Abstract
Synonymous mutations in messenger RNAs (mRNAs) can reduce protein–protein binding substantially without changing the protein’s amino acid sequence. Here, we use coarse-grain simulations of protein synthesis, post-translational dynamics, and dimerization to understand how synonymous mutations can influence the dimerization of two E. coli homodimers, oligoribonuclease and ribonuclease T. We synthe-size each protein from its wildtype, fastest- and slowest-translating synonymous mRNAs in silico and calculate the ensemble-averaged interaction energy between the resulting dimers. We find synonymous mutations alter oligoribonuclease’s dimer properties. Relative to wildtype, the dimer interaction energy becomes 4% and 10% stronger, respectively, when translated from its fastest- and slowest-translating mRNAs. Ribonuclease T dimerization, however, is insensitive to synonymous mutations. The structural and kinetic origin of these changes are misfolded states containing non-covalent lasso-entanglements, many of which structurally perturb the dimer interface, and whose probability of occurrence depends on translation speed. These entangled states are kinetic traps that persist for long time scales. Entanglements cause altered dimerization energies for oligoribonuclease, as there is a large association (odds ratio: 52) between the co-occurrence of non-native self-entanglements and weak-binding dimer conformations. Simulated at all-atom resolution, these entangled structures persist for long timescales, indicating the conclusions are independent of model resolution. Finally, we show that regions of the protein we predict to have changes in entanglement are also structurally perturbed during refolding, as detected by limited-proteolysis mass spectrometry. Thus, non-native changes in entanglement at dimer interfaces is a mechanism through which oligomer structure and stability can be altered.
Keywords: misfolding, function, kinetics, mRNA, non-equilibrium, protein folding, co-translational folding
Introduction
Oligomerization, the process of assembling multiple macromolecules into dimers and higher-order oligomers, is necessary for most proteins to function.1 These functional oligomeric assemblies require the correct type, number, conformational state, and orientation of each constituent protein monomer.2,3 For example, the monomers composing the active tetrameric forms of β-galactosidase4 and hemoglobin5 do not function efficiently on their own. An analysis of 452 human enzymes found roughly one-third (141) to be monomeric, one-third to be homodimers (125), and the remaining third to be heterodimers or higher order oligomers.6 Just as the native structures of proteins represent their minimum free energy structure at equilibrium, thermodynamics is also thought to dictate the structural ensemble of oligomeric complexes. From this thermodynamic perspective, the initial conditions and history associated with a system have no long-term effect on its behavior, meaning that the influence of translation-elongation kinetics should be irrelevant to the structures a dimer adopts.
Contrary to this prediction, experiments have revealed that changes to the speed of protein translation can perturb post-translational oligomerization and protein function over biologically long timescales, indicating a role of kinetics and changes in co-translational processes. For example, when the sub-optimal codon usage in the frq gene encoding the FRQ circadian clock protein in N. crassa is “optimized” by replacing rare codons with common synonymous codons that tend to be translated faster, it binds 60% less to the WC-2 protein even after controlling for soluble expression level changes. This decrease in affinity effectively abolishes N. crassa’s circadian rhythm measured over the course of multiple days.7 Thus, synonymous mutations can change the structure and function of protein complexes and cause phenotypic changes in organisms.
Recent studies8,9 have suggested a mechanism by which synonymous mutations can alter monomeric protein enzyme structure and function, and how these changes can persist in the presence of the proteostasis machinery – such as chaperones and the proteasome – that evolved to fix or remove misfolded proteins. These studies indicate that long-lived misfolded states are self-entangled, leading to reduced structure and function. Many of these entangled structures resemble the native state and thus can evade chaperones, avoid aggregation, and fail to be degraded, allowing them to remain soluble but less functional on timescales ranging from seconds to months or longer. The partitioning of nascent proteins into such soluble but self-entangled conformations has the potential to explain how changes to translation kinetics can disrupt oligomer formation for long time periods.
Here, we use coarse-grain and all-atom molecular dynamics simulations to understand the structural origin of altered dimerization when synonymous mutations are introduced into a protein’s mRNA template. Because FRQ is an intrinsically disordered protein10 whose binding interface and structure are unknown, we instead study the dimerization of two globular, cytosolic E. coli homodimers – oligoribonuclease and ribonuclease T – after synthesis from their wildtype, fastest-translating synonymous variant, and slowest-translating synonymous variant mRNA sequences. We chose these proteins on the basis that they are small, related, single-domain homodimers with high-resolution X-ray structures available of the full-length protein. Ribonuclease T folds relatively quickly and the speed of translation has no discernible influence on its ability to dimerize. On the other hand, the dimerization of oligoribonuclese (which shares the same RNase H-like topology as ribonuclease T) does depend on the mRNA variant from which it is synthesized. In the following, we identify a molecular origin for this phenomenon, show the results are robust to changes in model resolution, explain why the mechanism we identify is likely to be widespread across the proteome, and find Limited Proteolysis Mass Spectrometry data on global refolding reactions are consistent with the computationally observed entangled states.
Methods Summary
Full details of the coarse-grain model, coarse-grain and all-atom simulations, data analyses, and experiments are provided in the Supplementary Information. Briefly, we employed a previously published Gō-like coarse-grain methodology in which each amino acid is represented by a single interaction site. Ribosomal RNA is represented with one bead each for the ribose, phosphate, and pyrimidine nucleobases and two beads for purine nucleobases.9,11-13 Each protein monomer is synthesized one amino acid at a time inside the ribosomal exit tunnel. The dwell time at each nascent chain length during simulated translation is taken from the Fluitt-Vilijoen model14 for E. coli. The mean dwell times for each codon within this model were rescaled to produce an overall average elongation rate equivalent to 20 aa/s adjusted to account for the accelerated timescale of dynamic processes in the coarse-grain model.13 This procedure results in an overall in silico mean translation time of 12.6 ns. Predicted fastest- and slowest-translating synonymous variant mRNAs were generated for oligoribonuclease and ribonuclease T by replacing each codon in their wildtype sequences with the codon predicted by the Fluitt-Viljoen model to be the fastest- or slowest-translating synonymous codon, respectively (see Supplementary Information for more details). After translation is complete, the full-length nascent chain is released from the ribosome and 5 μs of post-translational dynamics simulated with the ribosome removed. Annealing simulations were carried out for random pairs of final structures from post-translational dynamics to generate dimer structures for interaction energy calculations. Hamiltonian (umbrella sampling) Replica Exchange15,16 simulations were run for representative annealed structures and WHAM17,18 was employed to construct the potential of mean force of dimerization as a function of the interface center-of-mass distance.
To identify non-covalent lasso entanglements, we utilize Gaussian linking numbers.19 Entangled structures were clustered by a leader algorithm.20 To assess whether entanglements correlate with the decrease in binding energy, we compute the conditional probability (odds ratio) of these two events co-occurring. Fisher’s Exact Test was applied to evaluate the significance. Metastable state analyses were performed using the Markov state model and clustering algorithms implemented in the PyEmma package.21 The solvent-accessible surface area of segments of residues was computed using Eq. 16 from ref.9.
For all-atom simulations, the backbone and sidechain atoms of selected structures were rebuilt with PD222 and Pulchra.23 The back-mapped structures were solvated in TIP3P24 water and neutralized by Na+ and Cl− counter-ions before adding 0.15-M sodium chloride to mimic the salt concentration inside the cell.25 Simulations were performed with GROMACS 201826 using the AMBER99SB-ildn forcefield.27 These computational techniques are summarized in Figure 1.
Figure 1. Simulating protein dimerization and entanglement at multiple resolutions.
(a) Side (left) and top (right) views of the coarse-grained 50S E. coli ribosome cutout (filled spheres) used in our simulations superimposed over the entire all-atom 50S subunit (transparent) from PDB ID 3R8T. Ribosomal RNA and protein are displayed in grey and green, respectively. The approximate location of the ribosome exit tunnel is indicated by a blue star in the top view. (b) Side view of a 181-residue oligoribonuclease ribosome nascent chain complex just prior to its release from the ribosome. Note that one side of the exit tunnel was cut away for visualization only. (c) Schematic of simulation protocol. One hundred nascent protein conformations are generated for Monomer A (purple) and Monomer B (grey). Each monomer is synthesized one amino acid at a time using coarse-grain protein and ribosome models (represented here by the purple/grey lines and green/grey shapes, respectively). After synthesis, the monomer is released from the ribosome and its bulk dynamics then simulated for 5 μs. Random combinations of the final structures from bulk dynamics are then selected from the sets of 100 Monomer A and 100 Monomer B trajectories and their lowest-energy dimer configurations determined by temperature annealing. (d) Initial coarse grain and resulting all-atom structures of oligoribonuclease monomer before and after back-mapping. (e) Same as (d) but for a dimeric oligoribonuclease structure.
For experiments, E. coli K12 cells were grown in two different sets of MOPS media then cultured cell pairs were mixed together before lysis. Native extracts were prepared by cryo-milling cells into 20 mM Tris-HCl (pH 8.2), 100 mM NaCl, 2 mM MgCl2, a portion of which was subject to global unfolding (in 6 M guanidinium chloride) and refolding (by 100-fold dilution). Limited proteolysis with proteinase K for 1 min was used to structurally probe proteins in their native and refolded forms. The Proteome Discoverer Software Suite was used to sequence peptides and perform label free quantification (LFQ).
Results
Synonymous mutations alter oligoribonuclease’s post-translational structural ensemble and ability to dimerize.
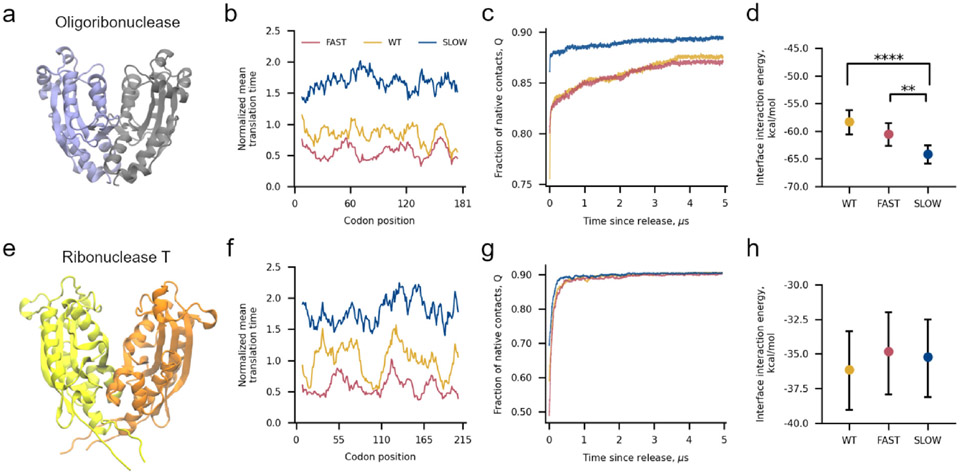
To test if oligoribonuclease’s ability to dimerize is perturbed by synonymous mutations we simulated its synthesis from mRNAs corresponding to its wildtype coding sequence, a slow-translating synonymous mRNA sequence composed of non-optimal codons, and a fast-translating synonymous mRNA sequence composed of optimal codons (Figure 2a, b). After synthesis, we simulated the release of oligoribonuclease from the ribosome followed by 5 μs of post-translational dynamics (the equivalent of approximately 20 s in real time13). We find that oligoribonuclease exhibits structural differences in its post-translational folding dynamics dependent on whether it was translated from the wildtype, fast-translating, or slow-translating mRNA sequences (Figure 2c). The slow-translating mRNA produces a monomer structural ensemble with a higher average fraction of native contacts (Q = 0.89, 95% CI: [0.87, 0.90], computed from bootstrapping 106 times, where Q only considers intra-monomer contacts) relative to the wild-type mRNA (Q = 0.86, 95% CI: [0.85, 0.88], 106 bootstraps) 5 μs after ribosome release. The ensemble of minimum energy dimeric structures from temperature annealing simulations (Figure 1c), reveals that this increase in native structure results in a more favorable dimer interface interaction energy of −64.1 kcal/mol (95% CI [−65.7, −62.4], 106 bootstraps) for the slow mRNA synthesis products compared to dimers produced from the wildtype mRNA of −58.3 kcal/mol (95% CI [−60.3, −55.9], 106 bootstraps). The difference between these interaction energies is significant (p = 2x10−5, permutation test 106 iterations). The average dimer interaction energy of −60.5 kcal/mol (95% CI: [−62.5, −58.4], 106 bootstraps) for the fast-translating mRNA is also different in comparison to the slow-translating mRNA (Figure 2d). These results demonstrate that oligoribonuclease’s post-translational dimerization affinity is affected by changes in translation-elongation speed.
Figure 2.
Altering translation kinetics affects the binding affinity of the oligoribonuclease homodimer. (a) 3D structure of oligoribonuclease from PDB ID 1YTA with Monomers A and B colored light purple and grey, respectively. (b) Mean translation time of codon positions, normalized by the average codon translation time across the 64 codons, within the fast-translating mutant (FAST, red), wildtype (WT, yellow), and slow-translating mutant (SLOW, blue) mRNA sequences used for oligoribonuclease simulations smoothed with a 15-codon moving average. (c) Moving average of fraction of intra-monomer native contacts as a function of time since oligoribonuclease’s release from the ribosome computed over all 200 trajectories (100 Monomer A + 100 Monomer B) for each mRNA. Individual time series were first smoothed by taking the mode within a sliding 15-ns window and then averaged together across all 200 monomer trajectories. (d) Average interface interaction energy between Monomers A and B computed over 200 different random pairs of monomers after annealing as described in Methods and Figure 1c. Error bars are 95% confidence intervals computed from bootstrapping 106 times. Brackets and asterisks indicate statistical significance of comparisons between means determined from permutation tests with 106 samples. (e) 3D structure of ribonuclease T from PDB ID 2IS3 with monomers A and B colored yellow and orange, respectively. (f) Normalized mean translation time of codon positions in ribonuclease T mRNAs used in our simulations. (g) Fraction of native contacts versus time computed from 200 Ribonuclease T trajectories for each mRNA template used. (h) Same as (d) but for interactions between monomers of ribonuclease T. One, two, three, or four asterisks indicate p ≤ 0.05, p ≤ 0.01, p ≤ 0.001, or p ≤ 0.0001, respectively. Fraction of native contact time series computed during simulated translation are available in Figure S8.
Dimerization of ribonuclease T is not influenced by synonymous mutations.
Ribonuclease T’s post-translational structural ensemble (Figure 2e-g) and dimerization interaction energy are not dependent on the translation schedule of the mRNA that encodes it (Figure 2h). No statistically significant differences are found between the average interface interaction energies of the dimer ensembles generated from the wildtype, fast-translating, or slow-translating mRNAs.
Oligoribonuclease, but not ribonuclease T, frequently populates self-entangled states that involve interface residues.
Recent computational studies predict that misfolded proteins often contain entanglements that form long-lived, native-like kinetic traps.8,9 In entangled protein structures, a segment of residues forms a loop (closed by a native contact) through which another segment of residues threads (Figure 3a).28 Mathematically, entanglements within a protein structure can be detected as a change in the Gauss linking number, g(i, j), of the native contacts relative to the folded state (see Methods and Eq. (4)-(6)). This metric of protein structure is topologically invariant29 and describes how segments of the protein are intertwined together in space (see Figure S7).
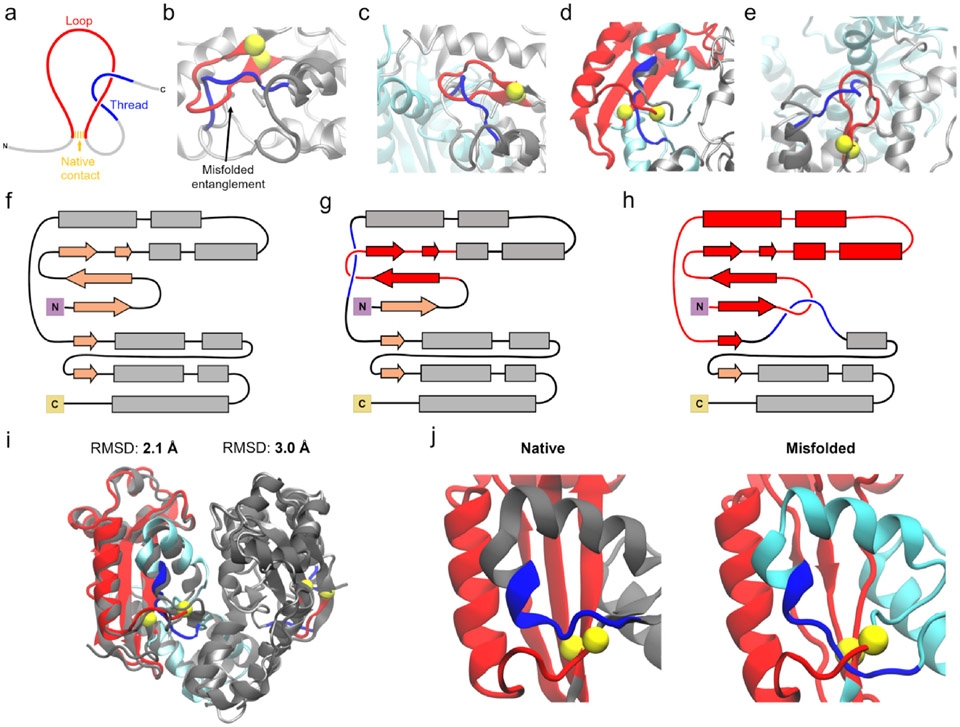
Figure 3. Entanglements in oligoribonuclease perturb its dimer interface.
(a) Entanglements occur when a threading segment (blue) passes through a loop (red) formed by another segment of residues and closed by a native contact (yellow). (b) Structure of oligoribonuclease Monomer B in which residues 96–102 thread through the loop closed by the native contact between residues 31 and 41. Portions of the misfolded structure not involved in the entanglement are displayed in gray, while the location of the threading segment in the native state is shown in dark grey. This structure corresponds to M3 (see Supplementary Data File 1). (c) Same as (b) except in the context of a dimeric complex after annealing (structure D4), with Monomer A displayed in cyan. (d) Structure of oligoribonuclease dimer in which residues 125–129 of Monomer A thread through the loop closed by the contact between residues 9 and 103 (structure D5). (e) Structure of oligoribonuclease Monomer B with the same entanglement as (b) but in the context of a dimer in which Monomer A is also entangled (structure D5). (f) Secondary structure diagram of the native state of a monomer of oligoribonuclease. (g) Secondary structure diagram of the entanglement shown in (b), (c), and (e). (h) Same as (g) but for the entanglement shown in (d). (i) Alignment of the Monomer A and Monomer B structures shown in (d) and (e) to the native state structure indicates they are overall native-like with ≤3-Å Cα Root Mean Square Deviation (RMSD) from the crystal structure after backmapping. (j) Left: native state dimer interface of Monomer A. Right: interface view of the entangled structure of Monomer A from (d). Residues 125–129 of Monomer A thread through the loop from residues 9 to 103, disrupting the formation of a β-sheet that forms part of the dimer interface. Throughout all panels, loops, threads, and the native contact that closes the loop are colored red, blue, and yellow, respectively. Misfolded conformations of Monomers A and B are show in cyan and silver, respectively, and native conformations of the threading segment (as in (b)-(e)) or the overall structure (as in (i) and (j)) are shown in dark grey. Coarse-grain structures were back-mapped to all-atom resolution to generate visualizations.
We identified various entanglements in structures of oligoribonuclease that cause disruptions relative to the native state (Methods). Representative structures of two frequently occurring entanglements, in which residues 96–102 or residues 125–129 thread through a loop, are displayed in Figures 3b-e, and schematic representations are shown in Figures 3g and h. Entanglement can occur in an isolated monomer (Figure 3b), in one monomer that forms part of a dimeric complex (Figure 3c), or in both monomers within a dimer (Figures 3d,e). Each of these entangled structures is very similar to the native structure; for example, the entangled dimer structure shown in Figures 3d and 3e has ≤ 3-Å Cα RMSD from the native state (Figure 3i). The high similarity of these entangled conformations to the native state suggests that they may remain soluble and evade proteostasis quality controls.9 Despite being well-folded overall, entanglements can structurally perturb the dimer interface. In the case of the entanglement of residues 125–129 in Monomer A, misplacement of a loop segment disrupts the formation of a β-sheet at the dimer interface (Figure 3j). This suggests that changes in dimer interaction energy could be caused by entanglements perturbing the dimer binding interface.
Synonymous mutations alter the population of entangled oligoribonuclease structures.
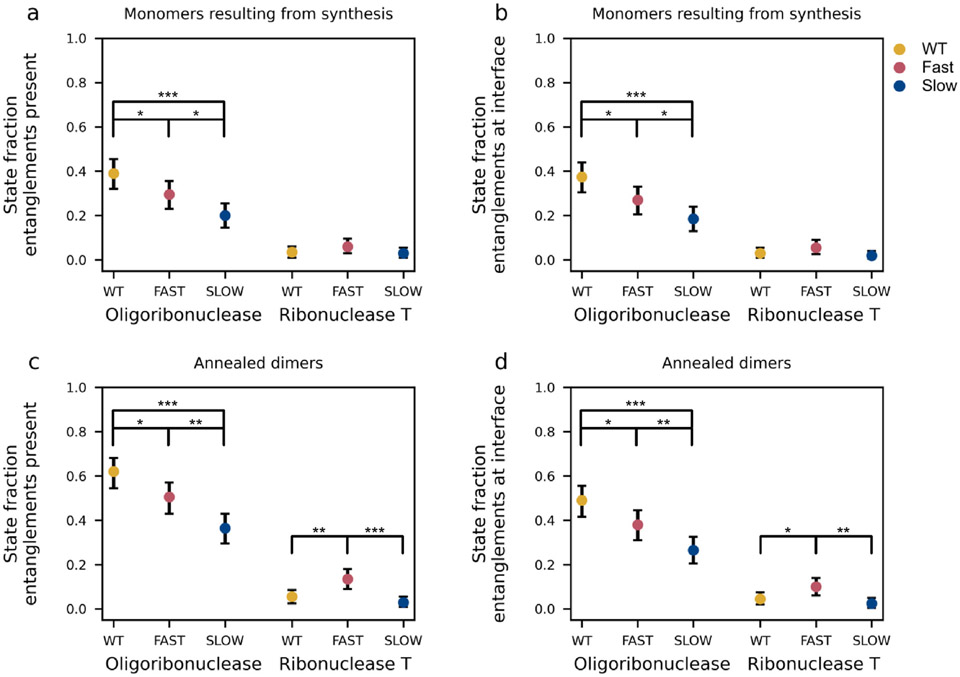
To quantify the influence of synonymous mutations on the likelihood of entanglement in oligoribuclease and ribonuclease T we computed the fraction of monomers (out of 200 independent trajectories) and dimers (out of 200 annealed structures) that exhibit a non-native change in entanglement for both oligoribonuclease and ribonuclease T from their wild-type, fast- and slow-translating mRNAs (Figure 4). We find that ribonuclease T exhibits relatively little entanglement (fraction entangled < 0.15) in both its monomeric and dimeric forms regardless of the translation-rate schedule used during its synthesis. Statistically significant differences are present depending on the translation schedule used (see, for example, Figure 4c), however, the magnitude of these population differences is small (less than 10.5%). This suggests why ribonuclease T’s dimer interaction energy is insensitive to synonymous mutations – any corresponding population changes in misfolded states are modest and have little effect on this protein’s ability to dimerize.
Figure 4. Changes in the population of self-entangled structures correlate with differences in dimer interaction energies.
(a) Fraction of monomer structures of oligoribonuclease and ribonuclease T generated by coarse-grain synthesis, ejection, and post-translational dynamics simulations using the wildtype (WT), fast-translating mutant (FAST), and slow-translating (SLOW) mutant mRNAs that have a gain in entanglement relative to the native state somewhere in their structure. (b) Same as (a) but limited to the specific set of entanglements involving residues at the dimer interface. (c) Same as (a) but computed for the dimer structures generated by annealing random pairs of monomers. (d) Same as (c) but limited to the specific set of entanglements involving interface residues. All error bars are 95% confidence intervals computed from bootstrapping 106 times. Brackets and asterisks indicate the statistical significance of comparisons between means determined from permutation tests with 106 samples. One, two, or three, asterisks indicate p ≤ 0.05, p ≤ 0.01, p ≤ 0.001, respectively.
In contrast, for oligoribonuclease, the population of conformations that display a non-native change in entanglement is larger in magnitude and more sensitive to changes in translation speed. For the wildtype, fast, and slow translation schedules, the fractions of dimer conformations with an overall entanglement are, respectively 0.62 (95% CI: [0.54, 0.68]), 0.51 (95% CI: [0.43, 0.57]), and 0.37 (95% CI: [0.29, 0.43], 106 bootstraps) (Figure 4c). This trend is anticorrelated with the average dimer interaction energies (Figure 2d) where values of −58.3, −60.5, and −64.1 kcal/mol, respectively, are found for the wildtype, fast, and slow mRNA templates. These results suggest that changes in the population of entangled states with perturbed interfaces, arising from changes in translation speed, cause the binding affinity between monomers to be altered.
Misfolded entangled states often involve the dimerization interface of oligoribonuclease.
Next, we asked how frequent it was for misfolded states to have the entanglement located at the dimer interface (see Methods and Figure 4). We find ribonuclease T has relatively low levels of entanglement at the dimer interface of misfolded structures, with probabilities of less than 0.15 (Figures 4b and d). In comparison, oligoribonuclease displays more frequent interface entanglements in both monomer and dimer misfolded structures (Figures 4b and d). Specifically, misfolded dimer structures have probabilities of entanglements located at the interface of 0.49 (95% CI: [0.41, 0.55]), 0.38 (95% CI: [0.31, 0.45]), and 0.26 (95% CI: [0.21, 0.32], 106 bootstraps) for the wildtype, fast-translating, and slow-translating mRNAs, respectively (Figure 4d). Thus, at least for oligoribonuclease, non-native entanglements are relatively frequent at the dimer interface.
Entanglements greatly reduce the likelihood of strong dimer interactions.
To test whether entanglements are associated with decreased average binding energy between monomers, we created a two-by-two contingency table categorizing annealed dimer structures as strongly or weakly bound and as having an entanglement present or not. A contingency table allows us to compute the conditional probability of these two events co-occurring, the odds ratio (effect size) that entanglement and weak binding are associated, and Fisher’s Exact Test tells us whether the association is significant. Statistically significant odds ratios other than 1 would establish an association between these two phenomena.
To classify structures based on their binding energy, we define dimer complexes with interface interaction energy less than or equal to the Xth percentile value from the wildtype interaction energy distribution to be strong binding and all others to be weak binding. Then, as a test of robustness, we systematically vary this threshold, X, in increments of + 5% from 5% to 95% and compute the odds ratio and p-value for each splitting of the data (Figure S1, Supplementary Data File 2).
For oligoribonuclease we find the odds ratio, where it is defined (plotted portions of Figures S1a and c), is significantly greater than 1 for a range of thresholds. For example, at a 50% threshold, the odds ratio is 51.9 and is statistically significant (p-value = 1.4 × 10−9, Fisher’s Exact test; see Supplementary Data File 2) for the slow-translating mRNA variant. This effect size and the consistent significance of these odds ratios demonstrates there is a very strong association between the presence of entanglements and the occurrence of dimers with weak dimerization energies.
Conversely, ribonuclease T’s odds ratio is never statistically different than 1.0 (Figures S1b and d). This indicates, as inferred earlier, that the modest population of entangled structures for ribonuclease T has no association with strong or weak dimerization occurring.
Equilibrium potentials of mean force of dimerization.
A concern is these results reflect only potential energy changes upon dimerization, not free energy changes. Equilibrium binding-and-unbinding simulations are computationally expensive even when employing enhanced sampling and coarse-grained protein models. Therefore, we could only compute the potential of mean force for dimerization for a small number of conformations. Specifically, we selected a representative dimer structure whose interaction energy is closest to the median value within the set of 200 annealed structures generated for a given synonymous mRNA. We ran Hamiltonian replica-exchange sampling to compute the potentials of mean force (Eq. 3) for the process of dimerization. For oligoribonuclease we find that the slow variant has a greater free energy of dimerization than the wildtype variant, indicating slow synthesis produces more stable dimers than the wildtype sequence (Figure S2a). For ribonuclease T we find no difference in the free energy of dimerization between the wildtype and slow variants indicating invariance of the binding strength to synonymous mutations (Figure S2b). Thus, these results, taken together with our earlier results reporting changes in the interaction potential energy and the statistically significant enrichment of entanglements at the interface of oligoribonuclease, are consistently showing an association between binding strength and near native changes in self-entanglement.
Entangled states are long-lived kinetic traps.
Entangled states that are long lived can have long-term impacts on protein structure and function. Therefore, we quantified the lifetime of these states in our simulations. While all monomers of ribonuclease T fold by 0.8 μs after release from the ribosome, some oligoribonuclease molecules fail to fold during the 5-μs post-translational simulations regardless of the mRNA sequence used. When synthesized from its wildtype, fast-translating, and slow-translating variants, respectively, 13% (95% CI [8%, 17%]), 14% (95% CI [9%, 19%]) and 10% (95% CI [6%, 14%], 106 bootstraps) of its monomers do not fold correctly (see Figure S3 and Methods). We used a kinetic curve-fitting procedure to estimate that these misfolded populations of oligoribonuclease require between 6 and 14 μs to fold (the equivalent of approximately 30–60 s of real time13). This indicates that these entangled states are kinetically trapped.
Entanglements persist in all-atom molecular dynamics for up to one microsecond.
To test whether some of our results generated using a Cα coarse-grain representation are resolution dependent we back-mapped representative entangled conformations of the oligoribonuclease dimer and monomer to atomistic resolution and simulated their aqueous dynamics for 500 ns. We ran three statistically independent trajectories for five different dimer starting structures and three different monomer structures selected to represent the entangled conformations most frequently populated and lowest in energy (see Methods and Supplementary Data File 1). In each case, the entanglement present in the coarse-grain model persists at all-atom resolution for the duration of the 500-ns simulation. In addition to these 500-ns simulations, we also extended the simulations for one randomly selected dimer and monomer structures to 1 μs and find the entangled states persist. This is evidenced by the time series of ⟨G⟩ for four representative entanglements, one in a monomer and three in dimers, displayed in Figure S4. Thus, the entangled structures we observe in our coarse-grained simulations can persist in all-atom models.
Entangled states are consistent with experimental signatures of altered structure populated during refolding.
Previous work of ours has found that many E. coli proteins cannot fully refold from denatured states,30 that one reason for this behavior is that upon attempted refolding, some proteins populate entangled misfolded states,9 and that proteins which are more dependent on cotranslational folding tend to be less competent to spontaneously refold correctly.8,31 Moreover, for some “nonrefolding” proteins, an unfolding-refolding cycle can elicit the population of misfolded states, including those with self-entanglements. To test our predicted misfolded structures we utilized previously published limited proteolysis-mass spectrometry (LiP-MS)9,30 data obtained from proteome-wide refolding studies conducted on E. coli extracts. In these experiments, the proteolysis profiles of native proteins (obtained directly from cell extracts) are compared to those from proteins that have been chemically denatured and then refolded through a dilution jump. We have previously shown that misfolded states populated co-translationally can also be populated during refolding,8,32 making it reasonable to compare our misfolded states to these data. Proteolytic fragments that exhibit large changes in their populations between the refolded and native samples are indicative of regions of the protein that have altered protease susceptibility upon refolding. Entanglements can alter protease accessibility. Thus, LiP-MS data provide a means to test the computationally predicted changes in entanglement. In the following, we focus on peptides from oligoribonuclease and ribonuclease T. We consider only peptides that exhibit at least a 2-fold change in abundance between the refolded (R) and native (N) samples (i.e., ) and are statistically different between the refolded and native sample (−log10 p ≥ 2, i.e. p < 0.01).30,31 Here, we limit our analysis to long-lived misfolded states by considering only statistically significant changes detected 120 min after the dilution jump (see Supplementary Data File 4 of ref. 9). At that time point oligoribonuclease has two peptides that show significant changes, consisting of residues 84–94 and 166–175, while ribonuclease T exhibits no significant peptides (out of five peptides detected from that protein; see Table S2). The same LiP-MS results for oligoribonuclease were also found in a previous study performed in a cytosol-like medium,31 and they are moreover qualitatively consistent with our prediction that oligoribonuclease is more likely to populate entangled states than ribonuclease T (Figure 4).
To make a detailed structural comparison between our predicted misfolded states and these LiP-MS data we identify metastable states in the two-dimensional Log-probability surface defined by the order parameters Q and G (Figures 5a-d, see Methods). Both proteins contain several metastable states with a high fraction of native contacts involving a change in entanglement, indicating these states are well folded but with structural perturbations introduced by entanglements (Figure 5). Two of the metastable states identified for oligoribonuclease (0 and 1 in Figures 5a and b) display increases in the solvent-accessible surface area of residues 84–94 relative to the native state reference simulations and together make up 10–16% of the conformations in the overall structural ensemble (Table S3, Figure 5e) produced by translation of the wildtype, fast, and slow mRNAs. Similarly, for residues 166–175, two metastable states (1 and 5) show a statistically significant increase in the solvent-accessible surface area and make up 65–80% of the structural ensembles across wildtype, fast, and slow (Figure 5f, Table S3). Additional metastable state plots for all synonymous mutants are shown in Figures S5 and S6. Thus, metastable states within our entangled state ensemble are consistent with the experimentally observed increase in protease accessibility of both proteolytic peptide fragments.
Figure 5.
Predicted changes in solvent accessible surface area are consistent with LiP-MS refolding experiments. (a) and (c) are the −Log(P) surfaces spanning the fraction of native contacts, Q, and change in entanglement, G, for oligoribonuclease and ribonuclease T, respectively, across WT, fast-translating, and slow-translating variants (see Figures S5 and S6 for results separated by variant). (b) and (d) are the resulting metastable states generated by Markov State models for the data in (a) and (c); the native-like states are 5 and 7, respectively. (e) Structures of oligoribonuclease in the native state and two entangled states with residues 84–94 highlighted in mauve. Percentages are the change in solvent-accessible surface area, relative to the average from the native state simulations, of residues 84 through 94 computed from the ensemble of structures arising from the wildtype translation schedule (see Table S3 for confidence intervals). For entangled conformations, the loop, thread, and contact closing the loop are shown in red, blue, and yellow, respectively (as in Figure 3). (f) Same as (e) but for residues 166–175. Note that State 5 does not contain an entanglement.
Ribonuclease T, on the other hand, only displays a single misfolded metastable state across all three variants with appreciable population (State 7, see Figures 5c and d, Table S4). Of the five sites identified for ribonuclease T, LiP-MS found no significant change in protease susceptibility after refolding; in agreement, our simulations show small changes in solvent-accessible surface area relative to the native state at these sites (Table S4), with all values close to zero. This observation is consistent with our prediction that this protein would refold efficiently, leading to a similar protease accessibility between the native and refolded samples.
Discussion
Our results provide a structural explanation for how changes in translation speed caused by synonymous mutations can alter the ability of soluble proteins to dimerize over long time scales. For the homodimer oligoribonuclease, synonymous mutations change the proportion of protein molecules that partition into soluble, misfolded, self-entangled conformations. These entangled conformations weaken the ensemble-averaged binding energy between the monomers over long time scales. In comparison to oligoribonuclease, ribonuclease T is largely insensitive to synonymous mutations that alter translation speed, with far fewer states with entangled dimer interfaces (≥15%) populated and those that are entangled exhibiting little population dependence on translation speed. We find that the key entangled states identified for oligoribonuclease persist at all-atom resolution for long timescales. On the other hand, ribonuclease T’s dimer binding energy does not change with the introduction of synonymous mutations. Finally, we used structure-based Markov State Models to compare metastable misfolded conformations with LiP-MS analysis of proteome-wide refolding, finding consistency with experimental data that indicate structural perturbation at residues 84–94 and 166–175 of oligoribonuclease. Taken together, these computational and experimental results suggest that oligoribunuclease frequently populates conformations that contain non-native entanglements, and that these long-lived kinetically trapped states fail to fold on biologically relevant timescales.
A commonly held assumption in the nascent protein folding field is that slower translation will result in more co-translational protein folding.33-37 Therefore, one would predict that any changes in dimer interaction energy would follow the trend that the slow-translating mRNA will result in the strongest binding, followed by the wild-type and fast-variant mRNAs of oligoribonuclease. This is not what we observe – we find, respectively, that slow, fast, and wild-type mRNA variants result in increasingly weaker dimer affinities. This result is explained by both kinetic and simulation models showing the influence of translation kinetics on co-translational protein folding is, for some proteins, non-monotonic. Faster translation can result in an increased yield of correctly folded protein by translating quickly through protein segments that are prone to misfolding.38,39 Is it surprising that the wildtype sequence produces the most entangled oligoribonuclease in our simulations? The evolutionary principle of parsimony, that evolution does not further optimize features that are already ‘sufficient’, suggests that the wildtype sequence is “good enough” under normal physiological conditions. That is, it produces enough folded, functional protein that its codon sequence need not be optimized to generate 100% folded, functional protein.40,41,42,43 Indeed, it is generally accepted that faults during primary biosynthesis lead to a substantial fraction of newly synthesized proteins to be immediately degraded.44 Thus, not all wildtype mRNA sequences have been maximally optimized for protein folding efficiency, with many mRNAs leading to enough functional protein so as not to be problematic for the cell. An alternative explanation based on our previous work is that although oligoribonuclease did not fully refold from the denatured state spontaneously,30 it could do so with the assistance of GroEL/ES.31 Hence, it could be that entangled monomers of oligoribonuclease that emerge from the ribosome become substrates for chaperone-mediated folding just like those that initially improperly folded after dilution from denaturant. It should be noted that other entangled states are challenging for chaperones to identify,31,32 which might place a greater onus on establishing folding correctly during translation in those situations.
Oligomer assembly can begin early in the life of a protein, with some nascent chains co-translationally dimerizing between adjacent ribosomes.1 It is unknown how many different proteins engage in such co-translational assembly, though it appears to be preferred by homodimers in specific folds, particularly coiled coils.1 Both ribonuclease T and oligoribonuclease have a RNase-H-like fold, which was not found by Bertolini and co-workers to be a high-confidence co-co assembling candidate. Therefore, in this study we chose to consider their dimerization after their release from the ribosome only. Additionally, the motivating experiments on FRQ assessed only post-translational dimerization. Based on our results, we speculate that co-translational interface interaction energies are likely to follow similar mechanisms as we have identified in this post-translational study. Investigating how synonymous mutations influence co-translational dimerization is an interesting avenue of future research for systems that are likely to assemble co-translationally. However, we point out that post-translational assembly could be the preferred pathway for a protein like oligoribonuclease which may require chaperone assistance between primary biosynthesis and dimerization due to the higher frequency of entanglement formation during cotranslational folding.
In summary, our results indicate that for some proteins synonymous mutations can modulate the amount of nascent protein that misfolds into non-native self-entangled conformations with reduced dimer interface interaction energies. For oligoribonuclease, slowing down or speeding up translation relative to its wildtype translation schedule leads to a reduction in entanglement, especially at the interface, leading to more stable dimers on average. Ribonuclease T, however, folds quickly and is less prone to misfolding, and is therefore largely unaffected by synonymous mutations. Finally, LiP-MS experiments and Markov state modeling of our post-translational data demonstrate that oligoribonuclease has specific patterns of misfolding that may correlate with entanglements. Taken in combination with a recent large-scale study of entanglement in the E. coli proteome9 and an in-depth analysis of the influence of entanglement on enzymatic activity,8 our results support an emerging view that near-native, entangled misfolded states are likely to be a common phenomenon that influences a wide range of protein functions.
Supplementary Material
Acknowledgements
L.P.D. acknowledges the Department of Science and Technology at Ho Chi Minh City for their support (Grant 07/2020/HD-KHCNTT). S.D.F. acknowledges support from the NIH Director’s New Innovator Award (DP2GM140926) and from the National Science Foundation Division of Molecular and Cellular Biology (MCB-2045844). M.S.L. acknowledges support by Narodowe Centrum Nauki in Poland (Grant 2019/35/B/ST4/02086) and by PLGrid Infrastructure and the supercomputer centre TASK in Gdansk, Poland. E.P.O. acknowledges funding from the National Institutes of Health (R35-GM124818) and the National Science Foundation (MCB-1553291). This work used the Extreme Science and Engineering Discovery Environment (XSEDE45), which is supported by National Science Foundation grant number ACI-1548562.
Footnotes
CRediT authorship contribution statement
Pham Dang Lan: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Validation, Visualization, Writing – original draft, Writing – review & editing. Daniel Allen Nissley: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. Ian Sitarik: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Validation, Visualization, Writing – original draft, Writing – review & editing. Quyen V. Vu: Data curation, Formal analysis, Investigation, Methodology. Yang Jiang: Investigation, Methodology, Validation, Visualization. Philip To: Data curation, Formal analysis, Investigation, Methodology. Yingzi Xia: Data curation, Formal analysis, Investigation, Methodology. Stephen D. Fried: Conceptualization, Investigation, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing. Mai Suan Li: Conceptualization, Investigation, Methodology, Project administration, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. Edward P. O’Brien: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
DECLARATION OF COMPETING INTEREST
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmb.2024.168487.
DATA AVAILABILITY
Programs and sample analyses are available on GitHub at https://github.com/obrien-lab-psu/Synonymous_mutations_can_alter_protein_dimerization.
References
- 1.Bertolini M, Fenzl K, Kats I, Wruck F, Tippmann F, Schmitt J, Auburger JJ, Tans S, et al. , (2021). Interactions between nascent proteins translated by adjacent ribosomes drive homomer assembly. Science 371 10.1126/science.abc7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goodsell DS, Olson AJ, (2000). Structural symmetry and protein function. Annu. Rev. Biophys. Biomol. Struct 29, 105–153. [DOI] [PubMed] [Google Scholar]
- 3.Levy ED, Pereira-Leal JB, Chothia C, Teichmann SA, (2006). 3D complex: a structural classification of protein complexes. PLoS Comput. Biol 2, 1395–1406. 10.1371/journal.pcbi.0020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Juers DH, Matthews BW, Huber RE, (2012). LacZ β-galactosidase: structure and function of an enzyme of historical and molecular biological importance. Protein Sci. 21, 1792–1807. 10.1002/pro.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schechter AN, (2008). Hemoglobin research and the origins of molecular medicine ASH 50th anniversary review hemoglobin research and the origins of molecular medicine. Blood 112, 3927–3938. 10.1182/blood-BLOOD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marianayagam NJ, Sunde M, Matthews JM, (2004). The power of two: protein dimerization in biology. Trends Biochem. Sci 29, 618–625. 10.1016/j.tibs.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 7.Zhou M, Guo J, Cha J, Chae M, Chen S, Barral JM, Sachs MS, Liu Y, (2013). Non-optimal codon usage affects expression, structure and function of clock protein FRQ. Nature 495, 111–115. 10.1038/nature11833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang Y, Neti SS, Sitarik I, Pradhan P, To P, Xia Y, Fried SD, Booker SJ, O’Brien EP, (2022). How synonymous mutations alter enzyme structure and function over long timescales. Nature Chem. 10.1038/s41557-022-01091-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nissley DA, Jiang Y, Trovato F, Sitarik I, Narayan KB, To P, Xia Y, Fried SD, O’Brien EP, (2022). Universal protein misfolding intermediates can bypass the proteostasis network and remain soluble and less functional. Nat. Commun 13 10.1038/s41467-022-30548-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wright PE, Dyson HJ, (2015). Intrinsically disordered proteins in cellular signaling and regulation. Nature Rev. Mol. Cell Biol 16, 18–29. 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nissley DA, Vu Q.v., Trovato F, Ahmed N, Jiang Y, Li MS, O’Brien EP, (2020). Electrostatic interactions govern extreme nascent protein ejection times from ribosomes and can delay ribosome recycling. J. Am. Chem. Soc 10.1021/jacs.9b12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Brien EP, Christodoulou J, Vendruscolo M, Dobson CM, (2012). Trigger factor slows co-translational folding through kinetic trapping while sterically protecting the nascent chain from aberrant cytosolic interactions. J. Am. Chem. Soc 10.1021/ja302305u. [DOI] [PubMed] [Google Scholar]
- 13.Leininger SE, Trovato F, Nissley DA, O’Brien EP, (2019). Domain topology, stability, and translation speed determine mechanical force generation on the ribosome. Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.1813003116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fluitt A, Pienaar E, Viljoen H, (2007). Ribosome kinetics and Aa-TRNA competition determine rate and fidelity of peptide synthesis. Comput. Biol. Chem 10.1016/j.compbiolchem.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sugita Y, Okamoto Y, (1999). Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Letter 10.1016/S0009-2614(99)01123-9. [DOI] [Google Scholar]
- 16.Sugita Y, Kitao A, Okamoto Y, (2000). Multidimensional replica-exchange method for free-energy calculations. J. Chem. Phys 113, 6042. 10.1063/1.1308516. [DOI] [Google Scholar]
- 17.Nguyen HL, Lan PD, Thai NQ, Nissley DA, O’Brien EP, Li MS, (2020). Does SARS-CoV-2 bind to human ACE2 more strongly than does SARS-CoV? J. Phys. Chem. B 124, 7336–7347. 10.1021/acs.jpcb.0c04511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar S, Rosenberg JM, Bouzida D, Swendsen RH, Kollman PA, (1992). THE weighted histogram analysis method for free-energy calculations on biomolecules. I. The Method. J. Comput. Chem 13, 1011–1021. 10.1002/JCC.540130812. [DOI] [Google Scholar]
- 19.Kauffman L, Balachandran AP, (1992). Knots and physics. Phys. Today 10.1063/1.2809632. [DOI] [Google Scholar]
- 20.Jain AK, Murty MN, Flynn PJ, (1999). Data clustering: a review. ACM Comput. Surveys 10.1145/331499.331504. [DOI] [Google Scholar]
- 21.Scherer MK, Trendelkamp-Schroer B, Paul F, Pérez-Hernández G, Hoffmann M, Plattner N, Wehmeyer C, Prinz JH, Noé F, (2015). PyEMMA 2: a software package for estimation, validation, and analysis of Markov models. J. Chem. Theory Comput 11, 5525–5542. 10.1021/acs.jctc.5b00743. [DOI] [PubMed] [Google Scholar]
- 22.Moore BL, Kelley LA, Barber J, Murray JW, MacDonald JT, (2013). High-quality protein backbone reconstruction from alpha carbons using gaussian mixture models. J. Comput. Chem 10.1002/jcc.23330. [DOI] [PubMed] [Google Scholar]
- 23.Rotkiewicz P, Skolnick J, (2008). Fast procedure for reconstruction of full-atom protein models from reduced representations. J. Comput. Chem 10.1002/jcc.20906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML, (1983). Comparison of simple potential functions for simulating liquid water. J. Chem. Phys 10.1063/1.445869. [DOI] [Google Scholar]
- 25.Ando T, Skolnick J, (2010). Crowding and hydrodynamic interactions likely dominate in vivo macromolecular motion. Proc. Natl. Acad. Sci. U.S.A 107, 18457–18462. 10.1073/pnas.1011354107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.James M, Murtola T, Schulz R, Smith JC, Hess B, Lindahl E, (2015). GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2, 19–25. 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 27.Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE, (2010). Improved side-chain torsion potentials for the amber ff99sb protein force field. Proteins Struct. Funct. Bioinf 78, 1950–1958. 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baiesi M, Orlandini E, Seno F, Trovato A, (2017). Exploring the correlation between the folding rates of proteins and the entanglement of their native states. J. Phys. A Math. Theor 10.1088/1751-8121/aa97e7. [DOI] [Google Scholar]
- 29.Rolfson D. Knots and links; 1976. [Google Scholar]
- 30.To P, Whitehead B, Tarbox HE, Fried SD, (2021). Nonrefoldability is pervasive across the E. Coli proteome. J. Am. Chem. Soc 143, 11435–11448. 10.1021/jacs.1c03270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.To P, Xia Y, Lee SO, Devlin T, Fleming KG, Fried SD, (2022). A proteome-wide map of chaperone-assisted protein refolding in a cytosol-like milieu. Proc. Natl. Acad. Sci. U.S.A, 119. 10.1073/pnas.2210536119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Halder R, Nissley DA, Sitarik I, Jiang Y, Rao Y, Vu QV, Li MS, Pritchard J, O’Brien EP, (2023). How Soluble misfolded proteins bypass chaperones at the molecular level. Nat. Commun 14 10.1038/S41467-023-38962-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komar AA, Lesnik T, Reiss C, (1999). Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Letter. 10.1016/S0014-5793(99)01566-5. [DOI] [PubMed] [Google Scholar]
- 34.Siller E, DeZwaan DC, Anderson JF, Freeman BC, Barral JM, (2010). Slowing bacterial translation speed enhances eukaryotic protein folding efficiency. J. Mol. Biol 396, 1310–1318. 10.1016/j.jmb.2009.12.042. [DOI] [PubMed] [Google Scholar]
- 35.Spencer PS, Siller E, Anderson JF, Barral JM, (2012). Silent substitutions predictably alter translation elongation rates and protein folding efficiencies. J. Mol. Biol 422, 328–335. 10.1016/j.jmb.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang G, Hubalewska M, Ignatova Z, (2009). Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nature Struct. Mol. Biol 16, 274–280. 10.1038/nsmb.1554. [DOI] [PubMed] [Google Scholar]
- 37.Nissley DA, Sharma AK, Ahmed N, Friedrich UA, Kramer GG, Bukau B, O’Brien EP, (2016). Accurate prediction of cellular co-translational folding indicates proteins can switch from post- to co-translational folding. Nat. Commun 10.1038/ncomms10341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’brien EP, Vendruscolo M, Dobson CM, (2014). Kinetic modelling indicates that fast-translating codons can coordinate cotranslational protein folding by avoiding misfolded intermediates. Nat. Commun 5, 2988. 10.1038/ncomms3988. [DOI] [PubMed] [Google Scholar]
- 39.Trovato F, O’Brien EP, (2017). Fast protein translation can promote co- and posttranslational folding of misfolding-prone proteins. Biophys. J 112, 1807–1819. 10.1016/j.bpj.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ratjen F, Bell SC, Rowe SM, Goss CH, Quittner AL, Bush A, (2015). Cystic fibrosis. Nature Rev. Dis. Primers 1, 15010. 10.1038/nrdp.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ward CL, Omura S, Kopito RR, (1995). Degradation of CFTR by the ubiquitin-proteasome pathway. Cell 83, 121–127. 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 42.Varga K, Jurkuvenaite A, Wakefield J, Hong JS, Guimbellot JS, Venglarik CJ, Niraj A, Mazur M, Sorscher EJ, Collawn JF, Bebok Z, (2004). Efficient Intracellular processing of the endogenous cystic fibrosis transmembrane conductance regulator in epithelial cell lines. J. Biol. Chem 279, 22578–22584. 10.1074/jbc.M401522200. [DOI] [PubMed] [Google Scholar]
- 43.Zhang L, Button B, Gabriel SE, Burkett S, Yan Y, Skiadopoulos MH, Dang YL, Vogel LN, et al. , (2009). CFTR delivery to 25% of surface epithelial cells restores normal rates of mucus transport to human cystic fibrosis airway epithelium. PLoS Biol. 7 10.1371/journal.pbio.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schubert U, Antón LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR, (2000). Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404, 770–774. 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- 45.Towns J, Cockerill T, Dahan M, Foster I, Gaither K, Grimshaw A, Hazlewood V, Lathrop S, Lifka D, Peterson GD, Ralph Roskies J, Ray Scott N-W-D, (2014). XSEDE: accelerating scientific discovery. Comput. Sci. Eng 16, 62–74. 10.1109/MCSE.2014.80. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Programs and sample analyses are available on GitHub at https://github.com/obrien-lab-psu/Synonymous_mutations_can_alter_protein_dimerization.