Abstract
Staphylococcus aureus is gaining worldwide attention because of its substantial impact on public health. The current study aimed to characterize S. aureus strains isolated from wild birds in the Kasur district of Punjab, Pakistan from 2021 to 2022. A total of one hundred samples were collected from five wild bird species. The samples were enriched, inoculated on selective agars and cultured for 24 hr at 37.00 ˚C. All isolates were verified by matrix-assisted laser desorption ionization–time-of-flight mass spectrometry (MALDI-TOF MS) and polymerase chain reaction (PCR) after Gram staining. Positive isolates were screened for phenotypic (Kirby-Bauer disk diffusion and minimum inhibitory concentration s), genotypic antibiotic resistance, and virulence genes. These samples yielded 30 (30.00%) S. aureus isolates, confirmed by polymerase chain reaction utilizing the 16S rRNA gene. Staphylococcus aureus was more prevalent in cloacal samples (16.00%) than oral samples (14.00%). Various S. aureus isolates showed varying degrees of resistance to three different antibiotics. Oxacillin (56.66%; n = 17) and tetracycline (33.33%; n = 10) showed the highest resistance rates with the lowest susceptibility (43.33%; n = 13). In contrast, vancomycin, rifampicin, linezolid, and daptomycin were 100% susceptible. Further disc diffusion study revealed resistance to tetracycline (33.33%), erythromycin (16.66%), and gentamicin (10.00%). The tetK gene was found in 33.33% of wild bird samples, while the ermA gene was found in 16.66% of samples. The aacA-D gene was only found in three (10.00%) isolates. None of the isolates tested positive for virulence genes. In conclusion, S. aureus is carried by wild birds in this area, posing a potentail threat to both humans and animals.
Key Words: accA-D, Antibiotic resistance, ermA, Staphylococcus aureus, tetK
Introduction
Birds are a globally distributed group of animals known for their diverse habitats and significant ecological contributions among vertebrates. Their ecological roles are remarkable, encompassing pest control, seed dispersal, flower pollination, scavenging carcasses, and exerting profound influences on the environment when compared to other species.1 Wild birds occupy pivotal positions in the transmission of zoonotic infections, exerting influences on local animal health, wildlife populations, and humans.2 They serve as prominent reservoirs of bacterial and viral diseases, with repercussions extending to birds, humans, and other animal species.3,4 Functioning as carriers, wild birds disseminate a variety of disease-causing microorganisms within and across populations, including isolated island ecosystems.5 The extensive interactions between wild animals and humans hold immense importance for both veterinary and public health. It is worth noting that a bird's microflora includes strains of bacteria that possess not only disease-causing attributes but also exhibit resistance to multiple drugs, in addition to harboring virulence genes.6
Wild birds are known for their potential to harbor and transmit a wide range of diseases, including more than 40 that can affect both animals and humans.7 Indeed, many wild bird species, including urban-dwelling birds like pigeons, are frequently found in areas such as garbage sites, manure-rich environments, untreated sewage water, and dumps.8 Pigeons, in particular, are commonly seen in both rural and urban settings, often in proximity to human populations. Unfortunately, they can serve as carriers of numerous zoonotic pathogens, which can be transmitted to humans, wildlife, and domestic animals through various means, including their feathers, feces, or secretions.9,10 Some wild birds, especially migratory species, cover extensive distances during their journeys, spanning hundreds of kilometers. In doing so, they can come into contact with diverse environments and populations, facilitating the transmission of disease-causing bacterial strains to humans and other animals.11 During their migratory periods, these birds assume a crucial role in the epidemiology of human zoonotic diseases by potentially introducing and spreading pathogens across regions. Monitoring and understanding these interactions between wild birds, their environments, and the diseases they may carry are vital for public health and wildlife conservation efforts.12 Staphylococcus aureus, is a Gram-positive facultative bacterium. It is notorious for being a source of hospital-acquired infections and has been isolated from a wide range of animals, including cattle, wildlife, and domesticated animals.13 Staphylococcus species are commonly found in the microflora of both humans and animals. The genus Staphylococcus encompasses two main groups: coagulase-negative staphylococci and coagulase-positive staphylococci.14Top of Form Staphylococcus aureus is an opportunistic bacterium that can cause a variety of infectious diseases in both animals and humans.15 Its presence has significant implications for the overall ecosystem, livestock production, and public health due to its potential to spread and cause disease.16 Antibiotic usage has increased by 35.00% over the past decade on a global scale. Countries such as Brazil, Russia, India, China, and South Africa have also experienced this trend. Projections indicate that antibiotic consumption is expected to surge by 67.00% over the next 15 years. This emphasizes the growing concern of antibiotic resistance and the need for responsible antibiotic use to mitigate its impact on human and animal health.17
Many scientists have worked hard to figure out why harmful bacteria become resistant to antibiotics because it is a big health problem that also affects the economy. Studies have shown that wild birds play a significant role in spreading these antibiotic-resistant bacteria, which makes the global problem of drug-resistant infections even worse. These avian species play a pivotal role in disseminating antibiotic-resistant strains across diverse environments and among different hosts.18 However, despite the global significance of this issue, there have been limited studies conducted in Pakistan to address this specific concern. The emergence, progression, and dissemination of antibiotic-resistant bacterial pathogens represent a critical and pressing concern within the context of global health. This investigation sought to ascertain the presence of S. aureus, a potential human pathogen, in wild avian populations and examine its exhibited levels of resistance across diverse antibiotic categories. Furthermore, this study conducted a comparative assessment of the precision of two pheno-typic methodologies for assessing the antibiotic resistance profiles of S. aureus: the Kirby Bauer Disc diffusion method and the innovative minimum inhibitory concentration (MIC) method, which was executed using Sensititre™ARIS HiQ™ System for antimicrobial susceptibility testing (Thermo Scientific, Waltham, USA). In addition, an examination of the tetK, ermA, and aacA genes, responsible for conferring resistance to tetracycline, erythromycin, and gentamicin, was conducted. The present investigation serves as a foundational resource for implementing the MIC technique in evaluating of antibiotic resistance trends among notable bacterial pathogens. This contributes to the holistic and integrated One Health approach.
Materials and Methods
Study area and sample collection. The sampling procedures were carried out as approved by the ethical review committee of the University of Veterinary and Animal Sciences (No. DR/37). The study was conducted in the Kasur district, located in the Punjab province of Pakistan. A total of 100 samples were gathered from five different wild bird species, which included House Crow (Corvus splendens), Red-vented Bulbul (Pycnonotus cafer), Common Myna (Acridotheres tristis), Wild Pigeon (Columba livia), and Bank Myna (Acridotheres ginginianus). These samples comprised oral and cloacal swabs, which were collected using sterile Phosphate-Buffered Saline (HiMedia Laboratories Pvt. Ltd, Mumbai, India). Then, the collected samples were transported to the One Health Laboratory at the University of Veterinary and Animal Sciences in Pattoki for further bacteriological analysis.
Isolation and identification of S. aureus . To analyze the collected samples, cloacal and oral swabs were initially enriched in Nutrient Broth (Oxoid, Basingstoke United Kingdom) and incubated overnight at 37.00 ˚C. Following incubation, a 30.00 μL sample was taken and streaked onto Mannitol salt agar (Oxoid) using a sterile wire loop. These plates were then incubated for 24 hr at 37.00 ˚C. Colonies with a characteristic yellow color, indicating mannitol fermentation, were carefully selected using a sterile wire loop. Gram staining was then used to further identify these distinctive yellow colonies on mannitol salt agar. Confirmation of the bacterial isolates was carried out using the Bruker matrix-assisted laser desorption ionization (MALDI) Biotyper (Bruker Daltonics GmbH, Bremen, Germany), a specialized microbial identification system located at the Pasteur Research Pole, School of Public Health, Faculty of Medicine, University of Hong Kong. Once the bacterial isolates were confirmed, they were subcultured onto Tryptic Soy Agar plates from HiMedia and subsequently incubated at 37.00 ˚C for 24 hr. Pure isolates were stored in glycerol stocks at 4.00 ˚C for future analysis and reference.
Polymerase chain reaction (PCR)-based confirmation of S. aureus . To extract pure DNA, Favorgen (FavorPrep, Taipei, Taiwan) TM DNA extraction mini kit was employed following the manufacturer's instructions. The quantification of DNA was assessed using a Nanodrop spectrophotometer. Confirmation of S. aureus isolates was achieved through the PCR method.19 Specifically, 16S rRNA S. aureus primers were utilized, with the following sequences: Forward Primer - 5’-GATTTGATCCTGGCTCAGGA-3’ and Reverse Primer - 5’-ACGTCAGATGTGCACAGTTACTTA-3’, resulting in a 484-base pair (bp) amplification product.19 The PCR reaction was prepared using Taq PCR Master Mix (BioShop Ontario, Canada). This reaction was created with a total volume of 25.00 µl, composed of 12.00 µL of master mix, 0.50 µL of the forward primer, 0.50 µL of the reverse primer, 3.00 µL of template DNA, and 9.00 µL of injection water. A thermocycler (T100TM; Bio-Rad, Hercules, USA) was employed, and the program followed standard conditions, consisting of 35 cycles with the following temperature settings: 94.00 ˚C for 10 min, 60.00 ˚C for 1 min, and 72.00 ˚C for 1 min. After this amplification process, a 10-min incubation step at 72.00 ˚C was performed. To verify the PCR product, gel electrophoresis was conducted using a 2.00% agarose gel obtained from bioWORLD (Dublin, USA). The desired bands were visualized and captured using a Gel Documentation system from Bio-Rad.
Antimicrobial susceptibility (disc diffusion method). The susceptibility of the bacterial isolates to specific antimicrobial drugs was evaluated using the disk-diffusion method. The following antimicrobial drugs were tested, each with its respective disk containing a defined amount (in µg per disk): tetracycline (30.00 µg), erythromycin (15.00 µg), and gentamicin (10.00 µg), all sourced from Thermo Scientific OxoidTM. To prepare the bacterial isolates for testing, a freshly made suspension was created in a saline solution, and its turbidity was assessed using a 0.50 McFarland standards solution.Subsequently, this bacterial suspension was streaked onto Muller Hinton Agar (Oxoid) plates. The prepared plates were then placed in an incubator (Thermo Scientific, Waltham, USA) set at 37.00 ˚C and left to incubate overnight to allow for susceptibility testing. After incubation, the diameter of the inhibition zone surrounding each drug-containing disk was carefully measured using a calibrated scale with a precision of 0.50 mm. The results obtained from these measurements were analyzed following the guidelines outlined by the Clinical and Laboratory Standards Institute, which provides standardized procedures for assessing antimicrobial susceptibility.
Minimum inhibitory concentration. A comprehensive analysis was conducted on the MICs for antimicrobial susceptibility using a total of 16 antibiotics. The antibiotics were as follow: chloramphenicol, oxacillin, ciprofloxacin, linezolid, penicillin, daptomycin, rifampicin, nitrofurantoin, gentamicin, tetracycline, levofloxacin, clindamycin, moxifloxacin, erythromycin, trimethoprim-sulfamethoxazole and vancomycin. These MIC assessments were carried out using the Thermo Scientific Sensititre system, specifically the ARIS HiQ AST System (Waltham, USA). The analysis took place at the Pasteur Research Pole, School of Public Health, Faculty of Medicine, University of Hong Kong.
Detection of antimicrobial resistance and virulence genes. The PCR tests were used to analyze virulence and antibiotic resistance genes using Taq PCR Master Mix. A total of 12.00 µL of master mix, 0.50 µL of forward primer, 0.50 µL of reverse primer, 3.00 µL of template DNA, and 9.00 µL of injection water were combined to create a final volume of 25.00 µL. The primer sequences and annealing temperatures for the targeted genes are provided in Table 1.20,21
Table 1.
Gene types along with 5’ to 3’ primer sequence and product size base pair (bp).
| Target genes | Primer Sequences (5'-3') | Amplicon size (bp) | References |
|---|---|---|---|
| tetK (tetracycline) | GTAGCGACAATAGGTAATAGT GTAGTGACAATAAACCTCCTA |
360 | 20 |
| tetM (tetracycline) | AGTGGAGCGATTACAGAA CATATGTCCTGGCGTGTCTA |
158 | 20 |
| ermA (erythromycin) | AAGCGGTAA ACC CCTCTGA TTCGCAAATCCC TTCTCAAC |
190 | 20 |
| aacA-D (gentamicin) | TAATCCAAGAGCAATAAGGGC GCCACACTATCATAACCACTA |
227 | 20 |
| etA | CTAGTGCATTTGTTATTCAA TGCATTGACACCATAGTACT |
119 | 21 |
| etB | ACGGCTATATACATTCAATT TCCATCGATAATATACCTAA |
200 | 21 |
Results
Prevalence of S. aureus in wild birds. Out of the 100 cloacal and oral samples collected from wild birds, it was found that the overall prevalence of S. aureus was 30 samples, constituting 30.00% of the total samples.
This confirmation was achieved through both matrix-assisted laser desorption ionization–time-of-flight mass spectrometry (MALDI-TOF MS) and the 16S rRNA gene analysis, as depicted in Figure 1. Among the various wild bird species studied, the two species that exhibited the highest frequency of S. aureus were C. livia, with one individual showing an 8.00% prevalence, and C. splendens, where two individuals had an 8.00% prevalence. Pycnonotus cafer presented a prevalence of 7.00%. Conversely, the lowest prevalence of S. aureus was observed in A. ginginianus, with four individuals having a 4.00% prevalence, and A. tristis, with five individuals displaying a 3.00% prevalence. It is noteworthy that cloacal samples from wild birds exhibited a higher prevalence (16.00%) of S. aureus compared to oral samples, which had a prevalence of 14.00%. This indicates that S. aureus was more commonly found in the cloacal region of the sampled wild birds (Table 2).
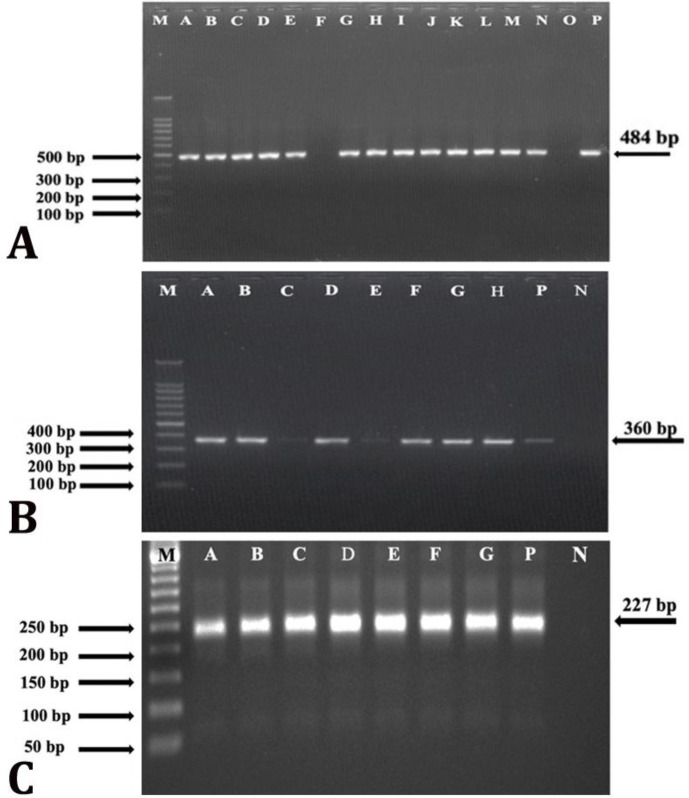
Fig. 1.
Polymerase chain reaction (PCR) amplification. A) The PCR-based detection of Staphylococcus aureus by 16S rRNA gene (484 bp). Lane M: 100 bp marker; Lane A to N: S. aureus isolates; Line O: Negative control; Line P: Positive control, B) The PCR-based detection of tetK gene (360 bp). Lane M: 100 bp marker; Lane A to H: S. aureus isolates positive for tetK gene. Lane P: Positive control; Lane N: Negative control, C) The PCR-based detection of the aacA-D gene (227 bp). Lane M: 50 bp marker; Lane A to G: S. aureus isolates positive for the aacA-D gene. Lane P: Positive control; Lane N: Negative control.
Table 2.
Prevalence of Staphylococcus aureus in oral and cloacal samples of wild bird species 20 samples from each bird.
| Serial No. | Wild birds species | Total number of S. aureus isolates | Oral (%) | Cloacal (%) |
|---|---|---|---|---|
| 1 | Corvus splendens | 8 | 3 (37.50) | 5 (62.50) |
| 2 | Pycnonotus cafer | 7 | 4 (57.14) | 3 (42.85) |
| 3 | Acridotheres tristis | 3 | 1 (33.33) | 2 (66.66) |
| 4 | Columba livia | 8 | 4 (50.00) | 4 (50.00) |
| 5 | Acridotheres ginginianus | 4 | 2 (50.00) | 2 (50.00) |
Antimicrobial resistance by disc diffusion method. The results of the disc diffusion method revealed that the S. aureus isolates obtained from wild birds exhibited multidrug resistance, as they displayed varying levels of resistance to three distinct antimicrobial drugs. Specifically, the antimicrobial resistance profile indicated a higher prevalence of tetracycline resistance among the S. aureus isolates from wild birds, either phenotypically or genetically, at a rate of 33.33%. This resistance was further broken down by bird species, with C. splendens having the highest prevalence at 40.00%, followed by A. ginginianus at 20.00%, P. cafer at 20.00%, A. tristis at 10.00% and C. livia at 10.00%. Additionally, the analysis revealed a high erythromycin resistance rate of 16.66% among the S. aureus isolates. This resistance was observed in C. splendens at 40.00%, A. ginginianus at 20.00%, C. livia at 20.00%, and P. cafer at 20.00%. In contrast, gentamicin demonstrated the lowest resistance level, with only 10.00% of the S. aureus isolates exhibiting resistance (Table 3).
Table 3.
Phenotypic and genotypic antimicrobial resistance of Staphylococcus aureus isolated from cloacal and oral samples of wild birds (n = 30).
| Antimicrobial class | Antimicrobial agents | Zone diameter (mm) | Wild birds S. aureus | Prevalence of resistance genes (%) | ||||
|---|---|---|---|---|---|---|---|---|
| S | I | R | S | I | R | |||
| Tetracyclines | Tetracycline (30.00 µg) | 19.00 | 15.00 - 18.00 | ≤ 14.00 | 10 | 10 | 10 | 10 (33.33) |
| Macrolides | Erythromycin 15.00µg | 23.00 | 14.00 - 22.00 | ≤ 13.00 | 9 | 16 | 5 | 5 (16.66) |
| Aminoglycosides | Gentamicin 10.00µg | 15.00 | 13.00 - 14.00 | ≤ 12.00 | 23 | 4 | 3 | 3 (10.00) |
S= susceptible; I= intermediate; and R= resistant.
Minimum inhibitory concentration based anti-microbial resistance. Based on the MIC analysis, the resistance patterns of S. aureus isolates from wild birds were as follows: the highest resistance was observed with oxacillin, affecting 56.66% of the isolates, and tetracycline, affecting 33.33% of the isolates. Among fluoroquinolones, ciprofloxacin and levofloxacin displayed the lowest resistance rates at 3.33% each, while moxifloxacin exhibited 0.00% resistance. Ciprofloxacin and moxifloxacin were found to be intermediate in resistance at 33.30%. The lowest susceptibility was recorded with oxacillin, at a rate of 43.33%. Conversely, vancomycin, rifampicin, linezolid, and daptomycin demonstrated the highest susceptibility, with a prevalence of 100%. The isolates exhibited varying degrees of susceptibility to other antibiotics: moxifloxacin (96.42%), levofloxacin (96.42%), trimethoprim-sulfa-methoxazole (93.33%), ciprofloxacin (93.33%), chloramphenicol (86.66%), clindamycin and nitrofurantoin (83.33%), gentamicin (76.66%), and erythromycin (66.66%). Oxacillin, tetracycline, and erythromycin were identified as the antibiotics exhibiting the most widespread resistance patterns among multidrug-resistant S. aureus bacteria found in wild birds (Table 4).
Table 4.
Antibiogram of minimum inhibitory concentration (MIC) of S. aureus isolated from cloacal and oral samples of wild birds.
| Serial No. | Antimicrobial class | Antimicrobial agents | MIC breakpoint (µg mL -1 ) | Wild birds S. aureus (%) | ||||
|---|---|---|---|---|---|---|---|---|
| S | I | R | S | I | R | |||
| 1 | Phenicols | Chloramphenicol | ≤ 8.00 | 16.00 | ≥ 32.00 | 26 (86.66) | 0 (0.00) | 4 (13.33) |
| 2 | Fluoroquinolone | Ciprofloxacin | ≤ 1.00 | 2.00 | ≥ 4.00 | 28 (93.33) | 1 (3.33) | 1 (3.33) |
| 3 | Lincosamides | Clindamycin | ≤ 0.50 | 1.00 - 2.00 | ≥ 4 .00 | 25 (83.33) | 0 (0.00) | 5 (16.66) |
| 4 | Lipopeptides | Daptomycin | ≤ 1.00 | - | - | 30 (100) | 0 (0.00) | 0 (0.00) |
| 5 | Macrolides | Erythromycin | ≤ 0.50 | 1.00 - 2.00 | ≥ 8.00 | 20 (66.66) | 1 (3.33) | 9 (30.00) |
| 6 | Aminoglycosides | Gentamycin | ≤ 4.00 | 8.00 | ≥ 16.00 | 23 (76.66) | 4 (13.33) | 3 (10.00) |
| 7 | Fluoroquinolone | Levofloxacin | ≤ 1.00 | 2.00 | ≥ 4.00 | 29 (96.66) | 0 (0.00) | 1 (3.33) |
| 8 | Oxazolidinones | Linezolid | ≤ 4.00 | - | ≥ 8.00 | 30 (100) | 0 (0.00) | 0 (0.00) |
| 9 | Fluoroquinolone | Moxifloxacin | ≤ 0.50 | 1.00 | ≥ 2.00 | 29 (96.42) | 1 (3.57) | 0 (0.00) |
| 10 | Nitrofurantoins | Nitrofurantoin | ≤ 32 | 64.00 | ≥ 128 | 25 (83.33) | 1 (3.33) | 4 (13.33) |
| 11 | Penicillin | Oxacillin | ≤ 2.00 | - | ≥ 4.00 | 13 (43.33) | 0 (0.00) | 17 (56.66) |
| 12 | Beta lactam | Penicillin | ≤ 0.12 | - | ≥ 0.25 | 25 (83.33) | 0 (0.00) | 5 (16.66) |
| 13 | Ansamycin | Rifampicin | ≤ 1.00 | 2.00 | ≥ 4.00 | 30 (100) | 0 (0.00) | 0 (0.00) |
| 14 | Tetracyclines | Tetracycline | ≤ 4.00 | 8.00 | ≥ 16.00 | 20 (66.66) | 0 (0.00) | 10 (33.33) |
| 15 | Folate Pathway Antagonists | Trimethoprim-sulfamethoxazole | ≤ 2.38 | - | ≥ 4/76 | 28 (93.33) | 0 (0.00) | 2 (6.66) |
| 16 | Glycopeptides | Vancomycin | ≤ 2.00 | 4.00 - 8.00 | ≥ 16.00 | 30 (100) | 0 (0.00) | 0 (0.00) |
S= susceptible; I= intermediate; and R= resistant.
Antimicrobial resistance and virulence gene detection. All isolates were screened for four antibiotic-resistance genes, and the results were as follows: tetK was observed in 33.33% of the isolates, with varying prevalence among different bird species. Specifically, C. splendens had a prevalence of 40.00%, A. ginginianus 20.00%, P. cafer 20.00%, A. tristis 10.00%, and C. livia 10.00% (Fig. 1). The tetM was found in only one isolate from C. splendens (3.22%). The ermA gene, associated with erythromycin resistance, was present in 16.66% of the samples. This resistance gene was distributed among different bird species, with C. splendens at 40.00%, A. tristis at 20.00%, C. livia at 20.00%, and P. cafer at 20.00% displaying this gene. The aacA-D gene, responsible for gentamicin resistance, was detected in 10.00% of the isolates. This gene was identified in isolates from C. splendens at (66.66%) and A. tristis at 33.33% (Fig. 1). The virulence genes etA and etB were tested in all 30 isolates. However, no isolate exhibited these virulence genes.
Discussion
Staphylococcus aureus is a significant contributor to the microbiota in humans and animals, and is recognized as one of the most prominent emerging pathogens worldwide. Understanding the pathogenic potential and transmission dynamics of S. aureus, especially in scenarios involving wild birds in their natural habitat, captivity, or when their feces contaminate the environment is crucial. The prevalence of S. aureus in such situations is highly relevant.22,23 Previous research in Pakistan has mainly focused on assessing the prevalence of S. aureus in livestock and poultry birds. Similar studies have been conducted globally, with a particular focus on poultry birds, in countries like Belgium, Nigeria, North Algeria, the United States of America, and Egypt. However, there is a significant gap in knowledge regarding the incidence of this pathogenic bacterium in wild bird populations in Pakistan.24-27 Understanding its prevalence in wild birds is essential for a comprehensive assessment of its impact on both animal and human health in the region. S. aureus is known to form commensal relationships with humans and various other animals, often being considered a natural component of their microbiota. The diversity in the pathogenicity and ecological preferences of these bacteria underscores their significance in epidemiological investigations. In our study, S. aureus was detected in 30.00% of the sampled birds. It is important to note that prevalence rates for S. aureus can vary in different geographic regions and bird species. For example, Ahmed et al. conducted a similar study in Egypt and found a slightly lower prevalence of S. aureus, at 24.30%. They collected tracheal and cloacal swabs from 125 pet birds, including species like the Red-rumped parrot (Psephotus haematonotus), Budgerigar (Melopsittacus undulatus), and Rosy-faced lovebird (Agapornis roseicollis).28 Additionally, research conducted by Szczuka et al. in Poland reported a prevalence of 100% for Staphylococci in domestic pigeons.29 These variations in prevalence rates may be influenced by factors such as geographic location, bird species, and the specific sampling methods employed. Such research provides valuable insights into the prevalence and distribution of S. aureus in different bird populations around the world.
Within the One Health framework, the presence of wild birds acting as reservoirs for infections displaying increased antibiotic resistance raises public health concerns. Our investigation found that 30.00% of wild birds tested positive for S. aureus. This aligns with a study by Silva et al. in central and north Portugal, where a prevalence rate of 20.90% for S. aureus isolation was reported in tracheal and cloacal swabs of nocturnal wild birds.30 A recent study by El-Mahallawy et al. determined that 70.00% of S. aureus isolates were obtained from various wild birds in Giza, Egypt, indicating a higher prevalence rate compared to our study.31 Our study also yielded results consistent with other research but with a lower occurrence of S. aureus in cloacal samples from wild birds obtained from street markets in Rio de Janeiro, Brazil.32 Previous studies have also identified S. aureus in the feces of corvids, marine animals, and migratory birds.33 In an Italian study focusing on wild birds of prey, Dipineto et al. found that 26 out of the samples, amounting to 35.60%, tested positive for S. aureus, slightly surpassing the prevalence observed in our research.34 Conversely, a study in Spain, which examined 324 samples from wild birds, identified coagulase-positive Staphylococcus in 27 cases (8.33%). Among these, only 15 were definitively identified as S. aureus, indicating a lower occurrence compared to our study.35 In contrast to our findings, Russo et al. investigated wild birds from 32 different species in Naples Federico, Italy, and discovered that only six out of 163 birds (3.68%) tested positive for S. aureus. This prevalence rate is notably lower when compared to studies conducted in other countries.36Top of Form
Antibiotic resistance is a growing concern worldwide particularly among S. aureus bacterial pathogens. Developing nations, such as Pakistan, are facing emerging threats from S. aureus and Methicillin-Resistant S. aureus. Unfortunately, our understanding of the prevalence of these bacterial diseases in these regions is severely limited compared to the global situation due to the absence of effective monitoring systems and diagnostic techniques. In our investigation, we observed varying levels of resistance among S. aureus isolates to three different antibiotics. Tetracycline and erythromycin are commonly used antimicrobial medications to treat staphylococcal infections in birds. Interestingly, our research indicated that these drugs had higher resistance rates against S. aureus compared to gentamicin. A similar trend of increased tetracycline resistance, as seen in our findings, was reported in poultry birds in Bangladesh, where an 80.60% resistance rate was documented.37 Moreover, Bagheri et al., in their study of drug resistance in companion birds in Iran, identified resistance rates of 9.00% for gentamicin and 30.00% for erythromycin in S. aureus, which aligns with our findings.38
The antimicrobial resistance rates observed in our study among wild birds are a cause for concern, as these birds typically do not face the selection pressure of antimicrobial drugs. Our research highlighted variations in resistance patterns among S. aureus isolates to the three antibiotics tested using the disc diffusion technique. Notably, tetracycline exhibited a significantly high resistance rate of 33.33% compared to all other antibiotics. Conversely, gentamicin displayed the lowest resistance rate at 3.00%. Recent data indicates that S. aureus isolates from migratory wild birds exhibited a resistance rate of 22.60% to tetracycline. Consistent with our study, an earlier research on wild birds in Brazil by Matias et al. concluded that gentamicin had a resistance rate of only 4.60% against S. aureus, while tetracycline showed the highest resistance rate at 11.90%.32 In our investigation, we observed a resistance rate of 16.66% to erythromycin. However, in Northeast Ohio, a much higher resistance rate of 61.50% was observed among S. aureus isolates from goose fecal samples, indicating greater resistance to erythromycin than our findings.39 In contrast to our results, recent research in Italy by Russo et al. found that 66.00% of S. aureus isolates in wild birds were resistant to both tetracycline and erythromycin.36 On the other hand, Silva et al. reported that 50.00% of the Staphylococcus strains examined were sensitive to these antibiotics.30 It is important to note that the excessive and inappropriate use of therapeutic drugs, primarily in human medicine but also in animals, including the use of antibiotics as growth promoters in cattle, significantly contributes to the rise of antibiotic resistance. Some research suggests that multidrug-resistant diseases can spread among animals and from animals to humans.40 The natural environment of wild birds can expose them to antibiotic-resistant bacteria. They may consume food or water sources contaminated with antibiotic-resistant bacteria.41 Antibiotic-resistant bacteria can be transferred from humans, livestock, and other animals to birds.42 Even without antibiotic treatment, horizontal gene transfer can cause antibiotic resistance in wild birds. Like other bacteria S. aureus can share genetic material. Genes that cause antibiotic resistance can survive in the environment and be picked up by bacteria over time.43 Wild birds' normal behaviors, such as foraging and preening, may expose them to antibiotic-resistant bacteria like S. aureus. The findings of our study underscore the potential role of wild birds as a source of zoonotic bacteria and potentially harmful genes, emphasizing the need for further research and surveillance in this area.
We have concluded that wild birds have the potential to spread antibiotic-resistant bacteria, posing significant risks to the health of both humans and animals. Wild birds can travel long distances, contributing to the spread of antibiotic-resistant microorganisms across various ecosystems. This study highlights that wild birds can act as reservoirs for S. aureus with both phenotypic (observable characteristics) and genotypic (genetic) antibiotic resistance. The cloacal samples of wild birds in this investigation had the highest prevalence with tetracycline and oxacillin showing the greatest resistance. Among all the resistance genes, tetK and emrA were the most common. None of the isolates tested positive for virulence genes. This study emphasizes the importance of diligent monitoring and understanding the mechanisms of antibiotic resistance in wildlife. Assessing the potential consequences of resistance gene spread on both human and animal populations can be achieved by monitoring the presence of pathogenic and antibiotic-resistant microbes in wild birds across different geographical locations.
Acknowledgments
We would like to thank the field personnel from the Department of Wildlife and Ecology at the University of Veterinary and Animal Sciences in Lahore, Pakistan, for their assistance with sample collection.
Conflict of interest
There is no conflict of interest.
References
- 1.Whelan CJ, Şekercioğlu ÇH, Wenny DG. Why birds matter: from economic ornithology to ecosystem services. J Ornithol . 2015;156(Suppl 1):227–238. [Google Scholar]
- 2.Robinson RA, Lawson B, Toms MP, et al. Emerging infectious disease leads to rapid population declines of common British birds. PLoS One. 2010;5(8):e12215. doi: 10.1371/journal.pone.0012215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amin MAM, Ali MNM, Awadallah MAI, et al. Prevalence of Enterobacteriaceae in wild birds and humans at Sharkia province; with special reference to the genetic relationship between E. coli and Salmonella isolates determined by protein profile analysis. J Am Sci. 2013;9(4):173–183. [Google Scholar]
- 4.Badouei MA, Blackall PJ, Koochakzadeh A, et al. Prevalence and clonal distribution of avian Escherichia coli isolates harboring increased serum survival (iss) gene. J Appl Poult Res. 2016;25(1):67–73. [Google Scholar]
- 5.Altizer S, Bartel R, Han BA. Animal migration and infectious disease risk. Science. 2011;331(6015):296–302. doi: 10.1126/science.1194694. [DOI] [PubMed] [Google Scholar]
- 6.Blyton MD, Pi H, Vangchhia B, et al. Genetic structure and antimicrobial resistance of Escherichia coli and cryptic clades in birds with diverse human associations. Appl Environ Microbiol. 2015;81(15):5123–5133. doi: 10.1128/AEM.00861-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaukler SM, Linz GM, Sherwood JS, et al. Escherichia coli, Salmonella, and Mycobacterium avium subsp paratuberculosis in wild European starlings at a Kansas cattle feedlot. Avian dis. 2009;53(4):544–551. doi: 10.1637/8920-050809-Reg.1. [DOI] [PubMed] [Google Scholar]
- 8.Fogarty LR, Haack SK, Wolcott MJ, et al. Abundance and characteristics of the recreational water quality indicator bacteria Escherichia coli and enterococci in gull feces. J Appl Microbiol. 2003;94(5):865–878. doi: 10.1046/j.1365-2672.2003.01910.x. [DOI] [PubMed] [Google Scholar]
- 10.Suphoronski SA, de Freitas Raso T, Weinert NC, et al. Occurrence of Salmonella sp and Escherichia coli in free-living and captive wild birds from 2010-2013 in Guarapuava, Paraná, Brazil. Afr J Microbiol Res. 2015;9(29):1778–1782. [Google Scholar]
- 11.Brugman VA, Horton DL, Phipps LP, et al. Epidemiological perspectives on West Nile virus surveillance in wild birds in Great Britain. Epidemiol Infect. 2013;141(6):1134–1142. doi: 10.1017/S095026881200177X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foti M, Mascetti A, Fisichella V, et al. Antibiotic resistance assessment in bacteria isolated in migratory Passeriformes transiting through the Metaponto territory (Basilicata, Italy) Avian Res. 2017;8:26 . [Google Scholar]
- 13.Gómez P, Lozano C, Camacho MC, et al. Detection of MRSA ST3061-t843-mecC and ST398-to11-mecA in white stork nestlings exposed to human residues. J Antimicrob Chemother. 2016;71(1):53–57. doi: 10.1093/jac/dkv314. [DOI] [PubMed] [Google Scholar]
- 14.França A, Gaio V, Lopes N, et al. Virulence factors in coagulase-negative Staphylococci. Pathogens. 2021;10(2):10. doi: 10.3390/pathogens10020170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chin D, Goncheva MI, Flannagan RS, et al. Coagulase-negative staphylococci release a purine analog that inhibits Staphylococcus aureus virulence. Nat Commun. 2021;12(1):1887 . doi: 10.1038/s41467-021-22175-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rossi CC, Pereira MF, Giambiagi-deMarval M. Underrated Staphylococcus species and their role in antimicrobial resistance spreading. Genet Mol Biol 2020. 43(1 suppl 2):e20190065. doi: 10.1590/1678-4685-GMB-2019-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Boeckel TP, Brower C, Gilbert M, et al. Global trends in antimicrobial use in food animals. Proc Natl Acad Sci USA. 2015;112(18):5649–5654. doi: 10.1073/pnas.1503141112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carroll D, Wang J, Fanning S, et al. Antimicrobial resistance in wildlife: implications for public health. Zoonoses Public Health. 2015;62(7):534–542. doi: 10.1111/zph.12182. [DOI] [PubMed] [Google Scholar]
- 19.Atyah MA, Zamri-Saad M, Siti-Zahrah A. First report of methicillin-resistant Staphylococcus aureus from cage-cultured tilapia (Oreochromis niloticus) Vet Microbiol. 2010;144(3-4):502–504. doi: 10.1016/j.vetmic.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 20.Strommenger B, Kettlitz C, Werner G, et al. Multiplex PCR assay for simultaneous detection of nine clinically relevant antibiotic resistance genes in Staphylococcus aureus. J Clin Microbiol. 2003;41(9):4089–4094. doi: 10.1128/JCM.41.9.4089-4094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson WM, Tyler SD, Ewan EP, et al. Detection of genes for enterotoxins, exfoliative toxins, and toxic shock syndrome toxin 1 in Staphylococcus aureus by the polymerase chain reaction. J Clin Microbiol. 1991;29(3):426–430. doi: 10.1128/jcm.29.3.426-430.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaudhary S, Batoo AS, Hussain K, et al. Prevalence of pododermatitis caused by Staphylococcus aureus in poultry birds of Jammu. Int J Livest Res. 2018;8(3):192–195. [Google Scholar]
- 23.Habib F, Malhi KK, Kamboh AA, et al. Antimicrobial susceptibility profile of Staphylococcus aureus isolates recovered from various animal species. J Anim Health Prod. 2015;3(4):99–103. [Google Scholar]
- 24.Persoons D, Van Hoorebeke S, Hermans K, et al. Methicillin-resistant Staphylococcus aureus in poultry. Emerg Infect Dis. 2009;15(3):452–453. doi: 10.3201/eid1503.080696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otalu Jr O, Kabir JJ, Okolocha EC, et al. Detection of methicillin-resistant Staphylococcus aureus in chicken carcasses and live birds in Zaria, Nigeria. FUTA J Res Sci. 2015;11(1):132–138. [Google Scholar]
- 26.Onaolapo JA, Igwe JC, Bolaji RO, et al. Antibiotics susceptibility profile of Staphylococcus aureus isolated from poultry birds in Kaduna, Nigeria. J Clin Microbiol Antimicrob. 2017;1(1):1–6. [Google Scholar]
- 27.Bakheet AA, Amen O, Habaty SHAL, et al. Prevalence of Staphylococcus aureus in broiler chickens with special reference to beta-lactam resistance genes in the isolated strains. Alex J Vet Sci. 2018;57(2):25–33. [Google Scholar]
- 28.Ahmed HA, Awad NFS, Abd El-Hamid MI, et al. Pet birds as potential reservoirs of virulent and antibiotic-resistant zoonotic bacteria. Comp Immunol Microbiol Infect Dis. 2021:75. doi: 10.1016/j.cimid.2020.101606. [DOI] [PubMed] [Google Scholar]
- 29.Szczuka E, Wesołowska M, Krawiec A, et al. Staphylococcal species composition in the skin microbiota of domestic pigeons (Columba livia domestica) PLoS One. 2023;18(7):e0287261. doi: 10.1371/journal.pone.0287261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silva V, Lopes AF, Soeiro V, et al. Nocturnal birds of prey as carriers of Staphylococcus aureus and other Staphylococci: diversity, antimicrobial resistance and clonal lineages. Antibiotics. 2022;11(2):240. doi: 10.3390/antibiotics11020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El-Mahallawy HS, Hamza DA, Ahmed ZS. Fecal carriage of S aureus and the mecA gene in resident wild birds and its zoonotic potential. J Appl Vet Sci. 2022;7(3):35–40. [Google Scholar]
- 32.Matias CAR, Pereira IA, Rodrigues DP, et al. Staphylococcus spp isolated from wild birds apprehended in the local illegal trade in Rio de Janeiro, Brazil, and relevance in public health. Lett Appl Microbiol. 2018;67(3):292–298. doi: 10.1111/lam.13035. [DOI] [PubMed] [Google Scholar]
- 33.Hubálek Z. An annotated checklist of pathogenic microorganisms associated with migratory birds. J Wildl Dis. 2004;40(4):639–659. doi: 10.7589/0090-3558-40.4.639. [DOI] [PubMed] [Google Scholar]
- 34.Dipineto L, Bossa LM, Pace A, et al. Microbiological survey of birds of prey pellets. Comp Immunol Microbiol Infect Dis. 2015;41:49–53. doi: 10.1016/j.cimid.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 35.Ruiz-Ripa L, Gómez P, Alonso CA, et al. Frequency and characterization of antimicrobial resistance and virulence genes of coagulase-negative staphylococci from wild birds in Spain Detection of tst-carrying S. sciuri isolates. Microorganisms. 2020;8(9):1317. doi: 10.3390/microorganisms8091317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russo TP, Minichino A, Gargiulo A, et al. Prevalence and phenotypic antimicrobial resistance among ESKAPE bacteria and enterobacterales strains in wild birds. Antibiotics (Basel) 2022;11(12):1825. doi: 10.3390/antibiotics11121825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ali Y, Islam MA, Muzahid NH, et al. Characterization, prevalence and antibiogram study of Staphylococcus aureus in poultry. Asian Pac J Trop Biomed. 2017;7(3):253–256. [Google Scholar]
- 38.Bagheri SS, Peighambari SM, Soltani M, et al. RAPD-PCR and drug resistance pattern of Staphylococcus aureus isolates recovered from companion and wild birds. Iran J Appl Anim Sci. 2019;13(4):356–364. [Google Scholar]
- 39.Thapaliya D, Dalman M, Kadariya J, et al. Characterization of Staphylococcus aureus in goose feces from state Parks in Northeast Ohio. EcoHealth. . 2017;14(2):303–309. doi: 10.1007/s10393-017-1227-z. [DOI] [PubMed] [Google Scholar]
- 40.Marshall B, Ochieng J. Levy S. Commensals: an underappreciated reservoir of antibiotic resistance. Probing the role of commensals in propagating antibiotic resistance should help preserve the efficacy of these critical drugs. Microbe. 2009;4:231–238. [Google Scholar]
- 41.Ramey AM, Ahlstrom CA. Antibiotic-resistant bacteria in wildlife: perspectives on trends, acquisition and dissemination, data gaps, and future directions. J Wildl Dis. 2020;56(1):1–15. [PubMed] [Google Scholar]
- 42.Carter DL, Docherty KM, Gill SA, et al. Antibiotic-resistant bacteria are widespread in songbirds across rural and urban environments. Sci Total Environ. 2018;627:1234–1241. doi: 10.1016/j.scitotenv.2018.01.343. [DOI] [PubMed] [Google Scholar]
- 43.Wu J, Huang Y, Rao D, et al. Evidence for environmental dissemination of antibiotic resistance mediated by wild birds. Front. Microbiol. 2018;9:Article 745. doi: 10.3389/fmicb.2018.00745. [DOI] [PMC free article] [PubMed] [Google Scholar]