Abstract
Background
Bioenergetic remodeling of core energy metabolism is essential to the initiation, survival, and progression of cancer cells through exergonic supply of adenosine triphosphate (ATP) and metabolic intermediates, as well as control of redox homeostasis. Mitochondria are evolutionarily conserved organelles that mediate cell survival by conferring energetic plasticity and adaptive potential. Mitochondrial ATP synthesis is coupled to the oxidation of a variety of substrates generated through diverse metabolic pathways. As such, inhibition of the mitochondrial bioenergetic system by restricting metabolite availability, direct inhibition of the respiratory Complexes, altering organelle structure, or coupling efficiency may restrict carcinogenic potential and cancer progression.
Scope of Review
Here, we review the role of bioenergetics as the principal conductor of energetic functions and carcinogenesis while highlighting the therapeutic potential of targeting mitochondrial functions.
Major Conclusions
Mitochondrial bioenergetics significantly contribute to cancer initiation and survival. As a result, therapies designed to limit oxidative efficiency may reduce tumor burden and enhance the efficacy of currently available antineoplastic agents.
Keywords: Mitochondria, Cancer, Bioenergetics, Energy transformation, Cell survival
1. Background
Cancer cells require energy to grow, perform work, transform, and replicate. Like all eukaryotic cells, adenosine triphosphate (ATP) in cancer cells serves as the principal form of chemical energy, which is made available through a variety of energy transformation processes. Over the past 100 years, multiple theories have emerged attempting to explain how and why bioenergetic adaptation occurs in cancer. However, it is now generally accepted that nutrient availability and need influence cancer cell metabolism heterogeneity and that a dynamic interplay between cancer cells and the tumor microenvironment (TME) enable metabolic flexibility to support tumor growth, cancer progression, and evasion of death [[1], [2], [3]].
Mitochondria are essential to cancer survival and progression by providing the energy and metabolites required to initiate and sustain growth, as well as modulate signaling process to confer plasticity in changing environments [4]. Bioenergetics govern the dynamic, multifaceted biology of mitochondria and more broadly, the cell, and are critical to cancer cell initiation, survival, and progression. A primary mechanism whereby mitochondria aid in cell survival is through the synthesis of ATP resulting from cellular respiration. However, the specific influence of mitochondrial respiratory function to survival of cancer cells or tumors remains incompletely understood.
Here, we review cancer cell metabolism with a specific focus on mitochondrial bioenergetics as a principal conductor of carcinogenesis and cancer progression. More specifically, we highlight how conserved bioenergetic processes are hijacked and/or used to support neoplastic formation, expansion, and cellular resilience. We discuss recent advances in the perturbation of energetic processes as it pertains to carcinogenic potential and the development of targeted therapies for cancer treatment.
2. Bioenergetics as the conductor of mitochondrial functions
2.1. Mitochondrial structure is critical for function
Mitochondria evolved to be highly conserved organelles of eukaryotic cells due to their remarkable ability to enable energy transformation and support biosynthetic demand [5,6]. In mammals, mitochondrial bioenergetics encompasses the energy conversion processes that occur on, within, and across the mitochondrial inner membrane (mtIM) (Figure 1). The interweaving of the biophysical and structural properties of a mitochondrion with core electrochemical functions is elegantly demonstrated by the coupling of substrate oxidation to ATP synthesis. Energetic coupling is achieved via a chemiosmotic mechanism which utilizes electrochemical potential differences across biological membranes. In this process, primary hydrogen ion pumps of the electron transfer system generate a large protonmotive force (pmF) across the mtIM, which then drives hydrogen ions (H+) back through a secondary pump on the mtIM into the matrix to synthesize ATP from adenosine diphosphate (ADP) and inorganic phosphate (Pi) [7]. The pmF consists of two components: (1) a small chemical potential difference resulting from the pH difference (ΔpH) and (2) a dominant electric potential difference (ΔΨp+) resulting from the distribution of all charged ions, including hydrogen ions, across the mtIM. The electron transfer system enables mitochondria to couple the oxidation of reduced fuel substrates with phosphorylation of ADP to ATP mediated by the pmF.
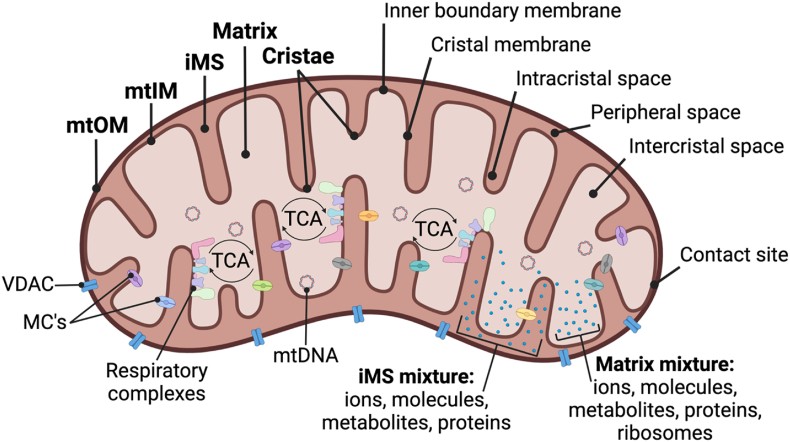
Figure 1.
Structural overview of the mitochondrion.
On an ultrastructural level, mitochondrion contain an outer membrane (mtOM), inner membrane (mtIM), intermembrane space (iMS) between the double membranes, and the matrix within the mtIM. The mtIM contains numerous invaginations, referred to as cristae, and is further distinguished as 1) the membrane near the mtOM (the inner boundary membrane) or 2) the membrane of the cristae (the crystal membrane). The cristae structure allows for increased surface area and a distinct microenvironment of the intracristal space, separate from the remaining peripheral space of the IMS, and the intercristal space, separate from other parts of the matrix. Despite having distinct structures, contact sites exist between the mtOM and the inner boundary membrane of the mtIM. The mtOM is decorated with numerous voltage-dependent anion channels (VDACs) that are relatively nonspecific for small proteins (<10 kDa). The mtOM is permeable to ions and most metabolites but prevents the leaking of IMS proteins, such as cytochrome c. In contrast, the mtIM is highly selective to ionic exchange and contains metabolite carriers (MC's) and channels that regulate the passage of metabolites and ions between the IMS and the matrix. Protons are pumped out of the matrix and into the IMS generating an ∼0.5–0.7 pH difference between the sides, and thus the IMS carries a positive charge and the matrix a negative charge. The mtIM area is roughly five times larger than the mtOM but the size, shape, and organization of the mtIM varies between mitochondria. The mtIM consists of two types of proton pumps: the primary proton pumps of the respiratory Complexes and the secondary FoF1 ATP synthase proton pump. The primary pumps catalyze the transfer of electrons from reducing equivalents, driving protons from the matrix to the IMS, making the matrix relatively more negative than the IMS, and electrically polarizing the mtIM. Polarization of the mtIM establishes the protonmotive force pmF. The respiratory Complexes, including the catalytic components of the ATP synthase, are anchored to the crystal membrane of the mtIM. The matrix also contains discrete microenvironments concentrated with a milieu of proteins, including enzymes of the TCA cycle, metabolites, ribosomes, granules, and mitochondrial DNA (mtDNA).
2.2. Oxidative phosphorylation and electron transfer: energy conversion process of the mitochondrial inner membrane
Forward reactions of electron transfer pathways are dictated by the extramitochondrial ATP to ADP ratio (i.e., cellular energy charge) and the chemical force of phosphorylation (ΔpFATP), affected by both Pi and pH. Respiration relies on shuttling redox equivalents between the tricarboxylic acid (TCA) cycle and the electron transfer system (ETS) to drive oxidative phosphorylation (OXPHOS) (Figure 2). Various substrates transported into the mitochondrial matrix enter the TCA cycle and undergo stepwise carbon rearrangement, addition, or subtraction to form metabolic intermediates. The TCA cycle is commonly visualized as the progressive transformation of oxaloacetate into eight intermediates (citrate, cis-aconitate, d-isocitrate, α-ketoglutarate, succinyl-CoA, succinate, fumarate, and malate) and the transfer of electrons to produce reducing equivalents [8]. The TCA cycle is regulated by product inhibition through allosteric regulation, with critical redox nodes regulated by isocitrate, α-ketoglutarate, succinate, and malate. The stoichiometry of substrates, nucleotides, and redox cofactors determine where the TCA cycle initiates and progresses [9,10]. For example, TCA cycle intermediates are believed to be saturating at rest and during strenuous activity in skeletal muscle and as such, efficiency is high [11]. Electron shuttles (nicotinamide adenine dinucleotide, NAD, and flavin adenine dinucleotide) are reduced as they accept the chemical energy released by the stepwise rearrangements of the TCA cycle and small amounts of ATP are generated by substrate phosphorylation.
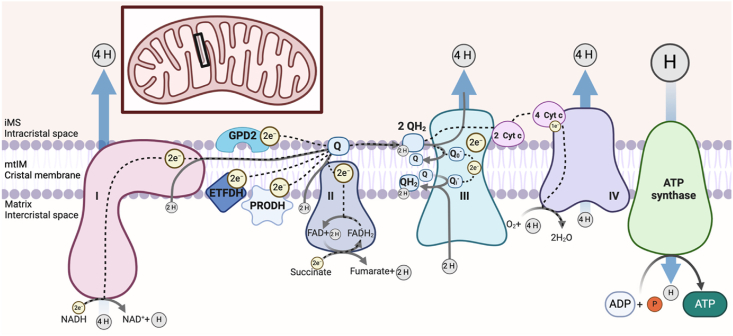
Figure 2.
Overview of the electron transfer system and pathways of oxidative phosphorylation.
The cristal membrane of the mitochondrial inner membrane (mtIM) is the primary site of oxidation and reduction reactions within both the electron transfer system (ETS) and oxidative phosphorylation (OXPHOS). The respiratory sequence initiates with oxidation of 3 NADH molecules at Complex I and succinate at Complex II. Of note, depending on the substrate, complete oxidation can yield more than 1 full turn of the tricarboxylic acid (TCA) cycle. Depending on the anaplerotic/cataplerotic status of cycle intermediates, a substrate may not undergo complete oxidation and thus a partial TCA cycle may occur. NADH oxidation to NAD+ +H+ transfers 2 electrons (e−) per NADH through the large Complex resulting in the transfer of 4 protons (H+) from the matrix into the intermembrane space (iMS) through Complex I. In contrast, Complex II oxidizes succinate to fumarate, transferring 2e− to bound FAD, yielding FADH2, which subsequently transfers the electrons to the Q-junction without hydrogen ion pumping. The electrons are passed from Complexes I and II to Q, an electron carrier, which is reduced to QH2, utilizing protons from the matrix. Electrons are transferred through the nonpolar region of the phospholipid mtIM bilayer to Complex III and protons are released into the iMS. Other membrane-bound structures contribute to net electron transfer in a tissue and organism-specific manner, including glycerol-3-phosphate dehydrogenase (GPD2), the electron transferring flavoprotein dehydrogenase (ETFDH), and proline dehydrogenase (PRODH). The complete transfer of 2 e− from Q through Complex III to cytochrome c requires 2 QH2 molecules and 2 cytochrome c molecules in a two-step process. 4 H+ from both QH2 molecules are pumped into the IMS and 2 H+ from the matrix are used to reform 1 QH2, resulting in a net proton gradient change of 6 H+. Each cytochrome c then transports an electron to Complex IV through the sequential reduction of a copper and heme group, requiring a total of 4 reduced cytochrome c molecules and allowing an oxygen molecule to bind and receive a total of 4e−. Additionally, four H+ from the matrix are pumped through Complex IV into the IMS and four H+ from the matrix are used to form two H2O, resulting in a net proton gradient change of eight H+. The established electrochemical proton gradient between the matrix and the IMS pushes protons through ATP synthase, down the gradient, back into the matrix. With the proton movement, ATP synthase spins and phosphorylates ADP with inorganic phosphate (Pi) to make ATP. However, with increased concentrations of ATP or if respiration is compromised and the pmF falls, the ATP synthase reaction can reverse and hydrolyze ATP to pump protons back out of the matrix to re-establish the pmF. Ultimately the efficiency of the OXPHOS system is influenced by electrochemical gradients, substrate and oxygen concentration, and the integrity of the respiratory Complexes.
In the mitochondria ATP are generated through the electron transfer-linked OXPHOS of ADP. The mtIM-bound ETS is made up of carriers and several distinct multi-protein Complexes (Figure 2) including but not limited to: Complex I (CI; NADH:ubiquinone oxidoreductase, Type 1 NADH dehydrogenase), Complex II (CII; succinate:ubiquinone oxidoreductase, succinate dehydrogenase), Complex III (CIII; coenzyme Q:cytochrome c-oxidoreductase, cytochrome bc1 Complex), and Complex IV (CIV; cytochrome c oxidase) [12]. Complex I is the largest respiratory Complex, forming an ‘L’ shape with a molecular mass of about 1 MDa and contains 44 subunits [13], seven of which are encoded by mitochondrial DNA (mtDNA) [14]. Complex II is ∼100 kDa with four subunits, all encoded by the nuclear genome in mammalian mitochondria [15]. Complex III is a 496 kDa dimeric protein and each monomer contains 11 subunits, one of which is encoded by mtDNA [16]. Complex IV is a 204 kDa protein containing 14 subunits, three of which are encoded by mtDNA [17]. Additional Complexes such as glycerol-3-phosphate dehydrogenase (GPD2), electron-transferring flavoprotein dehydrogenase (ETFDH), and proline dehydrogenase (PRODH) also transfer electrons to coenzyme Q (Q). Electron transfer from Complex III to IV requires the carrier, cytochrome c. Electron transfer through Complexes I, III, and IV involve the translocation of hydrogen ions from the matrix to the IMS, generating the protonmotive force pmF. Overall, respiratory capacity is impacted by the activity of the respiratory Complexes and the formation of mitochondrial supercomplexes.
2.3. Coupling efficiency
Mitochondrial respiration couples substrate oxidation to ATP production, but the degree of coupling varies greatly and highly influences the bioenergetic capacity of the cell. In this regard, bioenergetic efficiency is defined as the resulting fraction of useful energy converted to work. Under physiological conditions, 50–80% of substrate oxidation is coupled to ATP production. Thus, mitochondrial uncoupling is the disruption of the energy transducing proton circuit, dissociating electron transfer from ATP synthesis. Within a circuit, protons routinely flow from the IMS back into the matrix. Basal leak is the routine physiological exchange of protons that occurs along the respiratory system and accounts for up to 10% of oxygen consumption [18]. Proton leak may also be induced by endogenous and/or synthetic disruption of the electrochemical gradient to lower coupling efficiency. What mechanistically constitutes a biologic and/or synthetic mitochondrial chemical uncoupler remains discrepant and has evolved over time. In 1963, Edward Slater defined uncouplers as agents that (1) stimulate non-phosphorylating electron transfer (2), limit ATP synthesis without inhibition of electron transfer (3), stimulate ATP hydrolysis, and (4) inhibit exchange reactions catalyzed by the ATPase [19]. In 1979, Peter Heytler refined this definition to include both “classic” or “true” and “nonclassic” uncouplers [20]. Classic uncouplers were defined as moderately weak lipophilic acids with a pKa from about 4 to 8 that freely exchange protons across the mtIM by cycling between protonated and deprotonated forms of the acidic group on the uncoupler [20]. In contrast, nonclassic uncouplers were more loosely defined with variable mechanisms of action to dissipate the ΔΨp+, such as accumulation of a positive charge in the typically negative matrix or compounds that disrupt the protein structure or bind functional groups of membrane proteins [20]. Conversely, disruption of the ΔΨp+ may occur from dyscoupling activity, which uncouples nutrient oxidation from ATP production without significant reduction of the protonmotive force [21]. More recently, it was proposed that prototype mitochondrial uncouplers induce proton leak by activating UCP1 and ADP/ATP carriers rather than a protonophoric shuttling mechanism [22]. Importantly, uncoupling can result in tissue-dependent responses, including differences in respiratory kinetics [23] or toxicity, based on how different cell types can respond to the mitochondrial stress of uncoupling. ADP and oxygen availability also impact phosphorylation efficiency. When ADP is limited, the ADP/O2 flux ratio diminishes, but when oxygen is limited, the ADP/O2 flux ratio is maintained but noncoupled respiration under ADP is reduced [24].
2.4. Diversity of energy transformation
All cells are intimately linked by their need to produce and consume ATP, the molecular energy currency. Cells can produce upwards of 50 million molecules of ATP per cell per second [25] and contain over 6 billion ATP molecules at any given moment [26] to support biological work including ion transport, protein synthesis, contractility, and substrate phosphorylation. The dynamic utilization of energetic precursors such as lipids, carbohydrates, and proteins allows cells to meet their energetic and metabolic needs in the face of fluctuating physical environments (Figure 3). The two primary systems of ATP production are glycolysis and mitochondrial respiration. Notably, macromolecules can contribute to various points of this system (1): from lipids, the glycerol backbone can enter glycolysis and the fatty acids can undergo oxidation to form acetyl-CoA (2); from carbohydrates, simple sugars are broken down into glucose which can enter glycolysis; and (3) from proteins, almost every amino acid can be converted into pyruvate, acetyl-CoA, or a TCA cycle intermediate.
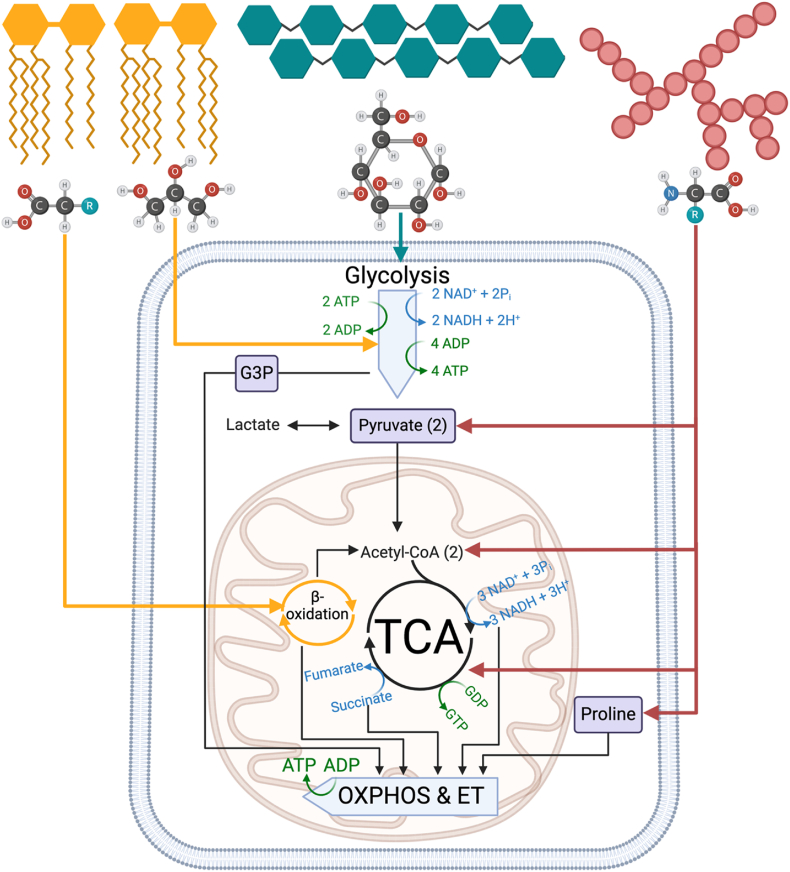
Figure 3.
Overview of catabolic pathways leading to mitochondrial ATP production.
Multiple pathways of substrate oxidation and reduction support energy transfer. Lipids (yellow) are broken down into glycerol backbone and fatty acids; carbohydrates (teal) are broken down into simple sugars and then glucose; and proteins (red) are broken down into monomeric amino acid units. Fatty acids, pyruvate, and many amino acids are transported into the mitochondria and undergo oxidation to fuel the tricarboxylic acid (TCA) cycle. Various metabolic intermediates such glycerol-3-phosphate (G3P) or proline can be transported into the mitochondria to fuel electron transport directly through the oxidative phosphorylation (OXPHOS) system independent of TCA cycle.
Glycolysis occurs in the cytoplasm and contributes to some ATP production while generating biosynthetic molecules for proteins, lipids, and nucleic acids [27]. Glycolysis nets two ATP per molecule of glucose in the stepwise conversion to two molecules of pyruvate and is divided into three phases: 1) the energy investment phase which consists of ATP hydrolysis to trap glucose in the cell by hexokinase-mediated conversion of glucose to glucose-6-phosphate, followed by phosphofructokinase-1-mediated conversion of fructose-6 phosphate to fructose-1,6-bisphosphate; 2) the cleavage of 6-carbon fructose-1,6-bisphosphate to form two 3-carbon molecules of glyceraldehyde-3-phosphate, and 3) the pay-off phase by reduction of NAD+ to NADH through glyceraldehyde-3-phosphate oxidation which confers high-energy reducing equivalents for downstream production of ATP and pyruvate [28]. In contrast, mitochondrial respiration produces ATP through a series of highly interconnected exergonic reactions: 1) TCA cycle regeneration of NADH and succinate [8] and 2) OXPHOS through oxidation of NADH and succinate and phosphorylation of ADP [12]. OXPHOS is generally an efficient metabolic process, producing over 30 ATP per molecule of glucose or glycogen dependent on the prevailing cellular milieu, bioenergetic status, and presence of oxygen molecules in the mitochondria [29].
Glycolysis and mitochondrial respiration synergistically contribute to cell survival, commonly depicted through their connection to pyruvate and lactate. If molecular oxygen is available for OXPHOS, pyruvate from glycolysis can enter the mitochondrial matrix to be oxidized to acetyl CoA which reacts with oxaloacetate to generate citrate and CoA and enter the TCA cycle to form reducing equivalents [8]. Conversely, pyruvate can remain in the cytoplasm, undergo fermentation, and be reduced to lactate by lactate dehydrogenase A [30]. Excess lactate is excreted into the extracellular space through monocarboxylate transporters (MCTs) and is then available for use by lactate-consuming cells that can further contribute to the pyruvate and coenzyme redox buffering exchange [30]. Another common denominator of both glycolysis and OXPHOS is the oxidizing coenzyme, NAD+ [31]. Glycolysis requires NAD+ for the pay-off phase, and fermentation of pyruvate also serves to recycle NADH back to NAD+, sustaining glycolysis [30]. OXPHOS produces NAD+ in the oxidation of NADH, and the conversions of three different TCA cycle intermediates also require NAD+ to generate NADH which, in turn, provides electrons for OXPHOS [12]. Thus, NAD+ is constantly synthesized, degraded, and recycled in both the cytoplasm and within the mitochondria in discrete redox cycles separated by compartmentalization. The cytoplasmic and mitochondrial NAD+/NADH pools are distinct but connected through redox shuttles and NAD biosynthetic pathways [32]. Notably, the mitochondrial NAD pool is defended even upon extreme cytoplasmic depletion, continuing to support cell viability and ATP until the mitochondrial membrane is compromised [32].
Mitochondria are central to metabolic flexibility by bestowing the energetic plasticity requisite to maintaining the cellular energy charge. For example, mitochondria can use succinate amongst other substrates as electron donors, while glycolysis does not have an alternative to NAD+ utilization. Glycolysis is fueled through glucose and glycerol, while mitochondrial respiration can be fueled by pyruvate as well as a variety of intermediates obtained through succinate, amino acids, mitochondrial glycerol-3-phosphate (G3P) oxidation, and fatty acid oxidation (FAO) [33,34]. Most amino acids can be converted into TCA cycle intermediates and fuel OXPHOS while others are converted into pyruvate, acetyl-CoA, or acetoacetate (which can be converted into acetyl-CoA) and eventually contribute to the TCA cycle as well [35]. G3P is a metabolic intermediate linking glycolysis, lipogenesis, and OXPHOS through the glycerophosphate shuttle [34]. GPD2 oxidizes G3P to dihydroxyacetone phosphate while reducing FAD to FADH2, enabling electron flow to coenzyme Q [34]. FAO occurs in the mitochondria and greatly contributes to energy transformation. Since fatty acid chains have more electrons around the carbons compared to other macronutrients, complete oxidation yields the most ATP per gram of nutrient, resulting in over 100 ATP molecules depending on the fatty acid length [36]. Once inside the cytoplasm, a fatty acid molecule is activated to form fatty acyl-CoA, in a two-step process utilizing ATP. Acyl-CoA is enzymatically shuttled into the mitochondrial matrix and subsequently shortened in a recurring sequence that produces one NADH, FADH2, and acetyl CoA molecule, per round of oxidation [36]. The resulting acetyl CoA molecules can then join with oxaloacetate to enter the TCA cycle to form citrate and the reduced coenzymes are available for OXPHOS, thus generating ATP. In summary, mitochondria evolved to support cell survival and expansion by developing an intricately regulated system of chemiosmosis coupled to the reduction of molecular oxygen. The efficiency of this process is mediated by intrinsic and extrinsic factors that ultimately dictate establishment of the pmF and OXPHOS capacity.
3. Mitochondria are central to metabolic adaptation in cancer
3.1. The Warburg legacy and prevailing metabolic theories in cancer
In the 1920's, Otto Warburg observed that cancer cells use glucose and secrete lactate, even in the presence of oxygen [[37], [38], [39], [40]]. These observations stood in stark contrast to the widely accepted Pasteur Effect, in which glycolysis is observed to be inhibited by oxygen. Shortly after Warburg's findings, Carl and Gerty Cori published a three-part series corroborating Warburg's finding, demonstrating that glycolysis was increased in tumors compared to non-tumor tissue [[41], [42], [43]]. In an effort to improve generalizability, Herbert Crabtree characterized the respiration of eight types of mouse tumors and a total of 56 different tumors [44]. Using Warburg's manometric technique, he observed that only ∼33% of tumors displayed the Warburg effect under all experimental conditions, and some displayed glucose-induced suppression of OXPHOS [44]. Crabtree also observed respiratory differences between tumor growth sites, hypothesizing that metabolic status may vary based upon tumor stage and types, introducing the concept of metabolic heterogeneity and the influence of nutrient availability [44]. Strikingly, Crabtree had previously measured a short-term reversible metabolic shift from OXPHOS to glycolysis induced by glucose or fructose independent of oxygen in Saccharomyces cerevisiae [45] and later coined the observations the “Crabtree effect”. Crabtree supported Warburg's theory but was critical of generalized application.
In 1956, Warburg further postulated that “damaged respiration” was responsible for the adaptive metabolism of cancer cells [46]. Warburg's hypothesis, though controversial and somewhat overstated, shaped decades of research, yielding profound mechanistic insight into cancer metabolism. In fact, Warburg's observation of the presence of aerobic glycolysis in tumors was later coined the “Warburg effect” in a 1972 publication by Efraim Racker [47]. In support of Warburg, high rates of glucose utilization have been observed in some cancer cells, where glycolysis contributes ∼50% of ATP synthesis compared to the ∼20% or less observed in most non-cancer cells [48]. In contrast, numerous investigations have demonstrated variable contributions of glycolysis to ATP, intact mitochondrial function independent of glycolysis, as well as metabolic flexibility dependent on substrate availability rather than pathway dependence [49,50]. In a meta-analysis of 31 cancer cell lines of various origins and 16 non-cancerous lines, OXPHOS contributed to ∼83% of the ATP pool in cancer cells and ∼80% in the non-cancer cells [48]. There is now a broad consensus that a high glycolytic rate in cancer is neither 1) universally caused by mitochondrial dysfunction nor 2) essential for cancer cell growth. Furthermore, the upwards of 50% of ATP synthesis attributable to glycolysis indicates that mitochondrial bioenergetic activity remains central to cancer cell energy metabolism.
Additional terminology and models have emerged to classify the presentation and origin of tumor energy metabolism. The cancer cell symbiosis model outlines how cancer cells relying on aerobic glycolysis are able to “feed” adjacent cancer cells or other cells in the TME utilizing OXPHOS via nutrient transport [51]. This model can be expanded beyond the energetic phenotypes of cells within the TME to capture the intercellular communication that occurs to ultimately promote tumor survival and growth. The TME is complex, consisting of a mixture of infiltrating and resident cells such as stromal and matrix cells, adaptive and myeloid immune cells, and vascular cells, that support tumor development and aid in evasion of cell death [52]. The Reverse Warburg Effect implicates tumor stroma as the primary source of glycolysis rather than cancer cells per se, which has led to the hypothesis that cancer cells induce aerobic glycolysis in cancer-associated fibroblasts and produce lactate and pyruvate for the cancer cells [53]. This concept is currently under active investigation across cancer types. In breast cancer, a hierarchy of TME cell-cell interactions was driven by cancer-associated fibroblasts that establish dominant autocrine circuit loops as well as paracrine circuits with tumor-associated macrophages [54]. Reflecting the diversity of the cell types within the TME, tumor metabolism is vastly heterogenous and may be highly influenced by non-cancer cells such as myeloid cells that are notably glycolytic compared to cancer cells which are known to be very glutamine dependent [55]. Indeed, cancer cells frequently violate assumptions of the Warburg effect. For example, cancer cells often display high OXPHOS gene expression with or without high glycolytic gene expression [56], and others demonstrate the ability to upregulate OXPHOS to enhance plasticity in response to changing environments [57], and to support metastasis [58]. Furthermore, cancer cells that display increased glycolysis do not share a single mechanism for the effect, and multiple control points have been elucidated for both increased glycolysis and limited mitochondrial metabolism. These include the overexpression of glucose transporters and high activity of glycolytic enzymes, as well as the inhibition of pyruvate dehydrogenase and TCA cycle enzymes, inhibition of substrate transporters, and some defects in the OXPHOS machinery [59].
Apart from glucose utilization, other branches of substrate metabolism also contribute to a cancer cell's ability to adapt. The glutamine theory postulates that increased glutamine uptake in cancer cells drives ATP synthesis through the TCA cycle and OXPHOS [60]. Since this initial observation, the influence of the glutamine pathway has been widely investigated and glutamine transporters have been found to be upregulated in many cancers, specifically SLC1A5, SLC7A5, and SLCA14 [61]. Furthermore, glutamine-driven OXPHOS was found to be the major means of ATP production in both normoxic and hypoxic cancer cells [62]. Energetic redundancy confers plasticity in non-cancerous and cancerous cells alike, however, the rapidly proliferative environment of cancer cells facilitates unique bioenergetic adaptations to further support survival. Originally, lactate was branded as a waste product in cancer cells, something to dispose of to avoid toxicity. The Cori's investigation of tumor carbohydrate metabolism hinted otherwise; they consistently noted considerable amounts of glycogen in tumors that disappeared with glucose restriction [41]. Four years later, the Cori's moved away from cancer research and published their Nobel Prize winning work demonstrating that lactic acid from muscle can be taken up by the liver to form glycogen, which in turn, can be broken down later to glucose for use in the muscle, termed the Cori Cycle [63]. It has since been demonstrated that cancer cells maintain the ability to produce lactate which gets shuttled out of the cell as well as the ability to take up extracellular lactate [64]. Additionally, intracellular lactate can be used for energy transformation through conversion into pyruvate and then undergo gluconeogenesis or be utilized in the TCA cycle [65]. The lactogenesis hypothesis posits that lactate is used both as a fuel for cancer cells as well as for carcinogenic signaling [66]. It further argues that lactate oncogenic signaling is required to achieve most hallmarks of cancer, such as angiogenesis, immune escape, and metastasis [66].
Additionally, some tumors grow in the absence of glucose, and have increased phosphoenolpyruvate carboxykinase (PEPCK) expression and abbreviated gluconeogenesis [67]. Other tumors display enhanced glucose uptake shuttling through the pentose phosphate pathway (PPP) [68] and/or upregulated FAO [69]. Furthermore, FAO intermediates and enzymes have been found to be upregulated in many cancers, such as triple negative breast cancer (TNBC) [70] and lung cancer [71]. FAO is associated with worse prognosis and exogenous fatty acids are essential mediators of metastasis [72], with free fatty acids from adipocytes enhancing invasiveness of pancreatic cancer cells [73]. Furthermore, some cancer cells have increased expression and unique isoform expression of mitochondrial fatty acid shuttle proteins, required for import into the mitochondria, along with increased FAO [74]. Key enzymes in fatty acid synthesis, such as fatty acid synthase (FASN), are often elevated in cancer. To this end, the FASN inhibitor, TVB-2640, demonstrated safety and potential as a therapeutic in the first clinical trial for treatment of advanced cancer [75]. Aside from fatty acid oxidation, fatty acids can be used to generate other lipids, such as sphingolipids. Increased fatty acid 2-hydroxylase (FA2H) and decreased ceramides promote pulmonary metastases, whereas supplementation with ceramides decrease cancer cell mitochondrial respiration and metastases [76]. Similarly, the role of G3P oxidation is also gaining interest in cancer metabolism research and GPD2 expression and activity is increased in multiple cancer types [77]. Taken together, the past 100 years of research on cancer metabolism has revealed that cancer is a heterogeneous disease and each cancer has a unique metabolic signature [78]. Even within a single tumor, constituent cells vary with distinct metabolic phenotypes from one cell to another. Furthermore, the energy producing pathways have been shown to be altered in different cancer subtypes and play distinct roles in the phases of cancer progression: with proliferating cells demonstrating glycolytic phenotypes, differentiating cells displaying increased OXPHOS, and metastatic cells exhibiting metabolic flexibility [79,80].
3.2. Mitochondrial function and cancer: A perspective
The elucidation of the vast functions of mitochondria over the last century has illuminated the complexity of Warburg's theory that aberrant glycolysis was attributable to mitochondrial “dysfunction”. Notable adaptations in mitochondria from cancer cells that highly influence function include mutations in genes encoding mitochondrial proteins [4], ultrastructural modifications [81], morphological changes [82], elevated ROS generation [83], increased mitochondrial membrane potential [84], and augmented energy metabolism [85,86]. Functional impairments are not necessarily reflective of overall organelle health and viability. For example, a genetic mutation in Complex I limits enzymatic activity and associated ATP production. However, this limitation may be overcome through compensatory utilization of other coupling pathways, such as Complex II, thus net ATP production is sustained. In this case, the variety of the pathways to ATP production ensures maintenance of energy homeostasis. Furthermore, the assumption that upregulation of glycolysis indicates that mitochondrial respiration is null or not substantially contributing to ATP production is presumptive, as very few cancer cells have been found to have more than 50% of ATP generated through glycolysis [48]. Indeed, the endergonic conversion of glucose to glucose-6-phosphate by hexokinase, would likely require OXPHOS, the sole exergonic source of ATP production, if it were ever limited. In the field of mitochondrial cancer research there has been a focus on mitochondrial function beyond ATP production and there is a need for more research on cancer mitochondrial respiration across cancer types, over the cancer continuum, and in response to therapies.
For cancer cells, the dynamic changes in mitochondrial function may be an adaptive response to an upstream influence and thus physiologically expected given the signaling stimuli. In fact, the mitochondrial plasticity found in cancer cells also exists in non-cancer cells [87]. Several tumor-related genes and proteins regulate metabolic and cell survival pathways that converge through mitochondria. For example, the P53 tumor suppressor gene, the most frequently mutated gene in cancer, allows for metabolic stress adaptation by enhancing the expression of mitochondrial proteins [88]. The constitutively expressed c-MYC transcription factor targets hundreds of nuclear encoded mitochondrial genes and thus heavily impacts the mitochondrial transcriptome [89]. Additionally, overexpression of hypoxia-inducible factor 1-alpha (HIF-1α) upregulates glycolytic metabolism under low oxygen conditions and suppresses mitochondrial respiration and biogenesis [90]. K-Ras, which is constitutively active in many cancers can also downregulate mitochondrial respiration and upregulate mitochondrial fission [91]. Furthermore, dysregulation of the central energy sensor AMP-activated protein kinase (AMPK), through gain or loss of AMPK activity, uncouples proliferation from mitochondrial energy sensing and allows for unhindered proliferation in the presence of oncogenic growth signaling [92]. A major downstream effector of the PI3K/AKT pathway is the mammalian target of rapamycin (mTOR), which supports tumor growth through mTOR Complex 1 (mTORC1) and mTOR Complex 2 (mTORC2) signaling to couple nutrient and growth factor sensing to cellular growth and promotion of mitochondrial biogenesis [93].
Mitochondrial quality control is intricately linked to tumor metabolic state, tumor heterogeneity, tissue type, microenvironment, and tumor stage [94]. Dominant mtDNA mutations that become established in a clonal cell population have been identified in many cancers. An analysis of paired tumor vs non-tumor tissue using The Cancer Genome Atlas (TCGA) data from 226 individuals demonstrated a wide range in frequency (13–63%) of somatic mutations in mtDNA across cancer type, with 65% of truncating mutations occurring in NADH dehydrogenase 5 [95]. Interestingly, mtDNA show very similar mutational signatures regardless of cancer tissue origin [96], likely due to the impaired function of mitophagy to identify and remove damaged and dysfunctional mitochondria resulting in turnover of fully functional mitochondria. Another analysis of TCGA data revealed variation in mtDNA copy number across 22 tumor types with decreases in mtDNA content demonstrated in breast, bladder, kidney, esophageal, head/neck, and liver cancer compared to control non-tumor tissue [97]. Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC1α) plays a crucial role in mitochondrial integrity as the master regulator of mitochondrial biogenesis, responding to stress. PGC1α expression largely corresponds to a cancer cell's metabolic status, with low expression indicating increased glycolysis and high expression indicating increased OXPHOS and potential for metabolic flexibility aiding tumor metabolic plasticity [98]. Interestingly, PGC1α expression is required for metastasis in some cancers [99] while it inhibits epithelial-to-mesenchymal transition (EMT) in others [100]. A recent study demonstrated that mtDNA content dictated colorectal cancer (CRC) cell survival and metastases with increases in the mtDNA copy number that would be required for CRC progression [101].
Imbalanced mitochondrial dynamics leading to network fragmentation, impaired OXPHOS activity, and release of metabolites and signaling molecules exacerbate tumor growth [102]. Specifically, hyper-fragmentation of mitochondrial networks mediates lung and breast cancer [103]. Furthermore, dynamin related protein 1-mediated mitochondrial fission promotes tumorigenesis in breast, gastric, ovarian, non-small cell lung, and pancreatic cancer as well as hepatocellular carcinoma and melanoma [104]. Metastatic breast cancer cells are more fragmented than non-metastatic cells and fission was found to be required for migration and invasion [105] and for survival of TNBC [106]. An increased ratio of fission to fusion corresponds with increased malignancy in ovarian cancer cells [107]. Interestingly, increased rates of mitochondrial fission in TNBC cells suppresses metastasis [108] and large, fused mitochondrial networks are found to support drug resistance in lung cancer cells [109]. Thus, it appears that hyper fission is required for some metastatic processes, but that induced fusion can improve cancer cell survival and induced fission can be detrimental to cancer cell survival. In addition to mitochondrial morphology, mitochondrial dynamic proteins coordinate and maintain mitochondrial bioenergetics. Mitochondrial Fission Factor (MFF) interacts with Voltage-Dependent Anion Channel-1 (VDAC1) to protect mitochondrial membrane integrity and support primary and metastatic prostate cancer progression [110].
Vulnerabilities may occur through rapidly dividing cells and mitochondria; thus, cancer cells can exploit mitophagy to evade cell death and promote survival. Since mitophagy is a quality control mechanism, it has been shown to have both anti- and pro-tumorigenic properties based on cancer subtype, context, and tumor stage [111]. In non-cancer cells, mitophagy clears damaged mitochondria that generate tumor-promoting ROS or other tumorigenic mitochondrial signals. Impairments in mitochondrial quality control, such as deficiencies in PTEN-induced kinase 1 and parkin-mediated mitophagy, appear to contribute to cancer development by causing genomic instability, accumulation of dysfunctional mitochondria, and alteration of bioenergetic processes [112]. Once a tumor is established, mitophagy can be utilized for survival against therapy-induced toxicity; for example, Pink1-mediated mitophagy contributes to survival of multi-drug resistance cancer cells, which is suppressed by gene silencing of Pink [113] or pharmacological inhibition of fission [114]. Like mitochondrial dynamics, for some tumors the impact of mitophagy depends on the point of tumorigenesis. For example, limiting mitophagy can promote tumorigenesis while enhancing mitophagy can drive a tumor to progress to metastasis [115]. Conversely, a mitophagy defect in mammary tumors has been shown to promote metastasis and predict poor prognosis [116]. These data suggest that there is not a singular pathway for disease progression, but that mitochondrial regulation remains critical in the diversity of the disease.
Oncogenic pathways are impacted by mitochondrial signaling networks that regulate cell death and oxidative stress. Mitochondria play a key role in the intrinsic apoptosis signaling cascade that regulates cytochrome c release through mitochondrial outer membrane permeabilization (MOMP) into the cytosol and activation of caspases [117]. Cancer cells can evade apoptosis by inhibiting genes that promote MOMP [118] and through mitochondrial hyper-fragmentation cause resistance to apoptotic stimuli [119]. Indicators of mitochondrial integrity, including ΔΨp+ and MOMP, also function as important signaling mechanisms. Increased ΔΨp+ is observed in many cancers and may also be an indicator of mitochondrial health, as a decrease in ΔΨp+ increases apoptotic susceptibility [120]. Decreasing ΔΨp+ in estrogen-receptor (ER) positive MCF-7 breast cancer cells, which displayed increased ΔΨp+ compared with a non-cancer control [84], TNBC MDA-MB-231 cells [121], and adult granulosa tumor KGN cells [122] has also been shown to promote apoptosis.
Mitochondria contribute to redox balance and oxidative stress regulation and are a source of ROS including superoxide, hydroxyl, and H2O2 [123]. ROS may be elevated in cancer, but the extent of the increase and the correlation with prognosis varies by tumor type. For example, evaluation of ROS metabolism using transcriptomics revealed eight distinct clusters depicting differences in ROS accumulation, oxidative stress, scavenging, biosynthesis, and subcellular sources (mitochondria vs cytoplasm) [124]. Cluster VII tumors are strongly associated with mitochondrial-derived ROS and are mostly hepatocellular carcinomas (97%) whereas cluster IV, majorly composed of pancreatic adenocarcinomas (98%) are associated with increased ROS scavenging [124]. Excess ROS production promotes lipid peroxidation, protein unfolding, and DNA damage, ultimately promoting carcinogenesis [125]. Localization of ROS production within the mitochondria may alter carcinogenic potential. For example, H2O2 released into the IMS contributes to proliferation and survival of colorectal cancer cells [126]. ROS can also function as signaling molecules that regulate cell proliferation, cytosolic networks, cellular metabolism, and many tumor promoting pathways [125]. Cancers with heightened ROS also often display increased ROS scavenger activity which mediates cancer cell survival [127]. The concomitant upregulation of antioxidant pathways prevents ROS-mediated cytotoxicity, thus the increased but tightly regulated ROS production allows tumors to maintain tumor-promoting ROS levels, supporting proliferation, angiogenesis, and metastasis [128]. Furthermore, phospholipid biosynthesis-mediated ROS production is purported to support tumor growth of pancreatic ductal adenocarcinoma [129].
Mitochondria also produce over 300 distinct metabolites generated through OXHPOS, TCA cycle, long-chain fatty acid β-oxidation, amino acid metabolism, and the urea, one-carbon cycle [130]. Mutations in mitochondrial substrate transporters and enzymes can result in accumulation of oncometabolites-metabolites that support cancer progression through nuclear gene transcription via chromatin modification as well as cytosolic signaling pathways [131]. Mitochondrial metabolites such as 2-hydroxyglutarate, fumarate, and succinate [132], have been shown to alter DNA or histones through methylation and acetylation changes [133]. An untargeted metabolomics profiling of human breast tumors led to the discovery of MYC-induced accumulation of the oncometabolite 2-hydroxyglutarate (2HG), possibly generated through glutamine utilization, in breast tumors which were associated with poor clinical prognosis [134]. Importantly, this discovery demonstrates the relevance of oncometabolite characterization and the potential use for cancer biomarkers. Recently, lactate was also identified as a potential oncometabolite, sustaining transcriptional regulation in breast cancer cells [135].
Somatic alterations to mtDNA and Complex function are some of the most common mutations across tumor types and specific patterns have been linked to patient prognosis [96,136,137]. For example, over 50% of tumors carry some form of somatic mtDNA mutation [138]. Advances in single-cell sequencing has improved the sensitivity and specificity of identifying mtDNA mutations, although the clonal relationships of mtDNA mutations still remain limited due to low heteroplasmy [139,140]. Currently the functional impacts of mtDNA mutations on the initiation and progression of cancer are still being elucidated. Primary prostate cancer tumors were found to have decreased NADH-linked respiratory capacity and increased succinate oxidative capacity compared to benign tissue, which was associated with mtDNA mutations in Complex I and worse prognosis [141]. Further, exogenous succinate was found to promote tumorigenesis in prostate cancer cells [142]. In addition, intestinal mtDNA mutation results in increased tumor proliferation and size, but not number in mice [142]. Improvements in mitochondrial base editing have recently arisen enabling the potential for more sophisticated mechanistic investigations of the roll of mtDNA mutations in cancer in the years to come [143].
Rewiring of bioenergetic function occurs in a tumor-dependent manner to support uncontrolled expansion. For example, Complex I impairment drives ROS-dependent activation of AKT, increasing glycolytic flux, cell survival, and carcinogenic potential in transformed human fibroblast cells [144]. In melanomas, Complex II, not Complex I, contributes to tumorigenicity via succinate-mediated increases in tumor antigen presentation [145]. In multiple cancer cell lines in vitro and in xenograft models, GPD2 mediates ferroptosis defense to promote cancer growth [146]. Conversely, GPD2 loss does not impact respiration but promotes cell proliferation via upregulation of cytosolic glycerol-3-phosphate in kidney cancer [147]. ETFDH expression is decreased in human hepatocellular carcinomas and associated with poor survival [148]. PRODH is highly upregulated by p53 and mediates non-small cell lung cancer progression, promoting epithelial to mesenchymal transition [149]. In breast cancer, patients often exhibit deficiencies in coenzyme Q10 (CoQ10), which blocks antioxidant defense and enhances proliferative potential [150]. Additionally, mitochondrial supercomplex formation and stabilization was recently found to promote breast and endometrial tumorigenesis [151] as well as pancreatic cancer growth [152]. Further study of mitochondrial Complex assembly and function are warranted to better understand the impact across cancer types as well as the mechanism for tolerating hypoxic conditions. Recently, it was observed that primary solid tumors have decreased TCA cycle flux compared with control tissue, whereas metastasis increased TCA flux [153]. This heterogeneity in mitochondrial alterations requires improved characterization of mitochondrial alterations with functional ramifications. In summary, essential mitochondrial functions are leveraged to sustain carcinogenesis and evade death in a tissue- and cell-type specific manner. Thus, understanding the degree by which a specific cancer type utilizes mitochondria for processes such as macromolecule synthesis and energy transfer is required to ultimately determine targetability and therapeutic potential.
Emerging concepts of cancer bioenergetics include spatial resolution of mitochondrial heterogeneity within cells, tumors, and the TME, and also in response to environmental changes. Mitochondria exist as dynamic organelles that continuously undergo remodeling, turnover, and networking. These dynamics have recently been illuminated with new techniques that visualize mitochondrial form and function [154,155]. For example, three-dimensional ultrastructural analysis revealed that distinct compartments of mitochondrial networks within tumors dictated the bioenergetic capacity of non-small cell lung cancer tumors [156]. Strides have also been made in the field of volume electron microscopy, where subcellular components, such as the nucleus and mitochondria, of liver tumors are visualized within their native 3-dimensional organization [157]. Mitochondrial mediated spatial regulation of tumor metabolism has also been elucidated through the lens of fluxomics. For example, cancer cells may rewire subcellular metabolic fluxes to enable flexibility by driving glutamine reduction under hypoxic conditions or by enabling pyrimidine biosynthesis and reversing TCA activity oxalacetate [158]. Recently, methods using computational deconvolution have improved resolution of spatial fluxes within intact cancer cells removing the need to fractionate cells [159]. Mitochondria transfer between cells, first observed via (ras homolog family member T1) RHOT1 mediated tunneling nanotubes (nano-scale channels between cells) over 10 years ago, is now an emerging area of study for TME, metabolic rewiring, and therapy. Recently, transfer of mtDNA via extracellular vesicles was identified as a novel mechanism of lung cancer metastasis [160]. Similarly, cancer cells appear to hijack and transfer nearby immune cell mitochondrion to promote cell growth and proliferation [161]. These imaging techniques combined with functional approaches to evaluate both the tumor and environment hold the promise of revolutionizing the fields of onco-anatomy and cancer bioenergetics.
4. Perturbing mitochondrial functions as a cancer therapeutic strategy
Cancer cells utilize evolutionarily conserved mitochondrial functions to sustain carcinogenesis and evade death signals based upon the fluctuating TME and subcellular metabolic landscape (Figure 4). Given the numerous nodes of cancer initiation and support, mitochondria serve as targets for a wide range of promising anti-cancer agents (Table 1). Several key TCA cycle enzymes, such as pyruvate dehydrogenase (PDH), isocitrate dehydrogenase (IDH), and α-KG dehydrogenase (KGDHC) and metabolite transporters, such as the mitochondrial pyruvate carrier (MPC), are being investigated for their anti-cancer potential in clinical trials [162]. CPI-613, a potential PDH and KGDHC inhibitor, which failed in a phase III clinical trial of pancreatic cancer [163] as well as acute myeloid leukemia in combination with chemotherapy (NCT03504410) is now being tested in combination therapies such as chemoradiation (NCT05325281).
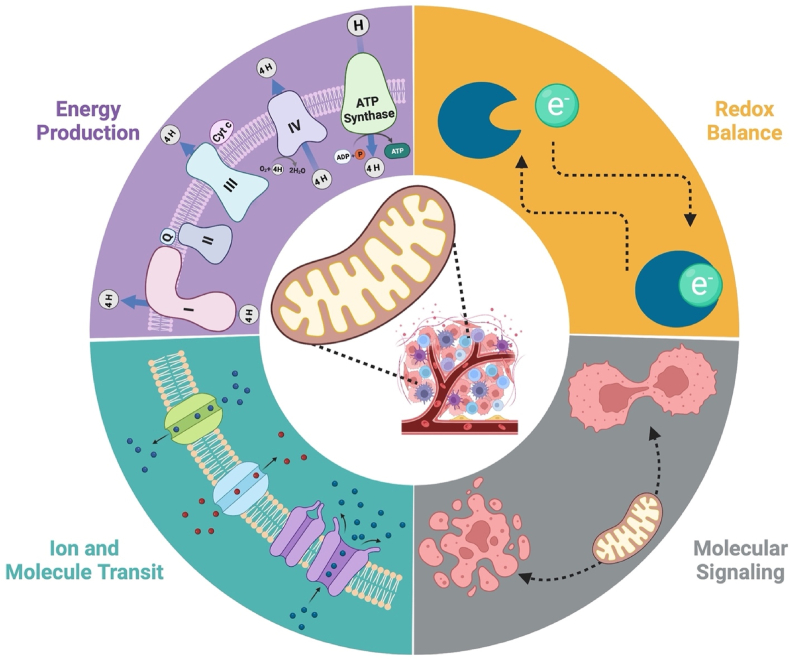
Figure 4.
Mitochondrial bioenergetic regulation of cancer cell survival and death evasion.
Mitochondria confer cancer survival and death evasion through the leveraging of highly plastic, conserved, and interconnected biological processes orchestrated through a dynamic bioenergetic system. Energy transformation through oxidative phosphorylation (OXPHOS) and electron transfer (ET) are dictated by coupling efficiency, the subcellular ADP/ATP ratios, and the availability of reducing equivalents. Redox networks within the mitochondria provide reducing equivalents requisite for energy transformation as well as managing mitochondrial reactive oxygen species (ROS) produced. The mitochondria require ion and molecule transport into the organelle to sustain mitochondrial morphology, driving forces, and functions. Additionally, mitochondria provide molecules and ions to the cancer cell. These metabolic byproducts can highly influence cell fate through molecular signaling cascades generated from the mitochondria.
Table 1.
Mitochondrial targets of anticancer agents.
| Biological process | Mitochondrial Target | Drug name | Level of evidence |
|---|---|---|---|
| TCA cycle enzymes | PDH and KGDHC inhibitor | CPI-613 (Devimistat) |
|
| IDH inhibitor | AG-221and AG-881 |
|
|
| IDH inhibitor | AGI-5198 |
|
|
| Metabolite transporters | MPC inhibitor | UK-5099 |
|
| HSP90 inhibitor | Gamitrinib |
|
|
| OXPHOS/ET | Complex II inhibitor Uncoupler |
Sorafenib |
|
| Complex I and ATP synthase | Tamoxifen |
|
|
| Complex I inhibitor | Metformin |
|
|
| Complex I inhibitor | IACS-010759 |
|
|
| Complex II inhibitor | α-tocopherol succinate |
|
|
| Iron chelator that inhibits complexes I, II, and IV | VLX600 |
|
|
| Methylation-controlled J protein that negatively regulates complex I | MCJ mimetics (MITOx20, MITOx30) |
|
|
| Uncoupler | NEN |
|
|
| Uncoupler | Nitazoxanide |
|
|
| Uncoupler DNA topoisomerase II inhibitor |
Ellipticine and derivatives |
|
|
| Uncoupler | CRMP |
|
|
| Uncoupler | BAM15 |
|
|
| Uncoupler | SR4 |
|
|
| Uncoupler | FR58P1a |
|
|
| Uncoupler Tyrosine kinase inhibitor |
Tyrphositn A9 |
|
|
| Uncoupler EAPC inhibitor |
ESI-09 |
|
|
| Antioxidants | Blocks GSH synthesis | Buthionine sulfoximine |
|
| Trx inhibitor | PX-12 |
|
|
| GPD2-dependent ROS formation inhibitor | KM04416 |
|
Inhibiting specific mitochondrial functions such as OXPHOS, electron transfer, biosynthesis, and/or metabolite signaling are being investigated extensively for antineoplastic activity [164]. Some FDA-approved anti-cancer therapies target mitochondrial function to inhibit cancer progression, such as Sorafenib, a mitochondrial Complex II inhibitor [165] and Tamoxifen, which inhibit Complex I and ATP synthase [166]. Metformin, a potential Complex I inhibitor, which is being tested in clinical trials for colorectal, breast, and prostate cancer [162] recently failed in a phase III clinical trial in breast cancer [167]. The small-molecule complex I inhibitor, IACS-010759, has a narrow therapeutic index and dose limiting toxicities and recently failed in phase I clinical trials in acute myeloid leukemia and sold tumors [168]. Antioxidant agents targeting glutathione (GSH) and thioredoxin (Trx) also demonstrate anti-cancer potential in current clinical trials [169]. Transfer of electron flow from Complex II to III with α-tocopherol succinate restricts proliferation and induces apoptosis in cancer cells [170]. Gamitrinib, a mitochondrially localized ATPase inhibitor of HSP90, limits carcinogenic potential in multiple tumor and cancer cell types [171,172], has displayed promising preclinical safety in dogs, and has advanced to a Phase I clinical trial [173]. VLX600, an iron chelator reported to inhibit Complexes I, II, and IV, similarly compromises tumor growth in models of colorectal cancer [171] was successful in a Phase I trial in patients with advanced solid tumors [174]. Additionally, chemoresistant cells that rely on mitochondrial ATP for transporter-mediated drug efflux are sensitized to chemotherapy using a methylation-controlled J protein that negatively regulates complex I and decreases respiration [175]. Inhibitors specifically targeting GPD2-dependent ROS formation are also being investigated [176,177], however, little is known about whether G3P oxidation alters cancer cell function and/or survival and further investigation is required. Collectively, these studies demonstrate that limiting mitochondrial function favorably augments cancer cell growth, but that specific inhibition has not yet yielded an efficacious anticancer therapeutic.
Dose limiting toxicity and lactic acidosis are major obstacles for mitochondrial inhibitors [178]. Compensatory and coordinated upregulation of other respiratory components or glycolysis to evade death and ensure survival are another limitation of inhibiting a specific component of the ETS. For example, pyruvate kinase interacts with mitofusin 2 on the outer mitochondrial membrane to signal demand for glycolytic flux [179]. Similarly, limiting the supply of NADH and FADH2 increases glycolytic ATP production [180]. Thus, the ideal mitochondrially targeted antineoplastic agent would sufficiently restrict ATP production and limit cytosolic ATP content below what is required to fuel hexokinase and phosphofructokinase, such that glycolysis could no longer proceed. One means of achieving such effect is to use combinatorial inhibition of upstream regulators of glycolysis and electron transfer. To this end, inhibition of Complex I via metformin synergizes with the prototype glycolysis inhibitor 2-deoxy-d-glucose to drive tumor regression [181].
Conversely, manipulating bioenergetic efficiency has the advantage of targeting the energy transformation process without the direct inhibition that may be leveraged in a cancer-specific way. Several mitochondrial uncouplers have demonstrated clinical tolerability and efficacy in the context of other metabolic diseases such as obesity and steatotic liver disease [182]. The efficacy of mitochondrial uncouplers as antineoplastic agents has been demonstrated preclinically across cancer models, with several entering clinical trial evaluation [183]. Controlled-release mitochondrial protonophore (CRMP), a chemically modified derivative of native DNP, was highly tolerable and reduced tumor growth in multiple pre-clinical models of colon cancer [184]. F16, a mitochondrial-localized lipophilic cation, inhibits cell cycle progression and induces cell death dependent upon on its uncoupling activity [185]. ESI-09, an exchange protein directly activated by cAMP (EAPC) inhibitor, was identified as a novel uncoupler with anticancer effects under low glucose conditions, specifically targeting quiescent cells [186]. The newly identified uncoupler FR58P1a inhibits migration of TNBC cells via Sirt1/AMPK/β1-integrin pathway [187]. Congruently, BAM15, a highly potent mitochondrial uncoupler, limits proliferation and initiates apoptosis in vitro and in vivo models of breast cancer by inhibition of ATP production and sustained mitochondrial stress [121]. BAM15 has also been found to inhibit proliferation of acute myeloid leukemia [188] and synergize with mitogen-activated protein kinase inhibitors to induce cell death in melanoma cells [189].
A subset of FDA-approved drugs were discovered to have uncoupling activity via untargeted library screens [190]. Sorafenib, a tyrosine kinase inhibitor used in standard-of-care chemotherapy for hepatocellular carcinoma, displays uncoupling activity at low-doses [191]. Niclosamide ethanolamine (NEN), commonly used for the treatment of parasitic infections, is a highly tolerable and potent uncoupler in addition to its other mechanisms of action [192]. NEN has been clinically investigated for its anticancer efficacy but was not sufficiently enriched to exert a direct anti-proliferative effect, in prostate cancer [193]. Reformulated nicolsamide successfully achieved target plasma levels in a Phase 1b trial [153]. Additionally, there have been other clinical trials using NEN for treatment, in prostate cancer one Phase II trial is ongoing (NCT02807805), and the trial in colon cancer was terminated due to low accrual (NCT02687009). Nitazoxanide, another antiparasitic with uncoupling potency, was tolerable and efficacious in combination with chemotherapy in chemotherapy-resistant nasopharyngeal carcinoma [194]. Ellipticine, a DNA topoisomerase II inhibitor, and its derivatives have well documented antitumor activity mediated by uncoupling but have not proceeded past Phase II clinical trials in cancer due to high incidence of toxicity [195]. Tyrphostin A9, a tyrosine kinase inhibitor, is considered one of the most powerful uncouplers discovered to date with potency in the nM range and the highest mtIM cycling rates [196] and has displayed powerful potency and demonstrated preliminary anticancer efficacy [197]. 1,3bis (dichlorophenyl)urea (SR4) a chemical uncoupler, was also identified as a potential anticancer therapeutic in a drug screen using leukemia HL-60 cells [198] and has demonstrated preliminary efficacy in preclinical models of obesity and leukemia, lung, melanoma, and hepatocellular carcinoma cancer models [199]. Overall, there is experimental evidence that uncoupling agents restrict proliferation and induce apoptosis in models of cancer. However, in vitro proof-of-concept studies have limited generalizability since tumors scavenge and self-sacrifice for ATP, conferring a survival advantage beyond individual cells. Increased preclinical and clinical investigation of uncoupling agents are needed to identify compounds that are both tolerable and effective.
5. Conclusions
Mitochondria are essential to cancer cell survival and progression by providing the energy and macromolecules required to initiate and sustain growth. Limitations in uncovering the contribution of mitochondrial bioenergetics to cancer pathophysiology persist due to the fact that tumors are heterogeneous tissues, housing a multiplicity of cell types in a dynamic extracellular environment. Diagnostic probing of the mitochondrial bioenergetic system within and around tumors will help to elucidate metabolic vulnerabilities over the cancer continuum and in response to primary therapies. Furthermore, therapies that limit the efficiency of mitochondrial ATP production will likely play an important role in primary treatment of cancer as well as in overcoming therapeutic resistance.
Funding
This research was supported in part by the National Institute of Health grants U54GM104940 (JPK) and T32 AT004094 (ERMZ).
CRediT authorship contribution statement
Elizabeth R.M. Zunica: Writing – review & editing, Writing – original draft, Visualization, Funding acquisition, Conceptualization. Christopher L. Axelrod: Writing – review & editing, Writing – original draft, Visualization, Conceptualization. L. Anne Gilmore: Writing – review & editing, Supervision. Erich Gnaiger: Writing – review & editing. John P. Kirwan: Writing – review & editing, Supervision, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank previous and current members of the Integrated Physiology and Molecular Medicine Laboratory, in particular Charles L. Hoppel, for consideration and contribution to this work. All figures were created using Biorender.com.
Abbreviations
- ATP
adenosine triphosphate
- TME
tumor microenvironment
- ΔμH+
transmembrane chemical potential difference [kJ·mol−1] or [mV]
- H+
hydrogen ion, hydron
- Pi
inorganic phosphate
- ADP
adenosine diphosphate
- Δp
pmF; protonmotive force [kJ·mol−1] or [mV]
- ΔpH
transmembrane pH difference
- ΔΨp+
mt-transmembrane electric potential difference [mV] or [kJ·mol−1]
- Redox
reduction-oxidation
- NAD+/NADH
nicotinamide adenine dinucleotide reducing equivalents
- FAD/FADH2
flavin adenine dinucleotide reducing equivalents
- ROS
reactive oxygen species
- TCA
tricarboxylic acid
- ETS
electron transfer system
- OXPHOS
oxidative phosphorylation
- mtDNA
mitochondrial DNA
- MCTs
monocarboxylate transporters
- G3P
glycerol-3-phosphate
- FAO
fatty acid oxidation
- mtOM
mitochondrial outer membrane
- mtIM
mitochondrial inner membrane
- iMS
intermembrane space
- VDACs
voltage-dependent anion channels
- MCs
metabolite carriers
Data availability
No data was used for the research described in the article.
References
- 1.DeBerardinis R.J., Chandel N.S. Fundamentals of cancer metabolism. Sci Adv. 2016;2(5) doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diehl F.F., Sapp K.M., Vander Heiden M.G. The bidirectional relationship between metabolism and cell cycle control. Trends Cell Biol. 2023;34(2):136–149. doi: 10.1016/j.tcb.2023.05.012. [DOI] [PubMed] [Google Scholar]
- 3.Weinberg S.E., Chandel N.S. Targeting mitochondria metabolism for cancer therapy. Nat Chem Biol. 2015;11(1):9–15. doi: 10.1038/nchembio.1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wallace D.C. Mitochondria and cancer. Nat Rev Cancer. 2012;12(10):685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McBride H.M., Neuspiel M., Wasiak S. Mitochondria: more than just a powerhouse. Curr Biol. 2006;16(14):R551–R560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 6.Frey T.G., Renken C.W., Perkins G.A. Insight into mitochondrial structure and function from electron tomography. Biochim Biophys Acta Bioenerg. 2002;1555(1):196–203. doi: 10.1016/s0005-2728(02)00278-5. [DOI] [PubMed] [Google Scholar]
- 7.Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature. 1961;191:144–148. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- 8.Berg J.M.T.J., Stryer L. Biochemistry. 5th ed. W H Freeman; New York: 2002. Chapter 17, the citric acid cycle. [Google Scholar]
- 9.Gnaiger E. Heat dissipation and energetic efficiency in animal anoxibiosis: economy contra power. J Exp Zool. 1983;228(3):471–490. [Google Scholar]
- 10.Gnaiger E. Surviving hypoxia: mechanisms of control and adaptation. CRC Press; Boca Raton, Ann Arbor, London, Tokyo: 1993. Efficiency and power strategies under hypoxia. Is low efficiency at high glycolytic ATP production a paradox? pp. 77–109. [Google Scholar]
- 11.Gibala M.J., MacLean D.A., Graham T.E., Saltin B. Tricarboxylic acid cycle intermediate pool size and estimated cycle flux in human muscle during exercise. Am J Physiol. 1998;275(2):E235–E242. doi: 10.1152/ajpendo.1998.275.2.E235. [DOI] [PubMed] [Google Scholar]
- 12.Berg J.M.T.J., Stryer L. Biochemistry. 5th ed. W H Freeman; New York: 2002. Chapter 18, oxidative phosphorylation. [Google Scholar]
- 13.Vercellino I., Sazanov L.A. The assembly, regulation and function of the mitochondrial respiratory chain. Nat Rev Mol Cell Biol. 2021;23(2):141–161. doi: 10.1038/s41580-021-00415-0. [DOI] [PubMed] [Google Scholar]
- 14.Vartak R., Deng J., Fang H., Bai Y. Redefining the roles of mitochondrial DNA-encoded subunits in respiratory Complex I assembly. Biochim Biophys Acta. 2015;1852(7):1531–1539. doi: 10.1016/j.bbadis.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rutter J., Winge D.R., Schiffman J.D. Succinate dehydrogenase - assembly, regulation and role in human disease. Mitochondrion. 2010;10(4):393–401. doi: 10.1016/j.mito.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xia D., Yu C.-A., Zhou F., Esser L. In: Encyclopedia of biophysics. Roberts G., Watts A., editors. Springer Berlin Heidelberg; Berlin, Heidelberg: 2018. Ubiquinol-cytochrome c oxidoreductase (complex III) pp. 1–8. [Google Scholar]
- 17.Zong S., Wu M., Gu J., Liu T., Guo R., Yang M. Structure of the intact 14-subunit human cytochrome c oxidase. Cell Res. 2018;28(10):1026–1034. doi: 10.1038/s41422-018-0071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Demine S., Renard P., Arnould T. Mitochondrial uncoupling: a key controller of biological processes in Physiology and diseases. Cells. 2019;8(8) doi: 10.3390/cells8080795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slater E.C. In: Metabolic inhibitors. Hochster R.M., Quastel J.H., editors. Academic Press; 1963. Chapter 32 - uncouplers and inhibitors of oxidative phosphorylation; pp. 503–516. [Google Scholar]
- 20.Heytler P.G. Uncouplers of oxidative phosphorylation. Methods Enzymol. 1979;55 doi: 10.1016/0076-6879(79)55060-5. 462-42. [DOI] [PubMed] [Google Scholar]
- 21.Rottenberg H. Decoupling of oxidative phosphorylation and photophosphorylation. Biochim Biophys Acta. 1990;1018(1):1–17. doi: 10.1016/0005-2728(90)90103-b. [DOI] [PubMed] [Google Scholar]
- 22.Bertholet A.M., Natale A.M., Bisignano P., Suzuki J., Fedorenko A., Hamilton J., et al. Mitochondrial uncouplers induce proton leak by activating AAC and UCP1. Nature. 2022;606(7912):180–187. doi: 10.1038/s41586-022-04747-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Axelrod C.L., King W.T., Davuluri G., Noland R.C., Hall J., Hull M., et al. BAM15-mediated mitochondrial uncoupling protects against obesity and improves glycemic control. EMBO Mol Med. 2020;12(7) doi: 10.15252/emmm.202012088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gnaiger E., Méndez G., Hand S.C. High phosphorylation efficiency and depression of uncoupled respiration in mitochondria under hypoxia. Proc Natl Acad Sci USA. 2000;97(20):11080–11085. doi: 10.1073/pnas.97.20.11080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flamholz A., Phillips R., Milo R. The quantified cell. Mol Biol Cell. 2014;25(22):3497–3500. doi: 10.1091/mbc.E14-09-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rechsteiner M., Hillyard D., Olivera B.M. Turnover of nicotinamide adenine dinucleotide in cultures of human cells. J Cell Physiol. 1976;88(2):207–217. doi: 10.1002/jcp.1040880210. [DOI] [PubMed] [Google Scholar]
- 27.Lunt S.Y., Vander Heiden M.G. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 28.Berg J.M.T.J., Stryer L. Biochemistry. 5th ed. W H Freeman; New York: 2002. Section 16.1, glycolysis is an energy-conversion pathway in many organisms. [Google Scholar]
- 29.Mookerjee S.A., Gerencser A.A., Nicholls D.G., Brand M.D. Quantifying intracellular rates of glycolytic and oxidative ATP production and consumption using extracellular flux measurements. J Biol Chem. 2017;292(17):7189–7207. doi: 10.1074/jbc.M116.774471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rabinowitz J.D., Enerbäck S. Lactate: the ugly duckling of energy metabolism. Nat Metab. 2020;2(7):566–571. doi: 10.1038/s42255-020-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hopp A.K., Grüter P., Hottiger M.O. Regulation of glucose metabolism by NAD(+) and ADP-ribosylation. Cells. 2019;8(8) doi: 10.3390/cells8080890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stein L.R., Imai S-i. The dynamic regulation of NAD metabolism in mitochondria. Trends Endocrinol Metabol: TEM. 2012;23(9):420–428. doi: 10.1016/j.tem.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spinelli J.B., Haigis M.C. The multifaceted contributions of mitochondria to cellular metabolism. Nat Cell Biol. 2018;20(7):745–754. doi: 10.1038/s41556-018-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mráček T., Drahota Z., Houštěk J. The function and the role of the mitochondrial glycerol-3-phosphate dehydrogenase in mammalian tissues. Biochim Biophys Acta Bioenerg. 2013;1827(3):401–410. doi: 10.1016/j.bbabio.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 35.Berg J.M.T.J., Stryer L. Biochemistry. 5th ed. W H Freeman; New York: 2002. Section 23.5, carbon atoms of degraded amino acids emerge as major metabolic intermediates. [Google Scholar]
- 36.Berg J.M.T.J., Stryer L. Biochemistry. W H Freeman; New York: 2002. Biochemistry. Section 22.2, the utilization of fatty acids as fuel requires three stages of processing. [Google Scholar]
- 37.Warburg O., Minami S. Versuche an überlebendem carcinom-gewebe. Klin Wochenschr. 1923;2(17):776–777. [Google Scholar]
- 38.Warburg O. Uber den Stoffwechsel der Karzinomezellen. Biochem Z. 1924;152:309–344. [Google Scholar]
- 39.Warburg O., Wind F., Negelein E. The metabolism of tumors in the body. J Gen Physiol. 1927;8(6):519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warburg O. In: Über die Katalytischen Wirkungen der Lebendigen Substanz: Arbeiten aus dem Kaiser Wilhelm-Institut für Biologie · Berlin-Dahlem. Warburg O., editor. Springer Berlin Heidelberg; Berlin, Heidelberg: 1928. Über den heutigen Stand des Carcinomproblems; pp. 501–509. [Google Scholar]
- 41.Cori C.F., Cori G.T. The carbohydrate metabolism of tumors: I. The free sugar, lactic acid, and glycogen content of malignant tumors. J Biol Chem. 1925;64(1):11–22. [Google Scholar]
- 42.Cori C.F., Cori G.T. The carbohydrate metabolism of tumors: II. Changes in the sugar, lactic acid, and CO2-COmbining power of blood passing through a tumor. J Biol Chem. 1925;65(2):397–405. [Google Scholar]
- 43.Cori C.F., Cori G.T. The carbohydrate metabolism of tumors: III. The rate of glycolysis of tumor tissue in the living animal. J Cancer Res. 1928;12(4):301–313. [Google Scholar]
- 44.Crabtree H.G. Observations on the carbohydrate metabolism of tumours. Biochem J. 1929;23(3):536–545. doi: 10.1042/bj0230536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crabtree H.G. The carbohydrate metabolism of certain pathological overgrowths. Biochem J. 1928;22(5):1289–1298. doi: 10.1042/bj0221289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 47.Racker E. Bioenergetics and the problem of tumor growth. Am Sci. 1972;60(1):56–63. [PubMed] [Google Scholar]
- 48.Zu X.L., Guppy M. Cancer metabolism: facts, fantasy, and fiction. Biochem Biophys Res Commun. 2004;313(3):459–465. doi: 10.1016/j.bbrc.2003.11.136. [DOI] [PubMed] [Google Scholar]
- 49.Ashton T.M., McKenna W.G., Kunz-Schughart L.A., Higgins G.S. Oxidative phosphorylation as an emerging target in cancer therapy. Clin Cancer Res. 2018;24(11):2482–2490. doi: 10.1158/1078-0432.CCR-17-3070. [DOI] [PubMed] [Google Scholar]
- 50.Moreno-Sánchez R., Marín-Hernández A., Saavedra E., Pardo J.P., Ralph S.J., Rodríguez-Enríquez S. Who controls the ATP supply in cancer cells? Biochemistry lessons to understand cancer energy metabolism. Int J Biochem Cell Biol. 2014;50:10–23. doi: 10.1016/j.biocel.2014.01.025. [DOI] [PubMed] [Google Scholar]
- 51.Li F., Simon M.C. Cancer cells don't live alone: metabolic communication within tumor microenvironments. Dev Cell. 2020;54(2):183–195. doi: 10.1016/j.devcel.2020.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Visser K.E., Joyce J.A. The evolving tumor microenvironment: from cancer initiation to metastatic outgrowth. Cancer Cell. 2023;41(3):374–403. doi: 10.1016/j.ccell.2023.02.016. [DOI] [PubMed] [Google Scholar]
- 53.Pavlides S., Whitaker-Menezes D., Castello-Cros R., Flomenberg N., Witkiewicz A.K., Frank P.G., et al. The reverse Warburg effect: aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell Cycle. 2009;8(23):3984–4001. doi: 10.4161/cc.8.23.10238. [DOI] [PubMed] [Google Scholar]
- 54.Mayer S., Milo T., Isaacson A., Halperin C., Miyara S., Stein Y., et al. The tumor microenvironment shows a hierarchy of cell-cell interactions dominated by fibroblasts. Nat Commun. 2023;14(1):5810. doi: 10.1038/s41467-023-41518-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reinfeld B.I., Madden M.Z., Wolf M.M., Chytil A., Bader J.E., Patterson A.R., et al. Cell-programmed nutrient partitioning in the tumour microenvironment. Nature. 2021;593(7858):282–288. doi: 10.1038/s41586-021-03442-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bi G., Bian Y., Liang J., Yin J., Li R., Zhao M., et al. Pan-cancer characterization of metabolism-related biomarkers identifies potential therapeutic targets. J Transl Med. 2021;19(1):219. doi: 10.1186/s12967-021-02889-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Danhier P., Bański P., Payen V.L., Grasso D., Ippolito L., Sonveaux P., et al. Cancer metabolism in space and time: beyond the Warburg effect. Biochim Biophys Acta Bioenerg. 2017;1858(8):556–572. doi: 10.1016/j.bbabio.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 58.Delaunay S., Pascual G., Feng B., Klann K., Behm M., Hotz-Wagenblatt A., et al. Mitochondrial RNA modifications shape metabolic plasticity in metastasis. Nature. 2022;607(7919):593–603. doi: 10.1038/s41586-022-04898-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burns J.S., Manda G. Metabolic pathways of the Warburg effect in health and disease: perspectives of choice, chain or chance. Int J Mol Sci. 2017;18(12) doi: 10.3390/ijms18122755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kovačević Z., Morris H.P. The role of glutamine in the oxidative metabolism of malignant cells. Cancer Res. 1972;32(2):326–333. [PubMed] [Google Scholar]
- 61.Bhutia Y.D., Babu E., Ramachandran S., Ganapathy V. Amino Acid transporters in cancer and their relevance to "glutamine addiction": novel targets for the design of a new class of anticancer drugs. Cancer Res. 2015;75(9):1782–1788. doi: 10.1158/0008-5472.CAN-14-3745. [DOI] [PubMed] [Google Scholar]
- 62.Fan J., Kamphorst J.J., Mathew R., Chung M.K., White E., Shlomi T., et al. Glutamine-driven oxidative phosphorylation is a major ATP source in transformed mammalian cells in both normoxia and hypoxia. Mol Syst Biol. 2013;9:712. doi: 10.1038/msb.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cori C.F., Cori G.T. GLYCOGEN FORMATION IN THE LIVER FROM d- AND l-LACTIC ACID. J Biol Chem. 1929;81(2):389–403. [Google Scholar]
- 64.Martínez-Reyes I., Chandel N.S. Waste not, want not: lactate oxidation fuels the TCA cycle. Cell Metabol. 2017;26(6):803–804. doi: 10.1016/j.cmet.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 65.Hui S., Ghergurovich J.M., Morscher R.J., Jang C., Teng X., Lu W., et al. Glucose feeds the TCA cycle via circulating lactate. Nature. 2017;551(7678):115–118. doi: 10.1038/nature24057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.San-Millán I., Brooks G.A. Reexamining cancer metabolism: lactate production for carcinogenesis could be the purpose and explanation of the Warburg Effect. Carcinogenesis. 2017;38(2):119–133. doi: 10.1093/carcin/bgw127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grasmann G., Smolle E., Olschewski H., Leithner K. Gluconeogenesis in cancer cells - repurposing of a starvation-induced metabolic pathway? Biochim Biophys Acta Rev Cancer. 2019;1872(1):24–36. doi: 10.1016/j.bbcan.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Patra K.C., Hay N. The pentose phosphate pathway and cancer. Trends Biochem Sci. 2014;39(8):347–354. doi: 10.1016/j.tibs.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pascual G., Avgustinova A., Mejetta S., Martín M., Castellanos A., Attolini C.S., et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature. 2017;541(7635):41–45. doi: 10.1038/nature20791. [DOI] [PubMed] [Google Scholar]
- 70.Camarda R., Zhou A.Y., Kohnz R.A., Balakrishnan S., Mahieu C., Anderton B., et al. Inhibition of fatty acid oxidation as a therapy for MYC-overexpressing triple-negative breast cancer. Nat Med. 2016;22(4):427–432. doi: 10.1038/nm.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Padanad M.S., Konstantinidou G., Venkateswaran N., Melegari M., Rindhe S., Mitsche M., et al. Fatty acid oxidation mediated by acyl-CoA synthetase long chain 3 is required for mutant KRAS lung tumorigenesis. Cell Rep. 2016;16(6):1614–1628. doi: 10.1016/j.celrep.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Koundouros N., Poulogiannis G. Reprogramming of fatty acid metabolism in cancer. Br J Cancer. 2020;122(1):4–22. doi: 10.1038/s41416-019-0650-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Okumura T., Ohuchida K., Sada M., Abe T., Endo S., Koikawa K., et al. Extra-pancreatic invasion induces lipolytic and fibrotic changes in the adipose microenvironment, with released fatty acids enhancing the invasiveness of pancreatic cancer cells. Oncotarget. 2017;8(11):18280–18295. doi: 10.18632/oncotarget.15430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nagarajan S.R., Butler L.M., Hoy A.J. The diversity and breadth of cancer cell fatty acid metabolism. Cancer Metabol. 2021;9(1):2. doi: 10.1186/s40170-020-00237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Falchook G., Infante J., Arkenau H.-T., Patel M.R., Dean E., Borazanci E., et al. First-in-human study of the safety, pharmacokinetics, and pharmacodynamics of first-in-class fatty acid synthase inhibitor TVB-2640 alone and with a taxane in advanced tumors. eClinicalMedicine. 2021;34 doi: 10.1016/j.eclinm.2021.100797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou X., Huang F., Ma G., Wei W., Wu N., Liu Z. Dysregulated ceramides metabolism by fatty acid 2-hydroxylase exposes a metabolic vulnerability to target cancer metastasis. Signal Transduct Targeted Ther. 2022;7(1):370. doi: 10.1038/s41392-022-01199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lamb R., Harrison H., Hulit J., Smith D.L., Lisanti M.P., Sotgia F. Mitochondria as new therapeutic targets for eradicating cancer stem cells: quantitative proteomics and functional validation via MCT1/2 inhibition. Oncotarget. 2014;5(22) doi: 10.18632/oncotarget.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim J., DeBerardinis R.J. Mechanisms and implications of metabolic heterogeneity in cancer. Cell Metabol. 2019;30(3):434–446. doi: 10.1016/j.cmet.2019.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Berridge M.V., Herst P.M., Tan A.S. Metabolic flexibility and cell hierarchy in metastatic cancer. Mitochondrion. 2010;10(6):584–588. doi: 10.1016/j.mito.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 80.Georgakopoulos-Soares I., Chartoumpekis D.V., Kyriazopoulou V., Zaravinos A. EMT factors and metabolic pathways in cancer. Front Oncol. 2020;10 doi: 10.3389/fonc.2020.00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Arismendi-Morillo G. Electron microscopy morphology of the mitochondrial network in human cancer. Int J Biochem Cell Biol. 2009;41(10):2062–2068. doi: 10.1016/j.biocel.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 82.Trotta A.P., Chipuk J.E. Mitochondrial dynamics as regulators of cancer biology. Cell Mol Life Sci: CMLS. 2017;74(11):1999–2017. doi: 10.1007/s00018-016-2451-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sullivan L.B., Chandel N.S. Mitochondrial reactive oxygen species and cancer. Cancer Metabol. 2014;2:17. doi: 10.1186/2049-3002-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bonnet S., Archer S.L., Allalunis-Turner J., Haromy A., Beaulieu C., Thompson R., et al. A mitochondria-K+ channel axis is suppressed in cancer and its normalization promotes apoptosis and inhibits cancer growth. Cancer Cell. 2007;11(1):37–51. doi: 10.1016/j.ccr.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 85.Porporato P.E., Filigheddu N., Pedro J.M.B.-S., Kroemer G., Galluzzi L. Mitochondrial metabolism and cancer. Cell Res. 2018;28(3):265–280. doi: 10.1038/cr.2017.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vyas S., Zaganjor E., Haigis M.C. Mitochondria and cancer. Cell. 2016;166(3):555–566. doi: 10.1016/j.cell.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kreuzaler P., Panina Y., Segal J., Yuneva M. Adapt and conquer: metabolic flexibility in cancer growth, invasion and evasion. Mol Metabol. 2020;33:83–101. doi: 10.1016/j.molmet.2019.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Levine A.J. p53: 800 million years of evolution and 40 years of discovery. Nat Rev Cancer. 2020;20(8):471–480. doi: 10.1038/s41568-020-0262-1. [DOI] [PubMed] [Google Scholar]
- 89.Morrish F., Hockenbery D. MYC and mitochondrial biogenesis. Cold Spring Harbor Perspect Med. 2014;4(5):a014225. doi: 10.1101/cshperspect.a014225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Papandreou I., Cairns R.A., Fontana L., Lim A.L., Denko N.C. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metabol. 2006;3(3):187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 91.Palorini R., De Rasmo D., Gaviraghi M., Danna L.S., Signorile A., Cirulli C., et al. Oncogenic K-ras expression is associated with derangement of the cAMP/PKA pathway and forskolin-reversible alterations of mitochondrial dynamics and respiration. Oncogene. 2013;32(3):352–362. doi: 10.1038/onc.2012.50. [DOI] [PubMed] [Google Scholar]
- 92.Caino M.C., Chae Y.C., Vaira V., Ferrero S., Nosotti M., Martin N.M., et al. Metabolic stress regulates cytoskeletal dynamics and metastasis of cancer cells. J Clin Invest. 2013;123(7):2907–2920. doi: 10.1172/JCI67841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hoxhaj G., Manning B.D. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat Rev Cancer. 2020;20(2):74–88. doi: 10.1038/s41568-019-0216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Martinez-Outschoorn U.E., Pavlides S., Sotgia F., Lisanti M.P. Mitochondrial biogenesis drives tumor cell proliferation. Am J Pathol. 2011;178(5):1949–1952. doi: 10.1016/j.ajpath.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Larman T.C., DePalma S.R., Hadjipanayis A.G., Protopopov A., Zhang J., Gabriel S.B., et al. Spectrum of somatic mitochondrial mutations in five cancers. Proc Natl Acad Sci U S A. 2012;109(35):14087–14091. doi: 10.1073/pnas.1211502109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yuan Y., Ju Y.S., Kim Y., Li J., Wang Y., Yoon C.J., et al. Comprehensive molecular characterization of mitochondrial genomes in human cancers. Nat Genet. 2020;52(3):342–352. doi: 10.1038/s41588-019-0557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Reznik E., Miller M.L., Şenbabaoğlu Y., Riaz N., Sarungbam J., Tickoo S.K., et al. Mitochondrial DNA copy number variation across human cancers. Elife. 2016;5 doi: 10.7554/eLife.10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tan Z., Luo X., Xiao L., Tang M., Bode A.M., Dong Z., et al. The role of PGC1α in cancer metabolism and its therapeutic implications. Mol Cancer Therapeut. 2016;15(5):774–782. doi: 10.1158/1535-7163.MCT-15-0621. [DOI] [PubMed] [Google Scholar]
- 99.Andrzejewski S., Klimcakova E., Johnson R.M., Tabariès S., Annis M.G., McGuirk S., et al. PGC-1α promotes breast cancer metastasis and confers bioenergetic flexibility against metabolic drugs. Cell Metabol. 2017;26(5):778–787. doi: 10.1016/j.cmet.2017.09.006. e5. [DOI] [PubMed] [Google Scholar]
- 100.Luo C., Lim J.H., Lee Y., Granter S.R., Thomas A., Vazquez F., et al. A PGC1α-mediated transcriptional axis suppresses melanoma metastasis. Nature. 2016;537(7620):422–426. doi: 10.1038/nature19347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sun X., Zhan L., Chen Y., Wang G., He L., Wang Q., et al. Increased mtDNA copy number promotes cancer progression by enhancing mitochondrial oxidative phosphorylation in microsatellite-stable colorectal cancer. Signal Transduct Targeted Ther. 2018;3(1):8. doi: 10.1038/s41392-018-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Anderson G.R., Wardell S.E., Cakir M., Yip C., Ahn Y-r, Ali M., et al. Dysregulation of mitochondrial dynamics proteins are a targetable feature of human tumors. Nat Commun. 2018;9(1):1677. doi: 10.1038/s41467-018-04033-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chan D.C. Mitochondrial dynamics and its involvement in disease. Annu Rev Pathol. 2020;15(1):235–259. doi: 10.1146/annurev-pathmechdis-012419-032711. [DOI] [PubMed] [Google Scholar]
- 104.Boulton D.P., Caino M.C. Mitochondrial fission and fusion in tumor progression to metastasis. Front Cell Dev Biol. 2022;10 doi: 10.3389/fcell.2022.849962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhao J., Zhang J., Yu M., Xie Y., Huang Y., Wolff D.W., et al. Mitochondrial dynamics regulates migration and invasion of breast cancer cells. Oncogene. 2013;32(40):4814–4824. doi: 10.1038/onc.2012.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen L., Zhang J., Lyu Z., Chen Y., Ji X., Cao H., et al. Positive feedback loop between mitochondrial fission and Notch signaling promotes survivin-mediated survival of TNBC cells. Cell Death Dis. 2018;9(11):1050. doi: 10.1038/s41419-018-1083-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Grieco J.P., Allen M.E., Perry J.B., Wang Y., Song Y., Rohani A., et al. Progression-mediated changes in mitochondrial morphology promotes adaptation to hypoxic peritoneal conditions in serous ovarian cancer. Front Oncol. 2021;10 doi: 10.3389/fonc.2020.600113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Humphries B.A., Cutter A.C., Buschhaus J.M., Chen Y.-C., Qyli T., Palagama D.S.W., et al. Enhanced mitochondrial fission suppresses signaling and metastasis in triple-negative breast cancer. Breast Cancer Res. 2020;22(1):60. doi: 10.1186/s13058-020-01301-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhou X., Li R., Chen R., Liu J. Altered mitochondrial dynamics, biogenesis, and functions in the paclitaxel-resistant lung adenocarcinoma cell line a549/taxol. Med Sci Mon Int Med J Exp Clin Res: Int Med J Exp Clin Res. 2020;26 doi: 10.12659/MSM.918216. [e] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Seo J.H., Agarwal E., Chae Y.C., Lee Y.G., Garlick D.S., Storaci A.M., et al. Mitochondrial fission factor is a novel Myc-dependent regulator of mitochondrial permeability in cancer. EBioMedicine. 2019;48:353–363. doi: 10.1016/j.ebiom.2019.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vara-Perez M., Felipe-Abrio B., Agostinis P. Mitophagy in cancer: a tale of adaptation. Cells. 2019;8(5):493. doi: 10.3390/cells8050493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bernardini J.P., Lazarou M., Dewson G. Parkin and mitophagy in cancer. Oncogene. 2017;36(10):1315–1327. doi: 10.1038/onc.2016.302. [DOI] [PubMed] [Google Scholar]
- 113.Liu H. Application of immunohistochemistry in breast pathology: a review and update. Arch Pathol Lab Med. 2014;138(12):1629–1642. doi: 10.5858/arpa.2014-0094-RA. [DOI] [PubMed] [Google Scholar]
- 114.Yao N., Wang C., Hu N., Li Y., Liu M., Lei Y., et al. Inhibition of PINK1/Parkin-dependent mitophagy sensitizes multidrug-resistant cancer cells to B5G1, a new betulinic acid analog. Cell Death Dis. 2019;10(3):232. doi: 10.1038/s41419-019-1470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chang J.Y., Yi H.S., Kim H.W., Shong M. Dysregulation of mitophagy in carcinogenesis and tumor progression. Biochim Biophys Acta Bioenerg. 2017;1858(8):633–640. doi: 10.1016/j.bbabio.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 116.Chourasia A.H., Tracy K., Frankenberger C., Boland M.L., Sharifi M.N., Drake L.E., et al. Mitophagy defects arising from BNip3 loss promote mammary tumor progression to metastasis. EMBO Rep. 2015;16(9):1145–1163. doi: 10.15252/embr.201540759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang C., Youle R.J. The role of mitochondria in apoptosis. Annu Rev Genet. 2009;43:95–118. doi: 10.1146/annurev-genet-102108-134850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fernald K., Kurokawa M. Evading apoptosis in cancer. Trends Cell Biol. 2013;23(12):620–633. doi: 10.1016/j.tcb.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Renault Thibaud T., Floros Konstantinos V., Elkholi R., Corrigan K.-A., Kushnareva Y., Wieder Shira Y., et al. Mitochondrial shape governs BAX-induced membrane permeabilization and apoptosis. Mol Cell. 2015;57(1):69–82. doi: 10.1016/j.molcel.2014.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gottlieb E., Armour S.M., Harris M.H., Thompson C.B. Mitochondrial membrane potential regulates matrix configuration and cytochrome c release during apoptosis. Cell Death Differ. 2003;10(6):709–717. doi: 10.1038/sj.cdd.4401231. [DOI] [PubMed] [Google Scholar]
- 121.Zunica E.R.M., Axelrod C.L., Cho E., Spielmann G., Davuluri G., Alexopoulos S.J., et al. Breast cancer growth and proliferation is suppressed by the mitochondrial targeted furazano[3,4-b]pyrazine BAM15. Cancer Metabol. 2021;9(1):36. doi: 10.1186/s40170-021-00274-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.MacDonald J.A., Kura N., Sussman C., Woods D.C. Mitochondrial membrane depolarization enhances TRAIL-induced cell death in adult human granulosa tumor cells, KGN, through inhibition of BIRC5. J Ovarian Res. 2018;11(1):89. doi: 10.1186/s13048-018-0463-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zorov D.B., Juhaszova M., Sollott S.J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol Rev. 2014;94(3):909–950. doi: 10.1152/physrev.00026.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shen S., Yan Z., Wu J., Liu X., Guan G., Zou C., et al. Characterization of ROS metabolic equilibrium reclassifies pan-cancer samples and guides pathway targeting therapy. Front Oncol. 2020;10 doi: 10.3389/fonc.2020.581197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pizzimenti S., Toaldo C., Pettazzoni P., Dianzani M.U., Barrera G. The "two-faced" effects of reactive oxygen species and the lipid peroxidation product 4-hydroxynonenal in the hallmarks of cancer. Cancers. 2010;2(2):338–363. doi: 10.3390/cancers2020338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sabharwal S.S., Dudley V.J., Landwerlin C., Schumacker P.T. H(2)O(2) transit through the mitochondrial intermembrane space promotes tumor cell growth in vitro and in vivo. J Biol Chem. 2023;299(5) doi: 10.1016/j.jbc.2023.104624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gorrini C., Harris I.S., Mak T.W. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov. 2013;12(12):931–947. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- 128.Liou G.-Y., Storz P. Reactive oxygen species in cancer. Free Radic Res. 2010;44(5):479–496. doi: 10.3109/10715761003667554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chen Z., Ho I.L., Soeung M., Yen E.-Y., Liu J., Yan L., et al. Ether phospholipids are required for mitochondrial reactive oxygen species homeostasis. Nat Commun. 2023;14(1):2194. doi: 10.1038/s41467-023-37924-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chen W.W., Freinkman E., Wang T., Birsoy K., Sabatini D.M. Absolute quantification of matrix metabolites reveals the dynamics of mitochondrial metabolism. Cell. 2016;166(5):1324–1337. doi: 10.1016/j.cell.2016.07.040. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sciacovelli M., Frezza C. Oncometabolites: unconventional triggers of oncogenic signalling cascades. Free Radic Biol Med. 2016;100:175–181. doi: 10.1016/j.freeradbiomed.2016.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Godel M., Ortone G., Anobile D.P., Pasino M., Randazzo G., Riganti C., et al. Targeting mitochondrial oncometabolites: a new approach to overcome drug resistance in cancer. Pharmaceutics. 2021;13(5):762. doi: 10.3390/pharmaceutics13050762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Patananan A.N., Sercel A.J., Teitell M.A. More than a powerplant: the influence of mitochondrial transfer on the epigenome. Curr Opin Physiol. 2018;3:16–24. doi: 10.1016/j.cophys.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Terunuma A., Putluri N., Mishra P., Mathé E.A., Dorsey T.H., Yi M., et al. MYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosis. J Clin Invest. 2014;124(1):398–412. doi: 10.1172/JCI71180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.San-Millán I., Julian C.G., Matarazzo C., Martinez J., Brooks G.A. Is lactate an oncometabolite? Evidence supporting a role for lactate in the regulation of transcriptional activity of cancer-related genes in MCF7 breast cancer cells. Front Oncol. 2020;9:1536. doi: 10.3389/fonc.2019.01536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gorelick A.N., Kim M., Chatila W.K., La K., Hakimi A.A., Berger M.F., et al. Respiratory complex and tissue lineage drive recurrent mutations in tumour mtDNA. Nat Metab. 2021;3(4):558–570. doi: 10.1038/s42255-021-00378-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ji X., Guo W., Gu X., Guo S., Zhou K., Su L., et al. Mutational profiling of mtDNA control region reveals tumor-specific evolutionary selection involved in mitochondrial dysfunction. EBioMedicine. 2022;80 doi: 10.1016/j.ebiom.2022.104058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ju Y.S., Alexandrov L.B., Gerstung M., Martincorena I., Nik-Zainal S., Ramakrishna M., et al. Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. Elife. 2014;3 doi: 10.7554/eLife.02935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kwok A.W.C., Qiao C., Huang R., Sham M.H., Ho J.W.K., Huang Y. MQuad enables clonal substructure discovery using single cell mitochondrial variants. Nat Commun. 2022;13(1):1205. doi: 10.1038/s41467-022-28845-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Miller T.E., Lareau C.A., Verga J.A., DePasquale E.A.K., Liu V., Ssozi D., et al. Mitochondrial variant enrichment from high-throughput single-cell RNA sequencing resolves clonal populations. Nat Biotechnol. 2022;40(7):1030–1034. doi: 10.1038/s41587-022-01210-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Schopf B., Weissensteiner H., Schafer G., Fazzini F., Charoentong P., Naschberger A., et al. OXPHOS remodeling in high-grade prostate cancer involves mtDNA mutations and increased succinate oxidation. Nat Commun. 2020;11(1):1487. doi: 10.1038/s41467-020-15237-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sant'Anna-Silva A.C.B., Perez-Valencia J.A., Sciacovelli M., Lalou C., Sarlak S., Tronci L., et al. Succinate anaplerosis has an onco-driving potential in prostate cancer cells. Cancers (Basel) 2021;13(7) doi: 10.3390/cancers13071727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mok B.Y., de Moraes M.H., Zeng J., Bosch D.E., Kotrys A.V., Raguram A., et al. A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature. 2020;583(7817):631–637. doi: 10.1038/s41586-020-2477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sharma L.K., Fang H., Liu J., Vartak R., Deng J., Bai Y. Mitochondrial respiratory complex I dysfunction promotes tumorigenesis through ROS alteration and AKT activation. Hum Mol Genet. 2011;20(23):4605–4616. doi: 10.1093/hmg/ddr395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mangalhara K.C., Varanasi S.K., Johnson M.A., Burns M.J., Rojas G.R., Esparza Moltó P.B., et al. Manipulating mitochondrial electron flow enhances tumor immunogenicity. Science. 2023;381(6664):1316–1323. doi: 10.1126/science.abq1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wu S., Mao C., Kondiparthi L., Poyurovsky M.V., Olszewski K., Gan B. A ferroptosis defense mechanism mediated by glycerol-3-phosphate dehydrogenase 2 in mitochondria. Proc Natl Acad Sci USA. 2022;119(26) doi: 10.1073/pnas.2121987119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yao C.H., Park J.S., Kurmi K., Hu S.H., Notarangelo G., Crowley J., et al. Uncoupled glycerol-3-phosphate shuttle in kidney cancer reveals that cytosolic GPD is essential to support lipid synthesis. Mol Cell. 2023;83(8):1340–1349 e7. doi: 10.1016/j.molcel.2023.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wu Y., Zhang X., Shen R., Huang J., Lu X., Zheng G., et al. Expression and significance of ETFDH in hepatocellular carcinoma. Pathol Res Pract. 2019;215(12) doi: 10.1016/j.prp.2019.152702. [DOI] [PubMed] [Google Scholar]
- 149.Liu Y., Mao C., Wang M., Liu N., Ouyang L., Liu S., et al. Cancer progression is mediated by proline catabolism in non-small cell lung cancer. Oncogene. 2020;39(11):2358–2376. doi: 10.1038/s41388-019-1151-5. [DOI] [PubMed] [Google Scholar]
- 150.Thapa M., Dallmann G. Role of coenzymes in cancer metabolism. Semin Cell Dev Biol. 2020;98:44–53. doi: 10.1016/j.semcdb.2019.05.027. [DOI] [PubMed] [Google Scholar]
- 151.Ikeda K., Horie-Inoue K., Suzuki T., Hobo R., Nakasato N., Takeda S., et al. Mitochondrial supercomplex assembly promotes breast and endometrial tumorigenesis by metabolic alterations and enhanced hypoxia tolerance. Nat Commun. 2019;10(1):4108. doi: 10.1038/s41467-019-12124-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hollinshead K.E.R., Parker S.J., Eapen V.V., Encarnacion-Rosado J., Sohn A., Oncu T., et al. Respiratory supercomplexes promote mitochondrial efficiency and growth in severely hypoxic pancreatic cancer. Cell Rep. 2020;33(1) doi: 10.1016/j.celrep.2020.108231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Bartman C.R., Weilandt D.R., Shen Y., Lee W.D., Han Y., TeSlaa T., et al. Slow TCA flux and ATP production in primary solid tumours but not metastases. Nature. 2023;614(7947):349–357. doi: 10.1038/s41586-022-05661-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Glancy B. Visualizing mitochondrial form and function within the cell. Trends Mol Med. 2020;26(1):58–70. doi: 10.1016/j.molmed.2019.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Collinson L.M., Bosch C., Bullen A., Burden J.J., Carzaniga R., Cheng C., et al. Volume EM: a quiet revolution takes shape. Nat Methods. 2023;20(6):777–782. doi: 10.1038/s41592-023-01861-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Han M., Bushong E.A., Segawa M., Tiard A., Wong A., Brady M.R., et al. Spatial mapping of mitochondrial networks and bioenergetics in lung cancer. Nature. 2023;615(7953):712–719. doi: 10.1038/s41586-023-05793-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.de Senneville B.D., Khoubai F.Z., Bevilacqua M., Labedade A., Flosseau K., Chardot C., et al. Deciphering tumour tissue organization by 3D electron microscopy and machine learning. Commun Biol. 2021;4(1):1390. doi: 10.1038/s42003-021-02919-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lee W.D., Mukha D., Aizenshtein E., Shlomi T. Spatial-fluxomics provides a subcellular-compartmentalized view of reductive glutamine metabolism in cancer cells. Nat Commun. 2019;10(1):1351. doi: 10.1038/s41467-019-09352-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Stern A., Fokra M., Sarvin B., Alrahem A.A., Lee W.D., Aizenshtein E., et al. Inferring mitochondrial and cytosolic metabolism by coupling isotope tracing and deconvolution. Nat Commun. 2023;14(1):7525. doi: 10.1038/s41467-023-42824-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhou Z., Qu C., Zhou P., Zhou Q., Li D., Wu X., et al. Extracellular vesicles activated cancer-associated fibroblasts promote lung cancer metastasis through mitophagy and mtDNA transfer. J Exp Clin Cancer Res. 2024;43(1):158. doi: 10.1186/s13046-024-03077-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhang H., Yu X., Ye J., Li H., Hu J., Tan Y., et al. Systematic investigation of mitochondrial transfer between cancer cells and T cells at single-cell resolution. Cancer Cell. 2023;41(10):1788–1802. doi: 10.1016/j.ccell.2023.09.003. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sainero-Alcolado L., Liaño-Pons J., Ruiz-Pérez M.V., Arsenian-Henriksson M. Targeting mitochondrial metabolism for precision medicine in cancer. Cell Death Differ. 2022;29(7):1304–1317. doi: 10.1038/s41418-022-01022-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Philip P.A., Bahary N., Mahipal A., Kasi A., Lima C.M.S.P.R., Alistar A.T., et al. Phase 3, multicenter, randomized study of CPI-613 with modified FOLFIRINOX (mFFX) versus FOLFIRINOX (FFX) as first-line therapy for patients with metastatic adenocarcinoma of the pancreas (AVENGER500) J Clin Oncol. 2022;40(16_suppl):4023. [Google Scholar]
- 164.Vasan K., Werner M., Chandel N.S. Mitochondrial metabolism as a target for cancer therapy. Cell Metabol. 2020;32(3):341–352. doi: 10.1016/j.cmet.2020.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhang C., Liu Z., Bunker E., Ramirez A., Lee S., Peng Y., et al. Sorafenib targets the mitochondrial electron transport chain complexes and ATP synthase to activate the PINK1-Parkin pathway and modulate cellular drug response. J Biol Chem. 2017;292(36):15105–15120. doi: 10.1074/jbc.M117.783175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Unten Y., Murai M., Koshitaka T., Kitao K., Shirai O., Masuya T., et al. Comprehensive understanding of multiple actions of anticancer drug tamoxifen in isolated mitochondria. Biochim Biophys Acta Bioenerg. 2022;1863(2) doi: 10.1016/j.bbabio.2021.148520. [DOI] [PubMed] [Google Scholar]
- 167.Goodwin P.J., Chen B.E., Gelmon K.A., Whelan T.J., Ennis M., Lemieux J., et al. Effect of metformin vs placebo on invasive disease-free survival in patients with breast cancer: the MA.32 randomized clinical trial. JAMA. 2022;327(20):1963–1973. doi: 10.1001/jama.2022.6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yap T.A., Daver N., Mahendra M., Zhang J., Kamiya-Matsuoka C., Meric-Bernstam F., et al. Complex I inhibitor of oxidative phosphorylation in advanced solid tumors and acute myeloid leukemia: phase I trials. Nat Med. 2023;29(1):115–126. doi: 10.1038/s41591-022-02103-8. [DOI] [PubMed] [Google Scholar]
- 169.Singh A., Faccenda D., Campanella M. Pharmacological advances in mitochondrial therapy. EBioMedicine. 2021;65 doi: 10.1016/j.ebiom.2021.103244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Dong L.F., Jameson V.J., Tilly D., Cerny J., Mahdavian E., Marin-Hernandez A., et al. Mitochondrial targeting of vitamin E succinate enhances its pro-apoptotic and anti-cancer activity via mitochondrial complex II. J Biol Chem. 2011;286(5):3717–3728. doi: 10.1074/jbc.M110.186643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Dong L., Gopalan V., Holland O., Neuzil J. Mitocans revisited: mitochondrial targeting as efficient anti-cancer therapy. Int J Mol Sci. 2020;21(21):7941. doi: 10.3390/ijms21217941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Reisbeck L., Linder B., Tascher G., Bozkurt S., Weber K.J., Herold-Mende C., et al. The iron chelator and OXPHOS inhibitor VLX600 induces mitophagy and an autophagy-dependent type of cell death in glioblastoma cells. Am J Physiol Cell Physiol. 2023;325(6):C1451–C1469. doi: 10.1152/ajpcell.00293.2023. [DOI] [PubMed] [Google Scholar]
- 173.Hayat U., Elliott G.T., Olszanski A.J., Altieri D.C. Feasibility and safety of targeting mitochondria for cancer therapy - preclinical characterization of gamitrinib, a first-in-class, mitochondriaL-targeted small molecule Hsp90 inhibitor. Cancer Biol Ther. 2022;23(1):117–126. doi: 10.1080/15384047.2022.2029132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Mody K., Mansfield A.S., Vemireddy L., Nygren P., Gulbo J., Borad M. A phase I study of the safety and tolerability of VLX600, an Iron Chelator, in patients with refractory advanced solid tumors. Invest N Drugs. 2019;37(4):684–692. doi: 10.1007/s10637-018-0703-9. [DOI] [PubMed] [Google Scholar]
- 175.Giddings E.L., Champagne D.P., Wu M.-H., Laffin J.M., Thornton T.M., Valenca-Pereira F., et al. Mitochondrial ATP fuels ABC transporter-mediated drug efflux in cancer chemoresistance. Nat Commun. 2021;12(1):2804. doi: 10.1038/s41467-021-23071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Raimondi V., Ciccarese F., Ciminale V. Oncogenic pathways and the electron transport chain: a dangeROS liaison. Br J Cancer. 2020;122(2):168–181. doi: 10.1038/s41416-019-0651-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Singh G. Mitochondrial FAD-linked glycerol-3-phosphate dehydrogenase: a target for cancer therapeutics. Pharmaceuticals. 2014;7(2):192–206. doi: 10.3390/ph7020192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhang X., Dang C.V. Time to hit pause on mitochondria-targeting cancer therapies. Nat Med. 2023;29(1):29–30. doi: 10.1038/s41591-022-02129-y. [DOI] [PubMed] [Google Scholar]
- 179.Li T., Han J., Jia L., Hu X., Chen L., Wang Y. PKM2 coordinates glycolysis with mitochondrial fusion and oxidative phosphorylation. Protein & Cell. 2019;10(8):583–594. doi: 10.1007/s13238-019-0618-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Liemburg-Apers D.C., Schirris T.J.J., Russel F.G.M., Willems P.H.G.M., Koopman W.J.H. Mitoenergetic dysfunction triggers a rapid compensatory increase in steady-state glucose flux. Biophys J. 2015;109(7):1372–1386. doi: 10.1016/j.bpj.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Cheng G., Zielonka J., Dranka B.P., McAllister D., Mackinnon A.C., Jr., Joseph J., et al. Mitochondria-targeted drugs synergize with 2-deoxyglucose to trigger breast cancer cell death. Cancer Res. 2012;72(10):2634–2644. doi: 10.1158/0008-5472.CAN-11-3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Noureddin M., Khan S., Portell F., Jorkasky D., Dennis J., Khan O., et al. Safety and efficacy of once-daily HU6 versus placebo in people with non-alcoholic fatty liver disease and high BMI: a randomised, double-blind, placebo-controlled, phase 2a trial. Lancet Gastroenterol Hepatol. 2023;8(12):1094–1105. doi: 10.1016/S2468-1253(23)00198-X. [DOI] [PubMed] [Google Scholar]
- 183.Shrestha R., Johnson E., Byrne F.L. Exploring the therapeutic potential of mitochondrial uncouplers in cancer. Mol Metabol. 2021;51 doi: 10.1016/j.molmet.2021.101222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Wang Y., Nasiri A.R., Damsky W.E., Perry C.J., Zhang X.-M., Rabin-Court A., et al. Uncoupling hepatic oxidative phosphorylation reduces tumor growth in two murine models of colon cancer. Cell Rep. 2018;24(1):47–55. doi: 10.1016/j.celrep.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wang J., He H., Xiang C., Fan X.Y., Yang L.Y., Yuan L., et al. Uncoupling effect of F16 is responsible for its mitochondrial toxicity and anticancer activity. Toxicol Sci. 2018;161(2):431–442. doi: 10.1093/toxsci/kfx218. [DOI] [PubMed] [Google Scholar]
- 186.Maeda Y., Kikuchi R., Kawagoe J., Tsuji T., Koyama N., Yamaguchi K., et al. Anti-cancer strategy targeting the energy metabolism of tumor cells surviving a low-nutrient acidic microenvironment. Mol Metabol. 2020;42 doi: 10.1016/j.molmet.2020.101093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Urra F.A., Muñoz F., Córdova-Delgado M., Ramírez M.P., Peña-Ahumada B., Rios M., et al. FR58P1a; a new uncoupler of OXPHOS that inhibits migration in triple-negative breast cancer cells via Sirt1/AMPK/β1-integrin pathway. Sci Rep. 2018;8(1) doi: 10.1038/s41598-018-31367-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Gao Zx, Cui Zl, Zhou Mr, Fu Y., Liu F., Zhang L., et al. The new mitochondrial uncoupler BAM15 induces ROS production for treatment of acute myeloid leukemia. Biochem Pharmacol. 2022;198 doi: 10.1016/j.bcp.2022.114948. [DOI] [PubMed] [Google Scholar]
- 189.Serasinghe M.N., Gelles J.D., Li K., Zhao L., Abbate F., Syku M., et al. Dual suppression of inner and outer mitochondrial membrane functions augments apoptotic responses to oncogenic MAPK inhibition. Cell Death Dis. 2018;9(2):29. doi: 10.1038/s41419-017-0044-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Goedeke L., Shulman G.I. Therapeutic potential of mitochondrial uncouplers for the treatment of metabolic associated fatty liver disease and NASH. Mol Metabol. 2021;46 doi: 10.1016/j.molmet.2021.101178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Jian C., Fu J., Cheng X., Shen L.-J., Ji Y.-X., Wang X., et al. Low-dose Sorafenib acts as a mitochondrial uncoupler and ameliorates nonalcoholic steatohepatitis. Cell Metabol. 2020;31(5):892–908.e11. doi: 10.1016/j.cmet.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Tang Z., Acuña U.M., Fernandes N.F., Chettiar S., Li P.-K., DE Blanco E.C. Structure–activity relationship of niclosamide derivatives. Anticancer Res. 2017;37(6):2839–2843. doi: 10.21873/anticanres.11635. [DOI] [PubMed] [Google Scholar]
- 193.Schweizer M.T., Haugk K., McKiernan J.S., Gulati R., Cheng H.H., Maes J.L., et al. A phase I study of niclosamide in combination with enzalutamide in men with castration-resistant prostate cancer. PLoS One. 2018;13(6) doi: 10.1371/journal.pone.0198389. [e] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Niu X., Liu P., Zhou X., Ou D., Wang X., Hu C. Anti-epidermal growth factor receptor (EGFR) monoclonal antibody combined with chemoradiotherapy for induction chemotherapy resistant locally advanced nasopharyngeal carcinoma: a prospective phase II study. Transl Oncol. 2024;39 doi: 10.1016/j.tranon.2023.101797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Paoletti C., Le Pecq J.B., Dat-Xuong N., Juret P., Garnier H., Amiel J.L., et al. Antitumor activity, pharmacology, and toxicity of ellipticines, ellipticinium, and 9-hydroxy derivatives: preliminary clinical trials of 2-methyl-9-hydroxy ellipticinium (NSC 264-137) Recent Results Cancer Res. 1980;74:107–123. doi: 10.1007/978-3-642-81488-4_15. [DOI] [PubMed] [Google Scholar]
- 196.Terada H. Uncouplers of oxidative phosphorylation. Environ Health Perspect. 1990;87:213–218. doi: 10.1289/ehp.9087213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Park S.J., Park Y.J., Shin J.H., Kim E.S., Hwang J.J., Jin D.H., et al. A receptor tyrosine kinase inhibitor, Tyrphostin A9 induces cancer cell death through Drp1 dependent mitochondria fragmentation. Biochem Biophys Res Commun. 2011;408(3):465–470. doi: 10.1016/j.bbrc.2011.04.053. [DOI] [PubMed] [Google Scholar]
- 198.Figarola J.L., Weng Y., Lincoln C., Horne D., Rahbar S. Novel dichlorophenyl urea compounds inhibit proliferation of human leukemia HL-60 cells by inducing cell cycle arrest, differentiation and apoptosis. Invest N Drugs. 2012;30(4):1413–1425. doi: 10.1007/s10637-011-9711-8. [DOI] [PubMed] [Google Scholar]
- 199.Figarola J.L., Singhal J., Tompkins J.D., Rogers G.W., Warden C., Horne D., et al. SR4 uncouples mitochondrial oxidative phosphorylation, modulates AMP-dependent kinase (AMPK)-Mammalian target of rapamycin (mTOR) signaling, and inhibits proliferation of HepG2 hepatocarcinoma cells. J Biol Chem. 2015;290(51):30321–30341. doi: 10.1074/jbc.M115.686352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.