Summary
Glomerella leaf spot (GLS), caused by the fungus Colletotrichum fructicola, is considered one of the most destructive diseases affecting apples. The VQ‐WRKY complex plays a crucial role in the response of plants to biotic stresses. However, our understanding of the defensive role of the VQ‐WRKY complex on woody plants, particularly apples, under biotic stress, remains limited. In this study, we elucidated the molecular mechanisms underlying the defensive role of the apple MdVQ37‐MdWRKY100 module in response to GLS infection. The overexpression of MdWRKY100 enhanced resistance to C. fructicola, whereas MdWRKY100 RNA interference in apple plants reduced resistance to C. fructicola by affecting salicylic acid (SA) content and the expression level of the CC‐NBS‐LRR resistance gene MdRPM1. DAP‐seq, Y1H, EMSA, and RT‐qPCR assays indicated that MdWRKY100 inhibited the expression of MdWRKY17, a positive regulatory factor gene of SA degradation, upregulated the expression of MdPAL1, a key enzyme gene of SA biosynthesis, and promoted MdRPM1 expression by directly binding to their promotors. Transient overexpression and silencing experiments showed that MdPAL1 and MdRPM1 positively regulated GLS resistance in apples. Furthermore, the overexpression of MdVQ37 increased the susceptibility to C. fructicola by reducing the SA content and expression level of MdRPM1. Additionally, MdVQ37 interacted with MdWRKY100, which repressed the transcriptional activity of MdWRKY100. In summary, these results revealed the molecular mechanism through which the apple MdVQ37‐MdWRKY100 module responds to GLS infection by regulating SA content and MdRPM1 expression, providing novel insights into the involvement of the VQ‐WRKY complex in plant pathogen defence responses.
Keywords: Apple, SA, Glomerella leaf spot, VQ protein, WRKY
Introduction
Apple (Malus × domestica Borkh.) is a deciduous tree belonging to the Rosaceae family. It is extensively cultivated in temperate regions worldwide and is an economically important fruit tree. Its fruit has a unique flavour and high nutritional value and is cherished by people. However, in natural environments, various pathogens can infect perennial apple trees. Glomerella leaf spot (GLS) is a fungal epidemic caused by Colletotrichum fructicola that occurs in almost all apple producing areas (Munir et al., 2016; Oo et al., 2018; Velho et al., 2015; Zhang et al., 2008a). GLS infects fruit and leaves, resulting in significantly decline in fruit quality and yield (Munir et al., 2016; Oo et al., 2018; Velho et al., 2015; Zhang et al., 2019a). Chemical treatments are commonly used to control GLS; however, they pose new challenges to food safety, environmental protection, and planting costs (Shan et al., 2021; Zhang et al., 2019a). Breeding pathogen‐resistant varieties and selecting pathogen‐resistant apple resources through molecular breeding are the most effective strategies to combat apple fungal diseases (Shan et al., 2021; Zhang et al., 2018, 2019a). Therefore, identifying new candidate genes and elucidating the molecular mechanisms underlying their participation in GLS are crucial for the molecular breeding of disease‐resistant varieties.
Several studies have revealed that in effector‐triggered immunity (ETI), disease‐resistant proteins (R proteins) interact with effector proteins produced by pathogens in a gene‐for‐gene manner, leading to rapid and robust defence responses in plants (Dodds and Rathjen, 2010; Meng et al., 2018). Most plant R proteins are NBS‐LRRs that contain nucleotide binding sites (NB) and leucine‐rich repeats (LRRs) (Dangl and Jones, 2001). Based on the structural characteristics of the N‐terminus domain, NBS‐LRR proteins can be divided into two classes: TIR‐NBS‐LRR (TNL), which contains the Toll/interleukin‐1 receptor (TIR) domain, and CC‐NBS‐LRR (CNL), which contains the coiled‐coil (CC) domain (Mackey et al., 2002; Meng et al., 2018). Previous studies have shown that the TIR and CC domains of NBS‐LRR proteins are involved in downstream signal regulation and the initiation of plant defence responses (Mackey et al., 2002; Meng et al., 2018). Resistance to Pseudomonas syringae pv. maculicola 1 (RPM1) encodes a CC‐NBS‐LRR protein and was the first disease resistance gene to be identified using molecular markers in the study of natural variations in Arabidopsis (Bisgrove et al., 1994; Grant et al., 1995). In apples, the TIR‐NBS‐LRR class genes, MdNLR16, Md‐TN1‐GLS, and MdTNL1‐1, have been reported to be involved in response to Alternaria alternata f. sp mali (ALT1), Glomerella cingulata, and GLS infection, respectively (Lv et al., 2022; Meng et al., 2018; Zhang et al., 2019a). In addition, the hairpin RNA MdhpRNA277 produces mdm‐siR277‐1 and mdm‐siR277‐2, which target CC‐NBS‐LRR genes in response to ALT1 infection (Zhang et al., 2018). However, there are no reports on the involvement of CC‐NBS‐LRR proteins in GLS tolerance in apples.
Salicylic acid (SA), an endogenous signalling molecule in plants, plays a crucial role in plant–pathogen interactions, and its level is required for PAMP‐triggered immunity (PTI), ETI, and systemic acquired resistance (SAR) (Zhang and Li, 2019). SA content in plants is primarily determined by SA homeostasis, including SA biosynthesis (regulated by the isochorismate synthase [ICS] and phenylalanine ammonia lyase [PAL] pathways) and SA metabolism (primarily controlled by Downy Mildew Resistant6 [DMR6] and SAG108) (Zhang et al., 2017b). In apples, C. fructicola effector CfEC12 interacts with MdNIMIN2, a NIM1‐interacting (NIMIN) protein that putatively modulates NPR1 activity in response to SA signalling (Shang et al., 2024). Furthermore, applying exogenous SA can significantly increase apple resistance to GLS (Zhang et al., 2016). Some studies have shown that Arabidopsis WRKY TFs directly bind to the promoter of SA synthesis, metabolism, and signal transduction pathway‐related genes, including ISOCHORISMATE SYNTHASE1 (ICS1), AVRPPHB SUSCEPTIBLE 3 (PBS3), Plant Antitoxin Deficiency 3 (PAD3), ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1) and pathogenesis‐related (PR), to regulate plant response to pathogen infection (Birkenbihl et al., 2012; Shan et al., 2021; van Verk et al., 2011). Similar molecular mechanisms have been observed in apples. For example, MdWRKY15 and MdWRKY46 activate the transcription of the SA synthase genes MdICS1 and MdPBS3.1 by directly binding to their promoters to enhance SA accumulation and confer resistance to Botryosphaeria dothidea (Zhao et al., 2019, 2020). MdWRKY17 reduces SA accumulation by activating the SA degradation gene MdDMR6, thereby increasing the sensitivity to GLS (Shan et al., 2021).
VQ proteins are a class of transcriptional regulatory cofactors containing VQ domains, named after 10 highly conserved amino acid residues (FxxhVQxhTG) in the VQ domain (Jing and Lin, 2015). VQ proteins typically interact with other proteins, such as WRKY, Phytochrome‐Interacting Factors1 (PIF1), ABSCISIC ACID‐INSENSITIVE5 (ABI5), and MAPK, to perform biological functions. Currently, research on the interaction between VQ proteins and WRKY transcription factors is extensive, as they collaboratively participate in regulating specific biological processes and stress responses. (Hu et al., 2013; Jiang and Yu, 2016; Lei et al., 2018; Ma et al., 2023a, 2023b). The interaction between MdVQ10 and MdWRKY75 in apples regulates wound‐triggered leaf senescence (Zhang et al., 2023a). However, the molecular mechanisms through which woody plant VQ‐WRKY complexes regulate disease resistance remain unclear.
The aim of this study was to elucidate the molecular mechanisms underlying the defensive role of the apple MdVQ37‐MdWRKY100 module in response to GLS infection caused by the fungus C. fructicola. Moreover, we investigated the impact of MdWRKY100 overexpression and RNA interference on GLS resistance in apple plants, focusing on their influence on SA content and the expression of the CC‐NBS‐LRR resistance gene MdRPM1. The results of the study may elucidate the crucial role of the WRKY‐VQ complex in the response of apples to GLS and contribute to improving the molecular breeding of apple varieties with GLS resistance.
Results
MdWRKY100 positively regulates resistance to GLS in apples
Our previous studies have shown that MdWRKY100 positively regulates resistance to bitter rot and salt stress in apples (Ma et al., 2021; Zhang et al., 2019b). This study revealed that the MdWRKY100 transcript was significantly upregulated after inoculation with C. fructicola for 48 and 72 h (Figure S1). To further investigate the biological role of MdWRKY100 in GLS tolerance, we used the previously obtained MdWRKY100 overexpression (OE) line MdWRKY100‐OE 4 as well as the MdWRKY100 RNA‐interference (RNAi) line MdWRKY100‐RNAi 3. After C. fructicola inoculation for 4 or 6 days, the MdWRKY100‐OE 4 seedlings showed highly resistance to C. fructicola; however, the MdWRKY100‐RNAi 3 seedlings displayed the opposite phenotype compared to wild‐type (WT) seedlings (Figure 1a). Disease index analysis confirmed this phenotypic difference (Figure 1b). Free SA, total SA content, and PAL activity in the MdWRKY100‐OE 4 seedlings were significantly higher than those in the WT seedlings. In contrast, free SA, total SA content, and PAL activity in the MdWRKY100‐RNAi 3 seedlings were significantly lower than those in the WT seedlings (Figure 1c–e). These results suggest that apple MdWRKY100 enhances resistance against GLS by altering SA content and PAL activity.
Figure 1.

MdWRKY100 positively regulates apple defence against Colletotrichum fructicola. Phenotypes (a) and disease index (b) of apple seedlings of WT, MdWRKY100 overexpression line (MdWRKY100‐OE 4) and MdWRKY100 RNAi line (MdWRKY100‐RNAi 3) after inoculation with C. fructicola for 4 and 6 days. Scale bar: 1 cm. Error bars represent SE (n = 18). The experiments were repeated thrice with similar results. ** in each panel indicates significantly different values relative to WT at P < 0.01, using Student's t‐test. The PAL activity (c) and the contents of endogenous free SA (d) and total SA (e) in apple seedlings of WT, MdWRKY100‐OE 4, and MdWRKY100‐RNAi 3 after inoculation with C. fructicola for 6 days. Error bars represent SD based on 3 independent replicates. ** and * in each panel indicate significantly different values relative to WT at P < 0.01 and P < 0.05, using Student's t‐test, respectively.
MdWRKY100 binds and inhibits the MdWRKY17 expression, and activates the MdPAL1 and MdRPM1 expression
DNA affinity purification sequencing (DAP‐seq) was performed to map the genome‐wide MdWRKY100 DNA‐binding sites to further explore the molecular mechanism and regulatory pathway through which MdWRKY100 regulates GLS infection. The in vitro synthesized MdWRKY100 protein was used to conduct affinity purification of sheared genomic DNA of mature ‘Gala’ leaves, followed by deep sequencing. In total 8750 highly reliable MdWRKY100 binding sites were identified; 24.9% MdWRKY100 binding peaks was distributed in the promoter regions, and 50.2%, 13.1%, and 3.9% were in the intergenic, intronic, and exon regions, respectively (Figure S2). MEME suite analysis showed that the motif sequence with the most significant enrichment of MdWRKY100 binding sites was ‘TTGACT/C', which was a typical W‐box element (Figure 2a). By screening the functional annotation of MdWRKY100 binding sites, we found that a previously reported susceptibility transcription factor during C. fructicola infection, MdWRKY17, a key enzyme gene of SA synthesis, MdPAL1 (Figure S3), and CC‐NBS‐LRR resistance genes (Figure S4) may be the target genes of MdWRKY100 (Table S1). RT‐qPCR results showed that the expression levels of MdPAL1 and MdRPM1 were upregulated by C. fructicola infection (Figure 2b), whereas those of MdCNL1‐MdCNL3 were unaffected by C. fructicola infection, indicating that MdPAL1 and MdRPM1 may play important roles in the apple's response to C. fructicola infection. To verify the reliability of the DAP‐seq results, yeast one‐hybrid (Y1H) and electrophoretic mobility shift assay (EMSA) assays were carried out. In Y1H assays, co‐transformed yeast cells containing pGADT7 empty vector and pAbAi‐MdWRKY17‐Pro, pAbAi‐MdPAL1‐Pro, or pAbAi‐MdRPM1‐Pro could not grow on ‐Leu medium with 200 ng/mL AbA; however, co‐transformed yeast cells containing pGADT7‐MdWRKY100 and pAbAi‐MdWRKY17‐Pro, pAbAi‐MdPAL1‐Pro or pAbAi‐MdRPM1‐Pro could grow on ‐Leu medium with 200 ng/mL AbA (Figure 2c). In the EMSA assays, MdWRKY100 was bound to Pro‐MdWRKY17 (Figure S5), Pro‐MdPAL1 (Figure S6), and Pro‐MdRPM1 (Figure S7) probes (Figure 2d). When the W‐box (TTGAC) in the MdWRKY17, MdPAL1, and MdRPM1 promoters was mutated to TTTTT, these bindings disappeared, indicating that the binding was specific. These results indicate that MdWRKY100 directly binds to the MdWRKY17, MdPAL1, and MdRPM1 promoters. Furthermore, we examined MdWRKY17, MdPAL1, and MdRPM1 expression levels in the WT and MdWRKY100 transgenic apple lines. The relative expression levels of MdRPM1 and MdPAL1 in MdWRKY100‐OE 4 seedlings were significantly higher than those in WT seedlings. The relative expression level of MdWRKY17 in MdWRKY100‐OE 4 seedlings was significantly lower than that in WT seedlings (Figure 2e). The relative expression levels of MdWRKY17, MdPAL1, and MdRPM1 in the MdWRKY100‐RNAi 3 seedlings displayed opposite trends. These results suggest that MdWRKY100 directly binds to the promoters of MdWRKY17, MdPAL1, and MdRPM1 to inhibit MdWRKY17 expression and activate MdPAL1 and MdRPM1 expression.
Figure 2.
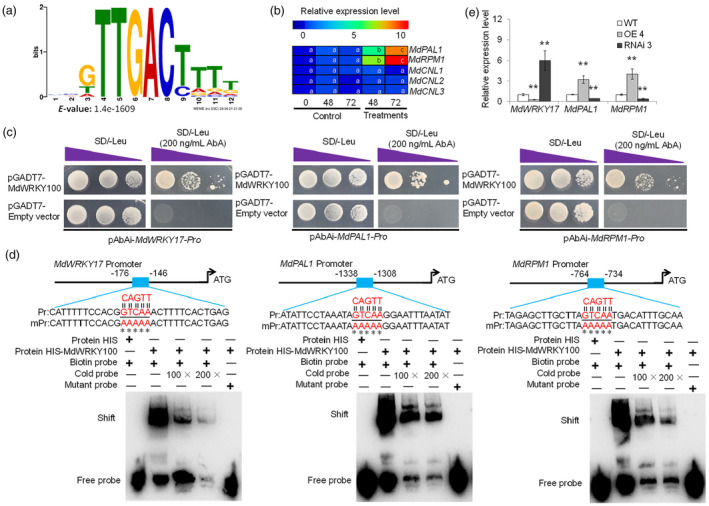
MdWRKY100 binds to the MdWRKY17, MdPAL1 and, MdRPM1 promoters. (a) Identified DNA‐binding motif of the MdWRKY100 protein by DAP‐seq. The e‐value of motif ‘GTTGACTTTT’ is 1.4e‐1609. Recombinant MdWRKY100 proteins were used to perform DAP‐seq. (b) Relative expression levels of MdPAL1, MdRPM1, and MdCNL1/2/3 after Colletotrichum fructicola infection. After qRT‐PCR data were re‐analysed, relative expression was calculated with respect to control samples (i.e., Control 0). Heat maps were generated using TIGR MeV v4.8.1 software. The bar at the top of the heat map presents relative expression values. Different letters in each panel indicate values are significantly different at P < 0.05, based on one‐way ANOVA and Duncan's test. (c) Y1H assay of MdWRKY100 binding to the MdWRKY17, MdPAL1, and MdRPM1 promoter fragments. The prey vector containing MdWRKY100 and the bait vectors pAbAi‐MdWRKY17‐Pro, pAbAi‐MdPAL1‐Pro, and pAbAi‐MdRPM1‐Pro were introduced into yeast strain Y1H, and the interaction between bait and prey enhanced Aureobasidin A (AbA) resistance. Yeast cells were grown on SD‐Leu media with 200 ng/mL AbA. The bait vector pAbAi‐MdWRKY17‐Pro plus pGADT7 empty vector, the bait vector pAbAi‐MdPAL1‐Pro plus pGADT7 empty vector, and the bait vector pAbAi‐MdRPM1‐Pro plus pGADT7 empty vector were also transformed into Y1H as negative controls. (d) EMSA confirming the in vitro binding of MdWRKY100 to the TTGAC motif of the MdWRKY17, MdPAL1, and MdRPM1 promoters. The labelled probe (Pr) containing the TTGAC motif was synthesized based on the sequence of the MdWRKY17, MdPAL1, and MdRPM1 promoters. In the mPr probe, TTGAC sequences are mutated to AAAAA. W‐box sequences are underlined. *Mutated sites. HIS alone was used as a negative control of the binding. (e) Expression patterns of MdWRKY17, MdRPM1, and MdPAL1 in MdWRKY100 transgenic lines. Error bars represent SD based on three biological replicates. ** in each panel indicates significantly different values relative to WT at P < 0.01, using Student's t‐test.
MdPAL1 and MdRPM1 positively regulate resistance to GLS in apples
To further investigate whether MdPAL1 is involved in resistance to GLS, we conducted transient overexpression and silencing MdPAL1 in the leaves of tissue‐cultured ‘Gala’ apple plantlets (Figure 3a). The transcription abundance of MdPAL1 and PAL activity increased in MdPAL1‐OE leaves and decreased in MdPAL1‐RNAi leaves 2 days after agroinfiltration compared with their controls (Figure 3b,c). We inoculated these leaves with C. fructicola for 5 days. We found that the overexpression of MdPAL1 significantly reduced the lesion area by increasing the free SA and total SA contents. In contrast, silencing MdPAL1 significantly increased the lesion area by reducing free SA and total SA contents compared with their controls (Figure 3d–f). These results indicate that MdPAL1 positively regulates apple resistance to GLS by increasing SA content.
Figure 3.
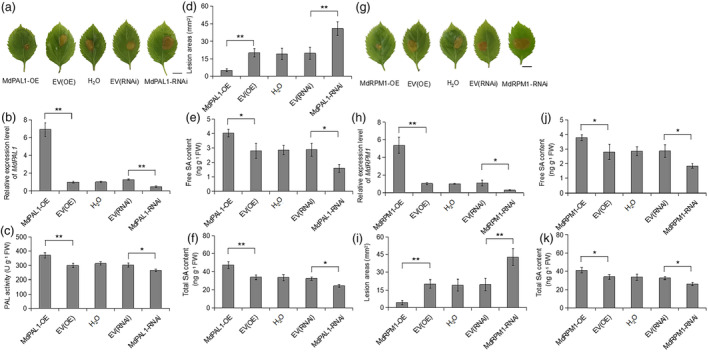
Transient overexpression and silencing of MdPAL1 and MdRPM1 in apple leaves (a) Disease symptoms on infiltrated leaves in which MdPAL1 was overexpressed or silenced at 48 h after inoculation with C. fructicola for 5 days. H2O; pK7GWIWG2D empty vector: EV(RNAi); pK7GWIWG2D‐MdPAL1 fusion vector: MdPAL1‐RNAi; pCambia2300 empty vector: EV(OE); pCambia2300‐MdPAL1 fusion vector: MdPAL1‐OE. Scale bar: 1 cm. (b) Relative expression levels of MdPAL1 in infiltrated leaves in which MdPAL1 was overexpressed or silenced at 48 h. Error bars represent SD based on three biological replicates. Statistics by Student's t‐test, **P < 0.01. (c) PAL activities in infiltrated leaves in which MdPAL1 was overexpressed or silenced at 48 h after inoculation with C. fructicola for 5 days. Error bars represent SD based on three biological replicates. Statistics by Student's t‐test, *P < 0.05 and **P < 0.01. (d) and (i) Lesion areas in infiltrated leaves after inoculation with C. fructicola for 5 days. Error bars represent SE (n = 18). The experiments were repeated thrice with similar results. Statistics by Student's t‐test, **P < 0.01. The contents of endogenous free SA (e) and total SA (f) in infiltrated leaves in which MdPAL1 was overexpressed or silenced at 48 h after inoculation with C. fructicola for 5 days. Error bars represent SD based on three biological replicates. Statistics by Student's t‐test, *P < 0.05 and **P < 0.01. (g) Disease symptoms on infiltrated leaves in which MdRPM1 was overexpressed or silenced at 48 h after inoculation with C. fructicola for 5 days. Scale bar: 1 cm. (h) Relative expression levels of MdRPM1 in infiltrated leaves in which MdRPM1 was overexpressed or silenced at 48 h. Error bars represent SD based on 3 biological replicates. Statistics by Student's t‐test, *P < 0.05 and **P < 0.01. The contents of endogenous free SA (j) and total SA (k) in infiltrated leaves in which MdRPM1 was overexpressed or silenced at 48 h after inoculation with C. fructicola for 5 days. Error bars represent SD based on three biological replicates. Statistics by Student's t‐test, *P < 0.05 and **P < 0.01.
We also investigated whether MdRPM1 participates in GLS resistance. Transient overexpression and silencing of MdRPM1 in the leaves of ‘Gala’ tissue‐cultured apple plantlets were performed (Figure 3g). The transcription abundance of MdRPM1 increased in MdRPM1‐OE leaves and decreased in MdRPM1‐RNAi leaves at 2 days after agroinfiltration compared with that in the controls (Figure 3h). Similarly, we inoculated these leaves with C. fructicola for 5 days and found that the overexpression of MdRPM1 significantly reduced the lesion area. In contrast, silencing of MdRMP1 significantly increased the lesion area compared to that of the controls (Figure 3i). Furthermore, the inoculation of C. fructicola in MdRPM1‐OE or MdRPM1‐RNAi leaves for 5 days altered endogenous free SA and total SA contents (Figure 3j,k). These results indicate that MdRPM1 positively regulates apple resistance to GLS.
MdWRKY100 physically interacts with MdVQ37
In our previous studies, we obtained two MdVQ37‐overexpressing transgenic lines and found that the overexpression of MdVQ37 reduced drought, salt, and heat tolerance (Dong et al., 2021, 2022a, 2022b). Previous studies have suggested that VQ proteins interact with group I and IIc WRKY transcription factors to regulate plant defence responses (Jing and Lin, 2015). Given that MdWRKY100 is a group I WRKY protein (Zhang et al., 2019b), we speculated that MdVQ37 may interact with MdWRKY100. Therefore, yeast two‐hybrid (Y2H) assay was used to detect the interaction between MdWRKY100 and MdVQ37. The full‐length MdWRKY100 cDNA with a deleted activation domain was fused to the pGBT9 bait vector (MdWRKY100▵1–227). As shown in Figure 4a, MdWRKY100▵1–227 protein interacted with MdVQ37 protein. To determine which regions of MdWRKY100 and MdVQ37 proteins are required for their interactions, we separated MdWRKY100 into its N‐WRKY (location 236–289), C‐WRKY (location 409–463), and C‐terminus domains (location 478–571) and divided MdVQ37 into its N‐terminus (location 1–31), VQ motif (location 32–41), and C‐terminus domain (location 91–181). The Y2H assay indicated that the deletion of the C‐terminus of MdVQ37 or the N‐WRKY and C‐terminus domains of MdWRKY100 did not affect these interactions. However, deleting the N‐terminus and VQ motif of MdVQ37 or the C‐WRKY domain of MdWRKY100 eliminated these physical interactions. These results demonstrate that the C‐WRKY domain of MdWRKY100 and the N‐terminus and VQ motif domains of MdVQ37 are critical for physical interactions in yeast.
Figure 4.
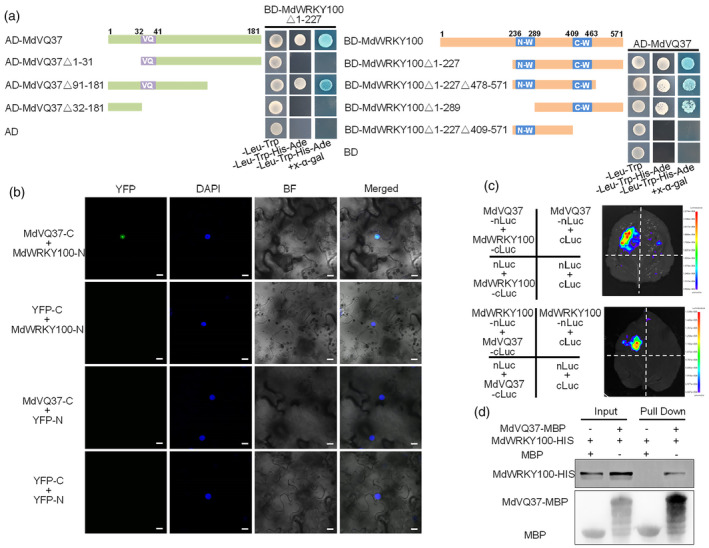
MdVQ37 physically interacts with MdWRKY100 in vitro and in vivo. (a) According to the domains of MdVQ37 (N‐terminus domain, VQ motif, and C‐terminus domain) and MdWRKY100 (N‐terminus domain, N‐WRKY, C‐WRKY, and C‐terminus domain), truncated MdVQ37 and MdWRKY100 with specific deletions were inserted into the pGAD424 prey vector and pGBT9 bait vector, respectively, and used to assess interactions. pGAD424 fusion prey vectors were co‐transformed with pGBT9 fusion bait vectors into yeast cells. Positive interactions were indicated by the ability of cells to grow on synthetic dropout medium with additive x‐α‐gal but lacking Leu, Trp, His, and Ade. Empty AD prey vector plus BD‐MdWRKY100▵1–227 fusion bait vector and Empty BD bait vector plus AD‐MdVQ37 fusion prey vector were used as negative controls. (b) Assay of bimolecular fluorescence complementation (BiFC), showing fluorescence in nuclear compartments of tobacco leaf epidermal cells that resulted from the complementation of the C‐terminus part of YFP fused to MdVQ37 (MdVQ37‐C) with N‐terminus part of YFP fused to MdWRKY100 (MdWRKY100‐N). No signal was observed from negative controls. Scale bar: 10 μm. (c) Split‐luciferase complementation assay showing the interaction of MdVQ37 and MdWRKY100. The construct combinations of MdVQ37 and MdWRKY100 were co‐transformed into N. benthamiana leaves. (d) Pull‐down assay for detecting the interaction between MdVQ37 and MdWRKY100. MdVQ37‐MBP or purified MBP was incubated with MdWRKY100‐His protein and purified using an MBP Purification Kit. Resultant protein samples were immunoblotted with anti‐MBP or anti‐HIS antibodies. The negative control was MBP.
We performed bimolecular fluorescence complementation (BiFC), split‐LUC, and pull‐down assays to verify the interaction between MdWRKY100 and MdVQ37 in vivo and in vitro. In the BiFC assays, when MdWRKY100‐N was co‐infiltrated with MdVQ37‐C in tobacco leaves, a YFP signal was detected in the nucleus; however, YFP‐C + MdWRKY100‐N, MdVQ37‐C + YFP‐N, and YFP‐C + YFP‐N showed no fluorescence (Figure 4b). In the split‐LUC assays, strong LUC activity was observed when MdWRKY100‐cLuc was co‐injected with MdVQ37‐nLuc or when MdWRKY100‐nLuc was co‐injected with MdVQ37‐cLuc into tobacco leaves (Nicotiana benthamiana) (Figure 4c). In pull‐down assays, MdWRKY100‐HIS was captured by MdVQ37‐MBP but not by MBP (Figure 4d). These observations suggest that MdWRKY100 physically interacts with MdVQ37 in vivo and in vitro.
MdVQ37 negatively regulates resistance to GLS in apples
The RT‐qPCR results showed that MdVQ37 expression was induced after inoculation with C. fructicola for 48 and 72 h (Figure S8a). Sub‐cellular localization results indicated that MdVQ37 was localized in the nucleus (Figure S8b). To investigate the biological role of MdVQ37 in GLS tolerance, two MdVQ37‐overexpressing transgenic lines were used. Upon C. fructicola inoculation for 5 days, transgenic apple lines were found hypersensitive to C. fructicola compared to WT plants (Figure 5a). Compared to that in WT plants, the lesion area in MdVQ37‐overexpressing transgenic lines was significantly increased, whereas PAL activity, free SA content, and total SA content were significantly decreased (Figure 5b–e). These results suggest that apple MdVQ37 negatively regulates resistance to GLS by altering SA content and PAL activity.
Figure 5.

MdVQ37 overexpression reduces resistance to Colletotrichum fructicola infection in transgenic apples. Phenotypes (a) and lesion areas (b) of detached leaves of wild‐type (WT) and MdVQ37 overexpression lines (L1 and L2) after inoculation with C. fructicola for 5 days. Scale bar: 1 cm. Error bars represent SE (n = 18). The experiments were repeated thrice, and similar results were obtained. ** in each panel indicates significantly different values relative to WT at P < 0.01, using Student's t‐test. The PAL activity (c) and contents of endogenous free SA (d) and total SA (e) in the detached leaves of the WT and MdVQ37 overexpression lines (L1 and L2) after inoculation with C. fructicola for 5 days. Error bars represent the standard deviation (SD) based on three independent replicates. * and ** in each panel indicates significantly different values relative to WT at P < 0.05 and P < 0.01, respectively, using Student's t‐test.
MdVQ37 represses the transcriptional activity of MdWRKY100
Having demonstrated the MdWRKY100‐MdVQ37 interaction (Figure 4), further investigations were conducted to understand whether MdVQ37 could affect the transcriptional function of MdWRKY100 using a transient expression system. To test this possibility, the MdWRKY17, MdRPM, and MdPAL1 promoters were inserted into the pGreenII 0800‐LUC vector and co‐transformed into tobacco leaves with pGreenII 62‐SK‐MdWRKY100 or a combination of pGreenII 62‐SK‐MdWRKY100/pGreenII 62‐SK‐MdVQ37 (Figure 6a). We found that MdWRKY100 induced the accumulation of MdRPM1 and MdPAL1 transcripts, with increased transactivation by almost 2.8‐fold and 2.2‐fold, respectively; however, MdWRKY100 inhibited the accumulation of MdWRKY17 transcripts (Figure 6b). Notably, the combination of MdVQ37 and MdWRKY100 downregulated the transcript levels of MdRPM1 and MdPAL1 and upregulated the transcript levels of MdWRKY17 in vivo (Figure 6b). Furthermore, we examined the expression levels of MdWRKY100, MdWRKY17, MdRPM1, and MdPAL1 in the WT and MdVQ37 transgenic apple lines. The relative expression levels of MdWRKY100, MdRPM1, and MdPAL1 in MdVQ37 transgenic apple lines were significantly lower than those in the WT plants. The relative expression level of MdWRKY17 in MdVQ37 transgenic apple lines was significantly higher than that in the WT plants (Figure 6c). These results indicate that MdVQ37 acts as a negative mediator of MdWRKY100 and represses its transcriptional activity.
Figure 6.
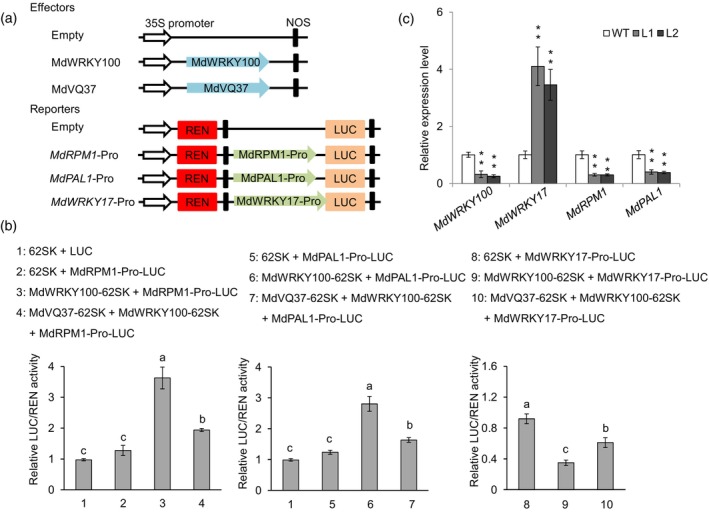
MdVQ37 inhibits the transcriptional activity of MdWRKY100. (a) Schematic showing the constructs used in the transient expression assays. The MdVQ37 and MdWRKY100 genes were cloned into the pGreenII 62‐SK vector to generate the effector constructs. The reporters were generated by recombining the promoter fragments of MdRPM1, MdPAL1, and MdWRKY17 into the pGreenII 0800‐LUC vector. (b) Dual‐luciferase reporter assay results. Different combinations of reporter and effecter plasmids were co‐infiltrated into the leaves of N. benthamiana. LUC/REN activity detection to MdVQ37 and MdWRKY100 co‐transformation to repress the transcriptional activity of MdWRKY100. Empty vector was used as the reference. Error bars represent SD based on three biological replicates. Different letters indicate significant differences between different combinations, according to one‐way ANOVA and Duncan's test (P < 0.05). (c) Expression patterns of MdWRKY100, MdWRKY17, MdRPM1, and MdPAL in MdVQ37 overexpression lines. Error bars represent SD based on three biological replicates. * and ** in each panel indicates significantly different values relative to WT at P < 0.05 and P < 0.01, respectively, using Student's t‐test.
Discussion
GLS is one of the most destructive diseases affecting apple production and restricting apple income in China. Multiple studies have shown that the WRKY transcription factor, as an important regulator of host–pathogen interactions, is an important component of the plant defence response signalling network and can positively or negatively regulate the defence response to multiple plant pathogens (Jiang et al., 2017; Rushton et al., 2010). In Arabidopsis and tomato, WRKY transcription factors reportedly interact with VQ proteins to jointly regulate the response to pathogens jointly (Huang et al., 2022; Jing and Lin, 2015). With the advancements in research, it has been proven that the interaction mechanism between WRKY and VQ in Arabidopsis, soybean, and apple only exists in some members of the WRKY family in groups I and IIc (Cheng et al., 2012; Dong et al., 2018; Zhou et al., 2016). These studies suggest that the interaction between WRKY and VQ regulates plant response to pathogens, a conserved molecular mechanism. Therefore, exploring the physical interactions between the apple WRKY transcription factor and VQ proteins may provide novel insights into the molecular mechanisms underlying plant defence responses. In apples, MdWRKY100, which encodes a group I WRKY subscription factor, positively regulates the resistance to Colletotrichum gloeosporioides infection (Zhang et al., 2019b). Our previous study demonstrated that apple VQ proteins interact with group I WRKY transcription factors. Based on our acquisition of MdVQ37 overexpression apple transgenic plants, we speculated that the MdWRKY100 transcription factor may interact with MdVQ37. This speculation was confirmed through yeast two‐hybrid experiments, and the results showed that the C‐terminus WRKY domain of MdWRKY100 and the VQ motif of MdVQ37 were crucial for the interaction between MdWRKY100 and MdVQ37 (Figure 4a). Furthermore, we demonstrated through BiFC, LCI, and pull‐down experiments that MdWRKY100 and MdVQ37 interact both in vivo and in vitro (Figure 4b–d). To characterize the role of the MdWRKY100‐MdVQ37 complex in the defence response of apples against C. fructicola, we investigated the performance of overexpression and silencing of MdWRKY100, as well as the overexpression of MdVQ37 in response to C. fructicola infection. Phenotypic analysis showed that the MdWRKY100 overexpression lines showed enhanced resistance to C. fructicola, whereas the MdWRKY100 RNAi and MdVQ37 overexpression lines showed weakened resistance to C. fructicola by increasing SA content (Figures 1 and 5). These results demonstrate the regulatory role of the MdWRKY100‐MdVQ37 complex in the response of apples to C. fructicola infection by affecting the biosynthesis or degradation of SA.
WRKY transcription factor can specifically bind to the cis‐acting element W‐box on the target gene promoter to activate or inhibit the expression of the target gene and participate in regulating the response of plants to biotic stress (Jiang et al., 2017; Rushton et al., 2010). For example, AtWRKY57 competes with AtWRKY33 to regulate the expression of the downstream target genes JASMONATE ZIM DOMAIN 1 (JAZ1) and JAZ5, thereby affecting the JA‐mediated defence signalling pathway involved in the regulation of B. cinerea resistance (Jiang and Yu, 2016). OsWRKY51 binds to the W‐box element of the OsPR10a promoter and functions as a positive regulator of the defence response against Xanthomonas oryzae pv. oryzae (Hwang et al., 2016). Apple MdWRKY79 can bind to and activate the NLR resistance gene, MdNLR16, thereby enhancing resistance to Alternaria alternata (Meng et al., 2018). In this study, we identified MdWRKY100 as a positive regulator of GLS infection. To further explore the molecular mechanism through which MdWRKY100 regulates GLS infection, we conducted DAP‐seq sequencing and found that MdWRKY17, MdPAL1, and MdRPM1 were potential target genes of MdWRKY100 (Figure 2a). Furthermore, we demonstrated through Y1H, EMSA, transient expression, and RT‐qPCR experiments that MdWRKY100 could bind to the W‐box motif of these target gene promoters, thereby inhibiting the expression of MdWRKY17 and promoting the expression of MdPAL1 and MdRPM1 (Figures 2b–e and 6a,b). This result suggests that MdWRKY100 is involved in the response to GLS infection by regulating these three target genes.
Plant SA homeostasis is dependent on SA biosynthesis and metabolism. Under biotic stress, both an increase in SA biosynthesis and a decrease in metabolic levels can enhance plant resistance to biotic stress. During the SA biosynthesis process, PAL is an important enzyme for SA biosynthesis in Arabidopsis, tobacco, pepper, soybean, and wheat (Kim and Hwang, 2014; Mauch‐Mani and Slusarenko, 1996; Shadle et al., 2003; Shine et al., 2016). For example, the overexpression of pepper CaPAL1 increases PAL activity and SA content to enhance resistance to microbial pathogen infection in transgenic Arabidopsis plants (Kim and Hwang, 2014). The overexpression of GmPAL2.1 remarkably increased SA content to improve resistance to Phytophthora sojae infection in soybean plants (Zhang et al., 2017a). Silencing of wheat AevPAL1 reduces nematode resistance in roots by decreasing the accumulation of SA (Zhang et al., 2021). The rice TPR‐domain RNA‐binding protein, BSR‐K1, regulates OsPAL1 mRNA levels to confer broad‐spectrum disease resistance (Zhou et al., 2018). However, the role of PAL in GLS tolerance remains unclear. In this study, we discovered that MdPAL1 positively contributes to GLS resistance by increasing SA accumulation (Figure 3a–f), indicating that MdPAL1 plays an essential role in GLS infection in apples. Mutations in the tomato SA degradation gene SlDMR6 confer broad‐spectrum resistance to fungi, bacteria, and oomycetes (Thomazella et al., 2021). The CRISPR/Cas9‐mediated simultaneous mutation of three salicylic acid 5‐hydroxylase (OsS5H/OsDMR6) confers resistance to rice blast and bacterial blight diseases (Liu et al., 2023). Wheat‐susceptible gene TaWRKY76 directly activates TaDMR6 expression to inhibit SA accumulation and reduce aphid resistance (Zhang et al., 2023b). The apple susceptibility gene MdWRKY17 directly activates the MdDMR6 expression to promote SA degradation and weaken resistance to GLS (Shan et al., 2021). The present study demonstrated that MdWRKY100 inhibited the expression of MdWRKY17 and reduced the degradation of SA; however, MdWRKY100 promoted the expression of MdPAL1, increased the accumulation of SA, and comprehensively improved the resistance to GLS in apples (Figures 1, 2, 3 and 6a,b).
CC‐NBS‐LRR proteins have been proven to play crucial roles in plant responses to pathogen infection in multiple plant species (Du et al., 2021; Grant et al., 1995; Wang et al., 2020, 2021). For example, transgenic rice lines overexpressing OsRLR1 (RPM1 like resistance gene 1) enhanced resistance to rice blast fungi and Xanthomonas oryzae through direct interaction with OsWRKY19, which facilitated the function of OsWRKY19 in the activation of the OsPR10 gene (Du et al., 2021). The dominant rice resistance gene, OsRSR1 (disease resistance gene RPM1), was correlated with sheath blight resistance in a genome‐wide association study, and OsRSR1 was validated through overexpression and knockdown assays (Wang et al., 2021). Wheat TaRPM1 positively contributes to Puccinia striiformis f. sp. tritici resistance in high‐temperature wheat seedlings through the BSMV virus‐induced gene silencing (VIGS) system (Wang et al., 2020). In addition, silencing of a CC‐NBS‐LRR gene, GbRVd, in cotton through VIGS substantially downregulated the SA content, resulting in increased susceptibility to Verticillium dahlia (Yang et al., 2016). However, the role of apple CC‐NBS‐LRR protein, MdRPM1, in GLS tolerance remains unclear. The present study demonstrated that MdRPM1 positively regulates resistance to GLS in apples by affecting SA content through overexpression and RNAi assays, indicating that MdWRKY100 increases resistance to GLS by increasing SA content and upregulating MdRPM1 expression (Figures 1, 2, 3 and 6a,b).
Transcriptional regulators typically inhibit or promote the transcriptional activity of WRKY transcription factors to regulate the transcript levels of their target genes (Jing and Lin, 2015; Li et al., 2014; Pan et al., 2018; Wang et al., 2022). For example, Arabidopsis VQ18 and VQ26 physically interact with ABI5 and inhibit its transcriptional activity, thereby negatively modulating the ABA response during seed germination (Pan et al., 2018). VQ29 interacted with PIF1, and the interaction decreased the stability of protein PIF1 and repressed XYLOGLUCAN ENDOTRANSGLYCOSYLASE7 expression to mediate the inhibition of hypocotyl elongation during early seedling development in Arabidopsis (Li et al., 2014). SlVQ7 acts as a positive interacting partner of the SlWRKY37 transcription factor and increases the transcript levels of SlWRKY53 and SlSGR1 to regulate dark‐induced leaf senescence in tomatoes (Wang et al., 2022). In the present study, MdVQ37 acted as a negative interacting partner of MdWRKY100 and repressed its transcriptional activity of MdWRKY100 to negatively regulate GLS tolerance in apples (Figures 4, 5, 6).
In summary, we proposed a functional model of the MdWRKY100‐MdVQ37 complex in apples, which responds to GLS infection via two pathways (Figure 7). The overexpression of MdWRKY100 in transgenic apple plants activated the expression of the target gene MdRPM1 to improve GLS tolerance. In contrast, MdWRKY100 upregulated MdPAL1 expression and downregulated MdWRKY17 expression, leading to SA accumulation that induces resistance to GLS infection. In contrast, the overexpression of MdVQ37 in transgenic apple plants inhibited the transcriptional activity of MdWRKY100 through the interaction between its VQ motif and the C‐terminus WRKY domain of MdWRKY100, which reduced the expression of MdRPM1 and SA accumulation, thereby increasing the sensitivity to GLS infection.
Figure 7.
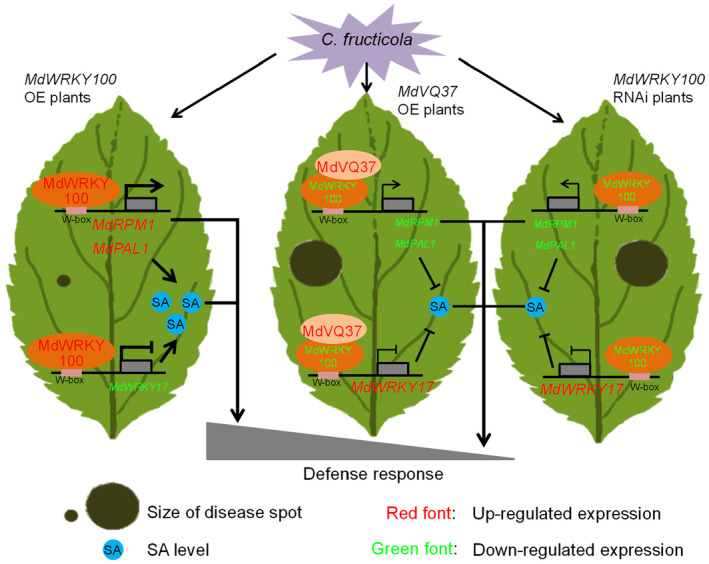
Model for MdWRKY100‐MdVQ37 module‐mediated GLS resistance in apples. On the one hand, the overexpression of MdWRKY100 improves C. fructicola tolerance, on the one hand by activating MdRPM1 expression, and on the other hand, the activation of MdPAL1 expression reduces MdWRKY17 expression, thereby increasing SA accumulation. However, the overexpression of MdVQ37 or RNAi silencing of MdWRKY100 enhances C. fructicola susceptibility to inhibit the transcriptional activity of MdWRKY100, along with decreases in MdRPM1 expression and SA accumulation.
Materials and methods
Plant materials and stress treatments
Apples (Malus domestica Borkh.) variety ‘Gala’ (GL‐3) was used for genetic transformation and expression analysis (Dai et al., 2013). The overexpression of MdVQ37 in transgenic apple lines (Dong et al., 2021) and overexpression of RNAi MdWRKY100 in transgenic apple lines (Zhang et al., 2019b) were performed in our previous studies. For the expression analysis of MdVQ37, MdWRKY100, MdRPM1, and MdPAL1 under C. fructicola infection, C. fructicola was cultured on potato dextrose agar medium at 25 °C for 7 d, and the fungal spores were collected and suspended in sterile water until the spore concentration was 106 cfu mL−1 for fungal inoculation. The detached, fully expanded leaves of 60‐day‐old GL‐3 plants were sprayed with spore suspension, incubated in a plastic box containing moist filter paper, and covered in a plastic box with vinyl film to maintain humidity at 25 °C. At 0, 48, and 72 h of treatment, detached leaves were collected for gene expression analysis.
To phenotypically assess MdVQ37 transgenic apple plants and transiently overexpressed or RNA‐interfered MdRPM1 and MdPAL1, the detached leaves from overexpression MdVQ37 transgenic apple lines and WT plants, as well as those from in vitro‐cultured GL‐3 and control seedlings with overexpression or RNA interference of MdRPM1 and MdPAL1 at 48 h were inoculated with a 10 μL spore suspension at the center, following the methodology described by Shan et al. (2021). After 5 days of treatment, the detached leaves were collected for phenotypic comparison and SA content analysis.
To phenotype MdWRKY100 transgenic apple plants, transgenic apple seedlings with overexpression or RNA interference of MdWRKY100 were sprayed with a spore suspension, incubated in Petri dishes containing moist filter paper at 25 °C, and then sampled at 0, 4, and 6 days after infection for detecting disease index and SA content.
Gene cloning and expression analysis
To clone MdVQ37, MdWRKY100, MdRPM1, and MdPAL1, their complete open reading frames (ORFs) were obtained from the fully expanded leaves of ‘Royal Gala’ apples via RT‐PCR using the specific primers listed in Table S1. The qRT‐PCR experiments were conducted according to the manufacturer's instructions using a QuantStudio 5 instrument (Applied Biosystems, Foster City, CA, USA) in conjunction with TransStart Top Green qRT‐PCR SuperMix (Transgen Biotech, Beijing, China), as previously described (Dong et al., 2018). The obtained data were calculated and analysed by the 2−▵▵CT method (Livak and Schmittgen, 2001), and the quantification results were normalized by MdMDH (Duan et al., 2023). Three biological replicates were used in each experiment.
Vector construction and genetic transformation
ORFs without the stop codon of MdRPM1 or MdPAL1 were ligased into the overexpression (OE) vector pCambia2300. Specific 300 bp fragments of MdRPM1 and MdPAL1 were introduced into the RNAi vector pK7GWIWG2D. Overexpression or RNA interference of MdRPM1 and MdPAL1 was performed by transiently transforming the leaves of in vitro‐cultured GL‐3 seedlings using Agrobacterium strain EHA105 carrying pCambia2300‐MdRPM, pCambia2300‐MdPAL1, pK7GWIWG2D‐MdRPM1, or pK7GWIWG2D‐MdPAL1, as described by Zhang et al. (2018) and Shan et al. (2021). The expression levels of MdRPM1 and MdPAL1 and PAL activity were determined 2 days after transient transformation, and GLS tolerance was recorded 5 days after infection, as described above. The experiment was independently repeated thrice.
Sub‐cellular localization assays
The CDS without the stop codons of MdVQ37 was cloned into the pCambia2300‐GFP vector and transformed into A. tumefaciens strain ‘GV3101’ by heat shock. Sub‐cellular localization assays were carried out according to the method previously reported (Duan et al., 2023).
Yeast two‐hybrid assays
Y2H assays were performed following the manufacturer's instructions (Clontech Laboratories, Mountain View, CA, USA). The full‐length MdWRKY100 protein showed strong self‐activation, and truncated fragments of MdWRKY100 were used to test these interactions. The truncated fragments of MdWRKY100 were introduced into the pGBT9 bait vector. The full‐length cDNA and truncated fragments of MdVQ37 were cloned into a pGAD424 prey vector. Different combinations of these constructs were transformed into the yeast strain ‘Y2H Gold’ using the lithium acetate method. The transformed yeast cells were cultured on yeast synthetic dropout medium lacking Trp and Leu (SD/−Trp‐Leu) and SD/−Trp‐Leu‐His‐Ade to observe yeast growth at 30 °C for 3 days. An assay of β‐galactosidase activity was performed using X‐α‐gal. Empty pGBT9 and pGAD424 vectors were used as negative controls (Dong et al., 2020).
Bimolecular fluorescence complementation assays
The BiFC assays were performed as described by Li et al. (2012). Briefly, the coding sequences of MdWRKY100 and MdVQ37 were amplified by PCR and inserted into pSPYNE‐35S and pSPYCE‐35S, respectively. The obtained fusion vectors, empty pSPYNE‐35S vector, and empty pSPYCE‐35S vector were transformed into A. tumefaciens strain ‘GV3101’ and co‐injected into tobacco leaves in different combinations. After 48 h of infection, yellow fluorescence in the infected leaves was observed using a confocal laser scanning microscope (LSM510 META; Zeiss, Oberkochen, Germany). 4′,6‐diamidino‐2‐phenylindole (DAPI) was used to stain cell nuclei.
Split‐luciferase complementation assay
Split‐luciferase completion assays were performed as described by Jing et al. (2022). Briefly, the coding sequences of MdWRKY100 and MdVQ37 were amplified by PCR and inserted into the pCAMBIA1300‐nLuc or pCAMBIA1300‐cLuc vectors. The obtained fusion vectors, empty pCAMBIA1300‐nLuc vector, and empty pCCAMBIA‐cLuc vector were transformed into A. tumefaciens strain ‘EHA105’ and co‐injected into tobacco leaves in different combinations. After 3 days of infection, 1 mM fluorescein (Promega) was sprayed onto the leaves, and the fluorescence signal in the infected leaves was observed using a CCD (Lumazone Pylon 2048 B).
Pull‐down assay
MdWRKY100‐HIS and MdVQ37‐MBP fusion proteins were produced in Escherichia coli strain BL21 and purified using HIS and MBP purification columns (Beyotime, Shanghai, China). Protein mixtures were captured using the MBP Purification Kit (Thermo Fisher Scientific). Eluted proteins were detected using anti‐HIS and anti‐MBP antibodies.
DAP‐seq sampling and data analysis
DNA purification and sequencing were performed by Bluescape Scientific Co., Ltd. (Hebei, China). The plant genomic DNA extraction kit (Tiangen Biotechnology) was used to extract genomic DNA from mature leaves of ‘Gala’ (Dong et al., 2023). Clean DNA beads (Bluescape Scientific Co., Ltd., Hebei, China) were used to purify the gDNA, and a NEXTFLEX Rapid DNA Seq Kit (PerkinElmer, Inc., Austin, TX, USA) was used to construct an affinity‐purified DNA library. The MdWRKY100 coding sequence was cloned into the pFN19K HaloTag T7 SP6 Flexi vector and expressed using a TNT SP6 Coupled Wheat Germ Extraction System (Promega, Madison, WI, USA). The expressed Halo‐MdWRKY100 fusion protein was purified and captured using Magne Halo‐tag beads (Promega, Madison, WI, USA). The MdWRKY100‐bound beads were incubated with adapter‐ligated gDNA libraries, and the technology was repeated twice. A negative control lacking MdWRKY100‐bound beads was incubated with adapter‐ligated gDNA libraries. The eluted DNA was sequenced in Illumina NavoSeq6000. The Bowtie2 software aligned the DAP‐seq reads with the apple GDDH13 v1.1 genome (Langmead and Salzberg, 2012). The MACS2 callpeak and IDR software were used to merge the peaks (P < 0.05) and score the reliability of the repeated peaks (Zhang et al., 2008b). MEME‐CHIP and Homer software were used to analyse the conserved motifs in the peak areas (Machanick and Bailey, 2011) and annotate the bound peaks (Heinz et al., 2010), respectively.
Yeast one hybrid assays
Y1H assay was performed according to the manufacturer's instructions (Clontech Laboratories, Mountain View, CA, USA). The coding sequence of MdWRKY100 was amplified using PCR and inserted into the pGADT7 vector. The promoters of MdWRKY17 and MdRPM1 were inserted into the pAbAi vector. The recombinant plasmids were transformed into the yeast strain Y1H. Transformed yeast cells were cultured in yeast synthetic dropout medium lacking Leu (SD/−Leu) and 200 ng/mL AbA to confirm positive interactions. Empty pGADT7 vector served as a negative control.
Electrophoretic mobility shift assay
MdWRKY100‐HIS was obtained as described for the pull‐down assay. EMSAs were performed using the LightShift Chemiluminescent EMSA kit (Thermo Scientific, Waltham, MA, USA) according to the manufacturer's instructions. Unlabeled probes were used as competitors, and HIS was used as a negative control, and the biotin‐labelled DNA probes are listed in Table S2.
Dual‐luciferase transient expression assay
The dual‐luciferase transient expression assay assays were performed as described by Yang et al. (2023). The promoters of MdWRKY17 and MdRPM1 were individually inserted into the pGreenII 0800‐LUC vector, and the ORF sequences of MdVQ37 and MdWRKY100 were cloned into the pGreenII 62‐SK vector. The empty pGreenII 62‐SK vector was used as a negative control. These vectors were transformed into Agrobacterium strain GV3101 and co‐injected into tobacco leaves in different combinations. After 2 days of incubation, LUC and Renilla luciferase (REN) activities were measured using a dual‐luciferase assay kit (Yeasen, Shanghai, China). Transcriptional activation of the target promoters was calculated as the ratio of LUC to REN. At least six transient assays were performed for each combination.
Measurement of SA contents
SA content was determined using 0.1 g leaf samples taken from WT and MdVQ37 overexpression apple plants after inoculation with C. fructicola for 5 days, leaf tissue collected from WT and MdWRKY100 transgenic apple seedlings after inoculation with C. fructicola for 6 days, and infiltrated leaves from MdPAL1‐OE, EV(OE), H2O, MdPAL1‐RNAi, and EV(RNAi) after inoculation with C. fructicola for 5 days. SA was extracted, purified, and analysed using liquid chromatography‐mass spectrometry according to a previously described method (Dong et al., 2021).
PAL activity assay
PAL activity was measured using a PAL activity kit (cas: PAL‐2‐Y) (Comin, Suzhou, China) according to the manufacturer's instructions. Briefly, 0.1 g of apple leaves was suspended in a 1 mL extraction buffer and centrifuged at 10000g for 10 min. The supernatant was collected as a crude extract for PAL activity assay.
Statistical analyses
All data were analysed using IBM SPSS (version 20) statistical software (IBM, Chicago, IL, USA). One‐way analysis of variance (ANOVA) was used to compare statistically significant differences between the control plants and transgenic lines using Duncan's test (P < 0.05) or Student's t‐test (P < 0.05, P < 0.01).
Accession numbers
Sequence data from this article can be found in the APPLE GENOME DATABASE (https://iris.angers.inra.fr/gddh13/) as follows: MdWRKY100 (MDP0000514115), MdVQ37 (MDP0000248043), MdWRKY17 (MD12G1181000), MdPAL1 (MD07G1172700), MdRPM1 (MD02G1199100), MdCNL1 (MD07G1056600), MdCNL2 (MD11G1069800), and MdCNL3 (MD08G1239100).
Conflict of interest
The authors declare that they have no competing interests.
Author contributions
F.M., Y.M. Y.T., X.Z., and Q.D. conceived and designed the experiments. Q.D., D.D., F.W., K.Y., Y.S., Y.W., and Z.J. performed the experiments. Q.D., D. W., C.X., P.J., H.L., S.G., G.Q., and K.M. analysed the data. Y.M. and Q.D. wrote the manuscript. Q.D., D.W., K.M., and Y.T. revised the manuscript. All authors read and approved the final version of the manuscript.
Conflict of interest
The authors declare no competing financial interests.
Supporting information
Figure S1 Relative expression level of MdWRKY100 after C. fructicola infection.
Figure S2 Genome‐wide analysis of MdWRKY100‐binding sites.
Figure S3 The phylogenetic tree of apple and Arabidopsis phenylalanine ammonia‐lyase proteins.
Figure S4 The phylogenetic tree of MdRPM1, MdCNL1, MdCNL2, MdCNL3, and Arabidopsis NBS‐LRR proteins.
Figure S5 The promoter sequence of MdWRKY17 gene.
Figure S6 The promoter sequence of MdPAL1 gene.
Figure S7 The promoter sequence of MdRPM1 gene.
Figure S8 Expression pattern and sub‐cellular localization of MdVQ37.
Table S1 MdWRKY100 DAP‐seq peaks located upstream of the transcription start site (TSS) of MdWRKY17, MdPAL1 and CC‐NBS‐LRR resistance genes.
Table S2 A list of primers used in this study.
Acknowledgements
This work was supported by grants from the Natural Science Foundation of Hebei Province (C2022204086 and C2023204128), the National Natural Science Foundation of China (31401852, 32072524 and 32272655), the Hebei Agriculture Research System (HBCT2024150206), and the Key Research and Development Program of Xingtai City (2022zz064).
Contributor Information
Xuemei Zhang, Email: zhangxuemei888@163.com.
Yi Tian, Email: tianyi@hebau.edu.cn.
Yue Ma, Email: yuema@syau.edu.cn.
Fengwang Ma, Email: fwm64@sina.com.
Data availability statement
The data that supports the findings of this study are available in the supplementary material of this article.
References
- Birkenbihl, R.P. , Diezel, C. and Somssich, I.E. (2012) Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol. 159, 266–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove, S.R. , Simonich, M.T. , Smith, N.M. , Sattler, A. and Innes, R.W.A. (1994) A disease resistance gene in Arabidopsis with specificity for two different avirulence genes. Plant Cell 6, 927–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y. , Zhou, Y. , Yang, Y. , Chi, Y.J. , Zhou, J. , Chen, J.Y. , Wang, F. et al. (2012) Structural and functional analysis of VQ motif‐containing proteins in Arabidopsis as interacting proteins of WRKY transcription factors. Plant Physiol. 159, 810–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, H.Y. , Li, W.R. , Han, G.F. , Yang, Y. , Ma, Y. , Li, H. and Zhang, Z.H. (2013) Development of a seedling clone with high regeneration capacity and susceptibility to Agrobacterium in apple. Sci. Hortic. 164, 202–208. [Google Scholar]
- Dangl, J.L. and Jones, J.D. (2001) Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. [DOI] [PubMed] [Google Scholar]
- Dodds, P. and Rathjen, J. (2010) Plant immunity: towards an integrated view of plant‐pathogen interactions. Nat. Rev. Genet. 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Dong, Q. , Duan, D. , Zheng, W. , Huang, D. , Wang, Q. , Li, X. , Mao, K. et al. (2021) MdVQ37 overexpression reduces basal thermotolerance in transgenic apple by affecting transcription factor activity and salicylic acid homeostasis. Hortic Res. 8, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, Q. , Duan, D. , Zheng, W. , Huang, D. , Wang, Q. , Yang, J. , Liu, C. et al. (2022a) Overexpression of MdVQ37 reduces drought tolerance by altering leaf anatomy and SA homeostasis in transgenic apple. Tree Physiol. 42, 160–174. [DOI] [PubMed] [Google Scholar]
- Dong, Q. , Duan, D. , He, J. , Zheng, W. , Huang, D. , Wang, Q. , Yang, J. et al. (2022b) Overexpression of MdVQ37 reduces salt stress tolerance in Malus domestica . Sci. Hortic. 300, 111077. [Google Scholar]
- Dong, Q. , Tian, Y. , Zhang, X. , Duan, D. , Zhang, H. , Yang, K. , Jia, P. et al. (2023) Overexpression of the transcription factor MdWRKY115 improves Drought and osmotic stress tolerance by directly binding to the MdRD22 promoter in apple. Hortic. Plant J. 10.1016/j.hpj.2023.05.005. (In press). [DOI] [Google Scholar]
- Dong, Q. , Zhao, S. , Duan, D. , Tian, Y. , Wang, Y. , Mao, K. , Zhou, Z. et al. (2018) Structural and functional analyses of genes encoding VQ proteins in apple. Plant Sci. 272, 208–2019. [DOI] [PubMed] [Google Scholar]
- Dong, Q. , Zheng, W. , Duan, D. , Huang, D. , Wang, Q. , Liu, C. , Li, C. et al. (2020) MdWRKY30, a group IIa WRKY gene from apple, confers tolerance to salinity and osmotic stresses in transgenic apple callus and Arabidopsis seedlings. Plant Sci. 299, 110611. [DOI] [PubMed] [Google Scholar]
- Du, D. , Zhang, C. , Xing, Y. , Lu, X. , Cai, L. , Yun, H. , Zhang, Q. et al. (2021) The CC‐NB‐LRR OsRLR1 mediates rice disease resistance through interaction with OsWRKY19. Plant Biotechnol. J. 19, 1052–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, D. , Yi, R. , Ma, Y. , Dong, Q. , Mao, K. and Ma, F. (2023) Apple WRKY transcription factor MdWRKY56 positively modulates drought stress tolerance. Environ. Exp. Bot. 212, 105400. [Google Scholar]
- Grant, M.R. , Godiard, L. , Straube, E. , Ashfield, T. , Lewald, J. and Sattler, A. (1995) Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269, 843–846. [DOI] [PubMed] [Google Scholar]
- Heinz, S. , Benner, C. , Spann, N. , Bertolino, E. , Lin, Y.C. , Laslo, P. , Cheng, J.X. et al. (2010) Simple combinations of lineage‐determining transcription factors prime cis‐regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y. , Chen, L. , Wang, H. , Zhang, L. and Yu, D. (2013) Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J. 74, 730–745. [DOI] [PubMed] [Google Scholar]
- Huang, H. , Zhao, W. , Li, C. , Qiao, H. , Song, S. , Yang, S. , Sun, L. et al. (2022) SlVQ15 interacts with jasmonate‐ZIM domain proteins and SlWRKY31 to regulate defense response in tomato. Plant Physiol. 190, 828–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, S.H. , Kwon, S.I. , Jang, J.Y. , Fang, I.L. , Lee, H. , Choi, C. , Park, S. et al. (2016) OsWRKY51, a rice transcription factor, functions as a positive regulator in defense response against Xanthomonas oryzae pv. oryzae . Plant Cell Rep. 35, 1975–1985. [DOI] [PubMed] [Google Scholar]
- Jiang, Y. and Yu, D. (2016) The WRKY57 transcription factor affects the expression of jasmonate ZIM‐domain genes transcriptionally to compromise Botrytis cinerea resistance. Plant Physiol. 171, 2771–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, J. , Ma, S. , Ye, N. , Jiang, M. , Cao, J. and Zhang, J. (2017) WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 59, 86–101. [DOI] [PubMed] [Google Scholar]
- Jing, Y. and Lin, R. (2015) The VQ motif‐containing protein family of plant‐specific transcriptional regulators. Plant Physiol. 169, 371–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing, Y. , Zhan, M. , Li, C. , Pei, T. , Wang, Q. , Li, P. , Ma, F. et al. (2022) The apple FERONIA receptor‐like kinase MdMRLK2 negatively regulates Valsa canker resistance by suppressing defence responses and hypersensitive reaction. Mol. Plant Pathol. 23, 1170–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D.S. and Hwang, B.K. (2014) An important role of the pepper phenylalanine ammonia‐lyase gene (PAL1) in salicylic acid‐dependent signalling of the defence response to microbial pathogens. J. Exp. Bot. 65, 2295–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B. and Salzberg, S.L. (2012) Fast gapped‐read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, R. , Ma, Z. and Yu, D. (2018) WRKY2/34–VQ20 modules in Arabidopsis thaliana negatively regulate expression of a trio of related MYB transcription factors during pollen development. Front. Plant Sci. 9, 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y.Y. , Mao, K. , Zhao, C. , Zhao, X.Y. , Zhang, H.L. , Shu, H.R. and Hao, Y. (2012) MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light‐induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol. 160, 1011–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Jing, Y. , Li, J. , Xu, G. and Lin, R. (2014) Arabidopsis VQ MOTIF‐CONTAINING PROTEIN29 represses seedling deetiolation by interacting with PHYTOCHROME‐INTERACTING FACTOR1. Plant Physiol. 164, 2068–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Yu, Y. , Yao, W. , Yin, Z. , Wang, Y. , Huang, Z. , Zhou, J. et al. (2023) CRISPR/Cas9‐mediated simultaneous mutation of three salicylic acid 5‐hydroxylase (OsS5H) genes confers broad‐spectrum disease resistance in rice. Plant Biotechnol. J. 21, 1873–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2−▵▵CT Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lv, L. , Liu, Y. , Bai, S. , Turakulov, K.S. , Dong, C. and Zhang, Y. (2022) A TIR‐NBS‐LRR gene MdTNL1 regulates resistance to Glomerella leaf spot in apple. Int. J. Mol. Sci. 23, 6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Y. , Xue, H. , Zhang, F. , Jiang, Q. , Yang, S. , Yue, P. , Wang, F. et al. (2021) The miR156/SPL module regulates apple salt stress tolerance by activating MdWRKY100 expression. Plant Biotechnol. J. 19, 311–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L. , Li, X. , Zhang, J. , Yi, D. , Li, F. , Wen, H. , Liu, W. et al. (2023a) MsWRKY33 increases alfalfa (Medicago sativa L.) salt stress tolerance through altering the ROS scavenger via activating MsERF5 transcription. Plant Cell Environ. 46, 3887–3901. [DOI] [PubMed] [Google Scholar]
- Ma, J. , Li, C. , Sun, L. , Ma, X. , Qiao, H. , Zhao, W. , Yang, R. et al. (2023b) The SlWRKY57‐SlVQ21/SlVQ16 module regulates salt stress in tomato. J. Integr. Plant Biol. 65, 2437–2455. [DOI] [PubMed] [Google Scholar]
- Machanick, P. and Bailey, T.L. (2011) MEME‐ChIP: motif analysis of large DNA datasets. Bioinformatics 27, 1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey, D. , Holt, B.F. , Wiig, A. and Dangl, J.L. (2002) RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1‐mediated resistance in Arabidopsis . Cell 108, 743–754. [DOI] [PubMed] [Google Scholar]
- Mauch‐Mani, B. and Slusarenko, A.J. (1996) Production of salicylic acid precursors is a major function of phenylalanine ammonia‐lyase in the resistance of Arabidopsis to Peronospora parasitica . Plant Cell 8, 203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, D. , Li, C. , Park, H. , González, J. , Wang, J. , Dandekar, A.M. , Turgeon, B.G. et al. (2018) Sorbitol modulates resistance to Alternaria alternata by regulating the expression of an NLR resistance gene in apple. Plant Cell 30, 1562–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munir, M. , Amsden, B. , Dixon, E. , Vaillancourt, L. and Gauthier, N.A.W. (2016) Characterization of Colletotrichum species causing bitter rot of apple in Kentucky orchards. Plant Dis. 100, 2194–2203. [DOI] [PubMed] [Google Scholar]
- Oo, M.M. , Yoon, H.Y. , Jang, H.A. and Oh, S.K. (2018) Identification and characterization of Colletotrichum species associated with bitter rot disease of apple in South Korea. Plant Pathol. J. 34, 480–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, J. , Wang, H. , Hu, Y. and Yu, D. (2018) Arabidopsis VQ18 and VQ26 proteins interact with ABI5 transcription factor to negatively modulate ABA response during seed germination. Plant J. 95, 529–544. [DOI] [PubMed] [Google Scholar]
- Rushton, P.J. , Somssich, I.E. , Ringler, P. and Shen, Q.J. (2010) WRKY transcription factors. Trends Plant Sci. 15, 247–258. [DOI] [PubMed] [Google Scholar]
- Shadle, G.L. , Wesley, S.V. , Korth, K.L. , Chen, F. , Lamb, C. and Dixon, R.A. (2003) Phenylpropanoid compounds and disease resistance in transgenic tobacco with altered expression of L‐phenylalanine ammonia‐lyase. Phytochemistry 64, 153–161. [DOI] [PubMed] [Google Scholar]
- Shan, D. , Wang, C. , Zheng, X. , Hu, Z. , Zhu, Y. , Zhao, Y. , Jiang, A. et al. (2021) MKK4‐MPK3‐WRKY17‐mediated salicylic acid degradation increases susceptibility to Glomerella leaf spot in apple. Plant Physiol. 186, 1202–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang, S. , Liu, G. , Zhang, S. , Liang, X. , Zhang, R. and Sun, G. (2024) A fungal CFEM‐containing effector targets NPR1 regulator NIMIN2 to suppress plant immunity. Plant Biotechnol. J. 22, 82–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine, M.B. , Yang, J.W. , El‐Habbak, M. , Nagyabhyru, P. , Fu, D.Q. , Navarre, D. , Ghabrial, S. et al. (2016) Cooperative functioning between phenylalanine ammonia lyase and isochorismate synthase activities contributes to salicylic acid biosynthesis in soybean. New Phytol. 212, 627–636. [DOI] [PubMed] [Google Scholar]
- Thomazella, D.P.T. , Seong, K. , Mackelprang, R. , Dahlbeck, D. , Geng, Y. , Gill, U.S. , Qi, T. et al. (2021) Loss of function of a DMR6 ortholog in tomato confers broad‐spectrum disease resistance. Proc. Natl. Acad. Sci. U. S. A. 118, e2026152118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velho, A.C. , Alaniz, S. , Casanova, L. , Mondino, P. and Stadnik, M.J. (2015) New insights into the characterization of Colletotrichum species associated with apple diseases in southern Brazil and Uruguay. Fungal Biol. 119, 229–244. [DOI] [PubMed] [Google Scholar]
- van Verk, M.C. , Bol, J.F. and Linthorst, H.J. (2011) WRKY transcription factors involved in activation of SA biosynthesis genes. BMC Plant Biol. 11, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Tian, W. , Tao, F. , Wang, J. , Shang, H. , Chen, X. , Xu, X. et al. (2020) TaRPM1 positively regulates wheat high‐temperature seedling‐plant resistance to Puccinia striiformis f. sp. tritici. Front . Plant Sci. 10, 1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, A. , Shu, X. , Jing, X. , Jiao, C. , Chen, L. , Zhang, J. , Ma, L. et al. (2021) Identification of rice (Oryza sativa L.) genes involved in sheath blight resistance via a genome‐wide association study. Plant Biotechnol. J. 19, 1553–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. , Gao, M. , Li, Y. , Zhang, J. , Su, H. , Cao, M. , Liu, Z. et al. (2022) The transcription factor SlWRKY37 positively regulates jasmonic acid‐ and dark‐induced leaf senescence in tomato. J. Exp. Bot. 73, 6207–6225. [DOI] [PubMed] [Google Scholar]
- Yang, J. , Ma, Q. , Zhang, Y. , Wang, X. , Zhang, G. and Ma, Z. (2016) Molecular cloning and functional analysis of GbRVd, a gene in Gossypium barbadense that plays an important role in conferring resistance to Verticillium wilt. Gene 575, 687–694. [DOI] [PubMed] [Google Scholar]
- Yang, J. , Guo, X. , Mei, Q. , Qiu, L. , Chen, P. , Li, W. , Mao, K. et al. (2023) MdbHLH4 negatively regulates apple cold tolerance by inhibiting MdCBF1/3 expression and promoting MdCAX3L‐2 expression. Plant Physiol. 1, 789–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. and Li, X. (2019) Salicylic acid: biosynthesis, perception, and contributions to plant immunity. Curr. Opin. Plant Biol. 50, 29–36. [DOI] [PubMed] [Google Scholar]
- Zhang, R. , Wang, S.F. , Cui, J.Q. , Sun, G.Y. and Gleason, M.L. (2008a) First report of bitter rot caused by Colletotrichum acutatum on apple in China. Plant Dis. 92, 1474. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Liu, T. , Meyer, C.A. , Eeckhoute, J. , Johnson, D.S. , Bernstein, B.E. , Nusbaum, C. et al. (2008b) Model‐based analysis of ChIP‐Seq (MACS). Genom. Biolol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Shi, X. , Li, B. , Zhang, Q. , Liang, W. and Wang, C. (2016) Salicylic acid confers enhanced resistance to Glomerella leaf spot in apple. Plant Physiol. Biochem. 106, 64–72. [DOI] [PubMed] [Google Scholar]
- Zhang, C. , Wang, X. , Zhang, F. , Dong, L. , Wu, J. , Cheng, Q. , Qi, D. et al. (2017a) Phenylalanine ammonia‐lyase2.1 contributes to the soybean response towards Phytophthora sojae infection. Sci. Rep. 7, 7242–7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Zhao, L. , Zhao, J. , Li, Y. , Wang, J. , Guo, R. , Sheng, G. et al. (2017b) S5H/DMR6 encodes a salicylic acid 5‐hydroxylase that fine‐tunes salicylic acid homeostasis. Plant Physiol. 175, 1082–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q. , Ma, C. , Zhang, Y. , Gu, Z. , Li, W. , Duan, X. , Wang, S. et al. (2018) A single‐nucleotide polymorphism in the promoter of a hairpin RNA contributes to Alternaria alternata leaf spot resistance in apple (Malus x domestica). Plant Cell 30, 1924–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Zhang, Q. , Hao, L. , Wang, S. , Wang, S. , Zhang, W. , Xu, C. et al. (2019a) A novel miRNA negatively regulates resistance to Glomerella leaf spot by suppressing expression of an NBS gene in apple. Hortic Res. 6, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, F. , Wang, F. , Yang, S. , Zhang, Y. , Xue, H. , Wang, Y. and Ma, Y. (2019b) MdWRKY100 encodes a group I WRKY transcription factor in Malus domestica that positively regulates resistance to Colletotrichum gloeosporioides infection. Plant Sci. 286, 68–77. [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Huang, Q. , Yi, L. , Song, X. , Li, L. , Deng, G. , Liang, J. et al. (2021) PAL‐mediated SA biosynthesis pathway contributes to nematode resistance in wheat. Plant J. 107, 698–712. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Xu, R. , Liu, Y. , You, C. and An, J. (2023a) MdVQ10 promotes wound‐triggered leaf senescence in association with MdWRKY75 and undergoes antagonistic modulation of MdCML15 and MdJAZs in apple. Plant J. 115, 1599–1618. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Fu, Y. , Liu, X. , Francis, F. , Fan, J. , Liu, H. , Wang, Q. et al. (2023b) SmCSP4 from aphid saliva stimulates salicylic acid‐mediated defence responses in wheat by interacting with transcription factor TaWKRY76. Plant Biotechnol. J. 21, 2389–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X. , Qi, C. , Jiang, H. , Zhong, M. , Zhao, Q. , You, C. , Li, Y. et al. (2019) MdWRKY46‐enhanced apple resistance to Botryosphaeria dothidea by activating the expression of MdPBS3.1 in the salicylic acid signaling pathway. Mol. Plant Microbe In. 32, 1391–1401. [DOI] [PubMed] [Google Scholar]
- Zhao, X. , Qi, C. , Jiang, H. , Zhong, M. , You, C. , Li, Y. and Hao, Y. (2020) MdWRKY15 improves resistance of apple to Botryosphaeria dothidea via the salicylic acid‐mediated pathway by directly binding the MdICS1 promoter. J. Integr. Plant Biol. 62, 527–543. [DOI] [PubMed] [Google Scholar]
- Zhou, X. , Liao, H. , Chern, M. , Yin, J. , Chen, Y. , Wang, J. , Zhu, X. et al. (2018) Loss of function of a rice TPR‐domain RNA‐binding protein confers broad‐spectrum disease resistance. Proc. Natl. Acad. Sci. U. S. A. 115, 3174–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y. , Yang, Y. , Zhou, X. , Chi, Y. , Fan, B. and Chen, Z. (2016) Structural and functional characterization of the VQ protein family and VQ protein variants from soybean. Sci. Rep. 6, 34663. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Relative expression level of MdWRKY100 after C. fructicola infection.
Figure S2 Genome‐wide analysis of MdWRKY100‐binding sites.
Figure S3 The phylogenetic tree of apple and Arabidopsis phenylalanine ammonia‐lyase proteins.
Figure S4 The phylogenetic tree of MdRPM1, MdCNL1, MdCNL2, MdCNL3, and Arabidopsis NBS‐LRR proteins.
Figure S5 The promoter sequence of MdWRKY17 gene.
Figure S6 The promoter sequence of MdPAL1 gene.
Figure S7 The promoter sequence of MdRPM1 gene.
Figure S8 Expression pattern and sub‐cellular localization of MdVQ37.
Table S1 MdWRKY100 DAP‐seq peaks located upstream of the transcription start site (TSS) of MdWRKY17, MdPAL1 and CC‐NBS‐LRR resistance genes.
Table S2 A list of primers used in this study.
Data Availability Statement
The data that supports the findings of this study are available in the supplementary material of this article.


