Abstract
To evaluate the impact of the diversity of antigen recognition by T lymphocytes on disease pathogenesis, we must be able to identify and analyze simultaneously cytotoxic T-lymphocyte (CTL) responses specific for multiple viral epitopes. Many of the studies of the role of CD8+ CTLs in AIDS pathogenesis have been done with simian immunodeficiency virus (SIV)- and simian-human immunodeficiency virus (SHIV)-infected rhesus monkeys. These studies have frequently made use of the well-defined SIV Gag CTL epitope p11C,C-M presented to CTL by the HLA-A homologue molecule Mamu-A*01. In the present study we identified and fine mapped two novel Mamu-A*01-restricted CTL epitopes: the SIVmac Pol-derived epitope p68A (STPPLVRLV) and the human immunodeficiency virus type 1 (HIV-1) Env-derived p41A epitope (YAPPISGQI). The frequency of CD8+ CTLs specific for the p11C,C-M, p68A, and p41A epitopes was quantitated in the same animals with a panel of tetrameric Mamu-A*01/peptide/β2m complexes. All SHIV-infected Mamu-A*01+ rhesus monkeys tested had a high frequency of SIVmac Gag-specific CTLs to the p11C,C-M epitope. In contrast, only a fraction of the monkeys tested had detectable CTLs specific for the SIVmac Pol p68A and HIV-1 Env p41A epitopes, and these responses were detected at very low frequencies. Thus, the p11C,C-M-specific CD8+ CTL response is dominant and the p41A- and p68A-specific CD8+ CTL responses are nondominant. These results indicate that CD8+ CTL responses to dominant CTL epitopes can be readily quantitated with the tetramer technology; however, CD8+ CTL responses to nondominant epitopes, due to the low frequency of these epitope-specific cells, may be difficult to detect and quantitate by this approach.
Although considerable data have accrued in studies of AIDS pathogenesis supporting a role for CD8+ cytotoxic T lymphocytes (CTL) in containing human immunodeficiency virus type 1 (HIV-1) replication, the importance of epitope specificity in antigen recognition by these cells remains poorly defined. Containment of HIV-1 replication in vivo has been correlated temporally with the generation of virus-specific CTL (5, 10, 23). In addition, a potent CTL response in chronically infected individuals is associated with low virus load and a stable clinical status (18, 21). However, little is known about the diversity of HIV-1 epitopes recognized by these CTL and the ramifications of diverse antigen recognition for disease pathogenesis. Some have suggested that the breadth of antigen recognition by CD8+ CTL may have a significant impact on success in controlling virus spread (23). To evaluate the impact of the diversity of antigen recognition by T lymphocytes on disease pathogenesis, we must be able to identify and analyze simultaneously CTL responses specific for multiple viral epitopes.
Recently it has become possible to define with quantitative precision distinct subpopulations of epitope-specific CD8+ CTL by soluble major histocompatibility complex (MHC) class I/peptide/beta-2-microglobulin (β2m) tetrameric complexes and flow cytometric analysis (2–4, 6, 8, 11, 17, 21). The application of this technique has so far been used primarily for the study of virus-specific CTL with specificity for dominant epitopes presented to T cells by MHC class I molecules. The tetramer technique for studying CTL should, however, be useful in evaluating CTL specific for multiple epitopes of the same virus. This might be accomplished through the use of a panel of tetrameric MHC class I/peptide/β2m complexes that define a variety of CTL epitopes restricted by either a single or multiple MHC class I molecules.
Simian immunodeficiency virus (SIVmac)- and simian-human immunodeficiency virus (SHIV) infection-infected rhesus monkeys develop a disease with remarkable similarities to HIV-1-induced disease in humans (13, 14, 24), providing powerful animal models in which to study AIDS pathogenesis. The evaluation of rhesus monkey CTL responses to SIVmac and SHIV has been facilitated by the use of well-defined viral CTL epitopes and their restricting MHC class I molecules. Specifically, SIVmac- and SHIV-specific CTL have been evaluated with considerable sensitivity in these animal models through the study of T-cell responses to the Gag epitope p11C,C-M presented by the HLA-A homologue molecule Mamu-A*01 (1, 11, 15, 16). The definition of additional CTL epitopes and the development of tetrameric staining approaches for their evaluation would considerably increase the utility of these animal models.
In the present study we have identified two additional Mamu-A*01-restricted CTL epitopes in SHIV-infected rhesus monkeys. We have generated tetrameric Mamu-A*01/peptide/β2m complexes that recognize CD8+ CTL specific for these epitopes and used them to evaluate the breadth of the Mamu-A*01-restricted CTL response in these monkeys.
MATERIALS AND METHODS
Animals.
Heparinized blood samples were obtained from rhesus monkeys (Macaca mulatta) experimentally infected with uncloned SIVmac strain 251, SHIV-89.6 (monkeys 206, 287, and 556), or SHIV-HXBc2 (monkeys L3 and L9). The experimental monkeys in the present study were infected with SHIV or SIVmac 3 to 72 months prior to their evaluation. The animals were maintained in accordance with the guidelines of the Committee on Animals for the Harvard Medical School (Cambridge, Mass.) and the Guide for the Care and Use of Laboratory Animals (20).
Selection of Mamu-A*01+ rhesus monkeys.
Rhesus monkeys were screened for the presence of the Mamu-A*01 allele by a PCR-based technique as previously described (9). EDTA-preserved whole blood from rhesus macaques was subjected to Ficoll diatrizoate density gradient centrifugation to isolate leukocytes, and the washed cell pellets were resuspended in 200 μl of phosphate-buffered saline. DNA extraction was then carried out with a QIAmp blood kit (Qiagen Inc., Chatsworth, Calif.). PCR was performed on 200 to 500 μg of extracted DNA with allele-specific primers in a 50 μl reaction mixture consisting of 60 mM Tris (pH 8.5), 2 mM MgCl2, 15 mM ammonium sulfate, 2 mM deoxynucleoside triphosphates (0.5 mM each), and 5 μl of Taq polymerase. Primers A*01/F (5′-GAC AGC GAC GCC GCG AGC CAA-3′) and A*01/R (5′-GCT GCA GCG TCT CCT TCC CC-3′) were used at final concentrations of 800 nM each. Two additional primers specific for a conserved MHC class II sequence (based on the macaque homologue of HLA-DRB3) were included in the reaction as internal positive controls. Primers 5′ MDRB (5′-GCC TCG AGT GTC CCC CCA GCA CGT TTC-3′) and 3′ MDRB (5′-GCA AGC TTT CAC CTC GCC GCT G-3′) were used at final concentrations of 680 nM each. PCR was carried out with a GeneAmp System 9600 thermocycler (Perkin-Elmer Inc., Norwalk, Conn.). Samples were denatured at 96°C for 2 min followed by 5 cycles of 25 s at 96°C and 60 s at 72°C; 21 cycles of 25 s at 96°C, 50 s at 67°C, and 45 s at 72°C; and 4 cycles of 25 s at 96°C, 60 s at 55°C and 80 s at 72°C. The PCR products were analyzed by 1% agarose gel electrophoresis. Ten microliters of each PCR reaction mixture was loaded per lane.
Potential Mamu-A*01-positive animals were identified by the presence of two bands, a 685-bp amplified product and a 260-bp band. DNA sequence analysis was then performed on all potential positive samples to confirm nucleotide sequence identity with the published Mamu-A*01 prototype sequence (16). Prior to being sequenced, the amplified DNA was treated with 1 U per reaction of shrimp alkaline phosphatase and 10 U of exonuclease I for 15 min at 37°C followed by 15 min at 80°C. The sequencing templates were then purified with a QIAquick PCR purification kit (Qiagen, Inc.). For each template, 70 ng of DNA was used for DNA sequencing together with 5 pmol of primer. Four PCR primers were used for sequencing: A*01/F and A*01/R, whose sequences are shown above, and B/1+ (5′-CTG CGC GGC TAC TAC AAC CA-3′) and G/1+ (5′-ATG TAA TCC TTG CCG TCG TA-3′). Sequencing was carried out at a central core sequencing facility on an ABI-373 stretch DNA-sequencing machine, using ABI AmpliTaq FS dye terminator chemistry (Perkin-Elmer, Inc.). All animals used in this study were genotypically Mamu-A*01 positive based on the above screening and were also Mamu-A*01 positive by functional CTL assay.
Peptide mapping of CTL epitopes.
The optimal SIVmac Pol and HIV-1 Env peptides presented by Mamu-A*01 to CTL were determined in functional effector cell assays. PBMC of Mamu-A*01+, infected rhesus monkeys were isolated by centrifugation over Ficoll-Hypaque (Ficopaque; Pharmacia). The PBMC were then specifically stimulated with antigen either by culture with autologous B lymphoblastoid cell line (B-LCL) cells infected with recombinant vaccinia virus or by the addition of 20- or 25-amino-acid peptides. When using recombinant vaccinia virus-infected B-LCL cells as stimulator cells, PBMC from infected, Mamu-A*01+ monkeys were maintained at a density of 2 × 106/ml and cocultured with an equal number of paraformaldehyde-fixed, autologous B-LCL cells infected with a recombinant vaccinia virus expressing HIV-1 gp160 (v299). When long peptides were used as stimulating antigens, PBMC from infected, Mamu-A*01+ monkeys were cultured with 50 μg of the indicated peptide (SIV Pol p68 [WQVTWIPEWDFISTPPLVRLVFNLV] or HIV-1 Env p41 [GKAMYAPPIEGQIRCSSNIT])/ml and maintained at a density of 5 × 106/ml. On day 3 of culture, the medium was supplemented with human recombinant interleukin 2 (rIL-2) (20 U/ml; provided by Hofmann-La Roche), and the cultures were maintained for an additional 7 days. The cells were then centrifuged over Ficoll-Hypaque and assessed as effectors in a standard 51Cr release assay at an effector-to-target (E/T) ratio of 10:1 as described below. The target cells were autologous B-LCL cells pulsed with decreasing concentrations (10 to 0.01 ng/ml) of the indicated peptides.
Cytotoxicity assay.
PBMC from infected, Mamu-A*01+ monkeys were cultured with 1.0 μg of the indicated optimal peptide (p11C,C-M [CTPYDINQM], p68A [STPPLVRLV], p41A [YAPPISGQI]) and maintained at a density of 2 × 106/ml. On day 3 of culture, the medium was supplemented with human rIL-2 (20 U/ml; provided by Hofmann-La Roche), and the cultures were maintained for an additional 7 days. The cells were then centrifuged over Ficoll-Hypaque and assessed as effectors in a standard 51Cr release assay at an E/T ratio of 10:1. The target cells were Mamu-A*01+ B-LCL cells pulsed during overnight 51Cr labeling either with 1.0 μg of the same peptide used to stimulate the cultured cells or with 1.0 μg of the control peptide p11B (ALSEGCTPYDIN) per ml. All wells were assayed in quadruplicate. The plates were incubated for 5 h in a humidified incubator at 37°C. Specific release was calculated as (experimental release − spontaneous release)/(maximum release − spontaneous release) × 100.
Mamu-A*01/peptide/β2m complex formation and staining of peptide-specific CD8+ T lymphocytes.
Mamu-A*01/p11C,C-M/β2m (SIVmac Gag), Mamu-A*01/p68A/β2m (SIVmac Pol), and Mamu-A*01/p41A/β2m (HIV-1 Env) complexes were prepared as previously described (11). Phycoerythrin (PE)-labeled ExtrAvidin (Sigma) was mixed stepwise with biotinylated Mamu-A*01/peptide complexes at a molar ratio of 1:4 to produce the tetrameric complexes. All antibodies used in this study were directly coupled to fluorescein isothiocyanate (FITC), PE-Texas red (ECD), or allophycocyanin (APC). The following monoclonal antibodies were used: anti-CD8α (Leu2a)-FITC (Becton Dickinson), anti-CD8αβ-ECD (Coulter), and anti-CD3-APC (FN18; kindly provided by D. M. Neville, Jr., National Institutes of Health, Bethesda, Md.).
The PE-coupled tetrameric Mamu-A*01/peptide/β2m complexes were used in combination with anti-CD8α-FITC, anti-CD8αβ-ECD, and anti-CD3-APC to stain 100 μl of fresh blood or 2 × 105 lymphocytes isolated by Ficoll-Hypaque density gradient centrifugation following in vitro peptide stimulation as previously described (11). Whole-blood samples were lysed with an Immunoprep reagent system and a Q-prep workstation (Beckman Coulter Inc.). Ten thousand gated events were collected, and samples were analyzed on a Coulter EPICS Elite ESP flow cytometer. Data analysis was performed with the EPICS Elite software (version 4.02; Beckman Coulter Inc.). Data presentation was performed by using WINMDI software version 2.7 (Joseph Trotter, La Jolla, Calif.) and PowerPoint 97 (Microsoft, Redmond, Wash.).
RESULTS
Identification of novel Mamu-A*01-restricted SIVmac and SHIV CTL epitopes.
To increase the power of the rhesus monkey model for exploring the role of CTL in AIDS immunopathogenesis, we sought to identify Mamu-A*01-restricted SIVmac- and SHIV-derived CTL epitopes in addition to the Gag p11C,C-M peptide. PBMC from Mamu-A*01+, SHIV-infected rhesus monkeys were stimulated in vitro with pools of overlapping 20- or 25-amino-acid peptides spanning the entire SIVmac Pol and HIV-1 Env proteins. The peptide-stimulated cultures were then assessed for CTL activity specific for autologous B-LCL cells pulsed with each of the individual peptides contained within the pool of stimulating peptides (data not shown). In studies of PBMC from three Mamu-A*01+, SHIV-infected monkeys, potential Mamu-A*01-restricted CTL epitopes were detected in an HIV-1 Env peptide (p41 [GKAMYAPPIEGQIRCSSNIT]; amino acids 393 to 412) (19) and in a SIVmac Pol peptide (p68 [WQVTWIPEWDFISTPPLVRLVFNLV]; amino acids 876 to 900) (7).
Identification of the optimal HIV-1 Env-derived Mamu-A*01-restricted CTL epitope.
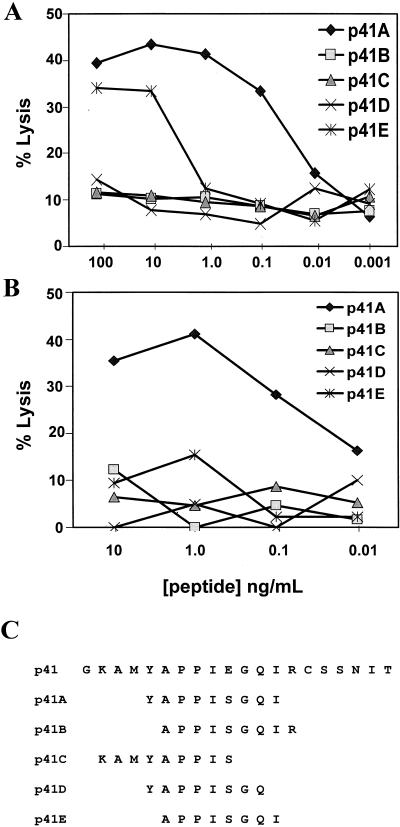
Since the peptide binding motif for Mamu-A*01 has recently been shown to have a proline residue at position 3 (1), we focused the fine mapping of these epitopes on 8- to 10-amino-acid peptides within these sequences that contain a proline at that position. For characterization of the HIV-1 Env-derived epitope, two different experimental approaches were used to generate HIV-1 Env-specific effector cells. In the first approach, paraformaldehyde-fixed, autologous B-LCL cells infected with a recombinant vaccinia virus (vv299) expressing the entire HIV-1 gp160 were used to stimulate PBMC from a SHIV-infected monkey. In the second approach, the 20-amino-acid peptide p41 was used to stimulate PBMC from a SHIV-infected monkey. Neither strategy should bias the specificity of the expanded effector cell population, since both depend upon intracellular antigen processing to generate the Mamu-A*01-binding peptide. HIV-1 Env-specific effector T cells generated by each of these approaches were used in a standard 51Cr release assay to assess lysis of autologous B-LCL cells pulsed with each of the peptides shown in Fig. 1C. Effector cells generated by both approaches exhibited preferential lysis of autologous B-LCL cells pulsed with the nine-amino-acid peptide p41A (YAPPISGQI) (Fig. 1A and B).
FIG. 1.
Mapping of the optimal Mamu-A*01-restricted HIV-1 Env CTL epitope. (A) PBMC from a SHIV-HXBc2-infected rhesus monkey (L28) were cultured for 10 days with paraformaldehyde-fixed, autologous B-LCL cells infected with a vaccinia virus (vv299) expressing HIV-1 gp160. The effector cells were assayed at an E/T ratio of 10:1 with autologous B-LCL targets cultured overnight with the indicated synthetic peptides. (B) PBMC from a SHIV-HXBc2-infected rhesus monkey (L3) were cultured for 10 days with the HIV-1 gp160 20-amino-acid p41 peptide (GKAMYAPPIEGQIRCSSNIT) at a final concentration of 50 μg/ml. The effector cells were assayed at an E/T ratio of 10:1 with Mamu-A*01+ B-LCL targets cultured overnight with the indicated synthetic peptides. (C) Sequences of p41-derived peptides with a proline (P) at position 3.
Identification of the optimal SIVmac Pol-derived Mamu-A*01-restricted CTL epitope.
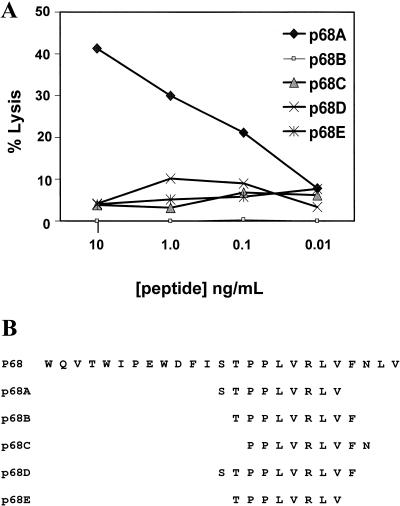
To map the SIVmac Pol-derived Mamu-A*01-restricted CTL epitope, PBMC from an SIVmac-infected monkey were stimulated in vitro with the 25-amino-acid p68 peptide. The resulting effector cells were then used to assess lysis of autologous B-LCL cells pulsed with a number of 8-, 9-, and 10-amino-acid peptides within the SIVmac Pol sequence containing a proline at position 3 (Fig. 2B). The nine-amino-acid peptide p68A (STPPLVRLV) was preferentially recognized by the Pol-specific effector cells (Fig. 2A).
FIG. 2.
Mapping of the optimal Mamu-A*01-restricted SIVmac Pol CTL epitope. (A) PBMC from an SIVmac-infected rhesus monkey (GL9) were cultured for 10 days with 10 μg of the SIVmac Pol 25-amino-acid p68 peptide (WQVTWIPEWDFISTPPLVRLVFNLV) per ml. The effector cells were assayed at an E/T Ratio of 10:1 with autologous B-LCL cells cultured overnight with the indicated synthetic peptides. (B) Sequences of p68-derived peptides with a proline (P) at position 3.
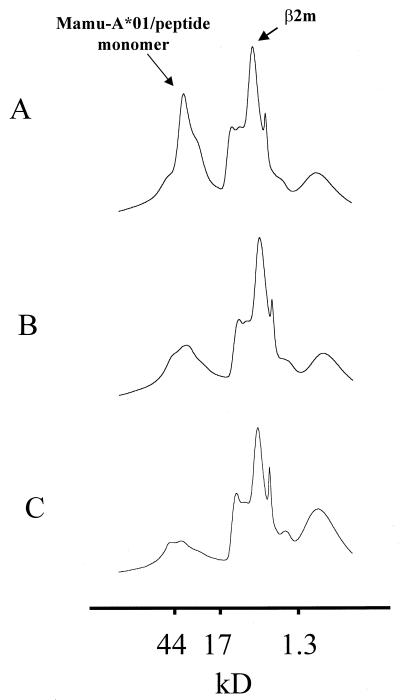
We then sought to confirm that p68A was the optimal Mamu-A*01-restricted SIVmac Pol epitope by assessing the interaction of selected peptides with the Mamu-A*01 molecule. Fluorescent dye-coupled tetrameric MHC class I/peptide/β2m complexes have recently been shown to bind subpopulations of epitope-specific CD8+ T cells, allowing for flow cytometric analysis of epitope-specific CTL. Critical to the development of these tetrameric staining reagents is the efficient in vitro folding of the soluble MHC class I monomers around specific peptides. The efficiency of this in vitro folding reaction can be monitored by assessing the conversion of the 31-kDa MHC class I heavy chain and the 12-kDa β2m to a 43-kDa MHC class I/peptide/β2m monomer (11). Peptides that bind with high affinity to the MHC class I heavy chain should induce efficient folding of the MHC class I/peptide/β2m monomers. Peptides that fail to bind efficiently to the MHC class I heavy chain should fail to produce high-molecular-weight MHC class I/peptide/β2m monomers.
To confirm the results of the functional SIVmac Pol peptide fine-mapping experiment shown in Fig. 2, we initiated small-scale in vitro folding reactions to assess the ability of the two nine-amino-acid peptides p68A and p68B to induce folding of the Mamu-A*01/β2m complex. Formation of the folded 43-kDa Mamu-A*01/peptide/β2m complex was monitored by gel filtration. As shown in Fig. 3A, the peptide p68A bound efficiently to Mamu-A*01, inducing the formation of a 43-kDa Mamu-A*01/peptide/β2m complex. In contrast to this, the peptide p68B failed to bind efficiently to Mamu-A*01 and failed to induce folding of the 43-kDa Mamu-A*01/peptide/β2m complex (Fig. 3B) above that seen in the absence of exogenous peptide (Fig. 3C).
FIG. 3.
Folding of soluble Mamu-A*01 and human β2m around SIVmac Pol-derived peptides in vitro. (A) Gel filtration profile of soluble Mamu-A*01 monomers refolded with human β2m and the SIVmac Pol-derived peptide p68A. (B) Gel filtration profile of soluble Mamu-A*01 monomers refolded with human β2m and the SIVmac Pol-derived peptide p68B. (C) Gel filtration profile of soluble Mamu-A*01 monomers refolded with human β2m in the absence of exogenous peptide.
CTL specific for SIVmac Pol p68A and HIV-1 Env p41A epitopes are detected in antigen-stimulated PBMC of some, but not all, SIVmac- and SHIV-infected Mamu-A*01+ rhesus monkeys.
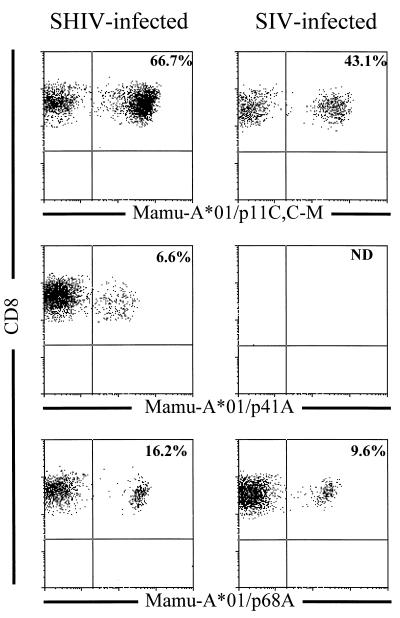
To assess the utility of the newly defined epitopes in monitoring CTL responses in SIVmac- and SHIV-infected, Mamu-A*01+ rhesus monkeys, PBMC from chronically infected animals were stimulated in vitro with 1.0 μg of the optimal SIVmac Gag p11C,C-M, SIVmac Pol p68A, or HIV-1 Env p41A peptide per ml for 10 days in the presence of rIL-2 and assessed for antigen-specific CTL by two independent assays: the detection of MHC class I/peptide/β2m tetramers binding CD8+ T lymphocytes and functional peptide-specific CTL activity (Table 1 and Fig. 4). In the PBMC of all SIVmac- and SHIV-infected, Mamu-A*01+ monkeys tested, high numbers of Mamu-A*01/p11C,C-M/β2m tetramer-binding cells were detected following in vitro peptide p11C,C-M stimulation; p11C,C-M-specific cells constituted from 17.4 to 66.7% of the peptide-stimulated CD8+ T cells. Consistent with the Mamu-A*01/p11C,C-M/β2m tetramer-binding data, high levels of functional p11C,C-M-specific CTL activity were detected in PBMC of all SIVmac- and SHIV-infected, Mamu-A*01+ monkeys tested.
TABLE 1.
CTL specific for SIVmac Gag p11C,C-M, SIVmac Pol p68A, and HIV-1 Env p41A epitopes detectable in PBMC of SHIV- and SIVmac-infected, Mamu-A*01+ rhesus monkeys after in vitro culture with peptidea
| Virus | Monkey | Gag peptide stimulated
|
Env peptide stimulated
|
Pol peptide stimulated
|
|||
|---|---|---|---|---|---|---|---|
| % Gag tetramer-binding CD8+ T cellsb | % p11C, C-M-specific lysisc | % Env tetramer-binding CD8+ T cells | % p41A-specific lysis | % Pol tetramer-binding CD8+ T cells | % p68A-specific lysis | ||
| SHIV | L3 | 17.4 | 43.3 | 5.4 | 15.4 | 0.5 | 14.9 |
| L9 | 36.0 | 46.5 | 1.5 | 6.9 | 1.5 | 0.6 | |
| 206 | 30.6 | 45.0 | 0.7 | 2.3 | 0.0 | 0.0 | |
| 287 | 66.7 | 66.2 | 6.6 | 32.7 | 16.2 | 57.6 | |
| 556 | 25.8 | 48.2 | 1.8 | 14.7 | 1.3 | 5.3 | |
| SIVmac | 138 | 8.7 | 61.5 | NDd | ND | 0.6 | 5.0 |
| 357 | 31.2 | 58.7 | ND | ND | 1.3 | 26.0 | |
| 403 | 56.6 | 59.7 | ND | ND | 4.4 | 6.1 | |
| 4DK | 43.1 | 43.1 | ND | ND | 9.6 | 37.2 | |
| GL9 | 44.8 | 47.9 | ND | ND | 5.9 | 30.7 | |
PBMC from the indicated rhesus monkeys were cultured in vitro for 10 days with 1.0 μg of the indicated optimal peptide per ml.
Results are reported as percent CD3+ CD8αβ+ PBMC binding the indicated Mamu-A*01/peptide tetrameric complex.
Data represent percent peptide-specific lysis at an E/T ratio of 10:1.
ND, not done.
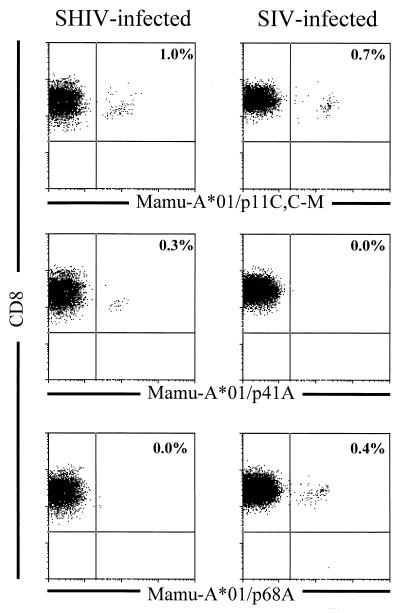
FIG. 4.
Tetramer binding to peptide-stimulated PBMC from SHIV- and SIVmac-infected, Mamu-A*01+ rhesus monkeys. PBMC from SHIV-89.6-infected monkey 287 and SIVmac-infected monkey 4DK were stimulated in vitro with 1.0 μg of the indicated optimal peptide (p11C, C-M [CTPYDINQM], p68A [STPPLVRLV], or p41A [YAPPISGQI]) per ml for 10 days in rIL-2-containing medium. The PE-coupled tetrameric Mamu-A*01/peptide/β2m complexes were used in combination with anti-CD8α-FITC, anti-CD8αβ-ECD, and anti-CD3-APC to stain 2 × 105 lymphocytes isolated by Ficoll-Hypaque density gradient centrifugation following this in vitro peptide stimulation. The percentage of CD8+ T lymphocytes staining positively with the Mamu-A*01/peptide/β2m complex is indicated in the upper right quadrant.
Mamu-A*01/p41A/β2m tetramers binding CD8+ T cells were detected in all the SHIV-infected animals tested following in vitro peptide p41A stimulation, with p41A-specific cells ranging from 0.7 to 6.6% of CD8+ T cells. However, greater than 10% functional Env-specific CTL activity could only be detected in the PBMC of three of five SHIV-infected animals. Animals L3 and L9 were chronically infected with SHIV-HXBc2, which encodes the HIV-1 Env p41A CTL epitope (YAPPISGQI) defined in the studies shown in Fig. 1. Animals 206, 287, and 556 were chronically infected with SHIV-89.6, which encodes a modified HIV-1 Env p41A CTL epitope (YAPPITGQI) containing an S-to-T amino acid change at position 6. Despite this difference in the HIV-1 Env CTL epitope, Mamu-A*01/p41A/β2m tetramer binding and p41A-specific CTL activity in the PBMC of these two groups of animals did not appear to differ significantly.
Mamu-A*01/p68A/β2m tetramer-binding cells were detected in four of five SHIV-infected animals and in all SIVmac-infected animals tested following in vitro peptide p68A stimulation. In the SHIV-infected animals, Mamu-A*01/p68A/β2m binding cells ranged from 0.5 to 16.2% of CD8+ T cells, and in the SIVmac-infected animals, Mamu-A*01/p68A/β2m binding cells ranged from 0.6 to 9.6% of CD8+ T cells. Functional p68A-specific CTL were detected in two of four SHIV-infected animals staining positive with the Mamu-A*01/p68A/β2m tetramer and in three of five SIVmac-infected animals.
These results demonstrate that SIVmac Gag p11C,C-M-specific CD8+ CTL are readily detected in all infected Mamu-A*01+ monkeys, while CD8+ CTL specific for the SIVmac Pol and HIV-1 Env epitopes are present at a lower frequency and are detected in only a fraction of the animals tested.
SIVmac Pol p68A and HIV-1 Env p41A tetramer-binding cells are only occasionally detected in freshly isolated PBMC of chronically infected Mamu-A*01+ rhesus monkeys.
To explore further the relative frequency of CTL specific for the Mamu-A*01-restricted SIVmac Pol and HIV-1 Env epitopes, we assessed freshly isolated whole blood of SIVmac- and SHIV-infected, Mamu-A*01+ monkeys for binding of the SIVmac Gag p11C,C-M, SIVmac Pol p68A, and HIV-1 Env p41A/Mamu-A*01 tetramers to CD8+ T cells (Table 2 and Fig. 5). As shown in Table 2, Mamu-A*01/p11C,C-M/β2m tetramer-binding CD8+ T cells were readily detected in freshly isolated peripheral blood of all SIVmac- and SHIV-infected animals at a frequency of 0.2 to 14.7% of CD8+ T cells. Mamu-A*01/p41A/β2m tetramer-binding CD8+ T cells were detected in freshly isolated peripheral blood of two of five SHIV-infected animals at a frequency of 0.1 to 0.3% of CD8+ T cells. The Mamu-A*01/p41A/β2m tetramer failed to bind to PBMC of the SIVmac-infected animals, confirming the specificity of the reagent. Mamu-A*01/p68A/β2m tetramer-binding CD8+ T cells were detected in freshly isolated peripheral blood of one of five SHIV-infected monkeys at a frequency of 0.1% of CD8+ T cells and one of six SIVmac-infected monkeys at a frequency of 0.4% of CD8+ T cells. It is interesting to note that the animals with measurable Mamu-A*01–p68A tetramer (287 and 4DK) and Mamu-A*01/p41A/β2m tetramer (L3 and 287) staining in freshly isolated PBMC were the animals with the highest level of tetramer staining following in vitro peptide stimulation.
TABLE 2.
Binding of freshly isolated CD8+ peripheral blood T cells from Mamu-A*01+, infected rhesus monkeys to Mamu-A*01–Gag, -Env, and -Pol peptide tetramers
| Virus | Monkeya | % Tetramer-binding CD8+ T cellsb
|
||
|---|---|---|---|---|
| SIVmac Gag p11C,C-M | HIV-1 Env p41A | SIVmac Pol p68A | ||
| SHIV | L3 | 1.0 | 0.3 | 0.0 |
| L9 | 0.7 | 0.0 | 0.0 | |
| 206 | 0.6 | 0.0 | 0.0 | |
| 287 | 2.7 | 0.1 | 0.1 | |
| 556 | 0.2 | 0.0 | 0.0 | |
| SIV | 138 | 3.8 | 0.0 | 0.0 |
| 357 | 1.1 | 0.0 | 0.0 | |
| 403 | 14.7 | NDc | 0.0 | |
| 4DK | 0.7 | 0.0 | 0.4 | |
| GL9 | 1.1 | 0.0 | 0.0 | |
Mamu-A*01+ rhesus monkeys chronically infected with SIVmac or SHIV (SHIV-89.6 or SHIV-HXBc2)
Percent freshly isolated CD3+ CD8αβ+ PBMC binding of the indicated MamuA*01/peptide tetrameric complex.
ND, not done.
FIG. 5.
Tetramer binding to freshly isolated PBMC from SHIV- and SIVmac-infected, Mamu-A*01+ rhesus monkeys. Fresh blood (100 μl) from SHIV-HXBc2-infected monkey L3 and SIVmac-infected monkey 4DK were stained with the indicated Mamu-A*01/peptide/β2m complex in combination with anti-CD8α-FITC, anti-CD8αβ-ECD, and anti-CD3-APC. The percentage of CD8+ T lymphocytes staining positively with the Mamu-A*01/peptide/β2m complex is indicated in the upper right quadrant.
DISCUSSION
The study of CTL is greatly facilitated by the elucidation of CTL epitopes and the MHC class I molecules that present these peptides to effector lymphocytes. Antigen-specific CTL populations are readily expanded in vitro by stimulation with epitope peptides, and target cells pulsed with these peptides can be used in 51Cr release assays to avoid the high levels of background lysis associated with the use of virus-infected target cells. To expand the utility of the SIV-macaque model for studies of AIDS pathogenesis and vaccine development, we sought to identify Mamu-A*01-restricted SIVmac and SHIV CTL epitopes in rhesus monkeys in addition to the previously reported SIVmac Gag p11C,C-M epitope (1, 16). Using PBMC from Mamu-A*01+, SHIV-infected rhesus monkeys stimulated in vitro with pools of overlapping peptides spanning the entire SIVmac Pol and HIV-1 Env proteins, we identified and fine mapped two novel Mamu-A*01-restricted CTL epitopes: the SIVmac Pol-derived epitope p68A (STPPLVRLV) and the HIV-1 Env-derived p41A epitope (YAPPISGQI).
Allen et al. recently defined a consensus motif for peptides that bind to the rhesus monkey HLA-A homologue molecule Mamu-A*01 (1). Sequence analysis of peptides eluted from purified Mamu-A*01 molecules revealed an enrichment of the signal for a proline residue in position 3, suggesting that proline at the third position of the CTL epitope is the anchor residue critical for the binding of a peptide to Mamu-A*01. They also identified threonine (T) at position 2, proline (P) at position 4, isoleucine (I) at position 6, asparagine (N) at position 7, and glutamine (Q) at position 8 as possible auxiliary anchor residues or preferred residues of Mamu-A*01-restricted CTL epitopes. The definition of the novel Mamu-A*01-restricted SIVmac Pol and HIV-1 Env epitopes in the present study confirms the predicted importance of proline as the position 3 anchor residue. Of the six residues reported in the consensus sequence by Allen et al. (1), p11C,C-M shows sequence identity with five. The SIVmac Pol p68A (threonine at P2, proline at P3, and proline at P4) and HIV-1 Env p41A (proline at P3, proline at P4, and glutamine at P8) each contain three residues with sequence identity to the consensus sequence (Table 3). There is reason to suppose that further epitope-mapping studies should allow the definition of Mamu-A*01-restricted CTL epitopes derived from SIVmac Env and viral auxiliary proteins.
TABLE 3.
Sequences of consensus and defined optimal Mamu-A*01-restricted CTL epitopes
| Epitope | Amino acid at position:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
| Consensusa | ∗b | T | P | P | ∗ | I | N | Q | ∗ |
| p11C,C-M | C | T | P | Y | D | I | N | Q | M |
| p41A | Y | A | P | P | I | S | G | Q | I |
| p68A | S | T | P | P | L | V | R | L | V |
Determined by peptide elution from Mamu-A*01 as reported by Allen et al. (1).
∗, any amino acid.
While all the Mamu-A*01+ rhesus monkeys evaluated had high-frequency SIVmac Gag p11C,C-M-specific CTL responses, only a fraction of these monkeys had detectable functional CTL specific for SIVmac Pol p68A and HIV-1 Env p41A, and these CTL were detected at relatively low frequencies by tetramer binding. These findings suggest that Gag p11C,C-M is a dominant epitope and both SIVmac Pol p68A and HIV-1 Env p41A are nondominant CTL epitopes. The reason for the difference in the frequency of CD8+ CTL specific for these epitopes is unclear. The ability of a viral peptide to elicit CTL is likely to be influenced by a number of factors: (i) the peptide-MHC class I affinity and rate of peptide dissociation, (ii) the amount of viral antigen expressed and the amount of antigen that can enter the MHC class I antigen processing pathway, (iii) the rate at which viral antigen is degraded, and (iv) the efficiency and selectivity of peptide transport into the endoplasmic reticulum by the TAP transporter (27). Some nondominant CTL epitopes appear to be naturally processed and presented but remain poorly immunogenic because of their low MHC class I binding affinities (22, 25, 26). It has been demonstrated that highly immunogenic CTL epitopes invariably exhibit MHC class I antigen binding affinities of 50 nM or less, while poorly immunogenic CTL epitopes often have MHC class I antigen binding affinities of 500 nM or more (25). The SIVmac Gag p11C,C-M epitope has been shown to bind to Mamu-A*01 with a 50% inhibitory concentration (IC50) of 4.3 nM (1), which is consistent with this peptide’s immunodominance. The fact that SIVmac Pol p68A- and HIV-1 Env p41A-specific CTL can be readily detected in some chronically infected, Mamu-A*01+ rhesus monkeys suggests that these epitopes can be naturally processed.
In a few instances Mamu-A*01/p41A/β2m and Mamu-A*01/p68A/β2m tetramer staining of CD8+ T cells was detected in the absence of demonstrable functional peptide-specific CTL activity. In other instances functional peptide-specific CTL were detected in PBMC that demonstrated very low Mamu-A*01/peptide/β2m tetramer binding. We have seen a quantitative correlation between functional CTL activity and the level of Mamu-A*01/peptide/β2m tetramer-binding CD8+ T cells only when greater than 5% of all CD8+ T cells bind the tetramer. A correlation between functional CTL activity and the level of Mamu-A*01/peptide/β2m tetramer binding has not been demonstrated when the tetramer positivity is as low as has been seen for the nondominant HIV-1 Env p41A and SIVmac Pol p68A epitopes. A failure to detect HIV-1 Env p41A- and SIVmac Pol p68A-specific functional CTL activity may be due to the inconsistency of functional CTL assays when low numbers of specific effector cells are used. It is also possible that an underlying abnormality in T-cell help in the infected monkeys results in a loss of detectable functional activity when low epitope-specific effector numbers are accessed.
Circulating Mamu-A*01/p11C,C-M/β2m tetramer-binding CD8+ T cells in the chronically infected monkeys ranged from 0.2 to 3.8% of all CD8+ CD3+ cells, with one monkey as high as 14.7%. The level of Mamu-A*01/p11C,C-M/β2m tetramer-binding CD8+ T cells in these chronically infected animals remained relatively stable during the course of their evaluation (data not shown). In the setting of primary SIVmac infection, we have recently demonstrated that tetramer-binding CD8+ CD3+ cells specific for the dominant p11C,C-M epitope appeared as early as day 11 postinfection, peaked on day 13 at 1.3 to 8.3%, and declined coincident with the fall in virus load (12). We do not yet know whether the kinetics of the emergence of the CTL response or its anatomic compartmentalization differs for CTL that recognize the dominant and nondominant epitopes.
Through the use of soluble MHC class I/peptide/β2m tetrameric complexes and flow cytometric analysis, it has become possible to define with quantitative precision distinct subpopulations of epitope-specific CD8+ CTL (2–4, 6, 8, 11, 17, 21). In the rhesus monkey model, the application of this technique has so far been restricted to the study of CTL with specificity for a single dominant epitope. In the current study, soluble MHC class I/peptide/β2m complexes were developed for multiple epitopes of the same virus presented to CTL by the same MHC class I molecule. This has allowed us to analyze the CD8+ CTL responses to both dominant and nondominant epitopes. The results indicate that CD8+ CTL responses to dominant CTL epitopes can be easily quantitated with this technology. However, CD8+ CTL responses to nondominant epitopes, due to the low frequency of these epitope-specific cells, may be difficult to quantitate with the tetramer technology.
ACKNOWLEDGMENT
This work was supported by National Institutes of Health grants AI42301, AI35166, and AI20729.
REFERENCES
- 1.Allen T M, Sidney J, del Guercio M F, Glickman R L, Lensmeyer G L, Wiebe D A, DeMars R, Pauza C D, Johnson R P, Sette A, Watkins D I. Characterization of the peptide binding motif of a rhesus MHC class I molecule (Mamu-A*01) that binds an immunodominant CTL epitope from simian immunodeficiency virus. J Immunol. 1998;160:6062–6071. [PubMed] [Google Scholar]
- 2.Altman J D, Moss P A H, Goulder P J R, Barouch D H, McHeyzer-Williams M G, Bell J I, McMichael A J, Davis M M. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 3.Busch D H, Pilip I M, Vijh S, Pamer E G. Coordinate regulation of complex T cell populations responding to bacterial infection. Immunity. 1998;8:353–362. doi: 10.1016/s1074-7613(00)80540-3. [DOI] [PubMed] [Google Scholar]
- 4.Callan M F, Tan L, Annels N, Ogg G S, Wilson J D, O’Callaghan C A, Steven N, McMichael A J, Rickinson A B. Direct visualization of antigen-specific CD8+ T cells during the primary immune response to Epstein-Barr virus in vivo. J Exp Med. 1998;187:1395–1402. doi: 10.1084/jem.187.9.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Z W, Kou Z C, Lekutis C, Shen L, Zhou D, Halloran M, Li J, Sodroski J, Lee-Parritz D, Letvin N L. T cell receptor V beta repertoire in an acute infection of rhesus monkeys with simian immunodeficiency viruses and a chimeric simian-human immunodeficiency virus. J Exp Med. 1995;182:21–31. doi: 10.1084/jem.182.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flynn K J, Belz G T, Altman J D, Ahmed R, Woodland D L, Doherty P C. Virus-specific CD8+ T cells in primary and secondary influenza pneumonia. Immunity. 1998;8:683–691. doi: 10.1016/s1074-7613(00)80573-7. [DOI] [PubMed] [Google Scholar]
- 7.Franchini G, Gurgo C, Guo H G, Gallo R C, Collalti E, Fargnoli K A, Hall L F, Wong-Staal F, Reitz M S., Jr Sequence of simian immunodeficiency virus and its relationship to the human immunodeficiency viruses. Nature. 1987;328:539–543. doi: 10.1038/328539a0. [DOI] [PubMed] [Google Scholar]
- 8.Gallimore A, Glithero A, Godkin A, Tissot A C, Pluckthun A, Elliott T, Hengartner H, Zinkernagel R. Induction and exhaustion of lymphocytic choriomeningitis virus-specific cytotoxic T lymphocytes visualized using soluble tetrameric major histocompatibility complex class I-peptide complexes. J Exp Med. 1998;187:1383–1393. doi: 10.1084/jem.187.9.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knapp L A, Lehmann E, Piekarczyk M S, Urvater J A, Watkins D I. A high frequency of Mamu-A*01 in the rhesus macaque detected by polymerase chain reaction with sequence-specific primers and direct sequencing. Tissue Antigens. 1997;50:657–661. doi: 10.1111/j.1399-0039.1997.tb02927.x. [DOI] [PubMed] [Google Scholar]
- 10.Koup R A, Safrit J T, Cao Y, Andrews C A, McLeod G, Borkowsky W, Farthing C, Ho D D. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuroda M J, Schmitz J E, Barouch D H, Craiu A, Allen T M, Sette A, Watkins D I, Forman M A, Letvin N L. Analysis of Gag-specific cytotoxic T lymphocytes in simian immunodeficiency virus-infected rhesus monkeys by cell staining with a tetrameric major histocompatibility complex class I-peptide complex. J Exp Med. 1998;187:1373–1381. doi: 10.1084/jem.187.9.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuroda M J, Schmitz J E, Charini W A, Nickerson C E, Lifton M A, Lord C I, Forman M A, Letvin N L. Emergence of CTL coincides with clearance of virus during primary simian immunodeficiency virus infection in rhesus monkeys. J Immunol. 1999;162:5127–5133. [PubMed] [Google Scholar]
- 13.Letvin N L, Daniel M D, Sehgal P K, Desrosiers R C, Hunt R D, Waldron L M, MacKey J J, Schmidt D K, Chalifoux L V, King N W. Induction of AIDS-like disease in macaque monkeys with T-cell tropic retrovirus STLV-III. Science. 1985;230:71–73. doi: 10.1126/science.2412295. [DOI] [PubMed] [Google Scholar]
- 14.Letvin N L, King N W. Immunologic and pathologic manifestations of the infection of rhesus monkeys with simian immunodeficiency virus of macaques. J Acquired Immune Defic Syndr. 1990;3:1023–1040. [PubMed] [Google Scholar]
- 15.Miller M D, Lord C I, Stallard V, Mazzara G P, Letvin N L. The gag-specific cytotoxic T lymphocytes in rhesus monkeys infected with the simian immunodeficiency virus of macaques. J Immunol. 1990;144:122–128. [PubMed] [Google Scholar]
- 16.Miller M D, Yamamoto H, Hughes A L, Watkins D I, Letvin N L. Definition of an epitope and MHC class I molecule recognized by gag-specific cytotoxic T lymphocytes in SIVmac-infected rhesus monkeys. J Immunol. 1991;147:320–329. [PubMed] [Google Scholar]
- 17.Murali-Krishna K, Altman J D, Suresh M, Sourdive D J, Zajac A J, Miller J D, Slansky J, Ahmed R. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity. 1998;8:177–187. doi: 10.1016/s1074-7613(00)80470-7. [DOI] [PubMed] [Google Scholar]
- 18.Musey L, Hughes J, Schacker T, Shea T, Corey L, McElrath M J. Cytotoxic-T-cell responses, viral load, and disease progression in early human immunodeficiency virus type 1 infection. N Engl J Med. 1997;337:1267–1274. doi: 10.1056/NEJM199710303371803. [DOI] [PubMed] [Google Scholar]
- 19.Myers G, Korber B, Wain-Hobson S, Smith R F, Pavlakis G N. Human retroviruses and AIDS 1993: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N. Mex: Los Alamos National Laboratory; 1993. [Google Scholar]
- 20.National Research Council. Guide for the care and use of laboratory animals. Washington, D.C: National Academic Press; 1996. [Google Scholar]
- 21.Ogg G S, Jin X, Bonhoeffer S, Dunbar P R, Nowak M A, Monard S, Segal J P, Cao Y, Rowland-Jones S L, Cerundolo V, Hurley A, Markowitz M, Ho D D, Nixon D F, McMichael A J. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science. 1998;279:2103–2106. doi: 10.1126/science.279.5359.2103. [DOI] [PubMed] [Google Scholar]
- 22.Oukka M, Riche N, Kosmatopoulos K. A nonimmunodominant nucleoprotein-derived peptide is presented by influenza A virus-infected H-2b cells. J Immunol. 1994;152:4843–4851. [PubMed] [Google Scholar]
- 23.Pantaleo G, Demarest J F, Soudeyns H, Graziosi C, Denis F, Adelsberger J W, Borrow P, Saag M S, Shaw G M, Sekaly R P, et al. Major expansion of CD8+ T cells with a predominant V beta usage during the primary immune response to HIV. Nature. 1994;370:463–467. doi: 10.1038/370463a0. [DOI] [PubMed] [Google Scholar]
- 24.Reimann K A, Tenner-Racz K, Racz P, Montefiori D C, Yasutomi Y, Lin W, Ransil B J, Letvin N L. Immunopathogenic events in acute infection of rhesus monkeys with simian immunodeficiency virus of macaques. J Virol. 1994;68:2362–2370. doi: 10.1128/jvi.68.4.2362-2370.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sette A, Vitiello A, Reherman B, Fowler P, Nayersina R, Kast W M, Melief C J, Oseroff C, Yuan L, Ruppert J, Sidney J, del Guerico M F, Southwood S, Kubo R T, Chesnut R W, Grey H M, Chisari F V. The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J Immunol. 1994;153:5586–5592. [PubMed] [Google Scholar]
- 26.van der Burg S H, Visseren M J, Brandt R M, Kast W M, Melief C J. Immunogenicity of peptides bound to MHC class I molecules depends on the MHC-peptide complex stability. J Immunol. 1996;156:3308–3314. [PubMed] [Google Scholar]
- 27.York I A, Rock K L. Antigen processing and presentation by the class I major histocompatibility complex. Annu Rev Immunol. 1996;14:369–396. doi: 10.1146/annurev.immunol.14.1.369. [DOI] [PubMed] [Google Scholar]