Abstract
Evidence is presented that the previously cloned type I duck interferon (DuIFN) cDNA encodes a homologue of mammalian interferon-alpha (IFN-α). Recombinant DuIFN-α was used to study the inhibition of duck hepatitis B virus (DHBV) replication in primary hepatocytes in order to determine the IFN-sensitive steps of the virus replication cycle. IFN-treated cells accumulated two- to threefold-lower amounts of viral RNA transcripts early during infection, when IFN was added before virus. This reduction was not due to inhibition of virus entry since initial covalently closed circular DNA levels were not decreased in IFN-treated cells. Interestingly, the inhibitory effect of IFN on viral RNA levels was not observed in cells infected with a mutant DHBV that fails to synthesize core protein, suggesting that an uncharacterized core protein-mediated enhancing effect is blocked by IFN. When IFN was added at 4 days postinfection, encapsidated viral RNA pregenomes disappeared from infected cells within 3 days. This depletion was not simply due to conversion of pregenomes to DNA since depletion was not blocked by phosphonoformic acid, an inhibitor of the viral reverse transcriptase. The intracellular concentration of intact nucleocapsids was reduced, suggesting that in the presence of IFN pregenome-containing capsids were selectively depleted in hepatocytes. Thus, two steps in DHBV replication that involve the viral core protein were inhibited by DuIFN-α.
Interferons (IFNs) are a group of polypeptides that are secreted from a variety of eukaryotic cells in response to external signals. They are classified into two groups, designated type I and type II. Type I IFNs were first recognized by their ability to render cells resistant to infection by a wide variety of viruses. Type II IFN, or IFN-γ, which exhibits antiviral and macrophage-activating activity, is produced primarily by T lymphocytes and natural killer cells during an immune response. IFNs exhibit antiviral activity through binding to specific cell surface receptors, thereby altering the expression of a specific set of responsive genes. The combined action of the corresponding gene products generates the antiviral state (reviewed in references 7, 10, and 29). The major type I IFNs are IFN-α and IFN-β, which are produced primarily in response to virus infection. Type I IFNs have been useful in the treatment of a number of chronic virus infections, including hepatitis B of humans (15).
The mechanism by which type I IFNs inhibit replication of viruses has been the focus of many studies (for a review, see reference 29), and several reports have demonstrated that IFN-α inhibits the replication of hepatitis B virus (HBV) in vitro (6, 20, 28, 35, 37) as well as in vivo. In addition, type I IFN expression is associated with suppression of HBV replication in vivo in a HBV transgenic mouse model in response to a variety of experimental treatments (2, 3, 12, 13). IFN-α has been shown to be produced by mononuclear cells and some fibroblasts in the liver during chronic HBV infection (18), and it may be active in modulating HBV replication.
Previously we reported that a recombinant type I IFN of ducks (DuIFN) inhibited the replication of the avian hepadnavirus duck hepatitis B virus (DHBV) in vitro and in vivo (14, 30). This animal model offers a unique opportunity to identify the steps of the hepadnavirus replication cycle which are sensitive to IFN. In this report, we first provide evidence that the cloned DuIFN is the counterpart of mammalian IFN-α and thus belongs to the same family of IFNs that has been shown to be effective in inducing remissions of chronic viral hepatitis B in patients.
We further show that treatment of primary duck hepatocytes with DuIFN-α leads to a decrease in total viral transcript levels produced early after infection. In addition, we found that treatment of infected hepatocytes results in the disappearance of the pool of pregenome-containing nucleocapsids, resulting in a gradual depletion of replicative viral DNA intermediates from the cell. IFN treatment had no apparent effect on the earliest viral replication steps or on virus release from infected cells.
MATERIALS AND METHODS
Animals.
Day-old ducklings were purchased from Metzer Farms (Redlands, Calif.). Blood was obtained from a leg vein and assayed for DHBV DNA by dot blot hybridization. Only DHBV-free ducklings were used for the experiments described here. All animal procedures were conducted according to guidelines published by the National Institutes of Health for the humane care and use of laboratory animals.
IFN induction in ducks.
The imidazoquinolinamine immunoenhancer S-28463 (27, 34) was obtained from M. A. Tomai (3M Pharmaceuticals). A solution containing S-28463 at a dose of 0.3 or 3 mg/kg of body weight was administered orally in a volume of 1 ml to ducklings at 3 weeks of age. Two ducklings were left untreated. Animals were sacrificed 2 h after drug administration, and tissues were snap-frozen in liquid nitrogen and stored at −80°C.
Primary duck hepatocytes and infection with DHBV.
Primary hepatocytes were prepared from 1- to 2-week-old DHBV-free ducklings by perfusion of the liver with collagenase as described previously (36). Cells were suspended in Leibowitz-15 medium (Gibco) supplemented with 5% fetal bovine serum, 0.5 g of glucose per liter, 15 mM HEPES, 10−5 M dexamethasone (Sigma), 1 mg of insulin (Sigma) per liter, 5 × 104 U of penicillin per liter, 50 mg of streptomycin per liter, and 10 ml of amphotericin B (Fungizone; Gibco) per liter. Hepatocytes were seeded into 60-mm-diameter dishes such that they reached confluence the following day, when the medium was exchanged for Leibowitz-15 medium supplemented with 1% dimethyl sulfoxide instead of fetal bovine serum (26). In some experiments, phosphonofurmic acid (PFA) was added to the medium at a concentration of 1 mM. No cytotoxic effects of PFA were evident microscopically in any of the experiments described here.
The DHBV strain used in this study was that sequenced by Mandart et al. (23). Virus particles containing a DHBV mutant genome, R8, defective in capsid protein production were obtained from supernatants of the chicken hepatoma cell line LMH (5, 19), cotransfected with an R8 expression vector and a capsid protein expression vector as previously described (16, 32). DHBV used to infect 2-day-old cultures was obtained from supernatants of the stable DHBV-transformed LMH cell line D2 (11). Virus particles were concentrated from supernatants by precipitation with 10% polyethylene glycol 8000 (33). Concentrated virus (ca. 1 × 108 to 2 × 108 viral genomes per 60-mm-diameter dish) was added to the hepatocyte culture medium, where it remained for 16 to 24 h before the inoculum was replaced with fresh medium.
Production of DuIFN-α in COS7 cells.
DuIFN-α was produced by using a eukaryotic expression vector as previously described (30). A fragment spanning the entire open reading frame of DuIFN-α was cloned into the plasmid pcDNA1 (Invitrogen, Carlsbad, Calif.), and the DNA was transfected into COS7 cells by calcium phosphate precipitation. At 72 h posttransfection, the culture supernatants were harvested and cleared of cell debris by centrifugation. The antiviral titer of these supernatants was determined by the cytopathic effect inhibition assay as described previously (30). Briefly, duck embryo fibroblasts were stimulated with twofold serial dilutions of DuIFN-α for 15 h before challenge with vesicular stomatitis virus at a multiplicity of 1. IFN titers are expressed as reciprocals of the dilutions that resulted in 50% protection against virus induced cell destruction.
Analysis of viral nucleic acids.
Selective extraction of covalently closed circular (cccDNA) and replicative intermediates was performed as previously described (32). Briefly, after lysis of hepatocytes in 1 ml of lysis buffer (50 mM Tris-HCl [pH 8.0], 1 mM EDTA, 150 mM NaCl, 1% sodium dodecyl sulfate [SDS]), cellular DNA and viral protein-bound DNA were precipitated by addition of 0.25 ml of 2.5 M KCl to form an insoluble protein-potassium dodecyl sulfate complex. After centrifugation, the cccDNA in the supernatant was isolated by phenol-chloroform (1:1) extraction and ethanol precipitation. The cccDNA fractions were digested briefly with 1 μg of RNase A per ml and analyzed by Southern blotting. Replicative intermediates were recovered from the pellet by dissolving in 1 ml of Tris-EDTA containing 0.5 mg of Pronase (Sigma) per ml. The dissolved pellet was incubated 1 h at 37°C to allow complete digestion of proteins to occur, and nucleic acids were recovered after extraction with phenol-chloroform and ethanol precipitation.
Assay of enveloped virus from infected hepatocyte cultures.
Viral DNA present in enveloped virus particles was assayed by the pronase-DNase I method (22). Briefly, 0.4 ml of culture supernatant was adjusted to 75 mM with Tris-HCl (pH 8.0), cell debris was removed by centrifugation, and the clarified supernatant was incubated at 37°C with 0.5 mg of pronase per ml for 1 h to degrade free nucleocapsids but not enveloped virus. Free viral DNA released from degraded nucleocapsids was removed by the addition of magnesium acetate to a final concentration of 6 mM and incubation with DNase I at a final concentration of 50 μg per ml. After 30 min at 37°C, the digestions were adjusted to 10 mM EDTA and 0.5% SDS and incubated at 37°C for 1 h to allow complete digestion of all proteins. DNA was recovered by phenol extraction and ethanol precipitation in the presence of 50 μg of carrier DNA per ml. Samples were digested briefly with 1 μg of RNase A per ml before agarose gel electrophoresis.
Analysis of viral RNA.
Encapsidated RNA was selectively extracted according to a method kindly provided by Stefan Wieland (The Scripps Research Institute, La Jolla, Calif.). Hepatocytes from a 60-mm-diameter dish were lysed in 600 μl of lysis buffer (50 mM Tris-HCl [pH 7.4], 1 mM EDTA, 150 mM NaCl, 1% Nonidet P-40), and the nuclei were removed by microcentrifugation. Free RNA was removed from one half of the cleared lysate by digestion with 4 U of micrococcus nuclease (Pharmacia) at 37°C for 15 min in the presence of 5 mM CaCl2. The protected encapsidated viral RNA and the remaining lysate were subjected to RNA extraction to yield encapsidated and total cytoplasmic RNA fractions.
RNA was extracted by the acid guanidinium isothiocyanate-phenol-chloroform method (4). Polyadenylated RNA was prepared from total RNA by using an Oligotex mRNA isolation kit from Qiagen Inc. (Santa Clarita, Calif.). RNA (10 μg per lane) was analyzed by electrophoresis through 1.2% agarose gels containing 10% (vol/vol) formaldehyde in electrophoresis buffer (20 mM morpholinepropanesulfonic acid, 5 mM sodium acetate, 1 mM EDTA [pH 7.0], 3.5% formaldehyde) and transferred to nylon membranes. A DNA fragment containing the DuIFN gene was labeled with [α-32P]dCTP by conventional nick translation and used as probe for the detection of IFN-α transcripts. Blots were washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 0.5% SDS at 65°C for 1 h and then in 0.1× SSC containing 0.5% SDS at 65°C for 30 min. A DNA fragment comprising the DHBV genome was labeled similarly and was used as probe for the detection of viral transcripts.
Extraction and assay of intact viral capsids.
Analysis of assembled capsids by agarose gel electrophoresis was performed as described previously (1). In brief, hepatocytes were lysed by addition of 0.5 ml of lysis buffer (50 mM Tris-HCl [pH 7.4], 1 mM EDTA, 0.2% Nonidet P-40) and incubation at 37°C for 10 min. Nuclei were removed by microcentrifugation, and aliquots (5 μl) were subjected to nondenaturing agarose gel electrophoresis, which separates assembled core proteins from unassembled proteins due to different migration properties. Proteins were transferred from the gel to nitrocellulose filters by capillary blotting with TNE (10 mM Tris-HCl [pH 7.4], 1 mM EDTA, 150 mM NaCl), and the filters were washed for 10 min in water and air dried. Capsids were detected by incubating the filters with a rabbit anti-core antibody followed by 125I-labeled goat anti-rabbit immunoglobulin G, as previously described (1).
RESULTS
The synthetic cytokine inducer S-28463 activates the DuIFN gene in vivo.
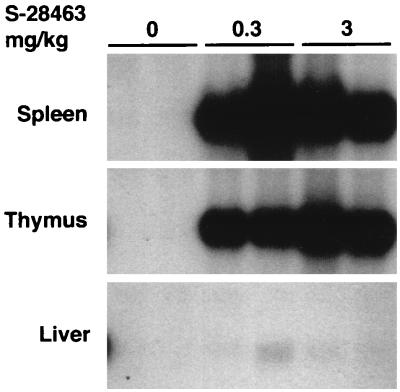
The imidazoquinolinamine drug imiquimod and its derivative S-28463 have been successfully used to induce IFN and other cytokines in mammals (27, 34). Since IFN-α genes, but not IFN-β genes, are selectively induced in mammals by this group of compounds (38), they have been recently used to classify the two subtypes of chicken type I IFNs, ChIFN1 and ChIFN2, as ChIFN-α and ChIFN-β, respectively (31). The recently cloned type I DuIFN shares roughly the same amino acid sequence homology with both type I ChIFNs (30), and therefore we used induction by S-28463 as a basis for its classification. We administered S-28463 to ducklings and assayed DuIFN transcripts in various tissues, using a radiolabeled DNA fragment from the type I DuIFN gene as Northern hybridization probe. DuIFN transcripts were detected at high levels in spleens and thymuses of ducks treated for 2 h with S-28463, whereas this RNA was not found in the corresponding organs of untreated control ducks (Fig. 1). Low amounts of induced transcript were also detected in the liver of one drug-treated duck. These results indicated that the cloned IFN gene represents the duck counterpart of mammalian genes encoding IFN-α and that cells of thymus and spleen are very effective producers of DuIFN-α in animals treated with S-28463.
FIG. 1.
Accumulation of DuIFN-α transcripts in organs of ducks treated with a synthetic cytokine inducer. Groups of two ducks were treated orally with 0.3 or 3 mg of S-28463 per kg or were left untreated. Animals were sacrificed 2 h after drug administration, and RNA was prepared from spleens, thymuses, and livers. Samples of 10 μg of total RNA were analyzed by Northern blotting using the cloned duck type I IFN as a probe.
DuIFN-α inhibited the accumulation of DHBV DNA in primary duck hepatocyte cultures in a concentration-dependent manner.
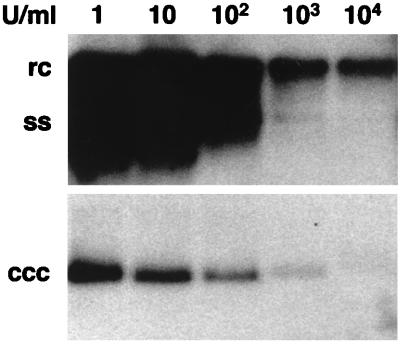
In initial experiments, we observed that productive infection of embryonic duck hepatocytes by DHBV was substantially reduced in the presence of recombinant DuIFN-α (100 U/ml) produced in Escherichia coli (30). This IFN also reduced or eliminated DHBV replication in infected ducklings after 10 days of treatment (14). To better characterize the mode of IFN action against DHBV replication, the first experiment in this study was aimed at evaluating the most effective concentration of DuIFN-α in infected primary duck hepatocytes. Since E. coli-produced recombinant DuIFN-α tended to precipitate upon storage (29a), DuIFN-α produced in COS7 cells was used for this and all following experiments. Hepatocyte cultures were treated with various doses of DuIFN-α for 3 days beginning at 1 day postinfection. At the end of treatment, the cells were harvested and assayed for viral replication as indicated by accumulation of two forms of intracellular viral DNA, cccDNA and DNA replicative intermediates. Dose-dependent inhibition of both cccDNA and replicative intermediate accumulation was seen at IFN concentrations of up to 103 U per ml (Fig. 2). Slightly lower viral DNA levels were observed with 104 U per ml, but we also found that cultures treated with this high IFN dose contained lower amounts of total nucleic acids (data not shown). To ensure that the observed effect in IFN-treated cultures was due to its antiviral effect and not to an antiproliferative or toxic effect, we decided to use 103 U of IFN per ml in all following experiments. Hepadnaviruses replicate their circular DNA genomes through a process of transcription and reverse transcription (reviewed in references 9 and 25). Reverse transcription is carried out within nucleocapsids by a viral polymerase. Nucleocapsids containing viral DNA are then assembled into envelopes and secreted from the cell. The major steps in replication that were assayed in these studies are depicted in Fig. 3.
FIG. 2.
The inhibitory effect of DuIFN-α on DHBV DNA accumulation in primary duck hepatocyte cultures is concentration dependent. Recombinant DuIFN-α was added to the cultures 1 day after infection. Culture medium containing the indicated concentrations of IFN was exchanged daily until day 4, when cells were harvested and analyzed for the presence of cccDNA and replicative intermediates. The migration positions of relaxed circular (rc), covalently closed circular (ccc), and single-stranded (ss) DHBV DNAs are indicated.
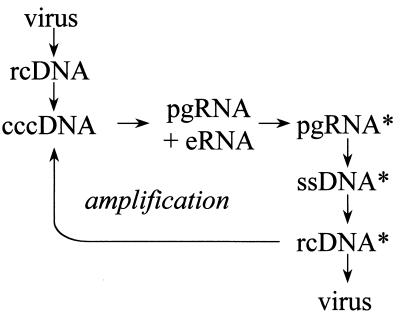
FIG. 3.
The replication pathway of hepadnaviruses. The pathway is shown from top to bottom. Infection is initiated by transfer of the viral DNA genome, an rcDNA molecule 3 kb in length, into the nucleus and conversion to cccDNA. cccDNA is the template for transcription of mRNAs for the envelope proteins (eRNA) and of genomic RNAs or pregenomes (pgRNA). Pregenomes are encapsidated by the viral capsid protein with the viral reverse transcriptase to form pregenome-containing capsids (pgRNA*) and are subsequently reverse transcribed within the nucleocapsids into single-stranded minus-strand DNA (ssDNA*). Pregenomes are degraded during the process of reverse transcription. Minus-strand DNA is used as the template for synthesis of the plus-strand DNA to generate rcDNA-containing capsids (rcDNA*). Early during infection, the capsids containing rcDNA molecules are utilized to produce additional molecules of cccDNA in the nucleus, a process referred to as cccDNA amplification. Later during infection, the number of cccDNA molecules stabilizes, and nucleocapsids containing rcDNA are exclusively assembled into envelopes and secreted from the cell as progeny virus particles.
DuIFN-α did not prevent the initial steps of the DHBV replication cycle.
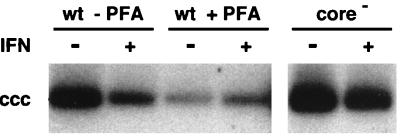
Early stages of DHBV virus replication include virus attachment, entry, and conversion of the genome to cccDNA. To identify the DuIFN-α sensitive steps in viral replication, we first examined whether IFN treatment had any effect on the initiation of infection. We compared the direct conversion of viral relaxed circular DNA (rcDNA) from the infecting virus to cccDNA during infection in untreated and DuIFN-α-treated cells. Synthesis of progeny cccDNA by amplification was inhibited either by treating the cells during infection with PFA, a specific inhibitor of the viral polymerase (8), or by infecting cells with virus particles that contained a mutated viral genome, R8, defective in the production of capsid protein due to a frameshift at codon 2 of the core open reading frame (16). These particles are unable to carry out viral DNA synthesis in infected cells, although viral cccDNA is produced from the infecting genome. At 2 days postinfection, cccDNA was extracted from the cells and analyzed by Southern blot hybridization (Fig. 4). In the absence of IFN, cells infected with both wild-type and mutant viruses contained viral cccDNA, indicating that conversion of some of the rcDNA genome of the infecting virus had occurred. During the first 2 days of infection, IFN treatment had little if any effect on the amount of cccDNA formed in either PFA-treated cells or cells infected by the mutant virus (Fig. 4). These results confirmed our previous findings, based on PCR analysis, that IFN treatment did not prevent the initial conversion of rcDNA from the input virus into cccDNA (30). Thus, viral attachment, penetration, and cccDNA formation were not influenced by DuIFN-α.
FIG. 4.
DuIFN-α does not inhibit cccDNA formation from input virus. DuIFN-α was added to the hepatocyte cultures 16 h prior to infection with either wild-type DHBV (wt) or a mutant virus, R8, defective in the production of core protein (core−). PFA (1 mM) was added to the indicated cultures on the day of infection. Medium was changed 1 day after infection and supplemented with DuIFN-α and PFA as indicated. All cells were harvested at 2 days postinfection and analyzed for cccDNA by Southern blotting.
DuIFN-α inhibited the accumulation of intracellular DHBV DNA but did not affect virus release.
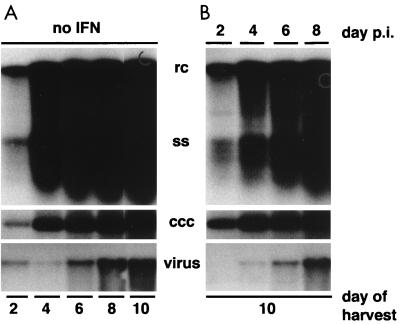
We next determined the kinetics of progeny virus production in DuIFN-α-treated cells. For this experiment, we infected hepatocyte cultures with DHBV and assayed the levels of intracellular viral DNA and rate of release of enveloped virus in untreated cells at various times after infection. At these same time points, DuIFN-α was added to a parallel set of infected cultures in which incubation with IFN was continued until day 10 postinfection. The levels of intracellular viral DNA and the rate of virus release in these cultures were compared to the levels measured at the start of IFN treatment. The results of this experiment are shown in Fig. 5 and presented graphically in Fig. 6.
FIG. 5.
Effect of DuIFN-α on intracellular viral DNA accumulation and virus release. DuIFN-α was added to hepatocyte cultures at the indicated days after infection. Medium was changed every other day, and DuIFN-α was added where appropriate. All cultures were harvested 24 h after the last medium change. Viral DNA was extracted from cells and supernatants and analyzed by Southern blotting. Intracellular viral DNA loaded onto the gel was equivalent to 1/6 of a 60-mm-diameter dish; the amount of virus loaded was equivalent to 1/10 of the medium from one plate. Panels represent the amounts of the DNA replicative intermediates, relaxed circular (rc) and single-stranded (ss) DHBV DNA, covalently closed circular DNA (ccc), and enveloped virus (virus) isolated from cultures at the indicated times post postinfection (p.i.).
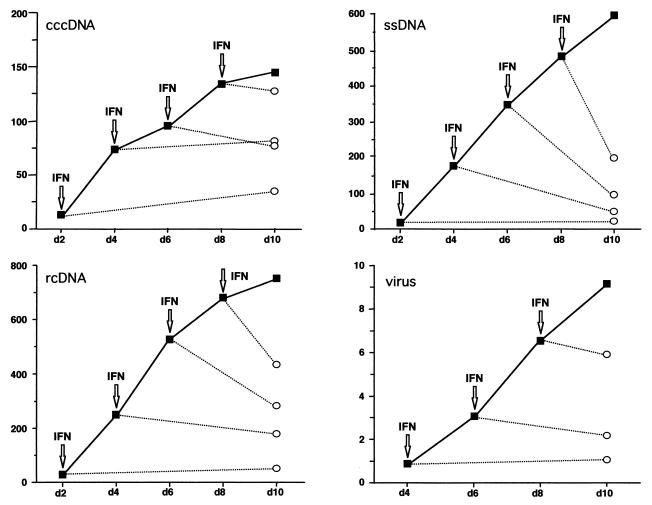
FIG. 6.
Effect of DuIFN-α on intracellular viral DNA accumulation and virus release. Southern blots from the experiment shown in Fig. 5 were analyzed quantitatively by phosphorimaging. Arrows indicate initial addition of DuIFN-α; open symbols indicate values of DuIFN-α-treated cultures; closed symbols indicate values of untreated cultures. Values on the y axis represent arbitrary units.
In the absence of DuIFN-α, accumulation of cccDNA and replicative intermediates continued throughout the 10-day duration of the experiment, and during this period there was a proportional increase in the rate of virion release (Fig. 5A). Initial addition of DuIFN-α at 2, 4, 6, or 8 days postinfection blocked any further increase in accumulation of intracellular viral DNA or release of virus, measured at day 10 postinfection (Fig. 5B). The ability of infected cells to maintain the rate of virus secretion in the presence of IFN indicated that steps involved in virus particle production, such as envelopment and export of enveloped virus particles, were not affected by treatment with DuIFN-α. Quantitative analysis of the intracellular viral DNA contents (Fig. 6) revealed that cccDNA levels remained unchanged during IFN treatment. In contrast, rcDNA and single-stranded DNA (ssDNA) were depleted from infected cells when IFN was added at day 4, 6, or 8 postinfection. The changes in the intracellular levels of each of the two types of replicative DNA intermediates are consistent with their continued conversion to more mature forms in the presence of IFN and, finally, their secretion from the cell. The observation that ssDNA levels decreased more than the levels of rcDNA suggests that the replenishment of ssDNA from encapsidated pregenomes was blocked in IFN-treated cells and that rcDNA lost due to virus secretion continued to be replenished from ssDNA. Together, these results indicate that IFN-sensitive steps in the viral life cycle lie between the formation of cccDNA and production of ssDNA.
RNA-containing viral nucleocapsids were depleted in DuIFN-α-treated cells.
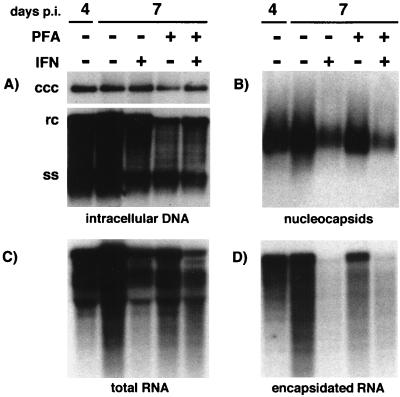
We next examined whether IFN treatment blocked steps occurring prior to reverse transcription. We started treatment of DHBV-infected hepatocytes beginning at 4 days postinfection to allow cccDNA amplification to occur and then maintained the cells in the presence of IFN for 3 days to look for evidence of depletion of viral transcripts or pregenome-containing capsids. PFA was added to a parallel set of cultures in order to test whether depletion of RNA-containing viral nucleocapsids would be due to maturation to ssDNA-containing capsids. The amounts of cccDNA, total RNA, encapsidated pregenome, replicative DNA intermediates, and total capsids were assayed at the start of treatment, at 4 days postinfection, and at 7 days postinfection. IFN treatment for 3 days caused only a modest decrease in replicative intermediates (Fig. 7A) and in total viral RNA (Fig. 7C). In contrast, encapsidated full-length pregenome RNA was almost completely eliminated from IFN-treated cells (Fig. 7D). The reduction in the encapsidated pregenome was accompanied by a corresponding reduction in the amount of total capsids (Fig. 7B), presumably reflecting the selective loss of those capsids that contained pregenomic RNA, but not of those in which DNA synthesis was already occurring. The loss of pregenome-containing capsids was not prevented by the presence of PFA during IFN treatment, suggesting that depletion of these capsids was not due to their conversion to DNA-containing capsids. In the absence of IFN, pregenome-containing capsids were maintained at a fairly constant level during PFA treatment. They were not observed to accumulate as one might have expected if conversion of pregenomic RNA to ssDNA by reverse transcription is blocked. The lack of effect of PFA on pregenome-containing capsids in this experiment was not due to a lack of inhibition of reverse transcription, as PFA effectively inhibited cccDNA amplification (Fig. 8),, a process that occurs through the reverse transcription pathway.
FIG. 7.
RNA-containing capsids are depleted from DuIFN-α-treated infected hepatocyte cultures. DHBV-infected hepatocytes were treated continuously with DuIFN-α and PFA (1 mM) beginning at 4 days postinfection (p.i.). The medium was changed daily. At 7 days after infection, the cells were harvested and analyzed for replicative intermediates and cccDNA (A), nucleocapsids (B), total RNA (C), and encapsidated RNA (D). The amount of viral DNA loaded corresponds to 1/5 of a 60-mm-diameter dish, the amount of nucleocapsids loaded corresponds to 1/100 of a dish, and the amounts of total and encapsidated RNA each correspond to 1/10 of a dish.
FIG. 8.
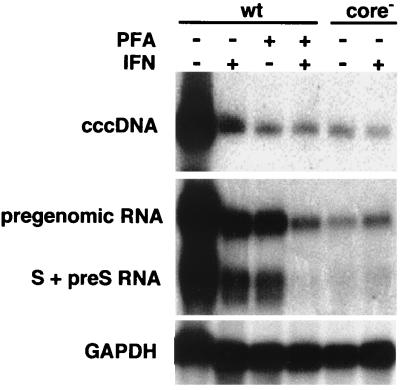
Effect of DuIFN-α on viral transcript levels. DuIFN-α was added to hepatocyte cultures 16 h prior to infection with either wild-type DHBV (wt) or a mutant virus, R8, defective in the production of core protein (core−). PFA (1 mM) was added to the indicated cultures on the day of infection. The medium was changed 1 day after infection and supplemented with DuIFN-α and PFA and was then renewed every other day until all cultures were harvested at 4 days postinfection. Parallel cultures were analyzed for cccDNA by Southern blotting (upper panel) and for polyadenylated viral RNA by Northern blotting (middle panel) using radiolabeled DHBV DNA as probe. Northern blots were rehybridized using the chicken glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA as probe (lower panel). cccDNA loaded onto the gel was equivalent to 1/4 of a 60-mm-diameter dish, and RNA loaded was equivalent to 9/10 of a dish.
DuIFN-α treatment reduced the level of viral transcripts during initiation of infection.
In an earlier experiment (Fig. 4), we showed that IFN treatment did not inhibit the conversion of rcDNA from the infecting virus into cccDNA. To test whether IFN affected the production of viral RNA from the initial cccDNA molecule, we incubated hepatocytes with or without IFN for 16 h, before they were infected either with wild-type virus or with the capsid protein deficient-mutant R8. Amplification of cccDNA was inhibited in the wild-type virus-infected cells by the addition of PFA at the time of infection, and IFN treatment was continued. At 4 days postinfection, the levels of viral pregenomic and envelope mRNAs and cccDNA were determined. Presence of PFA, IFN, or both agents and absence of the capsid protein (cells infected with the R8 mutant virus) all inhibited amplification of viral cccDNA (Fig. 8, upper panel). In cells in which PFA treatment inhibited new cccDNA synthesis, IFN treatment resulted in two- to threefold-reduced levels of pregenomic viral RNA present at 4 days postinfection (Fig. 8, middle panel). In contrast, IFN treatment of cells infected by the capsid-deficient mutant had no effect on RNA levels. Comparison of the ratio of viral RNA to cccDNA in wild-type virus-infected cells with the ratio in cells infected with the mutant R8 virus suggests that the expression of capsid protein enhanced the accumulation of viral transcripts and that IFN inhibited this enhancement. Capsid protein did not cause enhanced RNA accumulation solely by sequestration of RNA into capsids, since envelope mRNAs, which are not encapsidated, also were present in higher amounts in cells expressing capsid protein. The inhibition of viral RNA accumulation by IFN was also not due solely to reduced encapsidation of viral pregenomes because accumulation of S and pre-S RNAs was also inhibited. These results suggest a role of core protein in the establishment of viral transcript pools.
DISCUSSION
IFN-mediated inhibition of DHBV replication was observed to occur at two steps. The earliest effect of IFN was manifested as a reduction in the amount of polyadenylated viral RNAs transcribed from the input viral genome. This effect of IFN appeared to be due to inhibition of an apparent enhancing effect of the viral capsid protein, because infection with a capsid-deficient mutant virus produced similarly low levels of viral RNAs that were not affected further by IFN. A role for newly synthesized capsid protein in the early accumulation of nonencapsidated viral transcripts has previously not been described. Our data suggest that this novel function of the capsid protein is a target of DuIFN-α. These results contrast with previous reports in which human type I IFNs were reported to cause suppression of reporter gene expression from HBV promoters in the absence of the HBV core protein (28, 35).
In agreement with results reported previously for IFN-α inhibition of HBV replication in stably transformed cells (6), the strongest effect of IFN treatment resulted in a reduction of the levels of encapsidated viral RNA. More specifically, we observed the virtual depletion of capsids that contained full-length pregenomes from DuIFN-α-treated hepatocytes. The absence of this immediate precursor to viral DNA can probably account for the inhibition of subsequent accumulation of intracellular viral DNA forms and their gradual decline after IFN addition (Fig. 5 and 6). DNA synthesis per se was not apparently inhibited by IFN treatment, since intracellular viral DNA became enriched in the more mature species and virus continued to be secreted at an undiminished rate. We believe that this continued maturation and secretion of virus accounts for the reduction in intracellular viral DNA that we observed; however, the effect of IFN on DNA synthesis was not measured directly, and so we cannot be certain of this interpretation. The lack of strong inhibitory effects of IFN on virus production when IFN was added after infection had already been established is consistent with previous reports of modest inhibition of HBV by IFN in cells replicating HBV in vitro (6, 20, 21, 37).
However, the disappearance of pregenome-containing viral nucleocapsids from IFN-treated cells cannot be attributed to their conversion into more mature forms because, first, the reduction of pregenome-containing capsids was accompanied by a reduction in the total amount of capsids. A concomitant reduction in total capsids would not be expected to occur if the pregenome-containing capsids were simply converted to DNA-containing capsids because the total number of capsids would not change. Second, the addition of an inhibitor of viral DNA synthesis, PFA, which is expected to block conversion of pregenome-containing capsids into DNA-containing capsids, had no effect on the IFN-induced depletion of either encapsidated pregenomes or total capsids. These results suggest that in the presence of IFN, pregenome-containing capsids were depleted in the cells by a novel mechanism.
Curiously, the addition of PFA alone did not result in the accumulation of pregenome-containing capsids even when pregenomic RNA was still abundant in treated cells. Pregenomic RNA serves as the mRNA for translation of both capsid protein and P protein, the two proteins required for encapsidation of RNA and subsequent DNA synthesis (17). Since viral DNA synthesis is not required for RNA packaging (39), we expected that pregenomes would continue to be transcribed and encapsidated in the presence of PFA. The lack of accumulation of pregenome-containing capsids suggests that in the presence of PFA, these capsids are eliminated by an alternative pathway that does not require viral DNA synthesis, e.g., by destabilization or secretion from the cell. Whether such an alternative pathway operates in the absence of PFA is not known. If this alternative pathway exists, then the disappearance of pregenome-containing capsids from IFN-treated cells could be due to inhibition of encapsidation, which would result in the gradual elimination of those capsids that were already formed before the onset of IFN treatment. Alternatively, IFN may directly induce the destabilization or secretion of pregenome-containing capsids.
Both steps in DHBV replication that were found to be sensitive to IFN-induced inhibition involve the capsid protein, i.e., a capsid protein-dependent enhancement of viral RNA accumulation by an unknown mechanism, and encapsidation or turnover of pregenome-containing capsids. Thus, inhibition of both steps might be due to a single effect of IFN treatment on the capsid protein. The nature of such a hypothetical effect is not known at present.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants R37 CA40489 (to F.V.C.) and CA42542 (to J.S.) from the National Institutes of Health and a grant from the Deutsche Forschungsgemeinschaft (DFG) (to P.S.). U.S. was partially supported by a fellowship from the DFG (Schu 1152/1-1).
We are grateful to Stefan Weiland for advice on the measurement of encapsidated pregenome RNA. We thank Jacquelyn Moorhead and Amy Brown for excellent technical assistance and Jennifer Newmann for assistance with manuscript preparation.
Footnotes
Manuscript no. 12063-MEM from The Scripps Research Institute.
REFERENCES
- 1.Calvert J, Summers J. Two regions of an avian hepadnavirus RNA pregenome are required in cis for encapsidation. J Virol. 1994;68:2084–2090. doi: 10.1128/jvi.68.4.2084-2090.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cavanaugh V J, Guidotti L G, Chisari F V. Inhibition of hepatitis B virus replication during adenovirus and cytomegalovirus infections in transgenic mice. J Virol. 1998;72:2630–2637. doi: 10.1128/jvi.72.4.2630-2637.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cavanaugh V J, Guidotti L G, Chisari F V. Interleukin-12 inhibits hepatitis B virus replication in transgenic mice. J Virol. 1997;71:3236–3243. doi: 10.1128/jvi.71.4.3236-3243.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chomczynski P, Sacchi N. Single step method of RNA isolation by acid guanidinium thiocyanate phenol chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 5.Condreay L D, Wu T T, Aldrich C E, Delaney M A, Summers J, Seeger C, Mason W S. Replication of DHBV genomes with mutations at the sites of initiation of minus- and plus-strand DNA synthesis. Virology. 1992;188:208–216. doi: 10.1016/0042-6822(92)90751-a. [DOI] [PubMed] [Google Scholar]
- 6.Davis M G, Jansen R W. Inhibition of hepatitis B virus in tissue culture by alpha interferon. Antimicrob Agents Chemother. 1994;38:2921–2924. doi: 10.1128/aac.38.12.2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeMaeyer E, DeMaeyer-Guignard J. Interferons and other regulatory cytokines. New York, N.Y: Wiley-Interscience; 1988. [Google Scholar]
- 8.Fourel I, Saputelli J, Schaffer P, Mason W S. The carbocyclic analog of 2′-deoxyguanosine induces a prolonged inhibition of duck hepatitis B virus DNA synthesis in primary hepatocyte cultures and in the liver. J Virol. 1994;68:1059–1065. doi: 10.1128/jvi.68.2.1059-1065.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ganem D. Hepadnaviridae and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2703–2737. [Google Scholar]
- 10.Gilmour K C, Reich N C. Signal transduction and activation of gene transcription by interferons. Gene Expr. 1995;5:1–18. [PMC free article] [PubMed] [Google Scholar]
- 11.Gong S S, Jensen A D, Wang H, Rogler C E. Duck hepatitis B virus integrations in LMH chicken hepatoma cells: identification and characterization of new episomally derived integrations. J Virol. 1995;69:8102–8108. doi: 10.1128/jvi.69.12.8102-8108.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guidotti L G, Borrow P, Hobbs M V, Matzke B, Gresser I, Oldstone M B, Chisari F V. Viral cross talk: intracellular inactivation of the hepatitis B virus during an unrelated viral infection of the liver. Proc Natl Acad Sci USA. 1996;93:4589–4594. doi: 10.1073/pnas.93.10.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guidotti L G, Matzke B, Chisari F V. Hepatitis B virus replication is cell cycle independent during liver regeneration in transgenic mice. J Virol. 1997;71:4804–4808. doi: 10.1128/jvi.71.6.4804-4808.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heuss L T, Heim M H, Schultz U, Wissmann D, Offensperger W B, Staeheli P, Blum H E. Biological efficacy and signal transduction through STAT proteins of recombinant duck interferon in duck hepatitis B virus infection. J Gen Virol. 1998;79:2007–2012. doi: 10.1099/0022-1317-79-8-2007. [DOI] [PubMed] [Google Scholar]
- 15.Hoofnagle J H. Therapy of viral hepatitis. Digestion. 1998;59:563–578. doi: 10.1159/000007532. [DOI] [PubMed] [Google Scholar]
- 16.Horwich A L, Furtak K, Pugh J, Summers J. Synthesis of hepadnavirus particles that contain replication-defective duck hepatitis B virus genomes in cultured HuH7 cells. J Virol. 1990;64:642–650. doi: 10.1128/jvi.64.2.642-650.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang M J, Summers J. Infection initiated by the RNA pregenome of a DNA virus. J Virol. 1991;65:5435–5439. doi: 10.1128/jvi.65.10.5435-5439.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jilbert A R, Burrell C J, Gowans E J, Hertzog P J, Linnane A W, Marmion B P. Cellular localization of alpha interferon in hepatitis B virus infected liver tissue. Hepatology. 1986;6:957–961. doi: 10.1002/hep.1840060524. [DOI] [PubMed] [Google Scholar]
- 19.Kawaguchi T, Nomura K, Hirayama Y, Kitagawa T. Establishment and characterization of a chicken hepatocellular carcinoma cell line. Cancer Res. 1987;47:4460–4464. [PubMed] [Google Scholar]
- 20.Korba B E. In vitro evaluation of combination therapies against hepatitis B virus replication. Antiviral Res. 1996;29:49–51. doi: 10.1016/0166-3542(95)00915-9. [DOI] [PubMed] [Google Scholar]
- 21.Lampertico P, Malter J S, Gerber M A. Development and application of an in vitro model for screening anti hepatitis B virus therapeutics. Hepatology. 1991;13:422–426. [PubMed] [Google Scholar]
- 22.Lenhoff R J, Summers J. Construction of avian hepadnavirus variants with enhanced replication and cytopathicity in primary hepatocytes. J Virol. 1994;68:5706–5713. doi: 10.1128/jvi.68.9.5706-5713.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandart E, Kay A, Galibert F. Nucleotide sequence of a cloned duck hepatitis B virus genome: comparison with woodchuck and human hepatitis B virus sequences. J Virol. 1984;49:782–792. doi: 10.1128/jvi.49.3.782-792.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mason W S, Seal G, Summers J. Virus of Pekin ducks with structural and biological relatedness to human hepatitis B virus. J Virol. 1980;36:829–836. doi: 10.1128/jvi.36.3.829-836.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nassal M, Schaller H. Hepatitis B virus replication an update. J Viral Hepat. 1996;3:217–226. doi: 10.1111/j.1365-2893.1996.tb00047.x. [DOI] [PubMed] [Google Scholar]
- 26.Pugh J C, Summers J W. Infection and uptake of duck hepatitis B virus by duck hepatocytes maintained in the presence of dimethyl sulfoxide. Virology. 1989;172:564–572. doi: 10.1016/0042-6822(89)90199-2. [DOI] [PubMed] [Google Scholar]
- 27.Reiter M J, Testerman T L, Miller R L, Weeks C E, Tomai M A. Cytokine induction in mice by the immunomodulator imiquimod. J Leukoc Biol. 1994;55:234–240. doi: 10.1002/jlb.55.2.234. [DOI] [PubMed] [Google Scholar]
- 28.Romero R, Lavine J E. Cytokine inhibition of the hepatitis B virus core promoter. Hepatology. 1996;23:17–23. doi: 10.1002/hep.510230103. [DOI] [PubMed] [Google Scholar]
- 29.Samuel C E. Antiviral actions of interferon. Interferon regulated cellular proteins and their surprisingly selective antiviral activities. Virology. 1991;183:1–11. doi: 10.1016/0042-6822(91)90112-o. [DOI] [PubMed] [Google Scholar]
- 29a.Schultz, U. Unpublished data.
- 30.Schultz U, Kock J, Schlicht H J, Staeheli P. Recombinant duck interferon: a new reagent for studying the mode of interferon action against hepatitis B virus. Virology. 1995;212:641–649. doi: 10.1006/viro.1995.1522. [DOI] [PubMed] [Google Scholar]
- 31.Sick C, Schultz U, Munster U, Meier J, Kaspers B, Staeheli P. Promoter structures and differential responses to viral and nonviral inducers of chicken type I interferon genes. J Biol Chem. 1998;273:9749–9754. doi: 10.1074/jbc.273.16.9749. [DOI] [PubMed] [Google Scholar]
- 32.Summers J, Smith P M, Horwich A L. Hepadnavirus envelope proteins regulate covalently closed circular DNA amplification. J Virol. 1990;64:2819–2824. doi: 10.1128/jvi.64.6.2819-2824.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Summers J, Smith P M, Huang M J, Yu M S. Morphogenetic and regulatory effects of mutations in the envelope proteins of an avian hepadnavirus. J Virol. 1991;65:1310–1317. doi: 10.1128/jvi.65.3.1310-1317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomai M A, Gibson S J, Imbertson L M, Miller R L, Myhre P E, Reiter M J, Wagner T L, Tamulinas C B, Beaurline J M, Gerster J F, et al. Immunomodulating and antiviral activities of the imidazoquinoline S 28463. Antiviral Res. 1995;28:253–264. doi: 10.1016/0166-3542(95)00054-p. [DOI] [PubMed] [Google Scholar]
- 35.Tur-Kaspa R, Teicher L, Laub O, Itin A, Dagan D, Bloom B R, Shafritz D A. Alpha interferon suppresses hepatitis B virus enhancer activity and reduces viral gene transcription. J Virol. 1990;64:1821–1824. doi: 10.1128/jvi.64.4.1821-1824.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tuttleman J S, Pugh J C, Summers J W. In vitro experimental infection of primary duck hepatocyte cultures with duck hepatitis B virus. J Virol. 1986;58:17–25. doi: 10.1128/jvi.58.1.17-25.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ueda K, Tsurimoto T, Nagahata T, Chisaka O, Matsubara K. An in vitro system for screening anti hepatitis B virus drugs. Virology. 1989;169:213–216. doi: 10.1016/0042-6822(89)90057-3. [DOI] [PubMed] [Google Scholar]
- 38.Weeks C E, Gibson S J. Induction of interferon and other cytokines by imiquimod and its hydroxylated metabolite R 842 in human blood cells in vitro. J Interferon Res. 1994;14:81–85. doi: 10.1089/jir.1994.14.81. [DOI] [PubMed] [Google Scholar]
- 39.Wei Y, Tavis J E, Ganem D. Relationship between viral DNA synthesis and virion envelopment in hepatitis B viruses. J Virol. 1996;70:6455–6458. doi: 10.1128/jvi.70.9.6455-6458.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]