Significance
The STING-mediated IFN response is well known, STING signaling beyond IFN and innate immunity, particularly in the context of T cell biology and adaptive immunity, remains largely unexplored. Here, we have identified that STING trafficking activates MAPK-CREB signaling to induce the expression of IL-2 and TGF-β2, thereby facilitating Treg differentiation. In the Trex1−/− autoimmune disease mouse model, the prevailing view suggests that the disease is driven by constitutive STING-mediated IFN response, leading to T cell autoactivation. However, our findings indicate that CD4 T cell intrinsic STING activation induces Treg differentiation, potentially counterbalancing the autoimmunity associated with Trex1 deficiency. Overall, the STING-MAPK-CREB pathway represents an IFN-independent activity of STING with the potential to profoundly impact T cell effector functions.
Keywords: STING, type-I interferon, T cells, Innate immunity, autoimmunity
Abstract
The Type-I interferon (IFN-I) response is the major outcome of stimulator of interferon genes (STING) activation in innate cells. STING is more abundantly expressed in adaptive T cells; nevertheless, its intrinsic function in T cells remains unclear. Intriguingly, we previously demonstrated that STING activation in T cells activates widespread IFN-independent activities, which stands in contrast to the well-known STING-mediated IFN response. Here, we have identified that STING activation induces regulatory T cells (Tregs) differentiation independently of IRF3 and IFN. Specifically, the translocation of STING from the endoplasmic reticulum to the Golgi activates mitogen-activated protein kinase (MAPK) activity, which subsequently triggers transcription factor cAMP response element-binding protein (CREB) activation. The activation of the STING–MAPK–CREB signaling pathway induces the expression of many cytokine genes, including interleukin-2 (IL-2) and transforming growth factor-beta 2 (TGF-β2), to promote the Treg differentiation. Genetic knockdown of MAPK p38 or pharmacological inhibition of MAPK p38 or CREB markedly inhibits STING-mediated Treg differentiation. Administration of the STING agonist also promotes Treg differentiation in mice. In the Trex1−/− autoimmune disease mouse model, we demonstrate that intrinsic STING activation in CD4+ T cells can drive Treg differentiation, potentially counterbalancing the autoimmunity associated with Trex1 deficiency. Thus, STING–MAPK–CREB represents an IFN-independent signaling axis of STING that may have profound effects on T cell effector function and adaptive immunity.
A fundamental principle of the innate immune system is its remarkable ability to distinguish between “self” and “nonself.” Detecting foreign nucleic acids is a crucial strategy employed by the innate immune system to recognize invading pathogens. In mammals, pathogen-derived DNA is sensed in the cytosol by DNA sensor Cyclic GMP–AMP synthase (cGAS) (1), which produces a second messenger called 2′3′-Cyclic guanosine monophosphate-adenosine monophosphate (2′3′-cGAMP) to activate the signaling adaptor stimulator of interferon genes (STING) (1–4). Binding 2′3′-cGAMP to STING induces its translocation from the Endoplasmic Reticulum (ER) to the ER-Golgi intermediate compartment (ERGIC) and the Golgi, where it recruits kinase TBK1 and transcription factor IRF3 to induce the expression of type I interferons (IFN-I) and IFN-stimulated genes (5). While the primary outcome of STING activation is the induction of IFN-I, it has recently become evident that the STING pathway evolved over 600 Mya, predating the evolution of IFN-I (6). STING signaling beyond IFN-I and innate immunity remains largely unknown.
STING is highly expressed in T cells. Unlike the dominant IFN-I response of STING in macrophages, STING activation in T cells predominantly induces IFN-independent activities, such as nuclear factor kappa B (NF-κB), nuclear factor of activated T cells (NFAT), unfolded protein response (UPR), IL-2 pathways, TCR signaling, and cell death, among others (7). STING activation suppresses mTOR signaling, thus inhibiting T cell proliferation (8). STING impairs Rorγt-mediated Il17a transcription, thereby restraining Th17 differentiation (9). STING activation has been shown to induce Th1- and Th9-mediated antitumor immunity (10). It has been recently reported that STING promotes the expansion of regulatory B cells, a process dependent on IRF3 but independent of IFNAR, ultimately attenuating NK cell-mediated anti-tumor immunity (11). One of the major IFN-independent activities of STING is STING-mediated T cell death (7, 12–14), which was also shown in gain-of-function StingN153S/+ mice (14). StingN153S/+ mice suffer from T cell cytopenia and cell death, a condition that is genetically independent of cGAS, IRF3, or IFNAR (15–17). The StingN153S/+ mutation (N154S in human STING) causes STING to constitutively translocate from the ER to the Golgi, resulting in altered ER calcium homeostasis as well as elevated ER stress and UPR, leading to T cell death (14). STING-mediated T cell death is also physiologically important in tumor immune evasion (7). In a preclinical study of STING agonist therapy, high doses of ADU-S100 treatment led to a complete loss of CD8+ T cells in the tumors (18). We have also demonstrated that CD8+ T cells experience significant cell death, which is partly attributable to STING activation by tumor-derived cGAMP in implanted syngeneic mouse tumors (7). Thus, IFN-independent activities of STING in T cells are clearly physiologically important and merit further study.
Emerging evidence suggests that the cAMP response element-binding protein (CREB) is a key transcription factor that regulates T cell functions (19). The CRE has been identified in the promoters and enhancers of many T cell-specific genes, including Tcra, Tcrb, Cd3d, Cd8a, Il2, and Tgfβ2, thereby regulating their expression in T cells (20–25). A transgenic mouse engineered to express a dominant-negative form of CREB under the T cell-specific promoter, exhibited a significant impairment in proliferation and IL-2 production, ultimately leading to apoptotic cell death in T cells (26). Moreover, it has been reported that CREB stabilizes and induces FOXP3 expression, promoting the maintenance of the regulatory T cell (Treg) phenotype (27). CREB has been shown to be activated by several different serine-threonine kinases, including cAMP-dependent protein kinase A (PKA), protein kinase C (PKC), and Mitogen-activated protein kinases (MAPKs) (28).
The unique feature of STING activation requires its trafficking from ER to Golgi (5). It has been proposed that STING trafficking introduces a second dimension that regulates STING signaling (5). In fact, STING trafficking is required to initiate several IFN-independent activities, including autophagy, calcium signaling, and UPR. For instance, STING-mediated autophagy requires STING to translocate from the ER to ERGIC, a known source of autophagic membrane (29). STING-mediated UPR and calcium flux require a STING ER exit (14). Interestingly, both STING-mediated UPR and autophagy activities are mapped to the same motif (an α-helix containing amino acid 322 to 343), which is essential for STING trafficking (14, 29). In the current study, we have demonstrated that STING trafficking from the ER to the Golgi is sufficient to activate MAPK signaling, which in turn activates the transcription factor CREB to induce the expression of TGF-β2 and IL-2, thereby facilitating Treg differentiation. STING agonist administration or CD4+ T cell intrinsic STING activation caused by Trex1 deficiency also drives Treg differentiation in vivo. Thus, the STING–MAPK–CREB axis represents an IFN-independent signaling pathway that may have a profound impact on T cell effector function and adaptive immunity.
Results
STING Activation Triggers Treg Differentiation Independently of IRF3 and IFN-I.
STING is more abundantly expressed in T cells, and its activation in T cells triggers widespread IFN-independent activities that have not been previously appreciated (7). One of the major IFN-independent activities of STING is the induction of cell death (7). To investigate how different subsets of T cells respond to STING-mediated cell death, we polarized naïve CD4+ T cells isolated from Foxp3YFP reporter mice into Th0 and Treg cells. We observed significant Treg differentiation, as indicated by a substantial proportion (~39%) of T cells becoming Foxp3YFP+ cells (SI Appendix, Fig. S1A). We then treated Th0 and Treg cells with the STING agonist 5,6-dimethylxanthenone-4-acetic acid (DMXAA) to induce cell death. We found that the CD4+ T cells polarized under Treg conditions were less susceptible to STING-mediated cell death compared to those polarized under Th0 conditions (SI Appendix, Fig. S1B). Interestingly, under Treg polarization conditions, FOXP3+ cells (~39%) resisted STING-mediated cell death, while FOXP3- cells (~61%) were sensitive to STING-mediated cell death (SI Appendix, Fig. S1 A and C), suggesting that FOXP3+ Tregs are not responsive to STING-mediated cell death. To substantiate these findings, we treated naïve CD4 T cells from both WT and Sting−/− mice with our optimized Treg polarization condition, achieving a notable Treg differentiation efficiency of approximately 85% (SI Appendix, Fig. S1D). We then treated these terminally differentiated Tregs with DMXAA to assess their sensitivity to STING-mediated cell death. Our results revealed that FOXP3+ cells in WT CD4+ T cells exhibited complete resistance, whereas FOXP3- cells in the same cell population were susceptible to STING-mediated cell death. As a control, CD4+ T cells lacking STING exhibited no response to DMXAA-induced cell death, regardless of FOXP3 expression (SI Appendix, Fig. S1 D and E). We next evaluated how natural Tregs respond to STING-mediated cell death. We treated the Tregs from spleens with different doses of DMXAA and then assessed their response to cell death. We found that FOXP3- cells in the CD4+ T cell population committed rapid cell death, whereas FOXP3+ cells in the same population exhibited a much slower cell death rate (SI Appendix, Fig. S1 F and G).
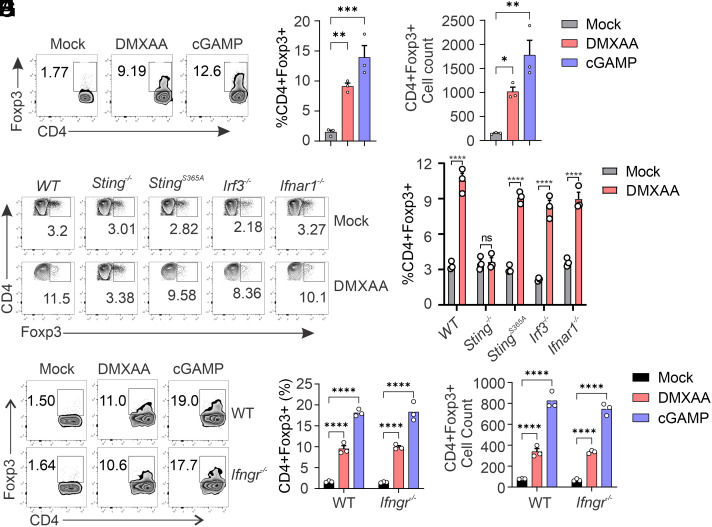
To understand the roles of STING in Tregs, we investigated Treg differentiation. TGF-β and IL-2 are the cytokines widely used to polarize naïve CD4+ T cells into Tregs. We treated WT and Sting−/− naïve CD4+ T cells with TGF-β and IL-2 to induce Treg differentiation. We observed that TGF-β-induced Treg differentiation is comparable between WT and Sting−/− CD4+ T cells (SI Appendix, Fig. S2 A and B). Furthermore, STING agonist stimulation doesn’t alter Treg differentiation (SI Appendix, Fig. S2 A and B), suggesting that STING is not required for TGF-β-mediated Treg differentiation. Interestingly, when we treated naïve CD4+ T cells from WT mice with DMXAA or cGAMP alone, it significantly induced Treg differentiation (Fig. 1 A–C), suggesting that STING activation is sufficient to induce Treg differentiation. The classic paradigm of STING signaling involves the activation of downstream IRF3 and IFN-I signaling. To test whether STING-mediated Treg differentiation is dependent on the IRF3-IFN pathway, we induced Treg differentiation in StingS365A/S365A, Irf3−/−, and Ifnar1−/− CD4+ T cells. We previously demonstrated that the StingS365A/S365A mutation precisely ablates STING-mediated IFN-I signaling while retaining STING-mediated IFN-independent activities (7). Our results indicate that DMXAA treatment induces a similar percentage of Treg differentiation in StingS365A/S365A CD4+ T cells compared to that in WT CD4+ T cells (Fig. 1 D and E). Furthermore, the deficiency of IRF3 or IFNAR1 does not affect STING-mediated Treg differentiation (Fig. 1 D and E).
Fig. 1.
STING activation induces Treg differentiation independently of IRF3 and IFN. (A–C) DMXAA or cGAMP treatment induces Treg differentiation. Wild-type (WT) naïve CD4 T cells were treated with mock, DMXAA (2.5 μg/mL), or cGAMP (10 μg/mL) for 3 d, and Treg differentiation was analyzed by intracellular staining of Foxp3 and FACS analysis. Representative FACS plots are shown in (A), Treg percentages (B) and cell numbers (C) are shown in bar graphs. (D and E) IRF3 or IFNAR1 deficiency does not affect STING-mediated Treg differentiation. WT, Sting−/−, StingS365A/S365A, Irf3−/−, and Ifnar1−/− naïve CD4 T cells were treated with mock or DMXAA (2.5 μg/mL) for 3 d, and Treg differentiation was analyzed by intracellular staining of Foxp3 and FACS analysis. Representative FACS plots are shown in (D), and statistics are shown in (E). (F–H) IFNGR1 deficiency does not affect STING-mediated Treg differentiation. WT and Ifngr1−/− naïve CD4 T cells were treated with mock, DMXAA (2.5 μg/mL), or cGAMP (20 μg/mL) for 3 d, and Treg differentiation was analyzed. Representative FACS plots are shown in (F), Treg percentages (G), and cell numbers (H) are shown in bar graphs. Error bars: SEM; ****P < 0.0001; *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant. Two-way ANOVA test. Data shown are representative of at least two independent experiments.
Type II IFN (IFNγ) is well known for its role in regulating Treg development. Interestingly, recent research has shown that IFNγ is implicated in T cell-mediated autoinflammation in STING-associated vasculopathy with onset in infancy (SAVI) mice (30). SAVI mice lacking IFNγR1 partially restore T cell death and proliferation deficiency, as well as improving Treg development (30). To investigate whether STING-mediated Treg differentiation depends on Type II IFN (IFNγ), we examined Treg differentiation in Ifngr1−/− CD4+ T cells. We found that deficiency of IFNγR1 does not alter DMXAA- or cGAMP-mediated Treg differentiation (Fig. 1 F–H). Collectively, these data suggest that STING activation is sufficient to induce Treg differentiation independently of IRF3, Type-I, and Type-II IFN.
STING Induces TGF-β2 and IL-2 Expression to Facilitate Treg Differentiation.
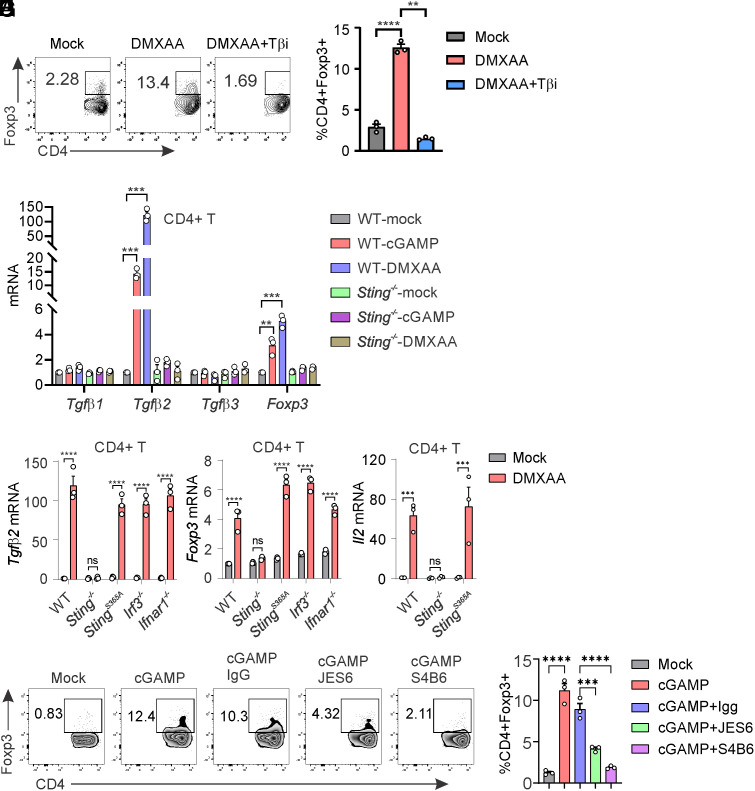
It is well established that TGF-β signaling can polarize naïve CD4+ T cells into Tregs. We sought to investigate whether STING-mediated Treg differentiation is dependent on TGF-β. We found that DMXAA-induced Treg differentiation is entirely blocked by TGF-β inhibitor treatment (Fig. 2 A and B), suggesting that STING-mediated Treg differentiation is indeed dependent on TGF-β. The TGF-β family is composed of three members, including TGF-β1, TGF-β2, and TGF-β3, which share similar biochemical properties in inducing TGF-β signaling (31). We observed that both cGAMP and DMXAA stimulation significantly induced TGF-β2 expression, but not TGF-β1 or TGF-β3 expression, in WT but not Sting−/− CD4+ T cells (Fig. 2C). Concurrently, we observed significant FOXP3 expression in response to STING activation in CD4+ T cells (Fig. 2C). Furthermore, STING activation induced similar levels of TGF-β2 and FOXP3 expression in StingS365A/S365A, Irf3−/−, and Ifnar1−/− CD4+ T cells compared to that of WT CD4+ T cells (Fig. 2 D and E), suggesting that STING-mediated TGF-β2 and FOXP3 expression is independent of IRF3 and IFN-I. IL-2 is another important cytokine that is required for Treg differentiation. We also found that STING activation dramatically induces IL-2 expression in WT and StingS365A/S365A CD4+ T cells but not in Sting−/− CD4+ T cells (Fig. 2F). To investigate whether IL-2 is required for STING-mediated Treg differentiation, we introduced anti-IL2 neutralizing antibodies. We found that the neutralization of IL2 efficiently blocked STING-mediated Treg differentiation (Fig. 2 G and H). Taken together, our results suggest that STING activation in CD4+ T cells induces TGF-β2 and IL-2 expression to facilitate Treg differentiation.
Fig. 2.
STING induces TGF-β2 and IL-2 expression to promote Treg differentiation. (A and B) TGF-β inhibitor treatment inhibits STING-mediated Treg differentiation. WT naïve CD4+ T cells were treated with DMXAA (2.5 μg/mL) in the presence or absence of TGF-β inhibitor SB 431542 (5 μM) for 3 d. Treg differentiation was analyzed by intracellular staining of Foxp3 and FACS analysis. Representative FACS plots are shown in (A), and statistics are shown in (B). (C) STING induces Tgfβ2 and Foxp3 expression. WT and Sting−/− CD4+ T cells were treated with mock, DMXAA (2.5 μg/mL), or cGAMP (10 μg/mL) for 3 d. Tgfβ1, Tgfβ2, Tgfβ3, and Foxp3 expression were detected by qRT-PCR. (D and E) The deficiency of IRF3 or IFNAR1 does not affect STING-mediated Tgfβ2 and Foxp3 expression. WT, Sting−/−, StingS365A/S365A, Irf3−/−, and Ifnar1−/− CD4+ T cells were treated with mock or DMXAA (2.5 μg/mL) for 3 d. Tgfβ2 (D) and Foxp3 (E) expression were detected by qRT-PCR. (F) STING activation induces Il2 expression. WT, Sting−/−, and StingS365A/S365A CD4+ T cells were treated with mock or DMXAA (2.5 μg/mL), and Il2 expression was detected by qRT-PCR. (G and H) IL-2 neutralization inhibits STING-mediated Treg differentiation. WT naïve CD4+ T cells were treated with cGAMP (10 μg/mL) in the presence or absence of IgG control, anti-IL2 neutralizing antibody JES6-1A12 (10 μg/mL) or S4B6-1 (10 μg/mL) for 3 d. Treg differentiation was analyzed. Representative FACS plots are shown in (G), and statistics are shown in (H). Error bars: SEM; **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not significant. Two-way ANOVA test. Data shown are representative of at least two independent experiments.
STING Activates CREB to Induce TGF-β2 and IL-2 Expression.
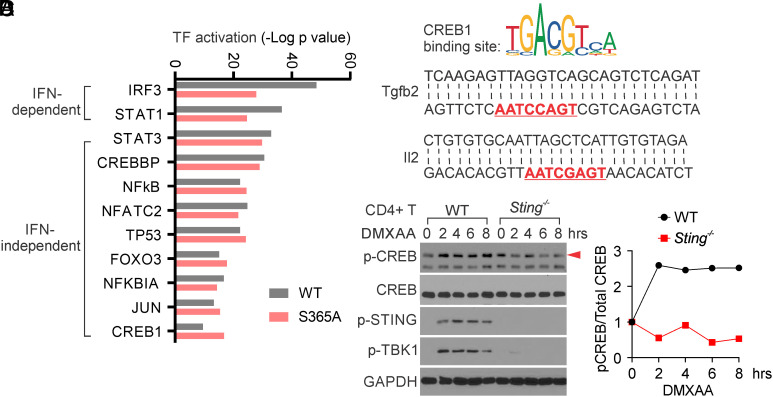
To investigate how STING transcriptionally induces TGF-β2 and IL-2 expression, we aimed to identify the transcription factors activated by STING to facilitate this process. Previously, we performed RNA-sequencing to compare STING-induced gene expression in WT, Sting−/−, and StingS365A/S365A T cells, and identified many STING-mediated IFN-independent gene expressions (7). Here, we performed ingenuity pathway analysis (IPA) to reanalyze our RNA-sequencing data and identified a dozen STING-mediated transcription factor activation (Fig. 3A). Among these candidates, IRF3 and STAT1 were two transcription factors more potently activated in WT compared to StingS365A/S365A T cells (Fig. 3A), suggesting their activation in a STING-mediated IFN-dependent manner. Indeed, IRF3 and STAT1 are two key transcription factors playing a crucial role in the STING-mediated IFN-I pathway. Interestingly, we also identified a group of transcription factors activated by STING in both WT and StingS365A/S365A T cells (Fig. 3A), indicating their activation in a STING-mediated IFN-independent manner. Among these transcription factors, NFκB was known to be activated by STING independently of IFN-I. Notably, we observed that CREB1, a critical transcription factor known to transcriptionally induce TGF-β2 and IL-2 expression (20, 24, 25, 32, 33), was also activated by STING independently of IFN-I (Fig. 3A). Our RNA-sequencing results also identified many CREB-associated gene expressions, including TGF-β2 and IL-2, in both WT and StingS365A/S365A T cells, but not in Sting−/− T cells, in response to STING activation (SI Appendix, Fig. S3). We scanned the Tgfβ2 and Il2 promoter sequences and identified critical CREB1 binding motifs in both gene promoter sequences (Fig. 3B). To verify whether STING activation can activate CREB, we isolated CD4+ T cells and stimulated them with DMXAA. We indeed found that DMXAA treatment induces the rapid activation of CREB in WT but not Sting−/− CD4+ T cells, as early as 2 h post-STING activation (Fig. 3 C and D). Together, our data suggest that STING activation promotes CREB activation to induce Tgfβ2 and Il2 expression independently of IFN-I.
Fig. 3.
STING activates CREB1 to induce TGF-β2 and IL-2 expression in T cells. (A) IPA prediction of transcription factor activation by STING. WT, Sting−/−, and StingS365A/S365A T cells were treated with mock or DMXAA, followed by RNA-sequencing analysis (7). The RNA-seq data were analyzed by IPA to predict transcription factor activation by STING. (B) CREB1 binding site is predicted to exist in the Tgfβ2 and Il2 promoter sequence. Tgfβ2 and Il2 promoter sequences were scanned by JASPAR (https://jaspar2020.genereg.net/) and found to contain a CREB1 binding motif, which is highlighted in red. (C and D) STING activates CREB1. WT and Sting−/− CD4+ T cells were treated with DMXAA for the indicated hours, and CREB and STING signaling were detected by Western blotting (C). Densitometry analysis is performed and normalized to the mock sample (D). The red arrow indicates the p-CREB band. Data shown are representative of at least two independent experiments.
STING Trafficking Is Required to Activate MAPK–CREB Signaling.
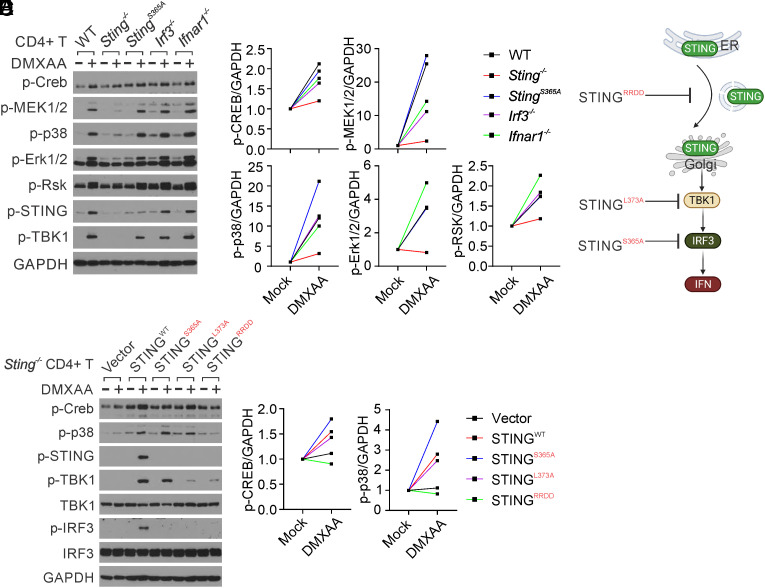
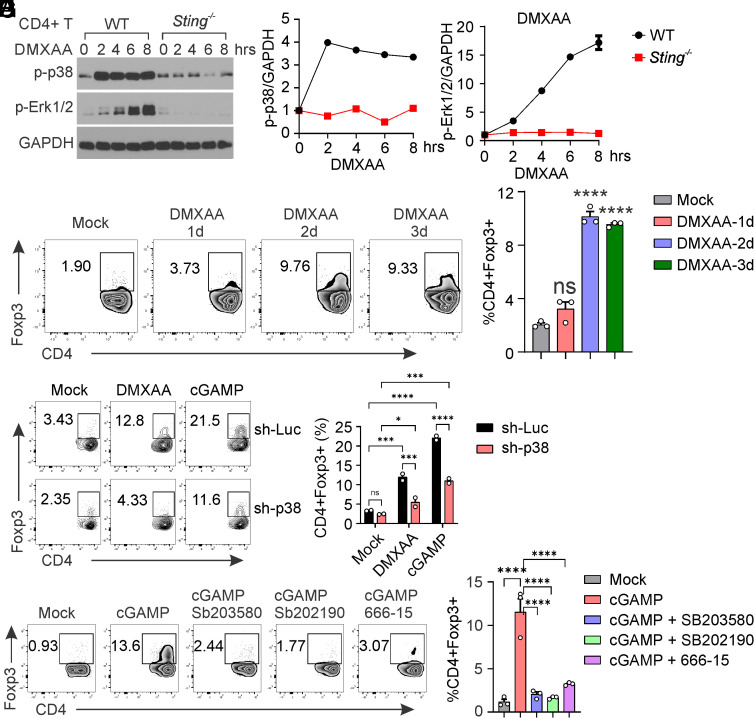
We then investigated how STING activates CREB. We found that STING does not directly interact with CREB in CD4+ T cells, regardless of STING activation (SI Appendix, Fig. S4A). CREB is a crucial transcription factor known to be activated by several serine-threonine kinases, including PKA, PKC, and MAPK (28). We examined these signaling pathways in CD4+ T cells following STING activation. STING agonist stimulation dramatically activated MAPK signaling, including MEK1/2, p38, ERK, and RSK, in WT but not Sting−/− CD4+ T cells (Fig. 4 A and B). Moreover, the StingS365A/S365A mutation or deficiency of IRF3 or IFNAR1 did not affect STING-activated MAPK–CREB signaling (Fig. 4A), suggesting that STING activates MAPK–CREB signaling independently of IRF3 and IFN-I. To further precisely identify the essential STING signaling activity necessary for MAPK–CREB activation, we reconstituted Sting−/− CD4+ T cells with specific STING mutants, including with STINGRRDD (STING trafficking deficient) (14, 29), STINGL373A (TBK1 binding & NFκB deficient) (34) and STINGS365A (IRF3 binding & IFN deficient) (7, 35, 36), each designed to halt STING signaling at a specific stage (Fig. 4C). After reconstitution, STINGWT restored DMXAA-induced phosphorylation of CREB, p38, STING, TBK1, and IRF3; STINGS365A was deficient in DMXAA-induced IRF3 phosphorylation but still capable of inducing TBK1, CREB, and p38 phosphorylation; and STINGL373A abrogated both DMXAA-induced TBK1 and IRF3 phosphorylation but still able to induce CREB and p38 phosphorylation (Fig. 4 D and E). In contrast, the STINGRRDD mutant, which we have previously shown to be deficient in STING trafficking (14), was unable to activate CREB and p38 (Fig. 4 D and E). Taken together, our results suggest that STING trafficking from the ER to the Golgi is required to activate MAPK–CREB signaling independently of IFN-I.
Fig. 4.
STING trafficking activates MAPK–CREB signaling in T cells. (A and B) Naïve CD4 T cells from WT, Sting−/−, StingS365A/S365A, Irf3−/−, and Ifnar1−/− mice were activated with anti-CD3 + anti-CD28 antibodies for 2 d, followed by treatment with mock or DMXAA (2.5 μg/mL) overnight. Cell lysates were then collected for the detection of MAPK–CREB signaling. Densitometry analysis is performed and normalized to the mock sample (B). (C) The schematic representation of STING mutations designed to selectively block STING signaling at specific stages. (D and E) Reconstitution of Sting−/− CD4+ T cells with STING mutants. Sting−/− CD4+ T cells were reconstituted with STING WT or various mutants, followed by stimulation with mock or DMXAA (2.5 μg/mL) overnight. MAPK–CREB and STING signaling were assessed by Western blotting. Densitometry analysis is shown in (E). The data presented are representative of at least three independent experiments.
MAPK p38-CREB Signaling Is Required for STING-Mediated Treg Differentiation.
To explore the correlation between STING-mediated MAPK activation and Treg differentiation, we examined the kinetics of these processes during STING activation. We found that MAPK p38 exhibited rapid activation as early as 2 h post-STING activation, with this signaling persisting for the subsequent 8 h. In contrast, ERK1/2 activation occurred more gradually following STING activation (Fig. 5 A–C). We then investigated Treg differentiation at various time points during STING activation. Interestingly, we observed that STING failed to induce Treg differentiation on day 1 but elicited robust Treg differentiation on days 2 and 3 post-STING activation (Fig. 5 D and E). These findings suggest that STING-mediated MAPK activation does not directly trigger Treg differentiation. Instead, it appears to facilitate Treg differentiation through downstream effects.
Fig. 5.
MAPK–CREB is required for STING-mediated Treg differentiation. (A–C) WT Naïve CD4 T cells were activated with anti-CD3 + anti-CD28 antibodies for 2 d, followed by treatment with mock or DMXAA (2.5 μg/mL) for indicated hours. Cell lysates were then collected for the detection of MAPK–CREB signaling. Densitometry analysis was performed and normalized to the mock sample (B and C). (D and E) WT naïve CD4+ T cells were stimulated with mock or DMXAA (2.5 μg/mL) for 1 to 3 d, followed by Treg analysis. Representative FACS plots are shown in (D), and statistics are shown in (E). (F and G) Knockdown of p38 inhibits STING-mediated Treg differentiation. WT naïve CD4+ T cells were transduced with lentiviral-mediated sh-luciferase control or sh-p38, followed by stimulation with mock, DMXAA (2.5 μg/mL), or cGAMP (20 μg/mL) for 3 d. Representative FACS plots are shown in (F), and statistics are shown in (G). (H and I) p38 or CREB inhibitor treatment inhibits STING-mediated Treg differentiation. WT naïve CD4+ T cells were treated with mock or cGAMP (20 μg/mL) in the presence or absence of p38 inhibitors sb203850 (10 μM) or sb202190 (10 μM), or CREB inhibitor 666-15 (1 μM), for 3 d. Treg differentiation was analyzed by intracellular staining of Foxp3 and FACS analysis. Representative FACS plots are shown in (H), and statistics are shown in (I). Error bars: SEM; *P < 0.05, ***P < 0.001, ****P < 0.0001; ns, not significant. Two-way ANOVA test. Data shown are representative of two independent experiments.
To investigate whether MAPK is required for STING-mediated Treg differentiation, we used lentiviral-based shRNA to knock down p38 expression in CD4+ T cells, then assessed its impact on Treg differentiation. Our results indicated that knocking down p38 (SI Appendix, Fig. S4B) significantly reduced DMXAA- or cGAMP-mediated Treg differentiation (Fig. 5 F and G). Additionally, pharmacological inhibition of p38 also dramatically inhibited STING-mediated Treg differentiation (Fig. 5 H and I). CREB is the transcription factor known to be activated by p38 (28). We also found that knocking down p38 diminished CREB activation in response to STING signaling (SI Appendix, Fig. S4B). Inhibition of CREB similarly greatly reduced STING-mediated Treg differentiation (Fig. 5 H and I). Taken together, our results suggest an essential role of MAPK–CREB signaling in STING-mediated Treg differentiation.
STING Activation Promotes Treg Induction in Mice.
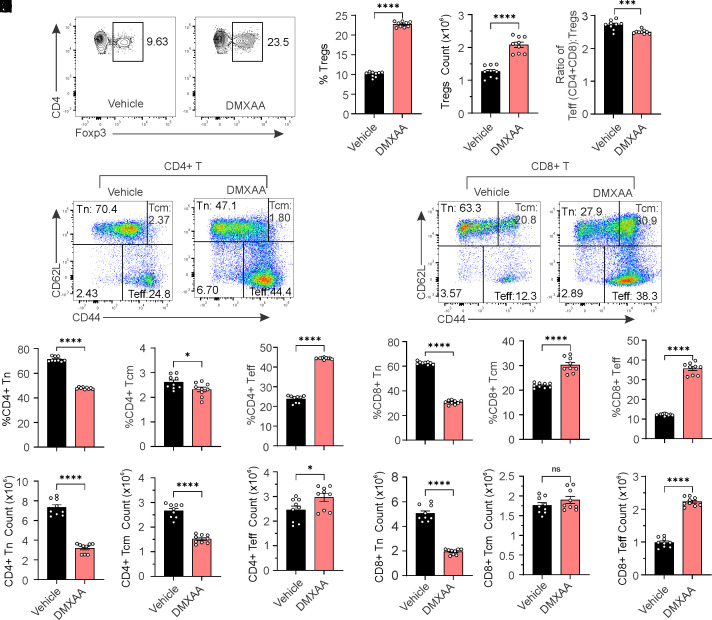
We next sought to investigate whether STING activation can induce Treg differentiation in mice. We treated mice with the STING agonist DMXAA and then assessed Tregs and other immune cell populations. Our results suggest that DMXAA treatment significantly increased the frequency and numbers of Treg cells in the periphery, while reducing the effector-to-Treg ratio (Fig. 6 A–D). Furthermore, we analyzed naïve, central memory, and effector T cells in both CD4+ and CD8+ T cell populations. STING activation led to a significant reduction in naïve and central memory T cells but promoted effector T cells in frequency and number in CD4+ T cells (Fig. 6 E–K). Similarly, STING activation reduced naïve T cells but enhanced central memory and effector T cells in CD8+ T cells (Fig. 6 L–R). Together, our results indicate that STING promotes Treg differentiation in mice.
Fig. 6.
STING activation promotes Treg differentiation in mice. Six-week-old mice were i.p. injected with vehicle or DMXAA (10 mg/kg) for 10 consecutive days. Splenocytes were harvested and stained for CD4, CD8, CD44, CD62L, and Foxp3. (A–C) Analysis of Treg cells. Representative FACs plots are shown in (A). Treg cell percentages and numbers are shown in (B and C). (D) The ratio of effector T cells to Treg cells. (E–K) Analysis of naïve, central memory, and effector CD4+ T cells. Representative FACs plots are shown in (E). The percentage (F–H) and cell number (I–K) of naïve, central memory, and effector CD4+ T cells are shown in bar graphs. (L–R) Analysis of naïve, central memory, and effector CD8+ T cells. Representative FACs plots are shown in (L). The percentage (M–O) and cell number (P–R) of naïve, central memory, and effector CD8+ T cells are shown in bar graphs. Error bars: SEM; *P < 0.05, ***P < 0.001, ****P < 0.0001; ns, not significant. Two-way ANOVA test.
T Cell Intrinsic STING Activation Induces Treg Differentiation in a Trex1−/− Autoimmune Disease Mouse Model.
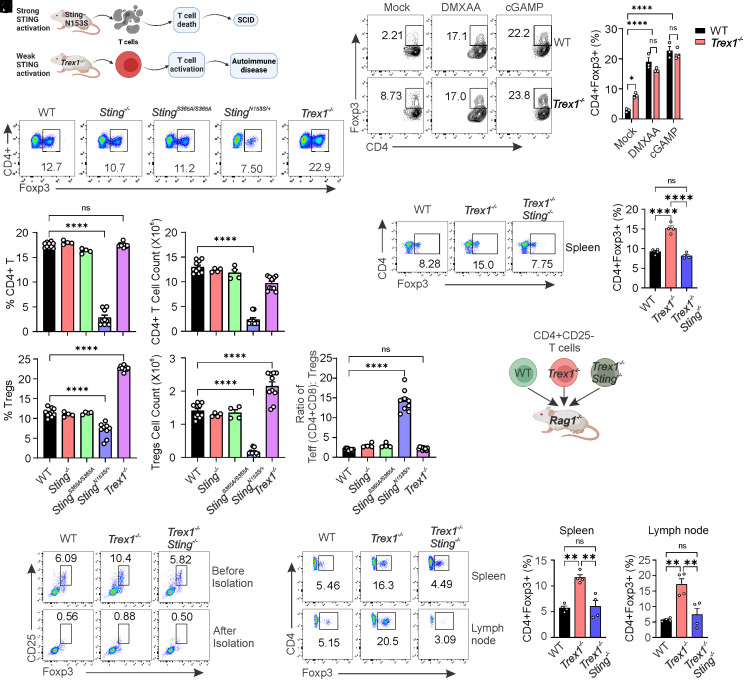
Chronic STING activation is associated with autoimmune and autoinflammatory diseases in humans (37). Various mouse models, including gain-of-function StingN153S/+ and Trex1−/−, have been developed to investigate STING-associated autoimmune diseases (15, 17, 38–44). Although disease development in StingN153S/+ and Trex1−/− mice is caused by constitutive STING activation (15, 17, 38–44), the downstream signaling driving disease pathogenesis differs. The gain-of-function StingN153S/+ mutation induces T cell death and cytopenia independently of IFN-I, leading to Severe Combined Immunodeficiency (SCID) (14, 17, 41). In contrast, Trex1 deficiency (Trex1−/−) results in constitutive STING-mediated IFN-I signaling, leading to T cell activation and autoinflammation (43, 44) (Fig. 7A). We sought to use these mouse models to investigate the impact of T cell intrinsic STING activation on Treg differentiation. Initially, we analyzed total CD4+ T cells and Tregs in both frequence and number and found no significant changes in Sting−/− and StingS365A/S365A mice compared to those in WT mice, suggesting that STING at the resting stage is not required for Treg development (Fig. 7 B–F). Next, we explored the impact of activated STING on Treg differentiation. The gain-of-function StingN153S/+ mutation induced T cell death (14), resulting in reduced CD4+ T cells and Tregs as expected (Fig. 7 B–F). However, CD4+ T cells showed no significant changes in Trex1−/− mice (Fig. 7 C and D), indicating that STING activation primed by Trex1 deficiency is not sufficient to induce CD4+ T cell death. In contrast, Treg cells significantly increased in Trex1−/− mice compared to WT mice (Fig. 7 E and F). We also observed that the effector-to-Treg ratio was significantly increased in StingN153S/+ mice, but not in Trex1−/− mice (Fig. 7G).
Fig. 7.
CD4+ T cell-intrinsic STING activation induces Treg differentiation in Trex1−/−autoimmune mice. (A) A schematic representation of constitutively activated STING mouse models. (B–G) CD4+ T cell and Treg analysis of WT, Sting−/−, StingS365A/S365A, StingN153S/+, and Trex1−/− mice. Splenocytes from the indicated mice were stained for Foxp3 and CD4. Representative FACS plots are shown in (B), statistics for CD4+ T cells and Tregs are shown in (C–F), and the ratio of effector T cells to Tregs is shown in (G). (H and I) WT and Trex1−/− naïve CD4 T cells were treated with mock, DMXAA (2.5 μg/mL), or cGAMP (20 μg/mL) for 3 d, and Treg differentiation was analyzed. Representative FACS plots (H) and statistics (I) are shown. (J and K) Treg analysis of Trex1−/− and Trex1−/− Sting−/− mice. Splenocytes were stained for Foxp3 and CD4. Representative FACS plots are shown in (J), and statistics of Tregs are shown in (K). (L) A schematic representation of adoptive transfer of naïve CD4+CD25- T cells into Rag1−/− mice. (M) Tregs were depleted in the isolated naïve CD4+ T cells from WT, Trex1−/−, and Trex1−/− Sting−/− mice. (N–P) Treg analysis of adoptively transferred CD4+ T cells. Naïve CD4+ T cells were adoptively transferred into Rag1−/− mice for 1 mo, and Tregs were analyzed in spleens and lymph nodes. Representative dot plots are shown in (N), and Treg percentages are shown in (O and P). Error bars: SEM; **P < 0.01, ****P < 0.0001; ns, not significant. Two-Way ANOVA test. Data shown are representative of at least two independent experiments.
We then analyzed naïve, central memory, and effector T cell subsets in both CD4+ and CD8+ T cells from these mice. The frequency and number of these subsets were comparable between WT, Sting−/−, and StingS365A/S365A mice (SI Appendix, Fig. S5). However, in StingN153S mice, the gain-of-function STING mutation significantly reduced the frequency and number of naïve T cells in both CD4+ and CD8+ T cells compared to WT mice (SI Appendix, Fig. S5 B, E, I, and L). While the percentage of central memory T cells showed a slight increase, their absolute numbers were notably decreased in both CD4+ and CD8+ T cell populations in StingN153S mice compared to WT mice (SI Appendix, Fig. S5 C, F, J, and M). The StingN153S mutation also resulted in an increased frequency, but not number, of effector T cells in both CD4+ and CD8+ T cells compared to WT mice (SI Appendix, Fig. S5 D, G, K, and N). Trex1-deficient mice exhibited a similar reduction in naïve T cell frequency and number (CD4+ and CD8+) as StingN153S mice (SI Appendix, Fig. S5). However, Trex1 deficiency had a distinct effect, expanding both central memory and effector T cells within both CD4+ and CD8+ subsets (SI Appendix, Fig. S5).
We next investigated whether STING activation driven by Trex1 deficiency also promotes Treg differentiation in vitro. Naïve CD4+ T cells from WT and Trex1−/− mice were treated with mock, DMXAA, or cGAMP to induce Treg differentiation. We found that intrinsic STING activation in Trex1−/− CD4+ T cells without ligand stimulation is sufficient to drive Treg differentiation (Fig. 7 H and I). Moreover, further STING activation by an agonist can boost Treg differentiation of both WT and Trex1−/− CD4+ T cells to a similar level (Fig. 7 H and I). Consistently, we also observed increased expression of Tgfb2 and Foxp3 in Trex1−/− CD4+ T cells compared to WT CD4+ T cells (SI Appendix, Fig. S4C).
To investigate whether the increased Tregs induction in Trex1−/− mice is dependent on STING, we generated Trex1−/−Sting−/− mice. The increased Treg cells in Trex1−/− mice were returned to the WT level upon STING deletion in Trex1−/−mice (Trex1−/−Sting−/−, Fig. 7 J and K), suggesting that constitutive STING activation drives Treg differentiation in Trex1−/− mice.
To further investigate whether this is a T cell-intrinsic or extrinsic effect of STING, we conducted T cell adoptive transfer experiments. We isolated naïve CD4+ T cells from WT, Trex1−/− and Trex1−/−Sting−/− mice and then adoptively transferred these cells into Rag1−/− mice to allow for their homeostatic proliferation and differentiation in the lymphopenic environment in vivo (Fig. 7L). We confirmed the complete depletion of Tregs in our isolated naïve CD4+ T cells before adoptive transfer (Fig. 7M). After 1 mo, T cells were isolated from both the spleen and lymph nodes, and Treg differentiation was analyzed. We observed a significant increase in Treg differentiation in mice that received adoptive transfer of Trex1−/− naïve CD4+ T cells compared to those that received WT naïve CD4+ T cells (Fig. 7 N–P). Interestingly, the elevated Treg differentiation observed in mice that received Trex1−/− naïve CD4+ T cells returned to WT levels upon STING deletion in Trex1−/− CD4+ T cells (Trex1−/−Sting−/−), both in the spleens and lymph nodes (Fig. 7 N–P). Collectively, our data suggest that intrinsic STING activation in CD4+ T cells induces Treg differentiation in vivo.
Discussion
STING is a crucial innate immune sensor that recognizes cGAMP produced by cGAS to induce IFN-I expression. Extensive research has focused on the STING-mediated IFN response in innate cells. Despite being more abundantly expressed in adaptive T cells, its intrinsic functions in T cells remain unclear. We previously demonstrated that STING activation in T cells induces widespread IFN-independent activities, including the activation of a cell death pathway (7). In our current study, we uncovered an IFN-independent signaling pathway of STING. We found that STING trafficking can activate MAPK signaling, leading to CREB transcription activation, which in turn induces the expression of TGF-β2 and IL-2 to facilitate Treg differentiation (SI Appendix, Fig. S6). Administration of a STING agonist or T cell-intrinsic STING activation primed by Trex1 deficiency promotes Treg differentiation in mice. Thus, the STING–MAPK–CREB axis represents an IFN-independent signaling pathway that may profoundly impact T cell effector function and adaptive immunity.
The unique characteristic of STING signaling is that STING activation requires its translocation from the ER to the Golgi. STING trafficking has been demonstrated to mediate several IFN-independent activities, including NFκB signaling, autophagy, calcium signaling, and UPR (14, 29, 45). A recent discovery revealed STING’s new function as a direct proton channel, mediating proton efflux from the Golgi (46, 47). Here, we found that STING trafficking is sufficient to activate MAPK–CREB signaling independently of IRF3 and IFN. CREB is a transcription factor known for its role in cell proliferation, differentiation, and survival (28). Given that STING trafficking is conserved across evolution, even in single-cell organisms (48), it is likely that STING may regulate cell growth and development in those lower organisms through the regulation of MAPK–CREB signaling.
How does STING trafficking activate the MAPK pathway? We previously demonstrated that STING activation induces ER stress in T cells (14). It has been reported that ER stress could activate Apoptosis signal-regulating kinase 1 (ASK1), an essential kinase that directly activates MAPK signaling (49). Additionally, we presented evidence that the STING ER exit induces calcium flux (14), which can also trigger MAPK activation (50). As T cell receptor (TCR) stimulation can activate MAPK signaling (51), it is possible that STING can enhance this process, and the STING signaling can synergize with TCR to induce MAPK activation.
Emerging evidence underscores the pivotal roles of CREB in immune responses, promoting the proliferation, survival, and regulation of T and B lymphocytes (19). CREB has been demonstrated to transcriptionally regulate many genes closely associated with T cell effector function, such as TCRα, TCRβ, CD3d, CD8a, IL-2, CD25/IL-2Ra, and IL-2Rg, suggesting that CREB may play a broad role in T cell function (20–23). In this study, we examine the transcriptional regulation of IL-2 and TGF-β2 by CREB in Treg differentiation. Our transcriptional analysis, comparing STING-induced gene expression in WT, Sting−/−, and StingS365A/S365A T cells, identified many differentially expressed genes (DEGs) associated with T cell effector function (7). Future validation of which DEGs are induced by CREB will provide insights into the roles of STING–MAPK–CREB signaling in the induction of other cytokine gene expressions, which may potentially influence T cell lineage development beyond Tregs. A recent study also showed that STING activation induces FOXP3 expression through the activation of SMAD3 and STAT5 (52). It is possible that multiple transcriptional programs might be activated by STING to regulate FOXP3 expression beyond CREB.
STING has been found to activate both type I (IFNα and IFNβ) and type II IFN (IFNγ) responses (30). Interestingly, SAVI disease has been defined as a type II interferonopathy, as deletion of IFNγR1 has been shown to improve multiple clinical features in SAVI mice, including lung disease and T cell cytopenia, including Tregs (30). Since IFNγ plays a crucial role in Treg development, it is possible that STING might regulate Treg differentiation through IFNγ. However, our data indicate that deletion of IFNγ does not affect STING-mediated Treg differentiation. Instead, knockdown of MAPK p38 or pharmacological inhibition of MAPK p38 or CREB markedly inhibits STING-mediated Treg differentiation. Thus, the STING–MAPK–CREB signaling axis represents an intrinsic T cell signaling pathway distinct from the STING-IFNγ signaling pathway in regulating Treg differentiation.
Throughout our study, we used different ligands, DMXAA and cGAMP, to activate STING. We also included STING knockout and other genetically modified STING as controls to exclude the STING-independent off-target effects of DMXAA. We demonstrated that administering the STING agonist DMXAA induces Treg development in WT mice. Although DMXAA is widely used as a STING agonist both in vitro and in vivo, it may have unknown targets. For example, we observed very mild MEK1/2 and p38 activation in STING knockout cells in response to DMXAA treatment (Fig. 4), albeit to a much lesser extent than in WT cells. Therefore, DMXAA-mediated Treg development (or other effects) may depend on STING along with another unknown target. To further clarify this finding, we analyzed Treg development in STING gain-of-function mice, including Trex1−/− and StingN153S/+ mice. We found that a gain-of-function STING mutation (StingN153S/+) induces Treg cell death, while constitutive activation of STING in Trex1−/− T cells promote Treg expansion. This finding further confirms that STING activation can promote Treg development in mice. To clarify the differential effects of STING activity on Treg development, we propose two possibilities. One possible explanation is that this may be related to STING signaling intensity. It is likely that intensive STING activation caused by STING gain-of-function mutation induces Treg cell death, while moderate STING activation caused by Trex1 deficiency promotes Treg differentiation. Another possible scenario may be related to how STING gets activated. The StingN153S/+ mutation-induced STING activation is independent of ligand binding, whereas STING activation in Trex1−/− T cells relies on STING ligand cGAMP binding. The distinct ways in which STING gets activated may determine which downstream signaling pathways would be activated.
Treg expansion in Trex1−/− mice depends on STING signaling, as this phenomenon disappears when STING is further deleted in Trex1−/−mice. This is likely a T cell intrinsic effect, as the adoptive transfer of Trex1−/− naïve CD4+ T cells into Rag1−/− mice also results in expansive Treg differentiation. Loss-of-function mutations in the human Trex1 gene cause autoimmune and autoinflammatory diseases, including Aicardi–Goutières syndrome (AGS) and systemic lupus erythematosus (SLE) (53, 54). Trex1−/− mice develop autoimmune phenotypes that recapitulate several clinical features in human patients. Trex1−/−-mediated autoimmunity initiates in nonhematopoietic cells but progresses via T lymphocytes (43). It has been proposed that STING-mediated IFN signaling caused by Trex1 deficiency promotes antigen presentation to activate T cell-mediated autoimmunity (43). However, the relative contribution of T cell intrinsic STING activation to T cell auto-activation is not well understood. STING activation in T cells promotes Th1 and Th9 differentiation (10), which may contribute to autoimmunity in Trex1−/− mice. However, we have shown that Treg cell percentage and number significantly increase in Trex1−/− mice due to T cell intrinsic STING activation. This finding leads us to propose the hypothesis that intrinsic STING activation in Tregs promotes Treg development, which may act as a rheostat to counter T cell-mediated autoimmunity in Trex1−/− mice. The conditional deletion of STING in different subtypes of T cells in Trex1−/−mice should provide more critical insights into how intrinsic STING functions in different subsets of T cells contribute to Trex1-associated autoimmunity. In fact, it has been reported that STING potently suppresses inflammation in a model of systemic lupus erythematosus (SLE) (55). Thus, the multifaceted functions of STING in pro- or anti-T cell activation may be critical to maintaining a homeostatic immune environment.
In summary, we have identified an IFN-independent signaling axis of STING in T cells, whereby STING trafficking triggers MAPK–CREB signaling to facilitate Treg differentiation both in vitro and in vivo. STING–MAPK–CREB signaling represents an IFN-independent pathway that may have a profound effect on T cell effector function beyond Treg differentiation.
Materials and Methods
Mice and Cells.
The Sting−/−, StingN153S/+, StingS365A/S365A, and Trex1+/− mice were obtained from Nan Yan (UT Southwestern Medical Center). Irf3−/− mice were obtained from Ganes Sen and Laura Nagy (Lerner Research Institute) with permission from Tadatsugu Taniguchi. Ifnar1−/−, Ifngr1−/−, Rag1−/−, and Foxp3YFP reporter mice were purchased from Jackson Labs. All mice were maintained in the Lerner Research Institute Biological Resources Unit and were used in accordance with protocols approved by the Institutional Animal Care and Use Committee at the Cleveland Clinic Foundation. HEK293T and MEFs were maintained in DMEM with 10% (vol/vol) FBS, 2 mM L-glutamine, 10 mM HEPES, and 1 mM sodium pyruvate (complete DMEM) with the addition of 100 U/mL penicillin and 100 mg/mL streptomycin and cultured at 37 °C with 5% CO2.
Antibodies.
The following antibodies were used for flow cytometry: anti-CD8α (53 to 6.7), anti-CD4 (L3T4), anti-CD3ε (145-2C11), anti-CD25 (PC61.5), anti-CD69 (H1.2F3), anti-CD44 (IM7), and anti-Foxp3 (MF-14) all from BioLegend. The splenocytes or T cells were stained with the indicated panel of antibodies according to the manufacturers’ instructors, and then analyzed by BD FACSymphony™ Flow Cytometer. Purified anti-CD3ε (145-2C11) and anti-CD28 (37.51; both from BioXCell) were used at the appropriate concentration for T cell activation. The following antibodies were used for immunoblotting analysis: anti-STING (D2P2F; Cell Signaling), anti-pSTING (D8F4W; Cell Signaling), anti-TBK1 (D1B4; Cell Signaling), anti-pTBK1 (D52C2; Cell Signaling), anti-IRF3 (D83B9, Cell Signaling), anti-pIRF3 (4D4G, Cell Signaling), Phospho-CREB (87G3, Cell Signaling), Phospho-p38 (D3F9; Cell Signaling), Phospho-SAPK/JNK (81E11; Cell Signaling), Phospho-MEK1/2 (41G9; Cell Signaling), Phospho-p44/42 MAPK (Erk1/2) (D13.14.4E; Cell Signaling), Phospho-p90RSK (D3H11; Cell Signaling), total CREB (48H2, Cell Signaling), total p38 (D13E1, Cell Signaling), and Phospho-MSK1 (Thr581; Cell Signaling).
RNA Isolation and qRT-PCR.
Total RNA was isolated with a TRI reagent according to the manufacturer’s instructions (Sigma-Aldrich). cDNA was synthesized with an iScript cDNA synthesis kit (Bio-Rad). The iTaq Universal SYBR Green Supermix (Bio-Rad) and an CFX96 Touch Real-Time PCR Detection System (Bio-Rad) were used for qRT-PCR analysis (primer sequences listed in SI Appendix, Table S1). Gapdh were used as housekeeping genes for data normalization.
T Cell Adoptive Transfer.
T cell adoptive transfer was performed as described previously (7). Briefly, the spleen was isolated, crushed, and passed through a 70-μM strainer to get the single cell suspension. Red blood cells were lysed by ACK lysis buffer. Naïve CD4+ T cells were isolated with the T Cell Isolation Kit (Miltenyi, #130-104-453) according to the manufacturer’s instructions. T cell purity was examined by staining CD4 and CD25 expression and flow cytometry analysis. 5 × 106 naïve CD4+ T cells were then adoptively transferred into Rag1−/− mice by i.v. injection. One month later, spleens and lymph nodes were harvested and analyzed for Treg differentiation in vivo by staining CD4 and FOXP3 expression.
T Cell Activation and Differentiation In Vitro.
Treg differentiation was performed according to the procedure described previously (56). 1 × 106 CD4+ T cells were isolated and activated in 24-well plates precoated with 10 mg/mL anti-CD3 (145-2C11, BioXCell) and 10 mg/mL anti-CD28 mAb (37.51, BioXCell). For Treg differentiation, 5 ng/mL TGF-β (BioLegend, #781802), 20 ng/mL IL-2 (Peprotech, #212-12), 5 μg/mL anti-IFNγ (BioLegend, #505834), and 5 μg/mL anti-IL4 (BioLegend, #504122) were added to the culture for 3 d. The FOXP3 intracellular staining kit (ThermoFisher, #00-5523-00) was used to quantify Treg differentiation according to the manufacturer’s instructions. For STING-mediated Treg differentiation, STING agonist DMXAA (2.5 μg/mL, Invivogen, #tlrl-dmx) or 2′3′-cGAMP (10 μg/mL, Invivogen, #tlrl-nacga23-02), 5 μg/mL anti-IFNγ (BioLegend, #505834), and 5 μg/mL anti-IL4 (BioLegend, #504122) were added to the culture for 3 d. The Treg differentiation were quantified by FOXP3 intracellular staining.
Viral Transduction of T Cells.
For ectopic gene expression, we cloned STING into the MSCV-puromycin (Addgene #68469) by Gibson HIFI assembly (NEB #E2621L). The point mutants of STING were made using the Quikchange II Site-Directed Mutagenesis Kit (Agilent Technologies, Inc.). To produce recombinant retroviruses, HEK293T cells were grown to a confluency of 80 to 90% for transfection. Retroviral constructs were transfected with pCL-Eco (Addgene#12371) at a 3:1 ratio in HEK293T cells using Lipofectamine 2000 (ThermoFisher) per the manufacturer’s instructions. Viruses were collected at day 2 to 3 after transfection and stored at −80 degrees. For retroviral transduction, CD4+ T cells were activated in 24-well plates precoated with 10 mg/mL anti-CD3 (145-2C11, BioXCell) and 10 mg/mL anti-CD28 mAb (37.51, BioXCell) for 24 h, and then spin-inoculated at 800 g with indicated recombinant virus in the presence of 5 mg/mL polybrene (Sigma-Aldrich) and 10 mM HEPES buffer (GIBCO) for 90 min. Transduced cells were selected by adding 4 mg/mL puromycin in the culture medium for 2 d.
For shRNA-mediated RNAi, p38 shRNA was cloned into the pLKO.1 lentivector, which carries puromycin resistance genes for transduction selection. HEK293T cells were grown to a confluency of 80 to 90% for transfection. For lentivirus production, the p38 shRNA lentivector, pMD2.G (Addgene #12259), and psPAX2 (Addgene #12260) were transfected at a 10:5:9 ratio in HEK293T cells. Lentivirus collection and T cell transduction were performed as described above.
DMXAA Administration of the Mice.
Six-week-old female WT mice were intraperitoneally injected with either vehicle or DMXAA (10 mg/kg) for 10 consecutive days. Splenocytes were harvested and subjected to cell surface staining for CD4, CD8, CD44, CD62L, and Zombie Aqua. Subsequently, FOXP3 staining was performed using the eBioscience™ Foxp3/Transcription Factor Staining Buffer Set (cat# 00-5523-00) according to the manufacturer’s instructions. The stained cells were then analyzed using a BD FACSymphony™ Flow Cytometer.
Statistical Analysis.
Data are presented as the mean ± SEM. Prism 10 (GraphPad) was used for statistical analysis. Statistical tests performed are indicated in figure legends. *P < 0.05; **P < 0.01; ***P < 0.001; and ****P < 0.0001.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Nan Yan (UT Southwestern Medical Center, Dallas) for Sting−/−, StingN153S/+, StingS365A/S365A, and Trex1−/−mice. We thank Ganes Sen and Laura Nagy (Lerner Research Institute, Cleveland) for Irf3−/−mice. We thank Robert Fairchild for critical reading of the manuscript. We thank Madelyn Dirrim for scientific editing of our manuscript. We thank members of the Wu laboratory for helpful discussions. This work was supported by the NIH (R37CA288747 to J.W., DP2-AI170515 to X.W.), a MRA Young Investigator Award (1034214 to J.W.), a V Foundation for Cancer Research Award (V2023-016 to J.W.), a VeloSano Award, and an American Cancer Society IRG.
Author contributions
W.L. and J.W. designed research; W.L., C.S., T.L., H.T., and J.W. performed research; W.L., C.S., T.L., H.T., X.W., and J.W. analyzed data; and W.L., C.S., T.L., H.T., X.W., and J.W. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Sun L., Wu J., Du F., Chen X., Chen Z. J., Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu J., et al. , Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ishikawa H., Barber G. N., STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishikawa H., Ma Z., Barber G. N., STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461, 788–792 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeltema D., Abbott K., Yan N., STING trafficking as a new dimension of immune signaling. J. Exp. Med. 220, e20220990 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Margolis S. R., Wilson S. C., Vance R. E., Evolutionary origins of cGAS-STING signaling. Trends Immunol. 38, 733–743 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Wu J., Dobbs N., Yang K., Yan N., Interferon-independent activities of mammalian STING mediate antiviral response and tumor immune evasion. Immunity 53, 115–126.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imanishi T., et al. , Reciprocal regulation of STING and TCR signaling by mTORC1 for T-cell activation and function. Life Sci. Alliance 2, e201800282 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Damasceno L. E. A., et al. , STING is an intrinsic checkpoint inhibitor that restrains the T(H)17 cell pathogenic program. Cell Rep. 39, 110838 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benoit-Lizon I., et al. , CD4 T cell-intrinsic STING signaling controls the differentiation and effector functions of T(H)1 and T(H)9 cells. J. Immunother. Cancer 10, e003459 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li S., et al. , STING-induced regulatory B cells compromise NK function in cancer immunity. Nature 610, 373–380 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larkin B., et al. , Cutting edge: Activation of STING in T cells induces type I IFN responses and cell death. J. Immunol. 199, 397–402 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gulen M. F., et al. , Signalling strength determines proapoptotic functions of STING. Nat. Commun. 8, 427 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu J., et al. , STING-mediated disruption of calcium homeostasis chronically activates ER stress and primes T cell death. J. Exp. Med. 216, 867–883 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warner J. D., et al. , STING-associated vasculopathy develops independently of IRF3 in mice. J. Exp. Med. 214, 3279–3292 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouis D., et al. , Severe combined immunodeficiency in stimulator of interferon genes (STING) V154M/wild-type mice. J. Allergy Clin. Immunol. 143, 712–725.e5 (2019), 10.1016/j.jaci.2018.04.034. [DOI] [PubMed] [Google Scholar]
- 17.Luksch H., et al. , STING-associated lung disease in mice relies on T cells but not type I interferon. J. Allergy Clin. Immunol. 144, 254–266.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sivick K. E., et al. , Magnitude of therapeutic STING activation determines CD8(+) T cell-mediated anti-tumor immunity. Cell Rep. 29, 785–789 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Wen A. Y., Sakamoto K. M., Miller L. S., The role of the transcription factor CREB in immune function. J. Immunol. 185, 6413–6419 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Solomou E. E., Juang Y. T., Tsokos G. C., Protein kinase C-theta participates in the activation of cyclic AMP-responsive element-binding protein and its subsequent binding to the -180 site of the IL-2 promoter in normal human T lymphocytes. J. Immunol. 166, 5665–5674 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Anderson S. J., Miyake S., Loh D. Y., Transcription from a murine T-cell receptor V beta promoter depends on a conserved decamer motif similar to the cyclic AMP response element. Mol. Cell Biol. 9, 4835–4845 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayall T. P., Sheridan P. L., Montminy M. R., Jones K. A., Distinct roles for P-CREB and LEF-1 in TCR alpha enhancer assembly and activation on chromatin templates in vitro. Genes Dev. 11, 887–899 (1997). [DOI] [PubMed] [Google Scholar]
- 23.Gao M. H., Kavathas P. B., Functional importance of the cyclic AMP response element-like decamer motif in the CD8 alpha promoter. J. Immunol. 150, 4376–4385 (1993). [PubMed] [Google Scholar]
- 24.Rodon L., et al. , Active CREB1 promotes a malignant TGFbeta2 autocrine loop in glioblastoma. Cancer Discov. 4, 1230–1241 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Wotton D., A CREB1-TGFbeta2 self-sustaining loop in glioblastoma. Cancer Discov. 4, 1123–1125 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Barton K., et al. , Defective thymocyte proliferation and IL-2 production in transgenic mice expressing a dominant-negative form of CREB. Nature 379, 81–85 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Kim H. P., Leonard W. J., CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: A role for DNA methylation. J. Exp. Med. 204, 1543–1551 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayr B., Montminy M., Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2, 599–609 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Gui X., et al. , Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature 567, 262–266 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stinson W. A., et al. , The IFN-gamma receptor promotes immune dysregulation and disease in STING gain-of-function mice. JCI Insight 7, e155250 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J., Zhao X., Wan Y. Y., Intricacies of TGF-beta signaling in Treg and Th17 cell biology. Cell Mol. Immunol. 20, 1002–1022 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hughes-Fulford M., et al. , Early immune response and regulation of IL-2 receptor subunits. Cell Signal. 17, 1111–1124 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Zhou W., et al. , Modulation of morphogenesis by noncanonical Wnt signaling requires ATF/CREB family-mediated transcriptional activation of TGFbeta2. Nat. Genet. 39, 1225–1234 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao B., et al. , A conserved PLPLRT/SD motif of STING mediates the recruitment and activation of TBK1. Nature 569, 718–722 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yum S., Li M., Fang Y., Chen Z. J., TBK1 recruitment to STING activates both IRF3 and NF-kappaB that mediate immune defense against tumors and viral infections. Proc. Natl. Acad. Sci. U.S.A. 118, e2100225118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamashiro L. H., et al. , Interferon-independent STING signaling promotes resistance to HSV-1 in vivo. Nat. Commun. 11, 3382 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Motwani M., Pesiridis S., Fitzgerald K. A., DNA sensing by the cGAS-STING pathway in health and disease. Nat. Rev. Genet. 20, 657–674 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Siedel H., Roers A., Rosen-Wolff A., Luksch H., Type I interferon-independent T cell impairment in a Tmem173 N153S/WT mouse model of STING associated vasculopathy with onset in infancy (SAVI). Clin. Immunol. 216, 108466 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Motwani M., et al. , Hierarchy of clinical manifestations in SAVI N153S and V154M mouse models. Proc. Natl. Acad. Sci. U.S.A. 116, 7941–7950 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin G. R., et al. , Expression of a constitutively active human STING mutant in hematopoietic cells produces an Ifnar1-dependent vasculopathy in mice. Life Sci. Alliance 2, e201800215 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bouis D., et al. , Severe combined immunodeficiency in stimulator of interferon genes (STING) V154M/wild-type mice. J. Allergy Clin. Immunol. 143, 712–725.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Bennion B. G., et al. , A human gain-of-function STING mutation causes immunodeficiency and gammaherpesvirus-induced pulmonary fibrosis in mice. J. Virol. 93, e01806-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gall A., et al. , Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity 36, 120–131 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stetson D. B., Ko J. S., Heidmann T., Medzhitov R., Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134, 587–598 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu S., et al. , Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347, aaa2630 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Xun J., et al. , A conserved ion channel function of STING mediates noncanonical autophagy and cell death. EMBO Rep. 25, 544–569 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu B., et al. , Human STING is a proton channel. Science 381, 508–514 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woznica A., et al. , STING mediates immune responses in the closest living relatives of animals. Elife 10, e70436 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Darling N. J., Cook S. J., The role of MAPK signalling pathways in the response to endoplasmic reticulum stress. Biochim. Biophys. Acta 1843, 2150–2163 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Rosen L. B., Ginty D. D., Weber M. J., Greenberg M. E., Membrane depolarization and calcium influx stimulate MEK and MAP kinase via activation of Ras. Neuron 12, 1207–1221 (1994). [DOI] [PubMed] [Google Scholar]
- 51.Adachi K., Davis M. M., T-cell receptor ligation induces distinct signaling pathways in naive vs. antigen-experienced T cells. Proc. Natl. Acad. Sci. U.S.A. 108, 1549–1554 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ni H., et al. , T cell-intrinsic STING signaling promotes regulatory T cell induction and immunosuppression by upregulating FOXP3 transcription in cervical cancer. J. Immunother. Cancer 10, e005151 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Vries B., et al. , TREX1 gene variant in neuropsychiatric systemic lupus erythematosus. Ann. Rheum. Dis. 69, 1886–1887 (2010). [DOI] [PubMed] [Google Scholar]
- 54.Crow Y. J., et al. , Mutations in the gene encoding the 3’-5’ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat. Genet. 38, 917–920 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Sharma S., et al. , Suppression of systemic autoimmunity by the innate immune adaptor STING. Proc. Natl. Acad. Sci. U.S.A. 112, E710–E717 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chou W. C., et al. , AIM2 in regulatory T cells restrains autoimmune diseases. Nature 591, 300–305 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.