistant to serum neutralization.
We previously reported that in vitro, the pathogenic SHIVSF33A isolate recovered from Mnu25814 in the symptomatic stage (104 weeks postinfection) replicated more efficiently and exhibited greater cytopathicity than the parental molecular clone SHIVSF33 (28). To determine whether SHIVSF33A had also changed antigenically, the ability of (i) a pool of sera from HIV-1-infected individuals and (ii) sera collected over time from Mnu25814 to neutralize SHIVSF33A was evaluated and compared to the results for SHIVSF33. We found that whereas SHIVSF33 was highly sensitive to neutralization by both HIV-1-positive sera and sera collected from Mnu25814 at 32 weeks postinfection and thereafter, the pathogenic variant was resistant (Table 2). To assess when neutralization escape was established, isolates recovered over time from Mnu25814 were examined for their neutralization sensitivity. We observed that viruses recovered at 32 and 52 weeks postinfection were still sensitive to serum neutralization. Virus recovered at 72 weeks postinfection exhibited enhanced neutralization resistance, and by 96 weeks postinfection, resistance was fully established (Table 3).
TABLE 2.
Neutralization of SHIVSF33 and SHIVSF33A by autologous SHIV and heterologous HIV-1 seraa
| Sera | IC90 for:
|
|
|---|---|---|
| SHIVSF33 | SHIVSF33A | |
| Autologous SHIV at wk: | ||
| 12 | <1:20 | <1:20 |
| 32 | 1:50 | <1:20 |
| 52 | 1:500 | <1:20 |
| 72 | 1:1,000 | <1:20 |
| 96 | 1:1,000 | <1:20 |
| 104 | 1:500 | <1:20 |
| Polyclonal HIV-1 | 1:500 | <1:20 |
Neutralization of SHIVSF33 and SHIVSF33A by sera collected over time from Mnu25814 and by a pool of sera collected from HIV-1-infected individuals was determined as described in the text, and the IC90 was then determined. Data represent one of three independent neutralization experiments.
TABLE 3.
Neutralization of sequential isolates by sera collected over time from Mnu25814a
| Wk of serum collection | IC90 for isolates from wk:
|
|||
|---|---|---|---|---|
| 32 | 52 | 72 | 96 | |
| 12 | <1:20 | <1:20 | <1:20 | <1:20 |
| 32 | 1:50 | 1:50 | <1:20 | <1:20 |
| 52 | 1:500 | 1:500 | <1:20 | <1:20 |
| 72 | 1:500 | 1:500 | <1:20 | <1:20 |
| 96 | 1:1,000 | 1:500 | 1:50 | <1:20 |
| 104 | 1:500 | 1:250 | 1:50 | <1:20 |
Sequential isolates recovered from Mnu25814 were propagated in CEMX174 cells, their titers were determined, and they were subjected to neutralization by autologous sera collected over time as described in Materials and Methods. The IC90 was then determined. Data represent one of two independent studies.
Sequence changes in gp120 of SHIVSF33 over time.
In an attempt to identify the genetic basis for the change in the neutralization sensitivity of SHIVSF33A, the sequences of the V1, V2, and V3 regions of envelope gp120 of viral variants present in Mnu25814 over time were determined. These regions were selected since they have been shown to contain or modulate major neutralization target sites of HIV-1 (11). The results in Fig. 1 show only a minor change in the sequences of these regions for variants present at 52 and 72 weeks postinfection. By 91 weeks postinfection, however, considerable amino acid substitutions had accumulated in all three regions examined, and these changes were maintained at later times. This increase in sequence diversity between 72 and 91 weeks postinfection parallels, in addition to the change in the neutralization sensitivity of the virus (Table 3), increases in the viral load of Mnu25814 (Table 1) and the pathogenicity of the virus (28). Of interest are amino acid substitutions in all three regions that alter potential glycosylation sites. In the V1 region, an asparagine (N)-to-histidine (H) change at the base of the loop abolishes a potential glycosylation site. In the V2 region, an N-to-serine (S) change repositions a potential glycosylation site. An arginine (R)-to-threonine (T) change at the N terminus of the V3 loop creates a potential glycosylation site at this position of the V3 loop. Since carbohydrate side chains have been reported to modulate immune responses and to play a role in immune evasion, mutant viruses were generated to assess whether these potential glycosylation changes contribute to the ability of SHIVSF33A to escape antibody recognition.
FIG. 1.
Amino acid alignment of the V1, V2, and V3 domains of SHIVSF33, SHIVSF33A, and sequential isolates obtained from Mnu25814. Viral DNA sequences encoding the V1 to V5 regions of the HIV-1 env gene were amplified by nested PCR. The amplified products were cloned, and the predicted amino acid sequences of the V1, V2, and V3 domains of the variants were determined and compared to the corresponding sequences of the reference SHIVSF33 clone (33WT). The numbers in parentheses represent the numbers of clones displaying the indicated sequence divided by the total number of clones sequenced; the consensus sequence of the SHIVSF33A isolate (33A) is provided for comparison. Dots are used to indicate identity, and overlining denotes the positions of glycosylation changes within the various domains. 52wk, 72wk, 91wk, and 96wk represent the variants present in Mnu25814 at 52, 72, 91, and 96 weeks postinfection, respectively.
Changes in potential glycosylation sites do not alter the replication kinetics or cytopathicity of SHIVSF33.
Glycosylation mutants were generated on the genomic backbone of the parental clone SHIVSF33 by introducing N-to-H (N/H), N/S, and NR/YT substitutions into the V1, V2 and V3 domains, respectively (Fig. 1). Combinations of glycosylation mutations in the V1 and V2 (V1V2), V1 and V3 (V1V3), V2 and V3 (V2V3), and V1, V2, and V3 (V123) domains were also generated. In addition, since the NR/YT amino acid substitution in the V3 domain involved a −1 charge change in addition to generating a potential glycosylation site, additional V3 mutants were generated to address specifically the role of glycosylation. These V3 mutants contained R/T and NR/YA substitutions within the loop. The former substitution creates a potential N-linked site, and the latter is isogenic for the NR/YT substitution, except for the R/A substitution, which results in only a charge change.
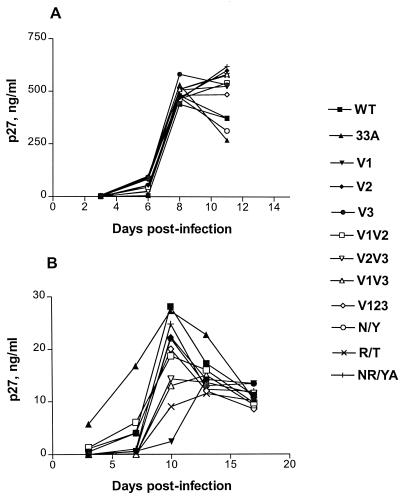
The abilities of the mutant viruses to replicate in RhPBMC and CEMX174 cells were examined and compared to that of parental SHIVSF33. We observed that, relative to SHIVSF33, the mutant viruses replicated with similar kinetics and to comparable titers in CEMX174 cells (Fig. 2A). The degree of cytopathicity induced by these mutant viruses was also similar to that induced by the wild-type virus (data not shown). In contrast, differences were seen for replication in RhPBMC (Fig. 2B). In agreement with a previous report, relative to SHIVSF33, SHIVSF33A replicated faster but to similar titers in this cell type (28). The kinetics of replication of the mutant viruses were comparable to those of wild-type SHIVSF33, but different levels of virus production were observed, with the V1 mutant virus replicating to the lowest titer and with the slowest kinetics. Nevertheless, these phenotypes of the V1 mutant virus depended on the batch of RhPBMC used (data not shown).
FIG. 2.
Replication of SHIVSF33 (WT), SHIVSF33A (33A), and glycosylation mutants in cells CEMX174 (A) and RhPBMC (B). CEMX174 cells (105) or RhPBMC (2 × 106) were infected with 100 TCID50 of each virus for 3 h at 37°C. The viral inocula were then removed, and the infected cells were maintained in culture media. At the times indicated, p27 antigen production in culture supernatants was determined.
V1 and V3 glycosylation mutations confer neutralization resistance.
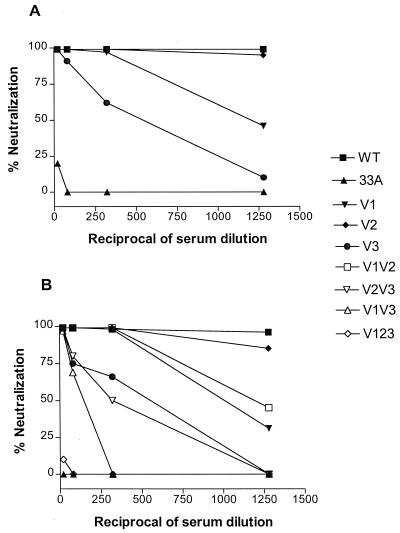
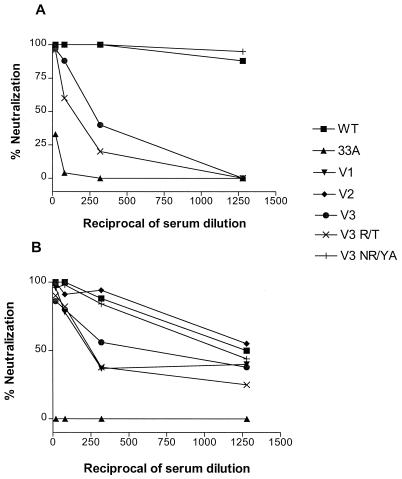
The neutralization sensitivities of the single V1 N/H, V2 N/S, and V3 NR/YT N-linked glycosylation mutants were first examined to determine whether any of the changes mediated evasion from immune recognition. Neutralization assays were performed with CEMX174 cells since, relative to the wild-type virus, the mutant viruses replicated with similar kinetics and to comparable titers in this cell type (Fig. 2A). The results in Fig. 3A show that either the removal of a potential N-linked glycosylation site in the V1 domain of SHIVSF33 or the creation of such a site in the V3 domain confers resistance to neutralization by sera collected from Mnu25814 at the symptomatic stage (96 or 104 weeks postinfection). The extent of neutralization resistance is higher for the V3 NR/YT mutant virus than for the V1 N/H virus. In contrast, repositioning of a potential glycosylation site within the V2 domain does not have an effect by itself. When the double and triple glycosylation mutant viruses were examined, we observed that a combination of the V1 and V3 mutations appeared to have an additive effect on conferring neutralization resistance, and this effect was enhanced by the presence of the V2 mutation in the V123 mutant virus (Fig. 3B). The V2 mutation, however, did not appear to have an effect in the context of the V1 or V3 mutation alone.
FIG. 3.
Neutralization of SHIVSF33 (WT), SHIVSF33A (33A), and single (A) or double and triple (B) glycosylation mutants by serum collected from Mnu25814 at 96 weeks postinfection. Serum neutralization was performed as described in Materials and Methods. The percent neutralization of each virus was determined and plotted against the reciprocal of serum dilutions used. Data represent one of three independent neutralization experiments, and similar findings were obtained with serum collected from Mnu25814 at 104 weeks postinfection.
A direct role of N-linked glycosylation in mediating neutralization escape is illustrated in the results summarized in Fig. 4A. We found that whereas the V3 NR/YT and R/T viruses are relatively resistant to neutralization by sera from Mnu25814, the V3 NR/YA virus is not. Taken together, the data show that the creation of an N-linked glycosylation site within the V3 loop of envelope gp120 alone is sufficient to confer neutralization resistance on the virus. Figure 4B shows that these changes in the V1 and V3 domains also confer partial resistance to neutralization by a pool of human polyclonal anti–HIV-1 sera. Again, the addition of an N-linked glycosylation site in the N terminus of the V3 loop rather than the charge change appears to be responsible for conferring partial neutralization escape from anti–HIV-1 sera.
FIG. 4.
Neutralization of V1, V2, and V3 glycosylation mutants by antisera to SHIV and HIV-1. Serum neutralization was performed with serum collected from Mnu25814 at 96 weeks postinfection (A) and with a pool of sera collected from HIV-1-infected individuals (B) as described in Materials and Methods. The percent neutralization was determined and plotted against the reciprocal of serum dilutions used. Data represent one of three independent neutralization experiments.
DISCUSSION
We have used the model of SHIV infection to define the nature of the selection process that occurs in vivo for HIV env gp120. We characterize genetically and antigenically the viral variants that evolved during the course of infection of rhesus macaques with molecularly cloned SHIV and establish the basis for their selection within the host. Longitudinal sequence analyses of viruses isolated from SHIVSF33-infected Mnu25814 demonstrate an accumulation of amino acid substitutions within HIV-1 envelope gp120 over time. The occurrence of the amino acid substitutions correlates with a rise in virus titers (Table 1) and with the change in antigenicity demonstrated here as well as the change in virus phenotype previously reported (28).
Whereas the parental SHIVSF33 clone is sensitive to neutralization by both human polyclonal HIV-1-positive sera and sera collected from Mnu25814 at all times postinfection, viral variants present in Mnu25814 late in infection are not (Tables 2 and 3). Using mutagenesis studies, we show that genetic changes in the V1 or V3 domain of gp120 mediate the change in the neutralization sensitivity of SHIVSF33 (Fig. 3 and 4). The degree of neutralization resistance conferred by changes in the V3 loop is higher than that conferred by changes in the V1 domain. Although we have not formerly proven that the genetic changes introduced in the V1 and V3 domains result in carbohydrate modifications, other studies have shown that similar sequence changes in HIV-1 envelope gp120 do lead to the anticipated biochemical changes (3, 17, 20, 24, 60). Our data obtained with additional V3 mutants offer further support of this notion (Fig. 4A). Thus, our findings establish a temporal relationship between sequence substitutions in the Env SU of HIV-1 and neutralization sensitivity for viruses that evolve during the course of an in vivo infection. Furthermore, we find that carbohydrates play an important role in conferring neutralization escape.
Significant resistance to serum neutralization is observed for the week-72 virus despite the lack of sequence changes in the V1 and V3 regions of the viral genome (Table 2 and Fig. 1). Since only four clones in these regions were sequenced, the possibility exists that resistant viruses were present as minor sequences and were not detected. The presence of such minor resistant variants within the mixed population of viruses isolated at week 72 would give the apparent appearance of neutralization resistance. The week-96 virus exhibits a neutralization resistance pattern identical to that of SHIVSF33A. The appearance of a fully resistant virus coincides with a time at which the titer of neutralizing antibodies against the parental SHIVSF33 clone is the highest (Table 2). Thus, the selection pressure for neutralization escape may have reached a maximum at that time. Interestingly, the acquisition of neutralization resistance is associated with an increase in the viral load of Mnu25814 (Table 1) and the pathogenicity of the virus in vivo (28).
The amino acid substitutions in the V3 domain that confer neutralization resistance create a potential glycosylation site (Fig. 1). It is possible that the presence of N-linked carbohydrates at this position of the loop shields the virus from immune recognition. Indeed, this N-linked glycosylation site within the V3 loop appears to be dispensable for virus replication and yet is highly conserved (24). The amino acid substitution in the V1 domain, however, results in the removal of a potential glycosylation site. Elimination of glycosylation sites in the V1 domain of HIV-1 and SIV has also been reported to affect the ability of monoclonal antibodies (MAbs) to bind and subsequently to neutralize viral infectivity (14, 18, 20, 47). Sequence changes in the V1 domain have been reported to alter V3 and CD4-binding-site recognition (7, 12, 20). It is conceivable, therefore, that the effect on neutralization mediated by the removal of the glycosylation site within the V1 domain of SHIVSF33 envelope gp120 occurs through modulation of the structures of the principal neutralizing epitopes.
The finding that the V3 glycosylation mutants are resistant to neutralization by autologous SHIV as well as heterologous HIV antisera suggests that the epitope(s) that is masked by the N-linked site in the V3 domain is shared between T-cell-line-adapted (TCLA) HIV-1SF33 and primary viruses that establish infections in vivo. This epitope could lie within the V3 loop itself. Indeed, broadly neutralizing anti-V3 loop MAbs that are directed against discontinuous conserved epitopes comprising the N-terminal side or both flanks of the V3 loop have been described (19, 35). Furthermore, the observation that the V3 NR/YA virus is still sensitive to serum neutralization (Fig. 4) is consistent with the notion that this masked epitope, if located within the V3 domain, is not linear in nature. Alternatively, the absence of the N-linked site in the V3 loop might lead to conformational changes that alter or expose a major neutralizing epitope in another region of the envelope. In view of the finding by Back et al. (3) that the removal of this highly conserved N-linked site in the amino terminus of the V3 loop of HXB2 envelope gp120 confers enhanced sensitivity to neutralization by both anti-V3 and anti-CD4 MAbs, epitopes located within the CD4-binding site could be affected. Anti-CD4-binding site antibodies are known to be broadly cross-neutralizing (54, 55).
It has been reported that whereas anti-V3 antibodies present in sera from HIV-1-infected individuals can effectively neutralize the TCLA MN strain, primary isolates are resistant (13, 51, 57, 58). Interestingly, the highly conserved N-linked glycosylation site that is located in the N terminus of the V3 loop but that is absent from envelope gp120 of HIV-1SF33 is also missing in the MN strain. Thus, the possibility exists that the reported (57) relative sensitivity and resistance of TCLA versus primary isolates to neutralization by anti-V3 antibodies are due to the absence or presence of this highly conserved N-linked site in the V3 loop. Studies with other TCLA viruses that contain this N-linked site or with molecularly cloned primary isolates that are genetically engineered to lack this conserved N-linked site should address this possibility.
The sera obtained from Mnu25814 late in infection (104 weeks postinfection) and at necropsy (132 weeks postinfection), although capable of neutralizing the parental SHIVSF33 clone, were unable to neutralize SHIVSF33A or variants recovered at 96 weeks postinfection (Table 1 and data not shown). Furthermore, sera from animals infected with cell-free SHIVSF33A do not neutralize the autologous virus (unpublished observations). These findings indicate that Mnu25814 mounted antibody responses that neutralized the parental SHIVSF33 envelope but not envelopes of viruses that evolved later in infection and suggest that SHIVSF33A does not appear to elicit a strong neutralizing response. Carbohydrate shielding of a neutralizing epitope(s) on the surface of SHIVSF33A might be responsible for the failure to elicit an effective response. Nevertheless, antibodies directed at putative shielded sites, in particular, those masked by the highly conserved N-linked site within the N terminus of the V3 loop, can be found in sera from HIV-infected individuals, since such sera neutralized parental SHIVSF33 and not the potential V3 glycosylation mutants constructed here. This observation suggests that the putative protected epitopes are immunogenic and are exposed to the immune system at some point during the natural process of HIV-1 infection. Alternatively, these epitopes may be present on immature viral proteins or debris, e.g., unprocessed gp160 that has been suggested to play a major role in eliciting immune responses (43). Identification of these epitopes should aid in the design of effective viral vaccines.
In summary, our studies on the genetic and antigenic changes in the env gene of SHIVSF33 variants identify neutralization escape as a major mechanism for viral adaptation in vivo. Our findings further support a role of carbohydrate side chains in mediating evasion from immunosurveillance and in modulating the immunogenicity of the envelope glycoprotein. Additional studies are required to define and compare the nature of the envelope glycoprotein epitopes recognized by antibodies present in SHIVSF33- and SHIVSF33A-infected macaques and to establish the role of neutralization resistance in the enhanced pathogenicity of SHIVSF33A infection.
ACKNOWLEDGMENTS
This work was funded by NIH grants AI41945 and CA72822.
We thank Leonidas Stamatatos and Lisa Chakrabarti for critical comments, Rei Tan for technical assistance, and Christina Chiaffarelli for preparation of the manuscript.
REFERENCES
- 1.Albert J, Abrahamsson B, Nagy K, Aurelius E, Gaines H, Nystrom G, Fenyo E M. Rapid development of isolate-specific neutralizing antibodies after primary HIV-1 infection and consequent emergence of virus variants which resist neutralization by autologous sera. AIDS. 1990;4:107–112. doi: 10.1097/00002030-199002000-00002. [DOI] [PubMed] [Google Scholar]
- 2.Arendrup M, Nielsen C, Hansen J-E S, Pedersen C, Mathiesen L, Nielsen J O. Autologous HIV-1 neutralizing antibodies: emergence of neutralization-resistant escape virus and subsequent development of escape virus neutralizing antibodies. J Acquired Immune Defic Syndr. 1992;5:303–307. [PubMed] [Google Scholar]
- 3.Back N K, Smit L, De Jong J J, Keulen W, Schutten M, Goudsmit J, Tersmette M. An N-glycan within the human immunodeficiency virus type 1 gp120 V3 loop affects virus neutralization. Virology. 1994;199:431–438. doi: 10.1006/viro.1994.1141. [DOI] [PubMed] [Google Scholar]
- 4.Back N K, Smit L, Schutten M, Nara P L, Tersmette M, Goudsmit J. Mutations in human immunodeficiency virus type 1 gp41 affect sensitivity to neutralization by gp120 antibodies. J Virol. 1993;67:6897–6902. doi: 10.1128/jvi.67.11.6897-6902.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benjouad A, Mabrouk K, Gluckman J C, Fenouillet E. Effect of sialic acid removal on the antibody response to the third variable domain of human immunodeficiency virus type-1 envelope glycoprotein. FEBS Lett. 1994;341:244–250. doi: 10.1016/0014-5793(94)80465-6. [DOI] [PubMed] [Google Scholar]
- 6.Bolmstedt A, Olofsson S, Sjogren-Jansson E, Jeansson S, Sjoblom I, Akerblom L, Hansen J E, Hu S L. Carbohydrate determinant NeuAc-Gal beta (1–4) of N-linked glycans modulates the antigenic activity of human immunodeficiency virus type 1 glycoprotein gp120. J Gen Virol. 1992;73:3099–3105. doi: 10.1099/0022-1317-73-12-3099. [DOI] [PubMed] [Google Scholar]
- 7.Boyd M T, Simpson G R, Cann A J, Johnson M A, Weiss R A. A single amino acid substitution in the V1 loop of human immunodeficiency virus type 1 gp120 alters cellular tropism. J Virol. 1993;67:3649–3652. doi: 10.1128/jvi.67.6.3649-3652.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burns D P, Collignon C, Desrosiers R C. Simian immunodeficiency virus mutants resistant to serum neutralization arise during persistent infection of rhesus monkeys. J Virol. 1993;67:4104–4113. doi: 10.1128/jvi.67.7.4104-4113.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burns D P, Desrosiers R C. Envelope sequence variation, neutralizing antibodies, and primate lentivirus persistence. Curr Top Microbiol Immunol. 1994;188:185–219. doi: 10.1007/978-3-642-78536-8_11. [DOI] [PubMed] [Google Scholar]
- 10.Burton D R. A vaccine for HIV type 1: the antibody perspective. Proc Natl Acad Sci USA. 1997;94:10018–10023. doi: 10.1073/pnas.94.19.10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burton D R, Montefiori D C. The antibody response in HIV-1 infection. AIDS. 1997;11(Suppl. A):S87–S98. [PubMed] [Google Scholar]
- 12.Carrillo A, Ratner L. Cooperative effects of the human immunodeficiency virus type 1 envelope variable loops V1 and V3 in mediating infectivity for T cells. J Virol. 1996;70:1310–1316. doi: 10.1128/jvi.70.2.1310-1316.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carrow E W, Vujcic L K, Glass W L, Seamon K B, Rastogi S C, Hendry R M, Boulos R, Nzila N, Quinnan G V J. High prevalence of antibodies to the gp120 V3 region principal neutralizing determinant of HIV-1MN in sera from Africa and the Americas. AIDS Res Hum Retroviruses. 1991;7:831–838. doi: 10.1089/aid.1991.7.831. [DOI] [PubMed] [Google Scholar]
- 14.Chackerian B, Rudensey L M, Overbaugh J. Specific N-linked and O-linked glycosylation modifications in the envelope V1 domain of simian immunodeficiency virus that evolve in the host alter recognition by neutralizing antibodies. J Virol. 1997;71:7719–7727. doi: 10.1128/jvi.71.10.7719-7727.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chamat S, Nara P, Berquist L, Whalley A, Morrow W J, Kohler H, Kang C Y. Two major groups of neutralizing anti-gp120 antibodies exist in HIV-infected individuals. Evidence for epitope diversity around the CD4 attachment site. J Immunol. 1992;149:649–654. [PubMed] [Google Scholar]
- 16.Cheng-Mayer C, Homsy J, Evans L A, Levy J A. Identification of human immunodeficiency virus subtypes with distinct patterns of sensitivity to serum neutralization. Proc Natl Acad Sci USA. 1988;85:2815–2819. doi: 10.1073/pnas.85.8.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fenouillet E, Gluckman J C, Bahaoui E. Role of N-linked glycans of envelope glycoproteins in infectivity of human immunodeficiency virus type 1. J Virol. 1990;64:2841–2848. doi: 10.1128/jvi.64.6.2841-2848.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer P B, Karlsson G B, Butter T D, Dwek R A, Platt F M. N-Butyldeoxynojirimycin-mediated inhibition of human immunodeficiency virus entry correlates with changes in antibody recognition of the V1/V2 region of gp120. J Virol. 1996;70:7143–7152. doi: 10.1128/jvi.70.10.7143-7152.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorny M K, VanCott T C, Hioe C, Israel Z R, Michael N L, Conley A J, Williams C, Kessler J A N, Chigurupati P, Burda S, Zolla-Pazner S. Human monoclonal antibodies to the V3 loop of HIV-1 with intra- and interclade cross-reactivity. J Immunol. 1997;159:5114–5122. [PubMed] [Google Scholar]
- 20.Gram G J, Hemming A, Bolmstedt A, Jansson B, Olofsson S, Akerblom L, Nielsen J O, Hansen J E. Identification of an N-linked glycan in the V1-loop of HIV-1 gp120 influencing neutralization by anti-V3 antibodies and soluble CD4. Arch Virol. 1994;139:253–261. doi: 10.1007/BF01310789. [DOI] [PubMed] [Google Scholar]
- 21.Gruters R A, Nefjes J J, Tersmette M, de Goede R E Y, Tulp A, Huisman H G, Miedema F, Ploegh H L. Interference with HIV-induced syncytium formation and viral infectivity by inhibitors of trimming glucosidase. Nature. 1987;330:74–77. doi: 10.1038/330074a0. [DOI] [PubMed] [Google Scholar]
- 22.Harouse J M, Tan R C, Gettie A, Dailey P, Marx P A, Luciw P A, Cheng-Mayer C. Mucosal transmission of pathogenic CXCR4-utilizing SHIVSF33A variants in rhesus macaques. Virology. 1998;248:95–107. doi: 10.1006/viro.1998.9236. [DOI] [PubMed] [Google Scholar]
- 23.Kinsey N E, Anderson M G, Unangst T J, Joag S V, Narayan O, Zink M C, Clements J E. Antigenic variation of SIV: mutations in V4 alter the neutralization profile. Virology. 1996;221:14–21. doi: 10.1006/viro.1996.0348. [DOI] [PubMed] [Google Scholar]
- 24.Lee W R, Syu W J, Du B, Matsuda M, Tan S, Wolf A, Essex M, Lee T H. Nonrandom distribution of gp120 N-linked glycosylation sites important for infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1992;89:2213–2217. doi: 10.1073/pnas.89.6.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levy J A. The transmission of HIV and factors influencing progression to AIDS. Am J Med. 1993;95:86–100. doi: 10.1016/0002-9343(93)90237-j. [DOI] [PubMed] [Google Scholar]
- 26.Li J, Lord C I, Haseltine W, Letvin N L, Sodroski J. Infection of cynomolgus monkeys with a chimeric HIV-1/SIVmac virus that expresses the HIV-1 envelope glycoproteins. J Acquired Immune Defic Syndr. 1992;5:639–646. [PubMed] [Google Scholar]
- 27.Li Y, Luo L, Rasool N, Kang C Y. Glycosylation is necessary for the correct folding of human immunodeficiency virus gp120 in CD4 binding. J Virol. 1993;67:584–588. doi: 10.1128/jvi.67.1.584-588.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luciw, P. A., C. P. Mandell, J. Li, T. A. Lowe, K. A. Schmidt, K. E. S. Shaw, and C. Cheng-Mayer. Fatal immunopathogenesis by SIV/HIV-1 (SHIV) containing the HIV-1SF33 env gene in juvenile and newborn rhesus macaques. Submitted for publication. [DOI] [PubMed]
- 29.Luciw P A, Pratt-Lowe E, Shaw K E S, Levy J A, Cheng-Mayer C. Persistent infection of rhesus macaques with T-cell-line-tropic and macrophage-tropic clones of simian/human immunodeficiency viruses (SHIV) Proc Natl Acad Sci USA. 1995;92:7490–7494. doi: 10.1073/pnas.92.16.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masuda T, Matsushita S, Kuroda M J, Kannagi M, Takatsuki K, Harada S. Generation of neutralization-resistant HIV-1 in vitro due to amino acid interchanges of third hypervariable env region. J Immunol. 1990;145:3240–3246. [PubMed] [Google Scholar]
- 31.Matthews T J, Weinhold K J, Lyerly H K, Langlois A J, Wigzell H, Bolognesi D P. Interaction between the human T-cell lymphotropic virus type IIIB envelope glycoprotein gp120 and the surface antigen CD4: role of carbohydrate in binding and cell fusion. Proc Natl Acad Sci USA. 1987;84:5424–5428. doi: 10.1073/pnas.84.15.5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDougal J S, Cort S P, Kennedy M S, Cabridilla C D, Feorino P M, Francis D P, Hicks D, Kalyanaraman V S, Martin L S. Immunoassay for the detection and quantitation of infectious human retrovirus, lymphadenopathy-associated virus (LAV) J Immunol Methods. 1985;76:171–183. doi: 10.1016/0022-1759(85)90489-2. [DOI] [PubMed] [Google Scholar]
- 33.McKeating J A, Gow J, Goudsmit J, Pearl L H, Mulder C, Weiss R A. Characterization of HIV-1 neutralization escape mutants. AIDS. 1989;3:777–784. doi: 10.1097/00002030-198912000-00001. [DOI] [PubMed] [Google Scholar]
- 34.Miedema F, Meyaard L, Koot M, Klein M R, Roos M T, Groenink M, Fouchier R A, Van’t Wout A B, Tersmette M, Schellekens P T, et al. Changing virus-host interactions in the course of HIV-1 infection. Immunol Rev. 1994;140:35–72. doi: 10.1111/j.1600-065x.1994.tb00864.x. [DOI] [PubMed] [Google Scholar]
- 35.Moore J P, Trkola A, Korber B, Boots L J, Kessler J A N, McCutchan F E, Mascola J, Ho D D, Robinson J, Conley A J. A human monoclonal antibody to a complex epitope in the V3 region of gp120 of human immunodeficiency virus type 1 has broad reactivity within and outside clade B. J Virol. 1995;69:122–130. doi: 10.1128/jvi.69.1.122-130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nara P L, Smit L, Dunlop N, Hatch W, Merges M, Waters D, Kelliher J, Gallo R C, Fischinger P J, Goudsmit J. Emergence of viruses resistant to neutralization by V3-specific antibodies in experimental human immunodeficiency virus type 1 IIIB infection of chimpanzees. J Virol. 1990;64:3779–3791. doi: 10.1128/jvi.64.8.3779-3791.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohgimoto S, Shioda T, Mori K, Nakayama E E, Hu H, Nagai Y. Location-specific, unequal contribution of the N glycans in simian immunodeficiency virus gp120 to viral infectivity and removal of multiple glycans without disturbing infectivity. J Virol. 1998;72:8365–8370. doi: 10.1128/jvi.72.10.8365-8370.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Overbaugh J, Luciw P A, Hoover E A. Models for AIDS pathogenesis: simian immunodeficiency virus, simian-human immunodeficiency virus and feline immunodeficiency virus infections. AIDS. 1997;11(Suppl. A):S47–S54. [PubMed] [Google Scholar]
- 39.Overbaugh J, Rudensey L M. Alterations in potential sites for glycosylation predominant during evolution of the simian immunodeficiency virus envelope gene in macaques. J Virol. 1992;66:5937–5948. doi: 10.1128/jvi.66.10.5937-5948.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pal R, Hoke G M, Sarngadharan M G. Role of oligosaccharides in the processing and maturation of envelope glycoproteins of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1989;86:3384–3388. doi: 10.1073/pnas.86.9.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palker T J, Clark M E, Langlois A J, Matthews T J, Weinhold K J, Randall R R, Bolognesi D P, Haynes B F. Type-specific neutralization of the human immunodeficiency virus with antibodies to env-encoded synthetic peptides. Proc Natl Acad Sci USA. 1988;85:1932–1936. doi: 10.1073/pnas.85.6.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park E J, Vujcic L K, Anand R, Theodore T S, Quinnan G V J. Mutations in both gp120 and gp41 are responsible for the broad neutralization resistance of variant human immunodeficiency virus type 1 MN to antibodies directed at V3 and non-V3 epitopes. J Virol. 1998;72:7099–7107. doi: 10.1128/jvi.72.9.7099-7107.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parren P W H I, Sattentau Q J, Burton D R. HIV-1 antibody—debris or virion? Nat Med. 1997;3:366–367. doi: 10.1038/nm0497-366d. [DOI] [PubMed] [Google Scholar]
- 44.Reitter J N, Means R E, Desrosiers R C. A role for carbohydrates in immune evasion in AIDS. Nat Med. 1998;4:679–684. doi: 10.1038/nm0698-679. [DOI] [PubMed] [Google Scholar]
- 45.Robert-Guroff M, Reitz M S J, Robey W G, Gallo R C. In vitro generation of an HTLV-III variant by neutralizing antibody. J Immunol. 1986;137:3306–3309. [PubMed] [Google Scholar]
- 46.Rudensey L M, Kimata J T, Benveniste R E, Overbaugh J. Progression to AIDS in macaques is associated with changes in the replication, tropism, and cytopathic properties of the simian immunodeficiency virus variant population. Virology. 1995;207:528–542. doi: 10.1006/viro.1995.1113. [DOI] [PubMed] [Google Scholar]
- 47.Rudensey L M, Kimata J T, Long E M, Chackerian B, Overbaugh J. Changes in the extracellular envelope glycoprotein of variants that evolve during the course of simian immunodeficiency virus SIVMne infection affect neutralizing antibody recognition, syncytium formation, and macrophage tropism but not replication, cytopathicity, or CCR-5 coreceptor recognition. J Virol. 1998;72:209–217. doi: 10.1128/jvi.72.1.209-217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sakuragi S, Shibata R, Mukai R, Komatsu T, Fukasawa M, Sakai H, Sakuragi J I, Kawamuri M, Ibuki K, Hayami M, Adachi A. Infection of macaque monkeys with a chimeric human and simian immunodeficiency virus. J Gen Virol. 1992;73:2983–2987. doi: 10.1099/0022-1317-73-11-2983. [DOI] [PubMed] [Google Scholar]
- 49.Schonning K, Jansson B, Olofsson S, Nielsen J O, Hansen J S. Resistance to V3-directed neutralization caused by an N-linked oligosaccharide depends on the quaternary structure of the HIV-1 envelope oligomer. Virology. 1996;218:134–140. doi: 10.1006/viro.1996.0173. [DOI] [PubMed] [Google Scholar]
- 50.Shibata R, Sakai H, Kiyomasu T, Ishimoto A, Hayami M, Adachi A. Generation and characterization of infectious chimeric clones between human immunodeficiency virus type 1 and simian immunodeficiency virus from an African green monkey. J Virol. 1990;64:5861–5868. doi: 10.1128/jvi.64.12.5861-5868.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spear G T, Takefman D M, Sharpe S, Ghassemi M, Zolla-Pazner S. Antibodies to the HIV-1 V3 loop in serum from infected persons contribute a major proportion of immune effector functions including complement activation, antibody binding, and neutralization. Virology. 1994;204:609–615. doi: 10.1006/viro.1994.1575. [DOI] [PubMed] [Google Scholar]
- 52.Stamatatos L, Cheng-Mayer C. An envelope modification that renders a primary, neutralization-resistant clade B human immunodeficiency virus type 1 isolate highly susceptible to neutralization by sera from other clades. J Virol. 1998;72:7840–7845. doi: 10.1128/jvi.72.10.7840-7845.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thali M, Charles M, Furman C, Cavacini L, Posner M, Robinson J, Sodroski J. Resistance to neutralization by broadly reactive antibodies to the human immunodeficiency virus type 1 gp120 glycoprotein conferred by a gp41 amino acid change. J Virol. 1994;68:674–680. doi: 10.1128/jvi.68.2.674-680.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thali M, Furman C, Ho D D, Robinson J, Tilley S, Pinter A, Sodroski J. Discontinuous, conserved neutralization epitopes overlapping the CD4-binding region of human immunodeficiency virus type 1 gp120 envelope glycoprotein. J Virol. 1992;66:5635–5641. doi: 10.1128/jvi.66.9.5635-5641.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tilley S A, Honnen W J, Racho M E, Hilgartner M, Pinter A. A human monoclonal antibody against the CD4-binding site of HIV1 gp120 exhibits potent, broadly neutralizing activity. Res Virol. 1991;142:247–259. doi: 10.1016/0923-2516(91)90010-z. [DOI] [PubMed] [Google Scholar]
- 56.Tremblay M, Wainberg M A. Neutralization of multiple HIV-1 isolates from a single subject by autologous sequential sera. J Infect Dis. 1990;162:735–737. doi: 10.1093/infdis/162.3.735. [DOI] [PubMed] [Google Scholar]
- 57.Vancott T C, Polonis V R, Loomis L D, Michael N L, Nara P L, Birx D L. Differential role of V3-specific antibodies in neutralization assays involving primary and laboratory-adapted isolates of HIV type 1. AIDS Res Hum Retroviruses. 1995;11:1379–1391. doi: 10.1089/aid.1995.11.1379. [DOI] [PubMed] [Google Scholar]
- 58.Vogel T, Kurth R, Norley S. The majority of neutralizing Abs in HIV-1-infected patients recognize linear V3 loop sequences. Studies using HIV-1MN multiple antigenic peptides. J Immunol. 1994;153:1895–1904. [PubMed] [Google Scholar]
- 59.Wilson C, Reitz M S J, Aldrich K, Klasse P J, Blomberg J, Gallo R C, Robert-Guroff M. The site of an immune-selected point mutation in the transmembrane protein of human immunodeficiency virus type 1 does not constitute the neutralization epitope. J Virol. 1990;64:3240–3248. doi: 10.1128/jvi.64.7.3240-3248.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu Z, Kayman S C, Honnen W, Revesz K, Chen H, Vijh-Warrier S, Tilley S A, McKeating J, Shotton C, Pinter A. Characterization of neutralization epitopes in the V2 region of human immunodeficiency virus type 1 gp120: role of glycosylation in the correct folding of the V1/V2 domain. J Virol. 1995;69:2271–2278. doi: 10.1128/jvi.69.4.2271-2278.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]