Abstract
Viral protein R (Vpr) of human immunodeficiency virus type 1 (HIV-1) transiently arrests cells in the G2 phase of the cell cycle and is a weak transcriptional transactivator. We found that Vpr increased HIV-1 long terminal repeat (LTR) activity in all cells examined but, when expressed at high levels, decreased HIV-1 LTR expression due to cytotoxic effects. Moreover, Vpr-mediated enhancement of HIV-1 LTR-driven transcription was observed in cycling primary human CD4+ T cells but not in terminally differentiated, noncycling primary human macrophages. In single-round infection experiments using primary human CD4+ T cells, proviral clones expressing either wild-type Vpr or Vpr mutants that retained the ability to cause a G2 arrest replicated to higher levels than proviruses lacking Vpr or expressing mutants of Vpr that did not cause an arrest. In support of the hypothesis that enhancement of HIV-1 LTR transcription by Vpr is an indirect effect of the ability of Vpr to delay cells in G2, counterflow centrifugal elutriation of cells into different phases of the cell cycle demonstrated that HIV-1 LTR expression was highest in G2. Finally, the ability of Vpr to upregulate viral transcription was dependent on a minimal promoter containing a functional TATA box and an enhancer.
Viral protein R (Vpr) is a 96-amino-acid virion-associated protein of human immunodeficiency virus type 1 (HIV-1) that localizes to the nucleus and the nuclear membrane of the infected cell (9, 14, 33, 45, 47). Vpr plays an important role in targeting the HIV-1 preintegration complex into the nucleus of nonproliferating cells such as terminally differentiated macrophages and hence is required for efficient viral replication in these cells (10, 17, 21, 37, 47). In addition, cycling cells infected with wild-type HIV encoding a functional Vpr arrest in the G2 phase of the cell cycle (5, 20, 24, 38). Vpr present in the virus particle is sufficient for this cell cycle arrest (35).
While HIV-1 Vpr alone is responsible for both the nuclear targeting and G2 arrest functions, HIV-2 and sooty mangabey-derived simian immunodeficiency virus encode for two proteins, Vpr and Vpx, that are responsible for the G2 cell cycle arrest and nuclear import functions, respectively (16, 46). Moreover, nuclear targeting of the preintegration complex and the G2 cell cycle arrest are two genetically separable functions of HIV-1 Vpr (47). Because the cell cycle arrest function of Vpr is conserved among diverse species of primate lentiviruses (34, 44) in a background of very high lentivirus mutation rates, we argue that the cell cycle arrest function must provide a selective advantage to the virus during its life cycle. We recently showed that HIV-1 mRNA levels were highest in the G2 phase of infected Jurkat T cells synchronized in the cell cycle and that the selective advantage for Vpr-induced G2 arrest could be explained by increased viral expression in G2-arrested cells (18).
In this report, we further examine the ability of Vpr to transactivate the HIV-1 long terminal repeat (LTR) and the relationship of LTR activation to the cell cycle. Using counterflow centrifugal elutriation analysis of Jurkat T cells, stably transfected with the luciferase gene under the control of the HIV-1 LTR, we confirm that HIV-1 LTR-driven luciferase expression (RNA and protein levels) is highest in the G2 phase of the cell cycle. We also demonstrate that Vpr is able to stimulate HIV-1 LTR-driven transcription in primary human CD4+ T cells which are subject to Vpr-mediated G2 cell cycle arrest. During a single round of infection, proviruses expressing Vpr alleles that are able to cause G2 arrest show increased replication in primary CD4+ T cells compared to Vpr alleles that are unable to cause a G2 arrest. Finally, we show that a minimal promoter containing a functional TATA box and an enhancer element is sufficient for the Vpr-mediated enhancement of basal transcription.
MATERIALS AND METHODS
Plasmids.
Vpr expression vectors that expressed HIV-1 Vpr from either the simian cytomegalovirus immediate-early (CMV IE) promoter (pCMV-Vpr) or the HIV-1 LTR (pHIV-Vpr) have been previously described (18). The HIV-1 LTR (−480 to +80 fragment) was cloned into both pGL-2 Basic (pwtLTR-luc1) (Promega, Madison, Wis.) and pGL-3 Basic (pwtLTR-luc2) (Promega) promoterless vectors for detection of HIV-1 LTR-driven luciferase gene activity (18). The −250 to +80 and the −81 to +80 LTR fragments were obtained by exonuclease III digests of HIV-1 LTR; the −171 to +80 and −161 to +80 LTR fragments were gracious gifts from Richard Gaynor, University of Texas, Southwestern; −480 to −20 LTR fragment was obtained by excision of the PvuII-HindIII fragment of the −480 to +80 LTR and religation of the resulting LTR fragment. These HIV-1 LTR sequences were then cloned into the pGL-2 Basic vector. HIV-1 LTRs (−480 to +80) containing mutations in selected transcription factor binding sites were cloned into Asp718-HindIII-cleaved pGL3-Basic. Mutations included a point mutant of the glucocorticoid response element (GRE−; +14 to +18 region of the LTR) (22), the Ets-1 and lymphoid enhancer binding factor 1 binding site mutants (Ets− and Lef−, respectively) (23, 42), E-box (HLH−) mutant, Sp1 transcription factor binding site mutant (Sp1−) (29), and mutations in both NF-κB transcription factor binding sites (NF-κB−) (30).
In addition, the human CMV IE promoter (gracious gift of Adam Geballe, Fred Hutchinson Cancer Research Center [FHCRC], Seattle, Wash.), the simian virus 40 (SV40) minimal promoter (Promega), the human dihydrofolate reductase (DHFR) promoter (gracious gift of Peggy Farnham, University of Wisconsin, Madison), and the mouse phosphoglycerate kinase 1 (PGK-1) promoter (gracious gift of Mark Groudine, FHCRC) were cloned into the HindIII site upstream of the luciferase gene in the pGL3-Basic plasmid.
PCR (PWO; Boehringer Mannheim, Indianapolis, Ind.) was used to amplify either the −161 to −20 or the −81 to −20 LTR fragment (primer sequences are available upon request). The HIV-1 LTR containing mutations in all three Sp1 binding sites has been described previously (29); mutations in the TATA element which abolished binding to TATA box binding protein (TBP) (49) were generated by PCR-based oligonucleotide-directed mutagenesis using primers 5′-CTAGCAAAATAGGCTGTCCC-3′ (sense) and 5′-CCGAAGCTTGCAGCTGCTCACATGCAG-3′ (antisense). Combinatorial amplification strategies allowed us to generate LTRs with mutations in either the NF-κB or Sp1 transcription factor binding sites in concert with the TATA box mutant, which were then subsequently cloned into the SmaI-HindIII-cleaved pGL-3 Basic vector.
To create a luciferase expression plasmid that could be stably expressed in vivo, the XhoI-SalI fragment from plasmid pwtLTR2 (containing the luciferase gene open reading frame under the direction of the full-length HIV-1 LTR) was cloned into SalI-digested pCEP-4 (Invitrogen, San Diego, Calif.) to create the plasmid HIV-1 LTR-luc/CEP4. This plasmid also contained an EBNA-1 expression cassette for maintenance of the plasmid and a hygromycin resistance gene for selection of luciferase-expressing cells.
Plasmids plucBS and pGAPDHBS were used to generate probes for RNase protection assays (RPA). Plasmid plucBS was derived from the −81 to +80 HIV-1 LTR-luciferase plasmid by PCR using primers 5′-CTAGCAAAATAGGCTGTCCC-3′ (sense) and 5′-TCCGGCGCCATGTTCACCTCG-3′ (antisense) and cloned into the SmaI-SacII site of pBluescript KS+ (Stratagene, Calif.). Plasmid plucBS was linearized with EcoRI, from which RNase protection probes were synthesized by T7 RNA polymerase, which produce a 152-nucleotide probe and, after RNase protection, a 110-nucleotide protected product. Plasmid pGAPDHBS was derived from pHcGAP (American Type Culture Collection, Rockville, Md.), which contains the human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA. Plasmid pHcGAP was cleaved with XbaI and HindIII, and the resulting 554-bp fragment was cloned into XbaI-HindIII-digested pBluescript KS+ (Stratagene). RNase protection probes were generated by cleaving plasmid pGAPDHBS with HindIII. Runoff transcripts synthesized by T7 RNA polymerase produced a 590-nucleotide probe and, after RNase protection, a 554-nucleotide protected product.
Cells.
Jurkat and H9 cells were grown and maintained in RPMI medium supplemented with 10% fetal bovine serum, while HeLa cells were grown and maintained in Dulbecco’s modified Eagle medium supplemented with 10% calf serum. The polyclonal Jurkat-LTRluc cell line was constructed by stable transfection of the plasmid HIV-1 LTR-luc/CEP4 into Jurkat cells by electroporation. Briefly, 5 × 106 cells were electroporated with 10 μg of plasmid at 250 V and 960 μF. At 48 h posttransfection, cells were placed in medium containing 0.2 mg of hygromycin B per ml.
Peripheral blood was obtained from the Puget Sound Blood Center as standard buffy coat preparations from healthy donors. Peripheral blood mononuclear cells (PBMC) were prepared by centrifugation on a Ficoll-Hypaque (Sigma, St. Louis, Mo.) density gradient. Primary human CD4+ T lymphocytes were derived by incubating 107 PBMC with fluorescein isothiocyanate-conjugated anti-CD4 monoclonal antibody (Leu-3a; Becton Dickinson) and sorting for CD4+ T cells with a fluorescence-activated cell sorter; 2.5 × 105 sorted CD4+ T lymphocytes were then stimulated in vitro with γ-irradiated (3,000 rads) allogeneic PBMC, anti-CD3 (OKT3; 30 μg/ml) monoclonal antibody, and interleukin-2 (50 U/ml) for 10 days. Peripheral blood monocytes were isolated from fresh PBMC by adherence to plastic at 37°C as described previously (47). Mature macrophages were derived by culturing the monocytes for 14 days in RPMI–10% pooled human serum in 12-well plates at a density of 5 × 105 cells/well. At this time, most of the cells had acquired the characteristic enlarged, spindle-shaped morphology.
Viruses and infections.
Construction of high-titer vesicular stomatis virus G protein (VSV-G) pseudotypes of Vpr+ and Vpr− HIV Δenv proviruses has been previously described (6). The H71R-Vpr and E24G-Vpr mutants were constructed by swapping the SalI-BamHI fragment from proviruses expressing the mutant vpr alleles as described previously (47) into the HIVΔenv proviral backbone. Viral titers were determined by the MAGI assay (25). Target cells were infected with equal multiplicities of infection (MOI) of virus stock in medium containing DEAE-dextran (20 μg/ml; Sigma) at 37°C for 2 h with intermittent agitation. Cells were washed extensively at the end of the incubation period and placed in fresh medium. Cells were harvested intermittently during the infection protocol and stained with fluorescein isothiocyanate-conjugated mouse anti-p24gag monoclonal antibody to determine the number of productively infected cells. In addition, cells were stained with propidium iodide (PI) as described before (5) to determine the cell cycle profile of the infected p24gag+ cells.
Transfections.
For transient transfections of Jurkat and H9 cell lines, 7.5 × 105 cells were washed twice in serum-free medium and resuspended in 0.5 ml of serum-free OPTI-MEM (GIBCO-BRL) containing 1.8 μg of total DNA and 3.6 μl of DMRIE-C (GIBCO-BRL) reagent per well of a 12-well plate. For transfection of HeLa cells, 2 × 105 cells were transfected in serum-free Dulbecco’s modified Eagle medium containing 2 μg of total DNA and 5 μl of Lipofectamine (GIBCO-BRL) reagent per well of a six-well plate. Medium containing 15% fetal bovine serum was added to each of the transfected wells 6 h posttransfection. Primary CD4+ T lymphocytes were electroporated in RPMI–10% pooled human serum at a concentration of 2 × 107 per ml, using 10 μg of total DNA in 0.4-cm gap cuvettes at 960 μF and 250 V (11). Peripheral blood monocyte-derived macrophages were transfected with Lipofectamine-Plus (GIBCO-BRL) reagent. Briefly, 5 × 105 cells were washed twice with serum-free medium and transfected with 0.8 μg of total DNA, 2.25 μl of Lipofectamine-Plus reagent, and 4 μl of Lipofectamine reagent. Three hours later, the transfection mix was replaced with fresh medium (RPMI/10% human serum). All transfections were performed in triplicate. Cells were harvested 48 h posttransfection and lysed in luciferase lysis buffer (Promega). Lysed extracts were used for luciferase assay with an automated injection apparatus (AutoLumat; EG&G Berthold). The protein concentration of the extracts was determined by the Bradford method (Pierce, Rockford, Ill.).
Counterflow centrifugal elutriation.
Jurkat-LTRluc cells (109) were harvested prior to use by centrifugation for 5 min at 300 × g and resuspended in 4 ml of elutriation media (Tris [pH 7.0], 5 mM EDTA, 1% fetal bovine serum). The concentrated cells were passed slowly through an 18-gauge needle three times prior to loading to ensure single-cell separation. Cells were separated into populations of progressively increasing cell size in a Beckman J-6B centrifuge equipped with the JE-6B elutriation rotor (Beckman Instruments, Inc., Palo Alto, Calif.) and Masterflex model 7550-60 digital pump (Cole-Parmer Instrument Co., Chicago, Ill.) (13). Initially, cells were loaded at a rotor speed of 1,600 rpm and pump speed of 30 ml/min. Thereafter, cell fractions were collected in 250-ml volumes at 5-ml/min intervals and rotor speed decrements of 20 rpm. Cell cycle profile of cells in each of the elutriated fractions was determined by PI staining as described previously (5). Total cellular RNA from each elutriated fraction (5 × 106) was isolated by using the Triazol (GIBCO-BRL) reagent as described previously (18). In addition, luciferase activity of each elutriated fraction (106) was determined by lysing cells in luciferase lysis buffer (Promega). Lysed extract was then used for luciferase assay with an automated injection apparatus (AutoLumat; EG&G Berthold).
RNase protection.
Total cellular RNA (2 to 10 μg) from cells in each elutriated fraction was dried under vacuum, resuspended in 10 μl of 1× hybridization buffer {80% formamide, 20 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid); pH 6.8], 20 mM sodium chloride, 0.5 mM EDTA} containing [α-32P]CTP-labeled RNA probe (2 × 104 cpm/μl), heated at 90°C for 1 min, and then incubated at 56°C for 12 to 16 h. The hybridization mixture was cooled to 37°C for 15 min, 100 μl of RNase digestion buffer (10 mM Tris-HCl [pH 7.6], 300 mM sodium chloride, 5 mM EDTA, 0.2 μg of RNase A/μl, 0.6 U of RNase T1/μl) was added, and the mixture was incubated at 30°C for 45 min. The RNase digestion was terminated by addition of sodium dodecyl sulfate and proteinase K to final concentrations of 0.1% and 100 μg/ml, respectively. The reaction mix was incubated for 15 min at 37°C and extracted once with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1). The RNA was precipitated in ethanol after the addition of ammonium acetate to 2 M and the addition of 30 μg of yeast tRNA. The RNA pellet was resuspended in 5 μl of 1× RNA loading buffer (95% formamide, 10 mM EDTA, 0.1% bromophenol blue, 0.1% xylene cyanol), heated for 2 to 3 min at 90°C, and resolved via denaturing polyacrylamide gel electrophoresis.
RESULTS
Vpr transactivates the HIV-1 LTR in transformed cell lines and in primary T cells in a dose-dependent fashion.
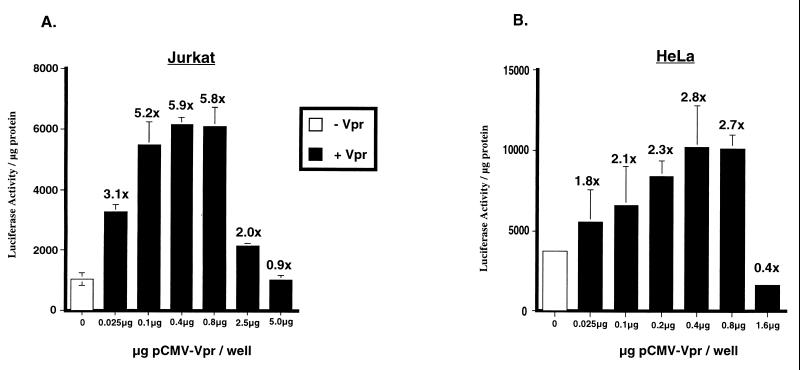
We and others have previously reported that HIV-1 Vpr mediates enhanced expression from the HIV-1 LTR in Jurkat T cells (9, 15, 18). In contrast, there are reports in the literature which suggest that Vpr is a repressor of transcription (4). In a transient transfection assay, we observed an increase in LTR activity in the presence of Vpr that was dependent on the amount of Vpr present in the cell and is evident from the dose-response curves generated in Jurkat and HeLa cells (Fig. 1A and B, respectively). Maximal induction was achieved when 0.6 μg of Vpr expression plasmid was used in Jurkat cells, and 0.2 μg of Vpr was used in HeLa cells. Use of higher levels of Vpr (greater than 1.0 μg) was deleterious to the cells, was associated with increased incidence of cell death (data not shown), and caused a precipitous drop in luciferase activity. The drop in luciferase activity correlates with the toxicity of Vpr in these cells and may explain why Vpr appears to be a repressor when expressed at high levels (4).
FIG. 1.
Dose-dependent increase in HIV-1 LTR-driven luciferase activity. Jurkat T (A) and HeLa (B) cells were transfected with a constant amount of plasmid pwtLTR-luc2 (0.625 and 0.2 μg, respectively) and indicated amounts of plasmid pCMV-Vpr. The empty CMV expression vector was used such that each sample received an equivalent amount of the expression plasmid. A representative experiment is shown for each cell line, with fold increase in luciferase activity obtained in the presence of Vpr over the basal level of the reporter alone indicated above the histograms. Experiment was repeated three independent times with similar results. Standard errors of means are denoted by error bars.
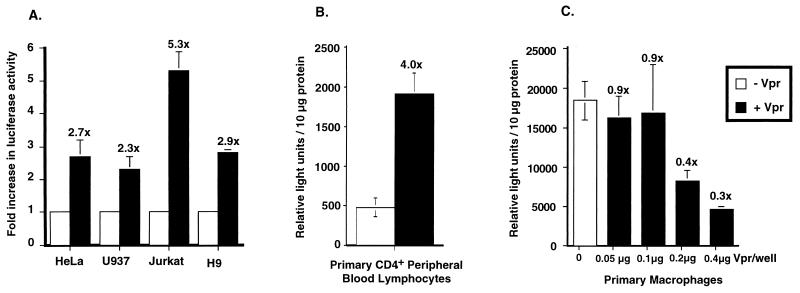
When introduced at optimum levels (Fig. 1), Vpr was an activator of HIV-1 LTR-driven transcription in each of the cell lines tested (Fig. 2A), resulting in a 2.5- to 5-fold increase in luciferase activity in the presence of Vpr. The ability of Vpr to transactivate the LTR was further examined in more relevant targets of HIV infection such as primary human CD4+ T cells and primary human macrophages. Buffy coats from healthy, HIV-seronegative donors were the source for both CD4+ T cells and terminally differentiated macrophages. Primary CD4+ T cells were electroporated with the luciferase expression plasmid (pwtLTR2) and a Tat expression plasmid, in the presence or absence of the Vpr expression plasmid (pHIV-Vpr). Tat expression plasmid was used in these transfections because the HIV-1 LTR-driven luciferase activity in primary cells in the absence of Tat was not significantly above background levels (data not shown). The presence of Vpr resulted in a three- to fourfold increase in LTR-driven luciferase activity (Fig. 2B).
FIG. 2.
Vpr mediates enhanced expression from HIV-1 LTR in transient transfections of transformed cell lines and primary human CD4+ T cells. (A) The following cell types were transiently transfected with pwtLTR-luc1 in the presence or absence of expression plasmid pCMV-Vpr by transfection protocols described in Materials and Methods: HeLa (epithelial), U937 (promonocytic), Jurkat (T cell), and H9 (T cell). HeLa cells were transfected with 0.2 μg of expression plasmids pwtLTR-luc2 and 0.2 μg of pCMV-Vpr. Jurkat, U937, and H9 cells were transfected with 0.625 μg of plasmid pwtLTR-luc2 and 0.625 μg of plasmid pCMV-Vpr (equivalent amount of empty Vpr expression vector was used as a control). The values of all transfections performed in the absence of Vpr are normalized to 1, to account for variations in transfection efficiency between different cells, and the fold increase in luciferase activity obtained in the presence of Vpr is noted above the shaded histograms. Values presented are the averages of three or more experiments. (B) Primary CD4+ T cells were electroporated with 5 μg of pwtLTR-luc2, 0.5 μg of pCMV-Tat, and 4.5 μg of pHIV-Vpr, while primary macrophages (C) were transfected with 0.2 μg of pwtLTR2 and indicated amounts of pHIV-Vpr. Primary cell transfections were performed on cells isolated from at least three independent donors. A representative experiment from three independent trials, each performed with different donor cells, is shown. Cells in all transfections were harvested 48 h posttransfection; lysates were assayed for luciferase activity as described in Materials and Methods and normalized to the protein content in each lysate. Standard errors of means are denoted by the error bars.
When similar transfections were carried out in terminally differentiated macrophages, cultured for 14 days prior to transfection, no increase in luciferase activity was observed in the presence of Vpr (Fig. 2C). In fact, cotransfections of the luciferase expression plasmid (pwtLTR2) with increasing amounts of the Vpr expression plasmid (pHIV-Vpr) resulted in a dose-dependent decrease in luciferase activity (Fig. 2C). These experiments in primary cells corroborate the transfection data obtained from transformed cell lines and provide evidence that Vpr can act as an activator of transcription in primary CD4+ T cells but not in differentiated macrophages.
Ability of Vpr to increase viral replication correlates with G2 arrest.
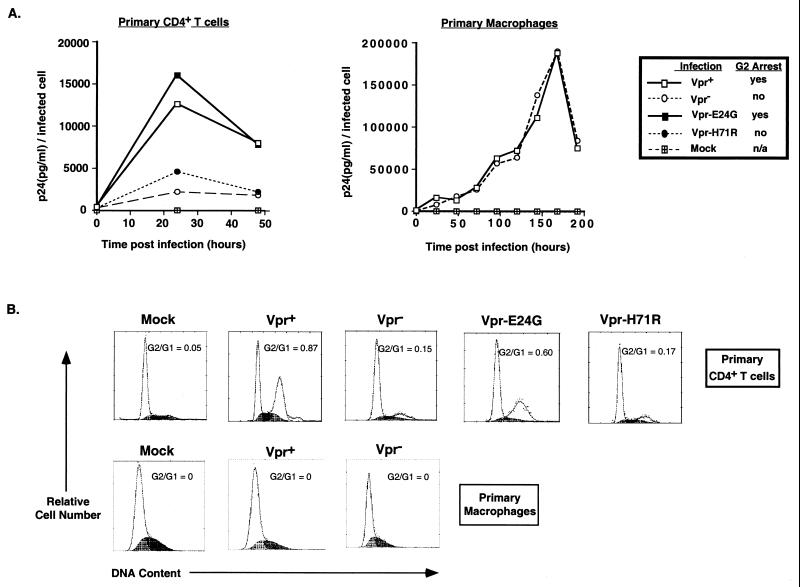
We also tested the effects of Vpr on viral replication in a single cycle of infection in primary human CD4+ T cells and terminally differentiated monocyte-derived macrophages. HIV Δenv proviruses, either lacking Vpr (Vpr−) or expressing wild-type Vpr (Vpr+), E24G-Vpr (able to cause G2 arrest), or H71R-Vpr (deficient for G2 arrest phenotype) were pseudotyped with VSV-G (6) and used to infect primary CD4+ T cells and macrophages at equal MOI. Infected cultures were periodically assessed for p24gag antigen release to measure viral replication; at the same time, the number of cells that were productively infected in the culture was determined by staining with a mouse anti-p24gag monoclonal antibody. Finally, the primary CD4+ T cells were harvested 48 h postinfection, and infected cells (p24gag positive) were assayed for cell cycle profile via PI staining.
Viruses that are able to cause G2 arrest in primary T cells (Vpr+ and E24G-Vpr) replicated to three- to fourfold-higher levels than the Vpr− or H71R-Vpr-expressing provirus neither of which is able to cause G2 arrest (Fig. 3). This increase in viral replication was seen within 24 h of infection, suggesting that Vpr has an early effect on viral replication. This correlates with the G2 arrest that is observed within 24 h of infection in cells infected with the HIV Δenv Vpr+ or E24G-Vpr provirus (data not shown). In contrast, the primary macrophages are primarily in the G1 phase of the cell cycle and are not induced to progress in the cell cycle as a consequence of infection with either the Vpr+ or Vpr− provirus (Fig. 3B). Even though the number of productively infected cells was higher in cultures infected with the HIV Δenv Vpr+ virus compared to cultures infected with HIV Δenv Vpr− virus (data not shown), there is no significant difference in the amount of viral production per infected cell at all times postinfection (Fig. 3A). Thus, the ability of Vpr to mediate enhanced expression from the HIV-1 LTR in CD4+ T cells, and to increase viral replication in a single round of infection, correlates with its ability to cause G2 arrest in CD4+ T cells (Fig. 2B and 3B).
FIG. 3.
Vpr-mediated G2 cell cycle arrest results in higher levels of viral replication in single round of infections of primary human CD4+ T cells. (A) Primary human CD4+ T cells were mock infected or infected with HIVΔenv VSV-G pseudotypes that expressed either wild-type Vpr (Vpr+), E24G-Vpr, H71R-Vpr, or no Vpr (Vpr−) at an MOI of 0.5. Primary human macrophages were mock infected or infected with HIVΔenv VSV-G pseudotypes that expressed either wild-type Vpr (Vpr+) or no Vpr (Vpr−) at an MOI of 0.5. The viruses contained a deletion in env to prevent multiple rounds of infection. Culture supernatants were removed at indicated times for measurement of p24gag secreted into the medium. The amount of p24gag (y axis) present in the culture supernatants is normalized to the number of infected cells as determined by staining for intracellular p24gag expression. (B) Primary human CD4+ T cells were harvested 48 h postinfection, while macrophages were harvested 192 h postinfection and analyzed for DNA content by PI staining of the nuclei (x axis, PI fluorescence intensity for DNA content; y axis, relative cell number). Flow cytometric profiles depicted are PI staining obtained for productively infected cells alone, as determined by staining for intracellular p24gag expression. The G2/G1 ratio for each of the infected cell population is indicated. A representative experiment from three independent trials, each performed with different donor cells, is shown.
LTR-driven expression is highest in the G2 phase of the cell cycle.
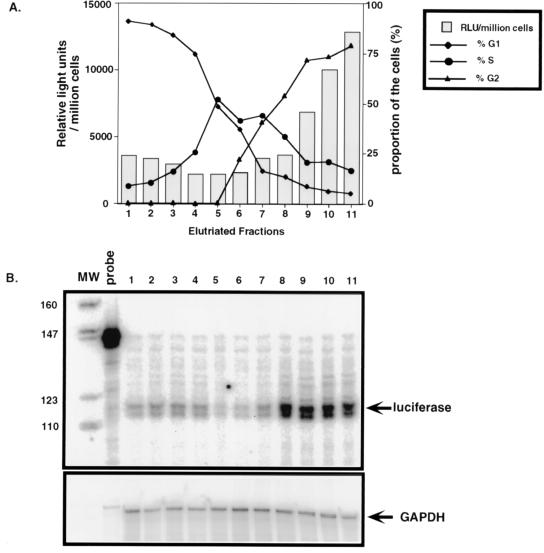
Our previous studies with cells chemically arrested in the cell cycle showed that the HIV LTR was most active in the G2 phase of the cell cycle (18). However, we wished to examine this in cells that were not drug treated. Therefore, a Jurkat cell line stably expressing luciferase under the control of the HIV-1 LTR was constructed (Jurkat-LTR-luc) and subjected to counterflow centrifugal elutriation, which separates cells on the basis of increasing cell size and mass (13). This allows the in vivo analysis of HIV-1 LTR-driven transcription in T cells in distinct phases of the cell cycle without the use of potentially disruptive cell cycle-arresting agents. Because separation is achieved in media, the procedure is noninvasive and does not perturb cell growth or metabolism, and the characteristics of the resultant populations closely resemble those of normal cycling cells (13).
Figure 4A shows the results of such an analysis. The majority of elutriated Jurkat-LTR-luc cells in fractions 1 to 3 are in G1, while cells in fractions 4 to 6 are in G1/S. Cells in fraction 7 are just beginning to enter G2, with the remaining fractions, 8 to 11, containing mostly G2/M cells. When cell lysates generated from equal numbers of cells in each phase of the cell cycle were assayed for luciferase activity, we found a reproducible three- to fourfold increase in luciferase activity in fractions containing cells mostly in G2 (fractions 8 through 11) compared to fractions containing cells mostly in G1 or S phase.
FIG. 4.
HIV-1 LTR is transcriptionally most responsive in the G2 phase of the cell cycle. (A) A total of 109 Jurkat-LTR-luc cells were separated by counterflow centrifugal elutriation into different phases of the cell cycle. The relative DNA content of each elutriated fraction was determined by PI staining of nuclei, and the proportion of cells in each phase of the cell cycle (G1, S, and G2/M phases) is indicated (right-hand y axis). Luciferase activity for each elutriated fraction determined from lysates of 106 cells from each fraction, expressed in relative light units (RLU), is represented by the histograms (left-hand y axis). (B) RPA of luciferase RNA from Jurkat-LTR-luc cells that were elutriated into different phases of the cell cycle. Total RNA isolated from 5 × 106 cells of each elutriated fraction was used for RPA. Each RPA reaction contained either 8 μg of total RNA hybridized to 2 × 105 cpm of the single-stranded luciferase RNA probe (top panel) or 2 μg of total RNA hybridized to 2 × 105 cpm of the single-stranded GAPDH RNA probe (bottom panel). The numbers on top of the lanes indicate the elutriated fraction number. MW, molecular weight standards. Sizes (in base pairs) are shown on the left.
To confirm that this increase in luciferase activity correlates with an increase in luciferase RNA levels, RPA with labeled antisense luciferase riboprobes was performed on total RNA isolated from cells in each elutriated fraction (Fig. 4B). As a control, expression of a cellular gene, GAPDH, was similarly assessed. The results of these assays demonstrate that luciferase RNA levels are markedly higher in the G2 phase (Fig. 4B, lanes 8 and 9) than in any other phase of the cell cycle (lanes 1 to 3). Levels of luciferase RNA were normalized to GAPDH RNA levels in these extracts, revealing a 3- to 3.5-fold increase in luciferase RNA in fractions containing cells predominantly in G2, compared to cells in G1, and a 4.5- to 5-fold increase over luciferase RNA levels seen in G1/S fractions. Therefore, the changes in luciferase protein levels (luciferase enzymatic activity (Fig. 4A) are roughly proportional to changes in the levels of luciferase RNA in these cells. Together these results indicate that transcripts driven by the HIV-1 LTR are most abundant in the G2 phase of the cell cycle.
LTR sequence requirements for Vpr-mediated transactivation.
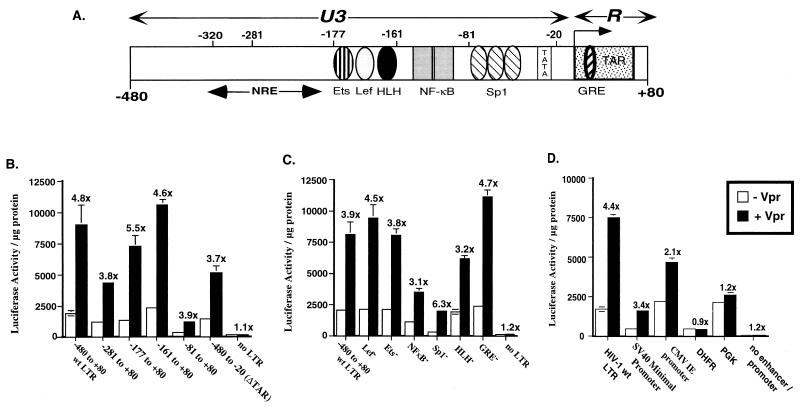
To examine the sequences of the LTR responsible for modulation by Vpr, we constructed a series of mutations (both deletions and point mutations) in the LTR and assayed their ability to be upregulated in the presence of Vpr in Jurkat cells. Initially, exonuclease III digests were performed on plasmid pwtLTR1 to generate a series of nested deletions in the LTR-luciferase clones. In addition, site-directed mutagenesis was carried out to abolish binding of individual transcription factors in the context of the full-length LTR. These LTR-luciferase plasmids were cotransfected into Jurkat cells in the presence or absence of Vpr. Removal of either the negative regulatory element (−320 to −177 of the LTR), the enhancer region (−177 to −81), or the TAR element (−20 to +80) had no effect on Vpr-mediated transactivation of HIV-1 LTR (Fig. 5B). Though decreases in the basal expression from the truncated LTR compared to the full-length LTR were observed, the −81 to +80 region of the LTR was sufficient for Vpr-mediated induction (fourfold increase [Fig. 5B]). In addition, mutations of individual transcription factor binding sites (Ets, Lef, NF-κB, Sp1, and GRE transcription factor binding sites) in the context of the full-length LTR had no effect on the ability of Vpr to stimulate LTR activation (Fig. 5C). These results suggest that a 61-bp LTR fragment (−81 to −20), containing the Sp1 transcription factor binding sites and the TATA box, is sufficient for the Vpr-mediated enhancement of HIV-1 LTR-driven expression.
FIG. 5.
Delineation of HIV-1 LTR sequence requirements for Vpr-mediated stimulation of the HIV-1 LTR. (A) Schematic representation of the HIV-1 LTR. Positions of the transcription factor binding sites that were assayed in the transfection experiments, the TATA box, and the TAR element are marked. NRE, negative regulatory element. Jurkat cells were cotransfected with 0.625 μg of expression plasmid pCMV-Vpr and 0.625 μg of either sequentially deleted LTRs (B); LTRs with mutations in the individual transcription factor binding sites including Lef, Ets, NF-κB, Sp1, helix-loop-helix (HLH) protein binding motif, and GRE (C); or viral (HIV-1 LTR, SV40 minimal promoter, and CMV IE promoter) and cellular (human DHFR and mouse PGK-1) promoters driving luciferase expression (D). Names of the plasmids in panel B identify the portion of the HIV-1 LTR included in each construct, and the nucleotide positions are denoted with respect to the +1 transcription start site. In all transfections, the empty CMV expression plasmid was cotransfected with each reporter plasmid as a control. Luciferase assays were performed on cell lysates harvested 48 h posttransfection, and results were expressed as luciferase activity in relative light units per microgram of protein (y axis). Fold increase in luciferase activity obtained in the presence of Vpr over the luciferase activity obtained in the absence of Vpr is indicated above the histograms, and error bars represent standard errors of mean. Reported values are means of transfections performed in triplicate. The experiment has been repeated at least three times with similar results. wt, wild type.
Vpr has been previously shown to transactivate several heterologous promoters (3, 9). Not surprisingly, Vpr was able to mediate enhanced expression from the SV40 minimal promoter/enhancer (3.4-fold increase) and, more modestly, the CMV IE promoter (2.1-fold increase), both of which contain, in addition to multiple transcription factor binding sites, an intact TATA box (Fig. 5D). However, this increase in expression in presence of Vpr was not observed following transient transfections with luciferase expression plasmids where luciferase expression was under the control of either the human DHFR promoter (DHFR-luc) or the mouse PGK-1 promoter (mPGK-luc) (Fig. 5D). As is typical of most housekeeping genes, DHFR and PGK-1 promoter regions are GC rich but lack a TATA box (2, 40). These results suggest that a functional TATA box might be a sequence requirement for the Vpr-mediated enhancement of expression.
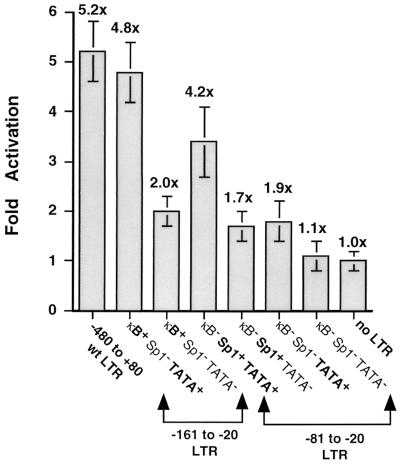
To investigate the requirement of the TATA element, a second series of mutations was generated in the −81 to +80 LTR-luciferase clone and subcloned into the pGL3 Basic vector (Fig. 6). PCR was used to selectively amplify the −161 to −20 LTR region, which contains the NF-κB and Sp1 transcription factor binding sites, and the TATA box or the −81 to −20 LTR region, which contains the TATA box and the Sp1 transcription factor binding sites. The TATA box (TATAA) was mutated by site-directed mutagenesis to TGTGA, a mutation that has previously been shown to abolish binding to TBP (49). Though deletion of the NF-κB binding sites alone (Sp1+ TATA+ clone; fourfold increase) and mutation of the Sp1 binding sites by themselves (κB+ Sp1− TATA+ clone; fivefold increase) had negligible effect on Vpr-mediated induction of LTR activity, mutations in the TATA box eliminated most of the enhancement (a twofold increase was observed) (Fig. 5E). Mutation of both NF-κB and Sp1 (κB− Sp1− TATA+) binding sites also abrogated most of the transcriptional enhancement (1.7-fold increase), suggesting that an enhancer element (either Sp1 or NF-κB) in addition to a functional TATA box is required for the Vpr-mediated upregulation of HIV-1 LTR-driven transcriptional activity. This suggests that binding of the TBP to the TATA box and subsequent assembly of the basal RNA polymerase II transcription apparatus on the HIV-1 LTR is one of the functional requirements for Vpr to exert its G2-specific effect on the HIV-1 LTR.
FIG. 6.
TATA box requirement for Vpr-mediated stimulation of the HIV-1 LTR. Jurkat cells were transfected with either the 141-bp fragment of the LTR (−161 to −20), containing the NF-κB and Sp1 transcription factor binding sites, and the TATA box or the 61-bp fragment of the LTR (−81 to −20), containing the Sp1 transcription factor binding sites and the TATA box, in the presence or absence of expression plasmid pCMV-Vpr. In all transfections, the empty CMV expression plasmid was cotransfected with each reporter plasmid as a control. The values of all transfections performed in the absence of Vpr are normalized to 1, and results are reported as fold increase in luciferase activity (indicated above the histograms) obtained in the presence of Vpr for each reporter plasmid (y axis). Reported values are means of at least three independent transfections, each performed in triplicate, and error bars represent standard errors of mean. wt, wild type.
DISCUSSION
Experiments presented in this study were designed to test the hypothesis that Vpr-mediated activation of the HIV-1 LTR is a consequence of the ability of Vpr to cause G2 arrest. First, counterflow centrifugal elutriation was used to separate cells in different phases of the cell cycle and analyze the cell cycle dependence of HIV-1 LTR-driven transcription. These experiments demonstrated that the HIV-1 LTR-driven expression is maximal in the G2 phase of the cell cycle. Furthermore, results from the transient transfections showed that the effect of Vpr on activation of the LTR is general and was observed both in transformed cell lines and in primary human CD4+ T cells. Importantly, in primary CD4+ T cells, the observed Tat induction in the presence of Vpr was greater than the enhancement observed in the absence of Vpr. Moreover, we provide evidence that the G2 arrest function of Vpr is important for high levels of viral replication in primary human CD4+ T cells. Finally, we show that a minimal promoter is all that is required to observe the Vpr-mediated augmentation of expression.
Previous reports have indicated that an interaction between Vpr and the Sp1 transcription factor is required for Vpr-mediated transcriptional enhancement of the HIV-1 LTR (3, 39, 48). Our results suggest that Sp1 binding to the LTR is not a necessary event for the Vpr-mediated augmentation of LTR activity (Fig. 5C and 6). It has also been reported that Vpr transactivation requires the NF-κB sites and the TATA box and that this induction is dependent on the ability of Vpr to stimulate p300/CBP coactivator function (15). It was also reported that Vpr-mediated induction of HIV-1 LTR activity in turn can be repressed by the adenovirus E1A gene product by its ability to bind and inhibit p300/CBP (15). Nonetheless, we find that neither mutation of both NF-κB binding sites in the context of a full-length LTR (NF-κB− construct [Fig. 5C]) nor deletion of the enhancer region which contains the NF-κB binding sites (−81 to +80 clone [Fig. 5B and 6]) has an effect on the ability of Vpr to stimulate transcription. These results are also consistent with previous reports in which HIV-1 LTRs spanning the region −80 to +80 and lacking NF-κB binding sites were observed to be sufficient for transactivation by Vpr in transient transfection assays (3, 39).
On the other hand, our results suggest that a minimal fragment of the HIV-1 promoter containing a functional TATA box motif and an enhancer element (either NF-κB or Sp1) is sufficient for LTR induction (Fig. 5 and 6) and that binding of the TBP to the TATA box is the necessary event for Vpr-mediated induction of HIV-1 LTR activity. In support of this hypothesis is the observation that the adenovirus E1A protein can repress core promoters alone by interacting with TBP directly, without the requirement for specific upstream enhancer elements (43). Thus, the previously observed dose-dependent increases in HIV transcription by p300/CBP protein (15) could be related to its ability to interact directly with the TFIID component of the RNA polymerase II basal transcription complex, which could result in enhanced binding of TBP to the TATA box (1, 26). Hence, there is no specificity for enhancer elements such as Sp1 and NF-κB in Vpr-mediated enhancement of HIV-1 LTR. Nonetheless, it is possible that LTR sequences in the immediate vicinity of the TATA box could provide the specificity for the Vpr-mediated enhancement of transcription.
The data from our studies with the cell line constitutively expressing luciferase under the control of the HIV-1 LTR confirm that LTR-driven expression is maximal in the G2 phase of the cell cycle (Fig. 4) and is largely, if not entirely, due to cell cycle-specific regulation of the basal transcriptional machinery. It is also possible that there is accumulation of luciferase RNA or that the luciferase RNA is more stable in G2. Previous studies have shown that Vpr arrests cells in the G2 phase of the cell cycle by preventing the activation of the p34cdc2 kinase (5, 20, 38). Interestingly, it has been reported that RNA polymerase II transcription in vitro by using a template containing only a TATA box and an enhancer element can be inhibited by the activated p34cdc2 kinase (27). Potential inhibitory targets of the mitotic p34cdc2 kinase include TBP, and other components of the basal transcription machinery, resulting in premature termination of polymerase II transcription as the cell enters mitosis (19, 41). It is tempting to speculate that Vpr by inhibiting p34cdc2 kinase activity is indirectly promoting RNA polymerase II transcription in vivo. Hence, the ability of Vpr to mediate increased expression from the HIV-1 LTR is likely indirect and, by Vpr arresting cells in the G2 phase of the cell cycle, provides an environment that is most conducive to HIV-1 promoter function.
The consequence of the Vpr-mediated G2 arrest and the subsequent LTR-driven transcription enhancement is clearly evident in the single round of infection experiments in primary human CD4+ T cells (Fig. 3A). In contrast, there was no observed increase in HIV-1 LTR-driven luciferase expression in transient transfections nor any significant increase in levels of viral replication in a single round of infection in terminally differentiated human macrophages in the presence of Vpr. We hypothesize that this result is a consequence of the fact that the macrophages are not cycling and hence are not subject to the G2 arrest function of Vpr (Fig. 3B). Rather, the nuclear import function of Vpr is crucial for establishment of productive infection in macrophages (17, 36, 37, 47).
The increase in p24gag levels observed within 24 h postinfection in the presence of Vpr suggests a role for the large amounts of virion associated Vpr during the early stages of viral life cycle. Since it has been reported that virion-associated Vpr can cause a G2 cell cycle arrest in T-cell lines within a few hours of infection (35), it is tempting to speculate that Vpr provides a cellular environment that is most conducive for efficient transcription from the HIV-1 LTR during the early stages of the viral life cycle to allow for production of essential viral proteins such as the Tat and Rev proteins. Arrest of the cell cycle in a state that fosters viral replication by increasing the availability of transcription factors, replication factors, and nucleotide pools is a common mechanism used by evolutionarily diverse pathogens, such as the herpesviruses (human CMV and Epstein-Barr virus) (8, 12), parvoviruses (Aleutian mink disease virus and minute virus of mice) (7, 32), and small DNA tumor viruses (SV40, human papillomavirus, and adenovirus) (28, 31). Our findings lead to the conclusion that Vpr-induced G2 arrest of the infected cell represents an important strategy for HIV-1 to promote its own replication.
ACKNOWLEDGMENTS
We thank P. Farnham (University of Wisconsin, Madison) for the DHFR promoter, R. Gaynor (University of Texas Southwestern Medical Center, Dallas) for the Sp1− mutant LTRs, A. Geballe (FHCRC, Seattle, Wash.) for the CMV IE promoter, K. Jones (Salk Institute, San Diego, Calif.) for the Lef, Ets, and GRE mutant LTRs, M. Groudine (FHCRC) for the mouse PGK-1 promoter, and S. Dewhurst (University of Rochester, Rochester, N.Y.) for the NF-κB− mutant LTR. We acknowledge L. Breeden (FHCRC) for the use of centrifugal elutriator; M. Linial, S. Bartz, W. C. Goh, M. Vodicka, and M. Kinsey for their help and insightful comments on the manuscript; and the FHCRC Flow Cytometry, Image Analysis, and Biotechnology laboratories for help with the figures.
This work was supported by NIH grant R01 AI30927.
REFERENCES
- 1.Abraham S E, Lobo S, Yaciuk P, Wang H H, Moran E. p300, and p300-associated proteins, are components of TATA-binding protein (TBP) complexes. Oncogene. 1993;8:1639–1647. [PubMed] [Google Scholar]
- 2.Adra C N, Boer P H, McBurney M W. Cloning and expression of the mouse pgk-1 gene and the nucleotide sequence of its promoter. Gene. 1987;60:65–74. doi: 10.1016/0378-1119(87)90214-9. [DOI] [PubMed] [Google Scholar]
- 3.Agostini I, Navarro J M, Rey F, Bouhamdan M, Spire B, Vigne R, Sire J. The human immunodeficiency virus type 1 Vpr transactivator: cooperation with promoter-bound activator domains and binding to TFIIB. J Mol Biol. 1996;261:599–606. doi: 10.1006/jmbi.1996.0485. [DOI] [PubMed] [Google Scholar]
- 4.Ayyavoo V, Mahboubi A, Mahalingam S, Ramalingam R, Kudchodkar S, Williams W V, Green D R, Weiner D B. HIV-1 Vpr suppresses immune activation and apoptosis through regulation of nuclear factor kappa B. Nat Med. 1997;3:1117–1123. doi: 10.1038/nm1097-1117. [DOI] [PubMed] [Google Scholar]
- 5.Bartz S R, Rogel M E, Emerman M. Human immunodeficiency virus type 1 cell cycle control: Vpr is cytostatic and mediates G2 accumulation by a mechanism which differs from DNA damage checkpoint control. J Virol. 1996;70:2324–2331. doi: 10.1128/jvi.70.4.2324-2331.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartz S R, Vodicka M A. Production of high-titer human immunodeficiency virus type 1 pseudotyped with vesicular stomatitis virus glycoprotein. Methods Enzymol. 1997;12:337–342. doi: 10.1006/meth.1997.0487. [DOI] [PubMed] [Google Scholar]
- 7.Beeck A O D, Anouja F, Mousset S, Rommelaere J, Caillet-Fauquet P. The nonstructural proteins of the autonomous parvovirus minute virus of mice interfere with the cell cycle, inducing accumulation in G2. Cell Growth Differ. 1995;6:781–787. [PubMed] [Google Scholar]
- 8.Cayrol C, Flemington E K. The Epstein-Barr virus bZIP transcription factor Zta causes G0/G1 cell cycle arrest through induction of cyclin-dependent kinase inhibitors. EMBO J. 1996;15:2748–2759. [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen E A, Dehni G, Sodroski J G, Haseltine W A. Human immunodeficiency virus vpr product is a virion-associated regulatory protein. J Virol. 1990;64:3097–3099. doi: 10.1128/jvi.64.6.3097-3099.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connor R I, Chen B K, Choe S, Landau N R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 11.Cron R Q, Schubert L A, Lewis D B, Hughes C C W. Consistent transient transfection of DNA into non-transformed human and murine T-lymphocytes. J Immunol Methods. 1997;205:145–150. doi: 10.1016/s0022-1759(97)00065-3. [DOI] [PubMed] [Google Scholar]
- 12.Dittmer D, Mocarski E S. Human cytomegalovirus infection inhibits G1/S transition. J Virol. 1997;71:1629–1634. doi: 10.1128/jvi.71.2.1629-1634.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donaldson K L, McShea A, Wahl A F. Separation by counterflow centrifugal elutriation and analysis of T- and B-lymphocytic cell lines in progressive stages of cell division cycle. J Immunol Methods. 1997;203:25–33. doi: 10.1016/s0022-1759(97)00011-2. [DOI] [PubMed] [Google Scholar]
- 14.Emerman M. HIV-1, Vpr and the cell cycle. Curr Biol. 1996;6:1096–1103. doi: 10.1016/s0960-9822(02)00676-0. [DOI] [PubMed] [Google Scholar]
- 15.Felzien L K, Woffendin C, Hottiger M O, Subbramanian R A, Cohen E A, Nabel G J. HIV transcriptional activation by the accessory protein, VPR, is mediated by the p300 co-activator. Proc Natl Acad Sci USA. 1998;95:5281–5286. doi: 10.1073/pnas.95.9.5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fletcher T M, III, Brichacek B, Sharova N, Newman M A, Stivahtis G, Sharp P M, Emerman M, Hahn B H, Stevenson M. Nuclear import and cell cycle arrest functions of the HIV-1 Vpr protein are encoded by two separate genes in HIV-2/SIV(SM) EMBO J. 1996;15:6155–6165. [PMC free article] [PubMed] [Google Scholar]
- 17.Gallay P, Stitt V, Mundy C, Oettinger M, Trono D. Role of the karyopherin pathway in human immunodeficiency virus type 1 nuclear import. J Virol. 1996;70:1027–1032. doi: 10.1128/jvi.70.2.1027-1032.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goh W C, Rogel M E, Kinsey C M, Michael S F, Fultz P N, Nowak M A, Hahn B H, Emerman M. HIV-1 Vpr increases viral expression by manipulation of the cell cycle: a mechanism for selection of Vpr in vivo. Nat Med. 1998;4:65–71. doi: 10.1038/nm0198-065. [DOI] [PubMed] [Google Scholar]
- 19.Gottesfeld J M, Forbes D J. Mitotic repression of the transcriptional machinery. Trends Biochem Sci. 1997;22:141–146. doi: 10.1016/s0968-0004(97)01045-1. [DOI] [PubMed] [Google Scholar]
- 20.He J, Choe S, Walker R, Di Marzio P, Morgan D O, Landau N R. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J Virol. 1995;69:6705–6711. doi: 10.1128/jvi.69.11.6705-6711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinzinger N K, Bukinsky M I, Haggerty S A, Ragland A M, Kewalramani V, Lee M A, Gendelman H E, Ratner L, Stevenson M, Emerman M. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones K A, Luciw P A, Duchange N. Structural arrangements of transcription control domains within the 5′-untranslated leader regions of the HIV-1 and HIV-2 promoters. Genes Dev. 1988;2:1101–1114. doi: 10.1101/gad.2.9.1101. [DOI] [PubMed] [Google Scholar]
- 23.Jones K A, Peterlin B. Control of RNA initiation and elongation at the HIV-1 promoter. Annu Rev Biochem. 1994;63:717–743. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- 24.Jowett J B, Planelles V, Poon B, Shah N P, Chen M L, Chen I S. The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J Virol. 1995;69:6304–6313. doi: 10.1128/jvi.69.10.6304-6313.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J Virol. 1992;66:3026–3031. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee W S, Kao C C, Bryant G O, Liu X, Berk A J. Adenovirus E1A activation domain binds the basic repeat in the TATA box transcription factor. Cell. 1991;67:365–376. doi: 10.1016/0092-8674(91)90188-5. [DOI] [PubMed] [Google Scholar]
- 27.Leresche A, Wolf V J, Gottesfeld J M. Repression of RNA polymerase II and III transcription during M phase of the cell cycle. Exp Cell Res. 1996;229:282–288. doi: 10.1006/excr.1996.0373. [DOI] [PubMed] [Google Scholar]
- 28.Moran E. DNA tumor virus transforming proteins and the cell cycle. Curr Opin Genet Dev. 1993;3:63–70. doi: 10.1016/s0959-437x(05)80342-9. [DOI] [PubMed] [Google Scholar]
- 29.Moses A V, Ibanez C, Gaynor R, Ghazal P, Nelson J A. Differential role of long terminal repeat control elements for the regulation of basal and Tat-mediated transcription of the human immunodeficiency virus in stimulated and unstimulated primary human macrophages. J Virol. 1994;68:298–307. doi: 10.1128/jvi.68.1.298-307.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nabel G, Baltimore D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature. 1987;326:711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- 31.Nevins J R. Adenovirus E1A: transcription regulation and alteration of cell growth control. Curr Top Microbiol Immunol. 1995;199:25–32. doi: 10.1007/978-3-642-79586-2_2. [DOI] [PubMed] [Google Scholar]
- 32.Oleksiewicz M B, Alexandersen S. S-phase-dependent cell cycle disturbances caused by Aleutian mink disease parvovirus. J Virol. 1997;71:1386–1396. doi: 10.1128/jvi.71.2.1386-1396.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paxton W, Connor R I, Landau N R. Incorporation of Vpr into human immunodeficiency virus type 1 virions: requirement for the p6 region of gag and mutational analysis. J Virol. 1993;67:7229–7237. doi: 10.1128/jvi.67.12.7229-7237.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Planelles V, Jowett J B, Li Q X, Xie Y, Hahn B, Chen I S. Vpr-induced cell cycle arrest is conserved among primate lentiviruses. J Virol. 1996;70:2516–2524. doi: 10.1128/jvi.70.4.2516-2524.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poon B, Grovit-Ferbas K, Stewart S A, Chen I S Y. Cell cycle arrest by Vpr in HIV-1 virions and insensitivity to antiretroviral agents. Science. 1998;281:266–269. doi: 10.1126/science.281.5374.266. [DOI] [PubMed] [Google Scholar]
- 36.Popov S, Rexach M, Ratner L, Blobel G, Bukrinsky M. Viral protein R regulates docking of the HIV-1 preintegration complex to the nuclear pore complex. J Biol Chem. 1998;273:13347–13352. doi: 10.1074/jbc.273.21.13347. [DOI] [PubMed] [Google Scholar]
- 37.Popov S, Rexach M, Zybarth G, Reiling N, Lee M A, Ratner L, Lane C M, Moore M S, Blobel G, Bukrinsky M. Viral protein R regulates nuclear import of the HIV-1 pre-integration complex. EMBO J. 1998;17:909–917. doi: 10.1093/emboj/17.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Re F, Braaten D, Franke E K, Luban J. Human immunodeficiency virus type 1 Vpr arrests the cell cycle in G2 by inhibiting the activation of p34cdc2-cyclin B. J Virol. 1995;69:6859–6864. doi: 10.1128/jvi.69.11.6859-6864.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sawaya B E, Khalili K, Mercer W E, Denisova L, Amini S. Cooperative actions of HIV-1 Vpr and p53 modulate viral gene transcription. J Biol Chem. 1998;273:20052–20057. doi: 10.1074/jbc.273.32.20052. [DOI] [PubMed] [Google Scholar]
- 40.Schilling L J, Farnham P J. Transcriptional regulation of the dihydrofolate reductase/rep-3 locus. Crit Rev Eukaryotic Gene Expr. 1994;4:19–53. doi: 10.1615/critreveukargeneexpr.v4.i1.20. [DOI] [PubMed] [Google Scholar]
- 41.Segil N, Guermah M, Hoffman A, Roeder R G, Heintz N. Mitotic regulation of TFIID: inhibition of activator-dependent transcription and changes in subcellular localization. Genes Dev. 1996;10:2389–2400. doi: 10.1101/gad.10.19.2389. [DOI] [PubMed] [Google Scholar]
- 42.Sheridan P L, Sheline C T, Cannon K, Voz M L, Pazin M J, Kadonaga J T, Jones K A. Activation of the HIV-1 enhancer by the LEF-1 HMG protein on nucleosome-assembled DNA in vitro. Genes Dev. 1995;9:2090–2104. doi: 10.1101/gad.9.17.2090. [DOI] [PubMed] [Google Scholar]
- 43.Song C, Loewenstein P M, Toth K, Green M. Transcription factor TFIID is a direct functional target of the adenovirus E1A transcription-repression domain. Proc Natl Acad Sci USA. 1995;92:10330–10333. doi: 10.1073/pnas.92.22.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stivahtis G L, Soares M A, Vodicka M A, Hahn B H, Emerman M. Conservation and host specificity of Vpr-mediated cell cycle arrest suggest a fundamental role in primate lentivirus evolution and biology. J Virol. 1997;71:4331–4338. doi: 10.1128/jvi.71.6.4331-4338.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Subbramanian R A, Kessous-Elbaz A, Lodge R, Forget J, Yao X J, Bergeron D, Cohen E A. Human immunodeficiency virus type 1 Vpr is a positive regulator of viral transcription and infectivity in primary human macrophages. J Exp Med. 1998;187:1103–1111. doi: 10.1084/jem.187.7.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tristem M, Marshall C, Karpas A, Hill F. Evolution of the primate lentiviruses: evidence from vpx and vpr. EMBO J. 1992;11:3405–3412. doi: 10.1002/j.1460-2075.1992.tb05419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vodicka M A, Koepp D M, Silver P A, Emerman M. HIV-1 Vpr interacts with the nuclear transport pathway to promote macrophage infection. Genes Dev. 1998;12:175–185. doi: 10.1101/gad.12.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang L, Mukherjee S, Jia F, Narayan O, Zhao L J. Interaction of virion protein Vpr of human immunodeficiency virus type 1 with cellular transcription factor Sp1 and trans-activation of viral long terminal repeat. J Biol Chem. 1995;270:25564–25569. doi: 10.1074/jbc.270.43.25564. [DOI] [PubMed] [Google Scholar]
- 49.Xiao H, Lis J T, Jeang K-T. Promoter activity of Tat at steps subsequent to TATA-binding protein protein recruitment. Mol Cell Biol. 1997;17:6898–6905. doi: 10.1128/mcb.17.12.6898. [DOI] [PMC free article] [PubMed] [Google Scholar]