Significance
Spinal and bulbar muscular atrophy (SBMA) is a slowly progressing neuromuscular disease caused by a polyglutamine (polyQ) expansion in the androgen receptor (AR), a ligand-activated transcription factor that, among many of its functions, regulates neurite growth and regeneration. Dysfunction of these processes has been implicated in SBMA; however, whether AR transcriptional functions contribute to this disease pathology is poorly understood. This study reveals that mechanisms independent of AR canonical transcriptional activity underly defects in neurite growth and maintenance in SBMA and identifies senescence as a previously recognized mechanism in SBMA pathology. These findings suggest that effective therapeutics for SBMA patients may not need to inhibit AR transcriptional function to be effective.
Keywords: SBMA, neurite, transcription, senescence
Abstract
Spinal and bulbar muscular atrophy (SBMA) is a slowly progressing neuromuscular disease caused by a polyglutamine (polyQ)-encoding CAG trinucleotide repeat expansion in the androgen receptor (AR) gene, leading to AR aggregation, lower motor neuron death, and muscle atrophy. AR is a ligand-activated transcription factor that regulates neuronal architecture and promotes axon regeneration; however, whether AR transcriptional functions contribute to disease pathogenesis is not fully understood. Using a differentiated PC12 cell model of SBMA, we identified dysfunction of polyQ-expanded AR in its regulation of neurite growth and maintenance. Specifically, we found that in the presence of androgens, polyQ-expanded AR inhibited neurite outgrowth, induced neurite retraction, and inhibited neurite regrowth. This dysfunction was independent of polyQ-expanded AR transcriptional activity at androgen response elements (ARE). We further showed that the formation of polyQ-expanded AR intranuclear inclusions promoted neurite retraction, which coincided with reduced expression of the neuronal differentiation marker β-III-Tubulin. Finally, we revealed that cell death is not the primary outcome for cells undergoing neurite retraction; rather, these cells become senescent. Our findings reveal that mechanisms independent of AR canonical transcriptional activity underly neurite defects in a cell model of SBMA and identify senescence as a pathway implicated in this pathology. These findings suggest that in the absence of a role for AR canonical transcriptional activity in the SBMA pathologies described here, the development of SBMA therapeutics that preserve this activity may be desirable. This approach may be broadly applicable to other polyglutamine diseases such as Huntington’s disease and spinocerebellar ataxias.
Spinal and bulbar muscular atrophy (SBMA; Kennedy’s disease) is a recessive, X-linked neurodegenerative disorder characterized by progressive bulbar and proximal limb muscle weakness, fasciculations, dysarthria, dysphagia, and mild androgen insensitivity, with onset between the third and fifth decades of life (1, 2). Due to a requirement for circulating testosterone, SBMA is a male-specific disease, with female carriers presenting only mild symptoms (3–5).
Pathologically, the disease is defined by degeneration of spinal cord and brainstem lower motor neurons and muscle atrophy, as well as sensory neuron deficits (1, 2, 6). SBMA is caused by a polyglutamine (polyQ)-encoding CAG repeat expansion of 38 repeats or greater in exon one of the androgen receptor (AR) gene (7). A cellular hallmark of disease is found in the misfolding and aggregation of mutant AR upon ligand binding, which presents as intranuclear inclusions composed of progressively proteolyzed mutant AR into insoluble N-terminal fragments in surviving motor neurons and muscle (8–11).
The androgen receptor is a member of the steroid hormone receptor family responsible for transcription of genes containing specific DNA-responsive elements. AR exists in an inactive aporeceptor complex bound to chaperones in the cytoplasm (12). Upon binding to its ligand, testosterone or dihydrotestosterone (DHT), AR undergoes a conformational change, translocates to the nucleus, and binds to androgen response elements (AREs) on DNA as a homodimer through its DNA-binding domain (DBD) (13). The first zinc finger of the AR DBD contacts the major groove of DNA at AREs and the second is responsible for AR homodimerization (13, 14). Mutations in the first zinc finger of the DBD inhibit AR binding to AREs but preserve androgen binding capabilities and are found in individuals with complete androgen insensitivity syndrome (15).
Androgens and AR serve important growth-promoting and trophic functions in motor neurons and muscle, the affected populations in SBMA. Androgens regulate cell morphology and dendritic length of highly and moderately androgen-sensitive motor neuron pools (16, 17). Furthermore, androgen administration enhances axon regeneration of both cranial and spinal motor neurons with evidence of functional recovery and attenuates neuronal loss following axotomy (18–23). In motor neuronal cell models, androgens exhibit a direct trophic effect and regulate neurite outgrowth in the presence of AR (24, 25).
Several lines of evidence point to the dysregulation of AR transcriptional function in SBMA. However, the role of this dysregulation in disease pathology is not completely resolved. There is a consensus in the field that polyQ-expanded AR has altered transcriptional activity compared to normal polyQ-length AR (26–31); however, the fact that SBMA patients show only partial androgen insensitivity indicates substantial retention of AR transcriptional activity (2). Moreover, in a Drosophila model of SBMA, mutation of the AR DNA-binding domain (A574D) eliminated the neurodegenerative phenotype (32). In contrast, promoting polyQ-expanded AR transcriptional activity through decreased SUMOylation in a mouse model of SBMA enhanced treadmill running and increased survival, simultaneously enhancing trophic support to muscle (30).
Defects in neurite outgrowth and retention have been identified in PC12 cells (33), NSC34 cells (34, 35), and patient iPSC-derived motor neurons (36) expressing polyQ-expanded AR. These studies revealed that increased polyQ lengths were associated with a decrease in neurites but differed in the requirement for hormone for the observed phenotypes, warranting further investigation. Additionally, despite the identification of candidates that may contribute to neurite loss in SBMA (33, 36), mechanisms remain poorly understood, and the role of AR transcriptional activity in this pathology has yet to be explored.
In this study, we utilized a differentiated PC12 cell model of SBMA to evaluate details of neurite loss, the role of polyQ-expanded AR transcriptional activity in this loss, and the mechanisms underlying this pathology. Using live-cell imaging to monitor neurite growth and retraction over time, we show that hormone-bound polyQ-expanded AR—independent of its ARE-binding-dependent transcriptional activity—inhibits neurite outgrowth, leads to neurite loss by inducing neurite retraction, and inhibits neurite regrowth following neurite retraction. Furthermore, we reveal that intranuclear inclusion formation serves as a catalyst for neurite retraction, which occurs alongside a dedifferentiation phenomenon defined by reduced β-III-Tubulin expression. Finally, we show that cells become senescent following neurite retraction, revealing a previously unappreciated targetable pathway for therapeutic intervention in SBMA.
Results
Hormone-Bound PolyQ-Expanded AR Inhibits Neurite Outgrowth, Independent of Canonical AR DNA Binding at AREs.
Prior studies identified an inhibitory effect of polyQ-expanded AR expression on neurite length (33–36). We therefore sought to determine the role of AR expression and transcriptional activity on neurite outgrowth in the presence and absence of androgens in PC12 cells. PC12 cells can be differentiated into neuronal cells upon treatment with nerve growth factor (NGF) (37), allowing the study of neurite outgrowth in the presence and absence of AR expression and DHT. The PC12 cells used in this study express full-length normal or polyQ-expanded AR under the control of a tetracycline-inducible promotor, where addition of doxycycline (Dox) induces AR expression. DHT treatment induces nuclear localization of AR and the formation of nuclear aggregates of polyQ-expanded AR (9).
To study the effect of AR expression and transcriptional activity on neurite outgrowth, we simultaneously treated PC12 cells with NGF to induce differentiation and Dox to induce equivalent expression of wild-type AR (AR10Q), DNA-binding competent polyQ-expanded AR (AR112Q), or DNA-binding-mutant polyQ-expanded AR (AR111Q V582F) (SI Appendix, Fig. S1 A and B). The V582F mutation is in the first zinc finger of the AR DNA-binding domain and modifies a residue that makes direct contact with DNA (38). This mutation eliminates ARE-dependent AR transcriptional activity while preserving androgen-binding capacity (15) (SI Appendix, Fig. S2). We assessed whether DNA-binding mutant AR111Q V582F maintained the capacity to bind to chromatin, which would suggest that it maintains transcriptional activity at sites other than AREs, possibly through tethering by other DNA-bound transcription factors (39–42). Through chromatin fractionation and western blot analysis, we observed, as anticipated, that AR binding to chromatin increased in the presence of DHT (SI Appendix, Fig. S3 A and B). We also found that AR111Q V582F bound to chromatin slightly more than AR112Q (SI Appendix, Fig. S3 C and D); furthermore, AR with enhanced DNA-binding and transcriptional activity mediated by mutation of AR SUMOylation sites (AR111Q K387/519R) (30) bound even more to chromatin than AR111Q V582F (SI Appendix, Fig. S3 C and D). Overall, the binding of AR111Q V582F to chromatin suggests it likely contributes to transcriptional activity at sites other than AREs.
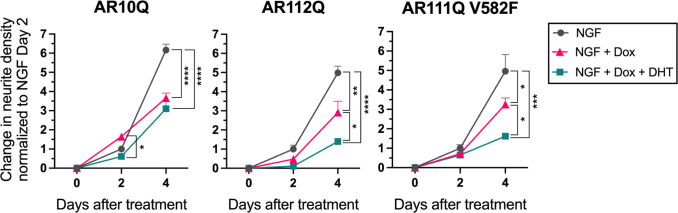
To determine the role of androgens in neurite outgrowth, we differentiated PC12 cells in the presence and absence of DHT, performed live-cell imaging on days 0, 2, and 4 of differentiation, and quantified neurite densities at these time points (SI Appendix, Fig. S4). After 4 d of differentiation, AR expression significantly reduced total neurite density, independent of AR polyQ-length and ARE-binding capacity (Fig. 1). Importantly, Dox treatment of PC12 cells that do not contain Dox-inducible AR had no effect on neurite density (SI Appendix, Fig. S5). In the presence of AR10Q, DHT treatment had no further effect on neurite density (Fig. 1, Left). However, in the presence of polyQ-expanded AR, DHT treatment further reduced neurite outgrowth (Fig. 1, Middle). Notably, this reduction in neurite density was independent of the ability of polyQ-expanded AR to bind DNA at AREs (Fig. 1, Right). Importantly, cell number was equivalent between treatment conditions for AR10Q, AR112Q, and AR111Q V582F cell lines, indicating that the effects of AR and DHT are specific to neurites and not due to differences in cell density (SI Appendix, Fig. S6). Overall, these analyses reveal that AR expression inhibits neurite outgrowth in PC12 cells, while DHT treatment further inhibits neurite outgrowth only for polyQ-expanded AR, independent of AR ARE-mediated transcriptional activity.
Fig. 1.
Hormone-bound polyQ-expanded AR inhibits neurite outgrowth, independent of AR DNA binding. Change in neurite density from 0 to 4 d of NGF, Dox, and DHT treatment, normalized to NGF day 2 for AR10Q, AR112Q, and AR111Q V582F cell lines. Data represent mean ± SEM from three wells per treatment condition, representative of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001, two-way mixed model ANOVA with Tukey’s multiple comparison test.
Neurite Loss in the Presence of PolyQ-Expanded AR Is Independent of AR Transcriptional Activity at AREs.
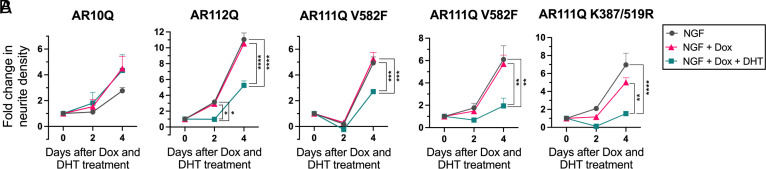
In neuronal models of SBMA, polyQ-expanded AR induces neurite loss (33–36). We sought to determine whether this neurite loss was dependent on polyQ-expanded AR ARE-mediated transcriptional activity. We differentiated PC12 cells to equivalent neurite densities of ~1,500 µm/mm2 (SI Appendix, Fig. S7A), chosen based on expression patterns of neuronal differentiation markers β-III-Tubulin, Synapsin-1, and GAP43 (37). We subsequently treated these differentiated cells with Dox to induce equivalent AR expression (SI Appendix, Fig. S1 A–C), DHT to induce AR nuclear localization, and AraC to eliminate any proliferating cells left in the culture. We performed live-cell imaging on days 0, 2, and 4 after Dox and DHT treatment and quantified neurite densities at these time points (SI Appendix, Fig. S7B). After 4 d of Dox and DHT treatment, AR10Q in the presence and absence of DHT had no significant effect on neurite density. However, hormone-bound polyQ-expanded AR expression in the presence of DHT significantly reduced neurite density, which was independent of AR ARE-mediated transcriptional activity (Fig. 2A). Since polyQ-expanded AR has altered transcriptional activity compared to WT AR (26–31), we wanted to confirm that the reduction in neurite density observed in the presence of hormone-bound polyQ-expanded AR was not due to a loss of AR transcriptional function. We therefore utilized PC12 cells expressing polyQ-expanded AR with enhanced transcriptional activity mediated by mutation of AR SUMOylation sites to enhance its DNA binding and activity (AR111Q K387/519R) (30). After 4 d of Dox and DHT treatment, AR111Q K387/519R in the presence of DHT induced a decrease in neurite density similar to that observed in the presence of AR111Q V582F (Fig. 2B). Taken together, these data indicate that neurite loss induced by hormone-bound polyQ-expanded AR in differentiated PC12 cells is independent of AR ARE-mediated transcriptional activity.
Fig. 2.
Hormone-bound polyQ-expanded AR induces neurite loss, independent of AR DNA binding. (A) Fold change in neurite density from 0 to 4 d of Dox and DHT treatment of differentiated AR10Q, AR112Q, and AR111Q V582F cells. (B) Fold change in neurite density from 0 to 4 d of Dox and DHT treatment of differentiated AR111Q V582F and AR111Q K387/519R cells. Data are mean ± SEM from four wells per treatment condition for the AR10Q cell line, and three wells per treatment condition for all other cell lines (sample size determined by power analysis), representative of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001, two-way mixed model ANOVA with Tukey’s multiple comparison test.
Hormone-Bound PolyQ-Expanded AR Induces Neurite Retraction in Differentiated PC12 Cells, Independent of AR DNA Binding at AREs.
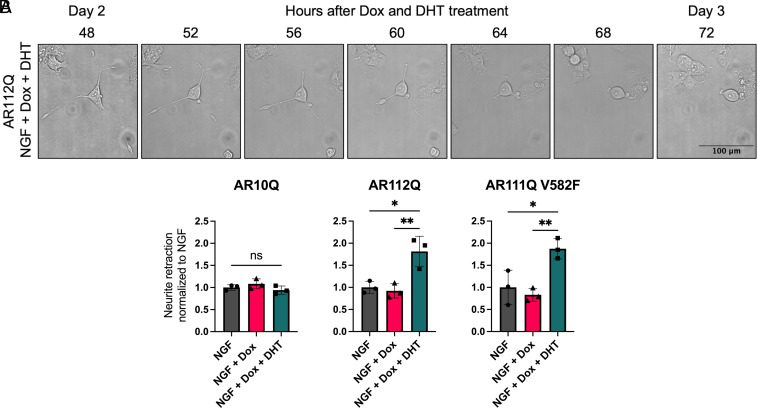
The loss of neurites from differentiated PC12 cells in the presence of hormone-bound polyQ-expanded AR (Fig. 2) suggested that these cells may undergo neurite retraction. To evaluate this question, we expressed AR10Q, AR112Q, or AR111Q V582F in the presence and absence of DHT in differentiated PC12 cells and performed live-cell imaging every 4 h to track and quantify neurite retraction of individual cells over time (SI Appendix, Fig. S8A). We found no difference in the percentage of cells with retracted neurites 2 d after Dox and DHT treatment for AR10Q, AR112Q, or AR111Q V582F (SI Appendix, Fig. S8B). However, at 3 d, DHT treatment of polyQ-expanded AR caused a significant increase in neurite retraction not observed for AR10Q-expressing cells; this increase was independent of polyQ-expanded AR DNA-binding capacity at AREs (Fig. 3 and SI Appendix, Fig. S9). These findings reveal that hormone-bound polyQ-expanded AR induces neurite retraction and that neurite retraction underlies the reduced neurite densities observed in Fig. 2.
Fig. 3.
Hormone-bound polyQ-expanded AR significantly increases neurite retraction, independent of AR DNA binding. (A) Representative phase-contrast images of a differentiated PC12 cell expressing hormone-bound AR112Q retracting its neurites over 24 h. (Scale bar, 100 µm.) (B) Quantification of neurite retraction normalized to NGF for AR10Q, AR112Q, and AR111Q V582F cell lines. Data represent mean ± SD from three wells per treatment condition, with an average of 41 cells analyzed per well, representative of three independent experiments. ns P > 0.05, *P < 0.05, and **P < 0.01, one-way ANOVA with Tukey’s multiple comparison test.
Cell Death Is Not the Primary Outcome Following Neurite Retraction in Differentiated PC12 Cells.
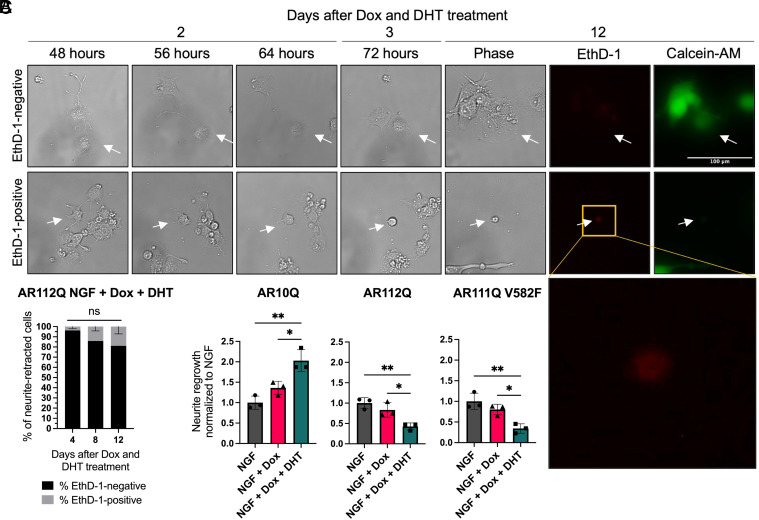
Upon observing an increase in neurite retraction in the presence of polyQ-expanded AR and DHT (Fig. 3), we questioned whether these cells underwent cell death following neurite retraction. We therefore performed a survival analysis of neurite-retracted cells (imaged as above from days 2 to 3 after Dox and DHT treatment) at 1, 5, and 9 d after neurite retraction (4, 8, and 12 d after Dox and DHT treatment). We chose the endpoint of 9 d (12 d of Dox and DHT treatment) because significant cell death is observed in undifferentiated PC12 cells expressing hormone-bound AR112Q under this condition (43). In the present study, polyQ-expanded AR was expressed at substantially lower levels than in these previous studies. We observed only a small percentage of cells undergoing cell death following neurite retraction (Fig. 4 A and B), indicating that most neurite-retracted cells expressing polyQ-expanded AR do not undergo cell death.
Fig. 4.
Hormone-bound polyQ-expanded AR does not induce significant cell death following neurite retraction but inhibits neurite regrowth, independent of AR DNA binding. (A and B) Survival analysis of hormone-bound AR112Q-expressing cells that retracted their neurites 3 d after Dox and DHT treatment. (A) Representative images of neurite-retracted hormone-bound AR112Q-expressing cells (white arrows) that were EthD-1-negative (Top) or EthD-1-positive (Bottom) 12 d after Dox and DHT treatment. The EthD-1-negative cell developed an enlarged and flattened morphology and stained positive for the live cell marker Calcein-AM. The gold box is an enlargement of the EthD-1 image. (Scale bar, 100 µm.) (B) Percentage of neurite-retracted hormone-bound AR112Q-expressing cells that were EthD-1-negative or EthD-1-positive 4 (n = 66), 8 (n = 44), and 12 (n = 29) d after Dox and DHT treatment. Data represent mean ± SEM from three wells for each time point. ns P > 0.05, 2-way ANOVA with Šídák’s multiple comparisons test. (C) Cells with retracted neurites 3 d after Dox and DHT treatment were tracked through live cell imaging for an additional 24 h. The percentage of cells that regrew their neurites by 4 d of Dox and DHT treatment was quantified and normalized to the NGF condition for each cell line. Data represent mean ± SD from three wells per treatment condition, with an average of 20 cells analyzed per well, representative of three independent experiments. *P < 0.05 and **P < 0.01, one-way ANOVA with Tukey’s multiple comparison test.
Hormone-Bound PolyQ-Expanded AR Inhibits Neurite Regrowth, Independent of AR DNA Binding at AREs.
We observed that neurite-retracted AR10Q-expressing cells maintain the capacity to regrow their neurites following retraction. We therefore quantified the percentage of neurite retracted cells that regrew neurites 4 d after Dox and DHT treatment and asked whether expressing AR in the presence and absence of DHT impacted this process. In the presence of AR10Q, DHT treatment significantly increased neurite regrowth (Fig. 4C). Conversely, DHT treatment significantly decreased neurite regrowth in the presence of polyQ-expanded AR, independent of AR DNA-binding capacity at AREs (Fig. 4C and SI Appendix, Fig. S10). These data support a previously identified role of androgens in axon regeneration (18–23) and reveal that this process is inhibited by DHT-bound polyQ-expanded AR, independent of its DNA binding at AREs.
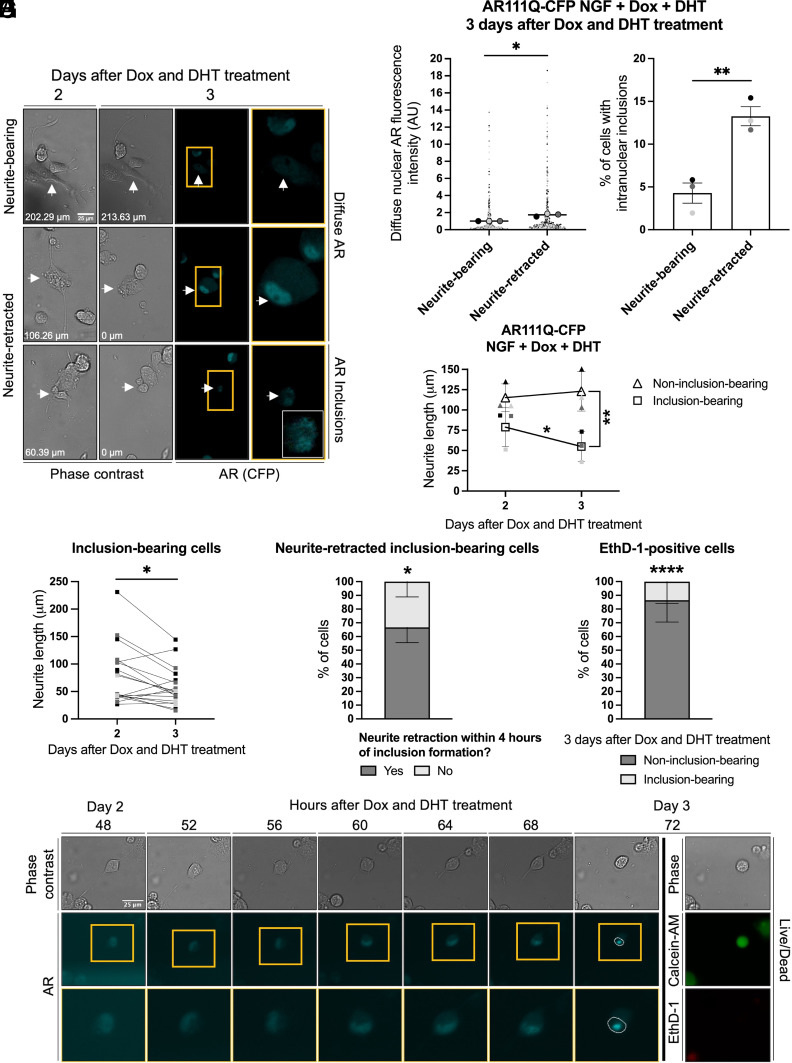
Neurite Retraction Is Associated with Increased Nuclear AR and AR Intranuclear Inclusions.
The presence of polyQ-expanded AR-containing intranuclear inclusions is a cytological feature of SBMA and other polyQ diseases. Thus, we sought to investigate the relationship between soluble AR, AR inclusions, and neurite retraction. To do so we utilized PC12 cells expressing AR111Q with a CFP-tag on the N terminus (AR111Q-CFP), which enabled detection of AR inclusion formation and neurite retraction through live-cell imaging (11). AR111Q-CFP was expressed at similar levels to AR112Q to ensure consistency in neurite retraction phenotypes (SI Appendix, Fig. S1D). Our analyses revealed that neurite-retracted cells had significantly greater diffuse nuclear AR (Fig. 5 A and B) and a significantly higher percentage of cells containing intranuclear inclusions (Fig. 5 A and C) compared to neurite-bearing cells 3 d after Dox and DHT treatment. We next asked whether there was a difference in total neurite lengths of individual neurite-bearing cells 2 and 3 d after Dox and DHT treatment between inclusion-bearing and non-inclusion-bearing populations. After 2 d of Dox and DHT treatment, there were no significant differences in neurite lengths between the two populations. However, by 3 d, inclusion-bearing cells had significantly shorter neurites than at 2 d, and significantly shorter neurites compared to non-inclusion-bearing cells (Fig. 5 D and E). Together, our analyses demonstrate that neurite retraction is associated with both increased diffuse AR and with intranuclear inclusions.
Fig. 5.
Increased diffuse nuclear AR and intranuclear inclusions are associated with neurite retraction. PC12 cells were differentiated, treated with Dox to induce expression of AR111Q-CFP and 10 nM DHT, and underwent live-cell imaging to analyze neurite retraction, nuclear AR fluorescence intensity, AR inclusions, and cell survival. (A) Representative phase-contrast images of neurite-bearing cells 2 d after Dox and DHT treatment with phase-contrast and fluorescence images (cyan, AR) of the same cells 24 h later. Total neurite lengths (µm) of the cells are shown. An enlarged and enhanced image with increased brightness and contrast of the CFP channel is included for the neurite-retracted, inclusion-bearing cell. (Scale bar, 25 µm.) (B) Diffuse nuclear AR fluorescence intensity (mean gray value; normalized to background) of neurite-bearing and neurite-retracted cells 3 d after Dox and DHT treatment. Each point represents a single cell (neurite-bearing, n = 405 cells; neurite-retracted, n = 353 cells), and each large circle (line at mean) represents the average fluorescence intensity from an individual experiment. Fluorescence intensities were normalized to the average fluorescence intensity in neurite-bearing cells. *P < 0.05, Welch’s t test. (C) Percentage of neurite-bearing and neurite-retracted cells with intranuclear inclusions 3 d after Dox and DHT treatment. Data represent mean ± SEM, and each point represents the average percentage from an individual experiment, with an average of 271 cells analyzed per experiment. **P < 0.01, independent samples t test. (D) Total neurite lengths (µm) of single inclusion-bearing (square, n = 18 cells) and non-inclusion-bearing (triangle, n = 244 cells) cells were quantified 2 and 3 d after Dox and DHT treatment. Small filled symbols represent the average neurite lengths quantified in an individual experiment; large open symbols represent mean ± SD from three independent experiments. *P < 0.05 and **P < 0.01, two-way repeated measures ANOVA with Šídák's multiple comparisons test. (E) Neurite lengths (µm) of single inclusion-bearing cells (n = 18 cells) 2 and 3 d after Dox and DHT treatment. Data are from three independent experiments. *P < 0.05, paired t test. (F) Percentage of cells that fully retracted their neurites within 4 h of forming AR inclusions. Data represent mean ± SD from three independent experiments, n = 27 cells. *P < 0.05, independent samples t test. (G) Percentage of EthD-1-positive cells 3 d after Dox and DHT treatment that were inclusion-bearing or non-inclusion-bearing. Data represent mean ± SD from two independent experiments, n = 71 cells. ****P < 0.0001, independent samples t test. (H) Representative images of a neurite-bearing hormone-bound AR111Q-CFP-expressing cell retracting its neurites (phase contrast image, Top row) and simultaneously developing an AR inclusion [cyan fluorescence image, Middle row (Bottom row = enlargement of image within gold box)] 3 d after Dox and DHT treatment. The white circle denotes the outline of the nucleus. At 3 d, the cell stained positive for the live cell marker Calcein-AM (green) but not the dead cell marker EthD-1 (red). (Scale bar, 25 µm.)
Given the observed correlation between intranuclear inclusions and neurite retraction, we wondered whether inclusion formation served as the catalyst for neurite retraction. We therefore utilized live-cell imaging to ask whether cells retracted their neurites within 4 h of forming an inclusion. Our analysis revealed that the majority (66.67%) of inclusion-bearing cells retracted their neurites within 4 h of forming an inclusion, suggesting that AR aggregation and inclusion formation induces neurite retraction (Fig. 5 F and H). Finally, we wondered whether the population of EthD-1-positive cells previously identified (Fig. 4B) consisted of inclusion-bearing or non-inclusion-bearing cells. Of the EthD-1-positive cells, the vast majority (86.49%) were non-inclusion-bearing (Fig. 5 G and H). Together, these analyses reveal that AR intranuclear inclusion formation serves as a catalyst for neurite retraction but does not induce cell death in this population of cells.
Differentiated PC12 Cells with Retracted Neurites Have Reduced β-III-Tubulin Expression, Independent of AR-DNA Binding at AREs.
Upon differentiation, PC12 cells express increased levels of neuronal proteins, including the neuronal-specific tubulin β-III-Tubulin (37). We therefore wondered whether cells with retracted neurites express reduced levels of β-III-Tubulin, which would indicate that they are undergoing dedifferentiation. While DHT treatment of AR10Q did not alter β-III-Tubulin levels, DHT treatment of AR112Q caused a significant reduction in β-III-Tubulin protein 2 and 4 d after Dox and DHT treatment (SI Appendix, Fig. S11 A and B). Furthermore, DHT treatment significantly reduced Tubb3 gene expression (encoding β-III-Tubulin protein) for AR10Q, AR112Q, and AR111Q K387/519R 4 d after Dox and DHT treatment; this did not reach statistical significance for AR111Q V582F (SI Appendix, Fig. S11C). We postulated that cells with retracted neurites are the specific cell population with decreased β-III-Tubulin. Indeed, we found that hormone-bound AR112Q-expressing cells with retracted neurites had significantly reduced cytoplasmic β-III-Tubulin expression compared to neurite-bearing cells (SI Appendix, Fig. S11D). We asked whether this reduction in β-III-Tubulin was dependent on AR DNA binding at AREs. Similar to AR112Q, we found that hormone-bound AR111Q V582F-expressing cells with retracted neurites had reduced cytoplasmic β-III-Tubulin expression (SI Appendix, Fig. S11E). Together, our analyses reveal that hormone-bound polyQ-expanded AR, independent of its transcriptional activity at AREs, reduces β-III-Tubulin expression in cells with retracted neurites, suggesting that these cells undergo dedifferentiation alongside neurite retraction.
Dedifferentiation upon Neurite Retraction Is Not Accompanied by Cell Cycle Reentry.
Previous work suggested that neurite retraction in the presence of polyQ-expanded AR is accompanied by mitotic reentry due to reduced APC/CCdh1 activity and aberrant reaccumulation of a Dbox-GFP Cyclin B1 reporter (33). We therefore investigated whether cell-cycle reentry occurred alongside reduced β-III-Tubulin expression in our model. Since changes in Cdh1 expression regulate APC/C activity (44), we quantified nuclear and cytoplasmic Cdh1; we observed equivalent Cdh1 expression for AR10Q and AR112Q in the presence and absence of DHT (SI Appendix, Fig. S12 A and B). We then compared nuclear Cdh1 expression between neurite-bearing and neurite-retracted cells in the presence of DHT-bound AR112Q and again observed no differences (SI Appendix, Fig. S12C). To evaluate APC/CCdh1 activity, we quantified Cyclin B1 expression to determine whether there was aberrant reaccumulation of the protein. We observed negligible nuclear or soma Cyclin B1 expression in neurite-bearing or neurite-retracted cells expressing hormone-bound AR112Q (SI Appendix, Fig. S12 D–F). Taken together, these data indicate that cell cycle reentry does not occur in neurite retracting cells in our model.
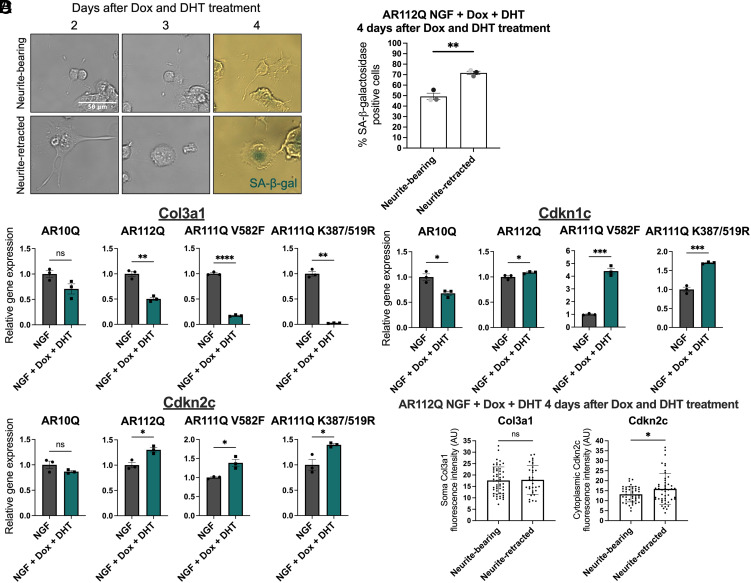
Hormone-Bound PolyQ-Expanded AR Induces Senescence in Differentiated PC12 Cells.
We observed that cells developed an enlarged and flattened morphology upon neurite retraction (Fig. 4A), which is characteristic of senescent cells (45). Since neurite retraction does not induce cell death (Fig. 4B) or cell-cycle reentry (SI Appendix, Fig. S12), we postulated that cells with retracted neurites become senescent. In addition to altered morphology, senescent cells have increased expression of pH-dependent senescence-associated β-galactosidase (SA-β-gal) and altered expression of senescence-associated genes (45). We found that the majority of neurite-retracted cells were SA-β-gal-positive 4 d after Dox and DHT treatment, significantly more than neurite-bearing cells (Fig. 6 A and B), revealing that cells become senescent following neurite retraction. To investigate whether there were broader changes in senescence-associated genes in the presence of AR112Q and DHT in differentiated PC12 cells, we screened for changes in 95 senescence-associated genes. We identified 15 genes with a 1.5-fold increase or 0.5-fold decrease in the presence of DHT in at least 1 of 3 independent experiments (SI Appendix, Fig. S13A). Upon validation of these 15 genes to evaluate the role of AR polyQ-length and transcriptional activity, we revealed that Cd44, Col1a1, Serpinb2, and Sparc were significantly decreased in the presence of DHT 4 and 8 d after Dox and DHT treatment, independent of AR polyQ-length and transcriptional activity at AREs (SI Appendix, Fig. S13 B and C). In contrast, we observed a polyQ length-dependent decrease in Col3a1 and increase in Cdkn1c and Cdkn2c expression 8 d after Dox and DHT treatment; this effect was independent of polyQ-expanded AR DNA binding at AREs (Fig. 6C). Overall, these data reveal that DHT treatment induces senescence in differentiated PC12 cells in a polyQ length-dependent, but ARE-mediated AR transcriptional activity-independent, manner.
Fig. 6.
Hormone-bound polyQ-expanded AR induces senescence in differentiated PC12 cells. (A) Representative images of neurite-bearing and neurite-retracted AR112Q-expressing cells stained for SA-β-galactosidase (SA-β-gal, blue) 4 d after Dox and DHT treatment. (Scale bar, 50 µm.) (B) Percentage of SA-β-galactosidase-positive neurite-bearing and neurite-retracted cells 4 d after Dox and DHT treatment. Data represent mean ± SEM and each point represents the average percentage from an individual experiment, with an average of 103 cells analyzed per experiment. *P < 0.05, independent samples t test. (C) qRT-PCR analysis was performed to evaluate relative expression of the senescence-associated genes Col3a1, Cdkn1c, and Cdkn2c in differentiated PC12 cells expressing AR10Q, AR112Q, AR111Q V582F, or AR111Q K387/519R 8 d after Dox and DHT treatment. Data represent mean ± SEM from three replicates per cell line. ns P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001, independent samples t test or Welch’s t test. (D) Quantification of soma Col3a1 and cytoplasmic Cdkn2c protein (mean gray value; normalized to background) in neurite-bearing and neurite-retracted cells expressing hormone-bound polyQ-expanded AR 4 d after Dox and DHT treatment. Each point represents a single cell (Col3a1: neurite-bearing, n = 53; neurite-retracted, n = 32; Cdkn2c: neurite-bearing, n = 45; neurite-retracted, n = 48). Data represent mean ± SD from three individual wells. ns P > 0.05, independent samples t test; *P < 0.05, Welch’s t test.
Since significantly more neurite-retracted cells than neurite-bearing cells were SA-β-gal-positive, we hypothesized that neurite-retracted cells displayed differential expression of senescence-associated genes. Based on immunocytochemistry analysis, we found no significant differences in Col3a1 expression between neurite-bearing and neurite-retracted cells expressing hormone-bound polyQ-expanded AR. However, Cdkn2c was significantly increased in neurite-retracted cells (Fig. 6D), matching the change observed in our RNA analysis. Thus, cells with retracted neurites display morphological, enzymatic, and gene expression changes consistent with the induction of senescence.
Pathways Related to Neurite Growth and Loss and Senescence Are Largely Unaltered upon Reduction or Enhancement of PolyQ-Expanded AR Transcriptional Activity at AREs.
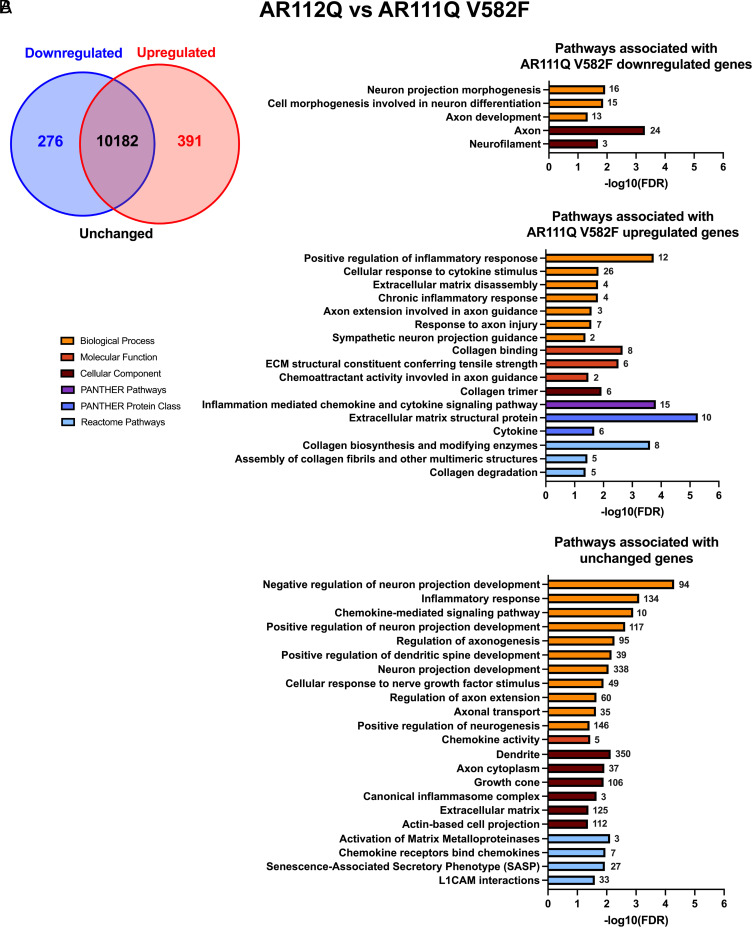
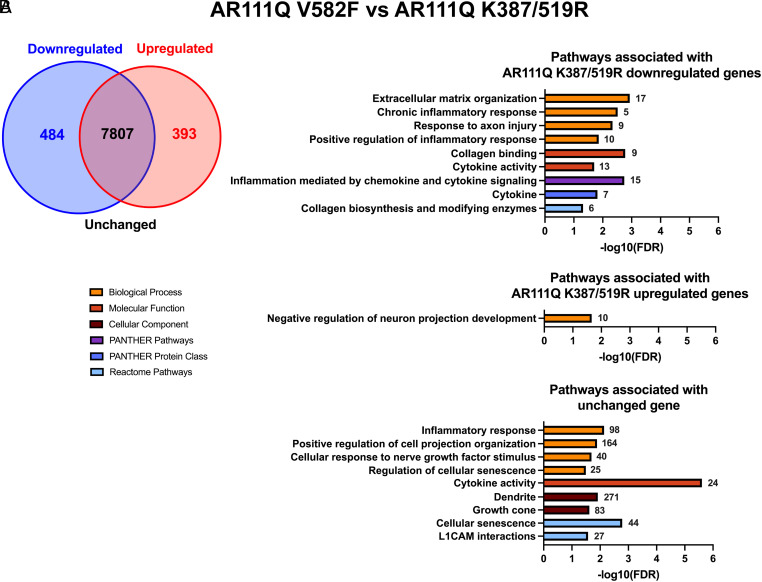
Our data revealing that neurite outgrowth (Fig. 1), neurite loss (Figs. 2 and 3), neurite regrowth (Fig. 4C), and senescence (Fig. 6) were unaltered by reducing or enhancing AR transcriptional activity at AREs suggest that these pathologies do not require this canonical AR function. To further confirm that these phenotypes are truly independent of AR transcriptional activity at AREs, we wanted to determine whether pathways related to neurite growth/loss and senescence are unaltered when AR transcriptional activity at AREs is manipulated. To do so, we performed RNA-seq transcriptomic analysis on differentiated PC12 cells expressing hormone-bound AR112Q, AR111Q V582F, and AR111Q K387/519R to compare samples with baseline (AR112Q) vs. reduced (AR111Q V582F) transcriptional activity at AREs and samples with reduced (AR111Q V582F) vs. enhanced (AR111Q K387/519R) transcriptional activity at AREs. Through principal component analysis, we confirmed that there is high variance between cell lines and low variance within replicate samples (SI Appendix, Figs. S14A and S15A). Down-regulated, up-regulated, and unchanged genes between AR112Q and AR111Q V582F samples, and between AR111Q V582F and AR111Q K387/519R samples, were identified and used in gene ontology (GO) enrichment analyses to identify significant pathways represented by these genes (SI Appendix, Figs. S14 B and C, S15 B and C, and S16). A total of 667 genes (276 down-regulated and 391 up-regulated) were differentially expressed in AR111Q V582F compared to AR112Q cells, with 10182 genes unchanged (Fig. 7A); a total of 877 genes (484 down-regulated and 393 up-regulated) were differentially expressed in AR111Q K387/519R compared to AR111Q V582F cells, with 7807 genes unchanged (Fig. 8A).
Fig. 7.
Enriched pathways related to neurite growth and senescence represented by differential and unchanged genes between AR112Q and AR111Q V582F cells. (A) Venn diagram depicting the number of down-regulated, up-regulated, and unchanged genes in AR111Q V582F samples compared to AR112Q samples. (B) Gene ontology (GO) analysis was conducted using down-regulated (absolute log2 fold change ≤−1), up-regulated (absolute log2 fold change ≥1), and unchanged (adjusted P > 0.05) gene sets and significant pathways related to neurite growth and senescence were selected and graphed by −log10(FDR). The number of down-regulated, up-regulated, or unchanged genes associated with each pathway is shown next to each bar.
Fig. 8.
Enriched pathways related to neurite growth and senescence represented by differential and unchanged genes between AR111Q V582F and AR111Q K387/519R cells. (A) Venn diagram depicting the number of down-regulated, up-regulated, and unchanged genes in AR111Q K387/519R samples compared to AR111Q V582F samples. (B) Gene ontology (GO) analysis was conducted using down-regulated (absolute log2 fold change ≤−1), up-regulated (absolute log2 fold change ≥1), and unchanged (adjusted P > 0.05) gene sets, and significant pathways related to neurite growth and senescence were selected and graphed by −log10(FDR). The number of down-regulated, up-regulated, or unchanged genes associated with each pathway is shown next to each bar.
We were particularly interested in pathways related to neurites and senescence and therefore asked whether genes that contribute to these pathways are largely down-regulated, up-regulated, or unchanged when AR transcriptional activity at AREs is altered. While pathways related to neuron projection development/extension and the SASP were identified from down-regulated, up-regulated, and unchanged gene sets, significant pathways were associated more often with unchanged genes, with a greater number of genes encompassing these pathways (Figs. 7B and 8B). Some noteworthy pathways represented by unchanged genes include neuron projection development (338 genes, AR112Q vs. AR111Q V582F) and cellular response to nerve growth factor stimulus (49 genes, AR112Q vs. AR111Q V582F; 40 genes, AR111Q V582F vs. AR111Q K387/519R). Additionally, the SASP (27 genes) was identified as a pathway associated with unchanged genes between AR112Q and AR111Q V582F samples, while cellular senescence (44 genes) was identified between AR111Q V582F and AR111Q K387/519R samples, further confirming that the induction of senescence in differentiated PC12 cells is independent of AR transcriptional activity at AREs. These data as a whole reveal that neurite growth/loss and senescence are independent of AR transcriptional activity at AREs in differentiated PC12 cells.
Discussion
SBMA is a devastating neuromuscular disease in which patients experience muscle weakness due to degeneration of lower motor neurons and muscle atrophy (1, 2). After the causative mutation for SBMA—a CAG repeat-expansion in the AR gene—was identified, studies revealed that the presence of androgens and nuclear localization of polyQ-expanded AR are required for disease pathology (3, 4, 43, 46). Whether the functional state of nuclear, hormone-bound polyQ-expanded AR, namely its transcriptional activity, is required for disease manifestation remains unclear. Understanding the role of AR transcriptional activity in disease mechanisms is crucial for the development of efficacious therapeutics for patients suffering from SBMA. Recent clinical trials using androgen deprivation, which notably inhibits AR transcriptional functions, showed limited efficacy (47–49), suggesting that the loss of AR trophic support to neurons and muscles upon androgen deprivation may be ultimately detrimental in the presence of polyQ-expanded AR. Furthermore, studies of neuronal models of SBMA revealed a loss of AR’s neurite growth-promoting functions, demonstrating a loss of neurites in the presence of polyQ-expanded AR (33–36). However, the mechanisms contributing to neurite loss in SBMA remained largely unknown.
In this study, we sought to determine the role of polyQ-expanded AR transcriptional activity in, and the mechanisms contributing to, neurite loss in SBMA. We utilized PC12 cells, which can be differentiated into neuronal cells (37), to study the processes of neurite outgrowth, neurite retraction, and neurite regrowth. Through live-cell imaging, we tracked single cells over time and performed immunocytochemical analyses to reveal molecular differences in morphologically distinct cell populations (i.e., neurite-bearing vs. neurite-retracted cells). Furthermore, the PC12 cells contain Doxycycline-inducible AR (9), allowing the selective expression of AR at specific time points in the differentiation process in order to study the role of AR in distinct morphological processes. Overall, our study revealed that hormone-bound polyQ-expanded AR inhibits neurite outgrowth, enhances neurite retraction, and inhibits neurite regrowth independent of its ability to bind AREs and identified senescence as a key mechanism involved in neurite retraction.
We first showed that DHT treatment inhibited neurite outgrowth in the presence of polyQ-expanded AR, independent of its transcriptional ability at AREs, while hormone had no effect on neurite outgrowth in the presence of wild-type AR. The inhibition of polyQ-expanded AR on neurite outgrowth is consistent with previous findings of reduced axonogenesis in spinal cord slice cultures from SBMA mice (50). The lack of growth-enhancing effect of DHT in the presence of normal polyQ-length AR, however, seemingly contradicts prior studies (24, 25, 51, 52). However, these differences could be explained by enhanced quantification methods used in our study compared to those utilized in earlier studies (24, 25); moreover, in contrast with earlier reports investigating low levels of cytoplasmic, transcriptionally inactive AR (51, 52), our studies described here examined the effect of AR that is nuclear and transcriptionally active in response to hormone. As observed in prior studies (33–36), we confirmed in our model that polyQ-expanded AR leads to a decrease in neurites in differentiated PC12 cells. We further showed that this decrease was dependent on the presence of DHT and was independent of polyQ-expanded AR transcriptional activity at AREs since DHT led to a decrease in neurite density when transcriptional activity was both eliminated and enhanced at AREs. Using live-cell imaging, we further revealed that individual cells undergo neurite retraction, which was enhanced by DHT-bound polyQ-expanded AR compared to the same cell line treated with NGF alone, independent of its transcriptional activity at AREs, revealing a toxic property of mutant AR. Finally, we revealed that some cells regrew their neurites following neurite retraction, a phenomenon that was enhanced by DHT-bound wild-type AR but inhibited by DHT-bound polyQ-expanded AR, independent of its transcriptional activity at AREs. Androgen-bound AR enhances axon regeneration after injury (18–23). Our results in vitro support these in vivo findings and reveal the loss of this function of hormone-bound polyQ-expanded AR that can have direct implications for functional outcomes of SBMA patients. The presence of glycolytic-to-oxidative fiber-type switching observed in mouse models of SBMA indicates denervation and reinnervation of muscle fibers (53–56). Our findings of reduced neurite regrowth suggest that the reinnervation process could be decreased in SBMA patients, leading to further functional decline.
Taken together, these morphological findings on the effect of hormone-bound polyQ-expanded AR on the growth and maintenance of neurites clearly reveal the lack of involvement of AR transcriptional activity at AREs—reducing or enhancing transcriptional activity at AREs was neither beneficial nor detrimental to the phenotypes observed. We confirmed that the neurite growth pathologies we identified are independent of AR transcriptional activity at AREs through RNA-seq analysis of differentiated PC12 cells with baseline, reduced, or enhanced transcriptional activity. The fact that multiple neurite projection–related pathways associated with numerous unchanged genes were identified as significant indicates that, as a whole, these pathways are not altered by changes to AR transcriptional activity at AREs. Our findings contrast with prior work in a Drosophila model of SBMA which demonstrated that AR DNA binding is required for SBMA phenotypes (32). It may be that the AR A574D mutation used to block AR DNA binding in that study differs in its effects from the mutated amino acid that we utilized, V582F, a residue that makes direct contact with DNA (38). Furthermore, it is plausible that AR transcriptional activity at AREs or at other sites through tethering contributes to other pathologies in SBMA not investigated here. Nonetheless, our findings may have direct therapeutic implications, as they suggest that AR ARE-mediated transcriptional functions do not contribute to neurite growth and retention pathologies. Therefore, therapeutics that preserve rather than inhibit ARE-dependent AR transcriptional activity at the neuromuscular junction may lead to enhanced functional outcomes for patients—as the trophic support supplied to muscle and motor neurons would be maintained—and should be considered for future development.
The fact that neurite loss was independent of AR transcriptional activity at AREs led us to hypothesize that another property of polyQ-expanded AR, such as its misfolding and aggregation, contributes to neurite loss. This idea was supported by the finding that hormone-bound polyQ-expanded AR did not impact neurite retraction from 0 to 2 d of Dox and DHT treatment (SI Appendix, Fig. S8), but did from 2 to 3 d (Fig. 3), a time-course similar to that of intranuclear inclusion formation in this model (9). As hypothesized, we observed a direct relationship between intranuclear inclusions and neurite retraction; cells were significantly more likely to fully retract their neurites upon forming an inclusion. Supporting this finding, we found that more cells with retracted neurites contained intranuclear inclusions than neurite-bearing cells; however, we also observed that cells with retracted neurites had significantly greater diffuse nuclear AR, which is seemingly contradictory. Previous studies of a Huntington’s disease neuronal model demonstrated that cells that form mutant HTT inclusion bodies have increased levels of diffuse huntingtin prior to inclusion formation (57). This suggests that in our model, cells with increased diffuse nuclear AR may be in the process of forming an inclusion, thus explaining why cells with retracted neurites have both increased diffuse AR and inclusions. Additionally, we found that cells with intranuclear inclusions had shorter neurites than non-inclusion-bearing cells, likely representing cells in the process of neurite retraction. Of interest is whether manipulations that reduce AR intranuclear inclusions, such as knockdown of the deubiquitinase USP7 (58) or inhibition of the AR N/C interaction (59), reduce the neurite retraction phenotype observed here.
We focused our molecular analyses on the cells with retracted neurites since they exhibited a clear morphological pathology. First, we observed a decrease in β-III-Tubulin expression in cells with retracted neurites; this was independent of AR DNA-binding capacity at AREs. β-III-Tubulin expression is low in undifferentiated PC12 cells and increases during the differentiation process (37); thus, the loss of β-III-Tubulin in neurite-retracted cells suggests that these cells are undergoing dedifferentiation. β-III-Tubulin is a neuronal-specific tubulin with roles in neurogenesis, axon guidance, and axon regeneration (60). Interestingly, DRG neurons from Tubb3−/− mice showed reduced neurite outgrowth and delayed axon regeneration following sciatic nerve crush (61). This finding suggests that reduced β-III-Tubulin expression could underly the reduction in neurite regrowth following neurite retraction in the presence of hormone-bound polyQ-expanded AR in our model.
In contrast to previous work (33), we found that neurite-retracted cells do not reenter the cell cycle upon neurite retraction. However, there are important distinctions between the differentiation models used in these two studies that may account for this difference. The NGF-mediated differentiation of PC12 cells used here resulted in neuronal cells that extend long neurites and express neuronal proteins. In contrast, the prior study used DHT-mediated differentiation, resulting in cells with relatively short neurites; neuronal markers were not assessed. Therefore, the cell cycle reentry observed might reflect partial, rather than terminal, differentiation, resulting in distinct sequelae from those observed in our study.
We found that senescence, not cell death, is the primary fate of cells with retracted neurites. In replicating cells, senescence is defined as a permanent cell cycle arrest; however, senescence also occurs in postmitotic cells including neurons, where it is thought to contribute to the aging process and neurodegenerative diseases such as Alzheimer’s disease and Parkinson’s disease (62, 63). Indeed, through qRT-PCR and RNA-seq, we revealed that hormone-bound polyQ-expanded AR induces senescence independent of its transcriptional activity at AREs. We revealed a broad change in the expression of senescence-associated genes, including collagens (Col1a1 and Col3a1), glycoproteins (Sparc and Cd44), and cell cycle regulators (Cdkn1c and Cdkn2c), suggesting the potential involvement of the extracellular matrix (ECM) in the induction of senescence and neurite retraction. These genes were shown to be both up- and down-regulated in senescent cells, differing between tissue type and role in senescence (64, 65); thus, both up- and downregulation of a gene can indicate the induction of senescence. Interestingly out of the differentially expressed genes identified in the presence of DHT, Sparc, Serpinb2, Cd44, Col3a1, and Col1a1 were both up- and down-regulated as part of the senescence-associated secretory phenotype (SASP), which consists of the secretion of inflammatory cytokines, immune factors, growth factors, and proteases (65). It has been suggested that protein aggregates can trigger senescence through induction of a proinflammatory state (66). Our data demonstrating a relationship between polyQ-expanded AR inclusion formation and neurite retraction, and senescence and neurite retraction, corroborate this idea, which warrants further study.
In summary, our study reveals a toxic function of polyQ-expanded AR in neurite growth and maintenance that is hormone-dependent and AR transcriptional activity-independent and identifies senescence as a pathway involved in this pathology. Whether the senescence pathway is activated in in vivo models of SBMA and in SBMA patients is an open question. Future studies will address this question and investigate the use of senolytics (67) as a therapeutic strategy for SBMA. Finally, neurite defects have recently been observed in other polyQ diseases (68, 69). The relationship we identified between inclusion formation and neurite retraction suggests that mechanisms revealed herein may apply to other polyQ diseases, presenting an avenue for treatment that is independent of the disease-causing gene.
Materials and Methods
Details of the Materials and Methods used in this study, including PC12 cell culture and differentiation; Construction of AR111Q V582F cell line; Analysis of AR and β-III Tubulin protein levels; MMTV-Luciferase assay; Chromatin fractionation assay; Live-cell imaging; Neurite density, neurite retraction, and neurite regrowth analyses (70); Survival analyses; Senescence-associated β-galactosidase analysis; Fluorescence intensity and neurite analyses in AR111Q-CFP cells; Immunocytochemical analysis; RNA extraction and qRT-PCR analysis; RNA extraction and RNA-Seq transcriptome analysis; and Statistical analyses, are provided in SI Appendix, Materials and Methods.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank the members of the Merry lab for helpful discussions and Tamar Berger for assistance with analysis of AR-V582F transcriptional activity. This work was supported by the NIH grant number R01 NS106302.
Author contributions
J.K. and D.E.M. designed research; J.K. performed research; J.K. and D.E.M. analyzed data; Y.L. created cell line central to study; and J.K. and D.E.M. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
The RNA-Seq data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (71) and are accessible through GEO Series accession number GSE267802 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE267802).
Supporting Information
References
- 1.Kennedy W. R., Alter M., Sung J. H., Progressive proximal spinal and bulbar muscular atrophy of late onset. A sex-linked recessive trait. Neurology 18, 671–680 (1968). [DOI] [PubMed] [Google Scholar]
- 2.Harding A. E., et al. , X-linked recessive bulbospinal neuronopathy: A report of ten cases. J. Neurol. Neurosurg. Psychiatry 45, 1012–1019 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chevalier-Larsen E. S., et al. , Castration restores function and neurofilament alterations of aged symptomatic males in a transgenic mouse model of spinal and bulbar muscular atrophy. J. Neurosci. 24, 4778–4786 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katsuno M., et al. , Testosterone reduction prevents phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Neuron 35, 843–854 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Soraru G., et al. , Spinal and bulbar muscular atrophy: Skeletal muscle pathology in male patients and heterozygous females. J. Neurol. Sci. 264, 100–105 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Sobue G., et al. , X-linked recessive bulbospinal neuronopathy. A clinicopathological study. Brain 112, 209–232 (1989). [DOI] [PubMed] [Google Scholar]
- 7.La Spada A. R., Wilson E. M., Lubahn D. B., Harding A. E., Fischbeck K. H., Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature 352, 77–79 (1991). [DOI] [PubMed] [Google Scholar]
- 8.Li M., et al. , Nuclear inclusions of the androgen receptor protein in spinal and bulbar muscular atrophy. Ann. Neurol. 44, 249–254 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Walcott J. L., Merry D. E., Ligand promotes intranuclear inclusions in a novel cell model of spinal and bulbar muscular atrophy. J. Biol. Chem. 277, 50855–50859 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Li M., et al. , Nonneural nuclear inclusions of androgen receptor protein in spinal and bulbar muscular atrophy. Am. J. Pathol. 153, 695–701 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heine E. M., et al. , Proteasome-mediated proteolysis of the polyglutamine-expanded androgen receptor is a late event in spinal and bulbar muscular atrophy (SBMA) pathogenesis. J. Biol. Chem. 290, 12572–12584 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pratt W. B., Welsh M. J., Chaperone functions of the heat shock proteins associated with steroid receptors. Semin. Cell Biol. 5, 83–93 (1994). [DOI] [PubMed] [Google Scholar]
- 13.van Royen M. E., van Cappellen W. A., de Vos C., Houtsmuller A. B., Trapman J., Stepwise androgen receptor dimerization. J. Cell Sci. 125, 1970–1979 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Lallous N., Dalal K., Cherkasov A., Rennie P. S., Targeting alternative sites on the androgen receptor to treat castration-resistant prostate cancer. Int. J. Mol. Sci. 14, 12496–12519 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lobaccaro J. M., et al. , Molecular modeling and in vitro investigations of the human androgen receptor DNA-binding domain: Application for the study of two mutations. Mol. Cell Endocrinol. 116, 137–147 (1996). [DOI] [PubMed] [Google Scholar]
- 16.Little C. M., Coons K. D., Sengelaub D. R., Neuroprotective effects of testosterone on the morphology and function of somatic motoneurons following the death of neighboring motoneurons. J. Comp. Neurol. 512, 359–372 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurz E. M., Sengelaub D. R., Arnold A. P., Androgens regulate the dendritic length of mammalian motoneurons in adulthood. Science 232, 395–398 (1986). [DOI] [PubMed] [Google Scholar]
- 18.Yu W. H., Yu M. C., Acceleration of the regeneration of the crushed hypoglossal nerve by testosterone. Exp. Neurol. 80, 349–360 (1983). [DOI] [PubMed] [Google Scholar]
- 19.Yu W. H., Administration of testosterone attenuates neuronal loss following axotomy in the brain-stem motor nuclei of female rats. J. Neurosci. 9, 3908–3914 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kujawa K. A., Emeric E., Jones K. J., Testosterone differentially regulates the regenerative properties of injured hamster facial motoneurons. J. Neurosci. 11, 3898–3906 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kujawa K. A., Jacob J. M., Jones K. J., Testosterone regulation of the regenerative properties of injured rat sciatic motor neurons. J. Neurosci. Res. 35, 268–273 (1993). [DOI] [PubMed] [Google Scholar]
- 22.Kujawa K. A., Kinderman N. B., Jones K. J., Testosterone-induced acceleration of recovery from facial paralysis following crush axotomy of the facial nerve in male hamsters. Exp. Neurol. 105, 80–85 (1989). [DOI] [PubMed] [Google Scholar]
- 23.Perez J., Kelley D. B., Trophic effects of androgen: Receptor expression and the survival of laryngeal motor neurons after axotomy. J. Neurosci. 16, 6625–6633 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brooks B. P., et al. , A cell culture model for androgen effects in motor neurons. J. Neurochem. 70, 1054–1060 (1998). [DOI] [PubMed] [Google Scholar]
- 25.Marron T. U., et al. , Androgen-induced neurite outgrowth is mediated by neuritin in motor neurones. J. Neurochem. 92, 10–20 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Irvine R. A., et al. , Inhibition of p160-mediated coactivation with increasing androgen receptor polyglutamine length. Hum. Mol. Genet. 9, 267–274 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Lieberman A. P., Harmison G., Strand A. D., Olson J. M., Fischbeck K. H., Altered transcriptional regulation in cells expressing the expanded polyglutamine androgen receptor. Hum. Mol. Genet. 11, 1967–1976 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Todd T. W., et al. , Nemo-like kinase is a novel regulator of spinal and bulbar muscular atrophy. Elife 4, e08493 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scaramuzzino C., et al. , Protein arginine methyltransferase 6 enhances polyglutamine-expanded androgen receptor function and toxicity in spinal and bulbar muscular atrophy. Neuron 85, 88–100 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chua J. P., et al. , Disrupting SUMOylation enhances transcriptional function and ameliorates polyglutamine androgen receptor-mediated disease. J. Clin. Invest. 125, 831–845 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prakasam R., et al. , LSD1/PRMT6-targeting gene therapy to attenuate androgen receptor toxic gain-of-function ameliorates spinobulbar muscular atrophy phenotypes in flies and mice. Nat. Commun. 14, 603 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nedelsky N. B., et al. , Native functions of the androgen receptor are essential to pathogenesis in a Drosophila model of spinobulbar muscular atrophy. Neuron 67, 936–952 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bott L. C., et al. , The polyglutamine-expanded androgen receptor responsible for spinal and bulbar muscular atrophy inhibits the APC/C(Cdh1) ubiquitin ligase complex. Sci. Rep. 6, 27703 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simeoni S., et al. , Motoneuronal cell death is not correlated with aggregate formation of androgen receptors containing an elongated polyglutamine tract. Hum. Mol. Genet. 9, 133–144 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Schindler M., et al. , Disruption of nongenomic testosterone signaling in a model of spinal and bulbar muscular atrophy. Mol. Endocrinol. 26, 1102–1116 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheila M., et al. , Phenotypic and molecular features underlying neurodegeneration of motor neurons derived from spinal and bulbar muscular atrophy patients. Neurobiol. Dis. 124, 1–13 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Karliner J., Merry D. E., Differentiating PC12 cells to evaluate neurite densities through live-cell imaging. STAR Protoc. 4, 101993 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaffer P. L., Jivan A., Dollins D. E., Claessens F., Gewirth D. T., Structural basis of androgen receptor binding to selective androgen response elements. Proc. Natl. Acad. Sci. U.S.A. 101, 4758–4763 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vlahopoulos S., et al. , Recruitment of the androgen receptor via serum response factor facilitates expression of a myogenic gene. J. Biol. Chem. 280, 7786–7792 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Norris J. D., et al. , The homeodomain protein HOXB13 regulates the cellular response to androgens. Mol. Cell 36, 405–416 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J., et al. , C/EBPalpha redirects androgen receptor signaling through a unique bimodal interaction. Oncogene 29, 723–738 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sawada T., et al. , Androgen-dependent and DNA-binding-independent association of androgen receptor with chromatic regions coding androgen-induced noncoding RNAs. Biosci. Biotechnol. Biochem. 85, 2121–2130 (2021). [DOI] [PubMed] [Google Scholar]
- 43.Montie H. L., et al. , Cytoplasmic retention of polyglutamine-expanded androgen receptor ameliorates disease via autophagy in a mouse model of spinal and bulbar muscular atrophy. Hum. Mol. Genet. 18, 1937–1950 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zachariae W., Schwab M., Nasmyth K., Seufert W., Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science 282, 1721–1724 (1998). [DOI] [PubMed] [Google Scholar]
- 45.Beck J., Horikawa I., Harris C., Cellular senescence: Mechanisms, morphology, and mouse models. Vet. Pathol. 57, 747–757 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Takeyama K., et al. , Androgen-dependent neurodegeneration by polyglutamine-expanded human androgen receptor in Drosophila. Neuron 35, 855–864 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Banno H., et al. , Phase 2 trial of leuprorelin in patients with spinal and bulbar muscular atrophy. Ann. Neurol. 65, 140–150 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Katsuno M., et al. , Efficacy and safety of leuprorelin in patients with spinal and bulbar muscular atrophy (JASMITT study): A multicentre, randomised, double-blind, placebo-controlled trial. Lancet Neurol. 9, 875–884 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Yamamoto T., et al. , An open trial of long-term testosterone suppression in spinal and bulbar muscular atrophy. Muscle Nerve 47, 816–822 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Ogura Y., et al. , Mid1 is associated with androgen-dependent axonal vulnerability of motor neurons in spinal and bulbar muscular atrophy. Cell Death Dis. 13, 601 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Di Donato M., Bilancio A., Auricchio F., Castoria G., Migliaccio A., Androgens and NGF mediate the neurite-outgrowth through inactivation of RhoA. Cells 12, 373 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Di Donato M., et al. , Cross-talk between androgen receptor/filamin A and TrkA regulates neurite outgrowth in PC12 cells. Mol. Biol. Cell 26, 2858–2872 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chivet M., et al. , Polyglutamine-expanded androgen receptor alteration of skeletal muscle homeostasis and myonuclear aggregation are affected by sex. Age and muscle metabolism. Cells 9, 325 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramzan F., et al. , Distinct etiological roles for myocytes and motor neurons in a mouse model of Kennedy’s disease/spinobulbar muscular atrophy. J. Neurosci. 35, 6444–6451 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rocchi A., et al. , Glycolytic-to-oxidative fiber-type switch and mTOR signaling activation are early-onset features of SBMA muscle modified by high-fat diet. Acta Neuropathol. 132, 127–144 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Molotsky E., Liu Y., Lieberman A. P., Merry D. E., Neuromuscular junction pathology is correlated with differential motor unit vulnerability in spinal and bulbar muscular atrophy. Acta Neuropathol. Commun. 10, 97 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arrasate M., Mitra S., Schweitzer E. S., Segal M. R., Finkbeiner S., Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431, 805–810 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Pluciennik A., et al. , Deubiquitinase USP7 contributes to the pathogenicity of spinal and bulbar muscular atrophy. J. Clin. Invest. 131, e134565 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zboray L., et al. , Preventing the androgen receptor N/C interaction delays disease onset in a mouse model of SBMA. Cell Rep. 13, 2312–2323 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Duly A. M. P., Kao F. C. L., Teo W. S., Kavallaris M., BetaIII-Tubulin gene regulation in health and disease. Front. Cell Dev. Biol. 10, 851542 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Latremoliere A., et al. , Neuronal-specific TUBB3 is not required for normal neuronal function but is essential for timely axon regeneration. Cell Rep. 24, 1865–1879.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sikora E., et al. , Cellular senescence in brain aging. Front. Aging Neurosci. 13, 646924 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baker D. J., Petersen R. C., Cellular senescence in brain aging and neurodegenerative diseases: Evidence and perspectives. J. Clin. Invest. 128, 1208–1216 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Levi N., Papismadov N., Solomonov I., Sagi I., Krizhanovsky V., The ECM path of senescence in aging: Components and modifiers. FEBS J 287, 2636–2646 (2020). [DOI] [PubMed] [Google Scholar]
- 65.Basisty N., et al. , A proteomic atlas of senescence-associated secretomes for aging biomarker development. PLoS Biol. 18, e3000599 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Golde T. E., Miller V. M., Proteinopathy-induced neuronal senescence: A hypothesis for brain failure in Alzheimer’s and other neurodegenerative diseases. Alzheimers Res. Ther. 1, 5 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee S., et al. , A guide to senolytic intervention in neurodegenerative disease. Mech. Ageing Dev. 200, 111585 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Capizzi M., et al. , Developmental defects in Huntington’s disease show that axonal growth and microtubule reorganization require NUMA1. Neuron 110, 36–50.e5 (2022). [DOI] [PubMed] [Google Scholar]
- 69.Buijsen R. A. M., et al. , Spinocerebellar ataxia type 1 characteristics in patient-derived fibroblast and iPSC-derived neuronal cultures. Mov. Disord. 38, 1428–1442 (2023). [DOI] [PubMed] [Google Scholar]
- 70.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Edgar R., Domrachev M., Lash A. E., Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
The RNA-Seq data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (71) and are accessible through GEO Series accession number GSE267802 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE267802).