Abstract
We recently reported the identification of sequences in the chicken genome that show over 95% identity to the novel envelope gene of the subgroup J avian leukosis virus (S. J. Benson, B. L. Ruis, A. M. Fadly, and K. F. Conklin, J. Virol. 72:10157–10164, 1998). Based on the fact that the endogenous subgroup J-related env genes were associated with long terminal repeats (LTRs), we concluded that these LTR-env sequences defined a new family of avian endogenous viruses that we designated the ev/J family. In this report, we have further characterized the content and expression of the ev/J proviruses. The data obtained indicate that there are between 6 and 11 copies of ev/J proviruses in all chicken cells examined and that these proviruses fall into six classes. Of the 18 proviruses examined, all share a high degree of sequence identity and all contain an internal deletion that removes all of the pol gene and various amounts of gag and env gene sequences. Sequencing of the gag genes, LTRs, and untranslated regions of several ev/J proviruses revealed a high level of identity between isolates, indicating that they have not undergone significant sequence variation since their introduction into the avian germ line. Although the ev/J gag gene showed a relatively weak relationship (46% identity and 61% similarity at the amino acid level) to that of the avian leukosis-sarcoma virus family, it retains several sequences of demonstrated importance for virus assembly, budding, and/or infectivity. Finally, evidence was obtained that at least some members of the ev/J family are expressed and, if translated, could encode Gag- and Env-related polypeptides.
We recently reported the identification of a new family of avian endogenous viruses designated the ev/J family (8). This family was identified during our investigation into the origin of the novel subgroup J envelope gene that was found several years ago in unusual strains of avian leukosis virus (ALV) (called ALV-J) in England (46). Prior to the description of subgroup J viruses, five major subgroups (A to E) of chicken retroviruses had been identified (reviewed in reference 34). The subgroup specificity of chicken retroviruses is determined by sequences within the portion of env that encodes the surface glycoprotein (called SU or gp85) (11, 12, 21). Sequence comparisons have demonstrated that the subgroup A to E SU proteins are between 80 and 85% identical and that subgroup specificity is determined by sequence variations that cluster in the variable and hypervariable regions of SU (11, 12, 21, 57). The SU-coding region of the subgroup J env gene, in contrast, shows only 40% identity to those of the subgroup A to E viruses (4, 5, 9, 58). Previous reports had suggested that the subgroup J envelope gene might have been derived, at least in part, from endogenous sequences (5). This suggestion was based on the fact that the subgroup J env gene included regions that showed a high degree of identity to the env gene of a previously identified avian endogenous virus called E51, a member of the ancient endogenous virus family (EAV) (see below and references 13, 14 and 22). However, the subgroup J env gene also included regions that were not related to E51 or to any other sequence in the GenBank database, indicating that if E51-type proviruses contributed to the generation of the subgroup J env gene, there must have been other elements that contributed as well. Through the use of subgroup J-specific primers and probes, we identified endogenous sequences in the chicken genome and in chicken cDNA libraries that showed over 95% identity to the entire subgroup J env gene (8). Further studies demonstrated that these J-related env sequences were associated with long terminal repeat (LTR)-like sequences, indicating that they identified a new family of endogenous proviruses, which we designated ev/J (8). Based on these studies, we concluded that the subgroup J envelope gene of ALV-J strains was acquired in one recombination event between a member of the exogenous avian leukosis-sarcoma virus (ALSV) family and ev/J sequences. The studies described in this report were undertaken to more thoroughly characterize the content and expression of this family of endogenous proviruses.
Three major families of endogenous viruses have been identified in chickens. The best characterized are the evs (for endogenous viruses). Twenty-one evs have been identified in White Leghorn chickens, and while most lines of chickens contain several evs, animals that lack all copies of this family of endogenous viruses have been bred (2, 3, 16, 33, 53). These animals are referred to as ev-0 lines. All evs that include an env gene exhibit a common subgroup specificity designated E. Sequence comparisons reveal that the E subgroup is as highly related to the A to D subgroups of exogenous viruses as these exogenous virus subgroups are to each other, showing an overall level of identity of between 85 to 90%. Some evs, such as ev-3 and ev-6, are expressed and produce functional envelope glycoproteins, while others such as ev-1, are classified as transcriptionally silent (6, 28, 29). In addition, several nondefective evs, such as ev-2, that can give rise to infectious virus have been identified, demonstrating that they include functional gag and pol genes as well as functional cis-acting sequences that mediate RNA packaging and processing. Sequence comparisons have revealed that the gag and pol genes of evs are over 90% identical to those of the exogenous ALSVs, demonstrating that the ev family of proviruses is closely related to the ALSV exogenous viruses. This fact is further supported by the ability to detect ev sequences in Southern blots of chicken genomic DNA by using probes from the exogenous virus gag, pol, and env genes under high-stringency hybridization conditions.
The other two classes of avian endogenous viruses, which have been less extensively characterized than the evs, are the ancient viruses (EAVs) (13, 14, 22) and the avian retrotransposons (ART-CH) (27, 41). Members of each of these families, which are only distantly related to both the evs and the ALSVs, are estimated to be present at approximately 50 copies per genome, include expressed copies, and are highly deleted. For example, the ART-CH proviruses that have been examined are relatively small elements (the longest is approximately 3 kbp) and include short regions (up to 100 bp) that show homology to the pol and env genes of the ev and ALSV families. The ART-CH viruses can also contain gag-related sequences, the longest example of which includes coding sequences for matrix (MA), p10, and most of capsid (CA) but lacks coding sequences for nucleocapsid (NC) and protease (PR). Sequence comparisons show that the ART-CH and ALSV Gag proteins have an overall level of identity of approximately 35% and a level of similarity of approximately 48%.
The EAVs are a diverse group and contain different classes, including EAV-0- and E51-type proviruses; these elements differ significantly in LTR sequence composition (13, 14, 22). All members are highly deleted, and none that include an intact pol or env coding region have been identified. Little has been reported on possible gag-related sequences in EAVs. The EAVs are classified as ancient elements, since EAV proviruses have been detected in genomic DNAs of distantly related avian species such as red jungle fowl (the progenitor of the domestic chicken), indicating that EAV proviruses were established in the germ line prior to speciation. While there is evidence that at least some EAVs are expressed, there is no evidence for production of virus-related proteins.
We previously reported the ev/J env gene sequences that were identified in PCR products, genomic phage clones, and cDNA clones generated from White Leghorn chickens (8). Data obtained from these analyses clearly demonstrated that the ev/J env gene was distinct from that of any of the previously described avian endogenous families but was over 95% identical to the env genes of various ALV-J isolates. Based on these data, we proposed that the ev/J env gene was the sole source of the subgroup J env gene of ALV-J viruses. The studies described in this report were designed to further investigate the ev/J family. Sequence analyses of ev/J isolates demonstrate a high degree of identity between isolates in all regions sequenced, indicating that little variation has occurred since their introduction into the chicken germ line. Data that demonstrate that at least some members of ev/J family are expressed into RNA and are capable of encoding Gag- and Env-related polypeptides were also obtained.
MATERIALS AND METHODS
Isolation of ev/J genomic and cDNA clones and sizing of genomic inserts by PCR.
To identify genomic sequences that included ev/J proviruses, a chicken genomic library (17) was screened with ALV-J SU- and TM-specific probes as previously described (8). Seventeen clones were positive with both probes; these isolates were subsequently purified and subjected to partial sequence analysis as described in Results. To determine the lengths of the proviral inserts in the different isolates, approximately 7.5 × 103 PFU was used directly in PCRs with 10 pmol of the ev/J LTR F and ev/J LTR R primers (Table 1) in a 50-μl volume with 0.1 U of Pwo polymerase (Boehringer Mannheim) according to the manufacturer’s recommendations. Cycle conditions were 94°C for 5 min followed by 35 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 3 min, after which time samples were incubated at 72°C for 7 min before being held at 4°C. Reaction products (5 μl of a total of 50 μl) were analyzed by agarose gel electrophoresis.
TABLE 1.
Oligonucleotide primers
| Oligonucleotide | Orientation in provirus | Coordinatesa | Sequence (5′-3′) |
|---|---|---|---|
| ev/J LTR primer F | 5′-3′ | −53 to −31ev/J | TTCGTGATTGGAGGAAACACTTG |
| ev/J LTR primer R | 3′-5′ | −83 to −54ev/J | GTTACACTTGGCACACAAAGGTGGCATAAC |
| ev/J 5′ | 5′-3′ | 289 to 305ev/J | CAATGGACCAAGTCATT |
| env HR1R | 3′-5′ | 5755 to 5774HP | GTTCCGTTCTTGATCATCTG |
| ev/J 3′ gag F | 5′-3′ | 2211 to 2231ev/J | ACACCATTGGTGGCGCGTGTC |
| TM R | 3′-5′ | 6999 to 7015HP | CCCGTCACATCGCGTTC |
Coordinates are according to the numbering for either the ev/J 3A clone (ev/J) or the HPRS-103 strain of ALV-J (HP).
To isolate ev/J cDNAs, a cDNA library was screened with ALV-J-specific probes as previously described (8). This library, obtained from Sharon Soodeen-Karamath and Ann Gibbins at the University of Guelph, had been constructed from RNA isolated from 48-h-old White Leghorn chicken embryos. Sequence analysis of clones obtained from this library revealed the presence of LTR-like sequences that were used to generate ev/J-specific LTR primers (Table 1). These LTR primers were used to amplify ev/J-specific proviruses directly from genomic DNA as described below.
Isolation of ev/J proviruses from genomic DNA by PCR.
The ev/J LTR F and R primers were used in PCRs to directly amplify ev/J proviruses from line 0 chicken genomic DNA. These reaction mixtures included 20 pmol of the ev/J LTR F and R primers (Table 1), 400 ng of genomic DNA, and 1.0 U of Pwo polymerase (Boehringer Mannheim) in a total volume of 100 μl with the buffer supplied by the manufacturer. Cycle conditions were 95°C for 5 min followed by 40 cycles of 95°C for 1 min, 60°C for 1 min, and 72°C for 12 min, after which time samples were incubated at 72°C for 10 min before being held at 4°C. The reaction products were pooled, and an aliquot was analyzed on a 1% agarose gel. The primary reaction product was approximately 2.2 kbp, and subsequent cloning and sequencing of this product identified class VI of ev/J proviruses. The larger product was always present in much smaller amounts, and reaction conditions were not modified to optimize increased production of this product. It is likely that this larger, approximately 4-kbp product represented examples of the class I through V ev/J proviruses.
PCRs to detect additional ev/J classes.
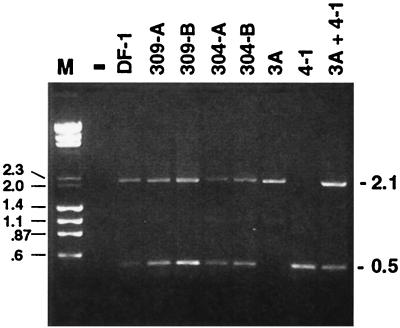
Genomic DNA was purified from two meat-type chicken lines (304 and 309) and from the line 0 White Leghorn chicken embryo fibroblast (CEF) cell line DF-1, and samples were subsequently analyzed by PCR with the ev/J 5′ and the env HR1R primers. Preliminary experiments were conducted to determine the optimal template concentrations that yielded discrete products. In all cases, only the 0.5- and 2.1-kbp bands shown in Fig. 3 were observed. As a result of this optimization, reactions were conducted with between 100 and 500 ng of purified DNA together with 20 pmol of each primer, 0.1 U of Pwo polymerase, and buffer supplied by the manufacturer in a 100-μl reaction mixture. The cycle conditions were 94°C for 5 min followed by 30 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min, after which time samples were incubated at 72°C for 7 min before being held at 4°C. Fifteen microliters of each reaction mixture was subjected to electrophoresis in a 1.45% agarose gel.
FIG. 3.
Genomic DNAs from chickens of distinct genetic backgrounds yield indistinguishable ev/J-specific PCR products. Genomic DNAs from White Leghorn (DF-1) or from two samples of two different meat-type (304 and 309) chickens were analyzed by PCR with the ev/J 5′ primer located in the 5′ UTR and the env HR1R primer (Table 1). In parallel reactions, DNAs from ev/J isolates 3A (class I) and 4-1 (class VI) were also amplified with the same primers either alone or together as indicated above the lanes. As shown, reactions that included the 3A or the 4-1 clone generated the expected products of 2.1 and 0.5 kbp respectively. When both DNAs were included in one reaction, there was roughly equal amplification of each DNA, indicating no size bias in amplification, at least between these two sizes. Bands that migrated identically to those obtained with cloned DNA templates were also seen in reaction products generated with chicken genomic DNAs isolated from the White Leghorn sample (DF-1) and from both samples of the two meat-type lines (304 and 309). Lane M, marker that included both lambda DNA digested with HindIII and φX174 DNA digested with HaeIII. The lane marked − contained a sample from a PCR mixture that lacked template DNA; similar negative results were obtained when either polymerase or primers were omitted from reactions (data not shown).
Reverse transcription-PCR (RT-PCR) assays.
Total RNA was isolated from a continuous line of line 0 fibroblasts (DF-1 cells) by the method of Chomczynski and Sacchi (15). Two hundred micrograms of total RNA was incubated twice with 150 U of RNase-free DNase I (Stratagene) for 2 h at 37°C and subsequently extracted with phenol-chloroform and precipitated by the addition of ethanol. Five micrograms of total, DNase I-treated RNA was then reverse transcribed with either oligo(dT) (85 pmol) or the ev/J LTR R primer (2 pmol) by using the SuperScript II kit (GIBCO-BRL) according to the manufacturer’s instructions in a 20-μl reaction volume. Samples were then treated with 2 U of RNase H (GIBCO-BRL) for 30 min at 37°C. Two-microliter portions of these reaction mixtures were then used in PCRs with the ev/J 3′ gag F and TMR primers (Table 1). The cycle conditions were as follows: 94°C for 2 min; 10 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 2 min; 25 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 2 min with 20 s of elongation for each cycle; and then incubation of samples at 72°C for 7 min before holding at 4°C. Reaction products were analyzed by agarose gel electrophoresis.
RESULTS
Structure of ev/J proviruses in phage genomic clones.
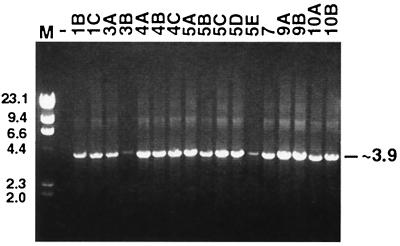
To obtain cloned copies of ev/J proviruses resident in the chicken genome, SU and TM subgroup J-specific env gene probes were used individually to screen a chicken genomic library as described previously (8). This screening led to the identification of 17 phage clones that were positive with both env gene probes. To determine the sizes of proviral inserts in each clone, the 17 phage clones were subjected to PCR with primers specific for ev/J LTR sequences (Table 1) as described in Materials and Methods. As shown in Fig. 1, a discrete product of approximately 3.9 kbp was amplified with all phage templates. These data indicated that all isolates included two viral LTRs, supporting the hypothesis that the subgroup J env-related sequences detected previously were part of LTR-containing proviruses. These data also demonstrated that all inserts were of approximately equivalent length, suggesting that all ev/J proviruses might be similar in content. In any case, the relatively small sizes of the amplified products indicated that these proviruses did not contain a full complement of gag, pol, and env gene-related information, which requires a coding capacity of approximately 7 kbp.
FIG. 1.
All ev/J proviruses isolated from a genomic library are similar in length and contain two LTRs. The 17 ev/J-containing phage clones isolated from a chicken genomic library were used in PCRs with the ev/J LTR F and ev/J LTR R primers (Table 1) as described in Materials and Methods. Reaction products were subjected to electrophoresis in a 1% agarose gel (lanes 3 through 19), and the results obtained demonstrate that all isolates generated one band of approximately 3.9 kbp. Lane M, lambda HindIII-digested marker DNA. Numbers above the sample lanes indicate the clone designation of each isolate. The lane marked − contained a sample from a PCR mixture that lacked phage DNA; similar negative results were obtained when either polymerase or primers were omitted from reaction mixtures (data not shown). Numbers on the left and right indicate kilobase pairs.
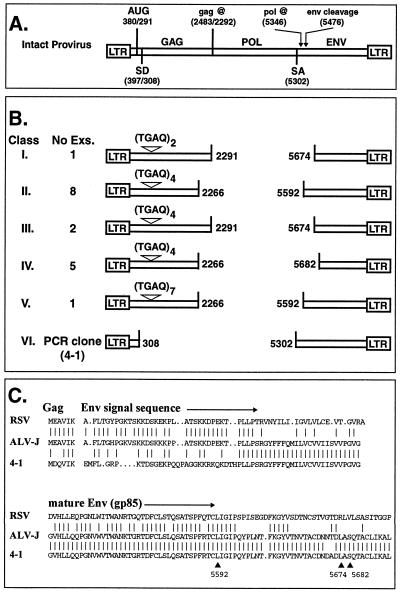
To further characterize the content of ev/J proviruses, additional PCRs and sequence analyses were conducted, and the results obtained were compared with those generated with wild-type proviruses. An intact provirus contains the gag, pol, and env genes flanked by two LTRs (Fig. 2A) and encodes two classes of RNA transcripts (reviewed in reference 60). The first transcript is a full-length RNA that can serve either as virion RNA or as mRNA for synthesis of the Gag and Gag-Pol polyproteins by using an AUG initiation codon near the 5′ end of the message. In avian retroviruses, the Gag mRNA includes the MA-, p10-, CA-, NC-, and PR-coding regions in one reading frame; a frameshift is required to generate the Gag-Pol precursor polyprotein. The second mRNA is a spliced message that is generated by processing full-length transcripts at the splice donor and acceptor sites indicated in Fig. 2A; this mRNA directs synthesis of the Env polyprotein. Translation of Env initiates at the same AUG near the 5′ end of the RNA transcript that is used to initiate Gag and Gag-Pol translation; the primary env gene translation product therefore includes six amino acids from the gag coding region. These 6 amino acids together with an additional 56 (in Rous sarcoma virus [RSV]) or 58 (in ALV-J) amino acids from the env coding region form the signal sequence at the amino terminus of the Env primary translation product (Fig. 2C). This sequence is removed by a site-specific cleavage event that is one step required to generate the mature gp85 (SU) and gp37 (TM) glycoproteins.
FIG. 2.
Structures of ev/J proviruses. (A) Structure of a wild-type provirus. Shown is the basic structure of a wild-type provirus, which contains the gag, pol, and env genes flanked by two LTRs. The AUG codon at nucleotide 380/291 is used to initiate the Gag and Gag-Pol polyproteins from full-length mRNA (the numbering system for the gag gene corresponds to that defined for RSV [number before the slash] and that predicted for the ev/J 3A clone [number after the slash]). Gag translation terminates at a UAG codon at 2483/2292 (2292 is the first nucleotide of the triplet that would contain the stop codon; sequence analysis demonstrates that clone 3A actually ends at nucleotide 2291). Also shown are the splice donor (SD) and splice acceptor (SA) sites used to generate the subgenomic env mRNA. In RSV, translation of the Env protein initiates at the AUG codon at nucleotide 380, which is retained in the env mRNA, and therefore the Env primary translation product includes six amino acids from Gag at its amino terminus (see also panel C). After translation, the amino-terminal 62-amino-acid signal sequence (6 amino acids derived from Gag and an additional 56 amino acids from Env) is cleaved during maturation of the Env polyprotein. This cleavage site is noted as occurring at nucleotide 5476 (numbering according to that established for the subgroup J env gene of HPRS-103 [4]), which is the number of the nucleotide that encodes the last amino acid in the signal sequence peptide. (B) Structure of ev/J proviral clones. The 17 ev/J proviruses isolated from the phage genomic library were divided into five classes (I to V) based on the identity of their deletion endpoints in gag and env and on the number of TGAQ repeats in Gag. The gag deletion endpoints are numbered according to the ev/J 3A clone, while the env deletion endpoints are numbered according to the prototypical subgroup J ALV HPRS-103. Also included is the number of examples (Exs.) of each class that were identified. Also shown is the structure of the ev/J provirus isolated by direct PCR amplification of line 0 genomic DNA (see Materials and Methods). This clone (4-1) defined a distinct class of provirus (class VI) that appears to be a faithful copy of a functional env mRNA. (C) Amino acid sequence of the primary Env translation products of RSV and of ALV-J together with that predicted for the 4-1 clone. Vertical lines between the sequences indicate identical amino acids; dots indicate gaps in the sequence. The arrows and numbers below the 4-1 sequence mark the deletion endpoints in env for the ev/J class.
Preliminary sequence data suggested that all of the cloned ev/J proviruses contained a deletion in pol (data not shown). To verify this, the proposed gag-env deletion junction regions of all 17 phage clones were subjected to sequence analysis, and the results obtained are shown diagrammatically in Fig. 2B. As indicated, sequence analyses confirmed that all of the ev/J proviruses analyzed contained an internal deletion that removed the entire pol gene and various amounts of gag and env. Two breakpoints were identified within gag (at nucleotides 2291 and 2266, using the numbering system for the ev/J 3A clone with the beginning of the LTR R region set as +1), and three different breakpoints were identified in env (at nucleotides 5592, 5674, and 5682, using the numbering system for the HPRS-103 strain of ALV-J [4]; env breakpoints are also shown in Fig. 2C). The gag and env gene breakpoints were found in different combinations giving rise to three distinct classes of proviruses (2291 with 5674, 2266 with 5592, and 2266 with 5682; additional classes shown in Fig. 2B represent a difference within the gag gene that is described below). Fourteen of the 17 isolates contained the same gag gene deletion breakpoint at nucleotide 2266. This position is within the viral PR gene (see below), and, if transcribed and translated, the provirus would direct synthesis of a protein that contained a 9-amino-acid truncation from the carboxy terminus of PR. As with other avian retroviruses, the PR-coding region of the ev/J proviruses is in the same reading frame as the remainder of the gag gene.
Three isolates contained a gag gene breakpoint at nucleotide 2291. This breakpoint leaves the gag coding region fully intact but results in the removal of the Gag translation stop codon. Thus, if ev/J proviruses with this structure are transcribed and translated, they could give rise to a full complement of gag gene products in the form of an unprocessed polyprotein; it is unclear, however, whether this product could be processed into the individual gag gene products.
The three env gene breakpoints were at nucleotides 5592, 5674, and 5682. These sites are all positioned downstream of both the splice acceptor site used to generate the env mRNA in wild-type viruses (nucleotide 5302) and the cleavage site within the Env polyprotein that generates the mature gp85 protein (encoded by nucleotide 5476 [Fig. 2C]). None of these isolates would therefore be capable of encoding a functional Env protein. However, all of the env gene deletion endpoints are in frame with the gag gene sequences, and therefore, if transcribed and translated, these proviruses could encode fusion proteins that contained Gag- and Env-related information.
Structure of a PCR clone.
A second approach to isolate ev/J clones was to amplify ev/J proviruses from total genomic DNA isolated from ev-0 cells by using the ev/J-specific LTR primers listed in Table 1. This approach generated one major product of approximately 2.2 kbp and a second, minor product of approximately 4.0 kbp (data not shown). The size of the larger product is the same as that of the class I through V proviruses isolated from genomic clones, indicating that this PCR product derives from proviruses of these classes. However, since the reaction conditions used in these preliminary PCRs produced only a relatively small amount of this product, no clones corresponding to the 4.0-kbp proviruses were further analyzed. Cloning and sequencing of the major 2.2-kbp product led to the identification of the sixth class of ev/J proviruses shown in Fig. 2B. A complete sequence analysis of a clone of this class (isolate 4-1) demonstrated that it is an apparently accurate DNA copy of an ev/J-encoded Env mRNA. This was determined by finding that the gag-env deletion endpoints in clone 4-1 were at nucleotides 308 and 5302, positions that correspond to the locations of the proviral splice donor and acceptor sites, respectively. The predicted sequence of the amino-terminal end of the Env primary translation product of clone 4-1 together with the corresponding regions from RSV and ALV-J is shown in Fig. 2C. As shown, clone 4-1 shows a four-of-six amino acid identity with RSV and ALV-J within the Gag portion of the predicted protein sequence but is only weakly related (approximately 45% identical) to the other two viruses over the first 30 amino acids of the env coding region. Downstream of this region of low identity, however, the 4-1 clone (Fig. 2C), as well as all other ev/J env genes sequenced (reference 8 and data not shown), shows over 95% identity to the subgroup J env genes of ALV-J viruses. The finding that the env gene homology between ev/J proviruses and ALV-J undergoes a transition from low to high identity within the region that encodes the signal sequence of the Env protein suggests that the recombination event that led to the acquisition of the subgroup J envelope gene likely occurred at or immediately downstream of the region encoding the PLLP sequence (residues 31 to 34 of the predicted amino acid sequence of the 4-1 env region) shared by all isolates (Fig. 2C).
Failure to detect pol sequences associated with ev/J proviruses in animals of different genetic background as determined by PCR.
The data shown above demonstrate that all ev/J proviruses isolated from the genomic library and by PCR lacked pol gene-related information. This was an unusual finding, since although other endogenous virus families are frequently defective due to deletions and mutations, we are aware of no other examples that so specifically delete one region of the genome. We were therefore interested to investigate whether we might be able to detect ev/J proviruses that included pol-related sequences in total genomic DNAs isolated from chickens with distinct genetic backgrounds. The chicken genomic DNA used to generate the phage genomic library described above was from a White Leghorn animal that contained the endogenous viruses ev-1, -3, and -6, while the DNA used to generate the ev/J PCR products was from a cell line derived from a line 0 White Leghorn animal (DF-1 cells). We therefore obtained DNAs from two chicken lines that are distinct from White Leghorns. The first was from an experimental meat-type line called White Rock (304 samples in Fig. 3), and the second was from a commercial line of meat-type birds (309 samples in Fig. 3). These DNAs, in parallel with DNA from the White Leghorn DF-1 cell line, were used in PCRs to attempt to identify ev/J proviruses that might include additional, pol-related information. The primers used in this analysis are listed in Table 1 and were the ev/J 5′ primer and the env HR1R primer. The regions corresponding to these oligonucleotides have been sequenced in at least four ev/J isolates, and each shows only a one-nucleotide variation among all clones (data not shown). They were therefore chosen as good candidates to screen for ev/J proviruses.
If all ev/J proviruses in the genome lack pol-related information and correspond in structure to the classes of proviruses shown in Fig. 2, we would expect the ev/J 5′ and HR1R primers to generate PCR products of approximately 2.1 kbp (classes I through V) and 0.5 kbp (class VI); generation of larger products would suggest the presence of proviruses that might also include pol gene information. As shown in Fig. 3, PCRs with DNAs from the line 0 DF-1 fibroblasts as well as two samples each of the commercial (309) and experimental meat-type (304) birds generated products that were indistinguishable from those generated in control amplifications that included a representative of the pol deletion class VI (isolate 4-1) and the class I-V (isolate 3A) proviruses. The mixed amplification reaction that included both the 3A and 4-1 clones demonstrated that both of these ev/J proviruses were amplified efficiently when present together, indicating that, at least for proviruses in these size ranges, there was not a preferential amplification of the smaller provirus. These data therefore suggest that at least the majority of ev/J proviruses present in the cells tested lack pol-related information. This conclusion is further supported by the fact that we have consistently failed to generate PCR products by using a pol primer that is 100 to 95% conserved in at least seven other types of avian retroviruses (data not shown). Thus, the data obtained to date indicate that the lack of pol-related information is a common property of the ev/J proviruses we have analyzed that are present in chickens with various genetic backgrounds.
gag genes.
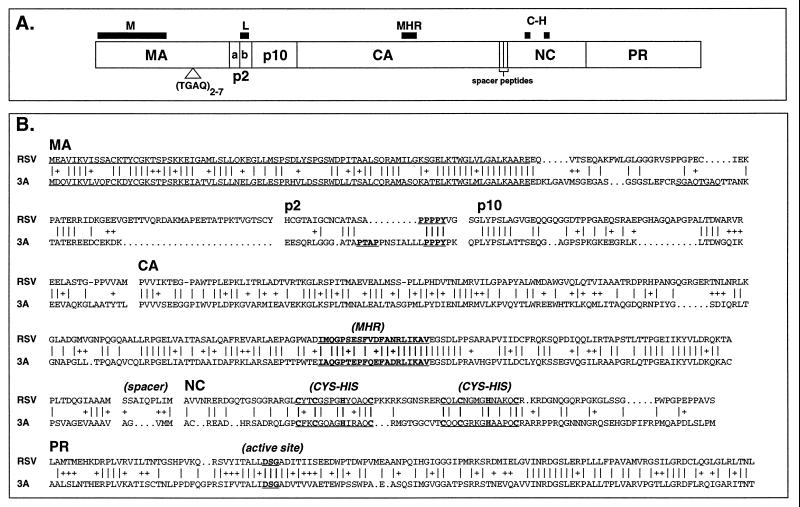
Previous analyses have demonstrated that the gag genes of the ev (subgroup E) family of endogenous viruses show an extremely high level of identity (95 to 98%) to those of the exogenous ALSV viruses. In contrast, another family of avian endogenous viruses, the avian retrotransposons (ART-CH), contain gag genes that are highly deleted and that show only a limited homology (35% identity and 48% similarity at the amino acid level) to the ALSV gag genes. The gag genes of the EAV family of avian endogenous viruses have not been sequenced. To determine the nature of the ev/J gag gene, three ev/J isolates were chosen for sequence analysis, and the results obtained were compared with those published for the RSV Gag protein (Fig. 4). The clones chosen for analyses were clone 3A (class I), clone 1C (class IV), and a PCR amplification product of 5E (class II). This analysis revealed that all isolates were devoid of stop codons in gag yet all were truncated. The 3A clone contained an intact coding region but lacked the gag translational stop codon, while the 1C and 5E clones terminated at nucleotide 2266 and therefore lacked the carboxy-terminal nine amino acids of PR. Comparison of the predicted amino acid sequences of the ev/J Gag proteins from the three sequenced ev/J proviruses showed a 92% level of identity between isolates, indicating a low degree of variation since their introduction into the chicken germ line. Comparison with other avian virus gag genes showed overall 59 and 50% levels of similarity and identity, respectively, to the highly deleted Gag sequence of the ART family of avian endogenous viruses (data not shown) and a 46% level of identity and a 61% level of similarity to the RSV Gag protein (Fig. 4B). As described in more detail below, however, the degree of identity to the RSV gag gene varied between and within each gene, and several regions in Gag that are required for assembly, budding, and/or infectivity are retained in ev/J proviruses.
FIG. 4.
Predicted amino acid sequence of the ev/J Gag protein compared with that of RSV. (A) Line drawing of the RSV Gag protein and locations of several sequences required for Gag function. The M, L, MHR, and C-H sequences are conserved motifs within retroviral Gag proteins, and their proposed functions are described in the text. The TGAQ sequence marked in Gag shows the approximate location of the repeated motif encoded by sequences within the ev/J gag gene. (B) Predicted amino acid sequence of the ev/J 3A clone compared with that of RSV. Vertical lines between the sequences indicate identical residues, + signs indicate conservative changes, and dots indicate gaps in sequence. The M, L, MHR, and other motifs mentioned in the text are underlined. The boundaries of predicted coding regions within the ev/J 3A clone are arbitrary and based on the best fit with RSV.
MA.
Studies with the RSV Gag protein have demonstrated that the first 86 amino acids of MA are required for efficient particle formation and for association of the Gag polyprotein with the plasma membrane (40, 59); this region is therefore referred to as the M region (Fig. 4A). The remainder of MA, however, is dispensable for budding and infectivity (40, 59, 63). Comparison of the predicted sequence of the first 86 amino acids of the ev/J MA-coding region with that of RSV shows a 58% level of identity and 72% level of similarity, while the remaining portion of MA shows only 24 and 34% levels of identity and similarity, respectively, between these proteins. The high degree of conservation between the amino-terminal portion of the ev/J MA protein and that of RSV suggests that the ev/J protein might have retained the membrane association function normally associated with this protein.
Sequence analysis of the 3A, 1C, and 5E clones revealed one major region of variability between these isolates, which localized towards the carboxy-terminal portion of the MA-coding region (beginning at amino acid 110 of the ev/J MA protein; underlined in Fig. 4B). In particular, it was found that these proviruses contained either two (3A) or four (1C and 5E) copies of a 12-bp sequence that encoded a 4-amino-acid repeat of TGAQ (or, as in the first repeat of the 3A clone in Fig. 4, SGAQ). We therefore sequenced this portion of all 17 ev/J clones and found three classes of clones: the two and four repeats noted above as well as one isolate that contained seven repeats. The most common number of repeats was four (15 of 17 isolates; see Fig. 2B). There was only one example each of the two-copy repeat (clone 3A, class I) and of the seven-copy repeat (clone 3B, class V). We are currently investigating the potential significance and stability of this sequence.
p2a and p2b.
During processing of the Gag polyprotein, the initial cleavage of MA includes the p2 region (Fig. 4A), which is removed in two subsequent proteolytic processing events to generate p2a and p2b (47, 48). In RSV the p2b region contains a proline-rich sequence (PPPPY; underlined in Fig. 4B) that is required late in infection for release of budding particles from the cell (44, 62, 64); this sequence is therefore referred to as the L sequence. A distinct but functionally equivalent L sequence (PTAP) has been identified in the p6 protein located in the carboxy-terminal region of the human immunodeficiency virus (HIV) Gag polyprotein (26, 30, 44). Examination of the ev/J sequence revealed the presence of a PPPY (clone 3A; Fig. 4B) or PPPPY (clones 1C and 5E; data not shown) sequence within the predicted p2b region, indicating that the ev/J Gag protein has retained this functionally important sequence. Interestingly, we also noted the presence of the HIV-specific PTAP sequence immediately upstream of the PPPPY sequence in all three ev/J proviruses that were sequenced in this region (Fig. 4B and data not shown). The finding of both versions of the L sequence in the ev/J Gag protein suggests that this family of proviruses might have two functional copies of this motif.
p10.
Comparison of the predicted amino acid sequence of the ev/J p10 protein with that of RSV p10 reveals 43 and 59% levels of identity and similarity, respectively, between these two proteins. Since discrete motifs that might be required for viral assembly and/or infectivity in p10 have not been defined, it is unclear whether functionally important sequences might be included in this region of the ev/J gag gene.
CA.
The predicted CA-coding region of ev/J proviruses shows 51 and 69% levels of identity and similarity, respectively, to the RSV CA-coding region. Within the CA protein of RSV is the major homology region (MHR), a 20-amino-acid sequence that is the most highly conserved motif among all retroviruses (18, 45). Although the function of this region is not clearly understood, it has been proposed that the MHR is required for postbudding events that generate a core that is structurally sound and infectious (18). The MHR contains three residues that are invariant: the glutamine at position 3, the glutamate at position 7, and the arginine at position 15. There are also several other highly conserved residues within the MHR, including a glycine at position 4, a proline at position 5, a serine or proline at position 8, a phenylalanine or tyrosine at position 12, and a phenylalanine or leucine at position 16. Comparison of the ev/J sequence with those of other viruses shows that it retains the invariant and the highly conserved residues noted above as well as several other residues within and immediately flanking the MHR. This high degree of conservation again suggests the retention of function of at least this portion of CA.
NC.
The NC proteins of RSV and HIV are of demonstrated importance in mediating Gag-RNA and Gag-Gag interactions and therefore have been classified as containing I (for interaction) domains (reviewed in reference 55). Sequence analyses of NC proteins have revealed that they contain clusters of basic amino acids as well as Cys-His motifs that may be required for efficient RNA-Gag interactions during packaging and/or virion maturation (1, 23–25, 37, 38, 50). While the exact relationship between these motifs and NC function is still under investigation, the conservation of these motifs in different retroviruses suggests that they are functionally significant. A comparison of the predicted amino acid sequence of the ev/J NC protein with that of RSV NC revealed that they show a relatively low level of similarity (36 and 42% levels of identity and similarity, respectively). However, the ev/J NC protein contains the cysteine and histidine residues that define the conserved zinc fingers in RSV, although the spacing between the two motifs is three amino acids shorter in the ev/J Gag protein. This region of the ev/J NC protein is also rich in basic residues, a feature of proposed importance for NC function.
PR.
The final region of the gag gene encodes the viral PR. Comparison of the ev/J and RSV PR-coding regions shows a relatively high degree of identity at the amino acid level (48% identity and 71% similarity). Importantly, the predicted ev/J PR retains the amino acids (DSG) that define the active site of the enzyme (10, 56) as well as a cluster of residues surrounding the active site.
UTRs and LTRs.
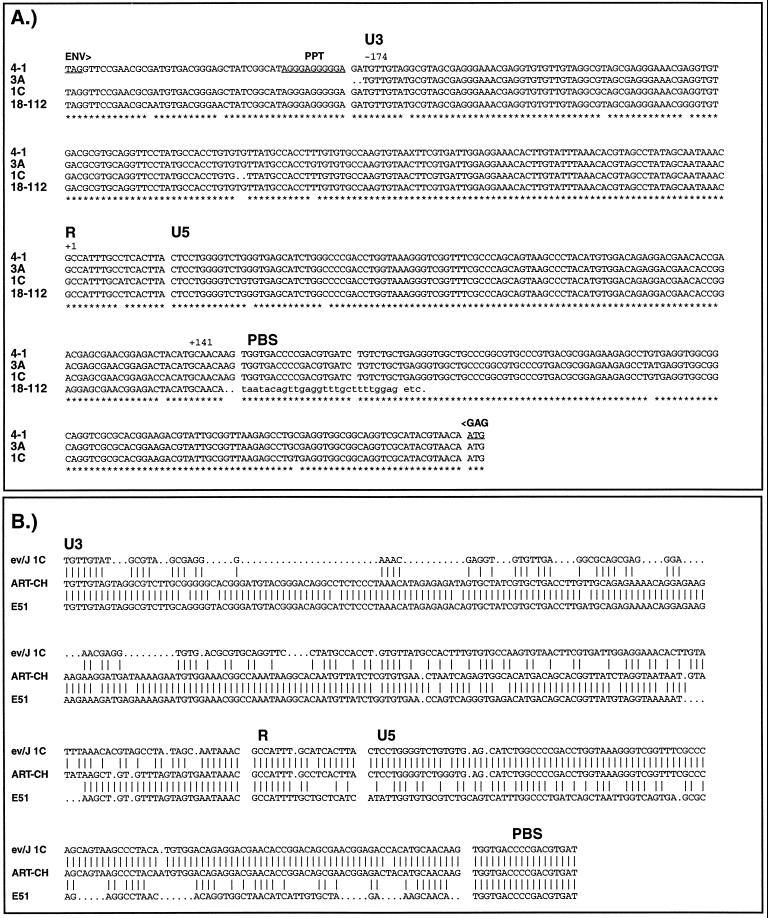
Sequences important in regulating transcription, translation, and RNA stability and processing have been localized to retroviral LTRs and to 5′ and 3′ untranslated regions (UTRs) immediately adjacent to viral LTRs. These regions in selected ev/J proviruses were therefore sequenced (Fig. 5A) and included the 5′ and 3′ LTRs and UTRs of clone 1C, the 5′ LTR and 5′ UTR of clone 3A, and the 5′ and 3′ UTRs of clone 4-1. Since the 4-1 clone was generated by using ev/J LTR primers, it was only possible to obtain partial sequence information from each LTR; the sequence of the 4-1 LTR shown in Fig. 5A is therefore a composite of information obtained from each end. Also included in Fig. 5A is the 3′ LTR from the 18-112 ev/J cDNA previously described (8). As shown, this cDNA was derived from an RNA that extended through the entire 3′ LTR into flanking cellular DNA and that therefore was not properly polyadenylated at the end of the R region. This was also seen with the other ev/J cDNAs sequenced (data not shown), suggesting that ev/J LTRs do not efficiently mediate polyadenylation of viral transcripts.
FIG. 5.
Sequences of ev/J LTRs and UTRs. (A) Shown are the nucleotide sequences of the LTRs and UTRs of genomic clones 3A (class I), 1C (class III), and 4-1 (class VI) and cDNA clone 18-112. The 3A sequence is that of the 5′ LTR and 5′ UTR. The sequences of the 1C 5′ and 3′ LTRs were identical except for the 2 bp missing from each end at the cell-LTR junction, so only one sequence is shown together with the associated 5′ and 3′ UTRs. The 4-1 provirus (class VI) was isolated by PCR amplification from genomic DNA with ev/J-specific LTR primers, and the sequence shown is therefore a composite sequence derived by combining information from the partial 5′ and 3′ LTRs. The Env translational stop codon (TAG) and the Gag translational initiation codon (ATG) are underlined. The lowercase letters at the end of the 18-112 sequence represent cellular DNA sequences. PPT, polypurine tract; PBS, primer binding site. Asterisks below the sequences represent nucleotides that are conserved among all isolates. Note that there are a total of only 12 differences out of 315 between any of the LTRs. (B) Sequence comparison of the ev/J 1C LTR with those of the ART-CH and EAV E51 proviruses. Vertical lines between sequences indicate identity, while dots indicate gaps in the sequence.
As indicated, the ev/J LTRs are 315 bp long and showed an extremely low degree of variability (maximum variation of 3.5%); sequencing of the 1C 5′ and 3′ LTRs demonstrated that they were identical. Two nucleotides were missing from all LTRs at their junctions with cell DNA, indicating that their insertions into the genome were via the normal integration mechanism. These data indicate that ev/J proviruses have undergone little variation since their introduction into the germ line and suggest that their acquisition was a relatively recent event. This is in contrast to data obtained with the ancient family of endogenous viruses (the EAV family), whose members, such as E51, EAV-0, E13, and E33, show an extremely high degree of variation within the LTR (13, 14). Sequence analysis of the U3 regions of ev/J LTRs revealed minimal similarity to U3 regions of exogenous viruses and to ev LTRs (data not shown) and only limited similarity to the U3 regions of the ART-CH and EAV E51 proviruses, which are 90% identical to each other (Fig. 5B). The lack of similarity between ev/J and other retroviral LTRs within U3 is in contrast to data obtained with the ev/J R and U5 regions, which are over 95% identical to the analogous regions from the ART-CH proviruses (which show only 55% identity to the E51 LTR in these regions). This high degree of similarity between ev/J and ART-CH elements extends into the 5′ UTR through the first 85 amino acids of MA (where the two proteins are 88% identical [data not shown]). All ev/J proviruses sequenced contain a tRNATrp primer binding site identical to that found in ALSV, ART-CH, and EAV proviruses (Fig. 5B and data not shown). Examination of the ev/J 3′ UTR (Fig. 5A) shows that there are only 34 nucleotides between the end of the env coding region and the polypurine tract. This is in contrast to the 5′ UTRs of ALSVs, which are between 200 and 350 bp in length and which contain an approximately 150-bp sequence called the direct repeat that is required for efficient RNA transport to the cytoplasm (42, 43) and RNA stability and/or Gag assembly (51, 52). There is no evidence for a direct repeat in ev/J proviruses.
ev/J proviruses are expressed.
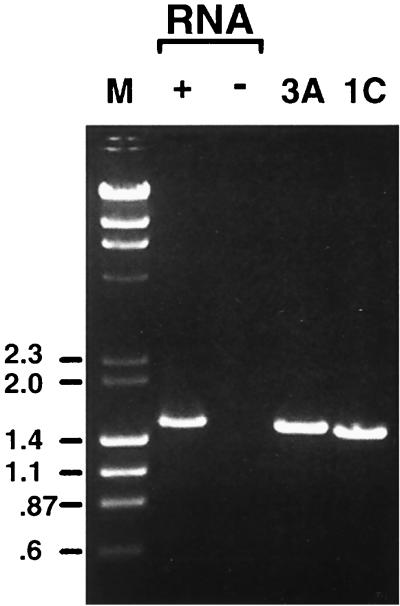
As mentioned above, ev/J-encoded cDNAs were isolated from a library made from chicken embryonic RNA, indicating that at least some ev/J proviruses are expressed in embryonic tissues. However, we were repeatedly unsuccessful in detecting ev/J-encoded RNAs in Northern blots under conditions where relatively weakly expressed messages such as c-myc were readily detectable (data not shown). One possible explanation for this result might be that ev/J-encoded RNAs are heterogeneous in length due to the fact that they are not efficiently polyadenylated within the viral LTR (see the sequence of the 18-112 cDNA in Fig. 5A). Therefore, RT-PCRs were conducted to evaluate ev/J expression with RNA isolated from CEFs. For these experiments, a variety of primers from the ev/J gag and env genes and from the ev/J LTRs were used and yielded positive results; those obtained with the ev/J 3′ gag and TM R primers are shown in Fig. 6. As shown, RT-PCRs that included cDNA generated from line 0 RNA produced a prominent band of approximately 1.5 kbp (lane 2), the size expected for transcripts encoded by the ev/J I through V proviral classes. No product was detected in samples in which oligo(dT) had been omitted from the cDNA synthesis reaction mixture (lane 3) or when either primers or polymerase was omitted from reaction mixtures (data not shown). Also included in Fig. 6 are PCR products generated with the ev/J 3′ gag and TM R primers from the cloned 3A and 1C ev/J proviruses. The size difference between the 3A and 1C PCR products is expected based on the different gag and env deletion endpoints in these clones, which should result in products that differ by 33 nucleotides. The RT-PCR product generated from line 0 CEF RNA is larger than the product generated from either the 3A or 1C clone, indicating that it was derived primarily from proviruses of the largest ev/J classes (II and V). This result was verified by direct sequence analysis of the RT-PCR product (data not shown).
FIG. 6.
ev/J proviruses are transcribed. RNA isolated from line 0 CEFs was reverse transcribed by using the SuperScript II kit (GIBCO-BRL) in either the presence (+) or absence (−) of an oligo(dT) primer, and products were subsequently amplified by PCR with primers specific for the 3′ end of the ev/J gag gene and for the subgroup J-specific TM region of the env gene (primers ev/J 3′ gag F and TM R [Table 1]). Parallel PCRs were conducted with the ev/J 3′ gag F and TM R primers with cloned 3A and 1C ev/J proviruses as templates. As shown, a prominent amplification product was generated in the cDNA-containing sample (lane 2) but was absent in samples in which oligo(dT) (lane 3), or reverse transcriptase or primers (data not shown) were omitted from reactions. The product generated from the 3A clone was slightly larger than that generated from the 1C clone, a result that would be expected due to the larger size of the class I versus class III proviruses (see Fig. 2). The RT-PCR product from the CEF RNA was larger than the class I product, indicating that it was derived from an RNA(s) encoded by class II and/or class V proviruses, a conclusion supported by direct sequence analysis of the RT-PCR product (data not shown). Lane M, markers (in kilobase pairs).
DISCUSSION
The data presented in this report further document the structure and expression of the ev/J family of avian endogenous viruses that were identified recently (8). Based on Southern blot analyses (reference 8 and data not shown), we estimate that there are between 6 and 11 ev/J proviruses in the genome of White Leghorn chickens. Screening of approximately three genome equivalents of a chicken genomic library led to the identification of 17 phage clones that hybridized with both SU and TM ALV-J-specific probes, a number in agreement with approximately six ev/J proviruses per genome. In addition, sequence analyses of ev/J proviruses identified by direct PCR amplification of chicken genomic DNA and from a phage genomic library identified six different classes of ev/J proviruses. The five classes identified in the genomic library were not equally represented. Instead, two classes (class II and class IV) were found eight and five times, respectively, while classes I, II, and V were represented by only one, two, and one isolate, respectively. We are therefore currently investigating which of the different isolates of ev/J proviruses within each class arose from the same provirus and which represent proviruses inserted at different locations in the cellular genome. In either case, the present data indicate that the ev/J proviral copy number is significantly less than the 50 copies per genome estimated for the EAV (13) and ART-CH (27) families of endogenous viruses.
Six classes of ev/J proviruses were identified. The first five classes, isolated from genomic phage clones, were strikingly similar in structure. All contained two LTRs, large regions with homology to ALSV gag and env genes, and an internal deletion that removed the entire pol coding region. The major differences noted between the proviral classes were the locations of the gag and env gene deletion breakpoints and the numbers of TGAQ repeats within the MA portion of Gag. In an attempt to determine if other regions of the 17 clones might contain significant variation, PCRs were conducted with a variety of primers that spanned the entire ev/J provirus (data not shown). Analyses of the products generated failed to reveal regions with detectable clone-to-clone variation; we estimate that differences of between at least 50 and 100 bp could have been detected, depending on the primer pair used. These data indicate that the ev/J family of proviruses are likely not ancient viruses similar to the EAV family of proviruses, which are an extremely diverse family of proviruses that were acquired prior to speciation of the domestic chicken (13). The sixth class of ev/J provirus, generated as a PCR product from genomic DNA, also lacked the pol gene and appeared to be a precise copy of an Env mRNA. These data therefore indicate that deletion of the pol gene is a common feature of all ev/J proviruses that we have analyzed, although it is possible that there are additional proviruses that we have failed to detect either in genomic libraries or by PCR. Based on the present data, a member of the identified ev/J family of proviruses is therefore not a likely candidate as the source of the reverse transcriptase activity that has been reported in uninfected line 0 CEFs (61).
Sequence analysis revealed that the gag and env gene deletion endpoints in the class I through V proviruses were not random, a finding that would have been expected if the ev/J proviruses analyzed suffered a pol gene deletion after insertion into the genome. Therefore, it is likely that the analyzed members of this family contained a pol deletion prior to their establishment in the avian germ line. This hypothesis necessitates that their replication and integration were dependent on the contribution of a helper virus that provided Pol function. It further suggests that the ev/J proviruses encode RNAs that can be packaged (see below).
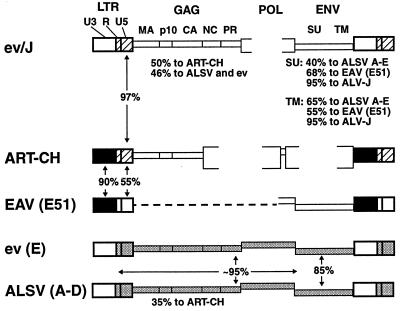
Comparisons of the LTRs and UTRs of several of the avian endogenous virus families revealed an interesting relationship between the ev/J family and the ART-CH family of proviruses (Fig. 7). Specifically, it was found that while the U3 regions of the ev/J and ART-CH LTRs were not highly related, the R and U5 regions of these proviruses were virtually identical (only four mismatches out of 140 bp). This relationship was the opposite of that found between the ART-CH and EAV E51 LTRs, which are virtually identical throughout the U3 region but much more poorly related within R and U5. The finding that the level of identity among these LTRs switches at the U3-R boundary raises the possibility that these LTRs might have arisen by recombination during reverse transcription, where the first step in replication is copying the R-U5 region from one viral strand before the first-strand transfer to a 3′ end that includes the same (or a related) R sequence. This possibility, together with the regions of similarity that have been noted between the ev/J env gene and that of the EAV E51 provirus, indicates that the avian endogenous viruses arose from related viruses and/or that their genomes have undergone (and possibly still are undergoing) genetic exchange.
FIG. 7.
Comparison of avian retrovirus genomes. Shown are the genomes of the indicated exogenous and endogenous avian retroviruses. Brackets indicate regions that are deleted in the proviruses, while the dashed line in the EAV provirus indicates regions that have not been sequenced. Regions shaded similarly indicate those that have high sequence identity; the percent identity of either the nucleotide sequence (for the LTRs) or the predicted amino acid sequences (for Gag- and Env-coding regions) is included either between or under the proviruses. The absence of shading indicates regions that show less than 70% identity to analogous regions from the other virus groups.
The 5′ and 3′ UTRs of avian viruses include sequences of suggested or demonstrated importance for translation regulation, regulation of splicing, RNA transport, and RNA stability. Examination of the 5′ UTR of ev/J proviruses indicates that it contains one open reading frame; the three open reading frames in the RSV 5′ UTR are of proposed importance in regulating whether full-length RNA molecules serve as virion RNA or mRNA (19, 20, 39, 54). However, the 5′ UTR of ev/J proviruses is considerably shorter than the corresponding region from RSV and in particular is missing sequences that include the RSV packaging signal (1, 7, 35, 36). Analysis of the ev/J 5′ UTR with the Mulfold folding program (data not shown) indicates that the sequence in this region is distinct from that of RSV and is unable to fold into a similar secondary structure that is of proposed importance in packaging (7). This might suggest that the ev/J-encoded RNAs would be inefficiently packaged into virions encoded by helper virus. However, the ev/J 5′ UTR is virtually identical to that of ART-CH proviruses, whose transcripts are reportedly efficiently transferred to NIH 3T3 cells that were infected by RSV produced from tumors (41). In addition, we have proposed that ev/J proviruses were the source of the subgroup J envelope gene (8), an event that would require genetic exchange between ev/J and an infectious ALV-type virus. If this is the case, the most likely mechanism for genetic exchange would be recombination during reverse transcription, a process that requires that ev/J-encoded RNA was packaged with the ALV-type genome to generate a heterozygous virus. This may have been a rare event, however, and the functionality of the ev/J packaging signal is therefore being evaluated experimentally.
The relationship of the ev/J gag gene to those of other avian retroviruses is depicted in Fig. 7. However, the relationship between these genes appears to be more complex when the individual proteins encoded by the gag gene are compared. Specifically, we found that the predicted amino acid sequence of the ev/J Gag protein indicates that it retains a number of motifs that are required for Gag protein function. This conservation of important regulatory regions makes it tempting to speculate that they have also retained function. This possibility is being investigated directly by inserting the ev/J gag gene from clone 3A, which is missing only the Gag translational stop codon, into the RCAS vector system (31, 32, 49) to evaluate its ability to function in virus assembly, budding, and entry.
RT-PCR experiments demonstrated that at least some ev/J proviruses are expressed in CEFs. In these studies, evidence was obtained only for expression of RNA that corresponded in length to transcripts that could be encoded by the class II and class V proviruses. This result might indicate that only a member(s) of one or both of these classes is expressed in CEFs. Alternatively, this result might reflect the fact that a total of eight class II genomic clones were isolated, making this the most abundant class of proviruses found. If this reflects the true representation of class II proviruses in the genome, the higher level of class II-encoded RNA might simply be the result of the greater abundance of class II proviruses in the genome. This question is under investigation.
Studies are also under way to investigate whether any of the ev/J mRNAs are translated. This will be important to determine in order to understand whether expression of Gag- or Env-related antigens might play a role in pathogenesis by exogenous viruses. We are particularly interested in investigating whether there might be a strain-to-strain difference in expression of ev/J proviruses and/or of Env-related antigens that might influence the ability of different strains to control infection by the subgroup J ALV strains that include an env gene derived from the ev/J proviruses.
ACKNOWLEDGMENTS
B.L.R. and S.J.B. contributed equally to this work.
We thank Sharon Soodeen-Karamath and Ann Gibbins at the University of Guelph for providing the cDNA library from which the ev/J-encoded cDNAs were isolated and Doug Foster at the University of Minnesota for his kind gift of DF-1 cells.
This work was supported by Public Health Service grants GM41571 and CA80097 from the National Institute of General Medical Sciences and by a grant from the Leukemia Research Fund. S.J.B. was supported by training grant CA09138 from the National Institutes of Health.
REFERENCES
- 1.Aronoff R, Hajjar A M, Linial M L. Avian retroviral RNA encapsidation: reexamination of functional 5′ RNA sequences and the role of nucleocapsid Cys-His motifs. J Virol. 1993;67:178–188. doi: 10.1128/jvi.67.1.178-188.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Astrin S M, Buss E G, Hayward W S. Endogenous viral genes are non-essential in the chicken. Nature. 1979;282:339–341. doi: 10.1038/282339a0. [DOI] [PubMed] [Google Scholar]
- 3.Astrin S M, Robinson H L, Crittenden L B, Buss E G, Wyban J, Hayward W S. Ten genetic loci in the chicken that contain structural genes for endogenous avian leukosis viruses. Cold Spring Harbor Symp Quanti Biol. 1980;44:1105–1109. doi: 10.1101/sqb.1980.044.01.119. [DOI] [PubMed] [Google Scholar]
- 4.Bai J, Howes K, Payne L N, Skinner M A. Sequence of host-range determinants in the env gene of a full-length, infectious proviral clone of exogenous Avian Leukosis Virus HPRS-103 confirms that it represents a new subgroup (designated J) J Gen Virol. 1995;76:181–187. doi: 10.1099/0022-1317-76-1-181. [DOI] [PubMed] [Google Scholar]
- 5.Bai J, Payne L N, Skinner M A. HPRS-103 (exogenous avian leukosis virus, subgroup J) has an env gene related to those of endogenous elements EAV-0 and E51 and an E element found previously only in sarcoma viruses. J Virol. 1995;69:779–784. doi: 10.1128/jvi.69.2.779-784.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker B, Robison H, Varmus H E, Bishop J M. Analysis of endogenous avian retrovirus DNA and RNA: viral and cellular determinants of retrovirus gene expression. Virology. 1981;114:8–22. doi: 10.1016/0042-6822(81)90248-8. [DOI] [PubMed] [Google Scholar]
- 7.Banks J D, Yeo A, Green K, Cepeda F, Linial M L. A minimal avian retroviral packaging sequence has a complex structure. J Virol. 1998;72:6190–6194. doi: 10.1128/jvi.72.7.6190-6194.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benson S J, Ruis B L, Fadly A M, Conklin K F. The unique envelope gene of the subgroup J avian leukosis virus derives from ev/J proviruses, a novel family of avian endogenous viruses. J Virol. 1998;72:10157–10164. doi: 10.1128/jvi.72.12.10157-10164.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benson S J, Ruis B L, Garbers A L, Fadly A M, Conklin K F. Independent isolates of the emerging subgroup J avian leukosis virus derive from a common ancestor. J Virol. 1998;72:10301–10304. doi: 10.1128/jvi.72.12.10301-10304.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blundell T, Sibanda B L, Pearl L. Three-dimensional structure, specificity and catalytic mechanism of renin. Nature. 1983;304:273–275. doi: 10.1038/304273a0. [DOI] [PubMed] [Google Scholar]
- 11.Bova C A, Manfredi J P, Swanstrom R. env genes of avian retroviruses: nucleotide sequence and molecular recombinants define host range determinants. Virology. 1986;152:343–354. doi: 10.1016/0042-6822(86)90137-6. [DOI] [PubMed] [Google Scholar]
- 12.Bova C A, Olsen J C, Swanstrom R. The avian retrovirus env gene family: molecular analysis of host range and antigenic variants. J Virol. 1988;62:75–83. doi: 10.1128/jvi.62.1.75-83.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyce-Jacino M T, O’Donoghue K, Faras A J. Multiple complex families of endogenous retroviruses are highly conserved in the genus Gallus. J Virol. 1992;66:4919–4929. doi: 10.1128/jvi.66.8.4919-4929.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyce-Jacino M T, Resnick R, Faras A J. Structural and functional characterization of the unusually short long terminal repeats and their adjacent regions of a novel endogenous avian retrovirus. Virology. 1989;173:157–166. doi: 10.1016/0042-6822(89)90231-6. [DOI] [PubMed] [Google Scholar]
- 15.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 16.Coffin J M, Tsichlis P N, Conklin K F, Senior A, Robinson H L. Genomes of endogenous and exogenous avian retroviruses. Virology. 1983;126:51–72. doi: 10.1016/0042-6822(83)90461-0. [DOI] [PubMed] [Google Scholar]
- 17.Conklin K F, Groudine M. Varied interactions between proviruses and adjacent host chromatin. Mol Cell Biol. 1986;6:3999–3407. doi: 10.1128/mcb.6.11.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Craven R C, Leure-duPree A E, Weldon R A, Jr, Wills J W. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J Virol. 1995;69:4213–4227. doi: 10.1128/jvi.69.7.4213-4227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donze O, Damay P, Spahr P F. The first and third uORFs in RSV leader RNA are efficiently translated: implications for translational regulation and viral RNA packaging. Nucleic Acids Res. 1995;23:861–868. doi: 10.1093/nar/23.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donze O, Spahr P F. Role of the open reading frames of Rous sarcoma virus leader RNA in translation and genome packaging. EMBO J. 1992;11:3747–3757. doi: 10.1002/j.1460-2075.1992.tb05460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dorner A J, Stoye J P, Coffin J M. Molecular basis of host range variation in avian retroviruses. J Virol. 1985;53:32–39. doi: 10.1128/jvi.53.1.32-39.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunwiddie C T, Resnick R, Boyce-Jacino M, Alegre J N, Faras A J. Molecular cloning and characterization of gag-, pol-, and env-related gene sequences in the ev- chicken. J Virol. 1986;59:669–675. doi: 10.1128/jvi.59.3.669-675.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dupraz P, Oertle S, Meric C, Damay P, Spahr P F. Point mutations in the proximal Cys-His box of Rous sarcoma virus nucleocapsid protein. J Virol. 1990;64:4978–4987. doi: 10.1128/jvi.64.10.4978-4987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorelick R J, Chabot D J, Ott D E, Gagliardi T D, Rein A, Henderson L E, Arthur L O. Genetic analysis of the zinc finger in the Moloney murine leukemia virus nucleocapsid domain: replacement of zinc-coordinating residues with other zinc-coordinating residues yields noninfectious particles containing genomic RNA. J Virol. 1996;70:2593–2597. doi: 10.1128/jvi.70.4.2593-2597.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorelick R J, Henderson L E, Hanser J P, Rein A. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: evidence for specific RNA recognition by a “zinc finger-like” protein sequence. Proc Natl Acad Sci USA. 1988;85:8420–8424. doi: 10.1073/pnas.85.22.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gottlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gudkov A V, Komarova E A, Nikiforov M A, Zaitsevskaya T E. ART-CH, a new chicken retroviruslike element. J Virol. 1992;66:1726–1736. doi: 10.1128/jvi.66.3.1726-1736.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayward W S. Size and genetic content of viral RNAs in avian oncovirus-infected cells. J Virol. 1977;24:47–63. doi: 10.1128/jvi.24.1.47-63.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayward W S, Braverman S B, Astrin S M. Transcriptional products and DNA structure of endogenous avian proviruses. Cold Spring Harbor Symp Quant Biol. 1980;44:1111–1121. doi: 10.1101/sqb.1980.044.01.120. [DOI] [PubMed] [Google Scholar]
- 30.Huang M, Orenstein J M, Martin M A, Freed E O. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hughes S H, Greenhouse J J, Petropoulos C J, Sutrave P. Adaptor plasmids simplify the insertion of foreign DNA into helper-independent retroviral vectors. J Virol. 1987;61:3004–3012. doi: 10.1128/jvi.61.10.3004-3012.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hughes S H, Kosik E, Fadly A M, Salter D W, Crittenden L B. Design of retroviral vectors for the insertion of foreign deoxyribonucleic acid sequences into the avian germ line. Poultry Sci. 1986;65:1459–1467. doi: 10.3382/ps.0651459. [DOI] [PubMed] [Google Scholar]
- 33.Hughes S H, Vogt P K, Stubblefield E, Robinson H, Bishop J M, Varmus H E. Organization of endogenous and exogenous viral and linked nonviral sequences. Cold Spring Harbor Symp Quant Biol. 1980;44:1077–1089. doi: 10.1101/sqb.1980.044.01.116. [DOI] [PubMed] [Google Scholar]
- 34.Hunter E. Viral entry and receptors. In: Coffin J M, Hughes S H, Varmus H, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 71–119. [PubMed] [Google Scholar]
- 35.Katz R A, Terry R W, Skalka A M. A conserved cis-acting sequence in the 5′ leader of avian sarcoma virus RNA is required for packaging. J Virol. 1986;59:163–167. doi: 10.1128/jvi.59.1.163-167.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knight J B, Si Z H, Stoltzfus C M. A base-paired structure in the avian sarcoma virus 5′ leader is required for efficient encapsidation of RNA. J Virol. 1994;68:4493–4502. doi: 10.1128/jvi.68.7.4493-4502.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meric C, Gouilloud E, Spahr P F. Mutations in Rous sarcoma virus nucleocapsid protein p12 (NC): deletions of Cys-His boxes. J Virol. 1988;62:3328–3333. doi: 10.1128/jvi.62.9.3328-3333.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meric C, Spahr P F. Rous sarcoma virus nucleic acid-binding protein p12 is necessary for viral 70S RNA dimer formation and packaging. J Virol. 1986;60:450–459. doi: 10.1128/jvi.60.2.450-459.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moustakas A, Sonstegard T S, Hackett P B. Alterations of the three short open reading frames in the Rous sarcoma virus leader RNA modulate viral replication and gene expression. J Virol. 1993;67:4337–4349. doi: 10.1128/jvi.67.7.4337-4349.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nelle T D, Wills J W. A large region within the Rous sarcoma virus matrix protein is dispensable for budding and infectivity. J Virol. 1996;70:2269–2276. doi: 10.1128/jvi.70.4.2269-2276.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nikiforov M A, Gudkov A V. ART-CH: a VL30 in chickens? J Virol. 1994;68:846–853. doi: 10.1128/jvi.68.2.846-853.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ogert R A, Beemon K L. Mutational analysis of the Rous sarcoma virus DR posttranscriptional control element. J Virol. 1998;72:3407–3411. doi: 10.1128/jvi.72.4.3407-3411.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ogert R A, Lee L H, Beemon K L. Avian retroviral RNA element promotes unspliced RNA accumulation in the cytoplasm. J Virol. 1996;70:3834–3843. doi: 10.1128/jvi.70.6.3834-3843.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parent L J, Bennett R P, Craven R C, Nelle T D, Krishna N K, Bowzard J B, Wilson C B, Puffer B A, Montelaro R C, Wills J W. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1995;69:5455–5460. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patarca R, Haseltine W A. A major retroviral core protein related to EPA and TIMP. Nature. 1985;318:390. doi: 10.1038/318390a0. [DOI] [PubMed] [Google Scholar]
- 46.Payne L N, Brown S R, Bumstead N, Howes K, Frazier J A, Thouless M E. A novel subgroup of exogenous avian leukosis virus in chickens. J Gen Virol. 1991;72:801–807. doi: 10.1099/0022-1317-72-4-801. [DOI] [PubMed] [Google Scholar]
- 47.Pepinsky R B, Mattaliano R J, Vogt V M. Structure and processing of the p2 region of avian sarcoma and leukemia virus gag precursor polyproteins. J Virol. 1986;58:50–58. doi: 10.1128/jvi.58.1.50-58.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pepinsky R B, Papayannopoulos I A, Campbell S, Vogt V M. Analysis of Rous sarcoma virus Gag protein by mass spectrometry indicates trimming by host exopeptidase. J Virol. 1996;70:3313–3318. doi: 10.1128/jvi.70.5.3313-3318.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petropoulos C J, Hughes S H. Replication-competent retrovirus vectors for the transfer and expression of gene cassettes in avian cells. J Virol. 1991;65:3728–3737. doi: 10.1128/jvi.65.7.3728-3737.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rein A, Harvin D P, Mirro J, Ernst S M, Gorelick R J. Evidence that a central domain of nucleocapsid protein is required for RNA packaging in murine leukemia virus. J Virol. 1994;68:6124–6129. doi: 10.1128/jvi.68.9.6124-6129.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simpson S B, Guo W, Winistorfer S C, Craven R C, Stoltzfus C M. The upstream, direct repeat sequence of Prague A Rous sarcoma virus is deficient in mediating efficient Gag assembly and particle release. Virology. 1998;247:86–96. doi: 10.1006/viro.1998.9233. [DOI] [PubMed] [Google Scholar]
- 52.Simpson S B, Zhang L, Craven R C, Stoltzfus C M. Rous sarcoma virus direct repeat cis elements exert effects at several points in the virus life cycle. J Virol. 1997;71:9150–9156. doi: 10.1128/jvi.71.12.9150-9156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skalka A, DeBona P, Hishinuma F, McClements W. Avian endogenous proviral DNA: analysis of integrated ev 1 and a related gs- chf- provirus purified by molecular cloning. Cold Spring Harbor Symp Quant Biol. 1980;44:1097–1104. doi: 10.1101/sqb.1980.044.01.118. [DOI] [PubMed] [Google Scholar]
- 54.Sonstegard T S, Hackett P B. Autogenous regulation of RNA translation and packaging by Rous sarcoma virus Pr76gag. J Virol. 1996;70:6642–6652. [PMC free article] [PubMed] [Google Scholar]
- 55.Swanstrom R, Wills J W. Synthesis, assembly, and processing of viral proteins. In: Coffin J M, Hughes S H, Varmus H, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 56.Tang J. Evolution in the structure and function of carboxyl proteases. Mol Cell Biochem. 1979;26:93–109. doi: 10.1007/BF00232887. [DOI] [PubMed] [Google Scholar]
- 57.Taplitz R A, Coffin J M. Selection of an avian retrovirus mutant with extended receptor usage. J Virol. 1997;71:7814–7819. doi: 10.1128/jvi.71.10.7814-7819.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Venugopal K, Smith L M, Howes K, Payne L N. Antigenic variants of J subgroup avian leukosis virus: sequence analysis reveals multiple changes in the env gene. J Gen Virol. 1998;79:757–766. doi: 10.1099/0022-1317-79-4-757. [DOI] [PubMed] [Google Scholar]
- 59.Verderame M F, Nelle T D, Wills J W. The membrane-binding domain of the Rous sarcoma virus Gag protein. J Virol. 1996;70:2664–2668. doi: 10.1128/jvi.70.4.2664-2668.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vogt V M. Retroviral virions and genomes. In: Coffin J M, Hughes S H, Varmus H, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 27–70. [PubMed] [Google Scholar]
- 61.Weissmahr R N, Schupbach J, Boni J. Reverse transcriptase activity in chicken embryo fibroblast culture supernatants is associated with particles containing endogenous avian retrovirus EAV-0 RNA. J Virol. 1997;71:3005–3012. doi: 10.1128/jvi.71.4.3005-3012.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wills J W, Cameron C E, Wilson C B, Xiang Y, Bennett R P, Leis J. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J Virol. 1994;68:6605–6618. doi: 10.1128/jvi.68.10.6605-6618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wills J W, Craven R C, Weldon R A, Jr, Nelle T D, Erdie C R. Suppression of retroviral MA deletions by the amino-terminal membrane-binding domain of p60src. J Virol. 1991;65:3804–3812. doi: 10.1128/jvi.65.7.3804-3812.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xiang Y, Cameron C E, Wills J W, Leis J. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J Virol. 1996;70:5695–5700. doi: 10.1128/jvi.70.8.5695-5700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]