Summary
Background
The 5-year survival rate of oesophageal squamous cell carcinoma (ESCC) is approximately 20%. The prognosis and drug response exhibit substantial heterogeneity in ESCC, impeding progress in survival outcomes. Our goal is to identify a signature for tumour subtype classification, enabling precise clinical treatments.
Methods
Utilising pre-treatment multi-omics data from an ESCC dataset (n = 310), an enhancer methylation-eRNA-target gene regulation network was constructed and validated by in vitro experiments. Four machine learning methods collectively identified core target genes, establishing an Enhancer Demethylation-Regulated Gene Score (EDRGS) model for classification. The molecular function of EDRGS subtyping was explored in scRNA-seq (n = 60) and bulk-seq (n = 310), and the EDRGS's potential to predict treatment response was assessed in datasets of various cancer types.
Findings
EDRGS stratified ESCCs into EDRGS-high/low subtypes, with EDRGS-high signifying a less favourable prognosis in ESCC and nine additional cancer types. EDRGS-high exhibited an immune-hot but immune-suppressive phenotype with elevated immune checkpoint expression, increased T cell infiltration, and IFNγ signalling in ESCC, suggesting a better response to immunotherapy. Notably, EDRGS outperformed PD-L1 in predicting anti-PD-1/L1 therapy effectiveness in ESCC (n = 42), kidney renal clear cell carcinoma (KIRC, n = 181), and bladder urothelial carcinoma (BLCA, n = 348) cohorts. EDRGS-low showed a cell cycle-activated phenotype with higher CDK4 and/or CDK6 expression, demonstrating a superior response to the CDK4/6 inhibitor palbociclib, validated in ESCC (n = 26), melanoma (n = 18), prostate cancer (n = 15) cells, and PDX models derived from patients with pancreatic cancer (n = 30).
Interpretation
Identification of EDRGS subtypes enlightens ESCC categorisation, offering clinical insights for patient management in immunotherapy (anti-PD-1/L1) and CDK4/6 inhibitor therapy across cancer types.
Funding
This study was supported by funding from the National Key R&D Program of China (2021YFC2501000, 2020YFA0803300), the National Natural Science Foundation of China (82030089, 82188102), the CAMS Innovation Fund for Medical Sciences (2021-I2M-1-018, 2022-I2M-2-001, 2021-I2M-1-067), the Fundamental Research Funds for the Central Universities (3332021091).
Keywords: Oesophageal squamous cell carcinoma (ESCC), Enhancer methylation, eRNA, Immunotherapy, CDK4/6 inhibitor therapy
Research in context.
Evidence before this study
Immunotherapy and CDK4/6 inhibitor therapy offer promising strategies for tackling ESCC. However, only a fraction of patients derive benefit from these innovative treatments, indicating that current subtyping methods insufficiently guide treatment decisions.
Added value of this study
We developed an Enhancer Demethylation-Regulated Gene Score (EDRGS) model, categorising pre-treatment patients with ESCC into EDRGS-high and EDRGS-low groups. EDRGS-high individuals exhibit a poorer prognosis with an immune-hot but immune-suppressive phenotype, suggesting a more favourable response to immunotherapy. EDRGS-low patients display a cell cycle-activated phenotype, indicating an improved response to CDK4/6 inhibitors.
Implications of all the available evidence
EDRGS surpassed PD-L1 in predicting the effectiveness of anti-PD-1/L1 therapy across ESCC, KIRC, and BLCA cohorts. Additionally, it outperformed CDK4/6 in forecasting the response to CDK4/6 inhibitors in ESCC, pancreatic cancer, melanoma, and prostate cancer cell lines or patient-derived xenograft (PDX) models. The EDRGS indicator could significantly impact treatment decisions, enhancing patient prognosis not only in ESCC but also in other cancer types.
Introduction
Oesophageal cancer (EC) ranks as the sixth most common cause of cancer-related death worldwide.1 Globally, oesophageal squamous cell carcinoma (ESCC) is the dominant histological type, representing around 87% of cases.2 Currently, conventional treatments based on histopathological criteria do not adequately inform treatment decisions due to the heterogeneity and variable clinical outcomes in ESCC. Advances in sequencing technology and multi-omics studies enhanced our understanding of the molecular pathology of ESCC.3, 4, 5 However, accurate cancer subtype classifications guiding clinical care and precision oncology are not fully determined, leading to insufficient clinical management in patients with ESCC and minimal survival improvements.
Most attempts to subtype ESCC were based on single-omics profiling to separate cohorts, which yielded numerous markers such as TP53, RB1, CDKN2A, and NFE2L2 gene mutations6, 7, 8 in genetic level; PGK1 activity5 and CLK1 activity9 in proteomic level; hypermethylation of WNT2 promoter10 in epigenetic level. Additionally, multiple clusters7,11 were revealed to be associated with prognosis in different omics levels. However, these groupings are able to guide drug development and predict prognosis, but are limited in informing ESCC treatment response. Although recent multi-omics-based molecular subtypings3,4 have led to a more comprehensive categorisation of ESCCs, and enhanced the understanding of the biology and treatment of ESCC, they do not alter clinical decision-making in ESCC yet. Hence, it is imperative to establish a subtyping system to improve patient selection for existing treatments, ultimately improving overall outcomes for ESCC.
Recently, targeted therapies and immunotherapy have been employed in the treatment of ESCC, with drugs targeting EGFR, VEGF, HER-2, CDK4/6, and PD-1/L1.12,13 Immunotherapy has made significant advancements in the treatment of patients with recurrent or metastatic ESCC.14 Therefore, Food and Drug Administration (FDA) has approved nivolumab (PD-1 monoclonal antibody) as adjuvant therapy for resected oesophageal cancer with residual disease post neoadjuvant chemoradiotherapy. However, the current clinical reliance on PD-L1 expression to assess immunotherapy efficacy remains a subject of international debate regarding its predictive value for patients with ESCC.15,16 Although CDK4/6 inhibitors have not yet been included in the ESCC treatment guidelines, several studies have reported CDK4/6 inhibitor therapy may offer another promising treatment option for patients with ESCC.13,17,18 Nevertheless, efficacy markers for CDK4/6 inhibitor therapy are still under investigation. Thus, there is an urgent need to uncover more reliable markers to select patients for immunotherapy and CDK4/6 inhibitor therapy.
Growing evidence has emphasised that an abnormal enhancer-driven transcriptional program is a fundamental driver of oncogenesis and tumour persistence.19, 20, 21 Enhancers are traditionally recognised as cis-regulatory elements that regulate gene expression. Recent research has found that enhancers can also be transcribed into non-coding RNAs, known as enhancer RNAs (eRNA). These eRNAs act as activators of enhancer activity and thus enhance the expression of target genes.22,23 As for the regulation of enhancers, it's worth noting that the predominant regulatory pattern observed in pan-cancer is the activation of enhancers through demethylation, resulting in the upregulation of eRNAs and target genes.24, 25, 26 It has been discovered that the regulatory interplay among C/EBPβ enhancer methylation, eRNA, and target gene, contributes to hepatocarcinogenesis through global transcriptional reprogramming.27 While enhancer methylation, eRNA, and target gene regulatory relationship is widespread in various cancers and serves as a crucial transcriptional regulatory pattern, it has not been systematically studied in ESCC yet.
In this study, we constructed an enhancer methylation-eRNA-target gene tripartite regulatory network based on treatment-naïve WGBS and RNA-seq data from 155 pairs of ESCC samples. An EDRGS model was constructed based on 12 core target genes identified from this regulatory network to classify ESCCs accurately. Importantly, EDRGS could not only indicate patient prognosis in various cancer types but also serve as an efficacy marker for immunotherapy (anti-PD-1/L1) or CDK4/6 inhibitor therapy in patients with ESCC or other over nine tumour entities.
Methods
eRNA quantification
We downloaded data for all eRNAs from the Human enhancer RNA Atlas (https://hanlaboratory.com/HeRA/), counted reads for eRNAs using samtools and normalized the expression values by the reads per million (RPM) method in RNA-Seq on 155 pairs of tumour and adjacent normal tissue samples in HRA003107 dataset.4 We considered only eRNAs with an RPM ≥1 as detectable eRNAs.
Enhancer methylation quantification
WGBS data in HRA003107 dataset4 was used to determine the enhancer methylation level. Methods for assessing the methylation levels at each site according to the approach introduced in the previous article.4 We directly calculated the average eRNA region CpG methylation level as the enhancer methylation level.
eRNA target gene identification
We identified eRNA target genes based on distance (≤1 MB) and significant co-expression correlations (Spearman correlation, R ≥ 0.3 and FDR <0.05).
Identification of a panel of core target genes
SVM, WGCNA, LASSO, and random forest methods were applied in the RNA-seq data of 155 pairs of tumour and adjacent normal tissue samples in HRA003107 dataset.4 In SVM approach, 148 target genes were used as the features for classification and five-fold cross-validation to evaluate the classification performance. In WGCNA method, we started by constructing a gene co-expression network, followed by employing modularization techniques to identify gene modules with similar expression patterns. In the LASSO method, a 10-fold cross-validation approach was employed, and the tuning parameter (λ) was selected based on minimizing the squared error. The random forest method was trained with parameters set to ntree = 1000 and mtry = 24. In the generalized linear model, the predictors were the transcriptome levels of target genes, the response variable was the sample type (tumour or normal), the random error distribution was the binomial distribution, and the link function was the logit link. The area under the ROC curve (AUC) assessed the panel's performance on both training (HRA003107)4 and test datasets (TCGA-ESCC and GSE53622). The expression values of the 12 target genes from the external validation sets were used as input for the model trained on the training set to obtain model predictions. Subsequently, the obtained predictions were compared with the actual sample types (normal or tumour) in the validation sets to calculate the AUC. We performed bootstrap resampling to assess whether there were significant differences among evaluated AUCs.
Construction and validation of the EDRGS
Through Stepwise Cox regression analysis, selecting based on the best subset selection criteria, we identified the final set of 12 target genes from the candidate pool to construct the multivariable Cox regression model. The multivariable Cox regression model incorporates the expression levels of 12 target genes as independent variables, with overall survival serving as the output variable. The proportional hazards assumption was accessed by Schoenfeld residuals method and the linearity assumption for quantitative predictors was accessed by Martingale residuals method. The coefficients of Cox model were estimated by Efron approximation method. EDRGS was calculated by multiplying gene expression values with their Cox model coefficients and summing them. Taking the median EDRGS as the cutoff value, EDRGS subtyping was established.
Survival analysis
The start time for survival analysis is defined as the time of diagnosis of ESCC, and the end time for survival analysis is determined by the time of death or, alternatively, the latest follow-up visit if death has not occurred. The median follow-up time is 797 days with an interquartile range (IQR) of 452.5–948.5 days in the training dataset. The censoring proportion of the training dataset is 2.58% (4/155), with reasons for censoring including but not limited to loss to follow-up and other instances where complete follow-up information could not be obtained. Prognostic power of EDRGS was assessed using Kaplan–Meier survival curves with log-rank tests.
Functional enrichment analysis
Initially, we employed the ‘DESeq2’ R package for differential expression analysis between high and low EDRGS groups. Subsequently, we conducted Gene Set Enrichment Analysis (GSEA) with the ‘clusterProfiler’ R package.
Immune cell infiltration analysis
We utilized ‘GSVA’ R package to conduct single-sample Gene Set Enrichment Analysis (ssGSEA) in the datasets of HRA003107.
Definition of different EDRGS-subtyping like ESCC cell line
Gene expression data from five datasets (GDSCv1, GDSCv2, PRISM, CGP, and CTRP2) for ESCC cell lines were collected.28, 29, 30, 31 Using the Nearest Template Prediction (NTP) method in the ‘CMScaller’ R package, ESCC cell lines were categorised into different EDRGS groups based on transcriptome similarities with EDRGS-phenotype patients with ESCC.
Evaluation of drug sensitivity
Pablociclib sensitivity data for ESCC cell lines were obtained from GDSCv1 and GDSCv2.29 Data on palbociclib sensitivity for melanoma, prostate cancer cells, and PDX models derived from patients with pancreatic cancer were sourced from GSE113268, GSE99675, and GSE113922. The AUC was used to assess the performance of EDRGS and CDK4/6 on these datasets.
Cell lines
The KYSE180 (RRID: CVCL_1349), KYSE410 (RRID: CVCL_1352), KYSE450 (RRID: CVCL_1353), and KYSE510 (RRID: CVCL_1354) cell lines were provided by Dr. Yutaka Shimada.32 The TE-8 (RRID: CVCL_F786) and 293T (RRID: CVCL_0063) cell lines were collected from the American Type Culture Collection. The TE-1 (RRID: CVCL_1759) cell line was purchased from the Cell Bank of Chinese Academy of Sciences. All above ESCC cells were cultured in RPMI-1640 medium supplemented with 10% foetal bovine serum, and 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% foetal bovine serum. All cells were cultured aseptically at 37 °C and 5% CO2 and passed through 0.25% trypsin. All cell lines were validated by short tandem repeat (STR) DNA fingerprinting and confirmed to be free of mycoplasma contamination.
Lentivirus production and generation of stable cell lines
The shRNA sequences were inserted into the pSIH1-puro vector (#26597; Addgene, Cambridge, MA, USA) (RRID: Addgene_26597). The shRNA sequences were as follows: shTP63e#1: GGACCTTGACGAATCACTTCC; shTP63e#2: GGAAACTACATAGGGTGAAGC. Lentivirus production and infection were conducted as previously described.33
Cell proliferation and cell viability assays
The cell proliferation rate and cell viability were assessed as previously described.34 Cell Counting Kit-8 reagent (CK04; Dojindo Laboratories, Kumamoto, Japan) was used to assess the cell proliferation rate and cell viability after drug treatment according to the manufacturer's instructions. The absorbance at 450 nm on days 2–5 was normalized to that measured on day 1, and the relative cell viability (a percentage compared to that in the control group) was calculated.
Live cell analysis
An IncuCyte Live Cell Imaging System (Essen Bioscience, Hertfordshire, UK) was used for live cell imaging. Cells were seeded in 96-well microplates, and the palbociclib (HY-50767, MedChemExpress, NJ, USA) was added after cell adhesion. The plates were placed in the IncuCyte system, taking snapshots of two zones per well every 3 h over a 72-h period. Quantitative cell confluency was performed using the included commercial software.
RT‒qPCR
RNA preparation and RT‒qPCR were performed according to previously described methods.34 The primers for the RT‒qPCR assay were as follows: TP63e-F: GCTGGGACCTTGACGAATCA; TP63e-R: CCTGCTGCCTCTATCCGAAG. TP63-F: GGACCAGCAGATTCAGAACGG; TP63-R: GGACCAGCAGATTCAGAACGG. β-actin-F: CTGGAACGGTGAAGGTGACA; β-actin-R: AAGGGACTTCCTGTAACAATGCA. For CPI-455 (HY100412; MedChemExpress, NJ, USA) and 5-AzaC (HY10586, MedChemExpress, NJ, USA) treatment, after seeding KYSE450 and KYSE180 cells for 12 h, the drugs were added to the cells according to the response concentration, and after 48 h, RNA was collected and qPCR was performed in triplicates to detect the expression levels of TP63 and TP63e.
Statistics
All statistical analyses (including Spearman correlation, Wilcoxon rank-sum test, Fisher's exact test, Chi-square test, one-way ANOVA test, two-way ANOVA test and unpaired Student's t test) were performed in R (version 4.2.3). Our trainset cohort HRA003107 consists of a total of 155 pairs of oesophageal squamous cell carcinoma and adjacent non-cancerous samples. We did not perform pre-selection of samples based on clinical information such as the patient's TNM stage, sex, age, etc. The clinical information for the samples is presented in Supplementary Table S1.
Ethics
We used deidentified data from the parent studies for this secondary analysis. As such, the study was considered non-human subjects research and did not require institutional review board approval. All study procedures for the parent studies, which were described in detail previously,4 were approved by the institutional review boards of participating medical centres. Participants or their surrogates provided written informed consent.
Role of funders
This study was supported by funding from the National Key R&D Program of China (2021YFC2501000, 2020YFA0803300), the National Natural Science Foundation of China (82030089, 82188102), the CAMS Innovation Fund for Medical Sciences (2021-I2M-1-018, 2022-I2M-2-001, 2021-I2M-1-067), the Fundamental Research Funds for the Central Universities (3332021091). The funders did not have any role in the study design, data collection, data analyses, interpretation, or the writing of the report.
Results
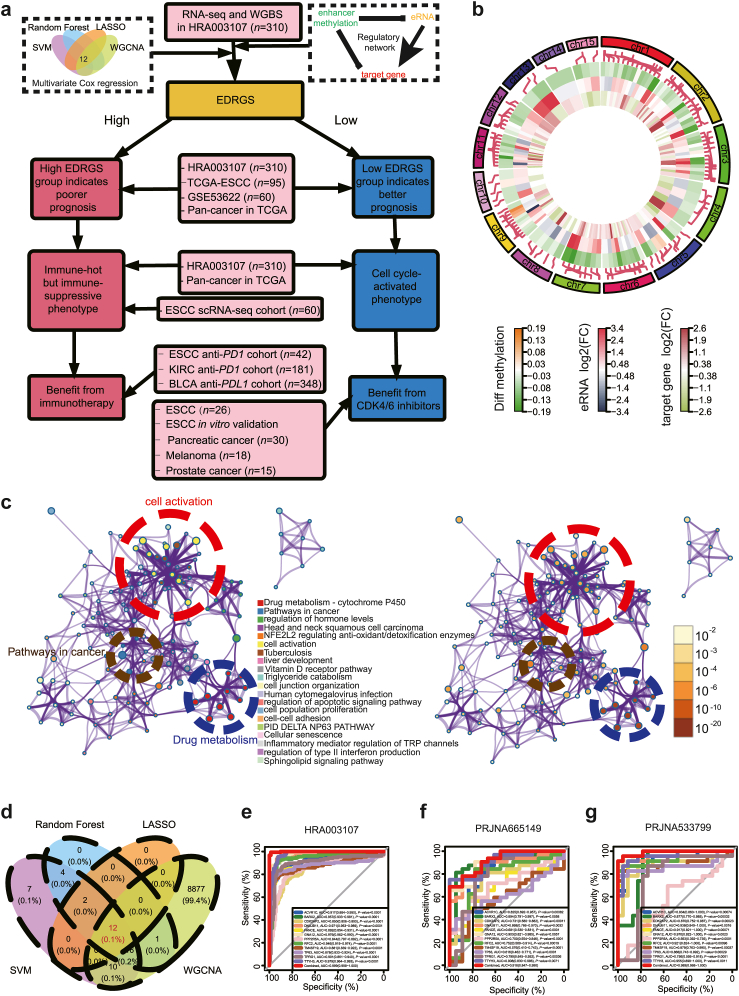
Construction of the enhancer methylation-eRNA-target gene tripartite regulatory network
Previous research has indicated that demethylation-induced enhancer activation followed by eRNA positively-regulated target genes is a dominant regulatory pattern in pan-cancer.26 To identify the core signature in ESCC, we created a tripartite regulatory network by analysing enhancer methylation's negative correlation with eRNAs and target genes, as well as eRNA's positive correlation with target genes based on HRA003107 dataset (Supplementary Fig. S1a). Hi-C data provides additional evidence on the target genes regulated by enhancers.35 Upon analysing Hi-C data by previous methods,36 we observed that 92.8% of enhancers in this regulatory network exhibited genomic interactions with their corresponding target genes (Supplementary Fig. S1b), confirming the reliability of this identified regulatory network.
The entire layout of the article is presented in Fig. 1a. In this tripartite regulatory network, we found that enhancer regions in tumour samples were primarily in a state of low methylation and high expression of eRNA and target gene compared to adjacent non-cancer samples (Fig. 1b). Considering that both methylated enhancers and eRNAs mainly depend on target genes to execute their functions, we conducted functional enrichment analysis of 148 target genes via the Metascape website37 (http://metascape.org) and identified that the tripartite network primarily encompasses functions related to drug metabolism, cancer-related pathways, and cell activation (Fig. 1c).
Fig. 1.
Enhancer methylation-eRNA-target gene regulation network in the ESCC cohort. a The comprehensive flowchart of the article. b The differences in enhancer methylation, eRNA expression, and target gene expression between tumour and adjacent normal tissues. c The functional enrichment of target genes in the tripartite network. d Core target gene screening via SVM, WGCNA, LASSO, and random forest. e The ROC curve of the diagnostic model in the HRA003107 cohort. f The ROC curve of the diagnostic model in the PRJNA665149 cohort. g The ROC curve of the diagnostic model in the PRJNA533799 cohort.
We employed four machine learning techniques, namely SVM, WGCNA, LASSO, and random forest, and thus consistently identified 12 target genes in the regulatory network of HRA003107 dataset that can accurately classify tumours from adjacent normal samples with an AUC of 0.999 [0.998–1] (Sensitivity = 0.994; Specificity = 0.974; Fig. 1d–e, Supplementary Fig. S2a–f). Next, we demonstrated high classification performance in another two independent ESCC datasets, with AUCs of 0.918 [0.847–0.99], and 0.989 [0.968–1] (Sensitivity = 0.906; Specificity = 0.667 in PRJNA665149 dataset; Sensitivity = 0.739; Specificity = 1 in PRJNA533799 dataset; Fig. 1f–g). In order to further confirm that the identified core genes were indeed regulated by enhancer methylation-eRNA regulatory pattern, we conducted in-silico and experimental validation in our subsequent research.
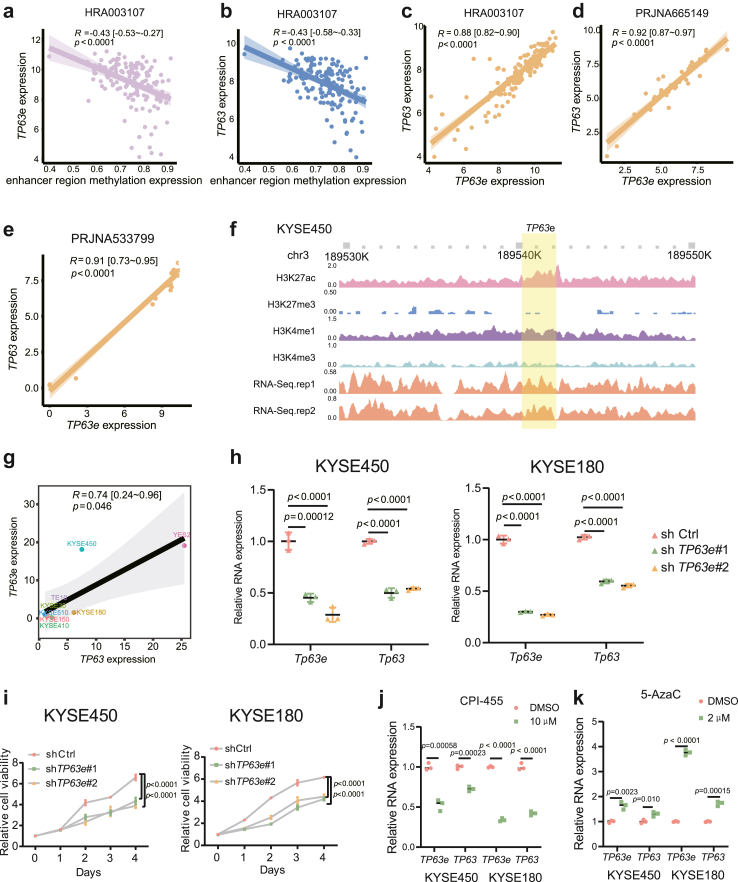
Validating the enhancer methylation-eRNA-target gene relationship
TP63 is one of the 12 core target genes identified earlier, and it is also a critical transcription factor in ESCC.38 Therefore, TP63 was used as an example to demonstrate the relationship between enhancer methylation, eRNA, and target genes. Indeed, we found a significant negative correlation between enhancer methylation of TP63 and the expression of both enhancer RNA of TP63 (TP63e) and TP63 in the HRA003107 cohort (Fig. 2a–b). Furthermore, a significant positive correlation was observed between the expression of TP63e and TP63 in PRJNA665149 and PRJNA533799 cohorts, which is in line with the outcomes from the HRA003107 cohort (Fig. 2c–e). These findings indicate a close association between TP63 and TP63e expression and highlight that the hypomethylation of the TP63 enhancer region might promote the upregulation of both TP63 and TP63e.
Fig. 2.
The relationship between TP63 enhancer methylation, TP63e expression, and TP63 expression. aTP63 enhancer methylation levels and TP63e expression relationship in the HRA003107 cohort (Spearman correlation, R = −0.43 [−0.53∼−0.27], p < 0.0001; the shaded area represents the 95% confidence region). bTP63 enhancer methylation levels and TP63 expression relationship in the HRA003107 cohort (Spearman correlation, R = −0.43 [−0.58∼−0.33], p < 0.0001; the shaded area represents the 95% confidence region). cTP63e expression and TP63 expression relationship in the HRA003107 cohort (Spearman correlation, R = 0.88 [0.82–0.90], p < 0.0001; the shaded area represents the 95% confidence region). dTP63e expression and TP63 expression relationship in the PRJNA665149 cohort (Spearman correlation, R = 0.92 [0.87–0.97], p < 0.0001; the shaded area represents the 95% confidence region). eTP63e expression and TP63 expression relationship in the PRJNA533799 cohort (Spearman correlation, R = 0.91 [0.73–0.95], p < 0.0001; the shaded area represents the 95% confidence region). f Identification of the TP63e segment using ChIP-seq and RNA-seq in KYSE450 cells. gTP63e expression and TP63 expression relationship in 8 ESCC cell lines by RT‒qPCR (Spearman correlation, R = 0.74 [0.24–0.96], p = 0.046; the shaded area represents the 95% confidence region). hTP63e and TP63 expression after knocking down TP63e in KYSE450 and KYSE180 cells (the data are presented as the mean ± SD values; error bars refer to Standard Deviation, SD; one-way ANOVA test, n = 3.). i The viability of KYSE450 and KYSE180 cells after knocking down TP63e (the data are presented as the mean ± SD values; error bars refer to Standard Deviation, SD; two-way ANOVA test, n = 5.). jTP63e and TP63 expression levels in KYSE450 and KYSE180 cells after CPI-455 treatment (the data are presented as the mean ± SD values; error bars refer to Standard Deviation, SD; unpaired Student's t test, n = 3). kTP63e and TP63 expression levels in KYSE450 and KYSE180 cells after 5-AzaC treatment (the data are presented as the mean ± SD values; error bars refer to Standard Deviation, SD; unpaired Student's t test, n = 3).
The determination of TP63e segment was based on our previous work,39 which involved detecting enrichment of H3K27ac, a high H3K4me1/H3K4me3 ratio, low H3K27me3 levels, and expressed transcripts in KYSE450 cells (Fig. 2f). Next, we validated the positive correlation between the expression of TP63e and TP63 by RT‒qPCR using a panel of 8 ESCC cell lines (Spearman correlation, R = 0.74 [0.24–0.96], p = 0.046, Fig. 2g). To investigate the impact of eRNA on the target gene, we knocked down TP63e, revealing a significant downregulation in TP63 expression and concurrent inhibition of KYSE450 and KYSE180 cell proliferation (Fig. 2h–i). Then, to investigate the impact of methylation on the expression of TP63 and TP63e, we treated KYSE450 and KYSE180 cells separately with the histone demethylase inhibitor CPI-455 and the DNA methyltransferase inhibitor 5-AzaC. After treatment with CPI-455, the expression of TP63 and TP63e was significantly reduced in KYSE450 and KYSE180 cells (Fig. 2j). Following the 5-AzaC treatment, the expression of TP63 and TP63e was significantly increased (Fig. 2k).
These results document that TP63 enhancer methylation inhibits the expression of both TP63 and TP63e, and TP63e upregulates the expression of TP63, thereby validating the reliability of the relationship between enhancer methylation, eRNA, and the target genes we have discovered.
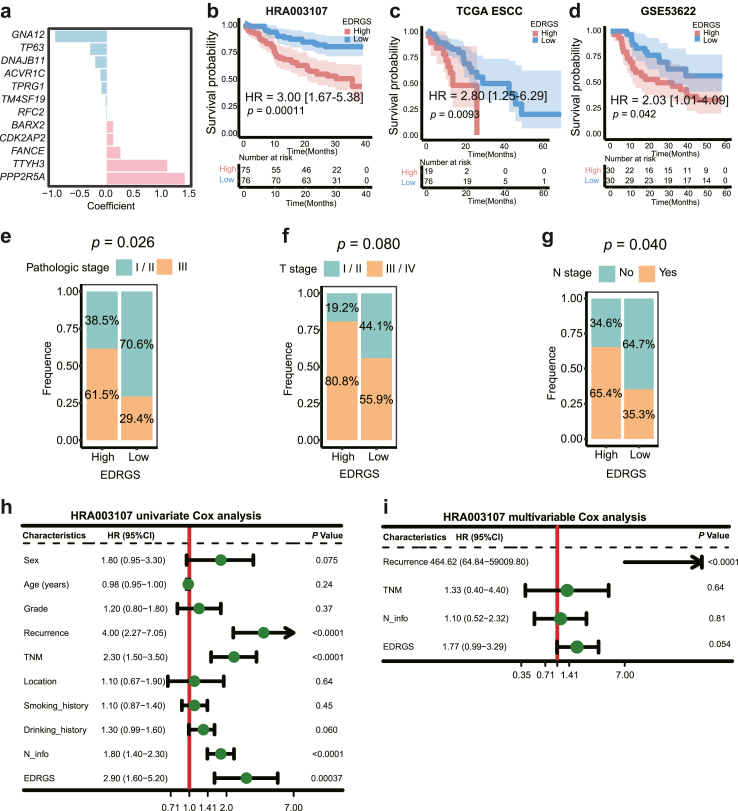
Construction of EDRGS and its prognostic implications in ESCC
After confirming the reliability of the relationship between enhancer methylation, eRNA, and target gene, a prognostic index, EDRGS, was constructed by multivariable Cox regression analysis in 12 core target genes for overall survival (OS) within the HRA003107 cohort, and their corresponding weights were displayed in Fig. 3a (Supplementary Table S2). The proportional hazards assumption and the linearity assumption for the multivariable Cox regression model are both satisfied (Supplementary Fig. S3a and b). Taking the median EDRGS as the cutoff value, high EDRGS patients consistently exhibited a worse OS than patients in the low EDRGS group in both the overall patient population (n = 151, p = 0.00011, log-rank test; Fig. 3b) and across various subgroups of clinical variables in the HRA003107 dataset (Supplementary Fig. S4). Similarly, high EDRGS patients showed a worse OS than patients in the low EDRGS group in TCGA (n = 95, p = 0.0093, log-rank test; Fig. 3c) and GSE53622 (n = 60, p = 0.042, log-rank test; Fig. 3d) ESCC datasets. Moreover, the proportion of patients in the EDRGS-high group who eventually succumbed to the disease or experienced recurrence was significantly higher than that in the EDRGS-low group (Supplementary Fig. S5a–d). In an ESCC scRNA-seq dataset40 (n = 60), we also observed that patients with a high EDRGS tended to have a more advanced pathological stage, T stage, and N stage (Fig. 3e–g). Besides, EDRGS was found to be a significant independent prognostic factor in patients with ESCC, according to univariate and multivariable Cox regression analyses (Fig. 3h–i). This was validated in both the TCGA-ESCC and GSE53622 datasets (Supplementary Fig. S5e–h).
Fig. 3.
A prognostic index for ESCC based on 12 core target genes. a The weights of 12 core target genes in the EDRGS model. b Patients with a high EDRGS had a worse prognosis than patients in the EDRGS-low group in the HRA003107 dataset (HR = 3.00 [1.67–5.38], p = 0.00011, log-rank test; the shaded area represents the 95% confidence region). c Patients with a high EDRGS had a worse prognosis than patients in the EDRGS-low group in the TCGA-ESCC dataset (HR = 2.80 [1.25–6.29], p = 0.0093, log-rank test; the shaded area represents the 95% confidence region). d Patients with a high EDRGS had a worse prognosis than patients in the EDRGS-low group in the GSE53622 dataset (HR = 2.03 [1.01–4.09], p = 0.042, log-rank test; the shaded area represents the 95% confidence region). e The pathologic stage in different EDRGS subgroups in the ESCC scRNA-seq dataset (p = 0.026, Chi-square test). f The T stage in different EDRGS subgroups in the ESCC scRNA-seq dataset (p = 0.080, Chi-square test). g The N stage in different EDRGS subgroups in the ESCC scRNA-seq dataset (p = 0.040, Chi-square test). h Univariate Cox analysis of clinicopathologic factors and EDRGS in the HRA003107 dataset. i Multivariable Cox analysis of the factors significant in the univariate Cox analysis in the HRA003107 dataset.
The above results indicate that EDRGS, as an independent risk factor for ESCC, has prompted us to investigate whether there are differences in molecular characteristics between different EDRGS subtypes.
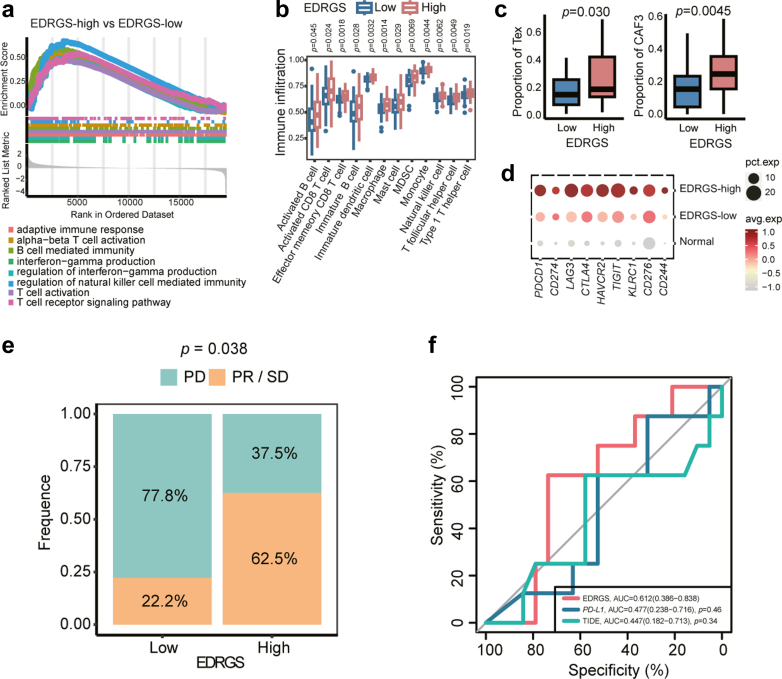
The EDRGS indicated differences in the tumour immune microenvironment and immunotherapy response
To unveil the molecular features among EDRGS subtypes in ESCCs, GSEA was performed in bulk RNA-seq (HRA003107, n = 310). Immune-related pathways, such as adaptive immune response, T-cell activation, and interferon-gamma production, were enriched in the EDRGS-high samples (Fig. 4a; FDR <0.05, Supplementary Table S3), suggesting an immune-hot phenotype from the aspect of gene expression patterns. We then further explored cell fractions in different EDRGS subtypes with scRNA-seq (GSE160269, n = 60) and bulk RNA-seq (HRA003107, n = 310). Based on ssGSEA, we found that compared to the EDRGS-low group, the EDRGS-high group had a higher level of immune cell infiltration, including activated CD8+ T cells, effector memory CD8+ T cells, and natural killer cells, etc (Fig. 4b), consistently implying an immune-hot phenotype. Furthermore, scRNA-seq data indicated a higher proportion of Tex and CAF3 cells (a subtype of cancer-associated fibroblasts that has been defined as associated with cancer progression) in patients with a higher EDRGS score (Fig. 4c). The expression of multiple immune checkpoint genes, such as PD-1, LAG3, TIM3 and TIGIT, was highly concordant between bulk RNA-seq (Supplementary Fig. S6a) and scRNA-seq (Fig. 4d) with a significant elevation in EDRGS-high subtype. These results indicated an immune-suppressive phenotype.
Fig. 4.
EDRGS-high phenotype indicated a better response to immunotherapy. a Immune-related pathways were enriched in the EDRGS-high group of the HRA003107 dataset. b Immune cell infiltration level estimated by ssGSEA in different EDRGS subgroups in the HRA003107 dataset (Wilcoxon rank-sum test; the definition for outliers is when the data falls below Q1–1.5 × IQR or exceeds Q3 + 1.5 × IQR; Q1: the 25th percentile; Q3: the 75th percentile; IQR = Q3–Q1). c The EDRGS-high group exhibited higher levels of Tex and CAF3 infiltration in the ESCC scRNA-seq dataset (Wilcoxon rank-sum test; the definition for outliers is when the data falls below Q1–1.5 × IQR or exceeds Q3 + 1.5 × IQR; Q1: the 25th percentile; Q3: the 75th percentile; IQR = Q3–Q1). d Immune checkpoint gene expression in the normal, EDRGS-low and EDRGS-high groups of ESCC scRNA-seq dataset. e The EDRGS-high group of patients with ESCC showed a better response to anti-PD-1 therapy in an ESCC anti-PD-1 therapy cohort (p = 0.038, Fisher's exact test). f ROC curves of the EDRGS, the expression of PD-L1 and TIDE score in an ESCC anti-PD-1 therapy cohort.
The findings from both bulk RNA-seq and scRNA-seq analyses suggest that the EDRGS-high group demonstrates an immune-hot but immune-suppressive phenotype, implying a favourable response to immunotherapy. Therefore, we predicted the response to immunotherapy in ESCC samples using TIDE score, an immunotherapy efficacy prediction index validated in melanoma,41 with a lower value indicating a higher likelihood of benefiting from immunotherapy. In alignment with our assumptions, patients in the EDRGS-high group showed significantly lower TIDE score41 (Supplementary Fig. S6b). To validate our findings, we assessed the reproducibility of the response prediction in an anti-PD-1 single-agent treatment cohort (n = 42).14 This validation also showed that the EDRGS-high group exhibited a more favourable response to treatment with anti-PD-1 antibody (p = 0.038, Fisher's exact test, Fig. 4e). What's even more significant is that when compared to two well-known methods for predicting immunotherapy response, namely the expression of PDL1 and TIDE score,41 EDRGS exhibited the best performance (Fig. 4f). Previous studies have indicated that higher somatic tumour mutational burden (TMB) was associated with better immunotherapy response,42,43 a trend consistent with what we observed in the EDRGS high group. Therefore, the influence of TMB on the predictive ability of EDRGS needs to be considered. The correlation analysis indicated that EDRGS and TMB were not significantly correlated (Spearman correlation, R = −0.12, p = 0.14; Supplementary Fig. S6c), and there was no significant difference in TMB values between the EDRGS low and high groups (Wilcoxon rank-sum test, p = 0.50; Supplementary Fig. S6d). These results indicate that EDRGS can potentially guide immunotherapy (anti-PD-1) for ESCCs.
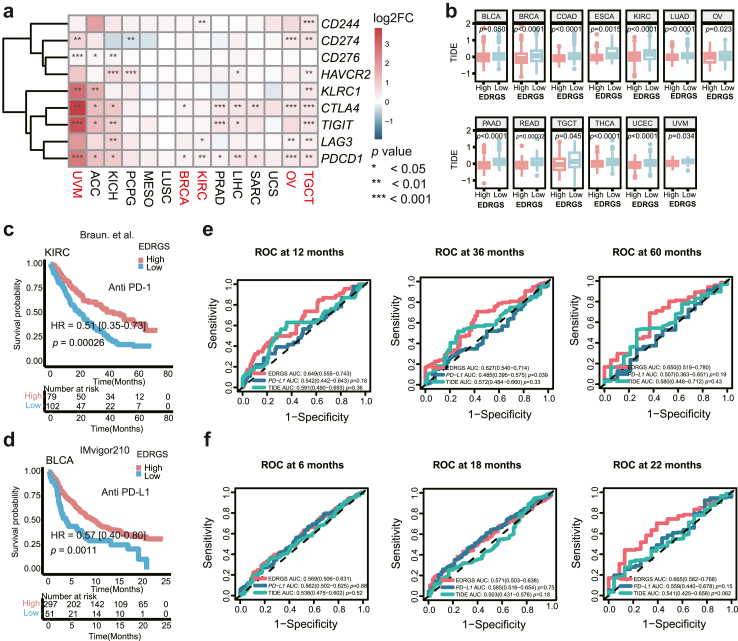
The EDRGS in a pan-cancer cohort and its association with immunotherapy response
To explore EDRGS's applicability in predicting prognosis and indicating immunotherapy response across cancer types, we applied it to a pan-cancer cohort of 33 cancer types using TCGA datasets. The EDRGS was significantly related to a worse prognosis in nine cancer types (Supplementary Fig. S7a–i). This suggests that EDRGS is relevant beyond ESCC and is especially prevalent in digestive tract malignancies and squamous cell carcinoma. Moreover, in five cancer types, namely, uveal melanoma (UVM), breast invasive carcinoma (BRCA), KIRC, ovarian serous cystadenocarcinoma (OV), and testicular germ cell tumours (TGCT), higher expression of immune checkpoint genes was observed in the EDRGS-high group (Fig. 5a). In line with ESCC, 13 other cancer types presented significantly decreased TIDE scores41 in the EDRGS-high group, indicating EDRGS's increased potential for an immunotherapy response (Fig. 5b). To validate the ability of the EDRGS to predict PD-1/PD-L1 treatment response, we assessed reproducibility in the PD-1 treatment cohort (n = 181, Braun et al.44) and PD-L1 treatment cohort (n = 348, IMvigor210). Consistently with ESCC, patients in the EDRGS-high group had a prominent survival advantage over patients in the EDRGS-low group in both cohorts embodying KIRC and BLCA (Fig. 5c–d). Furthermore, we observed that the EDRGS exhibited the strongest performance in KIRC cohort for the 12-, 36-, and 60-month follow-up periods, outperforming the accuracy of PD-L1 expression level and TIDE score41 (Fig. 5e). In BLCA cohort, best performance of EDRGS was observed at the 6- and 22-month follow-up points (Fig. 5f). In summary, patients with higher EDRGS may exhibit a better response to immunotherapy (anti-PD-1/L1) across various cancer types.
Fig. 5.
EDRGS-high phenotype indicated a better response to immunotherapy in various cancer types. a The expression of immune checkpoint genes in the EDRGS-high group compared to the EDRGS-low group across multiple cancer types (Wilcoxon rank-sum test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). b The TIDE score in different EDRGS subgroups across multiple cancer types (Wilcoxon rank-sum test; the definition for outliers is when the data falls below Q1–1.5 × IQR or exceeds Q3 + 1.5 × IQR; Q1: the 25th percentile; Q3: the 75th percentile; IQR = Q3–Q1). c Patients with a high EDRGS had a better response to anti-PD-1 therapy in the Braun et al.44 cohort (HR = 0.51 [0.35–0.73], p = 0.00026, log-rank test). d Patients with a high EDRGS had a better response to anti-PD-L1 therapy in the IMvigor210 cohort (HR = 0.57 [0.40–0.80], p = 0.0011, log-rank test). e ROC curves of EDRGS, PD-L1 expression level and TIDE score at 12-, 36-, and 60-month follow-up in Braun et al.44 cohort. f ROC curves of EDRGS, PD-L1 expression level and TIDE score at 6-, 18-, and 22-month follow-up in the IMvigor210 cohort.
EDRGS-low phenotype indicated a better response to CDK4/6 inhibitors
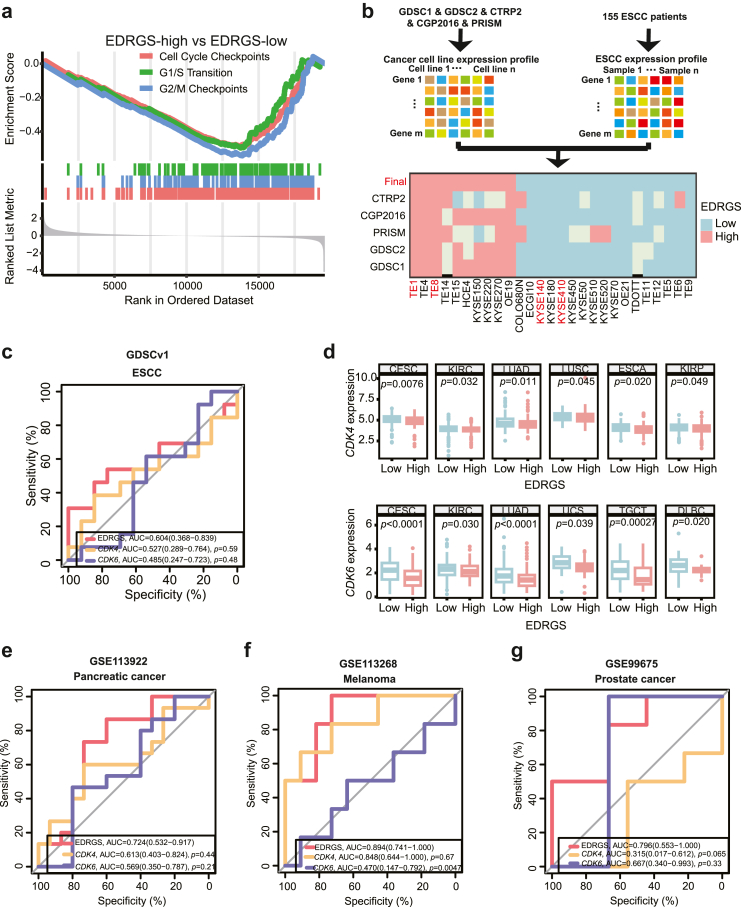
To reveal the molecular characteristics within the EDRGS-low group, GSEA was performed in bulk RNA-seq (HRA003107, n = 310). Cell cycle-related pathways, such as cell cycle checkpoints, G1/S transition and G2/M checkpoints, were enriched (Fig. 6a, Supplementary Table S3). Moreover, cell cycle-related genes, including CDK1, CDK2 and CDK4, showed significantly higher expression in the EDRGS-low group than in the EDRGS-high group (Supplementary Fig. S8a). Based on the functional characteristics of the EDRGS-low group, we hypothesized that patients in the EDRGS-low group were more likely to benefit from CDK4/6 inhibitor-palbociclib. It is the first CDK4/6 inhibitor approved as a cancer therapy and selectively inhibits the cyclin-dependent kinases CDK4 and CDK6.45 Due to the lack of transcriptomics data from patients with ESCC undergoing CDK4/6 inhibitor therapy, we were constrained to validate our hypothesis solely at the cellular level.
Fig. 6.
EDRGS-low group indicated a better response to CDK4/6 inhibitors. a Cell cycle-related pathways were enriched in the EDRGS-low group of HRA003107 dataset. b ESCC cell lines are classified into EDRGS-high or EDRGS-low-like cells. c ROC curves of the EDRGS, the expression of CDK4 and CDK6 in ESCC cells of the GDSCv1 dataset. d The expression of CDK4 and CDK6 in the EDRGS-high group compared to the EDRGS-low group across multiple cancer types (Wilcoxon rank-sum test; the definition for outliers is when the data falls below Q1–1.5 × IQR or exceeds Q3 + 1.5 × IQR; Q1: the 25th percentile; Q3: the 75th percentile; IQR = Q3–Q1). e ROC curves of the EDRGS, the expression of CDK4 and CDK6 in the pancreatic cancer patients-derived models treated with palbociclib (GSE113922). f ROC curves of the EDRGS, the expression of CDK4 and CDK6 in melanoma cell palbociclib treatment cohort (GSE113268). g ROC curves of the EDRGS, the expression of CDK4 and CDK6 in prostate cancer cell palbociclib treatment cohort (GSE99675).
For cellular-level validation, it was necessary to define ESCC cells exhibiting an EDRGS-high or EDRGS-low phenotype. To do so, we categorized ESCC cells into EDRGS-high/low-like groups using the NTP method, considering their transcriptome resemblances with patients with ESCC (Fig. 6b). Given CDK4/6's role in current clinic CDK4/6 inhibitor efficacy, we compared CDK4 or CDK6 expression across EDRGS groups. The results indicate higher CDK6 expression in EDRGS-low cells, with no significant difference in CDK4 expression in GDSCv1 and GDSCv2 datasets (Supplementary Fig. S8b–e), implying a relationship between EDRGS and the efficacy of CDK4/6 inhibitor therapy. To further validate our hypothesis, we compared the predictive performance of EDRGS and the expression of CDK4 or CDK6 in ESCC cell lines regarding sensitivity to palbociclib. Interestingly, EDRGS showed better performance than CDK4/6 expression supported by the GDSCv1 (n = 26) and GDSCv2 (n = 23) datasets (Fig. 6c, Supplementary Fig. S8f). Finally, we experimentally validated that cell lines KYSE410 and KYSE510 from the EDRGS-low group exhibited a significantly better response to palbociclib than cell lines TE1 and TE8 from the EDRGS-high group (Supplementary Fig. S8g). Given that DNA damage repair (DDR) can induce cell cycle arrest,46,47 potentially influencing patients' response to CDK4/6 inhibitors, we examined the impact of DDR on the predictive capability of EDRGS. The results indicated that EDRGS was not significantly correlated with either the overarching DDR pathway or its six individual major pathways48 (Supplementary Fig. S9a–g). Additionally, there were no discernible differences in the scores of each DDR pathway between the low and high EDRGS groups (Supplementary Fig. S9h–n). These results suggest that patients with EDRGS-low phenotype can potentially benefit from CDK4/6 inhibitors in the treatment of ESCC.
In order to investigate whether patients with low EDRGS may benefit from CDK4/6 inhibitors across cancer types, we first examined the expression of CDK4 and CDK6 between EDRGS-high and EDRGS-low groups in the pan-cancer cohort. In line with ESCC, the EDRGS-low group consistently exhibited a significant increase in the expression of CDK4 and/or CDK6 in multiple cancer types (Fig. 6d), suggesting a connection between EDRGS and the effectiveness of CDK4/6 inhibitor therapy in other cancer types. Next, with the aim of assessing the predictive capacity of EDRGS for the efficacy of CDK4/6 inhibitor treatment, we compared it with that of CDK4 or CDK6 expression across three independent datasets. Excitingly, we found that EDRGS exhibited the best classification performance compared to both CDK4 and CDK6 expression in the PDX models of pancreatic cancer treated with palbociclib (GSE113922, n = 30, Fig. 6e), melanoma cell palbociclib treatment cohort (GSE113268, n = 18, Fig. 6f), and prostate cancer cell palbociclib treatment cohort (GSE99675, n = 15, Fig. 6g).
In summary, patients characterized by EDRGS-low phenotype demonstrate improved response to CDK4/6 inhibitors in a diverse range of cancer types.
Discussion
A tremendous shift in cancer treatment of ESCC, from broad-spectrum treatment to targeted drugs, has provided promising outcomes in clinical practice or ongoing trials.12,13 However, response and survival benefit typically differs by population.14,17,18 A more comprehensive classification is needed to personalize patient therapy and thus improve response and outcomes. Here, we established a prognostic marker, EDRGS, which characterized ESCCs into two molecular subtypes reproducibly in a multicohort retrospective study. Specifically, not only in ESCC cohorts, patients in the EDRGS-high group demonstrated significantly improved responses to immunotherapy in 13 cancer types, whereas patients in the EDRGS-low group showed markedly better responses to CDK4/6 inhibitors in nine cancer types. This subtyping classification provides a conceptual framework to understand the heterogeneity of ESCC better and exhibits a potential to guide the selection of immunotherapy (anti-PD-1/L1) and CDK4/6 inhibitor therapy for patients across cancer types.
In this study, the EDRGS was constructed after establishing a 12-target-genes panel and performing multivariable Cox regression analysis. We are confident that the analysis process is solid enough to warrant a comprehensive evaluation thereafter. Accumulating evidence suggests that enhancer methylation-eRNA-target gene regulatory network is a predominant regulatory pattern observed in pan-cancer.26,27 However, it has not been systematically studied in ESCC yet. Therefore, this gene regulatory relationship was employed to initiate the discovery of the classifier. Out of 148 genes regulated by this tripartite network, a robust 12-target-genes panel to classify normal from tumorous tissues was identified. Training data from pre-treatment 155 pairs of adjacent non-cancerous and cancer tissues ensured an unbiased approach. This robust classification is further supported by the consistent identification of these 12 markers from four independent feature selection methods and reproducible results in two independent datasets. In all the comparisons, AUC values were consistently above 0.91, with no significant differences observed. This suggests that there was no major overfitting or underfitting problem generated during classifier calculation. Furthermore, the reliability of this regulatory network was confirmed by experimental validation. TP63, previously identified as a master regulator in ESCC,38 was also found in this 12-gene panel. Consistent with our bioinformatic analysis, experimental results indicated that TP63 was regulated by the TP63e and methylation. Finally, the results of multivariable Cox regression analysis were highly concordant between discovery and another two independent validation cohorts, consistently showing the poorer prognosis in the EDRGS-high group. This series of results underscore the high credibility of the EDRGS.
The EDRGS-high subtype corresponded to significantly elevated expression of immune checkpoints and increased infiltration of CAF3, accompanied by increased T cell infiltration and IFNγ signaling, implying an immune-suppressive but immune-hot phenotype. Though the phenotype of EDRGS was immune-suppressive, it could be reversed by the treatment of immune inhibitors.49,50 Additionally, consistent with previous studies,51,52 tumours with an inflamed phenotype tend to be more responsive to immune checkpoint inhibitors. Not surprisingly, our results suggest that EDRGS-high cases might respond to immunotherapy. No association between EDRGS and TMB further indicated that EDRGS is a immunotherapy efficacy prediction method that is independent of TMB. Immunotherapy has yielded very encouraging outcomes in a series of clinical trials when employed as the primary or secondary treatment for advanced oesophageal cancer.14,53 Nevertheless, merely 33.3% of patients with ESCC benefit from the immunotherapy.53 While PD-L1 expression may serve as a biomarker to reveal which patients are responsive to immunotherapy, it falls significantly short of accurately stratifying oesophageal cancer patients for immunotherapy. Additionally, there is still debate regarding the predictive value of PD-L1 in patients with oesophageal cancer.15,16 Inspiringly, compared to the expression of PD-L1 and TIDE score,41 which were already developed to predict immunotherapy response, the EDRGS model exhibited higher accuracy when predicting responders of anti-PD-1 therapy in a clinical trial with single-agent PD-1 therapy, indicating potential benefits for EDRGS-high subtype. Therefore, EDRGS can potentially act as a biomarker for immunotherapy (anti-PD-1) in patients with primary ESCC.
The EDRGS-low subtype was characterized by relatively higher expression of CDK4 and/or CDK6 and associated with cell cycle-related pathways that enhance cyclin D-CDK4/6 complexes kinase activity.13 In recent years, CDK4/6 inhibitors have achieved great progress in treating metastatic ER-positive, HER2-negative breast cancer and extensive-stage small-cell lung cancer.54,55 Despite insufficient clinical evidence in ESCC, research involving cell lines and PDX models indicates a promising future for clinical application in ESCCs.13,56 Encouragingly, the clinical trial NCT04433494, studying the efficacy of CDK4/6 inhibitors in the treatment of ESCC, is currently underway. In our study, the EDRGS-low subtype was linked to the pathways that govern the cell cycle. Consistent with prior research,13,17 we found that the ESCC cells with higher expression of CDK4/6 were more sensitive to palbociclib. We then proposed that individuals categorized as EDRGS-low subtype could potentially benefit from CDK4/6 inhibitor therapy. No association between EDRGS and DDR further indicated that EDRGS is a method for predicting the efficacy of CDK4/6 inhibitors, independent of DDR as a covariate. Compared to the expression of CDK4 or CDK6, EDRGS demonstrated the highest efficacy in predicting the response to CDK4/6 inhibitor treatment, supported by the validation in the datasets from PDX models of pancreatic cancer and cancer cells of melanoma and prostate cancer. Despite the promising efficacy of EDRGS, it still requires further validation with additional data from PDO/animal models and clinical trials in ESCC.
The impact of the EDRGS on patient survival and response prediction was identified and validated across multiple cancer types. In recent decades, the development of more universal diagnostic and therapeutic strategies in oncology has been facilitated by multiple pan-cancer biomarkers, such as BRCA1/BRCA257 and HER2.58 Therefore, it is necessary to discover the broader applicability of biomarkers across different cancer types. It was speculated that the identified EDRGS model in ESCC had the potential to be applied across a broad spectrum of cancers, given the knowledge that enhancer methylation-eRNA-target gene regulatory network is a common regulatory pattern observed in pan-cancer26 as well as 12 identified target genes were associated with cell activation and drug metabolism which was crucial for cancer development and treatment. Indeed, consistent with the observation in ESCC, we then found that EDRGS exhibited as an adverse prognostic factor in another nine cancer types. This result further supports the robustness of our EDRGS model. More importantly, the EDRGS-high group exhibited a better response to immunotherapy in another 13 cancer types, while the EDRGS-low group showed an improved response to CDK4/6 inhibitors in another nine cancer types. These findings suggest that EDRGS has the possibility to indicate patient prognosis and guide the selection of adjuvant therapies, not limited to ESCC alone.
Despite the compelling results found in our study, there are still some limitations to be noted. First, in the Cox regression model (Fig. 3i, Supplementary Fig. S4), the observed large or infinite hazard ratio (HR) estimates are meaningless artifacts resulting from data sparsity, suggesting the presence of sparse-data bias,59,60 which affects the evaluation of effect measures. This bias results from few outcome events per variable (EPV) or categorical covariates with very low or high prevalence, as examined in Supplementary Table S1 and Supplementary Fig. S4. Despite our application of Firth's penalized maximum likelihood bias reduction method to enhance the accuracy of risk prediction, sparse-data bias remains a significant limitation of the study. Second, while we adjusted for key covariates such as recurrence and TNM, we must acknowledge the potential impact of other variables, such as demographic factors, which could also affect prognosis assessment and warrant thorough consideration. Third, although our work suggests that the EDRGS model may be useful for the prognosis evaluation and the prediction of treatment response, it should be noted that the cohort for validation was limited. In predicting the efficacy of immunotherapy for ESCCs, the sample size of the anti-PD-1 therapy clinical trial was constrained, primarily because a combination regimen is a more prevalent choice for patients as opposed to single-agent anti-PD-1 therapy. In predicting the response of the CDK4/6 treatment for ESCCs, only cell-line data was available. In the pan-cancer study, the analysis was mainly based on a retrospective study collected from TCGA. Fourth, EDRGS expression was quantified at the transcriptome level, while the protein expression level doesn't always correspond to the RNA expression, and proteins such as mutation-derived neoantigens are directly involved in tumour immunity. Finally, despite the model showing good robustness, it may still be affected by several other factors, such as differences in genetic background, the stage at the time of diagnosis, and the treatment modalities. Further validation in a larger cohort, persuasive wet-lab experiments, and clinical studies are still needed to confirm the classifier's accuracy and future clinical use.
In conclusion, we constructed an ESCC enhancer methylation-eRNA-target gene regulation network and developed an EDRGS model that can potentially predict both the prognosis and the efficacy of immunotherapy and CDK4/6 inhibitor therapy, offering invaluable insights for the clinical management of pharmaceuticals in ESCC.
Contributors
W. Gao: Data curation, data analysis, investigation, methodology, writing-original draft, writing-review and editing. S. Liu: Wet lab validation, writing-review and editing. Y. Wu: Writing-original draft, writing-review and editing. W. Wei: Writing-review and editing, supervision. Q. Yang: Data curation, writing-review. W. Li: Data curation, writing-review. H. Chen: Conceptualization, supervision, writing-review and editing. A. Luo: Conceptualization, supervision, writing-review and editing. Y. Wang: Providing clinical insights and supervising the study during the revision stage. Z. Liu: Conceptualization, supervision, writing-review and editing. W. Gao, S. Liu, and Y. Wu collected data and directly accessed and verified the underlying data reported in the manuscript. All authors critically revised the manuscript and approved the final version.
Data sharing statement
Expression profile data analysed in this study were obtained from UCSC Xena (https://xena.ucsc.edu/) at TCGA ESCC dataset and Gene Expression Omnibus (GEO) at GSE53622, GSE113922, GSE113268 and GSE99675. The RNA-seq, WGBS and clinical information (sex, TNM stage and prognosis) data of 310 ESCC samples in HRA003107 (restricted access data, requests should be directed to https://ngdc.cncb.ac.cn/), along with the RNA expression profile of the ESCC immunotherapy cohort, were obtained from our previous study,4 with informed consent obtained from the patients. The IMvigor210 cohort was obtained from ‘IMvigor210CoreBiologies’ R package and Braun et al.44 cohort was obtained from the supplementary data. Code can be accessed at https://github.com/GAOwy123/EDRGS.
Declaration of interests
The authors have declared that no conflict of interest exists.
Acknowledgements
The funding for this research was provided by the National Key R&D Program of China (2021YFC2501000, 2020YFA0803300), the National Natural Science Foundation of China (82030089, 82188102), the CAMS Innovation Fund for Medical Sciences (2021-I2M-1-018, 2022-I2M-2-001, 2021-I2M-1-067), the Fundamental Research Funds for the Central Universities (3332021091), all awarded to Zhihua Liu.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2024.105177.
Contributor Information
Yanfeng Wang, Email: wangyf@cicams.ac.cn.
Zhihua Liu, Email: liuzh@cicams.ac.cn.
Appendix A. Supplementary data
References
- 1.Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Uhlenhopp D.J., Then E.O., Sunkara T., Gaduputi V. Epidemiology of esophageal cancer: update in global trends, etiology and risk factors. Clin J Gastroenterol. 2020;13(6):1010–1021. doi: 10.1007/s12328-020-01237-x. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research N Analysis Working Group, Asan U., Agency B.C.C., et al. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541(7636):169–175. doi: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Z., Zhao Y., Kong P., et al. Integrated multi-omics profiling yields a clinically relevant molecular classification for esophageal squamous cell carcinoma. Cancer Cell. 2023;41(1):181–195.e9. doi: 10.1016/j.ccell.2022.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Li L., Jiang D., Zhang Q., et al. Integrative proteogenomic characterization of early esophageal cancer. Nat Commun. 2023;14(1):1666. doi: 10.1038/s41467-023-37440-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song Y., Li L., Ou Y., et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 2014;509(7498):91–95. doi: 10.1038/nature13176. [DOI] [PubMed] [Google Scholar]
- 7.Cui Y., Chen H., Xi R., et al. Whole-genome sequencing of 508 patients identifies key molecular features associated with poor prognosis in esophageal squamous cell carcinoma. Cell Res. 2020;30(10):902–913. doi: 10.1038/s41422-020-0333-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng R., Yin Y., Wei Y., et al. Mutant p53 activates hnRNPA2B1-AGAP1-mediated exosome formation to promote esophageal squamous cell carcinoma progression. Cancer Lett. 2023;562 doi: 10.1016/j.canlet.2023.216154. [DOI] [PubMed] [Google Scholar]
- 9.Li Y., Yang B., Ma Y., et al. Phosphoproteomics reveals therapeutic targets of esophageal squamous cell carcinoma. Signal Transduct Target Ther. 2021;6(1):381. doi: 10.1038/s41392-021-00682-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao W., Lee H., Wu W., et al. Multi-faceted epigenetic dysregulation of gene expression promotes esophageal squamous cell carcinoma. Nat Commun. 2020;11(1):3675. doi: 10.1038/s41467-020-17227-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang L., Liou Y.L., Wan Z.R., et al. Aberrant DNA methylation of PAX1, SOX1 and ZNF582 genes as potential biomarkers for esophageal squamous cell carcinoma. Biomed Pharmacother. 2019;120 doi: 10.1016/j.biopha.2019.109488. [DOI] [PubMed] [Google Scholar]
- 12.Yang Y.M., Hong P., Xu W.W., He Q.Y., Li B. Advances in targeted therapy for esophageal cancer. Signal Transduct Target Ther. 2020;5(1):229. doi: 10.1038/s41392-020-00323-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J., Li Q., Yuan J., et al. CDK4/6 inhibitor-SHR6390 exerts potent antitumor activity in esophageal squamous cell carcinoma by inhibiting phosphorylated Rb and inducing G1 cell cycle arrest. J Transl Med. 2017;15(1):127. doi: 10.1186/s12967-017-1231-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang J., Xu B., Mo H., et al. Safety, activity, and biomarkers of SHR-1210, an anti-PD-1 antibody, for patients with advanced esophageal carcinoma. Clin Cancer Res. 2018;24(6):1296–1304. doi: 10.1158/1078-0432.CCR-17-2439. [DOI] [PubMed] [Google Scholar]
- 15.Liu J., Yang Y., Liu Z., et al. Multicenter, single-arm, phase II trial of camrelizumab and chemotherapy as neoadjuvant treatment for locally advanced esophageal squamous cell carcinoma. J Immunother Cancer. 2022;10(3):e004291. doi: 10.1136/jitc-2021-004291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang W., Xing X., Yeung S.J., et al. Neoadjuvant programmed cell death 1 blockade combined with chemotherapy for resectable esophageal squamous cell carcinoma. J Immunother Cancer. 2022;10(1):e003497. doi: 10.1136/jitc-2021-003497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin W.J., Su Y.G., Ding X.L., Zhao R., Zhao Z.J., Wang Y.Y. CDK4/6 inhibitor enhances the radiosensitization of esophageal squamous cell carcinoma (ESCC) by activating autophagy signaling via the suppression of mTOR. Am J Transl Res. 2022;14(3):1616–1627. [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou J., Wu Z., Wong G., et al. CDK4/6 or MAPK blockade enhances efficacy of EGFR inhibition in oesophageal squamous cell carcinoma. Nat Commun. 2017;8 doi: 10.1038/ncomms13897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asangani I.A., Dommeti V.L., Wang X., et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510(7504):278–282. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mack S.C., Pajtler K.W., Chavez L., et al. Therapeutic targeting of ependymoma as informed by oncogenic enhancer profiling. Nature. 2018;553(7686):101–105. doi: 10.1038/nature25169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loven J., Hoke H.A., Lin C.Y., et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153(2):320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lam M.T., Cho H., Lesch H.P., et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498(7455):511–515. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melo C.A., Drost J., Wijchers P.J., et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell. 2013;49(3):524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 24.Benetatos L., Vartholomatos G. Enhancer DNA methylation in acute myeloid leukemia and myelodysplastic syndromes. Cell Mol Life Sci. 2018;75(11):1999–2009. doi: 10.1007/s00018-018-2783-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fleischer T., Tekpli X., Mathelier A., et al. DNA methylation at enhancers identifies distinct breast cancer lineages. Nat Commun. 2017;8(1):1379. doi: 10.1038/s41467-017-00510-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan X., Li X., Sun J., et al. Enhancer methylation dynamics drive core transcriptional regulatory circuitry in pan-cancer. Oncogene. 2022;41(26):3474–3484. doi: 10.1038/s41388-022-02359-x. [DOI] [PubMed] [Google Scholar]
- 27.Xiong L., Wu F., Wu Q., et al. Aberrant enhancer hypomethylation contributes to hepatic carcinogenesis through global transcriptional reprogramming. Nat Commun. 2019;10(1):335. doi: 10.1038/s41467-018-08245-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corsello S.M., Nagari R.T., Spangler R.D., et al. Discovering the anti-cancer potential of non-oncology drugs by systematic viability profiling. Nat Cancer. 2020;1(2):235–248. doi: 10.1038/s43018-019-0018-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iorio F., Knijnenburg T.A., Vis D.J., et al. A landscape of pharmacogenomic interactions in cancer. Cell. 2016;166(3):740–754. doi: 10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rees M.G., Seashore-Ludlow B., Cheah J.H., et al. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat Chem Biol. 2016;12(2):109–116. doi: 10.1038/nchembio.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geeleher P., Cox N.J., Huang R.S. Clinical drug response can be predicted using baseline gene expression levels and in vitro drug sensitivity in cell lines. Genome Biol. 2014;15(3):R47. doi: 10.1186/gb-2014-15-3-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimada Y., Imamura M., Wagata T., Yamaguchi N., Tobe T. Characterization of 21 newly established esophageal cancer cell lines. Cancer. 1992;69(2):277–284. doi: 10.1002/1097-0142(19920115)69:2<277::aid-cncr2820690202>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 33.Nan Y., Liu S., Luo Q., et al. m(6)A demethylase FTO stabilizes LINK-A to exert oncogenic roles via MCM3-mediated cell-cycle progression and HIF-1alpha activation. Cell Rep. 2023;42(10) doi: 10.1016/j.celrep.2023.113273. [DOI] [PubMed] [Google Scholar]
- 34.Nan Y., Luo Q., Wu X., et al. DLGAP1-AS2-Mediated phosphatidic acid synthesis activates YAP signaling and confers chemoresistance in squamous cell carcinoma. Cancer Res. 2022;82(16):2887–2903. doi: 10.1158/0008-5472.CAN-22-0717. [DOI] [PubMed] [Google Scholar]
- 35.Ron G., Globerson Y., Moran D., Kaplan T. Promoter-enhancer interactions identified from Hi-C data using probabilistic models and hierarchical topological domains. Nat Commun. 2017;8(1):2237. doi: 10.1038/s41467-017-02386-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Z., Lee J.H., Ruan H., et al. Transcriptional landscape and clinical utility of enhancer RNAs for eRNA-targeted therapy in cancer. Nat Commun. 2019;10(1):4562. doi: 10.1038/s41467-019-12543-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou Y., Zhou B., Pache L., et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang Y.Y., Jiang Y., Li C.Q., et al. TP63, SOX2, and KLF5 establish a core regulatory circuitry that controls epigenetic and transcription patterns in esophageal squamous cell carcinoma cell lines. Gastroenterology. 2020;159(4):1311–1327.e19. doi: 10.1053/j.gastro.2020.06.050. [DOI] [PubMed] [Google Scholar]
- 39.Yuan J., Jiang Q., Gong T., et al. Loss of grand histone H3 lysine 27 trimethylation domains mediated transcriptional activation in esophageal squamous cell carcinoma. NPJ Genom Med. 2021;6(1):65. doi: 10.1038/s41525-021-00232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X., Peng L., Luo Y., et al. Dissecting esophageal squamous-cell carcinoma ecosystem by single-cell transcriptomic analysis. Nat Commun. 2021;12(1):5291. doi: 10.1038/s41467-021-25539-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang P., Gu S., Pan D., et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24(10):1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palmeri M., Mehnert J., Silk A.W., et al. Real-world application of tumor mutational burden-high (TMB-high) and microsatellite instability (MSI) confirms their utility as immunotherapy biomarkers. ESMO Open. 2022;7(1) doi: 10.1016/j.esmoop.2021.100336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samstein R.M., Lee C.H., Shoushtari A.N., et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51(2):202–206. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Braun D.A., Hou Y., Bakouny Z., et al. Interplay of somatic alterations and immune infiltration modulates response to PD-1 blockade in advanced clear cell renal cell carcinoma. Nat Med. 2020;26(6):909–918. doi: 10.1038/s41591-020-0839-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ettl J. Palbociclib: first CDK4/6 inhibitor in clinical practice for the treatment of advanced HR-positive breast cancer. Breast Care. 2016;11(3):174–176. doi: 10.1159/000447417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clay D.E., Fox D.T. DNA damage responses during the cell cycle: insights from model organisms and beyond. Genes. 2021;12(12):1882. doi: 10.3390/genes12121882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Magnander K., Elmroth K. Biological consequences of formation and repair of complex DNA damage. Cancer Lett. 2012;327(1-2):90–96. doi: 10.1016/j.canlet.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 48.Zhao N., Zhang Z., Wang Q., et al. DNA damage repair profiling of esophageal squamous cell carcinoma uncovers clinically relevant molecular subtypes with distinct prognoses and therapeutic vulnerabilities. eBioMedicine. 2023;96 doi: 10.1016/j.ebiom.2023.104801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu X., Tian X., Ji L., et al. A tumor microenvironment-specific gene expression signature predicts chemotherapy resistance in colorectal cancer patients. NPJ Precis Oncol. 2021;5(1):7. doi: 10.1038/s41698-021-00142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mao X., Xu J., Wang W., et al. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: new findings and future perspectives. Mol Cancer. 2021;20(1):131. doi: 10.1186/s12943-021-01428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dongye Z., Li J., Wu Y. Toll-like receptor 9 agonists and combination therapies: strategies to modulate the tumour immune microenvironment for systemic anti-tumour immunity. Br J Cancer. 2022;127(9):1584–1594. doi: 10.1038/s41416-022-01876-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hegde P.S., Chen D.S. Top 10 challenges in cancer immunotherapy. Immunity. 2020;52(1):17–35. doi: 10.1016/j.immuni.2019.12.011. [DOI] [PubMed] [Google Scholar]
- 53.Huang J., Xu J., Chen Y., et al. Camrelizumab versus investigator's choice of chemotherapy as second-line therapy for advanced or metastatic oesophageal squamous cell carcinoma (ESCORT): a multicentre, randomised, open-label, phase 3 study. Lancet Oncol. 2020;21(6):832–842. doi: 10.1016/S1470-2045(20)30110-8. [DOI] [PubMed] [Google Scholar]
- 54.Weiss J., Goldschmidt J., Andric Z., et al. Effects of trilaciclib on chemotherapy-induced myelosuppression and patient-reported outcomes in patients with extensive-stage small cell lung cancer: pooled results from three phase II randomized, double-blind, placebo-controlled studies. Clin Lung Cancer. 2021;22(5):449–460. doi: 10.1016/j.cllc.2021.03.010. [DOI] [PubMed] [Google Scholar]
- 55.Hoste G., Punie K., Wildiers H., et al. Palbociclib in highly pretreated metastatic ER-positive HER2-negative breast cancer. Breast Cancer Res Treat. 2018;171(1):131–141. doi: 10.1007/s10549-018-4827-6. [DOI] [PubMed] [Google Scholar]
- 56.Su D., Zhang D., Jin J., et al. Identification of predictors of drug sensitivity using patient-derived models of esophageal squamous cell carcinoma. Nat Commun. 2019;10(1):5076. doi: 10.1038/s41467-019-12846-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sokol E.S., Pavlick D., Khiabanian H., et al. Pan-cancer analysis of BRCA1 and BRCA2 genomic alterations and their association with genomic instability as measured by genome-wide loss of heterozygosity. JCO Precis Oncol. 2020;4:442–465. doi: 10.1200/PO.19.00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Won E., Janjigian Y.J., Ilson D.H. HER2 directed therapy for gastric/esophageal cancers. Curr Treat Options Oncol. 2014;15(3):395–404. doi: 10.1007/s11864-014-0292-6. [DOI] [PubMed] [Google Scholar]
- 59.Greenland S., Mansournia M.A., Altman D.G. Sparse data bias: a problem hiding in plain sight. BMJ. 2016;352 doi: 10.1136/bmj.i1981. [DOI] [PubMed] [Google Scholar]
- 60.Mansournia M.A., Geroldinger A., Greenland S., Heinze G. Separation in logistic regression: causes, consequences, and control. Am J Epidemiol. 2018;187(4):864–870. doi: 10.1093/aje/kwx299. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.