Summary
Pseudouridine (Ψ), the isomer of uridine, is ubiquitously found in RNA, including tRNA, rRNA, and mRNA. Human pseudouridine synthase 3 (PUS3) catalyzes pseudouridylation of position 38/39 in tRNAs. However, the molecular mechanisms by which it recognizes its RNA targets and achieves site specificity remain elusive. Here, we determine single-particle cryo-EM structures of PUS3 in its apo form and bound to three tRNAs, showing how the symmetric PUS3 homodimer recognizes tRNAs and positions the target uridine next to its active site. Structure-guided and patient-derived mutations validate our structural findings in complementary biochemical assays. Furthermore, we deleted PUS1 and PUS3 in HEK293 cells and mapped transcriptome-wide Ψ sites by Pseudo-seq. Although PUS1-dependent sites were detectable in tRNA and mRNA, we found no evidence that human PUS3 modifies mRNAs. Our work provides the molecular basis for PUS3-mediated tRNA modification in humans and explains how its tRNA modification activity is linked to intellectual disabilities.
Keywords: tRNA modification, pseudouridine synthase, PUS1, PUS3, anticodon stem loop, cryo-EM structure, Pseudo-seq, transcriptome
Graphical abstract

Highlights
-
•
Single-particle cryo-EM structures reveal how human PUS3 recognizes tRNAs
-
•
Two distinct interfaces in PUS3 are key for tRNA binding and positioning
-
•
PUS1- but not PUS3-dependent Ψ sites are found in RNAs other than tRNAs
-
•
Patient-derived mutations link PUS3’s tRNA modification activity to human disease
Lin, Kleemann et al. provide a comprehensive structure-function analysis of human PUS3, which catalyzes the conversion of uridine to pseudouridine (Ψ). PUS3 forms a homodimer to selectively bind and specifically modify tRNAs. No PUS3-dependent Ψs were detected in mRNA, linking the associated human diseases to tRNAs.
Introduction
RNA consists of four different nucleotides (A, U, G, and C) that can be modified to over 170 different chemical moieties.1 Pseudouridine (Ψ), also known as 5-ribosyluracil, is the most abundant RNA modification, affecting approximately 0.5% of all uridines.2 The formation of Ψ requires the cleavage of the N1–C1′ glycosidic bond of uridine, followed by a rotation of the base and its re-attachment to the ribose via C5, thus forming a C–Cʹ-glycosidic bond. This isomerization creates the opportunity for an additional hydrogen bond to form with neighboring nucleotides at the Hoogsteen edge of the base and increases base stacking, influencing the conformational landscape of RNAs3,4,5 and providing thermostability to structured RNAs.6 tRNAs are hotspots of RNA modifications and each tRNA molecule carries a plethora of modifications distributed throughout the molecule.7 Both cytosolic and mitochondrial tRNAs are decorated with Ψ at numerous positions.8 Ψ residues at these sites are critical for maintaining the tertiary structure of tRNAs, while simultaneously providing the flexibility required to dynamically fit into the restricted space at the A and P sites of translating ribosomes.9
The conversion of uridines to Ψs is catalyzed by pseudouridine synthases (PUSs).10 PUS enzymes are found in all domains of life and are categorized into six superfamilies: TruA, TruB, TruD, RluA, RsuA, and PUS10. Most PUS are “stand-alone” enzymes that autonomously bind target RNAs and catalyze the isomerization reaction without additional factors, with the exception of DKC1 (dyskerin pseudouridine synthase 1; TruB family), which acts as part of a multi-subunit complex. All PUS enzymes share a highly similar core structure despite a relatively low sequence conservation and utilize a catalytic reaction mechanism that is highly conserved across bacteria and eukaryotes, including mammals.11,12,13,14 Nevertheless, eukaryotic PUS enzymes harbor unique N and C termini that contain flexible loops, helices, and additional domains to facilitate substrate selectivity.12,14,15,16,17,18,19 The specific molecular mechanisms of how the different PUS enzymes execute site specificity across a diverse set of target RNAs, including tRNAs, remain elusive. In yeast, individual PUS enzymes are not essential, but their inactivation results in reduced growth rates under stress conditions.20 In humans, homozygous as well as compound heterozygous genomic mutations in the coding region of specific PUS enzymes have been linked to severe diseases, including a wide spectrum of neurodevelopmental and intellectual disorders.21,22,23,24,25,26
Recently, several high-throughput methods have been developed to map Ψ sites in cellular RNAs.27,28,29,30,31,32,33 The implementation of these transcriptome-wide technologies has shown that several PUS enzymes do not only target tRNAs but also introduce Ψ into specific sites in a subset of mRNAs and long noncoding RNA (lncRNA).18,31,33 Currently, only PUS10 has not been reported to modify mRNA targets, and PUS3 was only associated with a relatively small number of mRNA targets compared with other PUS enzymes in yeast and humans.18,31,33 In contrast to the conserved and well-defined positions of Ψ in tRNAs, Ψ sites are distributed across the entire length of mRNA transcripts,31,32 including exons, introns, and 3′/5′ untranslated regions (UTRs). Furthermore, the patterns of Ψ in cellular transcriptomes are tissue- and cell-type-specific and can be dynamically regulated in response to environmental conditions.29,33,34,35,36 The presence of Ψ in mRNA appears to affect splicing rates,29 extend mRNA half-life,37 suppress premature termination codons,38 and change ribosomal dynamics.39 Therefore, Ψ is not only critical for tRNA integrity and mRNA translation but also plays a major role at the post-transcriptional regulation of mRNA biogenesis. Without a full understanding of the spectrum of modifications that can be found on an RNA species, we cannot understand transcriptome-wide regulatory mechanisms.
PUS3, a member of the same TruA superfamily as PUS1, differs from PUS1 in many ways. PUS3 and PUS1 catalyze Ψ at different positions on tRNA, and clinically relevant mutations in both proteins have been associated with different diseases.25,40 Despite our general understanding of how bacterial TruA enzymes work,14,19,41 we still do not understand how these closely related PUS enzymes achieve distinct target selectivity for tRNA and other RNA species. In particular, we do not know the underlying mechanisms of PUS3, such as substrate recognition and selectivity, which has direct consequences for understanding the disease phenotype. Here, we determined the structures of unbound and tRNA-bound human full-length PUS3 using single-particle cryogenic electron microscopy (cryo-EM). Our structural and biochemical models illustrate how the specificity for tRNA substrates is accomplished by the homodimeric PUS3 enzyme, which has evolved to simultaneously bind the anticodon stem loop (ASL) and the T-arm of a single properly folded tRNA by a concerted action of the two monomers. Our transcriptome-wide analysis of Ψ sites in HEK293 cells confirms that PUS1 modifies both tRNAs and mRNAs. However, we found no evidence in human cells that PUS3 targets other RNA classes than tRNAs, including mRNAs, rRNAs, lncRNA, or small nuclear RNAs (snRNAs). This selectivity is consistent with our structure-function analysis and the specific binding of various tRNAs, including intron-containing pre-tRNAs. Finally, we characterize two main classes of pathogenic PUS3 variants: those that reduce protein stability and others that cause defects in tRNA binding and/or modification activity. In summary, our data suggest that the role of PUS3 in cells and human diseases is mainly (if not exclusively) related to its tRNA modification activity.
Results
The apo and tRNA-bound structures of human PUS3
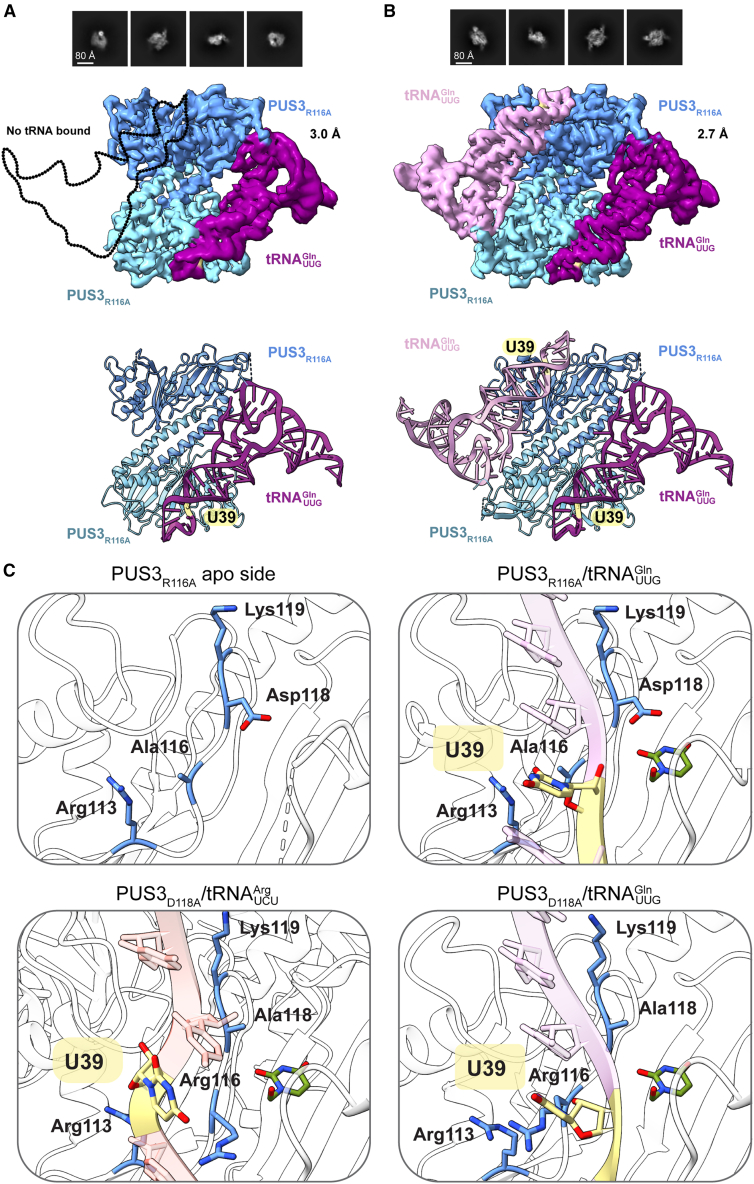
Human PUS3 harbors a conserved central PUS core domain14 (Figure 1A) flanked by extended eukaryote-specific N and C termini (Figure S1A). To understand how PUS3 forms a homodimeric complex25 and recognizes RNA substrates, we expressed full-length wild-type (WT) PUS3 (PUS3WT) and a catalytically inactive variant (PUS3D118A)25 in insect cells. The obtained gel filtration profiles indicate the formation of stable dimers for both proteins. In microscale thermophoresis (MST), the purified PUS3WT shows comparable affinities for all tested in vitro-transcribed (IVT) human tRNAs (Kd = 0.7–1.5 μM), including target tRNAs (e.g., , and ) and non-target tRNAs (e.g., ) (Figures 1A, S1B, and S1C). PUS3WT, but not PUS3D118A, converts U39 into Ψ39 in vitro but does not modify uridines in other positions of the tRNAs (Figures 1B and S1D), demonstrating that our purified PUS3WT is functional and displays the expected target specificity.
Figure 1.
Overview of the PUS3D118A apo and tRNA-bound cryo-EM structures
(A) Domain architecture of human PUS3 (top), protein purification of PUS3WT and PUS3D118A (left), and tRNA binding analyses using (tQ) or G39 (tQU39G). Calculated Kd values are shown.
(B) Detection of Ψ39 (left) using CMC-based primer extension. cDNA corresponding to the presence of Ψ (Ψ) and the primer (‣) are labeled.
(C) Representative 2D classes (top, scale bar: 80 Å), cryo-EM map (middle), and fitted model of PUS3D118A (PDB: 9F9Q). Dimensions and individual monomers are indicated (∗).
(D) Representative 2D classes (top, scale bar: 80 Å), cryo-EM map and atomic model of the PUS3- complex (PDB: 8OKD).
PUS3D118A is more stable than PUS3WT,25 so we collected a cryo-EM dataset (Figure S1E) and reconstructed a map at a global resolution of 6.5 Å (Gold-standard Fourier Shell Correlation/GSFSC0.143; Table 1). In the absence of an experimentally determined structural model, we employed AlphaFold242 to predict the model of a human PUS3 homodimer from its primary sequence. Despite the intermediate resolution, the quality of the cryo-EM map allowed us to fit the predicted model into the density (PDB: 9F9Q; Figure 1C). The PUS3 dimer resembles the overall architecture of its bacterial ortholog TruA from Thermus thermophilus (PDB: 1VS3).14 However, we noticed a striking difference in how the two monomers are held together compared with the bacterial homologs. The dimer interface (∼2,420 Å2) of PUS3 is exclusively formed by an anti-parallel coiled-coil domain, which is formed by a long C-terminal helix (aa 338–369) from each of the PUS3 monomers. Sequence alignments of PUS3 from various species (Figure S2A) show little sequence conservation in the C-terminal region, except for human and mouse, which is expected for coiled-coil domains. However, structural modeling and secondary structure predictions indicate the presence of a similar α helix also in PUS3 proteins of other eukaryotic organisms. Sequences of eukaryotic PUS1 and bacterial PUS3 homologs do not display a helical motif, which corroborates our observation that dimerization is achieved differently between eukaryotic and bacterial PUS3 enzymes. In detail, the interface between the two helices is formed by Leu348, His355, Thr359, and Leu369 (Figure S2B). The core structure of PUS3 appears rigid and consists of a canonical RNA-binding fold with a βαββαβ topology (Figures 1C and S2C). Surface charge analysis of the structure identified two positively charged surface regions of PUS3 that might be suitable to accommodate RNA substrates (Figure S2D). We did not detect densities for most N-terminal (aa 1–51) and C-terminal (aa 370–481) residues, but also did not observe major degradation products, suggesting that the termini are highly flexible.
Table 1.
Cryo-EM data collection, refinement, and validation statistics
| PUS3D118A; PDB: 9F9Q; EMD-16917; EMPIAR-16917 | PUS3D118A_tRNAGln; PDB: 8OKD; EMD-16926; EMPIAR-11511 | PUS3D118A_tRNAArg; PDB: 9ENE; EMD-19832; EMPIAR-12004 | PUS3D118A_pre-tRNAArg; PDB: 9ENF; EMD-19833; EMPIAR-12005 | PUS3R116A-2x tRNAGln; PDB: 9ENB; EMD-19830; EMPIAR-12003 | PUS3R116A-1x tRNAGln; PDB: 9ENC; EMD-19831; EMPIAR-12003 | |
|---|---|---|---|---|---|---|
| Data collection and processing | ||||||
| Magnification | 105,000× | 105,000× | 105,000× | 105,000× | 105,000× | 105,000× |
| Voltage (keV) | 300 | 300 | 300 | 300 | 300 | 300 |
| Electron exposure (e−/Å2) | 40 | 40 | 40 | 40 | 40 | 40 |
| Defocus range (µM) | −3 | −0.9 to −1.5 | −0.9 to −1.5 | −0.9 to −1.5 | −0.9 to −1.5 | −0.9 to −1.5 |
| Pixel size (Å) | 0.86 | 0.86 | 0.86 | 0.86 | 0.8456 | 0.8456 |
| Symmetry imposed | C2 | C2 | C2 | C2 | C2 | C1 |
| Initial particle images (no.) | 334,282 | 482,246 | 3,192,624 | 725,923 | 3,452,930 | 3,010,115 |
| Final particle images (no.) | 63,173 | 147,307 | 74,761 | 381,809 | 569,272 | 265,234 |
| Map resolution (Å) | 6.5 | 3.1 | 3.2 | 3.2 | 2.7 | 3.4 |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 |
| Map resolution range (Å) | 6.0 > 10 Å | 2.5 to >10 Å | 2.9 to >10 Å | 2.9 to >10 Å | 2.4–8.3 Å | 3.0 to >10 Å |
| Refinement | ||||||
| Initial model used | AlphaFold2 | AlphaFold2 | AlphaFold2 | AlphaFold2 | AlphaFold2 | |
| Model resolution (Å) | 3.3 | 3.1 | 3.1 | 3.1 | 3.9 | |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | |
| Model resolutionange (Å) | – | – | – | – | – | |
| Model composition | ||||||
| Non-hydrogen atoms | 7,964 | 8,490 | 8,245 | 8,268 | 6,162 | |
| Protein residues | 582 | 658 | 530 | 636 | 563 | |
| Nucleotide residues | 150 | 146 | 182 | 144 | 72 | |
| Ligands | – | – | – | Mg:6 | – | |
| B factors (Å2) | ||||||
| Protein | 135.64 | 170.54 | 150.66 | 176.01 | 212.43 | |
| Nucleotide | 211.14 | 283.81 | 454.72 | 257.95 | 289.89 | |
| Ligands | – | – | – | 106.04 | – | |
| Root-mean-square deviations (RMSDs) | ||||||
| Bond lengths (Å) | 0.002 | 0.004 | 0.003 | 0.005 | 0.004 | |
| Bond angles (°) | 0.502 | 0.638 | 0.607 | 0.512 | 0.512 | |
| Validation | ||||||
| MolProbity score | 1.76 | 1.80 | 2.16 | 1.56 | 1.69 | |
| Clashscore | 8.12 | 7.49 | 15.33 | 4.96 | 5.41 | |
| Poor rotamers (%) | 0 | 0.17 | 0 | 0.18 | 0 | |
| Ramachandran | ||||||
| Favored (%) | 95.44 | 94.24 | 92.35 | 95.65 | 93.99 | |
| Allowed (%) | 4.56 | 5.76 | 7.65 | 4.35 | 6.01 | |
| Disallowed (%) | 0 | 0 | 0 | 0 | 0 | |
| CC volume | 0.8 | 0.86 | 0.61 | 0.87 | 0.85 | |
We mixed PUS3D118A with the strongest-binding tRNA, (Figure S1C), to reconstitute a PUS3D118A-tRNA complex for structural analysis. Most two-dimensional (2D) classes showed PUS3 homodimers with two bound tRNA molecules while 6% of the particles corresponded to PUS3 dimers bound to a single tRNA molecule. After additional rounds of three-dimensional (3D) classification, the particle set with the best-defined features was refined at 3.1 Å global resolution (GSFSC0.143; PDB: 8OKD, Figures 1D and S2E; Table 1) after applying C2 symmetry. The structure reveals that PUS3 homodimers can accommodate two tRNAs simultaneously, as seen in the TruA-tRNA complex (Figure S2F). The bound tRNA adopts a canonical L-shaped architecture highly similar to the crystal structure of unbound yeast tRNAPhe (PDB: 1EHZ). The quality of the map allowed us to trace the phosphate backbone and nucleotides along most of the tRNA. However, the discriminator base (C73), the CCA-tail, and parts of the ASL (position 33–37) are only visible in lowpass-filtered maps, suggesting local flexibility. Furthermore, 3D-variability analyses did neither show different modes of tRNA binding nor reveal the position of the N and C termini. The overall structures of the unbound and tRNA-bound PUS3 dimer show no significant changes, indicating that PUS3 does not undergo major rearrangements when bound to tRNA (Figure S2G).
PUS3 uses two specific contact points to bind and position tRNAs
Each tRNA molecule in our structure contacts both PUS3 monomers and is held by the PUS3 dimer at two main contact points: the elbow region (T-arm) and the ASL. The variable loop and the acceptor stem are facing away from the PUS3 protein and do not make direct contacts (Figure 2A). In detail, the elbow region of tRNA is located on a platform formed by two helices (α1: aa 88–106 and α2: aa 157–169) that contain several basic residues (Lys99, Arg101, and Arg166) pointing toward the elbow, where the interaction between G19 and C56 takes place. At the other end of the tRNA molecule, the ASL is accommodated by the second PUS3 subunit positioning U39 in the catalytic cleft, poised for pseudouridylation. There, a cluster of positively charged residues (Arg113, Arg116, and Lys119) in the cleft binds to the phosphate backbone of the ASL. The L1 and L8 finger loops contact the major and minor grooves of the ASL, respectively (Figure 2B), similar to the TruA-tRNA complex (PDB: 2NR0).19 In the tRNA-bound state, the PUS3-dependent U39 residue is buried in the stem loop (stacking between A38 and C40) and positioned in close proximity to the catalytic residue Asp118 of PUS3. As U39 is still in the “flipped-in” conformation, our structural snapshot most likely represents a pre-reaction intermediate.19 The entire loop region (aa 110–119) that contains Asp118 is well-ordered, and Arg116 make contacts with the backbone of the ASL. Collectively, PUS3 specifically recognizes the tRNAs via their elbow and ASL regions without requiring additional RNA modifications at other positions.
Figure 2.
Substrate recognition of PUS3
(A) Close-up view of the elbow contact region (top), the ASL contact region (middle), and the sequence conservation (bottom).
(B) Cartoon representation of binding to PUS3D118A (aa 52–59 [pink], L1 [yellow], L8 [green], and U39 [wheat]).
(C and D) MST analyses of PUS3 mutants using (C) and PUS1 and PUS3 binding toward full-length tRNA (FL) and tRNA-ASL (D). Data are represented as mean ± SEM. Calculated Kd values are shown.
(E) Detection of Ψ39 in by PUS3 variants using CMC-based primer extension. cDNA corresponding to the presence of Ψ is labeled (Ψ). Average intensities of cDNA (bottom). (n = 3; n.s., no significance; ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001.) Data are represented as mean ± SEM.
Because the tRNA elbow contacts one monomer while the ASL contacts the other, we asked whether recognizing the tRNA substrate requires both binding regions and, therefore, dimerization of PUS3. We mutated the basic residues in the elbow contact region or in the ASL contact region to measure the individual contribution of each binding surface using MST. First, all tested preparations show comparable thermostability profiles, confirming that any change in affinity is not caused by an overall destabilization of the protein (Figures S3A–S3D). Most mutants in the catalytic site (PUS3R113A, PUS3R116A, and PUS3R159A) and the elbow contact point (PUS3K50A/K52A/R53A, PUS3K99A/R101A, and PUS3R50A/K52A/R53A/K99A/R101A) exhibit decreased tRNA binding affinities (Figure 2C), whereas the PUS3K119A mutant adjacent to Asp118 shows stronger binding compared with the WT. Mutations in the elbow contact region display strongly reduced binding affinity (Kd > 10 μM), confirming the contribution of these residues for tRNA binding to PUS3. To further corroborate this observation, we measured the binding affinity of PUS3 to a 17-mer RNA-hairpin corresponding to the ASL-sequence of (Figure 2D). PUS3 fails to bind to the ASL alone, which is consistent with data for the bacterial EcTruA dimer.14 In contrast, human PUS1, which functions as a monomer, binds to full-length tRNA and ASL with similar affinities (Kd of 0.53 ± 0.09 and 1.25 ± 0.12 μM). These results demonstrate that PUS3 requires both contact points to recognize and bind tRNAs with high affinity, whereas PUS1 recognizes tRNAs and short RNA hairpins with similar affinity.
Identification of residues affecting dimer formation and tRNA modification activity
Next, we used an in vitro pseudouridylation assay to analyze the catalytic activity of all purified PUS3 variants to modify U39 in (Figure 2E). We expected to observe catalytic defects for the active site mutants PUS3R116A and PUS3D118A, given their high conservation across the TruA superfamily. We also included additional active site mutants (PUS3R113A and PUS3K119A) that are less conserved, and mutants within the elbow contact site that are distal to the active site, which we did not expect to have a major effect on the catalytic activity of the enzyme after tRNA binding is achieved. As anticipated, only mutants in the central active site residues (PUS3R116A and PUS3D118A) showed significant defects in tRNA modification activity, whereas the remaining variants retained full activity. Of note, most elbow contact mutants still display pseudouridylation activity comparable with PUS3WT, while the PUS3R50A/K52A/R53A/K99A/R101A mutant shows lower activity, despite a fully intact active site. Our results are consistent with previous observations for the bacterial homolog EcTruA and the human PUS1 protein, confirming a highly conserved catalytic mechanism of pseudouridylation.19,40
Our structural comparison between bacterial TruA and human PUS3 reveals the formation of fundamentally different dimerization interfaces (Figure S2A). To further understand the functional necessity of PUS3 dimerization, we mutated central residues along the helix and generated PUS3L362R, PUS3H355T/T359A, and PUS3L362R/L366R/L369R mutants. Furthermore, we generated a variant that lacks the previously uncharacterized N terminus (PUS3Δ1-57) to investigate the role of the flexible N terminus of PUS3. We were able to purify small amounts of PUS3H355T/T359A and PUS3Δ1–57 but did not obtain soluble protein of the PUS3L362R and PUS3L362R/L366R/L369R. Purified PUS3H355T/T359A is soluble and forms stable homodimers, while PUS3Δ1–57 forms soluble oligomers (Figures S3A–S3C). Both PUS3H355T/T359A and PUS3Δ1-57 catalyze pseudouridylation at levels comparable with PUS3WT (Figure 2E). In summary, the formation of the central coiled-coil motif is crucial for the solubility and stability of PUS3. We posit that the identified hydrophobic residues on the side of the C-terminal helix facing the second monomer are key for dimer formation. As we have not been able to recombinantly purify a monomeric version of human PUS3, we conclude that dimerization of PUS3 contributes to its stability.
The tRNA elbow binding/recognition mode of PUS3
As the elbow region is a common structural feature of tRNAs, we wondered whether the elbow contact region of PUS3 facilitates the binding and positioning of other tRNAs as well. Therefore, we analyzed complexes between PUS3 and mature and the intron-containing pre-. First, PUS3 can bind to both tRNAs with similar affinities and efficiently catalyzes pseudouridylation (Figures 3A and 3B). Next, we determined the cryo-EM structures of PUS3D118A bound to human and pre-. Both structures reached overall resolutions of 3.2 (PDB: 9ENE) and 3.0 Å (PDB: 9ENF), respectively (Figures 3C, 3D, S3E, and S3F), and show that mature as well as the intron-containing bind to PUS3 in an almost identical conformation as . Of note, the intron-containing ASL of pre-tRNAArg is partially resolved (in low-pass filtered maps) and shows a ∼75° kink that points away from the PUS3 active site (Figure 3D). In summary, PUS3 uses both contact points (active site and elbow contact region) to recognize its target tRNAs as well as corresponding pre-tRNAs. This suggests that PUS3 employs both contact points to precisely define and position the target uridine in the active site in relative distance to the elbow region.19 Of note, to our knowledge, the structure of human PUS3 with pre- represents the first structural snapshot of a tRNA modifying enzyme bound to an intron-containing tRNA, suggesting that PUS3 acts before the tRNA splicing endonuclease (TSEN) complex, which removes tRNA introns.43,44,45,46
Figure 3.
Cryo-EM structures of PUS3 in complexes with or pre-
(A) MST analyses of PUS3WT using and pre-. Calculated Kd values are shown.
(B) Schematic 2D representation of and pre- illustrating A38, U39, and the intron (green). Detection of Ψ39 using CMC-based primer extension. cDNA corresponding to the presence of Ψ (Ψ) and full-length transcripts (‣) are labeled.
(C and D) Cryo-EM structures of PUS3- (PDB: 9ENE) (C) or PUS3-pre- (PDB: 9ENF), scale bar: 80 Å (D). Selected 2D classes (top), cryo-EM maps (middle), and atomic models of the PUS3-tRNA complexes (bottom). Schematic tRNAs are shown in the center (intron: green). The distances from the elbow to the Ψ sites are indicated.
Structural comparison between the empty and tRNA-bound active site of PUS3
After establishing that the highly conserved Arg116 is as catalytically important as Asp118 (Figure 2B), we obtained the cryo-EM structure of the PUS3R116A-tRNA complex to characterize the conformation of Asp118 within the active site. Despite its weaker tRNA binding affinity, it exhibits slightly higher thermostability when bound to tRNA (Figure S3D). Surprisingly, we identified two distinct types of PUS3- complexes (Figures 4A, 4B, and S4) that contained one or two molecules, respectively. For the PUS3R116A homodimer with two tRNA molecules bound, we obtained a C2-symmetrized map at an overall resolution of 2.66 Å (PDB: 9ENB). The asymmetrically occupied PUS3R116A homodimer, with only one tRNA bound, resulted in a reconstruction at an overall resolution of 3.15 Å resolution (PDB: 9ENC). The reduced affinity of PUS3R116A for tRNAs (Kd of ∼3.1 μM for PUS3R116A) might explain the presence of both conformations (Figure 2C). The overall architecture and positioning of the bound tRNA in both PUS3R116A structures is almost identical to other PUS3-tRNA complexes (Figures S5A–S5D). Despite the lower resolution of PUS3R116A with one tRNA bound, the map quality of the unoccupied catalytic site is significantly higher than for any of the apo PUS3 structures we obtained (Figures 1C and S5E–S5H; Table S1). The ASL region is also less well resolved in the higher-resolution structure of PUS3R116A in complex with 2 tRNAs and is only visible in low-pass filtered maps, as in all other maps obtained. We performed additional masked local refinement and 3D variability analyses to show that the ASL can indeed undergo specific movements (Video S1). Whether these movements that are restricted by PUS3 are an essential part of the modification reaction or simply represent the remaining degrees of freedom of the bound tRNA molecule is an interesting question that remains to be analyzed in the future.
Figure 4.
Overview of the catalytic site of PUS3
(A and B) Structures of PUS3R116A-tRNA complexes bound to one tRNA (A; PDB: 9ENC) or two tRNAs (B; PDB: 9ENB). Selected 2D classes (top, scale bar: 80 Å), cryo-EM maps (middle), and atomic models of the PUS3-tRNA complexes (bottom).
(C) Close-up view of the catalytic sites, highlighting the U39-containing strand and key residues. The modeled position of the flipped-out U39 (PDB: 1K8W) is shown (green).
With several high-resolution cryo-EM structures of unbound and tRNA-bound PUS3 complexes at hand, we analyzed whether specific residues undergo structural rearrangements due to tRNA binding and accommodation of U39 in the active site (Figure 4C). U39 is stacked inside the anticodon stem and base pairs with G31 in all our structures, suggesting that the PUS3R116A and PUS3D118A mutants trap the complex in a pre-catalytic conformation. We further show the relative position of U39 after flipping to illustrate that there is sufficient space in the active site of PUS3 to accommodate the flipped-out U39. The Asp118 residue is positioned in close proximity to the anticipated position of the flipped-out U39 in the empty and tRNA-bound PUS3 molecules, corroborating the key role of this residue in the modification reaction and the transient flipping out of U39 during the modification reaction. Overall, we did not identify significant rearrangements of the catalytic site, but observed that the ASL is positioned slightly differently in each of the structures. We conclude that binding to PUS3 induces local flexibility in the tRNA around U39 to promote the subsequent steps of the modification reaction.
Transcriptome-wide mapping of potential PUS3 targets
Our structural and biochemical results revealed a target-binding mode for PUS3 that appears to be tRNA specific. However, PUS3-dependent Ψ sites have been reported in mRNAs.31 To address this discrepancy, we created PUS3 knockout (PUS3−/−) HEK293 cells using CRISPR-Cas9 and applied Pseudo-seq, a transcriptome-wide method to identify Ψ sites based on specific labeling of Ψs by N-cyclohexyl-N′-(2-morpholinoethyl)carbodiimide methyl-p-toluenesulfonate (CMC) (Figure 5A).33 We similarly generated PUS1 knockout cells (PUS1−/−) to directly compare the target selectivity of monomeric (PUS1) and dimeric (PUS3) TruA-family members in human cells. We obtained three independent clones of PUS1−/− and PUS3−/− cells in which the respective proteins are undetectable and the expression level of other PUS enzymes is not affected. Next, we confirmed that the known Ψ sites in (Ψ28 for PUS1 and Ψ39 for PUS3) are absent in the respective knockout cells (Figures 5B, S6A, and S6B). We noticed that the overall cell morphology appears more roundish in PUS3 knockout cells, whereas PUS1−/− cells look similar to WT cells (Figure 5C).
Figure 5.
Transcriptome-wide analysis of Ψ sites
(A) Scheme of Pseudo-seq library preparation and data analysis. The dashed line indicates the peak of Ψ-dependent reads. CMC-dependent reverse transcription (RT) stops 1 nucleotide upstream of the Ψ site in the shown SLC29A1 transcript.
(B) Western blot analyses of PUS1 and PUS3 expression in the indicated HEK293 cell lines. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is the loading control (left). Detection of Ψ in from total RNA using CMC-based primer extension (right). cDNA corresponding to the presence of Ψ28 (blue) and Ψ39 (green) are labeled.
(C) Bright-field images of wild-type (WT), PUS1−/−, and PUS3−/− HEK293 cells (scale bar: 50 µm).
(D) Venn diagram showing the overlap between Ψ sites identified bybisulfite-induced deletion-sequencing (BID-seq),31 nanopore sequencing,47 and this study.
(E) K-mer frequency of the most abundant k-mer sequences of Ψ sites detected in the WT dataset. Sequences reported to be motifs of PUS7 (blue) or PUS4 (yellow) are indicated.33,36
(F) Distribution of detected Ψ sites in the 5′ untranslated region (5′ UTR), the coding sequence (CDS), the 3′ UTR, and at stop codons.
(G) Representative examples of identified Ψ sites of high frequency (adenylate kinase 2, AK2) and low frequency (phosphogluconate dehydrogenase, PGD).
(H) Detection of Ψ sites in cellular 18S rRNA33 using CMC-based primer extension. cDNA corresponding to the presence of Ψ (Ψ) are labeled (left). Ψ scores of these sites in 18S rRNA (right).
(I) Plots indicating the Ψ score of known sites in noncoding RNAs.
To apply stringent Ψ-calling criteria, we performed duplicate experiments for each of the three independent cell clones. Using this data, we determined a Ψ score for each uridine in the transcriptome. All replicates showed a high correlation, highlighting the robustness and reproducibility of our libraries. We then generated receiver operating characteristic curve (ROC) curves based on the fraction of reads resulting from CMC-Ψ-stalled reverse transcription and the Ψ-score to establish stringent cutoff values. Using these stringent filtering criteria, we identified 367 Ψ sites in HEK293 mRNA of which 94 overlap with reported sites from bisulfite-induced deletion sequencing (BID-seq) and 61 overlap with reported sites from nanopore sequencing (nanopore-seq) (Figure 5D; Table S2).31,32 We assessed the quality of our candidate sites by analyzing the sequence motifs and the location in which they occur within the transcripts. Notably, a substantial fraction of the sites was found in motifs specific to PUS4/TRUB1 or PUS7, indicating true positive sites (Figure 5E). In agreement with the previously reported site distribution, the identified sites are mainly located in the coding sequence (CDS) and 3′ UTR (Figure 5F).33 Furthermore, our approach successfully detected high-abundance sites (e.g., adenylate kinase 2 [AK2)] as well as the low-abundance site in phosphogluconate dehydrogenase [PGD; Figures 5G and S6C]).32 We not only confirmed reported sites in mRNAs but also in rRNA, snRNA, or lncRNA (Figures 5H, 5I, and S6D).
Next, we aimed to identify Ψ sites that depend on the activity of PUS1 or PUS3 in human cells. We successfully confirmed a well-reported PUS1-dependent site in MT-ND4 mRNA18 in our Pseudo-seq data and by reconstituting the modification reaction using purified components in vitro (Figure 6A). In total, we identified 9 high-confidence Ψ sites that were not detectable in the absence of PUS1. The four sites in MT-CO1, SLC35B2, CBR1, and CLUH have been previously associated with PUS148 (Figures 6B, 6C, S6E, and S6F). As most known Ψ sites in mRNA depend on PUS4/TRUB1 or PUS7,31,33,36,48 a low number of PUS1-dependent sites in HEK293 mRNAs was expected from recent studies.31,48 Previous reports31,33 had described PUS3-dependent Ψ sites in the transcriptome. However, we did not identify a single PUS3-dependent site in our large datasets (Figures 6B and S6G). Therefore, we specifically examined the signals of previously reported PUS3-dependent Ψ sites in mRNA31 in our own dataset. However, we found no evidence that any of the 10 top-ranked PUS3-dependent Ψ sites depend on PUS3 or are actual Ψ sites (Figure 6D).
Figure 6.
Transcriptome-wide analysis of PUS1- or PUS3-dependent Ψ sites
(A) Ψ score of the PUS1-dependent Ψ site in MT-ND4 mRNA in the wild type (WT), PUS1−/−, and PUS3−/− datasets (left). The predicted RNA secondary structure of the PUS1 site is shown and the targeted uridine is highlighted (middle). Detection of Ψ sites on in vitro-transcribed MT-ND4 or using CMC-based primer extension (right). cDNA corresponding to the presence of Ψ (Ψ) and the primer (‣) are labeled.
(B) Total number of Ψ sites detected in WT, and Ψ sites that are PUS1 or PUS3 dependent. Venn diagram showing the overlap between PUS1-dependent Ψ sites identified by BID-seq,31 pseudouridine assessment via bisulfite/sulfite treatment/PRAISE,48 and this study.
(C) Ψ scores of the 5 PUS1-dependent sites overlapping with PRAISE data and this study.
(D) Ψ scores of reported PUS3-dependent Ψ sites based on BID-seq according to our datasets (red, below the Ψ score threshold; orange, above the Ψ score threshold but below the used fraction of stalling reads). Ψ score for DBNDD2, GTF3C6, and ZFAND3 in our datasets. Ψ sites are indicated, while a reported low-confidence site is labeled with a black dot.
Ψ sites may occur only in specific cell types or under specific growth conditions, and it is possible that previously reported PUS3-dependent Ψ sites are not present in HEK293 cells.32 Hence, we further examined PUS3 activity on the reported transcripts by in vitro pseudouridylation to remove the cellular context, as we have confirmed the activity and specificity of purified PUS1 on an IVT MT-ND4 mRNA fragment (Figure 6A). Using as positive control, we did not detect PUS3 modification activity with any of the eight transcripts in vitro (Figures S7A and S7B). Furthermore, we used the ViennaRNA Package49 to assess the 2D secondary structures of these eight mRNA fragments. In contrast to the preferred modification motif of PUS1 (Figure S6E),18 the proposed PUS3-dependent uridines reside in various structural contexts with no detectable positional preference or recognizable motif (Figure S7C). Even though it is possible that low-occupancy sites are more efficiently modified in other cell types or under different growth conditions, we deem this unlikely. Our Pseudo-seq analysis and in vitro assays are consistent with our structural analysis, suggesting that PUS3 is not responsible for modifying these mRNAs.
In summary, our Ψ mapping provides highly reliable and consistent results for PUS1-dependent sites in tRNAs and mRNAs, confirming and extending prior literature. In agreement with our structural results, PUS3 appears to exhibit a high selectivity and specificity for modifying tRNAs, and we did not detect PUS3-dependent sites in mRNA in human cells or in vitro.
The biochemical characterizations of pathogenic PUS3 mutants
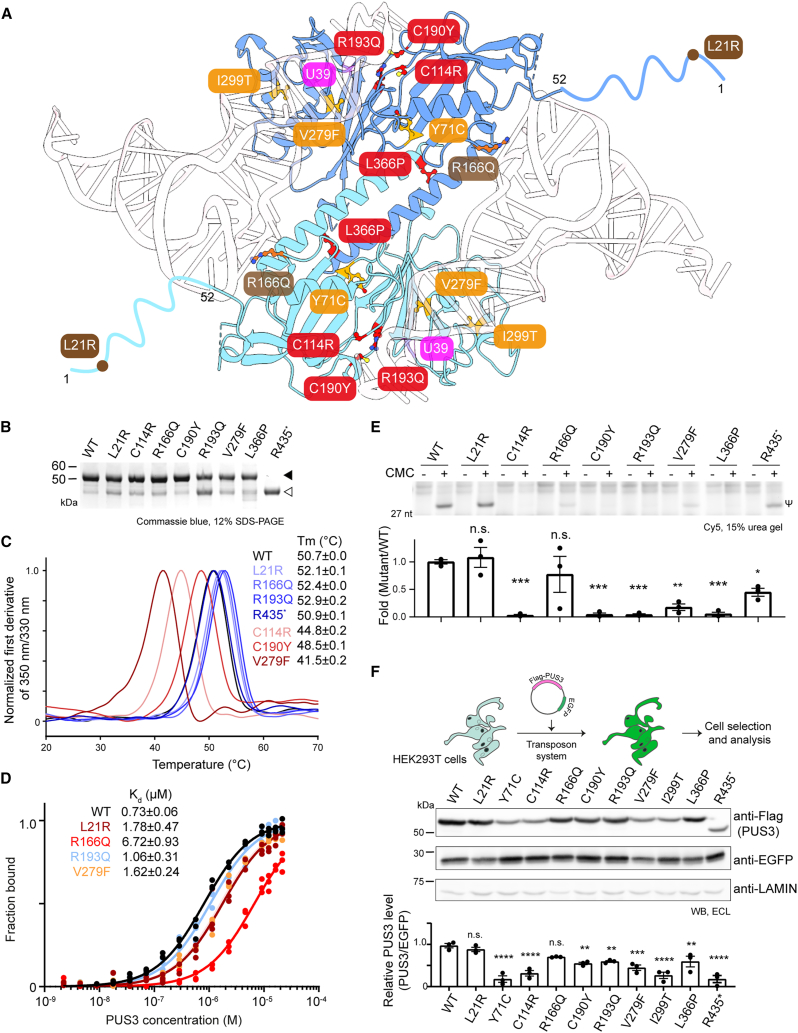
We previously found that two patient-derived variants of PUS3 (Y71C and I299T) cause intellectual disability by lowering PUS3 protein stability in vitro and in vivo.25 Recent studies also linked other clinically relevant variants of PUS3, including single amino acid substitutions, nonsense mutations, splice variants, and nucleotide substitutions in the start codon, to severe neurodevelopmental disorders.24,50 To understand how patient-derived mutations impact the structure and activity of human PUS3, we generated mutants carrying L21R, C114R, R166Q, C190Y, R193Q, V279F and L366P, and R435∗. All these missense mutation positions affect conserved residues of PUS3 (Figure S1) and can be mapped onto our structure of the human PUS3-tRNA complex (Figure 7A). Arg166 and Arg193 likely contact the tRNA substrate (Figure S7D), whereas the others are distributed across the entire protein and unlikely to be directly involved in tRNA binding. We produced all variants and measured their thermostability, tRNA binding and in vitro U39 tRNA modification activities (Figures 7B–7E). Most of the variants (PUS3L21R, PUS3R166Q, PUS3R193Q, and PUS3R435∗) displayed melting temperatures comparable with WT (between 50.7°C ± 0.02°C and 52.9°C ± 0.2°C), but the PUS3C114R, PUS3C190Y, and PUS3V279F mutants showed decreased stability (melting temperature [Tm] of 44.8°C ± 0.2°C, 48.5°C ± 0.1°C, and 41.5°C ± 0.2°C), which is consistent with decreased yields for these variants. We did not determine the Tm of the PUS3L366P mutant as it appears to form soluble oligomers (Figure S7E). We then characterized the mutants by comparing their ability to bind IVT human (Figure 7D). PUS3R193Q was the only variant that bound tRNA with similar affinities as the WT protein (Kd = 1.1 ± 0.3 μM). All other mutants had 2–3 times weaker tRNA binding affinities, with the PUS3R166Q mutant displaying the weakest affinity (Kd = 6.7 ± 0.9 μM). Comparing their catalytic activity toward tRNA, we found that the PUS3C114R, PUS3C190Y, PUS3R193Q, PUS3V279F, and PUS3L366P mutants completely failed to convert uridine to Ψ on , while PUS3R435∗ retained some activity and the rest of the mutants (PUS3L21R and PUS3R166Q) possessed activities comparable with WT (Figure 7E).
Figure 7.
Characterization of pathogenic PUS3 mutants in vitro and in cells
(A) Mapping pathogenic PUS3 variants. The residues are highlighted and color-coded (red, catalytic activity; orange, stability; brown, tRNA binding). U39 is highlighted.
(B) SDS-PAGE analysis showing purified PUS3 mutants (‣). The white triangle shows a PUS310–438 fragment confirmed by mass spectrometry.
(C) Protein thermostability analysis of PUS3 mutants. The melting temperatures (Tm) are listed in the inset.
(D) MST analyses of PUS3 mutants using showing the calculated Kd values.
(E) Detection of Ψ39 in by PUS3 mutants using CMC-based primer extension. cDNA corresponding to the presence of Ψ is labeled (Ψ). Average intensities of cDNA (n = 3; bottom).
(F) Scheme of Sleeping Beauty-mediated gene integration and protein expression in HEK293T cells. Western blot analysis of PUS3 and EGFP expression levels. LAMIN is the gel loading control. The averaged intensity of PUS3 is normalized against the intensity of EGFP. (n = 3; n.s.: no significance; ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001; ∗∗∗∗p ≤ 0.0001.) Data are represented as mean ± SEM.
Using human cells, we further asked whether the steady-state protein levels of the intrinsically unstable variants are affected. As we only had access to patient-derived cells for two of the variants, we overexpressed the recombinant mutants in HEK293T cells and monitored the expression levels. We utilized the Sleeping Beauty transposon system to integrate these variants, as well as the previously studied variants PUS3Y71C and PUS3I299T, into the genome to generate stable cell lines (Figure 7F). Consistent with our observations, we found that the steady-state levels of PUS3C114R, PUS3V279F, and PUS3R435∗ were as low as those of the mutants we previously characterized, PUS3Y71C and PUS3I299T. The levels of the other mutants, including PUS3C190Y, PUS3R193Q, and PUS3L366P, were slightly lower than the WT (about 50%–60% of WT). Meanwhile, the mRNA expression levels of the variants were all comparable with the WT (Figure S7F), pointing to altered protein stability as the main contributor to the observed differences in protein levels. In summary, our analyses show that certain patient-derived mutations of PUS3 lead to a loss of protein stability (PUS3C114R, PUS3C190Y, and PUS3V279F), whereas other variants affect tRNA binding and/or modification activity (PUS3L21R, PUS3R166Q, PUS3R193Q, PUS3L366P, and PUS3R435∗).
Discussion
PUS enzymes are ubiquitous across the tree of life and catalyze the most abundant RNA modification that regulates splicing rates,29 mRNA half-life,37 premature termination,38 and ribosomal dynamics,39 and yet the mechanisms underlying PUS substrate selection are largely unknown. Here, we present experimental evidence that PUS3 is unique among human PUS enzymes due to its dimerization, which is key to its mechanism of substrate recognition. Our structural, biochemical, and mutational analyses provide a framework for understanding the PUS3-mediated modification mechanism and its link to human diseases. Importantly, we show that PUS3 targets tRNAs, whereas PUS1 (like other PUS enzymes) modifies tRNAs as well as mRNAs.
In bacteria, members of the TruA, TruB, and TruD superfamilies target tRNAs, while members of the RluA and RsuA superfamilies preferentially act on rRNA. Many eukaryotic TruA, TruB, and TruD superfamily members relaxed their substrate selectivity and gained the ability to target mRNAs.33 Recently, Ψ sites have also been detected in the bacterial mRNA pool,51 but which bacterial PUS enzymes possess the ability to modify mRNAs remains unclear. Among all PUS superfamilies, the TruA family to which PUS1 and PUS3 belong, appears different from other PUS families for several reasons. Foremost, the hallmark of TruA is the formation of a homodimer, whereas other families act as monomers.14,41 Interestingly, TruA from Salmonella enterica exists as a monomer and forms a dimer in the presence of tRNA.52 Moreover, the elements that form the dimer interface in TruA are arranged differently in eukaryotic PUS3 homologs but still mediate the same feature of tRNA binding.13,14 For instance, our PUS3 structure displays that the C-terminal helix not only stabilizes the dimerization interface but also represents the only interaction point between the two monomers.14,53 In contrast, monomeric PUS enzymes like PUS4/TruB1 or PUS7 can bind to tRNAs as well as hairpin RNA substrates.53,54,55 Although PUS1 and PUS3 belong to the TruA family, both proteins target different sets of RNAs and different sites in tRNAs. They achieve their target selectivity and specificity by forming different oligomeric assemblies. PUS1 functions as a monomer and recognizes small, structured RNA motifs as well as a short and degenerate consensus-sequence motif,18,56 while PUS3 forms a homodimer that preferentially recognizes tRNAs and has relatively low affinity for other RNA motifs or sequences. This effect might have originally led to a diversification of TruA homologs by the establishment of PUS1.
All PUS proteins require a catalytic aspartate residue for executing the modification reaction,11 but other residues in the catalytic cleft or contacting sites are not as strictly conserved, contributing to differences in substrate recognition.41 For instance, the functionally important Arg residue in the catalytic RTDKGV-motif is only conserved in TruA, RluA, and RsuA families.14,19 Our structural and biochemical work highlights the functional role of Arg116 in human PUS3; however, the details of its contribution to the modification reaction need to be addressed in future studies.
The recent finding that mRNAs are chemically modified by specific cellular enzymes during RNA biogenesis has opened many exciting new research directions in cellular biochemistry. Transcriptome-wide searches for RNA modifications have identified a wealth of target sites, which represent the cellular epitranscriptome and include numerous Ψ sites.27,30,31,33,36 A key challenge is now to verify candidate sites and to distinguish functionally relevant modified sites that do not cause physiological consequences. As modification patterns are influenced by many additional factors like, e.g., cell type, growth conditions, or the metabolic state of the cells, it will be crucial to establish defined reference points. With the refinement of the detection methods, we generally observe a reduction of the numbers and a convergence toward high-confidence sites with functional relevance. Defining the target specificity of RNA modifying enzymes is an additional key step in this process. Hence, complementary studies that combine insights into the molecular mechanisms of target specificity with stringent calling criteria are essential to genuinely interpret and verify high-throughput mapping data reproducibly.28
Mutations in PUS enzymes are linked to several human diseases.10,57 For instance, mitochondrial myopathy and sideroblastic anemia (MLASA) is caused by mutations in PUS1, while numerous pathogenic variants of PUS7 cause several neurological and developmental disorders.23,40 Each pathogenic variant results in a loss of function, either by disrupting catalytic activity of the enzyme or by diminishing protein expression. Although patients commonly display intellectual disabilities, there is a wide spectrum of potential phenotypes that is patient-specific, suggesting a diverse range of affected cellular mechanisms that contribute to pathology.23,24,25,58,59 The body of available evidence suggests that this heterogeneous set of organismal-level phenotypes could be mediated through different subsets of RNA targets, many of which may have pleiotropic downstream effects themselves. PUS3 variants would be expected to affect tRNA-mediated protein synthesis across a wide range of mRNAs,24,25,50,59,60 whereas PUS1 or PUS7 may additionally regulate a specific subgroup of mRNAs directly at the level of RNA metabolism or translation.
In summary, our work provides a molecular framework for understanding the role of human PUS3 in health and disease. Foremost, the unique structure-dependent substrate selection by PUS3, which distinguishes it from similar PUS enzymes like PUS1, as shown in vitro and in vivo. Furthermore, we provide a structural basis for understanding the mechanism of its modification reaction and an in-depth characterization of several patient-derived variants. Our results pave the way to disentangle the complicated molecular relationships between different human PUS enzymes and to create a refined list of high-confidence Ψ sites in human cells. This knowledge will be critical to develop diagnostic markers for the linked disorders and to envision alternative treatment strategies.
Limitations of the study
Despite extensive sequencing and stringent search parameters in Pseudo-seq experiments, we did not identify high-confidence calls for PUS3-dependent Ψ sites in HEK293 cells. As HEK293 cells only express a subset of the human transcriptome, it is possible that PUS3-dependent Ψ sites are present in RNA transcripts of other cell types or under different growth conditions.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rabbit anti-PUS3 C-terminal domain | Abcam | Cat.#ab211270 |
| mouse anti-GAPDH | Sigma Aldrich | Cat.#MAB374; RRID:AB_2107445 |
| anti-FLAG M2 antibody | Sigma | Cat.#F3165; RRID:AB_259529 |
| anti B1-lamin | Invitrogen | Cat.#10H34L18; RRID:AB_2784553 |
| anti-GFP | Abcam | Cat.#ab290; RRID:AB_2313768 |
| HRP-conjugated secondary antibody | Cell Signaling; BD Pharmingen | Cat.#7074S; Cat.#554002; RRID:AB_395198 |
| Bacterial and virus strains | ||
| E. coli DH10Bac strain | Thermo Fisher Scientific | Cat.#10361012 |
| E. coli BL21 (DE3) CodonPlus-RIL strain | Agilent | Cat.#230245 |
| Chemicals, peptides, and recombinant proteins | ||
| HyClone SFM4 Insect cell culture media | Cytiva | Cat.# SH30913.02 |
| reduced GSH | BioShop | Cat.# GTH001 |
| PUS1 and PUS3 recombinant proteins | This study | N/A |
| LB Broth | BioShop | Cat.# LBL405 |
| Cy5-cytidine | Jena Bioscience | Cat.# NU-831-CY5 |
| RNase-free DNase I | Thermo Fisher Scientific | Cat.# 89836 |
| proteinase K | Merck | Cat.#P2308 |
| DEAE weak anion exchange column | Cytiva | Cat.# 17505501 |
| Superdex 75 Increase gel filtration column | Cytiva | Cat.#17-5174-01 |
| GSTPrep column | Cytiva | Cat.# 17528201 |
| HiLoad Superdex 200 pg preparative gel filtration column | Cytiva | Cat.#28989336 |
| premium capillaries (MST) | NanoTemper Technologies | Cat.# MO-K025 |
| glass capillary (DSF/DLS) | NanoTemper Technologies | Cat.# AN-041001 |
| QUANTIFOIL R2/1 copper grids (200 mesH) | Quantifoil | Cat.#N1-C15nCu20 |
| CMC | Sigma-Aldrich | Cat.#C106402 |
| SuperScriptIII | ThermoFisher Scientific | Cat.#18080044 |
| tracrRNA | IDT | Cat.#1075928 |
| TrueCut Cas9 Protein v2 | ThermoFisher Scientific | Cat.#A36498 |
| Lipofectamine CRISPRMAX Cas9 Transfection Reagent | ThermoFisher Scientific | Cat.#CMAX00001 |
| TRIzol | ThermoFisher Scientific | Cat.#15596018 |
| T4 PNK | NEB | Cat.#M0201L |
| T4 RNA ligase | NEB | Cat.#M0373L |
| AMV RT | Promega | Cat.#M5108 |
| CircLigase ssDNA ligase II | Epicentre | Cat.#CL9025K |
| cOmplete, EDTA-free Protease Inhibitor Cocktail | Sigma-Aldrich | Cat.#04693132001 |
| PageRuler prestained protein ladder | ThermoFisher Scientific | Cat.# 26619 |
| PVDF membranes (pore size 0.45 μmImmobilon-P or Immobilon-FL | ThermoFisher Scientific | Cat.# 88520 |
| SuperSignal™ West Pico PLUS Chemiluminescent Substrate | ThermoFisher Scientific | Cat.#34577 |
| M-MLV RT | Promega | Cat.#M1701 |
| RT-HS-PCR-Mix-SYBR-A | A&A Biotechnology | Cat.# 2017-100HS |
| NucleoBond AX100 column | MACHEREY-NAGEL | Cat.#740521 |
| Lipofectamine 3000 | ThermoFisher Scientific | Cat.#L300000 |
| Critical commercial assays | ||
| MEGAclearTM Transcription Clean-Up Kit | Thermo Fisher Scientific | Cat.#AM1908 |
| Poly(A)Purist™ MAG Kit | Thermo Fisher Scientific | Cat.#AM1922 |
| NEBNext® Multiplex Oligos for Illumina | NEB | Cat.#E7335S, E7500S, E7710S, E7730 |
| Universal RNA Purification Kit | EurX | Cat.# E3598 |
| Clean-Up RNA Concentrator kit | A&A Biotechnology | Cat.# 039-25C |
| Deposited data | ||
| Code used in this study | Zenodo | https://doi.org/10.5281/zenodo.11401096 |
| Deep-sequencing data generated in this study | GEO database (https://www.ncbi.nlm.nih.gov/geo/) | GSE255287 |
| Original imaging data | Mendeley Data | https://doi.org/10.17632/gdw8pr456d.2 |
| Experimental models: Cell lines | ||
| sf9 insect cell line | Expression Systems | Cat.# 94-001F |
| Hi5 insect cell line | ThermoFisher Scientific | Cat.# B85502 |
| Flp-In-T-REx-293 | ATCC | RRID:CVCL_U427 |
| HEK293T | ATCC | Cat.# CRL-3216 |
| Oligonucleotides | ||
| PUS3_crRNA:5′/AltR1/rUrCrUrGrCrUrCrArArGrUr ArCrArGrCrUrArGrUrGrUrUrUrUrArGrArGrCrUrAr UrGrCrU/AlrR2/3′; |
IDT | N/A |
| PUS1_crRNA:5′/AltR1/rArArUrArCrArGrCrCrUrGrAr CrCrGrGrArCrGrArGrUrUrUrUrArGrArGrCrUrArUrGr CrU/AltR2/-3′. |
IDT | N/A |
| (5′-AppGATATCGTCAAGATCGGAAGAGCACACGTC TGAA-ddC-3 |
IDT, Behrens et al.61 | N/A |
| RT primer: 5′-pRNAGATCGGAAGAGCGTCGTGTAGGGA AAGAG-iSp18-GTGACTGGAGTTCAGACGTGTGCTC-3 |
IDT, Behrens et al.61 | N/A |
| 5′-AATGATACGGCGACCACCGAGATCTACACTC TTTCCCTACACGACGCT∗C-3′ |
IDT, Behrens et al.61 | N/A |
| FLAG-PUS3: CAAGCCCATGGCTGACAACGATAC, CTTTCTTCAGACGCTGCACTTCC, GFP: AAGGG CATCGACTTCAAGG, TGCTTGTCGGCCATGATATAG |
Genomed | N/A |
| Recombinant DNA | ||
| Bac-to-Bac® Baculovirus Expression System | Kost et al.62 | N/A |
| pSB plasmid | Kowarz et al.63 | Addgene #60511 |
| transposase coding plasmid | Mates et al.64 | Addgene #34879 |
| Software and algorithms | ||
| MO. control software | NanoTemper Technologies | N/A |
| MO. AffinityAnalysis | Lin et al.65 | NanoTemper Technologies |
| Cryo-EM Single Particle Ab-Initio Reconstruction and Classification pipeline (CryoSPARC) | Punjani et al.66 | N/A |
| contrast transfer function (CTF) | Bepler et al.67 | N/A |
| blob picker and the TOPAZ particle picker | Bepler et al.67 | N/A |
| Alphafold2 | Jumper et al.42 | N/A |
| NAMDINATOR | Kidmose et al.68 | N/A |
| WinCoot | Emsley et al.69 | N/A |
| Phenix | Liebschner et al.70 | N/A |
| UCSF ChimeraX | Pettersen et al.71 | N/A |
| PR. PantaControl software | NanoTemper Technologies | N/A |
| PR. PantaAnalysis software | NanoTemper Technologies | N/A |
| CHOPCHOP | Labun et al.72 | https://chopchop.cbu.uib.no/ |
| TIDE | Brinkman et al.73 | https://tide.nki.nl/ |
| NIS-elements imaging software (v5.30.04) | Nikon | N/A |
| Cutadapt (v4.1) | Martin74 | https://cutadapt.readthedocs.io/en/stable/ |
| Bowtie (v2.5.0) | Langmead and Salzberg75 | https://github.com/BenLangmead/bowtie2 |
| Samtools (v1.15.1) | Danecek et al.76 | https://github.com/samtools/samtools |
| Bedtools (v2.30.0) | Quinlan and Hall77 | https://bedtools.readthedocs.io/en/latest/ |
| Bedops (v2.4.41) | Neph et al.78 | https://bedops.readthedocs.io/en/latest/ |
| Ensembldb (v2.22.0) | Rainer et al.79 | https://bioconductor.org/packages/release/bioc/html/ensembldb.html |
| AnnotationHub (v3.10.0) | Morgan and Shepherd80 | https://bioconductor.org/packages/release/bioc/html/AnnotationHub.html |
| Bio-Rad’s ImageLab (v5.1) | Bio-Rad | N/A |
| Other | ||
| Primerize method | Tian et al.81 | N/A |
| FPLC system | Cytiva | N/A |
| glow-discharger | Leica EM ACE 200 | N/A |
| Vitrobot Mark IV | Thermo Fisher | N/A |
| Titan Krios G3i | Thermo Fisher; Solaris, Poland | N/A |
| Gatan Quantum energy filter | Gatan | N/A |
| K3 Summit direct electron detector | Gatan | N/A |
| Prometheus PANTA | NanoTemper | N/A |
| ChemiDoc XRS+ System | Bio-Rad | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Sebastian Glatt (sebastian.glatt@uj.edu.pl).
Materials availability
Reagents generated in this study are available upon request.
Data and code availability
-
•
NGS raw and processed data have been deposited at GEO and are publicly available as of the date of publication. Accession number (GSE255287) is listed in the key resources table. The micrographs, atomic coordinates, and the cryo-EM maps of the described cryo-EM datasets have been deposited at the Electron Microscopy Public Image Archive, the Protein databank and the Electron Microscopy Data Bank and are publicly available as of the date of publication. The accession codes and DOI are listed in the Tables 1 and S1. Original gel images have been deposited at Mendeley and are publicly available as of the date of publication (https://doi.org/10.17632/gdw8pr456d.2)
-
•
All custom scripts described herein have been deposited on the Leidel Lab GitHub server and is publicly available (https://doi.org/10.5281/zenodo.11401096). Details are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Insect cell expression system
ORF-containing pFastBac constructs were transformed into a bacterial E. coli DH10Bac strain for production of the recombinant bacmid DNA. The sf9 and Hi5 cells were cultured in HyClone SFM4Insect cell culture media (SH30913, Cytiva) with 0.5% FBS in an incubator at 27 °C with 2% CO2.
Bacterial cell expression system
ORF-containing pETM30 constructs were transformed into a bacterial E. coli BL21 (DE3) CodonPlus-RIL strain. The cells were cultured in LB while shaking at 180 rpm at 37 °C.
Human cells lines
Flp-In™ T-REx™ 293 cells and HEK293T cells were grown in Dulbecco's Modified Eagle's Medium DMEM (D5671, Sigma) supplemented with 10% FBS (G3031P-500, Lucerna-Chem), 100 U penicillin, 100 μg streptomycin (P4333, Sigma), 2 mM Ala-Gln (G8541, Sigma) in a humidified incubator at 37 °C with 5% CO2.
Method details
Recombinant protein expression and purification
The codon-optimized sequence of a synthetic human PUS1 (Q9Y606) ORF or PUS3 (Q9BZE2) ORF fused with a glutathione-S-transferase (GST)-tag at N-terminus was cloned into the pETM30 or pFastBac vector, respectively. PUS1 and PUS3 mutants were generated using the standard mutagenesis PCR with primers that contain the mutation site sequence. PUS1 was recombinantly expressed in the strain BL21 (DE3) CodonPlus-RIL cells. Transformed cells were grown in LB broth and protein expressions were induced by isopropyl β-D-1-thiogalactopyranoside IPTG (1 mM) for overnight at 18 °C. PUS3 expression was performed using the Bac-to-Bac® Baculovirus Expression System62 according to the manual. In brief, the ORF-containing constructs were transformed into a bacterial E. coli DH10Bac strain for production of the recombinant bacmid DNA, which was transfected into sf9 insect cells. The recombinant baculoviruses were collected from sf9 cells and delivered to Hi5 cells to produce PUS3 recombinant proteins. The infected cells were cultured in HyClone SFM4Insect cell culture media with 0.5% FBS for three days, and then collected. PUS1 or PUS3 containing pellets were lysed in lysis buffer (50 mM HEPES, pH 7.5, 300 mM NaCl, 2 mM DTT, 5% glycerol, 2 mM MgCl2 containing protease inhibitors and DNase) followed by three cycles of freezing and thawing. The cell extract was sonicated, and the soluble fraction was separated from cell debris by centrifugation at 80,000 g for 90 min. The cleared supernatant was subjected to a GSTPrep column, and the column was washed with lysis buffer and washing buffer (same as lysis buffer but containing 1 M NaCl) and the protein of interest was then eluted in the elution buffer with 18 mM reduced GSH. The eluate was collected and incubated with GST-fused tobacco etch virus (TEV) protease overnight at 4 °C and followed by the removal of GST-fused TEV and the cleaved GST-tag via a second round of GST affinity chromatography. The flow through was concentrated and applied to a HiLoad Superdex 200 pg preparative gel filtration column, equilibrated with 20 mM HEPES, pH 7.5, 100 mM NaCl, 5 mM DTT, to obtain the pure and homogenous protein fractions. The purified protein was concentrated to 4 g/L and stored at -80 °C.
Production of RNAs
The DNA template for various tRNA and mRNA fragments was generated by PCR using the Primerize method81 and the T7 promoter sequence was introduced 5ʹ of the target sequence for a T7 RNA polymerase-driven in vitro transcription.65 An overnight T7-driven transcription reaction was performed at 37 °C with the following components: 20 mM Tris, pH 8.0, 5 mM DTT, 150 mM NaCl, 30 mM MgCl2, 2 mM spermidine, 20 mM NTPs, DNA template, RNasin (80 U/100 μl per IVT reaction), T7 RNA polymerase and pyrophosphatase (0.2 U/100 μL IVT reaction). For generating fluorescent RNAs, 1 mM of Cy5-cytidine (JenaBioscience) was added to the IVT reaction. RNase-free DNase I (Thermo Fisher Scientific) was added to digest the DNA template, followed by the addition of proteinase K (Merck) for 30 min and the reaction was stopped by the addition of 50 mM EDTA. The tRNA containing solution was subjected to a FPLC system using a DEAE weak anion exchange column (GE). RNA annealing was carried out by heating the RNA solution (prepared in annealing buffer containing 20 mM HEPES pH 7.5, 50 mM KCl, and 50 mM NaCl) to 80 °C for 2 min and slowly cooling to 25 °C with a ramp decrement (-0.4 °C/min) using a PCR program. A final concentration of 1 mM MgCl2 was added to the annealed RNA. The annealed RNAs were further purified using a Superdex 75 Increase gel filtration column (GE) in a buffer containing 20 mM HEPES, pH 7.5, 1 mM MgCl2, 150 mM NaCl and the fractions of interest were pooled, concentrated, and stored at -20 °C.
mRNA fragments were purified using MEGAclear™ Transcription Clean-Up Kit (AM 1908; Thermo Fisher Scientific) according to manufacturer’s protocol. The purified RNAs were run on a denaturing 10% UREA-PAGE for quality check, followed by temperature-gradient based annealing as described above. The mRNA was subsequently stored at -20 °C.
Microscale thermophoresis assay (MST)
For this assay, we used RNA that was labeled using Cy5-cytosine directly during in vitro transcription. 50 nM tRNA was mixed with serial dilutions of purified PUS3 with equal volume in 20 mM HEPES, pH 7.5, 100 mM NaCl, 2 mM MgCl2 and 2 mM DTT. The mixture was incubated at 4 °C for 30 min and then subjected to premium capillaries for measurement. The binding profiles were analyzed using MO. control software (NanoTemper Technologies) and the Kd values were calculated based on the measurement results (n=3) using MO. AffinityAnalysis (NanoTemper Technologies).65
Electron microscopy
QUANTIFOIL R2/1 copper grids (200 mesh) were cleaned using a glow-discharger (Leica EM ACE 200) at 8 mA for 60 seconds. PUS3 (0.3 g/l) with tRNA (10 μM) complex was prepared as mentioned above. Samples (2.5 μl) was plunge-frozen using a Vitrobot Mark IV (Thermo Fisher) set to 95% humidity and 4 °C with the following blotting parameters − 2 seconds for wait time, blot force 5 and 1 second for blotting time. Micrographs were acquired at 300 kV using a Titan Krios G3i (Thermo Fisher; Solaris, Poland) equipped with a Gatan BioQuantum energy filter and a K3 direct electron detector. Micrographs were collected (7353 micrographs for PUS3 and 8321 micrographs for PUS3-tRNA complex) with 0.86 Å pixel size and 0.5–3 μm under-focus for a total of 40 frames accumulating 40 e−/Å2 dose.
Image processing
Cryo-EM datasets were processed using Cryo-EM Single Particle Ab-Initio Reconstruction and Classification (CryoSPARC) pipeline.66 First, the collected movies were corrected with motion estimation and the averaged micrographs were then corrected with the contrast transfer function (CTF). Particles were picked using blob picker and the TOPAZ particle picker.67 Extracted particles were curated via 2D classification and ab-initio reconstructions. Junk particles were removed, and the rest particles were subjected to 3D classifications. The class with all required density was then refined with Non-Uniform refinement or Local refinement to improve the map quality.
Model building, refinement, and validation
An atomic model for the human PUS3 dimer was predicted using Alphafold2 as described previously.25 The model was first fitted into the map using NAMDINATOR68 and followed by manual curation in WinCoot.69 The obtained model models, except for the PUS3D118A apo structure, were then further refined and validated in Phenix.70 Figures were prepared using UCSF ChimeraX.71 Of note, the ASL as well as the CCA sequence at the 3ʹ end were omitted from the deposited atomic models, because the quality of the map was not sufficient to build a single representative model in these regions.
Protein size and dispersity measurements
Recombinant protein (10 μg) was prepared in 10 μl buffer (20 mM HEPES, pH 7.5, 150 mM NaCl and 5 mM DTT). The protein solution was subjected to a glass capillary and the measurement of hydrodynamic radius (rH) by dynamic light scattering (DLS) was performed using the Prometheus PANTA. PR. PantaControl software (NanoTemper Technologies) were used to control the experiment in modes: size analysis and thermal melting. Three independent measurements were performed, and datasets were analyzed and merged using the PR. PantaAnalysis software (NanoTemper Technologies).
Pseudouridylation and primer extension assays
PUS3 (0.3 μg) was mixed with tRNA substrates (6 μg) in a 25-μl reaction volume in reaction buffer containing 100 mM ammonium acetate, 100 mM NaCl, 20 mM Tris, pH 8.0, 5 mM MgCl2, 5 mM DTT.82 The reaction was performed at 37 °C for 5 min (in the condition with tRNA) or 1 h (in the condition with mRNAs) and the RNA was phenol-chloroform extracted, precipitated in ethanol and redissolved in H2O for the subsequent CMC treatment and reverse transcription. CMC with the methyl-p-toluenesulfonate salt (Sigma, Missouri, United States) was prepared at 0.2 M concentration in BEU buffer (50 mM bicine, pH 8.3, 4 mM EDTA, and 7 M urea).82 Twenty microliters of total RNA extract (60 μg), total tRNA extract (15 μg) or PUS3 treated tRNA (0.5-8 μg) were mixed with 100 μl of 0.2 M CMC at 37 °C for 30 min to form the Ψ-CMC conjugate. As a negative control, we retained an aliquot of each sample without adding CMC. Samples were then precipitated and treated with alkaline conditions (50 mM sodium bicarbonate, pH 10.7 and 2 mM EDTA) at 37 °C for 2 h to reverse the CMC conjugation on G and U nucleotides but not on Ψ. The RNA was precipitated and dissolved in H2O (15 μl). The obtained RNA was subjected to primer extension to detect the presence of Ψs.
Primer extension assays require BEU-treated and CMC-treated tRNA as the templates. The reverse transcription reaction was primed using tRNA-specific Cy5-labeled primer. Each primer was designed to be complementary to the 3’-end of each tRNA substrate (position 50-76 of tRNA). All ingredients were prepared in a 10-μl reaction volume. 80 ng tRNA was mixed with 1 μl 10 mM primer and 4.5 μl H2O, followed by heating at 80 °C for 3 min and 65 °C for 5 min and cooling down on ice for 1 minute. The sample mixture was then mixed with 2 μl of 5X reaction buffer, 0.5 μl of 100 mM DTT, 0.5 μl 10mM dNTPs, and 0.5 μl SuperScriptIII. The reverse transcription reaction was carried out at 50 °C for 15 min and followed by 85 °C for 5 min to stop the reaction. To remove proteins in the reaction, proteinase K (>700U) was added, and the sample was incubated at 37 °C for 30 min. The reverse transcribed products were resolved in a 15% urea denaturing gel run at 200 V for 60 min. Products with different sizes were visualized using a gel scanner.
Cell culture and CRISPR knockout generation
Flp-In™ T-REx™ 293 cells were grown in Dulbecco's Modified Eagle's Medium DMEM (D5671, Sigma) supplemented with 10% FBS (G3031P-500, Lucerna-Chem), 100 U penicillin, 100 μg streptomycin (P4333, Sigma), 2 mM Ala-Gln (G8541, Sigma) in a humidified incubator at 37 °C with 5% CO2. The online tool CHOPCHOP (https://chopchop.cbu.uib.no/) was used to predict the best target site in each gene of interest.72 The targeting CRISPR RNA (crRNA) was ordered from Integrated DNA Technologies (IDT), together with an ATTO550 labelled trans activating RNA (tracrRNA) (IDT, 1075928).
PUS3_crRNA:5′/AltR1/rUrCrUrGrCrUrCrArArGrUrArCrArGrCrUrArGrUrGrUrUrUrUrArGrArGrCrUrArUrGrCrU/AlrR2/3′; PUS1_crRNA:5′/AltR1/rArArUrArCrArGrCrCrUrGrArCrCrGrGrArCrGrArGrUrUrUrUrArGrArGrCrUrArUrGrCrU/AltR2/-3′.
crRNA:tracrRNA duplex formation, ribonucleoprotein (RNP) complex formation, and transfection was performed according to the IDT transfection protocol in Flp-In™ T-REx™ 293 cells which were seeded in 96-wells. The RNP was formed by using TrueCut Cas9 Protein v2 (Thermofisher, A36498), and the reverse transfection was performed using Lipofectamine CRISPRMAX Cas9 Transfection Reagent (Thermofisher, CMAX00001). Successful transfection was validated after 24 h by visualizing the ATTO550 tracrRNA inside cells using fluorescence microscopy. Two days after transfection, we diluted the cells to 10 cells/ml and seeded single cells into 96-well plates in 100 μl complete medium to obtain clonal cell lines. The genomic CRISPR-Cas9 target region of single cell colonies was analyzed by TIDE73 and mutants in which both alleles contained a frameshift mutation were used for further analysis. Live-cell images were taken from cultured cells with the DS-Fi3 camera attached to the Eclipse Ts2-FL microscope using NIS-elements imaging software (version 5.30.04, Nikon).
Pseudo-seq library preparation
Flp-In™ T-REx™ 293 cells were harvested with 90% confluency using 1 ml TRIzol per 10-cm dish and RNA was extracted according to the manufacturer protocol. Pseudo-seq libraries were prepared as described before33 with some adaptations. Poly-A RNA enrichment was performed with Poly(A)Purist™ MAG Kit (AM1922, Thermofisher) using 200 μg total RNA input. RNA was fragmented in 10 mM ZnCl2 for 55 s at 94 °C and quenched with 20 mM EDTA followed by ethanol precipitation. RNA was subsequently either CMC treated (0.4 M, +CMC) or mock treated (-CMC) in BEU buffer for 45 min at 40 °C and 1000 rpm. After ethanol precipitation, unspecific CMC labelling was removed by incubating the RNA in sodium carbonate buffer for 2 h at 50 °C and 1000 rpm followed by ethanol precipitation. RNA ends were repaired with T4 PNK (M0201L, NEB) for 2 h at 37 °C, precipitated and size selected (120-140 nt) by Urea-PAGE. A 3′ adenylated adapter (5′-AppGATATCGTCAAGATCGGAAGAGCACACGTCTGAA-ddC-3′)61 was then ligated to the RNA fragments using T4 RNA ligase (M0373L, NEB) for 4 h at 22 °C followed by ethanol precipitation. The reverse transcription was performed with AMV RT (M5108, Promega) and RT primer: 5′-pRNAGATCGGAAGAGCGTCGTGTAGGGAAAGAG-iSp18-GTGACTGGAGTTCAGACGTGTGCTC-3′. Truncated cDNAs (110-180 nt) were size selected using Urea-PAGE. Gel purified cDNA was circularized using CircLigase ssDNA ligase II (CL9025K, Epicentre) and PCR amplified with forward primer (5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCT∗C-3′) and barcode reverse primer (NEBNext® Multiplex Oligos for Illumina®, Index Primers Set 1-4, NEB E7335S, E7500S, E7710S, E7730S). PCR products were PAGE purified and sequenced on an Illumina NovaSeq 6000 using an SP and S2 flow cell in SR75 mode to yield 130 million reads per library (Table S3). We performed six replicates per condition (treated and untreated) in WT, PUS1-/- and PUS3-/- (36 libraries in total).
Analysis of Pseudo-seq datasets
After concatenating technical replicates (different sequencing lanes and sequencing runs of the same biological replicate) into a single FASTQ file, reads were adapter clipped and random nucleotides were trimmed using cutadapt version 4.1.74 Reads were then mapped to the protein coding transcriptome (MANE GRCh38 v1.0 ensemble rna) after mitochondrial transcripts from the human cDNA collection (Homo_sapiens.GRCh38.cdna.all.fa) were included using bowtie version 2.5.0.75,83 We observed that many reads had an non-templated nucleotide at the 5' end, likely due to the terminal deoxynucleotidyl transferase (TdT) activity of the AMV reverse transcriptase used in our study.84 For accurate mapping of Ψ sites, using the correct 5' end of the reads is crucial. Therefore, we developed a custom Python script to remove mismatching 5' nucleotides from all reads in all libraries. However, it's important to note that at a Ψ site, reads will ideally start at the +1 position (one nucleotide downstream) of the annotated T. If the cDNA contains a 5' T due to the TdT activity of the RT, this T would not be considered a mismatch. This situation would result in the start of the read being at the +0 position, making it impossible to identify this site as a Ψ site using these reads. To overcome this, we also removed all 5' Ts from all reads in all libraries. After thorough testing of the adjusted reads, we are confident that we successfully corrected for the TdT activity of the RT without losing information. This is supported by various quality control figures presented in the main text. The corrected reads were then mapped to the protein coding transcriptome as before. In addition, we mapped reads to rRNA extracted from NCBI (NR_145822.1, NR_145820.1, NR_145821.1 and NR_023363.1), snRNA (extracted with BioMart) and lncRNA (unique sequences of the high confidence set of LNCipedia Version 5.2)85 to investigate Ψ sites in these RNA classes and corrected the reads like for mRNA.
The analysis was performed based on calculating the ratio of 5' read ends (position at which the RT stalled) over the overall coverage at each transcript position as reported before.86 In brief, resulting SAM files were sorted and indexed using samtools version 1.15.1.76 Afterwards, the 5' coverage as well as the overall coverage at each transcript position was calculated using bedtools version 2.30.0 and bedops version 2.4.41.77,78 Both files of all replicates CMC treated and untreated were combined in one table for WT, PUS1-/- and PUS3-/- using GNU Awk 4.0.2. The average coverage, 5ʹ coverage/coverage, average 5ʹ coverage/coverage and the Ψ score was calculated using a custom R script (R version 4.3.2). The Ψ score was calculated by dividing the average 5ʹ coverage/coverage of the CMC treated dataset by the average 5ʹ coverage/coverage of the untreated dataset. Bedtools was used to call every nucleotide at each transcript position.
We aimed to generate ROC curves to select the optimal cutoffs for our analyses. However, there is a complication in doing so: Most known Ψ sites reside in highly abundant rRNA and snRNA. However, we used polyA-purified RNA to identify Ψ sites in mRNA targets that are less abundant and have different characteristics than rRNA and snRNA. Therefore, we did not use known rRNA and snRNA to define the cutoff as this may be misleading. Instead, we used an existing comparison of high-confidence Ψ sites based on four publications created by Tavakoli et al.: 1. RBS-Seq87 2. CeU-Seq,30 3. Pseudo-seq35 and 4. nanopore sequencing32 As these studies used different approaches, we expect that true positives are likely found in the overlap between these studies. However, the overlap between these studies is small, reflecting the general challenge to reliably assign Ψ sites in transcriptome-wide approaches as highlighted before.88,89
Ψ sites detected in 4/4 datasets: 5
Ψ sites detected in 3/4 datasets: 33
Ψ sites detected in 2/4 datasets: 183
Ψ sites detected in 1/4 datasets: 304
To be able to use a sufficient number of Ψ sites for a meaningful ROC curve, we chose 183 Ψ sites that are detected in at least 2 out of 4 datasets. This selection criterion provides a level of confidence in the true positive nature of these sites, even though we are aware that this list likely contains false positives.
We generated ROC curves considering candidate sites with 1 %, 5 %, 10 %, 2 0%, and 50 % of reads generated from synthesized cDNA stalling at a CMC-labelled Ψ (stalling fraction, 5ʹ read coverage/coverage) and Ψ score thresholds ranging from 10 to 0.1.
Filtering according to average coverage (50), 5ʹ coverage per CMC treated library (5 per library, 30 in total), average 5ʹ coverage/coverage of CMC treated dataset (0.2, stalling fraction), Ψ score (3.5) and a “T” one nucleotide upstream was performed with a custom python script. A Ψ site was determined to be PUS1/3 dependent if the filtering criteria were met in the wild-type and PUS1-/- or PUS3-/- datasets, respectively, but the stalling fraction was below 0.2 and the Ψ score below 3.5 and at least 50% of the score in WT in the PUS3-/- or PUS1-/- datasets. Transcript positions were transformed into genomic coordinates using the transcriptToGenome of ensembldb 2.22.0.79 For this the required resources EnsDb.Hsapiens.v108 were extracted from AnnotationHub version 3.10.0.80 The correlation plots were generated using log-transformed, library-size-normalized coverage data from each library. The k-mer sequences and transcript locations were extracted using custom Python scripts.
Western blot
Flp-In™ T-REx™ 293 and HEK293T cells were cultured as described above and lysed in modified RIPA buffer (65 mM Tris, pH 7.4, 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 1 mM EDTA, 0.1% SDS and 1% Triton-X100) supplemented with protease inhibitor cocktail cOmplete EDTA-free and phosphatase inhibitors by pipetting. The lysates were sonicated using Bioruptor® Plus for 10 cycles 20/20 s and centrifuged at 21,130 g for 10 min at 4 °C to remove debris. The supernatant was collected, and protein concentration was measured using a Bradford assay. Protein samples were mixed with Laemmli loading dye with β-mercaptoethanol and boiled for 5 min. Protein samples (30 μg) were resolved in 12% SDS-PAGE stain-free gel with PageRuler prestained protein ladder as a molecular weight marker. After electrophoresis, proteins in the gel were visualized using Bio-Rad’s ImageLab software (version 5.1) to verify equal protein loading. Proteins were transferred to PVDF membranes (pore size 0.45 μm, Immobilon-P or Immobilon-FL, ThermoFisher) using Trans-Blot Turbo Blotting System. Membranes were blocked for 1 h at room temperature in 5% non-fat dry milk in PBS-T (PBS with 0.01% Tween-20), followed by overnight incubation at 4°C with primary antibodies (rabbit anti-PUS3 C-terminal domain (#ab211270, Abcam); mouse anti-GAPDH (#MAB374, Sigma Aldrich); anti-FLAG® M2 antibody (F3165, Sigma); anti B1-lamin (10H34L18 Invitrogen); or anti-GFP (ab290, Abcam)) in 5% milk in PBS-T (1:500). Blots were washed three times 5 min with PBS-T, incubated 1 h at room temperature with HRP-conjugated secondary antibody (1:2000 in 5% milk) and washed three times 5 min with PBS-T. HRP signal was detected using SuperSignal™ West Pico PLUS Substrate (#34577, ThermoFisher) using the ChemiDoc XRS+ System.
RNA extraction and quantitative real-time PCR (qRT-PCR)
HEK293T cells were plated onto 6-well culture plates and at 90% confluency were scraped in 400 μl of lysis buffer and lysed by pipetting. RNA was isolated using Universal RNA Purification Kit (EurX) following the manufacturer’s protocol. The isolated total RNA was treated with DNase and concentrated using Clean-Up RNA Concentrator kit (A&A Biotechnology). The quantification and purity of the RNA were assessed with the NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific). RNA extract (1.0 μg) was reverse transcribed to cDNA using M-MLV RT (Promega) and oligo(dT)15 primers. The obtained cDNA was used for the quantification of recombinant FLAG-PUS3 variants mRNA levels using qPCR.
qPCR was performed using RT-HS-PCR-Mix-SYBR-A (A&A Biotechnology). Levels of the recombinant PUS3 mRNA were normalized to GFP expression and analyzed using the ΔΔCt method. The following primers were used: FLAG-PUS3: CAAGCCCATGGCTGACAACGATAC, CTTTCTTCAGACGCTGCACTTCC, GFP: AAGGGCATCGACTTCAAGG, TGCTTGTCGGCCATGATATAG. Expression levels of PUS3 variants were normalized against EGFP within the same transposition cassette. Relative PUS3 expression levels were calculated for each variant against wild-type PUS3.
Total tRNA extraction
HEK293T cells were cultured and collected from nine T75 flasks with 90% confluency. The cells were washed with PBS and lysed in 335 μl lysis buffer (10 mM Tris-HCl pH 7.5, 100 mM NaCl, 10 mM MgCl2, 1% Triton X-100, 0.5 mM DTT and 0.5% sodium deoxycholate). An equal volume of water was added to the lysate and the total RNA was isolated by three subsequent extractions of one volume of acid phenol-chloroform (Acid-Phenol:Chloroform, pH 4.5 (with IAA, 125:24:1)) and followed by final extraction with one volume of chloroform. For each extraction step, the mixture was vortexed thoroughly and followed by centrifugation at 4500 × g for 10 min at 4 °C. The upper aqueous phase was transferred to fresh tube and the extraction was performed again. The RNA, in the upper phase, was precipitated with 0.1 volume of 3 M sodium acetate (pH 6.3), 3 volumes of 96% EtOH, and 10 μg glycogen. The solution was incubated overnight at -80 °C and RNA was spun down at 7100 g for 30 min at 4 °C. The RNA pellet was washed in 70% EtOH and air-dried for 2 min. The pellet was then dissolved in RNase-free water and subjected to total tRNA isolation. A NucleoBond AX100 column was equilibrated with 10 ml equilibration buffer with Triton X-100 (10 mM Bis-Tris HCl, pH 6.3, 200 mM KCl, 15% EtOH and 0.15% Triton X-100). The total RNA (800–900 μg) was dissolved in 2 ml equilibration buffer without Triton X-100 and applied to the column. The column was washed twice with 12 ml wash buffer (10 mM Bis-Tris HCl, pH 6.3, 300 mM KCl, 15% EtOH). Bound tRNA was eluted with 12 ml elution buffer (10 mM Bis-Tris HCl, pH 6.3, 750 mM KCl, 1% EtOH) and followed by precipitation using 2.5 volumes of 96% EtOH and 10 μg glycogen. The solution was incubated overnight at -80 °C and tRNA was spun down at 7100 g for 30 min at 4 °C. The pellet was washed twice in 70% EtOH and air-dried for 2 min. The tRNA pellet was dissolved in 30 μl of RNase-free water. The quality of the extracted tRNA was checked via resolving the samples in a 10% urea gel, staining by ethidium bromide solution, and visualizing with a Bio-Rad ChemiDoc imaging system.
Stable cell line generation
FLAG-coding sequence with a Kozak sequence (5'-CCACGATGGAC-3') was cloned into the pSB plasmid (Addgene #60511)63 using the NcoI restriction site to create FLAG-PUS3. We used restriction enzyme cloning to shuffle the ORF of PUS3 variants from the pFast-Bac plasmid to the FLAG-pSB at NcoI and HindIII cloning sites. The plasmids were sequenced to confirm the in-frame construct. In this vector, the FLAG-PUS3 fusion gene is under control of a strong EF-1α promoter, whereas the eGFP-P2A-PuroR (enhanced green fluorescent protein sequence separated by self-cleaving P2A peptide from the puromycin resistance gene) was under control of a synthetic promotor in the same transposition cassette. HEK293T cells were cultured as mentioned above and co-transfected using Lipofectamine 3000 with FLAG-PUS3-pSB plasmid and transposase coding plasmid (Addgene #34879)64 at 70% confluency. We performed the transfection following the manufacturer’s protocol. The culture media was replaced 24 h after transfection. 2 μg/ml puromycin was added to select for cells with the cassette integrated into the genome, and the procedure was continued for 7 days with subsequent passages.
Quantification and statistical analysis
Graphed datasets are expressed as mean ± standard error of the mean (SEM) from three independent experiments. Statistical analysis was performed with GraphPad Prism software (version 7.05) using a one-way ANOVA (α = 0.05) with Dunnett’s multiple comparisons test. Statistically significant differences are indicated (∗∗p ≤ 0.01; ∗∗∗ p ≤ 0.001; ∗∗∗∗ p ≤0.0001).
Acknowledgments
We thank Jakub Nowak, Marcin Jaciuk, and Jie Wu for discussions and the Mass Spectrometry Core Facility (MCB) and Next Generation Sequencing Platform (University of Bern) for their support. The work was supported by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (101001394; S.G.), the Swiss National Science Foundation (310030_184947; S.A.L.), and from the National Science Centre (2019/35/D/NZ1/02397; T.-Y.L.). In addition, we thank the MCB SBCF (TEAMTECH_CORE_FACILITY/2017-4/6; FNP), SOLARIS (Polish Ministry and Higher Education; 1/SOL/2021/2), and PLGrid ACK Cyfronet (PLG/2023/016300) for access. The open-access publication of this article was funded by the Priority Research Area BioS under the program “Initiative of Excellence—Research University” at the Jagiellonian University in Krakow.
Author contributions
Conceptualization, T.-Y.L., L.K., S.A.L., and S.G.; methodology, T.-Y.L., L.K., A.C.-G., and J.J.; software, L.K.; validation, D.D., R.M., M.W., J.K., J.B., M.R., P.I., G.W., T.R., C.F., L.K., and J.J.; investigation, T.-Y.L., L.K., and J.J.; writing—original draft, T.-Y.L., J.J., and L.K.; writing—review and editing, T.-Y.L., L.K., S.A.L., and S.G.; supervision, T.-Y.L., S.A.L., and S.G.; funding acquisition, T.-Y.L., S.A.L., and S.G.
Declaration of interests
The authors declare no competing interests.
Published: July 11, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.molcel.2024.06.013.
Contributor Information
Ting-Yu Lin, Email: ting-yu.lin@durham.ac.uk.
Sebastian A. Leidel, Email: sebastian.leidel@unibe.ch.
Sebastian Glatt, Email: sebastian.glatt@uj.edu.pl.
Supplemental information
References
- 1.Boccaletto P., Stefaniak F., Ray A., Cappannini A., Mukherjee S., Purta E., Kurkowska M., Shirvanizadeh N., Destefanis E., Groza P., et al. MODOMICS: a database of RNA modification pathways. 2021 update. Nucleic Acids Res. 2022;50:D231–D235. doi: 10.1093/nar/gkab1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin T.Y., Mehta R., Glatt S. Pseudouridines in RNAs: switching atoms means shifting paradigms. FEBS Lett. 2021;595:2310–2322. doi: 10.1002/1873-3468.14188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Durant P.C., Davis D.R. Stabilization of the anticodon stem-loop of tRNA(Lys,3) by an A+-C base-pair and by pseudouridine. J. Mol. Biol. 1999;285:115–131. doi: 10.1006/jmbi.1998.2297. [DOI] [PubMed] [Google Scholar]
- 4.Davis D.R. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 1995;23:5020–5026. doi: 10.1093/nar/23.24.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vögele J., Duchardt-Ferner E., Kruse H., Zhang Z., Sponer J., Krepl M., Wöhnert J. Structural and dynamic effects of pseudouridine modifications on non-canonical interactions in RNA. RNA. 2023;29:790–807. doi: 10.1261/rna.079506.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kierzek E., Malgowska M., Lisowiec J., Turner D.H., Gdaniec Z., Kierzek R. The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic Acids Res. 2014;42:3492–3501. doi: 10.1093/nar/gkt1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krutyhołowa R., Zakrzewski K., Glatt S. Charging the code — tRNA modification complexes. Curr. Opin. Struct. Biol. 2019;55:138–146. doi: 10.1016/j.sbi.2019.03.014. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki T., Yashiro Y., Kikuchi I., Ishigami Y., Saito H., Matsuzawa I., Okada S., Mito M., Iwasaki S., Ma D., et al. Complete chemical structures of human mitochondrial tRNAs. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-18068-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charette M., Gray M.W. Pseudouridine in RNA: what, where, how, and why. IUBMB Life. 2000;49:341–351. doi: 10.1080/152165400410182. [DOI] [PubMed] [Google Scholar]
- 10.Rintala-Dempsey A.C., Kothe U. Eukaryotic stand-alone pseudouridine synthases–RNA modifying enzymes and emerging regulators of gene expression? RNA Biol. 2017;14:1185–1196. doi: 10.1080/15476286.2016.1276150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang L., Pookanjanatavip M., Gu X., Santi D.V. A conserved aspartate of tRNA pseudouridine synthase is essential for activity and a probable nucleophilic catalyst. Biochemistry. 1998;37:344–351. doi: 10.1021/bi971874+. [DOI] [PubMed] [Google Scholar]
- 12.McCleverty C.J., Hornsby M., Spraggon G., Kreusch A. Crystal structure of human Pus10, A novel pseudouridine synthase. J. Mol. Biol. 2007;373:1243–1254. doi: 10.1016/j.jmb.2007.08.053. [DOI] [PubMed] [Google Scholar]
- 13.Dong X., Bessho Y., Shibata R., Nishimoto M., Shirouzu M., Kuramitsu S., Yokoyama S. Crystal structure of the tRNA pseudouridine synthase TruA from Thermus thermophilus HB8. RNA Biol. 2006;3:115–122. doi: 10.4161/rna.3.3.3286. [DOI] [PubMed] [Google Scholar]
- 14.Foster P.G., Huang L., Santi D.V., Stroud R.M. The structural basis for tRNA recognition and pseudouridine formation by pseudouridine synthase I. Nat. Struct. Biol. 2000;7:23–27. doi: 10.1038/71219. [DOI] [PubMed] [Google Scholar]
- 15.Czudnochowski N., Wang A.L., Finer-Moore J., Stroud R.M. In human pseudouridine synthase 1 (hPus1), a C-terminal helical insert blocks tRNA from binding in the same orientation as in the Pus1 bacterial homologue TruA, consistent with their different target selectivities. J. Mol. Biol. 2013;425:3875–3887. doi: 10.1016/j.jmb.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan H., Agarwalla S., Moustakas D.T., Finer-Moore J., Stroud R.M. Structure of tRNA pseudouridine synthase TruB and its RNA complex: RNA recognition through a combination of rigid docking and induced fit. Proc. Natl. Acad. Sci. USA. 2003;100:12648–12653. doi: 10.1073/pnas.2135585100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guegueniat J., Halabelian L., Zeng H., Dong A., Li Y., Wu H., Arrowsmith C.H., Kothe U. The human pseudouridine synthase PUS7 recognizes RNA with an extended multi-domain binding surface. Nucleic Acids Res. 2021;49:11810–11822. doi: 10.1093/nar/gkab934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlile T.M., Martinez N.M., Schaening C., Su A., Bell T.A., Zinshteyn B., Gilbert W.V. mRNA structure determines modification by pseudouridine synthase 1. Nat. Chem. Biol. 2019;15:966–974. doi: 10.1038/s41589-019-0353-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hur S., Stroud R.M. How U38, 39, and 40 of many tRNAs become the targets for pseudouridylation by TruA. Mol. Cell. 2007;26:189–203. doi: 10.1016/j.molcel.2007.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lecointe F., Simos G., Sauer A., Hurt E.C., Motorin Y., Grosjean H. Characterization of yeast protein Deg1 as pseudouridine synthase (Pus3) catalyzing the formation of Ψ38 and Ψ39 in tRNA anticodon loop. J. Biol. Chem. 1998;273:1316–1323. doi: 10.1074/jbc.273.3.1316. [DOI] [PubMed] [Google Scholar]
- 21.Metodiev M.D., Assouline Z., Landrieu P., Chretien D., Bader-Meunier B., Guitton C., Munnich A., Rötig A. Unusual clinical expression and long survival of a pseudouridylate synthase (PUS1) mutation into adulthood. Eur. J. Hum. Genet. 2015;23:880–882. doi: 10.1038/ejhg.2014.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeharia A., Fischel-Ghodsian N., Casas K., Bykhocskaya Y., Tamari H., Lev D., Mimouni M., Lerman-Sagie T. Mitochondrial myopathy, sideroblastic anemia, and lactic acidosis: an autosomal recessive syndrome in Persian Jews caused by a mutation in the PUS1 gene. J. Child Neurol. 2005;20:449–452. doi: 10.1177/08830738050200051301. [DOI] [PubMed] [Google Scholar]
- 23.Shaheen R., Tasak M., Maddirevula S., Abdel-Salam G.M.H., Sayed I.S.M., Alazami A.M., Al-Sheddi T., Alobeid E., Phizicky E.M., Alkuraya F.S. PUS7 mutations impair pseudouridylation in humans and cause intellectual disability and microcephaly. Hum. Genet. 2019;138:231–239. doi: 10.1007/s00439-019-01980-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaheen R., Han L., Faqeih E., Ewida N., Alobeid E., Phizicky E.M., Alkuraya F.S. A homozygous truncating mutation in PUS3 expands the role of tRNA modification in normal cognition. Hum. Genet. 2016;135:707–713. doi: 10.1007/s00439-016-1665-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin T.Y., Smigiel R., Kuzniewska B., Chmielewska J.J., Kosińska J., Biela M., Biela A., Kościelniak A., Dobosz D., Laczmanska I., et al. Destabilization of mutated human PUS3 protein causes intellectual disability. Hum. Mutat. 2022;43:2063–2078. doi: 10.1002/humu.24471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Festen E.A.M., Goyette P., Green T., Boucher G., Beauchamp C., Trynka G., Dubois P.C., Lagacé C., Stokkers P.C.F., Hommes D.W., et al. A meta-analysis of genome-wide association scans identifies IL18RAP, PTPN2, TAGAP, and PUS10 as shared risk loci for Crohn’s disease and celiac disease. PLoS Genet. 2011;7 doi: 10.1371/JOURNAL.PGEN.1001283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lucas M.C., Pryszcz L.P., Medina R., Milenkovic I., Camacho N., Marchand V., Motorin Y., Ribas de Pouplana L.R., Novoa E.M. Quantitative analysis of tRNA abundance and modifications by nanopore RNA sequencing. Nat. Biotechnol. 2024;42:72–86. doi: 10.1038/s41587-023-01743-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helm M., Schmidt-dengler M.C., Weber M., Motorin Y., Gutenberg-universität J. General Principles for the Detection of Modified Nucleotides in RNA by Specific Reagents. Adv. Biol. 2021;5 doi: 10.1002/adbi.202100866. [DOI] [PubMed] [Google Scholar]
- 29.Martinez N.M., Su A., Burns M.C., Nussbacher J.K., Schaening C., Sathe S., Yeo G.W., Gilbert W.V. Pseudouridine synthases modify human pre-mRNA co-transcriptionally and affect pre-mRNA processing. Mol. Cell. 2022;82:645–659.e9. doi: 10.1016/j.molcel.2021.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X., Zhu P., Ma S., Song J., Bai J., Sun F., Yi C. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol. 2015;11:592–597. doi: 10.1038/nchembio.1836. [DOI] [PubMed] [Google Scholar]
- 31.Dai Q., Zhang L.S., Sun H.L., Pajdzik K., Yang L., Ye C., Ju C.W., Liu S., Wang Y., Zheng Z., et al. Quantitative sequencing using BID-seq uncovers abundant pseudouridines in mammalian mRNA at base resolution. Nat. Biotechnol. 2023;41:344–354. doi: 10.1038/s41587-022-01505-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tavakoli S., Nabizadeh M., Makhamreh A., Gamper H., McCormick C.A., Rezapour N.K., Hou Y.M., Wanunu M., Rouhanifard S.H. Semi-quantitative detection of pseudouridine modifications and type I/II hypermodifications in human mRNAs using direct long-read sequencing. Nat. Commun. 2023;14 doi: 10.1038/s41467-023-35858-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carlile T.M., Rojas-Duran M.F., Zinshteyn B., Shin H., Bartoli K.M., Gilbert W.V. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515:143–146. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lovejoy A.F., Riordan D.P., Brown P.O. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS One. 2014;9 doi: 10.1371/journal.pone.0110799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carlile T.M., Rojas-Duran M.F., Gilbert W.V. Transcriptome-wide identification of pseudouridine modifications using pseudo-seq. Curr. Protoc. Mol. Biol. 2015;112:4.25.1–4.25.24. doi: 10.1002/0471142727.mb0425s112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwartz S., Bernstein D.A., Mumbach M.R., Jovanovic M., Herbst R.H., León-Ricardo B.X., Engreitz J.M., Guttman M., Satija R., Lander E.S., et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159:148–162. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mauger D.M., Cabral B.J., Presnyak V., Su S.V., Reid D.W., Goodman B., Link K., Khatwani N., Reynders J., Moore M.J., et al. mRNA structure regulates protein expression through changes in functional half-life. Proc. Natl. Acad. Sci. USA. 2019;116:24075–24083. doi: 10.1073/pnas.1908052116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song J., Dong L., Sun H., Luo N., Huang Q., Li K., Shen X., Jiang Z., Lv Z., Peng L., et al. CRISPR-free, programmable RNA pseudouridylation to suppress premature termination codons. Mol. Cell. 2023;83:139–155.e9. doi: 10.1016/j.molcel.2022.11.011. [DOI] [PubMed] [Google Scholar]
- 39.Eyler D.E., Franco M.K., Batool Z., Wu M.Z., Dubuke M.L., Dobosz-Bartoszek M., Jones J.D., Polikanov Y.S., Roy B., Koutmou K.S. Pseudouridinylation of mRNA coding sequences alters translation. Proc. Natl. Acad. Sci. USA. 2019;116:23068–23074. doi: 10.1073/pnas.1821754116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bykhovskaya Y., Casas K., Mengesha E., Inbal A., Fischel-Ghodsian N. Missense mutation in pseudouridine synthase 1 (PUS1) causes mitochondrial myopathy and sideroblastic anemia (MLASA) Am. J. Hum. Genet. 2004;74:1303–1308. doi: 10.1086/421530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamma T., Ferré-D’Amaré A.R. Pseudouridine synthases. Chem. Biol. 2006;13:1125–1135. doi: 10.1016/j.chembiol.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 42.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hayne C.K., Butay K.J.U., Stewart Z.D., Krahn J.M., Perera L., Williams J.G., Petrovitch R.M., Deterding L.J., Matera A.G., Borgnia M.J., et al. Structural basis for pre-tRNA recognition and processing by the human tRNA splicing endonuclease complex. Nat. Struct. Mol. Biol. 2023;30:824–833. doi: 10.1038/s41594-023-00991-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sekulovski S., Sušac L., Stelzl L.S., Tampé R., Trowitzsch S. Structural basis of substrate recognition by human tRNA splicing endonuclease TSEN. Nat. Struct. Mol. Biol. 2023;30:834–840. doi: 10.1038/s41594-023-00992-y. [DOI] [PubMed] [Google Scholar]
- 45.Yuan L., Han Y., Zhao J., Zhang Y., Sun Y. Recognition and cleavage mechanism of intron-containing pre-tRNA by human TSEN endonuclease complex. Nat. Commun. 2023;14 doi: 10.1038/s41467-023-41845-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang X., Yang F., Zhan X., Bian T., Xing Z., Lu Y., Shi Y. Structural basis of pre-tRNA intron removal by human tRNA splicing endonuclease. Mol. Cell. 2023;83:1328–1339.e4. doi: 10.1016/j.molcel.2023.03.015. [DOI] [PubMed] [Google Scholar]
- 47.Begik O., Lucas M.C., Pryszcz L.P., Ramirez J.M., Medina R., Milenkovic I., Cruciani S., Liu H., Vieira H.G.S., Sas-Chen A., et al. Quantitative profiling of pseudouridylation dynamics in native RNAs with nanopore sequencing. Nat. Biotechnol. 2021;39:1278–1291. doi: 10.1038/s41587-021-00915-6. [DOI] [PubMed] [Google Scholar]
- 48.Zhang M., Jiang Z., Ma Y., Liu W., Zhuang Y., Lu B., Li K., Peng J., Yi C. Quantitative profiling of pseudouridylation landscape in the human transcriptome. Nat. Chem. Biol. 2023;19:1185–1195. doi: 10.1038/s41589-023-01304-7. [DOI] [PubMed] [Google Scholar]
- 49.Lorenz R., Bernhart S.H., Höner Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011;6 doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nøstvik M., Kateta S.M., Schönewolf-Greulich B., Afenjar A., Barth M., Boschann F., Doummar D., Haack T.B., Keren B., Livshits L.A., et al. Clinical and molecular delineation of PUS3-associated neurodevelopmental disorders. Clin. Genet. 2021;100:628–633. doi: 10.1111/cge.14051. [DOI] [PubMed] [Google Scholar]
- 51.Petrov D.P., Kaiser S., Kaiser S., Jung K. Opportunities and challenges to profile mRNA modifications in Escherichia coli. ChemBioChem. 2022;23 doi: 10.1002/cbic.202200270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arena F., Ciliberto G., Ciampi S., Cortese R. Purification of pseudouridylate synthetase I from salmonella typhimurium. Nucleic Acids Res. 1978;5:4523–4536. doi: 10.1093/nar/5.12.4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Phannachet K., Elias Y., Huang R.H. Dissecting the roles of a strictly conserved tyrosine in substrate recognition and catalysis by pseudouridine 55 synthase. Biochemistry. 2005;44:15488–15494. doi: 10.1021/bi050961w. [DOI] [PubMed] [Google Scholar]
- 54.Hoang C., Chen J., Vizthum C.A., Kandel J.M., Hamilton C.S., Mueller E.G., Ferré-D’Amaré A.R. Crystal structure of pseudouridine synthase RluA: indirect sequence readout through protein-induced RNA structure. Mol. Cell. 2006;24:535–545. doi: 10.1016/j.molcel.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 55.Czudnochowski N., Ashley G.W., Santi D.V., Alian A., Finer-Moore J., Stroud R.M. The mechanism of pseudouridine synthases from a covalent complex with RNA, and alternate specificity for U2605 versus U2604 between close homologs. Nucleic Acids Res. 2014;42:2037–2048. doi: 10.1093/nar/gkt1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sibert B.S., Fischel-Ghodsian N., Patton J.R. Partial activity is seen with many substitutions of highly conserved active site residues in human pseudouridine synthase 1. RNA. 2008;14:1895–1906. doi: 10.1261/rna.984508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mangum J.E., Hardee J.P., Fix D.K., Puppa M.J., Elkes J., Altomare D., Bykhovskaya Y., Campagna D.R., Schmidt P.J., Sendamarai A.K., et al. Pseudouridine synthase 1 deficient mice, a model for mitochondrial myopathy with sideroblastic anemia, exhibit muscle morphology and physiology alterations. Sci. Rep. 2016;6 doi: 10.1038/srep26202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De Paiva A.R.B., Lynch D.S., Melo U.S., Lucato L.T., Freua F., De Assis B.D.R., Barcelos I., Listik C., De Castro Dos Santos D., MacEdo-Souza L.I., et al. PUS3 mutations are associated with intellectual disability, leukoencephalopathy, and nephropathy. Neurol. Genet. 2019;5 doi: 10.1212/NXG.0000000000000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gulkovskyi R.V., Chernushyn S.Y., Livshits L.A. Novel gene PUS3 c.A212G mutation in Ukrainian family with intellectual disability. Biopolym. Cell. 2015;31:123–130. doi: 10.7124/bc.0008D6. [DOI] [Google Scholar]
- 60.Fang H., Zhang L., Xiao B., Long H., Yang L. Compound heterozygous mutations in PUS3 gene identified in a Chinese infant with severe epileptic encephalopathy and multiple malformations. Neurol. Sci. 2020;41:465–467. doi: 10.1007/s10072-019-04049-1. [DOI] [PubMed] [Google Scholar]
- 61.Behrens A., Nedialkova D.D. Experimental and computational workflow for the analysis of tRNA pools from eukaryotic cells by mim-tRNAseq. Star Protoc. 2022;3 doi: 10.1016/j.xpro.2022.101579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kost T.A., Condreay J.P., Jarvis D.L. Baculovirus as versatile vectors for protein expression in insect and mammalian cells. Nat. Biotechnol. 2005;23:567–575. doi: 10.1038/nbt1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kowarz E., Löscher D., Marschalek R. Optimized Sleeping Beauty transposons rapidly generate stable transgenic cell lines. Biotechnol. J. 2015;10:647–653. doi: 10.1002/biot.201400821. [DOI] [PubMed] [Google Scholar]
- 64.Mátés L., Chuah M.K.L., Belay E., Jerchow B., Manoj N., Acosta-Sanchez A., Grzela D.P., Schmitt A., Becker K., Matrai J., et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 2009;41:753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 65.Lin T.Y., Abbassi N.E.H., Zakrzewski K., Chramiec-Głąbik A., Jemioła-Rzemińska M., Różycki J., Glatt S. The Elongator subunit Elp3 is a non-canonical tRNA acetyltransferase. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-08579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. CryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 67.Bepler T., Morin A., Rapp M., Brasch J., Shapiro L., Noble A.J., Berger B. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods. 2019;16:1153–1160. doi: 10.1038/s41592-019-0575-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kidmose R.T., Juhl J., Nissen P., Boesen T., Karlsen J.L., Pedersen B.P. Namdinator - Automatic molecular dynamics flexible fitting of structural models into cryo-EM and crystallography experimental maps. IUCrJ. 2019;6:526–531. doi: 10.1107/S2052252519007619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liebschner D., Afonine P.V., Baker M.L., Bunkóczi G., Chen V.B., Croll T.I., Hintze B., Hung L.W., Jain S., McCoy A.J., et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., Morris J.H., Ferrin T.E. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Labun K., Montague T.G., Krause M., Torres Cleuren Y.N., Tjeldnes H., Valen E. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019;47:W171–W174. doi: 10.1093/nar/gkz365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brinkman E.K., Chen T., Amendola M., Van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42 doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. J. Comput. Biol. 2011;24:1138–1143. doi: 10.1089/cmb.2017.0096. [DOI] [Google Scholar]
- 75.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O., Whitwham A., Keane T., McCarthy S.A., Davies R.M. Twelve years of SAMtools and BCFtools. GigaScience. 2021;10 doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Quinlan A.R., Hall I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Neph S., Kuehn M.S., Reynolds A.P., Haugen E., Thurman R.E., Johnson A.K., Rynes E., Maurano M.T., Vierstra J., Thomas S., et al. BEDOPS: high-performance genomic feature operations. Bioinformatics. 2012;28:1919–1920. doi: 10.1093/bioinformatics/bts277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rainer J., Gatto L., Weichenberger C.X. Ensembldb: an R package to create and use Ensembl-based annotation resources. Bioinformatics. 2019;35:3151–3153. doi: 10.1093/bioinformatics/btz031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Morgan M., Shepherd L. 2023. AnnotationHub: Client to access AnnotationHub resources. [DOI] [Google Scholar]
- 81.Tian S., Yesselman J.D., Cordero P., Das R. Primerize: automated primer assembly for transcribing non-coding RNA domains. Nucleic Acids Res. 2015;43:W522–W526. doi: 10.1093/nar/gkv538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Adachi H., DeZoysa M.D., Yu Y.T. Detection and quantification of pseudouridine in RNA. Methods Mol. Biol. 2019;1870:219–235. doi: 10.1007/978-1-4939-8808-2_17. [DOI] [PubMed] [Google Scholar]
- 83.Morales J., Pujar S., Loveland J.E., Astashyn A., Bennett R., Berry A., Cox E., Davidson C., Ermolaeva O., Farrell C.M., et al. A joint NCBI and EMBL-EBI transcript set for clinical genomics and research. Nature. 2022;604:310–315. doi: 10.1038/s41586-022-04558-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen D., Patton J.T. Reverse transcriptase adds nontemplated nucleotides to cDNAs during 5′-RACE and primer extension. BioTechniques. 2001;30:574–581. doi: 10.2144/01303rr02. [DOI] [PubMed] [Google Scholar]
- 85.Volders P.J., Anckaert J., Verheggen K., Nuytens J., Martens L., Mestdagh P., Vandesompele J. Lncipedia 5: Towards a reference set of human long non-coding rnas. Nucleic Acids Res. 2019;47:D135–D139. doi: 10.1093/nar/gky1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Finet O., Yague-Sanz C., Hermand D. Epitranscriptomic mapping of RNA modifications at single-nucleotide resolution using rhodamine sequencing (Rho-seq) Star Protoc. 2022;3 doi: 10.1016/j.xpro.2022.101369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Khoddami V., Yerra A., Mosbruger T.L., Fleming A.M., Burrows C.J., Cairns B.R. Transcriptome-wide profiling of multiple RNA modifications simultaneously at single-base resolution. Proc. Natl. Acad. Sci. USA. 2019;116:6784–6789. doi: 10.1073/pnas.1817334116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zaringhalam M., Papavasiliou F.N. Pseudouridylation meets next-generation sequencing. Methods. 2016;107:63–72. doi: 10.1016/j.ymeth.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 89.Helm M., Motorin Y. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat. Rev. Genet. 2017;18:275–291. doi: 10.1038/nrg.2016.169. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
NGS raw and processed data have been deposited at GEO and are publicly available as of the date of publication. Accession number (GSE255287) is listed in the key resources table. The micrographs, atomic coordinates, and the cryo-EM maps of the described cryo-EM datasets have been deposited at the Electron Microscopy Public Image Archive, the Protein databank and the Electron Microscopy Data Bank and are publicly available as of the date of publication. The accession codes and DOI are listed in the Tables 1 and S1. Original gel images have been deposited at Mendeley and are publicly available as of the date of publication (https://doi.org/10.17632/gdw8pr456d.2)
-
•
All custom scripts described herein have been deposited on the Leidel Lab GitHub server and is publicly available (https://doi.org/10.5281/zenodo.11401096). Details are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.