Abstract
The production of the alphavirus virion is a multistep event requiring the assembly of the nucleocapsid core in the cytoplasm and the maturation of the glycoproteins in the endoplasmic reticulum and the Golgi apparatus. These components associate during the budding process to produce the mature virion. The nucleocapsid proteins of Sindbis virus and Ross River virus have been produced in a T7-based Escherichia coli expression system and purified. In the presence of single-stranded but not double-stranded nucleic acid, the proteins oligomerize in vitro into core-like particles which resemble the native viral nucleocapsid cores. Despite their similarities, Sindbis virus and Ross River virus capsid proteins do not form mixed core-like particles. Truncated forms of the Sindbis capsid protein were used to establish amino acid requirements for assembly. A capsid protein starting at residue 19 [CP(19–264)] was fully competent for in vitro assembly, whereas proteins with further N-terminal truncations could not support assembly. However, a capsid protein starting at residue 32 or 81 was able to incorporate into particles in the presence of CP(19–264) or could inhibit assembly if its molar ratio relative to CP(19–264) was greater than 1:1. This system provides a basis for the molecular dissection of alphavirus core assembly.
Sindbis virus (SINV) is a member of the Togaviridae family of enveloped, positive-strand RNA viruses and is the prototype of the Alphavirus genus (40). The alphavirus virion is approximately 710 Å in diameter and consists of four major components: the glycoprotein shell, the plasma membrane, the nucleocapsid core (NC), and the genomic RNA (3, 11, 30). The glycoprotein shell consists of 80 “spikes” arranged in a T=4 lattice. Each spike is composed of a trimer of heterodimers of the two glycoproteins, E1 and E2. The glycoprotein spikes penetrate a host cell-derived plasma membrane and interact directly with the NC. The NC is a 410-Å-diameter T=4 icosahedron consisting of 240 copies of the single nucleocapsid protein (CP) and a single genomic RNA of approximately 11,000 nucleotides.
The 264-amino-acid SINV CP is produced at the N terminus of the structural polyprotein, which is translated from a subgenomic mRNA (32). The CP cleaves itself from the polyprotein cotranslationally by using its endogenous serine protease activity to produce the mature CP (1, 14, 15). The structure of the C-terminal domain of the SINV CP, consisting of amino acids 107 to 264, has been determined to atomic resolution (4, 5). This region of the protein contains the proteinase domain (residues 114 to 264) and has an overall structure similar to that of chymotrypsin-like serine proteases. Adjacent to the proteinase domain is a region implicated in genome RNA recognition and consisting of amino acids 97 to 113 (12, 29, 43). The N-terminal 96 residues of the CP are poorly conserved among alphaviruses. This region consists of a large number of basic amino acids and is likely to be involved in nonspecific interactions with the genome RNA through charge neutralization. This region also contains a series of conserved hydrophobic and polar residues that are predicted to form an amphipathic α helix (residues 38 to 55) based on modeling of the primary sequence into a helical wheel plot (13a). The most striking feature of this putative helix is the presence of two conserved leucine residues at positions 45 and 52 and arranged on the same side of the helix. These residues may form an interaction motif between two CPs, in a manner similar to that of the large number of characterized leucine zipper proteins.
Attempts at a high-resolution structural solution of a complete alphavirus virion or of the NC by X-ray crystallography have been largely unsuccessful, due to poor diffraction of all crystals produced to date (9a, 16). However, cryoelectron microscopy (cryo-EM) and image reconstructions have provided a significant amount of data about the structure of the alphavirus particle and of the NC (3, 11, 30). The NC consists of a series of pentameric and hexameric capsomeres that project ∼40 Å from the core surface. Using a cryo-EM reconstruction of the related Ross River virus (RRV), Cheng et al. (3) fitted the atomic structure of monomeric SINV CP into the cryo-EM density and obtained a unique orientation of the CP in the core (3). These modeling studies indicated that residues 114 to 264 lie completely within the large surface projections (capsomeres) seen in the core and are involved in forming and/or maintaining those capsomeric structures. The results also predict that residues in the N-terminal region of the CP are involved in intercapsomere contacts, possibly through the proposed N-terminal leucine helix interaction.
The process of NC assembly is poorly understood. It is known from previous studies that immediately following translation and proteolysis, the CP is transiently associated with the large subunit of the ribosome (13, 36). The functional significance of this observation for core assembly is unknown; however, the core has also been shown to associate with ribosomes upon cell entry, and this association has been suggested to facilitate core disassembly (34, 43). Pulse-chase experiments indicate that within approximately 5 min of synthesis, the CP binds to genomic RNA and rapidly assembles into NCs (37). An in vitro assembly system for SINV core-like particles (CLPs) has been previously established with virus-purified CP (41). Assembly experiments were conducted in the presence of high concentrations of salt but were later modified with ionic conditions similar to those found in the cytoplasm (42). CLPs produced by these systems with viral genomic RNA closely resembled cytoplasmic cores purified from infected cells in size, shape, and composition. Furthermore, it was found that CLPs could be assembled from a variety of single-stranded nucleic acids, as well as several other polyanionic substrates. Although these assembly systems provided the first insights into the requirements for NC assembly, they had key limitations which hindered elucidation of an assembly pathway. The most notable of these limitations was the reliance on CP purified from assembled virus particles, thereby eliminating the ability to assay CP mutants that were defective in virus production. Although extensively investigated in vitro and in vivo, no specific intermediates in the NC assembly process have been identified (40).
We report a heterologous protein expression system, based in Escherichia coli, for the expression and purification of large quantities of the SINV CP from amino acid residues 19 to 264. An in vitro NC assembly system that uses the E. coli-expressed SINV CP and nucleic acid substrates is described. A similar expression, purification, and assembly system is also described for RRV. These in vitro systems are used to examine the nucleic acid requirements for assembly, to examine the regions of the SINV CP required for assembly, and to demonstrate the specificity of protein-protein and protein-nucleic acid interactions involved in core assembly.
MATERIALS AND METHODS
SINV CP cloning, expression, and purification.
CP cloning was performed with the pSBetB vector kindly provided by Ralf Mattes; this vector provides the E. coli argU gene for suppression of rare arginine codon usage (33). Two DNA oligonucleotides were synthesized to amplify the coding sequence (amino acid residues 1 to 264) of the SINV CP from the parental virus cDNA clone pToto64 (29). The 5′ oligonucleotide contained an NdeI restriction site spanning the capsid protein AUG start codon and additional overlapping nucleotides. The 3′ oligonucleotide was generated to produce a UAG termination codon immediately following the ultimate W264 residue, followed by a BamHI restriction site. Following DNA amplification with Pfu polymerase (Stratagene, La Jolla, Calif.), the product was digested with NdeI and BamHI and ligated into a similarly digested pSBetB vector. All cloning was carried out with E. coli MC1061. Positive clones were confirmed by restriction endonuclease digestion and DNA sequence analysis of the CP open reading frame and transformed into E. coli BL21(DE3) for protein expression. Initial protein expression and purification generated two copurifying capsid proteins. N-terminal protein sequencing was used to identify these proteins as amino acids 9 to 264 [CP(9–264)] and 19 to 264 [CP(19–264)] of the SINV CP. Attempts at preventing the degradation of the full-length protein to these CP fragments by use of a variety of protease inhibitors were not successful. Fast protein liquid chromatography (FPLC) elution gradients were optimized to separate the two CP species; the bulk of the purification yielded CP(19–264), with smaller yields of CP(9–264). In all instances, these CP species could be completely separated during purification.
For large-scale expression of CP(19–264), 1 liter of cells was grown in Luria-Bertani medium at 37°C with agitation until an A600 of 0.4 was reached, at which time the cells were induced with 1 mM (final concentration) isopropyl-β-d-thiogalactopyranoside (Inalco Pharmaceuticals, Milano, Italy). Following induction, cells were shifted to 28°C with agitation for 8 h to allow for maximal protein expression. The cells were pelleted in a GSA rotor (Sorvall, Wilmington, Del.) at 5,000 rpm for 10 min and resuspended in 20 ml of buffer A (100 mM NaH2PO4 [pH 6.8], 500 mM NaCl, 5 mM EDTA, 5% [vol/vol] glycerol) prior to lysis by two passages through a cold French pressure cell (SLM-Aminco, Urbana, Ill.) at 12,000 lb/in2. Unlysed cells and insoluble material were removed by centrifugation for 30 min at 14,000 rpm in an SS-34 rotor (Sorvall) at 4°C. The clarified cell lysate was loaded onto a Source 15S 10/10 (8.5-ml bed volume) FPLC column (Pharmacia Biotech, Uppsala, Sweden) equilibrated with buffer A. A linear salt gradient made from buffer A and buffer B (100 mM NaH2PO4 [pH 6.8], 2 M NaCl, 5 mM EDTA, 5% [vol/vol] glycerol) was used to elute the protein. Fractions containing CP were eluted at ∼1.1 M NaCl, concentrated with Centriprep-10 centrifugal concentrators (Amicon, Beverly, Mass.) at 4°C in an SS-34 rotor at 5,000 rpm, and exchanged into buffer C (25 mM HEPES [pH 7.4], 100 mM potassium acetate, 1.7 mM magnesium acetate). Concentrated proteins were applied to a Superdex 75 10/30 FPLC column (24-ml bed volume) (Pharmacia Biotech), and size exclusion chromatography was performed with buffer C. Protein purity was assessed by silver stain sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and found to be greater than 95%. Analysis of the purified protein on a Superdex 75 10/30 column with buffer C generated a single symmetric peak. Typical yields of CP(19–264) were approximately 4 to 10 mg/liter of cell culture. Truncated forms of CP were purified by a similar methodology or by previously published purification procedures, resulting in similar yields and purity (4). The truncations consisted of amino acids 32 to 264, 81 to 264, 106 to 264, and 114 to 264 of the SINV CP (Fig. 1A). All protein concentrations reported were determined by the Bio-Rad (Hercules, Calif.) protein assay.
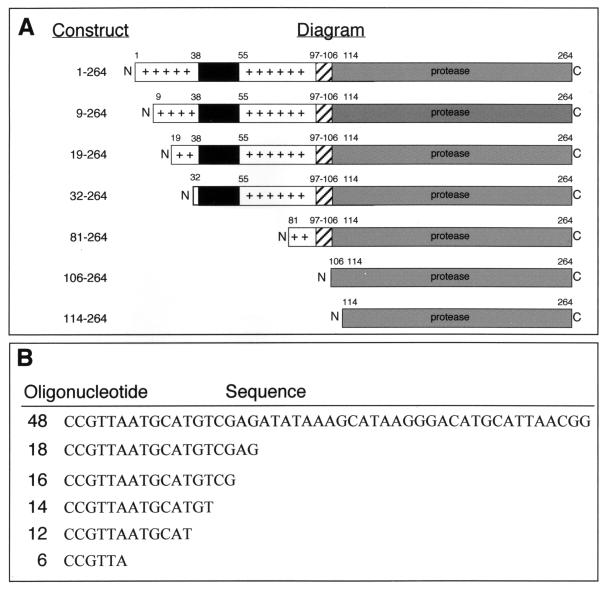
FIG. 1.
Schematic of SINV capsid constructs and synthetic DNA oligonucleotides used in the in vitro assembly studies. (A) Schematic of virus-encoded CP(1–264), E. coli-expressed and purified CP(9–264) and CP(19–264), and E. coli-expressed truncated CP(32–264), CP(81–264), CP(106–264), and CP(114–264). Important regions of the CP are indicated. These regions include the large blocks of positively charged residues at the amino terminus (residues 1 to 32 and 55 to 97; plus signs), the putative N-terminal α helix (residues 38 to 55; black bars), the specific RNA binding region (residues 97 to 106; hatched bars), and the serine protease region (residues 114 to 264; gray bars). All numbering corresponds to amino acids of the SINV CP. (B) Sequences of oligonucleotides used in assembly and minimum-length-requirement studies. The 48-base oligonucleotide is the standard assembly oligonucleotide. All sequences are shown 5′ to 3′.
RRV CP cloning, expression, and purification.
RRV CP cloning was performed essentially as described for SINV CP (21). Expression and test purification generated a single protein, which was identified as the RRV CP by N-terminal protein sequencing. For large-scale expression of RRV CP, 1 liter of cells was grown in Luria-Bertani medium at 37°C with agitation until an A600 of 0.8 was reached, at which time the cells were induced with 0.5 mM (final concentration) isopropyl-β-d-thiogalactopyranoside and allowed to grow for 8 h at 37°C. The cells were pelleted in a GSA rotor at 5,000 rpm for 10 min, resuspended in 20 ml of buffer D (100 mM NaH2PO4 [pH 6.8], 100 mM NaCl, 5 mM EDTA, 5% [vol/vol] glycerol), and lysed by two passages through a cold French pressure cell at 12,000 lb/in2. Insoluble material was removed by centrifugation for 30 min at 14,000 rpm in an SS-34 rotor at 4°C. The clarified cell lysate was loaded onto a Mono-S 5/5 (1-ml bed volume) FPLC column (Pharmacia Biotech) equilibrated with buffer D and eluted with a linear salt gradient of 100 mM to 2 M NaCl in buffer B. Fractions containing CP were concentrated with Centriprep-10 concentrators at 4°C in an SS-34 rotor at 4,000 rpm and exchanged into buffer C. Protein purity was assessed by silver stain SDS-PAGE and found to be greater than 90%. Typical yields of RRV CP(1–270) were approximately 8 to 12 mg/liter of cell culture.
In vitro assembly assays.
In vitro assembly assays were performed with purified SINV CP(19–264) and a nonspecific synthetic DNA oligonucleotide consisting of 48 bases (Fig. 1B). Equal volumes of prewarmed CP (400 μg/ml in buffer C) and oligonucleotide (240 μg/ml in buffer C) were mixed at 30°C and incubated for 30 min. Assembly reaction volumes were typically 100 μl. RRV assembly assays were performed under identical conditions. Assembly reactions with truncated forms of CP and other nucleic acid substrates were carried out in a similar fashion. Truncated oligonucleotides were synthetic DNA oligonucleotides comprising 3′-end truncations of the standard 48-base oligonucleotide used to establish the in vitro assembly systems (Fig. 1B). RNA assembly reaction substrates consisted of commercial preparations of ultrapure yeast tRNA (Boehringer Mannheim Biochemicals, Indianapolis, Ind.) or DNase-treated and purified in vitro transcripts of SINV genomic RNA from the full-length cDNA clone pToto64 (31). Viral RNAs were generated with and without a 5′ cap structure, and these two RNA species were analyzed separately.
Gradient assembly assay.
Following assembly, the presence of CLPs was assayed by sucrose gradient sedimentation. Assembly reaction mixtures (100 μl) were loaded onto 12-ml 25% freeze-thaw sucrose gradients prepared with buffer C. The samples were centrifuged at 4°C in an SW-41 rotor (Beckman, Palo Alto, Calif.) for 105 min at 38,000 rpm. Following sedimentation, gradients were fractionated into 1-ml samples for further assay.
For Western blot analysis, gradient fractions were separated by SDS-PAGE and transferred to nitrocellulose membranes. The membranes were exposed to a polyclonal anticapsid rabbit antibody and then to a secondary anti-rabbit goat immunoglobulin G antibody conjugated with horseradish peroxidase (Sigma, St. Louis, Mo.). Blots were subjected to chemoluminescence detection with an ECL kit (Amersham, Piscataway, N.J.) and exposed to X-ray film. Alternatively, samples were examined by an enzyme-linked immunosorbent assay with the polyclonal anticapsid rabbit antibody and the secondary anti-rabbit goat immunoglobulin G antibody conjugated with horseradish peroxidase.
Electron microscopy.
Following assembly and gradient sedimentation purification of CLPs, 3.5 μl of sample was placed on a prewashed, glow-discharged, 400-mesh copper grid coated with Formvar and carbon. Following 2 min of sample absorption and extensive washing with water, 7 μl of a 2% (wt/vol) uranyl acetate stain was applied. After 4 min of staining, grids were wick dried with Whatman no. 1 filter paper and allowed to air dry for a minimum of 20 min. Samples were then viewed on a Philips EM300 electron microscope with an acceleration voltage of 60 kV at a magnification of ×45,000. Images were captured on Kodak SO-163 EM film. Cytoplasmic and viral NCs were purified as previously described and prepared for microscopy as described above (29).
Agarose gel assembly assay.
Following assembly, CLPs were subjected to an electrophoretic mobility assay with agarose gels (17, 23). Assembled particles were electrophoresed at 120 V (constant voltage) on 0.8% agarose (wt/vol) gels in Tris-acetate (100 mM Tris [pH 8.1], 1.25 mM sodium acetate, 1 mM EDTA) electrophoresis buffer in the presence of ethidium bromide (5 μg/ml). Nucleic acid was directly visualized on a short-wavelength UV transilluminator; it has an altered mobility if packaged within a core particle. Additionally, gels were fixed in 40% (vol/vol) methanol with 10% (vol/vol) acetic acid, dried under vacuum, and stained with Coomassie brilliant blue R-250. Protein bands were visualized following a brief destaining in distilled water. The positions of assembled CLPs were determined by direct comparison with both unassembled CP and assembled CLPs that had been purified and examined by electron microscopy.
Truncated CP incorporation and inhibition assays.
Inhibition and incorporation of truncated forms of CP in core assembly reactions were performed under standard assembly reaction conditions. Briefly, CP(19–264) (final concentration, 400 μg/ml) and an increasing amount of truncated CP were mixed at 30°C in a total volume of 50 μl. Typically, the molar ratio of truncated CP to CP(19–264) varied from 0.1:1 to 40:1. The standard 48-base assembly oligonucleotide was then added at 240 μg/ml in a volume of 50 μl of buffer C. Samples were mixed thoroughly, incubated at 30°C for 30 min, and analyzed by the agarose gel assembly assay and gradient sedimentation-Western blotting.
Heterologous virus core assembly.
Heterologous mixed core assembly reactions were performed as a modification of the standard in vitro core assembly assays. Purified SINV CP and purified RRV CP were mixed to yield a final concentration of 400 μg/ml of each protein in buffer C. The standard 48-base assembly oligonucleotide was added to a final concentration of 240 μg/ml in a total reaction volume of 100 μl. Reactions were incubated for 30 min at 30°C and analyzed by the agarose gel assembly assay.
RESULTS
Protein expression and purification.
To establish an in vitro core assembly system, it was necessary to generate large quantities of wild-type CP. As was previously demonstrated, wild-type protein could be purified from virus particles, but this process represented a significant expenditure in time, effort, and materials (41). In addition, analysis of capsid mutants would be hindered if their replication efficiency were reduced compared with that of the wild-type virus. Therefore, the development of an E. coli-based expression system for the CP was a necessary prerequisite for the production of an in vitro assembly system that would be amenable to both biochemical and genetic manipulations.
Prokaryotic expression of CP initiating upstream of amino acid 81 has been impossible to produce in standard E. coli-based expression systems, most likely due to the multiple use of rare AGA and AGG arginine codons in the first 80 amino acids of the protein (data not shown). To overcome this problem, which is common for many eukaryotic proteins expressed in E. coli, a specialized expression system utilizing the pSBetB vector for the suppression of rare arginine codons was developed by Schenk and colleagues (33). This system allowed the production of several milligrams of CP per liter of bacterial culture, whereas earlier attempts at production of the protein with standard pET vectors failed to produce detectable protein, as assayed by Western blotting. However, SINV CP expression was found only at temperatures below 30°C. The expressed CP was found to have a narrow range of solubility in the initial cell lysates, with buffer A (see Materials and Methods) being an optimum buffer for lysis and initial chromatographic separations. With the purification method described above, quantities of 4 to 10 mg of SINV CP per ml could be produced at greater than 95% purity. Similar results were obtained with a modification of the SINV CP purification conditions for RRV CP. Typical yields of 8 to 10 mg of pure RRV CP per liter of cell culture were obtained.
Two SINV CPs were identified during the initial purification. The larger protein was identified by N-terminal protein sequencing as the CP starting at residue 9, the result of a cryptic Shine-Dalgarno translation initiation site. The second, smaller protein began at residue 19, as determined by N-terminal protein sequencing, and was probably generated by proteolysis during purification. With the purification strategy described above, it was possible to purify CP(19–264) independent of CP(9–264). For RRV, a single species, corresponding to the complete 270-amino-acid CP, was purified.
The E. coli-expressed CPs of SINV and RRV were examined for their oligomeric state following purification. Numerous crystallographic studies of the SINV CP have been carried out; the predominant form of the protein has been found as a dimer within the crystal (4, 5, 24). Although this crystallographic dimer was not observed by cryo-EM of native virions, many other viruses utilize structural protein dimers as building blocks (38, 39). Therefore, purified CPs were examined by analytical ultracentrifugation, gel permeation chromatography, and chemical cross-linking analyses. In all cases, only monomeric forms of the SINV and RRV CPs were found.
In vitro assembly system.
An in vitro assembly system based in part on the earlier in vitro assembly system developed with CP from disrupted virus particles (41) was developed with the E. coli-expressed protein and a variety of nucleic acid substrates. The establishment of assays that allowed for the detection of core assembly was necessary, since the original core assembly assays were performed by negative-stain electron microscopy, thereby requiring fairly large amounts of cores for detection. Sucrose gradient sedimentation analysis was initially used for the isolation of in vitro-assembled CLPs. Following centrifugation, an opaque band corresponding to assembled CLPs was present in reactions containing nucleic acid and protein. This band was not present when nucleic acid or protein was sedimented under identical conditions. Western blot analysis of gradient fractions with polyclonal anticapsid antibody indicated that CP sedimented with the opaque band when nucleic acid was present in assembly reactions and remained at the top of the gradient in reactions without nucleic acid. Its position in the gradient was consistent with the mobility of virus-isolated cores centrifuged under identical conditions. An example of an assembly reaction followed by gradient and Western blot analyses is shown in Fig. 2A. Electron microscopic analysis of the opaque band (Fig. 2A, fractions 7 and 8) by negative staining indicated the presence of CLPs (data not shown). At the bottom of the gradient, in fractions 11 and 12, there was also a significant amount of CP. However, in this case, electron microscopic analysis suggested the presence of CP aggregates with no CLPs present.
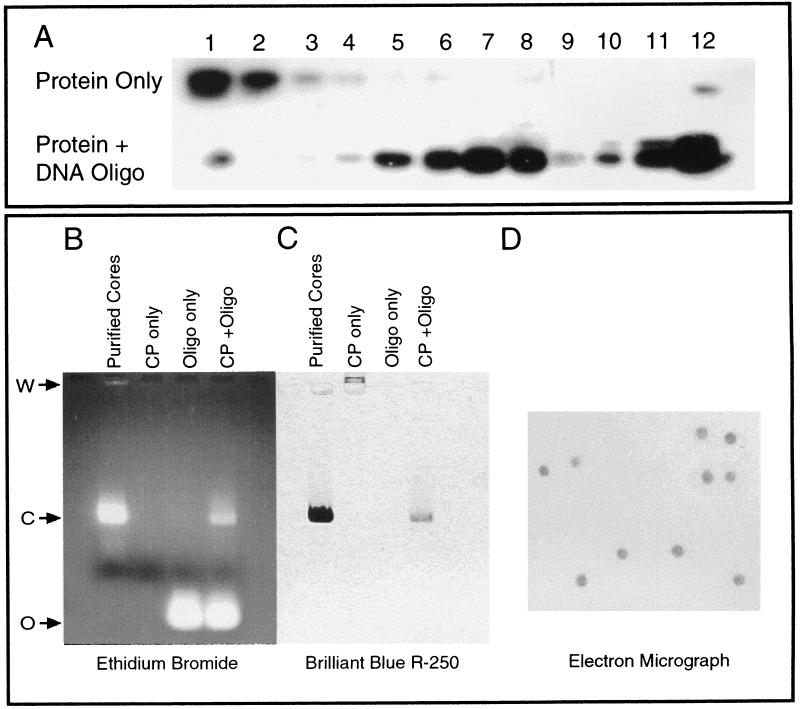
FIG. 2.
Analysis and detection of in vitro-assembled core particles. (A) In vitro core assembly with SINV CP(19–264) and the 48-base assembly oligonucleotide (Oligo) assayed by sucrose density gradient sedimentation followed by fractionation and Western blot analysis. The top gel represents assembly reactions performed without nucleic acid. The bottom gel represents assembly reactions containing both protein and nucleic acid. A peak in the lower gel gradient can be clearly seen in fractions 6 to 8. Numbers across the top correspond to gradient fractions from top (1) to bottom (12). (B) Agarose gel assay analysis of in vitro core assembly reactions stained for nucleic acid with ethidium bromide. First lane, purified in vitro-assembled CLPs from fraction 7 of the oligonucleotide-protein gel from panel A. Core (C) migration from the origin of electrophoresis (W) is indicated. Second lane, CP(19–264) alone; therefore no nucleic acid is apparent. Third lane, electrophoretic mobility of the 48-base assembly oligonucleotide alone (O). Fourth lane, standard in vitro assembly reaction with CP(19–264) and the 48-base oligonucleotide without purification or concentration. Both unincorporated oligonucleotide (O) and assembled core particles (C) are seen. The migration of core particles is identical to that of purified core particles in the first lane. (C) Agarose gel assembly reactions stained for protein with Coomassie brilliant blue R-250 stain. Lanes are identical to those in panel B. Note that cores in the first and fourth lanes contain protein which comigrates with nucleic acid incorporated in cores (B). CP alone (second lane) migrates in the direction opposite that of the CLPs from the origin of electrophoresis (W). (D) Uranyl acetate negative-stain electron micrograph of CLPs used in the first lanes of panels B and C. Magnification, ×45,000.
Large amounts of assembled CLPs could be produced with a wide range of protein and nucleic acid concentrations in a variety of assembly buffers at a neutral pH. Optimum assembly conditions were determined by examining the relative levels of core production and were found to be approximately 400 μg of CP per ml mixed with an equal volume of a 48-base nonspecific oligonucleotide at 120 to 250 μg/ml. Assembly was found to occur rapidly, with assembled CLPs detectable only a few minutes after mixing of the assembly components. Assembly reaction volumes from 10 to 400 μl were found to generate CLPs, with larger reaction volumes producing primarily large aggregates of protein and nucleic acid. Assembly reaction component concentrations could be varied extensively to produce larger or smaller amounts of particles, as long as the overall protein/nucleic acid ratio in the reaction remained constant. Assembly reaction concentrations of CP from 50 μg/ml to 5 mg/ml, the highest and lowest levels tested, were capable of producing core-like particles. Assembly was found to occur at 4, 25, 30, and 37°C. Both SINV and RRV CPs behaved in a similar manner, and particle assembly conditions were identical.
Agarose gel assay for core formation.
A more rapid assay for the detection of CLP formation was required to allow future assays of large numbers of mutant or truncated CPs. To fill this need, an agarose gel assay based on assembly studies performed for bacteriophages and small RNA plant viruses was developed to detect in vitro core assembly (17, 23). Following assembly, CLPs were subjected to an electrophoretic mobility assay with agarose gels. Assembled particles were electrophoresed on 0.8% agarose gels in Tris-acetate electrophoresis buffer in the presence of ethidium bromide (Fig. 2B). The mobility of the assembled NC in the gel system was determined by direct comparison with oligonucleotide alone and assembled CLPs which had previously been purified and examined by electron microscopy (Fig. 2D). Additionally, these agarose gels were fixed in 40% (vol/vol) methanol with 10% (vol/vol) acetic acid, dried under vacuum, and then stained with Coomassie brilliant blue R-250. The protein bands were visualized following brief destaining of the gel in distilled water (Fig. 2C). The mobility of the NC was determined by direct comparison with both unassembled CP and purified CLPs that had previously been examined by electron microscopy. In all instances, CP and nucleic acid present as assembled cores comigrated in the agarose gel assay and had a migration quite distinct from that of oligonucleotide or protein alone, thereby allowing for rapid determination of in vitro assembly experiment results.
Electron microscopy of native and in vitro-assembled NCs.
Negative-stain electron microscopy of in vitro-assembled DNA and RNA CLPs and purified viral and cytoplasmic cores was performed to confirm that the overall size and morphology of in vitro-assembled particles were similar to those of both cytoplasmic cores and cores isolated from virus. Figure 3 shows the negative-stain electron micrographs of these core particles and demonstrates that all of the cores have approximately the same size, shape, and general appearance, suggesting that in vitro-assembled CLPs are similar in appearance to native core particles. A slight size difference was seen between the viral and cytoplasmic cores and the in vitro-assembled particles. This difference was seen only in negative-stain electron microscopy, not in cryo-EM (data not shown), and suggests a staining artifact similar to that observed with Aura virus (46). When negative-stain electron microscopy and cryo-EM results for the various core preparations were examined, no visible size or morphology differences could be detected among the particles, suggesting the CLPs are very similar to native viral cores.
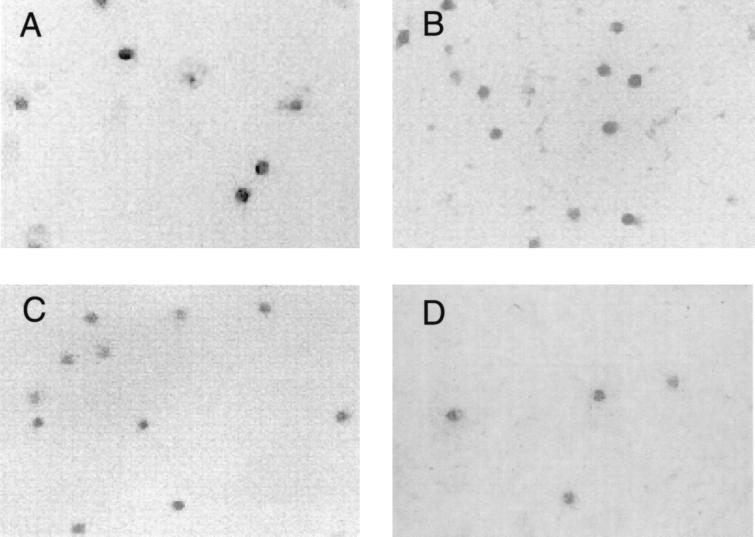
FIG. 3.
Negative-stain electron microscopy of SINV cores from in vivo and in vitro sources. (A) SINV cores isolated from purified wild-type Sindbis virions. (B) SINV core purified from the cytoplasm of infected BHK-21 cells. (C) In vitro-assembled SINV cores generated from CP(19–264) and the 48-base oligonucleotide. (D) In vitro-assembled SINV cores generated from CP(19–264) and viral genomic RNA. All electron micrographs were prepared as described in Materials and Methods and are presented at a magnification of ×45,000.
Nucleic acid requirements for in vitro core assembly.
It was previously shown that in vitro core assembly with virus-purified CP could be performed with viral RNA, tRNA, single-stranded DNA, and synthetic polyanions (41). To examine the nucleic acid requirements for in vitro core assembly with the E. coli-based CP assembly system, a series of nucleic acids were assayed for the ability to serve as substrates for core assembly. Table 1 provides a summary of the various nucleic acids tested in the in vitro core assembly assay. Assembly reactions were monitored by a combination of electron microscopy, agarose gel assay, and sucrose gradient-Western blot analysis in order to prevent any misinterpretation of results due to differential staining of the various lengths and types of nucleic acids by ethidium bromide in the agarose gel assay.
TABLE 1.
Nucleic acid requirements for assembly
| Nucleic acida | Typeb | Size | Assembly | Molar ratios tested (P:N)c | Optimum molar ratio (P:N)c |
|---|---|---|---|---|---|
| 48-nt oligod | ssDNA | 48 nt | Yes | 10:1–1:10 | 2:1 |
| 18-nt oligo | ssDNA | 18 nt | Yes | 10:1–1:20 | 1:1 |
| 16-nt oligo | ssDNA | 16 nt | Yes | 10:1–1:20 | 1:4 |
| 14-nt oligo | ssDNA | 14 nt | Yes | 10:1–1:20 | 1:20 |
| 12-nt oligo | ssDNA | 12 nt | No | 10:1–1:20 | |
| 6-nt oligo | ssDNA | 6 nt | No | 10:1–1:20 | |
| 77-nt oligo 1 | ssDNA | 77 nt | Yes | 10:1–1:10 | 2:1 |
| 77-nt oligo 2 | ssDNA | 77 nt | Yes | 10:1–1:10 | 2:1 |
| Oligo 1 + oligo 2 | dsDNA | 77 bp | No | 20:1–1:20 | |
| PCR A | dsDNA | 132 bp | No | 10:1–1:10 | |
| PCR B | dsDNA | 900 bp | No | 1:100–1:1,000 | |
| Plasmid | dsDNA | 4,400 bp | No | 1:100–1:1,000 | |
| tRNA | ssRNA | 81 nt | Yes | 10:1–1:10 | 2:1 |
| vRNA (no cap) | ssRNA | 11,704 nt | Yes | 1,000:1–0.1:1 | 400:1 |
| vRNA (cap) | ssRNA | 11,704 nt | Yes | 1,000:1–0.1:1 | 400:1 |
| None | No |
nt, nucleotide; oligo, oligonucleotide; PCR A, 132-bp double-stranded DNA fragment; PCR B, 900-bp double-stranded DNA fragment; vRNA, viral RNA.
ss, single stranded; ds, double stranded.
Molar ratios are given as moles of protein (P):moles of nucleic acid (N).
Standard assembly oligonucleotide.
To demonstrate that CLPs containing SINV genomic RNA could be produced, RNA was synthesized by in vitro transcription of a full-length cDNA clone, pToto64 (29). Viral RNA with and without a 5′ cap structure was assayed for the ability to generate CLPs. Viral RNA was found to serve as a substrate for in vitro core assembly and was specifically encapsidated when present in a mixture with tRNA or single-stranded DNA oligonucleotides (data not shown). The presence of a 5′ cap structure had no measurable effect on core assembly or the specificity of encapsidation. Both capped and uncapped viral RNAs were found to generate CLPs at an optimum molar ratio of 400 proteins per RNA (Table 2). Yeast tRNA was also found to serve as a substrate for in vitro CLP formation at capsid/nucleic acid molar ratios similar to those for the standard single-stranded DNA oligonucleotide, suggesting that both virus-specific RNA and nonspecific RNA could generate CLPs (Table 1).
TABLE 2.
Protein/nucleic acid ratios used in assembly
| Sample | Molar ratioa | Assembly |
|---|---|---|
| 48-base oligonucleotide | 20:1 | No |
| 10:1 | No | |
| 2:1 | Yesb | |
| 1:1 | Yes | |
| 0.1:1 | Yes (few) | |
| 0.01:1 | No | |
| Genomic RNA | >1,000:1 | No |
| 1,000:1 | Yes | |
| 500:1 | Yes | |
| 400:1 | Yesb | |
| 200:1 | Yes (few) | |
| 100:1 | No |
Protein:DNA oligonucleotide for the 48-base oligonucleotide and protein: RNA for genomic RNA.
Under the optimum assembly conditions for the indicated nucleic acid.
To determine if a minimum size of single-stranded DNA oligonucleotide was required for in vitro core assembly, synthetic DNA oligonucleotides composed of 3′ truncations of the standard 48-mer assembly oligonucleotide were generated in the following sizes: 18, 16, 14, 12, and 6 nucleotides (see Fig. 1B for their sequences). Assembly reactions were performed with these oligonucleotides at concentrations (240 μg/ml) or molar amounts (0.656 nmol/100-μl reaction) equal to those of the standard 48-mer control oligonucleotide, as well as a range of molar ratios of protein to nucleic acid of 10:1 to 1:20 (Table 1). It was found that 18-, 16-, and 14-base oligonucleotides were acceptable for assembling CLPs with optimum molar ratios of protein to nucleic acid of 1:1, 1:4, and 1:20, respectively, whereas the 12-mer and 6-mer oligonucleotides could not serve as substrates for CLP assembly at any ratios tested. The minimum size of single-stranded DNA oligonucleotide needed for detectable in vitro core assembly was found to be 14 nucleotides, and higher concentrations of shorter oligonucleotides were required for CLP assembly (16-mer and 14-mer oligonucleotides required greater than 1 mg/ml for assembly under standard conditions).
The ability of double-stranded DNA to incorporate into CLPs was examined with a set of complementary 77-base oligonucleotides which were annealed and incubated with CP(19–264) (400 μg/ml) over a range of concentrations (50 μg/ml to 1 mg/ml) corresponding to molar ratios of protein to nucleic acid of 10:1 to 1:10. No core formation was detected under any conditions assayed with the 77-bp double-stranded DNA fragment. Each of the two 77-base oligonucleotides at 240 μg/ml was competent for in vitro assembly under standard conditions when assayed independently as single-stranded DNA (Table 1). In order to further investigate the ability of double-stranded DNA to serve as a substrate for core assembly, both linear and circular double-stranded DNAs were assayed for CLP formation. A linear DNA fragment of 900 bp was assayed over a broad concentration range and found unable to form CLPs under any conditions assayed. A 132-bp PCR product assayed at molar ratios of 10:1 to 1:10 (protein to nucleic acid) was also found unable to generate CLPs in vitro. Similar negative assembly results were obtained with a circular plasmid DNA of 4,401 bp. Under no circumstances were CLPs detected when double-stranded DNA was used as a nucleic acid substrate for core assembly.
Attempts were also made to assemble CLPs in the absence of nucleic acid. Various buffers as well as a range of salt concentrations failed to produce any macromolecule that had a mobility similar to that of nucleic acid-containing CLPs in either the agarose gel assay or the sucrose centrifugation assay. In addition, electron microscopic analysis of nucleic acid-free assembly reactions failed to detect any CLPs or possible assembly intermediates.
Virus-specific capsid assembly reactions.
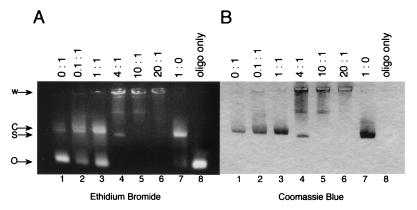
Several previous studies had demonstrated the flexibility of alphaviruses in utilizing structural and nonstructural proteins from more than one virus via recombinant heterotypic genomes (20, 26, 35, 44, 45). Chimeric viruses which exchanged the nonstructural and structural proteins, the capsid and the glycoproteins, and the individual glycoproteins were rescued. With the in vitro assembly system, it was possible to examine the specific nature of interactions involved in CP contacts within the assembly process by use of a mixture of purified viral CPs from SINV and RRV, thus determining whether phenotypically mixed particles could be formed in vitro. SINV and RRV CPs were mixed and added to the standard assembly oligonucleotide. In vitro-assembled CLPs of SINV and RRV migrated differently in the agarose gel assembly assay (Fig. 4), thereby allowing easy discrimination of CLP assembly in the gel assay during mixed-CP experiments. Mixed-protein assembly reactions failed to generate any detectable intermediate species with migration in the gel assay between those of SINV and RRV cores and representing a heterogeneous particle containing both species of CP (Fig. 4). In addition, immunoblotting of the agarose gel with an anticapsid antibody against the SINV CP which recognizes both SINV and RRV CPs failed to detect any intermediate species indicative of a mixed particle.
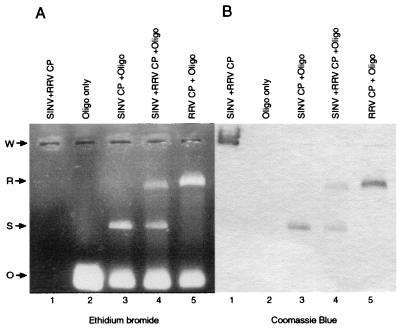
FIG. 4.
Heterologous in vitro core assembly. (A) Ethidium bromide-stained agarose gel of proteins only (lane 1), the 48-mer standard oligonucleotide (Oligo) only (lane 2), SINV assembly reaction (lane 3), RRV and SINV mixed assembly reaction (lane 4), and RRV assembly reaction (lane 5). The electrophoretic mobility of each species is indicated: O, oligonucleotide; S, SINV CLP; R, RRV CLP; W, origin of electrophoresis. Due to the difference in the migrations of SINV and RRV assembled CLPs, these species can readily be identified in mixed-protein experiments (compare lane 4 with the appropriate controls (lanes 3 and 5). (B) Gel from panel A stained with Coomassie brilliant blue R-250. No mixed particles (as shown by altered mobility) are detectable by protein staining or Western blotting (data not shown).
In vitro assembly with truncated CPs.
To address which regions of the CP of SINV were required for in vitro core assembly, a series of N-terminal truncations of the CP were cloned, expressed, purified, and examined for CLP formation. Assembly reactions were assayed by electron microscopy, an agarose gel assay, and gradient sedimentation Western blot assays. The proteins expressed are diagrammed in Fig. 1 and consist of amino acids 32 to 264, 81 to 264, 106 to 264, and 114 to 264. Additionally, small amounts of CP(9–264) were purified and assayed. The results of assembly assays with the truncated CP fragments are summarized in Table 3. CP(9–264) and the standard CP(19–264) were capable of generating CLPs in vitro when mixed with the 48-mer assembly oligonucleotide under standard reaction conditions. CP(32–264) did not assemble CLPs under standard assembly conditions or over a broad range of nucleic acid and protein concentrations (50 μg/ml to 1 mg/ml for protein and 50 μg/ml to 5 mg/ml for the 48-mer oligonucleotide). CP(81–264) has a deletion of the entire charged N-terminal portion of the CP and the putative N-terminal helix region. This construct was found to bind nucleic acid, as evidenced by band shifting of oligonucleotides in an agarose gel assembly assay (data not shown), but did not generate CLPs in vitro [the concentration ranges assayed were identical to those for CP(32–264)]. CP(106–264), which lacks the specific RNA binding capability found in CP(81–264), also failed to assemble CLPs in vitro. In addition, CLPs could not be generated with CP(114–264), which lacks both specific nucleic acid binding and protein-protein contact regions observed between monomers in crystal structures. Therefore, N-terminal truncations of CP past amino acid 19 do not permit the assembly of CLPs in vitro.
TABLE 3.
In vitro assembly with truncated capsid proteins
| Capsid protein | Assembly | Incorporationa | Inhibitiona |
|---|---|---|---|
| CP(9–264) | Yes | ||
| CP(19–264) | Yes | ||
| CP(32–264) | No | Yes | Yes |
| CP(81–264) | No | Yes | Yes |
| CP(106–264) | No | No | No |
| CP(114–264) | No | No | No |
Determined in the presence of CP(19–264).
Incorporation of truncated proteins into core particles.
As none of the CP truncations past amino acid 19 were capable of generating CLPs in vitro, although some had nucleic acid binding competence, the ability of these proteins to incorporate into particles during core assembly with CP(19–264) was investigated. Although the truncations may not be competent to complete the entire assembly process, many of the larger capsid proteins may still retain regions of the CP required to incorporate into core particles. Incorporation and identification of full-length and truncated proteins were assayed by both gradient sedimentation and an agarose gel assay followed by Western blot analysis. The small CP constructs, CP(114–264) and CP(106–264), were incapable of incorporating into core particles (Table 3). The larger CP constructs, CP(81–264) and CP(32–264), were capable of incorporating into CLPs when mixed with CP(19–264) at a molar ratio of 1:1 or less in the presence of the 48-mer assembly oligonucleotide. In Fig. 5, increased levels of CLP assembly in the presence of CP(81–264) are evidenced by increased levels of the CLP band (lanes 2 and 3). When the assembly products shown in lane 3 were analyzed by sucrose density centrifugation and Western blotting, the results indicated that the cores contained both CP(19–264) and CP(81–264) and demonstrated that the truncated protein could be incorporated into CLPs. Similar results were obtained with CP(32–264) (data not shown).
FIG. 5.
Incorporation and inhibition of CLP assembly by CP(81–264). Numbers above the gels represent molar ratios of CP(81–264) to CP(19–264). (A) Ethidium bromide-stained agarose gel demonstrating incorporation into and inhibition of standard assembly reactions with CP(81–264), CP(19–264), and the 48-base oligonucleotide. Lane 1, positive control reaction containing only CP(19–264) and the 48-base oligonucleotide under standard assembly reaction conditions. The positions of the unincorporated oligonucleotide (O), the CLP (C), and the origin of electrophoresis (W) are indicated. Lanes 2 through 6, increasing amounts of CP(81–264) added to the standard assembly reaction (at a constant volume). Lanes 2 and 3 show increased core assembly, as evidenced by the increased staining of the core band due to the incorporation of CP(81–264). Lanes 4 and 5 show abolished core assembly and the presence of an altered protein-nucleic acid migration position (S) corresponding to CP(81–264) bound to oligonucleotide (lane 7). A second, minor species of unknown origin is present in the agarose gel near the origin of electrophoresis in lanes 4 and 5. Lane 6 shows complete abolition of CLP formation. Lane 7, migration of CP(81–264) bound to oligonucleotide (S). The migration of this species is different from that of oligonucleotide alone (O) or oligonucleotide in CLPs (C). Lane 8, oligonucleotide (oligo) only. (B) Coomassie brilliant blue R-250 staining of the agarose gel shown in panel A.
As SINV CP(32–264) and CP(81–264) were capable of incorporating into core assembly reactions but were incapable of assembling CLPs alone, these truncations were investigated for their ability to inhibit core assembly. As the levels of CP(81–264) in the assembly reaction increased past a molar ratio of 1:1 relative to CP(19–264), no CLP assembly was seen, with a small amount of oligonucleotide shifted to the position of CP(81–264) nucleic acid binding (Fig. 5, lanes 4, 5, and 6); these results suggested inhibition of the assembly process. Therefore, it was found that both proteins could incorporate into core particles at low concentrations (molar ratios of up to 1:1) but inhibited core assembly at higher concentrations (4:1 and 40:1) (Table 3). In addition to the normal core migration position in the gel, additional bands corresponding to CP(81–264) bound to DNA (labeled S in Fig. 5) and another unidentified species were generated. The step at which CP(81–264) blocks assembly and the assembly intermediates involved in this process are currently under investigation.
DISCUSSION
The ability to generate a regular icosahedral NC structure from 240 copies of the alphavirus CP and the viral genomic RNA provides an interesting and challenging problem in macromolecular association. Numerous protein-protein and protein-nucleic acid interactions must occur in a complex fashion to generate the ordered assembly which is the NC (18). Investigation of this process in vivo by both genetic and biochemical analyses has allowed only limited insight into the process of core assembly (6, 7, 10). Mutations and in vivo core assembly assays have generated results that are difficult to interpret due to the complexity of the in vivo environment. Structural approaches with cryo-EM and X-ray crystallography have provided some insight as to the nature of core assembly by providing a picture of the final virion and its internal core (3, 11, 30). A system for the in vitro assembly of NCs was required to allow a more extensive and controlled examination of core formation. An in vitro assembly system was previously described by Wengler and colleagues and used CP isolated from virions (41, 42). Although this system was useful, its reliance on virus-purified CP restricted its use to the wild-type virus and possibly mutant viruses which grew well. To circumvent these restrictions, a prokaryotic expression system for the production and purification of large amounts of SINV and RRV CPs was developed, and an in vitro assembly system that uses these expressed proteins was established. Core formation assays were developed to allow examination of the NC assembly process. Examination of assembled core particles by electron microscopy, gradient sedimentation, and agarose gel assays suggested that in vitro-assembled cores were of a regular size and shape and appeared quite similar to native cytoplasmic cores or virus-purified cores. High-resolution analysis of the in vitro-assembled CLPs of both SINV and RRV by cryo-EM and image reconstruction is currently under way.
The in vitro assembly experiments presented were primarily designed to examine the requirements of NC assembly and to examine possible steps in the pathway. As the RRV and SINV assembly systems were nearly identical in protein and oligonucleotide requirements, a logical first step was to attempt to generate phenotypically mixed CLPs. It was expected that mixed cores, those that contain both SINV and RRV proteins, might be generated when assembly reactions were carried out with an equal mixture of the two CPs. The agarose gel assay developed to examine assembly provided a convenient assay for these experiments due to the difference in the migration of the cores of the two viruses following electrophoresis. One possible outcome of the mixing experiments would be a new species of core particle with a migration between those of the two parental core bands in the agarose gel assay. Surprisingly, no mixed-core-particle band was detected in these experiments. Furthermore, we did not detect any CLPs migrating between the two parental core bands, which would have been the result of both proteins mixing randomly into particles. This result suggests that the protein-protein interactions involved in the in vitro assembly of SINV and RRV CLPs are homotypic. It is interesting that these two homologous proteins, which share 68% sequence identity in their C-terminal domains (residues 97 to 264, SINV numbering) (9), have distinct protein-protein interactions in the core that are significant enough to prevent heterotypic mixing between the virus CPs. In contrast, chimeric viruses containing recombinant genomes have been shown to be viable when various combinations of the structural and nonstructural coding regions from two viruses are exchanged (20, 26, 35, 44, 45).
In order to understand which regions of the CP were required for core assembly, a series of amino-terminal truncations of the SINV CP were generated and assayed for their ability to generate CLPs in the in vitro assembly system. The initial SINV assembly experiments were conducted with the predominant species generated in the E. coli-based expression system, CP(19–264). The minor species, CP(9–264), was also assayed for in vitro assembly and was found to be identical in all assays to CP(19–264). These proteins, although used to establish the assembly system, represent truncations of the native wild-type CP. Therefore, the first 18 amino acids of the protein were dispensable for core assembly in vitro. Further truncation of the CP to residue 32 was sufficient to block CLP formation in the in vitro system. Truncation of the CP to amino acids 81 to 264, thereby completely eliminating the predicted N-terminal helix and the majority of basic amino acids from the N-terminal domain of the protein, prevented CLP formation but failed to block nucleic acid binding or incorporation of the protein together with CP(19–264) into CLPs. Deletion of the N-terminal 105 residues [CP(106–264)] or 113 residues [CP(114–264)] prevented assembly, incorporation, and nucleic acid binding. Previous results had demonstrated that these proteins do not bind nucleic acid, suggesting a requirement for nucleic acid in core assembly (29). Also, a direct correlation between the incorporation into CLPs of truncated CPs present at low ratios relative to CP(19–264) and the ability to inhibit core formation at high ratios was established.
Several in vivo studies have examined regions of the CP that play a role in assembly. Forsell et al. demonstrated that a deletion of the N-terminal domain of Semliki Forest virus (residues 1 to 112) prevented NC formation in vivo and abolished the production of virus particles, whereas significant deletions of the N-terminal basic domain could be made without a loss of core formation (10). In contrast, Owen and Kuhn isolated a SINV mutant which duplicated 80 residues of the positively charged N-terminal domain (residues 10 to 89) (29). From these results, it is likely that the residues of the N-terminal domain (residues 1 to 80) maintain flexible contacts involved in organizing the NC, permitting numerous permutations of the native amino acid sequence. As expected, however, certain N-terminal residues are more important for core assembly and virus production than others, particularly those in the N-terminal helix region (28a).
Examination of the nucleic acid requirements for core assembly in the in vitro system provides some interesting results not observed in previous in vitro assembly experiments. The ability to assemble CLPs in vitro with a variety of single-stranded nucleic acids but not with double-stranded DNA indicates that charge neutralization of the basic CP by polyanionic substrates is not the only role of nucleic acids in core assembly. The helical structure of DNA may prevent some nucleotide base-specific contacts required for protein-nucleic acid interactions in particle formation. The DNA helix also may pose problems for assembly due to the relative size and rigidity of the helix compared to those of single-stranded substrates. A second novel observation is the demonstration of specific recognition of the genomic viral RNA of SINV by the CP during in vitro assembly. This observation lends significant physiological relevance to the in vitro-assembled CLP. The third observation of interest is the presence of a minimum size of DNA oligonucleotide substrate required for core assembly. The demonstration of a minimum size of oligonucleotide substrate suggests a minimum CP nucleic acid binding “footprint” or possibly nucleic acid tethering between two adjacent CPs, as would be seen in a nucleic acid-bound protein dimer (2, 22).
The stoichiometry of nucleic acids of various lengths relative to a fixed amount of CP required for in vitro assembly demonstrates that shorter oligonucleotides must be present in larger molar amounts for assembly to occur. If core assembly required only a fixed number of specific nucleic acid binding sites to be occupied, equal amounts of short or long oligonucleotides would be required for assembly reactions. The requirement for larger amounts of shorter oligonucleotides suggests a “headful” encapsidation of nucleic acids (8, 25). These preliminary experiments do not exclude the possibility of both high-affinity specific site binding and low-affinity overall charge neutralization within the core. A more extensive kinetic analysis of nucleic acid binding and core composition is required to fully investigate this point.
The experiments presented here suggest the importance of nucleic acid in the process of NC assembly. The data suggest that nucleic acid is absolutely required for in vitro core formation. Furthermore, this hypothesis is supported by the inability to generate empty CLPs in vitro over a wide variety of conditions. If charge neutralization were the only role for nucleic acid in core assembly, then empty particle formation should be possible in high-ionic-strength buffers. Indeed, charge neutralization alone does not appear to be sufficient to drive core assembly, as double-stranded DNA fails to form core particles. The requirement of a minimum length of oligonucleotide substrate for in vitro assembly suggests the tethering of adjacent CPs together in a nucleic acid-bound dimer as the first step in core assembly. The minimum size may merely represent the minimum specific binding site footprint required for interactions. However, prior to the addition of nucleic acid, the CP is predominantly a monomeric protein, as determined by gel filtration chromatography and analytical ultracentrifugation analysis. Yeast two-hybrid genetic screens for interactions between CPs and between regions of the CP have failed to demonstrate homotypic interactions (40a), as have been seen in other viral systems (19, 27, 28). Following the addition of an appropriate nucleic acid substrate, a core particle of regular size and shape is rapidly generated. The lack of protein interactions in the absence of nucleic acid suggests that nucleic acid is involved in a preliminary step in the assembly process, most likely in the initial oligomerization of the CP. Subsequent steps are likely to involve protein sequences downstream of residue 81, since CP(81–264) can be incorporated and can inhibit the assembly of core particles by CP(19–264). Efforts are currently focused on the isolation and identification of the preliminary nucleic acid-bound protein species seen in core assembly by use of truncated and mutant CPs.
The development of an in vitro assembly system for SINV and RRV allows the process of alphavirus core assembly to be examined in great detail. Structural, genetic, and biochemical approaches that use the data and reagents from the in vitro assembly system can now be used to begin the molecular dissection of this core assembly process.
ACKNOWLEDGMENTS
We thank John Burgner for assistance with the analytical ultracentrifugation experiments. We also thank Tom Smith for valuable expertise in the development of protein purification conditions. Additionally, Ralf Mattes of the Institut für Industrielle Genetik, Stuttgart, Germany, kindly provided pSBetB vector DNA. Stimulating discussions with Chris Jones, Katherine Owen, Rushika Perera, and Michael Rossmann are also acknowledged.
This work was supported by Public Health Service grant GM56279 from the National Institutes of Health. Support from the Lucille P. Markey Foundation for structural studies at Purdue University is also acknowledged. T.L.T. was supported by an NIH biophysics training grant (GM98296). Support for A.E.H. and R.O. was provided by an NIH Public Health Service grant (AI 35212) to Michael Rossmann. R.O. also acknowledges the support of a Deutsche Forschungsgemeinschaft postdoctoral fellowship.
REFERENCES
- 1.Aliperti G, Schlesinger M J. Evidence for an autoprotease activity of Sindbis virus capsid protein. Virology. 1978;90:366–369. doi: 10.1016/0042-6822(78)90321-5. [DOI] [PubMed] [Google Scholar]
- 2.Chao K L, Lohman T M. DNA-induced dimerization of the Escherichia coli Rep helicase. J Mol Biol. 1991;221:1165–1181. doi: 10.1016/0022-2836(91)90926-w. [DOI] [PubMed] [Google Scholar]
- 3.Cheng R H, Kuhn R J, Olson N H, Rossmann M G, Choi H-K, Smith T J, Baker T S. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell. 1995;80:621–630. doi: 10.1016/0092-8674(95)90516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi H-K, Lee S, Zhang Y-P, McKinney B R, Wengler G, Rossmann M G, Kuhn R J. Structural analysis of Sindbis virus capsid mutants involving assembly and catalysis. J Mol Biol. 1996;262:151–167. doi: 10.1006/jmbi.1996.0505. [DOI] [PubMed] [Google Scholar]
- 5.Choi H K, Tong L, Minor W, Dumas P, Boege U, Rossmann M G, Wengler G. Structure of Sindbis virus core protein reveals a chymotrypsin-like serine proteinase and the organization of the virion. Nature (London) 1991;354:37–43. doi: 10.1038/354037a0. [DOI] [PubMed] [Google Scholar]
- 6.Coombs K, Brown D T. Organization of the Sindbis virus nucleocapsid as revealed by bifunctional cross-linking agents. J Mol Biol. 1987;195:359–371. doi: 10.1016/0022-2836(87)90657-7. [DOI] [PubMed] [Google Scholar]
- 7.Coombs K, Brown D T. Topological organization of Sindbis virus capsid protein in isolated nucleocapsids. Virus Res. 1987;7:131–149. doi: 10.1016/0168-1702(87)90075-x. [DOI] [PubMed] [Google Scholar]
- 8.Coren J S, Pierce J C, Sternberg N. Headful packaging revisited: the packing of more than one DNA molecule into a bacteriophage P2 head. J Mol Biol. 1995;249:176–184. doi: 10.1006/jmbi.1995.0287. [DOI] [PubMed] [Google Scholar]
- 9.Dalgarno L, Rice C M, Strauss J H. Ross River virus 26S RNA: complete nucleotide sequence and deduced sequence of the encoded structural proteins. Virology. 1983;129:170–187. doi: 10.1016/0042-6822(83)90404-x. [DOI] [PubMed] [Google Scholar]
- 9a.Fisher, B. R., R. J. Kuhn, and M. G. Rossmann. Unpublished results.
- 10.Forsell K, Suomalainen M, Garoff H. Structure-function relation of the NH2-terminal domain of the Semliki Forest virus capsid protein. J Virol. 1995;69:1556–1563. doi: 10.1128/jvi.69.3.1556-1563.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuller S D, Berriman J A, Butcher S J, Gowen B E. Low pH induces swiveling of the glycoprotein heterodimers in the Semliki Forest virus spike complex. Cell. 1995;81:715–725. doi: 10.1016/0092-8674(95)90533-2. [DOI] [PubMed] [Google Scholar]
- 12.Geigenmüller-Gnirke U, Nitschko H, Schlesinger S. Deletion analysis of the capsid protein of Sindbis virus: identification of the RNA binding region. J Virol. 1993;67:1620–1626. doi: 10.1128/jvi.67.3.1620-1626.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glanville N, Ulmanen J. Biological activity of in vitro synthesized protein: binding of Semliki Forest virus capsid protein to the large ribosomal subunit. Biochem Biophys Res Commun. 1976;71:393–399. doi: 10.1016/0006-291x(76)90295-3. [DOI] [PubMed] [Google Scholar]
- 13a.Gorbalenya, A. E., K. E. Owen, and R. J. Kuhn. Unpublished results.
- 14.Hahn C S, Strauss E G, Strauss J H. Sequence analysis of three Sindbis virus mutants temperature-sensitive in the capsid autoprotease. Proc Natl Acad Sci USA. 1985;82:4648–4652. doi: 10.1073/pnas.82.14.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hahn C S, Strauss J H. Site-directed mutagenesis of the proposed catalytic amino acids of the Sindbis virus capsid protein autoprotease. J Virol. 1990;64:3069–3073. doi: 10.1128/jvi.64.6.3069-3073.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrison S C, Strong R K, Schlesinger S, Schlesinger M J. Crystallization of Sindbis virus and its nucleocapsid. J Mol Biol. 1992;226:277–280. doi: 10.1016/0022-2836(92)90141-6. [DOI] [PubMed] [Google Scholar]
- 17.Heaton L A. Use of agarose gel electrophoresis to monitor conformation changes of some small, spherical plant viruses. Phytopathology. 1992;82:803–807. [Google Scholar]
- 18.Johnson J E, Speir J A. Quasi-equivalent viruses: a paradigm for protein assemblies. J Mol Biol. 1997;269:665–675. doi: 10.1006/jmbi.1997.1068. [DOI] [PubMed] [Google Scholar]
- 19.König S, Beterams G, Nassal M. Mapping of homologous interaction sites in the hepatitis B virus core protein. J Virol. 1998;72:4997–5005. doi: 10.1128/jvi.72.6.4997-5005.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuhn R J, Griffin D E, Owen K E, Niesters H G, Strauss J H. Chimeric Sindbis-Ross River viruses to study interactions between alphavirus nonstructural and structural regions. J Virol. 1996;70:7900–7909. doi: 10.1128/jvi.70.11.7900-7909.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn R J, Niesters H G M, Hong Z, Strauss J H. Infectious RNA transcripts from Ross River virus cDNA clones and the construction and characterization of defined chimeras with Sindbis virus. Virology. 1991;182:430–441. doi: 10.1016/0042-6822(91)90584-x. [DOI] [PubMed] [Google Scholar]
- 22.Kukolj G, Tolias P P, Autexier C, DuBow M S. DNA directed oligomerization of the monomeric Ner repressor from the Mu-like bacteriophage D108. EMBO J. 1989;10:3141–3148. doi: 10.1002/j.1460-2075.1989.tb08467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LeCuyer K A, Behlen L S, Uhlenbeck O C. Mutations of bacteriophage MS2 coat protein that alter its cooperative binding to RNA. Biochemistry. 1996;34:10600–10606. doi: 10.1021/bi00033a035. [DOI] [PubMed] [Google Scholar]
- 24.Lee S, Owen K E, Choi H K, Lee H, Lu G, Wengler G, Brown D T, Rossmann M G, Kuhn R J. Identification of a protein binding site on the surface of the alphavirus nucleocapsid protein and its implication in virus assembly. Structure. 1996;4:531–541. doi: 10.1016/s0969-2126(96)00059-7. [DOI] [PubMed] [Google Scholar]
- 25.Leffers G, Rao V B. A discontinuous headful packaging model for packaging less than headful length DNA molecules by bacteriophage T4. J Mol Biol. 1996;258:839–850. doi: 10.1006/jmbi.1996.0291. [DOI] [PubMed] [Google Scholar]
- 26.Lopez S, Yao J-S, Kuhn R J, Strauss E G, Strauss J H. Nucleocapsid-glycoprotein interactions required for assembly of alphaviruses. J Virol. 1994;68:1316–1323. doi: 10.1128/jvi.68.3.1316-1323.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luban J, Alin K B, Bossolt K L, Humaran T, Goff S P. Genetic assay for multimerization of retroviral gag proteins. J Virol. 1992;66:5157–5160. doi: 10.1128/jvi.66.8.5157-5160.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsumoto M, Hwang S B, Jeng K-S, Zhu N, Lai M M C. Homotypic interaction and multimerization of hepatitis C virus core protein. Virology. 1996;218:43–51. doi: 10.1006/viro.1996.0164. [DOI] [PubMed] [Google Scholar]
- 28a.Owen, K., R. Perera, and R. J. Kuhn. Unpublished results.
- 29.Owen K E, Kuhn R J. Identification of a region in the Sindbis virus nucleocapsid protein that is involved in specificity of RNA encapsidation. J Virol. 1996;70:2757–2763. doi: 10.1128/jvi.70.5.2757-2763.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paredes A M, Brown D T, Rothnagel R, Chiu W, Schoepp R J, Johnston R E, Prasad B V. Three-dimensional structure of a membrane-containing virus. Proc Natl Acad Sci USA. 1993;90:9095–9099. doi: 10.1073/pnas.90.19.9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rice C M, Levis R, Strauss J H, Huang H V. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J Virol. 1987;61:3809–3819. doi: 10.1128/jvi.61.12.3809-3819.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rice C M, Strauss J H. Nucleotide sequence of the 26S mRNA of Sindbis virus and deduced sequence of the encoded virus structural proteins. Proc Natl Acad Sci USA. 1981;78:2062–2066. doi: 10.1073/pnas.78.4.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schenk P M, Baumann S, Mattes R, Steinbiss H. Improved high-level expression system for eukaryotic genes in E. coli using T7 RNA polymerase and rare argtRNAs. BioTechniques. 1995;19:196–200. [PubMed] [Google Scholar]
- 34.Singh I, Helenius A. Role of ribosomes in Semliki Forest virus nucleocapsid uncoating. J Virol. 1992;66:7049–7058. doi: 10.1128/jvi.66.12.7049-7058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smyth J, Suomalainen M, Garoff H. Efficient multiplication of a Semliki Forest virus chimera containing Sindbis virus spikes. J Virol. 1997;71:818–823. doi: 10.1128/jvi.71.1.818-823.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Söderlund H. Kinetics of formation of Semliki Forest virus nucleocapsid. Intervirology. 1973;1:354–361. doi: 10.1159/000148864. [DOI] [PubMed] [Google Scholar]
- 37.Söderlund H, Ulmanen I. Transient association of Semliki Forest virus capsid protein with ribosomes. J Virol. 1977;24:907–909. doi: 10.1128/jvi.24.3.907-909.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sorger P K, Stockley P G, Harrison S C. Structure and assembly of turnip crinkle virus. II. Mechanism of reassembly in vitro. J Mol Biol. 1986;191:639–658. doi: 10.1016/0022-2836(86)90451-1. [DOI] [PubMed] [Google Scholar]
- 39.Speir J A, Munshi S, Wang G, Baker T S, Johnson J E. Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy. Structure. 1995;3:63–78. doi: 10.1016/s0969-2126(01)00135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40a.Tellinghuisen, T. L., and R. J. Kuhn. Unpublished results.
- 41.Wengler G, Boege U, Wengler G, Bischoff H, Wahn K. The core protein of the alphavirus Sindbis virus assembles into core-like nucleoproteins with the viral genome RNA and with other single-stranded nucleic acids in vitro. Virology. 1982;118:401–410. doi: 10.1016/0042-6822(82)90359-2. [DOI] [PubMed] [Google Scholar]
- 42.Wengler G, Wengler G, Boege U, Wahn K. Establishment and analysis of a system which allows assembly and disassembly of alphavirus core-like particles under physiological conditions in vitro. Virology. 1984;132:401–412. doi: 10.1016/0042-6822(84)90045-x. [DOI] [PubMed] [Google Scholar]
- 43.Wengler G, Würkner D, Wengler G. Identification of a sequence element in the alphavirus core protein which mediates interaction of cores with ribosomes and the disassembly of cores. Virology. 1992;191:880–888. doi: 10.1016/0042-6822(92)90263-o. [DOI] [PubMed] [Google Scholar]
- 44.Yao J, Strauss E G, Strauss J H. Molecular genetic study of the interaction of Sindbis virus E2 with Ross River virus E1 for virus budding. J Virol. 1998;72:1418–1423. doi: 10.1128/jvi.72.2.1418-1423.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yao J S, Strauss E G, Strauss J H. Interactions between PE2, E1, and 6K required for assembly of alphaviruses studied with chimeric viruses. J Virol. 1996;70:7910–7920. doi: 10.1128/jvi.70.11.7910-7920.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, W., B. R. Fisher, N. Olson, R. J. Kuhn, and T. S. Baker. Unpublished results.