Abstract
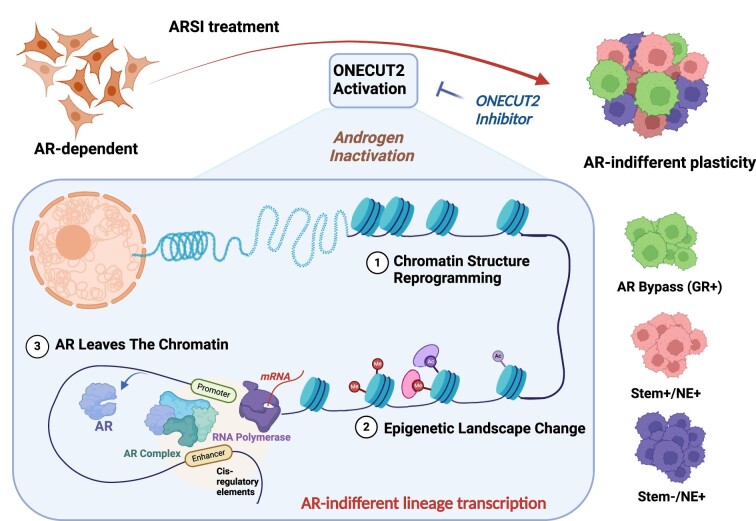
Androgen receptor- (AR-) indifference is a mechanism of resistance to hormonal therapy in prostate cancer (PC). Here we demonstrate that ONECUT2 (OC2) activates resistance through multiple drivers associated with adenocarcinoma, stem-like and neuroendocrine (NE) variants. Direct OC2 gene targets include the glucocorticoid receptor (GR; NR3C1) and the NE splicing factor SRRM4, which are key drivers of lineage plasticity. Thus, OC2, despite its previously described NEPC driver function, can indirectly activate a portion of the AR cistrome through epigenetic activation of GR. Mechanisms by which OC2 regulates gene expression include promoter binding, enhancement of genome-wide chromatin accessibility, and super-enhancer reprogramming. Pharmacologic inhibition of OC2 suppresses lineage plasticity reprogramming induced by the AR signaling inhibitor enzalutamide. These results demonstrate that OC2 activation promotes a range of drug resistance mechanisms associated with treatment-emergent lineage variation in PC and support enhanced efforts to therapeutically target OC2 as a means of suppressing treatment-resistant disease.
Graphical Abstract
Graphical Abstract.
Introduction
Prostate cancer (PC) is driven by the AR, a hormone-dependent nuclear receptor. AR-driven prostate adenocarcinoma can evolve to contain cell types with diminished luminal features, indicating lineage identity has been altered. This ‘lineage plasticity’ is thought to play a key role in tumor heterogeneity and development of lethal disease. Treatment-resistant phenotypes documented in PC include neuroendocrine (NE) differentiation (1), and activation of the glucocorticoid receptor (GR; NR3C1) (2). While NE transcriptional programs operate outside the AR axis and give rise to distinct histologic features, the GR assumes control of certain AR-regulated genes, resulting in preservation of the luminal phenotype of adenocarcinoma.
The HOX/CUT protein ONECUT2 (OC2) is a master transcription factor (TF) active in roughly 60% of mCRPC (3,4). OC2 promotes NEPC features, suggesting it plays a role as a driver of lineage plasticity and the emergence of drug resistance. Notably, OC2 can be directly targeted with a small molecule inhibitor (SMI) that suppresses established PC metastases in mice (3). Despite these insights, the mechanism of action of OC2 is not well defined. Here we describe a novel role for OC2 as a broadly acting lineage plasticity driver that operates across several distinct molecular pathways in adenocarcinoma as well as neuroendocrine tumors to promote lineage variation and drug resistance.
Materials and methods
Gene signature collection
We assembled a collection of gene signatures for this study based on previously published literature and the Molecular Signature Database (MSigDB) (5). The ‘Hallmark Androgen Response’ gene set from MSigDB was used as an AR signature. NE and Stem signatures were collected from published literature (6,7). Original papers and gene lists for each signature are included in Supplementary Table S2.
Digital spatial profiling (DSP) data analysis
To characterize OC2 expression in CRPC metastases (mCRPC), in situ hybridization (ISH) was performed with 152 tumor cores from 53 tumors from a recent study (8). The cores were ranked by OC2 in situ hybridization levels from high to low. IHC was performed for prostate-specific membrane antigen (PSMA). AR, NE and Stem signature scores were computed in each tumor core using the z-score method (9).
Single cell transcriptome data analysis
Preprocessing and data integration
Single-cell RNA-seq data from three published studies were used (10–12). For each dataset, doublets and triplets were removed using DoubletFinder (13). By default, 2000 highly variable genes were selected in each dataset, which were used in Principal Component Analysis (PCA). Cells with the number of genes <500 and >20% mitochondrial expression were removed (nFeature_RNA > 500 & percent.mi < 20%), resulting in a total of 36419 primary and 23607 CRPC cells. Systemic biases from the individual data were adjusted through canonical correlation analysis with the Seurat R package (version 4.3.0) (14). Batch effects were removed using SVA package ComBat function during multiple cohorts’ integration (15).
Dimensionality reduction
30 principal components were selected based on statistical significance from Elbow plot analysis. We then performed Uniform Manifold Approximation and Projection (UMAP) analysis with the 30 selected principal components to identify cell sub-populations (arXiv:1802.03426). Clustering was performed for integrated expression values using the Louvain community detection-based method with FindNeighbors function in Seurat package with the resolution is 0.8. To visualize the cell clusters, UMAP plots were used. The following markers were used for cell type annotation: Epithelial/Tumor: EPCAM, KRT8; T cells: CD3E, CD3D, TRBC1/2, TRAC; Myeloid cells: LYZ, CD86, CD68, FCGR3A; B cells and plasma cells: CD79A/B, JCHAIN, IGKC, IGHG3; Endothelial cells: CLDN5, FLT1, CDH1, RAMP2; Fibroblasts: DCN, C1R, COL1A1, ACTA2; Smooth muscle cells: TAGLN, CNN1; Mast cells: TPSAB1, resulting in 8 cell types consisting of 36 419 primary and 23 607 CRPC cells. In order for in-depth analysis of OC2 expressing cells, we selected OC2 expressing cells with a positive gene expression level. We then computed weighted z-score of the gene sets, including NE markers (CHGA, ASCL1 and BRN2), Androgen response genes, Stemness signature, and other PC-associated gene signatures for each cell type.
Single cell trajectory analysis
Pseudotime inference was performed to track the progression trajectory of OC2-expressing cancer cells by monocle3 R package (16). Epithelial cells from M0 stage in primary samples were selected as the start nodes. The learn_graph function was used to infer the trajectory of all OC2-expressed M0 and CRPC cells. Then, the plot_cells function was used to visualize the trajectory and pseudotime inference results. The signature score of each cell was defined by calculating z score.
TF activity inference and lineage identification
TF activity in a given sample or cell was computed using the decoupleR package (17). The VIPER method (18) was applied to infer TF activity based on mRNA expression of its targets including all targets with confidence levels from A to D (19). We identified cell type clusters defined by the TF activities using the K-means method in the ConsensusClustering package (20). Considering the known CRPC lineages and the Consensus Cumulative Distribution plot, the optimal number of clusters (K) was identified by surveying K values from 2 to 8.
Cell lines
LNCaP (#CRL-1740) was obtained from the American Type Culture Collection (ATCC) in September 2019 and authenticated using the Promega PowerPlex 16 system DNA typing (Laragen). LNCaP cells were grown in RPMI-1640 media (Gibco) supplemented with 10% FBS and penicillin/streptomycin. LAPC4 cells were from the Dr Stephen J. Freedland group. LAPC4 cells were grown in IMDM (Gibco) supplemented with 10% FBS and 1nM R1881 plus penicillin/streptomycin. Mycoplasma contamination was routinely monitored using the MycoAlert PLUS Mycoplasma Detection Kit (Lonza #LT07-118). The OC2 overexpression construct was generated by cloning the full-length OC2 cDNA (NM_004852) into the pLenti-C-Myc-DDK-IRES-Puro (Origene #PS10069) lentivirus system. The inducible OC2 was generated with pCDH-CuO-MCS-IRES-GFP (SystemBio #QM530A-2) and pCDH-EF1-CymR-T2A-RFP (SystemBio #QM300PA) system with Cumate (SystemBio # QM100A) turned-on treatment. OC2 knockdown was generated by validated shRNA clones TRCN0000013445 in the vector pLKO1 were purchased from Sigma. A nonmammalian shRNA control plasmid (TRC2-pLKO-puro non-target shRNA no. 1, Sigma) was used as a control. The AR-respond plasmid ARR3tk-eGFP/SV40-mCherry was purchased from Addgene (Addgene, #132360). Packing (psPAX2, Addgene #12260) and envelope (pMD2.G, Addgene #12259) plasmids were co-transfected into HEK293T cells to produce lentivirus. Cells were infected with lentivirus supplemented with 10 μg/ml polybrene, then selected by 2 ug/ml puromycin to generate the stable overexpression cells.
SCID mouse in vivo xenografts
All experimental protocols and procedures were approved by the Institutional Animal Care and Use Committee (IACUC009352) at Cedars-Sinai Medical Center. All relevant ethical regulations, standards, and norms were rigorously adhered to. For in vivo tumor growth assays, LNCaP control or OC2 OE cells were adjusted to 1 × 107 cells/ml in DPBS, followed by mixing with Matrigel at a 1:1 ratio (v/v). For each male SCID/Beige mouse (7-weeks-old; Charles River #CRL:250; CB17.Cg-PrkdcscidLystbg-J/Crl), a 100 μl mixture was subcutaneously injected into both flanks. Tumor length and width were measured with a caliper and tumor volumes were calculated using the formula of (length × width2)/2. At termination of the experiment, mice were euthanized, and tumor xenografts collected and sent to Cedars Sinai Pathology Laboratory for preparation of Formalin-fixed paraffin-embedded (FFPE) slides.
Tissue sample preparation
FFPE slides were obtained from TMA 95A, B and C, provided by the University of Washington collected from CRPC rapid autopsy patients who signed written informed consent under the aegis of the Prostate Cancer Donor Program at the University of Washington (IRB protocol no. 2341), to which the sections cut from all the positive and negative tissues and cell blocks were added on each slide. The positive controls included fetal Retina, Pancreas, Xenograft OC2 and HistoGel pre-wrapped cells block of organoid BS17077 with enforced OC2. The negative control consisted of organoid BS17077 with no vehicle and Xenograft Vec-Con1.
In situ hybridization
For in situ hybridization, the RNAscope® 2.5 HD detection reagent–Red kit (ACD, 322360) was used according to the manufacturer's instructions. The probes used in this study included the Hs-ONECUT2 probe (ACD, 473531) targeting ONECUT2 mRNA, the probe_DapB (ACD, 310043) as the negative control, and the Probe_PPiB (ACD, 313901) for the positive control.
Immunohistochemistry
FFPE slides were deparaffinized, and antigens unmasked using EnVision FLEX Target Retrieval Solution Low PH in DAKO PT link at 99°C for 20 min. Subsequently, the slides were loaded onto the Agilent Automated AS48 Link. Background reduction was achieved with EnVision Flex Peroxidase and Casein blocking agents. The primary antibodies used in this study included rabbit anti-human monoclonal antibodies of ONECUT2 (Sigma HPA057058) at 1:50 dilution, Androgen Receptor (Cell Marque, SP107) at 1:6400 dilution, Glucocorticoid Receptor (D6H2L) XP® (Cell Signaling, 1204) at 1:6000 dilution, and Synaptophysin (D8F6H) XP® at 1:200 dilution. Visualization was carried out with EnVision Flex HRP labeled Polymer (Agilent SM802), followed by development with Flex DAB + sub-chromogen and counterstaining with EnVision Flex hematoxylin. A peptide challenging assay was conducted using PrEst Antigen ONECUT2 (Sigma APrEST86051) with a 5:1 ratio of peptide to ONECUT2 antibody, pre-incubated for 1 h at room temperature, Flex mouse (cocktail IgG1, 2a,2b,3 and IgM) and immunoglobin fraction of serum from non-immunized rabbits to replace primary antibodies, as well as omitting the primary antibodies, were run with the same protocol parallel with experimental slides as negative controls.
Western blot analysis
Cell lysates were separated on 4–20% SDS-PAGE (Bio-Rad Laboratories) and transferred to nitrocellulose or PVDF membranes. The membranes were blocked with 5% non-fat dry milk and subsequently incubated with the pertinent primary antibody overnight. Anti-GR (D6H2L) (CST, #12041, 1:1000 dilution), Anti-AR (Activemotif, #39781, 1:2000 dilution), Anti-OC2 (Proteintech, #21916-1-AP, 1:1000 dilution), Anti-PSA (CST, #5365, 1:2000 dilution), Anti-Histone-H3 (CST, #9715, 1:2500 dilution), Anti-SMARCA4 (Abcam, #ab110641, 1:10000 dilution), Anti-SMARCA5 (Abcam, #ab72499, 1:2500 dilution), Anti-Myc-tag (CST, #2276, 1:1000 dilution), Anti-SYP (CST, #36406, 1:1000 dilution), Anti-REST (Sigma-Aldrich, #ZRB1455, 1:1000 dilution). Membranes were subsequently washed with TBST (0.1% Tween-20) and incubated with an HRP-conjugated secondary antibody (GE Healthcare Bio-Sciences). After washing with TBST, the protein bands were detected by the Chemidoc MP imaging system (Bio-rad).
Cell proliferation analysis
All procedures were performed according to the XTT cell viability kit protocol (CST, #9095). To assay viability, cells were plated at a density of 2000 cells/well in triplicate. 48 h after indicated treatment, viability was assessed at the absorbance of 450 nM. IC50 was generated by a non-linear regression function in GraphPad Prism 9.0. For proliferation assay, the cells were seeded in 2000/well and treated up to 96 h, then 450 nM absorbance was collected for further analysis.
RNA extraction and quantitative real-time polymerase chain reaction (RT-PCR)
Total RNA was extracted using Qiagen RNeasy Kit (Qiagen #74104). Messenger RNA was converted to the first-strand cDNA using iScript cDNA synthesis kit (Bio-Rad #1708891), followed by RT-PCR reaction using PowerUp SYBR Green PCR Master Kit (Applied Biosystems #A25742) in QuantStudio 5 Systems (Applied Biosystems #4309155). Primer sequences are provided in Supplementary Table S8. The comparative CT method was used to normalize gene expression to Actin/GAPDH.
Interactome analysis by immunoprecipitation-mass spectrometry (IP-MS)
For IP-MS to capture OC2 binding proteins, pLenti-C-Myc-DDK-IRES-Puro (Origene #PS10069) vector with myc-tagged OC2 or empty vector was stably transfected into LNCaP cells. OC2 overexpression cells will express OC2 with a fusion protein containing a myc-tag. The vector-control plasmid will not express the myc-tag protein. ChromoTek iST Myc-Trap Kit (Proteintech, #ytak-iST) was selected to perform the immunoprecipitation and prepare the proteomic sample generated from the host of alpaca. Cells were harvested and lysed. 1mg nuclear protein lysates were applied for immunoprecipitation of myc-tagged target and their binding proteins, followed with protein denaturation, reduction, alkylation, and peptide digestion and clean-up using ChromoTek iST Myc-Trap Kit. Eluted proteins were analyzed by gel-enhanced liquid chromatography-tandem mass spectrometry (GeLC-MS/MS) essentially as described (21). The resulting tryptic peptides in 10 μl solution was loaded onto a 2-cm trap column (Thermo Scientific) and separated on a 50-cm EASY-Spray analytical column (Thermo Scientific) heated to 55°C, using a 1-h gradient at the flow rate of 250 nL/min. The resolved peptides were ionized by an EASY-Spray ion source (Thermo Scientific), and mass spectra were acquired in a data-dependent manner (DDA) in an Orbitrap Fusion Lumos mass spectrometer (Thermo Scientific). MS1 scans were acquired in 240 000 resolution at m/z of 400 Th, with a maximum injection time of 250 ms and an ion packet setting of 4 × 105 for automatic gain control (AGC). Most intense peptide ions with charge state of 2–7 were automatically selected for MS/MS fragmentation by higher energy collisional dissociation, using 30% normalized collision energy. MS/MS spectra were acquired in the ion trap, using rapid ion trap scan at 1 × 104 AGC and 35 ms maximum injection time. Dynamic exclusion was enabled to minimize redundant MS2 acquisition.
The acquired RAW files were searched against the UniProt_Human database (released on 03/30/2018, containing 93316 protein sequences) with MaxQuant (v1.5.5.1) (22). The searching parameters include trypsin/P as the protease; carbamidomethyl (C) as fixed modification; oxidation (M), acetyl (protein N-term), and deamidation (NQ) as variable modifications; minimal peptide length as 7; up to two missed cleavages; mass tolerance for MS was 4.5 ppm and for MS/MS was 0.5 Da; identification of second peptides enabled; label free quantification (LFQ) enabled, with match-between-runs within 0.7 min. A standard false discovery rate of 0.01 was used to filter peptide-spectrum matches, peptide identifications, and protein identifications. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD050634.
RNA-sequencing and data processing
RNA concentration, purity, and integrity were assessed by NanoDrop (Thermo Fisher Scientific Inc.) and Agilent Bioanalyzer. 1ug Total RNA were shipped to BGI Americas for Pair-end 100bp sequencing using Strand Specific Transcriptome Library Construction Protocol and DNBSEQ platform. FastQC was applied to analyze Raw RNA-Seq fastq files. Trim Galore was used to remove the adapters. 150 bp paired-end reads were aligned to human reference genome (HG38) using STAR (-alignIntronMin 20 -alignIntronMax 1000000 -alignSJoverhangMin 8 -quantMode GeneCounts) method (23). Gene read counts matrix were used for further analysis. Differentially expressed genes were determined using edgeR packages (24). GSEA Preranked function was performed to identify significant biological functions (25). Genes were ranked by the log2 fold change between control and OC2 perturbed samples. ClusterProfiler R package was used to perform GO and KEGG pathway enrichment analysis based on differentially expressed gene set (26).
Cleavage under targets and release using nuclease (CUT & RUN) sequencing
All procedures were performed according to the manufacturer's protocol from Cell Signaling Technology (CST, #86652). Briefly, 100000 cells from both control and OC2 OE cells were resuspended in wash buffer (20 mM HEPES–NaOH pH 7.5, 150 mM NaCl, 0.5 mM spermidine, and protease inhibitor cocktail), concanavalin A-magnetic beads added, then rotated for 10 min at room temperature. Cell-bead conjugates were resuspended in 200 μl of digitonin buffer (wash buffer with 2.5% digitonin solution) containing 2ug of Anti-Tri-Methyl-Histone H3 (Lys27) (CST, #9733), 2ug of Anti-Tri-Methyl-Histone H3 (Lys4) (CST, #9751), 2ug of Anti-Acetyl-Histone H3 (Lys27) (D5E4) (CST, #8173), 2ug of Anti-Mono-Methyl-Histone H3 (Lys4) (D1A9) (CST, #5326), 5 ug of Anti-ONECUT2 (Proteintech, #21916-1-AP), 5ug of Anti-GR (D6H2L) (CST, #12041), 5ug of Anti-AR (Activemotif, #39781), 5 ug of Anti-FOXA1 (Activemotif, #398837) primary antibody, or rabbit IgG (CST, # 66362,), rotated overnight at 4 °C, resuspended in 250 μl of antibody buffer and 7.5 μl of the pAG-MNase enzyme (# 57813, CST), followed by the rotation at 4 °C for 1 h. After washing with digitonin buffer, ice-cold 150 μl of digitonin buffer containing CaCl2 was added and incubated on ice for 30 min followed by the addition of 150 μl of stop buffer containing 5 ng S. cerevisiae spike-in DNA used for sample normalization. After incubation at 37 °C for 15 min, samples were centrifuged at 16 000 g for 2 min at 4 °C. Tubes were placed on a magnetic rack, then supernatants were collected. DNA was purified using DNA purification buffers and spin columns (CST, #14209). The CUT & RUN library was generated with the DNA Library Prep Kit for Illumina (CST, #56795) combined with Multiplex Oligos for Illumina® (Dual Index Primers) (CST, # 47538). The adaptor was diluted 1:25 to avoid contamination. The PCR enrichment step run 15 cycles to amplify the adaptor-ligated CUT&RUN DNA.
CUT&RUN-seq data processing and downstream analysis
CUT&RUN-seq data of H3K27Ac, H3K27me3, H3K4me3, H3K4me1 and several TFs (ONECUT2, AR, FOXA1 and GR) was generated in LNCaP cell lines. Briefly, Trim Galore was utilized to remove contaminant adapters and read quality trimming. 150 bp paired-end reads were aligned to human reference genome (HG38) using Bowtie2 (v2.2.6) (27). As spike-in is commonly used as a control probe in DNA sequencing (28). We added the S. cerevisiae genome as spike-in control reference. Then, the mapping rates were used for calculating scale factor in each sample. Next, Picard MarkDuplicates tool was utilized to mark and remove PCR duplicates in each sample. ENCODE blacklisted regions on HG38 (29) were removed by using bedtools (30). Finally, a high accuracy peak calling method, SEACR (31), was used to identify significant peaks with the parameters: norm stringent. To visualize the signal in each sample, bamCompare function in DeepTools (v3.1.3) was used to generate Bigwig files with the parameters: -binSize 10 -numberOfProcessors 5 -normalizeUsing CPM -ignoreDuplicates -extendReads 200 (32). The scale factor was calculated from the above spike in step. Integrative Genomics Viewer (IGV) was used to visualize the signal in bigwig files (33). Due to the quality issue only one sample is generated for FOXA1 CUT&RUN in enzalutamide treatment. All other groups have replicates. We employed the GIGGLE method (34) to identify the TFs whose genome-wide binding profiles publicly available and produce by this study are highly enriched in the specific peaks.
Omni-ATAC sequencing
50000 viable LNCaP Vec Con and OC2 overexpressing cells were precipitated and kept on ice and subsequently resuspended ATAC Resuspension Buffer (RSB) (49.25ml nuclease-free water, 500 ul 1M PH7.5 Tris–HCl, 100 ul 5 M NaCl and 150 ul 1 M MgCl2). 50 ul of Transposition Master mix (25 ul 2× TD buffer, 1 ul Tagment DNA enzyme, 16.5 ul of PBS, 0.5 ul 1% Digitonin, 0.5 ul 10% Tween-20 and 6.5 ul Nuclease-free water) for 30 min reaction at 37°C with 1000 rpm mixing. DNA was then purified using Zymo DNA clean and Concentrator-5 kit. ATAC-seq libraries were prepared following the Buenrostro protocol (35). The Tagment DNA enzyme was gifted from Pattenden, Samantha Lab from UNC.
ATAC-seq data analysis
Trim Galore and Bowtie2 (v2.2.6) were used to do read quality trimming and mapping to the HG38 reference genome. PCR duplicates and blacklisted regions (HG38) were removed by the same methods described above. The Macs2 method was used to do peak calling based on the following parameters: -bdg -SPMR -nomodel -extsize 200 -q 0.01. Bigwig files were also generated by bamCompare function in Deeptools (v3.1.3) using the parameters: -binSize 10 -numberOfProcessors 5 -scaleFactorsMethod None -normalizeUsing CPM -ignoreDuplicates -extendReads 200. IGV software was used to do visualization. The HOMER software with findMotifsGenome.pl function was used to identify enriched motifs in selected regions. The parameters in findMotifsGenome.pl are hg38 -size 200 -len 8, 10, 12 (36). For visualization of the peak profiles from ATAC-seq and CUT&RUN-seq data, we used deepTools (v2.5.0) (32) to generate read abundance from all datasets around peak center (± 3 kb/ 5 kb), using ‘computeMatrix’. These matrices were then used to create heatmaps and profiles, using deep-Tools commands ‘plotHeatmap’ or ‘plotProfile’, respectively. For genome browser tracks, we used the pyGenometracks package (37) to generate plots for track visualization (Reference Genome, HG38).
Mutation constructs UGT2B15 promoter luciferase
Quick Change II XL site-directed mutagenesis kit (Agilent) manufacturers protocol was used to generate mutation constructs. PCR amplification of the UGT2B15 mutation constructs were cycled at 1 cycle 95°C 1 min, 18 cycles at 95°C 50 s 60°C 50 s 68°C 1 min/kb of plasmid length and 1 cycle at 68°C 7 min. The PCR product was then then placed on ice for 2 min to cool the reactions to ≤37°C. Then enzyme digestion of the PCR product was with the addition of 1 μl of the Dpn I restriction enzyme (10 U/μl).
UGT2B15mut_F_gttgtttctttctgtcatttctcatacttatatctgaggaaaagcaagccaagttaaaatataactgctaaaatttgaagtaaatacataata
UGT2B15mut_R_tattatgtatttacttcaaattttagcagttatattttaacttggcttgcttttcctcagatataagtatgagaaatgacagaaagaaacaac
Luciferase reporter assay
Cells were co-transfected with PGL4.10 luciferase backbone construct, and the pRL-SV40 vector (Promega #E2231) with Turbofectin 8.0 (Origene, #TF81001). After transfection overnight, cells were washed and treated for 6 h with CSRM-617 (10 uM). Luciferase activity was measured using the Dual-Luciferase Assay System (Promega, #1910).
DNA-protein affinity assay (DAPA)
10 million cells were harvested and nuclear protein and frozen at –80°C for each DAPA reaction. SRRM4 300 bp promoter region was amplified from gDNA with the primer (5′ TTTCTCCTCCCAAGACCTGCG, 5′ TCTGAGCTGGCTGAGCCTCT) using Phusion High-Fidelity PCR Kit (Thermoscientific #F553L). Then the DNA was biotinylated with BioNick DNA Labeling Kit (Invitrogen #18247015). Biotinylated 25 bp UGT2B15 WT and Mut probes were synthesized by IDT. Primers were diluted to 20 μM as working stocks for the DAPA reaction. Each primer was incubated with 20 μl of each primer (F/R) incubated in 60 μl of DAPA buffer (10 mM Tris Buffer, pH 7.4, 1 mM EDTA). The primer mix was incubated at 95°C for 3 min. The annealed primers were cooled at room temperature overnight and stored at -20°C. The nuclear protein fraction was isolated using the cytoplasmic and nuclear extraction protocol (Thermofisher # 78833). An aliquot of 250 μg of nuclear protein lysate was diluted in DAPA wash buffer (20 mM Hepes buffer, pH 7.4, 1 mM DTT, 0.1% Tween-20) up to a volume of 500 μl. The nuclear protein lysate was pre-cleared by adding 2 μl of poly D[IC] (Sigma-Aldrich) 25 μg/μl and 20 μl of High Capacity Strepavidin agarose beads (Thermofisher #20357). The pre-cleared protein lysate was incubated for 2 h at 4°C on an end-over-end rotator. The mixture was centrifuged at 3000×g at 4°C, and the protein lysate in the supernatant fraction was used for binding. The beads and buffer were boiled at 95°C for 5 min and used as a negative control sample. The pre-cleared lysate was bound to 250 ng of the biotynlated primer and an additional 2 μl of poly D[IC]. The protein-DNA mixture was incubated overnight at 4°C on an end-over-end rotator. The protein-DNA complex mixture was incubated with 20 μl of High Capacity Strepavidin agarose for 2 h at 4°C. The mixture was centrifuged a 3000×g for 1 min and the supernatant was removed. The pelleted beads were washed with three times with 500 μl DAPA wash buffer. All wash buffer was removed and then added 30 μl 4× lammeli buffer supplemented with 5% β-meracptoethanol. The beads and buffer were boiled at 95°C for 5 min then analyzed by western blot.
UGT2B15 WT probe(F) Biotinylated-TTTTAACTTGATTGATTTTTCCTCA
UGT2B15 WT probe (R)-TGAGGAAAAATCAATCAAGTTAAAA
UGT2B15 Mut probe (F) Biotinylated -TTTTAACTTGGCTTGCTTTTCCTCA
UGT2B15 Mut probe (R)-TGAGGAAAAGCAAGCCAAGTTAAAA
Surface plasmon resonance binding studies
Measurement of OC2 binding affinity for the UGT2B15 promoter DNA sequence was analyzed using surface plasmon resonance with Sartorius Octet SF3 instrument. The UGT2B15 promoter 20 bp sequence (5′-TAACTTGATTGATTTTTCCT-3′ for wild type and 5′-TAACTTGGCTGTCTTTTCCT-3′ for mutant) was immobilized to a streptavidin SADH sensor chip (Sartorius) in running buffer (10 mM HEPES pH 6.8, 250 mM NaCl, 0.005% Tween 20). The 20 bp reverse complement sequence (100 ng/ml), biotinylated at the 5′ end, was first immobilized to 75 response units (RU), followed by immobilization of the 20 bp forward sequence at 100 ng/ml to a final 150 RU, ensuring duplexed DNA immobilization. Recombinant OC2 DNA-binding domain protein (L330-W485) purified as previously published (4), was diluted in running buffer to 30, 40, 50, 70, 80 and 90 nM and injected over the immobilized, duplexed 20 bp DNA probe as well as reference channel. In the case of mutant UGT2B15, where no binding was observed, a subsequent positive control of wild-type UGT2B15 was performed on the same channel to confirm adequate experimental setup. The sensograms were fit with Octet SPR Analysis software (Sartorius) using a one binding site model with mass transport correction to calculate binding affinity (KD). All DNA oligos were purchased from IDT (synthesized in San Diego, CA, USA).
Genome-wide chromatin interaction analysis using Hi-C
Hi-C data was generated using the Arima-HiC kit, according to the manufacturer's protocols (38,39). The Juicer pipeline (version 1.6) was run on all samples using default parameters (40). The reads were aligned to the hg38 reference genome. Chromatin loop calls were identified using HiCCUPS (Hi-C Computational Unbiased Peak Search) algorithm with KR (Knight-Ruiz) normalization. TADs (Topologically Associating Domains) were identified using the Arrowhead algorithm at 10kb resolution. Eigenvalues were identified using the Eigenvector algorithm at 1Mb resolution across the genome. Then multiple .txt files containing eigenvalues were converted and merged into a single .wig file containing final A/B compartments calls. Enhancer-promoter loops were identified by integrating Hi-C loops with H3K27Ac binding peaks. The corresponding RNA expression change was shown by log2 fold change from RNA-Seq data (41).
ATAC-seq and RNA-seq data integration
To integrate gene expression (bulk RNA-seq) and chromatin accessibility profiles (ATAC-seq) induced by OC2, we first performed differential expression analysis using edgeR, and differential ATAC-seq peaks using the Differential ATAC-seq Toolkit (DAStk) (42) between control and OC2 OE. We then computed the z-scores for differential expression and differential motif enrichment for each TF based on the test P-values from edgeR and DAStk tools, respectively. The z-scores were combined into an overall z-score using the Stouffer's method (43) as the TF activity score. Finally, the significance level of the TF activity score was calculated by converting the combined z-score into a P-value using the standard normal cumulative distribution function. The positive and negative z-score represent the active and repressed status of TFs by the OC2 perturbation. For further consideration of TF activity. We estimated the contribution of downstream target genes to TF activity by calculating the fold enrichment score based on the TF-target interaction information from the Dorothea R packages (44). The TF fold enrichment was defined as the percentage of the number of differentially expressed target genes in the list belonging to all the TF target genes, divided by the corresponding percentage of the number of DEGs in all genes in the data. The fold enrichment score refers to the percent change in expression of the downstream targets. Significant TFs were selected with P-value <0.05 and fold enrichment score ≥1.5 (Supplementary Table S3).
Promoter H3K4 and H3K27 trimethylation analysis
Raw reads from H3K4me3 and H3K27me3 CUT&RUN-seq data were mapped to the hg38 human reference genome. Counts per million (CPM) mapped reads within ±2 kb of each transcription start site (TSS) was calculated in each sample. We averaged across multiple TSSs of one gene in each sample. Two-dimensional kernel density was constructed based on the H3K4me3 and H3K27me3 signal of all gene TSSs. This was visualized using the contour plot of the signals. The high/low cutoff for H3K4me3 was set at four standard deviations below the mean CPM value of the H3K4me3-high distribution (45). Activated, bivalent or repressed status of genes from control to over-expression OC2 samples can be identified in the contour plot. Each red dot represents a transcript from a gene promoter with OC2 binding.
Super-enhancer annotation
Rank Order of Super Enhancers (ROSE) was used to identify enhancers defined as H3K27Ac peaks 2 kb away from all TSSs (Supplementary Table S4-S7). After merging enhancer elements clustered within a distance of 12.5 kb, all super-enhancers in each sample were identified using a cutoff at the inflection point (tangent slope = 1) based on the ranking order of all typical-enhancers and super-enhancers (46). ROSE also provided the nearest genes to each typical-enhancer and super-enhancer, which can be used for functional analysis of these super-enhancers.
Statistics and reproducibility
Statistically significant data for in vitro and in vivo assays were assessed by unpaired two-tailed Student's t-test or Wilcoxon two-tailed rank-sum test, unless otherwise noted. Tests for differences in the case of more than two groups were performed by one-way ANOVA with Dunnett's post hoc test where appropriate, unless otherwise noted. GraphPad Prism and R (v.3.5, http://www.r-project.org/) were used for all statistical tests.
Results
OC2 is expressed in multiple CRPC lineages
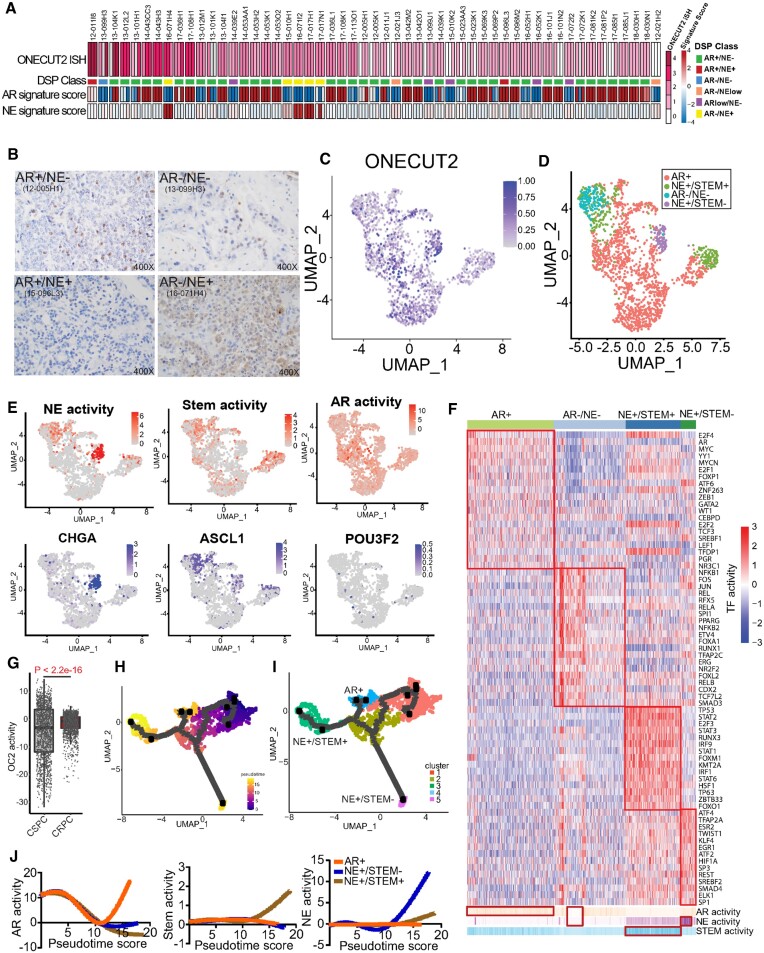
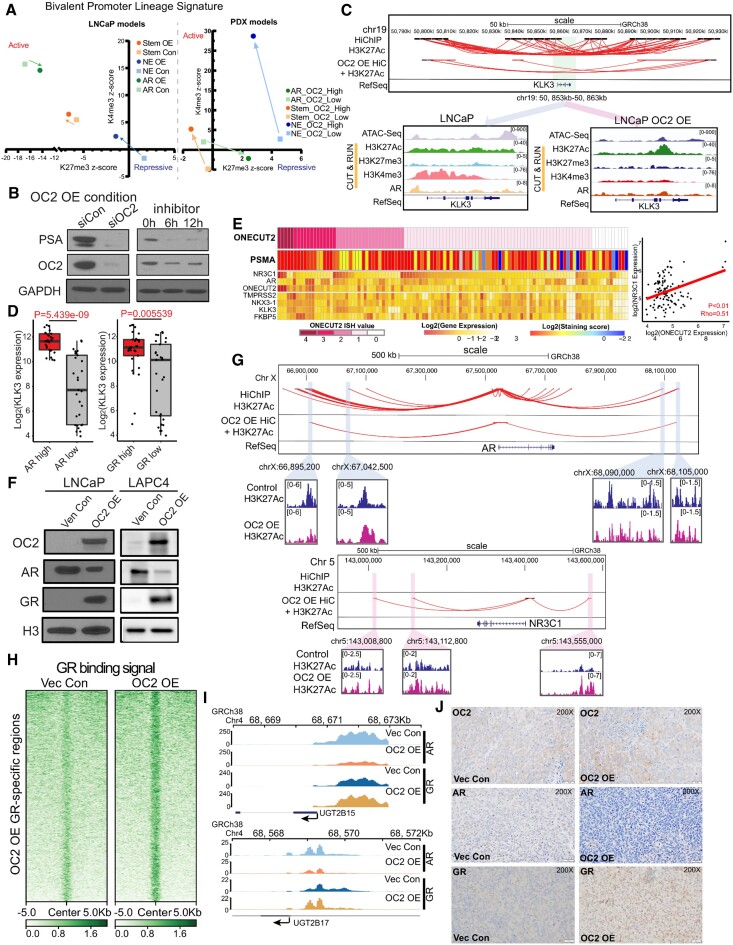
To characterize OC2 expression in CRPC metastases (mCRPC), we performed in situ hybridization (ISH) of 152 tumor cores from 53 tumors in a tissue microarray from a recent study (8). RNA-seq data from the same tissue was used to assign AR activity (AR+) and NE differentiation (NE+) status (6) (5). OC2 mRNA expression was detected in most specimens (52 out of 53), including AR–/NE+, double positive (AR+/NE+), double negative (AR–/NE–) and adenocarcinoma (AR+/NE–) phenotypes based on established publications (Figure 1A). All NEPC tumors exhibited relatively high OC2 expression, consistent with the described role of OC2 as an NE driver (3,4). Surprisingly, >80% of AR+/NE– tumors were OC2-positive, and high OC2 expression was also seen in AR+/NE+ and AR–/NE– specimens (Supplementary Figure S1A). Notably, 46 AR+/NE– cores, which represents 40% of the adenocarcinoma specimens, displayed high OC2 staining via ISH. High ISH staining correlated significantly with high immunohistochemical (IHC) score (Figure 1B, Supplementary Figure S1B, C). These findings indicate that OC2 expression is not restricted to NEPC but can occur widely within a range of CRPC phenotypes, including in AR-active tumors.
Figure 1.
OC2 is expressed in multiple CRPC lineages. (A) RNA scope in-situ hybridization of OC2 on UW-TMA95 series ((8)). NE and AR signature scores were calculated using RNA-seq data and established digital spatial profiling (DSP) class annotation from the matched samples collected from GSE147250. (Refer to methods).(B) Representative IHC staining of OC2 in CRPC specimens in established phenotypes. (C) UMAP plot illustrating OC2-expressing epithelial cells from 6 CRPC patients scRNA-seq datasets selected for further analysis. (D) UMAP plot illustrating OC2-expressing cells annotated to distinct lineages in CRPC. (E) UMAP plots illustrating OC2-expressing cells colored by three lineage signature activities (AR, NE, Stem) and expression levels of representative markers (CHGA, ASCL1 and POU3F2). (F) Unsupervised clustering of OC2 expressing epithelial cells from 6 CRPC scRNA-seq datasets using transcription factor (TF) activity from the high-confidence DoRothEA database. (Refer to methods). (G) CRPC patient samples show higher OC2 activity compared to CSPC patient samples (Refer to methods). (H) UMAP plot showing the pseudotime trajectory of OC2-expressing cells with progression from CSPC (M0 status) cells to CRPC. (I) UMAP plot showing development of three distinct lineages along the pseudotime trajectory. (J) AR, NE and Stem signatures were computed to track the development of three lineages.
To investigate further, we employed single-cell RNA sequencing (scRNA-seq) data from three independent studies comprising 14 patients with castration-sensitive PC (CSPC) and six with CRPC (10–12) (see Methods). Initially, OC2-expressing epithelial cells were selected from CRPC data (Figure 1C, Supplementary Figure S1E). Markers associated with distinct phenotypes in OC2-expressing epithelial cells and lineage signatures were mapped to the OC2-expressing cells (Figure 1D, E). Notably, the AR + phenotype was evident in most cells. The neural differentiation marker chromogranin A (CHGA) overlapped with the NE+/Stem- phenotype, while the NE driver ASCL1 overlapped with both NE+/Stem- and NE+/Stem+ phenotypes. Expression of the NE master regulator BRN2 (47) was not detected at the single-cell level (Figure 1E). These results, where NE + cells cluster into two different groups, are consistent with the results of the Brady et al. study (8) (Supplementary Figure S1D).
To address this heterogeneity, we performed a new phenotype annotation by computing 118 TF activity profiles based on mRNA expression of known targets, encompassing all high confidence targets (19) in the OC2-expressing epithelial cells. Hierarchical clustering identified four TF modules (17) (Supplementary Figure S1F), which correspond to distinct lineages based on standard markers representing AR, Stem and NE features. Four subtypes expressing OC2 were subsequently defined as displaying AR+, AR–/NE–, NE+/Stem+ and NE+/Stem– phenotypes (Figure 1F). Consistent with recent reports, the phenotype exhibiting Stem+ displays elevated interferon signaling (Supplementary Figure S1G), with high activity of IRF1/9 and STAT1/2/3/6 (48). Taken together, these results demonstrate that OC2-positivity coincides with distinct molecular phenotypes defined by divergent TF activity.
OC2 activity computed by gene signature (3) was significantly higher in CRPC compared to CSPC (Figure 1G). In CSPC, M1 metastatic disease in CSPC patients displayed higher OC2 activity compared to M0 patients (no detectable metastases). Similar results were observed in bulk RNA-seq data of primary tumors in M0/M1 patients (49) (Supplementary Figure S1H). These results suggest OC2 activity correlates with more advanced disease. To trace the OC2-expressing epithelial cell lineages, we first separated CSPC and CRPC cells in the combined scRNA-seq data. Pseudotime analysis revealed three branches developing during progression from CSPC (M0) to CRPC (Figure 1H, I). Established signatures were applied to assess pseudotime-dependent changes of AR+, NE+/Stem– and NE+/Stem+ lineages (Figure 1J). These suggest increases of NE+ and Stem+ activity at two terminal points (NE+/Stem+ and NE+/Stem– clusters). Consistent with this, OC2 activity is also high in the same terminal points (Supplementary Figure S1I-J). Prostate stem cell antigen (PSCA) is highly expressed at the start point (CSPC), while ASCL1 expression is enriched in NE+/Stem+ and NE+/Stem– clusters (Supplementary Figure S1J). PSCA is reported to be a marker for Stem-like Luminal L2 cells, described as a source of emergent lineage plasticity (50). These findings show OC2-expressing cells progressing along a trajectory from CSPC appear to adopt multiple routes to bypass AR dependence.
OC2 alters chromatin accessibility in the progression to CRPC
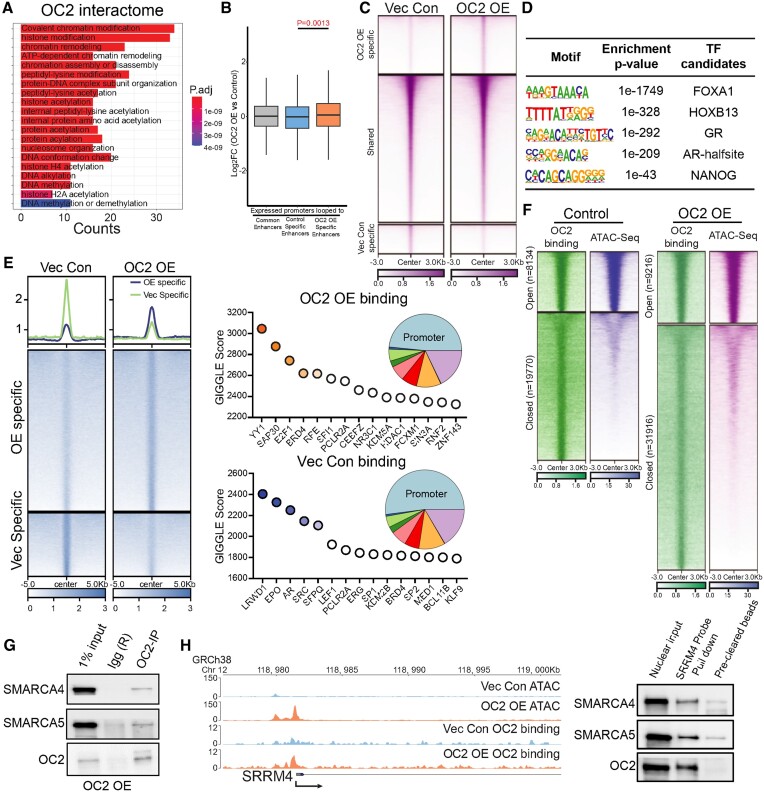
To uncover the mechanism underlying OC2-induction of heterogeneous lineages in the progression from CSPC to CRPC, we enforced expression of OC2 in AR-dependent LNCaP cells and investigated the OC2 interactome using Immunoprecipitation-Mass Spectrometry (IP-MS). The most abundant peptides in the OC2 interaction trap represented proteins involved in chromatin remodeling (Figure 2A; Supplementary Figure S2A). Hi-C chromosome capture (38,39) integrated with H3K27 acetylation (H3K27Ac) profiles demonstrated that OC2 OE increases enhancer-promoter interactions genome-wide, suggesting the newly formed looping structures contributed to OC2-directed gene expression changes (Figure 2B). The endogenous OC2 gene locus also showed specific enhancers looped to the OC2 promoter in OC2 OE LNCaP cells (Supplementary Figure S2B). Genome-wide chromatin accessibility, as assessed by ATAC-seq, increased in OC2 OE cells (Supplementary Figure S2C, D), with 40568 unique regions opened under this condition (Figure 2C). TF motif analysis of OC2-induced hyper-accessible regions revealed enrichment of FOXA1, HOXB13, GRE (glucocorticoid receptor element), AR-half-site and NANOG motifs (Figure 2D). To decipher the possible role of OC2 in chromatin remodeling, the OC2 cistrome was profiled with CUT&RUN-seq under the same conditions. Known TF binding motifs were ranked by similarity score to distinct OC2 binding sites. OC2 OE cells exhibited a distinct TF binding profile in comparison to control cells: OC2 showed co-occupancy with the AR, in non-perturbed cells, consistent with published data (3), whereas YY1 was the top co-occupied TF in OC2 OE cells (Figure 2E). Intriguingly, ∼75% of the OC2-bound regions consisted of closed chromatin. OC2 OE resulted in an increase in the number of OC2 binding peaks at closed chromatin. Accordingly, the ratio of closed/open peaks with OC2 binding increased from 2.5 (19 770 versus 8134) to 3.5 (31 916 versus 9216) with OC2 OE (Figure 2F), suggesting a chromatin remodeling function.
Figure 2.
OC2 interacts with CRCs and alters chromatin accessibility. (A) Gene Ontology analysis of the OC2 interactome proteins identified by IP-MS experiments (N = 2) showed enrichment of chromatin remodeling complexes. (B) HiC-seq was integrated with histone H3K27Ac CUT&RUN-seq to identify specific enhancers in LNCaP Vec Con and OC2 OE cells (N = 2). RNA-seq was then integrated to access the specific enhancer looped those upregulated gene promoter regions, suggesting the newly formed looping structures contributed to OC2-directed gene expression changes. (C) Normalized tag densities for ATAC-seq in LNCaP Vec Con and OC2 OE cells showed OC2 induced chromatin remodeling (N = 2). (D) Motif enrichment analysis for hyper-accessible regions in OC2 OE from ATAC-seq data. (E) Normalized tag densities for the OC2 cistrome in LNCaP Vec Con and OC2 OE cells through CUT&RUN-seq (Left). GIGGLE analysis for TF binding similarity in the OC2-specific cistrome in both conditions (Right) (N = 2). (F) Normalized tag densities for integrated analysis of ATAC-seq and OC2 CUT&RUN-seq. 75% of the OC2-bound regions consisted of closed chromatin. (G) IP-WB showed OC2 interacts with SMARCA4 and SMARCA5. (H) ATAC-seq suggested OC2 binds to the chromatin and induced chromatin opening in the SRRM4 promoter region (left). DNA affinity precipitation assay (DAPA) showed binding of SMARCA4 and SMARCA5 to the SRRM4 promoter region under OC2-enforced conditions (right) (Refer to methods).
Chromatin remodeling complexes (CRCs) modify chromatin architecture to allow TF access to condensed genomic DNA (51). Four CRC families exist in humans: Chromodomain Helicase DNA-binding (CDH), INO80, SWItch/Sucrose Non-Fermentable (SWI/SNF) and Imitation switch (ISWI) complexes. OC2 interactome data indicate that OC2 physically associates with multiple proteins in CRCs representing the SWI/SNF and ISWI complexes (Supplementary Figure S2E). The most abundant CRC peptides identified through IP-MS were those of SMARCA4 (SWI/SNF) and SMARCA5 (ISWI). INO80 and CDH families were either low in abundance or undetectable.
OC2 complexed with SMARCA4 and SMARCA5 (Figure 2G), demonstrating that both proteins directly interact with endogenous OC2. To explore the interaction of OC2 with CRCs at a physiologically relevant locus, the serine/arginine matrix 4 (SRRM4) promoter was selected. SRRM4 is an NE-associated splicing factor and lineage plasticity driver in CRPC (52). OC2 OE resulted in an increase in both SRRM4 mRNA level and chromatin accessibility at the endogenous SRRM4 promoter. OC2 bound to the SRRM4 promoter, consistent with OC2 as a direct regulator of this gene. OC2, SMARCA4 and SMARCA5 bound to a cloned segment of the SRRM4 promoter (Figure 2H). These findings are consistent with alteration of chromatin accessibility as an OC2 function.
OC2 activates multiple AR-independent lineage-defining factors
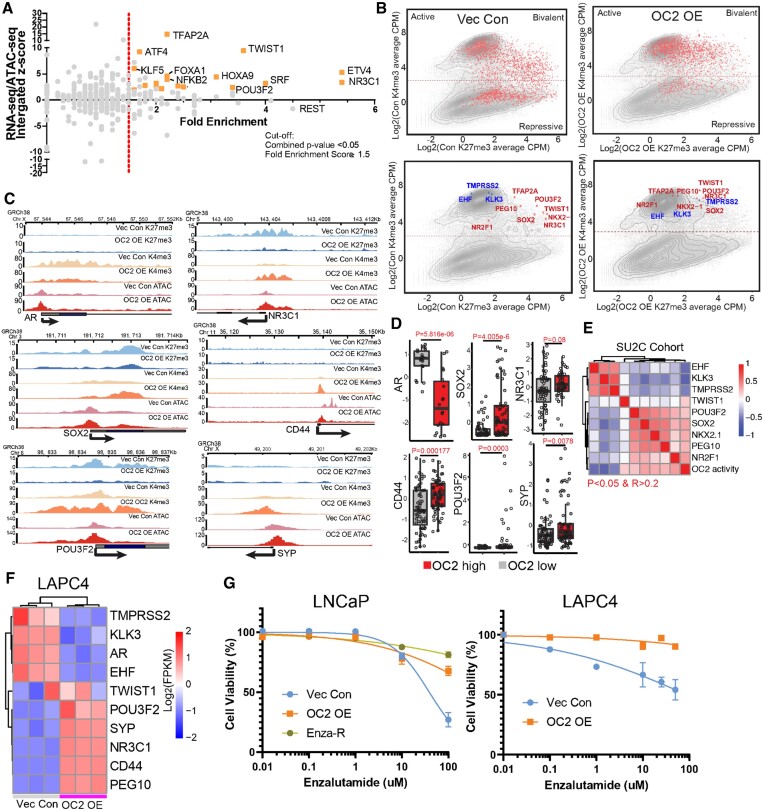
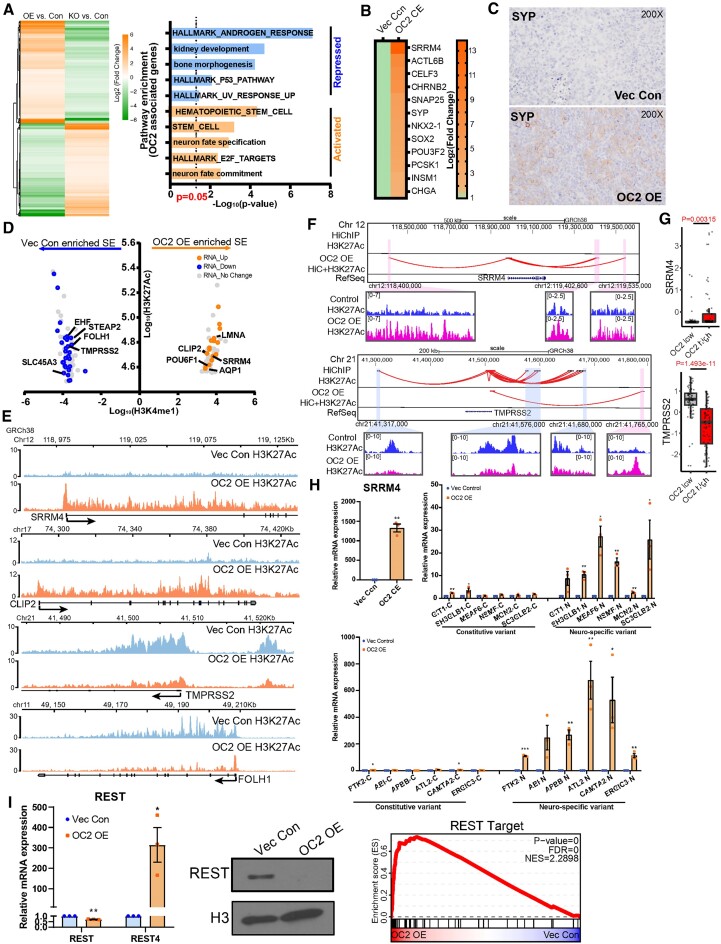
Multiple lineage-defining TF motifs were newly accessible under OC2 OE conditions (Figure 2D). To identify TF drivers and to characterize their transcriptional programs, we performed RNA-seq and ATAC-seq in response to OC2 OE. Integrative analysis of the transcriptional and chromatin landscape changes induced by enforced OC2 revealed a series of CRPC- and/or NEPC-associated TF genes, including NR3C1, ETV4, TWIST1, POU3F2, TFAP2A and KLF5, which were highly activated by OC2 OE (2,47,53–56) (Figure 3A, Supplementary Figure S3A–C). Importantly, these TFs were selected based on target gene enrichment (fold enrichment score), where the higher score represents a greater percentage of target gene changes (Supplementary Figure S3D).
Figure 3.
OC2 activates multiple AR-independent lineage-defining factors. (A) Integrated analysis for identifying OC2-driven candidate TFs based on RNA-seq, ATAC-seq and fold enrichment of downstream targets (refer to methods). Selected candidate genes were labeled in orange. Full candidates list is in Supplementary Table S3 (Cut-off: Fold enrichment score ≥1.5; Combined P < 0.05). (B) Log2 (CPM + 0.1) signal intensity of H3K4me3 and H3K27me3 within ±2 kb of the transcription start site (TSS) where OC2 binds. Each dot represents a unique transcript start site. The cutoff for H3K4me3 separation to indicate active and repression is based on two normal distributions of the signal. Selected genes are highlighted in the bottom row (Red in upregulated genes and Blue in downregulated genes). (C) H3K27me3, H3K4me3 and ATAC-seq signals in LNCaP Vec Con and OC2 OE cells for AR bypass genes AR and GR (NR3C1), NEPC genes (POU3F2 and SYP), Stem genes (SOX2 and CD44). (D) The expression levels of matched selected genes in the SU2C CRPC cohort based on stratification of OC2 activity. (E) Correlation of OC2 activity with candidate TF expression in the SU2C cohort. (F) OC2-driven TFs were also activated in LAPC4 Vec Con and OC2 OE cells (N = 3). (G) OC2 overexpression promotes enzalutamide resistance in LNCaP and LAPC4 cells. LogIC50: LNCaP (1.59 (Vec) versus 2.75 (OC2 OE) versus 4.46 (Enza-R)); LAPC4 (1.95 (Vec) versus 5.49 (OC2 OE))
We then investigated whether OC2-induced TF activation is associated with epigenetic modifications. OC2 binding ±2 kb around transcriptional start sites (TSS) were evaluated for H3K4me3 and H3K27me3 histone marks, which indicate active or repressed epigenomic states, respectively. OC2 OE resulted in a global shift from repressed or bivalent status toward transcriptional activity (Figure 3B, top). OC2-regulated TFs (marked red in the figure) exhibited an increase in the H3K4me3 activation mark and a decrease in the H3K27me3 repressive mark (Figure 3B, bottom). In contrast, AR downstream target genes (blue) moved in the opposing direction, indicating AR suppression by OC2 (Figure 3B, bottom). The cut-off of H3K4me3 signal is based on two-normal distribution (Supplementary Figure S3E). H3K27me3 and H3K4me3 marks were aligned with ATAC-seq profiles at genomic loci corresponding to AR+, NE+/Stem–, and NE+/Stem+ lineage-defining factors (Figure 3C), including those involved in NEPC (POU3F2, SYP), AR bypass (NR3C1, AR), as well as Stem-like features (SOX2, CD44) are shown epigenetically activated. We also checked the epigenetic profiles in 10 patient-derived-xenografts (PDXs); the top 3 PDXs (ranked by H3K4me3 signal) with OC2 activation were considered as OC2 high, while the bottom 3 samples were considered as OC2 low (Supplementary Figure S3F). Similar to LNCaP dataset, epigenetic activation of NEPC (POU3F2, SYP), AR bypass (NR3C1, AR), as well as Stem-like features (SOX2, CD44) was observed in OC2 high PDXs (Supplementary Figure S3G). Highly ranked TFs in response to OC2 OE were also highly ranked in OC2-high CRPC tumors from the updated SU2C cohort, which contains over 440 patients (57) (Figure 3D). Expression of OC2-regulated TFs correlated with OC2 activity computed by PC OC2 signature (3) in the SU2C CRPC cohort (Figure 3E). OC2-induced expression of AR-indifferent, lineage-defining factors was validated in an independent CSPC model, LAPC4 (Figure 3F). Enforced OC2 OE in both LNCaP and LAPC4 models resulted in an enzalutamide-resistant phenotype (Figure 3G). Taken together, these findings indicate that OC2 operates globally to activate numerous AR-indifferent, lineage-defining factors. Notably, OC2 OE alone, without ARSI challenge, resulted in the activation of multiple AR-indifferent lineages.
OC2 regulates AR target genes by upregulation of the glucocorticoid receptor
We also measured H3K27me3 and H3K4me3 histone marks to investigate the ability of OC2 to regulate the epigenetic status of the promoters of a panel of lineage signature genes in both the LNCaP model and PDX models. Upon OC2 activation, NE and Stem-like lineage signature scores exhibited epigenetic activation, whereas the AR-governed lineage was repressed (Figure 4A). OC2 OE suppressed prostate specific antigen (PSA/KLK3) mRNA and other AR targets. Conversely, OC2 knockdown using either shRNA or OC2 inhibitor CSRM-617 (3) restored AR signaling (Supplementary Figure S4A, B). However, surprisingly, residual PSA that remained was further suppressed, instead of being restored, by OC2 silencing (Figure 4B). A similar result was seen with the AR-target genes TMPRSS2, FKBP5, NKX3.1 and STK39 (Supplementary Figure S4C). This notable and unexpected finding suggests that OC2 may be capable of activating specific AR-dependent genes.
Figure 4.
OC2 activates AR-bypass pathways through GR. (A) Epigenetic status (H3K27me3 and H3K4me3 signal) of the promoters of a panel of lineage signature genes (AR/Stem/NE) represented by average z-score in LNCaP Vec Con and OC2 OE cells (left), and patient derived xenografts (PDX) with OC2 high and OC2 low phenotype (N = 3 for each). (B) OC2 knockdown with siRNA (10 uM) suppressed PSA in the OC2 OE condition, indicating OC2 in controls PSA-expressing lineage in OC2 OE cells. (C) Integrated analysis showed loss of enhancer looping to the KLK3 promoter with OC2 OE (Top). ATAC-seq and CUT&RUN-seq of H3K27Ac, H3K27me3, H3K4me3 and AR binding signals were aligned at KLK3 in LNCaP Vec Con and OC2 OE cells (Bottom). (D) KLK3 expression in AR and GR high versus low groups in SU2C cohorts. (E) mCRPC specimens (8) with evidence of AR activity, based on AR signature and positive PSMA staining, which show high OC2 expression, also exhibited either high AR or high GR expression (left). OC2 mRNA level is positively correlated with GR mRNA with Spearman's correlation coefficient (right). (F) OC2-induced GR upregulation with AR suppression in both LNCaP and LAPC4 cells. (G) Integrated HiC with H3K27Ac loops indicating enhancer regions near AR and NR3C1 (GR) promoter loci. (H) Normalized tag densities of CUT&RUN-seq at specific GR binding regions showed increased GR binding on the chromatin. (I) Loss of AR binding and gain of GR binding at the promoter loci of UGT2B15 and UGT2B17 in LNCaP OC2 OE cells. (J) OC2, AR and GR IHC staining in xenograft models created by subcutaneous injection of LNCaP Vec Con and OC2 OE cells.
The expected suppressive effect on PSA/KLK3 was seen in the integrated HiC and H3K27Ac data, where substantially fewer chromatin loops were evident in response to OC2 OE at the KLK3 promoter and surrounding regions (Figure 4C). This coincided with lower H3K4me3 signal (Figure 4C). Chromatin accessibility and AR binding peaks were also decreased, suggesting that PSA expression is no longer under the control of AR.
One of the top OC2-driven TFs (Figure 3A) is the GR(NR3C1), and GREs were identified among the most frequently co-occupied sites with OC2 OE (Figure 2E). GR drives expression of AR target genes in CRPC (2). KLK3 expression was higher in the GR-high subset of the SU2C CRPC cohort versus the GR-low subset (Figure 4D). Similarly, in the Brady et al. cohort (8), AR + specimens, validated by PSMA staining and showing high OC2 expression, exhibited a positive OC2/GR correlation (Figure 4E). In these specimens, AR and GR expression levels were inversely related, consistent with the ability of AR to suppress GR (58). High OC2 ISH values coincided with high OC2 mRNA in the Brady et al. cohort (Figure 4E). In both LNCaP and LAPC4 CSPC models, OC2 OE induced GR expression and suppressed AR in the nucleus (Figure 4F). Using an inducible system in LAPC4, GR expression was suppressed when OC2 expression was repressed (Supplementary Figure S4D), suggesting the GR is directly regulated by OC2. Consistent with this, endogenous OC2 bound to the NR3C1 promoter (Supplementary Figure S4E), and OC2 OE significantly increased chromatin accessibility at the promoter (Figure 3C). HiC revealed more 3-dimensional genomic contacts linked to the NR3C1 promoter in response to OC2, coinciding with elevated H3K27Ac signal in three putative enhancer regions, indicating enhancer reprogramming. Chromatin loops to the AR promoter were lost (Figure 4G). OC2 OE substantially increased GR binding genome-wide (N = 22751) as shown by CUT&RUN-seq (Figure 4H). AR binding to the AR-driven, androgen-inactivating genes UGT2B15 and UGT2B17, which are upregulated with androgen deprivation therapy or ARSI treatment (59,60), was reduced by OC2 OE, while GR binding was increased (Figure 4I). OC2 OE in LNCaP xenografts resulted in low AR levels and high GR levels (Figure 4J). Collectively, these results suggest that OC2-driven GR activation restores expression of AR target genes.
OC2 promotes NE features through super-enhancer reprogramming
OC2-associated genes in CSPC cells were identified in LNCaP OC2 OE cells, with the criterion they were oppositely regulated under OE and knockout conditions (fold change > 1.5 and adj.P < 0.05). NE and Stem-like pathways were prominent among the upregulated genes. Repressed pathways include androgen response and P53 hallmarks, suggesting emergence of lineage plasticity (Figure 5A). Enforced OC2 in LNCaP and LAPC4 cells activated an NE differentiation RNA signature (6) seen in aggressive PC (Figure 5B; Supplementary Figure S5A). Epigenetic activation of ASCL1 is also seen with OC2 OE cells (Supplementary Figure S5B). Consistent with this, a xenograft model derived from subcutaneous injection of OC2 OE cells exhibited positive SYP staining (Figure 5C). This OC2 activity signature, and a published NEPC signature (61), were applied to 18 patient-derived xenografts (6). Both signatures comparably identify NE tumors within the cohort, reinforcing the role of OC2 as a promoter of NE differentiation (Supplementary Figure S5C).
Figure 5.
OC2 promotes neuroendocrine features through super-enhancer reprogramming. (A) OC2 signature genes derived from AR-dependent LNCaP cells (Left). And the Gene Set enrichment analysis of OC2 signature genes (Right). (B) Enforced OC2 expression upregulates multiple NE signature genes in LNCaP cells. Non-expressing genes were removed from the heatmap. (C) SYP IHC staining in LNCaP Vec Con and OC2 OE cells xenograft models. (D) Genes proximal to super-enhancer regions identified as newly formed in OC2 OE cells versus control cells with corresponding change in RNA level. (E) Visualization of H3K27Ac signals around TMPRSS2 / FOLH1 (encoding PSMA) and SRRM4 / CLIP2 are shown. (F) Integrated HiC and H3K27Ac signal showed enhancers looped to promoter loci of TMPRSS2 and SRRM4. (G) Expression of TMPRSS2 and SRRM4 in SU2C cohorts stratified by OC2 activity. (H) Upregulation of the splicing factor SRRM4 promotes neural-specific splicing variants with OC2 overexpression (unpaired two-tailed Student's t-test, **P < 0.01, *P< 0.05). (I) Master neuronal suppressor REST was suppressed in the nucleus (middle) through SRRM4-mediated spicing variant REST4 (left), which promotes NE differentiation (right).
Genomic profiling of H3K27Ac was performed in OC2 OE and vector control cells to identify transcriptionally active regulatory elements. The H3K27Ac profile was substantially changed genome-wide in response to enforced OC2. Super-enhancers (SEs) are large chromatin domains that regulate lineage-specific gene expression (62,63). Genes proximal to SE regions identified as newly formed in OC2 OE versus control, in which there was a corresponding change in RNA expressed from these genes, are shown (Figure 5D, Figures S5D and S5E). The most highly upregulated gene with enforced OC2 was SRRM4 (Figure 5B), a splicing factor and NE-driver shown above to be directly regulated by OC2 (Figure 2H). SRRM4 is notable as an SE-driven gene when OC2 is activated because elevated H3K27Ac signal was seen across the entire genomic locus only under OC2 OE conditions (Figure 5E). Newly formed enhancer-promoter chromatin loops were seen at the SRRM4 locus with OC2 OE (Figure 5F). Notably, SEs associated with AR downstream targets in the control cells, including EHF, TMPRSS2, and FOLH1 (encoding PSMA), were repressed by OC2 OE (Figure 5E, Supplementary Figure S5F). Loss of 3-dimensional contacts near the TMPRSS2 promoter were also observed in the OC2 OE condition (Figure 5F, G). These findings confirm that AR is globally suppressed by OC2. The gene set reflecting presumptive OC2-associated, SE-activated or -repressed genes was consistently expressed in the OC2 high/low tumors in the SU2C CRPC cohort (Supplementary Figure S5G).
In accordance with the robust increase in SRRM4 expression with OC2 OE, SRRM4 activity was similarly increased under these conditions (Figure 5H, Supplementary Figure S5H). SRRM4 mediates neural-specific alternative splicing (52). One key downstream target gene is the RE1 silencing transcription factor (REST), a master repressor of neurogenesis. SRRM4-mediated splicing transforms REST into REST4, a transcriptionally inactive form, resulting in loss of the C-terminal repressor domain and diminished repressive function arising from cistrome competition (6). Elevated REST4 was seen in the OC2 OE condition, along with de-repression of a REST-target gene signature (NRSF_01 from MSIGDB (64)), and REST translocation into the nucleus was lost (Figure 5I). Depletion of OC2 using siRNA in OC2 OE failed to inhibit the expression of SRRM4 and its targets (Supplementary Figure S5I), suggesting that the SE formation mediated by OC2 is irreversible. Collectively, these findings indicate that OC2 regulates lineage plasticity, at least in part, through SE reprogramming.
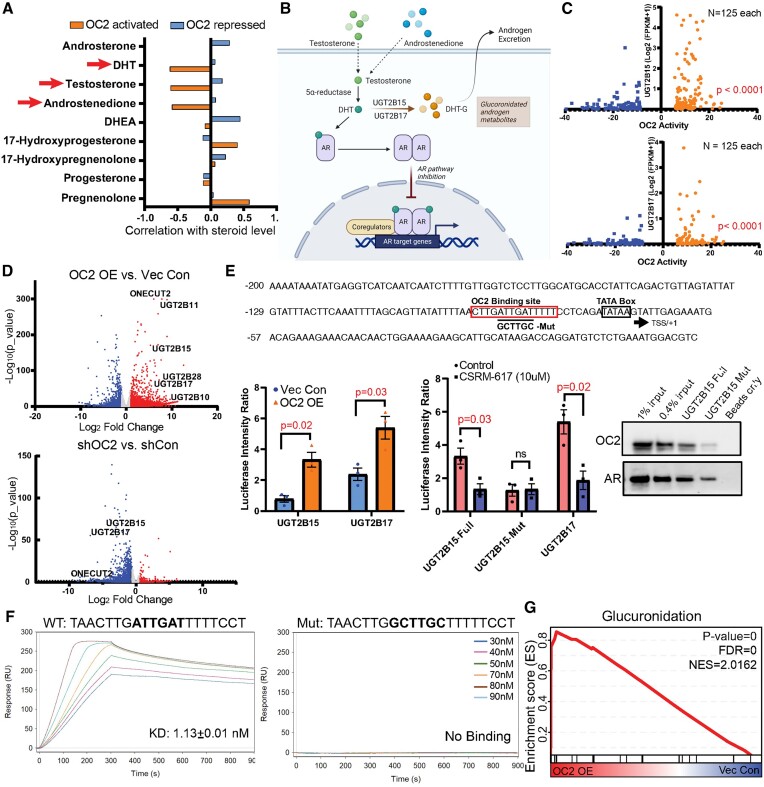
OC2 is a direct suppressor of active androgen
Mass spectrometry analysis and RNA-seq data from the same mCRPC tissue (65) showed that OC2 activity, determined by gene signature, inversely correlated with levels of dihydrotestosterone (DHT), testosterone and androstenedione (Figure 6A). These findings suggest a role for OC2 in androgen inactivation. DHT is inactivated in prostate cells by two glucuronidating enzymes, UGT2B15 and UGT2B17 (66,67) (Figure 6B). UGT2B15 and UGT2B17 expression was examined in patient tumors (57) with either high or low OC2 activity. Tumors with high OC2 exhibited increased levels of UGT2B15 and UGT2B17 overall (Figure 6C). Consistent with this, in LNCaP cells, UGT2B15 and UGT2B17 were upregulated by OC2 OE and suppressed when OC2 was silenced (Figure 6D).
Figure 6.
OC2 is a direct suppressor of active androgen. (A) The same mCRPC patient specimens showing OC2 activity inversely correlated with DHT, testosterone and androstenedione levels. (B) A graphic summary of UGT2B15 and UGT2B17 regulation of androgen glucuronidation to suppress the AR axis. (C) UGT2B15 and UGT2B17 are highly expressed in patients with high OC2 activity. (D) RNA-seq data showing OC2 overexpression induced UGT2B15/17 upregulation while knockdown of OC2 suppressed expression of both genes. (E) Loss of OC2 binding to the UGT2B15 promoter region when the predicted binding site is mutated. Luciferase reporter system showing OC2 upregulates UGT2B15 and UGT2B17. The luciferase signal with the OC2 binding site mutation was not affected by the OC2 inhibitor. (F) In a cell-free system using surface plasmon resonance, the OC2 DNA binding region exhibited high affinity to the wild-type UGT2B15 promoter while the mutated DNA sequence (90 nM) exhibited no binding signal. (G) Glucuronidation is highly activated in the OC2 OE condition.
OC2 bound to the UGT2B15 and UGT2B17 promoters in proximity to AR and FOXA1 (Supplementary Figure S6A). Motif scanning identified a putative OC2 binding site near the TATA box of UGT2B15 and UGT2B17 promoters (Supplementary Figure S6B). UGT2B15 and UGT2B17 promoter-luciferase constructs were significantly activated in OC2 OE versus control cells (Figure 6E). A 6-bp mutation was introduced at an OC2 site within the UGT2B15 promoter and incorporated into a promoter (UGT2B15 -2315/+24)-reporter system (Figure 6E). While the OC2 inhibitor CSRM-617 (3) suppressed wild-type UGT2B15 and UGT2B17 promoter activity, the mutant UGT2B15 promoter showed reduced activity in comparison to wild-type, and did not respond to the inhibitor (Figure 6E). OC2 bound to a 25bp probe corresponding to the OC2 motif in the UGT2B15 promoter, however OC2 binding was lost with the mutation, consistent with this site being an essential element for UGT2B15 regulation (Figure 6E). AR is a direct repressor of UGT2B15 and UGT2B17 expression (59,60). AR bound to both the wild-type and mutant UGT2B15 25 bp segment. Of note, the AR inhibitor enzalutamide only activated reporter activity from the wild-type but not the mutant construct, indicating that mutation of the OC2 binding site alters the response of this region to AR suppression (Supplementary Figure S6C).
OC2 binding to this region was validated using surface plasmon resonance. Recombinant, purified OC2 demonstrated high affinity (KD = 1.13 ± 0.01 nM) to the OC2 DNA binding region in the wild-type UGT2B15 promoter while showing no binding to the mutated fragment (Figure 6F). Comparison of the 5′ flanking region of several UGT2B genes demonstrated that the OC2-binding region is conserved across several family members (Supplementary Figure S6D). We also observed that enforced OC2 exerted broader effects on activation of glucuronidation genes (Figure 6G). These findings indicate that OC2 is a direct transcriptional activator of androgen-inactivating proteins that irreversibly deplete intracellular androgen. Because a low level of androgen is sufficient to promote lineage plasticity in PC cells (68), these findings identify another mechanism, distinct from those described above, whereby OC2 promotes the emergence of lineage variants.
OC2 inhibition suppresses a lineage plasticity program induced by enzalutamide
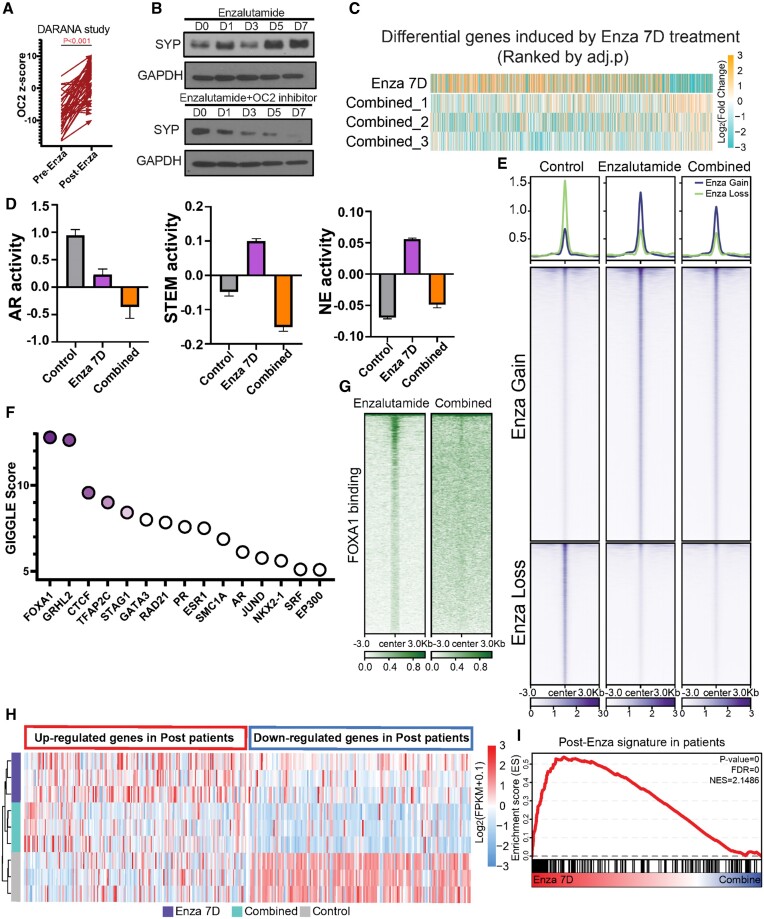
Enzalutamide induces lineage plasticity in AR-dependent PC cells through transcriptional reprogramming (69,70). OC2 was previously shown to be inactivated by REST (3). Enzalutamide-treated LNCaP cells suppressed REST and upregulated OC2 expression in a time-dependent manner (Supplementary Figure S7A). In patient cohort, OC2 activation is seen in response to ARSI therapy in pre- and post-enzalutamide matched samples (Figure 7A).
Figure 7.
OC2 inhibition suppresses lineage plasticity evoked by enzalutamide. (A) Pre- and post-enzalutamide treatment in the same patients showed OC2 activation following ARSI therapy (GSE197780). (B) OC2 inhibitor blocks enzalutamide-induced SYP expression in LNCaP cells. (Enzalutamide: 10uM; OC2 inhibitor: 10 uM). (C) Simultaneous combination (Combined) treatment of OC2 inhibitor with enzalutamide broadly suppressed enzalutamide-induced gene expression changes versus control. (D) AR, Stem and NE lineages were repressed by combined treatment with OC2 inhibitor. Heatmap of gene expression in these three lineages is shown. Non-expressing genes were removed from the heatmap. (E) Combined treatment with OC2 inhibitor greatly suppressed enzalutamide-induced chromatin accessibility changes (enzalutamide: 10 uM; OC2 inhibitor: 10 uM). (F) GIGGLE analysis identified FOXA1 as the most enriched TF in suppressed hyper-accessible regions of combination treatment versus enzalutamide alone. (G) FOXA1 CUT&RUN-seq showed combined treatment with OC2 inhibitor suppressed FOXA1-driven chromatin accessibility changes. (H-I) A gene signature derived from post-enzalutamide patient samples was consistent with perturbed genes seen in enzalutamide-treated LNCaP cells. SC treatment with OC2 inhibitor blocked enzalutamide-induced gene perturbation.
We applied two protocols to assess the outcomes of combining a direct OC2 inhibitor (10uM) with enzalutamide (10 uM): (i) a simultaneous protocol where both agents were applied to cells at the same time (hereafter referred to as the ‘combined’ condition); and (ii) a condition where the OC2 inhibitor was applied alone three days before the simultaneous administration of both agents (referred to as ‘pre-treatment’).
Combined treatment of LNCaP cells with enzalutamide and the OC2 inhibitor CSRM-617 suppressed SYP induction (Figure 7B) and broadly suppressed enzalutamide-induced gene expression changes (Figure 7C; Supplementary Figure S7B), indicating the potential of targeting OC2 to suppress lineage plasticity (Figure 7B). Combined treatment with OC2 inhibitor suppressed inflammatory response and EMT processes compared to enzalutamide alone (Supplementary Figure S7C). This finding aligns with a recent study demonstrating that an inflammatory response is required to induce lineage plasticity in PC cells (48). OC2 OE significantly released AR binding at SOX2 and POU3F2 (BRN2) loci, leading to activation of Stem and NE lineage genes (Supplementary Figure S7D). Enzalutamide treatment alone activated NE+/Stem + and NE+/Stem- lineages, while both were suppressed in the combined condition. AR activity was suppressed with enzalutamide, and combined treatment with the OC2 inhibitor further repressed AR (Figure 7D, Supplementary Figure S7E), consistent with the results shown in Supplementary Figure S4C. Similar activation of OC2 was also observed in the same patients before and after ADT treatment (Supplementary Figure S7G).
Enzalutamide used alone induced chromatin remodeling as reported (71) (Figure 7E). However, combined treatment with OC2 inhibitor greatly suppressed enzalutamide-induced chromatin accessibility changes. FOXA1 was identified as the most enriched TF at chromatin regions where OC2 inhibitor suppressed chromatin accessibility, suggesting that OC2 blocks FOXA1-mediated accessibility in enzalutamide-treated cells (Supplementary Figure S7F, Figure 7F). FOXA1 was shown to be involved in treatment-emergent NE transdifferentiation (45). FOXA1 binding intensity was dramatically lost with combined treatment, demonstrating that OC2 inhibition blocks enzalutamide-induced lineage plasticity, in part by suppressing FOXA1-mediated chromatin remodeling (Figure 7G).
A gene signature derived from post-enzalutamide patient samples was perturbed in enzalutamide-treated LNCaP cells, consistent with the patient data. Combined treatment blocked this enzalutamide-induced gene perturbation. (Figure 7H and I). These findings demonstrate that OC2 inhibition suppresses an enzalutamide-induced gene expression network seen in human PC.
The OC2 inhibitor alone activated AR signaling (Supplementary Figure S7H), suggesting that OC2 inhibition sensitizes cells to enzalutamide. To test this, using a second treatment protocol, LNCaP cells were pretreated with CSRM-617 for three days, followed by combined treatment with enzalutamide. This ‘pre-treatment’ was extremely potent in tumor cell killing compared to the combined condition (Supplementary Figure S7I). RNA expression profiling of the pre-treatment condition revealed predominant effects on cell cycle processes, consistent with the cell growth and survival results, suggesting that a therapeutic window exists for OC2 targeting even before ARSI therapy (Supplementary Figure S7J).
Discussion
In the present study, we characterized the mechanisms whereby OC2 drives lineage plasticity. We show that OC2, despite its NEPC driver function, can indirectly activate a portion of the AR cistrome. This is a unique activity that, to our knowledge, has not been previously described for any TF. We found OC2 to be widely expressed in CRPC tumors, and that it functions as a broadly acting lineage facilitator through its ability to alter chromatin accessibility, promote the formation of SE regions, and regulate gene expression by acting at bivalent (poised) promoters. OC2 suppresses the AR through multiple mechanisms, including promoting androgen inactivation through activation of glucuronidation genes that specifically and irreversibly disable androgens. We further show that these diverse mechanisms promoting plasticity include direct activation of NR3C1, the gene encoding the GR. The GR promotes disease progression in PC by assuming direct control of part of the AR cistrome under conditions of AR suppression, thus constituting a mechanism of resistance to AR-targeted therapy. Notably, activation of the GR occurs in adenocarcinoma, not NEPC. We show here that, through the intercession of the GR, OC2 can activate certain AR-regulated genes. Consequently, the ability of OC2 to promote lineage variation extends beyond NEPC to treatment-resistant adenocarcinoma.
We present evidence that OC2 directly activates multiple AR-independent, lineage-defining factors, including NR3C1, ETV4, TWIST1, BRN2 (POU3F2), KLF5 and TFAP2A. ETV4 mediates dosage-dependent prostate tumor initiation and promotes metastasis of prostate adenocarcinoma in response to PI3K and RAS co-activation (72). KLF5 opposes AR activities and drives the double-negative PC phenotype (56,73). TFAP2A activation indicates emergence of neural crest lineages (55). In contrast, classical AR downstream target genes (EHF, TMPRSS2, KLK3) were suppressed by OC2. Beyond direct epigenetic regulation within promoter regions, gene expression changes were also contributed by reprogrammed enhancers looped to these promoters. Consistent with the conclusion that OC2 has the capability to bind to closed chromatin and promote chromatin accessibility, we show that OC2 interacts with SMARCA4 and SMARCA5, which are important components of CRCs that modify chromatin architecture to allow TF access to condensed chromatin. The SWI/SNF complex is involved in the emergence of treatment induced NEPC. PROTACs targeting this complex dislodge SWI/SNF-mediated accessibility of oncoproteins to enhancers, and thus are a promising therapeutic approach for enhancer-addicted prostate cancers (74). ISWI complexes assist with nucleosome sliding. A recent study showed SMARCA5 contributes to the racial disparity in prostate cancer, suggesting the potentially distinct role of OC2 in prostate cancer development in patients of different race or ethnicity (75). Inactivation of CHD1, another chromatin remodeler, has been shown to promote the emergence of tumor heterogeneity and enzalutamide resistance (76). Notably, all four lineages driven by CHD1 loss (GR-driven, BRN2-driven, NR2F1-driven and TBX2-driven EMT) emerge with OC2 activation. Taken together, these results indicate that OC2 facilitates the development of multiple lineages through chromatin remodeling and epigenetic modification.
We also demonstrate that OC2 upregulates the UGT2B15 and UGT2B17 genes and is a direct transcriptional activator of UGT2B15. The UGT2B15/17 proteins irreversibly inactivate T and DHT. Analysis of human CRPC tumors revealed that tumors with high OC2 activation exhibited reduced androgen levels. These findings indicate that OC2 promotes androgen clearance, thus ensuring a low androgen environment. Reduced levels of androgen promote AR-indifferent lineage variation in PC cells (68). Consequently, OC2 facilitates lineage variation by activating gene expression programs suppressed by the AR. RNA expression in PC models, and analysis of human PC cohorts, indicate that OC2 exerts a broad effect on glucuronidation processes generally. Upregulation of glucuronidation has been reported to induce multi-drug resistance (77).
A small molecule inhibitor of OC2 represses the lineage plasticity program activated by enzalutamide, suggesting that OC2 inhibition may be a novel therapeutic strategy to prevent emergence of treatment-resistant variants. Our work suggests that OC2 inhibition could play an important role in blocking or delaying the emergence of CRPC, even in the context of castration-sensitive disease.
In summary, our results identify OC2 as a novel facilitator of multiple AR-indifferent lineages, operating by parallel routes to support the appearance of treatment-resistant adenocarcinoma as well as NEPC variants. Our work suggests that OC2 inhibition could play an important role in blocking or delaying the emergence of CRPC.
Supplementary Material
Acknowledgements
The authors thank Dr Gerhard Coetzee for helpful discussions.
Authors' contributions: C.Q., Y.Q., S.Y. and M.R.F. conceived and designed the study. C.Q. and Y.Q. carried out most experiments and analysis. M.R. contributed to the glucuronidation story and generated point-mutations. R.H. performed ISH staining and IHC stating. H.K. generated the ATAC-seq and RNA-seq integrated analysis pipeline. B.G. conducted SPR assay for OC2 binding to DNA. Y.Y. contributed to WB experiments. R.M.C., B.Z., and I.P.G contributed to antibody optimization and tissue staining. C.M., P.S.N. and L.B. provided patient microarray and spatial transcriptome dataset. W.Y. and S.Y. performed interactome analysis. E.A.M and S.Y. conducted androgen measurement in patients and statistical analysis. B.S.K. and H.Y. performed tissue microarray analysis. All authors interpreted results and edited the manuscript. C.Q., Y.Q., S.Y. and M.R.F. generated most of the figures and wrote the manuscript. All authors made intellectual contributions.
Contributor Information
Chen Qian, Departments of Urology and Biomedical Sciences, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA.
Qian Yang, Departments of Urology and Computational Biomedicine, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA.
Mirja Rotinen, Department of Health Sciences, Public University of Navarre, Pamplona, Navarra, Spain.
Rongrong Huang, Department of Pathology and Laboratory Medicine, UCLA, Los Angeles, CA 90095, USA.
Hyoyoung Kim, Departments of Urology and Computational Biomedicine, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA.
Brad Gallent, Departments of Urology and Biomedical Sciences, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA; Division of Medical Oncology, Department of Medicine, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA.
Yiwu Yan, Departments of Urology and Biomedical Sciences, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA.
Radu M Cadaneanu, Department of Surgical and Perioperative Care, VA Greater Los Angeles; Department of Urology and Jonsson Comprehensive Cancer Center, the David Geffen School of Medicine, UCLA, Box 951738, 10833 Le Conte Ave 66-188 CHS UCLA, Los Angeles, CA 90095, USA.
Baohui Zhang, Department of Surgical and Perioperative Care, VA Greater Los Angeles; Department of Urology and Jonsson Comprehensive Cancer Center, the David Geffen School of Medicine, UCLA, Box 951738, 10833 Le Conte Ave 66-188 CHS UCLA, Los Angeles, CA 90095, USA.
Salma Kaochar, Department of Medicine Section Hematology/Oncology Baylor College of Medicine, Houston, 77030 TX, USA.
Stephen J Freedland, Departments of Urology and Biomedical Sciences, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA.
Edwin M Posadas, Division of Medical Oncology, Department of Medicine, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA.
Leigh Ellis, Center for Prostate Disease Research, Mutha Cancer Center Research Program, Department of Surgery, Uniformed Services University of the Health Sciences and the Walter Reed National Military Medical Center; The Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., Bethesda, MD 20814, USA; Genitourinary Malignancies Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD 20892, USA.
Dolores Di Vizio, Departments of Urology, Pathology and Laboratory Medicine, and Biomedical Sciences, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA.
Colm Morrissey, Department of Urology, University of Washington, Seattle, WA 98195, USA.
Peter S Nelson, Divisions of Human Biology and Clinical Research, Fred Hutchinson Cancer Center, Seattle, WA 98109, USA.
Lauren Brady, Divisions of Human Biology and Clinical Research, Fred Hutchinson Cancer Center, Seattle, WA 98109, USA.
Ramachandran Murali, Departments of Urology and Biomedical Sciences, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA.
Moray J Campbell, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA.
Wei Yang, Department of Pathology and Cancer Center, Stony Brook University, NY 11794, USA.
Beatrice S Knudsen, Huntsman Cancer Institute, University of Utah, Salt Lake City, UT 84108, USA; Department of Pathology, University of Utah, Salt Lake City, UT 84108, USA.
Elahe A Mostaghel, Geriatric Research, Education and Clinical Center (GRECC), U.S. Department of Veterans Affairs Puget Sound Health Care System, Seattle, WA 98133, USA.
Huihui Ye, Department of Pathology, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA.
Isla P Garraway, Department of Surgical and Perioperative Care, VA Greater Los Angeles; Department of Urology and Jonsson Comprehensive Cancer Center, the David Geffen School of Medicine, UCLA, Box 951738, 10833 Le Conte Ave 66-188 CHS UCLA, Los Angeles, CA 90095, USA.
Sungyong You, Departments of Urology and Computational Biomedicine, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA.
Michael R Freeman, Departments of Urology and Biomedical Sciences, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA.
Data availability
We used a series of global data publicly available and generated new global data for this study (Supplementary Table S1). Initially, we obtained digital spatial profiling (DSP) transcriptome data from the Gene Expression Omnibus (GEO) database (GSE147250) (8). Single-cell transcriptome from PC patients, were integrated from three independent data sets (GSE141445, GSE157703 and GSE137829) (10–12), consisting of 6 CRPC and 14 primary samples. We also included tissue transcriptome data (GSE147493) of non-metastatic (M0) and metastatic (M1) PC needle biopsy samples from the Greater Los Angeles VA cohort (49) and the data of PC samples from various metastases sites in the Stand-Up-To-Cancer (SU2C) cohort (57), which was downloaded from cBioPortal (78). The global histone mark profiles, including H3K27Ac, H3K4me3 and H3K27me3 from LuCaP PC patient-derived xenograft (PDX) (45) were included for this study (GSE161948). Finally, drug-induced tissue transcriptome data from PC patients by enzalutamide treatment were obtained with the accession, GSE197780 (79). All global data generated for this study were deposited to GEO database with accessions (GSE244025). The proteomics data are available via ProteomeXchange with identifier PXD050634.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
UCLA Prostate Cancer SPORE [2P50CA092131, 1R01CA220327, 1R01CA271750]; Department of Defense [PC180541, PC190604, PC190482]; Institute for Prostate Cancer Research, the Pacific Northwest Prostate Cancer SPORE [P50CA97186, R01CA266452, R01 CA234715, R21 CA277368]. Funding for open access charge: NIH.
Conflict of interest statement. P.S. Nelson served as a paid advisor for Bristol Myers Squibb, Pfizer, and Janssen and received research support from Janssen for work unrelated to the present study.
References
- 1. Yamada Y., Beltran H. Clinical and biological features of neuroendocrine prostate cancer. Curr. Oncol. Rep. 2021; 23:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arora V.K., Schenkein E., Murali R., Subudhi S.K., Wongvipat J., Balbas M.D., Shah N., Cai L., Efstathiou E., Logothetis C. et al. Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell. 2013; 155:1309–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rotinen M., You S., Yang J., Coetzee S.G., Reis-Sobreiro M., Huang W.C., Huang F., Pan X., Yanez A., Hazelett D.J. et al. ONECUT2 is a targetable master regulator of lethal prostate cancer that suppresses the androgen axis. Nat. Med. 2018; 24:1887–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guo H., Ci X., Ahmed M., Hua J.T., Soares F., Lin D., Puca L., Vosoughi A., Xue H., Li E. et al. ONECUT2 is a driver of neuroendocrine prostate cancer. Nat. Commun. 2019; 10:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liberzon A., Birger C., Thorvaldsdottir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015; 1:417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Labrecque M.P., Coleman I.M., Brown L.G., True L.D., Kollath L., Lakely B., Nguyen H.M., Yang Y.C., da Costa R.M.G., Kaipainen A. et al. Molecular profiling stratifies diverse phenotypes of treatment-refractory metastatic castration-resistant prostate cancer. J. Clin. Invest. 2019; 129:4492–4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Smith B.A., Balanis N.G., Nanjundiah A., Sheu K.M., Tsai B.L., Zhang Q., Park J.W., Thompson M., Huang J., Witte O.N. et al. A Human adult stem cell signature marks aggressive variants across epithelial cancers. Cell Rep. 2018; 24:3353–3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brady L., Kriner M., Coleman I., Morrissey C., Roudier M., True L.D., Gulati R., Plymate S.R., Zhou Z., Birditt B. et al. Inter- and intra-tumor heterogeneity of metastatic prostate cancer determined by digital spatial gene expression profiling. Nat. Commun. 2021; 12:1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Levine D.M., Haynor D.R., Castle J.C., Stepaniants S.B., Pellegrini M., Mao M., Johnson J.M. Pathway and gene-set activation measurement from mRNA expression data: the tissue distribution of human pathways. Genome Biol. 2006; 7:R93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen S., Zhu G., Yang Y., Wang F., Xiao Y.T., Zhang N., Bian X., Zhu Y., Yu Y., Liu F. et al. Single-cell analysis reveals transcriptomic remodellings in distinct cell types that contribute to human prostate cancer progression. Nat. Cell Biol. 2021; 23:87–98. [DOI] [PubMed] [Google Scholar]
- 11. Dong B., Miao J., Wang Y., Luo W., Ji Z., Lai H., Zhang M., Cheng X., Wang J., Fang Y. et al. Single-cell analysis supports a luminal-neuroendocrine transdifferentiation in human prostate cancer. Commun. Biol. 2020; 3:778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ma X., Guo J., Liu K., Chen L., Liu D., Dong S., Xia J., Long Q., Yue Y., Zhao P. et al. Identification of a distinct luminal subgroup diagnosing and stratifying early stage prostate cancer by tissue-based single-cell RNA sequencing. Mol. Cancer. 2020; 19:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McGinnis C.S., Murrow L.M., Gartner Z.J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019; 8:329–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M. 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019; 177:1888–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang Y., Parmigiani G., Johnson W.E. ComBat-seq: batch effect adjustment for RNA-seq count data. NAR Genom Bioinform. 2020; 2:lqaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014; 32:381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Badia I.M.P., Velez Santiago J., Braunger J., Geiss C., Dimitrov D., Muller-Dott S., Taus P., Dugourd A., Holland C.H., Ramirez Flores R.O. et al. decoupleR: ensemble of computational methods to infer biological activities from omics data. Bioinform. Adv. 2022; 2:vbac016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alvarez M.J., Shen Y., Giorgi F.M., Lachmann A., Ding B.B., Ye B.H., Califano A. Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat. Genet. 2016; 48:838–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Garcia-Alonso L., Holland C.H., Ibrahim M.M., Turei D., Saez-Rodriguez J. Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res. 2019; 29:1363–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wilkerson M.D., Hayes D.N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010; 26:1572–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yan Y., Zhou B., Qian C., Vasquez A., Kamra M., Chatterjee A., Lee Y.J., Yuan X., Ellis L., Di Vizio D. et al. Receptor-interacting protein kinase 2 (RIPK2) stabilizes c-Myc and is a therapeutic target in prostate cancer metastasis. Nat. Commun. 2022; 13:669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008; 26:1367–1372. [DOI] [PubMed] [Google Scholar]
- 23. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012; 16:284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fardin P., Moretti S., Biasotti B., Ricciardi A., Bonassi S., Varesio L. Normalization of low-density microarray using external spike-in controls: analysis of macrophage cell lines expression profile. Bmc Genomics [Electronic Resource]. 2007; 8:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Amemiya H.M., Kundaje A., Boyle A.P. The ENCODE blacklist: identification of problematic regions of the genome. Sci. Rep. 2019; 9:9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Meers M.P., Tenenbaum D., Henikoff S. Peak calling by Sparse Enrichment analysis for CUT&RUN chromatin profiling. Epigenetics Chromatin. 2019; 12:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ramirez F., Ryan D.P., Gruning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dundar F., Manke T. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016; 44:W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011; 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Layer R.M., Pedersen B.S., DiSera T., Marth G.T., Gertz J., Quinlan A.R. GIGGLE: a search engine for large-scale integrated genome analysis. Nat. Methods. 2018; 15:123–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Buenrostro J.D., Wu B., Chang H.Y., Greenleaf W.J. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 2015; 109:21-29.1–21-29-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ramirez F., Bhardwaj V., Arrigoni L., Lam K.C., Gruning B.A., Villaveces J., Habermann B., Akhtar A., Manke T. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat. Commun. 2018; 9:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009; 326:289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014; 159:1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Durand N.C., Shamim M.S., Machol I., Rao S.S., Huntley M.H., Lander E.S., Aiden E.L. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 2016; 3:95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mariscal J., Vagner T., Kim M., Zhou B., Chin A., Zandian M., Freeman M.R., You S., Zijlstra A., Yang W. et al. Comprehensive palmitoyl-proteomic analysis identifies distinct protein signatures for large and small cancer-derived extracellular vesicles. J. Extracell. Vesicles. 2020; 9:1764192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tripodi I.J., Allen M.A., Dowell R.D. Detecting differential transcription factor activity from ATAC-Seq data. Molecules. 2018; 23:1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hwang D., Rust A.G., Ramsey S., Smith J.J., Leslie D.M., Weston A.D., de Atauri P., Aitchison J.D., Hood L., Siegel A.F. et al. A data integration methodology for systems biology. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:17296–17301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Garcia-Alonso L., Holland C.H., Ibrahim M.M., Turei D., Saez-Rodriguez J. Corrigendum: benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res. 2021; 31:745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Baca S.C., Takeda D.Y., Seo J.H., Hwang J., Ku S.Y., Arafeh R., Arnoff T., Agarwal S., Bell C., O’Connor E. et al. Reprogramming of the FOXA1 cistrome in treatment-emergent neuroendocrine prostate cancer. Nat. Commun. 2021; 12:1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., Young R.A. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013; 153:307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bishop J.L., Thaper D., Vahid S., Davies A., Ketola K., Kuruma H., Jama R., Nip K.M., Angeles A., Johnson F. et al. The master neural transcription factor BRN2 is an androgen receptor-suppressed driver of neuroendocrine differentiation in prostate cancer. Cancer Discov. 2017; 7:54–71. [DOI] [PubMed] [Google Scholar]
- 48. Chan J.M., Zaidi S., Love J.R., Zhao J.L., Setty M., Wadosky K.M., Gopalan A., Choo Z.N., Persad S., Choi J. et al. Lineage plasticity in prostate cancer depends on JAK/STAT inflammatory signaling. Science. 2022; 377:1180–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Miller E.T., You S., Cadaneanu R.M., Kim M., Yoon J., Liu S.T., Li X., Kwan L., Hodge J., Quist M.J. et al. Chromosomal instability in untreated primary prostate cancer as an indicator of metastatic potential. BMC Cancer. 2020; 20:398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Karthaus W.R., Hofree M., Choi D., Linton E.L., Turkekul M., Bejnood A., Carver B., Gopalan A., Abida W., Laudone V. et al. Regenerative potential of prostate luminal cells revealed by single-cell analysis. Science. 2020; 368:497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Clapier C.R., Iwasa J., Cairns B.R., Peterson C.L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 2017; 18:407–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li Y., Donmez N., Sahinalp C., Xie N., Wang Y., Xue H., Mo F., Beltran H., Gleave M., Wang Y. et al. SRRM4 Drives neuroendocrine transdifferentiation of prostate adenocarcinoma under androgen receptor pathway inhibition. Eur. Urol. 2017; 71:68–78. [DOI] [PubMed] [Google Scholar]
- 53. Li D., Zhan Y., Wang N., Tang F., Lee C.J., Bayshtok G., Moore A.R., Wong E.W.P., Pachai M.R., Xie Y. et al. ETV4 mediates dosage-dependent prostate tumor initiation and cooperates with p53 loss to generate prostate cancer. Sci. Adv. 2023; 9:eadc9446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shiota M., Yokomizo A., Tada Y., Inokuchi J., Kashiwagi E., Masubuchi D., Eto M., Uchiumi T., Naito S. Castration resistance of prostate cancer cells caused by castration-induced oxidative stress through Twist1 and androgen receptor overexpression. Oncogene. 2010; 29:237–250. [DOI] [PubMed] [Google Scholar]
- 55. Nouri M., Caradec J., Lubik A.A., Li N., Hollier B.G., Takhar M., Altimirano-Dimas M., Chen M., Roshan-Moniri M., Butler M. et al. Therapy-induced developmental reprogramming of prostate cancer cells and acquired therapy resistance. Oncotarget. 2017; 8:18949–18967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Che M., Chaturvedi A., Munro S.A., Pitzen S.P., Ling A., Zhang W., Mentzer J., Ku S.Y., Puca L., Zhu Y. et al. Opposing transcriptional programs of KLF5 and AR emerge during therapy for advanced prostate cancer. Nat. Commun. 2021; 12:6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Robinson D., Van Allen E.M., Wu Y.M., Schultz N., Lonigro R.J., Mosquera J.M., Montgomery B., Taplin M.E., Pritchard C.C., Attard G. et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015; 162:454. [DOI] [PubMed] [Google Scholar]
- 58. Xie N., Cheng H., Lin D., Liu L., Yang O., Jia L., Fazli L., Gleave M.E., Wang Y., Rennie P. et al. The expression of glucocorticoid receptor is negatively regulated by active androgen receptor signaling in prostate tumors. Int. J. Cancer. 2015; 136:E27–E38. [DOI] [PubMed] [Google Scholar]
- 59. Bao B.Y., Chuang B.F., Wang Q., Sartor O., Balk S.P., Brown M., Kantoff P.W., Lee G.S. Androgen receptor mediates the expression of UDP-glucuronosyltransferase 2 B15 and B17 genes. Prostate. 2008; 68:839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chouinard S., Barbier O., Belanger A. UDP-glucuronosyltransferase 2B15 (UGT2B15) and UGT2B17 enzymes are major determinants of the androgen response in prostate cancer LNCaP cells. J. Biol. Chem. 2007; 282:33466–33474. [DOI] [PubMed] [Google Scholar]
- 61. Beltran H., Prandi D., Mosquera J.M., Benelli M., Puca L., Cyrta J., Marotz C., Giannopoulou E., Chakravarthi B.V., Varambally S. et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med. 2016; 22:298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sengupta S., George R.E. Super-enhancer-driven transcriptional dependencies in cancer. Trends Cancer. 2017; 3:269–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lee H.K., Willi M., Shin H.Y., Liu C., Hennighausen L. Progressing super-enhancer landscape during mammary differentiation controls tissue-specific gene regulation. Nucleic Acids Res. 2018; 46:10796–10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Balanis N.G., Sheu K.M., Esedebe F.N., Patel S.J., Smith B.A., Park J.W., Alhani S., Gomperts B.N., Huang J., Witte O.N. et al. Pan-cancer convergence to a small-cell neuroendocrine phenotype that shares susceptibilities with hematological malignancies. Cancer Cell. 2019; 36:17–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mostaghel E.A., Marck B.T., Kolokythas O., Chew F., Yu E.Y., Schweizer M.T., Cheng H.H., Kantoff P.W., Balk S.P., Taplin M.E. et al. Circulating and intratumoral adrenal androgens correlate with response to abiraterone in men with castration-resistant prostate cancer. Clin. Cancer Res. 2021; 27:6001–6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhu Z., Chung Y.M., Sergeeva O., Kepe V., Berk M., Li J., Ko H.K., Li Z., Petro M., DiFilippo F.P. et al. Loss of dihydrotestosterone-inactivation activity promotes prostate cancer castration resistance detectable by functional imaging. J. Biol. Chem. 2018; 293:17829–17837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Barbier O., Belanger A. Inactivation of androgens by UDP-glucuronosyltransferases in the human prostate. Best Pract. Res. Clin. Endocrinol. Metab. 2008; 22:259–270. [DOI] [PubMed] [Google Scholar]
- 68. Beltran H., Hruszkewycz A., Scher H.I., Hildesheim J., Isaacs J., Yu E.Y., Kelly K., Lin D., Dicker A., Arnold J. et al. The role of lineage plasticity in prostate cancer therapy resistance. Clin. Cancer Res. 2019; 25:6916–6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Westbrook T.C., Guan X., Rodansky E., Flores D., Liu C.J., Udager A.M., Patel R.A., Haffner M.C., Hu Y.M., Sun D. et al. Transcriptional profiling of matched patient biopsies clarifies molecular determinants of enzalutamide-induced lineage plasticity. Nat. Commun. 2022; 13:5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mu P., Zhang Z., Benelli M., Karthaus W.R., Hoover E., Chen C.C., Wongvipat J., Ku S.Y., Gao D., Cao Z. et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science. 2017; 355:84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Davies A., Nouruzi S., Ganguli D., Namekawa T., Thaper D., Linder S., Karaoglanoglu F., Omur M.E., Kim S., Kobelev M. et al. An androgen receptor switch underlies lineage infidelity in treatment-resistant prostate cancer. Nat. Cell Biol. 2021; 23:1023–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Aytes A., Mitrofanova A., Kinkade C.W., Lefebvre C., Lei M., Phelan V., LeKaye H.C., Koutcher J.A., Cardiff R.D., Califano A. et al. ETV4 promotes metastasis in response to activation of PI3-kinase and Ras signaling in a mouse model of advanced prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E3506–E3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lundberg A., Zhang M., Aggarwal R., Li H., Zhang L., Foye A., Sjostrom M., Chou J., Chang K., Moreno-Rodriguez T. et al. The genomic and epigenomic landscape of double-negative metastatic prostate cancer. Cancer Res. 2023; 83:2763–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Xiao L., Parolia A., Qiao Y., Bawa P., Eyunni S., Mannan R., Carson S.E., Chang Y., Wang X., Zhang Y. et al. Targeting SWI/SNF ATPases in enhancer-addicted prostate cancer. Nature. 2022; 601:434–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Siddappa M., Hussain S., Wani S.A., White J., Tang H., Gray J.S., Jafari H., Wu H.C., Long M.D., Elhussin I. et al. African American Prostate cancer displays quantitatively distinct vitamin D receptor cistrome-transcriptome relationships regulated by BAZ1A. Cancer Res Commun. 2023; 3:621–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhang Z., Zhou C., Li X., Barnes S.D., Deng S., Hoover E., Chen C.C., Lee Y.S., Zhang Y., Wang C. et al. Loss of CHD1 promotes heterogeneous mechanisms of resistance to AR-targeted therapy via chromatin dysregulation. Cancer Cell. 2020; 37:584–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mazerska Z., Mroz A., Pawlowska M., Augustin E. The role of glucuronidation in drug resistance. Pharmacol. Ther. 2016; 159:35–55. [DOI] [PubMed] [Google Scholar]
- 78. Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012; 2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Linder S., Hoogstraat M., Stelloo S., Eickhoff N., Schuurman K., de Barros H., Alkemade M., Bekers E.M., Severson T.M., Sanders J. et al. Drug-induced epigenomic plasticity reprograms circadian rhythm regulation to drive prostate cancer toward androgen independence. Cancer Discov. 2022; 12:2074–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We used a series of global data publicly available and generated new global data for this study (Supplementary Table S1). Initially, we obtained digital spatial profiling (DSP) transcriptome data from the Gene Expression Omnibus (GEO) database (GSE147250) (8). Single-cell transcriptome from PC patients, were integrated from three independent data sets (GSE141445, GSE157703 and GSE137829) (10–12), consisting of 6 CRPC and 14 primary samples. We also included tissue transcriptome data (GSE147493) of non-metastatic (M0) and metastatic (M1) PC needle biopsy samples from the Greater Los Angeles VA cohort (49) and the data of PC samples from various metastases sites in the Stand-Up-To-Cancer (SU2C) cohort (57), which was downloaded from cBioPortal (78). The global histone mark profiles, including H3K27Ac, H3K4me3 and H3K27me3 from LuCaP PC patient-derived xenograft (PDX) (45) were included for this study (GSE161948). Finally, drug-induced tissue transcriptome data from PC patients by enzalutamide treatment were obtained with the accession, GSE197780 (79). All global data generated for this study were deposited to GEO database with accessions (GSE244025). The proteomics data are available via ProteomeXchange with identifier PXD050634.