Significance
Metastatic tumor cells must cope with the compression forces generated by interstitial migration and with the visco-elastic differences of various tissue microenvironments. Our study reveals that mechanical stress generated by confined migration overrides the ATR, ATM, and spindle assembly checkpoint (SAC) responses that protect nuclear and chromosome integrity. Thus, tumor cells migrating across constrictions exhibit aberrant chromosome segregation, chromosome instability, and accumulation of extra- chromosomal DNA circles. Confined migration also leads to amplification of the cMYC locus. Hence, mechanical stress has a causative role in generating cancer aneuploidy, genome instability, and oncogenic events, possibly explaining why cancer aneuploidy often does not correlate with mutations in the SAC genes and why c-MYC amplifications are strongly linked to metastatic tumors.
Keywords: mechanical stress, mitosis, aneuploidy, mitotic spindle, chromosome segregation
Abstract
Confined cell migration hampers genome integrity and activates the ATR and ATM mechano-transduction pathways. We investigated whether the mechanical stress generated by metastatic interstitial migration contributes to the enhanced chromosomal instability observed in metastatic tumor cells. We employed live cell imaging, micro-fluidic approaches, and scRNA-seq to follow the fate of tumor cells experiencing confined migration. We found that, despite functional ATR, ATM, and spindle assembly checkpoint (SAC) pathways, tumor cells dividing across constriction frequently exhibited altered spindle pole organization, chromosome mis-segregations, micronuclei formation, chromosome fragility, high gene copy number variation, and transcriptional de-regulation and up-regulation of c-MYC oncogenic transcriptional signature via c-MYC locus amplifications. In vivo tumor settings showed that malignant cells populating metastatic foci or infiltrating the interstitial stroma gave rise to cells expressing high levels of c-MYC. Altogether, our data suggest that mechanical stress during metastatic migration contributes to override the checkpoint controls and boosts genotoxic and oncogenic events. Our findings may explain why cancer aneuploidy often does not correlate with mutations in SAC genes and why c-MYC amplification is strongly linked to metastatic tumors.
Chromosomal instability (CIN) is characterized by a high rate of gains and losses of whole chromosomes, copy number variations (CNV), gross chromosomal rearrangements, and the activation of fragile sites. CIN can also result in the formation or overexpression of oncogenes by gene fusion as well as oncogene amplifications on extrachromosomal DNA (1–3). Aneuploidy can further enhance chromosome instability and promote replication stress (4). CIN contributes to drive intratumoral heterogeneity, disrupt cellular homeostasis, and promote cancer progression.
The spindle assembly checkpoint (SAC) is a surveillance system that monitors the attachment of chromosomes to the mitotic spindle preventing mitotic progression until all chromosomes are correctly positioned (5). Chemical or genetic conditions interfering with SAC lead to chromosome missegregation and aneuploidy, contributing to genomic instability and tumorigenesis (6–9). Though aneuploidy and CIN are hallmarks of tumors, paradoxically, genes encoding SAC proteins are rarely mutated in cancer cells (10, 11).
Cancer cells are constantly exposed to varying degrees of mechanical stress generated by the microenvironment, within primary tumor foci, or during metastatic invasion when cells infiltrate tissue vascular barriers (12). Nuclear deformations induced by interstitial migration result in nuclear envelope ruptures, replication stress, DNA damage, and cell cycle delays (13–18). Mechanical forces govern the functions of chromosomes (19) that can sense and transduce mechanical stress to the nuclear envelope and the nucleolus (20). Mechanical perturbations of the nuclear envelope trigger an Ataxia-Telangiectasia and Rad3-Related (ATR)-mediated, DNA damage–independent mechanotransduction response (20, 21), and the perturbations of the cytoskeleton activate an Ataxia-Telangiectasia Mutated (ATM)-dependent pathway that contributes to cytoskeleton and chromatin remodeling (22). Physical cues influence cell-cycle progression at multiple levels (23), and the nucleus itself can act as a mechanosensory organelle transducing mechanical stimuli into biochemical signals (24–26).
CIN and aneuploidy frequently emerge due to abnormal mitotic events, which raises questions about the potential involvement of mechanical stress during mitosis as a source of CIN. The mitotic process entails a finely tuned balance between mechanical forces generated by the mitotic spindle and kinetochore attachments, and molecular signals that ensure proper chromosome alignment and distribution. The mitotic spindle orientation can be influenced by cortical forces and requires a defined geometric space (27). A spindle apparatus experiencing mechanical stress generated by external forces induces SAC activation and mitotic delays (28–30). Acute confinement applied during prophase leads to premature nuclear internalization of cyclin B1 due to increased nuclear tension, resulting in aberrant chromosome segregation (31). Mitotic onset triggers distinct mechanical changes in the cell, including increased stiffness and a transition to a spherical shape. This phenomenon, known as mitotic cell rounding, is crucial for proper spindle assembly and positioning. Notably, artificial cell flattening during mitosis affects chromosome capture and bipolar spindle stability leading to SAC activation and delayed anaphase onset (32, 33). Moreover, Myosin II inhibition, a condition that reduces cell contractility, was shown to amplify confinement-induced chromosome losses due to segregation errors, independently on SAC (34).
A robust association exists between CIN and metastases. This is evidenced by the enhanced CIN observed in circulating tumor cells and metastases compared to primary tumors (35–39). We hypothesized that the mechanical stress generated by metastatic interstitial migration might be the cause of this elevated CIN. Here, we employed single-cell RNAseq (scRNA-seq) to analyze transcriptional and genomic alterations resulting from the migration of tumor cells across a single constriction within a customized microfluidic device. We demonstrate that confined migration in conditions that do not cause DNA damage becomes genotoxic for mitotic cells due to mitotic spindle defects, leading to chromosome missegregations, micronuclei formation, and aneuploidies. The mitotic defects induced by interstitial migration override the SAC, which remains functional in these cells. Indeed, the combination of SAC inhibition with mechanical stress synergistically exacerbates both the quantity and nature of abnormal mitotic events. Likely as a consequence of mechanical stress, tumor cells accumulate gene CNV including amplifications of the c-MYC locus, further promoting genetic heterogeneity.
Results
Interstitial Migration Delays Cell Cycle Progression.
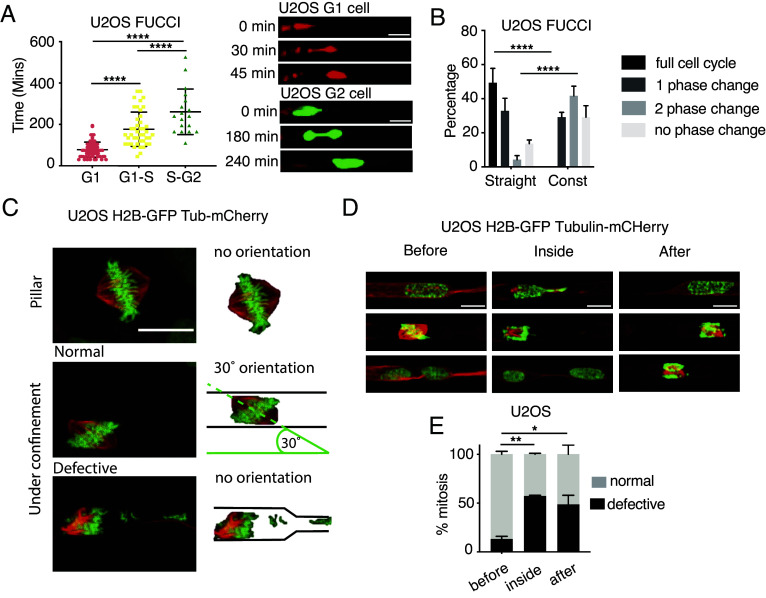
We investigated the cellular response to mechanical stress generated during confined migration using microfabricated devices containing channels with or without constrictions (16, 21) (SI Appendix, Fig. S1A). Previous studies employing these devices and bone osteosarcoma epithelioid U2OS cells showed that the ATR and ATM-mechanotransduction pathways protect cells from the mechanical stress induced during interstitial migration (21, 22). Notably, U2OS cells are well-established cell lines for studying cell cycle checkpoints as they have proficient ATR, ATM, and SAC pathways (40). Migration through constrictions with a limiting pore size generates DNA damage due to nuclear envelope ruptures (16, 21) and leakage of DNA repair factors out of the nucleus (15). To circumvent this, we specifically used larger 3,5 × 6 μm pore size (SI Appendix, Fig. S1A) for U2OS cells which shows an average nuclear volume of 343,347 μm3 (SI Appendix, Fig. S2B). This larger pore size ensures minimal DNA damage formation during cellular engagement at the pores. and mimics the pore size conditions encountered by tumor cells during metastasis (41–44) We monitored cell cycle of migrating U2OS cells using the FUCCI reporter system (45). S-G2 cell nuclei migrated significantly slower than G1 cell nuclei (Fig. 1A), likely due to the differences in their nuclear volume (46). Additionally, in both U2OS and HeLa cells, the speed of migration across constrictions increased linearly with the nuclear size, suggesting a direct relationship between cell cycle, size, and speed (SI Appendix, Fig. S1C). We then compared cell cycle progression in cells migrating across channels with or without constrictions. We imaged cells for 24 h and analyzed cell cycle progression of cells entering the channels in the first frame of the time lapse, regardless of their initial cell cycle stage. Migration across constrictions significantly delayed cell cycle progression, preventing cells from completing one full cycle in 24 h (Fig. 1B), while majority (approximately 80%) of cells passing through the straight channels progressed at least 2 cell cycle phases in 24 h. To investigate whether these cell cycle delays correlated with DNA damage occurrence, we stably expressed 53BP1-GFP, a protein that forms foci at DNA damage sites (47), in cells migrating across constrictions. In agreement with our previous observations (21), we found that the number of 53BP1-GFP foci did not significantly change before, during, or after migration across constrictions (SI Appendix, Fig. S1D).
Fig. 1.
Interstitial migration delays cell cycle progression, induces mitotic defects, and chromosome missegregation. (A) Cell cycle phase determines the speed of migration across constrictions. Analysis of the time to cross the constriction in U2OS cells labeled with FUCCI cell cycle marker. G1-phase cells (Red) pass-through the fastest while G2 cells (Green) take the longest. ****P value < 0.0001 one-way ANOVA. Bars show mean ± SD. (Scale bars: 20 μm.) (B) Mechanical constraints delay cell cycle progression. Cell cycle analysis in FUCCI-U2OS cells under various mechanically constrained environments (straight channels vs. channels with constrictions). ****P value < 0.0001 two-way ANOVA. Error bars are SEM. (C) Mitotic spindle organization in U2OS H2B-GFP Tubulin-mCherry migrating inside microchannels. The mitotic spindle orients following a 30° angle in cells undergoing mitosis inside straight channels; the presence of constrictions causes defective mitoses with altered spindle orientation and chromosome missegregations. (Scale bar: 20 μm.) (D and E) U2OS cells undergo defective mitoses while dividing across constrictions. (D) Examples of U2OS H2B-GFP Tubulin-mCherry cells dividing before, inside or after the constriction in channels. (Scale bars: 10 μm.) (E) Quantification of defective mitoses w.r.t their position in the channel. *P value < 0.05, **P value < 0.01 two-way ANOVA. Error bars are SEM.
Confined Migration Generates Aberrant Mitotic Events.
Nuclear compression during interstitial migration is particularly challenging for G2 cells (Fig. 1A). Most likely, the mechanical stress generated by confined migration is particularly challenging also in G2/M as chromosome segregation is affected when the spindle apparatus is exposed to compression forces (28–30); Moreover, mitotic cells per se experience mechanical stress, when chromatin condensation triggers nuclear envelope (NE) deformation (20). By labeling mitotic spindle using Tubulin-mCherry and the metaphase plate with H2B-GFP, we observed that cells dividing in channels prior to the constrictions exhibited a well-organized spindle pole oriented at approximately 30° relative to the channel’s longitudinal axis, and successfully completed mitosis (Fig. 1 C–E). In contrast, cells undergoing mitosis inside or immediately after exiting constrictions displayed disorganized spindles and metaphase plates (Fig. 1D). This often resulted in daughter cells with different nuclear size, indicative of unequal chromosome distribution during cytokinesis and micronuclei formation (Fig. 1 D and E). Furthermore, defective mitoses during interstitial migration in U2OS cells also correlated with increased cell death post mitosis (SI Appendix, Fig. S1E). Similar results were obtained in HeLa cells (SI Appendix, Fig. S1F). Altogether these results suggest that cell confinement during interstitial migration specifically impairs late cell cycle events, leading to abnormal mitosis characterized by chromosome missegregation, formation of lagging chromosomes and micronuclei.
The SAC Is Not Sufficient to Prevent Mitotic Defects during Confined Migration.
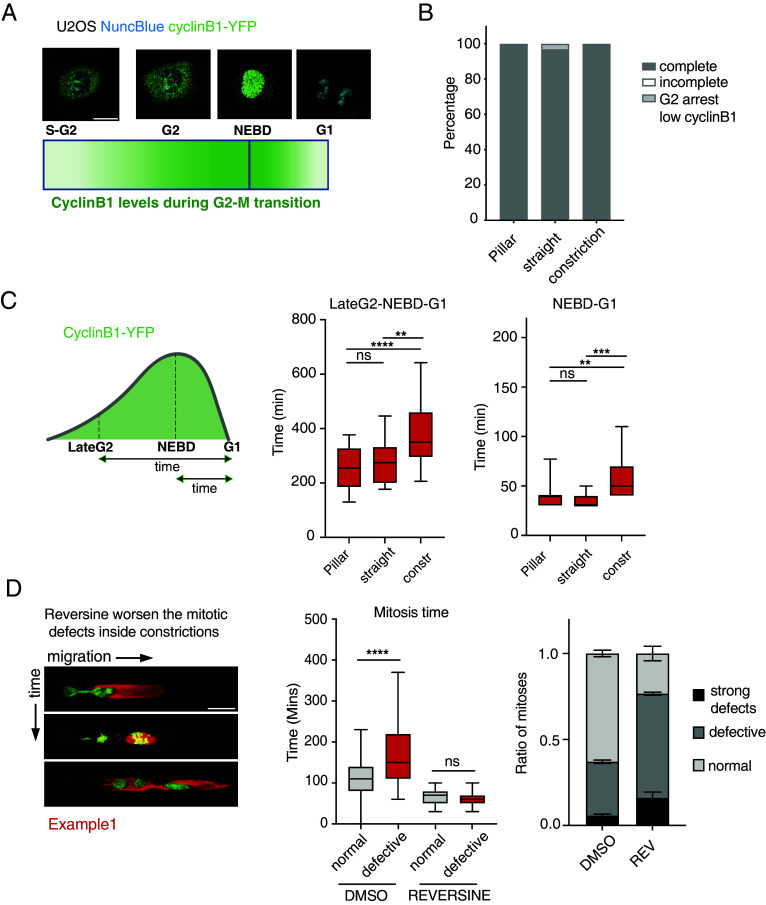
We observed that interstitial migration disrupts normal mitosis, a process typically expected to activate SAC. We therefore investigated whether U2OS cells have dysfunctional SAC or if the mechanical stress from confinement overrides the SAC response. During metaphase, the presence of unattached kinetochores activates the SAC, which delays cyclin B1 degradation by inhibiting APC/CCdc20 (5). Cyclin B1 is a G2/M cyclin that accumulates through late S-phase and G2, peaking at mitosis and is rapidly degraded during mitotic exit (48). We first confirmed that U2OS cells are SAC-proficient as they mounted strong mitotic arrest to increasing doses of nocodazole, a microtubule destabilizing drug (SI Appendix, Fig. S2A). We then analyzed the kinetics of G2-M transition during interstitial migration using endogenously tagged Cyclin B1 (Cyclin B1-YFP) (49) (Fig. 2A). Consistent with prior findings (Fig. 1 C and D), U2OS cells progressed through mitosis even while migrating through constrictions (Fig. 2B). We quantified Cyclin B1 kinetics in U2OS cells experiencing different levels of confinement within the microfluidic device: cells growing with no lateral constraints (pillar region), migrating through channels, and migrating through constrictions. Notably, migration through constrictions specifically prolonged the time between late G2 and G1 reentry as measured by cyclin B1 intensity (Fig. 2C). Furthermore, Cyclin B1 degradation during the nuclear envelope break down (NEBD)-G1 transition was also significantly delayed in these constricted cells (Fig. 2C). Cells undergoing abnormal mitoses exhibited a markedly longer mitotic duration compared to those with normal mitoses (Fig. 2D). Treatment with the SAC inhibitor Reversine (9) rescued the mitotic delay observed in constricted cells (Fig. 2D). However, Reversine treatment increased the fraction of cells undergoing defective mitoses of migrating cells (Fig. 2D and SI Appendix, Fig. S2B). Moreover, the combination of Reversine and mechanical stress resulted in even more severe mitotic defects, particularly in cells dividing across constrictions (SI Appendix, Fig. S2C). Taken together, these data indicate that U2OS cells undergo defective mitoses during interstitial migration despite possessing a functional SAC, this indicates that mechanical forces can bypass the SAC in these cells, leading to the formation of abnormal mitotic events.
Fig. 2.
Confined migration causes defective mitosis and aneuploidy in spite of a functional SAC. (A) Schematic representation of cyclinB1 expression during cell cycle along with example images from U2OS Cyclin B1-YFP cells where DNA was stained using the NuncBlue dye. Cyclin B1-YFP fluorescence signal increases at the end of S-phase and continues to show higher intensity though G2-phase until mitosis. Upon NEBD cyclin B1 enters the nucleus and is decayed as the sister chromatids separate into daughter cells. (Scale bar: 20 μm.) (B) Analysis of mitotic events in U2OS cells expressing Cyclin B1-YFP and migrating outside or inside microchannels. (C) Analysis of cyclin B1-YFP lifespan during mitosis in U2OS cells migrating inside channels. Cyclin B1-lifespan is increased in U2OS cells dividing inside constrictions (LateG2-G1) suggesting a mitotic stress. Mitosis inside constrictions also causes a mild delay in cyclin B1 degradation following NEBD in U2OS cells (NEBD-G1). ****P value < 0.0001, **P value < 0.01 ***P value < 0.001 one-way ANOVA. (D) Time-lapse images showing increased mitotic defects in U2OS H2B-GFP Tubulin-mCherry cells dividing inside microchannels (Left). Reversine abolishes the mitotic delay in U2OS H2B-GFP Tubulin-mCherry cells undergoing defective mitoses during interstitial migration suggesting that SAC is activated by chromosome missegregations caused by mechanical stress. Mitotic time is measured as the time-window between chromosome condensation in prophase and the generation of G1 daughter cells (Central graph). Reversine 0.5 μM treatment significantly worsens the mitotic defects in U2OS H2B-GFP Tubulin-mCherry cells dividing inside microchannels (Right graph). ****P value< 0.0001 one-way ANOVA. Error bars in the bar graph are SDs.
Interstitial Migration Induces Gene CNV.
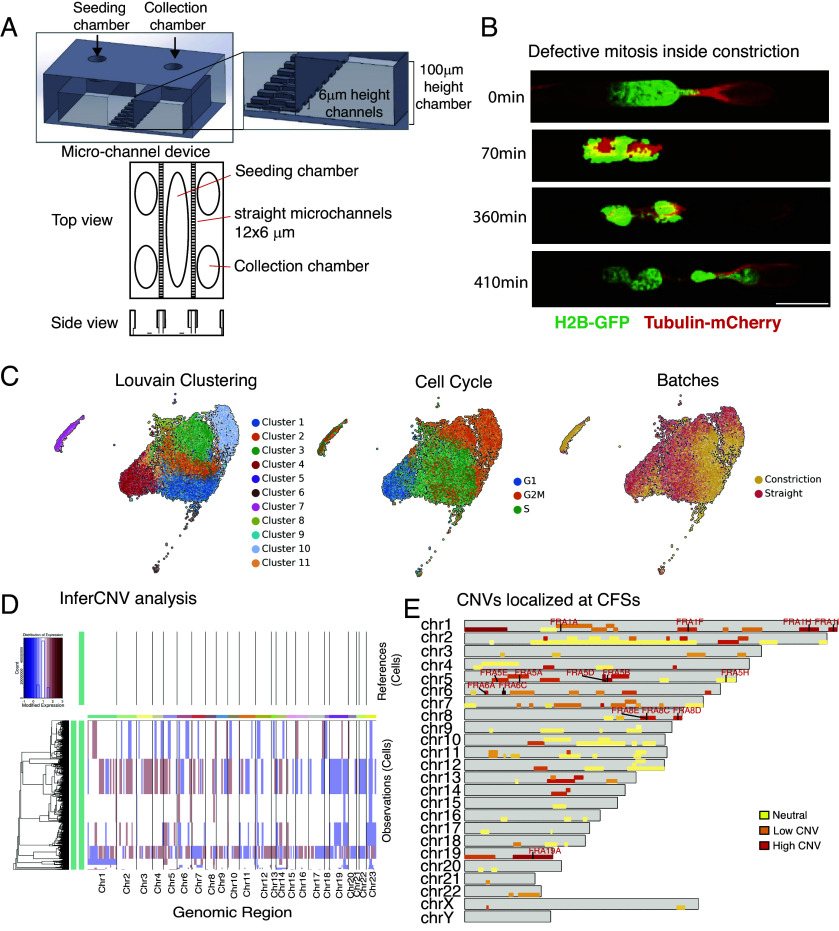
Building on our observations that mechanical stress from confined migration disrupts mitosis and potentially overrides the SAC, leading to chromosome missegregation, we hypothesized that this could contribute to CIN. We designed a custom-made microchannel device that allows us to collect cells after migrating across channels or channels with single constrictions, enabling subsequent analysis by scRNA-seq (Fig. 3A and SI Appendix, Fig. S3A). The device, made of PDMS (polydimethylsiloxane), has a central seeding chamber flanked by microchannels that connect the central chamber with collection chambers (Fig. 3A). The channels and constrictions of this device mirror the microfluidic system previously used to monitor U2OS migration by live cell imaging (SI Appendix, Figs. S1A and S3A). First, we confirmed that cells undergoing migration within the device displayed mitotic defects and micronuclei accumulation similar to those previously observed (Fig. 1 D and E) by live-cell imaging of H2B-GFP Tubulin-mCherry U2OS cells (Fig. 3B and SI Appendix, Fig. S3B).
Fig. 3.
Interstitial migration across one single constriction results in mitotic defects and copy number variation that affect also chromosome fragile sites (CFSs). (A) Schematic representation of the custom-made microchannel device system designed to collect cells migrated across straight channels or channels with 3,5 μm constrictions. (B) Example of U2OS H2B-GFP Tubulin-mCherry undergoing defective mitosis during migration across constriction inside the custom-made microchannel device. (Scale bar: 20 μm.) (C) Low dimensional UMAP embeddings of single-cell RNA-seq data showing automatic clustering based on transcriptomic profile of each cell (Left), cell cycle phase clustering (Middle) and identification of cells belonging to each batch, straight or constriction (Right). (D) InferCNV analysis of single-cell RNA-seq from U2OS clone migrated in the device with straight (reference) or constriction channels. Cells migrated across one single constriction show increase CNV at several genomic regions. (E) CNVs also localize at CFSs in U2OS cells migrated across constriction. Schematic representation showing all the individual chromosomes (gray bars) and the relative fragile sites (yellow to red bars). Color code refer to the frequency of CNV identified on each single fragile site (red: higher amount of CNV). The highlighted red names indicate fragile sites with the highest frequency of CNV across all CFSs.
To minimize genetic variation within the cell population, we isolated and karyotyped a single U2OS clone. These clonal cells were then subjected to migration in the microfluidic device: 20 devices contained straight channels without constraints and 20 incorporated a single constriction. After 72 h, cells were collected, incubated overnight, and processed for single-cell RNAseq.
We obtained the single-cell RNA transcriptome from 9,127 cells for cells migrating in channels without constriction (“straight”) and 7,360 cells for cells migrating in channels with constriction (“constriction”), with a median of identified genes ranging from 6,055 in straight to 7,128 in constriction condition. Cell cycle analysis identified comparable proportions of cells at different cell cycle stages across the two experimental conditions further supporting the notion that constricted migration affects duration of cell cycle transition rather than cell cycle phase itself (Fig. 3C and SI Appendix, Fig. S3B). In the constrictions sample, we found a small cell population characterized by a low number of transcripts, (less than 2,000), separated by the rest of the clusters, likely representing the micronuclear fraction (Fig. 3C, cluster 7).
We performed an InferCNV analysis on the sc-RNA transcriptomes to identify large-scale chromosomal CNV occurring in cells post constricted migration, using cells migrated across straight channels as reference, to account for described cell-line specific chromosomal alterations (https://depmap.org/portal/cell_line/ ACH-000364?tab=overview). We found that migration across a single constriction was sufficient to cause gain and loss of large chromosomal fragments or even entire chromosomes (Fig. 3D). Automatic clustering of inferred CNVs identified clusters of cells carrying similar genetic alterations. The putative micronuclear cluster (Fig. 3C) showed CNV on majority of chromosomes (Fig. 3D). We then cross-checked our inferred CNVs with the HumCFS database (50) to investigate whether the CNV regions contained chromosome fragile sites (CFSs). CFSs are large chromosomal regions that exhibit gaps and breaks on metaphase chromosomes upon replication stress. A graphical representation mapping the location of individual CFSs (according to HumCFS) on each human chromosome is included in SI Appendix, Fig. S3D. We found that 14 over 120 annotated CFSs localized at CNV regions in at least 50% of those cells migrated across constrictions (Fig. 3E); not all CNV regions contained CFSs (Fig. 3E and SI Appendix, Fig. S3 D and E). The 14 CFSs localized on five chromosomes chr1, chr5, chr6, chr8, and chr19 (Fig. 3E and SI Appendix, Fig. S3 D and E). Altogether, we observed that migration through a single constriction is sufficient to cause gross chromosomal rearrangements and CFSs expression.
Confined Migration Causes Transcriptional Deregulation of Oncogenic and Mechanosensory Pathways.
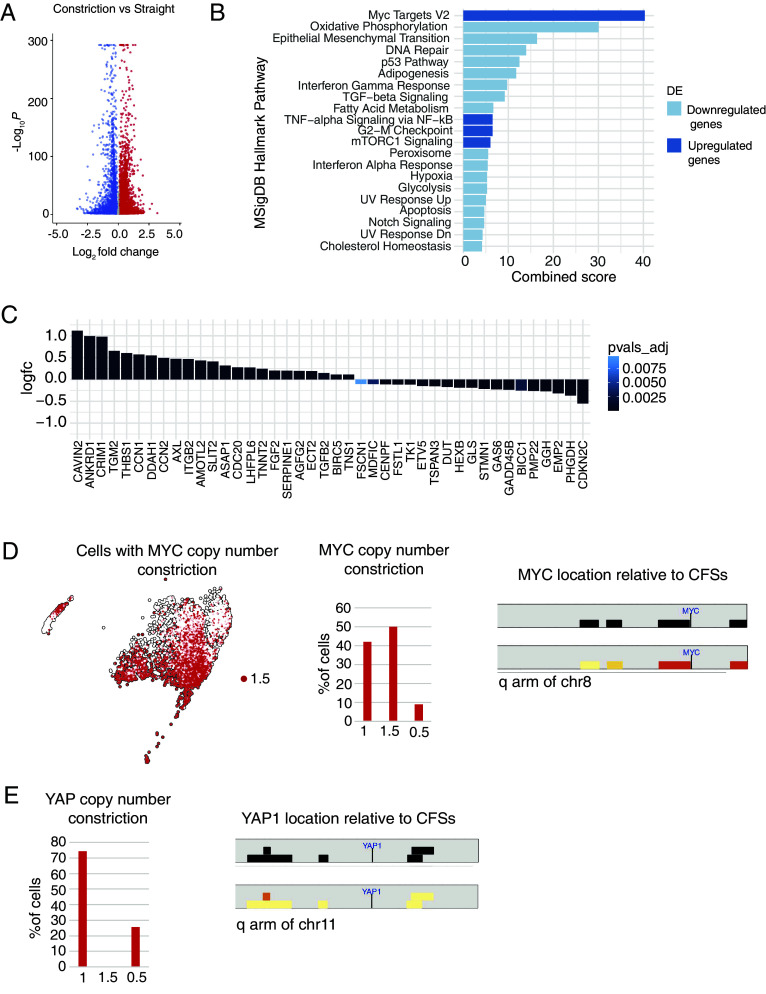
We performed a differential gene expression analysis to assess transcriptional deregulation across two experimental samples and employed the EnrichR tool to evaluate enrichment in specific pathways. Cells passing through constrictions displayed significant deregulation of several pathways (Fig. 4A). Among genes significantly up-regulated upon constriction, a signature for identified c-MYC targets scored in the top rank (Fig. 4B). In addition, tumor necrosis factor (TNF)-alpha signaling, together with mammalian target of rapamycin complex 1 (mTORC1) and G2-M checkpoint–related genes, were among the pathways highly enriched in up-regulated genes (Fig. 4B). We hypothesized that interstitial migration-induced cellular and nuclear deformations would activate the yes-associatedprotein (YAP)-mediated mechanotransduction pathway (51). As expected, several of the YAP signature target genes (52, 53) were induced in cells that migrated across constrictions (Fig. 4C).
Fig. 4.
Interstitial migration across one single constriction results transcriptional upregulation of pathways involving c-MYC, YAP, inflammation, and cell metabolism. (A) Volcano plot showing differential expression analysis comparing straight and constriction samples showing that cells migrated across constrictions have several genes deregulated at the transcriptional level. (B) Enrichr analysis of the transcriptionally up-regulated (blue) and down-regulated (light-blue) pathways in U2OS cells migrated across constrictions. c-MYC targets appear as the top-ranked transcriptionally up-regulated genes. (C) Graphs showing transcriptional upregulation of the YAP signature (52) in U2OS cells migrated across one constriction. (D) Identification of cells with gain of MYC copy number in the BBrowser map and bar-graph showing MYC CNV in cells migrated across constrictions (Left). Graphical representation highlighting the c-MYC locus on the q arm of chr8 located near two CFSs (FRA8E and FRA8C) that are affected by CNV following interstitial migration (Right). (E) Graphs showing absence of YAP copy number gain in U2OS cells migrated across one constriction (Left) and graphical representation highlighting the YAP1 locus on the q arm of chr11 located far from CFSs and in a region poorly affected by CNV following interstitial migration (Right).
Confined Migration Leads to Amplification of the c-MYC Locus.
We further analyzed the c-MYC gene locus in our InferCNV data and found that it was amplified in a substantial portion (~50%) of cells that migrated across constrictions (Fig. 4D). We noticed that the c-MYC gene is located closely to the CFSs FRA8E and FRA8C on Chr8, that were expressed upon interstitial migration (Fig. 4D).
Contrary to c-MYC, YAP gene copy numbers were not altered in cells that migrated across constrictions (Fig. 4E), suggesting that transcriptional upregulation of YAP target genes was likely due to YAP nuclear translocation following mechanical stress (54). Furthermore, YAP1 gene localized far from known CFSs and the chromosomal region surrounding the YAP1 gene showed minimal CNVs (Fig. 4E). We then quantified the c-MYC locus copy number in U2OS cells migrated across constrictions using fluorescence in situ hybridization (FISH). First, we probed the c-MYC locus in metaphase spreads of the U2OS clone used for the scRNA-seq experiment and retinal pigment epithelial-1 (RPE-1) cells (diploid nontransformed) as a reference (SI Appendix, Fig. S4A). The c-MYC FISH probe identified four copies of c-MYC in the terminal portion of an acrocentric chromosome in RPE-1 metaphases, as expected. The U2OS clone had higher cMYC copies in normal culture conditions, showing up to 10 copies in one metaphase spread, with minimal cell-to-cell variability (SI Appendix, Fig. S4A).
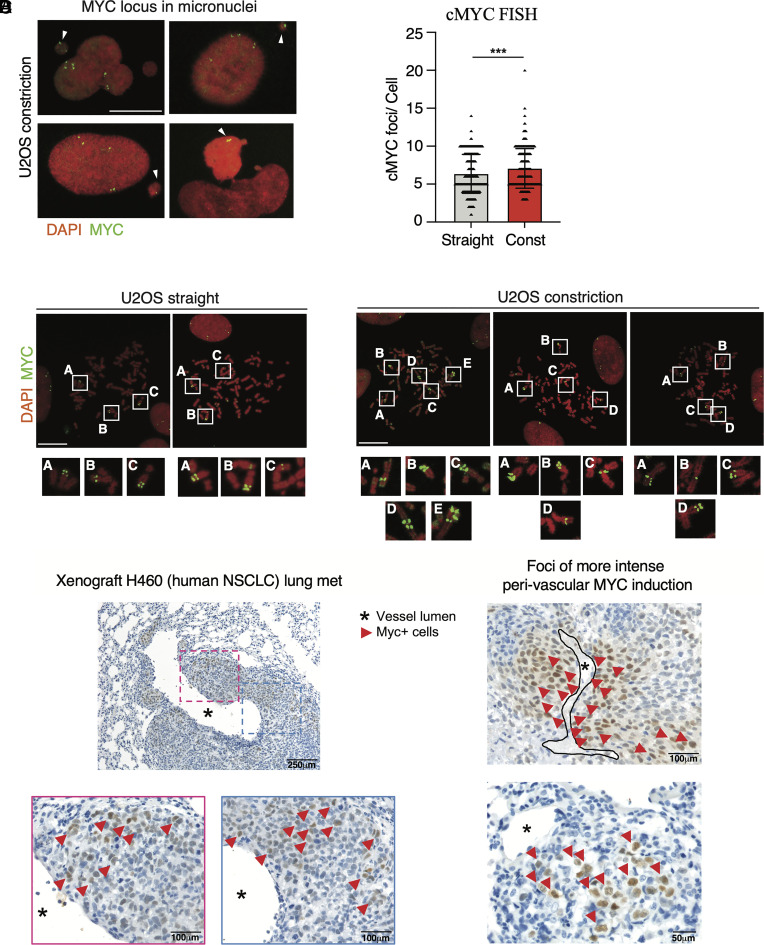
Using the microchannel device described above (Fig. 3A), we performed c-MYC FISH in metaphase spreads of U2OS that migrated across straight channels or channels with constrictions. We found that the c-MYC locus exhibited a perinuclear localization in interphase cells, independently of the constrictions (SI Appendix, Fig. S5B); moreover, the c-MYC locus was present in micronuclei generated during confined migration (Fig. 5A). Analysis of c-MYC foci/cell showed a significant increase of c-MYC copies in U2OS cells migrated across constrictions (Fig. 5 B and C).
Fig. 5.
Interstitial migration causes c-MYC locus amplifications. (A) FISH of c-MYC locus localized in micronuclei of U2OS cells migrated across constrictions. (Scale bar: 20 μm.) (B) c-MYC FISH showing increased number of c-MYC foci in U2OS cells migrated across channels with constrictions. ***P value < 0.001 unpaired t test, error bars are SDs. (C) Representative FISH images showing c-MYC amplifications found in metaphase spreads of U2OS cells migrated across constriction. The regular c-MYC locus configuration in U2OS cells migrated across straight channels is shown on the Left. (Scale bars: 20 μm.) (D) Immunohistochemistry images of lung metastasis derived from a xenograft model of human NSCLC stained for c-MYC. H460 cells metastasized in the lungs show increased heterogeneity for c-MYC protein expression.
Oncogene amplifications, including c-MYC, are linked to the generation of extrachromosomal circular DNA (55). We therefore quantified the number of extrachromosomal circular DNA (eccDNA)/cell in metaphase spreads of migrated cells. Passage through one constriction caused a significant increase in the number of eccDNA/cell (SI Appendix, Fig. S4E). Notably, the c-MYC locus was detected in a subset of eccDNAs, specifically in U2OS cells migrating across constrictions (SI Appendix, Fig. S5F).
Metastasized Foci Exhibit High c-MYC Expression in In Vivo Xenograft Models.
Our findings suggest that mechanical stress during interstitial migration particularly involving mitosis, disrupts chromosome segregations, which likely contributes to c-MYC amplifications and increased expression in tumor cells. To investigate the potential relevance of our findings in tumor settings in vivo, we analyzed c-MYC expression by immunohistochemistry (IHC) in a metastatic H460 lung cancer xenograft model with immune-compromised mice (56), and a syngeneic 4T1 metastatic murine breast cancer model (57). In both models, malignant cells populating metastatic foci in the lung and infiltrating the interstitial stroma around vessels showed local induction of c-MYC protein expression, giving rise to a heterogeneous fraction of cells expressing higher levels of c-MYC (Fig. 5D and SI Appendix, Fig. S5A).
Altogether these results suggest that mechanical stress encountered during interstitial migration causes mitotic defects and contributing to CIN, transcriptional deregulation, and c-MYC gene amplifications in metastatic secondary tumor cells.
Discussion
Interstitial Migration Causes Mitotic Defects and Micronuclei Formation.
We showed that mechanical stress imposed by interstitial migration, a model for metastatic migration (16, 58) enhances genotoxicity and fosters oncogene amplifications in cancer cells.
Confined migration can cause DNA damage when nuclei are exposed to compressive forces (15, 18). Both ATR and ATM kinases promote cell survival during this process (21, 22). ATR and ATM, besides controlling genome integrity, contain giant HEAT domain repeats ideal to sense mechanical stress (59, 60). While ATR safeguards nuclear envelope from compression forces (21), ATM is activated by cytoskeletal stress, mediating cytoskeleton, and chromatin remodeling (22). Even when compression does not cause obvious DNA damage signals, and when ATR and ATM are functional, interstitial migration can still generate genome rearrangements, micronuclei formation, and oncogene amplification. In spite of a functional SAC, cancer cells undergo abnormal cell divisions inside constrictions. Our observations imply that the compression forces imposed by interstitial migration can override the ATR, ATM, and SAC-mediated cell cycle checkpoints.
The aberrant chromosome segregation and mitotic apparatus exhibited while cell experience one single constriction generate daughter cells with unequal genetic material and may very well contribute to aneuploidy, hallmark of cancer. SAC genes are rarely mutated in tumors and, paradoxically, SAC proteins are often overexpressed in cancer cells (10), making the cause of metastatic cancer-associated CIN a major black box in cell biology. Here, we shed light on this outstanding issue by showing that confined migration contributes to aberrant chromosome segregation.
The mechanical stress imposed by constrictions likely perturbs mitosis by exerting pressure to the metaphase plate, disrupting the geometric organization of mitotic spindles. Nuclear compression during prophase may influence the coordination between chromosome condensation and NEBD, causing nuclear collapse and NE ruptures (16, 17), leakage of genomic DNA into the cytosol (21, 61, 62) and cyclin B1 premature internalization (31). These mitotic errors also generate micronuclei, thus triggering chromothripsis, intrachromosomal rearrangements, translocations, and the circular DNA formation (63). Accordingly, we observed accumulation of eccDNA molecules in U2OS cells migrating across constrictions. Compression during confined migration might also induce significant transcriptional, epigenetic, and topological alterations that may influence chromosome condensation.
Interstitial Migration Induces CNV.
We showed that confined migration through a single constriction is sufficient to induce genetic heterogeneity into a clonal population of tumor cells. Our analysis detected CNVs involving large portions of chromosomes, including the loss of entire chromosomes (chr1, chr19, chr22, and chr23). Taking into consideration that our in vitro conditions minimize: i) the effect of nuclear compression on DNA damage, ii) the number of passages across pores (one) iii) the initial genetic heterogeneity of the migrating tumor cell (single clone) iv) the external selective pressure, a logical expectation is that, in vivo, the mechanical stress generated by confined metastatic migration would have a more pronounced impact on CIN due to multiple rounds of nuclear squeezing and mitotic division occurring under external selective pressure.
We found that CFSs are affected by interstitial migration. A fraction of CFSs accumulate at CNV regions, particularly those located on chr1, chr5, ch6, chr8, and chr19. CFSs are large chromosomal regions exhibiting gaps and breaks on metaphase chromosomes upon replication stress. These sites are focal points for chromosomal rearrangements in cancer (63). CFSs are highly prone to copy number variation and often associated with oncogene amplification or deletion of tumor suppressors (64). CFSs are highly unstable even in precancerous stages, and play a relevant role in tumorigenesis (65). A common feature shared by all CFSs is their sensitivity to replication stress. Here, we showed that certain CFSs are sensitive to mechanical stress induced by interstitial migration. We speculate that both replication stress and confined migration generate similar mechanical stress at the chromatin level, leading to the expression of fragile sites: In the case of replication stress, the topological constraints exerted by stalled replication forks can be easily transmitted to the nuclear envelope (66); in the case of confined migration, nuclear compression generates mechanical stress at perinuclear chromatin. In this scenario, it is possible that those fragile sites expressed during interstitial migration might be localized close to the nuclear envelope. Intriguingly, those perinuclear chromosome regions have higher probability of micronucleation (67) and micronuclei are characterized by aberrant DNA replication events (68). Therefore, we suggest that confined migration-induced micronucleation contributes to the expression of perinuclear CFS.
Interstitial Migration Causes Transcriptional Deregulation.
Our scRNA-seq analysis focused on relatively stable changes in gene expression following interstitial migration, as we analyzed U2OS cells days after they traversed a single constriction. This approach captured gene expression patterns that persisted after the immediate response to mechanical stress. We found a significant upregulation of c-MYC target genes post constrictive migration. c-Myc activation promotes tumorigenesis and cancer aggressiveness by regulating cell proliferation, metabolism, protein synthesis, apoptosis, angiogenesis, immune evasion, extracellular matrix remodeling, and metastasis (69). These findings suggest that interstitial migration-induced c-MYC amplification likely triggers a c-MYC-dependent transcriptional program, further accelerating tumor progression.
Mechanical stress modulates transcription factors activity of armadillo/β-catenin, serum response factor, Myocardin-related transcription factor, YAP, and nuclear factor κB (NF-κB) (70–72). We found that several YAP-tafazzin (TAZ)-regulated genes were induced following migration across one constriction confirming that these cells displayed transcriptional features of mechanically challenged cells. Interestingly c-MYC and YAP were shown to integrate mitogenic and mechanical cues at the transcriptional level to promote cell proliferation and tumorigenesis (73). We found that TNF alpha signaling via NF-κB was up-regulated in cells that migrated across constrictions, likely indicating the activation of innate immunity pathway due to nuclear strains and increased NE ruptures. Additionally, we found significant transcriptional repression of interferon gamma and interferon alpha–related genes in these cells. These pathways are closely associated with cGAS–STING mediated inflammatory signaling, which is frequently repressed in aggressive tumors to promote immune system evasion (74).
Among the NF-κB regulated genes, interleukin-6 (IL6), gene was significantly up-regulated in cell migrated across constrictions. Notably, recent studies found that high levels of IL6 promote survival of highly chromosomal unstable tumors (75). We showed that tumor cells experience mitotic stress when dividing across constrictions and, accordingly, we found that genes functionally related to the G2-M checkpoint are up-regulated in those cells. Interestingly, several of those genes are classified as oncogenes, exhibiting overexpression in cancerous tissues and a known association with tumor aneuploidy (10). Moreover, some of these genes are targets of c-MYC, suggesting a potential role of c-MYC in their upregulation (76, 77). Therefore, cells that migrate across constrictions undergo chromosome missegregations and CIN that further promote oncogenic transformation. Of note, we also observed downregulation of genes regulated by p53, which is frequently mutated in aneuploid tumors (78).
Among the overexpressed pathways in constriction-migrated cells, we found mTORC1-related genes. Hyperactive mTOR is common in human tumors, sustaining tumor growth, proliferation, and metastasis; it also plays a role in regulating tumor mechanobiology (79, 80). Our findings align with published observations showing mTORC1 activation under different types of mechanical stimuli (81–83).
Our results suggest that interstitial migration induces a transcriptional reprogramming involving cell metabolism, oncogenes, G2/M transition, and inflammation that further promote tumorigenesis suggesting that mechanical stress during metastasis can contribute toward malignant transformation and cancer aggressiveness.
Interstitial Migration Causes Amplifications of c-MYC Oncogene.
CIN is associated with oncogenes amplifications, a phenomenon frequently observed in solid tumors (84). Among various oncogenes, c-MYC expression is dysregulated in up to 70% of human cancers, with amplification being the most common mechanism of c-MYC activation (85). c-MYC belongs to the MYC family of cellular oncogenes alongside N-MYC and L-MYC. These three genes are located on different chromosomes. Intriguingly, N-MYC and L-MYCare far less frequently amplified in tumors, despite their functional and structural homology to c-MYC (86). Both transcriptional upregulation and gene amplification of c-MYC are strongly linked with tumor metastasis (87–89).
Moreover, we provide in vivo evidence showing a significant heterogeneity of c-MYC expression in murine lung metastases. Consistent with previous observations, N-MYC or L-MYC did not exhibit transcriptional upregulation in our experimental model system, indicating that c-MYC is more frequently amplified among the MYC family of oncogenes.
The original U2OS clone, used for our migration experiments harbored stable c-MYC locus, not localized on extrachromosomal DNA molecules. c-MYC amplification was exclusively observed following migration across one single constriction. This finding strongly suggests that confined migration induces aberrant chromatin rearrangements, rather than acting through a selection mechanism favoring cells with preexisting amplified c-MYC. Moreover, we found the c-MYC locus in micronuclei of tumor cells that had migrated across constrictions. The c-MYC gene is located on chromosome 8 within the 8p24.21 region in human cells. Interestingly, this chromosomal region exhibits a pronounced tendency for amplification in human tumors, according to the Cancer Genome Atlas project (90). We showed that the CFSs FRA8C and FRA8E, that are localized in close proximity to the c-MYC locus, are among the CFSs with higher frequency of CNV in cells migrated across constrictions. Moreover, chromosome 8 showed a significantly higher probability of missegregation following mitotic errors caused by SAC inhibition (67). Frequent micronucleation of chromosome 8 has been attributed to its perinuclear position during interphase which is prone to segregation errors (67). Accordingly, the c-MYC locus is localized near the nuclear periphery during interphase also in U2OS cells. Peripheral chromosomes are prone to frequent missegregation, as they need to travel a longer distance to reach the metaphase plate. Additionally, endomembrane entrapment surrounding the mitotic spindle can contribute to segregation errors (91). We suggest that confined migration-induced mechanical stress could exacerbate this phenomenon by altering the membrane-free zone around the spindle, further entrapping peripheral chromosomes. c-MYC amplifications can occur on circular extrachromosomal elements (1, 92), suggesting that mechanical stress induced by confined migration causes micronucleation of the c-MYC locus, followed by chromothripsis events, thus directly promoting a route for intra- and extrachromosomal c-MYC amplifications.
Altogether our results suggest that the mechanical stress generated during confined and metastatic migration may have a causative role in enhancing CIN and oncogene amplification by overriding the ATR, ATM, and SAC cell cycle checkpoints.
Materials and Methods
Cell Culture and Drug Treatments.
HeLa cells were maintained in minimum essential medium with GlutaMAX (MEM; Life Technologies 41090-093) supplemented with 10% (vol/vol) fetal bovine serum (FBS) South America (SA) (Biowest), 0.1 mM nonessential amino acids (Microtech), sodium pyruvate 1 mM (Microtech), and penicillin–streptomycin (Microtech). 1% glutamine, 10% FBS SA-DMEM high glucose Lonza 12-614 was used for U2OS cells. hTERT-RPE-1 cells were maintained in DMEM-F12 (Gibco 31331-028) supplemented with 10% FBS SA, 1% glutamine, and penicillin–streptomycin. Cell culture was performed in 5% CO2, at 37 °C. Additional details are in SI Appendix, section S1.1.
Cell Migration Analysis in Microfabricated Channels for Live Cell Imaging.
Microchannels with and without constrictions were used in this work. For U2OS and HeLa cells, 12 μm width × 6 μm height channels with 3,5 μm wide constrictions were used.
Silicon molds of microchannels were from Matthieu Piel’s lab and PDMS channels were prepared as previously described (16, 21, 58); see details in SI Appendix, section S1.2. Cells loading was performed the day before (for HeLa) or 4 h before the beginning of time lapse (for U2OS cells). Time-lapse images were acquired (every 5 or 10 min, with z-stacks) on an Olympus Spinning Disk CSU based on an Olympus IX83 inverted microscope equipped with an Andor iXon Ultra camera and driven by CellSens software (Olympus). A 40× oil immersion objective was used to acquire images and cells were maintained in an environmental chamber at 37 °C in an atmosphere of 5% CO2. The images were processed using Image J and smoothed to reduce the background noise. Details are mentioned in SI Appendix, section S1.3 for details.
Live Cell Imaging and Cyclin B1 Intensity Quantifications Following Nocodazole Treatment.
For live cell analysis, U2OS-Cyclin B1-eYFP cells were seeded into 8-well μ-slides (Ibidi) 3 × 104 per well, synchronized with a double thymidine block (2.5 mM) and released in nocodazole (0, 50, 100, or 300 nM), approximatively 2 h before starting the imaging acquisition. See SI Appendix, section S1.4 for details.
Microchannel Device for Single-Cell RNA Sequencing, Genomic DNA Amplification, and cMyc-FISH.
The microfluidic chip was designed with one central chamber for cell seeding (6 × 20 mm in area) and two lateral chambers for cell collection (2 × 20 mm in area). Each region is filled with pillar arrays (50 μm in diameter and 100 μm in gap) to support. Cell seeding and cell collection regions are connected by 1,000 parallel constriction channels. The channel is 12 × 7 μm in cross-section and 100 μm in length. The constriction is 4 × 7 μm in cross-section and 12 μm in length. For control experiments, the microfluidic chip was designed to carry the same channels without constrictions. The silicon master was fabricated by Polifab using standard photolithography.
The PDMS microchannel device was prepared as previously described. A unique central cell-seeding chamber and two lateral cell collection chambers were created manually in the PDMS using a biopsy puncher (Rapid Core 15115-8 ID 3 mm). The PDMS was washed in pure ethanol for 10 min at room temperature (RT), followed by 10 min of sonication at maximum power to remove PDMS debris. PDMS channels were bound to bottom slide-glasses (2 devices/glass) using plasma treater followed by 45 min at 80 °C. Twenty devices with straight channels and 20 devices with constrictions-bearing channels were prepared for one experiment of migration and single-cell sequencing. The devices were stored at RT in a closed Petri until the day of experiment.
Cells Collection for Single-Cell RNA Sequencing of Cells Migrated Inside the Microfabricated Device.
Cells were loaded and migrated inside the microchannel device as described in SI Appendix, section S1.5. Cells were washed two times with Phosphate Buffered Saline (PBS), trypsin was added only in collection chamber and cells were incubated at 37 °C to allow proper trypsinization for approximately 5 min.
Cells were carefully resuspended to detach them from the device using a p200 pipette and collected in an Eppendorf tube 1.5 mL; cell culture medium was added to stop trypsin. Cells were centrifuged at 3,000 rpm for 2 min, the pellet was resuspended in cell culture medium and cells were seeded on a well of a 48-well plate.
Cell collection was performed on 10 devices at a time and cells were collected in the same Eppendorf tube. At the end of the procedure two samples were prepared, one for straight channels, one for channels with constriction. Cells were grown one more night at 37 °C on 48-well dish. Finally, cells were carefully washed four times with PBS (to remove eventual debris of PDMS that could interfere with single-cell sequencing procedure) trypsinized, centrifuged at 3,000 rpm for 2 min, washed ones in PBS, the cell pellet was resuspended in a 0.04% Bovine Serum Albumin (BSA) (extra pure) PBS solution. Samples from straight and constriction channels were resuspended in 20 μL and 10 μL of BSAPBS, respectively, in order to achieve a concentrated solution of cells in the range of 800 to 1,000 cells/microliter. One μL of cell solution was resuspended in final 12 μL of a Trypan Blue-PBS 1:1 solution and counted using a Burker counting chamber.
Samples were kept on ice until processed for single-cell sequencing. For sequencing details, see SI Appendix, section S1.6 and for additional information on scRNA-seq data processing and quality control see SI Appendix, section S1.7. Single-cell RNA sequencing data are available in ENA (PRJEB66123) (93).
Chromosomal Fragile Sites.
HumCFS is a comprehensive database for human chromosomal FS (50). It contains manually curated information on FS locations, genes, and miRNA in each FS, frequency of induction, and other supporting materials. The CNV predictions of each gene from the inferCNV observation matrix data were used to classify the respective FS into high/medium CNV or neutral based on the number of cells showing amplification/deletion. The FS were visualized using the bioconductor package, karyoploteR (94).
Note: The majority of the genes within a particular FS showed uniform CNV.
Metaphase Spreads on Cells Growing on Glass Slides.
Cells were seeded in 24-well plate at 30,000 cell/well concentration on the top of round slide glasses previously coated with fibronectin 10 μg/mL. The following day, cells were arrested in metaphase by adding KaryoMax™ Colcemid™ diluted in cell culture medium at a final concentration of 0.05 μg/mL for 4 h. Slides were then moved on a new 24-well plate and cells were treated with 1 mL of hypotonic solution for 10 min at RT. The hypotonic solution was gently removed, but not completely to avoid loss of round metaphases, followed by the addition of 1 mL ice-cold Carnoy’s fixative for 2 min, fixation was repeated twice.
Slides were then dried, moved on a new 24-well plate to be placed at 37° (without the cap) in a water bath, on the top of a thermo-block for 5 min. Fixed cells and metaphases on slides were then processed for FISH.
For metaphase spreads of migrated cells and reagents details, see SI Appendix, section S1.8 for details.
c-Myc FISH.
To visualize c-Myc by FISH we used the c-Myc FISH orange probe (MYC-20-OR, Empire Genomics) and followed the manufacture protocol for manual hybridization.
c-Myc FISH images were acquired on a Leica Sp8 confocal microscope. A 63× oil-immersion objective (NA 1.4) with a 2× zoom was used to acquire a single focal plane for DAPI and Cy3 channels and an image format of 2,048 × 2,048. The images were processed using Image J and the number of c-MYC foci were manually counted. See SI Appendix, section S1.9 for details.
Immunohistochemical Analyses.
Tissue sections of lung metastases from H460 xenografts (56) and 4T1 (95) mice were retrieved from archives of Tumor Immunology Laboratory and used for cMyc immunolocalization analyses.
Four-micrometer–thick tissue sections were deparaffinized, rehydrated, and unmasked using Novocastra Epitope Retrieval Solutions at pH6 in a thermostatic bath at 98 °C for 30 min. Subsequently, the sections were brought to room temperature and washed in PBS. After neutralization of the endogenous peroxidase with 3% H2O2 and Fc blocking by 0.4% casein in PBS (Novocastra), the sections were incubated with cMyc rabbit anti-mouse (clone Y69, 1:500, ab32072, Abcam) primary antibody. IHC staining was developed using the IgG (H&L)-specific secondary antibodies (Life Technologies, 1:500) and DAB (3,30-Diaminobenzidine, Novocastra) as substrate chromogen. Slides were analyzed under a Zeiss Axioscope A1 microscope and microphotographs were collected using a Zeiss Axiocam 503 Color digital camera with the Zen 2.0 Software (Zeiss).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Kristina Havas for critical reading of the manuscript, all the member of Marco Foiani laboratory for useful discussions, Hiroshi Arakawa, Ylli Doksani, Elia Zanella, Fabrizio Pennacchio, Domenico Delia, Grazisa Rossetti, Giulia Della Chiara, and Flora Ascione for technical suggestions. Arne Lindqvist for sharing the U2OS Cyclin B1-YFP cell line. Work in MF laboratory is supported by AIRC-5X1000-22759, AIRC-IG-21416.
Author contributions
G.B., G.R.K., A.C., and M.F. designed research; G.B., G.R.K., C.L., Q.L., R.B., F.O., D.P., M.P., V.C., C.T., and M.P. performed research; G.B., G.R.K., R.B., J.G., F.I., and L.D. analyzed data; M.F. acquired funding; G.R.K., S.P., G.S., A.C., and C.T. edited the manuscript; and G.B. and M.F. wrote the paper.
Competing interests
M.P. is of the board of Directors and stakeholder of CheckmAb s.r.l. and is a recipient of grants under a research agreement with Bristol-Myers Squibb and Macomics.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Giulia Bastianello, Email: giulia.bastianello@ifom.eu.
Marco Foiani, Email: marco.foiani@ifom.eu.
Data, Materials, and Software Availability
Single-cell RNAsequencing data have been deposited in ENA (PRJEB66123) (93).
Supporting Information
References
- 1.Turner K. M., et al. , Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature 543, 122–125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bach D. H., Zhang W., Sood A. K., Chromosomal instability in tumor initiation and development. Cancer Res. 79, 3995–4002 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitelman F., Johansson B., Mertens F., The impact of translocations and gene fusions on cancer causation. Nat. Rev. Cancer 7, 233–245 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Garribba L., et al. , Short-term molecular consequences of chromosome mis-segregation for genome stability. Nat. Commun. 14, 1353 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lara-Gonzalez P., Westhorpe F. G., Taylor S. S., The spindle assembly checkpoint. Curr. Biol. 22, R966–R980 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Schvartzman J. M., Duijf P. H., Sotillo R., Coker C., Benezra R., Mad2 is a critical mediator of the chromosome instability observed upon Rb and p53 pathway inhibition. Cancer Cell 19, 701–714 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shrestha R. L., et al. , CENP-A overexpression promotes aneuploidy with karyotypic heterogeneity. J. Cell Biol. 220, e202007195 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ippolito M. R., et al. , Gene copy-number changes and chromosomal instability induced by aneuploidy confer resistance to chemotherapy. Dev. Cell 56, 2440–2454.e6 (2021). [DOI] [PubMed] [Google Scholar]
- 9.Santaguida S., Tighe A., D’Alise A. M., Taylor S. S., Musacchio A., Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J. Cell Biol. 190, 73–87 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simonetti G., Bruno S., Padella A., Tenti E., Martinelli G., Aneuploidy: Cancer strength or vulnerability? Int. J. Cancer 144, 8–25 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tighe A., Johnson V. L., Albertella M., Taylor S. S., Aneuploid colon cancer cells have a robust spindle checkpoint. EMBO Rep. 2, 609–614 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gensbittel V., et al. , Mechanical adaptability of tumor cells in metastasis. Dev. Cell 56, 164–179 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Xia Y., et al. , Nuclear rupture at sites of high curvature compromises retention of DNA repair factors. J. Cell Biol. 217, 3796–3808 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfeifer C. R., Vashisth M., Xia Y., Discher D. E., Nuclear failure, DNA damage, and cell cycle disruption after migration through small pores: A brief review. Essays Biochem. 63, 569–577 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Irianto J., et al. , DNA damage follows repair factor depletion and portends genome variation in cancer cells after pore migration. Curr. Biol. 27, 210–223 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raab M., et al. , ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science 352, 359–362 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Denais C. M., et al. , Nuclear envelope rupture and repair during cancer cell migration. Science 352, 353–358 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shah P., et al. , Nuclear deformation causes DNA damage by increasing replication stress. Curr. Biol. 31, 753–765.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleckner N., et al. , A mechanical basis for chromosome function. Proc. Natl. Acad. Sci. U.S.A. 101, 12592–12597 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar A., et al. , ATR mediates a checkpoint at the nuclear envelope in response to mechanical stress. Cell 158, 633–646 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kidiyoor G. R., et al. , ATR is essential for preservation of cell mechanics and nuclear integrity during interstitial migration. Nat. Commun. 11, 4828 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bastianello G., et al. , Cell stretching activates an ATM mechano-transduction pathway that remodels cytoskeleton and chromatin. Cell Rep. 42, 113555 (2023). [DOI] [PubMed] [Google Scholar]
- 23.Gupta V. K., Chaudhuri O., Mechanical regulation of cell-cycle progression and division. Trends Cell Biol. 32, 773–785 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Venturini V., et al. , The nucleus measures shape changes for cellular proprioception to control dynamic cell behavior. Science 370, eaba2644 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Lomakin A. J., et al. , The nucleus acts as a ruler tailoring cell responses to spatial constraints. Science 370, eaba2894 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bastianello G., Foiani M., Mechanisms controlling the mechanical properties of the nuclei. Curr. Opin. Cell Biol. 84, 102222 (2023). [DOI] [PubMed] [Google Scholar]
- 27.Thery M., Jimenez-Dalmaroni A., Racine V., Bornens M., Julicher F., Experimental and theoretical study of mitotic spindle orientation. Nature 447, 493–496 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Cattin C. J., et al. , Mechanical control of mitotic progression in single animal cells. Proc. Natl. Acad. Sci. U.S.A. 112, 11258–11263 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Itabashi T., et al. , Mechanical impulses can control metaphase progression in a mammalian cell. Proc. Natl. Acad. Sci. U.S.A. 109, 7320–7325 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukherjee S., et al. , A gradient in metaphase tension leads to a scaled cellular response in mitosis. Dev. Cell 49, 63–76.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dantas M., Oliveira A., Aguiar P., Maiato H., Ferreira J. G., Nuclear tension controls mitotic entry by regulating cyclin B1 nuclear translocation. J. Cell Biol. 221, e202205051 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lancaster O. M., et al. , Mitotic rounding alters cell geometry to ensure efficient bipolar spindle formation. Dev. Cell 25, 270–283 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Cadart C., Zlotek-Zlotkiewicz E., Le Berre M., Piel M., Matthews H. K., Exploring the function of cell shape and size during mitosis. Dev. Cell 29, 159–169 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Hayes B. H., et al. , Confinement plus myosin-II suppression maximizes heritable loss of chromosomes, as revealed by live-cell ChReporters. J. Cell Sci. 136, jcs260753 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carter S. L., Eklund A. C., Kohane I. S., Harris L. N., Szallasi Z., A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat. Genet. 38, 1043–1048 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Pailler E., et al. , High level of chromosomal instability in circulating tumor cells of ROS1-rearranged non-small-cell lung cancer. Ann. Oncol. 26, 1408–1415 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gutenberg A., et al. , High chromosomal instability in brain metastases of colorectal carcinoma. Cancer Genet. Cytogenet. 198, 47–51 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Warth A., et al. , Chromosomal instability is more frequent in metastasized than in non-metastasized pulmonary carcinoids but is not a reliable predictor of metastatic potential. Exp. Mol. Med. 41, 349–353 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bakhoum S. F., et al. , Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 553, 467–472 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lawrence K. S., Chau T., Engebrecht J., DNA damage response and spindle assembly checkpoint function throughout the cell cycle to ensure genomic integrity. PLoS Genet. 11, e1005150 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolf K., et al. , Collagen-based cell migration models in vitro and in vivo. Semin. Cell Dev. Biol. 20, 931–941 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolf K., et al. , Physical limits of cell migration: Control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J. Cell Biol. 201, 1069–1084 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamauchi K., et al. , Development of real-time subcellular dynamic multicolor imaging of cancer-cell trafficking in live mice with a variable-magnification whole-mouse imaging system. Cancer Res. 66, 4208–4214 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Yamauchi K., et al. , Real-time in vivo dual-color imaging of intracapillary cancer cell and nucleus deformation and migration. Cancer Res. 65, 4246–4252 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Macurek L., et al. , Downregulation of Wip1 phosphatase modulates the cellular threshold of DNA damage signaling in mitosis. Cell Cycle 12, 251–262 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Panagiotakopoulou M., et al. , A nanoprinted model of interstitial cancer migration reveals a link between cell deformability and proliferation. ACS Nano 10, 6437–6448 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Schultz L. B., Chehab N. H., Malikzay A., Halazonetis T. D., p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 151, 1381–1390 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pines J., Hunter T., Cyclins A and B1 in the human cell cycle. Ciba Found. Symp. 170, 187–196; discussion 196–204 (1992). [PubMed] [Google Scholar]
- 49.Akopyan K., et al. , Assessing kinetics from fixed cells reveals activation of the mitotic entry network at the S/G2 transition. Mol. Cell 53, 843–853 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Kumar R., et al. , HumCFS: A database of fragile sites in human chromosomes. BMC Genomics 19, 985 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zanconato F., Cordenonsi M., Piccolo S., YAP/TAZ at the roots of cancer. Cancer Cell 29, 783–803 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cordenonsi M., et al. , The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell 147, 759–772 (2011). [DOI] [PubMed] [Google Scholar]
- 53.Zanconato F., et al. , Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat. Cell Biol. 17, 1218–1227 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dupont S., et al. , Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Yan Y., et al. , Current understanding of extrachromosomal circular DNA in cancer pathogenesis and therapeutic resistance. J. Hematol. Oncol. 13, 124 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bertolini G., et al. , A novel CXCR4 antagonist counteracts paradoxical generation of cisplatin-induced pro-metastatic niches in lung cancer. Mol. Ther. 29, 2963–2978 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miotti S., et al. , Antibody-mediated blockade of JMJD6 interaction with collagen I exerts antifibrotic and antimetastatic activities. FASEB J. 31, 5356–5370 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Heuze M. L., Collin O., Terriac E., Lennon-Dumenil A. M., Piel M., Cell migration in confinement: A micro-channel-based assay. Methods Mol. Biol. 769, 415–434 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Grinthal A., Adamovic I., Weiner B., Karplus M., Kleckner N., PR65, the HEAT-repeat scaffold of phosphatase PP2A, is an elastic connector that links force and catalysis. Proc. Natl. Acad. Sci. U.S.A. 107, 2467–2472 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perry J., Kleckner N., The ATRs, ATMs, and TORs are giant HEAT repeat proteins. Cell 112, 151–155 (2003). [DOI] [PubMed] [Google Scholar]
- 61.Frittoli E., et al. , Tissue fluidification promotes a cGAS-STING cytosolic DNA response in invasive breast cancer. Nat. Mater. 22, 644–655 (2022), 10.1038/s41563-022-01431-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miller K. N., et al. , Cytoplasmic DNA: Sources, sensing, and role in aging and disease. Cell 184, 5506–5526 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li S., Wu X., Common fragile sites: Protection and repair. Cell Biosci. 10, 29 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hellman A., et al. , A role for common fragile site induction in amplification of human oncogenes. Cancer Cell 1, 89–97 (2002). [DOI] [PubMed] [Google Scholar]
- 65.Bester A. C., et al. , Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell 145, 435–446 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bermejo R., et al. , The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell 146, 233–246 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Klaasen S. J., et al. , Nuclear chromosome locations dictate segregation error frequencies. Nature 607, 604–609 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Crasta K., et al. , DNA breaks and chromosome pulverization from errors in mitosis. Nature 482, 53–58 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meskyte E. M., Keskas S., Ciribilli Y., MYC as a multifaceted regulator of tumor microenvironment leading to metastasis. Int. J. Mol. Sci. 21, 7710 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mendez M. G., Janmey P. A., Transcription factor regulation by mechanical stress. Int. J. Biochem. Cell Biol. 44, 728–732 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Talwar S., Jain N., Shivashankar G. V., The regulation of gene expression during onset of differentiation by nuclear mechanical heterogeneity. Biomaterials 35, 2411–2419 (2014). [DOI] [PubMed] [Google Scholar]
- 72.Finch-Edmondson M., Sudol M., Framework to function: Mechanosensitive regulators of gene transcription. Cell Mol. Biol. Lett. 21, 28 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Panciera T., Azzolin L., Cordenonsi M., Piccolo S., Mechanobiology of YAP and TAZ in physiology and disease. Nat. Rev. Mol. Cell Biol. 18, 758–770 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Samson N., Ablasser A., The cGAS-STING pathway and cancer. Nat. Cancer 3, 1452–1463 (2022). [DOI] [PubMed] [Google Scholar]
- 75.Hong C., et al. , cGAS-STING drives the IL-6-dependent survival of chromosomally instable cancers. Nature 607, 366–373 (2022). [DOI] [PubMed] [Google Scholar]
- 76.Das S., Anczukow O., Akerman M., Krainer A. R., Oncogenic splicing factor SRSF1 is a critical transcriptional target of MYC. Cell Rep. 1, 110–117 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.den Hollander J., et al. , Aurora kinases A and B are up-regulated by Myc and are essential for maintenance of the malignant state. Blood 116, 1498–1505 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Taylor A. M., et al. , Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell 33, 676–689.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Menon S., Manning B. D., Common corruption of the mTOR signaling network in human tumors. Oncogene 27, S43–S51 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gargalionis A. N., Papavassiliou K. A., Basdra E. K., Papavassiliou A. G., mTOR signaling components in tumor mechanobiology. Int. J. Mol. Sci. 23, 1825 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ogasawara R., et al. , The role of mTOR signalling in the regulation of skeletal muscle mass in a rodent model of resistance exercise. Sci. Rep. 6, 31142 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.You J. S., et al. , The role of raptor in the mechanical load-induced regulation of mTOR signaling, protein synthesis, and skeletal muscle hypertrophy. FASEB J. 33, 4021–4034 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jacobs B. L., Goodman C. A., Hornberger T. A., The mechanical activation of mTOR signaling: An emerging role for late endosome/lysosomal targeting. J. Muscle Res. Cell Motil. 35, 11–21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schwab M., Oncogene amplification in solid tumors. Semin Cancer Biol 9, 319–325 (1999). [DOI] [PubMed] [Google Scholar]
- 85.Duffy M. J., O’Grady S., Tang M., Crown J., MYC as a target for cancer treatment. Cancer Treat Rev. 94, 102154 (2021). [DOI] [PubMed] [Google Scholar]
- 86.Dhanasekaran R., et al. , The MYC oncogene—The grand orchestrator of cancer growth and immune evasion. Nat. Rev. Clin. Oncol. 19, 23–36 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Maddipati R., et al. , MYC levels regulate metastatic heterogeneity in pancreatic adenocarcinoma. Cancer Discov. 12, 542–561 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Singhi A. D., et al. , MYC gene amplification is often acquired in lethal distant breast cancer metastases of unamplified primary tumors. Mod. Pathol. 25, 378–387 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Arriaga J. M., et al. , A MYC and RAS co-activation signature in localized prostate cancer drives bone metastasis and castration resistance. Nat. Cancer 1, 1082–1096 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tanaka H., Watanabe T., Mechanisms underlying recurrent genomic amplification in human cancers. Trends Cancer 6, 462–477 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ferrandiz N., Downie L., Starling G. P., Royle S. J., Endomembranes promote chromosome missegregation by ensheathing misaligned chromosomes. J. Cell Biol. 221, e202203021 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim H., et al. , Extrachromosomal DNA is associated with oncogene amplification and poor outcome across multiple cancers. Nat. Genet. 52, 891–897 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bastianello G., Foiani M., Mechanical stress during confined migration drives chromosomes mis-segregation and c-MYC amplifications. ENA. https://www.ebi.ac.uk/ena/browser/view/PRJEB66123. Deposited 14 September 2023.
- 94.Gel B., Serra E., karyoploteR: An R/Bioconductor package to plot customizable genomes displaying arbitrary data. Bioinformatics 33, 3088–3090 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Morello G., et al. , T cells expressing receptor recombination/revision machinery are detected in the tumor microenvironment and expanded in genomically over-unstable models. Cancer Immunol. Res. 9, 825–837 (2021). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Single-cell RNAsequencing data have been deposited in ENA (PRJEB66123) (93).