Summary
Soybean is a typical short‐day crop, and most commercial soybean cultivars are restricted to a relatively narrow range of latitudes due to photoperiod sensitivity. Photoperiod sensitivity hinders the utilization of soybean germplasms across geographical regions. When grown in temperate regions, tropical soybean responds to prolonged day length by increasing the vegetative growth phase and delaying flowering and maturity, which often pushes the harvest window past the first frost date. In this study, we used CRISPR/LbCas12a to edit a North American subtropical soybean cultivar named 06KG218440 that belongs to maturity group 5.5. By designing one gRNA to edit the nuclear localization signal (NLS) regions of both E1 and E1Lb, we created a series of new germplasms with shortened flowering time and time to maturity and determined their favourable latitudinal zone for cultivation. The novel partial function alleles successfully achieve yield and early maturity trade‐offs and exhibit good agronomic traits and high yields in temperate regions. This work offers a straightforward editing strategy to modify subtropical and tropical soybean cultivars for temperate growing regions, a strategy that could be used to enrich genetic diversity in temperate breeding programmes and facilitate the introduction of important crop traits such as disease tolerance or high yield.
Keywords: CRISPR/LbCas12a, E1, E1Lb, NLS, trade‐off, soybean maturity
Introduction
Soybean (Glycine max) is a major source of plant protein and oil for human food and animal feed (Carter et al., 2004; Graham and Vance, 2003; Hartman et al., 2011; Li et al., 2008). However, soybean is a photoperiod‐sensitive short‐day (SD) crop: SD conditions promote flowering, and long‐day (LD) conditions delay flowering (Garner and Allard, 1920). Photoperiod is the key meteorological factor that determines flower bud differentiation and adaptation to different ecological regions (Câmara et al., 1997) and represents the bottleneck of soybean production.
Twelve genes (E1‐E10, J) have been identified and isolated in the soybean photoperiod pathway (Bernard, 1971; Bonato and Vello, 1999; Buzzell and Voldeng, 1980; Cober et al., 1996, 2010; Kong et al., 2014; Lu et al., 2017; McBlain and Bernard, 1987; Ray et al., 1995; Watanabe et al., 2012; Yue et al., 2017). Four maturity genes, E1 to E4, play major roles in the control of flowering and maturity in soybean. (Jiang et al., 2014; Tsubokura et al., 2013, 2014; Xu et al., 2013). Among these four main genes, E1 is a legume‐specific transcription factor and functions as a flowering inhibitor (Xia et al., 2012; Zhang et al., 2016) and has a large impact on flowering time and time to maturity in soybean (Han et al., 2019; Upadhyay et al., 1994; Xu et al., 2015). E1 family genes are highly conserved in legumes and contain a putative bipartite nuclear location signal (NLS) composed of the KKRK and RRR basic domains at either end separated by 14 aa residues and a DNA‐binding B3 domain (Zhang et al., 2016).
Haplotype analysis has shown locus E1 to be under intense selection pressure during breeding (Xia, 2017). Seven alleles have been identified for E1, including 6 allelic natural recessive variations, e1‐as, e1‐nl, e1‐fs, e1‐re, e1‐p and e1‐b3a (Tsubokura et al., 2014; Xia et al., 2012; Zhai et al., 2015). The E1 and e1‐as alleles are commonly found in modern cultivars (Zhai et al., 2014). The geographic distribution of E1 alleles is mainly in subtropical or tropical soybean cultivars in lower latitude regions, whereas e1‐as is widely distributed in middle‐ or high‐latitude temperate regions. Three rare loss‐of‐function alleles (e1‐nl, e1‐fs and e1‐b3a) are found in very early‐mature soybean cultivars in high‐latitude cold regions (Jiang et al., 2014; Li et al., 2017; Liu et al., 2020; Xia et al., 2012; Zhai et al., 2015). Therefore, during soybean domestication from low latitudes to high latitudes, the E1 gene evolved from a functional photoperiod‐sensitive dominant allele in which the protein completely targeted the nucleus to the partially functional, reduced photoperiod‐sensitive allele e1‐as, in which the protein was distributed in both the nucleus and the cytoplasm, and finally to the nonfunctional, photoperiod‐insensitive alleles e1‐nl and e1‐fs, which cannot produce protein. The transition from later maturing to early maturing phenotypes in soybean is associated with selective sweeps at the E1 locus, and the change in subcellular localization patterns appears to represent a key mechanism underlying the differences in flowering time governed by E1, e1‐as, e1‐nl and e1‐fs. (Xia et al., 2012).
The expression of E1 is tissue‐specific, with high levels in fully expanded leaves and low levels in other tissues (Xia et al., 2012). E1 can up‐regulate the expression of the flowering‐inhibiting genes GmFT1a and GmFT4 (flowering locus T) and down‐regulate the expression of the flowering‐promoting genes GmFT2a and GmFT5a (flowering locus T) to determine the soybean developmental switch from vegetative to reproductive growth (Guo et al., 2015; Kong et al., 2010; Lin et al., 2021; Liu et al., 2018; Nan et al., 2014; Sun et al., 2011).
In addition to flowering time, E1 also controls post‐flowering stem growth and grain yield by regulating FT2a and FT5a expression. FT2a and FT5a both interact with FDL (FD‐like) to induce AP1 (APETALA 1) expression at axillary meristems and terminate stem growth by suppressing Dt1, which is an orthologue of Arabidopsis terminal flowering 1 (TFL1) (Takeshima et al., 2019). Additional research has revealed a basic regulatory mechanism including a negative feedback loop consisting of Dt1‐AP1 and competitive binding between Dt1 and FT5a to FDc1 to control the stem growth habit and flowering time in soybean, consequently affecting plant adaptation and yield (Yue et al., 2021).
There are two E1 homologues in soybean, E1La (Glyma04g24640.1) and E1Lb (Glyma18g22670.1) (Xia et al., 2012; Xu et al., 2015). The putative bipartite NLS and the helix–turn–helix structure in the N‐terminal region of the two E1‐L proteins are almost identical to those of E1. Virus‐induced gene silencing of E1La and E1Lb was found to additively promote flowering by up‐regulating FT2a and FT5a expression in the e1‐nl genotype background (Xu et al., 2015), indicating that E1 and E1La/b are not functionally redundant. It was also demonstrated that E1La and E1Lb together contribute to the repression of FT2a and FT5a, and each of them has independent effects on flowering time and time to maturity under a natural photoperiod (Dietz et al., 2021; Zhu et al., 2019).
Despite widespread awareness of the native alleles of E1(L) genes, the use of these alleles in temperate x tropical breeding programmes is hindered by linkage drag and stacking complexity. These challenges could be overcome using genome editing, but the application of E1 and E1Lb editing has not yet been reported in soybean. In this study, a subtropical soybean line was selected for genome editing. Leveraging the additive effect of E1 and its homologue E1Lb and depending on mutations such as frameshift knockout and NLS in‐frame deletion leading to partial function (e1‐as like), we created a series of new germplasms with shortened flowering time and time to maturity to adapt to different latitudinal zones. Our research provides a possible genome editing strategy to expand the latitudinal deployment of subtropical and tropical soybean cultivars, which will facilitate the introduction of useful traits in subtropical and tropical soybean germplasm to temperate regions, enriching the genetic diversity and possibly improving breeding outcomes in those regions.
Results
CRISPR/LbCas12a mutagenesis of E1 and E1Lb in a subtropical line
We transformed a subtropical soybean cultivar 06KG218440 (maturity group 5.5) with E1/E2/E3/E4/dt1 genotype (Figure S1) using Agrobacterium‐mediated genetic transformation. To create novel partial and dysfunctional alleles, a single CRISPR–Cas12a guide RNA (gRNA), TTTAGGACATCAAGGAGAAGATTCTGC, was designed to target the conserved NLS second basic domain of both E1 and E1Lb (Figure 1a).
Figure 1.
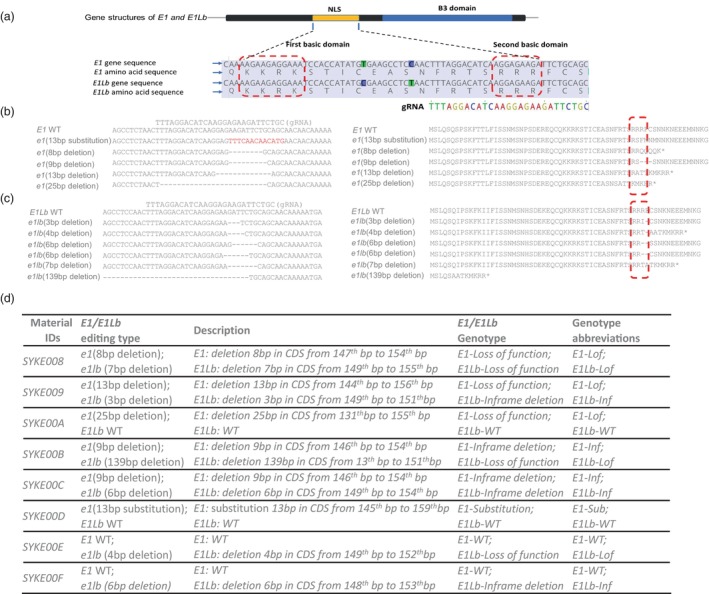
E1/E1Lb gRNA design, E1 mutations and E1Lb mutations, genotype of eight selected edited lines with different E1/E1Lb mutation combinations. (a) Gene structures of E1/E1Lb and gRNA target site. Gene structure of E1 and E1Lb, including putative bipartite NLS region and B3 domain. Two basic domains of NLS are conversed in both E1 and E1Lb. The gRNA was designed at second basic domain of NLS region in both E1 and E1Lb. The red box highlights the first and second basic domain of NLS. The coloured sequence is gRNA, and TTTA is the PAM of gRNA. (b) Gene sequence and amino acid sequence of the E1 mutations at gRNA target region in T2 homo edited lines. Minus represents deletion, and nucleotides in red indicate substitution. The amino acids in the red boxes show the changes that occurred at the second basic domain of NLS after editing. (c) Gene sequence and amino acid sequence of E1Lb mutations at gRNA target region in T2 homo edited lines. Minus represents deletion. The amino acids in the red boxes show the changes that occurred at the second basic domain of NLS after editing. (d) The genotype of the final eight selected edited lines in the T2 generation after segregation. Lof is the abbreviation for loss of function caused by non‐multiples three base pair deletion; Inf is the abbreviation for in‐frame deletion mutations that represents three or multiple three base pair deletion; and Sub is the abbreviation for a substitution mutation that represents base pair substitution.
The specific mutation types were determined by sequencing T2 generation plants after completely segregating out the Lbcas12a transgene. We detected various mutations in the NLS second basic domain of E1 and E1Lb, including frameshifts leading to premature stop codon, in‐frame deletions leading to 1–3 amino acid deletion and substitution leading to 5 amino acid substitution (Figure 1b,c). The frameshift mutations represent nonfunctional alleles such as e1‐fs, while in‐frame deletions and substitutions that removed or replaced one or two key arginine (R) amino acid(s) appear to create weak alleles with possible partial functions, such as e1‐as. Finally, eight T‐DNA‐free edited and non‐off‐target confirmed (Figure S2) lines with different combinations of E1 and E1Lb mutations were further studied (Figure 1d).
Functional analysis of the NLS of E1 and E1Lb
To analyse impact of various mutations in the NLS second basic domain, we compared flowering time of wild type, SYKE00D (E1‐Sub/E1Lb‐WT) and SYKE00A (E1‐Lof/E1Lb‐WT) under long‐day (16 h) greenhouse conditions. Wild type, SYKE00D and SYKE00A flowered at 57.2 ± 1.2, 48.2 ± 1.0 and 26.6 ± 1.4 days after emergence, respectively (Figure 2a). SYKE00D, which had the E1 substitution genotype, exhibited an intermediate flowering time between the E1‐WT and E1‐Lof genotypes (Figure 2a). Although an E1 in‐frame deletion mutation was not studied in the context of an E1Lb‐WT allele, we can compare SYKE008(E1‐Lof/E1Lb‐Lof) with SYKE00B(E1‐Inf/E1Lb‐Lof) and SYKE009(E1‐Lof/E1Lb‐Inf) with SYKE00C(E1‐Inf/E1Lb‐Inf) to understand the difference between E1 in‐frame deletion mutations and frameshifts. From these comparisons, it is clear that the E1 in‐frame deletion allele also has a weaker effect than E1‐Lof (Table S1). This indicates that the impact of the E1 substitution and E1 in‐frame deletion alleles, in which two R amino acids in the NLS second basic domain are replaced or removed, is equal. These appear to be weak alleles with partial function, such as e1‐as.
Figure 2.
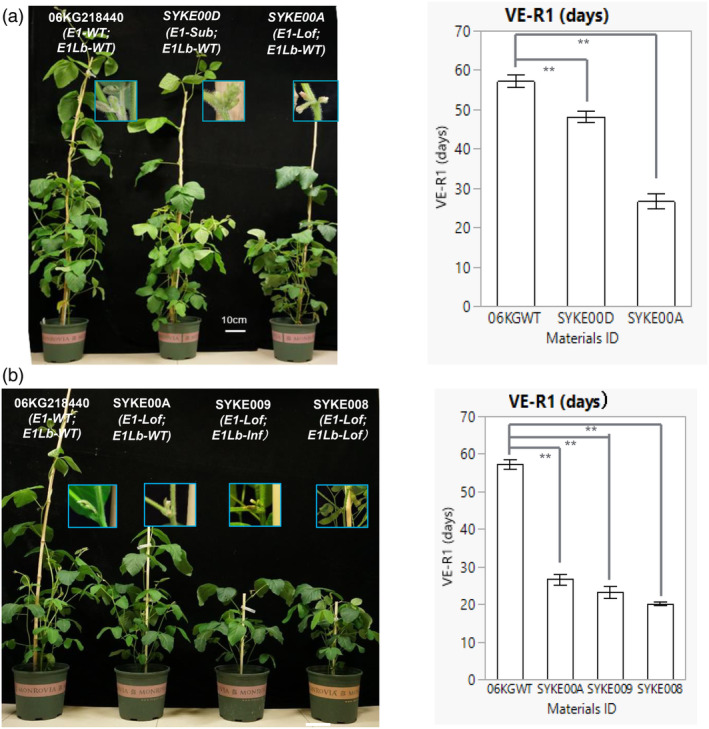
Effect of in‐frame deletion or substitution mutation of E1 and E1Lb on flowering time under long‐day (16 h light) condition in GH. (a) 38‐day‐old plants of 06KG218440 (E1‐WT, E1Lb‐WT); SYKE00D (E1‐Sub, E1Lb‐WT); and SYKE00A (E1‐Lof, E1Lb‐WT). Edited lines SYKE00A and SYKE00D showed obvious early flowering compared to 06KG218440, and flowering time of SYKE00D was intermediate between those of SYKE00A and 06KG218440. Each bar represents the mean of five plants. Error bars represent standard errors (**P < 0.01). (b) 30‐day‐old plants of E1Lb‐Inf and E1Lb‐Lof genotype of the E1‐Lof gene background after emergence. E1‐Lof combining either the E1Lb‐Inf (SYKE009) or the E1Lb‐Lof mutation (SYKE008) showed earlier flowering compared to both 06KG218440 and single E1‐Lof mutation (SYKE00A), but the E1Lb‐Inf mutation had less of an additive effect than E1Lb‐Lof mutation on early flowering. Detailed data for flowering times are provided in Table S1. Each bar represents the mean of five plants. Error bars represent standard errors (**P < 0.01). Flowering time is from VE (emergence) to R1 (first open flower in main stem). Sub is the abbreviation for substitution, Inf is the abbreviation for in‐frame deletion, and Lof is the abbreviation for loss of function. 06KGWT is the abbreviation for the 06KG218440 wild type.
Interestingly, in the E1‐WT background, compared to E1Lb‐WT, E1Lb‐Lof (SYKE00E) and E1Lb‐Inframe (SYKE00F) did not show difference on flowering time (Table S1). However, in the E1‐Lof background, SYKE008 (E1‐Lof/E1Lb‐Lof) flowered approximately 3 days earlier than SYKE009 (E1‐Lof/E1Lb‐Inf) (Figure 2b and Table S1). Similarly, in the E1‐Inframe genetic background, SYKE00B (E1‐Inf/E1Lb‐Lof) flowered approximately 9 days earlier than SYKE00C (E1‐Inf/E1Lb‐Inf) (Table S1). Thus, these data also indicated that in‐frame deletions in the NLS of E1Lb may produce a weak allele with partial function.
Protein distribution of E1 and E1Lb edited alleles
To test the hypothesis that mutations of the second basic domain of the NLS reduce the nuclear localization of E1 and E1Lb, we conducted a transient expression assay to determine the subcellular distributions of the mutant proteins.
We separately fused E1‐WT, E1‐Substitution, E1‐Inframe, E1‐Lof, E1Lb‐WT, E1Lb‐Inframe and E1Lb‐Lof to GFP driven by the 35S promoter. Confocal imaging indicated that E1‐WT‐GFP/E1Lb‐WT‐GFP localized primarily to the nuclei of tobacco epidermal cells (Figure 3a,b). In contrast, E1‐Substitution/E1‐Inframe/E1Lb‐Inframe GFP‐fused proteins were distributed in both the nucleus and the cytoplasm (Figure 3a,b). There was no signal for the E1‐Lof: GFP and E1Lb‐Lof: GFP constructs (Figure 3a,b), suggesting that the premature stop codons halted translation of these proteins. These results demonstrated that modifications to the second basic domain of NLS affect nuclear targeting in both E1 and E1Lb and suggest that the reduction of nuclear localization triggers the early‐flowering phenotype.
Figure 3.
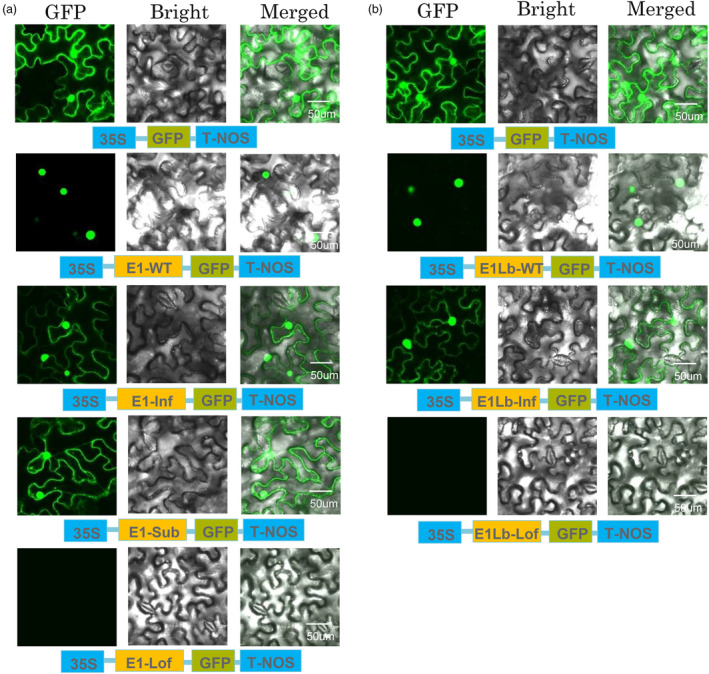
Subcellular localization of GFP and of the E1‐WT, E1‐Substitution, E1‐Inframe deletion, E1‐Loss of function, E1Lb‐WT, E1Lb‐Inframe deletion and E1Lb‐Loss of function fusion proteins. (a) Subcellular distribution of the E1 and E1 mutation proteins in tobacco epidermal cells. E1‐GFP, E1‐Sub‐GFP, E1‐Inf‐GFP and E1‐Lof‐GFP fusion proteins were produced transiently under the control of the CaMV 35S promoter and T‐Nos in tobacco epidermal cells and were observed under a confocal microscope. E1‐GFP protein was only distributed in nuclei, but E1‐Sub‐GFP and E1‐Inf‐GFP were distributed in both nuclei and cytoplasm. Moreover, no E1‐Lof‐GFP was detected. (b) Subcellular distribution of the E1Lb and E1Lb mutation proteins in tobacco epidermal cells. E1Lb‐GFP, E1Lb‐Inf‐GFP and E1Lb‐Lof‐GFP fusion proteins were produced transiently under the control of the CaMV 35S promoter and T‐Nos in tobacco epidermal cells and were observed under a confocal microscope. E1Lb‐GFP protein only distributed in nuclei, but E1‐Inf‐GFP distributed in both nuclei and cytoplasm. And no E1Lb‐Lof‐GFP was detected. The photographs were taken with a dark field for green fluorescence (left), with a bright field for the cell morphology (centre) and with a combination approach (merged, right). In a and b, the experiments were performed three times, with similar results obtained.
Flowering time and agronomic performance of E1 and E1Lb edited lines under greenhouse conditions
To achieve a range of early‐flowering time phenotype with our expectation, lines with various mutant combinations of E1 and E1Lb were selected and grown under both long‐day and short‐day conditions (12, 14 and 16 h light) in greenhouse.
The flowering times of single mutants with the E1 loss‐of‐function allele at 12, 14 and 16 h were 20.6, 25.8 and 26.6 d, respectively, which were significantly earlier than those of wild‐type plants, which were 26.6, 50 and 57 d, respectively (P < 0.01) (Table S1). The flowering times of single mutants with E1 substitution alleles were 23, 41.8 and 48.2 d, respectively, slightly earlier than those of wild‐type plants under 12, 14 and 16 h photoperiods (Table S1). Therefore, while E1 partial function reduced 06KG218440 photoperiod sensitivity, E1 loss of function made 06KG218440 completely photoperiod‐insensitive. When combining the E1 mutants with E1Lb mutant alleles, the photoperiod sensitivity and flowering time further decreased, and the effect of the E1Lb‐Inframe allele was weaker than that of the E1Lb‐Lof allele (Table S1). The edited lines with E1/E1Lb allelic combinations exhibited series of early flowering compared with wild‐type plants (Table S1) and showed same trend in three photoperiods. In addition, we observed that shorter flowering time in the E1/E1Lb allelic combinations seems to be correlated with decreased time to maturity and shorter plant height compared to wild type (Table S1).
Differential expression of FT genes determines the divergent phenotypes of E1 / E1Lb mutation combinations
In the molecular mechanism of soybean flowering time, there are two main flowering‐inhibiting genes, GmFT1a/FT4, and two main flowering‐promoting genes, GmFT2a and GmFT5a, that determine phase change during soybean development (Lin et al., 2021; Liu et al., 2018). To explore early‐flowering phenotype mechanism of edited lines, we performed qRT‐PCR on the FT genes under LD (14 h light) conditions. Expression of the flowering‐promoting genes FT2a and FT5a was activated in E1‐Lof edited lines but not in WT, and the FT5a expression level was much higher than that of FT2a (Figure 4a). In contrast, the expression of the flowering‐inhibiting genes FT1a and FT4 was suppressed in E1 edited lines, especially E1‐Lof (Figure 4b). Expression of the florigen gene AP1, which is downstream of FT2a/FT5a, was also increased in E1‐Lof edited lines. So, it indicated flowering time of the edited lines is closely correlated with the expression levels of these four FT genes and AP1 gene (Figure 4c).
Figure 4.
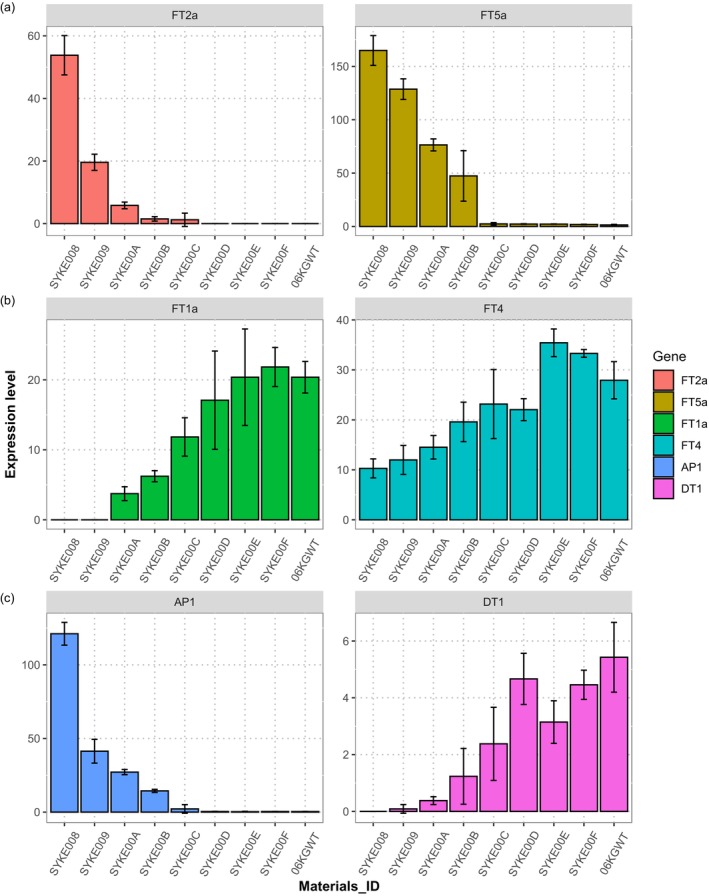
Variant of expression level of FT genes and DT1 gene in edited lines. (a) Expression levels of FT2a, FT5a in leaf of edited plants and 06KG218440 wild type. (b) Expression levels of FT1a, FT4 in leaf of edited plants and 06KG218440 wild type. (c) Expression levels of AP1, DT1 in stem tips of edited plants and 06KG218440 wild type. cDNA samples from 2nd extended leaf and stem tips of individual edited line and 06KG218440 WT at V3 stage under the 14‐h long‐day conditions, three biological replicates. GmCYP2 (Glyma.12G024700) was used as a control, whose expression level was set to 1 for all genes analysed to linearly adjust the corresponding gene expression levels in edited plants. Each bar represents the mean of three replications. Error bars represent standard errors. 06KGWT is abbreviations of 06KG218440 wild type.
We noted that the reduced stature of the greenhouse‐grown edited lines may be caused by premature stem growth termination. Therefore, the expression of DT1, the key gene controlling determinacy (Li et al., 2018), was analysed in each edited line. DT1 expression was strongly suppressed in the early‐flowering edited lines (Figure 4c). These results are reminiscent of those of a previous study that showed that FT5a inhibits Dt1 activity and directly up‐regulates AP1 (Yue et al., 2017).
E1 and E1Lb NLS‐edited subtropical lines grow normally in temperate field conditions
To determine the shifting of time to maturity in the E1 and E1Lb edited lines under field conditions, in 2022, we conducted efficacy trials at three different latitude regions: Yangling in Shaanxi Province, Shunyi‐Beijing City and Yongji‐Jilin Province.
Compared to the WT, the edited materials with different E1/E1Lb mutation combinations showed a series of shortened flowering times (Figure 5a) at all three field locations. With increasing latitude, the flowering time of each material was delayed, but photoperiod insensitivity was consistent with the trends under different photoperiod.
Figure 5.
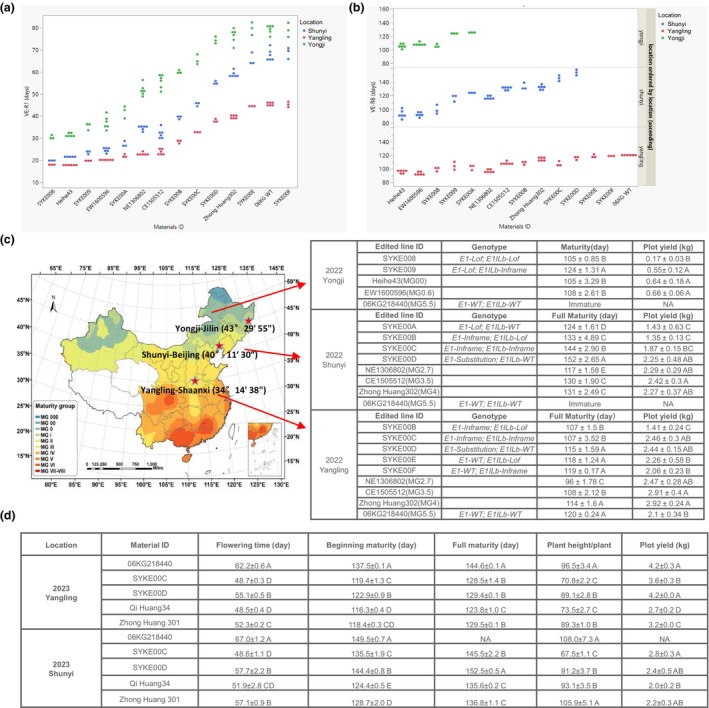
Flowering time, maturity and yield performance of edited lines in 2022 and 2023 field locations. (a, b) Flowering time and maturity time distribution of edited lines and reference lines in three locations (Yongji‐Jilin, Shunyi‐Beijing and Yangling‐Shaanxi) in 2022, and the reference lines as maturity group reference are Heihe43 (MG0), EW1600596 (MG0), NE1306802 (MG2.7), CE1505512 (MG3.5), Zhong Huang302 (MG4) and 06KG218440 (MG5.5). The edited lines showed series of shortened flowering time (a) and maturity (b) in three field locations and different adaptive in different latitude based on their maturity. (c, d) Maturity and yield comparison among edited lines and MG reference lines in their corresponding adaptive region in 2022 (c). Flowering time, maturity, plant height and yield comparison among two edited lines and local commercial lines in their adaptive region in 2023 (d). Data were mean ± SD. Significance of the difference was analysed by Student's t test, and a different letter indicates a significant difference between two lines. NA means no data because they are not fully mature.
Importantly, we found that only the E1 loss‐of‐function mutants enabled 06KG218440 to reach full maturity in the high‐latitude region of Jilin (Figure 5b). Combining E1‐Lof with E1Lb mutations further shortened 06KG218440 time to maturity, but this also caused the yield performance to decrease (Figure 5c).
In the more moderate latitude region of Beijing, E1‐Lof also significantly accelerated time to maturity. However, that allele also produced very poor yield (Figure 5c). The SYKE00D line, which has an E1 partially functional allele, matured before frost but 152 days after spring sowing (Figure 5c). Similarly, in Yongji, when E1 partial function alleles were combined with E1Lb mutations, maturity occurred earlier than single mutant, and plant height and yield also decreased (Figure 5c,d and Table S2). Compared to the reference lines in this maturity group (MG2‐3), SYKE00C(E1‐Inf/E1Lb‐Inf) and SYKE00D(E1‐Sub/E1Lb‐WT) showed similar yields without significant differences (Figure 5c). Meanwhile, E1Lb single mutations (SYKE00E and SYKE00F) and 06KG218440 could not reach full maturity in this region.
In the other moderate latitude region of Yangling, 06KG218440 was able to reach full maturity, but all the other materials matured earlier than it did after summer sowing (Figure 5b). At this latitude, SYKE00C(E1‐Inf/E1Lb‐Inf) and SYKE00D(E1‐Sub/E1Lb‐WT) seemed better adapted than 06KG218440: they matured 13 and 5 days earlier, respectively, and their yields were significantly higher than those of 06KG218440. However, their yield was still below that of the reference lines (Figure 5c).
To further confirm the yield potential of the two edited lines SYKE00C and SYKE00D in the temperature region, we reconducted a yield field trial with spring sowing in the 2023 Shunyi and Yangling locations. The results demonstrated that only SYKE00C matured successfully in both locations, and the time to maturity was very similar to those of the reference lines. Although SYKE00D also matured normally in both locations, the time to maturity was a little bit late before frost arrived in Shunyi. 06KG218440 was unable to reach full maturity in the Shunyi location in 2023 and very late mature in Yanling (Figure 5d). From a yield perspective, SYKE00C and SYKE00D showed higher plot yields than the local commercial reference lines Qi Huang 34 and Zhong Huang 301 at both sites, and at the Yangling site, the difference was significant (Figure 5d). Therefore, overall, when taking successfulness, maturity and yield into consideration, SYKE00C is more adaptive to cultivation in Shunyi, and SYKE00D is more adaptive to cultivation in Yangling.
Discussion
E1 and E1Lb play independent and interacting roles in flowering time, maturity and plant height
E1 has two homologues in soybean, E1La and E1Lb (Xia et al., 2012; Xu et al., 2015). Although it has been demonstrated that the E1L genes have a weaker effect on flowering time than E1, they clearly have an additive effect on flowering in the e1 genetic background (Xia et al., 2012). However, the interaction between the two genes E1 and E1Lb is not clear (Zhu et al., 2019).
To elucidate the relationship between E1 and E1Lb, we generated single and double mutants. From the phenotypic analysis of mutants in the greenhouse, E1 gene has a more impactful role on flowering time, time to maturity and plant height than does E1Lb: the phenotype of the E1Lb mutant was like that of the unedited WT control. However, E1E1Lb double mutants showed a significantly shorter flowering time than either of the single mutants. Similarly in field condition, the ability of E1Lb mutations to promote flowering and maturity appears to be abolished in a fully functional E1 background and only functions to accelerate flowering and maturity in the context of a partial function or dysfunctional E1. Therefore, the role of the E1Lb protein in repressing flowering appears to be redundant in the context of a functional E1. E1 and E1Lb redundantly and differentially regulate flowering time and plant height. These results also suggest that both genes regulate flowering time in at least a partially independent manner.
Function of the B3 domain and NLS of E1 and E1Lb
The function of the B3 domain is well understood, and two natural recessive alleles (e1‐fs and e1‐b3a) (Xia et al., 2012; Zhai et al., 2015) and one genome‐edited allele (Han et al., 2019) have truncated or deleted the B3 domain and significantly promoted flowering time. In our study, all frameshift mutations of E1 and E1Lb resulted in B3 domain deletion and had a significant effect on flowering time and time to maturity (Figure 3c), suggesting that the B3 domain of E1 and E1Lb is essential for their function.
An amino acid substitution in E1 from arginine to threonine occurs at exactly the first basic domain in the bipartite NLS, affecting nuclear targeting of the E1 protein (Xia et al., 2012). Here, we showed that partial loss‐of‐function mutations in E1 and E1Lb may be created from in‐frame deletions or substitution mutations in the second basic domain of the NLS. Edited lines with the E1/E1Lb in‐frame deletion or substitution genotype generally had flowering times intermediate between those of the WT allele and frameshift genotypes in both the greenhouse and the field. These results suggest that the second basic domain in the NLS of the E1 and E1Lb proteins is important for their function and implies that any arginine deletion or replacement in the second basic domain may have a similar effect on protein distribution as mis‐sense mutations in the first basic domain.
E1 and E1Lb editing create series of early‐flowering time phenotype
Shorten flowering time to different levels by using genome editing may require multiple gRNA to modify multiple FT genes (Cai et al., 2020; Kou et al., 2022; Zhao et al., 2022). Likewise, using a transgenic approach, one may create a range of flowering times simply by selecting different events with different levels of FT gene expression (Cao et al., 2015; Wang et al., 2016). In the present study, by creating novel weak allele via NLS editing and leveraging the additive effect of E1 gene and its homologue, one gRNA provided a combinatorial allelic series of both genes. QRT analysis on the various edited lines indicated that E1 and E1Lb editing combinations disrupt the balance of the soybean flowering ‘teeterboard’ under long‐day conditions, and the expression of the floral promoters (GmFT2a/GmFT5a) increased and that of the inhibitors (GmFT1a and GmFT4) decreased to different degrees compared to the wild type, leading to a series of shortened flowering times and times to maturity (Figure 6). In contrast to the complex approaches needed for creating allelic series such as editing regulatory regions (Liu et al., 2021; Song et al., 2022), this study also demonstrates a novel and straightforward way to regulate gene function by changing the protein distribution via NLS editing.
Figure 6.
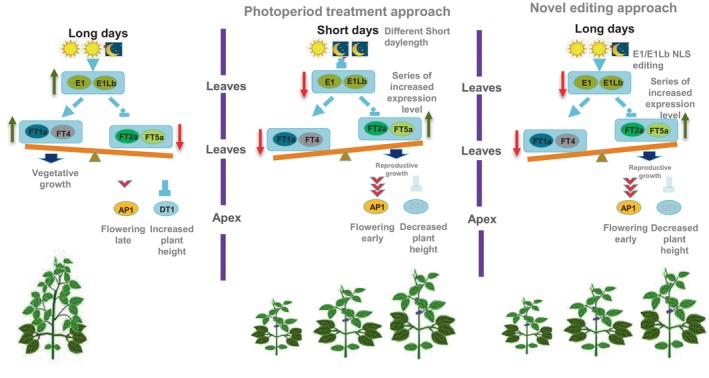
Molecular mechanism of E1/E1Lb editing manipulating soybean flowering and determinacy. E1/E1Lb editing breaks the equilibrium balance of soybean flowering regulation under long‐day conditions. The flowering‐promoting genes FT2a and FT5a up‐regulated and flowering‐inhibiting genes FT1a and FT4 down ‐ regulated to series of different expression level that is caused by E1/E1Lb NLS editing, consequently trigger AP1 gene expression increasing to a certain level and switching the soybean development to reproductive growth. Meanwhile, high FT5a expression represses key gene of soybean determinacy DT1, causing stem growth termination and plant height decreasing.
Balance maturity, growth habit and yield
Flowering time and stem growth have strong effects on the adaptability and grain yield in soybean (Cao et al., 2017; Heatherly and Smith, 2004). Early flowering and stem termination ensure that soybean quickly switches to reproductive growth and reaches reproductive maturity during the shorter frost‐free window in high‐latitude regions. However, this ability to flower quickly in LD conditions has to be balanced between maturity and yield potential: early maturity and stem termination can make soybean more adapted to high‐latitude regions but can also lead to low yield.
From our observations in the field trials, the earlier flowering time of all edited lines resulted in an earlier time to maturity, a shorter plant height and few nodes, leading to a smaller overall plant architecture. Moreover, the yield decreased.
It is important to monitor the link between flowering and stem indeterminacy during cultivar improvement. In our study, 06KG218440 was a determinate soybean cultivar with recessive dt1, and its expression level is low. Therefore, increased expression of FT5a caused by E1/E1Lb editing further suppresses DT1 expression, leading to earlier stem termination and low yield. However, if using an indeterminate cultivar with dominant DT1, the stem termination caused by early flowering may be relieved due to the relatively high expression of DT1. This may make flowering, maturity and yield more balanced. Moreover, weak alleles are another option, and in our study, an improved trade‐off between early maturity and yield was obtained in the weak alleles of E1 and E1Lb after NLS editing.
In summary, this study clarified the important role of the NLS second basic domain in the nuclear localization of E1 and E1Lb and provided a novel editing method to generate novel leaky alleles with partial function by editing the NLS region to change the protein distribution in the cell. Soybean E1 and its homologue E1Lb divergently but redundantly control flowering time, maturity and stem growth habit by regulating four FT genes and determinacy gene DT1. Our approach to develop different E1/E1Lb mutant combinations by editing led to a series of new genetic combinations with shortened flowering time and maturity. This approach may be used to enrich the soybean breeding pool and can expand the planting area of different soybean cultivars to higher latitude regions. The partial function alleles created by NLS editing should help solve trade‐off effects and provide a feasible method to move subtropical and tropical soybean germplasm to temperate regions.
Materials and methods
Plant materials and growth conditions
Soybean 06KG218440 (MG5.5) wild type and edited lines were grown in the greenhouse of Syngenta Beijing Innovation Center, China. The environmental setting was a day temperature of 28°C and a night temperature of 18°C. The supplemental light was a sodium lamp, and the average photon flux was 300 μmol/m2/s.
SgRNA design and construction of the CRISPR/Cas12a expression vector
The LbCas12a gene was codon‐optimized in Arabidopsis (Wolter and Puchta, 2019) and was driven by the Arabidopsis EF‐1 alpha A1 promoter plus the eFMV enhancer. The gRNA cassette including the GmUbi promoter and gRNA scaffolds was synthesized by GenScript (www.genscript.com) and cloned and inserted into a binary LbCas12a base vector that included an aadA gene driven by the soybean EF promoter as the plant selection marker.
Transformation of CRISPR/Cas12a in soybean and screening for mutations by sequencing analysis
Soybean (Glycine max, cultivar 06KG218440) seeds sterilized by chlorine gas were imbibed in germination medium at 25 °C in the dark. Imbibed seeds were used to prepare explants as described (Khan R. Method of transforming soybean, WIPO International Publication Number WO2004000006, 31 December 2003; Watts J. and Ganesan S. US Patent Number 9758792, 12 September 2017) by trimming off the hypocotyl and removing one cotyledon and leaf primordium. The shoot apical region and the cot‐node region were then gently wounded with the sharp end of a scalpel blade. The prepared explants were immediately infected with an Agrobacterium suspension of the Agrobacterium tumefaciens strain EHA101 containing a binary vector. With at least 2 h of inoculation, the explants were placed on cocultivation medium for 5 days at 23 °C in the dark.
After cocultivation, the explants were preferentially transferred to recovery medium without selection agent for approximately 7 days at 24 °C under a 16‐h light/8‐h dark regimen. The recovered explants with cotyledons were transferred to regeneration media along with glyphosate selection for approximately 3 weeks. The explants with developing multiple shoot clusters were transferred to elongation medium along with glyphosate selection for shoot elongation. Subcultures to fresh elongation medium were performed every 3 weeks until the elongated shoots (>3 cm) were long enough to be sampled for molecular analysis.
Mutation screening and identification of transgene‐free edited lines
Genomic DNA was isolated from young leaves for mutation analysis. The T‐DNA copy number was determined by TaqMan assay targeting Cas12a and aadA gene using ADH1 as an internal control (Table S3). The target genes E1 and E1Lb and the predicted off‐target gene were amplified using specific primers (Table S3). The PCR products were gel‐purified and sequenced by Sanger sequencing to characterize edited alleles. After obtaining the homozygous and T‐DNA free edits, the selected plants were used to produce seeds and for further trait evaluation in the greenhouse and field.
Flowering time and agronomic trait measurements in the greenhouse and field
T3 seeds derived from homozygous mutants without T‐DNA were used for additional phenotyping investigation in a greenhouse. Random complete block design was used for the phenotyping assay, including five blocks, one plant in each block and a total of five plants per genotype. The data were analysed by t test, and P < 0.05 was considered significant. Graphs were plotted using JMP.
Field experiments were conducted at three different latitude locations using T4 seeds, Yangling in Shaanxi Province, Shunyi‐Beijing City and Yongji‐Jilin Province in 2022. The experimental design was RCBD with three replicates (plots), and the materials included eight different E1/E1Lb combination edited lines, WT controls and 5 MG check reference lines. At the Shunyi and Yangling sites, there were two rows of plots, but at the Yongji site, there was a one‐row plot because of an insufficient seed quantity. At all three sites, the row length was 3 m, the row spacing was 75 cm, and the plant spacing was 10 cm. Ten uniformly growing plants per genotype were selected in each plot for agronomic trait investigation. Plot yield was calculated by total plants per genotype in two rows of each plot.
A total of 2023 field experiments were conducted at two different latitude locations using T5 seeds, Yangling in Shaanxi Province and Shunyi‐Beijing City. The experimental design was RCBD with 3–4 replicates (plots), and the plant materials included 2 different E1/E1Lb combination edited lines, WT controls and 2 local commercial lines. There were four‐row plots, the row length was 5.4 m, the row space was 75 cm, and the plant distance is 10 cm. Ten uniformly growing plants per genotype were selected in each plot for agronomic trait investigation. Total plants per genotype in two middle rows of each plot were used for yield evaluation. The data was analysed by variance analysis, and the significance of the difference was analysed by Student's t test using JMP software.
Gene expression analysis by quantitative real‐time PCR (qRT‐PCR)
Expression levels of GmFT2a, GmFT5a, GmFT1a, GmFT4, GmAP1 and GmDT1 in wild‐type plants and T3 homozygous mutants were analysed under 14‐h long‐day photoperiod conditions in a greenhouse. Fifteen days after emergence (DAE), at 10 am (4 h after light), the second trifoliate leaves and stem tips were sampled from plants with different genotypes. Total RNA was extracted following the protocol of Promega Magbeads. A plant genome extraction kit followed by DNA digestion with DNaseI (Promega) was then used. For reverse transcription, first‐strand cDNA synthesis was performed using MultiScribe Reverse Transcriptase (ABI). For qRT‐PCR, gene expression was examined using cDNA templates on an Applied Biosystems 7900 Real‐Time PCR System. The relative gene expression levels were assessed following a previously described method (Pfaffl, 2001). The mRNA level of cyclophilin (Glyma.12G024700) was used as a reference for normalization. The specific primers we used in this study are listed in Table S3. Three biological replicates were used for each gene.
Transient transfection assays
The subcellular localization of the edited GmE1 and GmE1Lb variants was conducted in Nicotiana benthamiana plants using Agrobacterium‐mediated transient transformation. Briefly, N. benthamiana plants were grown under long‐day conditions (16 h/light, 8 h/dark; 22–30 °C) in green chamber. The 4‐week‐old N. benthamiana leaves were infiltrated with A. tumefaciens strain GV3101 bearing GmE1/GmE1Lb‐mEGFP and control mEGFP plasmids, respectively. After 48‐h infiltration, the N. benthamiana leaves were collected and imaged by FLUOVIEW FV3000 Confocal Laser Scanning Microscope (Olympus Corporation, Tokyo, Japan). The open reading frames of GmE1, GmE1Lb and edited alleles were amplified from 06KG218440 and edited E2 plants, respectively, then fused into mEGFP binary vector at the N‐terminus of mEGFP (monomeric enhanced GFP, A206K) using NEBuilder HiFi DNA assembly cloning kit (NEB, E2621L).
Accession numbers
The gene sequences of Soybean were downloaded from the Phytozome database (https://phytozome‐next.jgi.doe.gov/) with the following accession numbers: E1(Glyma.06G207800), E1La(Glyma.04G156400), E1Lb(Glyma.04G143300), E2(Glyma.10G221500), E3(Glyma.19G224200), E4(Glyma.20G090000), GmFT2a(Glyma.16G150700), GmFT4(Glyma.08G363100), GmELF3(Glyma.04G050200), GmFT5a(Gyma.16G044100), GmFT1a(Glyma.18G298900), AP1(Glyma.16G091300) and DT1(Glyma.19G194300).
Conflict of interest statement
The authors declare no conflicts of interest.
Author contributions
Y.G. designed the research and wrote the manuscript. C.L. constructed the editing vector. Y.W. generated the transformants. Y.Z., S.W. and Y.G. performed the molecular analysis. Y.G. designed and conducted the greenhouse phenotyping. C.M. and Y.G. designed and conducted field trials. Y.C involved in project management and advised on the experiments. X.C supervised the project, advised on the experiments and revised the manuscript.
Supporting information
Figure S1 Alignment of the amino acid sequences encoded by the DT1 and dt1 alleles in 06KG218440.
Figure S2 Off‐target analysis of E1La and E1La sequencing in selected edited lines.
Figure S3 Plant architecture of eight selected edited lines with different genotypes with E1/E1Lb mutation combinations at the Shunyi site.
Table S1 Flowering time, maturity, yield, and agronomic traits of eight E1/E1Lb T3 edited lines under three different photoperiod treatments in a greenhouse.
Table S2 Flowering time, maturity, yield, and agronomic traits of eight E1/E1Lb edited lines at three latitude field locations in 2022.
Table S3 Primers and probes for the TaqMan assay, sequencing and QRT.
Acknowledgements
We are grateful to Syngenta fellow Kelliher Tim and Prof Qi Xie in State Key Laboratory of Crop Germplasm Innovation and Molecular Breeding for revising the manuscript. We thank Yintuan Wu, Hongchang Zhou and Hongchang Zhang for their efforts in field trial management in Shunyi, Yangling and Yongji. Syngenta fellow Jianping Xu, principal scientist Dawei Liang and Cohn Josh, programme manager Groenen Jose, Maurice and Yan Lu in regulatory affairs also provided strong support for experimental implementation.
Contributor Information
Yang Gao, Email: yang.gao-1@syngenta.com.
Xi Chen, Email: xi.chen@syngenta.com.
Data availability statement
The data that supports the findings of this study are available in the supplementary material of this article.
References
- Bernard, R.L. (1971) Two major genes for time of flowering and maturity in soybeans 1. Crop Sci. 11, 242–244. [Google Scholar]
- Bonato, E.R. and Vello, N.A. (1999) E6, a dominant gene conditioning early flowering and maturity in soybeans. Genet. Mol. Biol. 22, 229–232. [Google Scholar]
- Buzzell, R.L. and Voldeng, H.D. (1980) Research notes: inheritance of insensitivity to long day length. Soybean Genetics Newsletter 7, 26–29. [Google Scholar]
- Cai, Y. , Wang, L. , Chen, L. , Wu, T. , Liu, L. , Sun, S. , Wu, C. et al. (2020) Mutagenesis of GmFT2a and GmFT5a mediated by CRISPR/Cas9 contributes for expanding the regional adaptability of soybean. Plant Biotechnol. J. 18, 298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Câmara, G.M.S. , Sediyama, T. , Dourado‐Neto, D. and Bernardes, M.S. (1997) influence of photoperiod and air temperature on the growth, flowering and maturation of soybean (Glycine max (L.) Merrill). Sci. Agric. 54, 149–154. [Google Scholar]
- Cao, D. , Li, Y. , Lu, S. , Wang, J. , Nan, H. , Li, X. , Shi, D. et al. (2015) GmCOL1a and GmCOL1b function as flowering repressors in soybean under long‐day conditions. Plant Cell Physiol. 56, 2409–2422. [DOI] [PubMed] [Google Scholar]
- Cao, D. , Takeshima, R. , Zhao, C. , Liu, B. , Jun, A. and Kong, F. (2017) Molecular mechanisms of flowering under long days and stem growth habit in soybean. J. Exp. Bot. 68, 1873–1884. [DOI] [PubMed] [Google Scholar]
- Carter, T.E. , Nelson, R. , Sneller, C.H. and Cui, Z. (2004) Soybeans: Improvement, Production and Uses, 3rd edn. Madison: American Society of Agronomy. [Google Scholar]
- Cober, E.R. , Molnar, S.J. , Charette, M. and Voldeng, H.D. (2010) A new locus for early maturity in soybean. Crop Sci. 50, 524–527. [Google Scholar]
- Cober, E.R. , Tanner, J.W. and Voldeng, H.D. (1996) Genetic control of photoperiod response in early‐maturing, near‐isogenic soybean lines. Crop Sci. 36, 601–605. [Google Scholar]
- Dietz, N. , Combs‐Giroir, R. , Cooper, G. , Stacey, M. , Miranda, C. and Bilyeu, K. (2021) Geographic distribution of the E1 family of genes and their effects on reproductive timing in soybean. BMC Plant Biol. 21, 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner, W.W. and Allard, H.A. (1920) Effect of the relative length of day and night and other factors of the environment on growth and reproduction in plants. J. Agric. Res. 18, 553–606. [Google Scholar]
- Graham, P.H. and Vance, C.P. (2003) Legumes: importance and constraints to greater use. Plant Physiol. 131, 872–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, G. , Xu, K. , Zhang, X. , Zhu, J. , Lu, M. , Chen, F. , Liu, L. et al. (2015) Extensive analysis of GmFTL and GmCOL expression in Northern Soybean cultivars in field conditions. PLoS One 10, e0136601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, J. , Guo, B. , Guo, Y. , Zhang, B. , Wang, X. and Qiu, L.J. (2019) Creation of early flowering germplasm of soybean by CRISPR/Cas9 technology. Front. Plant Sci. 10, 1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman, G.L. , West, E.D. and Herman, T.K. (2011) Crops that feed the World 2. Soybean—worldwide production, use, and constraints caused by pathogens and pests. Food Secur. 3, 5–17. [Google Scholar]
- Heatherly, L.G. and Smith, J.R. (2004) Effect of soybean stem growth habit on height and node number after beginning bloom in the Midsouthern, USA. Crop Sci. 44, 1855–1858. [Google Scholar]
- Jiang, B. , Nan, H. , Gao, Y. , Tang, L. , Yue, Y. , Lu, S. , Ma, L. et al. (2014) Allelic combinations of soybean maturity Loci E1, E2, E3 and E4 result in diversity of maturity and adaptation to different latitudes. PLoS One 9, e106042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, F. , Liu, B. , Xia, Z. , Sato, S. , Kim, B.M. , Watanabe, S. , Yamada, T. et al. (2010) Two coordinately regulated homologs of FLOWERING LOCUS T are involved in the control of photoperiodic flowering in soybean. Plant Physiol. 154, 1220–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, F. , Nan, H. , Cao, D. , Li, Y. , Wu, F. , Wang, J. , Lu, S. et al. (2014) A new dominant gene E9 conditions early flowering and maturity in soybean. Crop Sci. 54, 2529–2535. [Google Scholar]
- Kou, K. , Yang, H. , Li, H. , Fang, C. , Chen, L. , Yue, L. , Nan, H. et al. (2022) A functionally divergent SOC1 homolog improves soybean yield and latitudinal adaptation. Curr. Biol. 32, 1728–1742.e1726. [DOI] [PubMed] [Google Scholar]
- Li, J. , Wang, X. , Song, W. , Huang, X. , Zhou, J. , Zeng, H. , Sun, S. et al. (2017) Genetic variation of maturity groups and four E genes in the Chinese soybean mini core collection. PLoS One 12, e0172106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. , Ding, Y. , Zhang, D. , Wang, X. , Tang, X. , Dai, D. , Jin, H. et al. (2018) Parallel domestication with a broad mutational spectrum of determinate stem growth habit in leguminous crops. Plant J. 96, 761–771. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Guan, R. , Liu, Z. , Ma, Y. , Wang, L. , Li, L. , Lin, F. et al. (2008) Genetic structure and diversity of cultivated soybean (Glycine max (L.) Merr.) landraces in China. Theor. Appl. Genet. 117, 857–871. [DOI] [PubMed] [Google Scholar]
- Lin, X. , Liu, B. , Weller, J.L. , Jun, A. and Kong, F. (2021) Molecular mechanisms for the photoperiodic regulation of flowering in soybean Journal of Integrative. Plant Biol. 63, 981–994. [DOI] [PubMed] [Google Scholar]
- Liu, L. , Gallagher, J. , Arevalo, E.D. , Chen, R. , Skopelitis, T. , Wu, Q. , Bartlett, M. et al. (2021) Enhancing grain‐yield‐related traits by CRISPR–Cas9 promoter editing of maize CLE genes. Nature Plants 7, 287–294. [DOI] [PubMed] [Google Scholar]
- Liu, L. , Song, W. , Wang, L. , Sun, X. , Qi, Y. , Wu, T. , Sun, S. et al. (2020) Allele combinations of maturity genes E1‐E4 affect adaptation of soybean to diverse geographic regions and farming systems in China. PLoS One 15, e0235397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W. , Jiang, B. , Ma, L. , Zhang, S. , Zhai, H. , Xu, X. , Hou, W. et al. (2018) Functional diversification of Flowering Locus T homologs in soybean: GmFT1a and GmFT2a/5a have opposite roles in controlling flowering and maturation. New Phytol. 217, 1335–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, S. , Zhao, X. , Hu, Y. , Liu, S. , Nan, H. , Li, X. , Fang, C. et al. (2017) Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat. Genet. 49, 773–779. [DOI] [PubMed] [Google Scholar]
- McBlain, B.A. and Bernard, R.L. (1987) A new gene affecting the time of flowering and maturity in soybeans. J. Hered. 78, 160–162. [Google Scholar]
- Nan, H. , Cao, D. , Zhang, D. , Li, Y. , Lu, S. , Tang, L. , Yuan, X. et al. (2014) GmFT2a and GmFT5a redundantly and differentially regulate flowering through interaction with and upregulation of the bZIP transcription factor GmFDL19 in soybean. PLoS One 9, e97669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M.W. (2001) A new mathematical model for relative quantification in real‐time RT‐PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, J.D. , Hinson, K. , Mankono, J.E.B. and Malo, M.F. (1995) Genetic control of a long‐juvenile trait in soybean. Crop Sci. 35, 1001–1006. [Google Scholar]
- Song, X. , Meng, X. , Guo, H. , Cheng, Q. , Jing, Y. , Chen, M. , Liu, G. et al. (2022) Targeting a gene regulatory element enhances rice grain yield by decoupling panicle number and size. Nat. Biotechnol. 40, 1403–1411. [DOI] [PubMed] [Google Scholar]
- Sun, H. , Jia, Z. , Cao, D. , Jiang, B. , Wu, C. , Hou, W. , Liu, Y. et al. (2011) GmFT2a, a soybean homolog of FLOWERING LOCUS T, is involved in flowering transition and maintenance. PLoS One 6, e29238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshima, R. , Nan, H. , Harigai, K. , Dong, L. , Zhu, J. , Lu, S. , Xu, M. et al. (2019) Functional divergence between soybean FLOWERING LOCUS T orthologues FT2a and FT5a in post‐flowering stem growth. J. Exp. Bot. 70, 3941–3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubokura, Y. , Matsumura, H. , Xu, M. , Liu, B. , Nakashima, H. , Anai, T. , Kong, F. et al. (2013) Genetic variation in soybean at the maturity locus E4 is involved in adaptation to long days at high latitudes. Agronomy 3, 117–134. [Google Scholar]
- Tsubokura, Y. , Watanabe, S. , Xia, Z. , Kanamori, H. , Yamagata, H. , Kaga, A. , Katayose, Y. et al. (2014) Natural variation in the genes responsible for maturity loci E1, E2, E3 and E4 in soybean. Ann. Bot. 113, 429–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay, A.P. , Ellis, R.H. , Summerfield, R.J. , Roberts, E.H. and Qi, A. (1994) Characterization of photothermal flowering responses in maturity isolines of soyabean [Glycine max (L.) Merrill] cv. Clark. Ann. Bot. 74, 87–96. [DOI] [PubMed] [Google Scholar]
- Wang, T. , Sun, M.‐Y. , Wang, X.‐S. , Li, W.‐B. and Li, Y.‐G. (2016) Over‐expression of GmGIa‐regulated soybean miR172a confers early flowering in transgenic Arabidopsis thaliana . Int. J. Mol. Sci. 17, 645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, S. , Harada, K. and Abe, J. (2012) Genetic and molecular bases of photoperiod responses of flowering in soybean. Breed. Sci. 61, 531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolter, F. and Puchta, H. (2019) In planta gene targeting can be enhanced by the use of CRISPR/Cas12a. Plant J. 100, 1083–1094. [DOI] [PubMed] [Google Scholar]
- Xia, Z. , Watanabe, S. , Yamada, T. , Tsubokura, Y. , Nakashima, H. , Zhai, H. , Anai, T. et al. (2012) Positional cloning and characterization reveal the molecular basis for soybean maturity locus E1 that regulates photoperiodic flowering. Proc. Natl. Acad. Sci. USA 109, E2155–E2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, Z.J. (2017) Research progress in whole‐genome analysis and cloning of genes underlying important agronomic traits in soybean. Chinese Bulletin Botany 52, 148–158. [Google Scholar]
- Xu, M. , Xu, Z. , Liu, B. , Kong, F. , Tsubokura, Y. , Watanabe, S. , Xia, Z. et al. (2013) Genetic variation in four maturity genes affects photoperiod insensitivity and PHYA‐regulated post‐flowering responses of soybean. BMC Plant Biol. 13, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, M. , Yamagishi, N. , Zhao, C. , Takeshima, R. , Kasai, M. , Watanabe, S. , Kanazawa, A. et al. (2015) The soybean‐specific maturity gene E1 family of floral repressors controls night‐break responses through down‐regulation of FLOWERING LOCUS T orthologs. Plant Physiol. 168, 1735–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue, L. , Li, X. , Fang, C. , Chen, L. , Yang, H. , Yang, J. , Chen, Z. et al. (2021) FT5a interferes with the Dt1‐AP1 feedback loop to control flowering time and shoot determinacy in soybean. J. Integr. Plant Biol. 63, 1004–1020. [DOI] [PubMed] [Google Scholar]
- Yue, Y. , Liu, N. , Jiang, B. , Li, M. , Wang, H. , Jiang, Z. , Pan, H. et al. (2017) A single nucleotide deletion in J encoding GmELF3 confers long juvenility and is associated with adaption of tropic soybean. Mol. Plant 10, 656–658. [DOI] [PubMed] [Google Scholar]
- Zhai, H. , Lü, S. , Wang, Y. , Chen, X. , Ren, H. , Yang, J. , Cheng, W. et al. (2014) Allelic variations at four major maturity E genes and transcriptional abundance of the E1 gene are associated with flowering time and maturity of soybean cultivars. PLoS One 9, e97636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, H. , Lü, S. , Wu, H. , Zhang, Y. , Zhang, X. , Yang, J. , Wang, Y. et al. (2015) Diurnal expression pattern, allelic variation, and association analysis reveal functional features of the E1 gene in control of photoperiodic flowering in soybean. PLoS One 10, e0135909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Zhai, H. , Wang, Y. , Tian, X. , Zhang, Y. , Wu, H. , Lü, S. et al. (2016) Functional conservation and diversification of the soybean maturity gene E1 and its homologs in legumes. Sci. Rep. 6, 29548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, F. , Lyu, X. , Ji, R. , Liu, J. , Zhao, T. , Li, H. , Liu, B. et al. (2022) CRISPR/Cas9‐engineered mutation to identify the roles of phytochromes in regulating photomorphogenesis and flowering time in soybean. The Crop Journal 10, 1654–1664. [Google Scholar]
- Zhu, J. , Takeshima, R. , Harigai, K. , Xu, M. , Kong, F. , Liu, B. , Kanazawa, A. et al. (2019) Loss of function of the E1‐like‐b gene associates with early flowering under long‐day conditions in soybean. Front. Plant Sci. 9, 1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Alignment of the amino acid sequences encoded by the DT1 and dt1 alleles in 06KG218440.
Figure S2 Off‐target analysis of E1La and E1La sequencing in selected edited lines.
Figure S3 Plant architecture of eight selected edited lines with different genotypes with E1/E1Lb mutation combinations at the Shunyi site.
Table S1 Flowering time, maturity, yield, and agronomic traits of eight E1/E1Lb T3 edited lines under three different photoperiod treatments in a greenhouse.
Table S2 Flowering time, maturity, yield, and agronomic traits of eight E1/E1Lb edited lines at three latitude field locations in 2022.
Table S3 Primers and probes for the TaqMan assay, sequencing and QRT.
Data Availability Statement
The data that supports the findings of this study are available in the supplementary material of this article.


