Abstract
Glycoprotein M (gM), the product of the UL10 gene of pseudorabies virus (PrV), is one of the few nonessential glycoproteins conserved throughout the Herpesviridae. In contrast to wild-type PrV strains, the UL10 gene product of the attenuated PrV vaccine strain Bartha (PrV-Ba) is not modified by N-glycans due to a mutation in the DNA sequence encoding the consensus N-glycosylation motif. To assay function of the UL10 protein in PrV-Ba, a UL10-deletion mutant (PrV-Ba-UL10−) was isolated. Surprisingly, in contrast to gM-deleted wild-type PrV, PrV-Ba-UL10− was severely impaired in plaque formation, inducing only foci of very few infected RK13, Vero, and PSEK cells and tiny plaques on MDBK cells. Since this effect was significantly more dramatic than in wild-type PrV, additional mutations known to be present in PrV-Ba were analyzed for their contribution to this phenotype. trans-complementation of the mutated PrV-Ba UL21 or gC protein by the wild-type version had no influence on the observed phenotype. In contrast, complementation of the gE/gI deletion rescued the phenotype. The synergistic effect of deletions in gE/gI and gM on plaque size was verified by construction of a gE/I/M triple mutant derived from wild-type PrV which exhibited the same phenotype. The dramatic effect of deletion of gM on plaque size in a gE/I− virus background was mainly attributable to a function of gM, and not of the gM/gN complex, as shown by analysis of a gE/I/N triple mutant. Interestingly, despite the strong effect on plaque size, penetration was not significantly impaired. In noncomplementing cells infected with the gE/I/M triple mutant, electron microscopy showed absence of secondary envelopment in the cytoplasm but occurrence of intracytoplasmic accumulations of nucleocapsids in association with electron dense material, presumably tegument proteins. These structures were not observed after infection of cells expressing either gE/I or gM. We suggest that gE/I and gM are required for late stages in virion morphogenesis prior to final envelopment in the cytoplasm.
During herpesvirus infection, at least two distinct types of processes involving membrane fusion occur (40, 50). Following attachment of free virions, penetration is induced by fusion of the virion envelope with the cellular cytoplasmic membrane, resulting in release of the nucleocapsid into the cytoplasm. Later in the infectious cycle, infectivity is directly transmitted from primary infected to neighboring noninfected cells. This process, which has been termed direct cell-to-cell spread and which is mainly responsible for the formation of plaques under agarose or semisolid methylcellulose media, is related to but distinct from penetration. So far, it is unclear whether membranes of infected and noninfected cells do in fact fuse, whether a modification of junctional regions allows transgress of infectivity, or whether virions are released into the intercellular space and then fuse with the neighboring cell.
In the alphaherpesviruses, essential glycoproteins B, H, and L (gB, gH, and gL) are required for both processes (40, 50). In herpes simplex virus type 1 (HSV-1), gD is also required for penetration and direct viral cell-to-cell spread (29), whereas in the related pseudorabies virus (PrV), gD is required for penetration of wild-type strains but not for direct viral cell-to-cell spread (44, 46). Moreover, recently a gD-deficient yet infectious PrV mutant which is able to penetrate in the absence of gD has been isolated (49).
In the process of direct viral cell-to-cell spread, gE and gI, which are nonessential for viral replication in tissue culture and form a noncovalently linked complex (19), also play a prominent role. Deletion of gE and/or gI leads to small-plaque phenotypes in several alphaherpesviruses (10, 12, 34, 42, 54, 55), whereby the size reduction is dependent on the individual parental virus strain and the type of host cell (3, 10, 33). gE and the gE/gI complex have also been shown to be involved in release of mature virions from target cells. The role that gE and gI play in direct cell-to-cell spread in vivo is even more dramatic, and both proteins are required for efficient transneuronal transsynaptic transfer of HSV-1 and PrV (1, 6, 11, 13, 26, 52). Thus, gE- and/or gI-deleted PrV strains are significantly attenuated in various model hosts and in PrV’s natural host, swine (35, 41).
Whereas gE and gI are conserved only within the alphaherpesviruses, homologs of gM have been found in all herpesvirus subfamilies. Despite this high conservation, gM is also a nonessential glycoprotein, and gM-deletion mutants of HSV-1 (2), equine herpesvirus 1 (EHV-1) (43), and PrV (7) have recently been isolated. Deletion of gM only slightly impairs penetration kinetics and plaque size but results in ca. 10- to 100-fold-decreased virus titers. gM forms a complex with the product of the UL49.5 homologous genes, as shown for PrV (21), bovine herpesvirus 1 (53), and Epstein-Barr Virus (27). Since the UL49.5 products are glycosylated in PrV and Epstein-Barr virus, they have been designated gN. In PrV, deletion of gN results in viable virus mutants which exhibit viral titers similar to those of wild-type virus (21). There is no effect on plaque size but a slight impairment in penetration. Thus, the effect of gM on penetration is most likely due to lack of the function of the gM/gN complex, whereas the effect on viral titers is attributable solely to absence of gM.
For several years we have been analyzing the attenuated PrV vaccine strain Bartha (PrV-Ba). PrV-Ba was obtained by multiple passages of a virulent field isolate in cultured chicken cells and embryonated eggs (4). Careful analysis revealed that PrV-Ba harbors three independent mutations which, in combination, result in its apathogenic phenotype. These include point mutations within the UL21 gene (23), a signal sequence mutation in the gC gene (48), and a deletion affecting the gE and gI genes located in the unique short region (31, 33, 45). We recently discovered that PrV-Ba also specifies a mutated UL10 gene, which in wild-type PrV encodes gM. In PrV-Ba point mutations abolish the single N-glycosylation consensus motif in UL10, and thus the PrV-Ba UL10 product is not N-glycosylated (8). In fact, so far we have no indication that the PrV-Ba UL10 protein is glycosylated at all. However, this mutation apparently does not affect pathogenicity of the virus since restoration of the UL21, gC, and gE/I defects in PrV-Ba results in a fully virulent virus (23).
Since PrV-Ba expressed a different UL10 protein, we wanted to analyze the effect of a UL10 deletion on the phenotype of PrV-Ba. To this end, we isolated and analyzed a UL10-deleted PrV-Ba variant, designated PrV-Ba-UL10−.
MATERIALS AND METHODS
Viruses and cells.
Virus mutants were derived from wild-type PrV strain Kaplan (PrV-Ka) (22) or the attenuated live vaccine strain PrV-Ba (4). PrV-1112 and PrV-B80 carry a gG–β-galactosidase (β-Gal) expression cassette at the gG locus (30, 38) and exhibit growth properties similar to those of their parental strains, PrV-Ka and PrV-Ba, respectively. Mutants with deletions of gE/I (PrV-gE/I−) and gM (PrV-gM−) have been described elsewhere (9, 35) (Fig. 1). Viruses were grown on porcine (PSEK), bovine (MDBK), rabbit (RK13), or African green monkey kidney (Vero) cells. VneoB4 cells are Vero cells that contain the viral genomic BamHI fragment 4 which contains the UL21 gene (23). Cells were propagated in Eagle’s minimum essential medium supplemented with 5 or 10% (RK13) fetal calf serum. Cotransfections were performed by calcium phosphate coprecipitation (17).
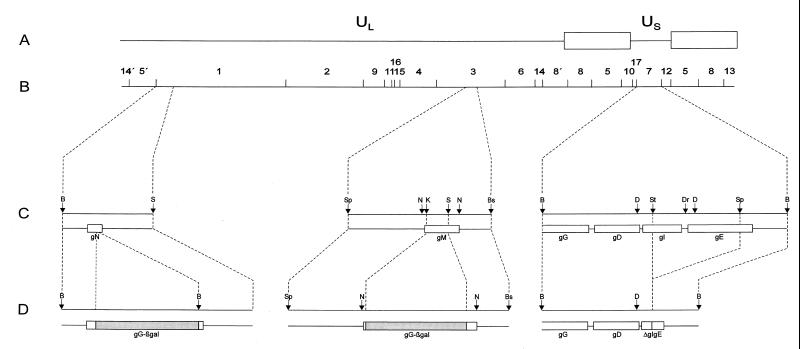
FIG. 1.
Mutations introduced into PrV. (A) Diagram of the PrV genome. It is divided into a unique long (UL) and unique short (US) region by inverted repeats (open rectangles) which bracket the latter. (B) BamHI restriction fragment map. (C) Locations of the glycoprotein genes which had been mutated as demonstrated in panel D. Relevant restriction sites: B, BamHI; Bs, BstEII; D, DdeI; Dr, DraI; N, NlaIII; K, KpnI; S, SalI; Sp, SphI; St, StuI.
Construction of gC-, gM-, and gE/I-expressing cell lines.
Cells which constitutively express gC were isolated after transfection of plasmid CMV-gC (16) into RK13 cells. Stable cell clones were selected in medium containing 500 μg of geneticin (Life Technologies, Eggenstein, Germany) per ml. Expression of gC was tested by indirect immunofluorescence and Western blotting using gC-specific monoclonal antibody B16-c8 (24). One cell clone, designated RK13-gC, was used for further testing.
For construction of a gM-expressing cell line, BamHI fragment 3 of viral genomic DNA was digested with NlaIII. A 1.2-kb NlaIII fragment which contains the UL10 open reading frame (Fig. 1) was cloned into vector pRc/CMV (InVitroGen, Leek, The Netherlands). The resulting plasmid, CMV-gM, was transfected into RK13 cells, and cell clones developing under medium containing 500 μg geneticin per ml were tested for gM expression by indirect immunofluorescence and Western blotting using an anti-gM peptide serum (7). One cell clone, RK13-gM, was used for further studies.
For isolation of a gE/I-expressing cell line, the following expression plasmids were constructed. Cloned BamHI fragment 7 was digested with DraI/BamHI, and the resulting 2.75-kb DraI/BamHI fragment, which comprises the complete gE gene, was cloned into expression vector pRc/CMV. The resulting plasmid was designated CMV-gE. For expression of gI, viral genomic BamHI fragment 7 was cleaved with DdeI. Insertion of a resulting 1.5-kb DdeI fragment, which contains the complete gI gene, into vector pRc/CMV resulted in plasmid CMV-gI. For construction of gE/I-expressing cell lines, plasmids CMV-gE and CMV-gI were cotransfected into RK13 cells. Cell clones were selected in medium containing 500 μg of geneticin per ml and tested for the expression of gE and gI by indirect immunofluorescence and Western blotting by using gE-specific monoclonal antibody, A9-b15-38 (24) and a gI-specific polyclonal antiserum (5), respectively. One of the resulting cell clones, RK13-gE/I, was used in this study.
Construction of mutants PrV-Ba-UL10−, PrV-gE/I/M−, and PrV-gE/I/N−.
For construction of PrV-Ba-UL10−, plasmid ΔUL10β (8) was cotransfected with DNA of PrV-Ba into RK13-gM cells. Virus progeny was screened for the appearance of a blue plaque phenotype under an agarose overlay containing 300 μg of Bluo-Gal (Life Technologies) per ml. Plaques were picked by aspiration and purified until all plaques stained blue. One randomly selected plaque isolate, PrV-Ba-UL10−, was further characterized by Southern and Western blotting. For isolation of a PrV-gE/I/M− mutant, DNA of PrV-gE/I− (35) was cotransfected with plasmid ΔUL10β into RK13-gM cells. β-Gal-positive plaques were isolated as described above. One plaque isolate, PrV-gE/I/M−, was chosen for further analysis. Correct recombination was verified by Southern blot analysis of mutant virus DNA using standard procedures.
Mutant PrV-gE/I/N− was isolated after cotransfection of gE/I− DNA and plasmid pU8341β (21). Progeny virus was assayed under Bluo-Gal agarose overlay, and blue plaques were purified. One plaque isolate was chosen for further analysis. The genotype of the mutant virus was verified by restriction analysis and Southern blotting.
Determination of plaque size.
Plaque size was measured after titration of virus mutants on the various cell lines after 2 days of incubation at 37°C under a methylcellulose medium. Thereafter, cells were fixed for 10 min in 2% formaldehyde–0.2% glutaraldehyde–0.02% Nonidet P-40–0.01% sodium deoxycholate in phosphate-buffered saline (PBS) and stained with a substrate solution for β-Gal containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 300 μg/ml; Roth, Karlsruhe, Germany)–16 mM potassium ferricyanide–16 mM potassium ferrocyanide–2 mM magnesium chloride–0.02% Nonidet P-40–0.01% sodium deoxycholate in PBS. Cells infected with mutant PrV-gE/I−, which does not express β-Gal, were fixed with 5% formaldehyde and stained with crystal violet. For each combination of virus and cell, 100 plaques were measured microscopically, and the average plaque size was determined. Values were calculated compared to PrV-B80 or PrV-1112, set at 100%. Average percentages as well as standard deviations were determined from three independent experiments.
Penetration kinetics.
For analysis of penetration kinetics (37), each well of RK13, Vero, and MDBK cells in six-well culture dishes was infected with approximately 500 PFU of PrV-1112, PrV-gE/I−, PrV-gM−, or PrV-gE/I/M− for 1 h at 4°C. Thereafter, the inoculum was removed and cells were overlaid with medium which had been prewarmed to 37°C to allow penetration. Immediately thereafter and after 5, 10, 20, and 40 min, extracellular virus was inactivated by treatment of the monolayer for 2 min with 40 mM citric acid–10 mM KCl–135 mM NaCl (pH 3.0). Cells were washed twice with PBS and overlaid with methylcellulose medium. After 2 days of incubation at 37°C, cells were fixed and stained. Plaques were counted, and percent penetration was calculated in comparison to a PBS-treated control.
One-step growth analysis.
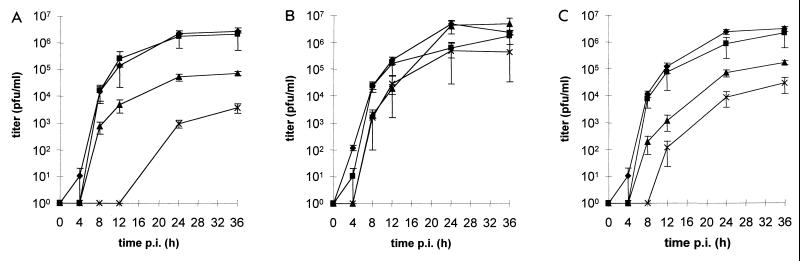
To monitor one-step growth normal, gM- or gE/I-expressing RK13 cells were infected with PrV-1112, PrV-gE/I−, PrV-gM−, or phenotypically gM-complemented PrV-gE/I/M− at a multiplicity of infection (MOI) of 5 for 1 h at 4°C. The triple mutant had to be grown on complementing cells to obtain virus stocks with appropriately high titers. Thereafter, the inoculum was substituted by prewarmed medium, and virus was allowed to penetrate for 90 min at 37°C. Then, remaining extracellular virus was inactivated by low-pH treatment. Cells and supernatants were harvested separately immediately thereafter (0 h) and after 4, 8, 12, 24, and 36 h of incubation at 37°C. Virus progeny was titrated on RK13 cells. Since no significant differences were observed in extra- and intracellular virus titers, the two were added and average values and standard deviations of two independent experiments were calculated.
Electron microscopy.
For ultrathin sectioning, nontransfected, gM-, or gE/I-expressing RK13 cells in petri dishes were infected at an MOI of 1 and fixed 16 h after infection for 60 min with 2.5% glutaraldehyde buffered in 0.1 M sodium cacodylate (pH 7.2), 300 mosmol (Merck, Darmstadt, Germany). They were then scraped off the plate, pelleted by low-speed centrifugation, and embedded in LMP agarose (Biozym, Oldendorf, Germany). Small pieces were postfixed for 60 min with 1% aqueous OsO4 (Polysciences Europe, Eppelheim, Germany) and stained with uranyl acetate. After stepwise dehydration in ethanol, the cells were cleared in propylene oxide, embedded in glycid ether 100 (Serva, Heidelberg, Germany), and polymerized at 59°C for 4 days. Ultrathin sections, counterstained with uranyl acetate and lead salts, were examined with an electron microscope (EM 400 T; Philips, Eindhoven, The Netherlands).
RESULTS
PrV-Ba-UL10− is severely impaired in direct viral cell-to-cell spread.
Although the wild-type PrV UL10 gene product had been shown to encode a nonessential glycoprotein (7), a UL10-deleted variant of PrV-Ba could be isolated only on cells which expressed gM and provided it in trans. This was in contrast to the situation in wild-type PrV, in which a gM-deletion mutant could easily be obtained on normal cells (7, 9). Analysis of PrV-Ba-UL10− led to the striking observation that this mutant, in contrast to the β-Gal-expressing UL10-positive PrV-B80, was able to induce only small foci of infected RK13, Vero, and PSEK cells (Fig. 2). Although not as dramatic, the size of plaques on MDBK cells was also markedly reduced. This defect could be fully compensated on gM-expressing RK13 cells, indicating that it was due to the absence of the UL10 protein and not attributable to any other mutation in the genome of the virus mutant. Thus, deletion of UL10 from PrV-Ba severely impaired its capacity for plaque formation, i.e., direct viral cell-to-cell spread.
FIG. 2.
Plaque morphology of mutants of PrV-Ba. The cell lines indicated on the left were infected under plaque assay conditions with either gG–β-Gal recombinant B80 or UL10–β-Gal recombinant Ba-UL10−. Two days after infection, cells were fixed and stained with X-Gal. Bar = 1 mm.
Absence of gE/gI from PrV-Ba is critical for the observed effect of deletion of UL10.
PrV-Ba harbors several mutations which have been shown to contribute to its avirulent phenotype. These include point mutations within the UL21 and UL44 (gC) genes (23, 48), as well as a large deletion in the unique short portion of the viral genome affecting the US7 (gI) and US8 (gE) genes (31, 45). To assay whether any of these known mutations contributes to the observed effect of deletion of UL10, cell lines expressing wild-type UL21, wild-type gC, and wild-type gE/gI were infected with PrV-Ba-UL10− and plaque size was analyzed. As shown in Fig. 3, only on gE/gI-expressing cells was PrV-Ba-UL10− able to form sizeable plaques. Neither the wild-type UL21 nor the UL44 gene rescued the phenotype. Thus, absence of gE/gI from the PrV-Ba genome was a prerequisite for the observed dramatic effect of deletion of UL10 on plaque size.
FIG. 3.
Plaque size of PrV-Ba-UL10− on different complementing cells. The cell lines indicated below the bars were infected under plaque assay conditions with PrV-Ba-UL10−. Two days after infection, cells were stained with X-Gal, and plaque diameters were measured microscopically and compared to the average diameter of plaques induced by the gG–β-Gal recombinant PrV-B80, which was taken as 100%. Bars indicate standard deviation.
Synergistic effect of deletion of gE/gI and gM from PrV-Ka on plaque size.
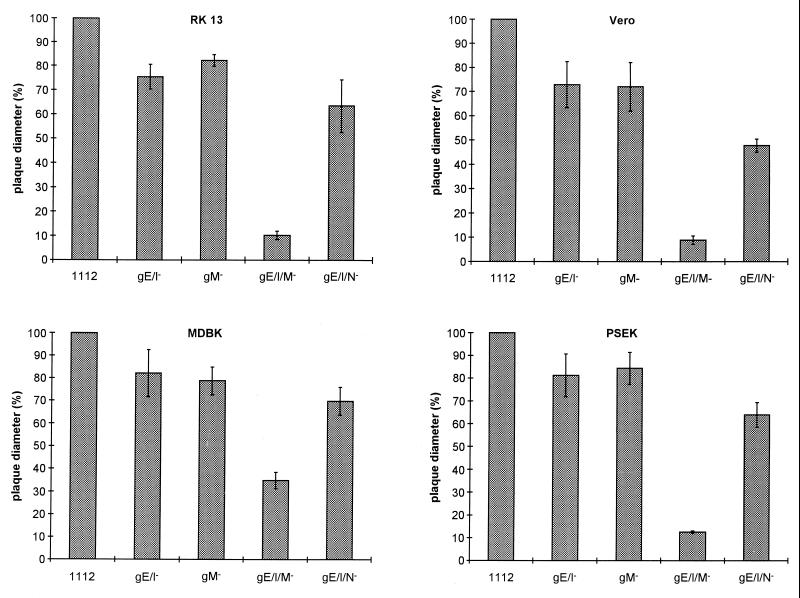
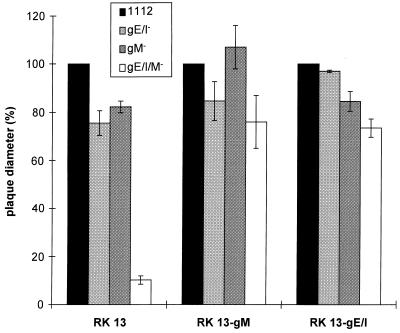
To analyze whether this was a phenomenon linked to PrV-Ba, the UL10 (gM) gene was deleted in a PrV-Ka mutant which already contained a deletion of the gE and gI genes. It is notable that deletion of gE and gI from PrV-Ka reduces plaque size by only ca. 20%, as was also observed after deletion of only gM (Fig. 4). In contrast, simultaneous absence of gE/gI and gM resulted in a decrease in plaque size by ca. 90%, resulting in only small foci of infected RK13, Vero, or PSEK cells. Small plaques were observed on MDBK cells; however, these plaques were also reduced in size by ca. 65% compared to wild-type PrV plaques. When plated on cells expressing gM or gE/gI, plaque size of the triple mutant gE/I/M was essentially restored to that of the single gM− or double gE/gI− mutant (Fig. 5).
FIG. 4.
Plaque size of different PrV mutants. RK13, Vero, MDBK, and PSEK cells were infected under plaque assay conditions with gG–β-Gal recombinant PrV-1112, PrV-gE/I−, PrV-gM−, PrV-gE/I/M−, or PrV-gE/I/N−. Two days after infection, cells were fixed and stained with X-Gal or crystal violet (for PrV-gE/I−). Plaque diameters were measured microscopically. The average diameter of plaques induced by PrV-1112 was counted as 100%, and diameters of plaques induced by the other mutant viruses were calculated accordingly. Bars indicate standard deviation.
FIG. 5.
Rescue of plaque formation by PrV-gE/I/M− in transcomplementing cells. The cell lines indicated at the bottom were infected with gG–β-Gal recombinant PrV-1112, PrV-gE/I−, PrV-gM−, or PrV-gE/I/M−. Diameters of plaques were measured 2 days after infection under plaque assay conditions. The average diameter of plaques induced by PrV-1112 was counted as 100%, and diameters of plaques induced by the other mutant viruses were calculated accordingly. Bars indicate standard deviation.
Absence of synergistic effect of gE/gI and gM deletions on penetration.
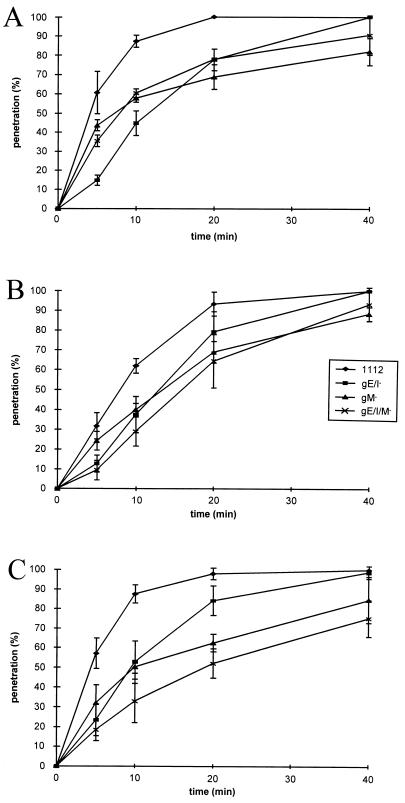
Deletion of gM has been shown to slightly reduce the rate of entry of PrV-Ka (7). To assay whether a combined deletion of gE/I and gM exhibits a stronger effect on entry, as has been observed for direct viral cell-to-cell spread, entry kinetics of PrV-1112, PrV-gE/I−, PrV-gM−, and PrV-gE/I/M− into RK13 (Fig. 6A), Vero (Fig. 6B), and MDBK (Fig. 6C) cells were examined. As shown in Fig. 6, penetration of the triple mutant into either cell line was not significantly different from that of the gM− or the gE/I− mutant. In fact, on RK13 and Vero cells, the mutant viruses exhibited almost identical phenotypes. These results show that the observed effect of the multiple deletion on plaque size is not paralleled by a similar effect on entry.
FIG. 6.
Penetration kinetics of PrV mutants. Rates of entry of gG–β-Gal recombinant PrV-1112, PrV-gE/I−, PrV-gM−, or PrV-gE/I/M− into wild-type RK13 (A), Vero (B), or MDBK (C) cells were determined as described elsewhere (37). Average values and standard deviation from three independent experiments are depicted.
Deletion of gM and not disruption of the gM/gN complex is mainly responsible for the observed synergistic effect on plaque size.
gM and gN of PrV form a disulfide-linked complex (21), and complex formation is required for virion localization of gN. To date, the function of this complex in virus replication is unclear. To assay whether the observed effect on plaque size is attributable to the gM/gN complex, we constructed a mutant virus which, in addition to deletions in the gE and gI genes, also contained a mutated gN gene which abolished expression of gN. As shown in Fig. 4, the gE/I/N− PrV mutant produced plaques which were only slightly smaller than those formed by the gE/I− and gM− mutants. Thus, the observed effect on plaque size by deletion of gM in a gE/I− virus background is due mainly to the loss of a gM function and not to absence of the gM/gN complex.
One-step growth analysis.
To assay the overall growth performance of the mutant viruses, one-step growth kinetics were established on noncomplementing, gM-expressing and gE/I-expressing RK13 cells. As shown in Fig. 7A, on normal RK13 cells the gE/I deletion mutant exhibited growth properties similar to those of PrV-Ka. In contrast, deletion of gM resulted in a ca. 50-fold decrease in viral titer, a defect which could be corrected on gM-expressing cells (Fig. 7B). The triple gE/I/M− mutant was severely impaired in replication on normal cells but grew with wild-type-like kinetics on gM-expressing cells. On gE/I-expressing cells (Fig. 7C), the triple mutant exhibited a growth which was somewhat impaired compared to PrV-gM− but significantly improved over replication on noncomplementing cells. PrV-gE/I− grew to similar titers as PrV-1112 on all cells.
FIG. 7.
One-step growth analysis of PrV-1112 (⧫), PrV-gE/I− (■), PrV-gM− (▴), or PrV-gE/I/M− (X) in nontransfected (A), gM-expressing (B), or gE/I-expressing (C) RK13 cells. At the indicated times after infection, supernatant and cells were harvested and titrated on RK13 cells, and titers were summed. Average values and standard deviation of two independent experiments are shown.
Electron microscopy.
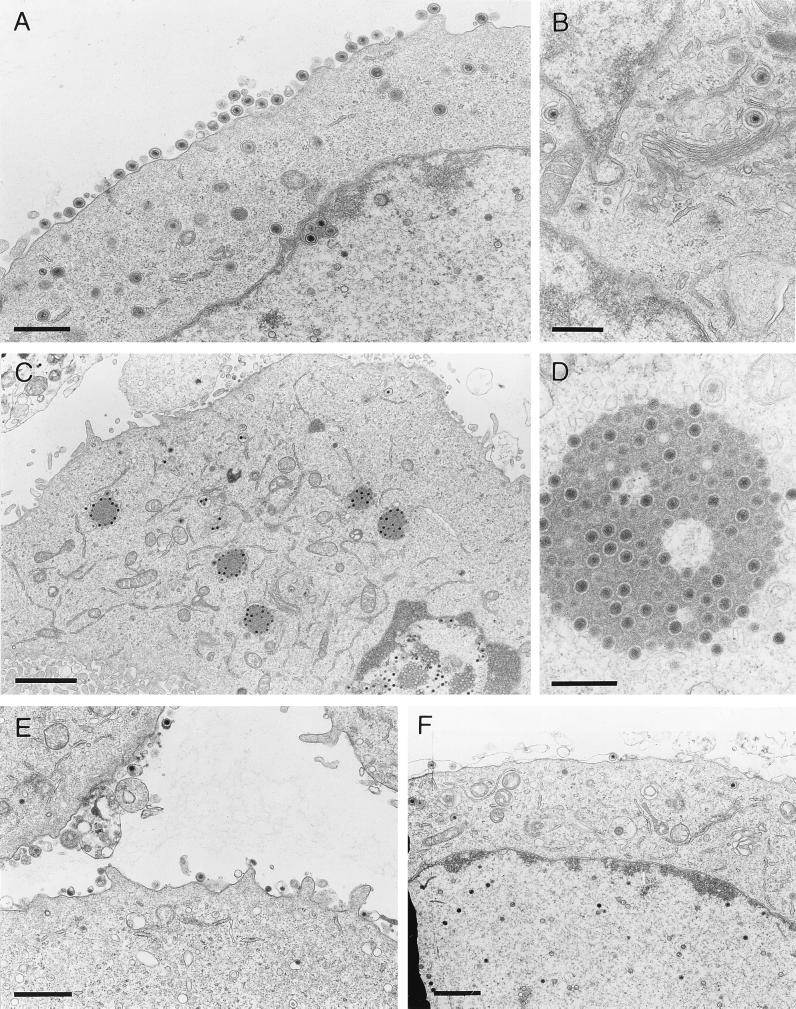
To pinpoint the defect in cells infected with the triple mutant, electron microscopic analyses were performed. To this end, noncomplementing as well as gE/I- or gM-expressing RK13 cells were infected at an MOI of 1 with the gE/I/M− PrV mutant. Sixteen hours after infection, cells were fixed, stained, and analyzed. As shown in Fig. 8A, after infection with wild-type virus numerous enveloped particles lined the outer surface of the cell, and all stages of intracellular virion maturation, i.e., capsid assembly in the nucleus, transit through the nuclear membrane with primary envelopment, and secondary envelopment in the trans-Golgi area (Fig. 8B), were observed. In contrast, in noncomplementing RK13 cells infected by the triple mutant, extracellular enveloped virions and intracytoplasmic enveloped particles were virtually absent. However, large accumulations of unenveloped capsids were observed in the cytoplasm in association with aggregated electron-dense material, presumably tegument proteins (Fig. 8C and D). These structures were not observed in the nucleus and were also absent after infection of RK13-gE/I (Fig. 8E) and RK13-gM (Fig. 8F) cells. In these cells, normal morphogenesis (18) appeared to be restored although fewer extracellular particles than in wild-type virus-infected cells were observed (Fig. 8A).
FIG. 8.
Electron microscopy. Nontransfected RK13 cells were infected with either wild-type PrV (A and B) or PrV-gE/I/M− (C and D) and analyzed 16 h after infection. (E and F) RK13-gE/I (E) and RK13-gM (F) cells after infection with the triple mutant. Bars represent 750 nm in panel A, 500 nm in panel B and D, 2 μm in panel C, and 1 μm in panels E and F.
DISCUSSION
One interesting feature of herpesviruses is that they encode proteins whose genes can be deleted without destroying viability of the virus in cell culture. These genes have been designated nonessential. Among the glycoproteins of the Alphaherpesvirinae, only gB, gD, gH, gL, and gK have been found to be indispensable for viral replication, whereas gC, gE, gG, gI, gJ, gM, and gN are nonessential for viral replication in cell culture (20, 50). Interestingly, glycoproteins which are conserved throughout the Herpesviridae and, therefore, are thought to execute particularly important functions for replication of herpesviruses can be found in both essential and nonessential proteins. Although it has always been argued that the nonessential proteins fulfill important roles in infection of the natural host which cannot be demonstrated in cell culture or by infection of model hosts, the fact that some of these proteins are highly conserved remained somewhat puzzling.
gE and gI are conserved within the Alphaherpesvirinae and form a noncovalently linked complex (19, 55). The gE/gI complex has been implicated in playing a role in direct viral cell-to-cell spread in that gE- or gI-deleted viruses sometimes exhibit a small-plaque phenotype. However, this is not always the case and appears to be dependent on the respective virus strain and host cell (3, 10, 35). More strikingly, gE/gI-deleted PrV and HSV exhibit specific defects in transneuronal spread of virions in several animal species, including PrV’s natural host, the pig (6, 10, 13, 25, 26, 52). Apparently, entry into primary neurons is not affected by the absence of the gE/gI complex, but transsynaptic transfer into second-order neurons is severely impaired or blocked. Although the precise role of the gE/gI complex in this process is unclear, in vitro studies indicated that this complex colocalizes with cell junctions, leading to the hypothesis that this interaction somehow alters sites of direct cell-to-cell contact to allow transcellular passage of virions (12).
gM belongs to those nonessential glycoproteins which are conserved throughout the Herpesviridae (28). Part of intracellular and virion gM can be found in a complex with another conserved nonessential protein, gN, the product of the UL49.5 gene (21). gM-deleted HSV-1, EHV-1, and PrV grow to 10- to 100-fold-lower titers in vitro and are attenuated in vivo (2, 7, 32, 43). Deletion of gM appears to have an effect on kinetics of penetration in EHV-1 and PrV and also reduces plaque size to various degrees. The effect on penetration, at least in PrV, appears to be a result of the disruption of the gM/gN complex since a gN-deleted PrV mutant exhibits a similar reduction in penetration kinetics but no alteration in plaque size (21).
As shown here, deletion of either gE/gI or gM has only a moderate effect on plaque size of PrV-Ka, which confirms previous results (8, 34). However, simultaneous inactivation of gE/gI and gM results in a severe deficiency in plaque formation, whereas penetration was not more affected by the combined deletions compared to gE/I- or gM-deleted viruses. This again shows the distinction between direct viral cell-to-cell spread and entry. Strictly speaking, a small-plaque phenotype as indication for impairment of direct viral cell-to-cell spread can be effected by deficiencies at various levels of viral replication. In particular, inactivation of a number of genes, e.g., UL3.5 or UL20, whose products are involved in virion morphogenesis and egress, results in strikingly reduced plaque size. In both cases, virion morphogenesis (18) is blocked at intracytoplasmic stages immediately prior to secondary envelopment (UL3.5 [14]) or after secondary envelopment before transfer to the cell surface (UL20 [15]). Simultaneous deletion of gE/gI and gM also appears to affect a step prior to secondary envelopment. However, whereas in the UL3.5− mutant cytoplasmic vesicles and capsids are juxtaposed, in the triple-deletion glycoprotein mutant capsids accumulate in the cytoplasm in association with electron-dense material which might represent tegument components. Unfortunately, no antibodies specific for PrV tegument proteins were available, and so final proof for this assumption is still lacking. It is interesting that around the capsids within these inclusions can be observed a clear halo which is indicative of a spatial separation between capsid and the amorphous proteinaceous material.
One-step growth analysis shows that the triple-deletion glycoprotein mutant replicates on noncomplementing cells with significantly lower efficiency than the gE/I and gM deletion mutants. This effect can be explained by the defect in virion morphogenesis resulting in less infectious virus being produced. It is noteworthy that this defect can also be compensated to the level of the gE/I- or gM-deleted mutants by replication on complementing, gE/gI- or gM-expressing cells. However, the decrease in titer in the gM− mutant as also observed after growth of the triple mutant on gE/I-expressing cells is not explained by our electron microscopic analyses. Thus, there might be another function of gM, in addition to its role in virion morphogenesis as described here, which affects overall growth performance of the virus.
Deletion of gE/gI specifically affects transneuronal spread of PrV and HSV-1, although the molecular basis for this phenotype is still unclear. Based on our results, one could speculate that the functional synergy of gE/gI and gM, as observed in cell culture, might not apply in all instances in vivo. Perhaps gM is unable to fulfill its role in virion egress or direct viral cell-to-cell spread in specialized cells such as neurons. Then, absence of gE/gI could result in a phenotype like the one demonstrated here in the triple-deletion mutant. This could also explain the necessity to phylogenetically conserve redundant functions to increase fitness of the virus in those circumstances. However, it is also conceivable that the observed deficiencies in transneuronal transfer (1, 6, 13, 26, 52) and virion morphogenesis (this report) are not correlated but reflect a multifunctional nature of the gE/gI complex.
At present it is unclear at exactly which step in virus maturation gE/gI and gM are involved. Possibly they are required for directing intracytoplasmic capsids to the site where final envelopment occurs, i.e., to the trans-Golgi region. This effect could also be indirect in that these glycoproteins primarily interact with tegument, which then recruits capsids to the site of envelopment. It is well known that in herpesviruses envelopment of tegument components occurs, leading to so-called light particles (L-particles) (47, 51). In this context, it is interesting that L-particles are readily observed when cells are infected with PrV mutants deficient in intranuclear capsid assembly, e.g., after inactivation of the UL28 gene (39). In contrast, in none of the mutants with defects in the intracytoplasmic egress pathway so far described for PrV, UL3.5− (14), UL20− (15), or gE/I/M− (this report), were L-particles produced. This finding argues that the products of these genes are required for the formation and release of enveloped structures with or without capsid.
In summary, we provide evidence for a synergistic effect of deletions of glycoproteins individually designated nonessential on a crucial step of virion morphogenesis. Neither of the two functional units gE/I or gM has so far been implicated in a late step in virus maturation preceding final envelopment of capsids in the cytoplasm. In the absence of either gE/I or gM, morphogenesis appears to proceed relatively normally, with only slightly reduced efficiency, as indicated by one-step growth analysis and confirmed by electron microscopy. In contrast, when both functional units are absent, virus maturation is inhibited and full capsids accumulate in the cytoplasm associated with electron-dense material, presumably tegument. We hypothesize that functions of so-called nonessential glycoproteins may in fact be at least partially redundant and may be uncovered only when more than one functional unit is inactivated. Nevertheless, these functions are important for the viral life cycle, which could explain conservation of the proteins involved.
ACKNOWLEDGMENT
This study was supported by grant DFG Me 854/4-1 from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Babic N, Klupp B G, Brack A, Mettenleiter T C, Ugolini G, Flamand A. Deletion of glycoprotein gE reduces the propagation of pseudorabies virus in the nervous system of the mouse after intranasal inoculation. Virology. 1996;219:279–284. doi: 10.1006/viro.1996.0247. [DOI] [PubMed] [Google Scholar]
- 2.Baines J D, Roizman B. The UL10 gene of herpes simplex virus 1 encodes a novel viral glycoprotein, gM, which is present in the virion and in the plasma membrane of infected cells. J Virol. 1993;67:1441–1452. doi: 10.1128/jvi.67.3.1441-1452.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balan P, Davis-Poynter N, Bell S, Atkinson H, Browne H, Minson T. An analysis of the in vitro and in vivo phenotypes of mutants of herpes simplex virus type 1 lacking glycoproteins gG, gE, gI or the putative gJ. J Gen Virol. 1994;75:1245–1258. doi: 10.1099/0022-1317-75-6-1245. [DOI] [PubMed] [Google Scholar]
- 4.Bartha A. Experimental reduction of virulence of Aujeszky’s disease virus. Mag Allatorv Lapja. 1961;16:42–45. [Google Scholar]
- 5.Brack, A., and B. G. Klupp. Unpublished results.
- 6.Card J P, Whealy M E, Robbins A K, Enquist L W. Pseudorabies virus envelope glycoprotein gI influences both neurotropism and virulence during infection of the rat visual system. J Virol. 1992;66:3032–3041. doi: 10.1128/jvi.66.5.3032-3041.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dijkstra J, Visser N, Mettenleiter T C, Klupp B G. Identification and characterization of pseudorabies virus glycoprotein gM as a nonessential virion component. J Virol. 1996;70:5684–5688. doi: 10.1128/jvi.70.8.5684-5688.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dijkstra J, Mettenleiter T C, Klupp B G. Intracellular processing of pseudorabies virus glycoprotein M (gM): gM of strain Bartha lacks N-glycosylation. Virology. 1997;237:113–122. doi: 10.1006/viro.1997.8766. [DOI] [PubMed] [Google Scholar]
- 9.Dijkstra J, Gerdts V, Klupp B G, Mettenleiter T C. Deletion of glycoprotein gM of pseudorabies virus results in attenuation for the natural host. J Gen Virol. 1997;78:2147–2151. doi: 10.1099/0022-1317-78-9-2147. [DOI] [PubMed] [Google Scholar]
- 10.Dingwell K, Brunetti C, Hendricks R, Tang O, Tang M, Rainbow A, Johnson D C. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J Virol. 1994;68:834–845. doi: 10.1128/jvi.68.2.834-845.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dingwell K, Doering L, Johnson D C. Glycoproteins E and I facilitate neuron-to-neuron spread of herpes simplex virus. J Virol. 1995;69:7087–7098. doi: 10.1128/jvi.69.11.7087-7098.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dingwell K, Johnson D C. The herpes simplex virus gE-gI complex facilitates cell-to-cell spread and binds to components of cell functions. J Virol. 1998;72:8933–8942. doi: 10.1128/jvi.72.11.8933-8942.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enquist L W, Husak P, Banfield B W, Smith G A. Infection and spread of alphaherpesviruses in the nervous system. Adv Virus Res. 1999;51:237–347. doi: 10.1016/s0065-3527(08)60787-3. [DOI] [PubMed] [Google Scholar]
- 14.Fuchs W, Klupp B G, Rziha H-J, Mettenleiter T C. Identification and characterization of the pseudorabies virus UL3.5 protein, which is involved in virus egress. J Virol. 1996;70:3517–3527. doi: 10.1128/jvi.70.6.3517-3527.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuchs W, Klupp B G, Granzow H, Mettenleiter T C. The UL20 gene product of pseudorabies virus functions in virus egress. J Virol. 1997;71:5639–5646. doi: 10.1128/jvi.71.7.5639-5646.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerdts V, Jöns A, Makoschey B, Visser N, Mettenleiter T C. Protection of pigs against Aujeszky’s disease by DNA vaccination. J Gen Virol. 1997;78:2139–2146. doi: 10.1099/0022-1317-78-9-2139. [DOI] [PubMed] [Google Scholar]
- 17.Graham F L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 18.Granzow H, Weiland F, Jöns A, Klupp B, Karger A, Mettenleiter T C. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J Virol. 1997;71:2072–2082. doi: 10.1128/jvi.71.3.2072-2082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson D C, Feenstra V. Identification of a novel herpes simplex virus type 1-induced glycoprotein which complexes with gE and binds immunoglobulin. J Virol. 1987;61:2208–2216. doi: 10.1128/jvi.61.7.2208-2216.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jöns A, Granzow H, Kuchling R, Mettenleiter T C. The UL49.5 gene of pseudorabies virus codes for an O-glycosylated structural protein of the viral envelope. J Virol. 1996;70:1237–1241. doi: 10.1128/jvi.70.2.1237-1241.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jöns A, Dijkstra J, Mettenleiter T C. Glycoproteins M and N of pseudorabies virus form a disulfide-linked complex. J Virol. 1998;72:550–557. doi: 10.1128/jvi.72.1.550-557.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaplan A S, Vatter A E. A comparison of herpes simplex and pseudorabies viruses. Virology. 1959;7:394–407. doi: 10.1016/0042-6822(59)90068-6. [DOI] [PubMed] [Google Scholar]
- 23.Klupp B G, Lomniczi B, Visser N, Fuchs W, Mettenleiter T C. Mutations affecting the UL21 gene contribute to avirulence of pseudorabies virus vaccine strain Bartha. Virology. 1995;212:466–473. doi: 10.1006/viro.1995.1504. [DOI] [PubMed] [Google Scholar]
- 24.Klupp, B. G., and E. Weiland. Unpublished results.
- 25.Knapp A C, Husak P J, Enquist L W. The gE and gI homologs from two alphaherpesviruses have conserved and divergent neuroinvasive properties. J Virol. 1997;71:5820–5827. doi: 10.1128/jvi.71.8.5820-5827.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kritas S K, Pensaert M, Mettenleiter T C. Role of gI, gp63 and gIII in the invasion and spread of Aujeszky’s disease virus (ADV) in the olfactory nervous pathway of pigs. J Gen Virol. 1994;75:2319–2327. doi: 10.1099/0022-1317-75-9-2319. [DOI] [PubMed] [Google Scholar]
- 27.Lake C, Molesworth S, Hutt-Fletcher L. The Epstein-Barr virus (EBV) gN homolog BLRF1 encodes a 15-kilodalton glycoprotein that cannot be authentically processed unless it is coexpressed with the EBV gM homolog BBRF3. J Virol. 1998;72:5559–5564. doi: 10.1128/jvi.72.7.5559-5564.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lehner R, Meyer H, Mach M. Identification and characterization of a human cytomegalovirus gene coding for a membrane protein that is conserved among human herpesviruses. J Virol. 1989;63:3792–3800. doi: 10.1128/jvi.63.9.3792-3800.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ligas M W, Johnson D C. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by β-galactosidase sequences binds to but is unable to penetrate into cells. J Virol. 1988;62:1486–1494. doi: 10.1128/jvi.62.5.1486-1494.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loewy A D, Bridgman P C, Mettenleiter T C. β-Galactosidase expressing recombinant pseudorabies virus for light and electron microscopic study of transneuronally labeled neurons. Brain Res. 1991;555:346–352. doi: 10.1016/0006-8993(91)90364-2. [DOI] [PubMed] [Google Scholar]
- 31.Lomniczi B, Watanabe S, Ben-Porat T, Kaplan A S. Genome location and identification of functions defective in the Bartha vaccine strain of pseudorabies virus. J Virol. 1987;61:796–801. doi: 10.1128/jvi.61.3.796-801.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacLean C A, Robertson L M, Jamieson F E. Characterization of the UL10 gene product of herpes simplex virus type 1 and investigation of its role in vivo. J Gen Virol. 1993;74:975–983. doi: 10.1099/0022-1317-74-6-975. [DOI] [PubMed] [Google Scholar]
- 33.Mettenleiter T C, Lukacs N, Rziha H-J. Pseudorabies virus avirulent strains fail to express a major glycoprotein. J Virol. 1985;56:307–311. doi: 10.1128/jvi.56.1.307-311.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mettenleiter T C, Schreurs C, Zuckermann F, Ben-Porat T. Role of pseudorabies virus glycoprotein gI in virus release from infected cells. J Virol. 1987;61:2764–2769. doi: 10.1128/jvi.61.9.2764-2769.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mettenleiter T C, Zsak L, Kaplan A S, Ben-Porat T, Lomniczi B. Role of a structural glycoprotein of pseudorabies in virus virulence. J Virol. 1987;61:4030–4032. doi: 10.1128/jvi.61.12.4030-4032.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mettenleiter T C, Lomniczi B, Sugg N, Schreurs C, Ben-Porat T. Host cell-specific growth advantage of pseudorabies virus with a deletion in the genome sequences encoding a structural glycoprotein. J Virol. 1988;62:12–19. doi: 10.1128/jvi.62.1.12-19.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mettenleiter T C. Glycoprotein gIII deletion mutants of pseudorabies virus are impaired in virus entry. Virology. 1989;171:623–625. doi: 10.1016/0042-6822(89)90635-1. [DOI] [PubMed] [Google Scholar]
- 38.Mettenleiter T C, Rauh I. A glycoprotein gX-βgalactosidase fusion gene as insertional marker for rapid identification of pseudorabies virus mutants. J Virol Methods. 1990;30:55–66. doi: 10.1016/0166-0934(90)90043-f. [DOI] [PubMed] [Google Scholar]
- 39.Mettenleiter T C, Saalmüller A, Weiland F. Pseudorabies virus protein homologous to herpes simplex virus type 1 ICP18.5 is necessary for capsid maturation. J Virol. 1993;67:1236–1245. doi: 10.1128/jvi.67.3.1236-1245.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mettenleiter T C. Initiation and spread of α-herpesvirus infections. Trends Microbiol. 1994;2:2–4. doi: 10.1016/0966-842x(94)90335-2. [DOI] [PubMed] [Google Scholar]
- 41.Mettenleiter T C. Pseudorabies (Aujeszky’s disease) virus: state of the art. Acta Vet Hung. 1994;42:153–177. [PubMed] [Google Scholar]
- 42.Mijnes J, Lutters B, Vlot A, van Anken E, Horzinek M C, Rottier P J M, de Groot R J. Structure-function analysis of the gE-gI complex of feline herpesvirus: mapping of gI domains required for gE-gI interaction, intracellular transport, and cell-to-cell spread. J Virol. 1997;71:8397–8404. doi: 10.1128/jvi.71.11.8397-8404.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Osterrieder N, Neubauer A, Brandmüller C, Braun B, Kaaden O-R, Baines J. The equine herpesvirus 1 glycoprotein gp21/22a, the herpes simplex virus type 1 gM homolog, is involved in virus penetration and cell-to-cell spread of virions. J Virol. 1996;70:4110–4115. doi: 10.1128/jvi.70.6.4110-4115.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peeters B, de Wind N, Hooisma M, Wagenaar F, Gielkens A, Moorman R. Pseudorabies virus glycoproteins gII and gp50 are essential for virus penetration, but only gII is involved in membrane fusion. J Virol. 1992;66:894–905. doi: 10.1128/jvi.66.2.894-905.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petrovskis E, Timmins J, Giermann T, Post L E. Deletions in vaccine strains of pseudorabies virus and their effect on synthesis of glycoprotein gp63. J Virol. 1986;60:1166–1169. doi: 10.1128/jvi.60.3.1166-1169.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rauh I, Mettenleiter T C. Pseudorabies virus glycoproteins gII and gp50 are essential for virus penetration. J Virol. 1991;65:5348–5356. doi: 10.1128/jvi.65.10.5348-5356.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rixon F J, Addison C, McLauchlan J. Assembly of enveloped tegument structures (L-particles) can occur independently of virion maturation in herpes simplex virus type 1 infected cells. J Gen Virol. 1992;73:277–284. doi: 10.1099/0022-1317-73-2-277. [DOI] [PubMed] [Google Scholar]
- 48.Robbins A, Ryan J, Whealy M, Enquist L W. The gene encoding the gIII envelope protein of pseudorabies virus vaccine strain Bartha contains a mutation affecting protein localization. J Virol. 1989;63:250–258. doi: 10.1128/jvi.63.1.250-258.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmidt J, Klupp B G, Karger A, Mettenleiter T C. Adaptability in herpesviruses: glycoprotein D-independent infectivity of pseudorabies virus. J Virol. 1997;71:17–24. doi: 10.1128/jvi.71.1.17-24.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spear P. Entry of alphaherpesviruses into cells. Semin Virol. 1993;4:167–180. [Google Scholar]
- 51.Szilagyi J F, Cunningham C. Identification and characterisation of a novel non-infectious herpes simplex virus-related particle. J Gen Virol. 1991;62:661–668. doi: 10.1099/0022-1317-72-3-661. [DOI] [PubMed] [Google Scholar]
- 52.Whealy M, Card J P, Robbins A K, Dubin J R, Rziha H-J, Enquist L W. Specific pseudorabies virus infection of the rat visual system requires both gI and gp63 glycoproteins. J Virol. 1993;67:3786–3797. doi: 10.1128/jvi.67.7.3786-3797.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu S X, Zhu P, Letchworth G J. Bovine herpesvirus 1 glycoprotein M forms a disulfide-linked heterodimer with the UL49.5 protein. J Virol. 1997;72:3029–3036. doi: 10.1128/jvi.72.4.3029-3036.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zsak L, Zuckermann F, Sugg N, Ben-Porat T. Glycoprotein gI of pseudorabies virus promotes cell fusion and virus spread via direct cell-to-cell transmission. J Virol. 1992;66:2316–2325. doi: 10.1128/jvi.66.4.2316-2325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zuckermann F, Mettenleiter T C, Schreurs C, Sugg N, Ben-Porat T. Complex between glycoproteins gI and gp63 of pseudorabies virus: its effect on virus replication. J Virol. 1988;62:4622–4626. doi: 10.1128/jvi.62.12.4622-4626.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]