Abstract
The efficiency of targeted knock-in for cell therapeutic applications is generally low, and the scale is limited. In this study, we developed CLASH, a system that enables high-efficiency, high-throughput knock-in engineering. In CLASH, Cas12a/Cpf1 mRNA combined with pooled adeno-associated viruses mediate simultaneous gene editing and precise transgene knock-in using massively parallel homology-directed repair, thereby producing a pool of stably integrated mutant variants each with targeted gene editing. We applied this technology in primary human T cells and performed time-coursed CLASH experiments in blood cancer and solid tumor models using CD3, CD8 and CD4 T cells, enabling pooled generation and unbiased selection of favorable CAR-T variants. Emerging from CLASH experiments, a unique CRISPR RNA (crRNA) generates an exon3 skip mutant of PRDM1 in CAR-Ts, which leads to increased proliferation, stem-like properties, central memory and longevity in these cells, resulting in higher efficacy in vivo across multiple cancer models, including a solid tumor model. The versatility of CLASH makes it broadly applicable to diverse cellular and therapeutic engineering applications.
Precise genomic engineering enables diverse molecular, cellular and therapeutic applications, including cell therapy1. Cell therapies that involve chimeric antigen receptor (CAR) transgene delivery via lentiviral or retroviral vectors into human T cells (CAR-Ts) have been approved by the US Food and Drug Administration (FDA) for the treatment of hematological cancers, such as B cell malignancies and multiple myeloma2–4. A rapid emergence of CAR-T and other forms of adoptive T cell therapies are underway, currently with over 1,100 active clinical trials (https://clinicaltrials.gov/) and numerous studies in pre-clinical stages5. There is a broad landscape of cell-based immunotherapies across a wide range of oncology indications6,7, all of which can benefit from next-generation cellular engineering approaches.
However, despite the recent success, there are still major hurdles for CAR-T therapy1. So far, no CAR-T therapy has been approved by the FDA for solid tumors. Even in hematological malignancies, despite high response rates, a large fraction of patients are either refractory or can quickly relapse8,9. Multiple fundamental challenges exist in the current forms of CAR-T therapy, including antigen loss, metabolic suppression in the tumor microenvironment, insufficient T cell trafficking to the cancer site, lack of effective cancer cell killing, severe toxicity especially in the form of cytokine release syndrome, suboptimal levels of T cell proliferation and, as often observed in the clinic, failure of CAR-T persistence6,7. A series of efforts have been attempted to enhance CAR-T function, such as re-structuring of the CAR’s promoter or signaling domains10,11; engineering of various CAR-T components, such as single-chain variable fragment (scFv) or transmembrane regions12,13; overexpression of boosting factors14; and co-administration of immunomodulating factors or viral vectors15, among many other endeavors. Several studies have tested the improvement of CAR-T cell function and persistence by changing co-stimulatory domains, lowering CAR binding affinity, deleting T-cell-dysfunction-related genes or by transient resting of CAR T cells16–20. Nevertheless, despite ongoing attempts, major hurdles remain that hinder the success of CAR-T therapy, and more efficient, higher-throughput engineering strategies are needed.
Most CAR-T and immune cell engineering approaches involve a single or small number of modifications based on rational designs or information from literature. These approaches can often miss the broader landscape of the immense combinatorial space to achieve superiority in cell therapy. One of the most efficient ways to build next-generation cell therapy and, thereby, to overcome current challenges is to engineer thousands of different variants and select the best ones for different features in an unbiased manner. This will require technologies with the capability of massively parallel engineering, as there are approximately 20,000 protein-coding genes and many non-coding elements in the human genome as well as numerous combinations. Gene editing in primary immune cells, such as human T cells, opens vast potential for engineering of next-generation cell-based therapeutics10. Although various T cell screens have been performed21–27, these CRISPR screening approaches to date predominantly rely on lentiviral, retroviral or transposon vectors, which are all random integration systems. These systems cannot control position effects and are naturally associated with risks of random insertional mutagenesis.
Genomic targeted knock-in is needed to precisely control such effects. A recent breakthrough demonstrated CRISPR–Cas9 gene editing for targeted delivery of a CAR transgene into a specific locus, such as TRAC28. Editing human T cells with Cas9 ribonucleoprotein (RNP) plus donor DNA templates also enables targeted knock-in of transgenes at a desired locus29. A recent study demonstrated pooled knock-in for T cells, with pools of a few dozen constructs30. However, targeted knock-in usually requires specifically designed constructs, and the scale has been limited so far in prior studies. The cellular toxicity associated with DNA electroporation naturally limits the scale of knock-in, especially in human T cells. Several technical barriers remain to overcome current limitations, including (1) how to build cellular knock-ins in a massive parallel manner; (2) how to achieve high-efficiency, stable and standardized knock-in pools with minimal cellular toxicity possible; and (3) how to ensure quantitative assessment between different knock-in variants in the same setting at high resolution. Accomplishing these technical potentials will open up diverse engineering and therapeutic development applications in T cells and many other cell types.
In this study, we developed a platform for highly efficient massively parallel knock-in engineering. This platform, called CRISPR-based library-scale AAV perturbation with simultaneous HDR knock-in (CLASH), enables rapid generation of customized scales of cellular knock-in pools in simple steps by mRNA electroporation with multi-functional pooled adeno-associated virus (AAV) transduction. The advantage of AAV in this platform is at least two-fold. First, it can efficiently transduce various type of cells, including primary human T cells, with minimal cytotoxicity. Second, it can be effectively designed to carry diverse cargos in silico, synthesized and packaged in vitro and used as homology-directed repair (HDR) templates in cellulo or in vivo. The mRNA carries a CRISPR effector enzyme, such as Cas12a/Cpf1, which performs the function of genome editing in a short pulse manner because the mRNA has a relatively short half-life in cells. Similarly to AAV, the mRNA component also has low cellular toxicity, which is particularly important when generating knock-in pools. We show that the CLASH system can efficiently create genomic knock-ins in a massive parallel manner via mRNA + AAV pool, enable engineering of different variants of knock-in constructs such as CARs with a standardized core component as well as variable components and can target specific sets of genes or variants to enhance the chance of evolving and selecting the promising cell therapy candidates for validation and downstream development. Unlike existing CRISPR screening approaches that use integration systems, CLASH is a massively parallel targeted integration system where all the engineered CARs and CRISPR RNAs (crRNAs) are precisely knocked-in into the same locus, thereby controlling positional effects and minimizing the risk of random insertional mutagenesis. We also define CLASH as a procedure to stably knock-in various constructs into cells by CRISPR effector mRNA electroporation and pooled AAV transduction. By directly applying CLASH in human primary T cells, we rapidly generated library scale of CAR-T pools with each candidate in the pool stably knocked-in into a defined desired locus in the genome, and we subjected them to unbiased selection for favorable features, such as anti-tumor function.
Results
Establishment of CLASH system
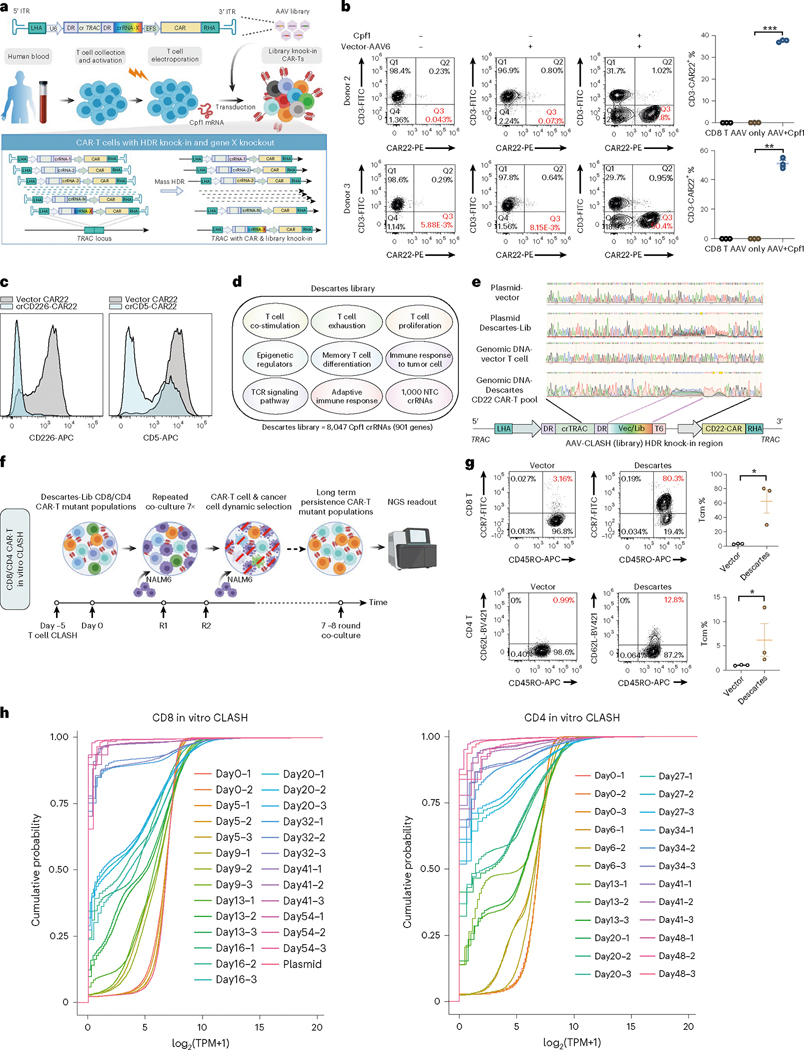
Taking advantage of AAV vectors, we encoded three components into the transgene: HDR arms for targeted knock-in; cargo expression cassette, such as a CAR; and Cas12a/Cpf1 crRNA expression cassette for genetic manipulation (Fig. 1a). Although CLASH/HDR can be used to target any genomic loci, we first designed it to target the TRAC locus for clinically relevant CAR knock-in. The base of the CAR expression cassette can be standardized so that all variants are directly comparable for their phenotypes. The crRNAs can be a single element or can be readily engineered in a pooled manner by simple molecular cloning. Due to the advantages of transactivating CRISPR RNA (tracrRNA) independence, multiple crRNAs can be engineered to be expressed under the same polymerase III promoter. We, therefore, engineered an AAV vector (CAR CLASH AAV vector, CLASH vector for short) expressing three components: (1) an anti-CD22 CAR construct with CD22-scFv, a transmembrane domain (TM) and a signaling domain (4–1BB, CD3z) (CAR22 for short); (2) a constitutive crRNA targeting the 5′ end of the first exon of TRAC to enable knock-in; and (3) a wild-card crRNA cassette separated with crTRAC by Cas12a/Cpf1 direct repeats (DRs) to test virtually any number of crRNAs against any set of genes. All of these components were flanked by the 5′ and 3′ TRAC HDR arms so that they can be simultaneously knocked-in into the same position (Fig. 1a). The CLASH AAV vector thereby offers three distinct functions in one setting: genomic knock-in, cargo transgene (CAR) expression and variable targeted mutagenesis.
Fig. 1 |. Establishment of CLASH system for massively parallel knock-in engineering.
a, Schematic of CLASH-mediated simultaneous CAR-T and Descartes library knock-in. b, Left, flow cytometry plots showing representative CAR22 knock-in into TRAC locus after CLASH vector-AAV6 transduction for 5 days (AAV6 g-MOI = 1 × 105). Right, quantification of CD3−CAR22+ percentages (infection replicates, n = 3). c, Representative flow cytometry analysis of CLASH knockout efficiency on surface makers (infection replicates, n = 3). d, Schematic of Descartes library design. Not drawn to scale. e, Sanger sequencing results of Descartes-Lib AAV plasmid and pooled Descartes-Lib CD22 CAR-T cells gDNA at TRAC locus. f, Schematic of the CD8 or CD4 CAR-T in vitro CLASH. CAR-T cells were co-cultured with NALM6 at E:T ratio = 0.2:1 for 7–8 rounds after electroporation. g, Left, representative flow cytometry plots of memory marker expression on CD8 or CD4 vector and Descartes-Lib CAR-T cells at endpoint of in vitro co-culture. Tcm = CD45RO+CCR7+ or CD45RO+CD62L+. Right, quantification of Tcm percentages (infection replicates, n = 3). h, Empirical CDF of crRNA representations in the genomic readouts of CD8 and CD4 in vitro Descartes-Lib CLASH knock-in CAR-T pool samples. TPM, transcripts per million. Statistics: in b and g, unpaired two-sided t-test was used to assess significance. *P < 0.05, **P < 0.01 and ***P < 0.001. Data are shown as mean ± s.e.m.
To enable massively parallel CAR knock-in human primary T cells, we developed and optimized a workflow for CLASH-mediated human CAR-T cell engineering (Fig. 1a and Supplementary Fig. 1a–c). We first delivered Cas12a/Cpf1 mRNA into human primary CD8 T cells by electroporation and then transduced with AAV6 encoding CLASH vector or library. To test CLASH’s CAR generation efficiency, the on-target integration of CAR into T cells was measured by fluorescence-activated cell sorting (FACS) at 5 days after transduction. By staining CD3 that forms a surface complex with T cell receptor (TCR)31, we detected TCR knockdown efficiency (in terms of CD3− T cells) of >60%, with on-target integration of CAR22 (CD3−CAR22+) at 37.4% and 51% in donor 2 and donor 3 CD8 T cells, respectively (Fig. 1b). To test the CLASH system’s targeting efficiency at the protein level, we designed crRNAs targeting T cell surface markers CD226 and CD5. Flow cytometry analysis showed that the CLASH vectors encoding crCD226 or crCD5 substantially knocked down CD226 and CD5 surface expression in CAR-T cells (Fig. 1c).
To achieve massively parallel CAR-T engineering with immunologically relevant targets, we designed a high-quality Cas12a/Cpf1 crRNA library, called Descartes, to diversify the wild-card crRNA position with targeted mutagenesis. The initial library targets 954 immune genes, chosen as a superset from gene sets that are implicated in T cell exhaustion32, epigenetic regulators33, T cell co-stimulation, memory T cell differentiation, TCR signaling pathway, adaptive immune response, immune response to tumor cell, T cell proliferation and an epigenetic regulator TET2 previously reported to be important for CAR-T function34 (Methods and Supplementary Dataset 1, CLASH dataset). All crRNAs were scored for selection using Seq-deepCpf1 (ref. 35) to enhance gene editing efficiency (Supplementary Fig. 1d,e). After filtering of genes with fewer than four effectively available crRNAs from the design, the final library of Descartes contained 8,047 crRNAs, targeting 901 genes with eight crRNAs per gene for most of the genes, plus 1,000 non-targeting controls (NTCs) (Fig. 1d and Supplementary Dataset 1). We cloned these libraries into the CLASH AAV vector and verified the library compositions by next-generation sequencing (NGS) using vector-specific primer readout (Supplementary Fig. 1f).
To test (1) whether the entire CLASH construct was integrated into the TRAC locus in the human T cell genome and (2) whether the scale of the knock-in was achieved for multiple constructs in the same pool of T cells, we used specific primers flanking the genomic regions outside the 5′ and 3′ HDR arm to amplify the genomic regions rather than the AAV donor and sequence the inserted region. Sanger sequencing results showed that, first, the designed knock-in regions were indeed in the genomic DNA (gDNA); and second, there is clear sequence degeneracy in the crRNA wild-card region, suggesting that diverse crRNAs exist in the targeted pool of human T cells (Fig. 1e and Supplementary Fig. 1h). With successful pooled knock-in observed, it was feasible to use this CLASH system for high-throughput generation of defined genomic-integrated CARs in custom-defined library scale in human T cells.
CLASH in vitro selection in long-term co-culture systems
CAR-T cell therapy is limited by poor T cell expansion and persistence, especially under chronic exposure to viral or tumor antigens that can result in T cell dysfunction17,36. Thus, we set out to harness the CLASH system to identify more persistent forms of CAR-T cells. We observed that repeated co-culture of CAR-T cells with antigen-specific tumor cells at a low effector:tumor (E:T) ratio significantly promoted T cell differentiation, leading to a reduced T central memory cell population (CD45RO+CD62L+) and decreased capacity to express IFNγ and TNFα with continuous exposure (Supplementary Fig. 1g,i). These results are consistent with a recent study showing that continuous antigen exposure recapitulates hallmark features of T cell dysfunction both in vitro and in CAR-T cell patients37. We, therefore, designed an in vitro long-term co-culture system to identify genes whose perturbation can increase the longevity and cytotoxicity of CAR-T cells under chronic antigen exposure (Fig. 1f). We first used the AAV Descartes library (Descartes-Lib) to rapidly generate pools of TRAC knock-in human CAR-T variants (Supplementary Fig. 1j). We also used the empty CLASH vector to generate control knock-in CAR-T cells that are identical except additional mutagenesis. After transduction with vector or Descartes-Lib AAV6, the control or pool mutant CAR-T cells were repeatedly co-incubated with NALM6 cells at E:T ratios of 0.2 for 7–8 rounds, and a fraction of them were collected at each round for gDNA prep and deep sequencing. We performed the long-term culture with three independent series, so that each CLASH Descartes CAR-T pool has a matched time series. We first performed this set of experiments using primary human CD8 T cells (‘CD8 in vitro CLASH’, ‘CD8 CLASH’ for short). We then repeated the whole set of experiment using primary human CD4 T cells (‘CD4 in vitro CLASH’, ‘CD4 CLASH’ for short), with slightly different timepoints of cancer stimulation, due to natural differences in the dynamics of CD8 versus CD4 CAR-T cell killing.
Initially (at day 0), the vector and Descartes-Lib transduced CAR-T cell pools showed similar immune phenotypes (Supplementary Fig. 1k,l). During tissue culture, we observed dynamic alterations of the cancer cell to CAR-T cell ratios after multiple rounds of co-culture, suggestive of selection within the pools. At the endpoint, vector CAR-T lost killing ability, whereas the Descartes-Lib CD8 CAR-T pool had markedly more efficient tumor cell clearance, with a concordant increase in IFNγ production and significant reduction in T cell exhaustion markers (PD-1 and LAG-3) (Supplementary Fig. 1m,n). Although the percentages of central memory T cell (Tcm) populations in CAR-T cells in vector and Descartes-Lib CAR-T cells were not different before stimulation, at the endpoint of selection the Descartes-Lib CAR-T pool showed a substantially higher fraction of Tcm populations compared to that of control CAR-T (Fig. 1g). The CD4 in vitro CLASH experiment also recapitulated this phenomenon, showing a common selection toward Tcm populations (Fig. 1g). The observation in the differences between pool mutant versus single-vector CAR control at the gross population level suggested that at least subsets of mutant variants in the Descartes-Lib pool were contributing to the shifts of various phenotypes of these CAR-T cells, demonstrating efficient selection by CLASH.
In vitro CLASH showed robust time-dependent selection
To determine the actual composition and dynamics of the variants over the whole timecourse, we developed a CLASH library readout method and performed NGS on all the experimental replicates at all collection timepoints over the entire timecourse of CAR-T: cancer cell co-culture (‘readout’). We performed the readouts for both CD8 in vitro CLASH and CD4 in vitro CLASH experiments and generated CLASH datasets delineating crRNA representation in the Descartes library across all samples, to provide a holistic picture of the CAR-T repertoires and how they change under defined selections (Supplementary Fig. 2a–c and Supplementary Dataset 1). Both CD8 and CD4 in vitro CLASH experimentsshowed the consistency of matched samples along the timecourse trajectory (Supplementary Fig. 2b). Over time, there was a gradual reduction in normalized counts for most crRNAs with a concomitant increase in a smaller proportion of the guides, suggesting selection for CAR-Ts with those guides (Supplementary Fig. 2c). In both CD8 and CD4 in vitro CLASH experiments, the diversity of the library dwindled over time, and the CAR-T library pool became increasingly dominated by a smaller fraction of crRNAs over time, as shown by the cumulative distribution function (CDF) plot (Fig. 1h), indicating a time gradient for this process.
To map out the whole set of variants of CAR-Ts in the Descartes pool, we performed a series of analyses to examine the enrichment at individual crRNA level and gene level on both CD8 and CD4 in vitro CLASH datasets. From an overall timecourse heat map of all crRNAs, in both CD8 and CD4 in vitro CLASH experiments, we observed that most crRNAs diminished over time and that most of the crRNAs became depleted at the fifth round of co-culture (Supplementary Fig. 2d,e). Distinct sets of crRNAs emerged as persistent clusters in varying degrees, with a different pattern in CD8 and CD4 in vitro CLASH experiments, as expected given the biological differences between these two cell types (Supplementary Fig. 2d,e). These observations demonstrated that both CD8 and CD4 T cells can be subjected to the pooled CAR knock-ins with CLASH and that selection for and against certain guides can be observed over time.
Timecourse enrichment analyses of in vitro CLASH
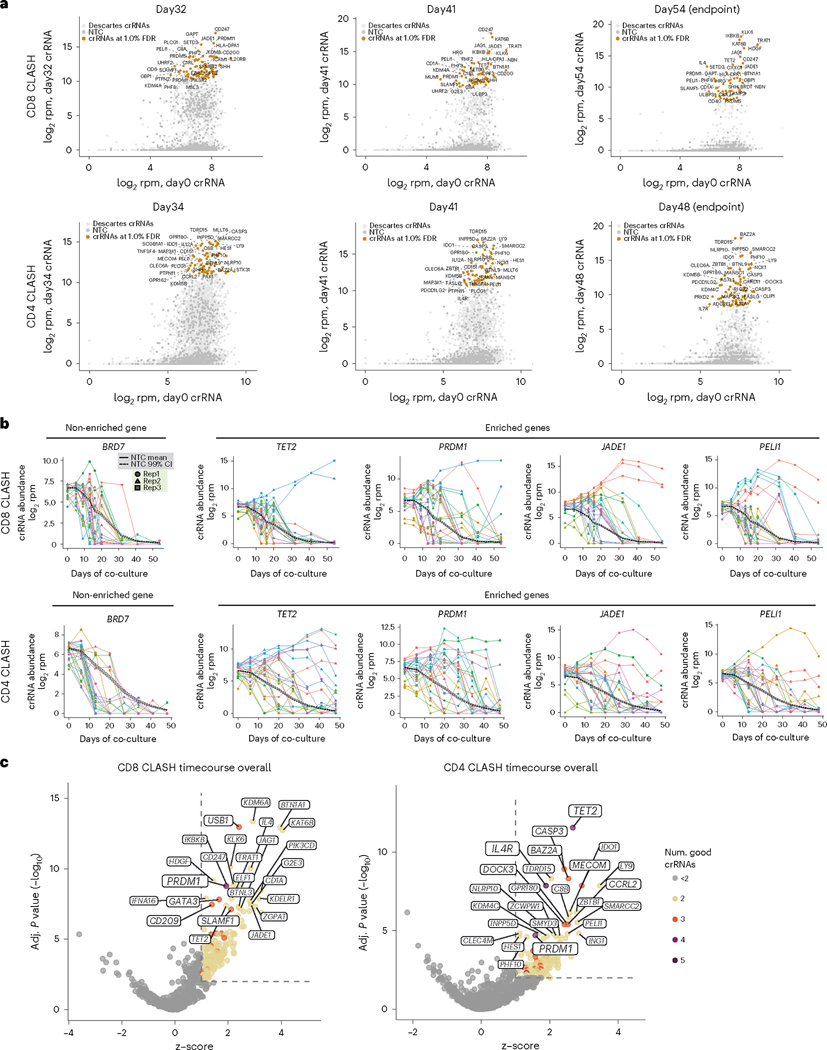
To identify which crRNAs and which genes’ perturbations in CAR-Ts in the Descartes pool lead to more persistent CAR-Ts with potentially enhanced anti-tumor function, we performed a series of timecourse screen analyses of enrichment on two fronts: (1) crRNA level and (2) gene level. We first performed crRNA enrichment analysis based on the bulk distributions of crRNAs in the library compared to the 1,000 NTCs (referred to as ‘bulk analysis’). As can be seen from the crRNA distributions, at the last three rounds of selection (‘last three rounds’ or ‘three latest timepoints’), although most crRNAs were depleted, a fraction of crRNAs were highly enriched (Fig. 2a). In CD8 in vitro CLASH at latest timepoints, highly enriched crRNAs (at a 1% false discovery rate (FDR); Methods) targeting genes such as JADE1, PELI1, CD247, HLA-DPA1, SLAMF1, PHF2, SETD3, IKBKB, PRDM5, TET2 and PRDM1 were observed (Fig. 2a). TET2 has been previously identified by the Carl June group as a key factor suppressing CAR-T expansion and persistence in vivo34 and, thus, represents as a gene for persistence that has been validated in the clinic. In CD4 in vitro CLASH, later timepoints showed highly enriched crRNAs (1% FDR) targeting genes such as PELI1, JADE1, NLPR10, IDO1, PHF10, CASP3, BTNL9 and TDRD15 (Fig. 2a).
Fig. 2 |. Timecourse in vitro CD8 and CD4 CLASH experiments identified genetic mutations that promote CAR-T cell persistence in long-term co-culture.
a, Bulk analysis of relative crRNA abundances in Descartes library at the three latest co-culture timepoints, compared with day 0 T cells. Points shown at crRNA level. Top 30 crRNAs (log2rpm, day X versus day 0) labeled with corresponding gene names. b, Timecourse analysis of crRNA abundance for selected significant genes. Dark solid line, average timecourse for the 1,000 NTC crRNAs. Dark dashed lines, upper and lower 99% CIs for NTCs. c, SAMBA analyses for CD8 and CD4 in vitro CLASH, taking the whole timecourse trajectory for each guide into account, using a model that regresses on time to identify overall significant genes (z > 1 and FDR-adjusted P < 0.01) (Methods). The top 25 genes by significance were labeled and sized by the number of ‘good crRNAs’ for that gene. BM, bone marrow.
We also examined the abundance of specific guides across the whole course of selection, including all individual crRNAs targeting the same gene for all three replicates (Fig. 2b, Supplementary Fig. 3a and Supplementary Dataset 1). We compared them to the trend of putative neutral guides—that is, the mean and 99% confidence interval (CI) of the 1,000 NTCs (Fig. 2b and Supplementary Fig. 3a). As a baseline, the NTCs rapidly decreased over time, with >95% depleted after co-culture for 30 days in CD8 CLASH and >90% depleted in CD4 CLASH (Fig. 2b and Supplementary Fig. 3a). We highlighted BRD7 as an example of a non-enriched gene showing similar timecourse behavior as NTCs, where the crRNAs for BRD7 also rapidly decreased over time (Fig. 2b and Supplementary Fig. 3a). For known T cell exhaustion or checkpoint inhibitor genes (such as PDCD1, BTLA and LAIR1), crRNAs were largely above the neutral upper 99% CI of NTC line; most crRNAs for such genes were depleted by the last round (Supplementary Fig. 3a), suggesting that these checkpoint genes were not necessarily the most important genes for regulating CAR-T persistence. In contrast, genes that scored as top hits (such as TET2, PRDM1, JADE1 and PELI1) were robustly selected for in both CD8 and CD4 CLASH, with two or more independent crRNAs maintaining high abundance at the three latest timepoints (Fig. 2b). These data reveal with greater granularity the guide-level behaviors of genetic perturbations determined as enriched at a gene level.
One limitation of bulk is that guide or gene enrichment can be evaluated only for each timepoint separately. However, among the other currently available pool screen analysis methods (such as RIGER, RSA, HiTSelect, BAGEL, CERES or MAGECK)38, none can integrate all factors, including time series, multiple guides targeting the same gene, control guides, FDRs and experimental groups. To define enriched genes using the entire timecourse trajectory using the entire set of information from the dataset (including all timepoints, all gene-targeting guides, all NTCs and all experimental groups), we used a custom generalized linear model (SAMBA) that factors in time in the design matrix (Methods), incorporating information from multiple guides to return statistical analysis at a gene level39. Using this approach, we identified genes that were significantly enriched in both CD8 and CD4 CLASH experiments (Supplementary Dataset 1 and Fig. 2c). This analysis returned a number of statistically significant genes when considering all crRNAs and all timepoints (‘overall significant genes’) (Supplementary Dataset 1). For example, at a criteria of FDR-adjusted q < 0.01 and a z-score cutoff of 1, there are 158 overall significant genes in CD8 CLASH, which reduce to 86 genes with a higher z-score cutoff (z-score > 1.5, FDR q < 0.01) (Fig. 2c and Supplementary Dataset 1). There are 136 overall significant genes in CD4 CLASH at q < 0.01 and z > 1, which reduce to 106 genes with higher z-score cutoff (z > 1.5, FDR q < 0.01) (Fig. 2c and Supplementary Dataset 1). There are 22 shared overall significant genes that scored in both CD8 CLASH and CD4 CLASH (z-score > 1, FDR q < 0.01), including TET2, PRDM1, KAT6B, PIK3CD, JADE1, BTNL3, PTPN11, PELI1, PRDM5, PTPN2, C7, ASXL1, MASP2, SHH, FASLG, RORC, KDM4C, TRAF2, DNMT3A, PIK3R2, SFMBT2 and TDRD15. Of note, PRDM1 and TET2 through the SAMBA analysis scored as top hits in both CD8 and CD4 in vitro CLASH experiments (Fig. 2c). Example divergent genes include GATA3, a top hit in CD8 in vitro CLASH (but not CD4 CLASH) that is a zinc-finger transcription factor driving dysfunction in CD8+ T cells40; and IL4R, a top hit in CD4 in vitro CLASH (but not CD8 CLASH) that is a critical receptor for the process of naive T cell differentiation into Th2 effector cells41. We visualized the overall in vitro timecourse of hit genes from SAMBA analyses (Supplementary Fig. 3b). These critical genes’ perturbations are enriched in the persistent CAR-T pools in the long-term co-culture setting. These data revealed the CD4 and CD8 CLASH hits from a systematic view, which further supported CLASH as an engineering screening platform.
Gene editing efficiency correlates with CLASH performance
To determine whether the gene editing ability of crRNAs correlate with their screen performance in the CLASH experiment, we used a mini AAV pool and performed a CLASH-MIPS (molecular inversion probe sequencing) experiment in T cells (Supplementary Fig. 4a and Supplementary Dataset 2, MIPS dataset). With biological triplicates, we jointly measured the crRNA abundance by genome-integrated crRNA library readout and the actual gene editing efficiency of individual crRNAs by MIPS and then compared the results with the crRNAs’ performance in CLASH experiments. Library readout successfully mapped the crRNA abundance of the minipool (Supplementary Fig. 4b). We compared the ability of individual crRNAs’ gene editing (measured by MIPS) to their screen performance in CLASH-Descartes experiment using the d32 (round 5) data as a balanced timepoint, when a substantial time period has elapsed for selection but where selection was not so strong that most crRNAs were lost (for example, last round). Considering all genes / all crRNAs measured, the overall gene editing efficiency (MIPS) was significantly correlated with mean screen performance (CLASH) (Supplementary Fig. 4c). This significant correlation also held regardless of whether the gene editing efficiency is normalized by crRNA abundance (Supplementary Fig. 4d) or not. These data suggested that the screen performance of crRNAs significantly correlates with their capacity for gene editing in T cells, and the crRNAs enriched in the screen largely represent true cutters and screen performers for the enriched genes.
CLASH-mediated in vivo selection
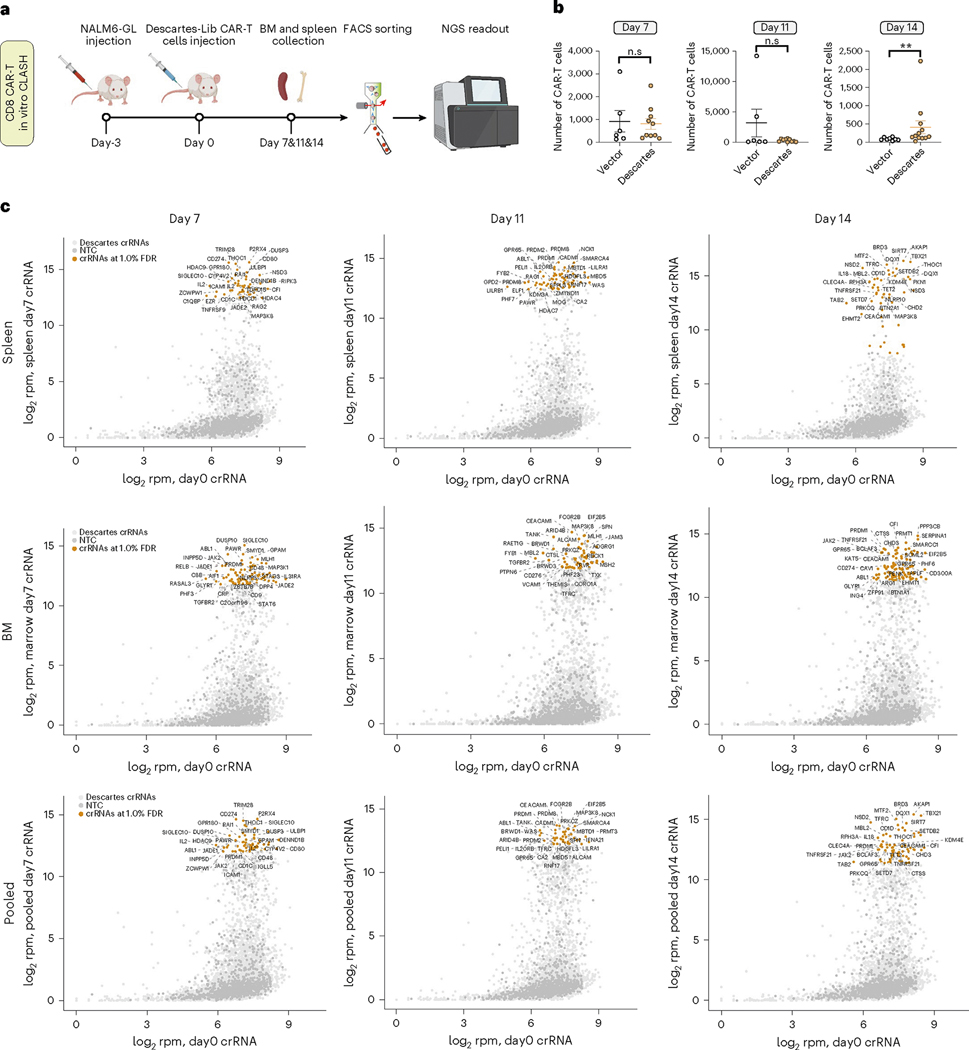
To further identify which CAR-T variants have better anti-tumor activity, we designed and performed a timecourse in vivo CLASH-Descartes experiment to identify genetic perturbations that can increase CAR-T persistence in a leukemia mouse model (Fig. 3a and Supplementary Dataset 1). We performed tumor induction by transplanting NALM6-GL cells into NSG mice. Three days after induction, we infused Descartes-Lib CAR-T variants into mice by adoptive transfer and collected bone marrow and spleen samples at day 7, day 11 and day 14. With the CAR-T infusion in this animal model, we found that, at these timepoints, cancer cells were largely cleared in bone marrow and spleen in both vector and Descartes-Lib groups, whereas more CAR-T cells survived in recipients of Descartes-Lib CAR-T group at day 14 (Fig. 3b and Supplementary Fig. 5a). We then read out the crRNA library representation of the Descartes library in these in vivo samples and analyzed the deep sequencing data to identify enriched crRNAs (Supplementary Fig. 5b–d). The CDF plot showed that the CAR-T library pool was dominated by a smaller fraction of crRNAs at day 7, day 11 and day 14, with strong selection already observed at day 7 (Supplementary Fig. 5b). We performed bulk analysis to identify enriched crRNAs and genes. Because the in vivo CLASH dataset has only three timepoints and a high level of dropouts due to the nature of strong selection, SAMBA is less suitable as it requires continuous distributions of data along the timecourse. From bulk analysis (FDR < 1%), we identified lists of crRNAs and genes enriched at each timepoint (Fig. 3c). Notable genes among these top hits included CEACAM1, a co-inhibitory receptor after T cell activation42. In addition, the T cell inhibitory receptor SIGLEC10 (ref. 43) and negative regulators of T cell proliferation and T cell differentiation, GPR65 (ref. 44) and TNFRSF21 (ref. 45), also scored as top genes. Of note, PRDM1 and TET2, important in the two in vitro CLASH experiments, were also identified as enriched genes in in vivo CLASH (Fig. 3c).
Fig. 3 |. In vivo CLASH in a mouse model of cancer with adoptive transfer of CAR-T pool.
a, Schematic of an in vivo CLASH-Descartes experiment in a mouse model of leukemia and CAR-T adoptive transfer. Three days after NALM6-GL injection, 2 × 106 vector or Descartes CAR-T cells were infused into mice. Bone marrow and spleen samples were collected at day 7 (n = 5 mice), day 11 (n = 5 mice) and day 14 (n = 6 mice) and sequenced by NGS. b, Quantification of the in vivo numbers of CAR-T cells at day 7, day 11 and day 14 (pooled spleen and bone marrow samples). Unpaired two-sided Mann–Whitney test was used to assess significance. Day 7 (n = 5 mice, 10 samples), day 11 (n = 5 mice, 10 samples) and day 14 (n = 6, 12 samples). Total tested live cells normalized to 1 million as inputs. **P < 0.01 and NS, not significant. Data are shown as mean ± s.e.m. c, Bulk analysis of relative crRNA abundances in Descartes library in CD8 in vivo CLASH experiment at day 7, day 11 and day 14 (spleen, bone marrow and pooled analyses) versus day 0 T cells. The top 30 crRNAs by log2rpm of the experimental condition (y axis) are labeled with corresponding gene names. Points are shown at the individual crRNA level.
Because solid tumor infiltration is an important topic for CAR-T cell therapy, we also tested the performance of CLASH in a solid tumor model (Supplementary Dataset 1). The CDF plot and clustering heat map again showed that, whereas the cellular CLASH pool of mutants clustered together, there is a strong shift of the library representation in tumor-infiltrating CAR-T pools from in vivo samples (Supplementary Fig. 5e,f). Evaluation of the tumor-infiltrating CAR-Ts as compared to the cellular pool revealed a number of top hits, such as ITK, PTPN22, PTPN6, LIG4 and TCF20, including several common top hits shared with the other in vitro and in vivo CLASH experiments, such as PRDM1, C1QC, DPF3 and ZCWPW2 (Supplementary Fig. 5g). Taken together, these data demonstrate the robustness of the in vivo CLASH and provide a ranked gene list of potential targets to improve CAR-T function.
Individual functional analysis of genes scored in CLASH
The naturally different biology of CD8 versus CD4 T cells, or in vitro versus in vivo, led to different selection. Nevertheless, we observed a number of shared genes between two or three different datasets (for example, PRDM1, JADE1, PELI1, EHMT1, KDM4E and NLRP10) (Figs. 2 and 3 and Supplementary Dataset 1). PRDM1 has been previously identified as a master regulator of normal CD8 T cells46. PELI1 is a negative regulator of T cells, and its natural function curtails autoimmunity47. NLRP10 has been linked to CD4+ T-cell-mediated IFNγ responses48. Several genes (for example, JADE1, EHMT1 and KDM4E) have relatively little attention in T cell studies.
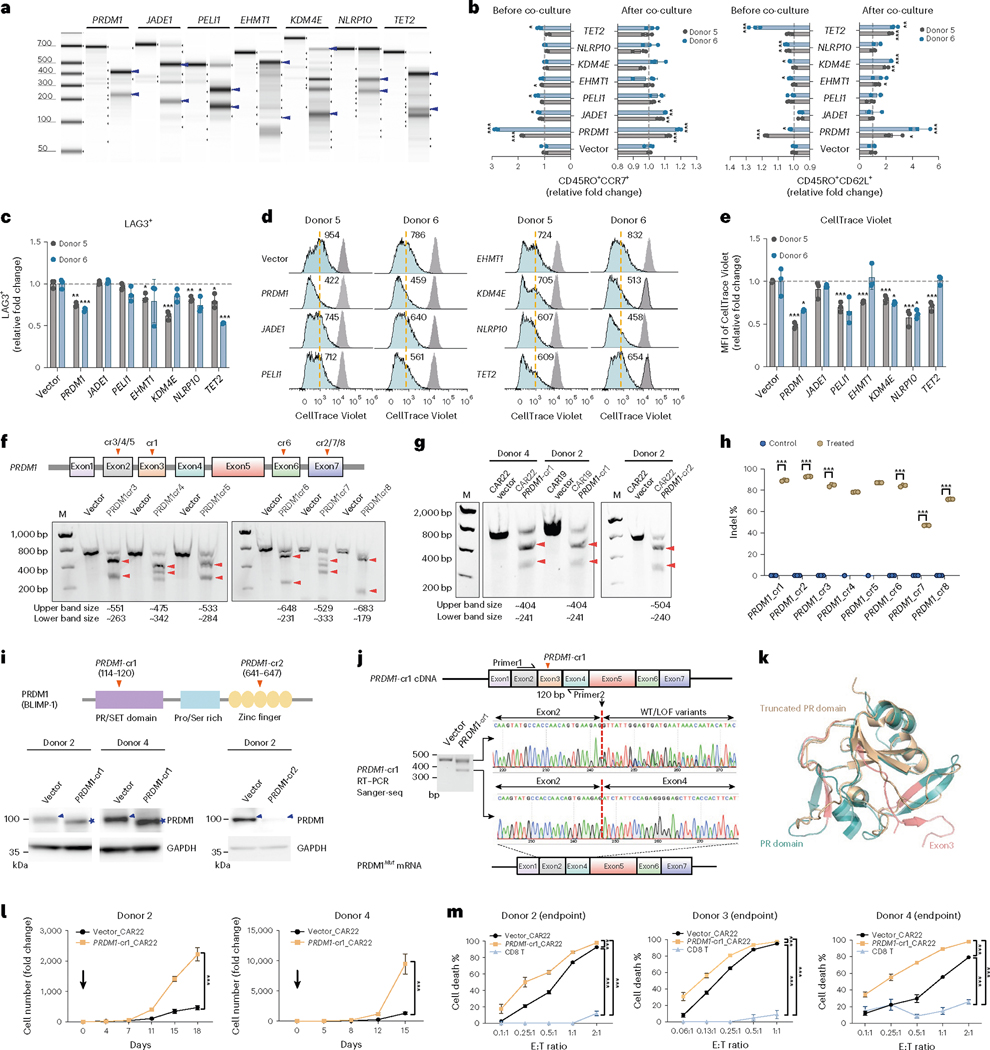
We further tested these candidate genes one by one in CAR-T cells. To validate whether these genetic variants of CAR-T cells have enhanced anti-tumor phenotypes, we first re-generated individual CAR-T variants, using single AAV-CLASH vectors encoding CAR22 and the top-rank crRNAs targeting each of these genes (PRDM1, JADE1, PELI1, EHMT1, KDM4E and NLRP10), along with TET2. We next measured the gene editing efficiency via T7E1 endonuclease assays. crRNAs for all these seven genes showed high efficiency of gene editing (Fig. 4a). We also tested the frequency of mono-allelic versus bi-allelic disruptions by flow sorting of single cells after gene editing mediated by the CLASH system (Supplementary Fig. 6a). Consistent with T7E1, more than 90% of monoclonal cells from crJADE1 and crPELI1 have gene disruptions (91.7% for crJADE1 and 90.0% for crPELI1) (Supplementary Fig. 6b,c). Among them, crJADE1 produced approximately 66.7% homozygous mutant and 25% heterozygous mutant, and crPELI1 produced approximately 55% homozygous mutant and 35% heterozygous mutant. (Supplementary Fig. 6b,c). We measured the Tcm population (CD45RO+CCR7+ and CD45RO+CD62L+) before or after co-culture with cognate cancer cells in two different CD3 T cell donors. Before co-culture, we observed that the Tcm populations were significantly increased in PRDM1 and NLRP10 mutant CAR-Ts as compared to vector control CAR-Ts. After stimulation with targeting cancer cells for 3 days, we found that PRDM1, JADE1, KDM4E and TET2 mutant CAR-Ts had higher Tcm populations in both T cell donors (Fig. 4b). Conversely, PRDM1 and TET2 mutants exhibited lower levels of activation marker CD69 and cytoplasmic granule toxins compared to vector CAR (Supplementary Fig. 6d,e). Accumulating evidence suggests that the balance between memory and effector T cell functions is required for T cell health and longevity49,50. We next tested whether these convergent genes also change other T cell phenotypes and functions. We quantified T cell exhaustion levels by CTLA4 and LAG3 staining and found that PRDM1-edited CAR-T cells have lower levels of CTLA4 after NALM6 stimulation (Fig. 4c). PRDM1, NLRP10 and TET2 CAR-T mutants exhibited lower levels of LAG3 compared to the vector (Fig. 4c). Interestingly, T cell proliferation was faster in PRDM1, PELI1, KDM4E and NLRP10 mutant CAR-Ts after two rounds of stimulation (Fig. 4d,e).
Fig. 4 |. Individual functional analysis of immune genes scored in CLASH.
a, T7E1 assay for individual crRNAs in CAR-T cells. Predicted cleaved bands, blue arrows. b–e, Flow analysis of mutant CAR-T cells in two donors (infection replicates, n = 3). b, Quantification of Tcm (CD45RO+CCR7+ or CD45RO+CD62L+). c, Quantification of exhaustion markers (LAG3+). d, Representative histograms of CellTrace Violet in mutant CAR-T cells, measured at day 0 (gray peak) or day 3 (cyan peak). Yellow dashed line, MFI for vector CAR-T. e, CellTrace Violet MFI quantification. f, T7E1 for PRDM1-cr3 to cr8 from donor 2 CAR-T cells. g, T7E1 for PRDM1-cr1and cr2 on another donor and different CAR-T construct. Note for a, f and g, gel is representative of three repeated experiments. h, Indel quantification of PRDM1-cr1 to cr8 in donor 2 CAR22 T cells (technical replicates, n = 3 for most samples, except cr4_control and c5_control (n = 1)). i, Top, schematics of PRDM1 protein primary structure. Bottom, PRDM1 protein western blot in vector and PRDM1 mutant CAR-T cells. Blot is representative of three repeated experiments. Blue arrowheads, WT protein. Blue stars, truncated mutant protein generated by PRDM1-cr1. j, Exon3 skipping after PRDM1-cr1 editing in CAR-T cells verified by Sanger sequencing of RT–PCR products. Note for f–j, cutting sites of crRNAs, vertical arrowheads on PRDM1 exon diagram. Predicted cleaved bands, horizontal arrowheads. Gel is representative of three repeated experiments. k, Protein structure alignment of PR domain (blue, PDB: 3DAL) and PR Δexon3 domain (gold, predicted via AlphaFold). Exon3 peptide, highlighted pink. l, Proliferation of PRDM1 mutant and control CD22 CAR-T cells with NALM6 stimulation (cell culture replicates, n = 3). m, Cytotoxicity of vector and PRDM1 mutant CAR-T cells for different donors (cell culture replicates, n = 3). Statistics: b, c and e, unpaired two-sided t-test; h, l and m, two-way ANOVA. *P < 0.05, **P < 0.01 and ***P < 0.001. Data are shown as mean ± s.e.m. LOF, loss of function.
Validation and characterization of PRDM1 mutations
Based on its robust performance in three CLASH experiments and initial in vitro validation results, PRDM1 represents a promising candidate of CAR-T engineering. We performed a series of experiments, including genetic dissection, biochemical characterization, transcriptomics, epigenetics and epigenomics and immunological profiling. We first measured all PRDM1 crRNAs’ gene editing efficiency individually in donor 2 anti-CD22 CAR-T cells (CD22 CARs) by T7E1 endonuclease assay and NGS. Most of the eight independent PRDM1 crRNAs, targeting different regions of the gene, demonstrated efficient gene editing, with two lead crRNAs validated in different donors (Fig. 4f–h). We found that PRDM1-cr2 led to strong reduction of the PRDM1 protein, yet, interestingly, PRDM1-cr1 led to a production of a smaller-sized protein recognized by the same PRDM1-specific antibody (Fig. 4i, blue stars). We performed immunoprecipitation using anti-PRDM1 antibody and peptide identification by mass spectrometry (IP-MS) and confirmed the absence of the peptides near the PRDM1-cr1 cutting sites in PRDM1-cr1-targeted mutant CAR-T cells, in all three replicates (Supplementary Fig. 9a,b). To determine the nature of the mutant variant of PRDM1 gene product produced by PRDM1-cr1, we designed two primers near the PRDM1-cr1 cutting site and used RT–PCR to identify the cDNA. Interestingly, we found that there was a 120-bp in-frame deletion in the lower DNA fragment that precisely corresponded to PRDM1 exon3 (Fig. 4j). These results demonstrated that CLASH PRDM1-cr1 generated a PRDM1 exon3 skipping (‘Δexon3’, ‘exon3 skip’) variant and produced a truncated PRDM1 protein in human primary T cells. We used AlphaFold2 to predict the structure of PRDM1 Δexon3 mutant truncated protein and overlayed with the PRDM1 PR domain structure (Protein Data Bank (PDB): 3DAL)51. The Δexon3 truncated protein abolished five β-strands in the PR domain of PRDM1 (Fig. 4k). We further tested the characteristics of this Δexon3 mutant. We observed a strong increase in antigen-specific proliferative capacity of Δexon3 mutant CAR-T cells in another two different donors (Fig. 4l). In the co-culture experiment endpoint, we found that Δexon3 mutant CAR-T cells, as compared to vector cells, had significantly higher cytotoxic effect against NALM6 cancer cells, across all E:T ratios, in CAR-Ts generated from three independent donors (Fig. 4m).
PRDM1 Δexon3 CAR-Ts have enhanced therapeutic efficacy
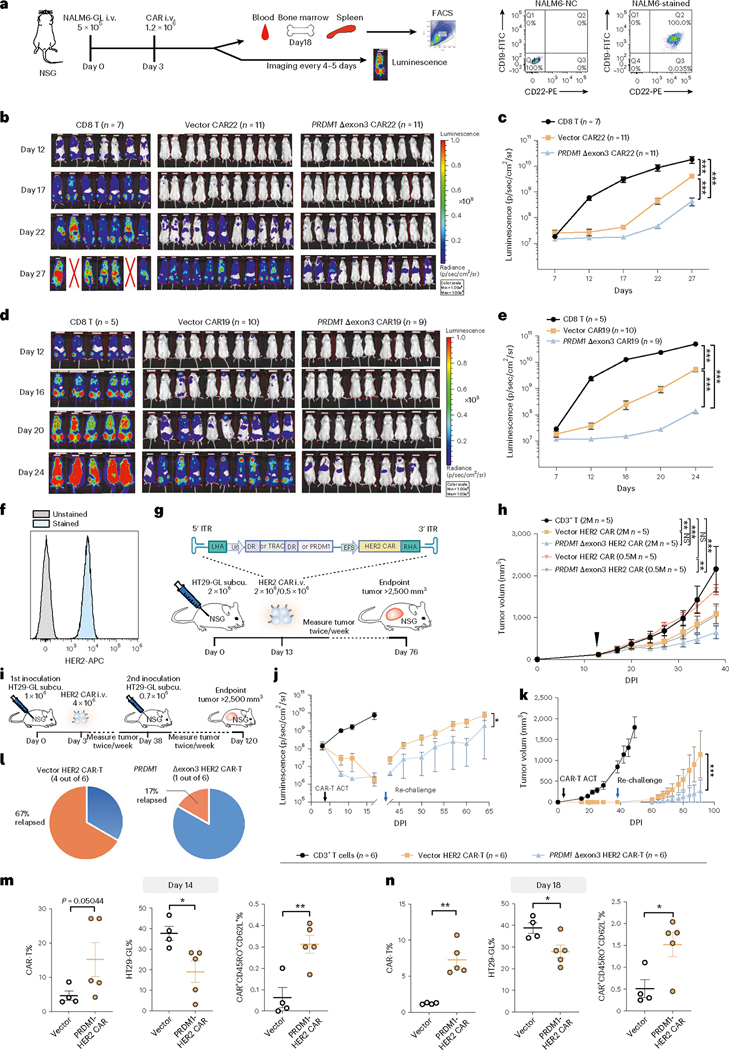
We further tested the pre-clinical therapeutic efficacy of PRDM1 Δexon3 CAR-T cells compared to their vector counterparts in vivo. In an NALM6-GL leukemia tumor model in immunodeficient NOD.Prkdc(SCID)/Il2rγ−/− (NSG) mice, we performed efficacy testing by adoptive transfer of anti-CD22 CAR-Ts, followed by monitoring of tumor burden (Fig. 5a). The NALM6 cells were first confirmed to be CD19;CD22 double-positive before being used for tumor induction (Fig. 5a). We performed In Vivo Imaging System (IVIS) imaging to follow the tumor burden in leukemic animals treated by untransduced CD8 T cells, vector-transduced anti-CD22 CD8 CAR-T cells and Δexon3 anti-CD22 CD8 CAR-T cells. As a result, we found that Δexon3 anti-CD22 CAR-T cells showed significantly stronger leukemia suppression than vector CAR-T cells, where substantial differences were seen in the tumor burden between vector and Δexon3 anti-CD22 CAR-T-cell-treated mice, starting 19 days after T cell adoptive transfer (22 days after tumor induction) (Fig. 5b,c). These data showed that Δexon3 CD22 CAR-Ts have stronger in vivo efficacy against cancer in a mouse model of leukemia. To validate the results, we constructed an independent AAV-CLASH vector with a different CAR construct against the CD19 antigen (anti-CD19 CAR-T, CD19 CAR and CAR19). We similarly generated Δexon3 CD19 CAR-T cells along with vector control CD19 CAR-T cells and performed in vivo efficacy testing using a similar cancer induction and adoptive transfer treatment regimen (Methods). Similar results were observed in CD19 CAR-T cells in vivo, where Δexon3 CD19 CAR-T cells showed significantly stronger leukemia suppression than vector CAR-T cells (Fig. 5d,e). We harvested the blood and organs from the animals treated in both groups and quantified the abundance of persistent CAR-Ts in vivo. Lower numbers of cancer cells and higher numbers of CAR-T cells were observed in bone marrow and spleen of recipients of Δexon3 CAR-T cells at 2 weeks after CAR-T infusion (Supplementary Fig. 7a–c). Consistent with in vitro findings, CAR-T cells from the bone marrow and spleen showed significantly higher levels of CD45RO+CD62L+ (Tcm) in mice treated with Δexon3 CAR-T cells as compared to the vector group (Supplementary Fig. 7d,e). These data suggest that Δexon3 CAR-T cells have enhanced efficacy, along with increased persistence and memory marker expression in vivo.
Fig. 5 |. PRDM1 Δexon3 CAR-Ts showed enhanced in vivo therapeutic efficacy in multiple mouse models of cancer.
a, Left, schematics of liquid cancer models. Right, FACS plot of CD19+CD22+ NALM6-GL cells. b,c, Bioluminescence imaging (b) and quantification (c) of NALM-6 leukemia-bearing NSG mice treated with the following: CD8 T (n = 7), vector CAR22 (n = 11) and PRDM1 Δexon3 CAR22 (n = 11). d,e, Bioluminescence imaging (d) and quantification (e) of NALM-6 leukemia-bearing NSG mice treated with the following: CD8 T (n = 5), vector CAR19 (n = 10) and PRDM1 Δexon3 CAR19 (n = 9). f, FACS histogram of HER2+ HT29-GL cells. g, Schematics of a HER2+ HT29 solid tumor model with two different doses of CAR-T treatments. Non-transduced CD3+ T (2 M, n = 5), vector HER2 CAR-T (2 M, n = 5; 0.5 M n = 5) and PRDM1 Δexon3 HER2 CAR-T (2 M, n = 5; 0.5 M, n = 5), all infused at day 13. h, Tumor growth curves of experiment in g. Black arrow, CAR-T treatment. i, Schematics of a rechallenge HT2 model with CAR-T treatment with the following: non-transduced CD3+ T (n = 6), vector HER2 CAR-T (n = 6) and Δexon3 HER2 CAR-T (n = 6). j–l, Quantification of whole-body bioluminescence (j) and tumor volume (k) over time, and percentage of relapsed animals, of experiment in g. Black arrow, CAR-T injection; blue arrow, tumor rechallenging. m,n, Quantification of cancer cells, tumor-infiltrating CAR-T cells and Tcm in tumor samples, from two cohorts with endpoints at day 14 (m) and day 18 (n) after infusion, respectively. Vector (n = 4) and PRDM1 Δexon3 CAR22 (n = 5). Statistics: c, e, h, j and k, two-way ANOVA; m and n, unpaired two-sided t-test. *P < 0.05, **P < 0.01 and ***P < 0.001 and NS, not significant. Data are shown as mean ± s.e.m. M, million.
To further explore the potential of Δexon3 CAR therapeutic effect in a solid tumor model, we generated a TRAC knock-in HER2-targeting CAR-T CLASH AAV vector and established two different mouse models using HER2+ HT-29-GL human colon cancer cells (Fig. 5f). In the first HT-29-GL solid tumor model (low-dose direct-treatment model), when the average tumor size reached 100 mm3, we infused two different does (0.5 million or 2 million CAR-T cells, single treatment) of vector or Δexon3 HER2 CAR-T cells into mice. In the 0.5-million CAR-T cell dose, the vector control HER2 CAR-T group had little therapeutic efficacy against HT-29-GL solid tumor (P > 0.05, endpoint mean tumor inhibition (MTI) = 22.8%); on the contrary, the Δexon3 HER2 CAR-T group had significant efficacy (P < 0.001, endpoint MTI = 51.2%) and was significantly stronger than control CAR-T (P < 0.01) (Fig. 5g,h). In the 2-million CAR-T cell dose, the vector control HER2 CAR-T group had little therapeutic efficacy, similarly to the ¼ dose (0.5 million) of Δexon3 HER2 CAR-T (P < 0.01, MTI = 49.4%); and the Δexon3 HER2 CAR-T group again demonstrated stronger efficacy (P < 0.001, MTI = 70%) (Fig. 5g,h).
In a second model (high-dose disease-relapse model), we infused 4 million vector or Δexon3 CAR-T cells into HT-29-GL tumor-bearing mice after transplantation for 3 days (Fig. 5i). All mice underwent complete tumor suppression by CAR-T cells 2 weeks after CAR-T adoptive transfer treatment (MTI = 100% for both vector and Δexon3 CAR-T groups) and remained completely tumor-free for the following 5 weeks of monitoring (Fig. 5j and Supplementary Fig. 8a). These mice were rechallenged with HT-29-GL cells 38 days after the initial transplantation (5 weeks after CAR-T infusion). Subsequently, mice in the vector control HER2 CAR-T group had rapid tumor growth; in contrast, mice in the Δexon3 HER2 CAR-T group had a prolonged period of tumor suppression and a significantly slower tumor growth from the rechallenge (P < 0.001) (Fig. 5j,k), consistent with the observation above that Δexon3 CAR-T cells are more persistent. Compared to four of six mice (67%) in the vector control HER2 CAR-T group that showed relapsed tumor growth, only one of six mice (17%) showed relapsed in the Δexon3 HER2 CAR-T treatment group, which was reflected in the overall survival (Fig. 5l and Supplementary Fig. 8a,b). We repeated the experiment with this HER2 CAR-T versus HT-29-GL solid tumor model, harvested the tumors from the animals treated by both Δexon3 and control HER2 CAR-T groups and quantified the abundance of persistent CAR-Ts (Supplementary Fig. 8c,d). Consistent with the results in leukemia model, lower numbers of cancer cells and higher numbers of CAR-T cells were observed in the tumors of recipients of Δexon3 CAR-T cells at both day 14 and day 18 after CAR-T infusion, against suggesting that Δexon3 mutant CAR-T cells are more potent and more persistent in vivo (Fig. 5m,n). Because in vivo persistence is linked to stem-like/memory properties, we analyzed this immunological characteristic and found that Δexon3 CAR-T cells as compared to control have significantly higher fraction of CD45RO+CD62L+ population (Fig. 5m,n). These in vivo data together demonstrated that PRDM1 Δexon3 HER2 CAR-T cells had significantly enhanced anti-tumor activity and performance in a solid tumor model.
PRDM1 Δexon3 loses interaction with histone H4
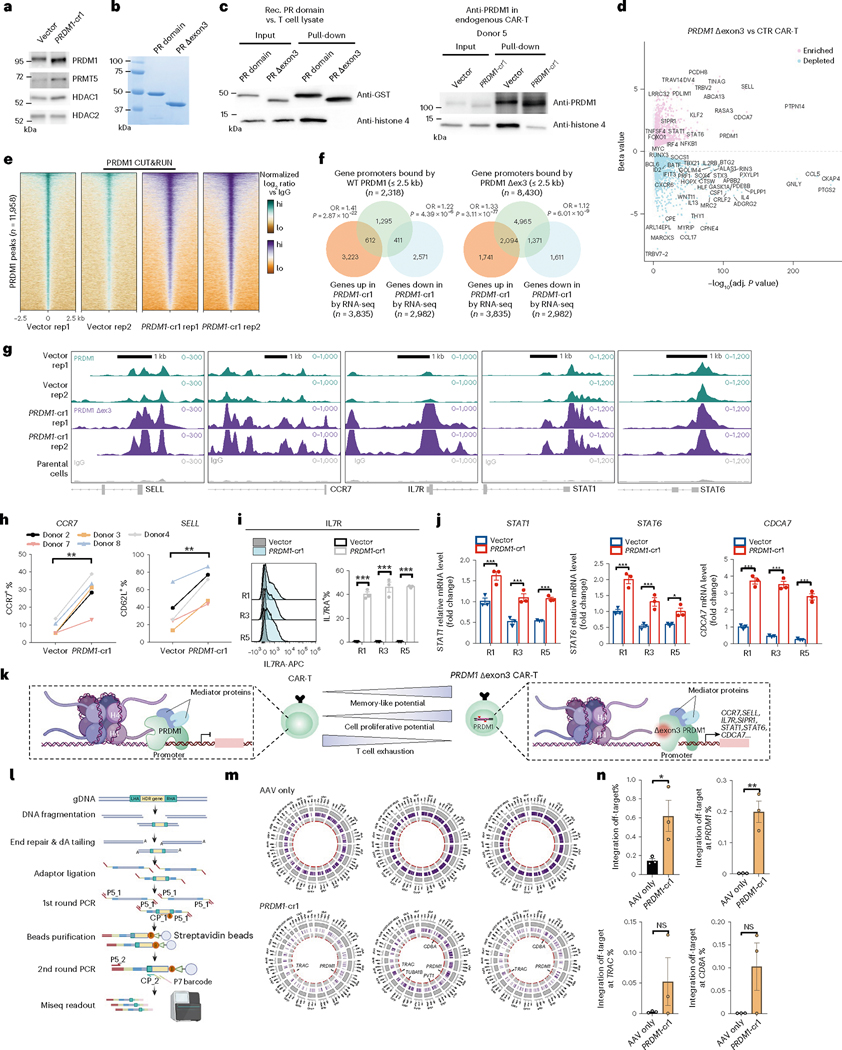
We then further investigated the mechanism of this unique PRDM1 Δexon3 mutant generated by the PRDM1-cr1 in CAR-T cells. PRDM1 exon3 is the main region responsible for encoding the N-terminal PR domain in PRDM1 (PRDI-BF1 or Blimp-1) protein. Disruption of the PR domain can result in a substantial loss of repressive function on multiple target genes52; however, despite various previous work that mostly involves full knockouts46,52–62, the underlying mechanism of the PR domain is, to date, still unknown. To unveil the underlying mechanism, we performed a series of genetic, biochemical and epigenetic experiments. Compared with the wild-type (WT) PR domain, the Δexon3 truncated protein abolished five β-strands, predicted to impair PR domain function (Fig. 4k). The PR domain of the PRDM gene family is a subclass of SET domains most closely related to the known histone methyltransferases63. Prior reports suggest that the PR domain of PRDM1 does not have the histone lysine N-methyltransferase activity64,65. PRDM1 collaboratively silences target gene expression by recruitment of chromatin-modifying co-repressors, such as G9a lysine methyltransferase64, histone deacetylases 1 and 2 (HDAC1/2)54 and protein arginine methyltransferase 5 (PRMT5)65. Interestingly, our co-immunoprecipitation (co-IP) western results showed that the interactions with PRMT5, HDAC1 and HDAC2 were retained in PRDM1 Δexon3 CAR-T cells (Fig. 6a). We, therefore, hypothesized that the PR domain of PRDM1 might have certain interactions with histones.
Fig. 6 |. Molecular characterization of PRDM1 Δexon3 mutant CAR-T cells and genome-wide AAV integration.
a, Co-IP western of PRDM1 binding proteins in CAR-T. b, Coomassie staining of recombinant human WT-PRDM1 PR domain and PR Δexon3 domain. c, Co-IP western revealed that PRDM1 PR domain interacts with histone H4 (HH4). Left, recombinant GST-tagged PR domain, but not Δexon3 mutant, binds HH4 (incubation with T cell lysate). Right, endogenous PRDM1, but not Δexon3, binds HH4 in CAR-T cells. Note: a–c, image is representative of three repeated experiments. d, Volcano plot of DEGs for PRDM1 Δexon3 versus control CAR-T (day 33, n = 3 biological replicates). Differential expression with Sleuth using Wald test. e, Heat map of all aligned CUT&RUN peaks of PRDM1 Δexon3 and WT-PRDM1 tracks. Data are normalized log2 ratios, comparing each sample to IgG average. f, Venn diagram of WT-PRDM1/PRDM1 Δexon3 bound genes and PRDM1 Δexon3 day 33 DEGs. PRDM1 peak ≤2.5 kb away from TSS was defined as WT-PRDM1 bound genes. Two-sided hypergeometric test, assuming 20,465 total human genes. g, Genome browser tracks of CUT&RUN on PRDM1 target genes’ segments. Note: e–g, vector (WT-PRDM1, two replicates), PRDM1 Δexon3 (two replicates) and averaged IgG control. h, CCR7+ and CD62L+ percentages in different healthy donors (n = 5) in vector or PRDM1-cr1 CAR-Ts (day 5). i, Flow cytometry analysis of IL7R (infection replicates, n = 3). j, qRT–PCR analysis of STAT1, STAT6 and CDCA7 (infection replicates, n = 3). k, Schematic of regulation mechanism of PRDM1 Δexon3 in CAR-T. l–n, Characterization of CLASH-PRDM1-cr1 CAR-T’s genome-wide AAV integration. l, Schematic of genome-wide AAV integration library preparation and readout. m, Circos plots of PRDM1-cr1 on-target/off-target integration sites. Purple bands, locations of all individual, unnormalized reads. Red dots, location and frequency of normalized integration reads. n, Quantification of m, for genome-wide sum frequencies of off-target events (top left) or representative genes (infection replicates, n = 3). Statistics: h, paired two-sided t-test; i and j, two-way ANOVA with Sidak’s multiple comparisons; n, unpaired two-sided t-test. *P < 0.05, **P < 0.01 and ***P < 0.001 and NS, not significant. Data are shown as mean ± s.e.m. NGS statistics are noted in specific panels. CTR, control; OR, odds ratio; Rec., recombinant; rep, replicate.
To identify the nature of the histone interactions of PRDM1 PR domain in an unbiased manner, we first biochemically generated and expressed recombinant WT and Δexon3 mutant PRDM1 PR domains (Fig. 6b) and bound them to a histone peptide microarray comprising 384 modified or unmodified histone peptides (Supplementary Fig. 9c). This histone microarray analysis revealed a number of highly specific interactions between WT PR domains and defined histone peptides, which were mostly abolished in the Δexon3 mutant PR domain (Supplementary Fig. 9d–g). This result also uncovered that histone H4 11–30 peptides have specific direct interactions with WT PRDM1 PR domain (Supplementary Fig. 9d–g). We next validated the interaction of PRDM1 PR domain with endogenous histone H4. We expressed the recombinant PR domain and the Δexon3 mutant PR domain tagged with N-terminal GST tag and immunoprecipitated them after incubation with primary T cell lysate. This co-IP western experiment validated that the WT PR domain has clean and strong binding with histone H4, whereas this interaction is fully abolished in the Δexon3 mutant/truncated PR domain (Fig. 6c). We then validated this result in a fully endogenous setting by co-IP western on WT (vector) and PRDM1 cr1 (Δexon3 mutant) CAR-T cells, from independent human donors (Fig. 6c and Supplementary Fig. 9h). Taken together, these data unveiled a previously undocumented observation that exon3 skipping in PRDM1 destroys the PR domain and leads to loss of histone H4 binding.
Because the Δexon3 mutant loses its ability to associate with histone H4, we hypothesized that this might alter its ability to regulate downstream gene expression. To further understand the transcriptional regulatory mechanism of PRDM1 specifically in cr1 targeted (Δexon3 mutant) CAR-T cells, we performed a biological triplicate timecourse transcriptomics experiment in Δexon3 CAR-T cells, along with vector control CAR-T cells and the untransduced human CD8 T cells as a baseline (Fig. 6d and Supplementary Fig. 10a). These transcriptome profiles reveal a systematic landscape of CAR-T gene expression with continuous cognate cancer antigen stimulation along the timecourse (Supplementary Dataset 4, Timecourse PRDM1 Δexon3 CAR-T mRNA-seq dataset). Distinct sets of differentially expressed genes (DEGs) were identified in the direct comparison between Δexon3 and vector control CAR-T cells (Supplementary Figs. 10b,c and 11), revealing a panel of highly significantly DEGs. PRDM1 Δexon3 CAR-Ts showed highly significant induction of genes such as PRDM1 itself, PCDH8, SELL/CD62L, PTPN14, RASA3, KLF2, STAT6, STAT1, IRF4, CDCA7 and NFKB1 and repression of genes such as CCL5, BATF, CXCR6, IL13, PRF1, IFIT13, ID2 and RUNX3 (Fig. 6d and Supplementary Fig. 10b,c).
Interestingly, the alterations of the gene expression showed a gradient pattern in both directions—that is, increasing expression of Δexon3 induced genes and decreasing expression of Δexon3 repressed genes along time (thus, along rounds of stimulation) (Supplementary Fig. 10b,c and Supplementary Dataset 4). To quantify and deconvolve the time factor effect, we performed a time series cluster analysis (Methods). This analysis revealed nine distinct clusters of genes with distinct behaviors along the timecourse between the Δexon3 mutant and control groups (Supplementary Fig. 11a,b and Supplementary Dataset 4). We performed Gene Ontology (GO) analysis of enriched biological processes of the gene sets in each cluster (Supplementary Fig. 11c and Supplementary Dataset 4). The DEGs, both upregulated and downregulated, showed high levels of overlaps between different timepoints, suggesting consistent gene expression signatures in Δexon3 CAR-T cells (Supplementary Fig. 11d,e). These differential expression results from the timecourse mRNA sequencing (mRNA-seq) revealed the collective signatures of T cell proliferation and apoptosis, T cell differentiation, signal transduction, inflammatory response and immune responses in Δexon3 CAR-T cells (Supplementary Fig. 11c,f,g).
Regulation mechanism of PRDM1 WT and Δexon3 mutant proteins
To directly investigate how PRDM1 regulates downstream genes in CAR-T cells and how Δexon3 mutant achieves high-performance anti-tumor immunity in CAR-Ts, we performed genome-wide mapping of chromatin binding using CUT&RUN, a modified approach of chromatin immunoprecipitation followed by sequencing (ChIP-seq)66, in human CAR-T cells using a specific endogenous PRDM1 antibody (Methods). The parallel CUT&RUN experiments also enable us to compare the genome-wide chromatin occupancy of WT PRDM1 (vector group) and PRDM1 Δexon3 (PRDM1 cr1 group). The CUT&RUN data mapped the overall genome-wide chromatin binding landscapes of PRDM1 WT and exon3 mutant proteins in human CAR-T cells (Supplementary Dataset 5, CUT&RUN dataset). When rank-ordered by occupancy over the 5-kb interval centered over WT PRDM1 binding sites, we observed that mutant PRDM1 Δexon3 bound similar genomic loci as WT PRDM1, where more than 90% of the WT PRDM1 peaks were co-bound by mutant PRDM1 Δexon3 (Fig. 6e and Supplementary Fig. 12a). Notably, the Δexon3 group peaks exhibited a broader range of signal intensity (Fig. 6e and Supplementary Fig. 12a). This finding suggested that, despite loss of histone H4 binding, the Δexon3 mutant still maintained chromatin binding capacity in CAR-T cells.
Integration of CUT&RUN and mRNA-seq showed that WT PRDM1 bound genes significantly overlapped with day 33 DEGs, including both upregulated genes (612 shared genes, P = 2.87 × 10−22) and downregulated genes (411 shared genes, P = 4.39 × 10−6) (Fig. 6f). This observation also held in PRDM1 Δexon3 bound genes, which also significantly overlapped with day 33 DEGs, including both upregulated genes (2,094 shared genes, P = 3.11 × 10−77) and downregulated genes (1,371 shared genes, P = 6.01 × 10−9) (Fig. 6f). The stronger signal of overlap in the upregulation side of DEGs is consistent with the role of PRDM1 being primarily a transcriptional repressor. These PRDM1 bound and PRDM1 Δexon3 DEGs contain potential direct targets of PRDM1 regulation in CAR-T cells. We then performed further analysis to investigate the targets of PRDM1 regulation in CAR-T cells. Functional annotation analysis of WT PRDM1 bound upregulated genes showed that they are enriched in cell cycle, cell division, antigen receptor-mediated signaling, chromatin organization and T cell differentiation (Supplementary Fig. 12b). PRDM1 bound downregulated genes are enriched in lymphocyte differentiation, T cell differentiation, cell–cell adhesion, apoptosis, cell death and tyrosine phosphorylation of STAT protein (Supplementary Fig. 12b). Among these potential direct targets, PRDM1 directly bound to genes that encode T cell memory factors, such as SELL/CD62L, CCR7 and IL7R genes, as well as master regulators for T cell proliferation and differentiation, such as CDCA7, STAT1 and STAT6 (Fig. 6g and Supplementary Fig. 12c). Interestingly, we also found that PRDM1 protein bound to the PRDM1 gene itself (Supplementary Fig. 12c), which further validated that PRDM1 is autoregulated55,67, here directly in human CAR-T cells.
To further test the hypothesis of PRDM1ʼs potential direct regulation on these bound genes at the functional level (protein or mRNA), we performed individual target measurements again in a timecourse of multi-round cancer stimulation. We measured the memory markers CCR7 and CD62L on different healthy donors 5 days after transduction in Δexon3 mutant (cr1) as compared to control CAR-T cells. Consistent with previous results, both markers were increased in Δexon3 in all five donors (Fig. 6h). In addition, IL7R was significantly upregulated across all three timepoints in Δexon3 CAR-T as compared to control (Fig. 6i). We then examined upstream regulators such as S1PR1, also a cluster 6 gene, which was found to exhibit a descending trend with continuous antigen exposure in control T cells; this effect was reversed in Δexon3 CAR-T cells (Supplementary Fig. 12d). In addition, STAT1, STAT6 and CDCA7, master regulators for T cells, were significantly increased in PRDM1 Δexon3 CAR-T cells as compared to control, which also gradually decreased over time with continuous antigen stimulation (Fig. 6j). On the other hand, SOCS1, inhibitor of T cell proliferation, was downregulated in a time-dependent manner in Δexon3 CAR-T cells as compared to vector control (Supplementary Fig. 12d). These data showed that the expression of these critical PRDM1 bound genes underlying major T cell pathways is significantly rewired in Δexon3 CAR-T cells. Consistent with the timecourse RNA-seq, flow cytometry analysis showed that Δexon3 CAR-T cells have decreased levels of TIM3, an immune checkpoint encoded by the HAVCR2 gene that also falls into cluster 2 of the RNA-seq timecourse (Supplementary Fig. 12e). We also examined additional surface checkpoints LAG3, 2B4/CD244 and CD39/ENTPD1 (refs. 68,69) and found that all these checkpoints were also significantly and consistently reduced on the T cell surface (Supplementary Fig. 12f), suggesting a robust dampening of exhaustion in Δexon3 CAR-T cells. These data together suggested that PRDM1 Δexon3 CAR-Ts generated by the PRDM1-cr1 have enhanced memory phenotypes, reduced T cell exhaustion, increased cell proliferation and are capable of maintaining longer-term effector function under continuous antigen exposure. These phenotypes are mediated mainly by the disruption of the interaction between the PR domain and histone H4 and the subsequent loss of targeted gene regulation abilities (Fig. 6k).
Genome-wide profiling of CLASH-mediated AAV integration
Because off-target integration is an important safety issue for CAR-T cell therapy involving AAV/CRISPR-mediated manipulations, we set out to map a genome-wide profiling of CLASH-PRDM1-mediated AAV integration. Previous studies have shown that the CRISPR/Cpf1 system has higher editing specificity compared to Cas9 nuclease by using GUIDE-seq, Digenome-seq and BLISS70–72. A recent study performed deep profiling and revealed the heterogeneity of integration outcomes in CRISPR knock-in experiments73. Considering these advances, to profile and quantify the CLASH-PRDM1-mediated genome-wide AAV integration, we modified a method based on GUIDE-seq74 and applied it to CLASH-PRDM1 CAR-T cells, with an AAV-only control without Cpf1 mRNA electroporation (Methods and Fig. 6l). Using ITR-based query sequences, we established a computational pipeline to identify chimeric off-target reads and their locations in the human genome. Using this approach, we performed whole-genome AAV integration mapping for PRDM1 Δexon3 CAR-T cells generated by the AAV-CLASH vector (Supplementary Dataset 6, Genome-wide AAV integration dataset).
Integrative Genomics Viewer (IGV)-based visualization of the normalized reads throughout the human genome showed a clean baseline level in the AAV-only control and a small number of detectable peaks for CLASH-PRDM1-cr1 samples (Supplementary Fig. 13a,b). Circos plot visualizations showed a similar pattern, with the distribution of off-target integration events throughout the human genome and relative frequencies, with the locations of peaks labeled at the center (Fig. 6m). These off-targets included the non-homologous end joining (NHEJ)-induced AAV integration after cpf1-mediated DNA cleavage at the PRDM1 locus. We found that the mean genome-wide sum frequencies of off-target integration events in CLASH-PRDM1-cr1 samples was 0.62% compared to 0.15% in samples receiving only the AAV vector without the Cpf1 mRNA (Fig. 6n). We observed certain detectable off-target integration events at genomic loci around CD8A, TUBA1B, PVT1, TRAC and PRDM1; the mean off-target integration frequency at the PRDM1 locus was 0.2%, 0.05% at the TRAC locus and 0.1% at the CD8A locus (Fig. 6n). These experiments measured the genome-wide AAV off-target integration events incurred with CLASH-PRDM1 CAR-T generation and estimated an overall genome-sum off-target integration rate at sub-percentage levels.
Discussion
Cell therapies including CAR-Ts are powerful therapeutics for cancer and potentially many other types of indications. To date, six clinical CAR-T products have been approved by the FDA2–4. A rapid emergence of CAR-T and other forms of adoptive cell therapies are in clinical or pre-clinical stages of development5. CAR-Ts can target diverse cancer antigens—for example, CD19 and CD22 for B cell malignancies2,6,75; B cell maturation antigen (BCMA) for multiple myeloma4,76; and mesothelin77, EGFRvIII78 or HER2 (ref. 79) for solid tumors. New CAR-T forms have recently emerged on a variety of targets, such as CD20, CD30, CD33, CD133, NKG2D, MUC1, claudin 18.2, claudin 6, GD2, GPC3 and others, at various stages of development15,80–86. These studies have unveiled a broad landscape of cell-based immunotherapies across a wide range of oncology indications6,7. However, cell therapy still faces many challenges1. For example, in vivo persistence is crucial for the durable therapeutic response in patients with cancer87. Persistence is also particularly crucial in allogeneic (‘off-the-shelf’) CAR-T products, as recent data from multiple front-running developers, such as Allogene, CRISPR Therapeutics and Horizon, showed that their allogeneic CAR-T candidates lack persistence in vivo. Thus, engineering better immune cells to overcome problems, such as efficacy, tumor infiltration, tumor microenvironment, toxicity and/or persistence, are crucial for future development of cellular therapeutics.
In this study, we established CLASH, a versatile platform for massive-scale knock-in engineering. In the CLASH system, in contrast to non-viral and DNA-based cDNA transgene knock-ins, the advantage of AAV vectors allows highly efficient, precise, knock-in, large-scale perturbations with high cell viability, by simply creating the viral vectors in a pooled manner. In contrast to regular CRISPR screens that have been applied on human and mouse primary T cells with lentiviral vectors and, more recently, by the transposon system21,22,24,25, CLASH is precisely targeted into a defined locus and, thereby, mitigates the positional effect or insertional mutagenesis.
By CLASHing human primary T cells with an AAV library, we directly generated a pool of differently mutated CAR-T cells and interrogated the immunologically relevant genetic perturbations that enhanced CAR-T anti-tumor function. Although we used a repeated multi-round co-culture assay for the in vitro screen, the screening condition can be set in different ways, such as hypoxia, nutrient deprivation and other approaches, to better mimic the tumor microenvironment. The in vitro and in vivo CLASH experiments revealed the timecourse dynamics of the mutants in critical genes that regulate anti-tumor immunology in CAR-T cells. Among these genes, TET2 as one of our top hits has been shown to improve the efficacy and persistence of CAR-T cells after disruption34, supporting the validity of the platform. PRDM1 emerged as another top candidate in our unbiased CLASH selections. Although PRDM1 was previously known as a critical transcriptional regulator for B cell and T cell differentiation46,57,59, its full knockouts showed important effects on T cell function and cell therapy46,52–62. In particular, recent findings suggested that modulating BLIMP1 knockout can enhance CAR-T cell adoptive transfer immunotherapy, further highlighting the importance of this gene and its therapeutic potential53. However, despite these previous works, the underlying mechanism of the critical PR domain remains unknown. For example, it is still unknown how PRDM1ʼs PR domain interacts with the histone; where PRDM1 binds in the chromosomes in human T cells, especially CAR-T cells; how these protein–protein and protein–DNA interactions affect downstream gene regulation; and what are the direct targets in CAR-T cells. Our CLASH generates Δexon3 mutant with a Cas12a/Cpf1 crRNA, which creates a specific PR domain truncation mutant rather than full null and, thereby, offers a unique window of opportunity to study the previously unchartered mechanisms directly in CAR-T-specific settings. We discovered that PRDM1 recognized a specific region of histone H4 (H4 11–30) and that this interaction was abolished in exon3 skip PRDM1 mutant CAR-T cells. We performed PRDM1 PR domain biochemistry and showed that the PRDM1 PR domain interacts with histone H4 methylation and acetylation and that loss of binding with histone H4 in Δexon3 mutant affects chromatin remodeling. Our CUT&RUN data revealed genome-wide chromatin binding profiles of PRDM1 WT and the exon3 skip mutant in CAR-T cells. PR domain disruption mediated by the engineered Δexon3 mutation caused a series of transcriptional rewiring of critical immune genes downstream of PRDM1 (PRDM1 bound and differentially expressed). Integration of CUT&RUN and timecourse RNA-seq data jointly revealed direct downstream targets such as CCR7, IL7R, SELL, STAT1 and STAT6, which we validated by individual pathway characterization. These genetic and epigenetic changes from exon3 skipping together lead to significantly altered immune characteristics in CAR-T cells, such as increased T cell proliferation, maintained cancer killing, reduced exhaustion and improved memory and persistence. These together translated into substantially better in vivo anti-tumor efficacy of Δexon3 mutant across several different CAR-Ts and respective cancer models, including a solid tumor model.
Although the current study focuses on human T cells in a CAR setting with a defined library, this study serves as a proof of principle to demonstrate CLASH’s feasibility, applicability and robustness. The future potential applications of CLASH can be broad. CLASH is not limited by one library and can be adapted for various libraries of other sizes. Although this study designed and used a focused library of ~8,000 crRNAs, it is possible to scale CLASH to other sizes of libraries, including genome-wide libraries. The places of mutagenesis or variation generation using AAV libraries can also be customized by design in addition to the crRNA pool. The features of selection can be determined by the users in the field, not limited to persistence or tumor infiltrations. In the future, CLASH can be applied to various other different settings to engineer other forms of transgenes in cell therapy. Finally, CLASH technology can, in principle, be applied to many other cell types, including other immune cells, stem cells, other primary cells or valuable cell lines and/or cells from other species in the future, simply by changing the AAV construct, library designs and selection schemes.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41587-022-01639-x.
Methods
Design and generation of CLASH AAV construct
To generate the CLASH AAV vector (pAAV-LHA-U6-DR-crTRAC-DR-BbsI-EFS-CAR-scFv-RHA or pXD60), the TRAC HDR arms and CD22BBz/CD19BBz CAR were amplified as previously reported88. The crRNA cassette, which contains one guide targeting the first exon of the TRAC locus and double BbsI cutting sites, was inserted after the left TRAC arm. Different fragments were cloned using Gibson assembly and traditional restriction cloning.
TRAC integration PCR
The gDNA was extracted after CLASH-vector AAV transduction for 9 days. The primer sequences are listed in a supplementary Excel table.
The PCR primers outside TRAC HDR arms were used:
TRAC HDR7FAGCAGCTCCTGCTTTCTGAG
TRAC HDR5RCACCTGGTGCATTCATGTGC
Using Phusion Flash High Fidelity Master Mix (Thermo Fisher Scientific), the thermocycling parameters for PCR were 98 °C for 2 minutes, 35 cycles of (98 °C for 1 second, 60 °C for 5 seconds, 72 °C for 1 minute) and 72 °C for 2 minutes.
The DNA was purified and sequenced by Sanger sequencing using primer:
HKO035gagggcctatttcccatgat
Non-viral T cell gene editing
Non-viral gene editing in T cells were carried out with the Cas9-RNP method in comparison with CLASH in parallel. In brief, for linearization of double-stranded DNA (dsDNA) HDR template, CLASH vector (pXD60) was digested by FD BpiI (Thermo Fisher Scientific) and purified with 1× SPRI (Beckman). RNPs were produced by complexing a two-component single guide RNA (sgRNA) to Cas9. The Cas9 guide RNA was designed to target the same sites as Cpf1 crRNA for TRAC using Benchling (Supplementary Table 1). Cas9 crRNA and tracrRNA were chemically synthesized (IDT) and resuspended in nuclease-free IDTE buffer at a concentration of 200 μM. The crRNA and tracrRNA were mixed at a 1:1 ratio and annealed as an sgRNA in nuclease-free IDTE buffer at 95 °C for 5 minutes and 37 °C for 10 minutes. RNPs were formed by the addition of SpCas9 nuclease (IDT) with 100 μM gRNA (1:2 Cas9-to-sgRNA molar ratio) at benchtop for 15 minutes. RNPs were mixed with 5 μg of linear dsDNA HDR template and immediately electroporated at the same condition with CLASH.
Design of Descartes Cas12a/Cpf1 crRNA library
A Cas12a/Cpf1 crRNA library, Descartes, was designed. In brief, a total of 954 immune genes were chosen as a superset from gene sets. Genes of interest were chosen as a superset from the following gene lists: T cell exhaustion32, epigenetic regulators33, T cell co-stimulation (GO: 0031295), memory T cell differentiation (GO: 0043379), TCR signaling pathway (GO: 0050852), adaptive immune response (GO: 0002250), immune response to tumor cell (GO: 0002418), T cell proliferation (GO: 0042098) and TET2. Guides were identified from the immune genes using CRISPOR software89 using consensus coding sequences (CCDSs), targeting only protein-coding exons. Genes with fewer than four available crRNAs were excluded. After this step, up to eight crRNAs were selected per gene, based on the top editing activity predicted from Seq-deepCpf1 scores35. The final library of Descartes contained 8,047 crRNAs, targeting 901 genes with eight crRNAs per gene for most genes, plus 1,000 NTCs. The genes and crRNAs of the Descartes library are provided in a Supplementary Dataset.
Library and individual crRNA cloning and preparation
The crRNA pools were synthesized by CustomArray. The Descartes library was amplified using a two-round PCR. The PCR product was purified by PCR purification kit (Qiagen). Descartes was cloned into the CLASH AAV plasmid by linearization with BbsI digestion and Gibson assembly. The Gibson assembly Descartes library products were transformed into high-efficiency competent cells (Endura) by electroporation. An estimated crRNA library coverage of ≥100× was observed after electroporation by colony counting. All the bacteria were harvested in a pool, and the plasmid library was purified using EndoFree Plasmid Maxi Kit (Qiagen). The representation of crRNAs in library plasmid was verified by NGS.
Packaging and purification of AAV6
The Descartes library, empty vector or individual gene targeting CLASH vectors were packaged by AAV6 serotype vectors to target human T cells. In brief, AAV6 serotype plasmid, packing plasmid pDF6 and AAV6 transgene vector plasmid were added at a ratio of 1.7:2:1, and then polyethyleneimine was added and mixed well by vortex. The solution was left at room temperature for 10–20 minutes, and then it was added dropwise into HEK293FT cells that were 80–90% confluent in 15 cm tissue culture dishes (Corning). Transfected cells were collected with PBS at 72 hours after transfection. For AAV purification, transfected cells were mixed with pure chloroform (1:10 volume) and incubated at 37 °C with vigorous shaking for 1 hour. Pure NaCl was added to meet a final concentration at 1 million, and then samples were centrifuged at 20,000g at 4 °C for 15 minutes. The aqueous layer was transferred to another tube, and the chloroform layer was discarded. Into the solution was added PEG8000 to 10% (w/v), followed by vigorous shaking to dissolve, and then the mixture was incubated at 4 °C for 1 hour. Samples were centrifuged at 20,000g at 4 °C for 15 minutes, and then the supernatant was discarded and the pellet was resuspended with Dulbecco’s PBS with MgCl2. The dissolved solution was treated with universal nuclease (Thermo Fisher Scientific), incubated at 37 °C for 30 minutes, added chloroform (1:1 volume), vortexed and centrifuged at 14,000g at 4 °C for 15 minutes. The aqueous layer was dumped into a 100-kDa molecular cutoff filter (Millipore) and centrifuged at 3,000g to concentrate the virus. Virus was tittered by quantitative PCR using custom TaqMan assays targeted to the U6 promoter.
Library-scale AAV transduction
Human primary peripheral blood CD8+ T cells, CD4+ T cells or human peripheral blood mononuclear cells (PBMCs) were purchased from STEMCELL Technologies. The CD8+ T cells from PBMCs were isolated by using the human CD8+ T Cell Isolation Kit (Miltenyi Biotec), and CD3+ T cells from PBMCs were isolated by using the Human Pan T Cell Isolation Kit (Miltneyi Biotec) according to the manufacturer’s protocol. T cells were cultured in X-VIVO media (Lonza) with 5% human AB serum and recombinant human IL-2 20 ng ml−1. Electroporation was performed after T cells thawed for 2 days. Cells were prepared at a density of ~2.5 × 106 cells per 100-μl tip reaction in electroporation Buffer R (Neon Transfection System Kits). A total of 20 reactions were set for the Descartes library electroporation. For each reaction, T cells were mixed with 10 μg of modified NLS-LbCpf1mRNA (TriLink) and electric shocked at program 24 (1,600 V, 10 ms and three pulses). After electroporation, the cells were transferred into 1 ml of pre-warmed X-VIVO media (with 5% human AB serum but without antibiotics) immediately. Indicated volumes of AAV6 at an estimated number of viral particles per cell (1 × 103–1 × 104) were added into the T cells after electroporation for 2–4 hours.
Due to the fact that they are empty of defective AAVs during packaging, rendering them non-infectious, the actual infectious viral particles are often substantially lower, especially for difficult-to-transduce cell types, such as T cells. The functional multiplicity of infection (f-MOI) is usually 3–4 orders of magnitude lower than that of genomic MOI (in genome copies) (g-MOI)90. In addition, a T cell only has two copies of genome, and, therefore, the CLASH HDR knock-in design caps the crRNA integration so that each cell can have no more than two different integrated crRNAs. At the transduction condition of the CLASH experiments (g-MOI = 1 × 103–1 × 104 viral particles per cell), the targeted effective multiplicity of infection (effective MOI, or integration MOI) was approximately ~0.1 in comparable screen terms.
Validation of massively parallel HDR knock-in
After electroporation for 5 days, the gDNA of massively parallelly engineered T cell pools was extracted by using QIAamp DNA Blood Mini Kit (Qiagen). In-Out PCR was used to amplify out the integrated fragments from gDNA. The PCR product was purified by PCR Purification Kit (Qiagen) and sequenced by the Keck Biotechnology Resource Laboratory (Yale).
CLASH timecourse dynamics of long-term CAR-T co-culture
T cells were infected with vector or Descartes AAV6 after electroporation with NLS-LbCpf1mRNA. After electroporation for 5 days, the percentage of positive CAR-T cells was determined by staining with CD3 and CAR-specific antibodies as previously described. In total, 2~3 × 106 positive CAR-T cells per replicate were used (8,047 crRNAs in Descartes library). The final coverage estimate is 250×~370× for CD8 and CD4 in vitro CLASH.
T cells were co-cultured with NALM6 at low E:T (0.2:1) ratio. After clearance of NALM6, new rounds of stimulation were performed until vector CAR-T cells were exhausted. After each round of stimulation, the T cells were harvested and frozen in liquid nitrogen. A total of eight rounds of co-culture were performed for CD8 in vitro CLASH and seven rounds for CD4 in vitro CLASH. The differences in stimulation timing and number of rounds were due to the effectiveness and actual times of cancer cell clearance by CD8 or CD4 CAR-T cell pools. gDNA was isolated by DNA Purification Kit (Qiagen) from the CD8 or CD4 CAR-T cell pools taken from the timepoints specified.
CLASH timecourse of in vivo CAR-T representation
NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were purchased from the Jackson Laboratory and bred in-house. For the leukemia mouse model, NSG mice between 6 weeks and 8 weeks of age were used and inoculated with 5 × 105 NALM6-GL cells by intravenous. Mice were randomly assigned to different groups before treatment. In total, 2 × 106 vector CAR-T cells or Descartes CAR-T cells (~250× coverage) were infused back to mice after 3 days. NSG mice were euthanized at days 7, 11 and 14. Spleen and bone marrow were collected immediately. For the solid tumor model, NSG mice were subcutaneously injected with 2 × 106 HER2+ HT-29-GL cells at day 0. At 20 days after transplantation, 3 × 106 Descartes HER2 CAR-T cells were infused into mice (library coverage ~370×). Tumors were collected after 12 days and digested with 100 U ml−1 of collagenase IV (Thermo Fisher Scientific) for 2 hours. Red blood cells were lysed by incubation with ACK (ammonium-chloride-potassium) Lysis Buffer (Thermo Fisher Scientific) for 2 minutes. After wash, the cell surface markers were labeled as described in the FACS part and measured by BD FACSAria II. CAR+ T cells were sorted, and the gDNA was extracted by using QuickExtract DNA Extraction Solution (Lucigen).
CLASH CAR-T crRNA library readout
The crRNA library readout was performed using a two-step PCR strategy, where the first In-Out PCR was used to amplify out the integrated fragments from gDNA, and the second PCR adds appropriate sequencing adapters to the products from the first PCR. The primer sequences are listed in a supplementary Excel table. For the first-round PCR, the thermocycling parameters were 98 °C for 1 minute, 20 cycles of (98 °C for 1 second, 60 °C for 5 seconds, 72 °C for 25 seconds) and 72 °C for 2 minutes. In each PCR reaction, we used 2 μg of total gDNA for in vitro and 5 μl of DNA extraction solution for in vivo. A total of 3–4 reactions were used to capture the full representation of the library. PCR products for each biological sample were pooled and used for amplification with barcoded second PCR primers. For the second-round PCR, the thermocycling parameters were 98 °C for 1 minute, 28 cycles of (98 °C for 1 second, 61 °C for 5 seconds, 72 °C for 10 seconds) and 72 °C for 2 minutes. Second PCR products were pooled and then normalized for each biological sample before combining uniquely barcoded separate biological samples. The pooled product was then gel purified from a 2% E-gel EX (Life Technologies) using the QIAquick Gel Extraction Kit (Qiagen). The purified pooled library was then sequenced with HiSeq or NovaSeq systems (Illumina).
CLASH CAR-T crRNA screen processing
Raw single-end FASTQ read files were filtered and demultiplexed using Cutadapt. Reads were demultiplexed for the barcodes included in the forward readout PCR primers. To identify and remove extra sequences immediately upstream of the crRNAs, the following settings were used: cutadapt -g TAATTTCTACTAAGTGTAGAT -e 0.1 -m 19–discard-untrimmed. The 20-bp crRNA sequences were then mapped to the designed Descartes library sequences using a bowtie index generated using the bowtie-build command in Bowtie 1.2.2. Mapping used the following settings: bowtie -v 1 -m 1; and the number of reads that had mapped to each crRNA within the library was quantified. This processing was used across the four key CLASH crRNA library readouts (CD8 in vitro, CD4 in vitro, CD8 in vivo and CD3 in vivo) as well as for the MIPS crRNA representation readout. Cells from the CD8 in vitro CLASH experiments were sequenced in two separate batches (early and late timepoints), and read counts for the different timepoints were merged into one table after separate readout processing for each batch.
Timecourse Cas12a/Cpf1 crRNA library representation analyses
Custom scripts were used to generate the crRNA library representation plots, which included library distribution box plots, CDF plots, correlation heat maps and overall timecourse abundance heat maps (collectively, ‘QC plots’). Pearson correlations for heat maps were calculated using the cor function in R, and empirical CDF was computed and plotted using stat_ecdf from ggplot. Then, to determine enriched guides, custom enrichment analysis was performed (‘bulk’ analysis)24. In brief, raw read counts were converted for each sample to a normalized reads per million total reads (rpm) and then log2-transformed. For bulk analysis, a crRNA was determined to be significant for a given timepoint if it was enriched using an FDR threshold of 1.0% based on the abundances of all NTCs—that is, a read count value above that of the 10th NTC out of a total 1,000 NTCs.
For the in vitro CLASH experiments, where the datasets consisted of 8–9 timepoints, an analytic approach was implemented using a generalized linear model (SAMBA) that considered the entire timecourse abundance trajectory across all timepoints, all sample groups and all gene-targeting and non-targeting guides39. This SAMBA-based screen analysis method incorporates data from all timepoints by fitting the data to a negative binomial generalized linear model and then performing a quasi-likelihood F-test. To narrow down a set of crRNAs that potentially contribute to the enrichment during any or all of the timepoints, initial crRNAs to be included in the SAMBA gene scoring analysis were defined as the top two guides per gene plus any additional guides above a 0.1 log fold change (FC) threshold (deemed ‘good crRNAs’ or ‘good guides’ for a gene). The crRNA statistics for all good guides (for example, adjusted P value, z-score or logFC) were then aggregated to obtain gene-level statistics. The design matrices were chosen to regress on time since day 0 and are described in greater detail in a Supplementary Dataset.
For visualizations, heat maps and statistical analyses, custom R scripts were used, making use of the following packages: ggplot, ggrepel, stringr, reshape2, VennDiagram, scales, strex, tidyr, stringr, data.table, gridExtra, viridis, pheatmap and ggpmisc.
MIPS library selection and cloning
The MIPS library contained 56 crRNAs, targeting seven genes, with approximately eight crRNAs per gene. The crRNAs were cloned as a minipool into the CLASH vector. The minipool in CLASH vector was used for CLASH experiment in human CD8 T cells (by mRNA electroporation and AAV6 transduction as above) to generate a CAR-T minipool. The size of the minipool was determined considering (1) the sensitivity of MIPS to measure genomic variants91; (2) the ability of MIPS to capture actual genome editing events in each of the specific crRNA target sites, with dilution of editing events in a pooled manner92; and (3) the relative gene editing challenges in T cells. The crRNA sequences are listed in a Supplementary Dataset. To generate the MIPS AAV vector, the pre-mixed crRNAs were cloned into the plasmid pXD60 by linearization with BbsI digestion and quick ligation. The MIPS library products were transformed into high-efficiency competent cells (Endura) by electroporation. An estimated crRNA library coverage of 300× (16,800 colonies) was observed after electroporation by colony counting. All the bacteria were harvested in pool. Plasmid library was purified using EndoFree Plasmid Maxi Kit (Qiagen). The representation of crRNAs in library plasmid was verified by NGS. MIPS library transduction is similar as described in the ‘Library-scale AAV transduction’ subsection. Three reactions were set for the MIPS library electroporation.
MIPS probe design
MIPS probes were designed according to previously published protocols (https://github.com/shendurelab/MIPGEN)93 and then processed by a customized selection algorithm. In total, 107 MIPS probes were designed using MIPgen. In brief, the 77 bp flanking the predicted cut site of each crRNA of all 56 unique crRNA were chosen as targeting regions, and the BED file with these coordinates was used as an input. These coordinates contained overlapping regions, which were subsequently merged into 31 unique regions. Each probe contains an extension probe sequence, a ligation probe sequence and a 6-bp degenerate barcode (NNNNNN) for PCR duplicate removal. A total of 107 MIPS probes were designed (sequences are listed in a supplementary Excel table), covering a total amplicon of 5,675 bp. The statistics for the MIP target size were as follows: minimum, 154 bp; maximum, 331 bp; mean, 183 bp; median, 163 bp. Each of the MIPS was synthesized using standard oligo synthesis (IDT), normalized and pooled.
MIPS target-capture sequencing
After 10 days of electroporation, CAR+ cells from the MIPS library and control group were sorted by FACS (BD), and then CAR-T cells gDNA was isolated by DNA Purification Kit (Qiagen). Experimental workflow was done by following standard protocols91. In brief, 50–100 ng of high-quality, non-fragmented gDNA was used for hybridization. After gap filling and ligation, circularized DNA molecules were used as template in PCR with universal primers complementary to the linker sequence. Then, sample-specific barcode sequences and Illumina adaptors were introduced during the PCR amplification step. After this amplification, DNA was purified and sequenced using 100-bp paired-end reads on an Illumina HiSeq 4000, NovaSeq or equivalent.
MIPS data analysis
For MIPS library crRNA representation in plasmid and samples, standard screen processing and mapping was performed as described above using a subset library with the MIPS crRNAs. For mutation-based MIPS target-capture sequencing analyses, raw pair-end FASTQ read files were first mapped to the reference hg38 Homo sapiens genome assembly and sorted using BWA and SAMtools. For coarse filtering, reads near target crRNA regions were selected (±1,000 bp) using BEDtools and then indexed and used for variant calling using SAMtools and VarScan version 2.4.1. with the parameters pileup2indel–min-coverage 1–min-reads2 1–min-var-freq 0.001–P value 0.05. Generated VCF files were then used for fine-mapping to crRNA cutting regions.
To define cutting regions, crRNAs were first collapsed based on whether their cutting sites were contiguous within or equal to 16 bp. For mapping, crRNA cutting regions were defined as ±8 bp from maximum/minimum of cutting sites for collapsed crRNAs (non-collapsed crRNAs will, therefore, have 16-bp windows, but collapsed crRNAs will be larger). For insertions, the variant position point in genome was kept as defined by VCF output. For deletions, the variant position point in genome was adjusted to reflect the center point of the deletion. Variants were mapped to crRNA cutting regions based on above. Further downstream analyses, including those comparing cutting efficiency with crRNA representation, were based on collapsed crRNA references. For MIPS crRNA representation in the library and associated cutting efficiency bar plots, crRNAs with near zero reads in library sequencing (PRDM-cr6 and PRDM-cr7 for MIP1, PRDM-cr1 for MIP3) were removed for visualization but were included in all other analyses and statistical tests. For analyses including MIPS cutting efficiency normalized by library representation, mean summed variant frequencies for each target region were divided by a normalization factor defined as: (mean library reads) / median (mean library reads). Statistical tests and visualizations were performed using custom R scripts.
CAR-T purification
CAR22+ T cells were purified by streptavidin microbeads (Miltenyi Biotec). In brief, 1 × 107 cells were suspended in 100 μl of labeling buffer and then incubated with 1 μg of Pierce Recombinant Biotinylated Protein L (Thermo Fisher Scientific) and 10 μl of FcR Blocking Reagent (Miltenyi Biotec) for 15 minutes at 4 °C. Cells were washed to remove unbound protein and labeled with 10 μl of streptavidin microbeads for 15 minutes on ice. After wash, the suspension was loaded onto MACS column for separation according to the manufacturer’s protocol (Miltenyi Biotec). Other types of CAR-T cells were sorted by BD FACSAria II.
Generation of CAR-T with individual gene perturbation
The individual crRNAs were cloned into the CLASH vector plasmid by linearization with BbsI digestion and Quick Ligation Kit (NEB). The ligated products were transformed into Stbl3 competent cells by heat shock at 42 °C for 90 seconds. A single clone was picked, and plasmid was isolated by Mini Prep Kit (Qiagen). The sequence of plasmid was identified by the Keck Biotechnology Resource Laboratory (Yale).
T7E1 assay
Five days after electroporation and AAV transduction, positive CAR-T cells were stained and sorted by BD FACSAria II. The gDNA was extracted by using QIAamp DNA Blood Mini Kit (Qiagen). PCR amplification of the genomic regions flanking the crRNAs was performed using the primers in a supplementary Excel table. Using Phusion Flash High Fidelity Master Mix (Thermo Fisher Scientific), the thermocycling parameters for PCR were 98 °C for 2 minutes, 35 cycles of (98 °C for 1 second, 60 °C for 5 seconds, 72 °C for 15 seconds) and 72 °C for 2 minutes. The PCR amplicons were then used for T7E1 assays according to the manufacturer’s protocol.
Nextera library prep and amplicon sequencing
The PCR products described as before in the T7E1 experiments were used for Nextera library preparation following the manufacturer’s protocols (Nextera XT DNA Library Preparation Kit, Illumina). In brief, 1 ng of purified PCR product was fragmented and tagged using the Nextera Amplicon Tagment Mix according to the manufacturer’s recommendations, followed by limited-cycle PCR with indexing primers and Illumina adaptors. After this amplification, DNA was purified and sequenced using 100-bp paired-end reads on an Illumina HiSeq 4000, NovaSeq or equivalent. For indel quantification, reads were mapped to expected amplicon sequences using BWA-MEM with the -M option. Then, 100-bp reads from the SAM file that fully mapped within a ±75-bp window of expected cut site within the amplicon were then identified (soft-clipped reads were discarded). Indel reads were then identified by the presence of ‘I’ or ‘D’ characters within the CIGAR string. Cutting efficiency was quantified as percentage of indels over total (indel plus WT) reads within the defined window.
Single-cell genotyping
After the gene editing and CAR introduction (individual gene CLASH), single positive CAR-T cells were sorted into round-bottom 96-well plates. PCR was performed after single CAR-T cells were cultured for 1 week. PCR amplification of the genomic regions flanking the crRNAs was performed using the same primers in theT7E1 assay. The thermocycling parameters for the first-round PCR were 98 °C for 2 minutes, 40 cycles of (98 °C for 1 second, 60 °C for 5 seconds, 72 °C for 15 seconds) and 72 °C for 2 minutes. PCR products were checked by using 2% E-Gel 96 Agarose Gels (Thermo Fisher Scientific). Second-round PCR was performed for the samples with positive PCR results, and the thermocycling parameters were 98 °C for 2 minutes, 34 cycles of (98 °C for 1 second, 60 °C for 5 seconds, 72 °C for 15 seconds) and 72 °C for 2 minutes. Final PCR products were cleaned by using 1× SPRI (Beckman) and sequenced by the Keck Biotechnology Resource Laboratory (Yale).
Kill assay (co-culture)
GFP and firefly luciferase gene were stably transduced into NALM6 cell line by using lentivirus. In total, 2 × 104 NALM6-GL cells were seeded in a 96-well plate. The engineered CAR-T, vector CAR-T or normal T cells were co-cultured with NALM6-GL at indicated E:T ratios for 24 hours. To test the luciferase expression in NALM6-GL, 150 μg ml−1 of d-luciferin (PerkinElmer) was added into each well. After 10 minutes, luciferase intensity was measured by a plate reader (PerkinElmer). Direct tumor cell killing was quantified by luminescence. Luminescence units were normalized to control (NALM6-GL without any effector cells, ).
The calculation formula is below:
CUT&RUN
Vector CAR-T cells and PRDM1cr1 exon3 skip mutant CAR-T cells were FACS isolated, and CUT&RUN was performed following the EpiCypher CUTANA user manual. In brief, 1 × 106 CAR-T cells were washed with cold PBS and resuspended in nuclei isolation buffer (100 mM HEPES-KOH pH 7.6, 100 mM KCl, 5 mM MgCl2, 0.5% NP40 supplemented with Roche Complete Protease Inhibitor) and incubated on ice for 10 minutes. Nuclei were washed twice with CUT&RUN wash buffer and incubated with activated concanavalin-A (ConA) beads for 10 minutes at room temperature. ConA-bead-bound nuclei were incubated overnight at 4 °C in CUT&RUN antibody buffer and antibodies, including anti-Blimp1/PRDM1 (CST9115), anti-H3K4me3 (positive control) and rabbit IgG (negative control) antibodies. After antibody incubation, ConA-bead-bound nuclei were washed once with Cell Permeabilization Buffer and then incubated with 2.5 μl of pAG-MNase for 10 minutes at room temperature. ConA-bound-nuclei were then washed twice with Cell Permeabilization Buffer. Targeted chromatin was digested and released by adding 1 μl of 100 mM CaCl2 and incubated on ice for 2 hours. The reaction was stopped by the addition of 33 μl of stop buffer. Then, 1 μl of Escherichia coli spike-in DNA was added to each sample as internal control. DNA fragments were purified by using DNA cleanup columns. CUT&RUN sequencing libraries were generated using NEBNext Ultra II DNA Library Prep Kit for Illumina and NEBNext Multiplex Oligos for Illumina (Index Primer Set 1 and 2). Library fragment size was analyzed by TapeStation (Agilent). The pass-QC libraries were sequenced on Illumina NovaSeq 150-bp paired-end reads.
CUT&RUN data processing
CUT&RUN FASTQ sequencing data were analyzed according to a previously published protocol94, with some modifications. Raw FASTQ files were first trimmed and cleaned using Trimmomatic95 and Kseq96. Cleaned FASTQ reads were then separately aligned to the human genome (hg38.p13) and the E. coli genome (GCF_000005845.2) using BWA-MEM97 with settings -M -t 8 -K 10000000. Peaks were called from the hg38 BAM alignment files using MACS2 callpeak98, comparing each experimental sample to each IgG control. The resulting peaks for a given sample were then merged with BEDtools99. These peaks were then further merged, combining the peaks from each biological condition (vector or PRDM1-cr1). To define genes that are bound by PRDM1, we annotated the peak calls from the vector-treated samples using HOMER100, assigning each peak to the closest transcription start site (TSS) (±2.5 kb). Overlaps between PRDM1 bound genes and DEGs were assessed by hypergeometric test, assuming 20,465 total human genes.
The number of aligned reads to each genome were then determined to calculate library scaling factors, as described in the EpiCypher CUTANA user manual. Specifically, the hg38-aligned read counts were scaled such that the reads aligned to the spike-in E. coli genome would be normalized across samples. The scaling factors were used to generate normalized BedGraph files, which were then converted into bigWig files. Replicate IgG control bigWig files were averaged by wiggleTools to create a single IgG control sample. Finally, signal visualization was performed using deepTools101 and Gviz102. CUT&RUN processed data and analyses are provided in a Supplementary Dataset.
Recombinant protein expression and purification
GST-tagged fusions of a human PRDM1 PR domain (amino acids 38–223) with C-terminal FLAG tag and an exon3 truncation version (amino acids 38–97 and 138–223) with C-terminal FLAG tag plasmid constructs (pGEX-6P-1) were transformed into an E. coli BL21(DE3) strain. Overnight cultures were diluted 1:100 in LB media supplemented with 100 μg ml−1 of ampicillin and grown at 37 °C at OD600 ~0.6–0.8. Protein expression was induced by the addition of 0.1 mM of isopropyl β-d-1-thiogalactopyranoside (IPTG), and cultures were grown further at 18 °C overnight. Expression of fusion proteins was verified by Coomassie blue staining. After cell harvest and lysis (20 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.1 mM TCEP) by sonication (6× pulses of 15 seconds each at half maximal power, on ice), the solution was clarified by centrifugation at 10,000g for 30 minutes at 4 °C and loaded onto a column containing 200 μl of Pierce glutathione agarose resin (Thermo Fisher Scientific, 16100). After incubation for 2 hours at 4 °C, the resin was washed five times with the lysis buffer, and fusion proteins were eluted with elution buffer (20 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.1 mM TCEP, 10 mM reduced glutathione).
Structure modeling by AlphaFold
The superposition of PRDM1 PR domain with truncated PR domain was aligned and visualized by Pymol 2.5.0. The crystal structure of PRDM1 PR domain (amino acids 38–223) is from PDB: 3DAL, and the structure of Truncated PR domain is predicted by AlphaFold2 (ref. 51) using MMseqs2 (https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb).
Histone peptide arrays
Probing protein binding with a histone peptide array platform (Active Motif, 13005) was carried out according to the manufacturer’s specifications with slight modifications, as described previously103. In brief, after a short blocking with 3% BSA in TBST (20 mM Tris-HCl pH 7.4, 150 mM NaCl and 2 mM KCl) for 1 hour at room tempterature, the peptide array slides were incubated with GST–PRDM1 PR domain FLAG proteins in the 3% BSA in TBST overnight at 4 °C with gentle agitation. After extensive washing with TBST, the slides were probed with rabbit monoclonal anti-FLAG (1:1,000, Cell Signaling Technology, 14793S), followed by goat anti-rabbit IgG horseradish peroxidase conjugate (1:1,000, Rockland, 611–1302). Data were analyzed by Array_Analyze_Software_v16.1 supplied by Active Motif.
Timecourse mRNA-seq experiment
T cells were infected with CLASH vector and PRDM1-cr1 (Δexon3) CAR22 AAV6 after electroporation with NLS-LbCpf1mRNA. After electroporation for 5 days, the percentage of positive CAR-T cells was determined by staining with CD3 and CAR-specific antibodies, as previously described. CAR-T cells were co-cultured with NALM6 at low E:T (0.2:1) ratio every 4–7 days and total 5 round was performed. CAR-T cells were harvested using TRIzol (Invitrogen) after each round stimulation. RNA was extracted using RNeasy Plus Mini Isolation Kit (Qiagen). mRNA library preparations were performed using a NEBNext Ultra RNA Library Prep Kit for Illumina, and samples were multiplexed using barcoded primers provided by NEBNext Multiplex Oligos for Illumina (Index Primers Set 1). Libraries were sequenced with NovasSeq systems (Illumina).
mRNA-seq processing
FASTQ files from mRNA-seq were analyzed using the Kallisto quant algorithm with setting -b 100 for transcript quantification104. Differential expression analysis was performed using Sleuth105. Differentially upregulated and downregulated genes were selected for DAVID analysis using a q-value cutoff of 1 × 10−3 (ref. 106). z-scores for timecourse heat map were calculated by log2-normalizion of gene counts followed by scaling by genes, and DEGs across the timepoints were determined by limma with contrasts set up to compare PRDM1 versus vector control CAR-T cells at each timepoint. Timecourse cluster analysis was performed with the R package maSigPro using ‘two.ways.forward’ as step method and otherwise default settings. Visualizations of DEGs, such as volcano plots and heat maps, were generated using standard R packages, such as ggplot2 and VennDiagram.
Flow cytometry
All antibodies for flow cytometry were purchased from BioLegend, and details are provided. All flow antibodies were used at 1:200 dilutions for staining unless otherwise noted. For surface staining, cells were stained with surface marker antibodies in the staining buffer of 2% FBS in PBS on ice for 30 minutes. Samples were washed twice with 2% FBS in PBS before analysis. For CAR staining, the CD22BBz CAR transduced T cells were incubated with 0.2 μg of CD22-Fc (R&D Systems) in 100 μl of staining buffer for 30 minutes and then stained with PE-IgG-Fc (BioLegend). CD19 CAR and HER2 CAR were detected by using PE-anti-FLAG (BioLegend). For intracellular cytokine staining analysis, CAR+ T cells and NALM6 were plated at a 1:1 E:T ratio in a 96-well plate (Corning) and 0.2 μl per test of Brefeldin A solution (1,000×, Clone BFA, BioLegend) for 5 hours. After incubation, intracellular cytokine staining was performed using the BD Cytofix/Cytoperm Fixation/Permeabilization Solution Kit according to the manufacturer’s instructions. All the antibodies are listed in the Reporting Summary.
CellTrace Violet assay
After two rounds of stimulation, T cells were collected and incubated with CellTrace Violet (1:2,000, stock concentration 5 mM) in PBS for 15 minutes at 37 °C. Five times the staining volume of T cell culture medium was added to the cell and incubated for 5 minutes. Half of cells were analyzed by Attune at VL1 channel as day 0. Another half of cells were resuspended in fresh T cell culture medium. After 3 days, the cells were measured again. The mean fluorescence intensity (MFI) of CellTrace Violet was used for doing quantification.
RT–PCR
RNA was extracted as described in RNA-seq. cDNA for qPCR was generated using M-MLV reverse transcriptase enzyme (Sigma-Aldrich) and Oligo dT (Thermo Fisher Scientific) following the manufacturer’s protocol. For the PRDM1 mutant sequence analysis, PCR was performed by using primers near the PRDM1-cr1 cutting site by using the cDNA as template (primer sequences are listed in a supplementary Excel table). For the RNA-seq validation, the cDNA was subjected to qPCR using TaqMan Real-Time PCR Master Mixes and TaqMan gene assay probes (Thermo Fisher Scientific). Samples were processed using Applied Bioscience Step One Plus Real Time machine, and relative mRNA expression was normalized to GAPDH controls. Relative mRNA expression was determined via the ΔΔCt method.
Western blot
Cells were lysed by ice-cold RIPA buffer (Boston BioProducts) containing protease inhibitors (Roche, Sigma-Aldrich) and incubated on ice for 30 minutes. Protein supernatant was collected after centrifuging at 13,000g at 4 °C for 30 minutes. Protein concentration was determined using the BCA Protein Assay Kit (Thermo Fisher Scientific). Protein samples were separated under reducing conditions on 4–20% Tris-HCl gels (Bio-Rad) and analyzed by western blotting using primary antibodies PRDM1/Blimp-1mouse mAb (R&D Systems, 1:1,000), followed by secondary anti-mouse HRP antibodies (Sigma-Aldrich, 1:10,000). Blots were imaged with an Amersham Imager 600.
Immunoprecipitation
PRDM1-deficient CD22 CAR-T cells or vector cells were lysed by ice-cold RIPA buffer with protease inhibitor cocktail (Roche) for 30 minutes. Immobilized BLIMP1/PRDM1 antibody (1:100, Cell Signaling Technology) was added on the NHS-activated agarose beads according to the manufacturer’s protocol (Thermo Fisher Scientific). Cell lysate was incubated with beads at 4 °C and rotated for 1 hour. The supernatant was discarded after spin-down for 1 minute at 3,000 r.p.m. Beads were washed four times with ice-cold TBS and boiled for 10 minutes with SDS loading buffer. Protein samples were loaded on 4–20% Tris-HCl gels (Bio-Rad).
Co-IP western
For validation of the interaction between recombinant PRDM1 PR domain and histone H4, 100 μl of Glutathione Resin (G-Biosciences, 786–280) was washed twice with PBS buffer (pH 7.4). Subsequently, 30 μl of PRDM1 PR domain with GST tag and PRDM1 PR truncated with GST tag (1 mg ml−1) in PBS buffer were added, and the resulting mixture was incubated for 2 hours. The resulting resin was washed with PBS five times and stored in PBS. The nuclei of T cells were isolated as mentioned above. The nuclei were lysed by ice-cold RIPA buffer with protease inhibitor cocktail (Roche) for 30 minutes. The supernatant was collected and incubated with pre-mixed beads. Bead suspensions were rotated at 4 °C for 6 hours and washed three times with TBST. Elution was carried out by 100 μl of PBS with 5 mM reduced glutathione (G-Biosciences, 786–588). The beads were boiled in 20 μl of 6× protein loading buffer (0.3 M Tris-HCl, pH 8.0, 30% glycerol, 100 mg ml−1 of SDS, 100 mg ml−1 of DTT and 100 μg ml−1 of bromophenol blue). Protein samples were collected and analyzed by western blotting using anti-GST mouse mAb (1:1,000, Cell Signaling Technology) and anti-histone H4 mouse mAb (1:1,000, Cell Signaling Technology).
For validation on fully endogenous histone H4, the nuclei of vector CAR-T cells and PRDM1-cr1 Δexon3 mutant CAR-T cells were isolated as described above. The nuclei were lysed by ice-cold RIPA buffer with protease inhibitor cocktail (Roche) for 30 minutes. The supernatant was collected and incubated with BLIMP1/PRDM1 antibody (Cell Signaling Technology) at 4 °C with rotation. At day 2, the lysate was incubated with Protein A/G Magnetic Beads (Thermo Fisher Scientific) for 6 hours. The beads were collected by placing the tube into a magnetic stand and washed four times with ice-cold TBS. The samples were boiled for 10 minutes with SDS loading buffer. Protein samples were collected and analyzed by western blotting using PRDM1/Blimp-1mouse mAb (1:1,000, Cell Signaling Technology) and anti-histone H4 mouse mAb (1:1,000, Cell Signaling Technology).
In-gel digestion and mass spectrometry
Protein-containing gel slices were digested with trypsin overnight. The resulting peptide mixtures were extracted from the gel and run directly on an Orbitrap Velos instrument (Thermo Fisher Scientific) with 120-minute liquid chromatography (Buffer A: 0.1% formic acid water; Buffer B: 0.1% formic acid MeCN; gradient: 0% to 95% buffer B; flow rate: 0.1 μl min−1) and tandem mass spectrometry (LC–MS/MS) using a standard TOP20 method procedure. In brief, MS1 m/z regions for 395–1,600 m/z ions were collected at 60,000 resolving power and used to trigger MS/MS in the ion trap for the top 20 most abundant ions. Active dynamic exclusion of 500 ions for 90 seconds was used during the LC–MS/MS method. Peptides were eluted with 300 nl min−1 flow rate using a NanoAcquity pump (Waters). Samples were trapped for 15 minutes with a flow rate of 2 μl min−1 on a trapping column 100-μm ID packed for 5 cm in-house with 5-μm Magic C18 AQ beads (Waters) and eluted with a gradient to 20 cm 75-μm ID analytical column (New Objective) packed in-house with 3-μm Magic C18 AQ beads (Waters).
Mass spectra were analyzed using Scaffold Q+/Q+S version 4.0 against the UniProt database. A mass deviation of 20 ppm was set for MS1 peaks, and 0.6 Da was set as the maximum allowed MS/MS peaks with a maximum of two missed. Maximum FDRs were set to 0.01 both on peptide and protein levels. Minimum required peptide length was five amino acids.
Genome-wide AAV integration library preparation
To perform genome-wide profiling of the CLASH-PRDM1 Δexon3 off-target integration events, a method modified from GUIDE-Seq74 was developed. In brief, CAR-T cells’ gDNA was extracted after AAV transduction for 9 days. The adapter was made by annealing the MiSeq common oligo with the sample barcode oligo (A01–A06) in TE buffer. The annealing program was set from 95 °C for 1 second and slow ramp-down (approximately −2 °C per minute) to 4 °C. The gDNA was fragmentated to ~1,500 bp by using an S220 focused ultrasonicator (Covaris). End-repair and dA-tailing were performed by using NEBNext UltraTM End Repair/dA-Tailing Module (NEB) according to the manufacturer’s protocol. The repaired DNA was ligated with annealed adaptor by using T4 DNA ligase at room temperature for 1 hour. The sample was cleaned by using 0.9× SPRI (Beckman). The genome-wide off-target integration library was performed using a two-step PCR strategy with streptavidin beads purification, where the first PCR was used to bait-prey the fragments with integrated CAR gene from gDNA by using biotinylated primer, and the biotinylated fragments were purified by using streptavidin beads (Thermo Fisher Scientific). Next, the second PCR added appropriate sequencing barcodes to the products from the first PCR. The Q5 was used for PCR (NEB). The thermocycling parameters for the two-round PCR were 98 °C for 30 seconds, seven cycles of (98 °C for 10 seconds, 70 °C (−1 °C per cycle), 72 °C for 1 minute), 13 cycles of (98 °C for 10 seconds, 63 °C for 30 seconds, 72 °C for 1 minute), 72 °C for 1 minute and hold at 4 °C. PCR products for each biological sample were normalized and pooled. DNA less than 1 kb was selected. The sample was sequenced by using custom sequencing primers with MiSeq (2× 300-bp pair-end). All oligo sequences are listed in a supplementary Excel table.
CLASH-PRDM1 Δexon3 genome-wide AAV integration data analysis
Paired-end reads were processed using Cutadapt 3.2, BWA 0.7.17 and SAMtools 1.12 to identify off-target integration events. R2 reads containing TRAC elements were first trimmed and selected using Cutadapt -G GTTTACTCGATATAAGGCCTTGA -e 0.2 -m 20–discard-untrimmed. Then, R2 reads containing ITR reads were trimmed and selected using Cutadapt -G AAAGGTCGCCCGACGCCCGG -e 0.2 -m 20 -O 15–discard-untrimmed. The ITR trimmed reads were then mapped to the H. sapiens genome assembly GRCh38 (hg38) using BWA. To see where mapped reads target in the genome without double-counting at positions, reads were converted to single base pair coordinates using the starting positions and then converted to BedGraph format. Reads were normalized for sequencing depth using the R2 reads containing TRAC elements. Visualizations were generated using Integrative Genomics Viewer 2.9.2 and R packages, including GenomicAlignments and ggbio.
In vivo CAR-T efficacy testing in mouse models of leukemia
NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were purchased from the Jackson Laboratory and bred in-house. Both male and female NSG mice between 6 weeks and 8 weeks of age were used. For the leukemia model, mice were intravenously inoculated with 5 × 105 NALM6-GL cells and randomly assigned to different groups before treatment. In total, 1.2 × 106 normal CD8 T cells, vector CAR-T cells or PRDM1 mutant CAR-T cells were infused back to mice after 3 days. Bioluminescent signal for each mouse was captured by using IVIS every 3–5 days. In brief, to generate bioluminescence signals, XenoLight d-luciferin (PerkinElmer) was intraperitoneally injected into mice at 150 mg kg–1, and images were analyzed by Living Image software. To test T cell phenotypes in vivo after treatment, NSG mice were euthanized at day 18. Peripheral blood, spleen and bone marrow were collected immediately. Red blood cells were lysed by incubated with ACK Lysing Buffer (Thermo Fisher Scientific) for 2 minutes. After wash, the cell surface markers were labeled as described in the FACS subsection and measured by BD FACSAria II.
In vivo CAR-T efficacy testing in solid tumor models
For the HER2 solid tumor CAR-T efficacy testing, two treatment models were adopted.
For the first model, NSG mice were subcutaneously injected with 2 × 106 HER2+ HT29-GL cells at day 0. Thirteen days after transplantation, 2 × 106 non-transduced CD3+ T, 2 × 106 / 0.5 × 106 vector HER2 CAR-T or PRDM1 cr1 Δexon3 mutant HER2 CAR-T cells were infused into mice. Tumors were measured twice a week. To evaluate the tumor-infiltrating CAR-T cell phenotypes in solid tumor models, NSG mice were subcutaneously injected with 4 × 106 HER2+ HT29-GL cells at day 0. At 11 days after transplantation, 2 × 106 vector HER2 CAR-T or PRDM1 cr1 Δexon3 mutant HER2 CAR-T cells were infused into mice. NSG mice were euthanized after CAR-T infusion for 14 days or 18 days in a separate cohort. Tumors were processed as described in the section on solid tumor screening. Cell surface markers were labeled as described in the FACS subsection and measured by BD FACSAria II.
For the second (rechallenge) model, 1 × 106 HT29-GL cells were subcutaneously injected into NSG mice. Three days after transplantation, 4 × 106 of non-transduced CD3+ T cells, vector HER2 CAR-T cells or PRDM1 Δexon3 mutant HER2 CAR-T cells were intravenously infused into mice. The mice were rechallenged with 0.7 × 106 HT29-GL cells at day 38. Tumors were measured twice a week. Mice were also imaged with IVIS once a week at the timepoints indicated in the figures.
Illustrations
Cartoons in Figs. 1a,f, 3a, 5g,i and 6k and Supplementary Figs. 4a, 6a, 9c and 10 were created with BioRender. A BioRender academic license/proof for using these artworks for publication is in place.
Standard statistical analysis
All statistical methods are described in figure legends and/or supplementary Excel tables. The P values and statistical significance were estimated for most analyses. For example, the unpaired, two-sided Mann–Whitney test was used to compare two groups. One-way ANOVA, two-way ANOVA, Dunnett’s multiple comparisons test and Tukey’s multiple comparisons test were used to compare multiple groups. Data between two groups were analyzed using a two-tailed unpaired t-test. Multiple t-test using the Holm–Sidak method was used for multiple group comparison. Different levels of statistical significance were assessed based on specific P values and type I error cutoffs (0.05, 0.01, 0.001 and 0.0001). Data analysis was performed using GraphPad Prism version 8 and R Studio. Source data and statistics are provided in a supplementary Excel table.
Supplementary Material
Acknowledgements
We thank M. Sznol, A. Bersenev, S. Seropian, I. Isufi, M. Müschen and D. Krause for discussions. We thank all members of the Chen laboratory as well as various colleagues in Yale Genetics, SBI, CSBC, MCGD, Immunobiology, BBS, YCC, YSCC and CBDS for assistance and/or discussions. We thank various Yale Core Facilities, such as YCGA, HPC, WCAC and KBRL, for technical support. S.C. is supported by NIH/NCI/NIDA (DP2CA238295, R01CA231112, U54CA209992-8697, R33CA225498 and 1RF1DA048811); DoD (W81XWH-20-1-0072 and W81XWH-21-1-0514); the Alliance for Cancer Gene Therapy; the Sontag Foundation (Distinguished Scientist Award); Pershing Square Sohn Cancer Research Alliance; Dexter Lu; the Ludwig Family Foundation; the Blavatnik Family Foundation; and the Chenevert Family Foundation. X.D. is supported by the Charles H. Revson Senior Postdoctoral Fellowship. J.J.P. is supported by an NIH Medical Scientist Training Program grant (T32GM136651). R.C. is supported by an NIH Medical Scientist Training Program grant (T32GM136651) and the National Research Service Award fellowship (F30CA250249). P.A.R. is supported by an NIH training grant (T32GM007499), the Lo Fellowship and NIH/NCI Diversity Supplement. S.S. is supported by a Mark Foundation for Cancer Research Emerging Leader Award, a Paul G. Allen Frontiers Group Distinguished Investigator Award and NIH/NIGMS R01GM122984.
Footnotes
Competing interests
A patent application has been filed by Yale University on CLASH (S.C., X.D., J.J.P. and Y.D. as inventors). S.C. is a founder of Cellinfinity Bio, which licensed the CLASH patent. S.C. is also a founder of EvolveImmune Tx, Chen Consulting and Chen Tech, unrelated to this study.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Code availability
Analytic codes used to generate figures that support the findings of this study will be available from the corresponding author upon reasonable request.
Additional information
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41587-022-01639-x.
Peer review information Nature Biotechnology thanks Li Tang, Hyongbum Henry Kim and Joseph Fraietta for their contribution to the peer review of this work.
Reprints and permissions information is available at www.nature.com/reprints.
Data availability
All data generated or analyzed during this study are included in this article and its supplementary information files. Specifically, source data and statistics for non-high-throughput experiments, such as flow cytometry, qPCR, protein experiments and other molecular or cellular assays, are provided in an Excel file of source data and statistics. Processed data for genomic sequencing (for example, CLASH, RNA-seq, amplicon sequencing, MIPS and AAV off-target) and other forms of high-throughput experiments are provided as processed quantifications in Supplementary Datasets. Genomic sequencing raw data are being deposited to the Gene Expression Omnibus (GEO), with the following accession numbers: GSE207143 for all CLASH screens; GSE219061 for MIPS, Nextera and AAV integration off-target; GSE207404 for RNA-seq; and GSE201997 for CUT&RUN107–110. CLASH vectors and libraries are available via material transfer agreements. All other data and materials that support the findings of this research are available either via public repositories or from the corresponding author upon reasonable request to the academic community. Source data are provided with this paper.
References
- 1.Lim WA & June CH The principles of engineering immune cells to treat cancer. Cell 168, 724–740 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Porter DL, Levine BL, Kalos M, Bagg A & June CH Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N. Engl. J. Med. 365, 725–733 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neelapu SS et al. Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N. Engl. J. Med. 377, 2531–2544 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raje N et al. Anti-BCMA CAR T-cell therapy bb2121 in relapsed or refractory multiple myeloma. N. Engl. J. Med. 380, 1726–1737 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang J, Pearce L, O’Donnell-Tormey J & Hubbard-Lucey VM Trends in the global immuno-oncology landscape. Nat. Rev. Drug Discov. 17, 783–784 (2018). [DOI] [PubMed] [Google Scholar]
- 6.June CH, O’Connor RS, Kawalekar OU, Ghassemi S & Milone MC CAR T cell immunotherapy for human cancer. Science 359, 1361–1365 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Weber EW, Maus MV & Mackall CL The emerging landscape of immune cell therapies. Cell 181, 46–62 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Porter DL et al. Chimeric antigen receptor T cells persist and induce sustained remissions in relapsed refractory chronic lymphocytic leukemia. Sci. Transl. Med. 7, 303ra139 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardner R et al. Acquisition of a CD19-negative myeloid phenotype allows immune escape of MLL-rearranged B-ALL from CD19 CAR-T-cell therapy. Blood 127, 2406–2410 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sadelain M, Rivière I & Riddell S Therapeutic T cell engineering. Nature 545, 423–431 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kosti P et al. Hypoxia-sensing CAR T cells provide safety and efficacy in treating solid tumors. Cell Rep. Med. 2, 100227 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ying Z et al. A safe and potent anti-CD19 CAR T cell therapy. Nat. Med. 25, 947–953 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneider D et al. Trispecific CD19-CD20-CD22–targeting duoCAR-T cells eliminate antigen-heterogeneous B cell tumors in preclinical models. Sci. Transl. Med. 13, eabc6401 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Lynn RC et al. c-Jun overexpression in CAR T cells induces exhaustion resistance. Nature 576, 293–300 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma L et al. Enhanced CAR-T cell activity against solid tumors by vaccine boosting through the chimeric receptor. Science 365, 162–168 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghorashian S et al. Enhanced CAR T cell expansion and prolonged persistence in pediatric patients with ALL treated with a low-affinity CD19 CAR. Nat. Med. 25, 1408–1414 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Savoldo B et al. CD28 costimulation improves expansion and persistence of chimeric antigen receptor–modified T cells in lymphoma patients. J. Clin. Invest. 121, 1822–1826 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weber EW et al. Transient rest restores functionality in exhausted CAR-T cells through epigenetic remodeling. Science 372, eaba1786 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu X et al. Genome-wide analysis identifies NR4A1 as a key mediator of T cell dysfunction. Nature 567, 525–529 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seo H et al. TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8+ T cell exhaustion. Proc. Natl Acad. Sci. USA 116, 12410–12415 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shifrut E et al. Genome-wide CRISPR screens in primary human T cells reveal key regulators of immune function. Cell 175, 1958–1971 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ting PY et al. Guide Swap enables genome-scale pooled CRISPR–Cas9 screening in human primary cells. Nat. Methods 15, 941–946 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Henriksson J et al. Genome-wide CRISPR screens in T helper cells reveal pervasive crosstalk between activation and differentiation. Cell 176, 882–896 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong MB et al. Systematic immunotherapy target discovery using genome-scale in vivo CRISPR screens in CD8 T cells. Cell 178, 1189–1204 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye L et al. In vivo CRISPR screening in CD8 T cells with AAV–Sleeping Beauty hybrid vectors identifies membrane targets for improving immunotherapy for glioblastoma. Nat. Biotechnol. 37, 1302–1313 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long L et al. CRISPR screens unveil signal hubs for nutrient licensing of T cell immunity. Nature 600, 308–313 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Z et al. In vivo CD8+ T cell CRISPR screening reveals control by Fli1 in infection and cancer. Cell 184, 1262–1280 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eyquem J et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 543, 113–117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roth TL et al. Reprogramming human T cell function and specificity with non-viral genome targeting. Nature 559, 405–409 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roth TL et al. Pooled knockin targeting for genome engineering of cellular immunotherapies. Cell. 181, 728–744 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Torikai H et al. A foundation for universal T-cell based immunotherapy: T cells engineered to express a CD19-specific chimeric-antigen-receptor and eliminate expression of endogenous TCR. Blood 119, 5697–5705 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wherry EJ et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity 27, 670–684 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Arrowsmith CH, Bountra C, Fish PV, Lee K & Schapira M Epigenetic protein families: a new frontier for drug discovery. Nat. Rev. Drug Discov. 11, 384–400 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Fraietta JA et al. Disruption of TET2 promotes the therapeutic efficacy of CD19-targeted T cells. Nature 558, 307–312 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim HK et al. Deep learning improves prediction of CRISPR–Cpf1 guide RNA activity. Nat. Biotechnol. 36, 239 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Shin H & Wherry EJ CD8 T cell dysfunction during chronic viral infection. Curr. Opin. Immunol. 19, 408–415 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Good CR et al. An NK-like CAR T cell transition in CAR T cell dysfunction. Cell 184, 6081–6100 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bodapati S, Daley TP, Lin X, Zou J & Qi LS A benchmark of algorithms for the analysis of pooled CRISPR screens. Genome Biol 21, 62 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Renauer P SAMBA: CRISPR Screen analysis with moderated Bayesian statistics and adaptive gene aggregation scoring. R package version 1.1.0. https://github.com/Prenauer/SAMBA (2022). [Google Scholar]
- 40.Singer M et al. A distinct gene module for dysfunction uncoupled from activation in tumor-infiltrating T cells. Cell 171, 1221–1223 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nelms K, Keegan AD, Zamorano J, Ryan JJ & Paul WE The IL-4 receptor: signaling mechanisms and biologic functions. Annu. Rev. Immunol. 17, 701–738 (1999). [DOI] [PubMed] [Google Scholar]
- 42.Gray-Owen SD & Blumberg RS CEACAM1: contact-dependent control of immunity. Nat. Rev. Immunol. 6, 433–446 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Bandala-Sanchez E et al. T cell regulation mediated by interaction of soluble CD52 with the inhibitory receptor Siglec-10. Nat. Immunol. 14, 741–748 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Tosa N et al. Critical function of T cell death-associated gene 8 in glucocorticoid-induced thymocyte apoptosis. Int. Immunol. 15, 741–749 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Liu J et al. Enhanced CD4+ T cell proliferation and Th2 cytokine production in DR6-deficient mice. Immunity 15, 23–34 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Rutishauser RL et al. Transcriptional repressor Blimp-1 promotes CD8+ T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity 31, 296–308 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang M et al. The ubiquitin ligase Peli1 negatively regulates T cell activation and prevents autoimmunity. Nat. Immunol. 12, 1002–1009 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vacca M et al. NLRP10 enhances CD4+ T-cell-mediated IFNγ response via regulation of dendritic cell-derived IL-12 release. Front. Immunol. 8, 1462 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joshi NS & Kaech SM Effector CD8 T cell development: a balancing act between memory cell potential and terminal differentiation. J. Immunol. 180, 1309–1315 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Louis CU et al. Antitumor activity and long-term fate of chimeric antigen receptor-positive T cells in patients with neuroblastoma. Blood 118, 6050–6056 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jumper J et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gyory I, Fejer G, Ghosh N, Seto E & Wright KL Identification of a functionally impaired positive regulatory domain I binding factor 1 transcription repressor in myeloma cell lines. J. Immunol. 170, 3125–3133 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Yoshikawa T et al. Genetic ablation of PRDM1 in antitumor T cells enhances therapeutic efficacy of adoptive immunotherapy. Blood 139, 2156–2172 (2022). [DOI] [PubMed] [Google Scholar]
- 54.Morgan MA, Mould AW, Li L, Robertson EJ & Bikoff EK Alternative splicing regulates Prdm1/Blimp-1 DNA binding activities and corepressor interactions. Mol. Cell. Biol. 32, 3403–3413 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martins G & Calame K Regulation and functions of Blimp-1 in T and B lymphocytes. Annu. Rev. Immunol. 26, 133–169 (2008). [DOI] [PubMed] [Google Scholar]
- 56.Shin H et al. A role for the transcriptional repressor Blimp-1 in CD8+ T cell exhaustion during chronic viral infection. Immunity 31, 309–320 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Crotty S, Johnston RJ & Schoenberger SP Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat. Immunol. 11, 114–120 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shin HM et al. Epigenetic modifications induced by Blimp-1 regulate CD8+ T cell memory progression during acute virus infection. Immunity 39, 661–675 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xin A et al. A molecular threshold for effector CD8+ T cell differentiation controlled by transcription factors Blimp-1 and T-bet. Nat. Immunol. 17, 422 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fu S-H, Yeh L-T, Chu C-C, Yen BL-J & Sytwu H-K New insights into Blimp-1 in T lymphocytes: a divergent regulator of cell destiny and effector function. J. Biomed. Sci. 24, 49 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Z et al. Hypermethylation of PRDM1/Blimp-1 promoter in extranodal NK/T-cell lymphoma, nasal type: an evidence of predominant role in its downregulation. Hematol. Oncol. 35, 645–654 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Zhu L et al. Blimp-1 impairs T cell function via upregulation of TIGIT and PD-1 in patients with acute myeloid leukemia. J. Hematol. Oncol. 10, 124 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang S Histone methyltransferases, diet nutrients and tumour suppressors. Nat. Rev. Cancer 2, 469–476 (2002). [DOI] [PubMed] [Google Scholar]
- 64.Győry I, Wu J, Fejér G, Seto E & Wright KL PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat. Immunol. 5, 299–308 (2004). [DOI] [PubMed] [Google Scholar]
- 65.Ancelin K et al. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat. Cell Biol. 8, 623–630 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Skene PJ & Henikoff S An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6, e21856 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Magnúsdóttir E et al. Epidermal terminal differentiation depends on B lymphocyte-induced maturation protein-1. Proc. Natl Acad. Sci. USA 104, 14988–14993 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mascanfroni ID et al. IL-27 acts on DCs to suppress the T cell response and autoimmunity by inducing expression of the immunoregulatory molecule CD39. Nat. Immunol. 14, 1054–1063 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Agresta L, Hoebe KH & Janssen EM The emerging role of CD244 signaling in immune cells of the tumor microenvironment. Front. Immunol. 9, 2809 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kleinstiver BP et al. Genome-wide specificities of CRISPR–Cas Cpf1 nucleases in human cells. Nat. Biotechnol. 34, 869–874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim D et al. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 34, 863–868 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Yan WX et al. BLISS is a versatile and quantitative method for genome-wide profiling of DNA double-strand breaks. Nat. Commun. 8, 1–9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Canaj H et al. Deep profiling reveals substantial heterogeneity of integration outcomes in CRISPR knock-in experiments. Preprint at https://www.biorxiv.org/content/10.1101/841098v1 (2019).
- 74.Tsai SQ et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR–Cas nucleases. Nat. Biotechnol. 33, 187–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fry TJ et al. CD22-targeted CAR T cells induce remission in B-ALL that is naive or resistant to CD19-targeted CAR immunotherapy. Nat. Med. 24, 20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mullard A FDA approves second BCMA-targeted CAR-T cell therapy. Nat. Rev. Drug Discov. 21, 249 (2022). [DOI] [PubMed] [Google Scholar]
- 77.Adusumilli PS et al. Regional delivery of mesothelin-targeted CAR T cell therapy generates potent and long-lasting CD4-dependent tumor immunity. Sci. Transl. Med. 6, 261ra151 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.O’Rourke DM et al. A single dose of peripherally infused EGFRvIII-directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Sci. Transl. Med. 9, eaaa0984 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ahmed N et al. HER2-specific chimeric antigen receptor-modified virus-specific T cells for progressive glioblastoma: a phase 1 dose-escalation trial. JAMA Oncol. 3, 1094–1101 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Posey AD Jr et al. Engineered CAR T cells targeting the cancer-associated Tn-glycoform of the membrane mucin MUC1 control adenocarcinoma. Immunity 44, 1444–1454 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brudno JN & Kochenderfer JN Chimeric antigen receptor T-cell therapies for lymphoma. Nat. Rev. Clin. Oncol. 15, 31 (2018). [DOI] [PubMed] [Google Scholar]
- 82.Kim MY et al. Genetic inactivation of CD33 in hematopoietic stem cells to enable CAR T cell immunotherapy for acute myeloid leukemia. Cell 173, 1439–1453 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang Y et al. CD133-directed CAR T cells for advanced metastasis malignancies: a phase I trial. Oncoimmunology 7, e1440169 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Weiss T, Weller M, Guckenberger M, Sentman CL & Roth P NKG2D-based CAR T cells and radiotherapy exert synergistic efficacy in glioblastoma. Cancer Res. 78, 1031–1043 (2018). [DOI] [PubMed] [Google Scholar]
- 85.Wei J, Han X, Bo J & Han W Target selection for CAR-T therapy. J. Hematol. Oncol. 12, 62 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Reinhard K et al. An RNA vaccine drives expansion and efficacy of claudin-CAR-T cells against solid tumors. Science 367, 446–453 (2020). [DOI] [PubMed] [Google Scholar]
- 87.Maude SL et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med. 371, 1507–1517 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dai X et al. One-step generation of modular CAR-T cells with AAV-Cpf1. Nat. Methods 16, 247–254 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Concordet JP & Haeussler M CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 46, W242–W245 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Francois A et al. Accurate titration of infectious AAV particles requires measurement of biologically active vector genomes and suitable controls. Mol. Ther. Methods Clin. Dev. 10, 223–236 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.O’Roak BJ et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 338, 1619–1622 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang G et al. Mapping a functional cancer genome atlas of tumor suppressors in mouse liver using AAV-CRISPR-mediated direct in vivo screening. Sci. Adv. 4, eaao5508 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Boyle EA, O’Roak BJ, Martin BK, Kumar A & Shendure J MIPgen: optimized modeling and design of molecular inversion probes for targeted resequencing. Bioinformatics 30, 2670–2672 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kong NR, Chai L, Tenen DG & Bassal MA A modified CUT&RUN protocol and analysis pipeline to identify transcription factor binding sites in human cell lines. STAR Protoc. 2, 100750 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bolger AM, Lohse M & Usadel B Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhu Q, Liu N, Orkin SH & Yuan GC CUT&RUNTools: a flexible pipeline for CUT&RUN processing and footprint analysis. Genome Biol 20, 192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li H Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://arxiv.org/abs/1303.3997 (2013). [Google Scholar]
- 98.Zhang Y et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Quinlan AR & Hall IM BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Heinz S et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ramirez F, Dundar F, Diehl S, Gruning BA & Manke T deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hahne F & Ivanek R Visualizing genomic data using Gviz and Bioconductor. Methods Mol. Biol. 1418, 335–351 (2016). [DOI] [PubMed] [Google Scholar]
- 103.Wang GG et al. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature 459, 847–851 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bray NL, Pimentel H, Melsted P & Pachter L Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 105.Pimentel H, Bray NL, Puente S, Melsted P & Pachter L Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods 14, 687 (2017). [DOI] [PubMed] [Google Scholar]
- 106.Sherman BT & Lempicki RA Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44 (2009). [DOI] [PubMed] [Google Scholar]
- 107.Dai X et al. Massively parallel knock-in engineering of persistent CAR-Ts [CLASH]. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE207143 (2022).
- 108.Dai X, Park J, Du Y & Chen S Massively parallel knock-in engineering of persistent CAR-Ts. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE219061 (2022).
- 109.Dai X, Park J & Chen S Massively parallel knock-in engineering of persistent CAR-Ts [XDRNA]. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE207404 (2022).
- 110.Dai X, Chow R, Gu J, Chen S Massively parallel knock-in engineering of persistent CAR-Ts. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE201997 (2022).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this article and its supplementary information files. Specifically, source data and statistics for non-high-throughput experiments, such as flow cytometry, qPCR, protein experiments and other molecular or cellular assays, are provided in an Excel file of source data and statistics. Processed data for genomic sequencing (for example, CLASH, RNA-seq, amplicon sequencing, MIPS and AAV off-target) and other forms of high-throughput experiments are provided as processed quantifications in Supplementary Datasets. Genomic sequencing raw data are being deposited to the Gene Expression Omnibus (GEO), with the following accession numbers: GSE207143 for all CLASH screens; GSE219061 for MIPS, Nextera and AAV integration off-target; GSE207404 for RNA-seq; and GSE201997 for CUT&RUN107–110. CLASH vectors and libraries are available via material transfer agreements. All other data and materials that support the findings of this research are available either via public repositories or from the corresponding author upon reasonable request to the academic community. Source data are provided with this paper.