Abstract
Background:
Carbapenemase-producing, carbapenem-resistant Pseudomonas aeruginosa (CP-CRPA) are extensively drug resistant bacteria. We investigated the source of a multistate CP-CRPA outbreak.
Methods:
Cases were defined as a U.S. patient’s first isolation of P. aeruginosa sequence type 1203 with the carbapenemase gene blaVIM-80 and cephalosporinase gene blaGES-9 from any specimen source collected and reported to CDC between January 1, 2022–May 15, 2023. We conducted a 1:1 matched case-control study at the post-acute care facility with the most cases, assessed exposures associated with case status for all case-patients, and tested products for bacterial contamination.
Results:
We identified 81 case-patients from 18 states, 27 of whom were identified through surveillance cultures. Four (7%) of 54 case-patients with clinical cultures died within 30 days of culture collection, and four (22%) of 18 with eye infections underwent enucleation. In the case-control study, case-patients had increased odds of receiving artificial tears compared to controls (crude matched OR: 5.0, 95% CI: 1.1, 22.8). Overall, artificial tears use was reported by 61 (87%) of 70 case-patients with information; 43 (77%) of 56 case-patients with brand information reported use of Brand A, an imported, preservative-free, over-the-counter (OTC) product. Bacteria isolated from opened and unopened bottles of Brand A were genetically related to patient isolates. FDA inspection of the manufacturing plant identified likely sources of contamination.
Conclusions:
A manufactured medical product serving as the vehicle for carbapenemase-producing organisms is unprecedented in the U.S. The clinical impacts from this outbreak underscore the need for improved requirements for U.S. OTC product importers.
Summary
The source of an extensively drug-resistant Pseudomonas outbreak causing significant morbidity and mortality was a nationally distributed, over-the-counter product. This investigation highlights the potential for manufactured drugs to be sources of multidrug-resistant organisms and the need for increased manufacturer oversight.
Introduction
Carbapenemase-producing, carbapenem-resistant Pseudomonas aeruginosa (CP-CRPA) are an emerging public health threat in the United States (U.S.). These bacteria are often extensively drug-resistant and are associated with higher mortality infections compared to non-CP-CRPA.1 In the U.S., carbapenemases are identified in approximately 2% of CRPA clinical isolates,2 though the proportion is higher in some other countries.1,3,4 CP-CRPA outbreaks in U.S. healthcare settings have primarily described transmission within a single healthcare facility or patient-sharing network;5,6,7 one multistate outbreak, linked to medical tourism, has been reported.8
A subset of CRPA identified in U.S. clinical laboratories are submitted to public health laboratories for carbapenem-resistance mechanism testing through the Antimicrobial Resistance Laboratory Network (AR Lab Network), with timely testing to inform infection control and public health decision-making.9 Since 2021, CDC has prioritized CP-CRPA isolates for whole genome sequencing (WGS) to understand the molecular epidemiology of circulating strains and to inform local (e.g., facility-level) outbreak investigations.
During September 2022, analysis of WGS data from a CP-CRPA outbreak at a post-acute care facility determined the strain had an unusual combination of genetic features. The strain was P. aeruginosa sequence type (ST) 1203, harboring the Verona Integron-mediated metallo-β-lactamase (VIM) variant blaVIM-80 and the Guiana extended-spectrum β-lactamase (GES) variant blaGES-9, neither of which had been previously reported in U.S. isolates. By mid-October, through retrospective and prospective review of sequenced isolates from the AR Lab Network, we identified 30 isolates from five states with the same genetic features that were closely genetically related and from cultures collected since May 2022. We describe the investigation into the outbreak source.
Methods
Initial investigation
We defined a case as a U.S. patient’s first isolation of P. aeruginosa ST1203 with blaVIM-80 and blaGES-9 from any specimen source collected and reported to CDC from January 1, 2022–May 15, 2023. In October 2022, CDC disseminated a national call for clinical laboratories to submit CRPA isolates not susceptible to ceftazidime or cefepime to the AR Lab Network for carbapenem resistance mechanism testing and to report VIM-producing CRPA isolates to public health for further characterization.
The initial investigation included cases identified through November 2022. We collected data on medical products used in healthcare facilities with cases and patient-level data on healthcare history, including indwelling devices and medications. To generate hypotheses about the outbreak source, we conducted a 1:1 matched case-control study at one post-acute care facility. Controls were identified from facility patients who had a negative screen for CP-CRPA and no clinical CRPA cultures and were matched to cases based on facility unit and overlapping dates of admission. Information on medical products, procedures, and devices were collected from medical records. We used conditional logistic regression to calculate crude and adjusted odds ratios; potential confounding variables were assessed a priori through directed acyclic graphs (see Supplemental Appendix for detailed methods).
Case Investigation
We collected data on all case-patients nationally using a standardized case report form, completed with medical records and, when possible, interviews with case-patients or their proxies. We classified exposure to artificial tears, a hypothesis that emerged from the case-control study, using tiered definitions to account for limitations in facility tracking of artificial tears brands received by individual patients and limited recall by outpatients. Exposure was confirmed if the brand of interest was documented in the medical record or reported by the case-patient or proxy. Exposure was probable if either (a) the case-patient received artificial tears in an inpatient facility that used the brand of interest concurrently with other brand(s) and did not document which specific brand(s) individual patients received, or (b) if a case-patient reported receiving a recall notice from a company or healthcare provider for artificial tears and could not remember the name of the recalled brand, but described a product compatible with the brand of interest. Exposure was possible if the case-patient received outpatient healthcare at a facility that was using the brand of interest during clinical care and/or providing the brand of interest to patients without documenting which brand(s) each patient received.
Data were collected and managed using REDCap electronic data capture tools hosted at CDC,10,11 and statistical analyses were conducted in R version 4.1.2.12 This activity was reviewed by CDC and conducted according to applicable federal law and CDC policy.13–16 This work did not receive any non-CDC funding support.
Traceback & Product Investigation
State and local health departments and the U.S. Food and Drug Administration (FDA) collected opened and unopened artificial tears products identified in the epidemiologic investigation from case-patient homes, healthcare facilities, distribution centers, and pharmacies. Products were cultured for the presence of bacteria and isolates underwent WGS. We conducted phylogenetic analysis on sequences from case-patients and products (see Supplemental Appendix for detailed methods).
Additional Control Measures
CDC and FDA disseminated public health communications about investigation findings. FDA conducted an onsite inspection of the manufacturer identified in the epidemiologic investigation in accordance with federal regulations.17
Results
Initial investigation
Most (n=26) cases confirmed prior to November 2022 belonged to three healthcare facility clusters in three states, which had been investigated as individual facility outbreaks before they were genetically linked by WGS analysis. One cluster was comprised of eye infections (n=4 patients) linked to an ophthalmology clinic, and two clusters occurred at post-acute care facilities (n=19 and n=3 patients), where cases were identified from urine, sputum, and surveillance rectal swab cultures collected in response to clinical cases. In November 2022, a fourth facility cluster was identified when an additional state reported seven patients who sought care at the same hospital for CRPA eye infections, three of whom used the same community health center. No history of healthcare outside the U.S. or interstate patient transfers were identified.
To generate hypotheses, we conducted a 1:1 matched case-control study with the first 16 cases identified in the largest facility cluster (Supplementary Appendix). Cases had 15 times greater odds of exposure to sterile water for inhalation than controls (crude matched OR: 15.0, 95% CI: 0.86, 262.6; aOR not calculated), and 5 times greater odds of exposure to artificial tears than controls (crude matched OR: 5.0, 95% CI: 1.1, 22.8; aOR: 4.7, 95% CI: 0.98, 22.5 [adjusting for mechanical ventilation], Supplementary Appendix, Table S1). Assessing these exposures across all facility clusters, only case-patients in two of four facilities used sterile water for inhalation, and the two facilities reported different sterile water brands. Case-patients in all four facility clusters used artificial tears; six different brands were each reported by at least two facilities. Given these findings and the eye infections associated with two clusters, we focused our investigation on artificial tears. Throughout the investigation, we explored the possibility of a contaminated ingredient in multiple products, including sterile water as an ingredient in artificial tears, and did not find sufficient evidence of manufacturing overlap for reported brands.
Case Investigation
As of May 15, 2023, we identified 81 cases from 18 states (Figure 1). Cultures were collected from May 1, 2022–April 6, 2023, with a median of 43 days (range: 14–310 days) between date of specimen collection and case confirmation (Figure 2). Isolates were extensively drug-resistant; 5 isolates tested at CDC by reference broth microdilution against an extended panel of antibiotics only demonstrated susceptibility to cefiderocol (Supplementary Appendix, Table S2). Overall, 54 (67%) cases were identified from clinical cultures of the eye (n=21, 39%), urinary tract (n=15, 28%), respiratory tract (n=13, 24%), blood (n=3, 5%), wound (n=1, 2%), and ear (n=1, 2%). Twenty-seven (33%) were identified from surveillance cultures of rectal swabs (n=26) and sputum (n=1). Case-patient clinical characteristics and outcomes varied across culture source (Table 1).
Figure 1.
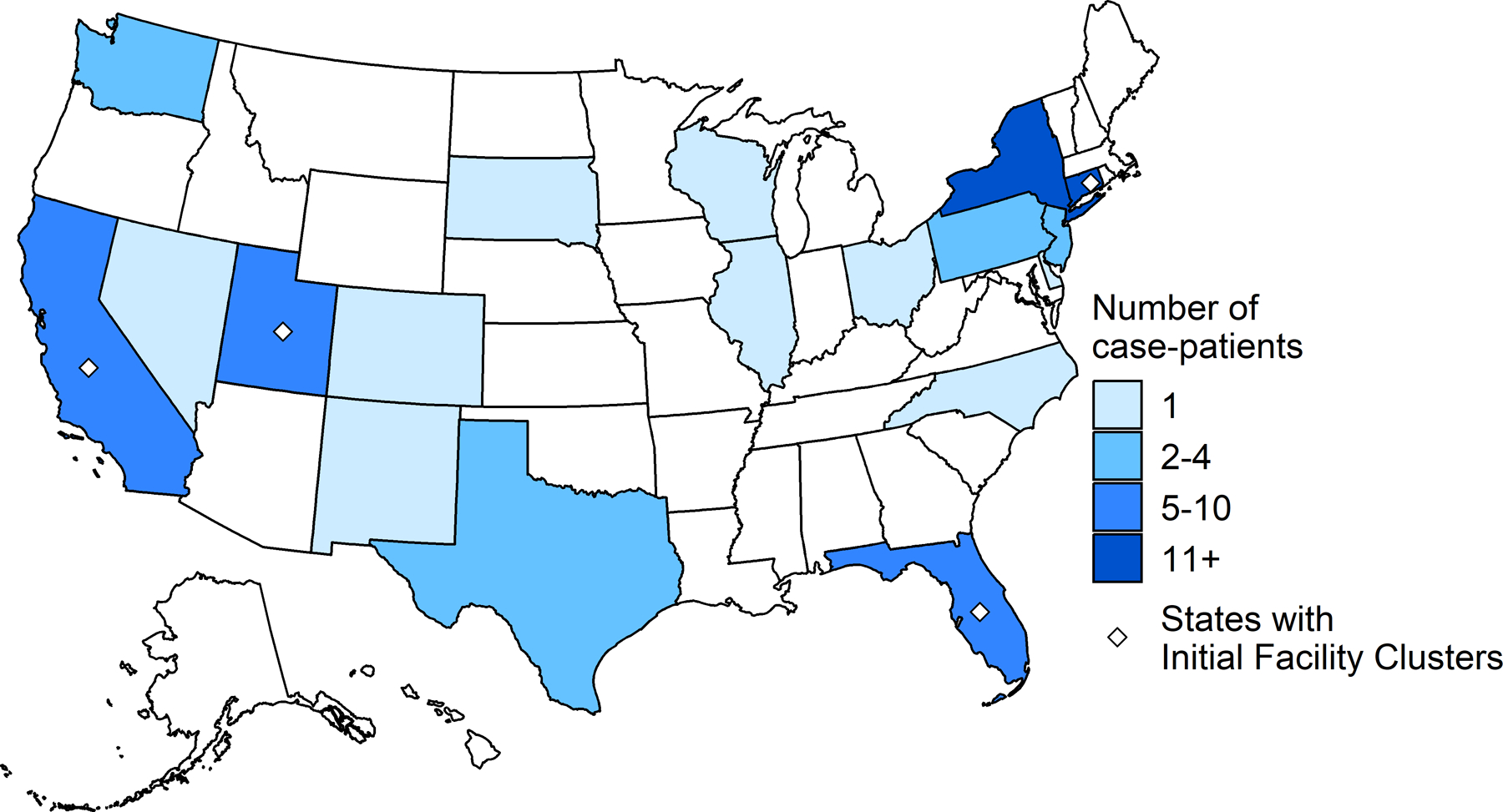
Number of patients with P. aeruginosa ST1203 with blaVIM-80 and blaGES-9, by state, January 1, 2022–May 15, 2023.
Figure 2.
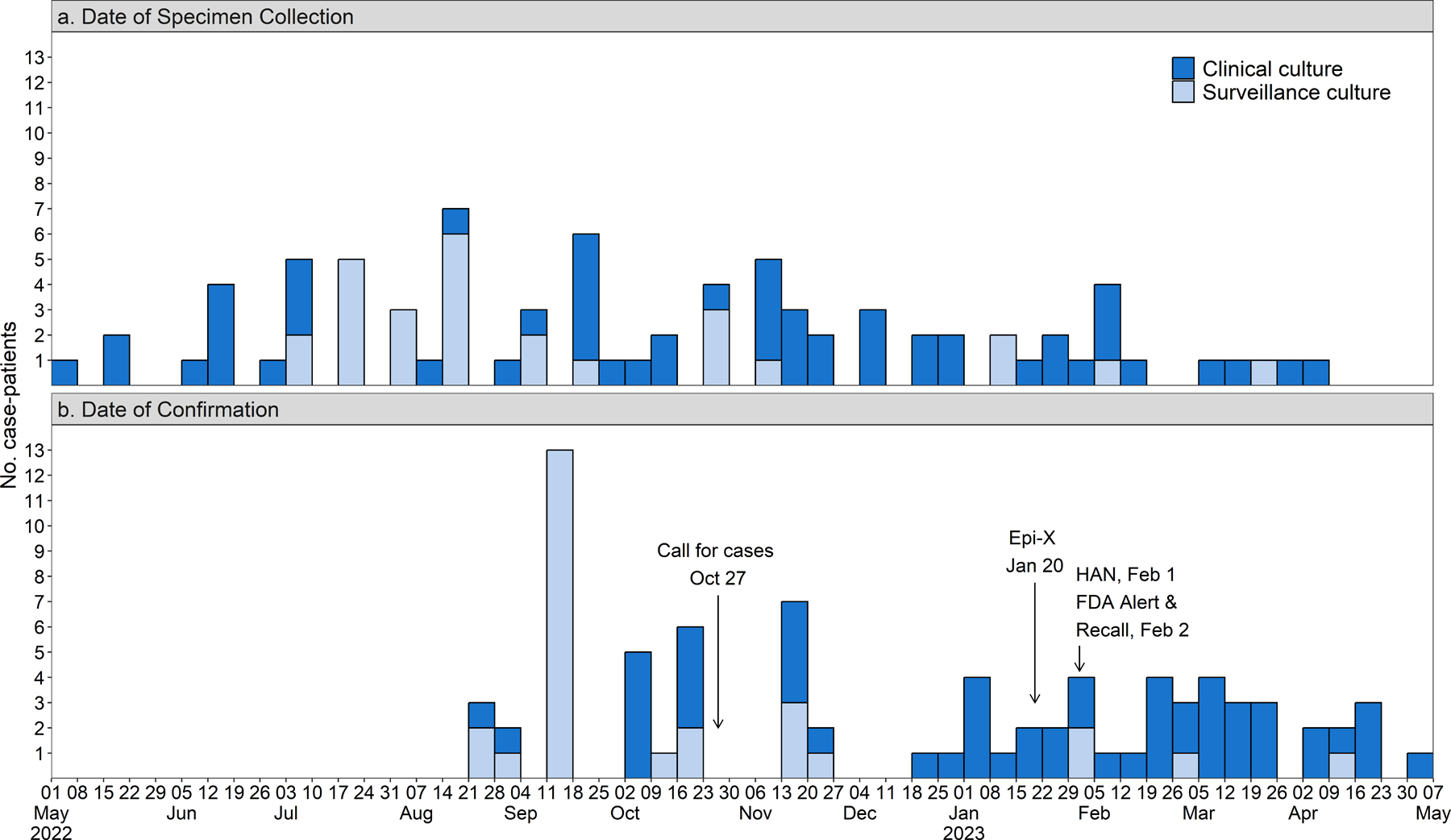
Patients with P. aeruginosa ST1203 with blaVIM-80 and blaGES-9 by (a) date of specimen collection and (b) date of confirmation, by specimen type. Key public health communications are noted in the bottom panel and were informed by the timing of case confirmation. Abbreviations include Epi-X: Epidemic Information Exchange and HAN: Health Alert Network.
Table 1.
Demographics, clinical characteristics, and outcomes of 81 patients with P. aeruginosa ST1203 with blaVIM-80 and blaGES-9, by culture source*
| Characteristic | All Case-Patients, N = 81 | Eye, N = 21 | Urinary, N = 15 | Respiratory, N = 13 | Blood, N = 3 | Other clinical, N = 2† | Surveillance, N = 27 |
|---|---|---|---|---|---|---|---|
| Demographics— n/N (%) | |||||||
| Age (years) Mean (SD) |
60 (24) | 75 (14) | 70 (22) | 44 (31) | 72 (5) | 67 (10) | 49 (19) |
| Median (range) | 63 (0–102) | 76 (42–102) | 70 (21–100) | 52 (0–83) | 74 (67–76) | 67 (60–74) | 51 (12–87) |
| Patient Sex—Male | 40/80 (50) | 9/21 (43) | 5/15 (33) | 6/12 (50) | 1/3 (33) | 1/2 (50) | 18/27 (67) |
| Setting of culture collection— n (%) | |||||||
| Inpatient Acute Care Hospital | 22 (27) | 3 (14) | 8 (53) | 8 (62) | 2 (67) | 1 (50) | 0 (0) |
| Long Term Acute Care Hospital | 27 (33) | 0 (0) | 1 (6.7) | 3 (23) | 0 (0) | 0 (0) | 23 (85) |
| Skilled nursing facility | 5 (6.2) | 0 (0) | 1 (6.7) | 0 (0) | 0 (0) | 0 (0) | 4 (15) |
| Emergency Department | 6 (7.4) | 2 (9.5) | 2 (13) | 1 (7.7) | 1 (33) | 0 (0) | 0 (0) |
| Outpatient clinic | 21 (26) | 16 (76) | 3 (20) | 0 (0) | 0 (0) | 1 (50) | 0 (0) |
| Underlying medical conditions— n (%)‡ | |||||||
| Charlson Comorbidity Index (CCI)—median (range) | 1 (0–12) | 1 (0–8) | 1 (0–8) | 2 (0–12) | 4 (2–6) | 3 (3–3) | 1 (0–8) |
| No CCI underlying conditions | 18 (22) | 5 (24) | 4 (27) | 2 (15) | 0 (0) | 0 (0) | 7 (26) |
| Diabetes | 19 (23) | 8 (38) | 3 (20) | 2 (15) | 0 (0) | 1 (50) | 5 (19) |
| Chronic lung disease | 16 (20) | 6 (29) | 3 (20) | 4 (31) | 1 (33) | 0 (0) | 2 (7.4) |
| Cerebrovascular disease | 14 (17) | 2 (9.5) | 4 (27) | 3 (23) | 1 (33) | 0 (0) | 4 (15) |
| Dementia | 13 (16) | 2 (9.5) | 1 (6.7) | 0 (0) | 1 (33) | 0 (0) | 9 (33) |
| Cancer | 13(16) | 2 (9.5) | 3 (20) | 2 (15) | 2 (67) | 2 (100) | 2 (7.4) |
| Any eye disease§ | 30 (37) | 19 (90) | 3 (20) | 1 (7.7) | 1 (33) | 1 (50) | 5 (19) |
| Glaucoma | 16 (20) | 13 (62) | 1 (6.7) | 0 (0) | 1 (33) | 0 (0) | 1 (3.7) |
| Cataracts | 14 (17) | 9 (43) | 2 (13) | 1 (7.7) | 1 (33) | 0 (0) | 1 (3.7) |
| Indwelling devices in place up to 3 months prior to first culture collection— n/N (%)‖ | |||||||
| No devices | 22/73 (30) | 14/15 (93) | 6/14 (43) | 0 (0) | 0 (0) | 1 (50) | 1 (3.7) |
| Tracheostomy | 39/75 (52) | 0 (0) | 3/14 (21) | 11 (85) | 0 (0) | 0 (0) | 25/26 (96) |
| Percutaneous endoscopic gastrostomy tube | 32/73 (44) | 0/15 (0) | 3/14 (21) | 7 (54) | 1 (50) | 0 (0) | 21 (78) |
| Invasive urinary catheter | 16/73 (22) | 0/15 (0) | 5/14 (36) | 5 (38) | 1 (50) | 0 (0) | 5 (19) |
| Mechanical Ventilation | 26/73 (36) | 0/15 (0) | 2/14 (14) | 9 (69) | 0 (0) | 0 (0) | 14 (52) |
| Outcomes for case-patients with incident clinical cultures—n/N (%) | |||||||
| Provider documented infection¶ | 42/50 (84) | 21 (100) | 8/11 (73) | 9 (69) | 3 (100) | 1 (50) | -- |
| New hospitalization** | 34/49 (69) | 11/18 (61) | 10/15 (67) | 10 (77) | 1 (50) | 0/1 (0) | -- |
| New intensive care unit admission** | 10/41 (24) | 0/14 (0) | 0/8 (0) | 7/10 (70) | 2 (100) | 1/1 (100) | -- |
| Death within 30 days of culture collection | 4/39 (10) | 1/17 (5.9) | 1/10 (10) | 0/9 (0) | 2 (100) | 0/1 (0) | -- |
Denominators are included for variables with missing data
Culture sources include wound (n=1) and ear (n=1)
The most frequent underlying medical condition for each specimen category is listed; full list in Supplementary Appendix File S6
Any eye disease reported includes: glaucoma (n=16), cataracts (n=14), macular degeneration (n=3), ocular hypertension (n=2), Sjorgren’s disease (n=2), blepharitis (n=1), retinal artery/vein occlusion (n=1), vitreomacular adhesion (n=1), undifferentiated anterior corneal surface disorder (n=1), lagophalmos (n=1), recurrent corneal erosion (n=1), filimentary keratitis (n=1), corneal stromal dystrophy (n=1), and ocular cicatricial pemphigoid (n=1).
Three most common devices reported among all case-patients, not including mechanical ventilation; full list in Supplementary Appendix File S6.
Associated infection with P. aeruginosa documented in the medical record
Hospitalization and admission to the intensive care unit (ICU) defined as new hospital or ICU admission, respectively, within 3 days prior to 2 weeks after culture collection. New admission to the ICU includes patients who were already hospitalized (i.e., admission more than 3 days prior to culture collection), but were transferred to ICU during timeframe of interest
Forty-two (84%) of 50 case-patients with the outbreak strain isolated from a clinical culture and available information had an associated infection documented in the medical record. These included eye infections ([n=21, 50%], comprising keratitis without progression [n=12/21, 57%], keratitis with progression to endophthalmitis [n=6/21, 28%], panophthalmitis [n=2/21; 9.5%], and unspecified eye infection [n=1/21, 5%]); urinary tract infection (8, 19%); respiratory tract infection (9, 21%); bacteremia (3, 7%); and otitis media with osteoradionecrosis (1, 2%). Four (10%) of 39 case-patients with clinical cultures who had outcome information available died within 30 days of incident specimen collection, and 4 (22%) of 18 case-patients with eye infections and available information underwent enucleation. An additional 14 were visually impaired in the affected eye 30 days after culture collection: 3 (23%) had moderate visual impairment (best corrected visual acuity [BCVA] ≤20/70 and >20/200) and 11 (77%) became legally blind (BCVA ≤20/200 or limited to hand motion or light perception). Among the 27 case-patients identified through surveillance cultures, 6 (22%) had subsequent clinical cultures with the outbreak strain (bloodstream [n=2], respiratory [n=3], urine [n=1]), which were collected a median 152 days (range: 22–252 days) after the incident surveillance culture.
The use of artificial tears was reported by 61 (87%) of 70 case-patients with information. Brand information was available for 56 (92%) patients, who collectively reported 39 different brands. Brand A, produced by Manufacturer Y, was reported in all four initial facility clusters and by 43 (77%) case-patients, who had confirmed (n=22), probable (n=14), or possible (n=7) use. One additional artificial tears product made by Manufacturer Y, Brand B, was reported by a single case-patient (Figure 3). No other brand was reported by more than 25% of patients with available information.
Figure 3.
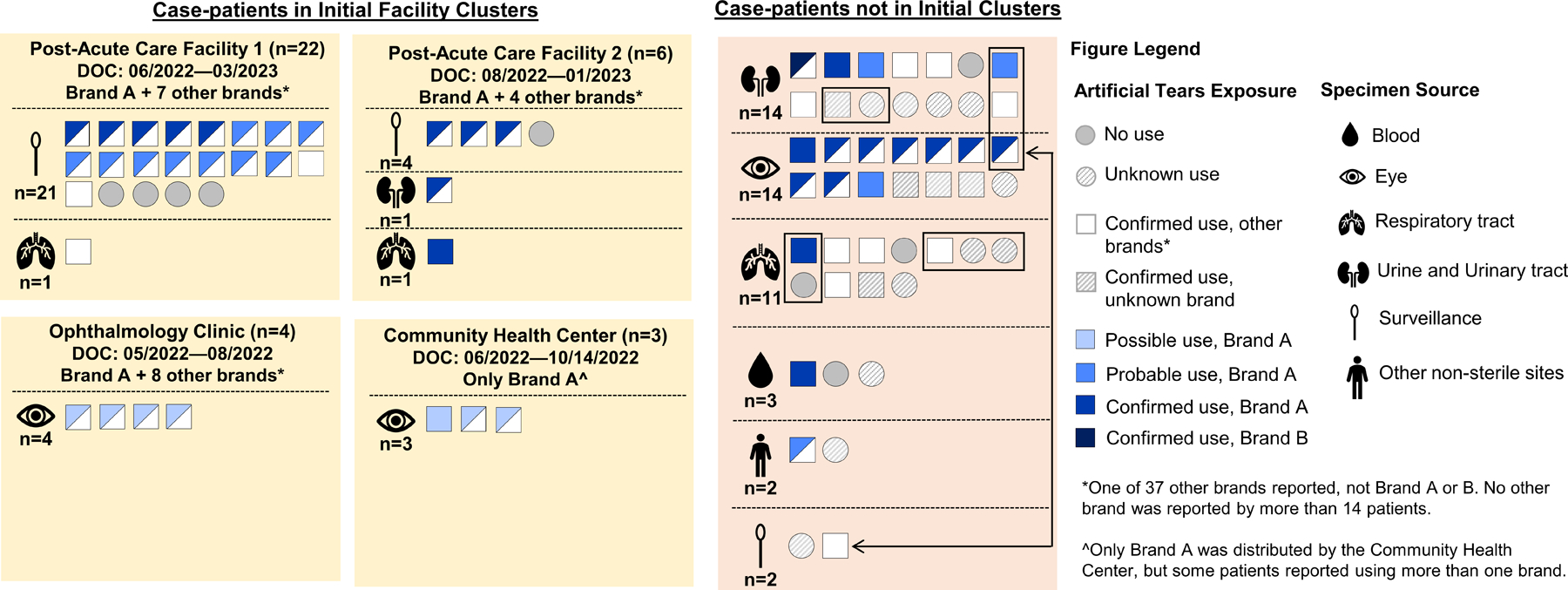
Artificial tears use among patients with P. aeruginosa ST1203 with blaVIM-80 and blaGES-9. Each square or circle represents an individual case-patient with use of artificial tears denoted by a shape and brand use indicated by color. Due to exposure uncertainty, we developed tiered definitions for brand exposure, which is shown only for Brands A and B; exposure to all other brands, regardless of exposure certainty, is shown in white. Exposure was confirmed if Brand A or B was documented in the medical record or reported by the case-patient or proxy. Exposure was probable if either (a) the case-patient received artificial tears in an inpatient facility that used Brand A or B concurrently with other brand(s) and did not document which brand(s) patients received, or (b) if a case-patient reported receiving a recall notice from a company or healthcare provider for artificial tears and could not remember the name of the recalled brand, but described a product compatible with Brand A or B. Exposure was possible if the case-patient received outpatient healthcare at a facility that was using Brand A or B during clinical care and/or providing Brand A or B to patients without documenting which brand(s) each patient received. Case-patients not in initial clusters who are epidemiologically linked are grouped with black rectangles or connected with a double-sided arrow.
Among 22 case-patients who did not report using artificial tears or reported a brand other than Brands A or B artificial tears, 12 (55%) overlapped temporally and/or spatially in healthcare facilities with other case-patients or were admitted to healthcare facilities where Brand A was used and may have acquired the outbreak strain through intra-facility transmission.
Traceback & Product investigation
Brands A and B artificial tears were preservative-free products produced by Manufacturer Y at the same site in India, packaged in multi-dose bottles, and sold to U.S. consumers over the internet and to U.S. healthcare facilities through medical product distributors.
CDC and FDA tested opened (n=17) and unopened (n=18) lots of Brand A artificial tears; 1–149 bottles per lot were tested. P. aeruginosa ST1203 with blaVIM-80 and blaGES-9 was recovered from 6 lots of opened Brand A, 2 lots of unopened Brand A, and a composite sample of 2 additional lots of unopened Brand A. Bacteria were isolated from five of seven (71%) unopened Brand B lots representing two different products, artificial tears (n=4 lots) and artificial eye ointment (n=1 lot), though P. aeruginosa was not recovered (see Supplemental Appendix Table S4 for bacterial species recovered from Brand B products).
Phylogenetic analysis of 108 isolates from case-patients (n=81) and opened (n=21) and unopened (n=6) Brand A bottles revealed that they varied by 0–110 single nucleotide polymorphisms (SNPs), derived from a clonal frame of 6.5 Mb (91.8%) of the reference genome. The majority (n=105) of the isolates only differed by 0–13 SNPs; two 0-SNP clusters were comprised of case-patient isolates from multiple states and Brand A product isolates from opened and unopened bottles (Figure 4). Outbreak isolates clustered distinctly from other P. aeruginosa ST1203 isolates from the U.S. and other countries (Figure 4).
Figure 4.
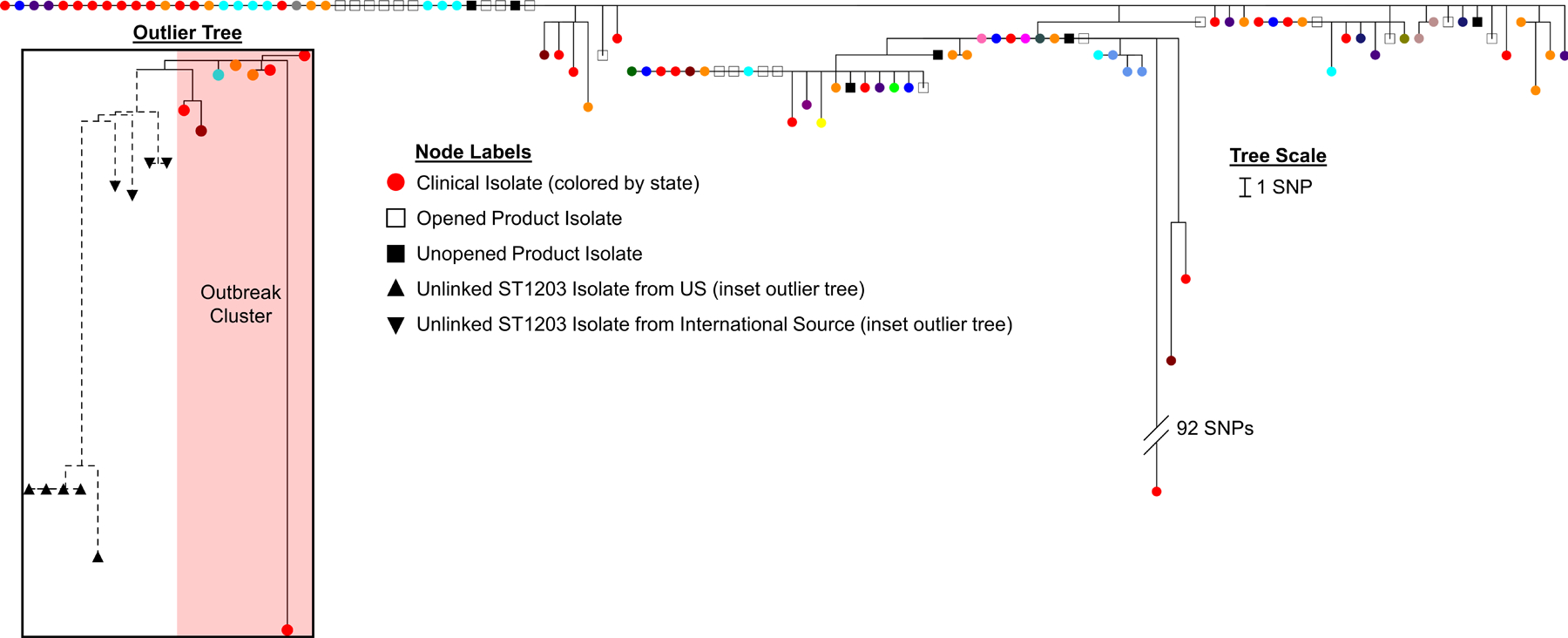
Phylogenetic tree of single nucleotide polymorphism (SNP) analysis of outbreak-related P. aeruginosa ST1203 with blaVIM-80 and blaGES-9. from 81 case-patient isolates (circular nodes, colored by state) and 27 isolates from opened and unopened Brand A artificial tears bottles (square nodes). The tree branch of a single isolate with a hypermutator genotype is truncated. Inset shows subset of outbreak isolates (circular nodes) clustering distinctly from other P. aeruginosa ST1203 isolates from both U.S. and international sources including India, Kenya, and Nepal (triangular nodes).
Additional Control Measures
On January 20, 2023, CDC recommended that clinicians and patients immediately discontinue use of Brand A Artificial Tears due to potential bacterial contamination via the Epidemic Information Exchange (Epi-X) network; additional CDC and FDA communications followed (Figure 2B). On February 2, 2023, Manufacturer Y issued a voluntary national recall for Brands A and B artificial tears.
FDA conducted an onsite inspection of Manufacturer Y beginning on February 20, 2023 and found their manufacturing process lacked appropriate procedures and practices to prevent microbiological contamination.18 On February 24, 2023, Brand B Artificial Eye Ointment was voluntarily recalled by Manufacturer Y based on the systemic nature of the inspection findings impacting all ophthalmic products at the facility and FDA testing of unopened Brand B Artificial Eye Ointment that yielded a preliminary non-sterility result. Manufacturer Y did not export additional products from the manufacturing site to the U.S. nor export Brands A and B to other countries.
Discussion
We used molecular epidemiology to determine that CP-CRPA from diverse specimen sources in different patient populations constituted a single outbreak. The combination of epidemiology and laboratory evidence, including the close genetic relatedness of isolates from case-patients across multiple states and from unopened Brand A product, indicates Brand A artificial tears was the outbreak source. Transmission of carbapenemase-producing organisms (CPOs) via a manufactured medical product represents a newly detected modality for spread in the U.S., with potentially long-term impacts on control of these emerging pathogens.
According to U.S. FDA regulations, over-the-counter (OTC) ophthalmic drug products sold in the U.S. must be sterile and multi-use products must contain preservatives.19,20 In our investigation, bacteria isolated from unopened Manufacturer Y products indicate they were not sterile, and FDA’s inspection of the manufacturer identified multiple deficient practices that were likely sources of contamination. This was the manufacturer’s first FDA inspection; under the current U.S. legislative system, foreign-based manufacturers can ship OTC products without having an FDA inspection.21 To address this gap, FDA proposed legislative changes for fiscal year 2024 to strengthen regulatory requirements for sterile manufacturing facility inspections before the distribution of OTC non-application drugs.22 Strengthening foreign manufacturers’ compliance with FDA’s Current Good Manufacturing Practices23,24 and improving requirements for U.S. importer accountabilities will ensure that products are made in accordance with U.S. regulations and may prevent future outbreaks.
The extent to which CPOs spread through contaminated medical products is unclear. Identification of this outbreak was aided by the outbreak strain’s unique genetic features; a contemporaneous CPO outbreak in Europe linked to a contaminated antibiotic was similarly recognized and tracked due to an unusual combination of resistance genes.25 Although the primary source of CPO transmission in the U.S. remains colonized and infected patients, these outbreaks raise the question of whether product-related outbreaks of CPOs occur more frequently than currently recognized but go undetected in the absence of unique genetic markers. This highlights a need to better characterize use of WGS to identify outbreaks of these organisms, including relatedness thresholds for identifying possible multistate clusters.
The use of a WGS-based case definition, rather than a more traditional epidemiology approach based on clinical presentation, place, and time, enabled us to determine isolates from diverse culture sources and healthcare settings were the same strain and likely had a common source. In contrast to previous outbreaks linked to contaminated ophthalmic products that exclusively described ocular infections,26–30 specimen sources in this outbreak were diverse, with only 26% of cases from eye specimens. Prior P. aeruginosa outbreaks linked to contaminated ophthalmic products may have had similar broad clinical impacts that went unrecognized.
Our investigation reinforces the potential amplification of known infection control and treatment challenges presented by CPOs when they are spread by a widely distributed product. First, carbapenemase identification is challenging because it requires specialized testing, which is available through the AR Lab Network if unavailable in clinical laboratories; however, low suspicion for CRPA to harbor a carbapenemase may deter isolate submission.9 Second, CPO transmission is difficult to control in healthcare facilities; interventions are labor-intensive, requiring high levels of adherence to infection prevention measures.31 Our data suggests that intra-facility transmission was the route of exposure for some case-patients who did not report the use of artificial tears, given the close genetic reladeness of case-patient isolates. Third, there are likely a large number of individuals colonized with the outbreak strain that have not been identified. Colonization can persist for months or years,32 and colonized patients can transmit to others and are at risk of infection with the colonizing organism. In our investigation, 22% of case-patients identified through surveillance cultures had subsequent clinical cultures, some more than six months after colonization was initially identified. The reservoir of colonized individuals created through widespread product exposure means that we expect to identify additional infections caused by the outbreak strain despite the product recall. Finally, outbreak isolates that underwent reference antimicrobial susceptibility testing were only susceptible to cefiderocol, demonstrating the limited treatment options available for CPOs.
This investigation had multiple limitations. The source of exposure for 10 case-patients who did not use Manufacturer Y products or share a healthcare facility with other case-patients is unknown, and 15 case-patients had unknown use of artificial tears or unknown brand. Exposure ascertainment was challenged by many factors, including the limited number of case-patient or proxy interviews completed for those who were in the community for part of the 90-day exposure period. Additionally, case-patients may not have remembered all artificial tears brands used; notably, six case-patients identified exposure to Manufacturer Y products only after the products were recalled. Issues of limited recall may have been exacerbated when case confirmation lagged specimen collection by several months, due to time-intensive laboratory testing and delays in submission of some isolates for WGS. While the 90-day exposure window attempted to balance periods of reasonable recall with potentially long colonization before case detection, the interval between product use and culture may have exceeded this period for some case-patients, leading to incomplete exposure data. Patients with eye infections may have been more likely to report ophthalmic products used compared to those with infections at other sites, introducing differential misclassification. Ascertainment of patient outcomes was also subject to limitations; although we used time periods relative to culture dates as proxies for assessing whether outcomes such as hospitalization and death were associated with clinical cases, it is possible case-patients were hospitalized for other reasons and case-patients died with, rather than due to, CP-CRPA infection.
The strengthening of U.S. public health infrastructure to detect and respond to antibiotic resistant threats enabled us to uncover a hidden outbreak with a novel source, prompting public health and regulatory actions that averted additional infections, vision loss, and death. Significant improvements in foreign-based ophthalmic manufacturer adherence to Current Good Manufacturing Practices may prevent drugs manufactured under suboptimal conditions from reaching U.S. patients. Additionally, U.S. importers and distributors bear legal responsibility for ensuring that products sold under their label are manufactured in accordance with U.S. regulations. The cumulative impact of this outbreak on the dissemination of extensively drug-resistant P. aeruginosa in the U.S. may only be fully realized over the coming years.
Supplementary Material
Acknowledgements
Members of the Multistate Pseudomonas Outbreak Investigation group include: Audrey Brezak, MPH; Allison C. Brown, PhD; Nicole Burton, PhD; Rebekah Carman, MPH; Dawn Chinn-Flournoy, DrPH; Jennifer Connolly, ICIP; Jennifer L. Dale, PhD; Carole Dieterly, DVM; Ashlie Dowdell; Jennifer Driscoll, MLS; Jalysa Erskine, MPH; Monica Giacomucci, MPH; Lajune Harris, MPH; Cam-Van Huynh, DDS; Melissa Judson, MPH; Manisha Juthani, MD; Eric N. Keller, MS; Lori Koenecke, MS; Alison Laufer Halpin, PhD; Stephen LaVoie, PhD; Victoria LeGarde, MPH; Jennifer MacFarquhar, MPH; Arif Mahmud, MD; Michael Mamerow, MS; Elise Mantell, PhD; Greta Michaelson, MHA; Jason Mitchell, MLT; Tisha Mitsunaga, DrPH; Marika Mohr, MS; Jennifer L. Morgan, MSN; Heather Moulton-Meissner, PhD; Judith Noble-Wang, PhD; Kelly Oakeson, PhD; Juliana Reyes, MS; Alessandro Rossi, PhD; Emily Schneider, MPH; Adrienne Sherman, MPH; Gloria Shin, PhD; Marla M Sievers, MPH; Megan Sredl, MPH; Ann-Catherine Stanton, MPH; Sheila vanTwuyver, BS.
We would like to thank the following people for their work on the investigation: Homero Aguilar, Amir Alavi, Rocio Balbuena, Jasna Braut-Taormina, Hollianne Bruce, Wiley Chambers, Kai-Shun Chen, Kristen Clancy, Michelle Cockrell, Karlos Crayton, Michael Cyrus, Marisa D’Angeli, Maria Diaz, Jan Dollete, Samuel Eskenazi, Lorene Fong, Rosalie Giardina, Jennifer Gogley, William Greendyke, Susan Hadman, Hollis Houston, Catherine Huck, Philip Istafanos, Amber Jean-Louis, Deborah Jones, Molly Kratz, Thao Kwan, Sammie La, Susan Lance, Megan Lasure, Cynthia Longo, Maria Machado, Gillian McAllister, Sherri McGarry, Susannah McKay, Derek L. Miller, Marissa Musk, Kelsey O’Yong, Elvis Patel, Arthur Pightling, Allison Rodriguez, Jeffery Rogers, Haydee Romero, Mona Satyam, Matthew Silverman, Gail Skolek, Amanda J. Smith, Anna Stahl, Luis Torres, Kavita K. Trivedi, and Nadine Wilmott.
We would also like to thank the following institutions: California Department of Public Health Microbial Diseases Laboratory and local public health laboratories, Florida Bureau of Public Health Laboratories, Florida Department of Health Healthcare-Associated Infections Prevention Team, Los Angeles County Public Health Laboratories, Maryland Department of Health Laboratories Administration Division of Microbiology and Division of Molecular Biology, NC State Laboratory of Public Health Molecular Diagnostics and Epidemiology Unit, Wadsworth Center AR Laboratory Team, and Wadsworth Center for Advanced Genomic Technologies Cluster.
Funding:
This work was supported by CDC.
Footnotes
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Visual abstract can be found here: AMultistateOutbreakofExtensivelyDrug-ResistantPseudomonasaeruginosaassociatedwithContaminatedArtificialTears,2022-2023|Tidbit(tidbitapp.io)
Conflict of interest: The authors declare no conflict of interest.
References
- 1.Reyes J, Komarow L, Chen L, et al. Global epidemiology and clinical outcomes of carbapenem-resistant Pseudomonas aeruginosa and associated carbapenemases (POP): a prospective cohort study. Lancet Microbe 2023;4(3):e159–e170. DOI: 10.1016/S2666-5247(22)00329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.CDC. Antibiotic Resistance & Patient Safety Portal (AR&PSP) AR Lab Network Data. U.S. Department of Health and Human Services, CDC. (https://arpsp.cdc.gov/).
- 3.Castanheira M, Deshpande LM, Costello A, Davies TA, Jones RN. Epidemiology and carbapenem resistance mechanisms of carbapenem-non-susceptible Pseudomonas aeruginosa collected during 2009–11 in 14 European and Mediterranean countries. J Antimicrob Chemother 2014;69(7):1804–14. (In eng). DOI: 10.1093/jac/dku048. [DOI] [PubMed] [Google Scholar]
- 4.Edelstein MV, Skleenova EN, Shevchenko OV, et al. Spread of extensively resistant VIM-2-positive ST235 Pseudomonas aeruginosa in Belarus, Kazakhstan, and Russia: a longitudinal epidemiological and clinical study. Lancet Infect Dis 2013;13(10):867–76. (In eng). DOI: 10.1016/s1473-3099(13)70168-3. [DOI] [PubMed] [Google Scholar]
- 5.Clegg WJ, Pacilli M, Kemble SK, et al. Notes from the Field: Large Cluster of Verona Integron-Encoded Metallo-Beta-Lactamase-Producing Carbapenem-Resistant Pseudomonas aeruginosa Isolates Colonizing Residents at a Skilled Nursing Facility - Chicago, Illinois, November 2016-March 2018. MMWR Morb Mortal Wkly Rep 2018;67(40):1130–1131. DOI: 10.15585/mmwr.mm6740a6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prestel C, Moulton-Meissner H, Gable P, et al. Dialysis Water Supply Faucet as Reservoir for Carbapenemase-Producing Pseudomonas aeruginosa. Emerg Infect Dis 2022;28(10):2069–2073. DOI: 10.3201/eid2810.220731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weng MK, Brooks RB, Glowicz J, et al. Outbreak investigation of Pseudomonas aeruginosa infections in a neonatal intensive care unit. Am J Infect Control 2019;47(9):1148–1150. (In eng). DOI: 10.1016/j.ajic.2019.03.009. [DOI] [PubMed] [Google Scholar]
- 8.Kracalik I, Ham DC, McAllister G, et al. Extensively Drug-Resistant Carbapenemase-Producing Pseudomonas aeruginosa and Medical Tourism from the United States to Mexico, 2018–2019. Emerg Infect Dis 2022;28(1):51–61. DOI: 10.3201/eid2801.211880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabour S, Huang JY, Bhatnagar A, et al. Detection and Characterization of Targeted Carbapenem-Resistant Health Care-Associated Threats: Findings from the Antibiotic Resistance Laboratory Network, 2017 to 2019. Antimicrob Agents Chemother 2021;65(12):e0110521. (In eng). DOI: 10.1128/aac.01105-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009;42(2):377–81. (In eng). DOI: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris PA, Taylor R, Minor BL, et al. The REDCap consortium: Building an international community of software platform partners. J Biomed Inform 2019;95:103208. (In eng). DOI: 10.1016/j.jbi.2019.103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2022. [Google Scholar]
- 13.44 U.S.C. § 3501 2018.
- 14.5 U.S.C. § 552a. 2018.
- 15.42 U.S.C. § 241(d) 2011.
- 16. 21 C.F.R. § 56.
- 17. 21 C.F.R. § 704.
- 18.FDA Form 483 Notice of Inspectional Observations. 2023.
- 19. 21 C.F.R. § 200.50.
- 20. 21 C.F.R. § 349.
- 21.Securing the U.S. Drug Supply Chain: Oversight of FDA’s Foreign Inspection Program. House Committee on Energy and Commerce, Subcommittee on Oversight and Investigations. https://www.fda.gov/news-events/congressional-testimony/securing-us-drug-supply-chain-oversight-fdas-foreign-inspection-program-121020192019.
- 22.FDA. “Evaluation of Non-Application Drug Manufacturers Before Marketing.” FDA’s FY 2024 Legislative Proposals. (https://www.fda.gov/media/166049/download). [Google Scholar]
- 23. 21 C.F.R. § 210.
- 24. 21 C.F.R. § 211.
- 25.Agergaard CN, Porsbo LJ, Sydenham TV, et al. Contaminated dicloxacillin capsules as the source of an NDM-5/OXA-48-producing Enterobacter hormaechei ST79 outbreak, Denmark and Iceland, 2022 and 2023. Euro Surveill 2023;28(9) (In eng). DOI: 10.2807/1560-7917.Es.2023.28.9.2300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang DC, Grant GB, O’Donnell K, et al. Multistate outbreak of Fusarium keratitis associated with use of a contact lens solution. JAMA 2006;296(8):953–63. DOI: 10.1001/jama.296.8.953. [DOI] [PubMed] [Google Scholar]
- 27.Sunenshine R, Schultz M, Lawrence MG, et al. An outbreak of postoperative gram-negative bacterial endophthalmitis associated with contaminated trypan blue ophthalmic solution. Clin Infect Dis 2009;48(11):1580–3. DOI: 10.1086/598938. [DOI] [PubMed] [Google Scholar]
- 28.Swaddiwudhipong W, Tangkitchot T, Silarug N. An outbreak of Pseudomonas aeruginosa postoperative endophthalmitis caused by contaminated intraocular irrigating solution. Trans R Soc Trop Med Hyg 1995;89(3):288. DOI: 10.1016/0035-9203(95)90545-6. [DOI] [PubMed] [Google Scholar]
- 29.Verani JR, Lorick SA, Yoder JS, et al. National outbreak of Acanthamoeba keratitis associated with use of a contact lens solution, United States. Emerg Infect Dis 2009;15(8):1236–42. DOI: 10.3201/eid1508.090225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bawankar P, Bhattacharjee H, Barman M, et al. Outbreak of Multidrug-resistant Pseudomonas Aeruginosa Endophthalmitis Due to Contaminated Trypan Blue Solution. J Ophthalmic Vis Res 2019;14(3):257–266. (In eng). DOI: 10.18502/jovr.v14i3.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.CDC. Interim Guidance for a Public Health Response to Contain Novel or Targeted Multidrug-resistant Organisms (MDROs). 2022. (https://www.cdc.gov/hai/pdfs/mdro-guides/Health-Response-Contain-MDRO-508.pdf).
- 32.Herrera S, Torralbo B, Herranz S, et al. Carriage of multidrug-resistant Gram-negative bacilli: duration and risk factors. Eur J Clin Microbiol Infect Dis 2023;42(5):631–638. (In eng). DOI: 10.1007/s10096-023-04581-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


