Abstract
Retroviral capsid assembly can occur by either of two distinct morphogenic processes: in type C viruses, the capsid assembles and buds at the plasma membrane, while in type B and D viruses, the capsid assembles within the cytoplasm and is then transported to the plasma membrane for budding. We have previously reported that a single-amino-acid substitution of a tryptophan for an arginine in the matrix protein (MA) of Mason-Pfizer monkey virus (MPMV) converts its capsid assembly from that of a type D retrovirus to that of the type C viruses (S. S. Rhee and E. Hunter, Cell 63:77–86, 1990). Here we identify a region of 18 amino acids within the MA of MPMV that is responsible for type D-specific morphogenesis. Insertion of these 18 amino acids into the MA of type C Moloney murine leukemia virus causes it to assemble an immature capsid in the cytoplasm. Furthermore, fusion of the MPMV MA to the green fluorescent protein resulted in altered intracellular targeting and a punctate accumulation of the fusion protein in the cytoplasm. These 18 amino acids, which are necessary and sufficient to target retroviral Gag polyproteins to defined sites in the cytoplasm, appear to define a novel mammalian cytoplasmic targeting/retention signal.
Electron microscopic (EM) studies have revealed two morphologically distinct pathways for capsid assembly in different retroviruses. In type C morphogenic viruses, such as murine leukemia virus (MuLV) and human immunodeficiency virus, Gag polyproteins are synthesized in the cytoplasm and then transported to the plasma membrane, where they are assembled into an icosahedral capsid. In contrast, the Gag polyproteins of type B and D viruses, such as mouse mammary tumor virus (MMTV) and Mason-Pfizer monkey virus (MPMV), are assembled into a capsid within the cytoplasm prior to transport to the cell membrane. Therefore, the newly synthesized Gag polyprotein appears to contain the necessary signals to direct its transport to or retention at the site of capsid self-assembly.
Our previous studies of MPMV demonstrated that a single-amino-acid substitution (mutant R55W) in the matrix protein (MA) could alter the capsid assembly site of type D viruses from a region within the cytoplasm to the inner surface of the plasma membrane (32). Others have reported that human immunodeficiency virus and Moloney murine leukemia virus (MoMuLV) with amino acid substitutions or deletions in MA undergo capsid assembly within cytoplasmic vacuoles rather than at the plasma membrane (3, 10, 12, 38). These studies strongly suggest that MA can play an essential role in determining the intracellular transport of Gag polyproteins to the site of capsid assembly. Moreover, they suggest that there are no inherent differences in the process of capsid assembly for different retroviruses; they differ only in the intracellular site to which Gag polyproteins are targeted and accumulated for assembly.
Based on mutagenic studies of MPMV MA, we proposed a model for intracytoplasmic assembly of type D retrovirus capsids (30–32). In MPMV-infected cells, Gag polyproteins, synthesized and modified with myristic acid in the cytosol, are transported to a defined site within the cytoplasm. There, the molecules presumably reach a sufficiently high concentration for efficient assembly of an immature capsid. The assembled capsids then become transport competent and are rapidly transported to the plasma membrane by an energy-dependent (40) intracytoplasmic transport machinery of the cell. Previous studies provide evidence for a cytoplasmic targeting/retention signal (CTRS) in MA that is responsible for intracytoplasmic capsid assembly in Type D retroviruses. This putative CTRS appears to act dominantly to retain Gag molecules at a site of intracytoplasmic capsid assembly. Without this CTRS, they would be individually transferred to a site at the plasma membrane where they would self-assemble concurrently with virus budding, as observed with MPMV mutant R55W (32) and, presumably, with type C viruses.
Interestingly, a region of 18 amino acids spanning residues 43 to 60 within MPMV MA is highly conserved between MPMV and MMTV (32). This region includes the arginine residue at position 55, which when mutated, can alter the capsid assembly site. Furthermore, recent nuclear magnetic resonance studies show that these 18 residues form an exposed loop on the surface of the MA protein (5). By contrast, this amino acid stretch appears to be partly deleted in MoMuLV MA: residues equivalent to 50 to 55 of MPMV MA are missing in MoMuLV MA, while flanking residues are highly conserved. Thus, we postulated that this short stretch of amino acids might function as a CTRS for type B- and D-specific intracytoplasmic capsid assembly.
In the present study, we have tested this hypothesis by determining whether this 18-amino-acid putative MPMV CTRS sequence can alter the site of MoMuLV capsid assembly. A mutant of MoMuLV, MLV/D-CTRS, was generated to contain the putative CTRS sequence of 18 amino acids from MPMV (D-CTRS) in the place of the 11 amino acids at the equivalent position in MoMuLV MA. Cells transfected with this mutant genome assemble immature capsids in the cytoplasm and not at the cellular membrane. Thus, these 18 amino acids are sufficient to direct proteins to the cytoplasm. Furthermore, MPMV MA tagged with the green fluorescent protein (GFP) accumulates at discrete spots in the cytoplasm. In contrast, mutant MA-GFP with the arginine substitution was found associated with the plasma membrane.
MATERIALS AND METHODS
DNAs and cells.
To generate mutants of MoMuLV, oligonucleotide-directed mutagenesis was carried out with various synthetic oligonucleotides as previously described (43). A 1.7-kbp SstI-XhoI fragment containing the entire coding sequence of MA from an infectious MoMuLV proviral DNA, pMOV3 (kindly provided by R. V. Srinivas, University of Alabama at Birmingham), was subcloned into vector M13mp18 to perform the mutagenesis. After mutagenesis, the mutated fragments were recloned into the MoMuLV expression vector pHMLV to replace the wild-type fragment, and the presence of the mutations was confirmed by dideoxy sequencing of the double-stranded DNA (36). The plasmid, pHMLV, contains an infectious MoMuLV proviral genome derived from pMOV3 and a hygromycin resistance gene under the control of the simian virus 40 early promoter.
To determine the subcellular localization of the putative CTRS-containing MA protein, the coding region of MPMV MA was amplified by PCR and then fused in frame to the N terminus of enhanced GFP from the jellyfish Aequorea victoria (pEGFP; Clonetech) to generate plasmid pMAD-CTRS/GFP. Plasmid pMAC-CTRS/GFP is a derivative of MPMV mutant R55W.
To establish cell lines containing integrated wild-type or mutant proviral DNA, semiconfluent monolayers of murine BALB/c 3T3 fibroblast SV-T2 cells (ATCC CCL-163.1) that are negative for MuLV were transfected with viral DNAs linearized with FspI as described previously (33). Cell colonies were selected in medium containing 400 mg of hygromycin B (GIBCO BRL)/ml and screened to determine whether they expressed viral structural proteins.
For the transient expression of viral proteins in COS-1 cells, the cells were transfected with either wild-type pHMLV or mutant DNA (3 mg/35-mm-diameter plate) by the modified calcium phosphate precipitation method as described by Chen and Okayama (4).
Radiolabeling and immunoprecipitation of virus proteins.
Cells were pulse-labeled for 20 to 30 min with [3H]leucine (0.8 mCi/ml; 157 Ci/mmol; DuPont) and chased for 4 h in complete growth medium as previously described (31). For fatty acid labeling, the cells were labeled with [9,10-3H]myristic acid (0.5 mCi/ml; DuPont) for 2 h (31).
Following lysis in lysis buffer A (1% Triton X-100, 1% sodium deoxycholate, 0.15 M NaCl, 0.05 M Tris [pH 7.5]), cell-associated viral proteins were immunoprecipitated with goat anti-MuLV antiserum (Division of Cancer Cause and Prevention, National Cancer Institute) or rabbit anti-Gag antibodies (31). Radiolabeled virus particles that were released into the culture medium were pelleted by centrifugation for 10 min at 80,000 rpm in a Beckman TLA 100 rotor at 4°C and lysed in lysis buffer B (0.1% sodium dodecyl sulfate [SDS], 1% Triton X-100, 1% sodium deoxycholate, 0.15 M NaCl, 0.05 M Tris [pH 7.5]). Virion-associated viral proteins were then immunoprecipitated with goat anti-MuLV antiserum.
The immunoprecipitated viral proteins were separated with a 12% resolving gel by SDS-polyacrylamide gel electrophoresis.
Fractionation of Gag polyprotein, capsid preparation, and Western blot analyses.
Gag polyproteins were fractionated into free and capsid-associated forms as previously described (32) with some modifications. Briefly, approximately 5 million SV-T2 cells of wild-type and mutant lines were lysed at room temperature for 1 h in 1 ml of ICAP buffer (1% Triton X-100, 0.25 M sucrose, 1.0 mM EDTA, 10 mM Tris [pH 7.5], 10 mg of DNase I/ml, 1 mg of leupeptin/ml, 1 mg of aprotinin/ml, 100 mg of phenylmethylsulfonyl fluoride/ml) supplemented with 500 mM NaCl to disrupt the Gag polyprotein association with cytoskeletal elements (8, 9). After removal of nuclei from the lysates by centrifugation for 5 min in a microcentrifuge at 4°C, the capsids were pelleted through a 35% sucrose cushion by centrifugation at 80,000 rpm for 15 min in a Beckman TLA120.2 rotor at 4°C. Viral proteins in each fraction were separately immunoprecipitated with rabbit anti-Gag antiserum, separated by SDS-polyacrylamide gel electrophoresis and visualized by immunoblotting with rabbit anti-Gag antibodies and peroxidase-conjugated donkey anti-rabbit antibodies with an enhanced chemiluminescence detection system (ECL; Amersham).
The sedimentable particles in the pellet fraction were further purified through a continuous 30 to 50% (wt/wt) linear sucrose gradient. The pellet fraction, obtained from approximately 60 million cells of the MLV/D-CTRS line as described above, was resuspended in 0.6 ml of ICAP buffer on ice for 2 h and then layered onto a 10-ml sucrose gradient. After centrifugation for 3 h at 36,000 rpm in a Beckman SW41 rotor at 20°C, fractions of 1 ml each were collected from the bottom, diluted with 1 ml of phosphate-buffered saline (PBS), and then ultracentrifuged at 80,000 rpm for 15 min in a Beckman TLA120.2 rotor at 4°C. The resulting pellets were analyzed to detect Gag polyproteins by Western blot assay with goat anti-MuLV antibodies followed by alkaline phosphatase-conjugated rabbit anti-goat antibodies.
Microscopic analyses.
For EM studies of capsid assembly, COS-1 cells were transfected with virus DNA, fixed for 1 h at room temperature with 1% glutaraldehyde, and washed in PBS. After postfixation with 1% osmium tetroxide, the cells were embedded in an epoxy resin mixture, sectioned, and stained with uranyl acetate and lead nitrate. All preparations were examined with a Philips 301 EM.
The in vivo subcellular localization of the fluorescence-tagged MA proteins was determined on a Bio-Rad MRC 1024 confocal microscope system with LaserSharp software. COS cells, transfected with pEGFP, pMAD-CTRS/GFP, or pMAC-CTRS/GFP and grown on glass coverslips, were washed with PBS and fixed with 4% paraformaldehyde in PBS for 20 min at room temperature. For GFP excitation, an argon laser at 488-nm wavelength was used.
RESULTS
Synthesis and processing of viral proteins in mutant virus-expressing cells.
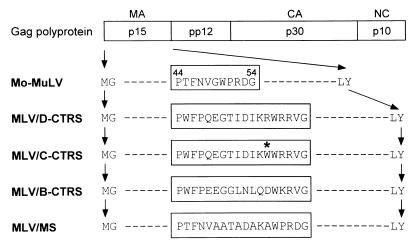
We generated four mutants of Mo-MuLV (Fig. 1). In MLV/D-CTRS, the putative CTRS sequence of 18 amino acids from MPMV (D-CTRS) substitutes for the 11 amino acids (residues 44 to 54) at the equivalent position in MoMuLV MA. MLV/C-CTRS contains the mutant CTRS, with tryptophan in place of arginine at position 55, derived from mutant R55W (C-CTRS). MLV/B-CTRS and MLV/MS have 18-amino-acid substitutions derived from the homologous, putative type B-specific CTRS sequence of MMTV (32) and from a multiple-alanine-substituted sequence, respectively. The alanines in MLV/MA substitute for each of the residues that are homologous between MPMV and MMTV.
FIG. 1.
Schematic representation of mutants of MoMuLV. The arrangement of the structural proteins within the gag-encoded polyprotein is schematically presented, with the partial amino acid sequences of MoMuLV p15 (MA) protein. In MLV/D-CTRS and MLV/C-CTRS, the 11 amino acids (residues 44 to 54) in MA, indicated in the box, are replaced with the 18 amino acids of the putative CTRS sequence of M-PMV (residues 43 to 60) and by the corresponding sequence from mutant MPMV R55W, respectively. The R55W substitution is marked with an asterisk. MLV/B-CTRS and MLV/MS have 18-amino-acid substitutions derived from the homologous, putative type B-specific CTRS sequence of MMTV (residues 44 to 61) and from a multiple-alanine-substituted sequence, respectively.
COS-1 cells were transiently transfected with wild-type DNA or MLV/D-CTRS or MLV/C-CTRS mutant DNA. Forty-eight hours after transfection, the cells were pulse labeled with [3H]leucine for 20 min and chased for 4 h in complete growth medium. Both cell-associated and released-virion-associated proteins were analyzed by immunoprecipitation with goat anti-MuLV antiserum (Fig. 2).
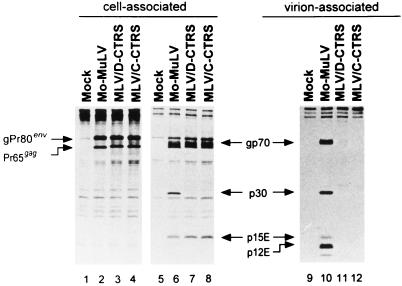
FIG. 2.
Immunoprecipitation of cell- and virion-associated viral proteins. To determine biosynthesis, processing, and release of viral proteins, proviral-DNA-transfected or mock-transfected COS cells were pulse labeled for 20 min with [3H]leucine (lanes 1 to 4) and chased for 4 h (lanes 5 to 8). Virus-specific proteins were immunoprecipitated from the lysed cells (cell-associated; lanes 1 to 8) or from the medium (virion-associated; lanes 9 to 12) with goat anti-MuLV antiserum.
Similar levels of two major precursor polyproteins, the Gag polyprotein Pr65gag and the envelope (Env) precursor gPr80env, were synthesized during the pulse labeling by cells expressing mutant as well as wild-type genomes (Fig. 2, lanes 1 to 4). The mutant Gag polyproteins are slightly larger by an amount consistent with the 7-amino-acid addition (18-amino-acid substitution for 11 amino acids [Fig. 2, lanes 3 and 4]).
During virus maturation, the MoMuLV Gag polyprotein is processed by the virus-encoded protease into four structural proteins: p15, pp12, p30, and p10 (1, 39). The Env precursor is cleaved by a cellular protease in the late Golgi compartment into two cell-associated glycoproteins, gp70 and p15E (11, 19). Following virus release, the transmembrane glycoprotein p15E is further processed by the viral protease into the virion-associated p12E (19, 29). As expected, the processing of Gag and Env precursors was clearly seen after a 4-h chase in wild-type MoMuLV-expressing cells (Fig. 2, lane 6); the intensities of the two precursor protein bands (gPr80env and Pr65gag) decreased with a concomitant appearance of gp70, p30, and p15E. In addition, extracellular virions that were released from the pulse-labeled cells during the 4-h chase could be detected from the culture fluids, as observed with bands of p12E along with mature viral proteins (Fig. 2, lane 10).
In cells expressing either mutant virus, normal processing of Env precursors was observed with bands of gp70 and p15E (Fig. 2, lanes 7 and 8). However, no p30 band could be detected, even though a significant proportion of the radiolabeled mutant Gag polyproteins was lost during the chase period. Furthermore, no viral proteins were detected in the culture medium (Fig. 2, lanes 11 and 12). Thus, both mutant Gag polyproteins with the putative D- and C-CTRS sequences were efficiently synthesized but were turned over (with an approximate 2-h half-life) without being efficiently processed or released into the culture medium.
Intracytoplasmic capsid assembly of the putative type D-specific CTRS-containing Gag polyproteins.
In MPMV-infected cells, completely assembled, intracytoplasmic immature capsids are stable and pelletable under mild nonionic detergent conditions, while mature capsids of virion particles as well as unassembled capsid precursors (either at the plasma membrane or within the cytoplasm) are solubilized (30, 32, 33). Exploiting these different sensitivities of the capsids, we carried out similar experiments to determine whether the mutant Gag polyproteins are preassembled into an immature capsid within the cytoplasm, as observed with MPMV. Since Gag polyproteins have been observed to form pelletable aggregates when they were overexpressed in COS cells, we established SVT2 cell lines with low-level expression of integrated wild-type or mutant proviral genomes.
Intracellular preassembled capsids were prepared as previously described (32). After lysis of SV-T2 cells in ICAP buffer supplemented with 500 mM NaCl to disrupt the Gag polyprotein association with cytoskeletal elements (8, 9), the capsids were pelleted through a 35% sucrose cushion. Soluble Gag polyproteins remained in the supernatant fraction while capsid-associated polyproteins were recovered in the pellet fraction. Viral proteins in each fraction were analyzed by immunoprecipitation and immunoblotting with rabbit anti-Gag antiserum (Fig. 3A).
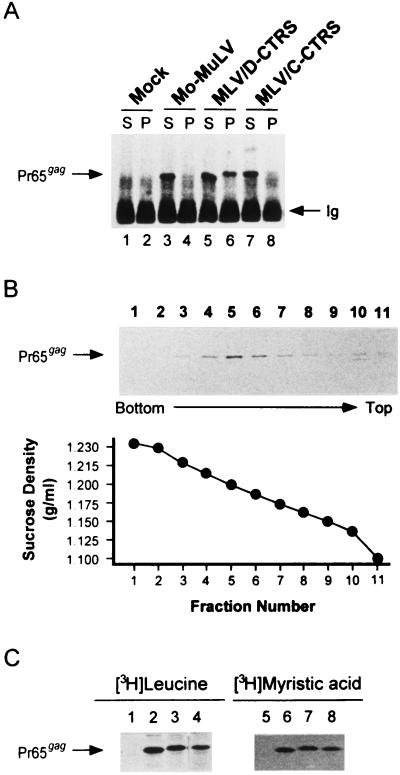
FIG. 3.
Determination of intracytoplasmic capsid formation and myristylation of mutant Gag polyproteins. (A) Western blot showing intracytoplasmic immature capsid assembly in mock-infected cells (lanes 1 and 2) and in SV-T2 cells expressing wild-type MoMuLV (lanes 3 and 4), MLV/D-CTRS (lanes 5 and 6), and MLV/C-CTRS (lanes 7 and 8). The cells were lysed in 1% Triton X-100-containing ICAP buffer and then fractionated into soluble (S; lanes 1, 3, 5, and 7) and pelletable (P; lanes 2, 4, 6, and 8) fractions by centrifugation. Gag polyproteins (Pr65gag) in each fraction were immunoprecipitated with rabbit anti-Gag antiserum and then visualized by Western blot analysis. Ig, heavy chains of rabbit immunoglobulin molecules. (B) The Gag polyprotein complexes in the pellet fraction of MLV/D-CTRS cells were purified through a continuous 30 to 50% (wt/wt) linear sucrose gradient. The fractions were collected from bottom to top, diluted with PBS, and ultracentrifuged to pellet high-molecular-weight particles. Particle-associated Gag polyproteins were detected by Western blot assay. The density of fraction 5 is 1.20 g/ml. (C) Myristylation of mutant Gag polyproteins was determined by labeling cells with [3H]leucine (lanes 1 to 4) or [3H]myristic acid (lanes 5 to 8). Radiolabeled Gag molecules of wild-type MoMuLV (lanes 2 and 6), MLV/D-CTRS (lanes 3 and 7), and MLV/C-CTRS (lanes 4 and 8), as well as mock-infected cells (lanes 1 and 5), were immunoprecipitated with rabbit anti-Gag antibodies.
Cell-associated Gag polyproteins (Pr65gag) of wild-type MoMuLV were found only in the soluble fraction (Fig. 3A, lane 3); none could be recovered from the pellet fraction (lane 4). This result is consistent with our earlier observations that the assembling capsid at the plasma membrane of type C morphogenic viruses is a relatively fragile structure. Thus, it is easily disrupted under these conditions (32).
In cells expressing MLV/D-CTRS, mutant Gag molecules were recovered in both fractions; approximately 20 to 30% of the total Gag molecules appeared to have been incorporated into stable, pelletable particles within the cytoplasm (Fig. 3A, lanes 5 and 6). Interestingly, no detergent-resistant particles were obtained with MLV/C-CTRS (Fig. 3A, lanes 7 and 8) that contains a mutated, presumably nonfunctional CTRS.
These detergent-resistant pelletable complexes obtained from MLV/D-CTRS-expressing cells appear to be different from the large oligomeric structures of Gag polyproteins observed in human immunodeficiency virus type 1-infected cells. The former were pelleted through a 35% sucrose cushion, equivalent to a density of 1.14 to 1.15 g/ml, while the latter have a density of 1.10 to 1.13 g/ml (21). Nevertheless, to confirm that these complexes resulted from highly ordered structures of Gag polyproteins with the type D-specific CTRS sequence, not from abnormal aggregates, pelletable material in MLV/D-CTRS cell lysates was further purified through a continuous 30 to 50% (wt/wt) linear sucrose gradient.
Most of the nonionic-detergent-resistant intracellular complexes of MLV/D-CTRS Gag polyproteins sedimented in fraction 5 of the gradient with a density of 1.20 g/ml (Fig. 3B), similar to that of naked immature retroviral capsids (20, 23, 35). Thus, these results, together with those mentioned above (Fig. 3A), strengthen the possibility that the type D-specific CTRS sequence causes Gag polyproteins to be retained within the cytoplasm for assembly of an immature capsid.
This dramatic change appears to result from the CTRS addition alone, not from a defect in myristylation of the proteins, in contrast to that observed with mutant simian immunodeficiency viruses, where nonmyristylated Gag precursors can assemble capsids within the cytoplasm of insect cells (6). Mutant Gag molecules of both MLV/D-CTRS and MLV/C-CTRS appear to be myristylated as efficiently as wild-type proteins (Fig. 3C) when the amounts of Gag polyproteins pulse labeled with [3H]leucine for 30 min (Fig. 3C, lanes 1 to 4) are compared to the amounts of those labeled with [9,10-3H]myristic acid for 2 h (lanes 5 to 8).
EM analyses.
The data described above strongly indicate that an insertion of 18 type D-specific amino acids into type C Gag polyproteins altered the viral morphogenesis to preassemble an immature capsid within the cytoplasm. However, these biochemical approaches could not distinguish between intracytoplasmic capsids that had assembled at a cytosolic site, as in type B and D viruses, and intracisternal capsids that had been released into cytoplasmic vacuoles, as in mutant type C viruses (3, 10, 12, 38). Thus, the effects of these amino acids on capsid assembly were more specifically evaluated by EM studies of COS cells transfected with wild-type or mutant proviral DNAs.
MoMuLV-infected cells show capsids assembling and budding at the plasma membrane, with numerous extracellular virions adjacent to the cells (Fig. 4A). Unlike MuLV-expressing NIH3T3 cells, in which it has been reported that capsids assemble at membranous intracellular sites (16), we have never observed infected COS cells assembling wild-type MuLV capsid structures within the cytoplasm. Thin-section electron micrographs of mutant MLV/C-CTRS-expressing COS cells show plasma membrane-associated structures at an early step in the assembly process (Fig. 4B). Thus, substitution per se of 18 amino acids (residues 43 to 60, derived from mutant MPMV R55W) for 11 amino acids (residues 44 to 54) in the MA protein of MoMuLV does not interfere with Gag polyprotein transport to and association with the plasma membrane. It does, however, appear to result in an assembly-release defect, since essentially no late budding structures were observed and the previously described pulse-chase experiments demonstrated a lack of virion release.
FIG. 4.
Electron micrographs of COS cells transfected with wild-type and mutant proviral genomes. (A) Cells with wild-type MoMuLV have assembling capsids at the plasma membrane (arrowhead) and release extracellular virions (arrow) with typical type C morphogenesis. (B) Cells with MLV/C-CTRS have plasma membrane-associated assembling capsid structures but no released viruses. (C and D) Cells with MLV/D-CTRS show capsids preassembled within the cytoplasm (C) and occasionally show capsids in the process of budding at the plasma membrane (arrow in panel D). Extracellular virions (arrowhead in panel D) can very rarely be seen. (E and F) In cells with MLV/B-CTRS, preassembled capsids are observed in the cytoplasm but not on the membrane (E) and a few capsids are in the process of budding at the plasma membrane (F). (G) In cells with MLV/MS, assembling capsids can be seen only at the plasma membrane. Original magnifications: ×75,000 (A, B, E, F, and G) and ×102,000 (C and D).
In contrast, MLV/D-CTRS-expressing cells show no capsids assembling at the plasma membrane (Fig. 4C). Instead, preassembled capsids are observed deep in the cytoplasm, not associated with intracellular membranes. Of note, we seldom detected intact capsids in the process of budding at the plasma membrane, and even more rarely did we detect extracellular virions (Fig. 4D).
The experiments described above demonstrate that a functional type D-specific CTRS enables type C Gag polyproteins to be targeted and retained at a cytoplasmic assembly site. Furthermore, we demonstrated that this morphogenic conversion is not simply the result of the conformational changes in Gag molecules caused by insertion of foreign amino acids into the MA domain, since it was not observed with the nonfunctional CTRS insertion. We further confirmed this conclusion by EM studies of two additional mutants of MoMuLV, MLV/B-CTRS and MLV/MS (Fig. 1). As with the mutant MLV/D-CTRS, MLV/B-CTRS viruses preassemble immature capsids in the cytoplasm but not on the membrane (Fig. 4E). A few capsids were also observed in the process of budding at the plasma membrane, but without detectable levels of virions released into the culture medium (Fig. 4F). By contrast, MLV/MS virus shows assembling structures associated with the plasma membrane (Fig. 4G), similar to those of MLV/C-CTRS. These results exclude the possibility that intracytoplasmic capsid assembly of D- or B-CTRS-containing Gag polyproteins was induced by a signal-independent, nonspecific process.
Intracellular localization of the putative CTRS-containing MA protein fused to GFP.
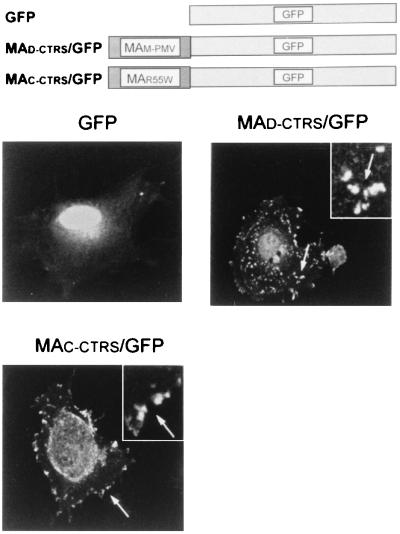
We further evaluated the subcellular localization of proteins containing the putative CTRS by using direct fluorescence to avoid nonspecific artifacts introduced by staining cells with antibodies in indirect immunofluorescence. Wild-type D-CTRS and mutant C-CTRS in the context of MPMV MA were fused to the GFP from the jellyfish A. victoria. GFP is widely used as a fluorescence reporter in studies of protein localization because many studies have shown that it does not alter protein trafficking. At 48 h after transfection, COS cells were imaged on a laser fluorescent confocal microscope for a better resolution of cellular structures (Fig. 5). The green staining of GFP alone, as expected, is distributed throughout the cell, with bright fluorescence at the nucleus (Fig. 5, panel GFP). Cells expressing the D-CTRS-containing the MA-GFP fusion protein have fluorescent deposits with irregular shapes distributed sporadically within the cytoplasm (panel MAD-CTRS/GFP). These fluorescent images are different from typical patterns of staining associated with vesicles or other cellular structures, suggesting that the fluorescence-tagged-MA protein with wild-type D-CTRS has accumulated at specific cytoplasmic locations. By contrast, this punctate cytoplasmic pattern was not observed with the MA-GFP chimera with C-CTRS; the green staining is associated with the plasma membrane, as well as with the nucleus (panel MAC-CTRS/GFP).
FIG. 5.
Subcellular distribution of MA protein fused to GFP in COS cells. The cells were transiently transfected with pEGFP, pMAD-CTRS/GFP, or pMAC-CTRS/GFP, and the fluorescence-tagged MA proteins were localized by laser fluorescent confocal microscopy. The green staining of GFP alone is distributed intensely in the nucleus and diffused faintly throughout the cytosol. In cells with the putative D-CTRS-containing MA-GFP fusion proteins (MAD-CTRS/GFP), fluorescent deposits can be seen sporadically within the cytoplasm (arrows) with an irregular body (inset). By contrast, GFP fused to mutant MA derived from mutant M-PMV R55W (MAC-CTRS/GFP) displays fluorescent patches associated with the plasma membrane (arrows) and a nuclear distribution.
DISCUSSION
In cells, newly synthesized proteins are directed from their sites of biosynthesis to their sites of activity by complex and highly regulated processes. To date, studies of many proteins have revealed the presence of signaling sequences within the primary structure of proteins. These sequences play an indispensable role in directing proteins to their destinations. The signals on secreted and membrane-spanning proteins and on proteins that direct them to the cellular organelles, such as nucleus, mitochondria, and chloroplasts, have been identified (2, 7, 15, 17, 18, 34). However, the intracytoplasmic trafficking by which proteins are targeted to other active sites within the cytoplasm is less well understood, even though it is likely that is also a highly directed and regulated process.
Previously, we proposed that the MA protein of MPMV, the prototype of the type D retroviruses, contains a specific, dominant signal that is essential to target Gag polyproteins to a site within the cytoplasm for type D-specific intracytoplasmic capsid assembly (32). When this signal was disrupted by a single-amino-acid substitution in MA (mutant R55W), Gag molecules no longer accumulated at the usual cytoplasmic sites. Instead, individual proteins were transported to the inner surface of the plasma membrane, where they self-assembled concurrently with virus budding, as observed with type C retroviruses. In this report, we have identified 18 amino acids, spanning residues 43 to 60 within MPMV MA, that act as a topogenic CTRS. When these 18 amino acids were inserted into the MA of type C MoMuLV, the capsid assembly process was altered to assemble an immature capsid in the cytoplasm. Interestingly, nuclear magnetic resonance has shown that these residues form a loop that is exposed on the surface of the MPMV MA protein (5). Thus, our findings imply that these 18 amino acids, presumably exposed on the surface of the MuLV Gag polyprotein, are sufficient to function as a CTRS to localize proteins within the cytoplasm. Moreover, these results confirm our previous results indicating that there are no inherent differences in the process of capsid assembly for different retroviruses; they differ only in the intracellular site to which Gag polyproteins are targeted and accumulated for assembly.
The MA protein of type C retroviruses has been suggested to play a critical role in transporting Gag polyproteins to the plasma membrane. Specifically, the basic residues, as well as myristic acid modification within the amino-terminal region of the protein, were found to form a bipartite plasma membrane-targeting signal (14, 28, 38, 41, 42). In the present study, we found that this signal appears to still be functional even with an amino acid insertion after the basic patch of mutant MuLVs. When a multiply substituted sequence (MLV/MS) or a nonfunctional CTRS (MLV/C-CTRS) was inserted, myristylated mutant Gag proteins were targeted to the plasma membrane and assembled capsids there (Fig. 3 and 4). This shows that the morphogenic conversion observed in mutant MuLV with the wild-type CTRS (mutant MLV/D-CTRS and MLV/B-CTRS) was not induced by the simple aggregation of Gag proteins at the site of biosynthesis. It is probable that the functional CTRS-containing Gag molecules accumulate at a defined site in the cytoplasm by specific signal-directed intracytoplasmic protein transport for capsid assembly. Consistent with our previous result with mutant MPMV R55W, where mutant Gag was efficiently and rapidly transported to the plasma membrane (32), these data suggest that the plasma membrane-targeting information in retroviral Gag must be overridden by the insertion of a dominant CTRS. This is supported by the results demonstrating that the MPMV MA protein accumulated in punctate patches within the cytoplasm, whereas mutant MA with the nonfunctional CTRS is found associated with the plasma membrane (Fig. 5). Furthermore, this pattern of distribution confirms our conclusion that the sequence of 18 amino acids in the MA protein of MPMV is critical to target and localize proteins to a specific site within the cytoplasm.
Although mutant Gag polyproteins were synthesized at a normal level and directed to an appropriate site, our biochemical assays demonstrated no detectable release of mutant virion particles (Fig. 2). In cells with mutant MLV/C-CTRS and MLV/MS, we observed only plasma membrane-associated structures at an early step in the assembly process. Furthermore, no capsids were detected assembling at the late stages. This implies that the insertion mutations described here do not interfere with protein transport to and association with the plasma membrane but do interfere with protein stability (causing the shorter half-life [Fig. 2]) and with the capsid assembly process. These effects could explain our finding of inefficient capsid assembly in the functional CTRS-containing MoMuLV: only a small portion of the total accumulated Gag molecules were associated with the nonionic detergent-resistant capsid (Fig. 3A), and few intracellular capsids were detected. It is also possible that efficient assembly of immature capsids in the cytoplasm may require other domains that are present in MPMV Gag polyproteins but missing in these MuLV chimeras. Indeed, MPMV mutants with deletions in the p12 domain that is present in type B and D viruses but absent from MuLV were severely impaired for capsid assembly (37).
In summary, the data presented here suggest the existence of a novel mechanism by which a cytoplasmic protein is specifically retained within the cytoplasm. Furthermore, this intracytoplasmic trafficking of proteins, as with proteins destined for the nucleus, mitochondria, and other organelles (7, 15, 17, 18, 22), appears to be specified by a short signal sequence within the primary structure of the protein. Numerous cellular proteins have been suggested to be receptor proteins that recognize signal sequences and mediate protein transport (13, 24–27). It seems likely, then, that specific eucaryotic CTRS receptor proteins direct and retain Gag polyproteins at specific cytoplasmic locations by using the host cell transport machinery. The discovery of this CTRS sequence may not only enhance our understanding of viral transport and assembly but also help elucidate the general process of intracytoplasmic protein trafficking in eucaryotic cells.
ACKNOWLEDGMENTS
We thank E. Arms at UAB for excellent technical assistance in the EM. We are grateful to members of our laboratory for discussions during the project and to J. Macke for substantive editing of the manuscript.
This work was supported by grant B-95003 to S.S.R. from the Samsung Biomedical Research Institute and by a grant to E.H. from the National Cancer Institute (R39 CA27834).
REFERENCES
- 1.Arcement L J, Karshin W I, Naso R B, Arlinghaus R B. “Gag” polyprotein precursors of Rauscher murine leukemia virus. Virology. 1977;81:284–297. doi: 10.1016/0042-6822(77)90145-3. [DOI] [PubMed] [Google Scholar]
- 2.Blobel G. Intracellular protein topogenesis. Proc Natl Acad Sci USA. 1980;77:1496–1500. doi: 10.1073/pnas.77.3.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cannon P M, Matthews S, Clark N, Byles E D, Iourin O, Hockley D J, Kingsman S M, Kingsman A J. Structure-function studies of the human immunodeficiency virus type 1 matrix protein, p17. J Virol. 1997;71:3474–3483. doi: 10.1128/jvi.71.5.3474-3483.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conte M R, Klikova M, Hunter E, Ruml T, Matthews S. The three-dimensional solution structure of the matrix protein from the type D retrovirus, the Mason-Pfizer monkey virus, and implications for the morphology of retroviral assembly. EMBO J. 1997;16:5819–5826. doi: 10.1093/emboj/16.19.5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delchambre M, Gheysen D, Thines D, Thiriart C, Jacobs E, Verdin E, Horth M, Burny A, Bex F. The GAG precursor of simian immunodeficiency virus assembles into virus-like particles. EMBO J. 1989;8:2653–2660. doi: 10.1002/j.1460-2075.1989.tb08405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dingwall C, Laskey R A. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 8.Edbauer C A, Naso R B. Cytoskeleton-associated Pr65gag and assembly of retrovirus temperature-sensitive mutants in chronically infected cells. Virology. 1984;134:389–397. doi: 10.1016/0042-6822(84)90306-4. [DOI] [PubMed] [Google Scholar]
- 9.Edbauer C A, Naso R B. Cytoskeleton-associated Pr65gag and retrovirus assembly. Virology. 1983;130:415–426. doi: 10.1016/0042-6822(83)90096-x. [DOI] [PubMed] [Google Scholar]
- 10.Facke M, Janetzko A, Shoeman R L, Krausslich H G. A large deletion in the matrix domain of the human immunodeficiency virus gag gene redirects virus particle assembly from the plasma membrane to the endoplasmic reticulum. J Virol. 1993;67:4972–4980. doi: 10.1128/jvi.67.8.4972-4980.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Famulari N G, Buchhagen D L, Klenk H D, Fleissner E. Presence of murine leukemia virus envelope proteins gp70 and p15(E) in a common polyprotein of infected cells. J Virol. 1976;20:501–508. doi: 10.1128/jvi.20.2.501-508.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freed E O, Orenstein J M, Buckler-White A J, Martin M A. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J Virol. 1994;68:5311–5320. doi: 10.1128/jvi.68.8.5311-5320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorlich D. Nuclear protein import. Curr Opin Cell Biol. 1997;9:412–419. doi: 10.1016/s0955-0674(97)80015-4. [DOI] [PubMed] [Google Scholar]
- 14.Gottlinger H G, Sodroski J G, Haseltine W A. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1989;86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gould S G, Keller G A, Subramani S. Identification of a peroxisomal targeting signal at the carboxy terminus of firefly luciferase. J Cell Biol. 1987;105:2923–2931. doi: 10.1083/jcb.105.6.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen M, Jelinek L, Jones R S, Stegeman-Olsen J, Barklis E. Assembly and composition of intracellular particles formed by Moloney murine leukemia virus. J Virol. 1993;67:5163–5174. doi: 10.1128/jvi.67.9.5163-5174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horwich A. Protein import into mitochondria and peroxisomes. Curr Opin Cell Biol. 1990;2:625–633. doi: 10.1016/0955-0674(90)90103-l. [DOI] [PubMed] [Google Scholar]
- 18.Kalderon D, Roberts B L, Richardson W D, Smith A E. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 19.Karshin W L, Arcement L J, Naso R B, Arlinghaus R B. Common precursor for Rauscher leukemia virus gp69/71, p15(E) and p12(E) J Virol. 1977;23:787–798. doi: 10.1128/jvi.23.3.787-798.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klikova M, Rhee S S, Hunter E, Ruml T. Efficient in vivo and in vitro assembly of retroviral capsids from Gag precursor proteins expressed in bacteria. J Virol. 1995;69:1093–1098. doi: 10.1128/jvi.69.2.1093-1098.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee Y-M, Yu X-F. Identification and characterization of virus assembly intermediate complexes in HIV-1-infected CD4+ T cells. Virology. 1998;243:78–93. doi: 10.1006/viro.1998.9064. [DOI] [PubMed] [Google Scholar]
- 22.Li H M, Chen L J. A novel chloroplastic outer membrane-targeting signal that functions at both termini of passenger polypeptides. J Biol Chem. 1997;18:10968–10974. [PubMed] [Google Scholar]
- 23.Lingappa J R, Hill R L, Wong M L, Hegde R S. A multistep, ATP-dependent pathway for assembly of human immunodeficiency virus capsids in a cell-free system. J Cell Biol. 1997;136:567–581. doi: 10.1083/jcb.136.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muckel E, Soll J. A protein import receptor of chloroplasts is inserted into the outer envelope membrane by a novel pathway. J Biol Chem. 1996;271:23846–23852. doi: 10.1074/jbc.271.39.23846. [DOI] [PubMed] [Google Scholar]
- 25.Otera H, Okumoto K, Tateishi K, Ikoma Y, Matsuda E, Nishimura M, Tsukamoto T, Osumi T, Ohashi K, Higuchi O, Fujiki Y. Peroxisome targeting signal type 1 (PTS1) receptor is involved in import of both PTS1 and PTS2: studies with PEX5-defective CHO cell mutants. Mol Cell Biol. 1998;18:388–399. doi: 10.1128/mcb.18.1.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pain D, Murakami H, Blobel G. Identification of a receptor for protein import into mitochondria. Nature. 1990;347:444–449. doi: 10.1038/347444a0. [DOI] [PubMed] [Google Scholar]
- 27.Pollard V W, Michael W M, Naldalny B, Slomi M C, Wang P, Dreyfuss G. A novel receptor mediated nuclear protein import pathway. Cell. 1996;88:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 28.Rein A, McClure M R, Rice N R, Luftig R B, Schultz D I. Myristylation site in Pr65gag is essential for virus particle formation by Moloney murine leukemia virus. Proc Natl Acad Sci USA. 1986;83:7246–7250. doi: 10.1073/pnas.83.19.7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rein A, Mirro J, Haynes J G, Ernst S M, Nagashima K. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J Virol. 1994;68:1773–1781. doi: 10.1128/jvi.68.3.1773-1781.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhee S S, Hunter E. Amino acid substitutions within the matrix protein of type D retroviruses affect assembly, transport and membrane association of a capsid. EMBO J. 1991;10:535–546. doi: 10.1002/j.1460-2075.1991.tb07980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhee S S, Hunter E. Myristylation is required for intracellular transport but not for assembly of D-type retrovirus capsids. J Virol. 1987;61:1045–1053. doi: 10.1128/jvi.61.4.1045-1053.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rhee S S, Hunter E. A single amino acid substitution within the matrix protein of a type D retrovirus converts its morphogenesis to that of a type C retrovirus. Cell. 1990;63:77–86. doi: 10.1016/0092-8674(90)90289-q. [DOI] [PubMed] [Google Scholar]
- 33.Rhee S S, Hunter E. Structural role of the matrix protein of type D retroviruses in Gag polyprotein stability and capsid assembly. J Virol. 1990;64:4383–4389. doi: 10.1128/jvi.64.9.4383-4389.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sabatini D D, Kreibich G, Morimoto T, Adesnik M. Mechanism for the incorporation of proteins in membranes and organelles. J Cell Biol. 1982;92:1–22. doi: 10.1083/jcb.92.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakalian M, Parker S D, Weldon R A, Jr, Hunter E. Synthesis and assembly of retrovirus Gag precursors into immature capsids in vitro. J Virol. 1996;70:3706–3715. doi: 10.1128/jvi.70.6.3706-3715.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5436–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sommerfelt M A, Rhee S S, Hunter E. Importance of p12 protein in Mason-Pfizer monkey virus assembly and infectivity. J Virol. 1992;66:7005–7011. doi: 10.1128/jvi.66.12.7005-7011.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soneoka Y, Kingsman S M, Kingsman A J. Mutagenesis analysis of the murine leukemia virus matrix protein: identification of regions important for membrane localization and intracellular transport. J Virol. 1997;71:5549–5559. doi: 10.1128/jvi.71.7.5549-5559.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Zaane D, Dekker-Michielsen M J, Bloemers H P J. Virus-specific precursor polypeptides in cells infected with Rauscher leukemia virus: synthesis, identification, and processing. Virology. 1976;75:113–129. doi: 10.1016/0042-6822(76)90011-8. [DOI] [PubMed] [Google Scholar]
- 40.Weldon R A, Jr, Parker W B, Sakalian M, Hunter E. Type D retrovirus capsid assembly and release are active events requiring ATP. J Virol. 1998;72:3098–3106. doi: 10.1128/jvi.72.4.3098-3106.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan X, Yu X, Lee T H, Essex M. Mutations in the N-terminal region of human immunodeficiency virus type 1 matrix protein block intracellular transport of the Gag precursor. J Virol. 1993;67:6387–6394. doi: 10.1128/jvi.67.11.6387-6394.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou W, Parent L J, Wills J W, Resh M D. Identification of a membrane binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zollar M J, Smith M. Oligonucleotide-directed mutagenesis: a simple method using two oligonucleotide primers and a single stranded DNA template. DNA. 1984;3:479–488. doi: 10.1089/dna.1.1984.3.479. [DOI] [PubMed] [Google Scholar]