Abstract
AlkB homolog 1 (ALKBH1) is a member of the AlkB family of dioxygenases that are dependent on Fe(II) and α-ketoglutarate. Mounting evidence demonstrates that ALKBH1 exhibits enzymatic activity against various substrates, including N6-methyladenosine (m6A), N1-methyladenosine (m1A), N3-methylcytidine (m3C), 5-methylcytosine (m5C), N6-methyladenine (N6-mA, 6mA), and H2A, indicating its dual roles in different biological processes and involvement in human diseases. Up to the present, there is ongoing debate regarding ALKBH1's enzymatic activity. In this review, we present a comprehensive summary of recent research on ALKBH1, including its substrate diversity and pathological roles in a wide range of human disorders, the underlying mechanisms of its functions, and its dysregulation. We also explored the potential of ALKBH1 as a prognostic target.
Keywords: ALKBH1, RNA demethylation, DNA demethylation, Cancer, N1-methyladenosine, N6-methyladenine
Highlights
-
•
ALKBH1 exhibits enzymatic activity against various substrates, including m6A, m1A, m3C, m5C, N6-mA and H2A.
-
•
Although various substrates of ALKBH1 have been reported, there is still controversy regarding its enzymatic activity.
-
•
ALKBH1 is involved in a wide range of human disorders acting multiple roles.
Abbreviations list
- α-ketoglutarate
(α-KG)
- single-stranded DNA
(ssDNA)
- fat mass and obesity-associated protein
(FTO)
- N6-methyladenosine
(m6A)
- N1-methyladenosine
(m1A)
- transfer RNA
(tRNA)
- N6, 2-O-dimethyladenosine
(m6Am)
- 3-methylthymine
(3-meT)
- 3-methyluracil
(3-meU)
- single-stranded RNA
(ssRNA)
- double-stranded DNA
(dsDNA)
- N3-methylcytidine
(m3C)
- N3-methylthymine
(m3T)
- N1-methylguanine
(m1G)
- 1-methyladenine
(1-meA)
- 3-methylcytosine
(3-meC)
- N6-methyladenine
(N6-mA, 6mA)
- N2,N2-dimethylguanosine
(m22G)
- DNA Methyltransferase 1
(DNMT1)
- tRNA selenocysteine
(tRNASec)
- 5-methylcytosine
(m5C)
- nucleotide recognition lid
(NRL)
- double-stranded β-helix
(DSBH)
- mitochondrial
(mt)
- N-6 adenine-specific DNA methyltransferase 1
(N6AMT1)
- mesenchymal stem cells
(MSCs)
- transcription factor 4
(ATF4)
- hypoxia-inducible factor 1 alpha
(HIF-1α)
- glycogen synthase 1
(GYS1)
- chemokine (C-X-C motif) ligand 14
(CXCL14)
- non-small cell lung cancer
(NSCLC)
- overall survival
(OS)
- Glioblastoma
(GBM)
- glioblastoma stem cell
(GSC)
- Gastric cancer
(GC)
- Tissue Microarray-Immunohistochemistry
(TMA-IHC)
- AMP-activated protein kinase
(AMPK)
- Colorectal cancer
(CRC)
- Head and neck squamous cell carcinoma
(HNSCC)
- Tongue squamous cell carcinoma
(TSCC)
- Hepatocellular carcinoma
(HCC)
- 5-hydroxymethylcytosine
(5 hmC)
- Pancreatic adenocarcinoma
(PAAD)
- pancreatic ductal adenocarcinoma
(PDAC)
- Sirtuin4
(SIRT4)
- Ovarian cancer
(OV)
- Triple negative breast cancer
(TNBC)
- Renal cell carcinoma
(RCC)
- G-protein-coupled receptor 137
(GPR137)
- chronic kidney disease
(CKD)
- vascular calcification
(VC)
- octamer-binding transcription factor 4
(Oct4)
- bone morphogenetic protein 2
(BMP2)
- Atherosclerosis
(AS)
- Cardiovascular diseases
(CVDs)
- myocardial infarction-associated transcript
(MIAT)
- Abdominal Aortic Aneurysm
(AAA)
- N-oxalylglycine
(NOG)
1. Introduction
AlkB family members are widely distributed in the biological kingdom and play vital roles in epigenetics as Fe(II)/α-ketoglutarate (α-KG)-dependent dioxygenases. The functions of AlkB proteins were initially unclear until the discovery in 2000 that they process DNA damage and act on lesions produced in single-stranded DNA (ssDNA), which they preferentially bind over duplex DNA [1]. In 1983, seven Escherichia coli mutants sensitive to methyl methane sulfonate but not UV radiation were identified, revealing a lower host cell reactivation capacity for phage lambda treated with methyl methane sulfonate [2]. Five mutations were identified in the same location as alkA (formerly known as alk) and might be identical to previously recognized alterations, while another mutation was discovered near nalA and designated alkB [2]. The phenotype of the alkB mutant differed from that of ada, as it exhibited a typical adaptive response to N-methyl-N′-nitro-N-nitrosoguanidine [2]. The AlkB gene has been recognized to be conserved from bacteria to mammals, with nine different mammalian ALKB homologs identified, including ALKBH1-8 and fat mass and obesity-associated protein (FTO) [3,4].
Since they all possess Fe2OG dioxygenase (Fe(II) and α-KG-dependent dioxygenase), they may all undergo oxidative dealkylation to remove alkyl adducts from nucleobases. However, their catalytic substrates differ. For instance, in the nucleus, FTO-mediated RNA demethylation occurs at internal N6-methyladenosine (m6A) in polyA RNA and N1-methyladenosine (m1A) in transfer RNA (tRNA), as well as m6A in U6 snRNA and N6,2-O-dimethyladenosine (m6Am) in U2 but also U1 snRNA. FTO also localizes to the cytoplasm, where it mediates the demethylation of internal m6A in polyA RNA and cap m6Am in polyA RNA, alongside tRNA m1A demethylation in the cytoplasm [[5], [6], [7]]. In addition, FTO is capable of oxidative demethylation of 3-methylthymine (3-meT) in ssDNA and 3-methyluracil (3-meU) in single-stranded RNA (ssRNA) [8]. ALKBH2 is reported to repair DNA alkylation lesions in double-stranded DNA (dsDNA) [9]. In contrast, ssDNA or ssRNA with N-methylated bases, such as m1A, N3-methylcytidine (m3C), N3-methylthymine (m3T), and N1-methylguanine (m1G), can be repaired by ALKBH3 to undo methylation damage and maintain the integrity of the DNA/RNA [10]. ALKBH3 is a demethylase for m1A in mRNA, 1-methyladenine (1-meA) and 3-methylcytosine (3-meC) in mammalian RNAs and N6-methyladenine (N6-mA, 6mA) in mammalian tRNA, and ALKBH3-directed tRNA demethylation has been shown to enhance protein translation efficiency [11,12]. Similarly, ALKBH6 prefers to bind to ssDNA or ssRNA. However, there is no obvious demethylase activity of ALKBH6 in numerous kinds of modified nucleic acids, such as m1A and m6A ssDNA/ssRNA [13]. In addition to its role in facilitating actin K84me1 and N6-meA demethylation in DNA, ALKBH4 also modulates DNA cytosine methylation by altering the expression level of the DNA methyltransferase 1 (DNMT1) protein [[14], [15], [16]]. Besides, Caenorhabditis elegans NMAD-1, the orthologous to human ALKBH4, is also reported as a demethylase for actin and prefers to regulate 6mA in the unpairing regions, and is thus possibly associated with dynamic chromosome regulation and meiosis regulation [17,18]. ALKBH5 has been identified as a demethylase that catalyzes the oxidative reversal of m6A in mRNA. Additionally, recent studies have reported that ALKBH5 demonstrates demethylation activity toward N6-mA in mRNA [19]. Finally, within nascent polycistronic mitochondrial RNA, ALKBH7 is responsible for demethylating N2,N2-dimethylguanosine (m22G) and m1A residues located within the mitochondrial Ile and Leu 1 pre-tRNA regions, respectively [20], while ALKBH8 is reported to modify tRNA selenocysteine (tRNASec) and possess tRNA methyltransferase activity [21,22].
However, of all the AlkB proteins, ALKBH1 demonstrates enzymatic activity against a variety of substrates and displays the highest sequence homology with E. coli AlkB, which is the focus of this review.
According to recent studies, ALKBH1 is reported to catalyze six substrates directly: m6A, m1A, m3C, 5-methylcytosine (m5C), N6-mA, and H2A [[23], [24], [25], [26], [27], [28]]. Importantly, the catalytic function of ALKBH1 with various substrates is related to various human disorders. Despite efforts, there continues to be disagreement concerning ALKBH1's enzymatic activity towards various substrates. In this review, we elaborate on the most recent developments in the investigation of ALKBH1 substrates over a wide range and their implications in human diseases. Most importantly, we propose solutions for the treatment of related diseases, making this review highly relevant and informative.
2. The biological role of ALKBH1
2.1. The structure of ALKBH1
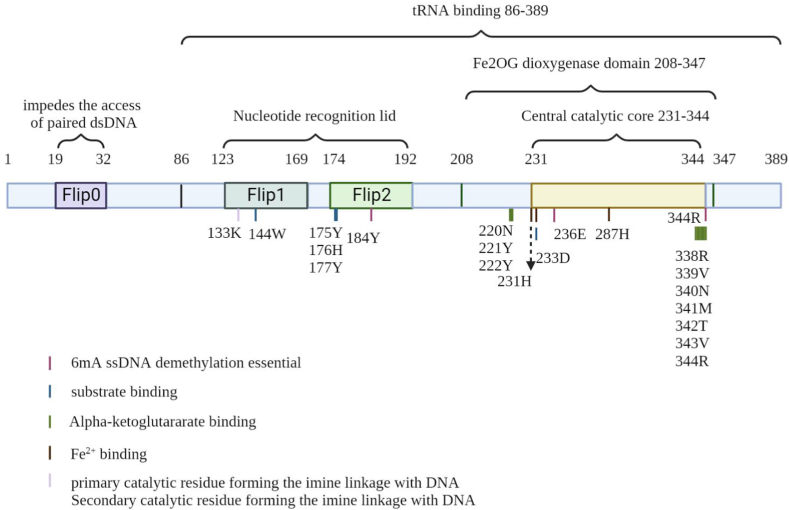
Full-length human ALKBH1 contains 389 amino acids, among which 19–369 amino acids have an enzymatically active center. ALKBH1 has a distinct N-terminal Flip0, a nucleotide recognition lid (NRL) with Flip1 and Flip2, and a central catalytic core [29]. The α-KG-dependent dioxygenase superfamily is distinguished by a highly conserved double-stranded β-helix (DSBH) structure in the catalytic core. The DSBH catalytic core has a typical eight-stranded (I–VIII) jelly-roll fold which is made up of a major sheet (βI, βVIII, βIII, βVI) and a minor sheet (βVII, βIV, βV and βII) to form a sandwich formation of Mn(II) and NOG at the active center [27]. The key domains of ssDNA selectivity in hALKBH1 are Flip0 and NRL. ALKBH1 has a conservative HxD … H metal ion-binding sequence and an R … R α-KG-binding sequence. α-KG is stabilized by three hydrogen bonds generated by the side chains of Asn220, Asn340, and Tyr222 and three salt bridges created by Arg338 and Arg344 [29]. In addition, Tian Li-Fei et al. found that the Tyr184-Arg344-Glu 236 stability triangle near the α-KG-binding site is indispensable for 6 mA ssDNA demethylation activity [29]. With the addition of α-KG, the entire structure of ALKBH1 shrinks. The NRL of ALKBH1 contains numerous structural differences from other members of the AlkB family. The ALKBH1 Flip1 region is distinct and long, providing a large binding area over the active site pocket, and is stabilized by extensive hydrophobic and hydrogen-bonding interactions by helical cluster formation among α3 and a ALKBH1-uniqued “INS” motif, which is a non-conserved long insertion between β(VII) and β(VIII) [27]. K158A/R159A/R160A/R162A and K167A/R169A mutants completely lose m6A demethylation activity, while K158A/R159A/R160A mutant activity is severely reduced [29]. Furthermore, Flip2 has two antiparallel β-sheets and a large loop with a high B factor. Key residues in the NRL region, such as Arg 169, Trp 170, Tyr177, and Trp179, have been identified as possible ALKBH1 determinants for 6 mA identification and demethylation. The unique composition and structure of Flip1 and Flip2 are believed to confer substrate selectivity on ALKBH1. Surprisingly, at the N-terminus, Flip0 (residues 19–32) collides with the unmethylated strand, which is required for ALKBH1 action and distinguishing single-stranded from paired double-stranded substrates. A hypothesized model for the binding of human ALKBH1 to ssDNA is proposed, in which one end of the nucleic acid strand interacts with Flip1, while the other end extends along the direction of the groove generated by Flip2 and Flip0 [29]. ALKBH1 exhibits a preference for DNA substrates with bubbles and bulges, as shown by previous studies [29,30]. It has been shown that residues Y184, H231, D233, H287, R338 and R344 in ALKBH1 catalyze the demethylation of m6A in RNA [23] (Fig. 1).
Fig. 1.
Functional domains of ALKBH1.
Recent study found that ALKBH1 preferred to catalyze the demethylation of N6-mA in bubbled or bulged DNA rather than single stranded DNA, and could not completely catalyze the demethylation of double stranded substrates [27,29,30]. Further experiments have shown that ALKBH1 can equally efficiently catalyze the demethylation of N6-mA in various local unpaired nucleic acids (such as bulge, R-loop, D-loop, and stem loop), revealing for the first time the high dependence of ALKBH1 on the substrate secondary structure. What is the structural basis for the unique substrate preference of ALKBH1, as other members of the AlkB family and Tet family have not shown a preference for local unpaired nucleic acids? The Flip1 of ALKBH1 is far from the enzyme active center and completely inverted, forming a sharp contrast with other members of the AlkB family and the Tet 2 “snap back” Flip1. The outward flipped Flip1 may cause ALKBH1 to fail to act on double stranded substrates, but it plays an important role in identifying the double stranded region of locally unpaired substrates. When ALKBH1 binds to the bulk substrate, its Flip1 still maintains a completely inverted conformation, causing it to be unable to actively invert N6-mA in the substrate, thus completely unable to catalyze demethylation of the double stranded substrate. Flip1 with outward flip and “α1” helix is located on both sides of the catalytic center and interacts with the double chain regions on both sides of the bulge. Among them, α1, the R24 and R159 residues on Flip1 are inserted into the small grooves of the double stranded region in the form of “Arginine finger”, respectively, to assist in presenting the N6-mA on the bulk to the catalytic center, highlighting the crucial role of the local unpaired substrate double stranded region in substrate recognition and clearly elucidating its preference for catalyzing local unpaired nucleic acid substrates rather than single stranded substrates. Further analyses found significant co localization between N6-mA and non paired regions of the genome, suggesting that the regulation of N6-mA may be closely related to the dynamic changes in the advanced structure of eukaryotic chromosomes. This will provide a new perspective for subsequent functional research of ALKBH1 and N6-mA [27].
ALKBH1 differs from other proteins in the AlkB family and has multiple new structural features. Its unique Flip0 region, located near the pocket of the active center, plays a crucial role in ALKBH1 substrate recognition and 6mA demethylase activity. Flip1, which is far from the active center and carries a positive charge, has a large number of alkaline amino acid residues on the side facing the active center, forming a larger positively charged region. Mutations in these alkaline residues cause ALKBH1 to lose its ability to bind to DNA. In addition, ALKBH1 binding Tyr184 and Glu 236 near the active center after α-KG moves towards the active center and forms a stable triangular structure with Arg344 through hydrogen bonding, ensuring the activity of ALKBH1 to execute 6mA demethylase function. In addition, Flip1 and Flip2 in the ALKBH1 structure contain a long loop, and their conformational changes, along with Tyr177 and Trp179 located on Flip2, are involved in the recognition and binding of DNA 6mA [29].
Collectively, there are three notably unique structural features to ALKBH1 arise outside the core structure: a far N-terminal α1 helix packed aside the catalytic center, the stretch-out of an extended Flip1, and a long insertion between β(VII) and β(VIII) (INS) that stabilizes Flip1 through helical cluster formation. Such features hold unique for ALKBH1 when comparing with all structures of its human paralogues determined so far, including ALKBH2, 3, 5, 7, 8 and FTO [27,29].
2.2. The substrate types of ALKBH1
ALKBH1 displays a wide distribution within cells, including the cytoplasm, nucleus, and mitochondria. In different regions, ALKBH1 plays different roles and has different enzymatic activities [28,[31], [32], [33]]. It is noteworthy that all potential substrates of ALKBH1, as we shown below, share a locally unpairing feature in which the modified nucleotide is prone to flipping out for demethylation.
2.2.1. H2A
Previous study has demonstrated that ALKBH1, which is found in the nucleus, has dioxygenase activity specific to histone H2A and plays a critical role in neural development by altering histone H2A methylation status [28]. Furthermore, the same group reports that ALKBH1 regulates the core transcriptional network of embryonic stem cells via histone H2A demethylation [34]. Prior research has postulated that ALKBH1 might modulate the methylation of histone H2A at K118 or K119 in embryonic stem cells [28] (Fig. 2A). However, despite claims that ALKBH1 has been reported to have H2A demethylation activity, this assertion has only been put forward by a single research team, necessitating further investigation for confirmation and reproducibility. Surprisingly, in vitro tests with recombinant ALKBH1 have failed to demonstrate reproducible histone H2A demethylation activity [35]. Moreover, Wu et al. conducted further investigations and eliminated the possibility of ALKBH1 functioning as a histone demethylase, as H2A K118/119 remains predominantly nonmethylated in wild-type or ALKBH1 knockout embryonic stem cells [36]. Nonetheless, there is still controversy over whether H2A is a dioxygenase substrate for ALKBH1, this area of research requires further investigation to clarify the role of ALKBH1 in histone H2A methylation.
Fig. 2.
Substrates of ALKBH1. The potential substrates of ALKBH1 include H2A, m1A, m6A, m3C, m5C and N6-mA. “?” indicates that there is currently controversy or a lack of good biochemical experiments in vitro.
2.2.2. m1A in tRNA
In 2016, He Chuan et al. discovered ALKBH1 as an RNA demethylase that causes methyl group removal from m1A in tRNA (Fig. 2B). Specifically, ALKBH1 targets m1A58 in tRNAs as its primary substrate [24]. The demethylation of target tRNAs by ALKBH1 not only causes diminished translation initiation and decreased use in protein synthesis but also induces tRNA destabilization and increased tRNA cleavage [24]. It is noteworthy that the influence of ALKBH1 on cell fate and tRNA cleavage is highly context-specific and dependent on the demethylation capability of the cellular stressors involved. Additionally, the cleavage of tRNAs following ALKBH1 modification and stress is not uniform across all tRNA species. Moreover, ALKBH1 knockdown fails to protect tRNAs from undergoing cleavage during demethylation stress [37]. Furthermore, RNA-mass analysis revealed a significant increase in the frequency of m1A modification in two mitochondrial (mt) tRNAs, namely, m1A16 in mt-tRNAArg and m1A58 in mt-tRNALys, in ALKBH1-knockout cells, indicating that ALKBH1, which is localized in mitochondria, exhibits demethylating activity toward mt-tRNAs [26]. Surprisingly, while an α1-truncated human ALKBH1 protein was reported to possess demethylation activity towards m1rA in tRNA [24], a subsequent study revealed that in vitro enzymatic assays hardly detected any activity towards m1rA in tRNA stem loop using the α1-truncated human ALKBH1 [27]. This finding casts doubt on the role of ALKBH1 in mediating the demethylation of m1A in tRNA, leaving this aspect shrouded in mystery.
2.2.3. m1A in mRNA
M1A is enriched in the 5′ UTR and near start codons of mRNA transcripts [11,38] (Fig. 2C). Furthermore, m1A-MAP displays prevalent m1A modification in mitochondrial-encoded transcripts [39]. M1A in mRNA responds dynamically to physiological conditions and has a positive correlation with protein production, indicating that m1A plays an important role in promoting methylated mRNA translation [38]. It is worth noting that the differential impact of m1A on translation efficiency depends on the mRNA region where it is present. For example, increased translation efficiency is associated with m1A in the 5′ UTR, especially at the mRNA cap. On the other hand, m1A in the CDS of mt-mRNA inhibits translation [39]. Liquid chromatography-tandem mass spectrometry analysis conducted by He Chuan et al., in 2016 revealed a modest increase in the overall m1A content of mRNA in ALKBH1−/− embryonic stem cells compared to ALKBH1 ± embryonic stem cells. Conversely, overexpression of ALKBH1 in HeLa cells resulted in a slight decrease in m1A in mRNA, suggesting that ALKBH1 might be responsible for mediating m1A demethylation in mRNA [24]. Additionally, depletion of ALKBH1 increased the m1A abundance in both METTL3 mRNA and MFAP2 mRNA [40,41]. Furthermore, ALKBH1 knockout cells exhibited a considerable decrease in mitochondrial protein synthesis relative to that in wild-type cells [26]. Future research need to look into the likelihood that ALKBH1 mediates the demethylation of m1A in mRNA.
2.2.4. m6A
M6A might be an ALKBH1 substrate. Li H. et al. reported that ALKBH1 plays a significant role in demethylating m6A in A549 cells, as depletion of ALKBH1 led to an increase in m6A levels [23] (Fig. 2D). Furthermore, in HEK293 cells, ALKBH1 was observed to localize to the mitochondria and exhibit m6A demethylase and AP lyase activities, as well as the ability to form a covalent adduct with AP-containing DNA, similar to that produced in E. coli [42]. These findings suggest that ALKBH1 has multifunctional roles in regulating DNA and RNA modifications in different cellular compartments. However, He Chuan et al. found that downregulation of ALKBH1 did not alter the m6A level in rRNA and did not noticeably affect the mRNA m6A level [24]. Thus, whether ALKBH1 is involved in the demethylation of m6A still needs further study.
2.2.5. m5C
ALKBH1 plays a crucial role in mitochondrial activity, as it is involved in the biogenesis of hm5Cm34 and f5Cm34 in ct-tRNALeu, as well as f5C34 in mt-tRNAMet, which are necessary for efficient translation [26,43]. ALKBH1 catalyzes the hydroxylation of m5C34, resulting in the formation of hm5C34, which is subsequently oxidized to f5C34 using O2 and 2-OG as substrates (Fig. 2E). Notably, ALKBH1 is required for mitochondrial translation and oxygen consumption, as ALKBH1-knockout cells show a significant reduction in both [26]. ALKBH1 also colocalizes with mtRNA granule markers, indicating its role in translation regulation rather than transcription [44]. Additionally, ALKBH1 deficiency in HEK293 cells is associated with increased mtDNA copy number and impaired mitochondrial function, further highlighting the critical role of ALKBH1 in mitochondrial function [26,42]. Furthermore, telencephalon-specific ALKBH1 conditional knockout mice exhibit hippocampal atrophy and impaired learning, emphasizing the importance of ALKBH1 in brain function [45]. However, these reports lacked robust biochemical experiments conducted in vitro.
2.2.6. m3C
In 2008, it was first discovered that ALKBH1 was a mitochondrial protein that demethylated m3C in ssDNA and ssRNA in vitro by oxidative demethylation, presumably carrying out base repair [31]. More recently, a study demonstrated that ALKBH1 could demethylate m3C in mammalian cell mRNA both in vitro and in vivo [25] (Fig. 2F). These findings broaden the understanding of ALKBH1's role in regulating RNA modifications; however, the studies lacked comprehensive biochemical assays performed in vitro.
2.2.7. N6-mA
Our knowledge of DNA N6-mA has been limited despite its discovery in prokaryotes and unicellular eukaryotes several decades ago [[46], [47], [48], [49], [50]]. However, recent advancements in deep sequencing have enabled the detection of N6-mA in high eukaryotes, revealing that [G/C]AGG[C/T] is the most prevalent motif at N6-mA modification sites across the human genome [[51], [52], [53]]. N6-mA density is markedly enriched within coding regions and is closely associated with gene transcriptional activation. Furthermore, N6-mA modification is a highly dynamic process that is governed by specific methyltransferases and demethylases. The most well-characterized methyltransferase is N-6 adenine-specific DNA methyltransferase 1 (N6AMT1), and ALKBH1 has been proposed as a candidate demethylase of DNA N6-mA [54] (Fig. 2G). Additionally, it has been established that the catalytic motif NPPY located in N6AMT1 and the critical residue D233 in ALKBH1 play indispensable roles in the enzymatic addition and removal of N6-mA [54]. However, several reports suggest that N6AMT1 does not serve as an N6-mA methyltransferase [55,56]. Besides, a study showed that METTL4 could mediate mammalian mtDNA 6mA methylation [57]. But another study demonstrates that the substrate catalyzed by METTL4 is RNA rather than DNA [58].
Additionally, knockout of ALKBH1 results in an increase in N6-methyladenine levels and leads to transcriptional silencing [36]. Mechanistically, ALKBH1 may function as a transcriptional activator by binding to regions of N6-mA enrichment and removing repressive N6-mA marks at selected genomic loci [59]. For example, ALKBH1 expression and decreased DNA N6-mA levels have been found to be essential for prompt sensory axon regeneration both in vivo and in vitro. ALKBH1 could aid in sensory axon regeneration by decreasing neuronal DNA N6-mA levels and consequently increasing the expression of neurodevelopmental genes [60]. Crucially, a study encompassing biochemical profiling and structural analyses of ALKBH1 offers compelling evidence that ALKBH1 serves as a crucial DNA demethylase, regulating genome N6-mA turnover of unpairing regions associated with dynamic chromosome regulation [27].
Moreover, it has been demonstrated that the induction of osteogenesis in human mesenchymal stem cells (MSCs) is accompanied by a significant increase in the expression level of ALKBH1. This augmentation of ALKBH1 expression plays a crucial role in regulating the levels of DNA N6-mA, which is imperative for osteogenic differentiation. Specifically, the reduction in N6-mA levels within the promoter region of activating transcription factor 4 (ATF4) via ALKBH1-mediated regulation is essential for ATF4 transcriptional activation, ultimately promoting the osteogenic differentiation of MSCs [61]. This phenomenon has been extensively documented in the literature [61] and has significant implications in the fundamental understanding of bone development and regeneration. Additional evidence suggests that ALKBH1 plays a critical role in the adipogenic differentiation of both human mesenchymal stem cells and 3T3-L1 cells, leading to excessive adipose accumulation in mice [62]. This effect is achieved through the demethylation of N6-mA residues in the hypoxia-inducible Factor 1 alpha (HIF-1α) and glycogen synthase 1 (GYS1) genes by ALKBH1, ultimately augmenting HIF-1 pathway activation and promoting adipogenic differentiation [62]. Moreover, during skeletal muscle development, the increase in the level of DNA N6-mA modification was accompanied by a strong decrease in ALKBH1 expression. In myogenesis, ALKBH1 is reported to inhibit the differentiation of skeletal muscle by regulating a core set of genes and multiple signaling pathways; for example, it increases chemokine (C-X-C motif) ligand 14 (CXCL14) expression and activates ERK signaling [63]. What's more, ALKBH1 has also been proposed as a 6 mA demethylase in human mtDNA, affecting oxidative phosphorylation [64].
Certain individuals hold the viewpoint that the existence of DNA N6-mA in mammals lacks concrete evidence and that the apparent identification of this epigenetic modification in mammalian DNA is a result of technical limitations such as antibody cross-reactivity, bacterial or RNA contamination, and other experimental confounding factors [65]. An article showed that the content of 6mA in mammalian genomes is very low and these 6mA are not catalyzed by methylases, but are metabolized and reused through methylolysis of m6A RNA, which is integrated into the genome by DNA polymerase [66]. Another study also reported that 6mA passed through the template independent poly of members of the DNA polymerase X family λ incorporating into the genome [67]. It is noteworthy that a recent article in 2022 showed that the observed 6mA in human glioblastomas is solely generated by DNA polymerase-mediated misincorporation. Additionally, in vitro experiments point toward that the generation of misincorporated DNA 6mA is associated with the cellular stresses-caused release of RNA N6-methyladenine (m6A) nucleoside, which is profoundly inhibited by hypoxia milieu. Furthermore, they confirmed ALKBH1, which has been proposed as a candidate demethylase of DNA 6mA in a number of reports, failed to erase the misincorporated DNA 6mA [51]. These criticisms underscore the need for rigorous experimental design and stringent validation of findings, particularly in the context of epigenetic modifications that are notoriously challenging to study. While the debate regarding the presence of N6-mA in mammalian DNA remains unresolved, further research efforts are required to definitively elucidate the nature and functional implications of this epigenetic mark in mammals.
Moreover, contemporary investigations have revealed an additional physiological function of ALKBH1. Specifically, two distinct murine studies have demonstrated that ALKBH1 gene knockout results in offspring production at a lower rate than the anticipated Mendelian ratio, with a greater proportion of male pups than female pups and a reduced body size compared to their wild-type littermates [68,69]. Furthermore, an earlier study indicated that ALKBH1 serves as an important mediator of placental trophoblast lineage differentiation [32].
Although various substrates of ALKBH1 have been mentioned, only the role of ALKBH1 as a demethylase for DNA N6-mA modification has been conclusively validated through structural biology and biochemical experiments, and this finding has been repeated by four independent research groups. This makes it tempting to believe that ALKBH1 relevant substrates predominantly would be DNA N6-mA rather than others.
3. ALKBH1 and cancer
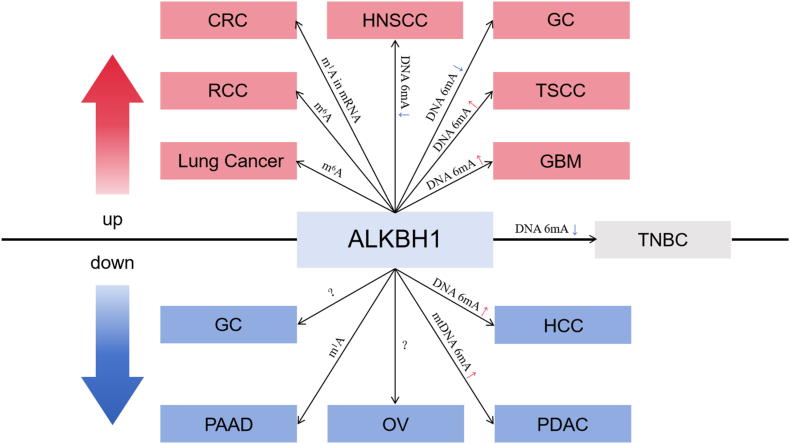
ALKBH1 is a multifaceted enzyme that acts on various substrates, and its dysregulation has been implicated in the pathogenesis of various cancers (Table 1). For instance, ALKBH1 has been shown to play a role in the demethylation of N6-mA in DNA, which has been associated with transcriptional silencing and tumor growth. Additionally, ALKBH1 is required for efficient mitochondrial activity and has been linked to mitochondrial dysfunction and cancer progression. Moreover, ALKBH1 is involved in the biogenesis of modified nucleotides in transfer RNAs, which has been shown to impact protein synthesis and contribute to cancer development. Given the diverse substrates and functions of ALKBH1 (Fig. 3), it is clear that this enzyme plays a critical role in cancer biology and represents a promising target for therapeutic intervention.
Table 1.
Roles of ALKBH1 in human cancer.
| Cancer type | Expression | Modification types | Modification expression in cancer | Biological functions | Molecular mechanisms | Targets | Reference |
|---|---|---|---|---|---|---|---|
| Lung cancer | up | m6A | No study | facilitated cell migration and invasion | No study | No study | 23 |
| RCC | up | m6A | No study | promotes RCC cell malignancy | ALKBH1 reduces m6A levels of GPR137 mRNA, which upregulates GPR137 mRNA levels, resulting in the increased GPR137 protein expression and the enhanced RCC cell biological actions consequently | GPR137 | 73 |
| CRC | up | m1A in mRNA | No study | Promote CRC metastasis | ALKBH1-mediated m1A demethylation of METTL3 mRNA promotes the metastasis of colorectal cancer by downregulating SMAD7 expression | METTL3 | 40 |
| HNSCC | up | DNA 6mA | down | Depletion of ALKBH1 inhibits HNSCC cells proliferation and colony formation of HNSCC patients-derived organoids | ALKBH1 enhances DDX18 expression by erasing DNA 6mA level and regulating its promoter activity | DDX18 | 75, 76 |
| GC | up | DNA 6mA | down | Not only enhances viability and migratory cell number, but also promotes the growth and metastasis of GC cells | ALKBH1-mediated 6mA demethylation in NRF1 transcriptional activity, leading to repressed AMPK signaling activation and a “Warburg” phenotype in GC cells | NRF1 | 81, 82 |
| GC | down | No study | No study | No study | No study | No study | 80 |
| GBM | up | DNA 6mA | up | Knockdown of ALKBH1 decreased proliferation, self-renewal, and tumor formation capacity. Upregulation of ALKBH1 did not elicit any significant changes in cell viability, self-renewal or in vivo tumor formation. | Depletion of ALKBH1 led to an increased N6-mA level in genomic DNA, which coordinated with H3K9me3, caused to transcriptional silencing of oncogenic pathways through decreasing chromatin accessibility. |
No study | 59 |
| TSCC | up | DNA 6mA | up | Knockdown of ALKBH1 promotes colony formation and cell migration | Knockdown of ALKBH1 results in increased 6mA levels | NF-κB pathway | 83 |
| HCC | down | DNA 6mA | up | ALKBH1 overexpression suppressed HCC cell viability, migration and invasion, enhanced apoptosis | When ALKBH1 was overexpressed, PARTICL was upregulated while PARTICL DNA 6mA modification was reduced | PARTICL | 88 |
| PAAD | down | m1A | No study | No study | ALKBH1 may participate in the occurrence and development of pancreatic cancer through mTOR and ErbB signaling pathway | mTOR and ErbB signaling pathway | 89 |
| PDAC | down | mtDNA 6mA | up | Knockdown of ALKBH1 blocks the transcription of mitochondrial DNA-coded genes, leads to mitochondrial dysfunction and inhibits pancreatic cancer cells | ALKBH1 can be downregulated by HRD1-SEL1L complex through ubiquitination when a tumor inhibitor, Sirtuin4 (SIRT4) overexpressed | mitochondrial DNA-coded genes | 90, 91 |
| OV | down | No study | No study | No study | No study | No study | 93 |
| TNBC | No change | DNA 6mA | down | No study | No study | No study | 94 |
Fig. 3.
The molecular mechanisms of ALKBH1 involved in cancers. Over-expression of ALKBH1 in cancers is in red; low expression of ALKBH1 in cancers is in blue; No change of ALKBH1 expression in cancer is in gray. ALKBH1 exhibits different enzymatic activity in different cancers. Arrows indicate changes in the expression of modifications in cancer compared with normal tissues. Over-expression of modification expression is in red, while low expression is in blue.
3.1. Lung cancer
Lung cancer is a major cause of mortality worldwide, ranking as the second most common cancer [70]. Non-small cell lung cancer (NSCLC), accounting for approximately 85 % of all lung cancer cases, has two main pathological subtypes, lung adenocarcinoma and lung squamous cell carcinoma [71]. In both NSCLC subtypes, ALKBH1 has been found to be upregulated in cancer tissues and cells, and higher levels of ALKBH1 mRNA are associated with lower overall survival (OS) rates in patients. Mechanistically, ALKBH1 promotes lung cancer by regulating m6A RNA demethylation, and depletion of ALKBH1 significantly reduces the invasion and migration abilities of lung cancer cells in vitro. In contrast, overexpression of ALKBH1 significantly facilitates the migration and invasion of lung cancer cells [23]. Furthermore, ALKBH1 mRNA levels are also significantly increased in NSCLC patients [72]. Collectively, these findings reveal that ALKBH1 may be as an ‘eraser’ of m6A, takes a great part in the invasion and metastasis of lung cancer.
3.2. Renal cell carcinoma (RCC)
Renal cell carcinoma is a malignant tumor with high mortality rates in adults worldwide. The latest study revealed that ALKBH1 is upregulated in RCC tissues and promotes RCC cell malignancy [73]. ALKBH1 promotes the viability and mobility of RCC cells. Mechanistic investigation further shows that ALKBH1 upregulates G-protein-coupled receptor 137 (GPR137) mRNA levels in RCC cells by decreasing GPR137 mRNA m6A levels, which enhances GPR137 protein expression and improves the biological functions of RCC cells as a result [73]. Moreover, ALKBH1 expression is correlated with GPR137 expression in RCC tissues. Taken together, the data indicate that ALKBH1 potentiates RCC cell biological actions through GPR137 in an m6A-dependent manner, which confirms ALKBH1's critical role in the regulation of RNA m6A and broadens its range of functions in cancer.
3.3. Colorectal cancer (CRC)
CRC is a common malignancy worldwide and a significant threat to human health, with high incidence and mortality rates [74]. The latest research revealed that ALKBH1, which is overexpressed in CRC, plays a crucial role in promoting metastasis and poor prognosis [40]. Through its m1A demethylation activity, ALKBH1 has been found to promote CRC metastasis both in vitro and in vivo when its expression level is increased. Furthermore, depletion of ALKBH1 leads to decreased METTL3 expression by methylating the m1A site in METTL3 mRNA, ultimately resulting in m6A demethylation in SMAD7 mRNA. More importantly, the downregulation of SMAD7 contributes to an aggressive phenotype, where the effects of ALKBH1 depletion or METTL3 depletion on CRC cell migration and invasion are reversed [40]. Collectively, these findings provide evidence to suggest that ALKBH1 may promote metastasis in CRC by destabilizing SMAD7 through METTL3. These results help to fill a gap in the current understanding of the role of ALKBH1 in CRC.
3.4. Head and neck squamous cell carcinoma (HNSCC)
HNSCC is a group of tumors that originate in various parts of the head and neck, including the oral cavity, hypopharynx, nasopharynx, oropharynx, or larynx. As the seventh most common malignancy worldwide, it is responsible for approximately 400,000 deaths per year [74]. In a significant proportion of HNSCC cases, ranging from 68 % to 90 %, there is a notable increase in the expression level of ALKBH1 [75]. Furthermore, a recent study suggests that ALKBH1 is highly expressed in HNSCC cells and patient tissues. Depletion of ALKBH1 inhibits HNSCC cell proliferation and colony formation in HNSCC patient-derived organoids [76]. In addition, ALKBH1 can enhance DDX18 expression by reducing the DNA 6mA level and controlling promoter activity. When DDX18 was overexpressed exogenously, ALKBH1 knockdown-induced cell proliferation arrest caused by DDX18 downregulation was attenuated [76]. This suggests that ALKBH1 may play a crucial role in the development and progression of HNSCC.
3.5. Glioblastoma (GBM)
GBM, the most prevalent and aggressive primary intrinsic brain tumor, exhibits markedly increased levels of N6-mA DNA modification [59]. This modification primarily represses gene expression in human GBM, contributing to heterochromatin formation and tumorigenesis. ALKBH1, identified as an N6-mA DNA demethylase, is essential for GBM stem cell growth, self-renewal, and tumor formation capacity and contributes to N6-mA colocalization with H3K9me3 genome wide [59]. Depletion of ALKBH1 facilitates heterochromatin formation in human GBM through N6-mA DNA modification, which results in transcriptional silencing of oncogenic pathways by decreasing chromatin accessibility. Moreover, knockdown of ALKBH1 decreased glioblastoma stem cell (GSC) proliferation and inhibited tumor formation. However, the upregulation of ALKBH1 did not elicit any significant changes in cell viability, self-renewal or in vivo tumor formation. Paradoxically, ALKBH1 was found to be overexpressed in GBM tissue relative to nontumor brain tissue and was strongly correlated with increased glioma grade and reduced survival rates. This apparent discrepancy can be attributed to the intricate and dynamic regulation of N6-mA, which is governed by both a demethylase and an unknown methyltransferase. Perturbing the dynamic regulation of N6-mA via ALKBH1 knockdown resulted in decreased proliferation, self-renewal, and tumor formation capacity, thereby underscoring the crucial role of N6-mA regulation in glioblastoma stem cell dependency [59].
On the contrary, Lyu et al. recently reported a significant reduction in genomic DNA 6mA content in human glioma, including glioblastoma. They demonstrated that the observed DNA 6mA in human glioblastoma and glioblastoma stem cells is extremely rare and is independent of DNA methyltransferase. Further in-depth study showed that the generation of the rare and non-epigenetic DNA 6mA solely attributed to the DNA polymerase-dependent misincorporation caused by DNA amplification. A study conducted in vitro suggests that the production of misincorporated DNA 6mA is linked to the release of RNA N6-methyladenine (m6A) nucleoside, which is significantly suppressed by a hypoxic environment. Their findings strongly suggest a marked decrease in misincorporated DNA 6mA content in glioma relative to normal brain tissues [51]. Moreover, more evidence highlights a positive correlation between misincorporated DNA 6mA content and OS rates among patients with glioma. Intriguingly, Lyu et al. reported both GBM and low-grade gliomas showed no change in overall survival (OS) rates in response to high expression of ALKBH1 mRNA. Nor did the misincorporated DNA 6mA exhibit any notable changes after ALKBH1 was depleted. These studies strongly suggest that there might be an unknown demethylase responsible for erasing the misincorporated DNA 6mA, but not ALKBH1(51).
Why do various studies' findings about GBM differ so much? We believe that the various detection techniques are the primary cause. There are currently 27 techniques available for identifying 6mA in various eukaryotes, particularly mammals [77]. However, in order to confirm or rule out the presence of 6mA, it is necessary to concurrently use multiple unrelated approaches to verify each other given the limitations of each detection and quantification technique for DNA 6mA in the further study. In addition, even though ALKBH1 has been proposed as a candidate demethylase of DNA 6mA in a number of reports, its structural features limit its ability to catalyze 6mA demethylation in ssDNA but not dsDNA [29]. Further experiments are necessary to confirm whether the biological effects of ALKBH1 are dependent on 6mA DNA demethylation.These discrepancies suggest the need for further studies to clarify the role of N6-mA and ALKBH1 in glioblastoma.
3.6. Gastric cancer (GC)
GC is a prevalent digestive malignancy worldwide that can be classified into different histological subtypes based on the Lauren classification, including intestinal, diffuse, and mixed [68,78]. Recent studies have shown that the m6A-related gene, ALKBH1, whose gene variants are associated with an increased risk of GC. For example, ALKBH1 rs1048147 is linked to the risk of mixed-type GC, while the GA and GA/GG variants of ALKBH1 rs1076496 increase the risk of GC in people aged ≥55 years but decrease it in those aged <55 years [79]. Moreover, aberrantly high mRNA expression of ALKBH1 is associated with poor OS in the KM and TCGA cohorts of GC patients [80], and ALKBH1mRNA is upregulated in GC tissues [73,81], which increases the numbers of viable and migratory cancer cells [81]. Additionally, miRNA-339–5p is reported to be the upstream regulator of ALKBH1, which binds to the 3′UTR of miRNA-339–5p. Overexpression of miRNA-339–5p leads to a decrease in the protein level of ALKBH1 in BGC-823 and SGC-7901 cells [81]. In contrast, the initial indication from tissue microarray-immunohistochemistry (TMA-IHC) that ALKBH1 protein expression is downregulated in GC tissues, subsequent analysis revealed a notable association between decreased ALKBH1 protein levels and more advanced tumor sizes (≥5 cm) and TNM stages in GC patients [80]. Surprisingly, a recent report showed that higher ALKBH1 protein expression was observed in primary gastric tumors than in matched adjacent non-tumor tissues. GC cells and tumors have lower 6mA levels than normal gastric cells [82]. In vitro and in vivo, ALKBH1 as an oncogene promotes the growth and metastasis of GC cells by acting a DNA 6mA demethylase [82]. Briefly, in a model of chemically induced tumorigenesis, GC tumor growth is suppressed in mice with ALKBH1 knockout. Mechanistically, NRF1 transcriptional activity is targeted by 6mA demethylation caused by ALKBH1 in GC, which results in repressed AMP-activated protein kinase (AMPK) signaling activation and a “Warburg” phenotype in GC cells [82]. In summary, these findings suggest that ALKBH1 may play multiple roles in the occurrence and development of gastric cancer.
3.7. Tongue squamous cell carcinoma (TSCC)
TSCC is a type of oral cancer that is becoming increasingly common, particularly among women and young adults. This aggressive cancer is characterized by its ability to metastasize to lymph nodes and other parts of the body [83]. Recent research has revealed that the genomic 6mA level is significantly increased in TSCC and that DNA 6mA promotes the proliferation and migration of tongue cancer cells. Additionally, the expression of ALKBH1 is increased in TSCC tissues compared to adjacent normal tissues. Knockdown of ALKBH1 results in increased 6mA levels and promotes colony formation and cell migration in TSCC cells. Further studies suggest that the NF-κB pathway may contribute to the enhanced migration of TSCC cells [83].
3.8. Hepatocellular carcinoma (HCC)
HCC is a primary liver cancer and the most common type, accounting for more than 80 % of hepatic cancer cases worldwide [84]. The incidence and mortality of HCC are continuously increasing, particularly in China, Southeast Asia, Mongolia, and Western and Eastern Africa [85]. Accumulating studies have shown that the covalent DNA modification 5-hydroxymethylcytosine (5 hmC) is closely associated with HCC development [86,87]. Recently, research has revealed another type of DNA modification, DNA 6mA, which is significantly increased in HCC tissues with a downregulation of ALKBH1 and upregulation of N6AMT1 [88]. ALKBH1 overexpression has been shown to suppress HCC cell viability, migration, and invasion while enhancing apoptosis. Interestingly, when ALKBH1 was overexpressed, PARTICL was upregulated, and PARTICL DNA 6mA modification was reduced, indicating that inhibiting DNA 6mA increased PARTICL expression. These findings suggest that PARTICL may be the key gene involved in the regulation of DNA 6mA in hepatic cancer cells [88]. Therefore, ALKBH1 may have a vital role in the progression and metastasis of HCC.
3.9. Pancreatic adenocarcinoma (PAAD)
PAAD is a highly lethal digestive system tumor. In fact, it is the seventh leading cause of cancer-related death worldwide, claiming approximately 430,000 lives per year according to the International Agency for Research on Cancer [74]. Emerging evidence suggests a strong association between m1A-modifying genes and the clinical progression of pancreatic cancer. Notably, the upregulation of select m1A-modifying genes positively correlates with copy number variation, as demonstrated by recent studies [89]. Notably, the diminished expression of the ALKBH1 gene has been linked to an unfavorable prognosis among patients diagnosed with PAAD, highlighting its potential clinical significance [89]. Mechanistically, it is believed that ALKBH1 may contribute to the development of pancreatic cancer through the mTOR and ErbB signaling pathways [89]. More recently, a study showed that the level of ALKBH1, a regulator of mtDNA 6mA demethylation modification, decreases gradually and the level of mtDNA 6mA increases during the malignant progression of pancreatic tumorigenesis [90]. Abnormal ALKBH1 expression and mtDNA 6mA mediate mitochondrial dysfunction to induce the occurrence and progression of pancreatic cancer. Moreover, Xiao Chai Hu Tang can increase ALKBH1 and mtDNA 6mA levels and regulate oxidative stress and the expression of mtDNA-encoded genes [90]. More importantly, another report indicated that in pancreatic ductal adenocarcinoma (PDAC), ALKBH1 can be downregulated by the HRD1-SEL1L complex through ubiquitination when a tumor inhibitor, Sirtuin4 (SIRT4), is overexpressed [91]. Knockdown of ALKBH1 blocks the transcription of mitochondrial DNA-coded genes, results in mitochondrial damage and inhibits the cell growth, migration, autophagy, and mitophagy of pancreatic cancer cells [91]. Above all, these results suggest that ALKBH1 may be a promising potent target in the prevention of pancreatic tumorigenesis.
3.10. Ovarian cancer (OV)
OV is a significant contributor to mortality from gynecological malignancies. Unfortunately, the vast majority of diagnoses occur in advanced stages, resulting in 5-year survival rates of only 30–40 % [92]. A recent study revealed that the AlkB family of Fe(II) and α-KG-dependent dioxygenases play important roles in ovarian serous carcinoma development. Notably, the GEPIA database showed that OV patients have a lower expression of ALKBH1. Further analysis using KM plotter indicated that high levels of ALKBH1 transcription are associated with shorter OS time in OV patients. Genetic alteration analysis using cBioPortal revealed that ALKBH1 mutations occurred in 16 % of the samples. The TIMER database demonstrated that the expression of ALKBH1 is positively linked to the infiltration of macrophages, neutrophils, and dendritic cells. Additionally, the global methylation levels of ALKBH1 were lower in OV patients than in healthy individuals according to the DiseaseMeth database. The downregulated methylation values might explain their difference in expression levels to some extent. These results indicate that ALKBH1 expression plays a crucial role in OV patient progression and could be useful in developing better diagnostic and treatment methods to improve their prognosis [93]. For another common malignant tumor in women, cervical cancer, ALKBH1 was found to significantly impact HeLa cancer cell survival, with silencing of ALKBH1 leading to decreased cell survival [75]. Additionally, silencing ALKBH8 led to elevated the protein levels of ALKBH1 in HeLa cells, while silencing ALKBH2 or ALKBH4 resulted in decreased protein levels of ALKBH1(75).
3.11. Triple-negative breast cancer (TNBC)
TNBC is a highly aggressive subtype of breast cancer that accounts for approximately 10–20 % of all breast cancer cases. Unfortunately, current clinical treatments for TNBC have proven to be relatively ineffective. In recent studies, researchers have found that TNBC tissues exhibit significantly lower levels of DNA 6mA than normal tissues [94]. However, in a set of 15 TNBC tissues analyzed, the expression level of the 6mA demethylase ALKBH1 did not show any significant change compared to the control tissues [94].
4. ALKBH1 and human noncancer diseases
4.1. Chronic kidney disease (CKD)
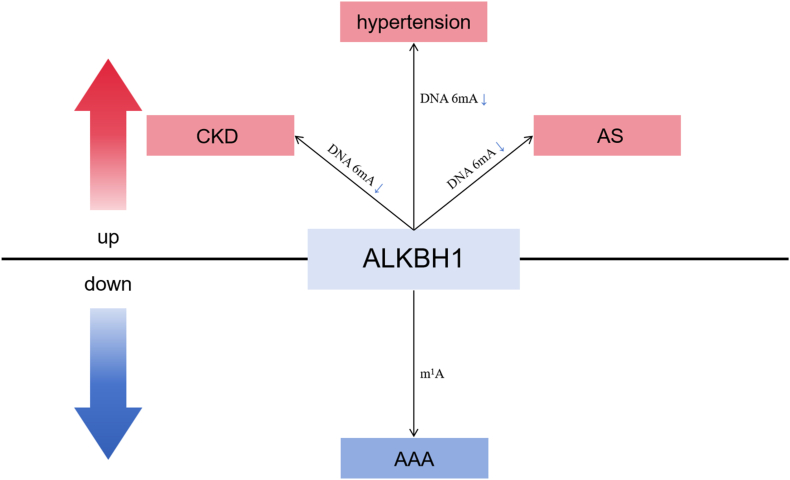
Cardiovascular complications are the leading cause of death in patients with CKD [95]. One prevalent complication is medial vascular calcification (VC), which is linked with hyperphosphatemia and contributes to subsequent cardiovascular morbidity and mortality [96,97]. In CKD, VC progressively increases with advanced stages of the disease. A recent study found that leukocytes and VSMCs exhibit a dynamic decrease in DNA 6mA levels along with calcification progression. The study further shows that upregulation of ALKBH1 mediates this observed 6mA reduction (Fig. 4). In vivo and in vitro experiments demonstrated that ALKBH1 overexpression aggravates VC progression and osteogenic reprogramming, while its depletion suppresses VC progression and osteogenic reprogramming. Mechanistically, ALKBH1-demethylated DNA 6 mA modification facilitates the binding of octamer-binding transcription factor 4 (Oct4) to the bone morphogenetic protein 2 (BMP2) promoter, which activates BMP2 transcription, leading to osteogenic reprogramming of VSMCs and subsequent VC progression (Table 2). The study suggests that targeting ALKBH1 might be a promising therapeutic method for reducing the burden of VC in CKD [98].
Fig. 4.
The molecular mechanisms of ALKBH1 involved in non-cancer diseases. Over-expression of ALKBH1 in diseases is in red; low expression of ALKBH1 in disease is in blue. ALKBH1 exhibits different enzymatic activity in different diseases. Arrows indicate changes in the expression of modifications in patients. Low expression of modification level is in blue.
Table 2.
Roles of ALKBH1 in human non-cancer diseases.
| Diseases type | Expression | Modification types | Modification expression in diseases | Biological functions | Molecular mechanisms | Targets | Reference |
|---|---|---|---|---|---|---|---|
| CKD | up | DNA 6mA | down | Aggravating medial vascular calcification (VC) progression and osteogenic reprogramming | Facilitating the binding of octamer-binding transcription factor 4 (Oct4) to the bone morphogenetic protein 2 (BMP2) promoter and activates BMP2 transcription | BMP2 | 98 |
| Hypertension | up | DNA 6mA | down | Angiotensin II-induced vascular remodeling | Regulating hypoxia-inducible factor 1α (HIF1α) | HIF1α | 100 |
| AS | up | DNA 6mA | down | Promoting the oncogenic role of the hypoxia-response gene myocardial infarction-associated transcript (MIAT) | Facilitating HIF1α binding and activation of MIAT | HIF1α | 107 |
| AAA | down | m1A | No study | No study | No study | No study | 111 |
4.2. Hypertension
Approximately a quarter of the adult population worldwide suffers from essential hypertension, which is the primary identifiable risk factor associated with death from severe complications [99,100]. The 6mA DNA level in leukocytes is low in patients with hypertension, but effective hypertension treatment has been shown to restore the 6mA DNA level in leukocytes to normal levels [100]. Decreased leukocyte 6mA DNA levels were found to be associated with age, systolic blood pressure, serum total cholesterol, and high-density lipoprotein levels [100]. Furthermore, hypertension mouse and rat models showed that elevated levels of ALKBH1, a demethylase of 6 mA, mediated the dynamic change in 6mA levels in leukocytes and vascular smooth muscle cells [100] (Fig. 4). ALKBH1-6mA directly and negatively regulates hypoxia-inducible factor 1α (HIF1α), which is involved in angiotensin II-induced vascular remodeling. By modulating HIF1α, knockdown of ALKBH1 impeded the angiotensin II-induced transformation, proliferation, and migration of vascular smooth muscle cells, as evidenced by the results of a recent study [100] (Table 2). These findings provide evidence that ALKBH1 plays a critical role in modulating the interaction between 6mA level alteration and HIF1α function in vascular remodeling, highlighting its potential as a novel therapeutic target.
4.3. Atherosclerosis (AS)
Cardiovascular diseases (CVDs) are responsible for 31 % of deaths worldwide, making them the leading cause of human mortality [101]. Among CVDs, AS is the major cause of morbidity and mortality worldwide [102,103]. Emerging evidence has suggested that leukocyte N6-methyladenine DNA levels are significantly reduced in AS patients and gradually decrease as plaque progresses. The decrease in DNA 6mA levels is closely associated with the upregulation of ALKBH1, the demethylase of DNA 6mA. ALKBH1 has been shown to promote the oncogenic role of the hypoxia-response gene myocardial infarction-associated transcript (MIAT) in glioblastoma [[104], [105], [106]] (Fig. 4). Additionally, oxidized low-density lipoprotein-induced ALKBH1 and m6A DNA demethylation facilitated HIF1α binding and activation of MIAT [107] (Table 2). These findings suggest that the ALKBH1-m6A regulatory axis may play a role in controlling plaque progression via MIAT, highlighting a potential therapeutic target for AS [107].
4.4. Abdominal aortic aneurysm (AAA)
AAA is a common vascular disease characterized by pathological dilatation and weakening of the abdominal aortic wall, with a high mortality rate of up to 81 % for men if ruptured [108,109]. Unfortunately, there are currently limited medical therapies available to mitigate aneurysm growth and prevent rupture [110]. However, recent research on the roles of epigenetic regulation, particularly RNA methylation, in AAA pathogenesis has revealed new therapeutic possibilities. One report has demonstrated a significant association between m1A regulation and human AAA pathogenesis and showed that ALKBH1 is downregulated in AAA [111] (Fig. 4) (Table 2). These findings suggest that targeting RNA methylation may be a promising approach for developing new therapies for AAA.
5. Inhibitors to ALKBH1
Based on the structure, the mechanism of inhibitor can be divided into three types [112]: binding to iron in the 2-OG biding site, such as binding to the substrate-binding site, and binding with an undetermined mode. Most early AlkB demethylase inhibitors, which containing a metal-binding site, are 2OG-competitive inhibitors, such as N-oxalylglycine (NOG), the cocrystal structure of which binding to ALKBH1 has been identified [27]. Inhibitors which bind to the substrate binding site only, however, do not possess any specific structural features like metal-chelation moiety. They only have a diversity in chemical structure, which makes the binding modes identified easily. The last one is mainly made up of natural products which have a broad range of biological targets and pharmacological activities. While this group of compounds may seem inspiring and interesting in the design of small molecular inhibitors, the identification process is also complicated, and the function of inhibitors is also unpredictable.
With the deeper exploration, many small molecular inhibitors targeting at AlkB family have been discovered. It is reported that ALKBH1 inhibitors have a number of highly conserved interactions with the 2OG-binding site and substrate-binding site (Table 3). However, in spite of the specific inhibitors of FTO, ALKBH3 and ALKBH5, high-selective inhibitor of ALKBH1 has not been identified [113]. And for some small molecular inhibitors,the selectivity is not high, and drug is still under development [30]. In addition to the small-molecule inhibitors, miRNA can also be a promising way to inhibit ALKBH1. It is reported that miRNA-339–5p suppresses the malignant development of gastric cancer by targeting ALKBH1, and other pathways are still to be identified [81].
Table 3.
Inhibitor of ALKBH1.
| Interactions between the inhibitors and ALKBH1 [112] | |||
|---|---|---|---|
| Binding Site | Mechanism | Inhibitors | Residues of ALKBH1 |
| 2-OG binding site | salt bridge interaction with –COOH | LipotF, citrate,2,4-PDCA,NOG,IOX1,IOX3 | Arg338 |
| hydrogen bonding with C O/-COOH/-OH | 2,4-PDCA,NOG,rhein, fluorescein | Arg 334 | |
| hydrogen bonding with C O/-COOH/-OH | citrate,NOG,entacapone | Asn220 | |
| hydrogen bonding with –COOH | LipotF,2,4-PDCA,NOG,IOX1,IOX3 | Asn340 | |
| hydrogen bonding with –COOH | LipotF,2,4-PDCA,NOG,IOX3 | Met 376 | |
| substrate-binding site | water-mediated bond | FB23 | Arg 234 |
| Hydrogen bonding with pyridyl nitrogen/-OH/nitrogen or oxygen atom of heterocyclic ring | LipotF, CHTB | ||
| π-π interactions with -C6H5 | rhein, CHTB,LipotF | His 231 | |
| hydrogen bonding with –OH | CHTB | Val 232 | |
| π-π interactions with -C6H5 | LipotF | Tyr177 | |
| hydrogen bonding with –OH | entacapone | Tyr184 | |
| hydrogen bonding with –OH | CDPCB | Phe 27 | |
| Van der Waals contact with -Cl | CDPCB | Leu 228 | |
| hydrophobic interaction with --C6H5 | LipotF | ||
6. Conclusion
Taken together, these findings highlight that ALKBH1 has unique catalytic substrates and functions that have been extensively studied. However, there is a need for further research to understand the relationship between the substrate diversity and specificity of ALKBH1 and its role in human diseases. Such research will provide crucial physiological insights and essential evidence for the screening and optimization of specific inhibitors for ALKBH1-related diseases. By understanding the underlying mechanisms of ALKBH1 in various diseases, we can develop targeted and effective therapies to combat these diseases. Therefore, future research in this area is of the utmost importance.
Financial disclosure
The authors have no funding to disclose.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
CRediT authorship contribution statement
Jing Zhong: Writing – original draft, Resources, Data curation. Zhengyang Xu: Writing – original draft, Software, Resources, Data curation. Ning Ding: Writing – review & editing, Resources, Data curation. Yanting Wang: Writing – review & editing, Validation, Resources. Wenwen Chen: Writing – review & editing, Writing – original draft, Supervision, Resources, Funding acquisition, Data curation.
Declaration of competing interest
The authors declare no conflict of interests.
Acknowledgements
The study was supported by the Natural Science Foundation of Zhejiang Province (LQ22C070003) and the National Natural Science Foundation of China (82303385).
Contributor Information
Jing Zhong, Email: 2512022@zju.edu.cn.
Zhengyang Xu, Email: 22218372@zju.edu.cn.
Ning Ding, Email: dingning_zju@zju.edu.cn.
Yanting Wang, Email: yantingw@zju.edu.cn.
Wenwen Chen, Email: 2520125@zju.edu.cn.
References
- 1.Dinglay S., Trewick S.C., Lindahl T., Sedgwick B. Defective processing of methylated single-stranded DNA by E. coli AlkB mutants. Gene Dev. 2000;14(16):2097–2105. [PMC free article] [PubMed] [Google Scholar]
- 2.Kataoka H., Yamamoto Y., Sekiguchi M. A new gene (Alkb) of Escherichia-coli that controls Sensitivity to methyl methane sulfonate. J. Bacteriol. 1983;153(3):1301–1307. doi: 10.1128/jb.153.3.1301-1307.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurowski M.A., Bhagwat A.S., Papaj G., Bujnicki J.M. Phylogenomic identification of five new human homologs of the DNA repair enzyme AlkB. BMC Genom. 2003;4(1):48. doi: 10.1186/1471-2164-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerken T., Girard C.A., Tung Y.C., et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318(5855):1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jia G., Fu Y., Zhao X., et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011;7(12):885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei J., Liu F., Lu Z., et al. Differential m(6)A, m(6)Am, and m(1)A demethylation mediated by FTO in the cell nucleus and cytoplasm. Molecular cell. 2018;71(6):973–985 e5. doi: 10.1016/j.molcel.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu Y., Jia G., Pang X., et al. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat. Commun. 2013;4:1798. doi: 10.1038/ncomms2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jia G., Yang C.G., Yang S., et al. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 2008;582(23–24):3313–3319. doi: 10.1016/j.febslet.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aas P.A., Otterlei M., Falnes P.O., et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421(6925):859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 10.Zhang C., Jia G. Reversible RNA modification N(1)-methyladenosine (m(1)A) in mRNA and tRNA. Dev. Reprod. Biol. 2018;16(3):155–161. doi: 10.1016/j.gpb.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X., Xiong X., Wang K., et al. Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat. Chem. Biol. 2016;12(5):311–316. doi: 10.1038/nchembio.2040. [DOI] [PubMed] [Google Scholar]
- 12.Ueda Y., Ooshio I., Fusamae Y., et al. AlkB homolog 3-mediated tRNA demethylation promotes protein synthesis in cancer cells. Sci. Rep. 2017;7 doi: 10.1038/srep42271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma L., Lu H., Tian Z., et al. Structural insights into the interactions and epigenetic functions of human nucleic acid repair protein ALKBH6. J. Biol. Chem. 2022;298(3) doi: 10.1016/j.jbc.2022.101671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li M.M., Nilsen A., Shi Y., et al. ALKBH4-dependent demethylation of actin regulates actomyosin dynamics. Nat. Commun. 2013;4:1832. doi: 10.1038/ncomms2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kweon S.M., Chen Y., Moon E., Kvederaviciute K., Klimasauskas S., Feldman D.E. An adversarial DNA N(6)-methyladenine-sensor network preserves polycomb silencing. Molecular cell. 2019;74(6):1138–11347 e6. doi: 10.1016/j.molcel.2019.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu K., Qi T.F., Miao W., Liu X., Wang Y. Quantitative proteomics revealed new functions of ALKBH4. Proteomics. 2022;22(7) doi: 10.1002/pmic.202100231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi Y., Yang H., Ding J. Caenorhabditis elegans NMAD-1 functions as a demethylase for actin. J. Mol. Cell Biol. 2023 Jun 1;15(1) doi: 10.1093/jmcb/mjad008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shang G., Yang M., Li M., Ma L., Liu Y., Ma J., Chen Y., Wang X., Fan S., Xie M., Wu W., Dai S., Chen Z. Structural basis of nucleic acid recognition and 6mA demethylation by Caenorhabditis elegans NMAD-1A. Int. J. Mol. Sci. 2024 Jan 5;25(2):686. doi: 10.3390/ijms25020686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng G., Dahl J.A., Niu Y., et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Molecular cell. 2013;49(1):18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L.S., Xiong Q.P., Pena Perez S., et al. ALKBH7-mediated demethylation regulates mitochondrial polycistronic RNA processing. Nat. Cell Biol. 2021;23(7):684–691. doi: 10.1038/s41556-021-00709-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evke S., Lin Q., Melendez J.A., Begley T.J. Epitranscriptomic reprogramming is required to prevent stress and damage from acetaminophen. Genes. 2022;13(3) doi: 10.3390/genes13030421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Songe-Moller L., van den Born E., Leihne V., et al. Mammalian ALKBH8 possesses tRNA methyltransferase activity required for the biogenesis of multiple wobble uridine modifications implicated in translational decoding. Mol. Cell Biol. 2010;30(7):1814–1827. doi: 10.1128/MCB.01602-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H., Zhang Y., Guo Y., et al. ALKBH1 promotes lung cancer by regulating m6A RNA demethylation. Biochem. Pharmacol. 2021;189 doi: 10.1016/j.bcp.2020.114284. [DOI] [PubMed] [Google Scholar]
- 24.Liu F., Clark W., Luo G., et al. ALKBH1-Mediated tRNA demethylation regulates translation. Cell. 2016;167(3):816–828 e16. doi: 10.1016/j.cell.2016.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma C.J., Ding J.H., Ye T.T., Yuan B.F., Feng Y.Q. AlkB homologue 1 demethylates N(3)-methylcytidine in mRNA of mammals. ACS Chem. Biol. 2019;14(7):1418–1425. doi: 10.1021/acschembio.8b01001. [DOI] [PubMed] [Google Scholar]
- 26.Kawarada L., Suzuki T., Ohira T., Hirata S., Miyauchi K., Suzuki T. ALKBH1 is an RNA dioxygenase responsible for cytoplasmic and mitochondrial tRNA modifications. Nucleic Acids Res. 2017;45(12):7401–7415. doi: 10.1093/nar/gkx354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang M., Yang S., Nelakanti R., et al. Mammalian ALKBH1 serves as an N(6)-mA demethylase of unpairing DNA. Cell Res. 2020;30(3):197–210. doi: 10.1038/s41422-019-0237-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ougland R., Lando D., Jonson I., et al. ALKBH1 is a histone H2A dioxygenase involved in neural differentiation. Stem Cell. 2012;30(12):2672–2682. doi: 10.1002/stem.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian L.F., Liu Y.P., Chen L., et al. Structural basis of nucleic acid recognition and 6mA demethylation by human ALKBH1. Cell Res. 2020;30(3):272–275. doi: 10.1038/s41422-019-0233-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao M.Z., Liu J.M., Xian C.L., Chen K.Y., Liu Z.Q., Cheng Y.Y. Therapeutic potential of ALKB homologs for cardiovascular disease. Biomed. Pharmacother. 2020;131 doi: 10.1016/j.biopha.2020.110645. [DOI] [PubMed] [Google Scholar]
- 31.Westbye M.P., Feyzi E., Aas P.A., et al. Human AlkB homolog 1 is a mitochondrial protein that demethylates 3-methylcytosine in DNA and RNA. J. Biol. Chem. 2008;283(36):25046–25056. doi: 10.1074/jbc.M803776200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan Z., Sikandar S., Witherspoon M., et al. Impaired placental trophoblast lineage differentiation in Alkbh1(-/-) mice. Dev. Dynam. : an official publication of the American Association of Anatomists. 2008;237(2):316–327. doi: 10.1002/dvdy.21418. [DOI] [PubMed] [Google Scholar]
- 33.Tsujikawa K., Koike K., Kitae K., et al. Expression and sub-cellular localization of human ABH family molecules. J. Cell Mol. Med. 2007;11(5):1105–1116. doi: 10.1111/j.1582-4934.2007.00094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ougland R., Jonson I., Moen M.N., et al. Role of ALKBH1 in the core transcriptional network of embryonic stem cells. Cell. Physiol. Biochem. : international journal of experimental cellular physiology, biochemistry, and pharmacology. 2016;38(1):173–184. doi: 10.1159/000438619. [DOI] [PubMed] [Google Scholar]
- 35.Ougland R., Rognes T., Klungland A., Larsen E. Non-homologous functions of the AlkB homologs. J. Mol. Cell Biol. 2015;7(6):494–504. doi: 10.1093/jmcb/mjv029. [DOI] [PubMed] [Google Scholar]
- 36.Wu T.P., Wang T., Seetin M.G., et al. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature. 2016;532(7599):329–333. doi: 10.1038/nature17640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rashad S., Han X., Sato K., et al. The stress specific impact of ALKBH1 on tRNA cleavage and tiRNA generation. RNA Biol. 2020;17(8):1092–1103. doi: 10.1080/15476286.2020.1779492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dominissini D., Nachtergaele S., Moshitch-Moshkovitz S., et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530(7591):441–446. doi: 10.1038/nature16998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X., Xiong X., Zhang M., et al. Base-Resolution mapping reveals distinct m(1)A methylome in nuclear- and mitochondrial-encoded transcripts. Molecular cell. 2017;68(5):993–1005 e9. doi: 10.1016/j.molcel.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen W., Wang H., Mi S., Shao L., Xu Z., Xue M. ALKBH1-mediated m1A demethylation of METTL3 mRNA promotes the metastasis of colorectal cancer by downregulating SMAD7 expression. Mol. Oncol. 2023;17(2):344–364. doi: 10.1002/1878-0261.13366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xue M., Mi S., Zhang Z., Wang H., Chen W., Wei W., Lou G. MFAP2, upregulated by m1A methylation, promotes colorectal cancer invasiveness via CLK3. Cancer Med. 2023 Apr;12(7):8403–8414. doi: 10.1002/cam4.5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muller T.A., Struble S.L., Meek K., Hausinger R.P. Characterization of human AlkB homolog 1 produced in mammalian cells and demonstration of mitochondrial dysfunction in ALKBH1-deficient cells. Biochem. Biophys. Res. Commun. 2018;495(1):98–103. doi: 10.1016/j.bbrc.2017.10.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haag S., Sloan K.E., Ranjan N., et al. NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. EMBO J. 2016;35(19):2104–2119. doi: 10.15252/embj.201694885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wagner A., Hofmeister O., Rolland S.G., et al. Mitochondrial Alkbh1 localizes to mtRNA granules and its knockdown induces the mitochondrial UPR in humans and C. elegans. J. Cell Sci. 2019;132(19) doi: 10.1242/jcs.223891. [DOI] [PubMed] [Google Scholar]
- 45.Kawarada L., Fukaya M., Saito R., Kassai H., Sakagami H., Aiba A. Telencephalon-specific Alkbh1 conditional knockout mice display hippocampal atrophy and impaired learning. FEBS Lett. 2021;595(12):1671–1680. doi: 10.1002/1873-3468.14098. [DOI] [PubMed] [Google Scholar]
- 46.Vanyushin B.F., Belozersky A.N., Kokurina N.A., Kadirova D.X. 5-methylcytosine and 6-methylamino-purine in bacterial DNA. Nature. 1968;218(5146):1066–1067. doi: 10.1038/2181066a0. [DOI] [PubMed] [Google Scholar]
- 47.Cummings D.J., Tait A., Goddard J.M. Methylated bases in DNA from Paramecium aurelia. Biochim. Biophys. Acta. 1974;374(1):1–11. doi: 10.1016/0005-2787(74)90194-4. [DOI] [PubMed] [Google Scholar]
- 48.Hattman S., Kenny C., Berger L., Pratt K. Comparative study of DNA methylation in three unicellular eucaryotes. J. Bacteriol. 1978;135(3):1156–1157. doi: 10.1128/jb.135.3.1156-1157.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pratt K., Hattman S. Deoxyribonucleic acid methylation and chromatin organization in Tetrahymena thermophila. Mol. Cell Biol. 1981;1(7):600–608. doi: 10.1128/mcb.1.7.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gorovsky M.A., Hattman S., Pleger G.L. (6 N)methyl adenine in the nuclear DNA of a eucaryote, Tetrahymena pyriformis. J. Cell Biol. 1973;56(3):697–701. doi: 10.1083/jcb.56.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lyu C., Niu Y., Lai W., et al. Rare and misincorporated DNA N(6)-methyladenine is a hallmark of cytotoxic stresses for selectively stimulating the stemness and proliferation of glioblastoma cells. Cell discovery. 2022;8(1):39. doi: 10.1038/s41421-022-00399-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Greer E.L., Blanco M.A., Gu L., et al. DNA methylation on N6-adenine in C. elegans. Cell. 2015;161(4):868–878. doi: 10.1016/j.cell.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang G., Huang H., Liu D., et al. N6-methyladenine DNA modification in Drosophila. Cell. 2015;161(4):893–906. doi: 10.1016/j.cell.2015.04.018. [DOI] [PubMed] [Google Scholar]
- 54.Xiao C.L., Zhu S., He M., et al. N(6)-Methyladenine DNA modification in the human genome. Molecular cell. 2018;71(2):306–318 e7. doi: 10.1016/j.molcel.2018.06.015. [DOI] [PubMed] [Google Scholar]
- 55.Xie Q., Wu T.P., Gimple R.C., Li Z., Prager B.C., Wu Q., Yu Y., Wang P., Wang Y., Gorkin D.U., Zhang C., Dowiak A.V., Lin K., Zeng C., Sui Y., Kim L.J.Y., Miller T.E., Jiang L., Lee-Poturalski C., Huang Z., Fang X., Zhai K., Mack S.C., Sander M., Bao S., Kerstetter-Fogle A.E., Sloan A.E., Xiao A.Z., Rich J.N. N6-methyladenine DNA modification in glioblastoma. Cell. 2018 Nov 15;175(5):1228–1243.e20. doi: 10.1016/j.cell.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu P., Nie S., Li B., Yang Z.Q., Xu Z.M., Fei J., Lin C., Zeng R., Xu G.L. Deficiency in a glutamine-specific methyltransferase for release factor causes mouse embryonic lethality. Mol. Cell Biol. 2010 Sep;30(17):4245–4253. doi: 10.1128/MCB.00218-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hao Z., Wu T., Cui X., Zhu P., Tan C., Dou X., Hsu K.W., Lin Y.T., Peng P.H., Zhang L.S., Gao Y., Hu L., Sun H.L., Zhu A., Liu J., Wu K.J., He C. N6-Deoxyadenosine methylation in mammalian mitochondrial DNA. Mol Cell. 2020 May 7;78(3):382–395.e8. doi: 10.1016/j.molcel.2020.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen H., Gu L., Orellana E.A., Wang Y., Guo J., Liu Q., Wang L., Shen Z., Wu H., Gregory R.I., Xing Y., Shi Y. METTL4 is an snRNA m6Am methyltransferase that regulates RNA splicing. Cell Res. 2020 Jun;30(6):544–547. doi: 10.1038/s41422-019-0270-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xie Q., Wu T.P., Gimple R.C., et al. N(6)-methyladenine DNA modification in glioblastoma. Cell. 2018;175(5):1228–1243 e20. doi: 10.1016/j.cell.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Q., Qian C., Feng H., et al. N6-methyladenine DNA demethylase ALKBH1 regulates mammalian axon regeneration. Neurosci. Bull. 2021;37(6):809–814. doi: 10.1007/s12264-021-00671-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou C., Liu Y., Li X., Zou J., Zou S. DNA N(6)-methyladenine demethylase ALKBH1 enhances osteogenic differentiation of human MSCs. Bone research. 2016;4 doi: 10.1038/boneres.2016.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu Y., Chen Y., Wang Y., et al. DNA demethylase ALKBH1 promotes adipogenic differentiation via regulation of HIF-1 signaling. J. Biol. Chem. 2022;298(1) doi: 10.1016/j.jbc.2021.101499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Diao L.T., Xie S.J., Yu P.J., et al. N(6)-methyladenine demethylase ALKBH1 inhibits the differentiation of skeletal muscle. Exp. Cell Res. 2021;400(2) doi: 10.1016/j.yexcr.2021.112492. [DOI] [PubMed] [Google Scholar]
- 64.Koh C.W.Q., Goh Y.T., Toh J.D.W., Neo S.P., Ng S.B., Gunaratne J., Gao Y.G., Quake S.R., Burkholder W.F., Goh W.S.S. Single-nucleotide-resolution sequencing of human N6-methyldeoxyadenosine reveals strand-asymmetric clusters associated with SSBP1 on the mitochondrial genome. Nucleic Acids Res. 2018 Dec 14;46(22):11659–11670. doi: 10.1093/nar/gky1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Douvlataniotis K., Bensberg M., Lentini A., Gylemo B., Nestor C.E. No evidence for DNA N (6)-methyladenine in mammals. Sci. Adv. 2020;6(12) doi: 10.1126/sciadv.aay3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Musheev M.U., Baumgärtner A., Krebs L., Niehrs C. The origin of genomic N6-methyl-deoxyadenosine in mammalian cells. Nat. Chem. Biol. 2020 Jun;16(6):630–634. doi: 10.1038/s41589-020-0504-2. [DOI] [PubMed] [Google Scholar]
- 67.Liu X., Lai W., Li Y., et al. N6-methyladenine is incorporated into mammalian genome by DNA polymerase. Cell Res. 2021;31:94–97. doi: 10.1038/s41422-020-0317-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nordstrand L.M., Svard J., Larsen E., et al. Mice lacking Alkbh1 display sex-ratio distortion and unilateral eye defects. PLoS One. 2010;5(11) doi: 10.1371/journal.pone.0013827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Muller T.A., Yu K., Hausinger R.P., Meek K. ALKBH1 is dispensable for abasic site cleavage during base excision repair and class switch recombination. PLoS One. 2013;8(6) doi: 10.1371/journal.pone.0067403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2022. CA A Cancer J. Clin. 2022;72(1):7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 71.Chen W., Zheng R., Zeng H., Zhang S., He J. Annual report on status of cancer in China, 2011. Chin. J. Cancer Res. 2015;27(1):2–12. doi: 10.3978/j.issn.1000-9604.2015.01.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang L., Feng X., Jiao Z., Gan J., Meng Q. Characterization of the prognostic and diagnostic values of ALKBH family members in non-small cell lung cancer. Pathol. Res. Pract. 2022;231 doi: 10.1016/j.prp.2022.153809. [DOI] [PubMed] [Google Scholar]
- 73.Li L., Zhu C., Xu Q., et al. ALKBH1 contributes to renal cell carcinoma progression by reducing N6-methyladenine of GPR137. Eur. J. Clin. Invest. 2023 doi: 10.1111/eci.13986. Advance online publication. [DOI] [PubMed] [Google Scholar]
- 74.Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 75.Pilzys T., Marcinkowski M., Kukwa W., et al. ALKBH overexpression in head and neck cancer: potential target for novel anticancer therapy. Sci. Rep. 2019;9(1) doi: 10.1038/s41598-019-49550-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guo C., Liu Z., Zhang H. DNA 6mA demethylase ALKBH1 regulates DDX18 expression to promote proliferation of human head and neck squamous cell carcinoma. Cell. Oncol. 2023;46(4):1097–1111. doi: 10.1007/s13402-023-00800-1. [DOI] [PubMed] [Google Scholar]
- 77.Feng X., He C. Mammalian DNA N6-methyladenosine: challenges and new insights. Mol Cell. 2023 Feb 2;83(3):343–351. doi: 10.1016/j.molcel.2023.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bijlsma M.F., Sadanandam A., Tan P., Vermeulen L. Molecular subtypes in cancers of the gastrointestinal tract. Nat. Rev. Gastroenterol. Hepatol. 2017;14(6):333–342. doi: 10.1038/nrgastro.2017.33. [DOI] [PubMed] [Google Scholar]
- 79.Li Y., Zhou D., Liu Q., Zhu W., Ye Z., He C. Gene polymorphisms of m6A erasers FTO and ALKBH1 associated with susceptibility to gastric cancer. Pharmacogenomics Personalized Med. 2022;15:547–559. doi: 10.2147/PGPM.S360912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li Y., Zheng D., Wang F., Xu Y., Yu H., Zhang H. Expression of demethylase genes, FTO and ALKBH1, is associated with prognosis of gastric cancer. Dig. Dis. Sci. 2019;64(6):1503–1513. doi: 10.1007/s10620-018-5452-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang C., Huang Y., Zhang J., Fang Y. MiRNA-339-5p suppresses the malignant development of gastric cancer via targeting ALKBH1. Exp. Mol. Pathol. 2020;115 doi: 10.1016/j.yexmp.2020.104449. [DOI] [PubMed] [Google Scholar]
- 82.Wang X., Wong C.C., Chen H., Fu K., Shi L., Su H., Guo S., Gou H., Hu X., Zhang L., Ji J., Yu J. The N6-Methyladenine DNA Demethylase ALKBH1 Promotes Gastric Carcinogenesis by Disrupting NRF1 Binding Capacity. 2023. [DOI] [PubMed] [Google Scholar]
- 83.Xi L., Yang Y., Xu Y., et al. The enhanced genomic 6 mA metabolism contributes to the proliferation and migration of TSCC cells. Int. J. Oral Sci. 2022;14(1):11. doi: 10.1038/s41368-022-00161-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sia D., Villanueva A., Friedman S.L., Llovet J.M. Liver cancer cell of origin, molecular class, and effects on patient prognosis. Gastroenterology. 2017;152(4):745–761. doi: 10.1053/j.gastro.2016.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ghouri Y.A., Mian I., Rowe J.H. Review of hepatocellular carcinoma: epidemiology, etiology, and carcinogenesis. J. Carcinog. 2017;16:1. doi: 10.4103/jcar.JCar_9_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu C., Liu L., Chen X., et al. Decrease of 5-hydroxymethylcytosine is associated with progression of hepatocellular carcinoma through downregulation of TET1. PLoS One. 2013;8(5) doi: 10.1371/journal.pone.0062828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thomson J.P., Ottaviano R., Unterberger E.B., et al. Loss of tet1-associated 5-hydroxymethylcytosine is concomitant with aberrant promoter hypermethylation in liver cancer. Cancer Res. 2016;76(10):3097–3108. doi: 10.1158/0008-5472.CAN-15-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lin Q., Chen J.W., Yin H., et al. DNA N6-methyladenine involvement and regulation of hepatocellular carcinoma development. Genomics. 2022;114(2) doi: 10.1016/j.ygeno.2022.01.002. [DOI] [PubMed] [Google Scholar]
- 89.Zheng Q., Yu X., Zhang Q., He Y., Guo W. Genetic characteristics and prognostic implications of m1A regulators in pancreatic cancer. Biosci. Rep. 2021;41(4) doi: 10.1042/BSR20210337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cai J., Shen W., Zhang G., Li X., Shen H., Li W., Tan C., Zhang T., Shi M., Yang Z., Li Y., Liu H., Zhao X. Xiao Chai Hu Tang alleviates the pancreatic tumorigenesis via improving the mtDNA N6-Methyladenine modification mediated mitochondrial dysfunction in Syrian hamster model. Phytomedicine. 2023 Jul 25;116 doi: 10.1016/j.phymed.2023.154840. [DOI] [PubMed] [Google Scholar]
- 91.Ping D., Pu X., Ding G., et al. Sirtuin4 impacts mitochondrial homeostasis in pancreatic cancer cells by reducing the stability of AlkB homolog 1 via deacetylation of the HRD1-SEL1L complex. Biochimica et biophysica acta. Gene regulatory mechanisms. 2023;1866(2) doi: 10.1016/j.bbagrm.2023.194941. Advance online publication. [DOI] [PubMed] [Google Scholar]
- 92.Nimmagadda S., Penet M.F. Ovarian cancer targeted theranostics. Front. Oncol. 2019;9:1537. doi: 10.3389/fonc.2019.01537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cai Y., Wu G., Peng B., et al. Expression and molecular profiles of the AlkB family in ovarian serous carcinoma. Aging. 2021;13(7):9679–9692. doi: 10.18632/aging.202716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sheng X., Wang J., Guo Y., Zhang J., Luo J. DNA N6-methyladenine (6mA) modification regulates drug resistance in triple negative breast cancer. Front. Oncol. 2020;10 doi: 10.3389/fonc.2020.616098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Blaha M.J., Budoff M.J., DeFilippis A.P., et al. Associations between C-reactive protein, coronary artery calcium, and cardiovascular events: implications for the JUPITER population from MESA, a population-based cohort study. Lancet. 2011;378(9792):684–692. doi: 10.1016/S0140-6736(11)60784-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Timmis A., Townsend N., Gale C., et al. European society of cardiology: cardiovascular disease statistics 2017. Eur. Heart J. 2018;39(7):508–579. doi: 10.1093/eurheartj/ehx628. [DOI] [PubMed] [Google Scholar]
- 97.Moe S.M., Chen N.X. Mechanisms of vascular calcification in chronic kidney disease. J. Am. Soc. Nephrol. : JASN (J. Am. Soc. Nephrol.) 2008;19(2):213–216. doi: 10.1681/ASN.2007080854. [DOI] [PubMed] [Google Scholar]
- 98.Ouyang L., Su X., Li W., et al. ALKBH1-demethylated DNA N6-methyladenine modification triggers vascular calcification via osteogenic reprogramming in chronic kidney disease. J. Clin. Invest. 2021;131(14) doi: 10.1172/JCI146985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mokdad A.H., Forouzanfar M.H., Daoud F., et al. Global burden of diseases, injuries, and risk factors for young people's health during 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2016;387(10036):2383–2401. doi: 10.1016/S0140-6736(16)00648-6. [DOI] [PubMed] [Google Scholar]
- 100.Guo Y., Pei Y., Li K., Cui W., Zhang D. DNA N(6)-methyladenine modification in hypertension. Aging. 2020;12(7):6276–6291. doi: 10.18632/aging.103023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li H.Z., Boulanger P. A survey of heart anomaly detection using ambulatory electrocardiogram (ECG) Sensors. 2020;20(5) doi: 10.3390/s20051461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tsao C.W., Aday A.W., Almarzooq Z.I., et al. Heart disease and stroke statistics-2022 update: a report from the American heart association. Circulation. 2022;145(8):e153–e639. doi: 10.1161/CIR.0000000000001052. [DOI] [PubMed] [Google Scholar]
- 103.Timmis A., Vardas P., Townsend N., et al. European Society of Cardiology: cardiovascular disease statistics 2021. Eur. Heart J. 2022;43(8):716–799. doi: 10.1093/eurheartj/ehab892. [DOI] [PubMed] [Google Scholar]
- 104.Galardi S., Savino M., Scagnoli F., et al. Resetting cancer stem cell regulatory nodes upon MYC inhibition. EMBO Rep. 2016;17(12):1872–1889. doi: 10.15252/embr.201541489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu Z., Wang H., Cai H., et al. Long non-coding RNA MIAT promotes growth and metastasis of colorectal cancer cells through regulation of miR-132/Derlin-1 pathway. Cancer Cell Int. 2018;18:59. doi: 10.1186/s12935-017-0477-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dong P., Xiong Y., Yue J., et al. Long noncoding RNA NEAT1 drives aggressive endometrial cancer progression via miR-361-regulated networks involving STAT3 and tumor microenvironment-related genes. J. Exp. Clin. Cancer Res. : CR. 2019;38(1):295. doi: 10.1186/s13046-019-1306-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wu L., Pei Y., Zhu Y., et al. Association of N(6)-methyladenine DNA with plaque progression in atherosclerosis via myocardial infarction-associated transcripts. Cell Death Dis. 2019;10(12):909. doi: 10.1038/s41419-019-2152-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Thompson R.W., Geraghty P.J., Lee J.K. Abdominal aortic aneurysms: basic mechanisms and clinical implications. Curr. Probl. Surg. 2002;39(2):110–230. doi: 10.1067/msg.2002.121421. [DOI] [PubMed] [Google Scholar]
- 109.Bains P., Oliffe J.L., Mackay M.H., Kelly M.T. Screening older adult men for abdominal aortic aneurysm: a scoping review. Am. J. Men's Health. 2021;15(2) doi: 10.1177/15579883211001204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Quaye K.B., Pack N., Wilson-Byrne T., Long C.A. Contemporary management of abdominal aortic aneurysms. Curr. Cardiol. Rep. 2022;24(4):431–438. doi: 10.1007/s11886-022-01662-z. [DOI] [PubMed] [Google Scholar]
- 111.Wu Y., Jiang D., Zhang H., et al. N1-Methyladenosine (m1A) regulation associated with the pathogenesis of abdominal aortic aneurysm through YTHDF3 modulating macrophage polarization. Frontiers in cardiovascular medicine. 2022;9 doi: 10.3389/fcvm.2022.883155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Perry G.S., Das M., Woon E.C.Y. Inhibition of AlkB nucleic acid demethylases: promising new epigenetic targets. J. Med. Chem. 2021 Dec 9;64(23):16974–17003. doi: 10.1021/acs.jmedchem.1c01694. [DOI] [PubMed] [Google Scholar]
- 113.Zhang Y., Wang C. Demethyltransferase AlkBH1 substrate diversity and relationship to human diseases. Mol. Biol. Rep. 2021 May;48(5):4747–4756. doi: 10.1007/s11033-021-06421-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.