Abstract
The rotavirus nonstructural protein NSP3 is a sequence-specific RNA binding protein that binds the nonpolyadenylated 3′ end of the rotavirus mRNAs. NSP3 also interacts with the translation initiation factor eIF4GI and competes with the poly(A) binding protein. Deletion mutations and point mutations of NSP3 from group A rotavirus (NSP3A), expressed in Escherichia coli, indicate that the RNA binding domain lies between amino acids 4 and 149. Similar results were obtained with NSP3 from group C rotaviruses. Data also indicate that a dimer of NSP3A binds one molecule of RNA and that dimerization is necessary for strong RNA binding. The dimerization domain of NSP3 was mapped between amino acids 150 and 206 by using the yeast two-hybrid system. The eukaryotic initiation factor 4 GI subunit (eIF-4GI) binding domain of NSP3A has been mapped in the last 107 amino acids of its C terminus by using a pulldown assay and the yeast two-hybrid system. NSP3 is composed of two functional domains separated by a dimerization domain.
Rotaviruses are members of the Reoviridae family, and their genome is composed of 11 molecules of double-stranded RNA which encode six structural proteins and six nonstructural proteins (8, 24). Viral transcriptase is activated as the virus enters the cell cytoplasm and synthesizes capped but nonpolyadenylated viral mRNAs. Viral mRNAs are either translated or used as templates for the synthesis of the genomic double-stranded RNAs. During this cycle, mRNAs are not tightly linked since coinfection of the same cell with different strains of virus results in gene reassortment at high frequency (37).
The viral mRNAs bear 5′ and 3′ untranslated regions of variable length which are bordered by two different sequences common to all genes. The 3′-end consensus sequence of group A rotaviruses is UGACC and is strictly conserved among all 11 segments. The 3′-end consensus sequence is different from one rotavirus serogroup to another; for example, in group C rotavirus, the 3′-end consensus sequence is UGGCU (4, 36). Reassortment between rotavirus strains of different serogroups has not been observed (52) and is thought to be restricted by these different consensus sequences.
We have reported that in group A rotavirus-infected cells, the nonstructural protein NSP3A is bound to the 3′ end of rotavirus mRNAs (31). In a previous report (32), we investigated the RNA sequence required for binding of NSP3A onto RNA in vitro and showed that the binding of the recombinant NSP3A to the 3′-end consensus sequence requires the last 5 bases UGACC. Mutations in the last 4 bases can greatly impair the interaction of NSP3A with RNA. NSP3 from group C rotavirus (NSP3C), expressed in baculovirus, binds to the 3′-end consensus sequence of group C rotavirus but not of group A rotavirus. These observations illustrate that NSP3 is a unique sequence-specific RNA binding protein.
Multimers of NSP3A expressed from rotavirus or baculovirus are observed only under nonreducing conditions (1, 24, 32); however, under the same conditions of analysis, the baculovirus-expressed NSP3C does not form multimers (33). Recently (31) we showed that NSP3A interacts in vivo with the translation initiation factor eukaryotic initiation factor 4, GI subunit (eIF-4GI). The translation of viral mRNA is enhanced at the expense of the translation of the cellular polyadenylated mRNA as NSP3A evicts the cellular poly(A) binding protein (PABP) from eIF-4GI. NSP3A may play a key role in the replication cycle of the rotavirus by allowing the efficient expression of the viral proteins in infected cells. Here we report on the identification of the RNA binding, multimerization, and eIF-4GI binding domains on the primary sequence of NSP3.
MATERIALS AND METHODS
Plasmid constructions.
Genes encoding NSP3 were obtained from the bovine RF strain of group A rotavirus (NSP3A; GenBank accession no. Z21639), devoid of the first three amino acids (containing residues 4 to 313), and from the porcine Cowden strain of group C rotavirus (NSP3C [402 amino acids]; GenBank accession no. M69115).
Recombinant proteins were expressed with a C-terminal His6 tag in a modified pET22b+ plasmid (Novagen). Plasmid pET22b+ containing the sequence of NSP3A cloned into NcoI (CCATGG)-XhoI had its leader sequence deleted (pET22LS−) by site-directed mutagenesis (22) (Table 1, oligonucleotide 1). N-terminal NSP3 deletion mutants were generated by site-directed mutagenesis by insertion of a NcoI site followed by linearization and recircularization of the plasmid. C-terminal deletions were obtained by insertion of a XhoI restriction site followed by linearization and recircularization of the plasmid (Table 1, oligonucleotides 2, 3, 4, 5, and 9) or by direct creation of a deletion inside the sequence (Table 1, oligonucleotides 6, 7, 8, and 10).
TABLE 1.
Oligonucleotides used for plasmid construction
| Mutation | Polarity | Oligonucleotide sequence (5′→3′) |
|---|---|---|
| Site-directed mutagenesis | ||
| pET22→pET22 LS− | − | CTGCTGTGTAGACTCCATGGTATATCTCCTTCTTAAAG |
| NSP3A→NSPA4–178 | − | CGATTCAAATCGTTTCTCGAGTTGTTCATATCT |
| NSP3A→NSP3A163–313 | − | CAACTTCCATGGTTTCTTCAAC |
| NSP3A→NSP3A206–313 | − | GTTTTGGAGAGAGTCCATGGTCTCGAGTACCTTTTTTGC |
| NSP3A→NSP3A238–LESM (NSP3A4–237, NSP3A242–313) | − | GACTGAAGACACCATGGACTCGAGTTTATTTTGTAG |
| NSP3A→NSP3A10–313 | − | AATCGAACTAGCCATGGTATATCTCCTTCTTAA |
| NSPA4–178→NSPA4–149 | − | GTGGTGCTCGAGACGTTCAATCTTATCTCT |
| NSPA4–178→NSPA4–130 | − | GTGGTGCTCGAGTGGAACTCCTTCACATT |
| NSPA4–178→NSPA4–178 MV54 | − | AACACCTGAATCGTCGACTACGTAATCAAA |
| NSPA4–178→NSPA4–178 KL106 | − | AGATGATAGCATAAGCTTAAGTTTGTTCAC |
| NSPA4–178→NSPA4–178 GS35 | − | TTCATTATAATCATAGGATCCGCCCATAAGTTC |
| NSPA4–178→NSPA4–178 QE172 | − | TTCAAATCGTTTTTCGAGCTCTTCATATCT |
| NSP3C→NSPC1–170 | − | ATCAATCATACACTCGAGTTCTTCTATCTT |
| NSPC1–170→NSPC1–140 | − | GTGGTGCTCGAGTACTGCATTTTCGTTTAA |
| PCR | ||
| NSPA163–313→NSPA163–290 | + | GTTGAAGAAGCCATGGAAGTTGAC |
| − | TAGCCGCTCGAGTATAATATTTCT | |
| Insertion of sequences | ||
| pGBT9→pGBT9-NS | + | AATTCGCCATGGATCCGTCGACTGACTGCA |
| − | GTCAGTCGACGGATCCATGGCG |
Constructs (Table 1, oligonucleotides 11 and 12) were obtained by PCR on the NSP3A163–313 sequence with oligonucleotides inserting NcoI or XhoI before cloning the PCR product into pET22LS−. Oligonucleotides used for point mutation are listed in Table 1. All the constructs were controlled by DNA sequencing.
Expression and purification of recombinant NSP3.
Plasmids were introduced into Escherichia coli BL21(DE3) by electroporation. Protein expression was induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM for 3 h at 37°C. His6-tagged proteins were purified on an Ni2+-Sepharose column (Pharmacia) after solubilization in lysis buffer A (20 mM Tris-HCl [pH 8], 0.5 M NaCl, 5 mM imidazole, 6 M urea). Renaturation was carried out at 4°C by step dialysis against renaturation buffer (50 mM Tris-HCl [pH 8], 0.5 mM oxidized gluthatione, 5 mM reduced gluthatione, 150 mM NaCl, 10% glycerol), with decreasing concentration of urea (from 3 M to 375 mM by twofold dilution steps over 12 h). After a final dialysis against renaturation buffer without urea, the insoluble proteins were removed by centrifugation (100,000 × g for 15 min). The concentration of the protein was estimated by comparison to a bovine serum albumin standard after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining. The semipurified baculovirus NSP3A (33) was used as a positive control.
Oligoribonucleotides.
Oligoribonucleotides (8 bases) were obtained by automated synthesis via dimethoxytrityl cyanoethyl RNA phosphoramidite chemistry with an Applied Biosystems 380A DNA synthesizer and purified as described previously (33). Purified oligoribonucleotides were used as probes for UV cross-linking and gel retardation assays after being labeled in vitro with T4 polynucleotide kinase and [γ-32P]ATP. Oligoribonucleotide A corresponds to the 3′-terminal sequence of group A rotavirus, 5′AUGUGACC. Oligoribonucleotide C corresponds to the 3′-terminal sequence of group C rotavirus, 5′ AUGUGGCU. Oligonucleotides bearing point mutations were called A/C for 5′AUGUGACU and C/A for 5′AUGUGGCC. A 5′-biotinylated oligoribonucleotide (GAUGUGACC; Eurogentec) was used for immobilization on an avidin chip in an optical biosensor (BIAcore 1000; Pharmacia) for analysis of the interaction with purified NSP3A proteins.
In vitro RNA-protein interactions.
Purified proteins (100 to 300 nM) were incubated with 5′-end-labeled oligoribonucleotides (3.5 nM) in 15 μl of RNA binding buffer (10 mM HEPES [pH 7.9], 40 mM KCl, 1 mM EDTA, 1 mM dithiothreitol [DTT], 5% glycerol) for 20 min at room temperature and UV cross-linked on ice 5 cm away from a 254-nm UV light source for 10 min (1 J/cm2). Complexes were then analyzed under reducing or nonreducing conditions by SDS-PAGE, and the gels were fixed, dried, and autoradiographed.
Identical reaction mixtures were prepared for gel retardation assays and performed at 30°C for 30 min before being loaded onto a nondenaturing polyacrylamide gel (8% acrylamide, 0.2% bisacrylamide) in 0.5× TAE buffer (27 mM Tris [pH 7.9], 13.2 mM sodium acetate [pH 7.9], 4 mM EDTA, 10% glycerol). Samples (plus traces of bromophenol blue) were loaded with the current on and electrophoresed at 4°C at 15 mA with recirculation of the 0.5× TAE running buffer. After the samples had entered the gel, the current was increased to 30 mA until the dye had migrated the two-thirds of the way along the gel. Wet gels were autoradiographed.
RNA binding was also studied in real time by surface plasmon resonance with a BIAcore apparatus (12, 26). The surface plasmon resonance is expressed in resonance units (RU: 1,000 RU corresponds to a change in adsorbed mass of 1 ng/mm2). Experiments were performed at 25°C in degassed and sterilized RNA binding buffer containing 0.05% surfactant P20. The 5′-biotinylated oligoribonucleotide was immobilized on an avidin sensor chip. Prior to the experiments, the purified proteins were dialyzed against RNA binding buffer and then injected with a continuous flow of RNA binding buffer. Immobilized RNA-protein complexes were washed with the same buffer. The interactions were visualized on a sensorgram in which RUs are plotted as a function of time and analyzed by “Global Fitting” or “Separate association/dissociation” version 3.0 (BIAcore).
Chemical cross linking was carried out following UV cross-linking of NSP3A to the labeled oligoribonucleotide, to stabilize RNA-bound multimers of NSP3A. Glutaraldehyde (0 to 0.5% [vol/vol] per thousand) was added to the samples for 10 min, at room temperature. Products were directly analyzed by SDS-PAGE under reducing conditions.
Yeast two-hybrid system.
The yeast two-hybrid test was used to analyze protein-protein interactions (10, 11). The plasmids that allowed the expression of fusion proteins were pGBT9 and pGAD424 (Clontech). pGBT9 contains the sequence for the GAL4 DNA binding domain, and pGAD424 contains the GAL4 activation domain. The polylinker of pGBT9 was modified by insertion (via a pair of annealed oligonucleotides [Table 1, oligonucleotides 13 and 14]) of NcoI and SalI restriction sites between the EcoRI and PstI sites to give pGBT9-NS. Sequences encoding NSP3A and some deletion mutants were obtained from pET22LS− after digestion with NcoI and XhoI and then cloned into pGBT9-NS digested with NcoI and SalI. Sequences cloned into pGBT9-NS were excised by digestion with EcoRI and PstI and inserted into pGAD424. The eIF-4GI+37–176 in pGAD has been described previously (31). Transfections were performed into Saccharomyces cerevisiae HF7C by the lithium acetate method as specified by the manufacturer (Clontech). Transformants were plated on medium lacking tryptophan and leucine (to monitor the efficiency of cotransfection) and on plates lacking tryptophan, leucine, and histidine (to detect an interaction between the proteins) and incubated at 30°C for 3 days.
Immunoprecipitations from rotavirus-infected cells.
The RF strain of group A rotavirus was propagated in monkey kidney cells (MA104) in Eagle’s minimum essential medium in the presence of trypsin (0.44 μg/ml; Sigma type IX). Infections were carried out in 60-mm-diameter dishes at a multiplicity of infection of 10 PFU/cell. Labelling was performed between 2 and 6 h postinfection with 200 μCi of Tran35S-label (1,000 Ci/mmol; 10 mCi/ml). The cells were washed with cold phosphate-buffered saline (PBS) and then incubated for 5 min with either cold PBS or cold PBS containing 40 mM N-ethylmaleimide acid (NEM) (45). Lysis was performed in 1 ml of lysis buffer B (50 mM Tris [pH 8], 125 mM NaCl, 20 mM EDTA, 0.1% SDS, 1% deoxycholate, 1% Triton X-100, 2 μg of aprotinin per ml) with or without 40 mM NEM. Cell debris was pelleted by centrifugation (12,000 × g for 5 min). NSP3A was immunoprecipitated by adding 1 μl of mouse monoclonal ascitic fluid ID3 (1) to 100 μl of supernatant, making the mixture up to 1 ml with lysis buffer B, and incubating it overnight at 4°C. Following the initial incubation, 30 μl of a suspension (1:1 [wt/vol] in lysis buffer B) of protein A-Sepharose was added and the incubation was continued for 1 h at 4°C with end-over-end rotation. Protein A-Sepharose beads were centrifuged, washed four times with 1 ml of lysis buffer B without NEM, and directly boiled in loading buffer with or without 150 mM β-mercaptoethanol (10 mM Tris-HCl [pH 6.8], 2% SDS, 10% glycerol) before analysis of bound proteins by SDS-PAGE. The gels were fixed in 20% ethanol–10% acetic acid, treated with Amplify (Amersham), dried, and subjected to autoradiography.
Protein interactions in vitro.
The His6-tag pull-down assay with the in vitro translation product of eIF-4GI+37–176 has been described previously (31). The pull-down assay allows the detection of an interaction between a purified His6-tagged protein and an in vitro transcription-translation product in rabbit reticulocyte lysate labeled with [35S]methionine. Experiments were performed with NSP3A rapidly batch purified from a small-scale culture. A 1-ml volume of an induced culture was centrifuged, and the pellet was solubilized overnight in lysis buffer A at 4°C. After centrifugation (100,000 × g for 15 min), the supernatant was applied to the Ni2+-charged resin in buffer A for 1 h at 4°C with gentle rotation. Renaturation of the immobilized proteins occurred during sequential incubations with decreasing concentration of urea in lysis buffer (10 min each with 4 M, 2 M, 1 M, and no urea respectively). The resin was then equilibrated in 50 mM Tris (pH 7.4)–150 mM NaCl–0.4% Nonidet P-40, 5 μg of leupeptin per ml–10 μg of aprotinin per ml and processed as previously described (31).
RESULTS
Rotavirus NSP3 expression in E. coli and RNA binding properties of NSP3A and NSP3C.
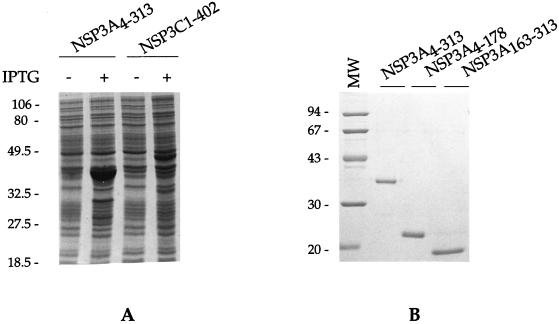
The E. coli T7 expression system developed by Studier (44) was used to express NSP3 from group A and C rotaviruses. The fusion of the first (NSP3C) or second (NSP3A) methionine to the Shine-Dalgarno sequence allows high-level expression of mature NSP3 upon induction by IPTG (Fig. 1A). The presence of a stretch of 6 histidine residues (His6 tag) at the C-terminal end of NSP3 allows the purification of the recombinant proteins on a metal chelate column.
FIG. 1.
Expression of NSP3 in E. coli and purification of deletion mutants. (A) The cDNA encoding NSP3 from group A and group C rotavirus was cloned in pET22b+LS− and introduced in E. coli BL21(DE3). Expression of the viral proteins was induced by addition of 1 mM IPTG to the cell culture for 3 h. Whole-cell proteins were resolved by SDS-PAGE and stained with Coomassie blue. (B) Recombinant proteins were purified on an Ni2+-Sepharose column and renatured by step dialysis. Proteins were resolved by SDS-PAGE and stained with Coomassie blue. MW, molecular weight (in thousands).
The recombinant proteins were solubilized in 6 M urea, purified by metal chelation chromatography, and then renatured by step dialysis against decreasing concentrations of urea. The purification conditions allow for recovery of one-third of the produced NSP3 as soluble material. Figure 1B depicts an example of the purified NSP3A used in the experiments described below.
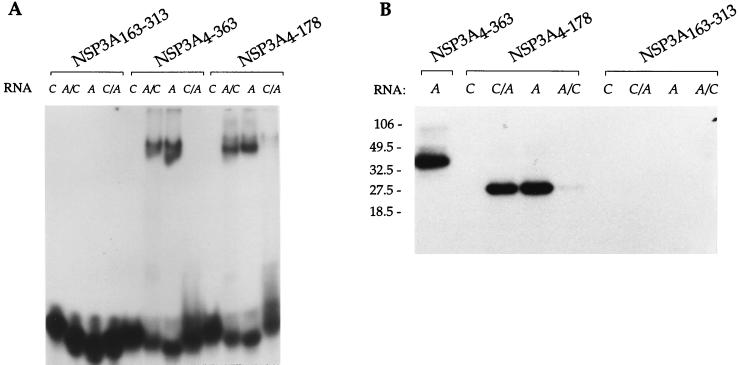
The E. coli recombinant proteins were initially tested for their ability to bind RNA specifically, as observed previously with baculovirus recombinant proteins (33). When tested by gel retardation (Fig. 2A) or UV cross-linking (Fig. 2B), NSP3A4–363 (which corresponds to amino acids 4 to 363 of NSP3A) presents the binding specificity of NSP3A. NSP3A4–363 binds strongly to the A and A/C RNA sequences, poorly to C/A RNA, and not at all to the group C 3′-end consensus sequence UGGCU. With NSP3C, the opposite result was obtained; NSP3C1–402 binds the C and C/A RNA but not the group A 3′-end consensus sequence UGACC (data not shown).
FIG. 2.
RNA binding properties of NSP3 and deletion mutants expressed in E. coli. Purified protein from the experiment in Fig. 1 were incubated with the different labeled oligoribonucleotides (RNA), and complexes were analyzed by gel retardation (A) or subjected to UV cross-linking and analysis by denaturing SDS-PAGE (B). RNA A corresponds to the 3′-terminal sequence of group A rotavirus (5′AUGUGACC). Oligoribonucleotide C corresponds to the 3′-terminal sequence of group C rotavirus (5′AUGUGGCU). The sequence of oligonucleotide A/C is 5′AUGUGACU and that of oligonucleotide C/A is 5′AUGUGGCC. The sizes of the molecular weight markers are indicated on the left in thousands.
The results indicate that the His6 tag at the C terminus did not impair the RNA binding properties of NSP3 and that the proteins produced in E. coli exhibited the same properties as the baculovirus recombinant NSP3.
C-terminal boundaries of the RNA binding domain of NSP3.
Sequence comparison between NSP3A and NSP3C reveals that the first half of the proteins is well conserved even though the C-terminal half is not (36, 38). We initially deleted each half of NSP3A and used the purified truncated proteins (Fig. 1B) in gel retardation and UV cross-linking assays. NSP3A4–178 presented the same RNA binding specificity as did the full-length protein in both the gel retardation assay (Fig. 2A) and UV cross-linking assay (Fig. 2B). The complement, NSP3A163–313, did not indicate RNA binding ability either by gel retardation (Fig. 2A) or by UV cross-linking (Fig. 2B).
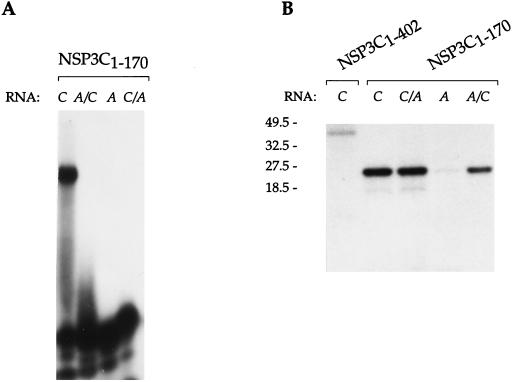
Larger C-terminal NSP3A deletions, including NSP3A4–149 and NSP3A4–130, were also tested. Binding of NSP3A4–130 to RNA was not detected in the gel retardation or UV cross-linking assay. Interaction of NSP3A4–149 with RNA was not detectable by the gel retardation assay, whereas NSP3A4–149 bound to RNA with the full NSP3A specificity when observed by UV cross-linking (data not shown). This behavior is characteristic of a weakened RNA binding property that leads to RNA-NSP3A4–149 complexes that are not stable enough to be detected by gel retardation. The first half of NSP3C (NSP3C1–170; Fig. 3) bound RNA with the same specificity as the full-length NSP3C did. The deletion mutant, NSP3C1–140, was not able to cross-link even to the group C oligoribonucleotide (data not shown).
FIG. 3.
RNA binding properties of NSP3C1–402 and NSP3C1–170 mutants expressed E. coli. Purified NSP3C and NSP3C1–402 were incubated with the different labeled oligoribonucleotides (RNA), and complexes were analyzed by gel retardation (A) or subjected to UV cross-linking and analysis by denaturing SDS-PAGE (B). The sizes of the molecular weight markers are indicated on the left in thousands.
The results indicate that the RNA binding properties of NSP3 are located in the first half of the protein and that the C-terminal boundary for the RNA binding domain is located between amino acids 130 and 149 for NSP3A and between amino acids 140 and 170 for NSP3C.
N-terminal boundaries of the RNA binding domain of NSP3A.
The N-terminal limit of the NSP3A RNA binding domain was also investigated with deletion mutants. As stated above, the deletion of the first 3 amino acids of NSP3A did not abolish its RNA binding properties (Fig. 2). Deletion of the first 9 amino acids (NSP3A10–313) or addition of a signal peptide of 22 amino acids at the N terminus of NSP3A and NSP3C totally abolished their RNA binding properties (data not shown). The N-terminal end of the RNA binding domain of NSP3A corresponds to the N terminus of the protein. Considering this result, we did not attempt to delimit the N-terminal boundary of NSP3C.
Point mutations in the RNA binding domain affect RNA binding.
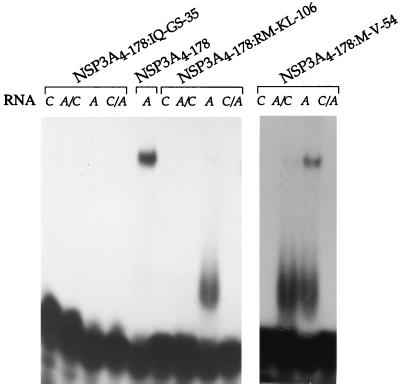
Initially, point mutations were introduced into NSP3A4–178 to identify which region of the RNA binding domain confers specificity toward the group A or group C RNA 3′-end sequence. Four positions of NSP3A were replaced by group C sequence: IQ to GS, M to V, RM to KL, and Q to E mutations were introduced at positions 35 to 36, 54, 106 to 107, and 172 of NSP3A4–178 (proteins NSP3A4–178:IQ-GS-35, NSP3A4–178:M-V-54, NSP3A4–178:RM-KL-106, and NSP3A4–178:Q-E172), respectively. UV cross-linking indicated that the mutants showed the NSP3A4–178 characteristics of RNA binding except for NSP3A4–178:IQ-GS-35, which did not cross-link to any RNA (data not shown). Gel retardation assay indicated that NSP3A4–178:Q-E-172 exhibited the RNA binding properties of NSP3A4–178 (data not shown) and that NSP3A4–178:IQ-GS-35 did not exhibit any RNA binding activity. For NSP3A4–178:M-V-54 and NSP3A4–178:RM-KL-106, the RNA-protein complex observed by gel retardation was less intense and a novel, poorly defined band migrating slightly above the free probe was observed (Fig. 4). This novel band probably represents a monomer of NSP3 bound on RNA. The NSP3A4–178:M-V-54 and NSP3A4–178:RM-KL-106 mutations introduced in NSP3 probably affect the dimerization of the RNA binding domain and not the contact between the protein and the RNA.
FIG. 4.
RNA binding properties of point mutants of NSP3A. Purified NSP3A proteins containing point mutations were incubated with different labeled oligoribonucleotides (RNA), and complexes were analyzed by gel retardation.
Point mutations did not alter the specificity of NSP3A binding, but some of them quantitatively modified its RNA binding ability and confirmed the presence of the RNA binding domain of NSP3 in its N-terminal region.
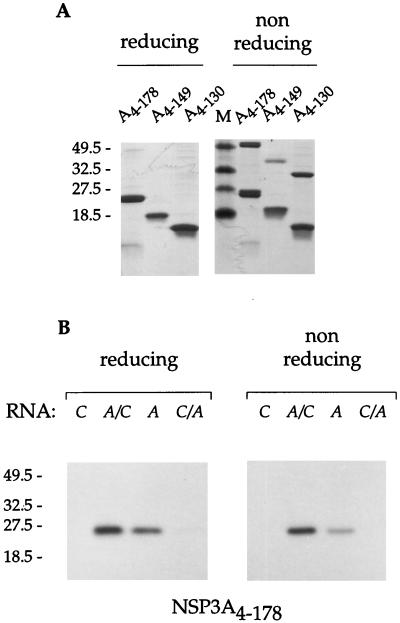
Disulfide bonds in the RNA binding domain impair RNA binding.
Two cysteine residues (positions 123 and 139) are conserved in the RNA binding domain of different strains of NSP3A (38). It has been reported that NSP3A, but not NSP3C (32), forms multimers when analyzed under nonreducing conditions (1, 23, 32, 33). Dimers of NSP3A4–178, NSP3A4–149, and NSP3A4–130 were observed when the proteins were analyzed under nonreducing conditions (Fig. 5A). When the same proteins were treated with as little as 20 mM DTT, dimers were no longer observed, suggesting that at least some of the cysteines are engaged in forming intermolecular disulfide bonds (data not shown). When NSP3A4–178 (Fig. 5B) and NSP3A4–149 (data not shown) were cross-linked to the labeled RNA A before analysis by PAGE under nonreducing conditions, only monomers of the proteins were labeled. Furthermore, treatment of the proteins with 20 mM DTT before UV cross-linking did not lower the quantity of NSP3A4–178 or NSP3A4–149labelled monomer (data not shown). Data suggest that the RNA binding properties of NSP3A are impaired when cysteine 123 and/or 139 is engaged in intermolecular disulfide bonds.
FIG. 5.
Disulfide bonds in the RNA binding domain of NSP3A impair RNA binding. (A) Purified NSP3A deletions were analyzed by SDS-PAGE under reducing or nonreducing conditions and stained with Coomassie blue. (B) The NSP3A4–178 deletion mutant was UV cross-linked to the radiolabeled oligoribonucleotides indicated and analyzed by SDS-PAGE under reducing or nonreducing conditions. Autoradiograms of the gels are shown. The sizes of the molecular weight markers are indicated on the left in thousands.
NSP3A forms dimers when bound on RNA.
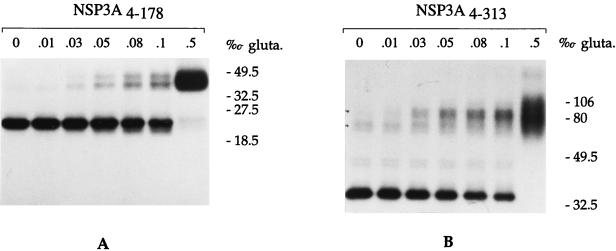
Chemical protein-protein cross-linking in the presence of DTT after UV cross-linking of NSP3A with the radiolabeled RNA A was used to determine if NSP3A forms multimers when bound on RNA. NSP3A4–363 and NSP3A4–178 were incubated under reducing conditions with RNA A and subjected to UV cross-linking (Fig. 6). Increasing concentrations of glutaraldehyde (0 to 0.5% [vol/vol] per thousand) were added, and the samples were incubated for 10 min at room temperature. The samples were analyzed by SDS-PAGE under reducing conditions. Analysis of NSP3A4–178 (Fig. 6A) revealed two close bands of about 45 kDa, which represent dimers of the protein cross-linked to the oligoribonucleotide. The increase of overall labeled NSP3 when the concentration of glutaraldehyde rose from 0.1 to 0.5% (vol/vol) per thousand is probably due to additional RNA-protein cross-linking induced by glutaraldehyde. Increasing the concentration of glutaraldehyde (up to 1%) or the incubation time did not lead to complexes of higher molecular weight. The presence of dimers of two slightly different sizes could reflect the presence of one or two cross-linked molecules of RNA per dimer. The presence of a dimer of NSP3A4–178 cross-linked to two RNAs in Fig. 6A could result from the dissociation of NSP3 dimers (after UV cross-linking to the RNA probe) followed by random reassociation and protein-protein cross-linking with glutaraldehyde. The reassociation of two NSP3A monomers, each with the RNA cross-linked to it, leads to the artifactual formation of an NSP3 dimer with two RNA molecules.
FIG. 6.
NSP3A binds RNA as a dimer. The purified RNA binding domain of NSP3 (A) or the full-length NSP3 (B) was UV cross-linked to the radiolabeled oligoribonucleotide A, incubated with increasing concentration of glutaraldehyde (gluta.), and analyzed by reducing SDS-PAGE. Autoradiograms are shown. The sizes of the molecular weight markers are indicated on the right in thousands.
The full-length NSP3 (Fig. 6B), when subjected to the same conditions, migrated as a diffuse band between 80 and 106 kDa. The diffuse band was interpreted as the result of heterogeneous intra- and intermolecular cross-linking of the NSP3A dimer. A degree of multimerization higher than dimers of the full-length NSP3A was not observed.
Quantitative study of the RNA-protein interaction.
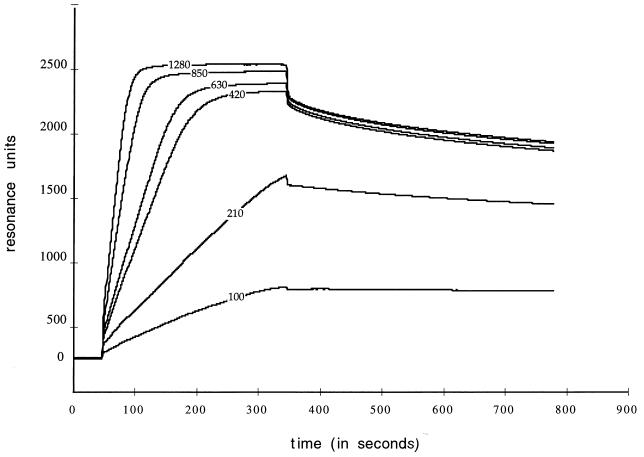
BIAcore analysis allows the detection of a real-time interaction between a molecule in the liquid phase and an immobilized ligand (12, 27). The signal observed (measured in RU) is directly proportional to the mass of bound molecules. BIAcore analysis was used to study the interaction between the recombinant purified NSP3A and an oligoribonucleotide corresponding to the 3′-end consensus sequence of rotavirus group A mRNAs. This oligoribonucleotide was biotinylated at its 5′ end (b-GAUGUGACC, 3.4 kDa), and 186 RU was immobilized on avidin chips. A signal of 2,010 RU was obtained at equilibrium with the highest concentrations of NSP3A4–178 (20.7 kDa) injected (Fig. 7). Since the ratio 186 RU/3.4 kDa = 55 is half the ratio 2,010 RU/20.7 kDa = 97, this result clearly shows that two molecules of NSP3A4–178 interacted with one molecule of oligoribonucleotide. Similar results were obtained after the injection of different concentrations of NSP3A4–178 on different amounts of immobilized RNA (data not shown). The ascending part of the curve could not be perfectly fitted with the association models tested, and it is probable that the association corresponds to a multistep phenomenon which is not accessible to analysis with the software utilized. The dissociation part of the curve was clearly separated into two phases. A first dissociation with an average rate kd1 = 1.2 × 10−2 s−1 was followed by a second dissociation with an average rate kd2 = 3.1 × 10−4 s−1. The data suggest that the two steps correspond to the dissociation of the protein dimer and the dissociation of the protein from the oligoribonucleotide. It is not possible to precisely attribute one constant to a particular dissociation.
FIG. 7.
Analysis of NSP3-RNA interaction in real time by BIAcore. The 5′-end-biotinylated RNA UGUGACC(186 RU) was immobilized on an avidin chip. NSP3A34–178 purified from E. coli at the concentration indicated (micromolar) was injected, and the signal (in RU) is plotted as a function of time.
The analysis of the real-time interaction between NSP3A4–178 and a biotinylated oligoribonucleotide confirms the results obtained by chemical cross-linking and shows that a dimer of NSP3A4–178 is bound to one molecule of oligoribonucleotide.
Absence of a disulfide bond between NSP3A molecules in infected cells.
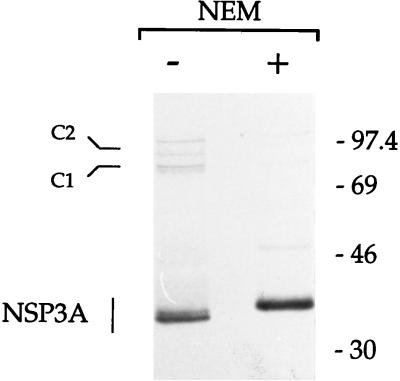
It has been reported that NSP3A expressed from rotavirus or baculovirus can form multimers of 100 and 80 kDa (C1 and C2, respectively) (24), which are visible only after electrophoresis under nonreducing conditions. The discrepancies between these results and the observation that disulfide bonds impaired binding to the RNA prompted a reexamination of the presence of disulfide bonds in NSP3A produced during rotavirus infection. Artifactual disulfide bonds have been observed during the preparation of cell lysates (45) and can be eliminated by NEM. Rotavirus-infected cells were incubated with NEM before being lysed by the method described by Svensson et al. (45), and NSP3A was immunoprecipitated. Following analysis by PAGE under nonreducing conditions (Fig. 8), much less C1 multimer was observed and the quantity of C2 multimer was reduced compared to that in lysates prepared in the absence of NEM (Fig. 8). NEM did not totally abolish the formation of C1 and C2 complexes, possibly because inaccessible cysteines buried in the protein structure were not exposed to NEM and immunoprecipitation washes were not performed in the presence of NEM (due to its toxicity). Note that, as described previously (45), the NEM treatment resulted in tightening of the VP6 (46-kDa) band. The monomer of NSP3A formed one band that migrated more slowly when immunoprecipitated from the lysates prepared in the presence of NEM. This phenomenon was also observed when the samples were analyzed under reducing conditions (data not shown) and could result from the presence of NEM molecules on each of the four cysteine residues of NSP3A.
FIG. 8.
Absence of disulfide bonds on NSP3A produced during rotavirus infection. NSP3 immunoprecipitated from rotavirus-infected cells lysates prepared in the presence (+) absence (−) of NEM was analyzed by nonreducing SDS-PAGE. An autoradiogram of the gel is shown. The molecular weight standards are indicated on the right in thousands. C1 and C2 refer to NSP3 oligomers described previously (23).
Dimerization domain of NSP3A by the two-hybrid test in yeast.
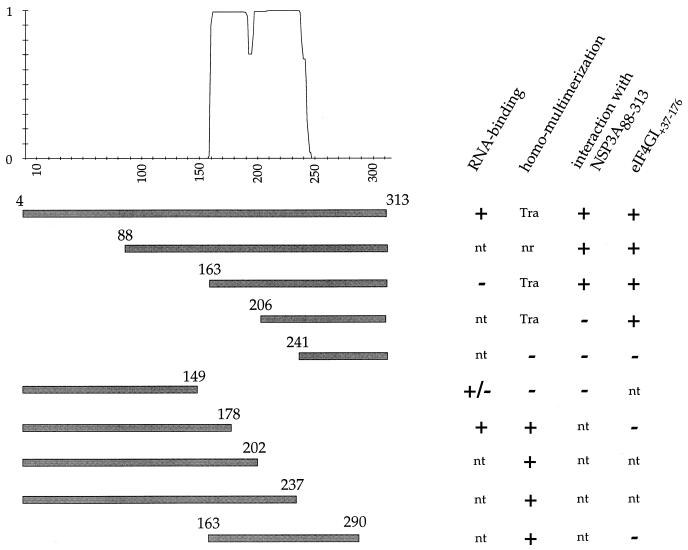
The yeast two-hybrid system has been developed to detect protein-protein interactions (10, 11) and may allow the detection of interactions not usually observed by traditional biochemical approaches (29). Furthermore, the composition of yeast internal medium is closer to the mammalian cell internal medium than to E. coli, and we (34) and others (17) have reported that the multimerization of NSP3A can be detected by this test. Considering that the dimerization of NSP3A does not appear to involve disulfide bonds, an attempt was made to map the dimerization domain of NSP3A. The use of Lupas’s algorithm (23) for the prediction of coiled coils in protein predicted a highly probable coil between amino acids 159 and 245 of NSP3A (Fig. 9) and lower probability of a coil at about residue 192. In addition to the mutants used for the RNA binding studies, new mutants were constructed (Fig. 9) and expressed as fusion proteins in the yeast two-hybrid vectors. Deletion mutants contained the entire coiled-coil (163 to 313, 88 to 313), the first (4 to 202) or the second (206 to 313) half, and no coiled-coil (241 to 313, 4 to 149) sequences. Since transactivation of the HIS3 reporter gene is, in some cases (9, 31) more sensitive than that of the lacZ reporter gene, the interaction of the mutants of NSP3A with themselves was studied by plating transfected cells on medium lacking histidine. For the NSP3A206–313 and NSP3A163–313 mutants, as with the full-length NSP3A, the homomultimerization cannot be assessed due to the spontaneous transactivation of the HIS3 reporter. With these mutants, the multimerization was studied by using NSP3A88–313 (Fig. 9) as partner.
FIG. 9.
RNA binding, dimerization, and eIF-4GI binding domains of NSP3A. The figure summarizes the results obtained for RNA binding (UV cross-linking and gel retardation; +/− indicates that the binding was observed by UV cross-linking but not by gel retardation) with the different purified NSP3A mutants studied. The dimerization (homomultimerization and interaction with NSP3A88–313) of the same mutants expressed in the yeast two-hybrid system is indicated. The interaction with eIF-4GI summarizes the results obtained by the pull-down and two-hybrid assays. The probability of coiled-coil structures in NSP3 predicted by the Lupas algorithm with a 28-amino-acid window (23) is indicated at the top of the figure. Numbers indicate amino acid positions. nt; not tested, Tra; transactivates, nr; not relevant.
The results of the two-hybrid test (Fig. 9) indicated that mutants containing amino acids 163 through 178 (i.e., the first half of the predicted coiled coil) are able to multimerize. Mutant NSP3A206–313, containing the second half of the predicted coiled coil (44 amino acids or six turns), was unable to interact with NSP3A88–313, but mutant NSP3A4–178, containing only the first 28 amino acids (four turns) of the coiled coil, dimerized.
Localization of the binding domain of NSP3A to eIF-4GI.
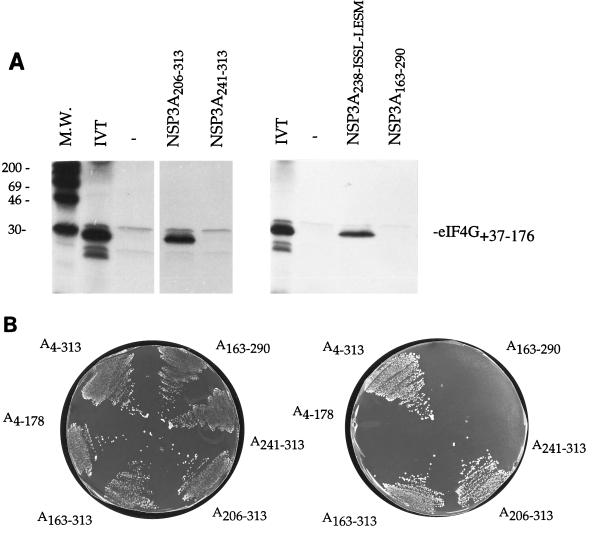
Recently (31), we reported an interaction between NSP3 and eIF-4GI (for reviews, see references 13 and 26) and delimited a 40-amino-acid stretch of eIF-4GI that was necessary and sufficient to interact with NSP3. The region of interaction of NSP3A on eIF-4GI was located in the N-terminal half of NSP3A and was determined by a pull-down assay (31). The pull-down assay was also used to identify this region more precisely on shorter fragments of NSP3A. Deletion mutants of NSP3A were tested with the radiolabelled translation product (Fig. 10A) of eIF-4GI. NSP3A206–313 was able to bind the radiolabelled product but not NSP3A241–313, NSP3A4–237, or NSP3A163–290. The full-length mutant protein NSP3A238-ISSL-LESM (amino acids 238 to 241: ISSL replaced by LESM), from which the two deletions NSP3A4–237 and NSP3A241–313 were expressed, were also tested. The mutations in NSP3A238-ISSL-LESM do not prevent the interaction with eIF-4GI. The sequences of NSP3A deletion mutants were transferred into the yeast two-hybrid plasmid pGAD424 and tested against pGBT9-eIF4G+37:176 (Fig. 10B). The result was entirely consistent with the pull-down assay; eIF4G+37:176 interacts with a region of NSP3A localized between amino acids 206 and 313.
FIG. 10.
Domain of NSP3 that interacts with eIF-4GI. The domain of NSP3A necessary for interaction with eIF-4GI were mapped by the pull-down (A) and two-hybrid (B) assays with deletion mutants. (A) Autoradiograms of the gels. Controls consist of beads incubated without recombinant protein (−). An aliquot of the in vitro-translated and labelled eIF4GI+37:176 fragment (IVT) used in this experiment is shown. The sizes of the molecular weight (M.W.) standards are indicated on the left in thousands. (B) HF7C yeast colonies, cotransfected with the indicated NSP3 fragment cloned in pGAD424 and the eIF-4GI fragment (+37:176) cloned in pGBT9, were streaked on medium plates lacking tryptophan and leucine (left) and on medium plates lacking tryptophan, leucine, and histidine (right).
DISCUSSION
RNA binding domain of NSP3A and NSP3C.
E. coli-expressed truncated proteins have facilitated mapping of the RNA binding domains of NSP3A and NSP3C. NSP3A4–178 and NSP3C1–170 are the shortest deletion mutants able to bind RNA with the specificity of the full-length proteins in either the gel retardation assay or the UV cross-linking assay.
Gene 7 encodes NSP3A in the RF strain of rotavirus and presents two in-frame methionines (M1 and M4). NSP3A, starting at M4, presents the RNA binding properties of the full-length NSP3A. Further deletion from the N terminus of NSP3A abolished the RNA binding, whether by deleting an important domain of the protein or by impairing the folding of the protein. Interestingly, when N-terminal fusions were produced in E. coli, no RNA binding was observed (data not shown), suggesting that no deletion or addition is allowed at the N terminus of NSP3A. Different arguments favor M4 as the initiation methionine of NSP3A in vivo. First, M4 has a better environment as an initiation codon (21) than M1 does. Comparison of NSP3A and NSP3C shows that NSP3C M1 is analogous to NSP3A M4. Finally, an avian gene (GenBank accession no. AB009626) and a human rotavirus gene (14) encode an NSP3 starting at M4. The first 3 amino acids are not necessary for the RNA binding properties of NSP3A whether or not the first methionine (M1) is used in vivo.
The C-terminal boundary of the RNA binding domain of NSP3A is less precise. A deletion of 30 amino acids at the C terminus of NSP3A4–178 (mutant NSP3A4–149) did not abolish the binding of NSP3A on the RNA but, rather, weakened it; RNA binding is not visible by gel retardation, but the protein can be cross-linked on the RNA.
Sequence comparison between NSP3A and NSP3C indicates that the first half of the protein (amino acids 81 to 137) is very well conserved (44% identity between amino acids 49 and 86 compared to an overall identity of less than 20%). Data indicates that the first 170 amino acids of NSP3C are also functionally homologous to those of NSP3A; indeed, NSP3C1–170 presents the same specificity for group C RNA as the baculovirus-expressed NSP3C does.
The comparison of the amino acid sequence of NSP3A and NSP3C reveals that the RNA binding half of the protein bears most of the homology. The conservation of some aromatic residues that could be involved in base stacking and of some basic amino acids that could interact with the phosphate backbone is of particular interest. More extensive point mutagenesis experiments may allow the determination of the protein sequence involved in the specific RNA binding properties of NSP3. Most often, RNA binding proteins interact with RNA targets which contain various secondary structures as stem-loops, pseudoknots, bulges, and three-dimensional shapes (20). NSP3 has the remarkable property of binding to a short RNA sequence which is unlikely to be able to make a stable secondary structure. The amino acid sequence of NSP3 does not reveal similarity to the ribonucleoprotein motif present on many RNA binding proteins (20, 28). No other RNA binding motifs (5) such as the arginine-rich motif (35), zinc finger (7), KH domain (41), or RGG and RS motifs (3, 53) are found on NSP3. The only homology of NSP3 to other RNA binding proteins is a short motif identified by van Staden et al. (50) between different Reoviridae RNA binding proteins; the bluetongue virus NS2, reovirus sigma NS, and rotavirus NSP3. Binding of recombinant bluetongue virus NS2 to RNA does not seem to be sequence specific (48, 54). Sigma NS purified from mammalian reovirus-infected cells reportedly prefers reovirus 3′-end mRNA (16, 42), while avian reovirus-expressed (51) and E. coli-expressed sigma NS (39) do not show any specificity. Limited mutations outside the conserved motif, as well as deletion of the entire motif (47), abolished RNA binding by NS2 (15, 54). It should be noted that the mutation of RM to KL at positions 106 and 107 (Fig. 4) in the so-called van Staden motif considerably diminishes the affinity of NSP3 for its RNA target. Thus, it does not seem that the presence of this motif is a signature of sequence-specific RNA binding proteins or that it is an obligatory part of the RNA binding domain; the role of this motif may be related to other functions of the proteins or to their dimerization.
Absence of disulfide bonds in vivo.
Before the experiments reported here were performed, the degree of multimerization of NSP3A in rotavirus-infected cells or in baculovirus was not clear (1, 24, 32, 33). Two forms of multimers held by disulfide bonds (C1 and C2) were observed in nonreducing SDS-PAGE; however, their calculated molecular weights did not allow a clear distinction between whether dimers or trimers of NSP3A existed. The observations that disulfide bonds with C123 and/or C139 were deleterious for the RNA binding properties of NSP3A and that NSP3C did not form dimers under nonreducing conditions (33) prompted a reevaluation of the presence of disulfide bonds in NSP3A produced in rotavirus-infected cells. The much smaller quantity of C1 and C2 recovered after immunoprecipitation of cell lysates prepared in the presence of NEM strongly suggests that these complexes are formed during cell lysis and are therefore artifacts of cell lysate preparation. Under these lysis conditions, formation of high-molecular-weight oligomers of NSP3A could result from multiple intermolecular disulfide bonds and may not reflect the true oligomerization state of NSP3A. Oligomers of NSP3 higher than dimers were not detected after chemical cross-linking of the full-length NSP3A.
NSP3 forms dimers.
The region of NSP3A from amino acids 150 to 240 is predicted to form a coiled coil as predicted by Lupas’s algorithm (Fig. 10). This region does not include the leucine zipper present in NSP3A from the SA11 strain (24). The coiled-coil region of NSP3A can be further divided into two parts. The first part, from amino acids 150 to 178, is able to multimerize by itself, as observed in the two-hybrid test (mutant NSP3A4–178 versus NSP3A4–149), while the second part (downstream of amino acid 200) does not dimerize. These data are supported by the observation that the NSP3A206–313 mutant is not able to interact with NSP3A88–313 in the two-hybrid test. Similar observations have been made with the smooth muscle myosin; fragments of the protein containing up to 15 heptad repeats failed to dimerize (49).
It seems reasonable to hypothesize that two aspects of NSP3A dimerization can occur. First, NSP3 dimerizes upon binding to the RNA and the region required for dimerization is the same (amino acids 3 to 149) as the RNA binding domain. The dimerization of this domain is strictly dependent on the presence of RNA, since it could not be observed by the two-hybrid test. Mutations introduced into the RNA binding domain (at positions 54 and 106) can weaken the dimerization of the RNA binding domain on RNA, as observed by the appearance of a transient monomeric NSP3-RNA complex (Fig. 4). The dimerization of NSP3A on the RNA may explain the cooperativity of the binding observed previously (33) and the complexity of the association of NSP3 with RNA (Fig. 7). Second, the region downstream of amino acid 149 is able to multimerize, as observed by the two-hybrid test, and is predicted by sequence analysis to form a coiled coil. The dimerization of this region is independent of the RNA binding domain dimerization, as observed in the two-hybrid test with a fragment of NSP3 lacking the first 149 amino acids. The region downstream of amino acid 206 does not seem able to multimerize spontaneously, despite the high probability that it forms a coiled coil (Fig. 9). Such a phenomenon has been observed for the dimerization of cortexillin (43), where a 14-amino-acid sequence at the beginning of the coil is absolutely necessary for the formation of a longer dimer. Interestingly, the cortexillin and the NSP3A region from 149 to 178 are both rich in charged amino acids, suggesting that the NSP3 region from 149 to 178, by spontaneously forming a coiled coil, triggers the formation of a longer coil, which extends to approximately amino acid 240.
The BIAcore study indicated that a dimer of NSP3A4–178 was interacting with each RNA molecule. This result was confirmed by chemical cross-linking of NSP3A4–178 and extended to the full-length NSP3 by the same method. The differences in RNA binding between NSP3A4–178 and NSP3A4–149 can be attributed to the lower ability of NSP3A4–149 to make stable dimers. This interpretation is reinforced by the observation that NSP3A4–178 is able to interact with itself but not with NSP3A4–149 in the two-hybrid test.
RNA binding and eIF-4GI binding domains are functionally and structurally independent.
The domain of interaction of NSP3 with eIF-4GI has been refined by the pull-down assay (31) and the two-hybrid assay. This region has been delimited to the C terminus of NSP3A between amino acids 206 and 363. This fragment was not able to dimerize by itself or with NSP3A88–313 despite the presence of the last 40 amino acids of the predicted coiled coil. The domain of interaction of NSP3A with eI4FGI is opposite to the RNA-binding domain, and the two domains can function independently. This property of NSP3 contrasts with the yeast and mammalian PABPs (19, 46). In the case of these two cellular proteins, the second RNA binding motif (of the four present on the protein) interacts with eIF-4GI. It has also been recently shown that the 40 amino acids of eIF-4GI that are necessary and sufficient for its interaction with NSP3A are necessary (30) or sufficient (19) for the interaction of eIF-4GI with the mammalian PABP. Despite binding on the same site on eIF-4GI, the PABP and NSP3 do not have strong sequence similarities even in their respective eIF-4GI binding domains. It should be emphasized that the binding of the viral and cellular proteins present some important differences. NSP3A binds strongly to eIF-4GI in the absence of RNA and is able to evict PABP from the translation initiation factor (31). Unlike mammalian PABP, the binding of yeast PABP is highly dependent on the presence of a poly(A)+ RNA. Crystalization of each protein with the 40-amino-acid domain of eIF-4GI will reveal the structural basis for these differences.
The independence of the RNA and eIF-4GI binding domains of NSP3 reflects its dual role. Interaction of NSP3 with eIF-4GI in the absence of viral mRNA will induce the shutoff of translation of cellular mRNAs. Exclusive interaction of NSP3 with viral mRNAs will protect them from degradation via the deadenylation pathway (2, 6). Simultaneous interaction of NSP3 with viral mRNAs and eIF-4GI will allow the efficient translation of the viral mRNAs (30).
It is interesting that NSP3 has been found associated in part with the cell cytoskeleton (24). The homology observed between the dimerization domain of NSP3 and the coiled-coil structures of cellular protein-forming cell filaments has sustained the idea that NSP3A could interact with some cellular structures. The association of NSP3 with the cell cytoskeleton could be mediated by its interaction with eIF-4GI. Translation and localization of cellular mRNAs has been linked to their association with the cell cytoskeleton (18) via the translation machinery or addressing protein (40).
The mapping of the different functional domains of NSP3 allows a schematic representation of NSP3 in the infected cell, bound by its C terminus on the eIF-4G and presenting the viral mRNAs to the translation machinery. The cocrystalization of NSP3 with eIF-4G and a viral mRNA could refine this rather coarse representation.
ACKNOWLEDGMENTS
We are deeply indebted to Jean Cohen for his continuous scientific and personal support and for critical reading of the manuscript. Thanks to Scott A. Kramer for checking the English. Thanks to Ali Mirazimi for advice on the NEM protocols and also to Pierre Lindenbaum and Patrice Vende for assistance with the two-hybrid experiments. We acknowledge the skillful technical assistance of N. Castagné.
M. Piron is supported by a fellowship from the French Ministère de l’Education Nationale de la Recherche et de la Technologie. T.D., J.G., and D.P. belong to the INRA staff. This work was funded in part by “Programme de Recherches Fondamentales en Microbiologie, Maladies Infectieuses et Parasitologie” of MENRT.
REFERENCES
- 1.Aponte C, Mattion N M, Estes M K, Charpilienne A, Cohen J. Expression of two bovine rotavirus non-structural proteins (NSP2, NSP3) in the baculovirus system and production of monoclonal antibodies directed against the expressed proteins. Arch Virol. 1993;133:85–95. doi: 10.1007/BF01309746. [DOI] [PubMed] [Google Scholar]
- 2.Beelman C A, Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- 3.Birney E, Kumar S, Krainer A R. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bremont M, Chabanne-Vautherot D, Vannier P, McCrae M A, Cohen J. Sequence analysis of the gene (6) encoding the major capsid protein (VP6) of group C rotavirus: higher than expected homology to the corresponding protein from group A virus. Virology. 1990;178:579–583. doi: 10.1016/0042-6822(90)90357-w. [DOI] [PubMed] [Google Scholar]
- 5.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 6.Caponigro G, Parker R. Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol Rev. 1996;60:233–249. doi: 10.1128/mr.60.1.233-249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clemens K R, Wolf V, McBryant S J, Zhang P, Liao X, Wright P E, Gottesfeld J M. Molecular basis for specific recognition of both RNA and DNA by a zinc finger protein. Science. 1993;260:530–533. doi: 10.1126/science.8475383. [DOI] [PubMed] [Google Scholar]
- 8.Estes M K. Rotaviruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1625–1655. [Google Scholar]
- 9.Estojak J, Brent R, Golemis E A. Correlation of two-hybrid affinity data with in vitro measurements. Mol Cell Biol. 1995;15:5820–5829. doi: 10.1128/mcb.15.10.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 11.Fields S, Sternglanz R. The two-hybrid system: an assay for protein-protein interactions. Trends Genet. 1994;10:286–292. doi: 10.1016/0168-9525(90)90012-u. [DOI] [PubMed] [Google Scholar]
- 12.Fisher R J, Fivash M. Surface plasmon resonance based methods for measuring the kinetics and binding affinities of biomolecular interactions. Curr Opin Biotechnol. 1994;5:389–395. doi: 10.1016/0958-1669(94)90047-7. [DOI] [PubMed] [Google Scholar]
- 13.Gallie D R. A tale of two termini: a functional interaction between the termini of an mRNA is a prerequisite for efficient translation initiation. Gene. 1998;216:1–11. doi: 10.1016/s0378-1119(98)00318-7. [DOI] [PubMed] [Google Scholar]
- 14.Gault, E., and A. Gabarg-Chenon. 1998. Personal communication.
- 15.Gillian A L, Nibert M L. Amino terminus of reovirus nonstructural protein sigma NS is important for ssRNA binding and nucleoprotein complex formation. Virology. 1998;240:1–11. doi: 10.1006/viro.1997.8905. [DOI] [PubMed] [Google Scholar]
- 16.Gomatos P J, Prakash O, Stamatos N M. Small reovirus particle composed solely of sigma NS with specificity for binding different nucleic acids. J Virol. 1981;39:115–124. doi: 10.1128/jvi.39.1.115-124.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez R A, Torres-Vega M A, Lopez S, Arias C F. In vivo interactions among rotavirus nonstructural proteins. Arch Virol. 1998;143:981–996. doi: 10.1007/s007050050347. [DOI] [PubMed] [Google Scholar]
- 18.Hesketh J E. mRNA targeting: signals in the 3′-untranslated sequences for sorting of some mRNAs. Biochem Soc Trans. 1996;24:521–527. doi: 10.1042/bst0240521. [DOI] [PubMed] [Google Scholar]
- 19.Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A) binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kenan D J, Query C C, Keene J D. RNA recognition: towards identifying determinants of specificity. Trends Biochem Sci. 1991;16:214–220. doi: 10.1016/0968-0004(91)90088-d. [DOI] [PubMed] [Google Scholar]
- 21.Kozak M. Interpreting cDNA sequences: some insights from studies on translation. Mamm Genome. 1996;7:563–574. doi: 10.1007/s003359900171. [DOI] [PubMed] [Google Scholar]
- 22.Kunkel T A, Bebenek K, McClary J. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- 23.Lupas’s algorithm. [Online.] http://www.isrec.isb-sib.ch/software/COILS_form.html. [26 April 1999, last date accessed.]
- 24.Mattion N M, Cohen J, Aponte C, Estes M K. Characterization of an oligomerization domain and RNA-binding properties on rotavirus nonstructural protein NSP3. Virology. 1992;190:68–83. doi: 10.1016/0042-6822(92)91193-x. [DOI] [PubMed] [Google Scholar]
- 25.Mattion N M, Cohen J, Estes M K. Rotavirus proteins. In: Kapikian A, editor. Viral infections of the gastrointestinal tract. New York, N.Y: Marcel Dekker, Inc.; 1994. pp. 169–249. [Google Scholar]
- 26.Merrick W C, Hershey J W B. The pathway and mechanism of eucaryotic protein synthesis. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 31–69. [Google Scholar]
- 27.Myszka D G. Kinetic analysis of macromolecular interactions using surface plasmon resonance biosensors. Curr Opin Biotechnol. 1997;8:50–57. doi: 10.1016/s0958-1669(97)80157-7. [DOI] [PubMed] [Google Scholar]
- 28.Nagai K, Oubridge C, Ito N, Avis J, Evans P. The NRP domain: a sequence-specific RNA-binding domain involved in processing and transport of RNA. Trends Biochem Sci. 1995;20:235–240. doi: 10.1016/s0968-0004(00)89024-6. [DOI] [PubMed] [Google Scholar]
- 29.Patel R C, Sen G C. PACT, a protein activator of the interferon-induced protein kinase, PKR. EMBO J. 1998;17:4379–4390. doi: 10.1093/emboj/17.15.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piron, M., and D. Poncet. Unpublished results.
- 31.Piron M, Vende P, Cohen J, Poncet D. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J. 1998;17:5811–5821. doi: 10.1093/emboj/17.19.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poncet D, Aponte C, Cohen J. Rotavirus protein NSP3 (NS34) is bound to the 3′-end consensus sequence of viral mRNAs in infected cells. J Virol. 1993;67:3159–3165. doi: 10.1128/jvi.67.6.3159-3165.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poncet D, Laurent S, Cohen J. Four nucleotides are the minimal requirement for RNA recognition by rotavirus non-structural protein NSP3. EMBO J. 1994;13:4165–4173. doi: 10.1002/j.1460-2075.1994.tb06734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poncet D, Lindenbaum P, L’Haridon R, Cohen J. In vivo and in vitro phosphorylation of rotavirus NSP5 correlates with its localization in viroplasms. J Virol. 1997;71:34–41. doi: 10.1128/jvi.71.1.34-41.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puglisi J D, Tan R, Calnan B J, Frankel A D, Williamson J R. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science. 1992;257:76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- 36.Qian Y A, Jiang B M, Saif L J, Kang S Y, Ojeh C K, Green K Y. Molecular analysis of the gene 6 from a porcine group C rotavirus that encodes the NS34 equivalent of group A rotaviruses. Virology. 1991;184:752–757. doi: 10.1016/0042-6822(91)90446-i. [DOI] [PubMed] [Google Scholar]
- 37.Ramig R F. Genetics of the rotaviruses. Annu Rev Microbiol. 1997;51:225–255. doi: 10.1146/annurev.micro.51.1.225. [DOI] [PubMed] [Google Scholar]
- 38.Rao C D, Das M, Ilango P, Lalwani R, Rao G S, Gowda K. Comparative nucleotide and amino acid sequence analysis of the sequence-specific RNA-binding rotavirus nonstructural protein NSP3. Virology. 1995;207:327–333. doi: 10.1006/viro.1995.1087. [DOI] [PubMed] [Google Scholar]
- 39.Richardson M A, Furuichi Y. Synthesis in Escherichia coli of the reovirus nonstructural protein sigma NS. J Virol. 1985;56:527–533. doi: 10.1128/jvi.56.2.527-533.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ross A F, Oleynikov Y, Kislauskis E H, Taneja K L, Singer R H. Characterization of a beta-actin mRNA zipcode-binding protein. Mol Cell Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siomi H, Choi M, Siomi M C, Nussbaum R L, Dreyfuss G. Essential role for KH domains in RNA binding: impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome. Cell. 1994;77:33–39. doi: 10.1016/0092-8674(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 42.Stamatos N M, Gomatos P J. Binding to selected regions of reovirus mRNAs by a nonstructural reovirus protein. Proc Natl Acad Sci USA. 1982;79:3457–3461. doi: 10.1073/pnas.79.11.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steinmetz M O, Stock A, Schulthess T, Landwehr R, Lustig A, Faix J, Gerisch G, Aebi U, Kammerer R A. A distinct 14 residue site triggers coiled-coil formation in cortexillin I. EMBO J. 1998;17:1883–1891. doi: 10.1093/emboj/17.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Studier F W. Use of bacteriophage T7 lysozyme to improve inducible T7 expression system. J Mol Biol. 1991;219:37–44. doi: 10.1016/0022-2836(91)90855-z. [DOI] [PubMed] [Google Scholar]
- 45.Svensson L, Dormitzer P R, von Bonsdorff C H, Maunula L, Greenberg H B. Intracellular manipulation of disulfide bond formation in rotavirus proteins during assembly. J Virol. 1994;68:5204–5215. doi: 10.1128/jvi.68.8.5204-5215.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tarun S Z, Jr, Sachs A B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 47.Theron J, Huismans H, Nel L H. Identification of a short domain within the non-structural protein NS2 of epizootic haemorrhagic disease virus that is important for single strand RNA-binding activity. J Gen Virol. 1996;77:129–137. doi: 10.1099/0022-1317-77-1-129. [DOI] [PubMed] [Google Scholar]
- 48.Theron J, Nel L H. Stable protein-RNA interaction involves the terminal domains of bluetongue virus mRNA, but not the terminally conserved sequences. Virology. 1997;229:134–142. doi: 10.1006/viro.1996.8389. [DOI] [PubMed] [Google Scholar]
- 49.Trybus K M, Freyzon Y, Faust L Z, Sweeney H L. Spare the rod, spoil the regulation: necessity for a myosin rod. Proc Natl Acad Sci USA. 1997;94:48–52. doi: 10.1073/pnas.94.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Staden V, Theron J, Greyling B J, Huismans H, Nel L H. A comparison of the nucleotide sequences of cognate NS2 genes of three different orbiviruses. Virology. 1991;185:500–504. doi: 10.1016/0042-6822(91)90808-o. [DOI] [PubMed] [Google Scholar]
- 51.Yin H S, Lee L H. Identification and characterization of RNA-binding activities of avian reovirus non-structural protein sigmaNS. J Gen Virol. 1998;79:1411–1413. doi: 10.1099/0022-1317-79-6-1411. [DOI] [PubMed] [Google Scholar]
- 52.Yolken R, Arango-Jaramillo S, Eiden J, Vonderfecht S. Lack of genomic reassortment following infection of infant rats with group A and group B rotaviruses. J Infect Dis. 1988;158:1120–1123. doi: 10.1093/infdis/158.5.1120. [DOI] [PubMed] [Google Scholar]
- 53.Zamore P D, Williamson J R, Lehmann R. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA. 1997;3:1421–1433. [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao Y, Thomas C, Bremer C, Roy P. Deletion and mutational analyses of bluetongue virus NS2 protein indicate that the amino but not the carboxy terminus of the protein is critical for RNA-protein interactions. J Virol. 1994;68:2179–2185. doi: 10.1128/jvi.68.4.2179-2185.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]