Abstract
Background
Common genetic variants with small effect sizes have been associated with rotator cuff tearing although very few rare, highly penetrant variants have been identified. The purpose of this pilot study was to identify dominant coding variants that segregated with affected individuals in pedigrees at high risk for rotator cuff tears (RCTs). We hypothesize that rare variants contribute to symptomatic RCTs and that they can be identified in related cases with a full-thickness tear requiring surgical management.
Methods
We used the Utah Population Database to identify pedigrees that exhibited a significant excess of individuals who had undergone surgical repair of a full-thickness RCT. We analyzed whole exome sequence analysis to identify rare coding variants in 9 independent affected cousin pairs (first or second cousins) who had undergone arthroscopic surgery for repair of a full-thickness RCT (mean age at diagnosis 68 years). Validation of association of the candidate variants with risk for rotator cuff tearing was accomplished utilizing data from the UK Biobank and a separate cohort of unrelated cases of full-thickness RCTs.
Results
A total of 82 rare (minor allele frequency <0.005) coding variants were identified as shared in at least one cousin pair affected with full-thickness rotator cuff tearing belonging to a high-risk pedigree, which included variants in RUNX1, ADAM12, TGFBR2, APBB1, PDLIM7, LTBP1, MAP3K4, and MAP3K1. Analysis of 39 of these variants with data available in the UK Biobank (3899 cases with rotator cuff injury and 11,697 matched controls; mean case age 59.9 years) identified a significant association with the APBB1 gene (OR = 2.37, P = .007, uncorrected). The PDLIM7 allele was found to be in significant excess in RCT cases in a separate cohort of Utah patients with full-thickness RCTs (10 carriers out of 458 independent, unrelated patients; minor allele frequency of 0.022) compared to a minor allele frequency of 0.0058 for the European (non-Finnish) control population rate (749 carriers out of 128612 tested) (chi-square test: 19.3 [P < .001]).
Discussion
The analysis of closely related individuals with confirmed full-thickness RCTs from high-risk pedigrees has identified 82 rare, shared candidate genetic predisposition coding variants. Association of the PDLIM7 allele with risk for tear was confirmed in an independent cohort of RCTs. Further analysis of the variant alleles is required for confirmation of these genes in rotator cuff tearing.
Keywords: Rotator cuff tear, Genetic variants, Pedigrees, High risk, UPDB, APBB1, PDLIM7
Rotator cuff disease has previously been shown to have a strong genetic predisposition.21 Several studies have utilized large genetic databases to identify common genetic variants associated with rotator cuff tearing.1,10,15,18, 19, 20,22 A few studies have had sufficient power to confirm variants associated with rotator cuff tearing in independent datasets.1,18 Multiple genes have been previously identified as being associated with rotator cuff tearing, including those associated with extracellular matrix production, tendon maturation or modulation, cell adhesion, growth factors critical to tendon healing, and function and hormonal receptors. 1,10,15,18, 19, 20,22 Identification of genes in rotator cuff tear (RCT) patients allows for understanding the pathogenesis of the disease which can be utilized potentially for prevention but also as a template for further work investigating genetic factors associated with tear progression and repair healing.
RCTs are common, complex injuries. Genetic variants, both common and rare, likely have an important role in the etiology of RCTs. Common variants with small risk are most easily identified using large genetic databases of unrelated individuals while rare, highly penetrant variants associated with large risk can be efficiently detected using a small number of related individuals. The Utah Population Database (UPDB) is a computerized genealogy of the Utah pioneer founders and descendants which has been updated with statewide birth certificates (father, mother, child) and linked to vital records (eg, death certificates), cancer, census, and medical data. The UPDB allows identification of families that have a higher preponderance of specific diseases including musculoskeletal disease like osteoarthritis and osteoporosis. We define high-risk pedigrees as those with a statistically significant excess number of cases compared to control pedigrees. Sequence data for multiple members of the same high-risk pedigree with the same disorder, like rotator cuff tearing, allows identification of shared rare genetic variants. The value of studying high-risk pedigrees is that the cases within likely have a stronger genetic contribution to their disease, and importantly, they are more likely to carry the same pathogenic variants at a much higher rate than occurs in the population, thereby enhancing signal detection and, by extension, relative efficiency. Only Azzara et al have reported on rare variants identified in four patients with rotator cuff tearing using exome sequencing.2
The purpose of this pilot study was to identify affected individuals with full-thickness RCTs who were members of high-risk RCT pedigrees, perform whole exome sequencing (WES) on selected sets of relatives, and then identify any rare, deleterious genetic variants shared among these individuals. We hypothesize that rare variants contribute to symptomatic RCTs and that they can be identified in related cases with a full-thickness tear requiring surgical management. Rare variants were tested for association with risk of rotator tear utilizing an independent genetic database, the UK Biobank. Select variants with a strong biologic mechanism associated with rotator cuff tearing were also evaluated for association with risk for RCT in an independent set of unrelated RCT cases by comparing the prevalence observed with the prevalence of these variants in a population dataset.
Materials and methods
The UPDB data include information on over 11 million individuals of which almost 3 million have at least 3 generations of genealogy data; some pedigrees extend to 12 generations. The individual genealogy data have been record-linked to electronic medical records from 2014 to 2021 representing over 2 million patients from the University of Utah, one of the largest healthcare providers in the state, allowing identification of genetic relationships among individuals diagnosed and treated for various diseases, including RCTs. The UPDB was queried for patients who had undergone an arthroscopic rotator cuff repair (RCR) using the Current Procedural Terminology (CPT) code 29827. Among the 1.2 million individual patients in the University of Utah Health Sciences Center with at least 3 generations of genealogy, there were 1070 living patients with a medical record containing a CPT4 procedure code indicative of RCR (CPT 29827). All ancestors of the 1070 RCR cases with linked genealogy data were analyzed to identify 1694 clusters of at least two related RCR cases (individuals could be part of multiple clusters) descending from a common founder or founder pair (pedigrees).
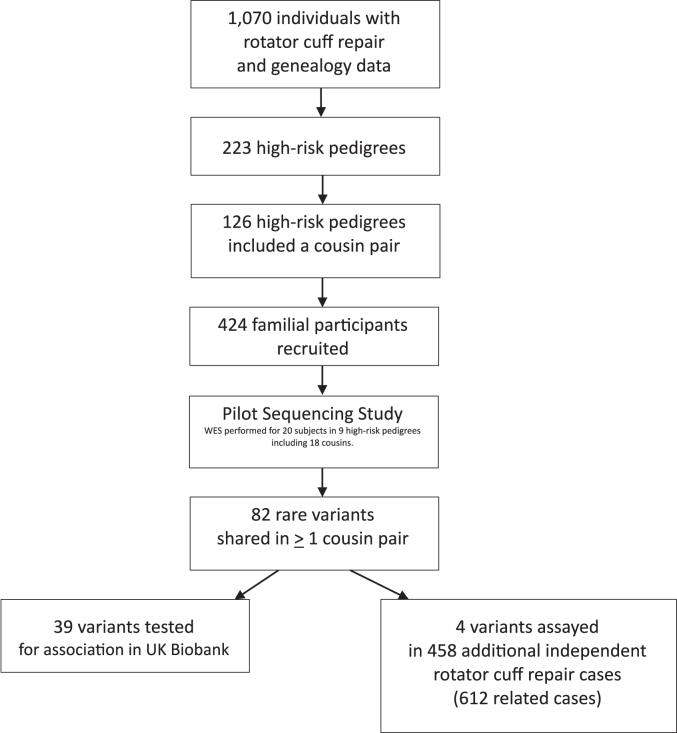
Because some clusters of RCR cases could have occurred by chance, we tested each of the clusters/pedigrees to determine whether there was a significant excess of observed-to-expected RCR cases among the descendants in each pedigree. The expected number of cases was estimated using internal cohort-specific rates for rotator cuff tearing estimated as follows. All individuals in the UPDB with at least 3 generations of genealogy data and linked hospital data were assigned to a cohort based on biological sex, birthplace (Utah or not), and 5-year birth year range. Cohort-specific rates of rotator cuff tearing were estimated as the number of cases in each cohort divided by the total number of UPDB individuals with at least 3 generations of genealogy and hospital data in each cohort. For each pedigree, the total number of descendants was counted by cohort. The total number of observed cases in a pedigree was counted; the expected number of RCR cases was estimated by summing the cohort-specific rate of rotator cuff tearing for all descendants. If a significant excess of observed to expected RCR cases was noted (P < .05), the pedigree was termed high risk for RCR. Two hundred twenty-three of the 1694 pedigrees were found to have a significant excess of cases; 126 of these high-risk pedigrees included a pair of affected cousins (first or second cousins). We targeted these related cousins who had undergone a RCR procedure for recruitment, as well as other affected relatives. We recruited, consented, and sampled 424 familial subjects who had a RCR. All patients had confirmation of a diagnosis of full-thickness rotator cuff tearing and repair through review of both the operative reports for each patient and review of each patient’s magnetic resonance imaging by a fellowship-trained shoulder and elbow surgeon (Fig. 1).
Figure 1.
Flow chart showing the selection of individuals included in this pilot study.
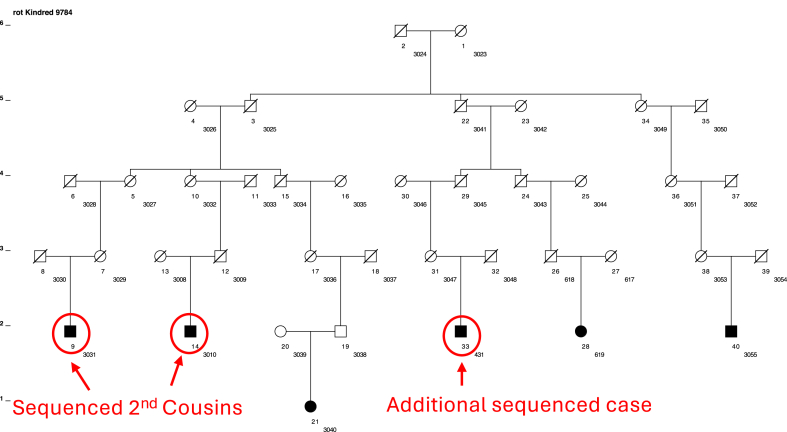
For this pilot study, 20 recruited members of 9 of the high-risk pedigrees were selected to sequence and analyze to identify rare, shared variants. An example studied pedigree is shown in Figure 2. Each pedigree included a sampled affected first- or second-cousin pair and any other sampled, affected relatives, for a total of 20 unique subjects. Six pedigrees had two sampled individuals with RCR, two pedigrees had 3 sampled individuals with RCR, and one pedigree had 4 sampled individuals with RCR. One individual was in two nonoverlapping pedigrees. Demographic information about patients was recorded from the medical record including age, sex, body mass index, handedness, presence of diabetes, and smoking status. Patients were also queried about other tendon problems as well as if they had any other family members with a RCT.
Figure 2.
Example of one of the 9 high-risk pedigrees with three individuals with rotator cuff repair recruited in which DNA was obtained and sequenced. Males are represented as squares, females as circles. Individuals with slash marks are deceased. This pedigree contains 2521 descendants from the founder pair; however, only the connecting relatives between cases are shown. There are 6 sampled and confirmed RCR cases (black circles). This pedigree has a significant excess of RCR with 6 confirmed cases compared to 2.59 expected RCR cases based on age- and sex-matched population rates (P = .048). The sequenced second cousins are indicated. There is one additional recruited RCR case as indicated. RCR, rotator cuff repair.
Preoperative magnetic resonance imagings were evaluated for coronal tear size (cm), sagittal tear size (mm), and supraspinatus fatty infiltration. Coronal tear size was measured on T2-weighted coronal sections with retraction measured as the distance from the lateral edge of the greater tuberosity to the tendon end. Sagittal tear size was determined by measuring the anteroposterior length of the exposed rotator cuff footprint of the lateral-most section of the greater tuberosity on the sagittal T2-weighted images. Tears were classified by size into small (<1 cm), medium (1 cm to 3 cm), large (3 cm to 5 cm), and massive (>5 cm) sizes based on sagittal tear size. Supraspinatus fatty infiltration was measured using the Fuchs classification as a modification of the Goutallier classification for computed tomography scans.11,12 Grading was on the most lateral sagittal T1-weighted image on which the scapular spine was in contact with the scapular body. The stages were identified as follows: stage 0, normal; stage 1, some fatty streaks; stage 2, fatty infiltration throughout with less fat than muscle; stage 3, fatty infiltration with equal muscle and fat; stage 4, fatty infiltration with more fat than muscle.
DNA for all subjects was isolated from cellular content collected from a saliva sample. WES was performed at Huntsman Cancer Institute High-Throughput Genomics Core. DNA libraries were prepared from 1.5 μg of DNA using the IDT xGEN Human Exome v2 capture kit. Samples were run on the Illumina NextSeq (Illumina, San Diego, CA, USA) instrument. Reads were mapped to the human genome GRCh38 reference using BWA-mem for alignment, and variants were called using Genome Analysis Toolkit (GATK) version 4.1.4.1 (Broad Institute, Cambridge, MA, USA) software following Broad Institute Best Practices Guidelines (25). Exome capture resulted in an average of 95% of target bases covered by greater than 10× coverage across the exome, with an average depth of 265X. Variants occurring outside the exome capture kit’s intended coverage area were removed. Variants were called and filtered using standard GATK VQSR filters (QD < 2.0, QUAL < 30 MQRankSum < −12.5, ReadPosRankSum < −8.0, FS > 60.0, SOR > 3.0, max-gaussians = 2, and truth-sensitivity-filter-level 99.0). Variants were annotated with Annovar (Biobase, Jinan, China), which contains predicted pathogenicity scores from 15 in-silico functional prediction algorithms.
Summary statistics were available from a previously performed genome-wide association study (GWAS) identifying candidate variants for rotator cuff injury defined by International Classification of Diseases9/10 diagnostic codes using case and control imputed genetic data from the UK Biobank.22 Validation was attempted for each of the candidate variants identified in the current analysis utilizing the summary statistics from the previously performed UK Biobank GWAS.
DNA from 612 patients (relationships assessed utilizing the UPDB genealogy) with full-thickness posterosuperior RCTs had been previously collected. These cases were used to validate a subset of variant genes selected based upon function. Some of these individuals were related to each other. While variant testing was performed for all 612 individuals, for those individuals who were part of a pedigree, we counted only one individual per pedigree to meet the independence assumption required for estimating the prevalence rate of the variant in the Utah rotator cuff population and chi-square association testing. For these 612 individuals, 286 individuals were part of 134 pedigrees and 324 individuals had no other identified relative in our dataset. Hence, our effective total number of independent cases was 458 participants with a RCT. Genetic variants in the selected genes were evaluated using Taqman assays in the 612 patients (458 effective independent cases) to estimate the percentage of variants in this population. All 612 subjects were assayed for the variants so that we could identify any new pedigrees that were segregating one of these variants. The prevalence of these variants in the RCT dataset was compared to the prevalence of these variants in a European (non-Finnish) population (gnomeAD). Chi-square analysis was utilized to test for differences in prevalence.
Results
All 20 patients from the 9 high-risk pedigrees had a full-thickness RCT that underwent arthroscopic repair. The average age at the time of the magnetic resonance imaging was 60 years (range: 49 to 72 years). Of the 20 patients, 13 were male and 7 were female. The average body mass index was 32 kg/m2 (range: 23 to 47 kg/m2). The RCT affected 10 of 20 patients on their dominant side. 25% (5 of 20) patients reported that they had a history of smoking. 40% (8 of 20) patients reported a prior history of other tendon disorders in addition to the RCT. 35% (7 of 20) patients reported a history of prior tendon surgeries in addition to their RCR surgery. 25% (5 of 20) patients reported a major trauma resulting in the RCT while 25% (5 of 20) patients reported a history of diabetes. 30% (6 of 20) patients reported a family member having a RCT or having rotator cuff surgery including one patient who had a twin brother having a RCT. The average anterior posterior tear width was 2.1 cm and the average tear retraction was 2 cm.
A total of 82 rare (minor allele frequency <0.005) candidate predisposition variants were identified as shared in at least one cousin pair affected with a full-thickness RCT from a high-risk pedigree (Table I). All variants occurred in exomic regions of the DNA. Of the rare variants identified with a likely contribution to RCTs or healing based on previous associations with extracellular matrix organization, muscle density/function or tendinopathy, one variant was identified in the gene Runt-related transcription factor 1 (RUNX1), one in the gene Disintegrin and metalloproteinase domain-containing protein 12 (ADAM12), one in the gene Transforming growth factor beta receptor 2 (TGFBR2), one in Amyloid Precursor Protein-Binding Protein (APBB1), two separate variants in two genes in the mitogen activated protein kinase (MAPK) pathway (MAP3K4, MAP3K1), one variant in PDZ and LIM domain protein 7 (PDLIM7), and one variant in latent transforming growth factor beta binding protein 1 (LTBP1).
Table I.
Chromosomal location and gene associated with variant single nucleotide polymorphisms.
| Gene_refGene | Chr | Start | End | Ref | Alt | Func_refGene |
|---|---|---|---|---|---|---|
| AIM2 | chr1 | 159062696 | 159062696 | - | T | exonic |
| GPATCH2 | chr1 | 217431290 | 217431290 | G | A | exonic |
| APOB | chr2 | 21042446 | 21042446 | T | C | exonic |
| ATAD2B | chr2 | 23757445 | 23757445 | C | T | exonic |
| LTBP1 | chr2 | 33134781 | 33134781 | C | G | exonic |
| FAM98A | chr2 | 33584927 | 33584927 | C | A | exonic |
| CCDC150 | chr2 | 196666795 | 196666795 | A | - | exonic |
| KIAA2012 | chr2 | 202190209 | 202190209 | - | A | exonic |
| CRYGD | chr2 | 208121934 | 208121934 | G | C | exonic |
| PER2 | chr2 | 238271374 | 238271374 | C | T | exonic |
| TGFBR2 | chr3 | 30650380 | 30650380 | A | - | exonic |
| RBM5 | chr3 | 50118455 | 50118456 | GA | - | exonic |
| ASTE1 | chr3 | 131014203 | 131014203 | T | - | exonic |
| RASA2 | chr3 | 141487100 | 141487100 | C | T | exonic |
| PPP1R2 | chr3 | 195527870 | 195527870 | T | - | exonic |
| IDUA | chr4 | 1004341 | 1004341 | C | T | exonic |
| FGFRL1 | chr4 | 1025267 | 1025268 | CA | - | exonic |
| COMMD8 | chr4 | 47453083 | 47453083 | T | C | exonic |
| PDZD2 | chr5 | 32058056 | 32058056 | C | A | exonic |
| PDZD2 | chr5 | 32074423 | 32074423 | C | T | exonic |
| AMACR | chr5 | 34007920 | 34007920 | C | G | exonic |
| NDUFS4 | chr5 | 53560667 | 53560667 | C | A | exonic |
| MAP3K1 | chr5 | 56882022 | 56882027 | CAACAA | - | exonic |
| RNF145 | chr5 | 159203621 | 159203621 | - | T | exonic |
| PDLIM7 | chr5 | 177492655 | 177492655 | G | A | exonic |
| TTBK1 | chr6 | 43282943 | 43282945 | GAG | - | exonic |
| NUP43 | chr6 | 149727030 | 149727030 | G | A | exonic |
| IYD | chr6 | 150389354 | 150389354 | G | A | exonic |
| MAP3K4 | chr6 | 161098319 | 161098324 | CTGCTG | - | exonic |
| MICALL2 | chr7 | 1448629 | 1448629 | G | A | exonic |
| ZNF727 | chr7 | 64078166 | 64078166 | T | C | exonic |
| CROT | chr7 | 87357531 | 87357531 | G | - | exonic |
| PRAG1 | chr8 | 8378000 | 8378000 | G | C | exonic |
| CLDN23 | chr8 | 8702666 | 8702666 | G | A | exonic |
| C8orf48 | chr8 | 13567218 | 13567218 | A | G | exonic |
| NAT1 | chr8 | 18222237 | 18222237 | C | T | exonic |
| SNTB1 | chr8 | 120548788 | 120548788 | G | A | exonic |
| HMCN2 | chr9 | 130362103 | 130362103 | C | T | exonic |
| AGPAT2 | chr9 | 136676978 | 136676978 | G | A | exonic |
| KIAA1217 | chr10 | 24494592 | 24494592 | A | G | exonic |
| ADAM12 | chr10 | 126155235 | 126155235 | T | C | exonic |
| OR52H1 | chr11 | 5544772 | 5544772 | G | A | exonic |
| APBB1 | chr11 | 6402643 | 6402643 | T | C | exonic |
| PITPNM1 | chr11 | 67500158 | 67500158 | C | T | exonic |
| TSKU | chr11 | 76795629 | 76795631 | CTG | - | exonic |
| CBL | chr11 | 119206523 | 119206525 | CAC | - | exonic |
| ZNF384 | chr12 | 6667904 | 6667909 | TGCTGC | - | exonic |
| FOXJ2 | chr12 | 8047962 | 8047964 | CAG | - | exonic |
| PZP | chr12 | 9154819 | 9154819 | G | A | exonic |
| KRT8 | chr12 | 52901210 | 52901210 | A | C | exonic |
| DHX37 | chr12 | 124980724 | 124980726 | CTC | - | exonic |
| LRCOL1 | chr12 | 132606198 | 132606200 | CAG | - | exonic |
| DOCK9 | chr13 | 98805114 | 98805114 | T | C | exonic |
| RTN1 | chr14 | 59607408 | 59607408 | T | A | exonic |
| HIRIP3 | chr16 | 29995177 | 29995177 | G | A | exonic |
| DOC2A | chr16 | 30006621 | 30006621 | T | G | exonic |
| E2F4 | chr16 | 67195891 | 67195893 | CAG | - | exonic |
| WWP2 | chr16 | 69908784 | 69908784 | A | G | exonic |
| ZFHX3 | chr16 | 72958393 | 72958393 | C | T | exonic |
| ELAC2 | chr17 | 13017084 | 13017084 | T | C | exonic |
| SUPT6H | chr17 | 28696909 | 28696909 | A | T | exonic |
| EFCAB5 | chr17 | 30080913 | 30080913 | G | T | exonic |
| CCT6B | chr17 | 34928049 | 34928049 | C | A | exonic |
| RAD51D | chr17 | 35116913 | 35116913 | T | C | exonic |
| SLFN11 | chr17 | 35363002 | 35363002 | G | C | exonic |
| KRT9 | chr17 | 41571748 | 41571748 | C | T | exonic |
| HDAC5 | chr17 | 44088559 | 44088559 | C | T | exonic |
| C17orf53 | chr17 | 44161924 | 44161924 | A | G | exonic |
| TSPOAP1 | chr17 | 58310043 | 58310045 | TCC | - | exonic |
| DPP9-AS1 | chr19 | 4682867 | 4682868 | AG | - | exonic |
| OR7E24 | chr19 | 9251065 | 9251065 | T | - | exonic |
| OR7E24 | chr19 | 9251064 | 9251064 | - | T | exonic |
| FAM98C | chr19 | 38408862 | 38408864 | AAG | - | exonic |
| FPR3 | chr19 | 51824240 | 51824240 | G | A | exonic |
| SLC9A8 | chr20 | 49850763 | 49850763 | - | T | exonic |
| LTN1 | chr21 | 28966884 | 28966884 | T | - | exonic |
| RUNX1 | chr21 | 34792308 | 34792308 | A | G | exonic |
| BRWD1 | chr21 | 39278806 | 39278806 | G | C | exonic |
| POTEH | chr22 | 15708022 | 15708022 | C | G | exonic |
| MED15 | chr22 | 20564628 | 20564628 | - | CAG | exonic |
| CHKB | chr22 | 50579775 | 50579775 | T | C | exonic |
| MED12 | chrX | 71140798 | 71140800 | CAG | - | exonic |
Of the 82 rare candidate predisposition variants, only 39 were observed in UK Biobank data, most likely due to the low allele frequency threshold used here. Analysis of these variants in the 3899 UK Biobank cases with rotator cuff injury and the 11,697 genetically matched controls (mean case age 59.9 years) identified one significant association in the APBB1 gene (OR = 2.37, P = .007, uncorrected). Power was calculated for the UK Biobank dataset. Based on 3899 cases, 11,697 matched population controls, and assuming a RCT prevalence of 20.7% in the general population,24 a significance level of 0.0013 (0.05/39 variants available for validation), and a disease allele frequency of 0.005, we had 80% power to detect an odds ratio of 1.74. The odds ratio we observed for the APBB1 gene is similar to the odds ratio predicted with sufficient power, although the significance criterion for the APBB1 gene (alpha = 0.0013) was not achieved. Although variants in the MAPK pathway (MAP3K4, MAP3K1), RUNX1, ADAM12, and TGFBR2 were identified as candidates, UK Biobank data were not available for them.
Due to their plausible biologic role in RCT pathogenesis, four variants were selected to test for significant association with RCTs in the set of independent Utah cases with a RCT: APPB1, ADAM12, PDLIM7, and LTBP1. The APBB1 (chr11:6423873-6423873 T > C, rs1800425) variant was observed in the original cousin pair as well as in 2 additional unrelated, heterozygous carriers who were not related to the cousin pair or to each other, resulting in a prevalence of 2/458 RCT cases (0.43%). The decision to choose only these four variants over others was based upon identifying the best plausible biologic role for RTC pathogenesis as determined by the team along with a limitation in resources to only assess a maximal number of four total variants.
The PDLIM7 (chr5:176919656-176919656 G > A, rs148209669) variant was observed in the original 3 related cousins plus 10 additional heterozygote carriers resulting in a prevalence of 10/458 RCT cases (2.18%). Linked genealogy data were available for 8 of these 10 cases and none of the 8 were related to the cousin pair nor to each other.
The ADAM12 (chr10:126155235-126155235 T > C, rs367875671) variant was observed in the original cousin pair plus two additional heterozygote carriers, resulting in a prevalence of 2/458 RCT cases (0.44%). Genealogy data were available for one of the additional carriers. The one additional carrier was 8 meioses (3rd cousins) distant from the original cousin pair.
The LTBP1 (chr2:33134781-33134781 C > G, rs201777796) variant was observed in the original cousin pair plus two additional heterozygote carriers, resulting in a prevalence of 2/458 RCT cases (0.44%). Linked genealogy data were available for one of the additional carriers. This individual with linked genealogy data was not related to the cousin pair.
The PDLIM7 variant had the highest prevalence of variant carriers in our RCT population therefore it was compared to the prevalence of variant carriers in a European control population. The PDLIM7 rare allele was found to be in significant excess in the Utah RCT cases, assuming 10 additional independent carriers of the PDLIM7 variant out of 458 independent, RCT subjects tested (minor allele frequency of 0.022 after excluding the three related cousin pair participants) compared to a minor allele frequency of 0.00582 for the European (non-Finnish) population rate (749 carriers out of 128,612 tested). The chi-square test result is 19.27 (P = .000011).
Discussion
The results of this study present the data from the identification of high-risk RCR pedigrees and genetic analysis of patients who have undergone RCR for a full-thickness RCT. We have identified 82 rare candidate predisposition variants proposed to be associated with tearing identified in 20 related patients from high-risk pedigrees. We attempted to validate these candidates using a previously performed GWAS that identified candidate variants for rotator cuff injury from the UK Biobank.22 Approximately half of the variants identified in the present study were available in the UK Biobank dataset, and one significant association in the APBB1 gene was identified. Among the rare variants identified, several of the genes have previously been associated with tendon pathology or healing, wound inflammation, or nicotine dependence. Each of these variants are potential future targets for further genetic analyses to confirm their association with rotator cuff pathology as well as basic science studies to identify their biologic role in the development of tendon pathology.
Several of the identified rare variants have a potential biologic role in the development of rotator cuff pathology. APBB1 is an adaptor protein in the nucleus and variants have been identified as being associated with nicotine dependence.5 RCTs and failure of repair healing are strongly linked to the usage of nicotine.3 ADAM12 is a disintegrin and metalloproteinase and is critical for cell-matrix interaction has been identified as being upregulated at the edge of torn rotator cuff tendons in comparison to normal intact rotator cuff tissue.4 Dysregulation of ADAM12 may predispose patients to increased risk for rotator cuff tearing. RUNX1 is a transcription factor involved in upregulating the Hippo-YAP pathway and has a critical role in directing cell proliferation, differentiation, and apoptosis.6 The Hippo pathway has been previously shown in a rat RCR model to impair tendon healing if downregulated.13 Elevated expression of RUNX1 has been shown to contribute to tendon-bone healing after ACL reconstruction using bone mesenchymal stem cells in an animal model.14 Impairment of RUNX1 function may influence tenocyte cell proliferation or apoptosis resulting in tearing or impaired tendon healing. PDLIM7 gene encodes a protein that is involved in the assembly of an actin filament-associated complex but, more importantly, is a target for ubiquitin-proteasome-mediated atrophy of skeletal muscle.8 Dysregulation of PDLIM7 may influence disuse atrophy associated with rotator cuff injury and be a risk factor for tear development or progression.
The TGF-β pathway has been shown to have a critical role in the etiology of tendon injury as well as repair through regulating collagen synthesis, cell differentiation, and cell adhesion.17 TGFBR2 has been shown to be upregulated in the supraspinatus tendon in an animal impingement model.9 LTBP1 is an extracellular matrix protein that binds latent TGF-beta and influences its availability in bone and connective tissue.7 Consequently, dysregulation of the TGF-β pathway is a potential pathogenic mechanism for the development of rotator tearing. Finally, the MAPK genes are enzymes critical to the MAPK pathway which is responsive to environmental stress and hypoxia resulting in upregulation of hypoxia-inducible factor 1 alpha (HIF-1alpha) leading to angiogenesis.16 Alteration of the MAPK pathway has also been shown to regulate fatty degeneration of the rotator cuff musculature in the setting of RCTs.23 Rare variants in genes within the MAPK pathway are likely candidates to result in both tendon tearing through impaired angiogenesis but also muscle degeneration in the setting of rotator cuff injury.
A majority of the genetic variants reported to be associated with RCTs are common variants. 1,10,15,18, 19, 20,22 These variants likely have a small but significant association with the development of RCTs. Only a limited number of studies have investigated rare variants. Similar to our study, Azzara et al performed WES using a family-based design to identify rare variants associated with RCTs.2 In contrast to us, they selected a single nuclear family with two related individuals with bilateral RCTs (ie, father and son) and compared variants shared by the affected relatives to two healthy relatives (ie, mother and 2nd son) without RCTs.2 They initially identified 91,000 rare, shared variants between the affected mother and son, which they reduced to 347 rare, shared variants within coding/splicing regions of the genome. Azzara et al further refined their list of 347 rare variants to 16 variants in 16 genes predicted as damaging and not associated with other known diseases.2 Ultimately, they selected three variants in three genes that were associated with connective tissue or tendons (COL23A1, EMILIN3, and HDAC10).2 We compared our list of 82 genes with rare, shared variants carried by a cousin pair with RCTs to their published list of 16 genes associated with bilateral RCTs; there was no overlap in the gene lists. This is likely due to differences in study design; Azzara et al focused on bilateral tears, which was not a requirement in our study. Our design was based on rare variant sharing in cousin pairs where all identified rare, shared variants were then evaluated for potential association with the risk of RCTs. The advantage of investigating shared rare variants in affected cousin pairs is that there is a substantial reduction in the number of variants to follow-up; first cousins share on average 12.5% of the genomes identical-by-descent, whereas second cousins share on average 6.25% of these genomes identical-by-descent. In contrast, parent–offspring relationships share an estimated 50% of their genomes identical-by-descent. Azzara et al using affected nuclear family members identified a much larger number of shared, rare variants, as would be expected. Our current study has identified new rare, highly penetrant candidate RCT predisposition alleles. Understanding new genetic variants associated with rotator cuff tearing allows further understanding of the biology of rotator cuff tearing and therefore opens avenues for potential biologic solutions to improve rotator cuff tendon healing. Biologic augmentations for repairs could be designed to directly counter the genetic impairment leading to injury. Further validation and functional testing of these identified variants in animal models is required as well as continued investigation of additional shared variants in the larger RCT pedigree resource we have identified.
The primary strength of this pilot study is the identification of rare, shared, candidate, predisposition variants for rotator cuff tearing using high-risk RCT pedigrees. The primary limitation of this pilot study was the small sample size of 20 patients. Although given the findings of the current study, we plan to study more families in the future. Additional limitations include that genealogy data does not always represent biological relationships and because the population represented in the UPDB is almost exclusively of Northern European ancestry, these findings are limited to similar populations and should not be extrapolated. Another limitation is recognizing that many of the variants/genes identified have limited available biology associated with them that is related to tendonopathy. While we have focused on genes with stronger biological plausibility related to RCTS based on current knowledge, the publication of our complete list of shared candidate variants allows continued investigation of these variants/genes as the biology of these genes becomes more fully established. Based upon the strong candidate variants/genes identified here, the current data support obtaining funding to perform analysis of the larger dataset.
Conclusion
Our analysis of closely related individuals with confirmed full-thickness RCTs from high-risk pedigrees has identified 82 rare, shared candidate genetic predisposition coding variants. Among these, rare alleles of APBB1, RUNX1, ADAM12, TGFBR2, PDLIM7, LTBP1 as well as several MAPK pathway gene variants were identified. Association of the PDLIM7 allele with risk for tear was confirmed in an independent cohort of RCTs. Further analysis of the variant alleles is required for confirmation of the role of these genes in RCT.
Acknowledgments
We thank the Pedigree and Population Resource of Huntsman Cancer Institute, University of Utah (funded in part by the Huntsman Cancer Foundation) for its role in the ongoing collection, maintenance and support of the Utah Population Database (UPDB). We thank the University of Utah Center for Clinical and Translational Science (CCTS) (funded by NIH Clinical and Translational Science Awards), the Pedigree and Population Resource, University of Utah Information Technology Services and Biomedical Informatics Core for establishing the Master Subject Index between the Utah Population Database and the University of Utah Health Sciences Center.
Disclaimers:
Funding: This study received support from the Veterans Administration Merit Review Grant (Number 1157449) US Department of Veterans Affairs.
Conflicts of interest: Robert Z. Tashjian is a paid consultant for Zimmer/Biomet, Stryker, Enovis and Mitek; has stock in Conextions and Genesis; receives intellectual property royalties from Stryker, Shoulder Innovations and Zimmer/Biomet; receives publishing royalties from the Journal of Bone and Joint Surgery and Springer and serves on the editorial board for the Journal of Bone and Joint Surgery. The other authors, their immediate families, and any research foundation with which they are affiliated have not received any financial payments or other benefits from any commercial entity related to the subject of this article.
Footnotes
University of Utah IRB approval was obtained (IRB #00029048).
References
- 1.Assunção J.H., Godoy-Santos A.L., Dos Santos M.C.L.G., Malavolta E.A., Gracitelli M.E.C., Ferreira Neto A.A. Matrix metalloproteases 1 and 3 promoter gene polymorphism is associated with rotator cuff tear. Clin Orthop Relat Res. 2017;475:1904–1910. doi: 10.1007/s11999-017-5271-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azzara A., Ambrogioni L., Cassano I., Lintas C., Longo G., Denaro V., et al. Genetic characterization in familial rotator cuff tear. Biology (Basel) 2022;11:1565. doi: 10.3390/biology11111565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishop J.Y., Santiago-Torres J.E., Rimmke N., Flanigan D.C. Smoking predisposes to rotator cuff pathology and shoulder dysfunction: a systematic review. Arthroscopy. 2015;31:1598–1605. doi: 10.1016/j.arthro.2015.01.026. [DOI] [PubMed] [Google Scholar]
- 4.Chaudhury S., Xia Z., Thakkar D., Hakimi O., Carr A.J. Gene expression profiles of changes underlying different-sized human rotator cuff tendon tears. J Shoulder Elbow Surg. 2016;25:1561–1570. doi: 10.1016/j.jse.2016.02.037. [DOI] [PubMed] [Google Scholar]
- 5.Chen G.B., Payne T.J., Lou X.Y., Ma J.Z., Zhu J., Li M.D. Association of amyloid precursor protein-binding protein, family B, member 1 with nicotine dependence in African and European American smokers. Hum Genet. 2008;124:393–398. doi: 10.1007/s00439-008-0558-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chuang L.S.H., Ito Y. The multiple interactions of RUNX with the hippo-YAP pathway. Cells. 2021;10:2925. doi: 10.3390/cells10112925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dallas S.L., Keene D.R., Bruder S.P., Saharinen J., Sakai L.Y., Mundy G.R., et al. Role of the latent transforming growth factor beta binding protein 1 in fibrillin-containing microfibrils in bone cells in vitro and in vivo. J Bone Miner Res. 2000;15:68–81. doi: 10.1359/jbmr.2000.15.1.68. [DOI] [PubMed] [Google Scholar]
- 8.D'Cruz R., Plant P.J., Pablo L.A., Lin S., Chackowicz J., Correa J., et al. PDLIM7 is a novel target of the ubiquitin ligase Nedd4-1 in skeletal muscle. Biochem J. 2016;473:267–276. doi: 10.1042/BJ20150222. [DOI] [PubMed] [Google Scholar]
- 9.Eliasberg C.D., Wada S., Carballo C.B., Nakagawa Y., Nemirov D.A., Bhandari R., et al. Identification of inflammatory mediators in tendinopathy using a murine subacromial impingement model. J Orthop Res. 2019;37:2575–2582. doi: 10.1002/jor.24434. [DOI] [PubMed] [Google Scholar]
- 10.Figueiredo E.A., Loyola L.C., Belangero P.S., Campos Ribeiro-Dos-Santos Â.K., Emanuel Batista Santos S., Cohen C., et al. Rotator cuff tear susceptibility is associated with variants in genes involved in tendon extracellular matrix homeostasis. J Orthop Res. 2020;38:192–201. doi: 10.1002/jor.24455. [DOI] [PubMed] [Google Scholar]
- 11.Fuchs B., Weishaupt D., Zanetti M., Hodler J., Gerber C. Fatty degeneration of the muscles of the rotator cuff: assessment by computed tomography versus magnetic resonance imaging. J Shoulder Elbow Surg. 1999;8:599–605. doi: 10.1016/s1058-2746(99)90097-6. [DOI] [PubMed] [Google Scholar]
- 12.Goutallier D., Postel J.M., Bernageau J., Lavau L., Voisin M.C. Fatty muscle degeneration in cuff ruptures. Pre- and postoperative evaluation by CT scan. Clin Orthop Relat Res. 1994;304:78–83. [PubMed] [Google Scholar]
- 13.Huang Y., Pan M., Shu H., He B., Zhang F., Sun L. Vascular endothelial growth factor enhances tendon-bone healing by activating Yes-associated protein for angiogenesis induction and rotator cuff reconstruction in rats. J Cell Biochem. 2020;121:2343–2353. doi: 10.1002/jcb.29457. [DOI] [PubMed] [Google Scholar]
- 14.Kang K., Geng Q., Cui L., Wu L., Zhang L., Li T., et al. Upregulation of Runt related transcription factor 1 (RUNX1) contributes to tendon-bone healing after anterior cruciate ligament reconstruction using bone mesenchymal stem cells. J Orthop Surg Res. 2022;17:266. doi: 10.1186/s13018-022-03152-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kluger R., Burgstaller J., Vogl C., Brem G., Skultety M., Mueller S. Candidate gene approach identifies six SNPs in tenascin-C (TNC) associated with degenerative rotator cuff tears. J Orthop Res. 2017;35:894–901. doi: 10.1002/jor.23321. [DOI] [PubMed] [Google Scholar]
- 16.Korbecki J., Simińska D., Gąssowska-Dobrowolska M., Listos J., Gutowska I., Chlubek D., et al. Chronic and cycling hypoxia: drivers of cancer chronic inflammation through HIF-1 and NF-κB activation: a review of the molecular mechanisms. Int J Mol Sci. 2021;22:10701. doi: 10.3390/ijms221910701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y., Liu X., Liu X., Peng Y., Zhu B., Guo S., et al. Transforming growth factor-β signalling pathway in tendon healing. Growth Factors. 2022;40:98–107. doi: 10.1080/08977194.2022.2082294. [DOI] [PubMed] [Google Scholar]
- 18.Miao K., Jiang L., Zhou X., Wu L., Huang Y., Xu N., et al. Role of matrix metalloproteases 1/3 gene polymorphisms in patients with rotator cuff tear. Biosci Rep. 2019;39 doi: 10.1042/BSR20191549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Motta Gda R., Amaral M.V., Rezende E., Pitta R., Vieira T.C., Duarte M.E., et al. Evidence of genetic variations associated with rotator cuff disease. J Shoulder Elbow Surg. 2014;23:227–235. doi: 10.1016/j.jse.2013.07.053. [DOI] [PubMed] [Google Scholar]
- 20.Roos T.R., Roos A.K., Avins A.L., Ahmed M.A., Kleimeyer J.P., Fredericson M., et al. Genome-wide association study identifies a locus associated with rotator cuff injury. PLoS One. 2017;12 doi: 10.1371/journal.pone.0189317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tashjian R.Z., Farnham J.M., Albright F.S., Teerlink C.C., Cannon-Albright L.A. Evidence for an inherited predisposition contributing to the risk for rotator cuff disease. J Bone Joint Surg Am. 2009;91:1136–1142. doi: 10.2106/JBJS.H.00831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tashjian R.Z., Kim S.K., Roche M.D., Jones K.B., Teerlink C.C. Genetic variants associated with rotator cuff tearing utilizing multiple population-based genetic resources. J Shoulder Elbow Surg. 2021;30:520–531. doi: 10.1016/j.jse.2020.06.036. [DOI] [PubMed] [Google Scholar]
- 23.Wilde J.M., Gumucio J.P., Grekin J.A., Sarver D.C., Noah A.C., Ruehlmann D.G., et al. Inhibition of p38 mitogen-activated protein kinase signaling reduces fibrosis and lipid accumulation after rotator cuff repair. J Shoulder Elbow Surg. 2016;25:1501–1508. doi: 10.1016/j.jse.2016.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamamoto A., Takagishi K., Osawa T., Yanagawa T., Nakajima D., Shitara H., et al. Prevalence and risk factors of a rotator cuff tear in the general population. J Shoulder Elbow Surg. 2010;19:116–120. doi: 10.1016/j.jse.2009.04.006. [DOI] [PubMed] [Google Scholar]