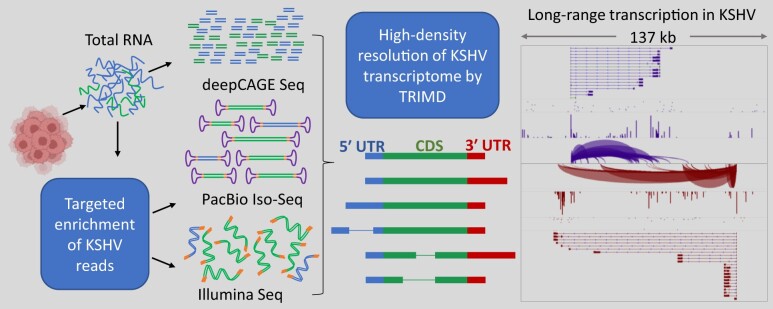
Abstract
Kaposi's sarcoma-associated herpesvirus is the etiologic agent of Kaposi's sarcoma and two B-cell malignancies. Recent advancements in sequencing technologies have led to high resolution transcriptomes for several human herpesviruses that densely encode genes on both strands. However, for KSHV progress remained limited due to the overall low percentage of KSHV transcripts, even during lytic replication. To address this challenge, we have developed a target enrichment method to increase the KSHV-specific reads for both short- and long-read sequencing platforms. Furthermore, we combined this approach with the Transcriptome Resolution through Integration of Multi-platform Data (TRIMD) pipeline developed previously to annotate transcript structures. TRIMD first builds a scaffold based on long-read sequencing and validates each transcript feature with supporting evidence from Illumina RNA-Seq and deepCAGE sequencing data. Our stringent innovative approach identified 994 unique KSHV transcripts, thus providing the first high-density KSHV lytic transcriptome. We describe a plethora of novel coding and non-coding KSHV transcript isoforms with alternative untranslated regions, splice junctions and open-reading frames, thus providing deeper insights on gene expression regulation of KSHV. Interestingly, as described for Epstein-Barr virus, we identified transcription start sites that augment long-range transcription and may increase the number of latency-associated genes potentially expressed in KS tumors.
Graphical Abstract
Graphical Abstract.
Introduction
Kaposi's sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus-8 (HHV-8) is endemic in sub-Saharan Africa and was originally identified in Kaposi's sarcoma (KS) lesions from human immunodeficiency virus (HIV) infected patients (1). Transmission of KSHV is mostly mediated by saliva and sexual routes. In addition to KS, which is a pleomorphic and highly vascularized tumor, KSHV is also etiologically associated with two lymphoproliferative diseases including primary effusion lymphoma (PEL) and a subset of multicentric Castleman's disease (MCD) in immunocompromised patients (2,3). Similar to other herpesviruses, KSHV has two replication phases, latent and lytic, and a fine balance of these phases has been found essential for establishment of KSHV-induced malignancies (4).
The double-stranded (ds) DNA of the KSHV genome is ∼165–170 kb long and consists of unique coding sequences and variable lengths of internal and terminal repeat regions. In order to fully understand the molecular pathogenesis of KSHV associated malignancies, it is crucial to annotate the highly dense transcriptome of the KSHV genome, which has added complexity due to active transcription of both DNA strands. The foundation of the KSHV annotation was based primarily on the detection of open reading frames (ORFs), demarcated by a canonical start codon (AUG) at the 5′ end and a stop codon (UAA, UAG or UGA) at the 3′ end of a protein coding sequence (5). Genome-wide mapping and individual studies exploring the 3′ untranslated regions of annotated KSHV ORFs using techniques such as rapid amplification of cDNA ends (RACE), and polyadenylation sequencing identified salient transcription termination sites for some annotated ORFs (6,7). Northern blots, tiled microarrays and high-throughput RNA sequencing (RNA-Seq) analysis of KSHV gene expression in different cell lines further improved our identification of coding and non-coding transcript features of KSHV (8,9). The most extensive short-read sequencing combined with ribosome profiling performed by Arias et.al to study the KSHV transcriptome, mapped the transcription start sites (TSSs) and polyadenylation sites for a significant number of pre-annotated KSHV ORFs along with identification of novel transcripts (10).
Short-read sequencing methods are, however, recently being found to be inadequate for analyzing complex transcriptomes and resolving multiple overlapping transcripts that share the same promoter or polyadenylation sites. The occurrence of bidirectional transcription, polycistronism and alternative splicing from closely spaced genes within the KSHV genome have collectively limited the short-read RNA-Seq studies from completely identifying and distinguishing individual KSHV transcripts. Third generation sequencing technologies that have the capability to produce substantially longer reads are currently recognized as more proficient for analyzing the complexity of coding and non-coding RNAs in herpesvirus genomes (11,12). However, long-read RNA-Seq methods have their own limitations, which mainly include polymerase errors and truncated ends in sequencing products (13).
Data integration from multiple sequencing platforms has recently emerged as the most powerful approach for studying complex transcriptomes of human viruses including Herpes Simplex Virus 1 (HSV1) and Human Cytomegalovirus (HCMV) with high definition (14–16). In addition to the identification of full-length novel transcripts, cross-platform transcriptome studies have also demonstrated transcriptional activity from unexpected genomic regions and frequent readthrough transcription in gamma-herpesviruses including Epstein-Barr virus (EBV) and Murine Herpesvirus 68 (MHV68) (17,18). This has been facilitated by the development of an RNA-Seq data analysis tool named Transcript Resolution by Integration of Multiplatform Data (TRIMD), that determines complete transcript structures by integrating data from three different sequencing platforms; including Illumina short-read RNA-Seq, deepCAGE (Cap Analysis for Gene Expression) sequencing and Pacific Biosciences (PacBio) Single-Molecule Real-Time (SMRT) long-read RNA-Seq (17). Despite various attempts to analyze the KSHV transcriptome, we lack a high-resolution annotation of the KSHV transcriptome that is inclusive of the complete transcript features and alternatively spliced isoforms. There is also limited evidence of KSHV intergenic transcription or long-range transcripts from the KSHV genome such as has been described for EBV (19,20). We therefore, pursued a re-evaluation of the KSHV transcriptome using third generation sequencing in combination with short-read RNA-Seq with the aim of significantly updating the KSHV annotation and enhancing our understanding of KSHV gene expression and regulation during lytic replication and host pathogenesis.
Another major caveat limiting previous studies from identifying novel transcript features of KSHV, is the very low proportion of viral to host transcripts in the total RNA isolated from KSHV infected cell lines. Many KSHV transcripts that are expressed at moderate to low levels can remain undetected from sequencing platforms due to the presence of high levels of cellular transcripts, which dominate the RNA pool. In order to achieve a high resolution KSHV transcriptome, we have also applied hybridization-based target enrichment approaches to increase the percentage of KSHV reads in both short- and long-read RNA-Seq data (21).
In our study, we have reported a detailed genome-wide transcript structure resolution of the KSHV lytic transcriptome performed by integrating RNA-Seq data from different sequencing platforms including SMRT long-read, deepCAGE and Illumina short-read sequencing from the lytically induced body-cavity-based lymphoma cell line, BCBL-1. Overall, our high-resolution analysis of the KSHV transcriptome identified numerous novel TSSs, polyadenylation sites and splice junctions representing a total of 994 unique KSHV transcripts. Moreover, most of the transcript features identified in previous studies were affirmed in our TRIMD analysis, indicating high accuracy and improved depth of our approach. Finally, we also validated interesting novel transcripts coming from newly identified promoter regions by RT-PCR and Nanopore single molecule sequencing.
Materials and methods
Cell culture and lytic reactivation of KSHV infected cells
Body-cavity-based lymphoma cell line 1 (BCBL-1) cells (22) were cultured in RPMI 1640 media supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, 1 mM sodium pyruvate, 100 U/ml penicillin and 100 mg/ml streptomycin at 37°C under 5% CO2. iSLK cells (23) were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin, 1 μg/ml puromycin, 250 μg/ml G418 and 250 μg/ml hygromycin at 37°C under 5% CO2. Lytic reactivation in BCBL-1 cells was induced with 12-O-tetradecanoylphorbol-13-acetate (TPA) (20 ng/ml) and sodium butyrate (2 mM), while lytic reactivation in iSLK cells was induced by using a combination of doxycycline (1 μg/ml) and sodium butyrate (1 mM).
RNA extraction
Total RNA was extracted from BCBL-1 or iSLK cells before and after lytic reactivation using TRIzol reagent (Invitrogen, Catalog No. 15596026) according to the manufacturer's protocol. For all sequencing purposes, RNA samples with a RIN>= 8 were used (as indicated by the Agilent BioAnalyzer or TapeStation analysis). If necessary, RNA samples were purified and concentrated using the ZYMO Research RNA Clean and Concentrator kit (Zymo, Catalog No. R1015).
deepCAGE sequencing
For 5′ deepCAGE sequencing, single-end 50 bp nAnT-iCAGE libraries (24) were prepared using RNA extracted from induced (48 h) BCBL-1 cells. These libraries were then loaded on the Illumina HiSeq 2500 instrument for sequencing. Library preparation and sequencing were performed by DNAform, Yokohama, Japan.
Probe design for target enrichment
For enrichment of KSHV specific RNAs from the total RNA pools, we used the myBaits Custom Target Enrichment v4 kit (Arbor Biosciences, myBaits Custom RNA-Seq). Biotinylated RNA baits, 70 nucleotides each, were custom designed and manufactured by Arbor Biosciences across the full-length KSHV genome (Genbank Accession: NC_009333.1) to provide a 2X coverage. These biotinylated RNA probes complementary to the complete KSHV reference genome were used for selective enrichment of KSHV specific RNAs. All the reagents and protocols for target enrichment were also supplied by Arbor Biosciences with the myBaits custom kit.
Illumina library preparation and enrichment for KSHV
Libraries for short read Illumina sequencing were prepared from uninduced and 48 h lytic reactivated BCBL-1 cells. Ribosomal RNA was depleted from total RNA using NEB rRNA depletion kit (NEB, Catalog No. E6310) and following the manufacturer's protocol in three parallel reactions, each with 1 μg total RNA as input. NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (NEB, Catalog No. E7760) was then used to prepare Illumina libraries from 50 ng of rRNA depleted RNA samples (RIN > 7). Prepared libraries were barcoded using index primers from NEB (NEB, Catalog No. 6440).
Two of the four 48 h induced Illumina libraries were hybridized to KSHV probes for enrichment of KSHV specific RNAs. Target enrichment was performed with KSHV probes using the manufacturer's protocol for the myBaits custom kit (Arbor Biosciences, Product name: myBaits Custom RNA-Seq). In brief, 350 ng of Illumina libraries (in 7 μl TE buffer) were used as input to hybridize with 5.5 μl (concentration not disclosed by company) of biotinylated RNA probes at 65°C overnight (16 h) to form cDNA-probe hybrids. The hybridized cDNA molecules were pulled-down using the streptavidin-coated magnetic beads, washed to remove any non-specific DNAs and eluted in 30 μl TE buffer. Enriched libraries were re-amplified using NEBNext Ultra II Q5 master mix (1X), IDT library amplification primers (500 nM), and 15 μl (of total 30 μl) enriched library as template in a PCR reaction of 50 μl, according to the guidelines for amplification in the enrichment protocol. Amplified enriched libraries were quality tested on the bioanalyzer for size and concentration before sequencing. All libraries were loaded in equal concentration to perform paired-end sequencing on a NovaSeq 6000 instrument to obtain 50 million reads per sample.
PacBio library preparation and enrichment for KSHV
For PacBio Single-Molecule Real-Time (SMRT) long-read RNA-Seq, full-length cDNAs were generated using 300 ng of native and polyadenylated RNA from 48 h induced BCBL-1 cells as templates. Synthesis and amplification of full-length cDNAs was performed with Low Input RNA: cDNA Synthesis and Amplification kit (NEB, Catalog No. E6421) as per the manufacturer's guidelines. Full-length cDNAs were hybridized with KSHV baits to capture the KSHV specific cDNAs. To enrich for KSHV RNAs, total or size-fractionated full-length cDNAs were hybridized with KSHV genome specific baits (as mentioned above for Illumina library hybridization) according to instructions of hybridization protocol for myBaits custom, Arbor Biosciences (Arbor Biosciences, myBaits Custom RNA-Seq). Instead of using the Illumina library specific blockers provided in hybridization kit, we added the Low Input RNA: cDNA Synthesis and Amplification kit primers (Supplementary Table S1) in the blocking reaction. It is necessary to add these additional primers in the blocking reaction to reduce probe binding to free primers and avoid non-specific pull-down during enrichment. Enriched cDNA libraries (∼40 ng) were then re-amplified using the library amplification kit (NEB, Catalog No. 6421). We performed size fractionation on both enriched and non-enriched cDNA libraries. SMRTbell adaptors (Iso-SeqTM) were added using reagents from the PacBio SMRTbell Template Prep Kit 1.0-SPv3 starting from 1 μg of cDNA. This procedure resulted in 250–300 ng of SMRT bell library (i.e. 25–30% yield). The final libraries were eluted in 15 μl of 10 nM Tris HCl, pH 8.0. Library fragment sizes were estimated by the Agilent TapeStation (genomic DNA tapes), and this data was used for calculating molar concentrations. Overall, eight SEQUEL SMRT cells were used: two with non-size-selected non-enriched RNA, two with non-size-selected enriched RNA, two with 1.5–3 kb fraction of enriched RNAs, two with > 3 kb fraction of enriched RNAs and one SEQUEL II SMRT cell was used for sequencing >3 kb fraction of non-Enriched RNA libraries. Between 9–10 pM of library was loaded (diffusion loading) onto the SMRT cell. For optimum read length and output, libraries were sequenced on LR SMRT cell, using 4 h pre-extension, 20 h movies and v3 chemistry reagents. All other steps for sequencing were done according to the recommended protocol by the PacBio SMRT Link Sample Setup and Run Design modules (SMRT Link 6.0). One SMRT cell run generated 500–650 thousand reads with an average polymerase read length of 50–75 kb.
Alignments of deepCAGE and Illumina RNA-Seq to the KSHV genome
Paired-end Illumina RNA-Seq data was mapped to the KSHV reference genome (NC_009333.1), using STAR (25) with default parameters. To align deepCAGE reads, we used STAR with parameters -outFiltermultimapNmax 100 and -outSAMprimaryFlag AllBestScore to identify start sites in repeat regions.
Processing of PacBio SMRT reads and alignment to the KSHV genome
For processing the PacBio SMRT RNA-Seq data into full-length transcript isoforms, we individually performed the steps recommended in the Iso-Seq3 pipeline developed by PacBio for isoform analysis. In brief, the subreads obtained from each sample were first processed into circular consensus reads (CCS), followed by trimming of sequencing adapters to obtain full-length (FL) reads. Next, FL reads with concatemers or without poly(A) tails were eliminated to refine into full-length non-concatemers (FLNC) reads. Finally, FLNCs obtained from all the Iso-Seq samples were clustered (unpolished by Quiver) into consensus full-length (CFL) sequences that were mapped to the KSHV reference genome (NC_009333.1) using GMAP (26).
Integration of RNA-Seq data for transcript structure validation using TRIMD
Identification of complete KSHV transcript structures was performed using the Transcriptome Resolution of Multiplatform Data (TRIMD) method (17) that verifies the sequence and features of Iso-Seq CFLs, from the PacBio SMRT-Seq platform, by using complementary high-throughput data from deepCAGE and Illumina RNA-Seq platforms. In brief, CFL start sites were calculated as the weighted average of CFL start coordinates in each cluster of non-softclipped Iso-Seq CFL 5′ ends mapping within 8 bp of each other. DeepCAGE start site clusters were identified using Paraclu (27) requiring a minimum of 30 tags/cluster, maximum/baseline density ratio of minimum 2 units and cluster length of 1–20 bp. Iso-Seq consensus start sites occurring in a minimum of 2 CFLs and mapping within 3 bases of deepCAGE consensus start sites were considered as validated. For identification of splice junctions, TRIMD parameters were set to only validate Iso-Seq CFL junctions (mapped by GMAP) that could be verified with a minimum of 3 Illumina reads (mapped by STAR). For validation of polyadenylation sites, CFL polyadenylation sites were first determined by calculating the weighted average of non-softclipped Iso-Seq CFL 3′ end clusters mapping within 8 bp of each other. Illumina RNA-Seq reads containing a run of at least 5 As (plus strand) or 5 Ts (minus strand), in their ends with at least 2 of these bases as softclipped were used for polyadenylation site identification. Consensus polyadenylation sites were then determined by calculating the weighted average of the cluster of these poly(A)-tailed Illumina reads that mapped within 8 bp of each other. Iso-Seq consensus polyadenylation sites detected in at least 5 CFLs and located within 10 bp upstream or 4 bp downstream of an Illumina consensus polyadenylation site were then considered as TRIMD-identified 3′ ends. Iso-Seq CFLs verified for their TSSs, splice junction(s) (if any) and polyadenylation sites, with supplementary data from one of the other two platforms, were determined to be the final TRIMD-identified transcripts.
Coding potential analysis and annotation of TRIMD-identified transcripts
To annotate the unique TRIMD-identified transcripts we first performed a BLAST search for these transcripts against all the previously annotated (NC_009333.1) ORFs using the NCBI_BLASTP tool. Transcripts revealing complete BLAST matches to known coding ORFs were annotated as transcript isoforms of the respective BLAST-hit gene, in ascending numerical order for TSS occurrence. For example, transcript isoforms for ORF4 are annotated as ORF4-01, ORF4-02 and so on. Transcripts identified as having more than one complete ORF as BLAST-hits were annotated for the longest matching ORF. For the remaining transcripts that reported no matches or incomplete matches to the previously annotated ORFs, we performed a computational analysis by TransDecoder (v5.5.0) to evaluate their coding potential and identify the single-best coding ORFs within these transcripts. We followed the parameters to retain all ORFs longer than 50 amino acids (-m 50) with a canonical start codon (–no_refine_starts). The novel identified ORFs were annotated according to their loci on the NC_009333.1 reference and sequence overlap with previously annotated genes. For example, novel ORFs with a sequence overlapping ORF4 were annotated as ORF4a, ORF4b and so on. ORFs identified with their sequences overlapping to the N’ terminus of one gene and C’ terminus of a different downstream gene with in-frame coding sequence were annotated as Fusion ORFs (fus.ORF). Transcripts that were not identified to have any coding potential with either BLAST or TransDecoder were annotated as non-coding to overlapping ORF (nc.ORF) or anti-sense transcripts (as.ORF).
cDNA preparation and Nested RT-PCR for transcript validation
Total RNAs (2.5 μg) isolated from each sample were treated with DNase I (NEB, Catalog No. M0303) to remove the genomic DNA. DNase treated RNAs were re-purified using phenol chloroform extraction and then used as a template for cDNA preparation with the Superscript IV (SSIV) First-Strand Synthesis System (ThermoFisher, Catalog No. 18091200) using the random hexamers protocol as described in the user manual for the SSIV kit.
For PCR validation of TRIMD-identified abundant transcripts (Score/Number of Iso-Seq CFLs > 10) 1 μl of cDNA (equivalent to 120 ng RNA) was PCR amplified (33X) with Phusion High-Fidelity PCR Master Mix (NEB, Catalog No. M0531) and primers designed across the novel identified exon-exon junction. For validation of rare TRIMD-identified transcripts (Score < 10), we performed a nested PCR, using 1 μl of the first PCR product as template and a new set of primers designed within the amplicon of the first PCR product and across the splice junction. Resulting RT-PCR fragments were validated by Sanger sequencing (Genewiz, Azenta Life Sciences) from both forward and reverse primers.
Preparing libraries for nanopore direct RNA, direct cDNA and PCR-cDNA sequencing
To prepare libraries for sequencing by Oxford Nanopore Technology (ONT), total RNA was isolated from BCBL-1 and WT-BAC16 infected iSLK cells after 0, 24 and 48 h of induction of lytic replication. Poly(A)-tailed RNA was selected from 75 μg of each RNA sample using the Dyna Beads mRNA purification kit (Invitrogen, Catalog No. 61006). To prepare direct-RNA sequencing libraries from poly(A)-selected BCBL-1 RNAs (50 ng each), we used the SQK-RNA002 library preparation kit by ONT and followed the guidelines of the kit for preparing libraries. Direct-cDNA sequencing libraries were prepared with 100 ng each of purified BCBL-1 mRNAs using the SQK-DCS109 kit by ONT according to the protocol described in the user manual. Libraries for PCR-cDNA sequencing were prepared using 1 ng of each of the purified mRNAs from uninduced and induced WT-BAC16 infected iSLK cells. The SQK-PCB109 kit by ONT was used for PCR-cDNA sequencing library preparation according to the guidelines for poly(A)-selected RNA.
Sequence alignment for Nanopore sequencing data
All the ONT reads were basecalled and demultiplexed using Guppy, an ONT basecaller. ONT reads were aligned to their respective genomes, i.e. NC_009333.1 for BCBL-1 cell libraries and GQ994935.1 for iSLK cell libraries, using minimap2 with the appropriate parameters (-ax splice –secondary = no -C 5). Alignment files were converted to Gene Transfer Format (GTF) files for each library, using samtools v1.10 and bedtools v2.29.2. Using SQANTI3 QC (28) we identified reads in cDNA libraries categorized as antisense and reoriented their alignments in the GTF files.
Converting TRIMD transcripts to GQ994935 coordinates
The sequences for TRIMD-identified transcripts on NC_009333.1 coordinates were extracted using gffread v.0.12.7. TRIMD transcript sequences were then aligned to the GQ994935.1 genome using minimap2 and converted to GTF files using samtools v1.10 and bedtools v2.29.2.
Comparing TRIMD-identified transcripts with Nanopore sequencing data
SQANTI3 QC (28) was used to match the reads that aligned to the KSHV genome in each of the nine ONT libraries to classify their splice junction patterns relative to the TRIMD-identified transcripts. Reads assigned as full-splice match (FSM) have an exact match of all of their junctions with one of the TRIMD-identified transcripts. The incomplete splice match (ISM) category includes reads with consecutive junction matches to a TRIMD-identified transcript but missing junctions on either the 5′ or 3′ ends. Novel in catalog (NIC) classification describes reads with all their junctions present in the TRIMD transcriptome, however in a novel combination, and novel not in catalog (NNIC) class have reads with splice junctions not identified in the TRIMD transcriptome. The number of TRIMD junctions and junction chains detected in each of the libraries was also counted by SQANTI3 QC.
Calculating relative abundance of unique ORFs during latent and lytic cycle
The nanopore sequencing reads were compared to the TRIMD-identified transcripts using SQANTI3 as described above and full-splice matched reads were counted for all transcripts. The respective counts were assigned to the unique ORFs exhibited by each transcript and cumulative counts were generated for each ORF to estimate the relative abundance of each ORF isoform in BCBL-1 and iSLK-WT-BAC16 infected cells before (0 h) and after (24 and 48 h) lytic reactivation. The ORFs that were not found to have novel isoforms and that remained undetected in all samples were not included in the counts table.
Pediatric KS tissue RNA-Seq data analysis
The raw sequencing data for pediatric KS tissue samples were obtained from the NCBI’s Sequence Read Archive (SRA BioProject PRJNA975091)(29). The raw sequence files were mapped to KSHV genome reference NC_009333.1 with STAR using the 994 TRIMD-identified transcripts as the annotation file. KSHV transcripts originating at nucleotide position (nt.) 119010 and nt. 124005 with Reads Per Kilobase of transcript per Million mapped reads (RPKM) values >3 in a minimum of three samples were analyzed further for detecting uniquely mapped reads across the splice-junction of these transcripts with their splice donor bases (GU) at nt. 118906 and nt. 123842, respectively. The number of reads mapping across the splice junction of transcripts were identified from the ‘SJ.out.tab’ output of the STAR alignment, that summarizes the high confidence splice junctions.
Results
Target enrichment of KSHV from BCBL-1 cells
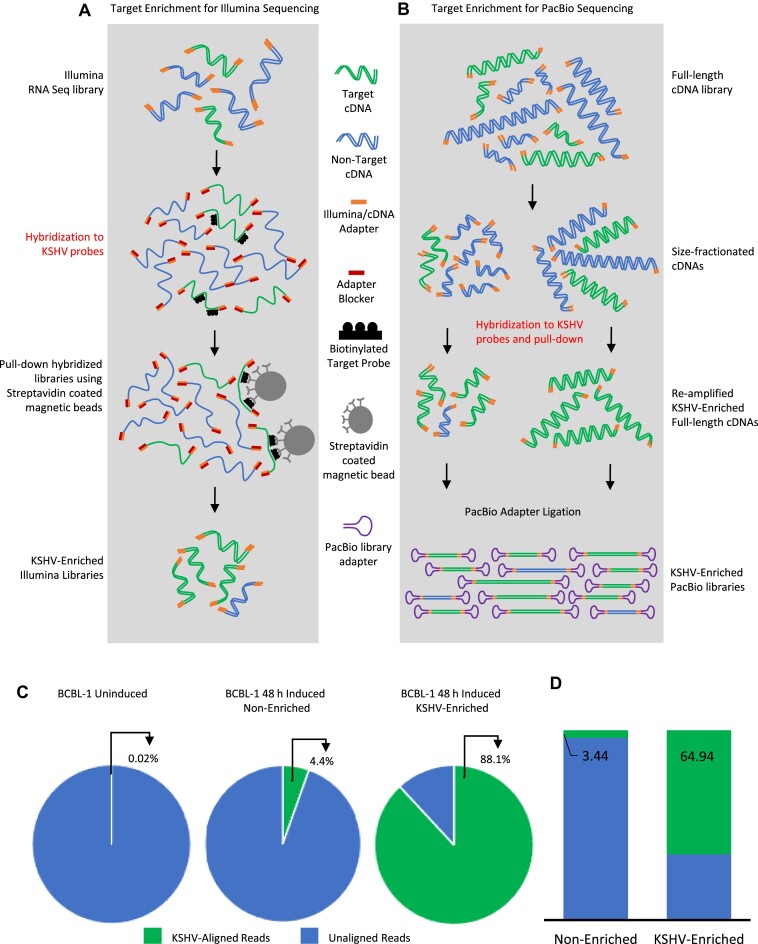
BCBL-1 cells derived from primary effusion lymphoma (PEL), naturally harbor KSHV and are a biologically relevant model for studying KSHV during both the latent and lytic phases of infection (22). In order to perform a high-resolution analysis of the KSHV transcriptome, our prerequisite was to start with a high percentage of KSHV transcripts in the total inputs for RNA sequencing, which could not be increased in BCBL-1 to >5% even after 48 h induction of lytic replication. The reactivation of BCBL-1 cells was confirmed by validating the increased expression of replication and transcription activator (RTA) gene (Supplementary Figure S1A). To further increase the percentage of KSHV reads in our RNA-Seq study, we performed hybridization-based target enrichment of KSHV specific RNAs in our libraries for both short and long read sequencing approaches (Figure 1A and B).
Figure 1.
Target enrichment and alignment breakdown. (A) Workflow for enrichment of Illumina RNA-Seq libraries using biotinylated RNA probes designed complementary to the complete KSHV genome. (B) Enrichment protocol for full-length cDNAs for KSHV before PacBio sequencing library preparation. (C, D) Percentage of total Illumina (C) and PacBio (D) sequencing reads mapped to the KSHV reference before and after enrichment with KSHV probes.
As the TRIMD pipeline utilizes long-reads as a scaffold for building annotated transcript structures; it necessitates sequencing them with more coverage and greater depth. Because the PacBio SMRT library molecules are circular with U-shaped adapters ligated to the ends of the multimerized cDNAs, these molecules cannot be efficiently enriched by target selection through hybridization. Hence, we developed an alternative library construction workflow whereby target selection was performed on complete cDNA libraries before the adapters were ligated. This approach prevented intramolecular hybridization of circular molecules affecting target hybridization efficiency (Figure 1B). The purified KSHV enriched full-length cDNAs were then re-amplified before adapter ligation. The KSHV-enriched and amplified circular SMRT library molecules were sequenced on both Sequel or Sequel II SMRT cells, providing deeper coverage.
We then compared the percentage of KSHV mapped reads before and after target enrichment, with all samples sequenced at a similar depth. In the Illumina RNA-Seq data, <1% of total reads from uninduced BCBL-1 cells mapped to the KSHV genome, which increased to 5.4% in the induced BCBL-1 samples (Figure 1C). Our target enrichment was successful as the percentage of KSHV mapped reads increased significantly to 88.1% in the induced BCBL-1 samples. Similarly, the percentage of SMRT reads mapping to the KSHV reference increased substantially from 3.4% to 65% after enrichment (Figure 1D, S1B). The coverage of long-reads across the entire KSHV genome was comparable indicating that the enrichment protocol did not introduce any bias in the selection of transcripts (Supplementary figure S1B). A summary of all read numbers pre- and post- target selection is provided in Supplementary Table S2.
We note that target selection was instrumental to our study since the percentage of viral reads in KSHV lytic replication is significantly lower than in models used for EBV and MHV68 TRIMD transcriptome analysis (17,18).
Integration of data from different RNA-Seq platforms using TRIMD
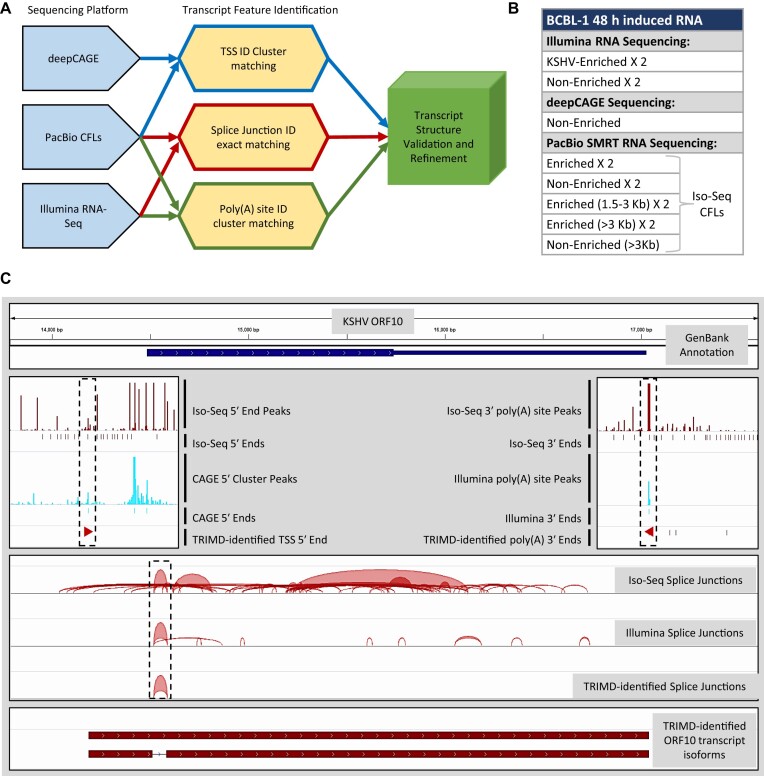
In order to perform global genome-wide resolution of the KSHV transcriptome we used TRIMD, a pipeline that integrates data from three different sequencing platforms for identifying complete transcript structures. PacBio SMRT reads processed into consensus full-length sequences (CFLs) formed the basis of transcript structure analysis by serving as a scaffold in the TRIMD pipeline for identification of transcript structure. TRIMD was used to validate CFL 5′ ends by integrating deepCAGE sequencing data, and to validate splice junctions and 3′ ends using the short-read RNA-Seq data. The CFLs with validated transcript ends and splice junctions (if present) with supporting data from more than one sequencing platform were finally classified as complete transcripts by TRIMD analysis (Figure 2A).
Figure 2.
TRIMD method for transcriptome resolution. (A) Workflow showing the steps of the TRIMD pipeline integrating data from multiple sequencing platforms for identifying complete transcript structures. (B) BCBL-1 48 h RNA samples used for each sequencing platform. (C) An example validation of KSHV ORF10 transcript isoforms using TRIMD representing individual feature validation in dotted blocks.
Briefly, deepCAGE sequencing was performed on non-enriched libraries, while Illumina and SMRT sequencing was performed on both, the KSHV-enriched and non-enriched RNA-Seq libraries (Figure 2B). SMRT reads obtained from all PacBio library samples were first processed into CFLs using the Iso-Seq pipeline. The processed sequences from all platforms were then individually mapped to the KSHV reference genome (NC_009333.1). Overall, 0.8 × 106 of 9.4 × 106 total reads from deepCAGE sequencing mapped to the KSHV genome. Of the total 144 × 106 reads from Illumina sequencing, 75.2 × 106 mapped to the KSHV genome, while 0.11 × 106 of the total 0.86 × 106 Iso-Seq CFLs mapped to the KSHV reference. We found that despite the size fractionation of full-length cDNAs followed by equal loading of all fractions on individual SMRT cells, the length-distribution of SMRT sequencing was biased towards smaller sequences with less than three percent of Iso-Seq CFLs >4 kb mapping to the KSHV genome (Supplementary figure S1C). We observed that the numbers of both short- and long-reads mapping to the viral genome robustly increased due to the target enrichment process when compared to the previously used inputs for TRIMD analysis of EBV and MHV68 (17,18). The KSHV mapped Iso-Seq CFLs also indicated a remarkable coverage of the genome by our long-read sequencing data (Supplementary figure S2). Finally, the KSHV mapped reads from all platforms were integrated using the TRIMD pipeline to identify complete transcript structures. An example for identification of complete ORF10 transcript isoforms from various CFLs that mapped to the ORF10 gene (Supplementary figure S3) is illustrated in Figure 2C. This example depicts the identification of transcript ends and a splice variant of ORF10 from integration of all three sequencing datasets using TRIMD (Figure 2C).
Genome-wide resolution of the KSHV lytic transcriptome
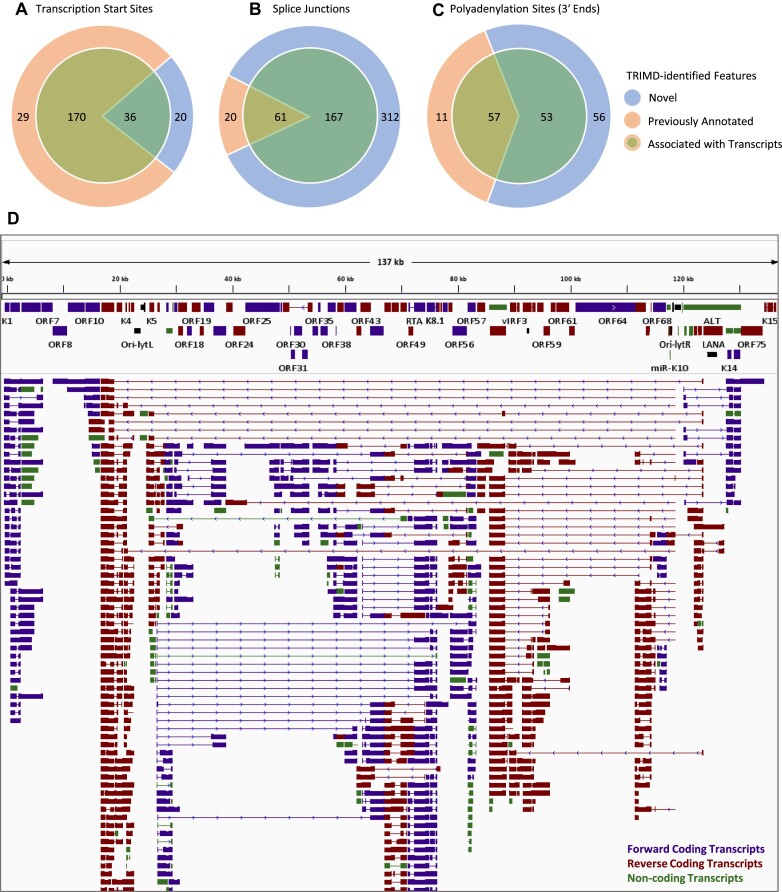
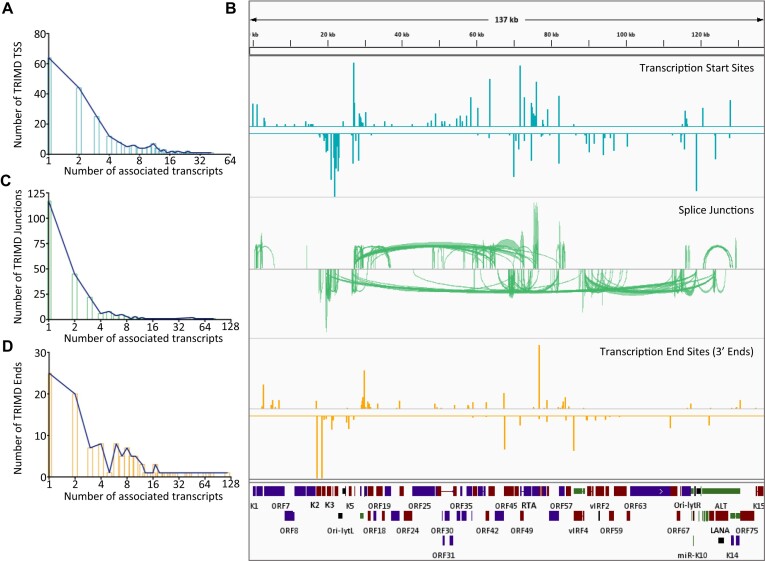
In addition to TRIMD’s fundamental verification of the Iso-Seq CFL features with supporting evidence from short-read sequencing data, we further improved the confidence of TRIMD-identified transcripts by following stringent parameters at different steps for identification of individual transcript features (see methods). Specifically, validation of transcript 5′ ends required their detection in at least two CFLs and identification of splice junctions in CFLs required validation by a minimum of three Illumina reads. In summary, our analysis resulted in identification of 255 TSSs with a total of 56 novel and 199 TSSs annotated previously by short-read sequencing methods or most recently by a direct-cDNA sequencing method (Figure 3A) (10,12). Further adding to the precision of our data, 129 of TSS loci identified by TRIMD coincide with the transcription start site clusters (TSCs) identified by the RAMPAGE method, which combines template-switching and cap-trapping for identifying TSCs (30). We have also identified a total of 560 splice junctions (85% novel) and 183 polyadenylation sites (59% novel) for KSHV lytic transcripts, with each feature supported by evidence from two sequencing platforms (Figure 3B-C). The splice junctions identified from our study are also inclusive of 193 of 387 splice junctions identified recently in a study focused on identifying ORF57-dependent RNA splicing in KSHV (31). A detailed comparison of TRIMD-identified transcript features to previous KSHV annotations and transcript feature studies is shown in Supplementary Table S3. While the stringent parameters followed in our analysis resulted in enhanced transcript resolution, it also resulted in the lack of detection for some previously annotated transcripts; for example the Antisense-to-Latency Transcript (ALT) transcript, which is more than 10 kb in length, or limited detection of features as identified recently by the direct-cDNA sequencing approach (8,12). In summary, the step-wise feature validation of Iso-Seq CFLs by TRIMD identified 994 unique transcripts representing the lytic transcriptome of KSHV in BCBL-1 cells (Figure 3D, Supplementary Table S3). The utility and stringency of our approach is further supported by the fact that the unique transcript structures are represented by 81% of 5′ ends, 60% of 3′ ends and 46% of all mapped splice-junctions, all validated individually by data from multiple sequencing platforms (Figure 3A-C).
Figure 3.
KSHV lytic transcriptome resolved by TRIMD. (A–C) Identification of novel and previously annotated TSSs (A), splice junctions (B) and polyadenylation sites (C) by TRIMD analysis of KSHV lytic transcriptome from 48 h induced BCBL-1 cells. The inner circles display the numbers of each feature represented in the complete full-length transcript isoforms validated by TRIMD. (D) An integrated genome viewer (IGV) snapshot of TRIMD-identified KSHV transcripts (lower panel) mapped to the KSHV reference genome (NC_009333.1) (top panel). The figure is limited to display the most comprehensive view on IGV (maximum zoom-out), and complete details of transcriptome are provided in Supplementary Table S3.
The identification of transcript isoforms with novel transcript features, as summarized in Supplementary Table S3, demonstrates that a large number of KSHV genes have multiple TSSs and exhibit alternatively spliced isoforms. Overall, we identified TSSs and polyadenylation sites for 77 of the 86 KSHV ORFs as annotated in GenBank, along with many new alternatively spliced transcript isoforms. The nine KSHV ORFs that could not be verified for complete transcript structures by TRIMD include ORF7, ORF17, ORF19, ORF20, ORF29, ORF63, ORF64, ORF75 and K15.
Overlapping transcripts share transcript features in KSHV
We further analyzed the distribution of transcript features in TRIMD-identified transcripts. From a total of 206 TSSs that were associated with TRIMD transcripts, 142 were shared by more than one transcript (Figure 4A). We observed that TSSs on the plus strand exhibited a higher prevalence within the initial 100 kb of the KSHV genome, preceding the right origin of lytic replication (ori-lytR) and the latency-associated region (Figure 4B). Conversely, TSSs on the minus strand were predominantly distributed near ori-lytR and the genome segment spanning from the RTA to the LANA encoding region. Overall, 29 loci on the KSHV genome were observed as highly dominant sites of transcription initiation, with >10 transcripts originating from each of these TSSs (Supplementary Table S4). Furthermore, we have identified three genomic loci with high promoter activity augmenting long-range transcription of multiple genes (Supplementary figure S4). These include, two TSSs in the latency-associated region at nucleotide positions (nt.) 119010 and 124005, which together give rise to more than 32 novel transcripts that are spliced into a variety of coding and non-coding genes located all across the KSHV genome, with some genes located at 40–100 kb from the respective TSS (Supplementary figure S4A-B). Another TSS at nt. 27048 associated with 24 transcripts including isoforms of distal genes, RTA, K8 and K8.1, is predicted to augment long-range transcription (Supplementary figure S4C).
Figure 4.
Transcript feature distribution. (A) Distribution of TSSs across TRIMD-identified KSHV transcripts with x-axis representing the number of validated TSSs and y-axis representing the number of transcripts arising from corresponding number of TSS. (B) (from top to bottom) Distribution of TSSs (blue), splice junctions (green) and 3′ends (yellow) over KSHV reference genome with all the top panels displaying the forward transcript features and bottom panels displaying the reverse transcript features. Height of bars for 5′ and 3′ ends and the density of junctions in splice-junctions track represent the relative occurrence of transcripts feature. (C, D) Distribution of splice junctions (C) and 3′ends (D) across TRIMD-identified KSHV transcripts with x-axis representing the number of validated features and y-axis representing the number of transcripts exhibiting the corresponding numbers of features.
Similarly, we examined the distribution of 228 splice junctions and observed that >100 splice junctions are shared amongst multiple KSHV transcripts (Figure 4C). Splice junctions that are shared by more than 20 transcripts each, are another interesting finding of our study revealing potential novel protein isoforms. We found that splice junctions on the minus strand were more abundant and with introns of varying lengths, including both short and extremely long ones, with some extending between 10 to 100 kb (Figure 4B, Supplementary figure S5). In contrast, the occurrence of splice junctions on the plus strand was less frequent, mostly observed between K3 and the RTA region (Figure 4B, Supplementary figure S5). The distribution pattern of polyadenylation sites (3′ ends) was comparable to the patterns observed for 5′ ends, in that only 25 of the 110 3′ ends represented in complete transcripts are unique to single transcripts, while 85 are shared between multiple transcripts (Figure 4D). The most prevalent plus strand 3′ end sites were located downstream of the RTA coding region, while on the minus strand the density of 3′ ends, was highest in the region around the K2 gene (Figure 4B).
Annotation of TRIMD-identified unique KSHV transcripts
Furthermore, by performing a BLAST search against annotated genes and computational analysis to predict the coding potential of TRIMD-identified transcripts (detailed in methods), we have annotated 85% of transcripts as monocistronic or polycistronic isoforms of 138 unique coding sequences (Supplementary figure S6A, Supplementary Table S3). These include 77 GenBank annotated and 61 novel predicted ORFs from our study. As observed for other transcript features, most of the ORFs were found to be shared between multiple transcripts, with 445 transcripts identified as monocistronic and 402 as bicistronic or polycistronic harbouring 2–6 potential coding sequences (Supplementary figure S6B). The remaining 149 TRIMD-identified transcripts (15%) could not be linked to any complete coding sequence and therefore were categorized as long non-coding RNAs (lncRNAs), substantially increasing the annotation of KSHV lncRNAs.
We note that a subset of transcripts annotated as coding with complete BLAST hits to coding ORFs were identified as non-coding by coding potential analysis tools due to longer 5′UTRs. However, to simplify the annotations, we continued to designate these transcripts as coding protein isoforms. Additionally, we observed that several TSSs associated with TRIMD-identified coding transcripts have been previously demonstrated to associate with translation of upstream ORFs including ORF6U, ORF11U, K5U, ORF21U, ORF28U, ORF34U, ORF38U, ORF47U, ORF61U, ORF71U and ORF75U detected using ribosomal profiling of KSHV mRNAs (10). Previous identification of ORF35.1 and ORF35.2 as upstream ORFs to ORF35 by Kronstad et al., provides thorough insights into the KSHV gene expression regulation by upstream translated ORFs that have the potential to suppress translation from proximal downstream ORFs and enhance translation of distal downstream ORFs (32). With reference to these studies, we predict that many of the TRIMD-identified polycistronic transcripts, may exhibit additional regulatory mechanism for translation of each ORF by the activity of potential upstream ORFs or small ORFs (smORFs) in the 5′ UTRs of these transcripts.
Identification of long-range KSHV transcripts
Our TRIMD-based transcriptome analysis identified long-range transcription across 40–100 kb large genomic regions, specifically from the latency associated regions on the right, spanning to lytic genes situated at the left end (Figure 3D). Thus far, long-range transcription has only been identified in EBV for expression of EBNA1 from the Cp promoter near the oriP locus (19,20,33). A recent study by Majerciak et.al. has previously reported limited evidence of long-range transcription in KSHV (31).
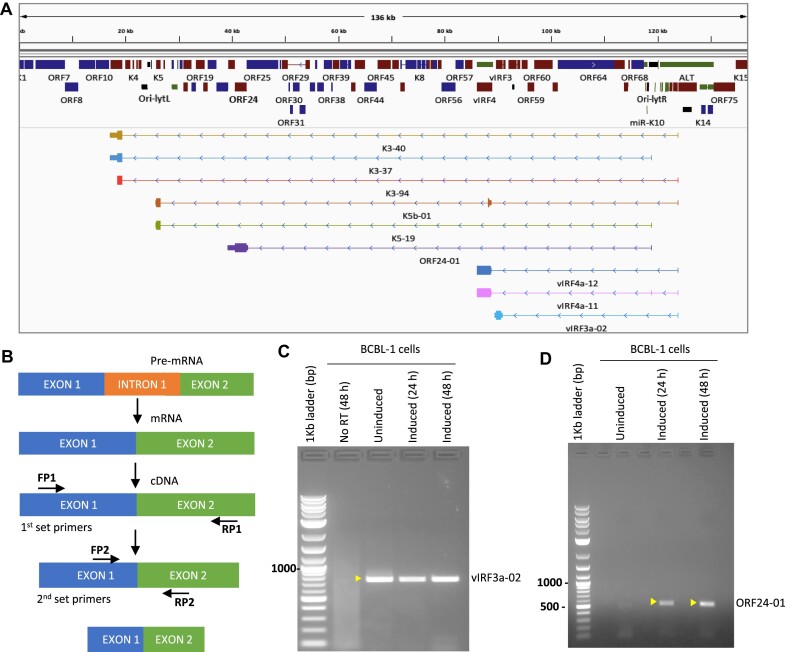
Surprisingly, our study has revealed three predominant TSSs at nt. 27048, 119010 and 124005 that are associated with long-range transcription which through both differential termination (readthrough of polyadenylation sites) and differential splicing give rise to more than 60 transcript isoforms encompassing nearly 20% of the KSHV ORFs (Supplementary figure S4, S5). Interestingly, among the transcripts transcribed from the two promoters in the latency regions we found transcripts encoding for KSHV E3 Ubiquitin ligases, K3 and K5, KSHV TATA-binding protein homolog, ORF24, and viral Interferon Regulatory Factor (vIRF1-4) proteins (Figure 5A). In order to further verify the molecular existence of the long-range transcripts, we performed strand-specific nested RT-PCRs to amplify newly identified splice junctions (Figure 5B) of both high and low expressed isoforms. We investigated the existence of these transcripts in BCBL-1 uninduced and induced cells and verified the novel splice variants for K3 and K5 transcripts, which were found to originate nearly 100 kb upstream from either of the two strong latency promoters (Supplementary Figure S7A–C). K3 and K5 long-range transcripts were detected only in induced cells. In contrast, we detected vIRF3a and vIRF4a isoforms in both uninduced and induced BCBL-1 cells, suggesting that these genes can contribute to both the latent and lytic phases of KSHV replication (Figures 5C, S7D). The existence of the ORF24 transcript isoform originating from near the left inverted repeat proximal to ori-lytR was also verified by nested PCR in uninduced and induced cells, albeit at much lower levels in uninduced cells (Figure 5D). The former result was surprising since ORF24 is a virally-encoded ortholog of a TATA-box binding protein that is required for transcription of late lytic genes containing noncanonical TATA boxes (34). To rule out that the transcript detected during latency in BCBL-1 cells was not due to spontaneous reactivation of a small number of cells, we repeated the RT-PCR validation of this transcript in iSLK cells that are strictly latent and still detected the ORF24 isoform in uninduced cells (Supplementary figure S7E).
Figure 5.
Identification and molecular validation of novel splice junctions. (A) An IGV panel displaying the TRIMD-identified splice variants of annotated KSHV genes potentially generated from long-range or readthrough transcription. (B) Flow diagram showing the nested PCR method used for validation of novel exon-exon junctions in KSHV transcripts. (C, D) Agarose gel electrophoresis for the nested PCR amplification of cDNA from uninduced and induced BCBL-1 cells, across novel identified exon-exon junction of (C) vIRF3 coding isoform (vIRF3a-02) that originates from near the LANA coding region at 124005 nt and (D) ORF24 transcript isoform (ORF24-01) that originates at 119010 nt (ori-lytR TSS).
The other set of long-range transcripts was identified in the forward orientation and were observed to splice primarily into RTA (ORF50), K8 and K8.1 transcript isoforms (Supplementary figure S4C). RT-PCR across the novel splice junction of the ORF50a transcript isoform, spanning the nearly 50 kb intron, further confirmed the existence of this junction in iSLK-WT-BAC16 infected cells before and after induction of lytic replication (Supplementary figure S7F). Furthermore, we also performed Sanger sequencing of all amplified PCR products to verify the exact sequence of novel identified exon-exon junctions ruling out any potential artefact stemming from our bioinformatic analysis or off-target PCR amplification (Supplementary file S2). The Sanger sequencing of exon-exon junctions from nested RT-PCR products validated the existence of transcript variants that all initiate between 40–100 kb upstream of their respective coding sequences.
Alternative coding sequences of annotated KSHV genes
It is noteworthy that many of the KSHV transcript isoforms, which we demonstrated in the former section including vIRF3a, vIRF4a and ORF50a, are also coding variants of the annotated KSHV ORFs. By utilizing the combination of NCBI-BLAST and TransDecoder for annotation (see methods), we identified coding isoforms for both early and late genes as listed in Supplementary Table S3. For instance, we identified six coding isoforms of ORF4, including three splice variants, with the C-terminus sequence conserved in all isoforms. (Supplementary figure S8A). Evidence for alternative splicing in the coding sequence of ORF4 has also been reported previously by transcription profiling of ORF4 in PEL cells (35). We tested for one of these splice junctions using strand-specific RT-PCR, as mentioned above, and found that the ORF4a/4d spliced isoforms are present only in the induced BCBL-1 cells while the unspliced ORF4 isoforms are present in both induced, and uninduced BCBL-1 cells, albeit at lower levels (Supplementary figure S8B). However, it remains elusive if the identified amplicons are derived from either full-length (ORF4/4b) or the N-terminus truncated (ORF4a/4d) ORF4 isoforms.
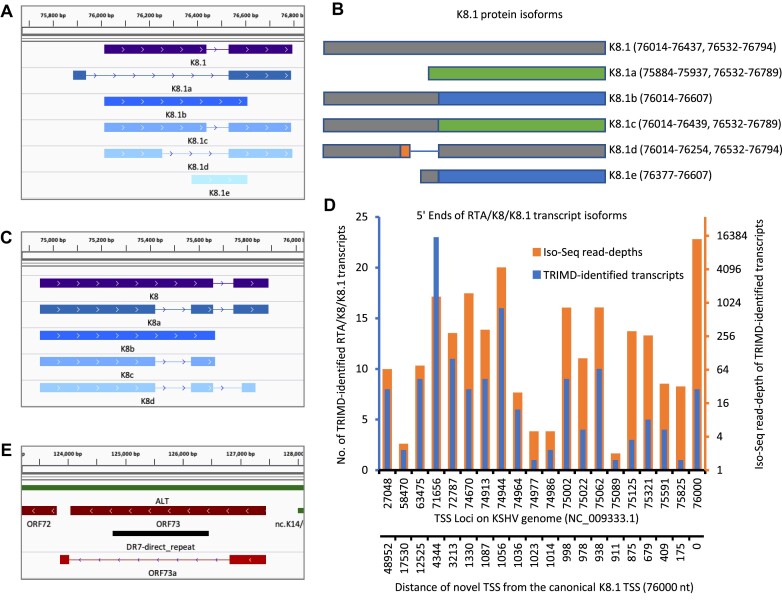
Our high-resolution transcriptome analysis has also identified six alternative coding isoforms of K8.1, a viral glycoprotein (36–38), which is a structural component of the KSHV virion that facilitates virus entry by adhering to cell surface receptors (Figure 6A). The identified K8.1 isoforms included all previously annotated isoforms suggesting high efficacy of our annotation (39,40). By performing multiple sequence alignment of all K8.1 protein isoforms, we have demarcated the identical and divergent domains of each coding isoform. We deciphered that four of the identified K8.1 coding isoforms including K8.1, K8.1b, K8.1c and K8.1d exhibit the same amino-acid sequence on their N-terminus, while only K8.1d and K8.1 share the same amino-acid sequences on their C-terminus (Figure 6B, S9A). The other two K8.1 isoforms completely lack the N-terminus amino-acids of the annotated K8.1 protein. The identification of >50 transcript variants and six coding isoforms of K8.1, suggests that the heterogenous expression of K8.1 isoforms may contribute to different entry mechanisms in different cell types or conditions.
Figure 6.
Alternate coding sequences of K8.1 and K8 proteins. (A) Alternate coding isoforms of K8.1 protein identified by TRIMD analysis. (B) Multiple sequence alignment of all K8.1 protein isoforms with each colour representing different amino acid sequences. Domains marked with same colour depict same sequence of amino acids in polypeptide chain. Numbers on the right indicate the nt. loci demarcating exon boundaries of the respective isoforms. (C) Alternate coding isoforms of K8 protein identified by TRIMD. (D) Distribution of K8.1, K8/K8.1 and RTA/K8 K8.1 encoding transcript isoforms over 19 individual TSSs with each orange bar depicting the total number of long reads (Iso-Seq read depth) founding the identification of complete transcripts originating from a TSS and the respective blue bars depicting the total numbers of complete TRIMD-identified transcripts identified from the same TSS loci. (E) Coding isoform of the LANA (ORF73) protein identified by TRIMD analysis.
Nevertheless, a downstream phenotypic analysis of the alternative coding sequences will be required to evaluate the functional importance of the identified K8.1 coding isoforms. In addition to coding isoforms of K8.1, TRIMD revealed alternative coding sequences for various other KSHV ORFs, including five coding isoforms of the K8-bZip protein that functions in viral replication and seven coding isoforms of the vIRF4 protein, which is a crucial protein required by KSHV for host immune modulation during its life cycle (Figures 6C, S9B). To gain more insights into all the mono- and polycistronic K8.1 transcripts, we further analyzed the distribution of TSSs associated with K8.1, K8/K8.1 and RTA/K8/K8.1 transcripts, and determined that TSSs for K8.1 transcription are distributed over 20 different loci on the KSHV genome, with two TSSs located >5 kb upstream and one TSS determined as the predominant forward strand TSS situated nearly 50 kb upstream of the K8.1 gene loci (Figure 6D). We observed the highest Iso-Seq read-depth of K8.1 transcripts from the canonical TSS (76 000 nt), as identified earlier by Tang et al. (40), and variable read-depths from other TSSs previously unidentified. The broad distribution of TSSs associated with K8.1 transcripts suggests that there is alternative usage of promoters by KSHV to encode essential genes.
We also detected a coding isoform of KSHV LANA (ORF73) protein, that exhibits the same N-terminal domain as LANA but has a different sequence of amino-acids on its C-terminal (Figure 6E). Previously, N-terminal truncated cytoplasmic isoforms of LANA have been characterized to inhibit cGAS-STING-dependent induction of interferon for promoting reactivation of KSHV (41). In contrast, the novel LANA isoform (ORF73a) identified from our study is a C-terminal truncated LANA generated by alternative splicing of a LANA precursor transcript, having the C-terminal and central repeat-region spliced out during mRNA processing.
Identification of fusion-ORFs, alternative UTRs for annotated ORFs and long non-coding antisense-to-ORF transcript isoforms
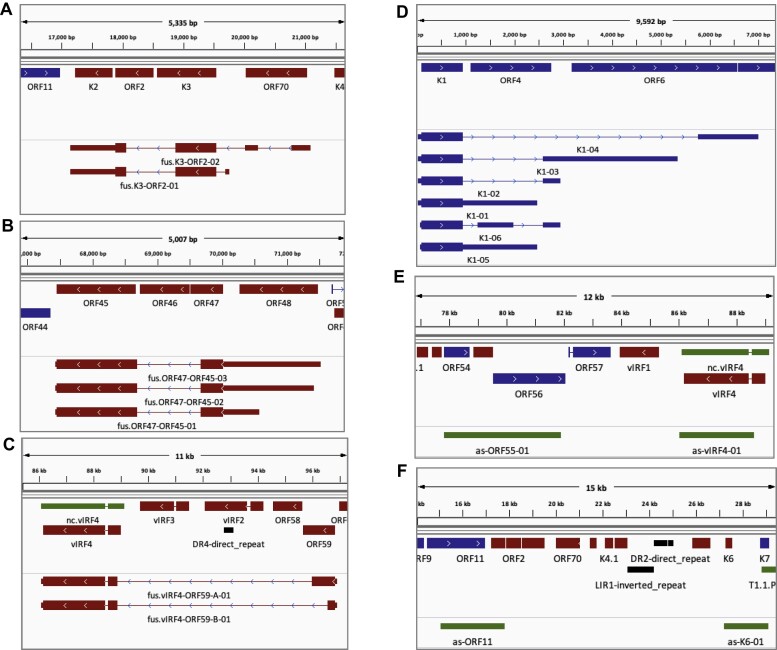
We further evaluated TRIMD unique transcripts for novel features other than alternative coding sequences. We observed that the complex KSHV transcriptome with multiple overlapping transcripts also encodes fusion-ORFs, defined as the in-frame coding sequences comprising of N-terminus from one protein and C-terminus from another protein. Specifically, we identified fusion ORFs for K3-ORF2, ORF47-ORF45 and two distinct fusion ORFs for ORF59-vIRF4 (Figure 7A–C). While these observations are potentially further expanding the KSHV proteome, any validation and functional studies of these potentially multifunctional proteins is beyond the scope of this report. Identification and validation of more than one TSS and termination sites for most coding genes is an important finding from our analysis. For instance, we identified four different poly-A sites of transcripts encoding K1 (Figure 7D). In addition to transcripts with novel ORFs and fusion-ORFs, we also identified non-coding transcripts antisense (as) to coding-ORFs including as-ORF55, as-vIRF4, as-ORF11 and as-K6 (Figure 7E, F). Our robust target enrichment approach for KSHV reads in sequencing data enhanced the identification of the transcript isoforms with alternative untranslated regions (UTRs), transcripts encoding for fusion ORFs and novel non-coding antisense transcripts not previously reported.
Figure 7.
Fusion ORFs, alternate TSS and anti-sense RNAs identified by TRIMD. (A-C) IGV panels displaying the TRIMD-identified fusion coding isoforms of (A) the K3 and ORF2, (B) ORF45 and ORF47, and (C) two alternate fusion isoforms of ORF59 and vIRF4. (D) IGV panel displaying six TRIMD-identified isoforms of K1 represented by four different polyadenylation sites and two different TSSs. (E-F) IGV panels displaying the non-coding RNAs identified by TRIMD that are found antisense to the coding KSHV ORFs including transcripts antisense to (E) ORF55 (as-ORF55-01), vIRF4 (as-vIRF4-01), (F) ORF11 (as-ORF11) and K6 (as-K6-01).
Comparing TRIMD-identified transcripts with nanopore sequencing of KSHV transcripts
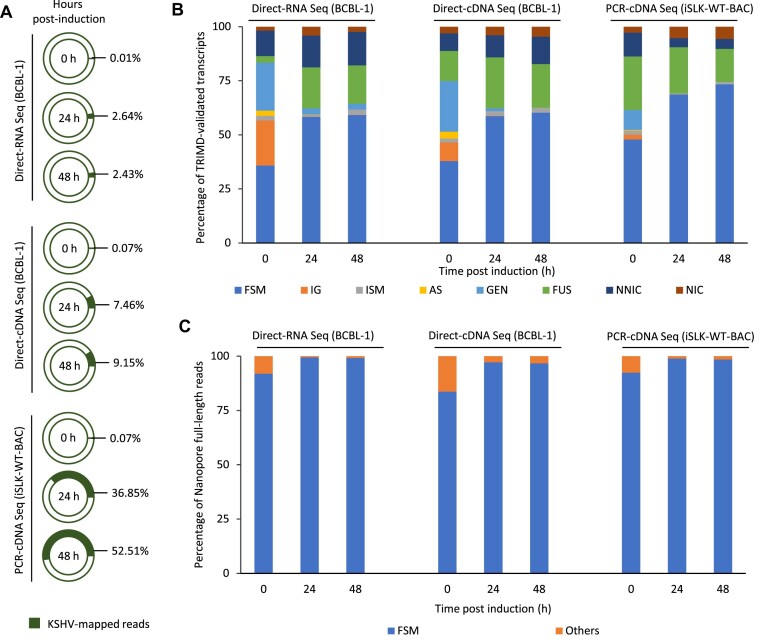
In order to further verify the TRIMD high-resolution transcriptome and to decipher long KSHV transcripts that could not be identified with our stringent TRIMD approach, we next performed an Oxford Nanopore Technology (ONT)-based high-throughput sequencing of KSHV transcripts in both BCBL-1 and iSLK-WT-BAC16 infected cells. As nanopore technology facilitates end-to-end sequencing of full-length transcripts, we used the nanopore direct-RNA and direct-cDNA platforms for sequencing BCBL-1 cells, and nanopore PCR-cDNA sequencing for obtaining reads from infected iSLK cells. While our TRIMD transcripts represent the lytic transcriptome of KSHV, we performed the nanopore sequencing of BCBL-1 and iSLK cells during both latent (0 h) and lytic phases at 24 and 48 h post induction, to enable a comparison of latent and lytic transcriptomes. In total, we obtained 2–4 million reads with direct-RNA seq, 0.8–1.6 million reads for direct-cDNA seq and 7–11 million reads with PCR-cDNA seq. We observed that the percentage of KSHV mapped reads was less than 1% in the uninduced cells for all sequencing platforms (Figure 8A). The mapped reads percentage increased to 2.4–2.6% and 7.4–9.1% in direct-RNA and direct-cDNA seq, respectively, of induced BCBL-1 cells. In contrast, we obtained 36.8–52.5% KSHV mapped reads from PCR-cDNA sequencing of lytically reactivated iSLK-WT-BAC16 cells (Figure 8A). The rationale behind performing PCR-cDNA sequencing for iSLK-WT-BAC16 cells was to obtain long-reads with excessive depth, as we have not utilized other sequencing platforms for analyzing the KSHV transcriptome from iSLK cells. The higher percentage of viral reads in iSLK cells is a result of transactivation by the RTA transgene under a tetracycline inducible promoter (23).
Figure 8.
Comparison of TRIMD-identified transcripts and ONT sequencing reads. (A) Percentage of total reads that aligned to KSHV as generated from the direct-RNA and direct-cDNA sequencing of BCBL-1 cells and PCR-cDNA sequencing of iSLK-WT-BAC16 cells, before and after induction. (B) TRIMD-identified transcripts classified into full splice match (FSM), incomplete splice match (ISM), intergenic (IG), antisense (AS), genic (GEN), fusion genes (FUS), novel in catalogue (NIC) and novel not in catalogue (NNIC) transcripts with reference to full-length KSHV reads obtained from different platforms of nanopore sequencing of BCBL-1 and iSLK-WT-BAC16 cells. (C) Full-length KSHV transcripts processed from nanopore sequencing platforms of BCBL-1 cells and iSLK-WT-BAC16 cells classified into full splice match (FSM) and Others (including ISM, GEN, FUS, NIC, NNIC and AS) with reference to TRIMD-identified lytic transcripts.
Next, we used SQANTI3 QC (28) for transcript classification, to categorize the unique transcripts identified by TRIMD with reference to full-length reads obtained from each of the nanopore sequencing data. We found that >50% of unique TRIMD transcripts could be identified as full splice match (FSM), including each splice junction, to ONT reads from induced samples, compared to <50% FSM observed for ONT reads from uninduced cells (Figure 8B). Additionally, we observed that 15–20% of unique TRIMD transcripts were classified as fusion by SQANTI3 QC, suggesting that these transcripts mapped across more than one ONT read. The classification of unique TRIMD transcripts, with ONT reads as reference, suggested that the integration of KSHV-enriched multiplatform sequencing data by the TRIMD pipeline has validated many more splice junctions compared to those identified by ONT long-read processing. Furthermore, we also classified the ONT full-length reads with reference to TRIMD unique transcripts. Interestingly, we found that >96% of ONT reads from all induced samples and 83–93% of ONT reads from all uninduced samples were classified as full-splice match (FSM) to the TRIMD-identified KSHV transcripts (Figure 8C). This further established that our approach for KSHV transcriptome analysis provides a much higher resolution of the complex bidirectional and overlapping KSHV transcriptome. Furthermore, the expansive set of unique splice junctions identified by our TRIMD analysis include almost all splice junctions that have been identified previously or in our data by ONT sequencing (12). However, the mapping of ONT reads to the KSHV reference genome also identified a number of long KSHV transcripts (>4 kb), which were not detected in our TRIMD approach.
We also analyzed the nanopore sequencing data to estimate the relative abundance of novel identified splice variants of annotated ORFs during latent and lytic cycles in both BCBL-1 and iSLK cells (Supplementary Table S5). The counts of ONT reads identified as full-splice-match by SQANTI3 to unique ORF isoforms demonstrate differential expression of coding variants during the latent and lytic phases of KSHV, thus providing preliminary evidence to explore the functional role of alternative coding isoforms.
Detection of TRIMD-identified long-range transcripts in pediatric KS tissue samples
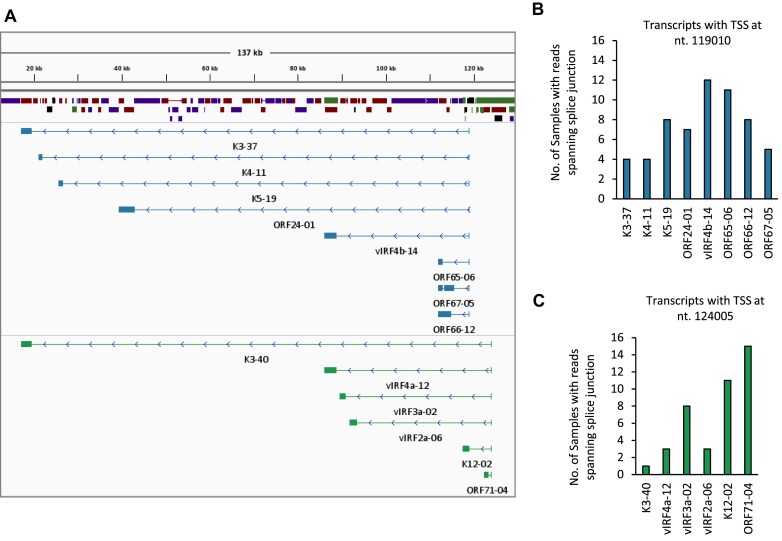
To further comprehend the significance of TRIMD-identified transcripts, we analyzed recently published RNA-Seq data of pediatric KS tissue samples (29). With our rationale to test the presence of long-range identified transcripts in KS samples, we mapped the RNA-Seq data from 16 samples to KSHV using the TRIMD-identified transcripts of interest as a reference (Figure 9A). We observed that four samples had reads spanning the splice junction of TRIMD-identified K3 and K4 transcript isoforms, with the TSSs of these transcripts in the latency associated region at nt. 119010, splice donor site at nt. 118906 and splice acceptor sites at 100 and 80 kb upstream, respectively (Figure 9B). Additionally, we detected splice junction covering-reads for K5, ORF24, vIRF4b, ORF65 and ORF66 long-range transcript isoforms in more than six samples, with 75% of total samples found positive for reads spanning the splice junction of vIRF4b-14 (Figure 9B). Similarly, we observed the presence of several long-range transcripts with TSS at nt. 124005 and splice donor at nt. 123842 in the KS tissue samples, with isoforms of vIRF3a, K12 and ORF71 detected in 8 or more samples (Figure 9C). The detection of TRIMD-identified transcripts in clinical samples of KS underscores the significance of our analysis.
Figure 9.
Pediatric KS tissue samples exhibiting long-range KSHV transcripts. (A) IGV panel displaying different short and long-range spliced transcripts originating from the two TSSs located in the latency region. (B, C) Number of pediatric KS tissue samples (SRA BioProject PRJNA975091) that were detected positive for RNA-Seq reads uniquely mapping across the splice junction (SJ) of TRIMD-identified long-range transcripts with (B) TSS at nt. 119010 and splice donor bases (GU) at nt. 118906 and (C) TSS at nt. 124005 and splice donor bases (GU) at nt. 123842.
Discussion
Our approach of target enriched transcript structure analysis of the KSHV transcriptome by integration of data from different sequencing platforms with TRIMD has enabled the de-complexification of KSHV transcripts at a very high-resolution. We report validation of 994 unique KSHV transcripts from induced BCBL-1 cells (Supplementary Table S3). It is noteworthy, that in addition to the identification of a plethora of novel KSHV transcripts, our unique set of KSHV transcripts also represents previously reported transcript features that were identified by single molecule or high-throughput analysis of KSHV transcripts. Overall, our lytic transcriptome is represented by 206 TSSs, 110 polyadenylation sites and 228 splice junctions, with a potential to encode for 147 unique ORFs exhibiting canonical start codons (AUG) (Figure 3). Prior investigations into the KSHV transcriptome have delineated the transcription initiation and termination sites for multiple KSHV ORFs (9,10,12,30). Although these studies have substantially contributed to our comprehension of KSHV transcripts, they exhibited differential efficiency in discerning the alternative UTRs and splice variants across distinct KSHV transcripts. The TRIMD method for transcript structure identification used in our study, overcomes these limitations by applying both Illumina and SMRT sequencing data to build transcript structures with high confidence.
The precise identification of TSS, 3′ ends of polycistronic transcripts, and alternative splice junctions in previously annotated and novel transcripts, has remained relatively underexplored due to the limited sequencing coverage. To investigate the lytic KSHV transcriptome, previous studies have used the TREX-BCBL-1 and iSLK-WT-BAC infected cells, which expresses the doxycycline-inducible RTA transactivator throughout the lytic cycle, leading to higher gene expression but also does not 100% recapitulate the kinetics of immediate early, early and late genes during the replication cycle (23,42). Our analysis on BCBL-1 induced with TPA and NAB yielded only 1–5% of KSHV mapped reads. The percentage of KSHV mapped reads significantly increased to 88.1% and 65% after target enrichment for the short and long read RNA-Seq platforms, respectively. Furthermore, target enrichment prior to integration of data from different sequencing platforms, strongly enhanced the validation of both high and low expressed transcripts by TRIMD, detection of which has remained limited in previous annotations (12). This approach can be utilized in the future for identification of KSHV transcriptomes in other cell lines and importantly in clinical KS tissue samples, that has remained highly challenging with small RNA-Seq platforms (43).
Similar to EBV and MHV68, KSHV displays a high degree of 3′ poly(A) signal readthrough transcripts, which result in numerous polycistronic transcripts with common 3′ ends. Previously, the genome wide 3′ RACE approach to map 3′ UTRs for all KSHV ORFs, reported only a subset of shared polyadenylation sites for genes clustered in close proximity (7). Furthermore, modified poly(A) sequencing of KSHV with Illumina platform identified a total of 55 polyadenylation sites, of which 20 were found associated to single genes and the others were associated to multiple genes (6). Our validation of novel 3′ UTRs has identified monocistronic transcript isoforms for various previously annotated as either bicistronic or polycistronic genes due to lack of identification of individual 3′ UTRs. For instance, the RTA transcript has previously been described as a polycistronic transcript with three ORFs encoding RTA, K8 and K8.1. In contrast, we have identified an early 3′ end for RTA transcription along with three other 3′ ends identified associated with K8, K8/K8.1 and RTA/K8/K8.1 transcripts, indicating that KSHV has the potential to generate both monocistronic and polycistronic transcripts for clustered genes. Similarly, we identified monocistronic isoforms of K1, ORF21, ORF56, ORF62, ORF72 and K14, and proximal polyadenylation sites for additional ORF clusters including K4.2/K4.1, ORF34/35/36 and ORF62/61/60 not previously identified (6,7,9,12). The use of transcript isoforms with different 3′ UTR lengths may suggest differential stability of mRNAs and potential control of gene expression by miRNA regulation.
Furthermore, within the set of TRIMD-identified KSHV transcripts, we observed various novel transcript isoforms that share the same TSS but distinct 3′ ends due to differential readthrough across poly(A) signals. The most prominent examples of such transcripts are the multiple short- and long-range transcripts that arise from three predominant TSSs near ori-lytR or ori-lytL (nt. 27048, 119010 and 124005) and terminate at different distant genomic loci that are alternatively spliced from these long pre-mRNAs (Supplementary figure S4). Transcription initiation from these TSSs in combination with alternative usage of polyadenylation sites strongly indicate the potential of KSHV to transcribe additional coding and non-coding RNAs from a single promoter. Importantly, we validated the expression and structure of some of these novel transcripts during both latent and lytic replication, which potentially increases the repertoire of latency-associated genes.
Compared to polyadenylation sites, the identification of TSSs for KSHV transcripts in previous studies has been more efficient with a high overlap with our data set (9,10,12,30). However, we have identified a total of 206 TSSs many of which are shared among different transcripts. In total, we have identified at least one TSS for nearly all GenBank annotated KSHV ORFs, along with many novel alternative TSSs for the majority of KSHV genes (Figure 3D, Supplementary Table S3, S4).
The occurrence of transcript isoforms with alternative TSSs further implies that KSHV utilizes advanced regulatory mechanisms to control the expression of coding and non-coding genes. Because each TSS can be regulated by different upstream promoters, the occurrence of cellular and viral TATA-binding proteins (TBPs) and their respective binding elements near the transcript 5′ ends can impact the gene expression from each site. Interestingly, we found that a few TSS validated in our data are more proximal to ORF24, the KSHV TATA-binding protein homolog, binding motif (TATTWAA) than the canonical TATA box, suggesting that ORF24 could mediate the recruitment of RNA PolII at these sites (34,44). TRIMD-identified several isoforms of ORF4, ORF9, ORF17.5, ORF26, ORF32, ORF38, ORF39, ORF45, K8.1, ORF59, ORF65, ORF67 which are examples of transcripts with 5′ ends more proximal to an upstream ORF24 binding motif compared to host TATA boxes.
The detection of transcript isoforms generated from multiple TSSs during lytic replication suggests that active transcription from nearly all putative TSSs is carried out by KSHV during lytic replication, with quantitative bias towards augmenting transcription from a few very strong promoters. For example, we have identified an alternative TSS upstream of the Kaposin (K12) ORFs, that was previously described to be driven from a common promoter expressing ORF71, ORFF72 and Kaposins during latency (45,46), which is active during lytic replication and gives rise to multiple transcripts.
5′ UTRs regulate translation efficiency as well as cellular localization of mRNAs. For example, it has been shown that longer 5′ UTRs harbor sequences for binding to regulatory proteins, such as poly(C)-motifs for nuclear retention (47,48). Consistent with this, MHV68 ORFs with long 5′ UTRs were significantly enriched for poly(C) binding protein (PCBP1, PCBP3) motifs, and ORF55 isoforms with long 5′ UTRs were exclusively present in the nucleus while ORF55 isoforms with the shortest 5′ UTRs were distributed between cytoplasmic and nuclear fractions (18). Therefore, we predict that KSHV transcripts with shorter 5′ UTRs are efficiently exported for translation. For instance, we have identified various mono and polycistronic transcripts containing the K3 (MIR1), ORF2 and K2 (vIL6) coding sequences, with many of these isoforms generated from different TSSs. A similar example is cited for RTA/K8/K8.1 transcripts that were associated with nearly 20 TSSs (Figure 6D). While most of these transcripts have coding frames for all three genes, the coding potential for individual ORFs is predicted as highest for transcripts that have shorter UTRs, meaning high proximity of TSSs and polyadenylation sites to start and stop codons, respectively (49,50). These also include a large number of newly identified non-coding RNAs some of which may exert their potential function in the nucleus.
We identified two predominant TSSs near the latency associated region and one predominant TSS between K5 and K6 that generate long-range readthrough transcripts, with different polyadenylation sites and alternative splicing of long introns (Supplementary Figure S4). Evidence of readthrough transcription has been reported earlier from EBV, MHV68 as well as HSV genomes, that increase the transcriptional diversity of these viruses, during both latency and lytic replication (17,18). Indeed, the first identified latency-associated gene augmented by long-range transcription is the EBV EBNA1 gene transcribed from a nearly 100 kb long pre-mRNA (19,20,33). Interestingly, we have identified multiple long-range transcripts from the KSHV latency region. Additionally, read through transcription of Kaposin and the miRNA locus augmented by the LANA TSS through multiple polyadenylation signals has previously been described for KSHV (45). This mechanism allows for differential expression levels of different latency-associated genes expressed from a single promoter. Two prominent transcriptional hubs situated in the latency region are found to generate numerous multiply-spliced mRNAs. Alternative splicing of these transcripts generates mRNAs for multiple KSHV genes including K3, K5, ORF24, vIRF3 and vIRF4 (Supplementary Figure S4A, B). The identification of these novel transcripts from TSSs that are located 40–100 kb upstream of their respective ORFs potentially expands the role of these lytic genes in the context of latency. Albeit, we note that some of these transcripts are expressed at low levels. As we have validated the presence of some of these transcripts like ORF24 during latency by RT-PCR and Sanger sequencing, it would be interesting to develop and perform more sensitive assays to identify additional rare transcript isoforms including K3 and K5 from these regions, in latent cell populations and KS-tissue samples. Interestingly, a previous study has reported K3- and K5-dependent MHC class I down-regulation in latently infected cells under conditions where their respective transcripts were not detectable (51).
Among the most valuable insights obtained from our target enriched TRIMD analysis of KSHV transcripts is the identification of expanded splicing events occurring throughout the KSHV genome. Splice variant isoforms for transcripts that classically represent different phases of KSHV replication including, immediate early (RTA, ORF45, K4.2), early (K8/K8.1), delayed early (K3, K5, ORF57, ORF37), late lytic (ORF6, ORF44) and latent (LANA, vFLIP, vCyclin, vIRF3) phases of KSHV replication have been identified at a very high resolution by our analysis (Figure 3D, Supplementary Table S3). We observed that differential splicing of mRNA precursors can generate a large number of unique KSHV transcripts that potentially can be translated into identical, overlapping, or completely distinct proteins; thereby expanding the KSHV proteome.
Interestingly, some alternative splicing events identified within the coding sequences of previously annotated ORFs are also predicted to encode for proteins that may or may not share overlapping sequences with currently annotated ORFs, thus classified as coding protein isoforms or completely novel ORFs. For instance, we found that alternative splicing of vIRF4 transcripts can generate seven different coding variants of vIRF4 that need to be further characterized at the protein level (Supplementary Figure S9B). The KSHV replication associated protein (RAP) or K-bZIP (K8), contributes to KSHV replication and gene expression, and induces G1 cell cycle arrest during the lytic cycle. K8 has been previously identified to be transcribed from two distinct promoters, with one for transcription during the immediate early phase and the second during the delayed early phase, both of which are further processed into several spliced transcript isoforms (52). Likewise, evidence of differential splicing of K8.1 mRNAs and its coding variants has been reported earlier by different groups (40). However, in addition to identification of multiple TSSs associated with RTA/K8/K8.1 and K8/K8.1 transcripts, we have identified a high number of splice variants of these transcripts that have potential to encode two alternative coding sequences of RTA, five isoforms of K8 and six isoforms for K8.1. This observation, in conjunction with previous reports, further highlights the sensitivity of our high-resolution transcriptome analysis. The alternative coding sequences identified in our study only represent the full-length coding sequences with canonical start codons. Various downstream analyses can be performed on these transcripts to further identify the potential roles of KSHV upstream ORFs (uORFs), short ORFs (sORFs), internal ORFs and ORFs with non-AUG start codons, as previously identified for KSHV by the ribosome profiling method (10).
Human herpesviruses, including KSHV, are known to have evolved many strategies to evade host immune responses during primary infection and establishment of cellular latency for lifelong persistence in their hosts. One of these mechanisms includes transcriptional modulation. While we have obtained different annotations for KSHV transcripts, the extensive complexity of the KSHV transcriptome was not fully appreciated in previous studies due to low coverage and the use of single sequencing platforms. To overcome these limitations, we combined multiple short and long-read sequencing techniques with state-of-the-art target enrichment from BCBL-1 cells and our stringent TRIMD data analysis pipeline (17). In addition to the identification of many unique transcript features, we have annotated these transcripts with respect to their coding potential suggesting the existence of many new protein isoforms. Furthermore, the detection of long-range KSHV transcripts in pediatric KS samples signifies the clinical relevance of KSHV transcriptome annotation. Our high-density annotations have laid a rich foundation for future studies of newly identified coding and long non-coding RNAs towards a better understanding about their functional roles during KSHV replication, latency and pathogenesis. Target enriched long-read sequencing of KS tissue samples can further enhance the identification of transcripts with splice junctions and long-range KSHV transcription, to gain more valuable insights into the pathogenesis of KS in future studies.
Supplementary Material
Acknowledgements
Author contributions: R.S., R.R., E.K.F. and S.A.T. conceptualized the project. R.S. performed the majority of molecular work with help from N.K. and A.F. T.O. and E.K.F. designed the TRIMD analysis pipeline. D.M. and R.S. developed the PacBio sequencing Target Enrichment platform. R.S. and N.K. with help from T.O. analyzed sequencing data. R.S. and R.R. wrote the manuscript. R.S., N.K. and R.R. revised and edited the manuscript.
Contributor Information
Ritu Shekhar, Department of Molecular Genetics and Microbiology, University of Florida, Gainesville, FL, USA.
Tina O'Grady, Department of Pathology, Tulane University, New Orleans, LA, USA.
Netanya Keil, Department of Molecular Genetics and Microbiology, University of Florida, Gainesville, FL, USA; UF Genetics Institute, University of Florida, Gainesville, FL, USA.
April Feswick, Department of Molecular Genetics and Microbiology, University of Florida, Gainesville, FL, USA.
David A Moraga Amador, UF Interdisciplinary Center for Biotechnology Research, University of Florida, Gainesville, FL, USA.
Scott A Tibbetts, Department of Molecular Genetics and Microbiology, University of Florida, Gainesville, FL, USA; UF Health Cancer Center, University of Florida, Gainesville, FL, USA; UF Genetics Institute, University of Florida, Gainesville, FL, USA.
Erik K Flemington, Department of Pathology, Tulane University, New Orleans, LA, USA.
Rolf Renne, Department of Molecular Genetics and Microbiology, University of Florida, Gainesville, FL, USA; UF Health Cancer Center, University of Florida, Gainesville, FL, USA; UF Genetics Institute, University of Florida, Gainesville, FL, USA.
Data availability
The data underlying this article are available in NIH data repository, Gene Expression Omnibus (GEO) and can be accessed with the unique accession GSE250435.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
National Institutes of Health/National Cancer Institute [P01 CA214091 to R.R, S.A.T., E.K.F.]; R.S. and R.R. also received pilot funding from the UF Health Cancer Center Next Generation Sequencing Shared Resource. Funding for open access charge: NIH/NCI [P01 CA214091].
Conflict of interest statement. None declared.
References
- 1. Chang Y., Cesarman E., Pessin M.S., Lee F., Culpepper J., Knowles D.M., Moore P.S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994; 266:1865–1869. [DOI] [PubMed] [Google Scholar]
- 2. Cesarman E., Chang Y., Moore P.S., Said J.W., Knowles D.M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 1995; 332:1186–1191. [DOI] [PubMed] [Google Scholar]
- 3. Soulier J., Grollet L., Oksenhendler E., Cacoub P., Cazals-Hatem D., Babinet P., d’Agay M.F., Clauvel J.P., Raphael M., Degos L. et al. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995; 86:1276–1280. [PubMed] [Google Scholar]
- 4. Broussard G., Damania B. Regulation of KSHV latency and lytic reactivation. Viruses. 2020; 12:1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rezaee S.A.R., Cunningham C., Davison A.J., Blackbourn D.J. Kaposi's sarcoma-associated herpesvirus immune modulation: an overview. J. Gen. Virol. 2006; 87:1781–1804. [DOI] [PubMed] [Google Scholar]
- 6. Majerciak V., Ni T., Yang W., Meng B., Zhu J., Zheng Z.M. A viral genome landscape of RNA polyadenylation from KSHV latent to lytic infection. PLoS Pathog. 2013; 9:e1003749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bai Z., Huang Y., Li W., Zhu Y., Jung J.U., Lu C., Gao S.J. Genomewide mapping and screening of Kaposi's sarcoma-associated herpesvirus (KSHV) 3' untranslated regions identify bicistronic and polycistronic viral transcripts as frequent targets of KSHV microRNAs. J. Virol. 2014; 88:377–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chandriani S., Xu Y., Ganem D The lytic transcriptome of Kaposi's sarcoma-associated herpesvirus reveals extensive transcription of noncoding regions, including regions antisense to important genes. J. Virol. 2010; 84:7934–7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bruce A.G., Barcy S., DiMaio T., Gan E., Garrigues H.J., Lagunoff M., Rose T.M. Quantitative analysis of the KSHV transcriptome following primary infection of blood and lymphatic endothelial cells. Pathogens. 2017; 6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Arias C., Weisburd B., Stern-Ginossar N., Mercier A., Madrid A.S., Bellare P., Holdorf M., Weissman J.S., Ganem D KSHV 2.0: A comprehensive annotation of the Kaposi's sarcoma-associated herpesvirus genome using next-generation sequencing reveals novel genomic and functional features. PLoS Pathog. 2014; 10:e1003847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moldovan N., Tombacz D., Szucs A., Csabai Z., Balazs Z., Kis E., Molnar J., Boldogkoi Z. Third-generation sequencing reveals extensive polycistronism and transcriptional overlapping in a baculovirus. Sci. Rep. 2018; 8:8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Prazsak I., Tombacz D., Fulop A., Torma G., Gulyas G., Dormo A., Kakuk B., Spires L.M., Toth Z., Boldogkoi Z. KSHV 3.0: a state-of-the-art annotation of the Kaposi's sarcoma-associated herpesvirus transcriptome using cross-platform sequencing. 2023; bioRxiv doi:23 September 2023, preprint: not peer reviewed 10.1101/2023.09.21.558842. [DOI] [PMC free article] [PubMed]
- 13. Amarasinghe S.L., Su S., Dong X., Zappia L., Ritchie M.E., Gouil Q. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 2020; 21:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mooney M., McWeeney S. Data integration and reproducibility for high-throughput transcriptomics. Int. Rev. Neurobiol. 2014; 116:55–71. [DOI] [PubMed] [Google Scholar]
- 15. Whisnant A.W., Jurges C.S., Hennig T., Wyler E., Prusty B., Rutkowski A.J., L’Hernault A., Djakovic L., Gobel M., Doring K et al. Integrative functional genomics decodes herpes simplex virus 1. Nat. Commun. 2020; 11:2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stern-Ginossar N., Weisburd B., Michalski A., Le V.T., Hein M.Y., Huang S.X., Ma M., Shen B., Qian S.B., Hengel H. et al. Decoding human cytomegalovirus. Science. 2012; 338:1088–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. O’Grady T., Wang X., Honer Zu Bentrup K., Baddoo M., Concha M., Flemington E.K. Global transcript structure resolution of high gene density genomes through multi-platform data integration. Nucleic Acids Res. 2016; 44:e145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. O’Grady T., Feswick A., Hoffman B.A., Wang Y., Medina E.M., Kara M., van Dyk L.F., Flemington E.K., Tibbetts S.A. Genome-wide transcript structure resolution reveals abundant alternate isoform usage from murine gammaherpesvirus 68. Cell Rep. 2019; 27:3988–4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sample J., Hummel M., Braun D., Birkenbach M., Kieff E. Nucleotide sequences of mRNAs encoding Epstein-Barr virus nuclear proteins: a probable transcriptional initiation site. Proc. Natl. Acad. Sci. U.S.A. 1986; 83:5096–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bodescot M., Perricaudet M., Farrell P.J. A promoter for the highly spliced EBNA family of RNAs of Epstein-Barr virus. J. Virol. 1987; 61:3424–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mamanova L., Coffey A.J., Scott C.E., Kozarewa I., Turner E.H., Kumar A., Howard E., Shendure J., Turner D.J. Target-enrichment strategies for next-generation sequencing. Nat. Methods. 2010; 7:111–118. [DOI] [PubMed] [Google Scholar]
- 22. Renne R., Zhong W., Herndier B., McGrath M., Abbey N., Kedes D., Ganem D Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 1996; 2:342–346. [DOI] [PubMed] [Google Scholar]
- 23. Myoung J., Ganem D Generation of a doxycycline-inducible KSHV producer cell line of endothelial origin: maintenance of tight latency with efficient reactivation upon induction. J. Virol. Methods. 2011; 174:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Murata M., Nishiyori-Sueki H., Kojima-Ishiyama M., Carninci P., Hayashizaki Y., Itoh M. Detecting expressed genes using CAGE. Methods Mol. Biol. 2014; 1164:67–85. [DOI] [PubMed] [Google Scholar]
- 25. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu T.D., Watanabe C.K. GMAP: A genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005; 21:1859–1875. [DOI] [PubMed] [Google Scholar]
- 27. Frith M.C., Valen E., Krogh A., Hayashizaki Y., Carninci P., Sandelin A. A code for transcription initiation in mammalian genomes. Genome Res. 2008; 18:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tardaguila M., de la Fuente L., Marti C., Pereira C., Pardo-Palacios F.J., Del Risco H., Ferrell M., Mellado M., Macchietto M., Verheggen K. et al. SQANTI: extensive characterization of long-read transcript sequences for quality control in full-length transcriptome identification and quantification. Genome Res. 2018; 28:396–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Caro-Vegas C., Peng A., Juarez A., Silverstein A., Kamiyango W., Villiera J., McAtee C.L., Mzikamanda R., Tomoka T., Peckham-Gregory E.C. et al. Pediatric HIV+ Kaposi sarcoma exhibits clinical, virological, and molecular features different from the adult disease. JCI Insight. 2023; 8:e167854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ye X., Zhao Y., Karijolich J. The landscape of transcription initiation across latent and lytic KSHV genomes. PLoS Pathog. 2019; 15:e1007852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Majerciak V., Alvarado-Hernandez B., Lobanov A., Cam M., Zheng Z.M. Genome-wide regulation of KSHV RNA splicing by viral RNA-binding protein ORF57. PLoS Pathog. 2022; 18:e1010311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kronstad L.M., Brulois K.F., Jung J.U., Glaunsinger B.A. Reinitiation after translation of two upstream open reading frames (ORF) governs expression of the ORF35-37 Kaposi's sarcoma-associated herpesvirus polycistronic mRNA. J. Virol. 2014; 88:6512–6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rogers R.P., Woisetschlaeger M., Speck S.H. Alternative splicing dictates translational start in Epstein-Barr virus transcripts. EMBO J. 1990; 9:2273–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Davis Z.H., Verschueren E., Jang G.M., Kleffman K., Johnson J.R., Park J., Von Dollen J., Maher M.C., Johnson T., Newton W. et al. Global mapping of herpesvirus-host protein complexes reveals a transcription strategy for late genes. Mol. Cell. 2015; 57:349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Spiller O.B., Robinson M., O’Donnell E., Milligan S., Morgan B.P., Davison A.J., Blackbourn D.J. Complement regulation by Kaposi's sarcoma-associated herpesvirus ORF4 protein. J. Virol. 2003; 77:592–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chandran B., Smith M.S., Koelle D.M., Corey L., Horvat R., Goldstein E. Reactivities of human sera with human herpesvirus-8-infected BCBL-1 cells and identification of HHV-8-specific proteins and glycoproteins and the encoding cDNAs. Virology. 1998; 243:208–217. [DOI] [PubMed] [Google Scholar]
- 37. Li M., MacKey J., Czajak S.C., Desrosiers R.C., Lackner A.A., Jung J.U. Identification and characterization of Kaposi's sarcoma-associated herpesvirus K8.1 virion glycoprotein. J. Virol. 1999; 73:1341–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu L., Renne R., Ganem D., Forghani B. Human herpesvirus 8 glycoprotein K8.1: expression, post-translational modification and localization analyzed by monoclonal antibody. J. Clin. Virol. 2000; 17:127–136. [DOI] [PubMed] [Google Scholar]
- 39. Chandran B., Bloomer C., Chan S.R., Zhu L., Goldstein E., Horvat R. Human herpesvirus-8 ORF K8.1 gene encodes immunogenic glycoproteins generated by spliced transcripts. Virology. 1998; 249:140–149. [DOI] [PubMed] [Google Scholar]
- 40. Tang S., Zheng Z.M. Kaposi's sarcoma-associated herpesvirus K8 exon 3 contains three 5'-splice sites and harbors a K8.1 transcription start site. J. Biol. Chem. 2002; 277:14547–14556. [DOI] [PubMed] [Google Scholar]
- 41. Zhang G., Chan B., Samarina N., Abere B., Weidner-Glunde M., Buch A., Pich A., Brinkmann M.M., Schulz T.F. Cytoplasmic isoforms of Kaposi sarcoma herpesvirus LANA recruit and antagonize the innate immune DNA sensor cGAS. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E1034–E1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nakamura H., Lu M., Gwack Y., Souvlis J., Zeichner S.L., Jung J.U. Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J. Virol. 2003; 77:4205–4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moorad R., Kasonkanji E., Gumulira J., Gondwe Y., Dewey M., Pan Y., Peng A., Pluta L.J., Kudowa E., Nyasosela R. et al. A prospective cohort study identifies two types of HIV+ Kaposi Sarcoma lesions: proliferative and inflammatory. Int. J. Cancer. 2023; 153:2082–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nandakumar D., Glaunsinger B. An integrative approach identifies direct targets of the late viral transcription complex and an expanded promoter recognition motif in Kaposi's sarcoma-associated herpesvirus. PLoS Pathog. 2019; 15:e1007774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pearce M., Matsumura S., Wilson A.C. Transcripts encoding K12, v-FLIP, v-cyclin, and the microRNA cluster of Kaposi's sarcoma-associated herpesvirus originate from a common promoter. J. Virol. 2005; 79:14457–14464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Grundhoff A., Ganem D Mechanisms governing expression of the v-FLIP gene of Kaposi's sarcoma-associated herpesvirus. J. Virol. 2001; 75:1857–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ghosh G., Samui S., Das S., Singh V., Pal D., Das S., Naskar J., Sinha Roy S., Basu U. Poly C Binding Protein 2 dependent nuclear retention of the utrophin-A mRNA in C2C12 cells. RNA Biol. 2021; 18:612–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Palazzo A.F., Lee E.S. Sequence determinants for nuclear retention and cytoplasmic export of mRNAs and lncRNAs. Front. Genet. 2018; 9:440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hinnebusch A.G., Ivanov I.P., Sonenberg N. Translational control by 5'-untranslated regions of eukaryotic mRNAs. Science. 2016; 352:1413–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mitschka S., Mayr C. Context-specific regulation and function of mRNA alternative polyadenylation. Nat. Rev. Mol. Cell Biol. 2022; 23:779–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Adang L.A., Tomescu C., Law W.K., Kedes D.H. Intracellular Kaposi's sarcoma-associated herpesvirus load determines early loss of immune synapse components. J. Virol. 2007; 81:5079–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yamanegi K., Tang S., Zheng Z.M. Kaposi's sarcoma-associated herpesvirus K8beta is derived from a spliced intermediate of K8 pre-mRNA and antagonizes K8alpha (K-bZIP) to induce p21 and p53 and blocks K8alpha-CDK2 interaction. J. Virol. 2005; 79:14207–14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in NIH data repository, Gene Expression Omnibus (GEO) and can be accessed with the unique accession GSE250435.