Abstract
The adenovirus type 5 mutant dl1520 was engineered previously to be completely defective for E1B-55K functions. Recently, this mutant (also known as ONYX-015) has been suggested to replicate preferentially in p53− and some p53+ tumor cell lines but to be attenuated in primary cultured cells (C. Heise, A. Sampson-Johannes, A. Williams, F. McCormick, D. D. F. Hoff, and D. H. Kirn, Nat. Med. 3:639–645, 1997). It has been suggested that dl1520 might be used as a “magic bullet” that could selectively lyse tumor cells without harm to normal tissues. However, we report here that dl1520 replication is independent of p53 genotype and occurs efficiently in some primary cultured human cells, indicating that the mutant virus does not possess a tumor selectivity. Although it was not the sole host range determinant, p53 function did reduce dl1520 replication when analyzed in a cell line expressing temperature-sensitive p53 (H1299-tsp53) (K. L. Fries, W. E. Miller, and N. Raab-Traub, J. Virol. 70:8653–8659, 1996). As found earlier for other E1B-55K mutants in HeLa cells (Y. Ho, R. Galos, and J. Williams, Virology 122:109–124, 1982), dl1520 replication was temperature dependent in H1299 cells. When p53 function was restored at low temperature in H1299-tsp53 cells, it imposed a modest defect in viral DNA replication and accumulation of late viral cytoplasmic mRNA. However, in both H1299 and H1299-tsp53 cells, the defect in late viral protein synthesis appeared to be much greater than could be accounted for by the modest defects in late viral mRNA levels. We therefore propose that in addition to countering p53 function and modulating viral and cellular mRNA nuclear transport, E1B-55K also stimulates late viral mRNA translation.
The adenovirus type 5 (Ad5) E1B-55K protein performs several functions that are important in viral replication. During the early phase of infection, E1B-55K counteracts E1A functions that would otherwise lead to the stabilization of p53 and the induction of apoptosis (17, 20, 54, 67, 68). In the late phase, E1B-55K functions in a complex with the E4-orf6 gene product (72, 74) to stimulate the cytoplasmic accumulation and translation of the viral late mRNAs (2, 3, 12, 35, 43, 50, 65, 84, 89). This is accompanied by the shutoff of host mRNA nuclear export and of host protein synthesis (2, 6).
E1B-55K interferes with p53 function during viral infection through at least two mechanisms. In the early phase of infection, E1B-55K binds the amino terminus of p53, inhibiting p53 transactivation function (45, 56, 86). E1B-55K possesses an intrinsic transcriptional repression domain that inhibits expression from a number of different promoters when targeted by fusion with the Gal4 DNA binding domain (88). This activity inhibits transcription initiation in vitro and is targeted to cellular promoters containing p53-specific binding sequences by direct interaction with DNA-bound p53 (56). A similar activity has now been described for the E4-orf6 protein. Like E1B-55K, E4-orf6 also binds p53 and antagonizes p53-mediated transactivation (23, 62). The repression at p53-responsive promoters mediated by these two adenovirus proteins has been proposed to interfere with E1A-induced apoptosis, enhancing both the transformation of nonpermissive cells and lytic replication in permissive cells (23, 60, 62, 86–88).
The E4-orf6 and E1B-55K proteins also regulate the function of p53 by affecting its half-life. The half-life of p53 is markedly reduced during infection, depending on the presence of both E1B-55K and E4-orf6. In the absence of either of these proteins, a dramatic increase in cellular p53 levels is observed (34, 66, 69, 81). The E1B-55K and E4-orf6 proteins appear to be the only viral proteins required to destabilize p53, as they have been shown to accelerate p53 degradation when transiently expressed (70, 81).
Late in infection, the E1B-55K protein performs an additional function in a complex with the E4-orf6 gene product (72, 74). Studies by Shenk and others have shown that E1B-55K and E4-orf6 together modulate the preferential cytoplasmic accumulation of the viral late mRNAs (3, 12, 35, 50, 64, 65, 84). The characterization of this late-phase E1B-55K function has been performed mainly with HeLa cells. During the infection of HeLa cells, E1B-55K mutants replicate viral DNA to much the same level as does the wild-type virus (2, 50, 64, 84). Far fewer viral late messages accumulate in the cytoplasm, however, and viral late protein synthesis is greatly reduced (3, 50, 64, 65, 84). The replication of E1B-55K− mutants is thus severely impaired in HeLa cells (5, 37, 41, 50, 64).
The exact mechanism through which E1B-55K and E4-orf6 modulate the preferential nuclear export of the viral late mRNAs is not well understood. It has been shown, however, that the E1B-55K/E4-orf6 complex can physically shuttle between the nucleus and cytoplasm (22). A model that has emerged, then, is one in which E1B-55K and E4-orf6 directly or indirectly bind mRNAs transcribed from adenovirus DNA during the late phase of infection and escort them through nuclear pores (22, 63). The mechanistic relationship, if any, between this late-phase E1B-55K/E4-orf6 function and the inhibition of transcriptional activation by p53 is not understood.
The E1B-55K− mutant dl1520 was constructed so as to express little if any E1B-55K function. dl1520 contains a stop codon following the second codon of the E1B-55K-coding region (a mutation that does not alter the sequence of the E1B-19K protein encoded in an overlapping reading frame) plus a deletion of the E1B-55K-coding region beyond the 3′ end of the E1B-19K-coding region (5). Given the requirement for E1B-55K in the inactivation of p53 during infection, Bischoff et al. hypothesized that an E1B-55K-defective virus might preferentially replicate in p53-negative human cells, in which this function of E1B-55K would not be required (10). Indeed, in support of this hypothesis, dl1520 was found to replicate similarly to wild-type Ad5 in p53− C33A cells but to be attenuated in p53+ U2OS or cultured primary human cells (10, 39). Furthermore, in a tumor xenograft model in nude mice, dl1520 was found to be effective in the treatment of p53− C33A human tumors but to be ineffective against U87 p53+ tumors (10, 39). Based on these studies, phase I and II clinical trials with dl1520 (renamed ONYX-015) in the treatment of human p53− head and neck carcinomas were initiated (10).
It was surprising to us that dl1520 replicated efficiently in p53− cell lines. While the E1B-55K function that neutralizes p53 would not be required in such cells, it was not clear why the requirement for E1B-55K in the nucleocytoplasmic trafficking and translation of the late mRNAs observed in HeLa cells would not also apply in p53− tumor cell lines. Indeed, contrary to the initial findings of Bischoff et al., recent papers have reported that the ability of dl1520 and other E1B-55K− mutants to replicate in various human tumor cells is independent of the p53 status of the host cell (31, 32, 36, 71, 82). Furthermore, in diametric opposition to the hypothesis that an E1B-55K mutant might replicate preferentially in p53− cells, one paper (36) proposed that p53 function is in fact required in adenovirus-infected cells for the generation of the cytopathic effect and the subsequent release of progeny virions. Goodrum and Ornelles alternatively propose that the replication and cytolytic properties of dl1520 are determined by the phase of the cell cycle during which infection occurs (31, 32). In their studies, the replication efficiency of E1B-55K− mutants did not correlate with the p53 status of the host but was instead determined by the proportion of cells in S phase at the time of infection (31, 32). The correlation between p53 expression and the replication of an E1B-55K− virus has thus remained controversial, and the host range determinant of E1B-55K− mutants is still unknown (48, 52).
To better understand the effects which p53 function may have on the replication of an E1B-55K− mutant, we examined dl1520 replication in a number of primary cultured human cells and immortalized cell lines. To evaluate the specific effects of p53 on dl1520 replication, the p53− H1299 cell line (59) and a stable transformant of H1299 expressing a temperature-sensitive p53 allele (A135V) (29) were studied. Our analyses of yield of infectious virions, viral DNA replication, late message accumulation, and late viral protein synthesis in these cells confirmed requirements for E1B-55K in the inactivation of p53 and the nuclear export of viral mRNAs during the late phase of infection. Additionally, however, these studies revealed that E1B-55K may further function in significantly stimulating the translation of viral late mRNAs.
MATERIALS AND METHODS
Cells and viruses.
293 (33), HeLa (75, 78), C33A (18, 75), U2OS (15, 21), Saos-2 (15, 21), RKO (4), H1299 (59), A549 (49), HepG2 (11, 26), Hep3B (11, 26), SK-OV-3 (85), WI38, IMR90, and human neonatal kidney (HNK) monolayer cultures were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS). The growth media for the RKO RC10.1, RKO RC10.3, RKOp53.13, and H1299-tsp53 cell lines were further supplemented with G418 at concentrations of 200 μg/ml for the RKO-derived transformants (79, 80) and 400 μg/ml for H1299-tsp53 (29). PC-3 (13) and OVCAR-3 (85) cells were grown in RPMI with 10 and 20% FCS, respectively. The 293, HeLa, C33A, U2OS, A549, WI38, IMR90, HepG2, SK-OV-3, and OVCAR-3 cell lines were obtained commercially from the American Type Culture Collection, and the HNK cells were obtained from Biowhittaker. The RKO, RKO RC10.1, RKO RC10.3, and RKOp53.13 cells were generously provided by Michael Kastan (Johns Hopkins University) and have been described previously (79, 80). The Hep3B cells were obtained from J. S. Economou (UCLA), the PC-3 cells were obtained from Phil Koeffler (UCLA), and the Saos-2 cell line was obtained from Arnold Levine (Princeton University). The H1299 and H1299-tsp53 cells were kind gifts of Nancy Raab-Traub (University of North Carolina, Chapel Hill) and have been described previously (29, 59).
The wild-type Ad5 and the E1B-55K-defective adenovirus mutant dl1520 were described by Barker and Berk (5). The dl1520 virus contains a deletion within the E1B-55K open reading frame from nucleotide 2496 to 3323 and a point mutation at nucleotide 2022 which terminates translation of the protein (5). All viruses were propagated in 293 cell suspension cultures.
Ad5 and dl1520 replication in primary cells and tumor cell lines.
Monolayers (60-mm-diameter dishes) of the indicated cell lines at 70 to 80% confluency were infected with the wild-type Ad5 and dl1520 virus at a multiplicity of infection (MOI) of 5. Infections were performed in 1 ml of DMEM supplemented with 2% FCS over 1 h at 37°C with intermittent rocking. At the end of the adsorption period, medium was added back for a final volume of 5 ml. At the indicated times, infected cultures and supernatants were harvested and freeze-thawed three times to release progeny virions. Viral yields were then determined by plaque assay on complementing 293 monolayer cultures (33).
p21 and hdm2 Western analysis.
p53 function in the H1299-tsp53 cells was verified by examining p21 and hdm2 levels by Western analysis as previously described (29). In brief, C33A, H1299, and H1299-tsp53 cells maintained at 39°C were shifted to 32°C. At various times after the temperature shift (12, 24, and 48 h), equivalent numbers of cells were lysed in sodium dodecyl sulfate (SDS) sample buffer (73). Whole-cell lysates from 2.5 × 105 cells were subsequently resolved by SDS–12% polyacrylamide gel electrophoresis (SDS–12% PAGE) and transferred to nitrocellulose. The p21 and hdm2 proteins were detected by using the WAF1 (Ab-1) and MDM2 (Ab-1) antibodies from Calbiochem according to the manufacturer’s specifications.
Viral DNA analysis.
Viral DNA was isolated from infected H1299 and H1299-tsp53 cells by a modified Hirt method (40). In brief, the H1299 and H1299-tsp53 cells were infected with Ad5 and dl1520 at an MOI of 5 as described above. At the indicated times postinfeciton, 1.4 × 106 cells were harvested and washed once with phosphate-buffered saline (PBS). The cells were then resuspended in 0.5 ml of 10 mM Tris-HCl (pH 7.0)–10 mM EDTA, lysed by the addition of an equal volume of 10 mM Tris-HCl (pH 7.0)–10 mM EDTA–1.2% SDS–2 mg of pronase per ml, and incubated at 37°C for 2 h. High-molecular-weight DNA was then precipitated overnight at 4°C by the addition of 0.25 ml of 5 M NaCl and cleared from the lysates by microcentrifugation. Low-molecular-weight DNA was then extracted from the supernatants by precipitation with isopropanol. The resultant pellet was resuspended in 10 mM Tris-HCl (pH 8.0)–1 mM EDTA and phenol-chloroform extracted twice. Viral DNA was then isolated by ethanol precipitation in the presence of 0.3 M sodium acetate. DNA from 3.5 × 105 cells was digested with the restriction endonuclease XhoI and resolved on a 0.8% agarose gel containing 0.25 μg of ethidium bromide per ml.
Preparation of cytoplasmic and nuclear RNAs.
H1299 and H1299-tsp53 cells (2 × 107 to 3 × 107) were infected with either wild-type or mutant virus at an MOI of 30. Cytoplasmic RNA was harvested by the method described by Berk and Sharp (8). Nuclear RNA was isolated by a method modified from that of Sambrook et al. (73). Briefly, Ad5- and dl1520-infected cells were lysed in a 0.65% Nonidet P-40–50 mM NaCl–10 mM Tris-HCl (pH 7.8)–1.5 mM MgCl2 disruption buffer. The nuclei were isolated by centrifugation and subsequently washed once with isotonic disruption buffer before being lysed in a 100 mM Tris-HCl (pH 8.0)–150 mM NaCl–10 mM EDTA–1% SDS buffer. Upon shearing of the cellular DNA, the lysates were treated with proteinase K (200 μg/ml) for 30 min at 37°C and phenol-chloroform extracted twice. Nucleic acids were then precipitated with ethanol in the presence of 250 mM NaCl and resuspended in a 50 mM Tris-HCl (pH 7.8)–1 mM EDTA buffer. Samples were then treated with RNase-free DNase I (100 μg/ml) (Gibco) in the presence of 10 mM MgCl2, 1 mM dithiothreitol, and 1,000 U of recombinant pancreatic RNase inhibitor (RNasin; Promega) per ml. EDTA and SDS were then added to the samples for final concentrations of 10 mM and 0.2%, respectively. Upon phenol-chloroform extraction, the RNAs were recovered through precipitation with ethanol and resuspended in 10 mM Tris-HCl (pH 7.0)–1 mM EDTA–0.1% SDS.
S1 analysis of RNA.
S1 analyses were performed with single-stranded DNA probes by the method described by Berk (7). The fiber oligonucleotide probe (Lifetech) spans the splice acceptor of the gene encoding the fiber protein (IV) and corresponds to Ad5 nucleotides 31115 to 31017. The probe was 5′ end labelled with [γ-32P]ATP by using T4 polynucleotide kinase. Each hybridization reaction mixture was comprised of 1 pmol of the probe and 50 μg of nuclear or cytoplasmic RNA derived from uninfected and Ad5- or dl1520-infected H1299 and H1299-tsp53 cells. RNA-DNA hybrids were allowed to form for 3 h at 63°C and were subsequently digested with 200 U of S1 nuclease (Boehringer-Mannheim) for 30 min at room temperature. S1-protected products were recovered by ethanol precipitation, and one-fifth of the reaction mixture was resolved by urea–6% PAGE. The gel was then fixed in 5% trichloroacetic acid (TCA) and dried before analysis with a Molecular Dynamics PhosphorImager.
Analysis of late viral proteins.
Viral late protein expression in Ad5- and dl1520-infected H1299 and H1299-tsp53 cells was evaluated by a method similar to that described by Yew et al. (87). In brief, 70 to 80% confluent H1299 and H1299-tsp53 monolayer cultures (60-mm-diameter dishes) were infected with wild-type Ad5 or dl1520 at an MOI of 30. At the indicated times postinfection, the infected cells were washed once each with PBS and methionine- and cysteine-deficient DMEM supplemented with 2% dialyzed newborn calf serum. The cells were then preincubated for 15 min in fresh methionine-deficient medium plus 2% dialyzed newborn calf serum to deplete intracellular pools of methionine and subsequently were metabolically labelled with 75 μCi of 35S-Tranlabel (ICN) for 1 h at the indicated temperatures. The labelled cells were then washed twice in PBS and lysed in a 0.5% Na-deoxycholate–0.5% Nonidet P-40–50 mM NaCl–25 mM Tris-HCl (pH 8.0)–1 mM phenylmethylsulfonyl fluoride disruption buffer for 30 min on ice. The lysates were then cleared of cellular debris through microcentrifugation and precipitated with 5% TCA to determine the acid-precipitable counts per microliter present in each lysate. An equal number of TCA-precipitable counts was then resolved on an SDS–10% polyacrylamide gel. The gel was then fixed in acetic acid–methanol–double-distilled water (10:30:60) and dried. The hexon and fiber proteins were quantitated with a Molecular Dynamics PhosphorImager.
Microscopy and image analysis.
WI38, HNK, SK-OV-3, and H1299 cells (60-mm-diameter dishes) at 70 to 80% confluency were infected with Ad5 at an MOI of 5 as described above. At 48 and 72 h postinfection, infected samples were observed with a Zeiss AXIOSKOP microscope with a 100× objective. Images were acquired with a Sony DKC-5000 camera by using Adobe Photoshop software.
RESULTS
The replication efficiency of the dl1520 virus does not correlate with cellular p53 status.
To further understand the effects which p53 function may have on the replication of an E1B-55K− Ad5 mutant, dl1520 replication was analyzed in a panel of cell lines of various p53 status by assaying for the production of PFU following infection. The Ad5 and dl1520 viruses were used to infect the cell lines indicated in Table 1 at an MOI of 5. The cell lines evaluated included a group with wild-type p53 sequences as well as a group in which p53 was either mutated, deleted, or inactivated by the presence of other viral proteins. In addition, Ad5 and dl1520 replication was assayed in a number of primary cultured human cell strains, including the WI38 and IMR90 primary human lung fibroblasts and HNK cells. Viral yields were determined at 72 h postinfection by plaque assay on complementing 293 cells (33). The relative replication efficiency of dl1520 within each cell line was then determined by calculating a ratio of viral yields upon infection with the dl1520 and Ad5 viruses (dl1520/Ad5 ratio).
TABLE 1.
Ad5 and dl1520 replication in primary and tumor cell linesa
| Cell line | p53 status (reference) | Virus yield (PFU/cell)
|
Reference(s) | ||
|---|---|---|---|---|---|
| Ad5 | dl1520 | dl1520/Ad5 | |||
| 293 | (33) | 7,020 | 11,000 | 1.9 ± 0.67 | 5; this study |
| HeLa | HPV transformed (76, 78) | 15,400 | 347 | 0.021 ± 0.012 | 5, 32, 71; this study |
| U2OS | Wild type (15, 21) | 2,030 | 21.3 | 0.012 ± 0.0039 | 10, 32, 71; this study |
| RKO | Wild type (4) | 886 | 137 | 0.30 ± 0.30 | 71; this study |
| HepG2 | Wild type (11, 26) | 4,280 | 1,790 | 0.40 ± 0.067 | This study |
| A549 | Wild type (49) | 11,000 | 3,340 | 0.24 ± 0.12 | 32, 71; this study |
| IMR90 | Wild type (15) | 2,110 | 550 | 0.36 ± 0.27 | This study |
| WI38 | Wild type (26) | 2,560 | 86.0 | 0.036 ± 0.0047 | This study |
| HNK | Wild type | 27,600 | 9,610 | 0.36 ± 0.11 | This study |
| C33A | R273C (76) | 4,830 | 2,420 | 0.54 ± 0.087 | 10, 39, 71; this study |
| OVCAR-3 | R248Q (85) | 7,720 | 734 | 0.10 ± 0.057 | This study |
| RKO p53.13 | A143V (80) | 1,180 | 260 | 0.24 ± 0.16 | This study |
| SK-OV-3 | Partial deletion (85) | 5,590 | 278 | 0.063 ± 0.021 | This study |
| Hep3B | Null (11, 26) | 7,000 | 1,040 | 0.14 ± 0.067 | This study |
| PC-3 | Insertion → frameshiftb (13) | 2,960 | 73.1 | 0.048 ± 0.018 | This study |
| H1299 | Null (59) | 25,500 | 10,600 | 0.25 ± 0.22 | 32, 71; this study |
| Saos-2 | Null (15, 21) | 226 | 9.00 | 0.050 ± 0.020 | 32; this study |
| RKO RC 10.1 | HPV E6 transformed (79, 80) | 843 | 39.7 | 0.062 ± 0.023 | This study |
| RKO RC 10.3 | HPV E6 transformed (79, 80) | 544 | 60.1 | 0.16 ± 0.063 | This study |
Seventy percent confluent monolayers (60-mm-diameter dishes) of cells were infected with Ad5 (wild type) or dl1520 virus (E1B-55K mutant) at an MOI of 5. Viral particles were harvested 72 h postinfection, and the number of PFU was determined by plaque assay on 293 cells (33). Values represent the averages from at least three independent experiments (± standard deviations).
An insertion at codon 138 results in a shift of reading frame and a new in-frame stop codon at codon 169.
The replication of the dl1520 virus varied widely from cell line to cell line. Viral replication occurred to near-wild-type levels in a subset of cell lines, including the p53-negative C33A (10, 18, 75) and Hep3B (11, 26) cells and the p53-positive A549 (32, 49) and HepG2 (11, 26) cells. In other cell lines, however, the replication of the mutant virus was severely restricted. In the U2OS (p53 positive [10, 15, 21]) and PC-3 (p53 negative [13]) cell lines, for example, dl1520 replication was reduced nearly 2 orders of magnitude compared to that of wild-type Ad5. The requirement for E1B-55K in adenovirus replication therefore appeared to vary from cell line to cell line and further did not correlate with the p53 genotype of the host cell.
The replication of dl1520 was further analyzed in WI38, IMR90, and HNK primary cell cultures. Whereas the replication of the mutant virus within the WI38 primary human fibroblasts was severely reduced compared to that of the wild-type Ad5 (dl1520/Ad5 ratio = 0.036), dl1520 replication within HNK cells was only modestly affected (dl1520/Ad5 ratio = 0.36). The observations made for tumor cell lines could therefore be extended to nonimmortalized cell strains: the ability of a cell to support the replication of the dl1520 virus appeared to be independent of its p53 status.
p53 function restricts dl1520 replication.
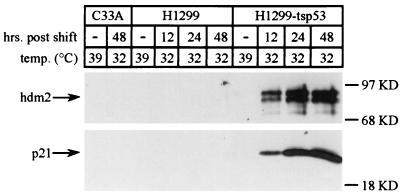
To examine the effects which p53 function may have on dl1520 replication apart from other genetic background differences between various immortalized tumor cell lines, a cell line expressing a temperature-sensitive allele of p53 was studied. The H1299-tsp53 cell line has been previously described (29) and was derived from the p53-negative H1299 cell line (59) by stable transformation with an expression cassette for the temperature-sensitive p53 point mutant A135V. The activity of this p53 mutant can be regulated by shifting the growth temperature of the cells from 39°C, where the A135V mutant does not function, to 32°C, where it does (57, 58). Infections at 32°C were performed 24 h after temperature shift from the growth temperature of 39°C. p53 function under these conditions was confirmed by examining the induction of two known transcriptional targets of p53, p21 and hdm2 (reviewed in reference 51), as previously described (29). Western blotting of cellular extracts (Fig. 1) showed that p21 and hdm2 were undetectable in the p53-negative C33A and H1299 cell lines under all conditions examined. However, in H1299-tsp53 cells, p21 and hdm2 expression was observed only at the reduced temperature where p53 A135V function is restored.
FIG. 1.
p53 function is induced in H1299-tsp53 cells at 32°C. The p53 statuses of H1299, H1299-tsp53, and C33A cells were confirmed by examining p21 and hdm2 expression at temperatures previously determined to be nonpermissive (39°C) and permissive (32°C) for p53 A135V function (29, 57, 58). H1299, H1299-tsp53, and C33A cells were shifted from their normal growth temperature (39°C) to 32°C. At the indicated times after the temperature shift, equal numbers of cells were lysed in SDS sample buffer (73) and resolved by SDS–12% PAGE. Upon transfer to a nitrocellulose support, Western analyses of p21 and hdm2 were performed by using the WAF1 (Ab-1) and MDM2 (Ab-1) antibodies (Calbiochem). The positions of p21 and hdm2 are denoted by arrows, and molecular mass markers in kilodaltons (KD) are indicated at the right of each panel.
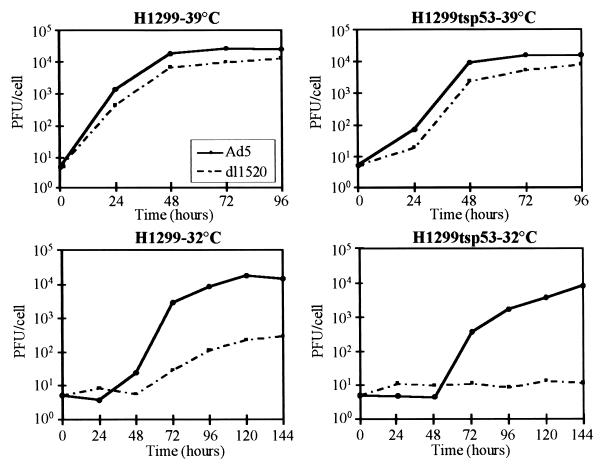
Viral yields from Ad5- and dl1520-infected H1299 and H1299-tsp53 cells were determined at 24-h intervals after infection by assaying for plaque formation on the complementing 293 cells (33) (Fig. 2). At 39°C, replication of wild-type Ad5 occurred efficiently in both H1299 and H1299-tsp53 cells, producing 2.5 × 104 and 1.5 × 104 PFU per cell, respectively, at 96 h postinfection. dl1520 replication was only marginally defective in both cell lines at 39°C, resulting in one-half of the viral yield of wild-type Ad5.
FIG. 2.
dl1520 replication is affected by both temperature and p53 function. Ad5 and dl1520 replication was monitored as a function of time postinfection in the H1299 and H1299-tsp53 cells at temperatures both nonpermissive (39°C) and permissive (32°C) for p53 A135V function. Infections at both temperatures were performed at an MOI of 5. Infections at 32°C were performed 24 h after the temperature shift from 39°C. Infected cells and supernatants were harvested at 24-h intervals after infection, and viral yields determined by assaying for plaque formation on 293 cells (33). Values are presented as the number of PFU derived per cell on a logarithmic scale and represent the averages from three independent experiments performed in duplicate.
dl1520 was much more defective for replication in H1299 cells at 32°C. The maximal dl1520 viral yield was reduced 50-fold compared to that of the wild-type Ad5 under these conditions. This cold-sensitive growth phenotype has been observed previously for the E1B-55K mutants hr6, hrcs13, and dl338 (41, 50). Furthermore, in H1299-tsp53 cells at 32°C, where p53 function was restored, virtually no dl1520 replication occurred, although Ad5 replicated to ∼104 PFU/cell. The low-temperature-dependent (Fig. 2, lower left panel) and additional p53-imposed (Fig. 2, lower right panel) defects in dl1520 replication suggest that E1B-55K has both p53-independent and p53-dependent functions in adenovirus replication at 32°C.
p53 function inhibits dl1520 viral DNA replication.
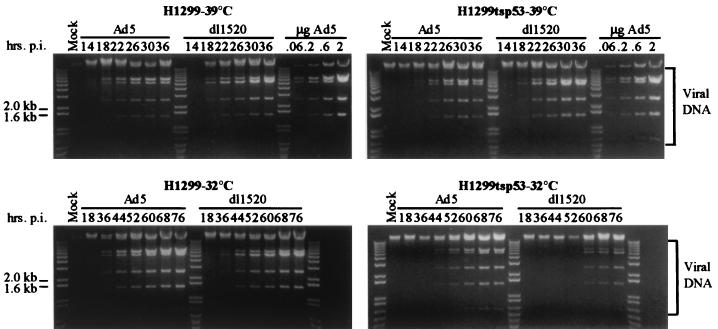
To understand the molecular basis underlying the p53-dependent replication defect of dl1520, viral DNA replication, late mRNA synthesis, and viral late protein synthesis were examined. Viral DNA replication was monitored as a function of time postinfection at both 39 and 32°C. Infections at 32°C were performed 24 h after the shift to the lower temperature. H1299 and H1299-tsp53 cells were infected with Ad5 and dl1520 at an MOI of 5, and viral DNA was isolated at various times postinfection. Isolated DNA was digested with XhoI and analyzed by agarose gel electrophoresis and ethidium bromide staining (Fig. 3).
FIG. 3.
Viral DNA replication is restricted by p53 in dl1520-infected cells. H1299 and H1299-tsp53 cells were mock infected or infected with wild-type Ad5 or dl1520 at an MOI of 5 as described in Materials and Methods. Infections were performed at both 39 and 32°C. Infections at 32°C were carried out 24 h after the shift from 39°C. Viral DNA was harvested from infected cells by a modified Hirt method (40) at the indicated times postinfection (p.i.). DNA isolated from an equal number of cells was digested with the restriction endonuclease XhoI and subsequently resolved by agarose gel electrophoresis and visualized by ethidium bromide staining. Known quantities of purified Ad5 DNA were included as standards for quantitation. The bands corresponding to the XhoI-digested viral DNA fragments are indicated by the brackets at right. The 1.0-kb plus ladder (Gibco BRL) is also included so that the sizes of the individual fragments may be delineated.
At 39°C, Ad5 and dl1520 replicated their DNAs efficiently in both H1299 and H1299-tsp53 cells (Fig. 3). Although production of dl1520 PFU was modestly reduced in both cell lines compared to that of wild-type Ad5 (Fig. 2), viral DNA synthesis for both dl1520 and Ad5 commenced at the same time and exhibited similar kinetics of accumulation. Additionally, when compared to that in Ad5-infected cells, the accumulation of viral DNA in dl1520-infected cells was not reduced.
At the lower temperature, viral DNA replication occurred over a prolonged period. However, viral DNA in Ad5-infected H1299 cells accumulated to levels comparable to that observed at 39°C. Furthermore, despite the 50-fold defect observed in the production of dl1520 PFU (Fig. 2), viral DNA in the dl1520-infected cells was not reduced compared to that found in Ad5-infected cells.
The presence of p53 function at the lower temperature did not affect viral DNA synthesis in Ad5-infected H1299-tsp53 cells (Fig. 3). Indeed, viral DNA accumulated to similar levels at 32 and 39°C. In contrast, there was an ∼8-h delay before viral DNA accumulated to detectable levels in dl1520-infected H1299-tsp53 cells compared to wild-type Ad5-infected cells, and viral DNA accumulation was reduced to approximately one-third of wild-type Ad5 levels. These results indicate that p53 function in the absence of E1B-55K restricts viral DNA replication. The additional p53-imposed defect in dl1520 viral yield observed in H1299-tsp53 cells at 32°C (Fig. 2) can therefore be accounted for in part by reduced dl1520 DNA replication.
Effects of p53 on dl1520 RNA metabolism.
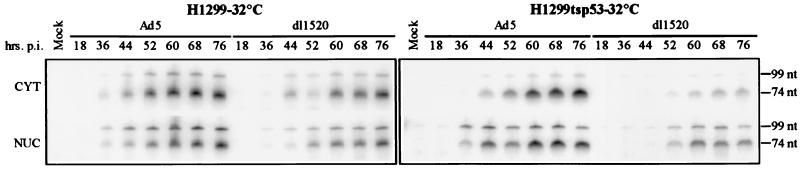
To further understand the biochemical parameters underlying the p53-imposed dl1520 replication defect, late message accumulation in Ad5- and dl1520-infected H1299 and H1299-tsp53 cells was monitored as a function of time postinfection at 32°C. Infections at 32°C were performed 24 h after the shift to the lower temperature and at an MOI of 30 to increase the synchrony of infection. S1 analyses were performed with nuclear and cytoplasmic RNAs isolated from the infected cells at various times postinfection. The probe utilized overlapped the splice acceptor of the gene encoding the fiber protein (IV) and permitted the detection of the mature fiber mRNA (74-nucleotide protected product) as well as incompletely processed species (99-nucleotide protected product). The fiber RNA was chosen for this analysis because in dl338-infected HeLa cells, the L5/fiber RNA was the most severely affected of all the major late species (50, 65). S1-protected products were resolved by PAGE (Fig. 4) and quantitated by PhosphorImager analysis (Table 2).
FIG. 4.
Effects of p53 on late RNA metabolism. H1299 and H1299-tsp53 cells were infected with Ad5 and dl1520 at an MOI of 30. Infections were performed at 32°C and at 24 h after the shift to the lower temperature. Nuclear (NUC) and cytoplasmic (CYT) RNAs were harvested at the indicated times postinfection (p.i.), and the accumulation of the fiber RNA was monitored by S1 analysis. Late fiber species were detected by utilizing a 32P-labelled oligonucleotide probe (Lifetech) overlapping the splice acceptor of the fiber gene. S1-protected products were resolved by urea-PAGE and subsequently visualized by PhosphorImager analysis. The 74- and 99-nucleotide (nt) protected species represent the mature and incompletely spliced forms of the fiber mRNA, respectively. Quantitation of the effects of p53 on nuclear and cytoplasmic RNA accumulation is shown in Table 2.
TABLE 2.
dl1520 macromolecular synthesis
| Cell line | Temp (°C) | p53 functiona | Viral DNAb | Fiber mRNAc
|
Late proteind
|
Virus yielde | ||
|---|---|---|---|---|---|---|---|---|
| NUC | CYT | Hexon | Fiber | |||||
| H1299 | 39 | − | +++ | NDf | ND | 36 ± 17 | 38 ± 13 | 46 ± 28 |
| H1299-tsp53 | 39 | − | +++ | ND | ND | 33 ± 7.9 | 40 ± 7.7 | 54 ± 12 |
| H1299 | 32 | − | +++ | 83 ± 20 | 52 ± 12 | 5.8 ± 1.7 | 7.9 ± 1.3 | 2.0 ± 1.0 |
| H1299-tsp53 | 32 | + | + | 29 ± 9.8 | 22 ± 10 | 0 | 0 | 0.20 ± 0.15 |
Determined by Western analysis of p21 and hdm2 expression levels.
Qualitative assessment of Hirt DNA isolated from dl1520-infected cells at 36 (39°C) and 68 (32°C) h postinfection. +++, viral DNA levels comparable to the wild-type Ad5 level; +, modest reduction.
Nuclear (NUC) and cytoplasmic (CYT) fiber mRNA levels in dl1520-infected cells as determined from S1 analyses at 68 h postinfection. Values represent the averages from three experiments (± standard deviations) and are presented as percentages of Ad5 values.
Late viral protein levels in dl1520-infected cells as measured by PhosphorImager analysis of in vivo [35S]methionine-cysteine-labelled cells. Values represent the averages from three independent experiments (± standard deviations). Measurements were taken at 36 (39°C) and 68 (32°C) h postinfection and are presented as percentages of the Ad5 values.
Percentage of PFU/cell released by dl1520-compared to Ad5-infected cells. Viral yields were determined by plaque assay on complementing 293 cells (33). Values represent the averages from three independent experiments (± standard deviations) and were determined at 96 (39°C) and 144 (32°C) h postinfection.
ND, not determined.
The accumulation of fiber message in the nuclei of dl1520-infected H1299 cells was not significantly reduced compared to that in Ad5-infected H1299 cells (Fig. 4). The processed fiber message accumulated to ∼80% of the wild-type level in the nucleus at 68 h postinfection (Table 2). In the cytoplasm of these cells, however, a moderate defect in the accumulation of mature fiber transcript was observed. Fiber mRNA was reduced to ∼50% of the wild-type level. In H1299 cells, therefore, there was a modest requirement for E1B-55K in modulating the nucleocytoplasmic transport and/or cytoplasmic stability of the fiber mRNA. A low level of incompletely processed primary transcript was also detected in the cytoplasm.
In the presence of p53 function, additional defects in the nuclear and cytoplasmic accumulation of fiber message in dl1520-infected cells were observed (Fig. 4). The nuclear accumulation of mature fiber message at 68 h postinfection was reduced to ∼30% of the wild-type level, and the cytoplasmic level was reduced to ∼20% (Table 2). Incompletely processed forms of the major late transcript were reduced to a similar extent in the nucleus, accumulating to ∼30% of the Ad5 level. The late cytoplasmic message defect under these conditions therefore appeared to be the product of two smaller defects, one at the level of viral RNA accumulation in the nucleus and one affecting the nuclear transport and/or stability of the fiber mRNA species in the cytoplasm.
Expression of viral late proteins.
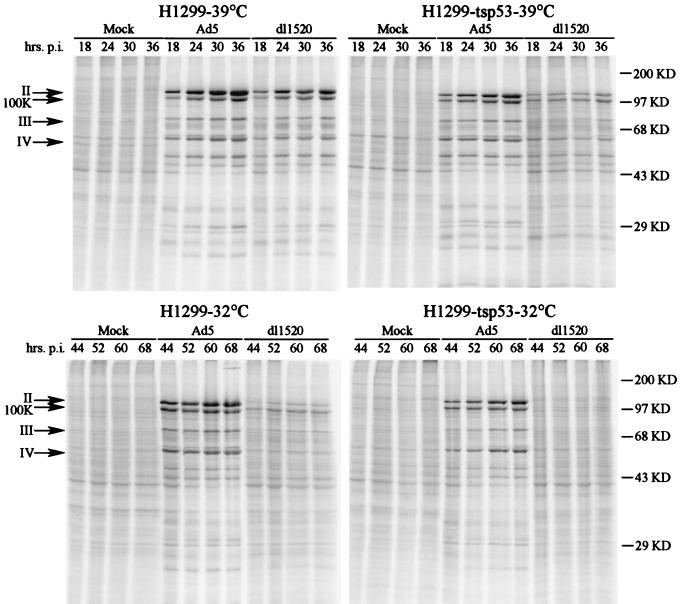
The impact of p53 on the ability of the dl1520 virus to direct the synthesis of viral late proteins was next examined. The H1299 and H1299-tsp53 cells were infected with Ad5 and dl1520 at an MOI of 30 at both 39 and 32°C and metabolically pulse-labelled with [35S]methionine and [35S]cysteine at various times postinfection (Fig. 5). The magnitude of dl1520 defects in viral late protein synthesis was subsequently determined by PhosphorImager analysis. Ad5 infection induced the synthesis of distinct viral late proteins, including the major structural proteins hexon (II), penton (III), and fiber (IV) and the 100K protein, which functions in virion assembly (Fig. 5). In Ad5-infected H1299 and H1299-tsp53 cells, there was a concomitant general shutoff of host cell translation, as evidenced by the decrease in labelling of most cellular polypeptides observed in mock-infected cells.
FIG. 5.
Protein expression during the late phase of infection in H1299 and H1299-tsp53 cells at 39 and 32°C. H1299 and H1299-tsp53 cells were infected with wild-type Ad5 or the E1B-55K mutant (dl1520) at 39 and 32°C. An MOI of 30 was used, and infections at 32°C were carried out at 24 h after the temperature shift. Late viral protein expression was monitored at various times postinfection (p.i.) by in vivo labeling with [35S]methionine-cysteine for a 1-h period. Lysates were TCA precipitated, and an equal number of precipitable counts was resolved by SDS–10% PAGE. The positions of the molecular mass markers are indicated in kilodaltons (KD) at the far right, and the bands corresponding to the major late proteins hexon (II), L4 100K protein, penton (III), and fiber (IV) are denoted by arrows on the left. The effects of temperature and p53 function on hexon and fiber synthesis were determined by PhosphorImager analysis and are shown in Table 2.
At 39°C, quantitative analysis of the hexon and fiber species revealed a moderate reduction in the level of late proteins synthesized in dl1520-infected cells. In both H1299 and H1299-tsp53 cells, hexon and fiber levels at 36 h postinfection were reduced to 30 to 40% of wild-type levels (Table 2). At the lower temperature, however, the magnitude of the viral late protein synthetic defect was much greater. The expression of the hexon and fiber proteins in H1299 cells at 68 h postinfection was reduced to 5.8 and 7.9% of the wild-type level, respectively. Furthermore, in the H1299-tsp53 cells at 32°C, where p53 function had been restored, an additional defect in late viral protein production was observed, resulting in undetectable levels of the hexon and fiber proteins. These severe reductions in the expression of late viral proteins were accompanied by near-complete defects in the shutoff of host translation. In general, the defects in late gene expression were consistent with those seen in the yield of progeny PFU.
Ad5 replication and induction of the cytopathic effect do not correlate with cellular p53 status.
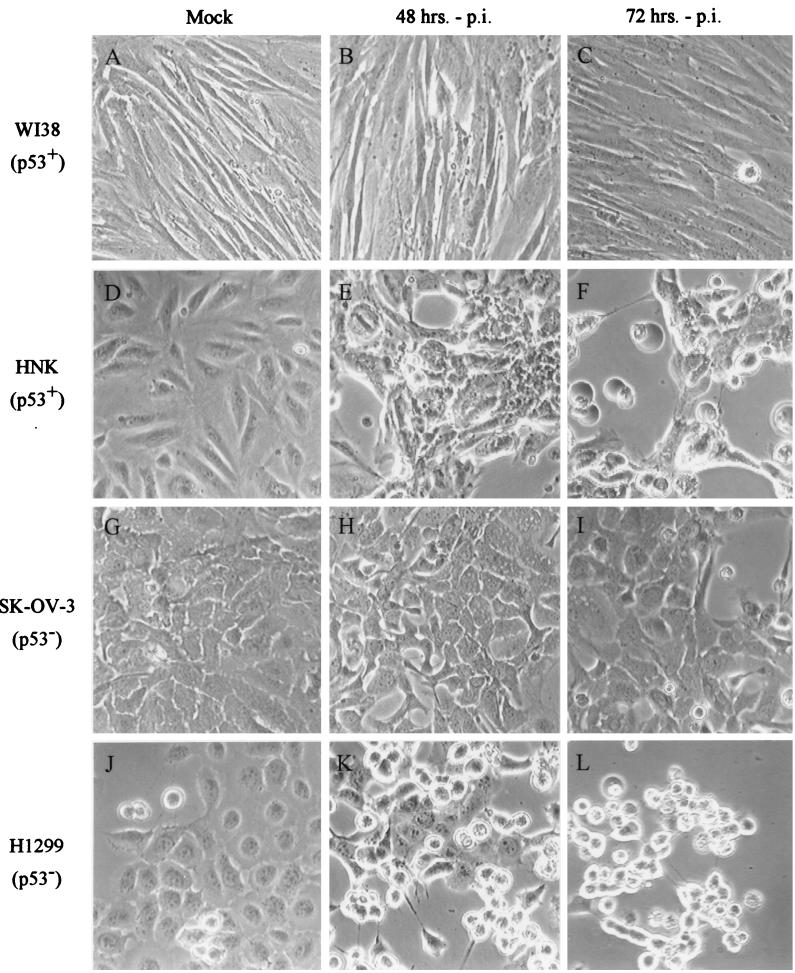
Recent studies by Hall et al. have suggested that p53-dependent cell death may be required for induction of the adenovirus cytopathic effect and release of progeny virions from infected cells (36). However, this suggestion was not consistent with our observations. p53+ WI38 (26) and HNK nonimmortalized cell strains and p53− SK-OV-3 (85) and H1299 (59) cell lines were infected with Ad5 and dl1520 at an MOI of 5 and examined by phase-contrast microscopy at 48 and 72 h postinfection (Fig. 6). The p53-positive HNK and p53-negative H1299 cells exhibited obvious cytopathic effects and cell lysis by 72 h postinfection (Fig. 6F and L), as evidenced by the detachment of the monolayer and the formation of round, light-refractile bodies. Both cell lines supported adenovirus replication efficiently, producing 27,600 and 25,500 PFU per cell by this time (Table 1). Neither the p53-negative SK-OV-3 (Fig. 6H and I) (85) nor the p53-positive WI38 (Fig. 6B and C) (26) cells exhibited significant cytopathology at the times examined. Both cell strains supported Ad5 replication efficiently, however, producing 5,590 and 2,560 PFU per cell, respectively, at 72 h postinfection (Table 1). Thus, Ad5 induction of the cytopathic effect in various cell lines and cell strains varies widely and does not correlate with the expression of wild-type p53 or even viral replication.
FIG. 6.
Adenovirus-induced cytopathic effect does not correlate with p53 genotype. Ad5- and dl1520-induced cytolysis was examined in the p53-positive WI38 (A to C) (26) and HNK (D to F) nonimmortalized cell strains and in the p53-negative SK-OV-3 (G to I) (85) and H1299 (J to L) (59) cell lines. Infections were performed at an MOI of 5 as described in Materials and Methods. Micrographs of infected cells were taken at 48 (B, E, H, and K) and 72 (C, F, I, and L) h postinfection (p.i.).
DISCUSSION
p53 status does not determine dl1520 host range.
In this study we have reexamined the correlation proposed by researchers at ONYX Pharmaceuticals between the p53 status of a cell and its ability to support the replication of dl1520 (5) (also known as ONYX-015 for commercial reasons [10, 39]). The replication of dl1520 varied widely in the cell lines and primary human cells that we examined, and the ability to support dl1520 replication did not correlate with the p53 genotype (Table 1). Our results are in agreement with other recent studies on this issue (31, 32, 71, 82a), including a subsequent paper from ONYX (39). However, this second ONYX paper (39) suggested that dl1520 might replicate preferentially in tumor cells independently of their p53 status compared to nontransformed primary cells (39). In contrast to this suggestion, our observation of efficient dl1520 replication in two primary cell strains, HNK and IMR90 (Table 1), indicates that the mutant virus does not replicate selectively in tumor cells.
In addition to the lack of correlation between p53 status and the ability to support the replication of dl1520, Rothmann et al. (71) recently reported that both p53-positive and p53-negative cells are susceptible to dl1520-mediated cytolysis. Consequently, cell killing by dl1520 cannot be predicted based on the p53 status of the host cell (71). Our studies further confirm these findings (Fig. 6) and contradict the recent suggestion that the induction of cytopathic effects by Ad5 requires p53-mediated apoptosis (36, 69).
The ONYX studies and the several recent studies in response to the original paper of Bischoff et al. (10), including this one, revealed a wide variation in the ability of dl1520 to replicate in various cell lines and primary cell strains (Table 1) (31, 32, 71). Earlier studies of dl1520 and other E1B-55K mutants have been performed primarily with HeLa cells, where viral replication is strictly dependent on E1B-55K (2, 5, 37, 64). In HeLa cells, cytoplasmic late viral mRNA levels and late protein synthesis are severely diminished following infection with E1B-55K mutants compared to wild-type Ad5 (2, 84, 87). It appears, however, that in many cell lines and primary cell strains, significant late mRNA transport and translation can occur in the absence of E1B-55K.
We examined late viral protein synthesis in several of the cell lines listed in Table 1 by performing pulse-labelling studies at 28 h postinfection (data not shown). In general, we observed diminished dl1520 compared to wild-type Ad5 late viral protein synthesis in cell lines that did not support dl1520 replication, as had been shown for dl1520 and other E1B-55K mutants in HeLa cells (3, 65, 84, 87). In cell lines where dl1520 replication was reduced only two- to fourfold compared to Ad5 replication, dl1520 late protein synthesis was similarly only modestly reduced compared to that of Ad5. At present, we do not understand why E1B-55K is strictly required for viral replication in some host cells and not others.
p53-dependent and p53-independent functions of E1B-55K.
As mentioned above, most earlier studies on the influence of E1B-55K on viral DNA replication and late viral RNA synthesis, nuclear export, and cytoplasmic stability have been performed with HeLa cells (3, 50, 64, 65, 84). However, experiments with HeLa cells make it difficult to determine which effects of E1B-55K are due to the inhibition of p53 function and which are due to p53-independent functions of E1B-55K. This is because HeLa cells contain wild-type p53 whose function is inhibited by endogenous human papillomavirus (HPV) type 18 E6 protein, which targets p53 for degradation by a ubiquitin-proteosome mediated mechanism (76). However, in HeLa cells infected by an adenovirus E1B mutant, the stabilization of p53 induced by E1A functions (17, 54, 67) overcomes HPV E6 mediated-p53 degradation, and p53 levels rise (16).
To better understand which molecular processes requiring E1B-55K function are dependent on E1B-55K’s ability to inactivate p53 function and which are independent of its effects on p53 activity, we performed experiments with p53− H1299 cells and H1299-tsp53 cells stably transformed with the temperature-sensitive p53 A135V allele (29). Although p53 function did not appear to be the sole host range determinant for dl1520 replication in our panel of cell lines, it did influence dl1520 replication in H1299 cells. As for other E1B-55K mutant viruses analyzed in HeLa cells (41, 50), the replication of dl1520 in H1299 cells was temperature dependent (32). At 39°C, dl1520 replication was only modestly impaired compared to that of Ad5 in both H1299 and H1299-tsp53 cells (where there is no p53 function) (Fig. 2). However, in H1299 cells at 32°C, the dl1520 yield was reduced 50-fold compared to that of Ad5 (Table 2). Furthermore, when a high level of p53 function was restored for 24 h prior to infection by shifting H1299-tsp53 cells to 32°C (conditions that lead to the induction of the p53-responsive genes for hdm2 and p21 Cki1 [Fig. 1]), a substantial additional defect in the dl1520 yield was observed. Under these conditions, virtually no dl1520 replication occurred, whereas the presence of E1B-55K allowed replication to an extent similar to that for wild-type Ad5 in the parental p53− H1299 cells. These findings indicate that E1B-55K performs at least two functions required for maximal replication in H1299-tsp53 cells at low temperature, one that is independent of p53 function and one that counteracts cellular responses that require p53 function. Similar findings have been reported by Goodrum and Ornelles (32). In their studies, however, the p53-imposed replication defect was not as severe as that observed in our experiments.
Biochemical analyses were performed to investigate the molecular mechanisms underlying the additional restriction to dl1520 replication in H1299 cells imposed by p53. In addition to addressing questions related to p53 function, these studies uncovered unanticipated differences in the block to E1B-55K mutant replication in H1299 versus HeLa cells. As was the case for E1B-55K mutant-infected HeLa cells (50, 84), viral DNA replication in dl1520-infected H1299 cells at 32°C (Fig. 3) was comparable to that of wild-type Ad5. However, whereas a substantial decrease in viral late cytoplasmic RNA levels was observed in HeLa cells at 32°C (50, 65), a result confirmed with dl1520 (data not shown), in H1299 cells at 32°C the dl1520 mutation caused only a modest, twofold decrease in cytoplasmic fiber RNA (Fig. 4; Table 2). A similar only-modest reduction of the hexon mRNA level also was observed in H1299 cells at 32°C (data not shown). Despite these moderate effects on the cytoplasmic accumulation of late viral RNAs, late viral protein synthesis was markedly decreased in H1299 cells at 32°C (Fig. 5; Table 2).
The additional effect of p53 function in H1299 cells was analyzed by shifting H1299-tsp53 cells to 32°C 24 h prior to infection. When p53 function was restored, dl1520 DNA replication was delayed and the DNA accumulated to only approximately one-third of the level of wild-type Ad5 (Fig. 3). Cytoplasmic fiber RNA was reduced to ∼20% of the Ad5 level, and nuclear fiber RNA was similarly reduced to ∼30% (Fig. 4; Table 2). The additional p53-imposed decrease in cytoplasmic fiber RNA levels was likely due to the p53-imposed reduction in viral DNA replication (Fig. 3) and subsequent decrease in RNA synthesis.
The severe inhibition of late viral protein synthesis observed in dl1520-infected H1299 cells at 32°C was still more severe when p53 function was restored in H1299-tsp53 cells at 32°C. Under these conditions, late viral protein synthesis was barely detectable (Fig. 5). This may have been a consequence of the further decrease in late viral mRNA coupled with the very low levels of viral late protein synthesis in H1299 cells at 32°C. Alternatively, p53 function may have induced an additional block to late viral protein synthesis in H1299-tsp53 cells at 32°C.
It is also formally possible that viral yields in dl1520-infected H1299-tsp53 cells at 32°C were reduced in response to the induction of p53-dependent apoptosis. We consider this final possibility unlikely, however, as the antiapoptotic function of E1B-19K (83) remains intact in the dl1520 virus (5), and the Hirt analyses of infected cells failed to reveal significant amounts of nuclear or viral DNA fragmentation indicative of apoptosis (Fig. 3).
E1B-55K promotes late mRNA translation in H1299 cells.
The defect in dl1520 late protein synthesis in H1299 cells at 32°C was reproducibly much more pronounced than the reduction in late mRNA levels (Fig. 4 and 5; Table 2). Hexon and fiber protein synthesis in these cells was reduced to 6 to 8% of the wild-type level in the face of more modest reductions in cytoplasmic mRNA (Table 2). We therefore propose that E1B-55K may perform an additional function in stimulating the translation of viral late proteins in H1299 cells. A similar activity in HeLa cells may have been masked by the severe reduction in cytoplasmic late RNA levels in those cells (50, 84). It is also possible that the pronounced decrease in late viral protein synthesis observed for E1B-55K mutants is due to the cumulative effects of modest decreases in the expression of other late viral proteins that stimulate late viral protein synthesis (38, 43).
In the late phase of adenovirus infection, the host translational machinery is redirected to the near-exclusive synthesis of late viral proteins (Fig. 5) (reviewed in reference 77). During this time, viral late messages represent the majority of RNAs associated with polyribosomes (77). This selective expression of viral proteins results from the preferential accumulation of viral late RNAs in the cytoplasm and, once there, on the tripartite leader associated with the major late mRNAs (9, 24, 53). The tripartite leader has been proposed to facilitate translation initiation in the presence of limiting concentrations of the cap-binding complex eIF-4F, which stimulates the interaction of capped mRNAs with the 40S ribosomal subunit during translation initiation (24, 25). In cells coinfected with adenovirus and poliovirus, for example, adenovirus late mRNA synthesis continues despite the inactivation of eIF-4F through cleavage of its p220/eIF-4G subunit by a poliovirus protease (14, 24).
The activity of the cap-binding complex is also regulated by the phosphorylation of the eIF-4E subunit of eIF-4F (44). Phosphorylation of this factor correlates with the ability of the eIF-4F complex to recruit the 40S ribosomal subunit to capped mRNAs (44). Reduced phosphorylation of eIF-4E occurs during the inhibition of translation during the heat shock response, and phosphorylation is increased upon the stimulation of cell growth with mitogens (46, 47).
eIF-4E phosphorylation is also reduced late in adenovirus infection (42). The subsequent impaired activity of eIF-4F has been proposed to shut off the synthesis of host proteins (42, 89). The translation of adenovirus late messages continues, however, due to the presence of the tripartite leader (24, 25). Intriguingly, in HeLa cells infected with the E1B-55K mutant dl338, eIF-4E remains largely phosphorylated (89). The high proportion of phosphorylated eIF-4E correlates with the failure of dl338 to shut off cellular protein synthesis at late times (89). Zhang et al. (89) demonstrated that the decrease in eIF-4E phosphorylation at late times in Ad5-infected cells is due to an inhibition of eIF-4E phosphorylation as opposed to an increase in the rate of dephosphorylation. They speculated that the inhibitor of the unknown eIF-4E kinase(s) might be a late viral protein and that the failure of dl338-infected cells to inhibit eIF-4E phosphorylation might be secondary to the dl338 defect in late viral mRNA transport (89). However, it remains possible that E1B-55K has a more direct role in inhibiting eIF-4E phosphorylation.
The intriguing and as-yet-unexplained ability of incubation at 39°C to compensate for E1B-55K mutations in cells where E1B-55K function is required (32, 37, 41) (Fig. 1) might be explained if the stimulation of late protein synthesis by E1B-55K results from the dephosphorylation of eIF-4E. We speculate that in Ad5-infected H1299 cells, eIF-4E is dephosphorylated during the late phase of infection, just as in HeLa cells (42), and that this process fails in dl1520-infected H1299 cells at 32°C just as it does in dl338-infected HeLa cells (89). This would account for the inability of dl1520 to shut off host protein synthesis at 32°C in H1299 cells. This could in turn result in the decreased synthesis of late viral proteins by preventing the preferential translation of late viral mRNAs with the tripartite leader when eIF-4F activity is limiting (24, 25). The inability of E1B-55K mutants to dephosphorylate eIF-4E may be compensated for at 39°C, as the heat shock response, if induced at 39°C, would result in the dephosphorylation of eIF-4E (47). Further experiments will be required to test this hypothesis.
Similarities between E1B-55K/E4-orf6 and HIV Rev in the control of mRNA nuclear export and translation.
The human immunodeficiency virus (HIV) Rev protein (reviewed in reference 61) has functions that in many ways parallel those of the E1B-55K/E4-orf6 complex. Like the E1B-55K/E4-orf6 complex, Rev modulates the preferential export of HIV late RNAs during the late phase of infection (27, 55). Both Rev and E4-orf6 possess leucine-rich nuclear export signals, which appear to confer these nuclear export functions (22, 28). Additionally, when overexpressed, E4-orf6 can inhibit the Rev-mediated nuclear export of unspliced RNAs, suggesting that these factors may bind a common saturable factor(s) involved in RNA transport or use overlapping export pathways (22).
Similar to the findings of the present study, the effect of Rev on the cytoplasmic accumulation of late viral messages varies with the host cell and does not absolutely correlate with protein expression (1, 19). In lymphoid cells, protein expression from some HIV late RNAs is severely restricted in the absence of Rev, despite the near-wild-type accumulation of the message in the cytoplasm (1). It has further been shown that polyribosomal association of these late RNAs is reduced in the absence of Rev, suggesting that Rev may affect ribosome loading onto the late RNAs (1, 19).
The parallels that may be drawn between the E1B-55K/E4-orf6 complex and HIV Rev are intriguing. It is also tempting to speculate that the abilities of the complex retroviruses and adenoviruses to usurp cellular RNA export and translational processes are somehow linked. Further studies to address this possibility and to better understand the mechanism by which E1B-55K affects the translation of late viral proteins are in progress.
ACKNOWLEDGMENTS
We thank Michael Kastan (Johns Hopkins University) for the RKO, RKO RC10.1, RKO RC10.3, and RKOp53.13 cell lines, J. S. Economou (UCLA) for the Hep3B cell line, Phillip Koeffler (UCLA) for the PC-3 cell line, and Arnold Levine (Princeton University) for the Saos-2 cell line. We are especially indebted to Kathi Fries and Nancy Raab-Traub (University of North Carolina, Chapel Hill) for providing the H1299 and H1299-tsp53 cell lines. We also gratefully acknowledge Carol Eng for providing excellent technical assistance and the members of Utpal Banerjee’s laboratory for assistance with microscopy.
This work was supported by Public Health Service National Research Service Award GM07185 predoctoral fellowship to J.N.H. and by Public Health Service grant CA 64799.
REFERENCES
- 1.Arrigo S J, Chen I S Y. Rev is necessary for translation but not cytoplasmic accumulation of HIV-1 vif, vpr, and env/vpu 2 RNAs. Genes Dev. 1991;5:808–819. doi: 10.1101/gad.5.5.808. [DOI] [PubMed] [Google Scholar]
- 2.Babiss L E, Ginsberg H S. Adenovirus type 5 early region 1B gene product is required for efficient shutoff of host protein synthesis. J Virol. 1984;50:202–212. doi: 10.1128/jvi.50.1.202-212.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babiss L E, Ginsberg H S, Darnell J E. Adenovirus E1B proteins are required for accumulation of late viral mRNA and for effects on cellular mRNA translation and transport. Mol Cell Biol. 1985;5:2552–2558. doi: 10.1128/mcb.5.10.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker S J, Markowitz S, Fearon E R, Willson J K, Vogelstein B. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science. 1990;249:912–915. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- 5.Barker D D, Berk A J. Adenovirus proteins from both E1B reading frames are required for transformation of rodent cells by viral infection and DNA transfection. Virology. 1988;56:107–121. doi: 10.1016/0042-6822(87)90441-7. [DOI] [PubMed] [Google Scholar]
- 6.Beltz G A, Flint S J. Inhibition of HeLa cell protein synthesis during adenovirus infection. J Mol Biol. 1979;131:353–373. doi: 10.1016/0022-2836(79)90081-0. [DOI] [PubMed] [Google Scholar]
- 7.Berk A J. Characterization of RNA molecules by S1 nuclease analysis. Methods Enzymol. 1989;180:334–347. doi: 10.1016/0076-6879(89)80110-7. [DOI] [PubMed] [Google Scholar]
- 8.Berk A J, Sharp P A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977;12:721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- 9.Berkner K L, Sharp P A. Effect of the tripartite leader on synthesis of a non-viral protein in an adenovirus 5 recombinant. Nucleic Acids Res. 1985;13:841–857. doi: 10.1093/nar/13.3.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bischoff J R, Kirn D H, Williams A, Heise C, Horn S, Muna M, Ng L, Nye J A, Sampson-Johannes A, Fattaey A, McCormick F. An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science. 1996;274:373–376. doi: 10.1126/science.274.5286.373. [DOI] [PubMed] [Google Scholar]
- 11.Bressac B, Galvin K M, Liang T J, Isselbacher K J, Wands J R, Ozturk M. Abnormal structure and expression of p53 gene in human hepatocellular carcinoma. Proc Natl Acad Sci USA. 1990;87:1973–1977. doi: 10.1073/pnas.87.5.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bridge E, Ketner G. Interaction of adenoviral E4 and E1b products in late gene expression. Virology. 1990;174:345–353. doi: 10.1016/0042-6822(90)90088-9. [DOI] [PubMed] [Google Scholar]
- 13.Carroll A G, Voeller H J, Sugars L, Gelmann E P. p53 oncogene mutations in three human prostate cancer cell lines. Prostate. 1993;23:123–134. doi: 10.1002/pros.2990230206. [DOI] [PubMed] [Google Scholar]
- 14.Castrillo J L, Carrasco L. Adenovirus late protein synthesis is resistant to the inhibition of translation induced by poliovirus. J Biol Chem. 1987;262:7328–7334. [PubMed] [Google Scholar]
- 15.Chandar N, Bilig B, McMaster J, Novak J. Inactivation of p53 gene in human and murine osteosarcoma cells. Br J Cancer. 1992;65:208–214. doi: 10.1038/bjc.1992.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiou S-K, Tseng C-C, Rao L, White E. Functional complementation of the adenovirus E1B 19-kilodalton protein with Bcl-2 in the inhibition of apoptosis in infected cells. J Virol. 1994;68:6553–6566. doi: 10.1128/jvi.68.10.6553-6566.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiou S-K, White E. p300 binding by E1A cosegregates with p53 induction but is dispensable for apoptosis. J Virol. 1997;71:3515–3525. doi: 10.1128/jvi.71.5.3515-3525.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crook T, Wrede D, Vousden K H. p53 point mutation in HPV negative human cervical carcinoma cell lines. Oncogene. 1991;6:873–875. [PubMed] [Google Scholar]
- 19.D’Agostino D M, Felber B K, Harrison J E, Pavlakis G N. The Rev protein of human immunodeficiency virus type 1 promotes polysomal association and translation of gag/pol and vpr/env mRNAs. Mol Cell Biol. 1992;12:1375–1386. doi: 10.1128/mcb.12.3.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Debbas M, White E. Wild-type p53 mediates apoptosis by E1A, which is inhibited by E1B. Genes Dev. 1993;7:546–554. doi: 10.1101/gad.7.4.546. [DOI] [PubMed] [Google Scholar]
- 21.Diller L, Kassel J, Nelson C E, Gryka M A, Litwak G, Gebhardt M, Bressac B, Ozturk M, Barker S J, Vogelstein B, Friend S H. p53 functions as a cell cycle control protein in osteosarcomas. Mol Cell Biol. 1990;10:5772–5781. doi: 10.1128/mcb.10.11.5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dobbelstein M, Roth J, Kimberly W T, Levine A J, Shenk T. Nuclear export of the E1b-55kDa and E4-34kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 1997;16:4276–4284. doi: 10.1093/emboj/16.14.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dobner T, Horikoshi N, Rubenwolf S, Shenk T. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science. 1996;272:1470–1473. doi: 10.1126/science.272.5267.1470. [DOI] [PubMed] [Google Scholar]
- 24.Dolph P J, Huang J, Schneider R J. Translation by the adenovirus tripartite leader: elements which determine independence from cap-binding protein complex. J Virol. 1990;64:2669–2677. doi: 10.1128/jvi.64.6.2669-2677.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dolph R J, Racaniello V, Villamarin A, Palladino F, Schneider R J. The adenovirus tripartite leader may eliminate the requirement for cap-binding protein complex during translation initiation. J Virol. 1988;62:2059–2066. doi: 10.1128/jvi.62.6.2059-2066.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farshid M, Tabor E. Expression of oncogenes and tumor suppressor genes in human hepatocellular carcinoma and hepatoblastome cell lines. J Med Virol. 1992;38:235–239. doi: 10.1002/jmv.1890380402. [DOI] [PubMed] [Google Scholar]
- 27.Felber B K, Hadzopoulou-Cladaras M, Cladaras C, Copeland T, Pavlakis G N. Rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc Natl Acad Sci USA. 1989;86:1495–1499. doi: 10.1073/pnas.86.5.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. The HIV-1 rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 29.Fries K L, Miller W E, Raab-Traub N. Epstein-Barr virus latent membrane protein 1 blocks p53-mediated apoptosis through the induction of the A20 gene. J Virol. 1996;70:8653–8659. doi: 10.1128/jvi.70.12.8653-8659.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reference deleted.
- 31.Goodrum F D, Ornelles D A. The early region 1B 55-kilodalton oncoprotein of adenovirus relieves growth restrictions imposed on viral replication by the cell cycle. J Virol. 1997;71:548–561. doi: 10.1128/jvi.71.1.548-561.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goodrum F D, Ornelles D A. p53 status does not determine outcome of E1B 55-kilodalton mutant adenovirus lytic infection. J Virol. 1998;72:9479–9490. doi: 10.1128/jvi.72.12.9479-9490.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graham F L, Smiley J, Russell W C, Nairn R. Characterization of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–72. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 34.Grand R J A, Grant M L, Gallimore P H. Enhanced expression of p53 in human cells infected with mutant adenoviruses. Virology. 1994;203:229–240. doi: 10.1006/viro.1994.1480. [DOI] [PubMed] [Google Scholar]
- 35.Halbert D N, Shenk T. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J Virol. 1985;56:250–257. doi: 10.1128/jvi.56.1.250-257.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hall A R, Dix B R, O’Carroll S J, Braithwaite A W. p53-dependent cell death/apoptosis is required for a productive adenovirus infection. Nat Med. 1998;4:1068–1072. doi: 10.1038/2057. [DOI] [PubMed] [Google Scholar]
- 37.Harrison T, Graham F, Williams J. Host-range mutants of adenovirus type 5 defective for growth in HeLa cells. Virology. 1977;77:319–329. doi: 10.1016/0042-6822(77)90428-7. [DOI] [PubMed] [Google Scholar]
- 38.Hayes B W, Telling G C, Myat M M, Williams J F, Flint S J. The adenovirus L4 100-kilodalton protein is necessary for efficient translation of viral late mRNA species. J Virol. 1990;64:2732–2742. doi: 10.1128/jvi.64.6.2732-2742.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heise C, Sampson-Johannes A, Williams A, McCormick F, Hoff D D V, Kirn D H. ONYX-015, an E1B gene attenuated adenovirus, causes tumor-specific cytolysis and antitumoral efficacy that can be augmented by standard chemotherapeutic agents. Nat Med. 1997;3:639–645. doi: 10.1038/nm0697-639. [DOI] [PubMed] [Google Scholar]
- 40.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 41.Ho Y S, Galos R, Williams J. Isolation of the type 5 adenovirus mutants with a cold-sensitive host range phenotype: genetic evidence of an adenovirus transformation maintenance function. Virology. 1982;122:109–124. doi: 10.1016/0042-6822(82)90381-6. [DOI] [PubMed] [Google Scholar]
- 42.Huang J, Schneider R J. Adenovirus inhibition of cellular protein synthesis involves inactivation of cap-binding protein. Cell. 1991;65:271–280. doi: 10.1016/0092-8674(91)90161-q. [DOI] [PubMed] [Google Scholar]
- 43.Imperiale M J, Akusjarvi G, Leppard K N. Post-transcriptional control of adenovirus gene expression. Curr Top Microbiol Immunol. 1995;199:139–171. doi: 10.1007/978-3-642-79499-5_6. [DOI] [PubMed] [Google Scholar]
- 44.Joshi-Barve S, Rychlik W, Rhoads R E. Alteration of the major phosphorylation site of eukaryotic protein synthesis initiation factor 4E prevents its association with the 48S initiation complex. J Biol Chem. 1990;265:2979–2983. [PubMed] [Google Scholar]
- 45.Kao C C, Yew P R, Berk A J. Domains required for in vitro association between the cellular p53 and the adenovirus 2 E1B 55K proteins. Virology. 1990;179:806–814. doi: 10.1016/0042-6822(90)90148-k. [DOI] [PubMed] [Google Scholar]
- 46.Kaspar R L, Rychlik W, White M W, Rhoads R E, Morris D R. Simultaneous cytoplasmic redistribution of ribosomal protein L32 mRNA and phosphorylation of eukaryotic initiation factor 4E after mitogenic stimulation of Swiss 3T3 cells. J Biol Chem. 1990;265:3619–3622. [PubMed] [Google Scholar]
- 47.Lamphear B J, Panniers R. Heat shock impairs the interaction of cap-binding protein complex with 5′ mRNA cap. J Biol Chem. 1991;266:2789–2794. [PubMed] [Google Scholar]
- 48.Lane D P. Killing tumor cells with viruses—a question of specificity. Nat Med. 1998;4:1012–1013. doi: 10.1038/2000. [DOI] [PubMed] [Google Scholar]
- 49.Lehman T A, Bennett W P, Metcalf R A, Welsh J A, Ecker J, Modali R V, Ullrich S, Romano J W, Appella E, Testa J R, Gerwin B I, Harris C C. p53 mutations, ras mutations, and p53-heat shock protein complexed in human lung carcinoma cell lines. Cancer Res. 1991;51:4090–4096. [PubMed] [Google Scholar]
- 50.Leppard K N, Shenk T. The adenovirus E1B 55 kD protein influences mRNA transport via an intranuclear effect on RNA metabolism. EMBO J. 1989;8:2329–2336. doi: 10.1002/j.1460-2075.1989.tb08360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Levine A J. p53, the gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 52.Linke S P. Has the smart bomb been defused? Nature. 1998;395:13–15. doi: 10.1038/25595. [DOI] [PubMed] [Google Scholar]
- 53.Logan J, Shenk T. Adenovirus tripartite leader sequence enhances translation of mRNAs late after infection. Proc Natl Acad Sci USA. 1984;81:3655–3659. doi: 10.1073/pnas.81.12.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lowe S W, Ruley H E. Stabilization of the p53 tumor suppressor is induced by adenovirus E1A and accompanies apoptosis. Genes Dev. 1993;7:535–545. doi: 10.1101/gad.7.4.535. [DOI] [PubMed] [Google Scholar]
- 55.Malim M H, Hauber J, Le S-Y, Maizel J V, Cullen B R. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 56.Martin M E D, Berk A J. Adenovirus E1B-55K represses p53 activation in vitro. J Virol. 1998;72:3146–3154. doi: 10.1128/jvi.72.4.3146-3154.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martinez J, Georgoff I, Martinez J, Levine A J. Cellular localization and cell cycle regulation by a temperature-sensitive p53 protein. Genes Dev. 1991;5:151–159. doi: 10.1101/gad.5.2.151. [DOI] [PubMed] [Google Scholar]
- 58.Michalovitz D, Halevy O, Oren M. Conditional inhibition of transformation and of cell proliferation by a temperature-sensitive mutant of p53. Cell. 1990;62:671–680. doi: 10.1016/0092-8674(90)90113-s. [DOI] [PubMed] [Google Scholar]
- 59.Mitsudomi T, Steinberg S M, Nau M M, Carbone D, D’Amico D, Bodner H K, Oie H K, Rinnoila R I, Mulshine J L, Minna J D, Gazdar A F. p53 gene mutations in non-small-lung cell cancer cell lines and their correlation with the presence of ras mutations and clinical features. Oncogene. 1992;7:171–180. [PubMed] [Google Scholar]
- 60.Moore M, Horikoshi N, Shenk T. Oncogenic potential of the adenovirus E4orf6 protein. Proc Natl Acad Sci USA. 1996;93:11295–11301. doi: 10.1073/pnas.93.21.11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nakielny S, Fischer U, Michael W M, Dreyfuss G. RNA transport. Annu Rev Neurosci. 1997;20:269–301. doi: 10.1146/annurev.neuro.20.1.269. [DOI] [PubMed] [Google Scholar]
- 62.Nevels M, Rubenwolf S, Spruss T, Wolf H, Dobner T. The adenovirus E4orf6 protein can promote E1A/E1B-induced focus formation by interfering with p53 tumor suppressor function. Proc Natl Acad Sci USA. 1997;94:1206–1211. doi: 10.1073/pnas.94.4.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ornelles D A, Shenk T. Localization of the adenovirus early region 1B 55-kilodalton protein during lytic infection: association with nuclear viral inclusions requires the early region 4 34-kilodalton protein. J Virol. 1991;65:424–439. doi: 10.1128/jvi.65.1.424-429.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pilder S, Leppard K, Logan J, Shenk T. Functional analysis of the adenovirus E1B 55K polypeptide. Cancer Cells. 1986;4:285–290. [Google Scholar]
- 65.Pilder S, Moore M, Logan J, Shenk T. The adenovirus E1B-55K transforming polypeptide modulates transport or cytoplasmic stabilization of viral and host cell mRNAs. Mol Cell Biol. 1986;6:470–476. doi: 10.1128/mcb.6.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Querido E, Marcellus R C, Lai A, Charbonneau R, Teodoro J G, Ketner G, Branton P E. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J Virol. 1997;71:3788–3798. doi: 10.1128/jvi.71.5.3788-3798.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Querido E, Teodoro J G, Branton P E. Accumulation of p53 induced by the adenovirus E1A protein requires regions involved in the stimulation of DNA synthesis. J Virol. 1997;71:3526–3533. doi: 10.1128/jvi.71.5.3526-3533.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rao L, Debbas M, Sabbatini D, Hockenberry D, Korsmeyer S, White E. The adenovirus E1A proteins induce apoptosis, which is inhibited by the E1B 19-kDa and Bcl-2 proteins. Proc Natl Acad Sci USA. 1992;89:7742–7746. doi: 10.1073/pnas.89.16.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ridgway P J, Hall A R, Myers C J, Braithwaite A W. p53/E1b58kDa complex regulates adenovirus replication. Virology. 1997;237:404–413. doi: 10.1006/viro.1997.8782. [DOI] [PubMed] [Google Scholar]
- 70.Roth J, Konig C, Weinzer S, Weigel S, Ristea S, Dobbelstein M. Inactivation of p53 but not p73 by adenovirus type 5 E1B 55-kilodalton and E4 34-kilodalton oncoproteins. J Virol. 1998;72:8510–8516. doi: 10.1128/jvi.72.11.8510-8516.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rothmann T, Hengstermann A, Whitaker N J, Sheffner M, Hausen H Z. Replication of ONYX-015, a potential anticancer adenovirus, is independent of p53 status in tumor cells. J Virol. 1998;72:9470–9478. doi: 10.1128/jvi.72.12.9470-9478.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rubenwolf S, Schutt H, Nevels M, Wolf H, Dobner T. Structural analysis of the adenovirus type 5 E1B 55-kilodalton-E4orf6 protein complex. J Virol. 1997;71:1115–1123. doi: 10.1128/jvi.71.2.1115-1123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 74.Sarnow P, Hearing P, Anderson C W, Halbert D N, Shenk T, Levine A J. Adenovirus early-region 1B 58,000-dalton tumor antigen is physically associated with an early-region 4 25,000-dalton protein in productively infected cells. J Virol. 1984;49:692–700. doi: 10.1128/jvi.49.3.692-700.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Scheffner M, Munger K, Byrne J C, Howley P M. The state of p53 and retinoblastoma genes in human cervical cancer cell lines. Proc Natl Acad Sci USA. 1991;88:5523–5527. doi: 10.1073/pnas.88.13.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 15 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 77.Schneider R J. Cap-independent translation in adenovirus infected cells. Curr Top Microbiol Immunol. 1995;203:117–129. doi: 10.1007/978-3-642-79663-0_6. [DOI] [PubMed] [Google Scholar]
- 78.Schwarz E, Freese U K, Gissmann L, Mayer W, Roggenbuck B, Stremlau A, zur Hausen H. Structure and transcription of human papillomavirus sequences in cervical carcinoma cells. Nature. 1985;314:111–114. doi: 10.1038/314111a0. [DOI] [PubMed] [Google Scholar]
- 79.Slebos R J C, Lee M H, Plunkett B S, Kessis T D, Williams B O, Jacks T, Hedrick L, Kastan M B, Cho K R. p53-dependent G1 arrest involves pRB-related proteins and is disrupted by the human papillomavirus 16 E7 oncoprotein. Proc Natl Acad Sci USA. 1994;91:5320–5324. doi: 10.1073/pnas.91.12.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Slichenmyer W J, Nelson W G, Slebos R J, Kastan M B. Loss of a p53-associated G1 checkpoint does not decrease cell survival following DNA damage. Cancer Res. 1993;53:4164–4168. [PubMed] [Google Scholar]
- 81.Steegenga W T, Riteco N, Jochemsen A G, Fallaux F J, Bos J L. The large E1B protein together with the E4orf6 protein target p53 for active degradation in adenovirus infected cells. Oncogene. 1998;16:349–357. doi: 10.1038/sj.onc.1201540. [DOI] [PubMed] [Google Scholar]
- 82.Turnell A S, Grand R J A, Gallimore P H. The replicative capacities of large E1B-null group A and group C adenoviruses are independent of host cell p53 status. J Virol. 1999;73:2074–2083. doi: 10.1128/jvi.73.3.2074-2083.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.White E. Regulation of p53-dependent apoptosis by E1A and E1B. Curr Top Microbiol Immunol. 1995;199:34–57. [PubMed] [Google Scholar]
- 84.Williams J, Karger B D, Ho Y S, Castaglia C L, Mann T, Flint S J. The adenovirus E1B 495R protein plays a role in regulating the transport and stability of the viral late messages. Cancer Cells. 1986;4:275–284. [Google Scholar]
- 85.Yaginuma Y, Westphal H. Abnormal structure and expression of the p53 gene in human ovarian carcinoma cell lines. Cancer Res. 1992;52:4196–4199. [PubMed] [Google Scholar]
- 86.Yew P R, Berk A J. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature. 1992;357:82–85. doi: 10.1038/357082a0. [DOI] [PubMed] [Google Scholar]
- 87.Yew P R, Kao C C, Berk A J. Dissection of functional domains in the adenovirus 2 early 1B 55K polypeptide by suppressor-linker insertional mutagenesis. Virology. 1990;179:795–805. doi: 10.1016/0042-6822(90)90147-j. [DOI] [PubMed] [Google Scholar]
- 88.Yew P R, Liu X, Berk A J. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes Dev. 1994;8:190–202. doi: 10.1101/gad.8.2.190. [DOI] [PubMed] [Google Scholar]
- 89.Zhang Y, Feigenblum D, Schneider R J. A late adenovirus factor induced eIF-4E dephosphorylation and inhibition of cell protein synthesis. J Virol. 1994;68:7040–7050. doi: 10.1128/jvi.68.11.7040-7050.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]