Abstract
Accurate RNA structure models are crucial for designing small molecule ligands that modulate their functions. This study assesses six standalone RNA 3D structure prediction methods—DeepFoldRNA, RhoFold, BRiQ, FARFAR2, SimRNA and Vfold2, excluding web-based tools due to intellectual property concerns. We focus on reproducing the RNA structure existing in RNA-small molecule complexes, particularly on the ability to model ligand binding sites. Using a comprehensive set of RNA structures from the PDB, which includes diverse structural elements, we found that machine learning (ML)-based methods effectively predict global RNA folds but are less accurate with local interactions. Conversely, non-ML-based methods demonstrate higher precision in modeling intramolecular interactions, particularly with secondary structure restraints. Importantly, ligand-binding site accuracy can remain sufficiently high for practical use, even if the overall model quality is not optimal. With the recent release of AlphaFold 3, we included this advanced method in our tests. Benchmark subsets containing new structures, not used in the training of the tested ML methods, show that AlphaFold 3′s performance was comparable to other ML-based methods, albeit with some challenges in accurately modeling ligand binding sites. This study underscores the importance of enhancing binding site prediction accuracy and the challenges in modeling RNA–ligand interactions accurately.
Graphical Abstract
Graphical Abstract.
Introduction
In recent years, there has been a significant surge in interest within the scientific community regarding RNA molecules, especially in the context of developing therapeutics targeting RNA. To effectively meet this challenge, accurate high-resolution three-dimensional structures of RNA are essential. However, experimental methods for resolving RNA structures remain difficult, costly, and time-consuming (1,2). As an alternative, computational prediction of 3D RNA structures, with or without supplemental experimental data such as distance restraints or RNA secondary structure, is being pursued. Still, this field is in a developmental stage similar to protein structure prediction before the advent of AlphaFold (3), and the potential of applying AlphaFold approach to RNA with precision comparable to that achieved for proteins is currently viewed with skepticism (4). Generally speaking, this is due to an insufficient number of known experimentally solved structures. As of 1 May 2024, there are only 6526 structures of RNA molecules available in the RCSB database (5), including 4688 RNA-protein complexes and excluding the molecules with DNA and NA-hybrids. For comparison, there are 212 000 entries with protein structures.
Many research groups are therefore focusing on the development of new bioinformatics methods for the prediction of RNA tertiary structures with high accuracy. These methods can be broadly classified into three categories: physics-based, knowledge-based, and Machine Learning (ML)-based (6–8). Physics-based methods, such as Molecular Dynamics (MD) (9–20), rely on the principles of physics to predict RNA’s 3D structure. All-atom physics-based approaches are computationally expensive as they extensively explore folding pathways to find stable conformations. Typically, they are applied to smaller RNA molecules and utilize additional restraints (11) or coarse-grained models due to the computational demands (21). One of the key challenges in these methods is the inherent limitations of the force fields they use, which is an area of ongoing development in the field (22,23). Knowledge-based methods make use of tools derived from known RNA structures, employing templates, fragments, or scoring functions to model RNA 3D structures (24–39). ML-based methods utilize a range of artificial intelligence techniques for the prediction of 3D structures (40–46). Both knowledge-based and ML-based approaches depend on existing experimentally solved RNA structures to derive principles governing RNA’s 3D structure, with their major limitation being the insufficient number of such experimentally solved structures available for reference.
Results from the RNA-Puzzles (47–51) and Critical Assessment of protein Structure Prediction (CASP) (52,53) competitions have revealed that user experience is a critical factor in accurately predicting RNA structures. Unfortunately, the expertise required for high-level RNA structure prediction is not widely available, as only a few research laboratories have access to bioinformaticians with extensive experience in this area. Considering this, our publication focuses on evaluating the performance of currently available standalone methods for RNA structure prediction when executed with default settings. These methods rely solely on sequence data and, where applicable, secondary RNA structure information as inputs. This evaluation aims to determine the effectiveness of these methods in the absence of extensive user experience in RNA structure prediction. Recent studies have demonstrated that the geometrical accuracy of automatically predicted RNA 3D models can be significantly enhanced by applying straightforward energy minimization routines (54,55). These findings suggest that while user expertise plays a critical role in the initial predictions, the correct topology and subsequent refinements via energy minimization also contribute to the accuracy of computational RNA structure prediction. Our evaluation excludes the consideration of web-based RNA structure prediction tools, owing to potential intellectual property issues associated with their use.
The benchmark dataset employed in our analysis was compiled from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB) database (56). It includes a diverse array of RNA structural elements, as detailed in the Methods section. In response to the growing interest in targeting RNA with small molecules, our test set specifically comprises RNAs whose structures have been resolved in complexes with small molecules and are available in RCSB. This selection is crucial, considering that high-resolution RNA structures are vital for virtual screening and subsequent compound development. This is particularly important as RNA molecules are quite adaptable, and their structure can change upon contact with small molecules. Since docking with flexible RNA backbone remains a significant challenge (57); therefore, the primary aim of our study is to evaluate the efficacy of available algorithms not just in predicting the overall 3D structure of RNA, but more importantly, in accurately reconstructing RNA–ligand binding sites.
While benchmarking initiatives such as CASP and RNA-Puzzles have significantly advanced RNA structure prediction by focusing on entire RNA molecules, this study narrows the scope to RNA–ligand complexes. These complexes often exhibit structural variations from their isolated RNA counterparts, underscoring the necessity of specialized prediction models. By concentrating on the ligand binding sites within RNA, which are crucial for RNA functionality and the development of RNA-targeted therapeutics, we offer a nuanced perspective that complements existing RNA structure modeling efforts.
In our study, we tested six different RNA structure prediction methods available as standalone programs in 2022: DeepFoldRNA (42), RhoFold (formerly, E2Efold-3D) (41), BriQ (38), FARFAR2 (26), SimRNA (37) and Vfold2 (39) (detailed in the Methods section). DeepFoldRNA and RhoFold, both machine-learning (ML)-based methods, were operated without secondary structure restraints. In contrast, the BRiQ program, which necessitates secondary structure restraints as mandatory input, was run exclusively with these restraints. The remaining three programs—FARFAR2, SimRNA and Vfold2—are statistical-based methods and were evaluated under both scenarios: with and without secondary structure restraints. The accuracy of the predicted RNA 3D structures was assessed using various metrics, including root mean square deviation (RMSD), template modeling score (TM), and interaction network fidelity (INF).
As an exception, we also examine the performance of the recently released AlphaFold3 (58) in predicting RNA structures. Although AlphaFold3 is a server with non-commercial status, we chose to evaluate its performance due to its potential to be a groundbreaking tool in RNA structure prediction. Our small test set for all machine learning (ML)-based methods consists of structures not included in their respective training sets, allowing us to assess the generalization capabilities of these ML-based approaches.
Materials and methods
Dataset of RNA structures
We curated a dataset of RNA structures from the RCSB PDB available until 31 May 2023, with a focus on structures exclusively containing RNA, excluding those featuring DNA, proteins, or nucleic acid hybrids. This dataset encompasses structures determined through X-ray crystallography (XRD) and nuclear magnetic resonance (NMR). In our selection process, we emphasized RNA structures solved with small-molecule ligands, deliberately excluding both metal ions and a variety of small molecules commonly present in crystallization buffers, such as buffering agents and stabilizers, which are not considered functional ligands in our study. This criterion was chosen to highlight RNA–ligand interactions of biological significance. Our selection was guided by the need to benchmark RNA 3D structure modeling methods, many of which are optimized for single-chain RNA structures. Consequently, we included only those structures that align with the capabilities of these methods. Additionally, we excluded structures with extensively modified residues, such as glycol nucleic acids (GNAs) and locked nucleic acids (LNAs), to maintain a focus on more typical RNA sequences. This meticulous selection process resulted in a dataset comprising 139 RNA structures (Supplementary Table S1). Of the 139 structures in our dataset, 107 were resolved using XRD with a resolution of 4 Å or better. Among these, 93 structures have a resolution finer than 3 Å, and 24 structures are within 2 Å. The remaining structures were determined using NMR. The secondary structures were extracted from the 3D structures using the x3dna-dssr program v1.9.10 (59). All secondary structures underwent manual inspection and cleaning to eliminate artefacts introduced by x3dna-dssr.
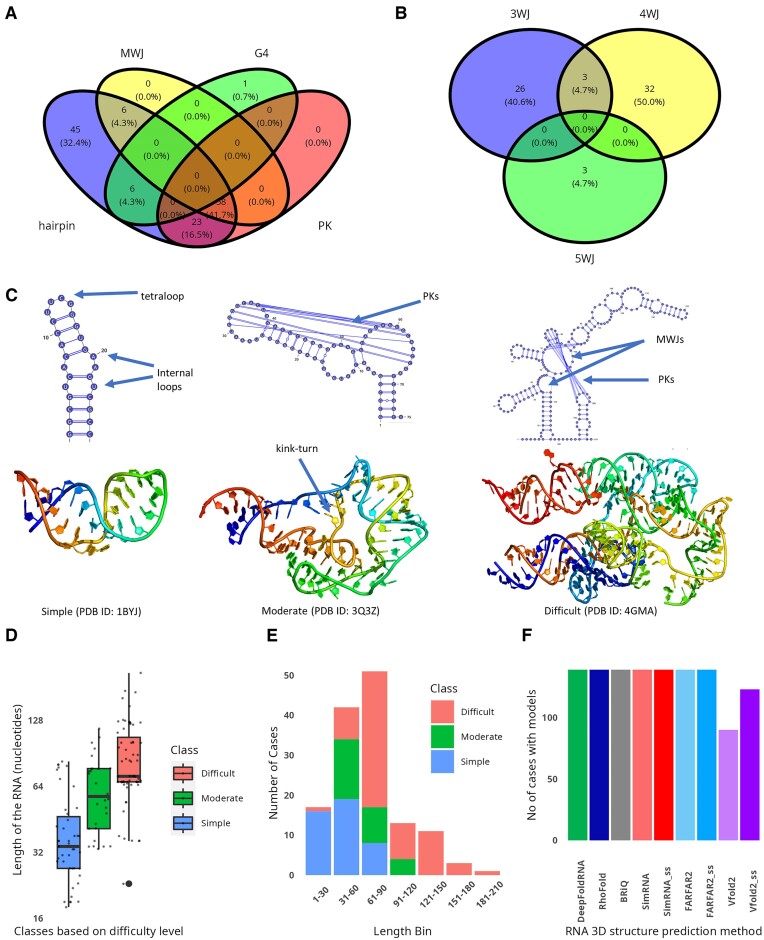
The dataset comprises of RNAs with diverse structural features such as simple hairpins (HP), HPs with pseudoknots (PK), multi-way junctions (MWJ), MWJs with PKs, G-quadruplexes (G4) and HPs with G4 (Supplementary Table S1, Figure 1A). It includes 45 simple HPs characterized by the presence of bulges or internal loops without additional structural elements. The dataset also contains 64 structures with MWJs: 26 with three-way junctions (3WJ), 32 with four-way junctions (4WJ), and 3 with five-way junctions (5WJ). Among these, 61 cases feature a single type of junction, while the remaining three cases exhibit a combination of both 3WJs and 4WJs (Figure 1B). Furthermore, there are 81 cases with PKs, out of which 58 include MWJs, and 23 consist solely of HPs. Additionally, the dataset includes seven cases with G4s, one of which is a short sequence containing only the G4 element, and the remaining six cases have the G4 as a part of HP structure.
Figure 1.
Dataset of reference and models of RNA structures. Venn diagrams showing the distribution of structural elements of RNA in the dataset: (A) hairpins, multiway junctions (MWJ), pseudoknots (PK) and G-quadruplex (G4) structures; (B) three-way junctions (3WJ), four-way junctions (4WJ) and five-way junctions (5WJ). Categories based on difficulty of modeling are ‘simple’, ‘moderate’, and ‘difficult’, with (C) example cases (PDB IDs: 1BYJ, 3Q3Z and 4GMA) and (D) RNA length distribution, and (E) RNA distribution in 30-nucleotide length bins. (F) Successful cases of modeling the RNA 3D structure. Each RNA 3D structure prediction method is represented in a distinct color. The models from simulations without and with using secondary structure restraints are shown in light and dark shades of the same color, respectively.
Based on the complexity of structural elements present in the RNA, we classified the dataset into three distinct categories: ‘simple’, ‘moderate’, and ‘difficult’. The ‘simple’ category consists of 45 HPs that feature only bulges or internal loops without additional structural elements. The ‘moderate’ category comprises 29 structures: 6 are MWJs with HPs, and 23 are HPs with PKs. The ‘difficult’ category encompasses 65 structures, which include 58 MWJs with PKs, and seven RNAs with G-quadruplexes. An example from the ‘simple’ category is the 16S RNA fragment (PDB ID: 1BYJ), which consists of two bulges and a tetraloop (Figure 1C, left) (60). The c-di-GMP-II riboswitch RNA (PDB ID: 3Q3Z) is a representative of the ‘moderate’ category, featuring an HP with loops interconnected with PKs (Figure 1C middle). In 3D, this RNA forms a PK helix with a kink-turn motif (61). From the ‘difficult’ category, the adenosylcobalamin riboswitch (PDB ID: 4GMA), which showcases both MWJs and PKs forming a complex structure, stands as an exemplary case (Figure 1C, right) (62).
The distribution of RNA lengths across different structural classes shows overlap (Figure 1D). Importantly, every RNA in our dataset exceeding 120 nucleotides, characterized by their complex structural elements, is categorized as ‘difficult’ (Figure 1E). Additionally, this classification encompasses a notable exception: a short RNA comprising 23 nucleotides, uniquely distinguished by the presence of a G-quadruplex (G4) as its sole structural element.
During the final stages of preparing this publication, AlphaFold 3 was released (58). To benchmark the performance of all ML-based methods (AlphaFold 3, DeepFoldRNA, and RhoFold), we developed two new datasets: Blind set 1 (B1) and Blind set 2 (B2) (see Supplementary Table S2).
AlphaFold 3 was last trained with data available up to October 31, 2021. DeepFoldRNA and RhoFold were last updated in March 2022 and October 2022, respectively.
Blind set 1 (B1) includes structures from the Protein Data Bank (PDB) released after November 1, 2022. This ensures that none of the structures in B1 were available during the training of AlphaFold 3, DeepFoldRNA, or RhoFold, providing an independent evaluation of these models. Verification was done using RNACentral's sequence search tool to confirm that no similar RNAs were released before 31 October 2022.
Blind set 2 (B2) includes everything in B1 plus additional structures released after 31 October 2021. These additional structures were also not used in the training of AlphaFold 3 but have homologous structures in the training sets of DeepFoldRNA, or RhoFold. This could slightly reduce the robustness of the comparisons by making the test set less challenging to DeepFoldRNA and RhoFold compared to B1. The purpose of B2 is to extend the evaluation of AlphaFold 3 by including a broader range of data.
Modeling of RNA 3D structures
We analyzed the performance of DeepFoldRNA, RhoFold, BRiQ, FARFAR2, SimRNA and Vfold2 programs with the sequence of RNA as a standard input for all methods.
DeepFoldRNA utilizes deep self-attention-based neural networks to predict spatial restraints, including distance maps and inter-residue orientations, which are converted into negative log-likelihood potentials (42). These potentials guide the L-BFGS folding simulations that generate full-length RNA structure models by minimizing with respect to backbone pseudo-torsion angles. Following initial folding simulations, DeepFoldRNA employs SimRNA to refine the generated RNA structure models. Despite DeepFoldRNA’s integration of SimRNA for model refinement, the core predictive mechanisms of DeepFoldRNA—especially its use of deep learning for restraint generation—are distinct from SimRNA’s simulation methods.
RhoFold generates MSA, processs it through a dual transformer-based model setup, comprising RNA Foundation Model (RNA-FM) and RhoFormer, to generate refined sequence embeddings (40). These embeddings are fed into an 8-layer structure module that iteratively predicts and refines the three-dimensional spatial configuration of RNA molecules. The system cycles predictions to enhance accuracy, ultimately outputting high-confidence, detailed RNA structures.
SimRNA is a Monte Carlo simulation tool for RNA structure prediction, uses a coarse-grained representation of RNA molecules to dynamically model their 3D structures (37). It includes several simulation modes, such as isothermal simulations, simulated annealing, and Replica Exchange Monte Carlo (REMC), to explore the RNA conformational space by overcoming energy barriers and sampling a diverse set of structural configurations. The SimRNA supports various restraints and constraints, such as secondary structure restraints and pairwise atom-atom distance constraints, which tailor the energy function (63).
FARFAR2 can construct detailed models of medium-sized RNAs and assembles RNA structures either de novo or from known segments (26). It features a fragment assembly protocol for modelling. This method offers various customization options such as user provided input template files, used of experimental data such as chemical shift and MOHCA-Seq (64).
BRiQ is an RNA structure prediction and refinement tool which leverages a nucleobase-centric sampling algorithm coupled with knowledge-based potential (38). This method focuses on enhancing the accuracy of RNA structural models by refining both local and global conformations around predicted or known base pairs.
Vfold2 pipeline offers an automated approach to predict RNA 3D structures from given sequences, leveraging a two-step modeling process (39). Initially, the pipeline uses the Vfold2D model (65) to predict the RNA’s 2D structure, setting a structural foundation that informs subsequent 3D modeling. For 3D structure prediction, the pipeline utilizes Vfold3D (32,66) and VfoldLA (33) models, which assemble A-form helices with loops and motif templates derived from existing RNA 3D structures, although predictions can be limited by the current scope of the template library.
Certain methods (FARFAR2, SimRNA and Vfold2) allowed for the incorporation of a given secondary structure, and these were tested both with and without this additional input. BRiQ, on the other hand, was run only with secondary structure restraints, as they are mandatory. The inclusion of secondary structure as an auxiliary input was decided upon because its prediction is relatively straightforward – there are well-established methods available as standalone and webservers (RNAFold (67), RNAStructure (68), IPKnots (69)) that forecast secondary structure with commendable accuracy (∼70%) (70,71). In the subsequent text, results for SimRNA, FARFAR2, and Vfold2, when run without incorporating secondary structure, will be denoted by their respective method names. However, in cases where these methods include secondary structure as an additional input, ‘_ss’ will be appended to the method name (for example SimRNA_ss). For modeling with secondary structures, ideal secondary structures based on the reference structure were employed.
For both DeepFoldRNA and RhoFold programs, we used only sequence as input for the modelling. The programs were set up locally along with the sequences from Rfam (72), RNACentral (73) and NCBI nucleotide (74) databases. Both programs generate multiple sequence alignments (MSA) to perform the prediction of secondary structures and other restraints used in the simulations. We also set up locally the external dependencies necessary to run the DeepFoldRNA program: PETfold (75), rMSA (76), SimRNA (37), QRNAS (77) and Spot-RNA-1D (78). The DeepFoldRNA program generated up to six models for each simulation while RhoFold produced a single model for each RNA.
The SimRNA method was run with and without secondary structure restraints. For each RNA, the program was run with eight independent REMC simulations starting with different random seeds, and with ten replicas per simulation. Each simulation was run for 16 million iterations. The simulations were performed following the protocols described in the book chapter on SimRNA (63) and the lowest energy structures from the top three clusters were selected for further analysis. This selection approach follows the study by Manfredonia et al. (79), which used SimRNA to investigate RNA binding sites, identifying these conformations as likely the most stable and biologically relevant within each cluster.
FARFAR2 was executed for one million cycles of Monte Carlo simulations, both with and without secondary structure restraints. It generated up to five representative models for benchmarking studies.
For the BRiQ method the simulations were only performed with secondary structure restraints. This method generated only one model per simulation and was used in further analysis.
The Vfold2 pipeline was run with and without secondary structure restraints. Without secondary structures, Vfold2 internally utilizes Vfold-2D to generate multiple alternative secondary structures, each leading to ensembles of 3D models labelled as npk1, npk2, pk1, pk2, etc., where ‘pk’ and ‘npk’ denote secondary structures generated with and without PK prediction, respectively. Up to five models from these ensembles were selected for further analysis. With secondary structure restraints, Vfold2 generated a single ensemble of models, from which up to the first five models were chosen.
We have provided the runtime and CPU information for one example case from each length bin in Supplementary Table S3. We performed all the simulations on CPUs. Unfortunately, we were unable to estimate the runtime for DeepFoldRNA and Vfold2 simulations from the available log files.
For both B1 and B2 datasets, we ran the AlphaFold 3 server (http://alphafoldserver.com/) with default settings. The predictions were run without any secondary structure restraints as the server does not support them. All other methods were run for all three datasets as described above in this section.
All models selected from each method underwent refinement using QRNAS, which was run with default settings for 5000 steps to optimize the structural geometry, including measures such as the clashscore. This refinement ensures that the quality of geometry is addressed and optimized as part of our pipeline.
Model evaluation metrics
The structural similarity between the native and the predicted 3D structures was measured using RMSD. Models were superimposed on the native structure using PyMOL (Schrödinger, Inc.) (80), and root mean square deviation values were calculated for all heavy atoms. In cases where reference structures solved by XRD had multiple biological assemblies in the asymmetric unit, superpositions were performed for all the assemblies. The reference assembly with the minimum RMSD to the model was selected for calculating other measures. For coordinates solved by NMR, all conformers were used for one-to-one comparison with the models. The reference conformer with the minimum RMSD to the model was then used for calculating other evaluation metrics. When RNA structure prediction programs generated more than one model, only the model with the lowest RMSD was retained for further calculation of performance measures, as described in this section.
The TM-score metric is used to assess the topological similarity of the model to the reference coordinates. The TM score is more sensitive to the global fold similarity than to the local structural variations and is length independent. The score has values in the range (0,1], where 1 indicates a perfect match between the model and reference structure. TM scores were calculated using the RNA-align program (81). For smaller RNAs the TM scores are unreliable (81), and based on our analysis, the threshold of RNA length at which the TM score accurately describes the predicted structures' accuracy is approximately 60 nt. Consequently, we excluded TM scores for RNAs below 60 nucleotides (Supplementary Figure S1) from our analysis.
The Interaction Network Fidelity (INF) is defined as the Matthews correlation coefficient (MCC) between the interactions of the reference structure and the predicted model, and it quantifies the overall agreement of a model with respect to the reference structure (82). A perfect model would have an INF value of 1, indicating an ideal match to the reference structure. Conversely, an INF value of 0 suggests a substantial deviation from the reference structure or a complete mismatch in the interactions in the predicted models. The INF_all values are used to assess the overall agreement of interactions between the predicted model and reference structure. The specific types of interactions can be assessed by INF_stack for stacking interactions, INF_wc for Watson-Crick interactions and INF_nwc for non-Watson-Crick interactions.INF values were calculated using the rna-tools program (83).
Evaluation of ligand binding interface
In this study, we employed the NACCESS (84), which is based on the Lee and Richards algorithm (85), to calculate the solvent accessible surface area (SASA) of RNA, and the complexes. Alternative methods to calculate SASA, such as AREAIMOL (86), FreeSASA (87), Surface Racer (88), and PDBASA (part of PDBREMIX https://github.com/boscoh/pdbremix), could be used and are reviewed in a book chapter on tools for analyzing protein-RNA interfaces, applicable to RNA-Ligand complexes (89). The structures of the unbound forms of RNA, used in this calculation, were taken from the complex. Atoms that lose SASA of at least 0.01 Å2 upon complexation were designated as interface atoms. The residues containing these interface atoms were designated as interface residues (90). Furthermore, the list of interface residues was expanded to incorporate their pairing partners, based on the secondary structure derived from the native RNA structure.
For the models generated by various methods, the interface RMSD (I-RMSD) values were calculated to assess their accuracy. Interface residues were extracted from both the native structures and the models and were then superposed using PyMOL (Schrödinger, Inc.) (80). The superposed structures were then used to calculate the I-RMSD for all heavy atoms.
Results and discussion
Overview of performance of RNA 3D modeling methods
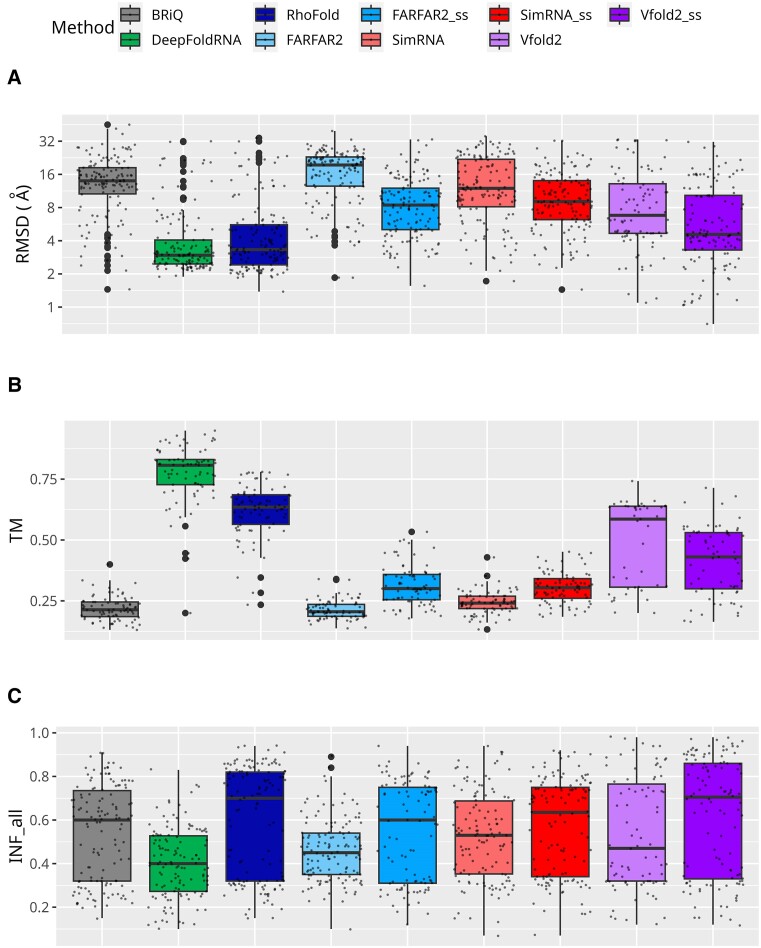
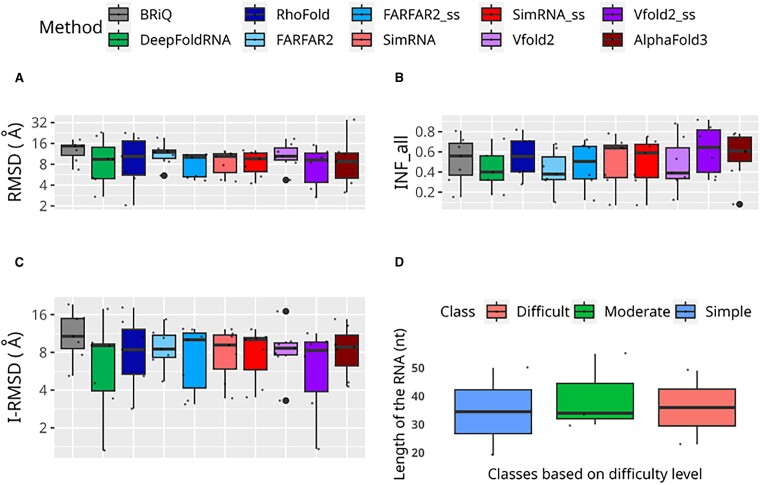
Among the six methods tested, Vfold2 was the only one that failed to generate a model for some RNA molecules. The Vfold2 and Vfold2_ss generated models for 90 and 123 out of 139 cases, respectively (Figure 1F). To evaluate available standalone programs, we utilized RMSD value which is one of the widely used measures to quantify the differences between the superposed coordinates of reference and model structures. Among all the methods tested, the models generated by ML-based method DeepFoldRNA have the lowest median (2.96 Å) and spread (Interquartile range, IQR 1.60 Å) of RMSD values (Figure 2A). Another ML-based method RhoFold has slightly higher RMSDs (median 3.32 Å) with a larger spread of values (IQR 3.17 Å). For FARFAR2_ss, SimRNA_ss and Vfold2_ss, the RMSD values are better than without introducing the secondary structural restraints into the modeling pipeline. Among all the non-ML based methods tested in this study, the lowest median RMSD value (4.56 Å) is observed for models generated by the Vfold2_ss. It is noteworthy that the models exhibiting the lowest RMSD values (0.70 Å) among all tested methods were generated by Vfold2_ss.
Figure 2.
Performance of RNA 3D modeling methods. Performance measures for RNA 3D structure prediction methods are displayed across the overall dataset: (A) RMSD, (B) TM and (C) INF_all. Logarithmic scale is used for the ordinates for RMSD. TM scores for below 60 nucleotides are not included. Each method is represented by a unique color. Models generated without secondary structure restraints are shown in lighter shades, while those with restraints are in darker shades of the same color.
Among all the methods tested in this study, the DeepFoldRNA method has the highest TM values with median 0.81 (Figure 2B). RhoFold has slightly lower TM values than DeepFoldRNA with median 0.64, however, the values are higher than all non-ML based methods tested in this study. Among the non-ML methods, the highest median values are observed for Vfold2 (0.59) and Vfold2_ss (0.43). Incorporating secondary structural restraints does not have a big impact on the median TM scores of FARFAR2 (0.30 vs 0.21), SimRNA (0.30 vs 0.24).
Despite achieving the lowest RMSD values and the highest TM values, the DeepFoldRNA method shows relatively low values for INF_all (median: 0.40, Figure 2C). However, the RhoFold method exhibits second best results of the median RMSD and TM values has the second highest INF_all median value (0.70) among all tested methods. Among all the programs tested, Vfold2_ss has the highest median value (0.71) INF_all. The models generated by BRiQ program have INF_all (median = 0.60) comparable to other non-ML methods run with secondary structures. Introduction of secondary structure restraints improves the INF_all for all methods. The median values do not change significantly for FARFAR2 (0.45 to 0.58) and SimRNA (0.53 to 0.63). However, the third quartile (Q3) values increase significantly for FARFAR2 (0.54 to 0.75). In contrast, Vfold2_ss outperforms Vfold2, with a median INF_all value of 0.71 compared to Vfold2’s 0.47. The results for INF_stack are very similar to those obtained for INF_all (Supplementary Figure S2A).
The INF_wc median values close to zero are observed for models generated using the DeepFoldRNA and FARFAR2 methods (Supplementary Figure S2B). Additionally, the models generated by both DeepFoldRNA and FARFAR2 show the lowest values for IQR for INF_wc indicating inferior quality of the Watson-Crick interaction predicted by these methods. Among the ML-based methods tested in this study, RhoFold has higher median INF_wc values (0.77) and shows significantly better prediction of Watson-Crick interactions compared to the DeepFoldRNA. Introducing secondary structure restraints in the modeling pipelines improves the median INF_wc values for all three methods, FARFAR2 (0.0 to 0.63), SimRNA (0.26 to 0.74) and Vfold2 (0.25 to 0.90). The models generated by BRiQ program have INF_wc (median = 0.76) comparable to other non-ML methods run with secondary structures. Among all the methods tested, the best median of INF_wc values are observed for the models generated by Vfold2 pipeline with secondary structure restraints.
The distribution of INF_nwc values across the models generated by all methods indicates a general shortfall in accurately predicting non-Watson-Crick interactions, as evidenced by the pervasive presence of median values at or near 0 (Supplementary Figure S2C). This suggests that there is room for significant improvement in this area for all the methods evaluated. Incorporating secondary structures into the prediction workflows slightly increases the Q3 values of INF_nwc across the three methods that were evaluated: FARFAR2 (0.09 to 0.27), SimRNA (0.17 to 0.36), and Vfold2 (0.44 to 0.62). Moreover, highest Q3 values are observed for Vfold2_ss (0.62) and RhoFold (0.55). Furthermore, the data points above the third quartile (Q3) for the RhoFold cluster range between 0.55 and 1 (median 0.67), whereas for Vfold2_ss, they are distributed between 0.62 and 1 (median 0.82). This indicates that among the models with the highest INF_nwc values, the results from Vfold2 are slightly superior.
The overview of RNA 3D modeling methods illuminates the varied capabilities and limitations of different computational approaches in RNA structure prediction. Both ML-based methods, DeepFoldRNA and RhoFold, consistently show superior performance in RMSD and TM score metrics (Figure 2A,B), demonstrating their strength in structural accuracy and global fold similarity. However, DeepFoldRNA lags in Interaction Network Fidelity, particularly in INF_all and INF_stack, where RhoFold and Vfold2_ss exhibit stronger results (Figure 2C, Supplementary Figure S2A-C). The analysis of INF_wc and INF_nwc further underscores the necessity for improvements, especially in accurately predicting non-Watson-Crick interactions. We should also point out that ML-based methods may show artificially inflated results due to the presence of same or homologous structures in the training sets. Therefore, we constructed blind test set B1 to evaluate the DeepFoldRNA and RhoFold methods and is detailed in a subsequent section. Overall, this study not only highlights the strengths of ML-based methods in certain aspects of RNA modeling but also identifies critical areas for enhancement. Among the non-ML-based methods, the Vfold2 method predicts models with lower RMSD (Figure 2A), higher TM scores (Figure 2B) and better INF values (Figure 2C, Supplementary Figure S2A-C), making the models more reliable in terms of global fold and local structures. The contrasting performances across various metrics and methods underscore the importance of selecting the appropriate tool for specific modeling challenges and pave the way for future advancements in RNA 3D structure prediction. Additionally, it is important to note that, results of template-based methods like Vfold2 and FARFAR2 can be dependent on the availability of appropriate templates. This underscores the importance of utilizing well-curated templates to enhance prediction accuracy.
Performance across different difficulty classes
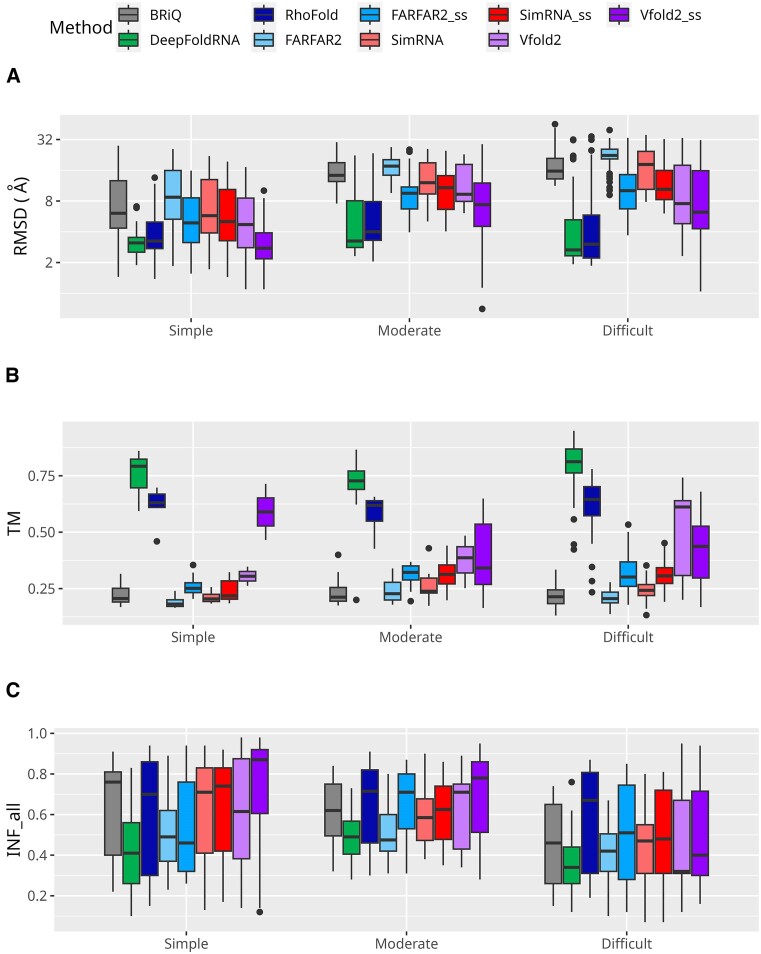
Based on the complexity of structural elements present in the RNA, we classified the dataset into three distinct categories: ‘simple’, ‘moderate’, and ‘difficult’ (see Materials and Methods section). Across ‘moderate’ and ‘difficult’ categories, the models generated by ML-based methods exhibit the lowest median and IQR RMSD values (Figure 3A). Among them DeepFoldRNA has slightly lower median values than RhoFold in ‘moderate’ and ‘difficult’ categories (3.25 Å and 2.66 Å versus 4.00 Å and 3.03 Å, respectively). In addition, in ‘simple’ category, models generated by Vfold2_ss (median 2.76 Å) exhibit better results to ML-based methods (medians of 3.11 Å and 3.25 Å for DeepFoldRNA and RhoFold, respectively). Moreover, among the non-ML-based methods, Vfold2_ss also has the lowest median values in ‘moderate’ (7.38 Å) and ‘difficult’ (6.21 Å) categories. It is noteworthy that the models with the lowest RMSD across all categories were generated using Vfold2 in ‘simple’ category (1.10 Å) and Vfold2_ss in both ‘moderate’ and ‘difficult’ categories (0.70 Å and 1.04 Å, respectively). In all three categories, the TM values are highest for the models generated by ML-based methods, however DeepFoldRNA slightly outperformed RhoFold in ‘simple’, ‘moderate’ and ‘difficult’ categories (medians median 0.79, 0.73 and 0.81 versus 0.63, 0.62 and 0.64, respectively) (Figure 3B). This difference between ML-based and non-ML-based methods is more profound in the ‘moderate’ and ‘difficult’ categories, compared to the ‘simple’ category. Unexpectedly, for the ‘difficult’ category, both ML-based methods and Vfold2 (including Vfold2_ss) exhibit significantly better results compared to the ‘moderate’ category, considering both median RMSDs (2.66, 3.03, 7.65 and 6.21 Å versus 3.25, 4.00, 9.32 and 7.38 Å, respectively) and TM scores (0.81, 0.64, 0.61 and 0.44 versus 0.73, 0.62, 0.39 and 0.34, respectively). While the improved performance of ML-based methods could be attributed to overtraining, this explanation appears less plausible for the results observed with Vfold2 and Vfold2_ss.
Figure 3.
Performance of RNA 3D modeling methods across difficulty classes. Performance measures for RNA 3D structure prediction methods are displayed across the different difficulty classes: (A) RMSD, (B) TM and (C) INF_all. Logarithmic scale is used for the ordinates for RMSD. TM scores for below 60 nucleotides are not included. Each method is represented by a unique color. Models generated without secondary structure restraints are shown in lighter shades, while those with restraints are in darker shades of the same color.
Similar to the observations for the overall dataset, the DeepFoldRNA method performs poorly in all categories for INF_all, INF_stack, INF_wc and INF_nwc (Figure 3C, Supplementary Figure S3A–C), while RhoFold has still quite good results among all methods in all categories, considering both IQR and median. The models from Vfold2_ss pipeline have the highest INF_all median values in ‘simple’ and ‘moderate’ categories (0.87, 0.78, respectively), while for ‘difficult’ categories all methods have relatively low medians of INF_all (Figure 3C). In general, it can be observed that methods incorporating secondary structure as an additional input yield superior performance compared to those without secondary structure. In the ‘simple’ category for models generated by the FARFAR2_ss method, we observe a slight decrease in the median values from 0.49 to 0.46, while the Q3 values increase from 0.62 to 0.76. The methods with the highest INF_all scores in the ‘simple’ category include Vfold2_ss, BriQ, SimRNA_ss, SimRNA and RhoFold (medians 0.87, 0.76, 0.74, 0.71 and 0.70, respectively). Notably, incorporating a secondary structure in SimRNA does not significantly impact the results here.
The highest median INF_stack values in the ‘simple’ (0.85) and ‘moderate’ (0.78) categories are observed in models generated by Vfold2_ss pipeline (Supplementary Figure S3A). In the ‘moderate’ categories, models generated by Vfold2_ss, RhoFold and exhibit similarly high INF_stack median value (0.75 and 0.78, respectively). For ‘difficult’ category RhoFold, FARFAR2_ss, SimRNA_ss, SimRNA and Vfold2_ss slightly outperform other methods with median INF_stack around 0.6. However, RhoFold has highest Q3 value (0.83), while models with the highest INF_stack were generated by Vfold2 (0.95) and Vfold2_ss (0.94), followed by RhoFold (0.89).
Unlike INF_stack, which shows minimal variation across methods, INF_wc values significantly differ between methods (Supplementary Figure S3B). In the ‘simple’ category, Vfold2_ss, BRiQ, SimRNA_ss, SimRNA, and RhoFold exhibit high median values—above 0.85. In contrast, DeepFoldRNA and FARFAR2 display unexpectedly low results, with medians around zero. The scenario is similar in ‘moderate’ category, except that FARFAR2_ss's median increases to 0.93, while SimRNA’s median decreases to 0.44. In the ‘simple’ and ‘moderate’ categories, the highest median values of INF_wc are observed for Vfold2_ss (1.0) and FARFAR2_ss (0.93), respectively. However, in the ‘difficult’ category, models generated by all methods have median values close to zero for INF_wc.
Across all ‘difficulty’ classes, INF_nwc values are consistently low for each method (Supplementary Figure S3C). These results are unsurprising given that one of the foremost challenges in both secondary and tertiary RNA structure prediction is the accurate prediction and reconstruction of non-canonical base pairs (91,92).
In summary, our comparative analysis across different complexity categories demonstrates a consistent trend in the superior performance of ML-based methods, particularly DeepFoldRNA, in terms of RMSD and TM values, across ‘simple’, ‘moderate’, and ‘difficult’ classes of RNA structures. Despite this, DeepFoldRNA exhibits limitations in accurately predicting interactions, as evidenced by its lower INF metrics in all categories. RhoFold, while trailing slightly behind DeepFoldRNA in RMSD and TM scores, shows a notably better performance in INF metrics, indicating a more balanced approach between structural accuracy and interaction fidelity. The Vfold2 method, particularly when augmented with secondary structure restraints, not only stands out among non-ML based methods but also demonstrates competitive performance in TM scores and INF metrics, rivaling that of ML-based approaches across various levels of structural complexity. Furthermore, SimRNA and SimRNA_ss demonstrate high precision in predicting interaction fidelity, which diminishes as the complexity of the RNA structure increases. These findings highlight the nuanced trade-offs inherent in different RNA modeling methods and underscore the importance of selecting appropriate tools based on the specific requirements of the structural complexity being addressed.
Performance across different lengths of RNA
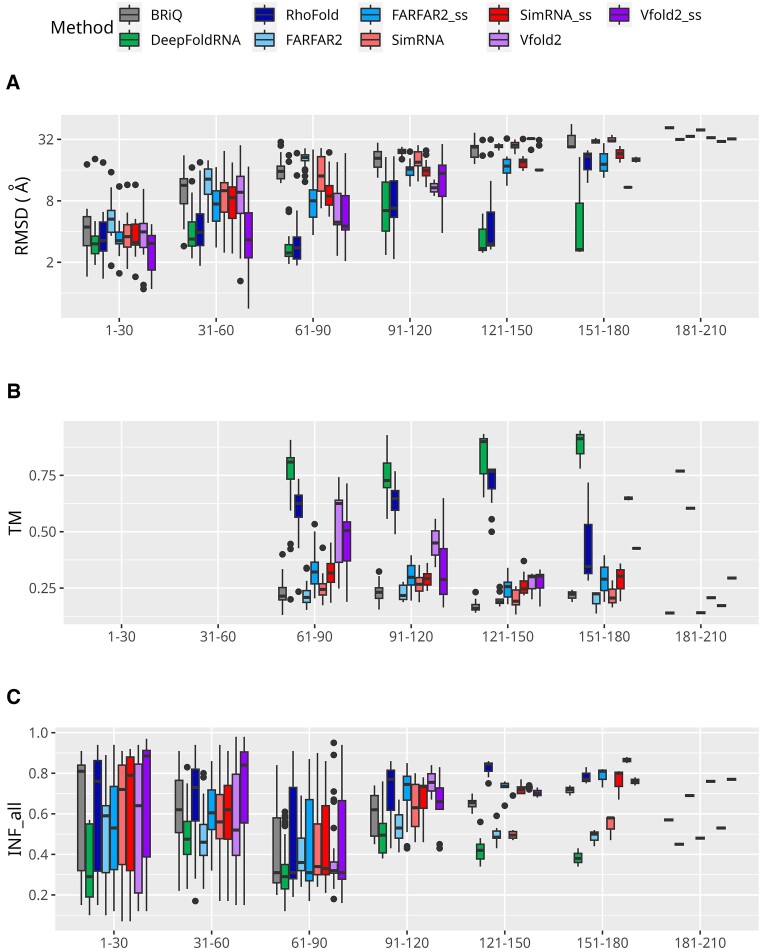
To investigate the impact of RNA length on the quality of model predictions, we divided the test set into intervals, each differing by 30 nucleotides. The distribution of RNA structures across these intervals is detailed in Supplementary Table S1. Notably, the final interval comprises only one structure (Figure 1E).
The length of the RNA has a significant effect on the quality of the models generated. This phenomenon is both natural and predictable, as the increase in RNA length leads to a higher proportion of complex structures. In the initial length interval of RNA (1–30 nt), predominantly ‘simple’ structures are observed. And in this category, all methods have the best results. In the subsequent interval (30–40 nt), the prevalence of ‘simple’ and ‘moderate’ structures becomes equal, accompanied by a minor presence of ‘difficult’ structures. Here all methods, except for ML-based methods and Vfold2_ss, significantly increase RMSD. In the later categories, the majority are classified as ‘difficult’ structures (Figure 1E). As expected, the general trend observed in this study is that the RMSD values increase as the length of the RNA increases (Figure 4A). This increase is more pronounced for non-ML based methods than for ML-based methods. Furthermore, for lengths below 60 nucleotides, a lot of models generated by Vfold2_ss have reasonable RMSD values (median 3.3 Å). In the length range 151–180, there only three cases and both Vfold2 and Vfold2_ss generated models only for two of them. No method accurately predicted the structure of the longest RNA molecule, from the category 181–210 nt.
Figure 4.
Performance of 3D modeling methods across RNA lengths. Performance measures for RNA 3D structure prediction methods are displayed according to RNA sequence length. Metrics include (A) RMSD, (B) TM and (C) INF_all, organized into sets of box plots representing 30-nucleotide length bins. The TM scores for RNAs below 60 nucleotides are not included. Logarithmic scale is used for the ordinates for RMSD. Each method is represented by a unique color. Models generated without secondary structure restraints are shown in lighter shades, while those with restraints are in darker shades of the same color.
The TM score displays values akin to those anticipated, considering the RMSD: methods characterized by a low median RMSD, including both ML-based approaches and Vfold2_ss, exhibit higher median TM score values (Figure 4A,B). Across all length bins, models from the DeepFoldRNA method have the highest TM score (>0.7) in all length categories. These differences observed in TM scores between models from DeepFoldRNA and other methods are higher for longer RNAs (Figure 2C). Both ML-based methods tested in this study outperform non-ML-based methods for longer RNAs. Among the non-ML-based methods tested, models from the Vfold2_ss program has the highest TM scores.
For the DeepFoldRNA program, the INF_all values are poor across all the length bins tested in this study, while RhoFold performs much better (Figure 4C). Regarding the INF metrics, it is difficult to observe trend correlating INF_all, INF_stack and INF_wc values with the length of the RNA modeled (Figure 4C, Supplementary Figure S4A–C). However, it appears that for all methods, models generated for RNA within the range of 60–90 nucleotides exhibit the poorest INF values, whereas those within the 1–30 nucleotides range display the highest INF values. The trend of low INF_nwc values persists irrespective of RNA length (Figure 3C).
To summarize, our analysis reveals a clear influence of RNA length on the quality of structural models generated by various computational methods. A key observation is the increasing RMSD values with RNA length, more so for non-ML based methods, indicating a decline in model accuracy for longer RNA sequences. While ML-based methods, particularly DeepFoldRNA, maintain better performance across all RNA lengths, their limitations become more apparent in longer RNAs, as reflected in the higher RMSD values. In terms of interaction network fidelity metrics, such as INF_all, INF_stack, INF_wc and INF_nwc, the absence of a consistent trend across varying RNA lengths suggests that these aspects of model quality are less dependent on RNA length. Overall, these findings underscore the need for continued refinement of RNA modeling methods, particularly for longer RNA sequences, to improve both structural accuracy and interaction fidelity.
Performance across different structural classes
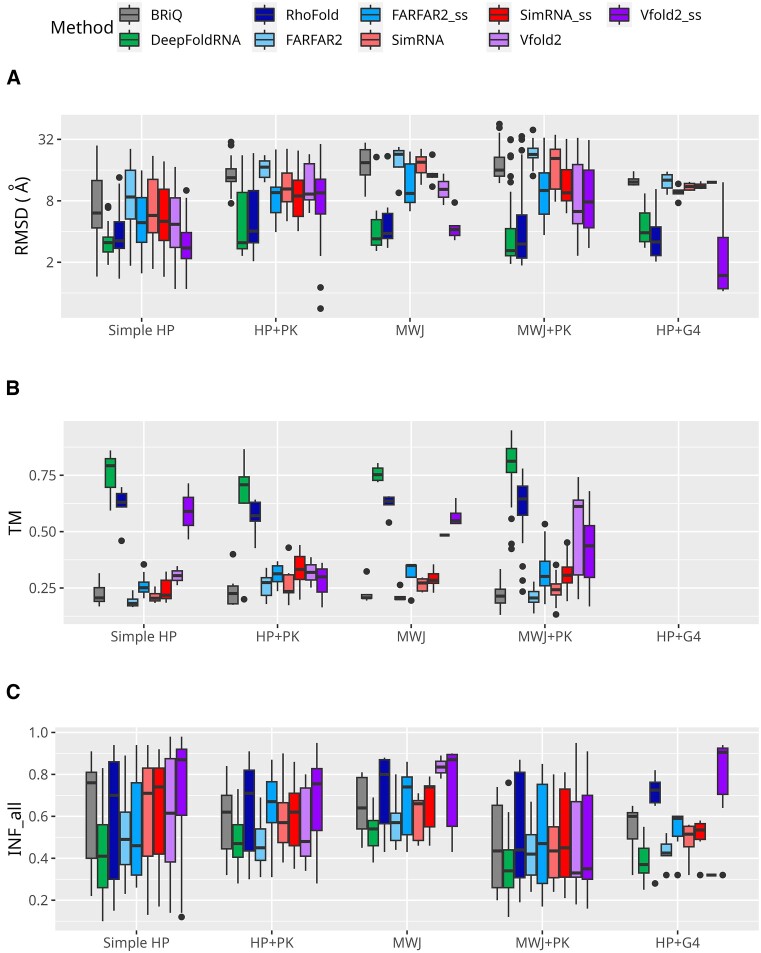
We also examined the performance of methods predicting different types of RNA structures. We divided the dataset into the following groups: simple HPs, HP + PK, MWJs, MWJ + PK, G4 and HP + G4 (see Materials and methods section).
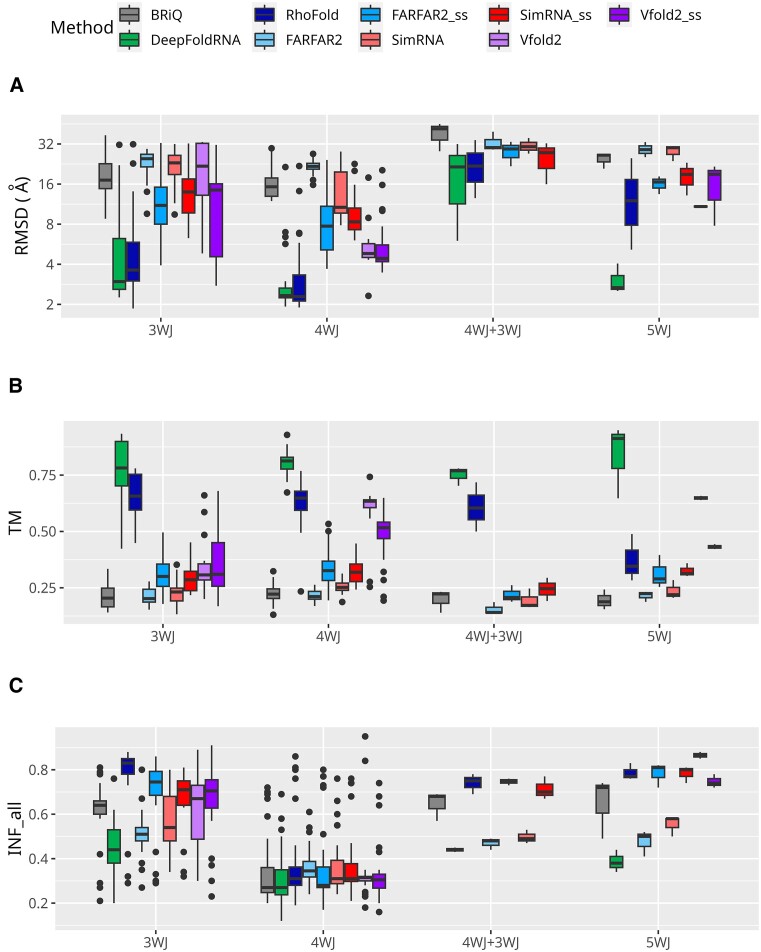
For all structural categories except simple G4, ML-based algorithms achieved quite good results with median RMSDs ranging between 2.60 and 4.04 Å (Figure 5A). Among non-ML methods, Vfold2_ss demonstrated the best performance across all categories. Notably, in categories such as simple HP and HP + G4, Vfold2_ss exhibited the lowest median RMSD value (2.76 Å and 1.50 Å, respectively). Short RNA with only G4 structure was incorrectly modeled either as a HP or a single-stranded structure by all tested methods. However, when G4 is part of a HP RNA, Vfold2 and RhoFold generally model it correctly. The iMango-III aptamer, an RNA structure incorporating a G4 in an HP (93), is a case in point (Supplementary Figure S5). If we examine different types of MWJs separately, we observe that, for simpler structures, ML-based methods, especially DeepFoldRNA, frequently propose reasonable models (RMSD < 6 Å) (Figure 6A). However, very complex structures consisting of a combination of 3WJ and 4WJ are not well-modeled by any of the tested methods. Non-ML-based methods struggle to predict the structures of MWJs, as evident in Figure 6A, where their medians significantly exceed those obtained with ML-based methods. For 4WJs, among non-ML methods, the lowest RMSD medians are observed for Vfold2_ss (4.42), Vfold2 (4.82), FARFAR2_ss (7.73) followed by SimRNA_ss (8.33). Notably, all methods achieved their best results in the 4WJ category. For the structures with 4WJ + 3WJ, Vfold2 did not produce any models.
Figure 5.
Performance of 3D modeling methods across RNA structural classes. Performance measures for RNA 3D structure prediction methods are displayed according to structural classes where HP, PK, MWJ and G4 stand for hairpins, pseudoknots, multi-way junctions and G-quadruplexes, respectively. Metrics include (A) RMSD, (B) TM and (C) INF_all. Logarithmic scale is used for the ordinates for RMSD. TM scores for below 60 nucleotides are not included and hence the HP + G4 category is not shown in TM plot. The simple G4 category is also excluded from the plot as there is only one case. Each method is represented by a unique color. Models generated without secondary structure restraints are shown in lighter shades, while those with restraints are in darker shades of the same color.
Figure 6.
Performance of 3D modeling methods for different multiway junctions. Performance measures for RNA 3D structure prediction methods are displayed for various MWJs, where 3WJ, 4WJ and 5WJ stands for three-way, four-way and five-way junctions, respectively. Metrics include (A) RMSD, (B) TM and (C) INF_all. Logarithmic scale is used for the ordinates for RMSD. TM scores for RNAs below 60 nucleotides are not included. Each method is represented by a unique color. Models generated without secondary structure restraints are shown in lighter shades, while those with restraints are in darker shades of the same color.
In all the categories, models generated by ML-based methods attain reasonable TM values (Figure 5B). In Simple HP, MWJ, and MWJ + PK categories, in addition to ML-based methods, TM score medians achieved by Vfold2 and Vfold2_ss are reasonable. Specifically, Vfold2_ss shows medians of 0.59 for Simple HP, 0.55 for MWJ, and Vfold2 posts a median of 0.44 in the MWJ + PK category. DeepFoldRNA demonstrates superior performance, producing models with higher TM scores than all other methods across all types of MWJs modeled in this study followed by RhoFold and Vfold2 with Vfold2_ss in the mentioned categories (Figure 6B). None of the models generated by non-ML-based methods attain a TM-score above 0.75. The TM score values for the G4 and HP + G4 categories are not considered as the RNAs are less than 60 nucleotides.
For all categories except MWJ + PK, the highest median INF_all scores are seen in models generated by Vfold2_ss (Figure 5C). For MWJ + PK, medians for all methods are very similar, lying within the range of 0.3 to 0.5. RhoFold, FARFAR2_ss, SimRNA_ss, Vfold2, Vfold2_ss and BRiQ consistently demonstrate significantly higher INF_all medians for 3WJ (0.83, 0.75, 0.71, 0.67, 0.71, and 0.64, respectively) and 5WJ (0.77, 0.81, 0.80, 0.87, 0.74 and 0.72, respectively) categories when compared to other methods. For the 4WJ + 3WJ category, RhoFold the best median INF_all values are observed for RhoFold (0.75), FARFAR2_ss (0.75), SimRNA_ss (0.70) and BriQ (0.68). For the 4WJ all methods yield very similar results, characterized by low medians (0.27 to 0.35) (Figure 6C).
For simple HPs, Vfold2_ss has the highest median INF_stack values (0.85), followed by SimRNA_ss (0.73), BriQ (0.72), SimRNA (0.70), Vfold2 (0.68), RhoFold (0.67) and FARFAR2 (0.62) (Supplementary Figure S6A). For HPs with PKs, MWJ and HPs with G4s, Vfold2_ss again has the highest INF_stack median (0.78) but is followed by FARFAR2_ss (0.73), RhoFold (0.72), SimRNA_ss (0.68), Vfold2 (0.63) and BriQ (0.63). For MWJ + PK category the highest median INF_stack is observed for SimRNA_ss (0.63), followed by FARFAR2_ss (0.62) and RhoFold (0.61). No methods exhibit reasonable INF_stack values for the simple G4 structure (0.12 to 0.24). However, the highest median INF_stack value is observed in HP + G4 category for Vfold2_ss (0.92). Among ML-based methods, DeepFoldRNA exhibits poorer INF_stack values compared to RhoFold in all categories. Among the different MWJs modeled in this study, 3WJs, 5WJ and 4WJ + 3WJ have better median INF_stack values (0.54 to 0.83, 0.43 to 0.86, and 0.52 to 0.77, respectively) than 4WJ (0.36 to 0.47) for all methods (Supplementary Figure S7A). DeepFoldRNA consistently has the lowest INF_stack medians values across all types of MWJs modeled.
Across all structural classes of RNAs, DeepFoldRNA exhibits poor INF_wc values compared to other methods (Supplementary Figure S6B). For simple HPs, HPs with PKs, and MWJs, models generated by Vfold2_ss, BriQ, SimRNA_ss, and RhoFold have high INF_wc values (medians above 0.85). In the case of HPs with G4 structures, Vfold2_ss, BRiQ, RhoFold and FARFAR2_ss, produce models with similarly high INF_wc values (medians above 0.9). However, for MWJs with PKs, while these methods show a broader range of INF_wc values, the median values are close to zero. Notably, the INF_wc median values for 3WJs (above 0.82) and 5WJs (above 0.88) obtained by BriQ, RhoFold, FARFAR2_ss, SimRNA_ss and Vfold2_ss are reasonable compared to all methods for 4WJs. These methods also have reasonable median INF_wc for 4WJ + 3WJs modeled in this study (above 0.8) except for Vfold2_ss which did not generate any models in this category (Supplementary Figure S7B).
For INF_nwc, the results are consistently poor across all methods. However, the models generated by Vfold2_ss and Vfold2 in the MWJ category, as well as the Vfold2_ss and RhoFold models in the HP + G4 category, exhibit INF_nwc medians above 0.5. (Supplementary Figure S6C). In the analysis of different MWJs within this study, RhoFold for 3WJ has a median of 0.58. For 5WJs, RhoFold, BRiQ, FARFAR2_ss, SimRNA_ss, and Vfold2 register median INF_nwc values exceeding 0.5 (Supplementary Figure S7C).
In summary, this comprehensive analysis of RNA structural classes reveals distinct patterns in the performance of various modeling methods. DeepFoldRNA consistently shows strong capabilities in generating models with low RMSD values across most classes, particularly excelling in simple HPs and various MWJ structures. However, its performance in terms of INF metrics, notably INF_wc and INF_nwc, is less robust compared to other methods. Vfold2, especially when supplemented with secondary structure restraints, emerges as a robust performer in both RMSD and TM score evaluations, particularly for HPs with G4 structures and in the INF metrics across several classes. RhoFold, while showing a broader range in some metrics, consistently ranks high in INF_stack values. Utilizing their default configurations, both SimRNA and FARFAR2 methodologies demonstrate higher RMSD metrics and reduced TM scores. Nonetheless, they exhibit a notable capacity for modeling local interactions with quite high precision, as evidenced by INF values. In addition, these methods provide a comprehensive array of options for the advanced user. Significantly, the integration of secondary structure data consistently enhances the quality of outcomes produced by SimRNA and FARFAR2. This enhancement suggests that the incorporation of experimental data into RNA structure modeling significantly augments the robustness of the resulting models. This enhancement is reflected in the CASP (52,94) and RNA-Puzzles (49,50) results, where the outputs from experienced groups significantly outperform those from servers (http://www.rnapuzzles.org/results/). These findings highlight the nuanced strengths and limitations of each method, underscoring the importance of method selection based on the specific structural characteristics of the RNA being modeled. These insights into the diverse performance across methods not only highlight their individual strengths and weaknesses but also underscore the critical aspects to consider when selecting an appropriate modeling approach for specific RNA structures.
Ligand binding interfaces
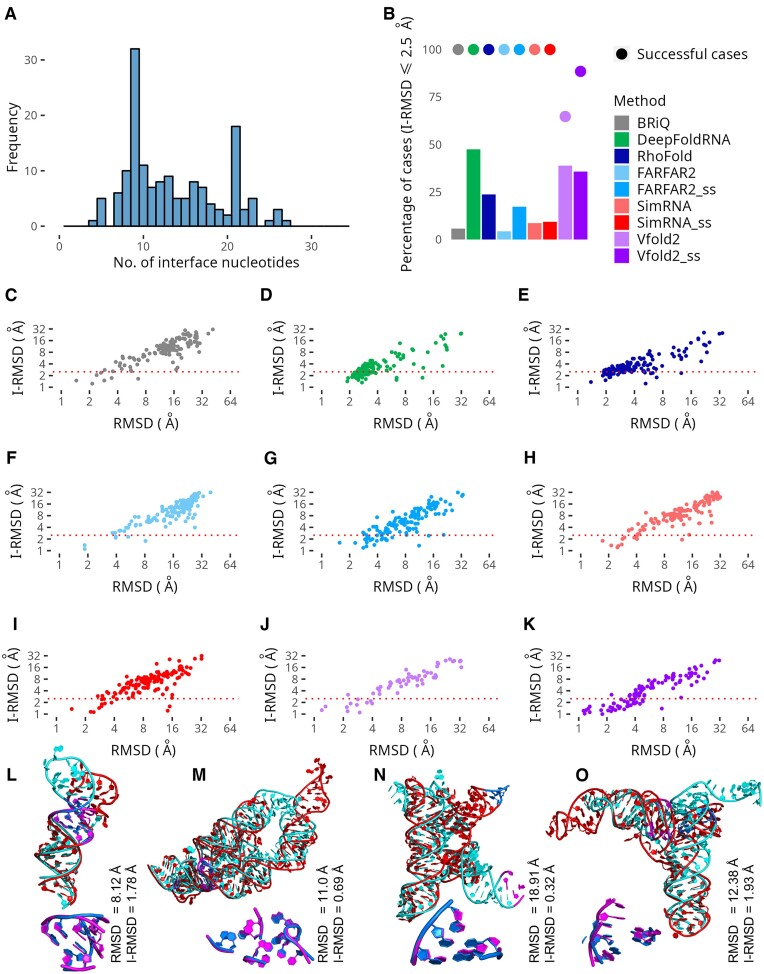
The quality of the binding site is crucial for the efficacy of virtual screening processes. Therefore, we evaluate the precision of predicting the binding pocket using various methods. The ligand binding interfaces were calculated as described in the Materials and Methods section. Analysis of the RNA–ligand interface size, as illustrated in Figure 7A, uncovers a bimodal distribution with two significant peaks representing interface sizes of nine and twenty-one nucleotides, respectively. The primary peak at nine nucleotides signifies the most prevalent interface size, while the secondary peak at 21 nucleotides highlights a distinct subset of RNA–ligand complexes, suggesting a broad structural diversity within RNA–ligand interactions. This diversity, underscored by the distribution's right-skewness (0.6597) and slight negative kurtosis (–0.3724), hints at varying levels of specificity and affinity across different binding interfaces.
Figure 7.
Ligand binding interfaces. (A) Distribution of ligand binding pocket size. (B) Bar plots showing the percentage of successful modeling cases achieving an interface RMSD (I-RMSD) of 2.5 Å or below. The percentage of successful modeling cases is also provided as points (•) on the plots for comparison. (C–K) Plot of I-RMSD vs RMSD for methods tested in this study. Each method is represented by a unique color in panels C–K. Models generated without secondary structure restraints are shown in lighter shades, while those with restraints are depicted in darker shades of the same color. Example cases of RNA models include: (L) FARFAR2 model of 7SK small nuclear RNA (PDB ID: 2KX8), (M) Vfold2 model of the lysine riboswitch (PDB ID: 3D0U), (N) BRiQ model of yybP-ykoY riboswitch (PDB ID: 4Y1M), and (O) SimRNA model of class III preQ1 riboswitch (PDB ID:4RZD). For these models, each panel displays two images: the upper image showing the superpositions of models (red) to the native structure (cyan), and the lower image highlighting the superpositions of corresponding ligand binding regions (in marine and magenta, respectively).
Further delving into the complexities of RNA–ligand interaction structures, Supplementary Figure S8 offers a detailed visualization of six diverse RNA–ligand complexes. These examples span a range of interface sizes from minimal contacts to extensive binding surfaces, providing a visual testament to the variability inherent in RNA–ligand complexes. Such insights are indispensable for understanding the nuanced mechanisms of RNA functionality and for advancing the design of RNA-targeted therapeutic interventions.
The assessment of ligand binding interface modeling is conducted through interface RMSD (I-RMSD) values. As the threshold for interface RMSD (I-RMSD) increases from ≤1 to ≤ 4 Å across different RNA 3D structure prediction methods, there is a general trend of increasing success rates, indicating a higher percentage of models achieving the specified accuracy (Supplementary Figure S9). The 2.5 Å threshold for interface RMSD (I-RMSD) is chosen to strike a balance between strict accuracy demands and the realistic achievability of model predictions. Across the entire dataset, a moderate to strong correlation is observed between I-RMSD and overall RMSD, as well as between I-RMSD and TM scores for most methods, corroborated by significant P-values that confirm statistical significance (Supplementary Table S4). However, this relationship changes when the analysis is restricted to models with an I-RMSD of 2.5 Å or less. In this subset, the correlations weaken substantially or disappear, suggesting that high-fidelity interface modeling may not always align with the overall model accuracy or TM scores (Figure 7C–K). This divergence suggests that the precision of binding site modeling might be influenced by factors not reflected in the metrics that assess the accuracy of the entire structure.
The number of models reaching the I-RMSD threshold (≤2.5 Å) varies considerably among the methods (Figure 7B). DeepFoldRNA, RhoFold, BRiQ, FARFAR2, and SimRNA consistently generated models for all 139 cases. In contrast, Vfold2 produces models for only 90 cases while Vfold2_ss for 123 cases (Figure 1F). Among all evaluated methods, DeepFoldRNA yields the highest percentage of models with the most accurate interface (47.50%), with Vfold2 and Vfold2_ss close behind at 38.9% and 35.8%, respectively. A considerable number of accurate interfaces were also generated by RhoFold (23.7%), and FARFAR2_ss (17.3%). The inclusion of secondary structure information does not significantly enhance the results obtained by SimRNA. However, both SimRNA and SimRNA_ss successfully reproduced the interaction sites for a greater number of RNA-small molecule complexes compared to FARFAR2 (8.63% and 9.35% versus 4.32%, respectively). The improvement associated with the use of secondary structure is primarily related to global folding patterns and Watson-Crick interaction (Figures 2–6, Supplementary Figures S2–S4 and S6–S7) rather than to the precision of the binding pocket accuracy, which often includes unpaired nucleotides. (Figure 7B).
In this study, we delve into the evaluation of modeling methods, focusing on their ability to predict interaction sites for small molecules accurately. As can be observed in Figure 7, panels C–K, each method yields complexes with poorly predicted global RNA folds, yet with precisely predicted interaction sites for small molecules, as indicated by high RMSD and low I-RMSD values. This increases the number of RNA models that can be used as starting points for virtual screening. The 7SK small nuclear RNA (PDB ID: 2KX8), is an example of ‘simple’ category with a simple HP structure, is 42 nucleotides long. The FARFAR2 model has an RMSD of 8.12 Å and an I-RMSD of 1.78 Å, indicating a significant level of accuracy in the binding region (Figure 7L). In the case of the lysine riboswitch (PDB ID: 3D0U), which spans 161 nucleotides and falls into the ‘difficult’ class featuring 5WJ structure with PKs, the modeling results present an intriguing scenario (Figure 7M). The model generated by Vfold2 shows an RMSD of 11.0 Å and a TM score of 0.64. Despite this, the model achieves an I-RMSD of 0.69 Å. The yybP-ykoY riboswitch (PDB ID: 4Y1M), with a length of 107 nucleotides, falls into the ‘moderate’ difficulty class, with a 4WJ. The BRiQ model (Figure 7N) exhibits precise modeling of the binding site, as evidenced by an I-RMSD of 0.36 Å, despite facing challenges in overall structure representation, with an RMSD of 18.91 Å and a TM score of 0.32. For class III preQ1 riboswitch (PDB ID: 4RZD), with a length of 102 nucleotides and categorized in the ‘difficult’ class, the modeling results offer an interesting perspective. This RNA, characterized by a 3WJ with PKs, SimRNA model with an RMSD of 12.38 Å and a TM score of 0.29 (Figure 7O). However, the I-RMSD is recorded at 1.93 Å, indicating less deviations at the local binding site structure compared to the global structure.
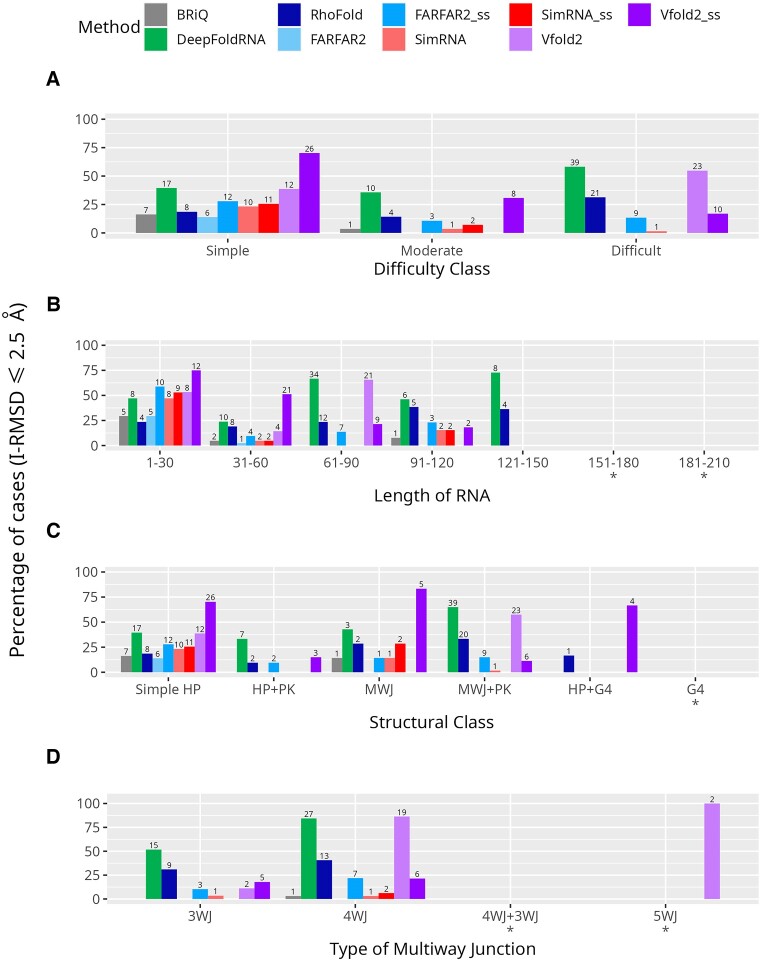
In the ‘simple’ category and for RNA sequences up to 30 nucleotides, most methods achieved models with an I-RMSD ≤ 2.5 for over 25% of the RNA samples. Beyond this category, the 25% threshold was typically surpassed by DeepFoldRNA or one of the Vfold2 variants. Specifically, in the MWJ category, SimRNA_ss and RhoFold successfully modeled the interaction site for over 25% of the structures. Furthermore, RhoFold exceeded the 25% threshold in subsequent categories: ‘difficult’, 91–120 nt, 121–150 nt, MWJ + PK, 3WJ and 4WJ (Figure 8A–D). This suggests that, as expected, most methods are adept at accurately reconstructing the interaction site with a small molecule for small RNAs with simpler structures. However, it is also evident that DeepFoldRNA and Vfold2 are capable of effectively handling more complex RNA structures. This variation underscores the need for nuanced and tailored approaches in RNA structure prediction, catering to the specific challenges posed by different RNA sizes and structural complexities. Perhaps incorporating evolutionarily stabilized regions of RNA could also improve the accuracy of predicted structures, especially in regions critical to their function and those that serve as binding sites for small molecules.
Figure 8.
Distribution of models with interface RMSD (I-RMSD) of 2.5 Å or below. Bar plots showing the percentage of successful modeling cases achieving an interface RMSD (I-RMSD) ≤ 2.5 Å across organized into (A) difficulty classes (B) 30-nucleotide length bins (C) structural classes, and (D) type of multiway junctions. Each method is represented by a unique color. Models generated without secondary structure restraints are shown in lighter shades, while those with restraints are depicted in darker shades of the same color. The number of models with I-RMSD ≤ 2.5 Å are shown above the bars and the classification with less than five cases are marked by asterisk (*) below the labels on abscissa.
In summary, our analysis of ligand binding interfaces underscores the complexity and diversity of RNA–ligand interactions, as evidenced by the bimodal nucleotide frequency distribution at these interfaces. A disconnect exists between the overall accuracy of structure prediction and the precision of binding interface modeling, highlighted by the variable correlation between interface RMSD (I-RMSD) and global structural accuracy metrics. Moreover, the varied performance across different RNA structure prediction methods—particularly regarding high-accuracy interface modeling—underscores the need for tailored approaches and the potential advantages of incorporating secondary structure restraints in specific instances. It should be noted that some methods, such as DeepFoldRNA and Vfold2 models, often yield models with precise interface surfaces when executed with default settings. However, it is frequently necessary to supply not only the secondary structure but probably also to employ more sophisticated techniques. For example, as it was found in last CASP, RNA-Puzzles and other experiments, partial modeling based on templates, addition of constraints from experimental methods, like SAXS/SANS/NMR, CryoEM, combined with the experienced use of the program, can lead to significantly improved outcomes (50,52,94–99). This analysis illuminates the intricate nature of RNA–ligand interactions and the challenges involved in accurately modeling these interfaces for bioinformatics and drug design applications.
Performance of ML-based methods on blind datasets
To obtain a more reliable comparison of ML-based methods, we included all tested methods along with the AlphaFold3 server on smaller test sets of RNA structures which are not present in their training data. Our evaluation utilized two datasets, B1 and B2 (see Materials and methods).
In the B1 dataset, which includes seven structures, none of the methods displayed outstanding median RMSD values (Figure 9A), indicating a moderate level of performance. AlphaFold 3 achieved the lowest median RMSD at 8.79 Å, with other methods showing median RMSDs ranging from 9.2 to 10.5 Å, except for FARFAR2 and BRiQ, which recorded higher values at 11.95 and 14.7 Å, respectively. Some methods, including DeepFoldRNA, RhoFold, and AlphaFold 3, showed better performance on simpler RNA structures with RMSDs below 4 Å. However, AlphaFold 3′s performance dipped in the B2 dataset, registering a median RMSD of 11.22 Å (Supplementary Figure S12A). This increase suggests difficulties in handling more complex RNA structures absent from its training data. This dataset includes two additional challenging cases (Figures 9D, Supplementary Figure S12D), highlighting a disparity in dataset familiarity among the methods, which may have affected AlphaFold 3 more significantly.
Figure 9.
Performance of 3D modeling methods on blind dataset 1 (B1) containing structures not used in training by the ML methods. Performance measures for RNA 3D structure prediction methods are displayed across the B1 dataset: (A) RMSD and (B) INF_all (C) I-RMSD. (D) Length distribution of RNAs on three difficulty classes of modeling - ‘simple’, ‘moderate’, and ‘difficult’.
Regarding INF_all, the B1 dataset showed the highest medians for Vfold2_ss, SimRNA, AlphaFold 3, SimRNA_ss, BRiQ, RhoFold and FARFAR2_ss with scores from 0.65 to 0.51 (Figure 9B). The B2 dataset presented similar trends (Supplementary Figure S12B), with AlphaFold 3 maintaining consistency across both datasets, suggesting its reliability in capturing interaction patterns despite varying structural accuracies. However, it should be noted that these values remain insufficient and do not significantly surpass the INF values obtained by other methods.
The median INF_stack values ranged from 0.5 to 0.67 for all tested methods across both datasets (Figures S10A and S13A), showcasing some consistency in the modeling capabilities of the methods. The highest interaction network fidelity involving Watson-Crick interactions (INF_wc) was observed in Vfold2_ss, followed by AlphaFold 3, BriQ, SimRNA_ss and SimRNA (Supplementary Figures S10B, S13B), reflecting a general competence in predicting stacking interactions. The median INF_nwc values are below 0.3 for all methods tested for both datasets (Figures S10C and S13C).
In I-RMSD, Vfold2_ss (8.34 Å), Vfold2 (8.63 Å) and FARFAR2 (8.51 Å) (Figure 9C) displayed the lowest median values in the B1 dataset, indicating their precision in modeling. The lowest I-RMSD values were observed for DeepFoldRNA (1.32 Å) and Vfold2_ss (1.36 Å) for PreQ1 type 1 (PDB ID: 8FB3) (100) and PreQ1 type III (PDB ID: 8FZA) riboswitches, respectively (101). AlphaFold 3 and RhoFold showed varying degrees of accuracy (median 8.83 Å versus 8.4 Å), with AlphaFold 3 not performing as well in some cases compared to RhoFold (lowest 4.27 Å versus 2.8 Å). For the B2 dataset, some methods like RhoFold (6.31 Å) and DeepFoldRNA (7.26 Å) performed better than in B1. AlphaFold3 has a median I-RMSD of 8.65 Å and a minimum of 3.16 Å, which is lower than the one observed in the B1 dataset. This lowest I-RMSD value is observed for the methyl transferase ribozyme with a 3WJ (PDB ID: 7V9E) (102), even though the overall structure is not modeled well (RMSD = 12.42 Å).
For modeling RNA structures that effectively interact with ligands, only DeepFoldRNA and Vfold2_ss managed to model one structure within the B1 dataset below the 2.5 Å I-RMSD cutoff (Figures 9C and Supplementary Figure S11, panels A-J), illustrating limited success in highly accurate interface modeling. In contrast, the B2 dataset (Supplementary Figure S14, panels A–J) also includes RhoFold among the methods that achieve interfaces within this cutoff (Supplementary Figure S14, panels A–J). Interestingly, AlphaFold 3 did not model any interfaces within this threshold in either dataset, indicating specific limitations in accurately modeling ligand interaction interfaces. This highlights a broader challenge in predicting complex RNA structures comprehensively.
In conclusion, while AlphaFold 3 shows potential in RNA structure prediction, it did not meet the high expectations across all tested metrics. The comparative analysis with DeepFoldRNA and RhoFold underscores areas for improvement and sets the stage for future methodological enhancements. Continuous updates and iterative testing against new RNA structures will be crucial for refining these models' predictive accuracy and advancing our understanding of RNA biology.
Conclusions
The various methods used in this study exhibit varying performance across distinct structural classes. The performance of machine learning (ML)-based methods for RNA structure prediction depends on the composition of the training and testing sets. For our testing set ML-based methods have been shown to be more effective than non-ML methods in delineating long and complex RNA structures. This performance gap indicates a potential for improvement as the training sets expand with newly solved structures (4). While the local resolution of predictions is not yet on par with ML-based global predictions, targeted efforts to refine this aspect are crucial. It is important to mention that as the details of the training sets used by ML-based methods are not publicly disclosed, we cannot exclude the possibility that structures from our test set may also appear in the training sets of the methods we are evaluating. Hence, the results obtained by ML-based methods may be overestimated. Currently, there is no universally reliable method for high-resolution RNA 3D structure prediction. A breakthrough in the prediction of RNA structures can be expected, as in the field of protein prediction, but before that several conditions must be met, including, above all, the availability of a larger number of known structures (4).
Our study provides valuable insights for selecting and applying various prediction methods. ML-based methods are effective for generating models that have good global folds. Many of these models meet the I-RMSD criterion, making them suitable for initial models, particularly in drug development targeting small molecules. However, for detailed structural features such as intra-molecular interactions and specific elements like G-quadruplexes, non-ML-based methods demonstrate superior accuracy. Consequently, a synergistic strategy that incorporates machine learning (ML) for global structural predictions and non-ML methods for detailed binding interface modeling could offer significant advantages. Notably, the inclusion of secondary structure restraints significantly improves the performance of non-ML-based methods in binding site modeling. The outcomes of recent RNA-Puzzles and CASP competitions demonstrate that an experienced user, who incorporates the structures of homologous RNA molecules and various experimental data, significantly enhances prediction accuracy compared to outcomes achieved using the default settings of the program (50,52,94–97,103). This enhancement may be important for modeling small molecule binding sites, which must be functional and, therefore, frequently demonstrate evolutionary conservation in both sequence and structure. Consequently, it becomes crucial to use an integrated modeling approach, incorporating various biochemical and biophysical data and utilizing diverse computational tools for improving the models.
Our findings indicate that models with low RMSD, predicted using all methods with default settings, exhibit accurately predicted binding sites for small molecules (Figures 7C-K). Interestingly, some models with higher RMSD (ranging from 8 to 16 Å) also display well-predicted interaction sites with small molecules, thereby expanding the pool of RNA models viable for virtual screening. Given the rapid advancements in the precision and capabilities of artificial intelligence-based methods, significant progress in RNA structure prediction is anticipated shortly. In conclusion, researchers without access to experienced structural bioinformaticians can now independently utilize one of the top-performing methods, such as Vfold2, DeepFoldRNA, or RhoFold. Nevertheless, experienced bioinformaticians may prefer methods like SimRNA or FARFAR2, which offer enhanced flexibility for integrating both expertise and experimental data (63,64).
Our evaluation of the newly released AlphaFold 3 on the B1 and B2 datasets, which are constrained by the limited number of available structures, reveals that it did not model any RNA interfaces within the stringent I-RMSD cutoff of ≤2.5 Å. This result, derived from a relatively small data sample, may not capture the full potential of AlphaFold 3 across varied scenarios. In contrast, methods like DeepFoldRNA and RhoFold were able to successfully model interfaces in some RNA structures within this cutoff, highlighting potential specific challenges AlphaFold 3 faces in accurately capturing these crucial interaction details, which are vital for a thorough understanding of RNA-small molecule interactions. Nonetheless, the limitations posed by the small size of the datasets suggest that these results should be viewed as preliminary. The ongoing development and integration of more comprehensive datasets in future evaluations are expected to provide deeper insights and may enhance the performance of AlphaFold 3. Such continuous advancements are critical for the progress of computational methods in RNA biology, indicating that while the tool shows promise, there is considerable scope for improvement and refinement.
It is crucial to remember that RNA structure prediction extends beyond foreseeing a singular, static structure. The dynamic nature of RNA, characterized by its rapid conformational shifts, plays a pivotal role in recognition processes, and significantly impacts ligand binding affinity. The energy differences between these various conformations can range from modest to dramatic, exerting a crucial influence on binding affinity. Recognizing and precisely quantifying RNA dynamics is indispensable for successfully designing RNA-based structures and exploring new avenues in RNA-targeted therapeutics (104). The advancement in RNA structure dynamics heavily depends on the customization of MD force fields, originally designed for proteins, to suit the distinctive characteristics of RNA. Notably, the aromatic ring stacking interactions, a hallmark of RNA’s molecular recognition, require the precise adjustment of electrostatic parameters in these force fields (22,104–109). Accurate modeling of these interactions is vital for a deeper understanding of how small molecules interact with RNA and for identifying the most effective locations for functional groups. Employing molecular dynamics with a force field enabling realistic depiction of RNA dynamics could be crucial in selecting alternative RNA models for performing virtual screening. In this scenario, selecting representative structures statistically from the molecular dynamics (MD) trajectory may substitute for structures determined by nuclear magnetic resonance (NMR), which represent the conformer binding small molecules. These are not always depicted by the cluster of the most frequent conformer (109).
While RNA structure prediction methods presently achieve sufficient accuracy for generating initial global structure models, they may fall short of the precision required for high-resolution modeling crucial in structure-based drug design. Achieving finer details necessary for designing effective small-molecule therapeutics targeting specific RNA motifs and binding sites demands a higher level of accuracy than currently observed. Future research, including comprehensive docking experiments, will play an important role in determining whether the accuracy of these methods is sufficient for practical applications.
Supplementary Material
Acknowledgements
Author contributions: C.N. ran programs, prepared the testing set, analyzed results, and wrote the manuscript; S.K. wrote the manuscript, and supervised the analysis; R.B. and J.N. prepared the reliable testing set; I.T. supervised the analysis and wrote the manuscript.
Contributor Information
Chandran Nithin, Molecure SA, 02-089 Warsaw, Poland; Laboratory of Computational Biology, Biological and Chemical Research Center, Faculty of Chemistry, University of Warsaw, 02-089 Warsaw, Poland.
Sebastian Kmiecik, Laboratory of Computational Biology, Biological and Chemical Research Center, Faculty of Chemistry, University of Warsaw, 02-089 Warsaw, Poland.
Roman Błaszczyk, Molecure SA, 02-089 Warsaw, Poland.
Julita Nowicka, Molecure SA, 02-089 Warsaw, Poland.
Irina Tuszyńska, Molecure SA, 02-089 Warsaw, Poland.
Data availability
All models obtained using the methods we described are hosted on Mendeley Data: http://dx.doi.org/10.17632/8yg88x7rdk.3.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
The study on RNA at Molecure SA is supported by project: ‘Breakthrough discovery services for new small molecule medicines targeting mRNA for incurable diseases’ [FENG.01.01-IP.02-1256/23], cofinanced by the European Union under the European Funds for a Modern Economy program, action 1.1 SMART path; C.N. and S.K. acknowledge funding from the National Science Centre, Poland [OPUS 2020/39/B/NZ2/01301]; C.N gratefully acknowledge Polish high-performance computing infrastructure PLGrid (HPC Centers: ACK Cyfronet AGH, CI TASK, WCSS) for providing computer facilities and support within computational grant no. PLG/2022/016043. Funding for open access charge: Molecure SA.
Conflict of interest statement. At the time of the study Chandran Nithin, Roman Błaszczyk, Julita Nowicka and Irina Tuszyńska were employees and shareholders of Molecure SA (formerly OncoArendi Therapeutics SA), which develops small molecules targeting RNA. The authors report no other conflicts of interest in this work.
References
- 1. Zhang J., Fei Y., Sun L., Zhang Q.C. Advances and opportunities in RNA structure experimental determination and computational modeling. Nat. Methods. 2022; 19:1193–1207. [DOI] [PubMed] [Google Scholar]
- 2. Ponce-Salvatierra A., Astha, Merdas K., Nithin C., Ghosh P., Mukherjee S., Bujnicki J.M Computational modeling of RNA 3D structure based on experimental data. Biosci. Rep. 2019; 39:BSR20180430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A. et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021; 596:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schneider B., Sweeney B.A., Bateman A., Cerny J., Zok T., Szachniuk M. When will RNA get its AlphaFold moment?. Nucleic Acids Res. 2023; 51:9522–9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burley S.K., Bhikadiya C., Bi C., Bittrich S., Chao H., Chen L., Craig P.A., Crichlow G.V., Dalenberg K., Duarte J.M. et al. RCSB Protein Data Bank (RCSB.org): delivery of experimentally-determined PDB structures alongside one million computed structure models of proteins from artificial intelligence/machine learning. Nucleic Acids Res. 2023; 51:D488–D508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang X., Yu S., Lou E., Tan Y.-L., Tan Z.-J. RNA 3D structure prediction: progress and perspective. Molecules. 2023; 28:5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dawson W.K., Bujnicki J.M. Computational modeling of RNA 3D structures and interactions. Curr. Opin. Struct. Biol. 2016; 37:22–28. [DOI] [PubMed] [Google Scholar]
- 8. Ghosh P., Nithin C., Joshi A., Stefaniak F., Wirecki T.K., Bujnicki J.M. Computational modeling methods for 3D structure prediction of ribozymes. Ribozymes. 2021; 861–881. [Google Scholar]
- 9. Krokhotin A., Dokholyan N.V.. Chen S.-J., Burke-Aguero D.H. Chapter three - computational methods toward accurate RNA structure prediction using coarse-grained and all-atom models. Methods in Enzymology, Computational Methods for Understanding Riboswitches. 2015; 553:Academic Press; 65–89. [DOI] [PubMed] [Google Scholar]
- 10. Ding F., Sharma S., Chalasani P., Demidov V.V., Broude N.E., Dokholyan N.V. Ab initio RNA folding by discrete molecular dynamics: from structure prediction to folding mechanisms. RNA. 2008; 14:1164–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Göç Y.B., Poziemski J., Smolińska W., Suwała D., Wieczorek G., Niedzialek D. Tracking topological and electronic effects on the folding and stability of guanine-deficient RNA G-quadruplexes, engineered with a new computational tool for De Novo Quadruplex folding. Int. J. Mol. Sci. 2022; 23:10990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sponer J., Bussi G., Krepl M., Banáš P., Bottaro S., Cunha R.A., Gil-Ley A., Pinamonti G., Poblete S., Jurecka P. RNA structural dynamics as captured by molecular simulations: a comprehensive overview. Chem. Rev. 2018; 118:4177–4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Krokhotin A., Houlihan K., Dokholyan N.V. iFoldRNA v2: folding RNA with constraints. Bioinformatics. 2015; 31:2891–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mustoe A.M., Al-Hashimi H.M., Brooks C.L. III Coarse grained models reveal essential contributions of topological constraints to the conformational free energy of RNA bulges. J. Phys. Chem. B. 2014; 118:2615–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang D., Li J., Chen S.-J. IsRNA1: de novo prediction and blind screening of RNA 3D structures. J. Chem. Theory Comput. 2021; 17:1842–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Malhotra A., Tan R.K., Harvey S.C. Modeling large RNAs and ribonucleoprotein particles using molecular mechanics techniques. Biophys. J. 1994; 66:1777–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tan R.K.Z., Petrov A.S., Harvey S.C. YUP: A molecular simulation program for coarse-grained and multiscaled models. J. Chem. Theory Comput. 2006; 2:529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li J., Chen S.-J. RNAJP: enhanced RNA 3D structure predictions with non-canonical interactions and global topology sampling. Nucleic Acids Res. 2023; 51:3341–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pasquali S., Derreumaux P. HiRE-RNA: a high resolution coarse-grained energy model for RNA. J. Phys. Chem. B. 2010; 114:11957–11966. [DOI] [PubMed] [Google Scholar]
- 20. Denesyuk N.A., Thirumalai D. Coarse-grained model for predicting RNA folding thermodynamics. J. Phys. Chem. B. 2013; 117:4901–4911. [DOI] [PubMed] [Google Scholar]
- 21. Li J., Chen S.-J. RNA 3D structure prediction using coarse-grained models. Front Mol. Biosci. 2021; 8:3135–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Salsbury A.M., Lemkul J.A. Recent developments in empirical atomistic force fields for nucleic acids and applications to studies of folding and dynamics. Curr. Opin. Struct. Biol. 2021; 67:9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liebl K., Zacharias M. The development of nucleic acids force fields: from an unchallenged past to a competitive future. Biophys. J. 2023; 122:2841–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Das R., Baker D. Automated de novo prediction of native-like RNA tertiary structures. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:14664–14669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Das R., Karanicolas J., Baker D. Atomic accuracy in predicting and designing noncanonical RNA structure. Nat. Methods. 2010; 7:291–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Watkins A.M., Rangan R., Das R. FARFAR2: improved De Novo Rosetta prediction of complex global RNA folds. Structure. 2020; 28:963–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Parisien M., Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008; 452:51–55. [DOI] [PubMed] [Google Scholar]
- 28. Popenda M., Szachniuk M., Antczak M., Purzycka K.J., Lukasiak P., Bartol N., Blazewicz J., Adamiak R.W. Automated 3D structure composition for large RNAs. Nucleic Acids Res. 2012; 40:e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhao Y., Huang Y., Gong Z., Wang Y., Man J., Xiao Y. Automated and fast building of three-dimensional RNA structures. Sci. Rep. 2012; 2:734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang J., Wang J., Huang Y., Xiao Y. 3dRNA v2. 0: an updated web server for RNA 3D structure prediction. Int. J. Mol. Sci. 2019; 20:4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang Y., Wang J., Xiao Y. 3dRNA: building RNA 3D structure with improved template library. Comput. Struct. Biotechnol. J. 2020; 18:2416–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cao S., Chen S.-J. Physics-based de novo prediction of RNA 3D structures. J. Phys. Chem. B. 2011; 115:4216–4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xu X., Chen S.-J. Hierarchical assembly of RNA three-dimensional structures based on loop templates. J. Phys. Chem. B. 2018; 122:5327–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhou L., Wang X., Yu S., Tan Y.-L., Tan Z.-J. FebRNA: an automated fragment-ensemble-based model for building RNA 3D structures. Biophys. J. 2022; 121:3381–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kamga Youmbi F.I., Kengne Tchendji V., Tayou Djamegni C. P-FARFAR2: a multithreaded greedy approach to sampling low-energy RNA structures in Rosetta FARFAR2. Comput. Biol. Chem. 2023; 104:107878. [DOI] [PubMed] [Google Scholar]
- 36. Chojnowski G., Zaborowski R., Magnus M., Mukherjee S., Bujnicki J.M. RNA 3D structure modeling by fragment assembly with small-angle X-ray scattering restraints. Bioinformatics. 2023; 39:btad527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Boniecki M.J., Lach G., Dawson W.K., Tomala K., Lukasz P., Soltysinski T., Rother K.M., Bujnicki J.M. SimRNA: a coarse-grained method for RNA folding simulations and 3D structure prediction. Nucleic Acids Res. 2016; 44:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xiong P., Wu R., Zhan J., Zhou Y. Pairing a high-resolution statistical potential with a nucleobase-centric sampling algorithm for improving RNA model refinement. Nat. Commun. 2021; 12:2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li J., Zhang S., Zhang D., Chen S.-J. Vfold-pipeline: a web server for RNA 3D structure prediction from sequences. Bioinformatics. 2022; 38:4042–4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li Y., Zhang C., Feng C., Pearce R., Lydia Freddolino P., Zhang Y. Integrating end-to-end learning with deep geometrical potentials for ab initio RNA structure prediction. Nat. Commun. 2023; 14:5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shen T., Hu Z., Peng Z., Chen J., Xiong P., Hong L., Zheng L., Wang Y., King I., Wang S., Sun S, Li Y E2Efold-3D: end-to-End deep learning method for accurate de novo RNA 3D structure prediction. 2022; arXiv doi:04 July 2022, preprint: not peer reviewed https://arxiv.org/abs/2207.01586.
- 42. Pearce R., Omenn G.S., Zhang Y. De novo RNA tertiary structure prediction at atomic resolution using geometric potentials from deep learning. 2022; bioRxiv doi:15 May 2022, preprint: not peer reviewed 10.1101/2022.05.15.491755. [DOI]
- 43. Townshend R.J.L., Eismann S., Watkins A.M., Rangan R., Karelina M., Das R., Dror R.O. Geometric deep learning of RNA structure. Science. 2021; 373:1047–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang W., Feng C., Han R., Wang Z., Ye L., Du Z., Wei H., Zhang F., Peng Z., Yang J. trRosettaRNA: automated prediction of RNA 3D structure with transformer network. Nat. Commun. 2023; 14:7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sha C.M., Wang J., Dokholyan N.V. Predicting 3D RNA structure from the nucleotide sequence using euclidean neural networks. Biophys. J. 2023; 123:1–11. [DOI] [PubMed] [Google Scholar]
- 46. Baek M., McHugh R., Anishchenko I., Baker D., DiMaio F. Accurate prediction of nucleic acid and protein-nucleic acid complexes using RoseTTAFoldNA. Nat. Methods. 2022; 21:117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cruz J.A., Blanchet M.-F., Boniecki M., Bujnicki J.M., Chen S.-J., Cao S., Das R., Ding F., Dokholyan N.V., Flores S.C. et al. RNA-puzzles: a CASP-like evaluation of RNA three-dimensional structure prediction. RNA. 2012; 18:610–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Miao Z., Adamiak R.W., Blanchet M.-F., Boniecki M., Bujnicki J.M., Chen S.-J., Cheng C., Chojnowski G., Chou F.-C., Cordero P. et al. RNA-puzzles Round II: assessment of RNA structure prediction programs applied to three large RNA structures. RNA. 2015; 21:1066–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Miao Z., Adamiak R.W., Antczak M., Batey R.T., Becka A.J., Biesiada M., Boniecki M.J., Bujnicki J.M., Chen S.-J., Cheng C.Y. et al. RNA-puzzles round III: 3D RNA structure prediction of five riboswitches and one ribozyme. RNA. 2017; 23:655–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Miao Z., Adamiak R.W., Antczak M., Boniecki M.J., Bujnicki J., Chen S.-J., Cheng C.Y., Cheng Y., Chou F.-C., Das R. et al. RNA-puzzles Round IV: 3D structure predictions of four ribozymes and two aptamers. RNA. 2020; 26:982–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gumna J., Antczak M., Adamiak R.W., Bujnicki J.M., Chen S.-J., Ding F., Ghosh P., Li J., Mukherjee S., Nithin C. et al. Computational pipeline for reference-free comparative analysis of RNA 3D structures applied to SARS-CoV-2 UTR models. Int. J. Mol. Sci. 2022; 23:9630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Das R., Kretsch R.C., Simpkin A.J., Mulvaney T., Pham P., Rangan R., Bu F., Keegan R.M., Topf M., Rigden D.J. et al. Assessment of three-dimensional RNA structure prediction in CASP15. Proteins Struct. Funct. Bioinf. 2023; 91:1747–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kryshtafovych A., Antczak M., Szachniuk M., Zok T., Kretsch R.C., Rangan R., Pham P., Das R., Robin X., Studer G. et al. New prediction categories in CASP15. Proteins Struct. Funct. Bioinf. 2023; 91:1550–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Popenda M., Zok T., Sarzynska J., Korpeta A., Adamiak R.W., Antczak M., Szachniuk M. Entanglements of structure elements revealed in RNA 3D models. Nucleic Acids Res. 2021; 49:9625–9632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Carrascoza F., Antczak M., Miao Z., Westhof E., Szachniuk M. Evaluation of the stereochemical quality of predicted RNA 3D models in the RNA-Puzzles submissions. RNA. 2022; 28:250–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000; 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Agu P.C., Afiukwa C.A., Orji O.U., Ezeh E.M., Ofoke I.H., Ogbu C.O., Ugwuja E.I., Aja P.M. Molecular docking as a tool for the discovery of molecular targets of nutraceuticals in diseases management. Sci. Rep. 2023; 13:13398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Abramson J., Adler J., Dunger J., Evans R., Green T., Pritzel A., Ronneberger O., Willmore L., Ballard A.J., Bambrick J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature. 2024; 630:493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lu X.-J., Bussemaker H.J., Olson W.K. DSSR: an integrated software tool for dissecting the spatial structure of RNA. Nucleic Acids Res. 2015; 43:e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yoshizawa S., Fourmy D., Puglisi J.D. Structural origins of gentamicin antibiotic action. EMBO J. 1998; 17:6437–6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Smith K.D., Shanahan C.A., Moore E.L., Simon A.C., Strobel S.A. Structural basis of differential ligand recognition by two classes of bis-(3′-5′)-cyclic dimeric guanosine monophosphate-binding riboswitches. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:7757–7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Johnson J.E. Jr, Reyes F.E., Polaski J.T., Batey R.T B12 cofactors directly stabilize an mRNA regulatory switch. Nature. 2012; 492:133–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wirecki T.K., Nithin C., Mukherjee S., Bujnicki J.M., Boniecki M.J. Modeling of three-dimensional RNA structures using SimRNA. Methods Mol. Biol. 2020; 2165:103–125. [DOI] [PubMed] [Google Scholar]
- 64. Watkins A.M., Das R.. Kawaguchi R.K., Iwakiri J. RNA 3D modeling with FARFAR2, online. RNA Structure Prediction. 2023; NY: Springer US; 233–249. [DOI] [PubMed] [Google Scholar]
- 65. Cheng Y., Zhang S., Xu X., Chen S.-J. Vfold2D-MC: a physics-based hybrid model for predicting RNA secondary structure folding. J. Phys. Chem. B. 2021; 125:10108–10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Xu X., Zhao P., Chen S.-J. Vfold: a web server for RNA structure and folding thermodynamics prediction. PLoS One. 2014; 9:e107504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lorenz R., Bernhart S.H., Höner zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L. ViennaRNA package 2.0. Algorithms Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Reuter J.S., Mathews D.H. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinf. 2010; 11:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sato K., Kato Y., Hamada M., Akutsu T., Asai K. IPknot: fast and accurate prediction of RNA secondary structures with pseudoknots using integer programming. Bioinformatics. 2011; 27:i85–i93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhao Y., Wang J., Zeng C., Xiao Y. Evaluation of RNA secondary structure prediction for both base-pairing and topology. Biophys. Rep. 2018; 4:123–132. [Google Scholar]
- 71. Justyna M., Antczak M., Szachniuk M. Machine learning for RNA 2D structure prediction benchmarked on experimental data. Brief. Bioinf. 2023; 24:bbad153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kalvari I., Nawrocki E.P., Ontiveros-Palacios N., Argasinska J., Lamkiewicz K., Marz M., Griffiths-Jones S., Toffano-Nioche C., Gautheret D., Weinberg Z. et al. Rfam 14: expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res. 2021; 49:D192–D200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. RNAcentral Consortium RNAcentral 2021: secondary structure integration, improved sequence search and new member databases. Nucleic Acids Res. 2021; 49:D212–D220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D. et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016; 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Seemann S.E., Gorodkin J., Backofen R. Unifying evolutionary and thermodynamic information for RNA folding of multiple alignments. Nucleic Acids Res. 2008; 36:6355–6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhang C., Zhang Y., Pyle A.M. rMSA: a sequence search and alignment algorithm to improve RNA structure modeling. J. Mol. Biol. 2023; 435:167904. [DOI] [PubMed] [Google Scholar]
- 77. Stasiewicz J., Mukherjee S., Nithin C., Bujnicki J.M. QRNAS: software tool for refinement of nucleic acid structures. BMC Struct. Biol. 2019; 19:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Singh J., Paliwal K., Singh J., Zhou Y. RNA backbone torsion and pseudotorsion angle prediction using dilated convolutional neural networks. J. Chem. Inf. Model. 2021; 61:2610–2622. [DOI] [PubMed] [Google Scholar]
- 79. Manfredonia I., Nithin C., Ponce-Salvatierra A., Ghosh P., Wirecki T.K., Marinus T., Ogando N.S., Snijder E.J., van Hemert M.J., Bujnicki J.M. et al. Genome-wide mapping of SARS-CoV-2 RNA structures identifies therapeutically-relevant elements. Nucleic Acids Res. 2020; 48:12436–12452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Schrödinger L.L.C. 2015; The PyMOL Molecular Graphics System, Version 2.5.
- 81. Gong S., Zhang C., Zhang Y. RNA-align: quick and accurate alignment of RNA 3D structures based on size-independent TM-scoreRNA. Bioinformatics. 2019; 35:4459–4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Parisien M., Cruz J.A., Westhof É., Major F. New metrics for comparing and assessing discrepancies between RNA 3D structures and models. RNA. 2009; 15:1875–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Magnus M. rna-tools.Online: a Swiss army knife for RNA 3D structure modeling workflow. Nucleic Acids Res. 2022; 50:W657–W662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hubbard S.J., Thornton J. NACCESS: Program for Calculating Accessibilities. 1992; Department of Biochemistry and Molecular Biology, University College of London. [Google Scholar]
- 85. Lee B., Richards F.M. The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol. 1971; 55:379. [DOI] [PubMed] [Google Scholar]
- 86. Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G.W., McCoy A. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. Sect. D. 2011; 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Mitternacht S. FreeSASA: an open source C library for solvent accessible surface area calculations. F1000Research. 2016; 5:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Tsodikov O.V., Record J.M.T., Sergeev Y.V Novel computer program for fast exact calculation of accessible and molecular surface areas and average surface curvature. J. Comput. Chem. 2002; 23:600–609. [DOI] [PubMed] [Google Scholar]
- 89. Mukherjee S., Nithin C.. Tripathi T., Dubey V.K. Chapter 11 - advanced computational tools for quantitative analysis of protein–nucleic acid interfaces. Advances in Protein Molecular and Structural Biology Methods. 2022; Academic Press; 163–180. [Google Scholar]
- 90. Mukherjee S., Nithin C., Divakaruni Y., Bahadur R.P. Dissecting water binding sites at protein–protein interfaces: a lesson from the atomic structures in the Protein Data Bank. J. Biomol. Struct. Dyn. 2019; 37:1204–1219. [DOI] [PubMed] [Google Scholar]
- 91. Matarrese M.A.G., Loppini A., Nicoletti M., Filippi S., Chiodo L. Assessment of tools for RNA secondary structure prediction and extraction: a final-user perspective. J. Biomol. Struct. Dyn. 2023; 41:6917–6936. [DOI] [PubMed] [Google Scholar]
- 92. Singh J., Paliwal K., Zhang T., Singh J., Litfin T., Zhou Y. Improved RNA secondary structure and tertiary base-pairing prediction using evolutionary profile, mutational coupling and two-dimensional transfer learning. Bioinformatics. 2021; 37:2589–2600. [DOI] [PubMed] [Google Scholar]
- 93. Trachman R.J., Autour A., Jeng S.C.Y., Abdolahzadeh A., Andreoni A., Cojocaru R., Garipov R., Dolgosheina E.V., Knutson J.R., Ryckelynck M. et al. Structure and functional reselection of the Mango-III fluorogenic RNA aptamer. Nat. Chem. Biol. 2019; 15:472–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Kretsch R.C., Andersen E.S., Bujnicki J.M., Chiu W., Das R., Luo B., Masquida B., McRae E.K.S., Schroeder G.M., Su Z. et al. RNA target highlights in CASP15: evaluation of predicted models by structure providers. Proteins Struct. Funct. Bioinf. 2023; 91:1600–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Baulin E.F., Mukherjee S., Moafinejad S.N., Wirecki T.K., Badepally N.G., Jaryani F., Stefaniak F., Amiri Farsani M., Ray A., Rocha de Moura T. et al. RNA tertiary structure prediction in CASP15 by the GeneSilico group: folding simulations based on statistical potentials and spatial restraints. Proteins Struct. Funct. Bioinf. 2023; 91:1800–1810. [DOI] [PubMed] [Google Scholar]
- 96. Chen K., Zhou Y., Wang S., Xiong P. RNA tertiary structure modeling with BRiQ potential in CASP15. Proteins Struct. Funct. Bioinf. 2023; 91:1771–1778. [DOI] [PubMed] [Google Scholar]
- 97. Sarzynska J., Popenda M., Antczak M., Szachniuk M. RNA tertiary structure prediction using RNAComposer in CASP15. Proteins Struct. Funct. Bioinf. 2023; 91:1790–1799. [DOI] [PubMed] [Google Scholar]
- 98. Luo B., Zhang C., Ling X., Mukherjee S., Jia G., Xie J., Jia X., Liu L., Baulin E.F., Luo Y. et al. Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing. Nature Catalysis. 2023; 6:298–309. [Google Scholar]
- 99. Mulvaney T., Kretsch R.C., Elliott L., Beton J.G., Kryshtafovych A., Rigden D.J., Das R., Topf M. CASP15 cryo-EM protein and RNA targets: refinement and analysis using experimental maps. Proteins Struct. Funct. Bioinf. 2023; 91:1935–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Schroeder G.M., Akinyemi O., Malik J., Focht C.M., Pritchett E.M., Baker C.D., McSally J.P., Jenkins J.L., Mathews D.H., Wedekind J.E. A riboswitch separated from its ribosome-binding site still regulates translation. Nucleic Acids Res. 2023; 51:2464–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Schroeder G.M., Kiliushik D., Jenkins J.L., Wedekind J.E. Structure and function analysis of a type III preQ1-I riboswitch from Escherichia coli reveals direct metabolite sensing by the Shine-Dalgarno sequence. J. Biol. Chem. 2023; 299:105208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Deng J., Wilson T.J., Wang J., Peng X., Li M., Lin X., Liao W., Lilley D.M.J., Huang L. Structure and mechanism of a methyltransferase ribozyme. Nat. Chem. Biol. 2022; 18:556–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Li J., Zhang S., Chen S.J. Advancing RNA 3D structure prediction: Exploring hierarchical and hybrid approaches in CASP15. Proteins. 2023; 91:1779–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Childs-Disney J.L, Yang X., Gibaut Q.M.R, Tong Y., Batey R.T., Disney M.D. Targeting RNA structures with small molecules. Nat. Rev. Drug Discov. 2022; 21:736–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Yuan Y., Fu S., Huo D., Su W., Zhang R., Wei J. Multipolar electrostatics for hairpin and pseudoknots in RNA: Improving the accuracy of force field potential energy function. J. Comput. Chem. 2021; 42:771–786. [DOI] [PubMed] [Google Scholar]
- 106. Li Z., Mu J., Chen J, Chen H.F. Base-specific RNA force field improving the dynamics conformation of nucleotide. Int. J. Biol. Macromol. 2022; 222:680–690. [DOI] [PubMed] [Google Scholar]
- 107. Jing Z., Ren P. Molecular Dynamics Simulations of Protein RNA Complexes by Using an Advanced Electrostatic Model. J. Phys. Chem. B. 2022; 126:7343–7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. He W., Naleem N., Kleiman D., Kirmizialtin S. Refining the RNA Force Field with Small-Angle X-ray Scattering of Helix-Junction-Helix RNA. J. Phys. Chem. Lett. 2022; 13:3400–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Ganser L.R., Lee J., Rangadurai A., Merriman D.K., Kelly M.L., Kansal A.D., Sathyamoorthy B., Al-Hashimi H.M. High-performance virtual screening by targeting a high-resolution RNA dynamic ensemble. Nat. Struct. Mol. Biol. 2018; 25:425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All models obtained using the methods we described are hosted on Mendeley Data: http://dx.doi.org/10.17632/8yg88x7rdk.3.