Summary
Alzheimer’s disease (AD) is a complex pathophysiological disease. Allowing for heterogeneity, not only in disease manifestations but also in different progression patterns, is critical for developing effective disease models that can be used in clinical and research settings. We introduce a machine learning model for identifying underlying patterns in Alzheimer’s disease (AD) trajectory using longitudinal multi-modal data from the ADNI cohort and the AIBL cohort. Ten biologically and clinically meaningful disease-related states were identified from data, which constitute three non-overlapping stages (i.e., neocortical atrophy [NCA], medial temporal atrophy [MTA], and whole brain atrophy [WBA]) and two distinct disease progression patterns (i.e., NCA WBA and MTA WBA). The index of disease-related states provided a remarkable performance in predicting the time to conversion to AD dementia (C-Index: 0.923 ± 0.007). Our model shows potential for promoting the understanding of heterogeneous disease progression and early predicting the conversion time to AD dementia.
Subject areas: neuroscience, machine learning
Graphical abstract

Highlights
-
•
Our model was trained on a longitudinal ADNI cohort, validated with the AIBL cohort
-
•
Our model identified 10 disease-related states in Alzheimer’s disease trajectory
-
•
The identified states constituted 3 unique stages and 2 progression patterns
-
•
The identified states excelled in predicting the time to conversion to AD dementia
Neuroscience; Machine learning
Introduction
Alzheimer’s disease (AD) is a multifactorial neurodegenerative disorder with a complex pathophysiology,1,2 which gradually affects subsequent events that include synaptic and neuronal loss, and cognitive decline.3,4 Generally, the progression of AD is over decades, in a non-linear manner, and with heterogeneous clinical representations and varied deterioration rates between patients due to complicated genetic and environmental factors interactions.5,6,7 There is an increasing interest in disease progression models of AD because of their potential application in understanding disease development mechanisms,8,9 guiding patient management, providing disease prognosis, and designing treatment strategies.10,11
Advances in cerebrospinal fluid (CSF) biomarker research and imaging modalities (e.g., positron emission tomography (PET)),12 combined with computational methods (e.g., machine learning and deep learning techniques), have substantially enhanced the ability to model AD progression. Specifically, existing models learn the variability of long-term disease progression from short-term observational data and can then predict the progression of patients from their historical data.10,13,14 Note that many of these studies were based on Jack’s model,15 where all subjects follow the same disease progression pattern but with different onset times as well as deterioration speeds. Subjects were temporally ordered according to a disease progression score16 aimed at quantifying disease severity and therapeutics’ effectiveness. Accordingly, computational models using a sigmoid-shaped curve,17 Gaussian process,18 or ordinary differential equations19 were developed to fit the progression of biomarkers and measure the disease progression score. Although previous work advanced AD research, most of these studies are subject to the limitation of cross-sectional design and lack the ability to model individualized disease trajectories to discover distinct disease progression patterns, which do exist, especially in the early course of the disease. Recently, machine learning models were proposed to cluster patients into several specific disease subtypes and then calculated the progression score to quantify the disease stage of each subtype.10,13,14 Instead of assuming that all individuals follow a common progression pattern, this modeling strategy considered that patients with each specific disease subtype adhere to a separate progression pattern. However, the differences and connections of progression between disease subtypes were not well investigated.
In this study, we alleviated methodological limitations of existing models to reach a better understanding of the heterogeneity in AD, and aimed to discover the underlying distinct disease progression patterns of AD from longitudinal multi-modal data, including cognitive scores (Mini-Mental State Examination, MMSE; Clinical Dementia Rating Scale Sum of Boxes, CDRSB; Alzheimer’s Disease Assessment Scale-Cognitive Subscale, ADAS-cog and so forth), structural magnetic resonance imaging (MRI) data, genetic features (APOE4) and CSF biomarkers20,21,22(amyloid-beta (Aβ) plaques that collect between neurons and disrupt cell function; and hyperphosphorylated tau - the microtubule-associated protein, which forms insoluble filaments that accumulate as neurofibrillary tangles in AD). Specifically, we employed a personalized Hidden Markov model on longitudinal data to assess the disease staging of patients who developed cognitive abnormalities and AD dementia during follow-up subsequently. Our model can identify biologically and clinically meaningful disease-related states and their transitions, which can be further clustered into non-overlapping disease stages as well as distinct disease progression patterns characterized by varied brain regional atrophies, different cognitive measures, and demographics, and therefore has the potential for guiding the disease management, boosting the prediction performance of progression, and improving the efficacy of clinical trials.
Results
Disease-related states, stages, and progression patterns
We employed Personalized-HMM to identify disease-related states with their transitions, and the 5-fold cross-validation on the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort resulted in a ten-state model (Figures 1A and 1B, Table S8) as well as a transition matrix (Table 1). Each state was primarily differentiated by the demographics (age, sex, education), genetics (Apolipoprotein E4 alleles, APOE4), cognitive function scores (MMSE, CDRSB, ADAS, and so forth) and atrophy brain regions (hippocampus, fusiform, entorhinal, and so forth) (Figure 1C; Figures S6–S8). By clustering transition trajectories of disease-related states of participants (Table 1), three non-overlapping disease stages A (labeled as neocortical atrophy [NCA]), B (labeled as medial temporal atrophy [MTA]), and C (labeled as whole brain atrophy [WBA]), which form two distinct disease progression patterns, i.e., stage A (NCA) stage C (WBA), and stage B (MTA) stage C (WBA), were identified (details in the section "Visualization of brain regional atrophy"). Stages A and B are parallel, succeeded with stage C. Intuitively, the probability of one disease-related state turning to itself was relatively high, and there were few transitions between stages A and B. The transition with a relatively high probability were A-I A-II (0.180), A-II A-III (0.219), A-III A-IV (0.213), A-IV C-I (0.253), B-I B-II (0.146), B-II C-I (0.111), C-I C-II (0.223), C-II C-III (0.312), and C-III C-IV (0.197). As explicitly described by cognitive scores, C-IV is the most severe state with almost exclusively transitions to its own state (C-IV C-IV:0.947). It thus can be regarded as the terminal state in AD progression. The evolution of disease-related states varied from one state to another over time (Figures 2E–2G).
Figure 1.
Overview of Alzheimer’s disease progression analysis
(A) The identified disease-related states and their transitions. The thickness of pointing lines between states denotes the probability of the transition. The thicker blue lines represent two main progression patterns.
(B) Percentage of CN, MCI, and dementia in number of follow-up visits by state.
(C) Corrected p-values for each of the pairwise state comparisons in terms of representative covariates, including Age, APOE4, MMSE, and Hippocampus. Analysis was performed using post-hoc Nemenyi test after all groups were found to be statistically significant using Kruskal-Wallis test, and the Benjamini-Hochberg false discovery rate correction was used to account for multiple testing.
Table 1.
Transition matrix between the identified disease-related states
| A-I | A-II | A-III | A-IV | B-I | B-II | C-I | C-II | C-III | C-IV | |
|---|---|---|---|---|---|---|---|---|---|---|
| A-I | 0.787 | 0.180 | 0.018 | 0.011 | 0.000 | 0.001 | 0.001 | 0.001 | 0.001 | 0.000 |
| A-II | 0.126 | 0.636 | 0.219 | 0.015 | 0.000 | 0.000 | 0.001 | 0.003 | 0.000 | 0.000 |
| A-III | 0.016 | 0.136 | 0.623 | 0.213 | 0.000 | 0.002 | 0.004 | 0.005 | 0.001 | 0.000 |
| A-IV | 0.002 | 0.002 | 0.058 | 0.597 | 0.002 | 0.001 | 0.253 | 0.072 | 0.013 | 0.000 |
| B-I | 0.000 | 0.000 | 0.000 | 0.002 | 0.823 | 0.146 | 0.023 | 0.004 | 0.002 | 0.000 |
| B-II | 0.000 | 0.000 | 0.000 | 0.001 | 0.058 | 0.753 | 0.111 | 0.074 | 0.003 | 0.000 |
| C-I | 0.000 | 0.000 | 0.001 | 0.029 | 0.001 | 0.015 | 0.592 | 0.223 | 0.130 | 0.010 |
| C-II | 0.000 | 0.003 | 0.003 | 0.024 | 0.003 | 0.018 | 0.191 | 0.407 | 0.312 | 0.039 |
| C-III | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.020 | 0.041 | 0.740 | 0.197 |
| C-IV | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.035 | 0.018 | 0.947 |
The values larger than 0.1 are marked with bold font.
Figure 2.
Participants grouping and disease state dynamics with clinical progression
(A) Number of participants grouped by diagnosis, Abeta status, and state.
(B) Number of participants grouped by diagnosis, pTau status, and state.
(C) Number of participants in subgroups in terms of Abeta and pTau measurements.
(D) Number of participants in states based on Abeta and pTau measurements.
Stream graphs visualize the proportions of disease-related states over time with respect to (E) CN MCI, (F) MCI AD and (G) AD. Data from 1192 ADNI participants.
Visualization of brain regional atrophy
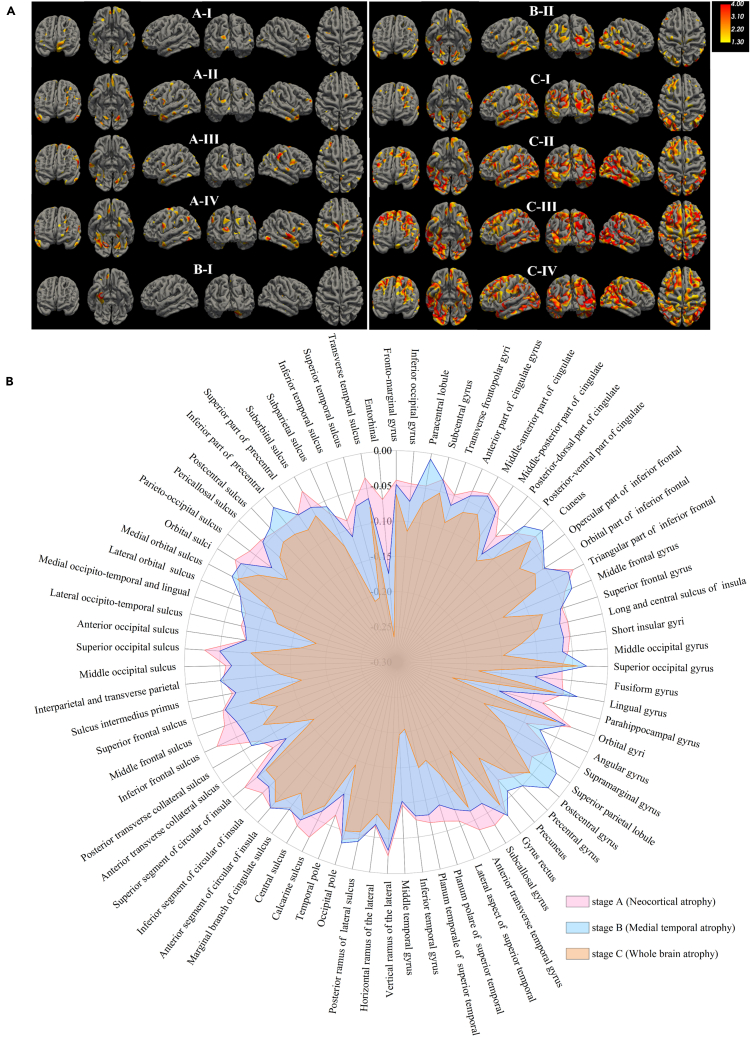
Participants labeled with different disease-related states showed distinct atrophy signatures, which were measured by voxel-based group comparison results between the CN group and the group of participants with the index of a specific state (Figure 3A). These disease-related states were highly reproducible when we trained the model on participants who were Abeta-positive in the ADNI dataset (Figure S10). Specifically, starting atrophy regions of the brain appeared to differ between subjects in different illness stages. In fact, we can visually interpret the imaging states as (i) A-I and B-I, preserved brain volume, exhibit no significant atrophies compared to CN; (ii) In stage A (A-I, A-II, A-III, A-IV), participants showed similar atrophy trajectories (multi-regional sporadic features, with atrophy in the parietal, occipital and frontal), and thus were labeled as neocortical atrophy (NCA) type; (iii) Compared with the healthy control group, there exist pronounced atrophies in entorhinal and fusiform gyrus of participants in stage B (B-I, B-II), which were essential brain regions that induce hippocampal atrophy. In this sense, patients in stage B can be labeled as medial temporal atrophy (MTA) type; (iv) Patients in stage C (C-I, C-II, C-III, C-IV) displayed severe atrophy over the whole brain region and thus were labeled as whole brain atrophy (WBA) type.
Figure 3.
Characterization of ten atrophy states and three progression stages of neurodegeneration
(A) Visualization of atrophy in brain regions of the identified disease-related states from the ADNI dataset, generated by comparing with healthy controls. A generalized linear model was utilized with a t-value threshold set at 1.3, with statistical significance attributed to activities where the t-value exceeded this threshold. Yellow indicates less atrophy, while red indicates more atrophy.
(B) Atrophy in brain regions corresponding to disease stages: A (Neocortical Atrophy, NCA), B (Medial Temporal Atrophy, MTA), and C (Whole Brain Atrophy, WBA).
Amyloid/tau & disease-related state
The hallmark pathology of AD includes the presence of β-amyloid neuritic plaques and tau protein-containing neurofibrillary tangles. We analyzed the correlations between the identified disease-related states and Abeta/pTau measures (Figures 2A and 2B, Tables S9 and S10): (i) Most of CN participants (including those with negative Abeta status (A-), positive Abeta status (A+), negative pTau status (T-), and positive pTau status (T+)) were expressed in the states A-I, A-II, and B-I. (2) A-II, A-III, B-I, and B-II corresponded to a large number of cognitively impaired but nondemented participants with A- or T-; and MCI participants with A+ or T+ mainly concentrated in the states A-III, A-IV, B-II, and C-I. (3) There were a comparable number of participants with AD dementia and labeled with C-I and C-II, who had similar distributions in amyloid status, predominately A+ (46.51% and 57.26%) and T+ (42.64% and 43.55%); participants within states C-III and C-IV were mostly A+ (91.91%, 100.00%) and T+ (79.77%, 87.10%) (Tables S9 and S10). Apparently, the index of disease-related states can be used to classify participants based on the Abeta and pTau measurements, providing insight into the disease progression and presence of pathology. Participants were grouped as normal (A-T-), as falling along the typical AD continuum (A+T+), as AD with dominant pathology (A+T-), or as suspected non-AD pathology (SNAP/A-T+) (Figure 2C). Those participants along the AD continuum further concentrated in the states A-IV, C-I, C-II, C-III, and C-IV. Participants with A+T+ tended to have more severe neurodegeneration than those with A+T-, as expected (Figure 2D and Table S11).
Characteristics of disease progression patterns
Two distinct disease progression patterns, i.e., A C, and B C, were identified from data (Figure 1A). The average age of participants at the initial state A-I of pattern A C was 68.25, while the average age of participants at state B-I was 77.96, indicating that pattern B C may correspond to a population with a later onset of AD dementia. Besides, the proportion of APOE4 gene loci in participants with pattern A C was higher than that of B C participants (A-I: APOE4, 41.2%; B-I: APOE4, 32.6%). The significant differences between the states of stage A and stage B were revealed in terms of cognitive score and core brain regions (Figure S9).
We analyzed participants' representative atrophy brain regions in the three discovered stages (Figure 3B). We identified disease-related states of each stage (Figures S11–S13) compared to healthy controls. Participants in stage A exhibited sporadic atrophy in multiple regions, including the parietal, occipital, and frontal lobes. In contrast, participants in stage B exhibited clear atrophy in brain regions such as the entorhinal and fusiform gyrus of the temporal lobe, which are essential locations for hippocampal atrophy. Significant differences in the shrinking brain regions between disease stages A and B indicate that the early course of AD is essentially heterogeneous. Participants progressing to stage C had accelerated atrophy rates in almost all selected regions, especially in the middle, inferior temporal lobe, hippocampus, and cingulate gyrus (Figure 3B and Figure S13). The hippocampus and temporal lobe were stage C’s most predominant atrophy regions, indicating that a reduction in hippocampal volume or atrophy of the temporal lobe is a hallmark imaging feature of AD.20,23,24,25
The discovered AD progression patterns had different progression trends. Regarding cognitive deterioration, participants with positive Abeta (A+) or pTau (T+) in stage A had an average more rapid decline over time than those in stage B (Table 2; Figures 4C–4F), but the deterioration trend forparticipants in stage A was non-linear and accelerated once these participants turned into state A-IV (Figure S11). Participants in stage C have significantly worse cognitive abilities than those in stages A and B (Figure S14). At baseline, the ratio of brain regions of participants in stage B was smaller than that of patients in stage A, and the volume atrophies of brain regions of participants in stage C was faster than those in stages A and B.
Table 2.
Statistical analysis of the identified disease-related stages and clinical indicators
| stage A | stage B | stage C | stage A (A+) | stage B (A+) | stage C (A+) | stage A (T+) | stage B (T+) | stage C (T+) | |
|---|---|---|---|---|---|---|---|---|---|
| Number of follow-up visits | 2723 | 1409 | 1692 | 419 | 229 | 433 | 366 | 164 | 370 |
| AGE (mean (SD)), y | 71.86 (7.24) | 78.80 (6.07) | 75.94 (7.45) | 72.18 (6.73) | 77.96 (5.56) | 74.93 (7.33) | 72.53 (6.91) | 78.98 (5.30) | 74.91 (7.49) |
| PTEDUCAT (mean (SD)), y | 16.02 (2.75) | 16.19 (2.65) | 15.21 (3.03) | 15.97 (2.78) | 16.62 (2.58) | 15.49 (2.87) | 15.99 (2.77) | 16.45 (2.42) | 15.40 (2.91) |
| Label (%) | |||||||||
| CN | 225 (8.3) | 161 (11.4) | 0 (0.0) | 24 (5.7) | 25 (10.9) | 0 (0.0) | 22 (6.0) | 25 (15.2) | 0 (0.0) |
| MCI | 2198 (80.7) | 1072 (76.1) | 439 (25.9) | 324 (77.3) | 185 (80.8) | 118 (27.3) | 274 (74.9) | 126 (76.8) | 101 (27.3) |
| Dementia | 300 (11.0) | 176 (12.5) | 1253 (74.1) | 71 (16.9) | 19 (8.3) | 315 (72.7) | 70 (19.1) | 13 (7.9) | 269 (72.7) |
| Male gender (vs. female %) | 1475 (54.2) | 996 (70.7) | 946 (55.9) | 240 (57.3) | 156 (68.1) | 249 (57.5) | 194 (53.0) | 101 (61.6) | 202 (54.6) |
| APOE4 (%) | |||||||||
| Non-carriers | 1355 (49.8) | 839 (59.5) | 531 (31.4) | 109 (26.0) | 117 (51.1) | 121 (27.9) | 108 (29.5) | 92 (56.1) | 100 (27.0) |
| 1 allele | 1036 (38.0) | 461 (32.7) | 871 (51.5) | 227 (54.2) | 93 (40.6) | 219 (50.6) | 192 (52.5) | 63 (38.4) | 187 (50.5) |
| 2 alleles | 332 (12.2) | 109 (7.7) | 290 (17.1) | 83 (19.8) | 19 (8.3) | 93 (21.5) | 66 (18.0) | 9 (5.5) | 83 (22.4) |
| CDRSB (mean (SD)) | 1.60 (1.72) | 1.86 (1.83) | 4.61 (2.79) | 2.08 (2.04) | 1.70 (1.67) | 4.37 (2.46) | 2.14 (2.00) | 1.62 (1.77) | 4.39 (2.48) |
| ADAS11 (mean (SD)) | 9.61 (5.90) | 9.75 (5.03) | 20.96 (8.93) | 11.16 (6.62) | 9.90 (4.71) | 20.28 (7.86) | 11.43 (6.59) | 9.98 (5.07) | 20.20 (7.95) |
| ADAS13 (mean (SD)) | 15.25 (8.47) | 15.73 (7.14) | 31.54 (10.55) | 17.95 (8.81) | 15.82 (6.56) | 30.94 (9.02) | 18.42 (8.61) | 15.70 (7.14) | 30.85 (9.05) |
| ADASQ4 (mean (SD)) | 5.01 (2.82) | 5.00 (2.33) | 8.72 (1.77) | 6.05 (2.70) | 4.93 (2.24) | 8.84 (1.37) | 6.32 (2.59) | 4.84 (2.33) | 8.89 (1.33) |
| MMSE (mean (SD)) | 27.51 (2.63) | 27.48 (2.58) | 22.38 (4.23) | 26.96 (2.90) | 27.31 (2.40) | 22.76 (3.82) | 26.83 (2.81) | 27.39 (2.56) | 22.76 (3.86) |
| RAVLT_immediate (mean (SD)) | 35.61 (12.12) | 33.61 (9.87) | 21.01 (8.55) | 32.75 (10.35) | 34.64 (9.21) | 21.72 (7.18) | 32.21 (10.12) | 35.31 (9.43) | 21.74 (7.18) |
| RAVLT_learning (mean (SD)) | 4.16 (2.67) | 4.12 (2.58) | 1.71 (1.83) | 3.69 (2.53) | 4.41 (2.55) | 1.73 (1.70) | 3.56 (2.48) | 4.65 (2.54) | 1.62 (1.66) |
| RAVLT_forgetting (mean (SD)) | 4.49 (2.53) | 4.54 (2.61) | 4.20 (1.85) | 5.13 (2.44) | 4.76 (2.39) | 4.45 (1.89) | 5.23 (2.42) | 4.62 (2.65) | 4.48 (1.88) |
| RAVLT_perc_forgetting (mean (SD)) | 59.47 (35.37) | 58.26 (41.46) | 91.49 (21.11) | 69.16 (37.96) | 58.13 (29.70) | 91.29 (21.33) | 72.22 (30.44) | 55.59 (31.46) | 92.39 (19.22) |
| TRABSCOR (mean (SD)) | 103.34 (62.57) | 125.26 (71.48) | 206.89 (94.56) | 115.13 (66.25) | 135.52 (70.89) | 203.92 (88.41) | 112.46 (65.88) | 127.82 (69.86) | 203.11 (88.34) |
| FAQ (mean (SD)) | 3.48 (5.49) | 4.61 (6.03) | 13.26 (8.21) | 5.14 (6.22) | 3.76 (5.09) | 12.61 (7.76) | 5.32 (6.29) | 3.31 (5.19) | 12.65 (7.72) |
| mPACCdigit (mean (SD)) | −5.05 (5.88) | −5.13 (5.13) | −15.66 (6.66) | −7.49 (5.88) | −5.70 (4.37) | −15.38 (5.17) | −7.90 (5.66) | −5.26 (4.86) | −15.50 (5.32) |
| mPACCtrailsB (mean (SD)) | −4.56 (5.56) | −5.12 (4.87) | −15.14 (6.43) | −6.71 (5.51) | −5.71 (4.03) | −14.85 (4.90) | −7.02 (5.45) | −5.29 (4.51) | −14.89 (5.08) |
| Ventricles/ICV (mean (SD)), % | 2.12 (0.94) | 3.60 (1.41) | 3.41 (1.41) | 2.30 (1.00) | 3.86 (1.36) | 3.29 (1.39) | 2.15 (0.95) | 3.44 (1.44) | 3.14 (1.32) |
| Hippocampus/ICV (mean (SD)), % | 0.47 (0.08) | 0.39 (0.06) | 0.36 (0.07) | 0.44 (0.08) | 0.39 (0.06) | 0.37 (0.07) | 0.44 (0.08) | 0.40 (0.06) | 0.37 (0.06) |
| WholeBrain/ICV (mean (SD)), % | 69.30 (5.11) | 62.57 (4.06) | 62.68 (4.83) | 68.74 (5.12) | 62.41 (3.73) | 63.39 (4.51) | 68.32 (5.14) | 62.73 (4.18) | 63.66 (4.54) |
| Entorhinal/ICV (mean (SD)), % | 0.24 (0.05) | 0.21 (0.05) | 0.18 (0.04) | 0.23 (0.05) | 0.21 (0.05) | 0.18 (0.04) | 0.22 (0.06) | 0.21 (0.05) | 0.18 (0.04) |
| Fusiform/ICV (mean (SD)), % | 1.20 (0.16) | 1.03 (0.13) | 0.98 (0.16) | 1.18 (0.17) | 1.02 (0.15) | 0.99 (0.16) | 1.17 (0.16) | 1.02 (0.15) | 1.00 (0.15) |
| MidTemp/ICV (mean (SD)), % | 1.33 (0.17) | 1.18 (0.15) | 1.08 (0.18) | 1.29 (0.17) | 1.17 (0.16) | 1.10 (0.17) | 1.28 (0.17) | 1.16 (0.17) | 1.10 (0.16) |
CN, Cognitive Normal; MCI, Mild Cognitive Impairment; APOE4, number of Apolipoprotein E4 alleles; CDRSB, Clinical Dementia Rating Scale Sum of Boxes; ADAS-cog, Alzheimer’s Disease Assessment Scale–Cognitive Subscale; MMSE, Mini-Mental State Examination; RAVLT, Rey’s Auditory Verbal Learning Test; LDELTOTAL, logical memory delayed recall total; TRABSCOR, Trail Making Test-B, FAQ, Functional Activities Questionnaire; mPACCdigit, Preclinical Alzheimer’s Cognitive Composite Scores; mPACCtrailsB, Preclinical Alzheimer’s Cognitive Composite trails B, MidTemp, Middle Temporal Gyrus; ICV: Intracerebral Volume.
Figure 4.
Predictive ability of stages
(A) Survival curves of participants progress from stage A/B to stage C.
(B) Survival curves of participants progress from stage A/B/C to AD dementia.
(C and D) Survival curves of participants progressing from stage A/B to stage C in terms of Abeta+ and pTau+, respectively.
(E and F) Survival curves of participants progressing from stage A/B/C to AD dementia regarding Abeta+ and pTau+, respectively.
(G and H) Survival curves of participants progressing from stage A/B to stage C regarding Abeta and pTau status, respectively.
(I and J) Survival curves of participants progressing from stage A/B/C to AD dementia regarding Abeta and pTau, respectively. The p-value derived by the log rank test indicates a statistically significant difference. The survival tables below the curves show the number of patients currently at risk, censored, or having an event (event representing progression from stage A/B to stage C or stage A/B/C to AD dementia) at each time point corresponding to the x axis.
Survival analysis
Kaplan-Meier analysis revealed that participants in stage A or stage B had a similar risk of progressing to stage C at every point of progression (Figure 4A). Participants in stage A and stage B had a 50% risk of developing AD dementia around 84 months. In contrast, the survival curve of stage C declined sharply (Figure 4B), indicating that participants in stage C either already had AD dementia or would rapidly develop AD dementia.
Survival curves stratified by Abeta and pTau biomarkers revealed that participants in stage A progressing to stage C or AD dementia were mainly dominated by Abeta+ or pTau+ populations. However, for participants in stage B, the influence of pTau status on the disease progression was relatively less significant (Figures 4D and 4F). As for participants in stage C, who were already at a higher risk of developing dementia, their deterioration rate was not affected by Abeta or pTau status.
Regarding the progression from any state to the final C-IV state or AD dementia (Figures S15 and S16), Kaplan-Meier analysis revealed that participants in states A-III or B-II had only a 25% risk of conversion to the terminal state C-IV within 96 months (Figure S15). In contrast, participants in stage A-IV had a higher probability of progressing to stage C-IV after 108 months. In fact, the probability of progression to the terminal stage C-IV for participants in the other states of Stage C was significantly higher than for those in any state of stage A/B. Intuitively, Kaplan-Meier curves of time to conversion to AD dementia show that participants in states of stage A or B had slow progression rates to AD dementia (Figure S16). In contrast, the probability of participants in stage C turning into AD dementia was relatively high.
Prediction performance using the index of disease-related states
We evaluated the performance using the index of the disease-related state as a predictor of time to conversion to AD dementia, compared with other well-known AD risk factors, i.e., APOE genotype, MMSE score, Abeta and pTau measures, and hippocampal volume. The predictor "index of disease-related states" achieved superior performance in the prediction of time to conversion to AD dementia (C-Index:0.923 ± 0.007) than the other well-known risk factors (Hippocampal, 0.774 ± 0.010; MMSE, 0.808 ± 0.006; Abeta, 0.7445 ± 0.013; pTau, 0.689 ± 0.006; APOE4, 0.607 ± 0.012) in the ADNI cohort (Figures 5A and 5B). Similar findings were observed in the AIBL dataset (Tables S12 and S13, Figures 5C and 5D; Figures S17–S19). Moreover, our model’s performance is substantiated by the results from the C-Index and ROC curves (Table S14 and Figure S20), consistently highlighting its exceptional predictive capability compared to conventional machine learning approaches. To address data imbalance, we restricted the number of individuals in the disease population and controlled the ratio of those progressing to AD dementia versus those not progressing. Our model utilized identified disease states as predictive biomarkers for AD progression, maintaining stable predictive performance across both balanced and imbalanced populations, with improved performance observed in the imbalanced group (Table S15 and Figure S21).
Figure 5.
Evaluations of time-to-conversion prediction
We evaluated the predictive performance of time-to-conversion to AD dementia using the C-Index across several clinical variables (Abeta, pTau, APOE4, Hippocampus, MMSE) using violin plots to display 5-fold cross-validated results. Significance levels are marked as ∗ (p < 0.05), ∗∗ (p < 0.01), and ∗∗∗ (p < 0.001), assessed by the one-sided Wilcoxon signed-rank test.
(A) Predictive ability of the identified states in the ADNI dataset.
(B) ROC curves for conversion time prediction to AD dementia in the ADNI dataset.
(C) Predictive ability of the identified states in the AIBL dataset.
(D) ROC curves for conversion time prediction to AD dementia in the AIBL dataset.
Discussion
The mechanism underlying the marked heterogeneity in AD progression still needs to be understood. In this study, we utilized, validated, and evaluated a machine learning model, i.e., personalized-HMM, to uncover the latent patterns from longitudinal data documented in the trajectories of patients with AD. Our model accounts for the heterogeneity of AD in its manifestations and progression. Specifically, by capitalizing on the rich real-world data from ADNI and AIBL, we were able to overcome the challenges associated with the lack of longitudinal data and deficiency in the performance of personalized disease progression analysis as well as time-to-event prediction. AD progression models were often inspired by the amyloid cascade hypothesis, crystallized into Jack’s hypothetical15 model of biomarker dynamics, which states that the main AD biomarkers become abnormal in a temporally ordered manner. Even though there is a large consensus that Amyloid-β plays a critical role in AD pathophysiology, growing evidence shows that AD progression comes from a multifactorial interaction of processes and that all combinations of biomarker abnormalities are frequently present in the cognitively normal population.26,27 Multiple biomarkers drive our model, do not assume the order in which the biomarkers become abnormal, and can stage the entire spectrum of AD in real-world data populations. These non-overlapping disease stages and their unique patterns of brain atrophy, cognitive measures, and demographic features would be responsible for the heterogeneity observed in biomarker trajectories and discrepancies between observations and the amyloid cascade hypothesis.
The most striking finding of this study is that our model identified ten meaningful disease-related states, which manifest differently in terms of participants’ demographics, genetics, cognitive score, and atrophy of brain regions. The identified states constitute 3 non-overlapping stages and 2 distinct disease progression patterns, hidden in the heterogeneous trajectories of patients with AD, explicitly offering a straightforward means for a clinician to stratify patients about the likelihood of progression within a particular time frame. More importantly, the discovered disease-related states, stages, and progression patterns are interpretable and versatile with respect to both the severity and heterogeneity of AD progression. This feature allows the investigation of complex and nonlinear relationships between the discovered states/stages/patterns and clinical outcomes of interest (e.g., cognitive score, clinical measures, and so forth), which might benefit informing clinical trial recruitment. Our model does not assume disease subtypes or directly infer disease stages but explicitly identifies disease-related states and their transitions from real-world data. This fundamental property of our model is different from the existing learning models, which assume patients with AD either follow a common progression pattern or were categorized as several disease subtypes and then separately modeled the progression of each disease subtype.8,13,15 Contrary to the finding of Yang et al.,10,11 which considered individuals who expressed similar disease manifestations in the initial phase of AD, we found that participants were initially in either stage A or B and then progressed to stage C, suggesting that although the advanced AD is clinically similar, heterogeneity exists in the early course of the disease. Our findings in the ADNI and AIBL cohorts coincide with the study results of another independent clinical cohort, the French MEMENTO cohort, which investigated the brain atrophy subtypes in participants of subjective cognitive complaints or MCI.28 The study proposed that the typical/diffuse atrophy subtype can be recognized as the continuum of the two subtypes, either the limbic-predominant or the hippocampal-sparing subtype. The clinical and anatomical heterogeneity broadly corresponds to regional Tau deposition as the major contributor.29,30 Our study offers insights into AD and, more importantly, how it progresses.
Applying personalized-HMM to MRI data enriched with AD pathology identified 3 non-overlapping stages of regional brain atrophy expressed in participants across the AD spectrum, which were highly reproducible in validation experiments. These stages range from mild to advanced atrophy and define two progression pathways. The 3 non-overlapping stages have clinically meaningful implications. Stage A exhibited sporadic atrophy in multiple regions, including the parietal, occipital, and frontal lobes. In contrast, participants in stage B exhibited apparent atrophy in brain regions such as the entorhinal and fusiform gyrus of the temporal lobe. The three non-overlapping stages had different trends in cognitive scores and the ratios of brain sub-regions over time (Figure S14). Specifically, the cognitive deterioration of participants in stage B was slightly faster than that of stage A after two years of follow-up. Significant differences in the shrinking brain regions between disease stages A and B indicate that the early course of AD is essentially heterogeneous. Stage C is a composite of advanced or ‘end-stage’ neurodegeneration stages. While entirely typical of cognitive decline and worsening regional brain atrophy, indicating a late-stage similarity of widespread brain atrophy across multiple pathologies.
Different demographics and specific atrophy regions of the brain can characterize the heterogeneity of disease progression. For instance, there are more old male participants in pattern B C (state B-I: 64.4% male, average age 77.96; B-II: 75% male, average age 79.10) than those in pattern A C (e.g., A-I: 36.6% male, average age 68.25; A-IV: 61.4% male, average age 73.35), suggesting a fundamental difference between the discovered patterns in terms of age and sex of participants whose cognitive functions start to decline and clinical syndromes appear. Literature indicates that male patients have a later disease onset than female patients,31,32 and this proposition was confirmed by our findings. In addition, survival curves (Figures 4D and 4F) show a significant association with pTau in pattern A C but not B C. This finding may be caused by potential sex-related differences, highlighting the possibility that accelerated tau proliferation in women significantly contributes to a greater risk of faster deterioration of the disease.33,34 Regarding the atrophy of brain regions, we observed that there were sporadic atrophies in many brain regions for participants in disease stage A. On the contrary, evident atrophies in the entorhinal and fusiform gyrus for patients in stage B were observed, suggesting that brain regions contributed differently to characterize the difference of AD progression. Both gray matter and white matter loss in the hippocampus were prominent in pattern B C compared to A C. These brain alterations were considered of high clinical relevance, resulting in—for instance—more likely to show typical clinical syndromes of symptomatic AD and vice versa.35,36,37
Participants in the two patterns had varied cognitive score trends over time and different atrophic rates in brain regions. On average, participants with positive Abeta or pTau in stage A had slightly faster cognitive decline than those in stage B. With in-depth analysis, we found that participants with early states (i.e., A-I, A-II, and A-III) in stage A had slow cognitive decline, but this trend became accelerated once participants turned into state A-IV, indicating that the progression of AD is non-linear in nature, even in the preclinically early stage of the disease. Our findings were consistent with the results of Scheltens et al., who found that the hippocampal-sparing subtype has a faster decline related to atypical AD variants.38 However, controversial results have shown that the hippocampal-sparing subtype has also been associated with less aggressive disease progression.39 The discrepancies highlight the importance of investigating the disease mechanism underlying such heterogeneity. Of note, the atrophy of brain regions and the deterioration rates of participants in both stages A and B were clearly different from each other, speculating that the AD-correlated micro-structural alternations in brain regions might indicate their involvement in these changes, and these imaging alternations might be results of pathophysiological changes in AD progression. These findings suggest that AD is a heterogeneous disorder with varied progression rates, especially in the early disease course.
The index of the identified disease-related states provides remarkable performance on predicting the time to conversion to AD dementia, in comparison with the other commonly used AD risk factors, including APOE genotype, MMSE score, Abeta and pTau measures, and hippocampal volume, underlining the potential significance of this predictive index for progression prognosis. Note that many studies have specifically focused on extracting essential biomarkers for the early prediction of AD onset. Although promising performance of conventional risk factors, e.g., APOE genotype, MMSE score, Abeta and pTau measures, and so forth have been demonstrated for prognostic analysis, the discriminative power of these relatively simple features is limited, especially when used for the prediction task of time to conversion to AD dementia. Our results show that the index of the identified disease-related states performed significantly better than conventional imaging, genetic, and clinical biomarkers on both ADNI and AIBL cohorts, suggesting that the identified states have significant potential for characterizing AD progression. The experimental results also illustrated the robust generalization performance across different cohorts.
Conclusion
We aimed to capture the full spectrum of AD progression and proposed a machine-learning model for identifying underlying disease-related states from longitudinal data. The discovered states constitute non-overlapping disease stages and exhibit distinct disease progression patterns. These findings have implications for predicting individuals likely to progress along a specific disease trajectory within a defined time frame. Consequently, they contribute a significant step toward addressing the persistent challenge of early Alzheimer’s disease prevention and individualized Alzheimer’s disease management.
Limitations of the study
Despite promising findings, some limitations of our study should be addressed. First, we trained and evaluated our model on ADNI data with an external validation in the AIBL cohort. The generalizability of the proposed model needs to be evaluated using large-scale experiments. To this end, external validation could be performed on other large datasets to verify whether the discovered patterns are consistent across diverse populations. This is left for future work. Another limitation is that our study did not consider the impact of clinical interventions on the trajectory of patients with AD. Clinical intervention may reverse the disease in patients with early MCI, and intervention in the later stage of the disease will also delay the progression of the disease. Treatment effects should be estimated in either a prospective or retrospective manner for better AD progression analysis and management.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Alzheimer's Disease Neuroimaging Initiative (ADNI) | SciCrunch Registry | RRID:SCR_003007; http://adni-info.org/ |
| Australian Imaging Biomarkers and Lifestyle Study of Aging (AIBL) | SciCrunch Registry | www.aibl.csiro.au |
| Software and algorithms | ||
| Python (version 3.7.13) | Python Software Foundation | RRID:SCR_008394; https://www.python.org/ |
| Pandas (version 1.3.5) | Python package | RRID:SCR_018214; https://pandas.pydata.org/ |
| Numpy (version 1.21.6) | Python package | RRID:SCR_018214; https://pandas.pydata.org/ |
| Pytorch (version 1.12.6) | Python package | RRID:SCR_018536; https://pytorch.org/ |
| Scikit-learn (version 0.22.2) | Python package | RRID:SCR_002577; http://scikit-learn.org/ |
| R (version 4.2.1) | R software | RRID:SCR_002577; http://scikit-learn.org/ |
| FreeSurfer (version 6.0) | FreeSurfer software | RRID:SCR_001847; http://surfer.nmr.mgh.harvard.edu/ |
| Origin (version 9.0) | Origin software | RRID:SCR_014212; http://www.originlab.com/index.aspx?go=PRODUCTS/Origin |
| GraphPad Prism (version 9.0) | GraphPad software | RRID: SCR_002798; http://www.graphpad.com/ |
| Gephi (version 0.10) | Gephi software | RRID:SCR_004293; http://gephi.org/ |
| Personalized_HMM_disease_progression | This Paper | https://github.com/ZJU-BMI/Personalized_HMM_disease_progression. |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Zhengxing Huang (e-mail: zhengxinghuang@zju.edu.cn).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
The data supporting the findings of this study were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) and the Australian Imaging Biomarkers and Lifestyle Study of Ageing (AIBL), which are available from the ADNI database (https://adni.loni.usc.edu) and AIBL database (https://aibl.csiro.au/) upon registration and compliance with the data use agreement.
-
•
The source code pertaining to both the personalized hidden Markov model and data analysis in this manuscript has been deposited on GitHub and is publicly available as of the date of publication; URLs are provided at https://github.com/ZJU-BMI/Personalized_HMM_disease_progression.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Our training cohort includes 1530 individuals (117 CN MCI, 1075 MCI AD, and 338 CN CN) from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) multi-center database,40,41 and the validation cohort includes 266 individuals (31 CN MCI, 56 MCI AD, and 179 CN CN) from the Australian Imaging, Biomarker & Lifestyle Flagship Study of Ageing (AIBL) database.42,43 Details of both cohorts are depicted in Tables S3–S5. Disease groups contain patients from CN to MCI or from MCI to AD Dementia (1192 participants, 5824 visiting records) in ADNI, and the CN population (CN CN, 388 participants, 1843 visiting records) was used as healthy controls. We included the demographics, cognitive function score, and T1 structural magnetic resonance imaging (MRI) scans of the ADNI cohort for model learning and used the AIBL dataset for evaluation.
Method details
Follow-up time intervals
Different time intervals do have an impact on the HMM model. If the time interval is short, the transition between hidden states may be more frequent, leading to a more accurate estimation of the state transition probability. Conversely, if the time interval is long, the transition between hidden states may be relatively sparse, leading to an inaccurate estimation of the state transition probability. Since the training model requires a three-dimensional data format, we need to perform time alignment on the included longitudinal research data. We conducted a statistical analysis on the follow-up frequency of 1192 individuals in the disease groups (CN-MCI, MCI-AD, AD-AD). Most of the population had follow-up frequencies between 4-6 visits, with only 38 individuals having more than eight follow-up visits (Table S1). Due to padding the data length with zeros at the end, which does not affect the outcome, for individuals with more than eight follow-up visits, some follow-up data points will be discarded to maintain consistent data lengths across the population. To minimize the increase in input data volume while retaining the original follow-up data of the population as much as possible, we chose to perform time alignment based on eight follow-up visits.
When performing time alignment, if the patient had less than eight visits, supplemented to eight visits, and the supplemental numerical feature was replaced with 0 (Padding zeros to the features is merely a transformation of the input data format and does not affect the predicted state of the model). If the patient had more than eight follow-up visits, the baseline and final follow-up visits were taken, and eight visits were randomly selected in the middle: guaranteeing 1192 patients (disease group), each with eight follow-up data. Then, we conducted a statistical analysis on the time interval between consecutive follow-ups of the study population in the disease group and plotted a histogram (Figure S2). According to the histogram distribution, almost all observed follow-ups occurred within six months to 1 year of the expected timing. This time interval is relatively uniform and stable, allowing a better estimation of the state transition probability.
Model variables description
The variables included in the study are listed in Table S2, and the abbreviations in parentheses will be used throughout the manuscript. During model training, the following variables were utilized: demographic indicators such as age and educational level (PTEDUCAT); cognitive assessment scales including CDRSB, ADAS11, ADAS13, ADASQ4, MMSE, RAVLT_immediate, RAVLT_learning, RAVLT_forgetting, RAVLT_perc_forgetting, LDELTOTAL, TRABSCOR, FAQ, mPACCdigit, and mPACCtrailsB; core brain region volumes (normalized by ICV) such as Ventricles, Hippocampus, WholeBrain, Entorhinal, Fusiform, and MidTemp; and covariate indicators including gender and APOE4. Beta-amyloid (Abeta) protein and phosphorylated tau protein (pTau) are used for result analysis.
The demographic and baseline clinical characteristics of the AIBL cohort are listed in Table S5. AIBL serves as an external validation dataset; however, in comparison with ADNI, several essential covariates (including ADAS-cog, RAVLT-cog, etc.) were not available in the AIBL cohort. To address this issue, we assumed that the samples in ADNI and AIBL followed an identical independent data distribution. We merged the data from ADNI and AIBL and then imputed the missing data of AIBL using a multiple regression algorithm. Afterward, we assigned the participants in AIBL to the states learned from the ADNI data and used the model derived from ADNI to predict the conversion time for AIBL patients.
Longitudinal data preparation
A sample's retained data includes clinical indicators (cognitive testing) and T1-weighted volumetric MRI scans. The sample was excluded if one modality of the data was missing. Participants were eligible for study inclusion if they had at least two times of T1-weighted volumetric MRI scans (Figure S1 for details on exclusion and inclusion criteria). To ensure that informative clinical variables were selected and the correlation between variables could be diluted, we only included clinical variables with a missing rate smaller than 30% and adopted multivariate imputation by chained equation (MICE) to impute the missing data, using R package MICE (version 3·14). Images were processed with the longitudinal stream of FreeSurfer 6.0 through a fully automated pipeline, as suggested in literature.44,45,46 For both ADNI and AIBL cohorts, 74 anatomical regions of interest (ROIs) (Table S6) of each hemisphere were identified in gray matter using the Destrieux (2009) brain template.47,48 We normalized the regional volumes by intracranial volume (ICV) to compensate for inter-individual differences in brain morphology and total head size. For visualization of disease states, tissue density maps, referred to as Destrieux, were computed. Individual images were first registered with a single subject brain template and segmented into GM and WM tissues. Destrieux maps encode, locally and separately, each tissue type and the volumetric changes observed during the registration.
Personalized hidden Markov model (Personalized-HMM)
A personalized hidden Markov model49,50 was trained and applied using longitudinal clinical measures and imaging signatures. The model assumes that a disease trajectory manifests as a series of disease-related states, not specified a priori but latent in data, in which each state is defined by a mixture of distribution of clinical measures. The objective of the model is to generate two primary matrices, a transition matrix and an observational matrix, which describe the probability of transitions between disease-related states and the distribution of clinical measures associated with each state, respectively. Hereby, we denote latent states as z, and corresponds to the trajectory of a target patient i and corresponds to a visiting record documented at time t in the trajectory. Analogously, observations are denoted by . The standard transition model is given as follows:
| (Equation 1) |
where indicates a categorical distribution. Of note, an HMM with a Gaussian observation model is specified as , where represents a multivariate Gaussian distribution with mean . Due to the heterogeneity of AD progression for individual patients, it is necessary to allow patients to deviate from the population at large. To this end, we introduced patient specific latent variables to modify the mean response of patient i:
| (Equation 2) |
This modeling strategy enables personalization in how states might manifest in an individual. The model places Gaussian priors on the personalized effects and .
| (Equation 3) |
By employing zero mean Gaussian priors with appropriately chosen variances , encoding prior belief that while heterogeneity among patients and between states exists, the scale of this heterogeneity is small and the personalized effects do not deviate too far.
Learning algorithm
We adopted the expectation-maximization algorithm51,52 to train the personalized-HMM model by maximizing a lower bound of the model likelihood using the training data. The model approximated the posterior distributions over the local latent variables () with tractable variational approximations and relied on point estimates for the global parameters (θ). A structured variational approximation was employed to retain the dependence between and , , and the temporal structure within :
| (Equation 4) |
where represents the variational free parameters. We used Gaussians with full covariances to parameterize the variational distributions, and , where denotes lower triangular matrix.
The model minimized the Kullback-Leibler divergence between the variational approximation and the posterior, and learned the model parameters θ by maximizing the corresponding evidence lower bound (ELBO):
| (Equation 5) |
where is the entropy.
| (Equation 6) |
where . We maximized the ELBO via coordinate ascent alternating between updates to variational parameters λ and model parameters θ. Once the model was learned, the appropriate sequence of disease-related states for a target patient trajectory can be inferred using the Viterbi algorithm.53,54 The model was trained using 5-fold cross validation and the number of hidden states was appropriately selected (Figures S3 and S4). Eventually, the interpretation of the disease-related states as well as their transition routes were captured through visualization.
Hidden state inference
Sample splitting of the training set and testing set of ADNI was shown in Figure S4. In detail, the training cohort (ADNI data) was divided into 5 parts (Tables S7 and S8). At each round of the study, 4/5 of the data (training) was used to learn the model parameters, and the remaining 1/5 of the data (validation) was used to assess the result. We select the best model by comparing the loss values of 5 models, and the model with the lowest and most stable loss value will be chosen as the final model. The number of states was chosen based on the performance of the validation data. This analysis was performed to decrease the risk of overfitting, as increasing the number of states should always improve performance on the training data. However, the model would eventually overfit and generalize poorly to the validation data. In this study, the number of states 3 to 14 were considered in the analysis. The results of the cross-validation study are summarized in Figure S5.
External model validation
External validation of the proposed model was performed using the data extracted from the Australian Imaging Biomarkers and Lifestyle Study of Ageing (AIBL) cohort. In total, there were 179 CN patients, 31 CN MCI patients, and 56 MCI Dementia patients extracted from the AIBL cohort as the external validation dataset (Table S5). The data preprocessing procedure was the same as that of ADNI. Compared with ADNI, several essential covariates (including ADAS-cog, RAVLT-cog etc.) were unavailable in the AIBL cohort. To remedy it, we assumed that samples in ADNI and AIBL followed the identically independent data distribution, merged data of ADNI and AIBL, and then imputed the missing data of AIBL by using a multiple regression algorithm. After that, we assigned participants in AIBL to the states learned from ADNI data and used the model learned from ADNI data to predict the conversion time of AIBL patients.
Defining the labels of the stages
The disease status was learned and obtained from the model using data from the disease group populations (CN-MCI, MCI-AD: 1192, all ADNI data), while the CN group population (CN-CN) served as a healthy control. This control group was utilized for visualizing brain region atrophy and calculating the atrophy rate of each brain region across different disease states. We employed Freesurfer, a brain imaging processing software, along with generalized linear models,55 to create brain region atrophy visualization maps for each state (Figure 3A). The labels for the three stages were named based on the calculation of the atrophy rates in each stage, and were further refined with input from professional clinical doctors. Below are the detailed steps for naming the labels of the three stages:
-
(a)
Brain region extraction: Prior to the commencement of the study, we categorized the population into three groups (CN-CN, CN-MCI, MCI-AD) based on their clinical progression trajectories. In determining disease states, our study focused exclusively on the non-healthy groups (CN-MCI, MCI-AD). We employed the healthy group (CN-CN) as a control for calculating the brain atrophy rate in each assigned state. All images were processed using FreeSurfer 6.0 through a fully automated pipeline. We identified 74 anatomical ROIs using the Destrieux brain template and normalized their volumes based on ICV.
-
(b)
Atrophy rate calculation: the calculation of brain atrophy rates for each state is based on cross-sectional data, and the atrophy rate calculation follows the formula below:
Here, "i" represents the number of individuals in each assigned state group; "j" represents the assigned state; "m" represents the number of individuals in the CN group; and "v" represents the grey matter volume of a brain region.
-
(c)
Expert guidance: Combine guidance from professional clinical doctors with consideration of brain regions relevant to the disease state and corresponding atrophy rates (Figures S11–S13).
-
(d)
Label naming: Based on the atrophy rate results, expert guidance, and clinical background knowledge, we determined appropriate labels (neocortical atrophy, medial temporal atrophy, and whole brain atrophy) for each stage.
These steps aim to better name the labels of the three stages based on objective data and professional guidance to describe brain atrophy in different disease states more accurately.
Quantification and statistical analysis
Throughout the manuscript, statistical significance was defined as a p-value of less than 0.05 after correcting for multiple comparisons where necessary, as described below. Estimates for statistical parameters, degrees of freedom where applicable, and the n for each statistical test are included throughout the results and supplemental information section, except for statistical tests involving comparisons of survival curves, where n values are included in the respective figure (Figures S15–S18).
Demographics analysis
The chi-square test was employed to compare categorical variables, such as sex and APOE4 status. One-way analysis of variance (ANOVA) was utilized to analyze differences in continuous variables, such as age and Mini-Mental State Examination (MMSE) scores. The significance level was set at p < 0.05 (Tables S4 and S5).
Disease-related states analysis
We compared groups stratified by different disease-related states using one-way analysis of variance (ANOVA) for continuous variables and chi-square tests for categorical variables (Tables S8 and S12). Corrected p-values were calculated for each pairwise state comparison with respect to cognitive assessments and core brain regions in Alzheimer's disease. These results are intended to support the interpretation of the distinguishing features of these states. Post-hoc analysis using the Nemenyi test was performed after all groups were found to be statistically significant by the Kruskal-Wallis test, and the Benjamini-Hochberg false discovery rate correction was applied to account for multiple testing (Figure 1C and Figures S6–S9).
Atrophy visualization analysis
We used a generalized linear model to visualize and statistically analyze cortical atrophy in the disease-related states compared to the healthy control group (Figure 3A and Figure S10). The t-value threshold was set at 1.3, with statistical significance attributed to activities where the t-value exceeded this threshold.
Survival and predictive analysis
Kaplan-Meier analysis was used to analyze the time to conversion to AD dementia/terminal state, and the log-rank test was used to assess statistical significance (Figure 4 and Figures S15–S18). Survival tables below the survival curves show the number of patients at risk, censored, or having an event (event representing progression from any state to AD dementia) at each time point corresponding to the x-axis. We evaluated the predictive performance of time-to-conversion using the C-Index and compared it with other clinical variables (Abeta, pTau, APOE4, hippocampus, and MMSE) using the one-sided Wilcoxon signed-rank test (Figure 5).
Software packages
All statistical analyses were conducted using R (version 4.2.1). Figures in the paper were created using tools such as Freesurfer, Origin, GraphPad, Gephi, and R software. The URLs for accessing these software datasets are listed in the key resources table.
Acknowledgments
This work was partially supported by the Key R&D Program of Zhejiang under Grant No. 2022C03134, National Nature Science Foundation of China under Grant No. 82272129, Natural Science Foundation of Xinjiang Autonomous Region No.2022D01C434, and the State Key Laboratory of Pathogenesis, Prevention, Treatment of Central Asian High Incidence Diseases Fund No.SKL-HIDCA-2022-23.
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U019 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO; Janssen Alzheimer Immunotherapy Research And Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
The Australian Imaging Biomarkers and Lifestyle (AIBL) study:www.aibl.csiro.au, is a consortium between Austin Health, CSIRO, Edith Cowan University, the Florey Institute (The University of Melbourne), and the National Aging Research Institute. Partial financial support was provided by the Alzheimer's Association (US), the Alzheimer's Drug Discovery Foundation, an anonymous foundation, the Science and Industry Endowment Fund, the Dementia Collaborative Research Centres, the Victorian Government’s Operational Infrastructure Support program, the McCusker Alzheimer's Research Foundation, the National Health and Medical Research Council, and the Yulgilbar Foundation. Numerous commercial interactions have supported data collection and analysis. In-kind support has also been provided by Sir Charles Gairdner Hospital, Cogstate Ltd., Hollywood Private Hospital, the University of Melbourne, and St. Vincent’s Hospital.
Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database:adni.loni.usc.edu. As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in the analysis or writing of this report. A complete listing of ADNI investigators can be found at http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf. Data used in the preparation of this article were obtained from the Australian Imaging Biomarkers and Lifestyle flagship study of aging (AIBL) funded by the Commonwealth Scientific and Industrial Research Organisation (CSIRO), which was made available at the ADNI database (www.loni.usc.edu/ADNI). The AIBL researchers contributed data but did not participate in the analysis or writing of this report. AIBL researchers are listed at www.aibl.csiro.au.
Author contributions
ZH, YCZ, and MP conceived and designed the study. SZ developed and validated the deep learning system supervised by ZH and YS with clinical input from YCZ, JY, ZYL, and FFZ. FY and JY did the statistical analysis. SZ and ZH also contributed to computational analysis and validations. FW, YS and YZ provided critical reading and suggestions. SZ, JY and ZH drafted the article with input from YCZ, MP, and JS. ZH and YCZ contributed equally to the work as senior authors. All authors subsequently critically edited the report. All authors read and approved the final report. The corresponding author and senior authors had full access to all data. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Declaration of interests
The authors declare that they have no competing interests.
Published: June 14, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.110263.
Contributor Information
Yi-Cheng Zhu, Email: zhuyc@pumch.cn.
Zhengxing Huang, Email: zhengxinghuang@zju.edu.cn.
Supplemental information
References
- 1.Sloane P.D., Zimmerman S., Suchindran C., Reed P., Wang L., Boustani M., Sudha S. The public health impact of Alzheimer’s disease, 2000-2050: potential implication of treatment advances. Annu. Rev. Public Health. 2002;23:213–231. doi: 10.1146/annurev.publhealth.23.100901.140525. [DOI] [PubMed] [Google Scholar]
- 2.Toledo J.B., Arnold S.E., Raible K., Brettschneider J., Xie S.X., Grossman M., Monsell S.E., Kukull W.A., Trojanowski J.Q. Contribution of cerebrovascular disease in autopsy confirmed neurodegenerative disease cases in the National Alzheimer’s Coordinating Centre. Brain. 2013;136:2697–2706. doi: 10.1093/brain/awt188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang X.X., Tian Y., Wang Z.-T., Ma Y.-H., Tan L., Yu J.-T. The epidemiology of Alzheimer’s disease modifiable risk factors and prevention. J. Prev. Alzheimers Dis. 2021;8:313–321. doi: 10.14283/jpad.2021.15. [DOI] [PubMed] [Google Scholar]
- 4.Suzzi S., Croese T., Ravid A., Gold O., Clark A.R., Medina S., Kitsberg D., Adam M., Vernon K.A., Kohnert E., et al. N-acetylneuraminic acid links immune exhaustion and accelerated memory deficit in diet-induced obese Alzheimer’s disease mouse model. Nat. Commun. 2023;14:1293. doi: 10.1038/s41467-023-36759-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korologou-Linden R., Bhatta L., Brumpton B.M., Howe L.D., Millard L.A.C., Kolaric K., Ben-Shlomo Y., Williams D.M., Smith G.D., Anderson E.L., et al. The causes and consequences of Alzheimer’s disease: phenome-wide evidence from Mendelian randomization. Nat. Commun. 2022;13:4726. doi: 10.1038/s41467-022-32183-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iturria-Medina Y., Adewale Q., Khan A.F., Ducharme S., Rosa-Neto P., O’Donnell K., Petyuk V.A., Gauthier S., De Jager P.L., Breitner J., Bennett D.A. Unified epigenomic, transcriptomic, proteomic, and metabolomic taxonomy of Alzheimer’s disease progression and heterogeneity. Sci. Adv. 2022;8 doi: 10.1126/sciadv.abo6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cullen N.C., Leuzy A., Janelidze S., Palmqvist S., Svenningsson A.L., Stomrud E., Dage J.L., Mattsson-Carlgren N., Hansson O. Plasma biomarkers of Alzheimer’s disease improve prediction of cognitive decline in cognitively unimpaired elderly populations. Nat. Commun. 2021;12:3555. doi: 10.1038/s41467-021-23746-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poulakis K., Pereira J.B., Muehlboeck J.-S., Wahlund L.-O., Smedby Ö., Volpe G., Masters C.L., Ames D., Niimi Y., Iwatsubo T., et al. Multi-cohort and longitudinal Bayesian clustering study of stage and subtype in Alzheimer’s disease. Nat. Commun. 2022;13:4566. doi: 10.1038/s41467-022-32202-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eldholm R.S., Barca M.L., Persson K., Knapskog A.-B., Kersten H., Engedal K., Selbæk G., Brækhus A., Skovlund E., Saltvedt I. Progression of Alzheimer’s disease: A longitudinal study in Norwegian memory clinics. J. Alzheimers Dis. 2018;61:1221–1232. doi: 10.3233/JAD-170436. [DOI] [PubMed] [Google Scholar]
- 10.Yang Z., Nasrallah I.M., Shou H., Wen J., Doshi J., Habes M., Erus G., Abdulkadir A., Resnick S.M., Albert M.S., et al. A deep learning framework identifies dimensional representations of Alzheimer’s Disease from brain structure. Nat. Commun. 2021;12:7065. doi: 10.1038/s41467-021-26703-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwak K., Giovanello K.S., Bozoki A., Styner M., Dayan E., Alzheimer’s Disease Neuroimaging Initiative Subtyping of mild cognitive impairment using a deep learning model based on brain atrophy patterns. Cell Rep. Med. 2021;2:100467. doi: 10.1016/j.xcrm.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jack C.R., Jr., Bennett D.A., Blennow K., Carrillo M.C., Dunn B., Haeberlein S.B., Holtzman D.M., Jagust W., Jessen F., Karlawish J., et al. NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 2018;14:535–562. doi: 10.1016/j.medj.2022.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young A.L., Marinescu R.V., Oxtoby N.P., Bocchetta M., Yong K., Firth N.C., Cash D.M., Thomas D.L., Dick K.M., Cardoso J., et al. Uncovering the heterogeneity and temporal complexity of neurodegenerative diseases with Subtype and Stage Inference. Nat. Commun. 2018;9:4273. doi: 10.1038/s41467-018-05892-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eshaghi A., Young A.L., Wijeratne P.A., Prados F., Arnold D.L., Narayanan S., Guttmann C.R.G., Barkhof F., Alexander D.C., Thompson A.J., et al. Author Correction: Identifying multiple sclerosis subtypes using unsupervised machine learning and MRI data. Nat. Commun. 2021;12:3169. doi: 10.1038/s41467-021-22265-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jack C.R., Jr., Knopman D.S., Jagust W.J., Petersen R.C., Weiner M.W., Aisen P.S., Shaw L.M., Vemuri P., Wiste H.J., Weigand S.D., et al. Tracking pathophysiological processes in Alzheimer’s disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol. 2013;12:207–216. doi: 10.1016/S1474-4422(12)70291-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jedynak B.M., Lang A., Liu B., Katz E., Zhang Y., Wyman B.T., Raunig D., Jedynak C.P., Caffo B., Prince J.L., Alzheimer's Disease Neuroimaging Initiative A computational neurodegenerative disease progression score: method and results with the Alzheimer’s disease Neuroimaging Initiative cohort. Neuroimage. 2012;63:1478–1486. doi: 10.1016/j.neuroimage.2012.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sebaugh J.L., McCray P.D. Defining the linear portion of a sigmoid-shaped curve: bend points. Pharm. Stat. 2003;2:167–174. doi: 10.1002/pst.62. [DOI] [Google Scholar]
- 18.Liu H., Ong Y.-S., Shen X., Cai J. When Gaussian process meets big data: A review of scalable GPs. IEEE Trans. Neural Netw. Learn. Syst. 2020;31:4405–4423. doi: 10.1109/TNNLS.2019.2957109. [DOI] [PubMed] [Google Scholar]
- 19.Lu L., Meng X., Mao Z., Karniadakis G.E. DeepXDE: A deep learning library for solving differential equations. SIAM Rev. Soc. Ind. Appl. Math. 2021;63:208–228. [Google Scholar]
- 20.Whitwell J.L., Dickson D.W., Murray M.E., Weigand S.D., Tosakulwong N., Senjem M.L., Knopman D.S., Boeve B.F., Parisi J.E., Petersen R.C., et al. Neuroimaging correlates of pathologically defined subtypes of Alzheimer’s disease: a case-control study. Lancet Neurol. 2012;11:868–877. doi: 10.1016/S1474-4422(12)70200-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whitwell J.L., Josephs K.A., Murray M.E., Kantarci K., Przybelski S.A., Weigand S.D., Vemuri P., Senjem M.L., Parisi J.E., Knopman D.S., et al. MRI correlates of neurofibrillary tangle pathology at autopsy: a voxel-based morphometry study. Neurology. 2008;71:743–749. doi: 10.1212/01.wnl.0000324924.91351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kantarci K., Avula R., Senjem M.L., Samikoglu A.R., Zhang B., Weigand S.D., Przybelski S.A., Edmonson H.A., Vemuri P., Knopman D.S., et al. Dementia with Lewy bodies and Alzheimer disease: neurodegenerative patterns characterized by DTI. Neurology. 2010;74:1814–1821. doi: 10.1212/WNL.0b013e3181e0f7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van de Pol L.A., Hensel A., Barkhof F., Gertz H.J., Scheltens P., van der Flier W.M. Hippocampal atrophy in Alzheimer disease: age matters. Neurology. 2006;66:236–238. doi: 10.1212/01.wnl.0000194240.47892. [DOI] [PubMed] [Google Scholar]
- 24.Barkhof F., Polvikoski T.M., van Straaten E.C.W., Kalaria R.N., Sulkava R., Aronen H.J., Niinistö L., Rastas S., Oinas M., Scheltens P., Erkinjuntti T. The significance of medial temporal lobe atrophy: a postmortem MRI study in the very old. Neurology. 2007;69:1521–1527. doi: 10.1212/01.wnl.0000277459.83543.99. [DOI] [PubMed] [Google Scholar]
- 25.Grubman A., Chew G., Ouyang J.F., Sun G., Choo X.Y., McLean C., Simmons R.K., Buckberry S., Vargas-Landin D.B., Poppe D., et al. A single-cell atlas of entorhinal cortex from individuals with Alzheimer’s disease reveals cell-type-specific gene expression regulation. Nat. Neurosci. 2019;22:2087–2097. doi: 10.1038/s41593-019-0539-4. [DOI] [PubMed] [Google Scholar]
- 26.Jack C.R., Jr., Wiste H.J., Weigand S.D., Therneau T.M., Knopman D.S., Lowe V., Vemuri P., Mielke M.M., Roberts R.O., Machulda M.M., et al. Age-specific and sex-specific prevalence of cerebral β-amyloidosis, tauopathy, and neurodegeneration in cognitively unimpaired individuals aged 50–95 years: a cross-sectional study. Lancet Neurol. 2017;16:435–444. doi: 10.1016/S1474-4422(17)30077-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ingala S., De Boer C., Masselink L.A., Vergari I., Lorenzini L., Blennow K., Chételat G., Di Perri C., Ewers M., van der Flier W.M., et al. Application of the ATN classification scheme in a population without dementia: Findings from the EPAD cohort. Alzheimers Dement. 2021;17:1189–1204. doi: 10.1002/alz.12292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Planche V., Bouteloup V., Mangin J.-F., Dubois B., Delrieu J., Pasquier F., Blanc F., Paquet C., Hanon O., Gabelle A., et al. Clinical relevance of brain atrophy subtypes categorization in memory clinics. Alzheimers Dement. 2021;17:641–652. doi: 10.1002/alz.12231. [DOI] [PubMed] [Google Scholar]
- 29.Ossenkoppele R., Schonhaut D.R., Schöll M., Lockhart S.N., Ayakta N., Baker S.L., O’Neil J.P., Janabi M., Lazaris A., Cantwell A., et al. Tau PET patterns mirror clinical and neuroanatomical variability in Alzheimer’s disease. Brain. 2016;139:1551–1567. doi: 10.1093/brain/aww027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petersen C., Nolan A.L., de Paula França Resende E., Miller Z., Ehrenberg A.J., Gorno-Tempini M.L., Rosen H.J., Kramer J.H., Spina S., Rabinovici G.D., et al. Alzheimer’s disease clinical variants show distinct regional patterns of neurofibrillary tangle accumulation. Acta Neuropathol. 2019;138:597–612. doi: 10.1007/s00401-019-02036-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang H., Oh C.-K., Amal H., Wishnok J.S., Lewis S., Schahrer E., Trudler D., Nakamura T., Tannenbaum S.R., Lipton S.A. Mechanistic insight into female predominance in Alzheimer’s disease based on aberrant protein S-nitrosylation of C3. Sci. Adv. 2022;8 doi: 10.1126/sciadv.ade07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma J., Zhang H., Liang F., Li G., Pang X., Zhao R., Wang J., Chang X., Guo J., Zhang W. The male-to-female ratio in late-onset multiple acyl-CoA dehydrogenase deficiency: a systematic review and meta-analysis. Orphanet J. Rare Dis. 2024;19:72. doi: 10.1186/s13023-024-03072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paul B.D. DUB’ling down uncovers an X-linked vulnerability in Alzheimer’s disease. Cell. 2022;185:3854–3856. doi: 10.1016/j.cell.2022.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buckley R.F., Scott M.R., Jacobs H.I.L., Schultz A.P., Properzi M.J., Amariglio R.E., Hohman T.J., Mayblyum D.V., Rubinstein Z.B., Manning L., et al. Sex mediates relationships between regional tau pathology and cognitive decline. Ann. Neurol. 2020;88:921–932. doi: 10.1002/ana.25878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Franjic D., Skarica M., Ma S., Arellano J.I., Tebbenkamp A.T.N., Choi J., Xu C., Li Q., Morozov Y.M., Andrijevic D., et al. Transcriptomic taxonomy and neurogenic trajectories of adult human, macaque, and pig hippocampal and entorhinal cells. Neuron. 2022;110:452–469.e14. doi: 10.1016/j.neuron.2021.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivashko-Pachima Y., Hadar A., Grigg I., Korenková V., Kapitansky O., Karmon G., Gershovits M., Sayas C.L., Kooy R.F., Attems J., et al. Discovery of autism/intellectual disability somatic mutations in Alzheimer’s brains: mutated ADNP cytoskeletal impairments and repair as a case study. Mol. Psychiatry. 2021;26:1619–1633. doi: 10.1038/s41380-019-0563-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johansson M., Stomrud E., Insel P.S., Leuzy A., Johansson P.M., Smith R., Ismail Z., Janelidze S., Palmqvist S., van Westen D., et al. Mild behavioral impairment and its relation to tau pathology in preclinical Alzheimer’s disease. Transl. Psychiatry. 2021;11:76. doi: 10.1038/s41398-021-01206-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheltens P., De Strooper B., Kivipelto M., Holstege H., Chételat G., Teunissen C.E., Cummings J., van der Flier W.M. Alzheimer’s disease. Lancet. 2021;397:1577–1590. doi: 10.1016/S0140-6736(20)32205-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferreira D., Verhagen C., Hernández-Cabrera J.A., Cavallin L., Guo C.-J., Ekman U., Muehlboeck J.-S., Simmons A., Barroso J., Wahlund L.-O., et al. Distinct subtypes of Alzheimer’s disease based on patterns of brain atrophy: longitudinal trajectories and clinical applications. Sci. Rep. 2017;7:46263. doi: 10.1038/srep46263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chu N.N., Gebre-Amlak H. Navigating Neuroimaging Datasets ADNI for Alzheimer’s Disease. IEEE Consumer Electron. Mag. 2021;10:61–63. doi: 10.1109/mce.2021.3056872. [DOI] [Google Scholar]
- 41.Hammers D.B., Kostadinova R., Unverzagt F.W., Apostolova L.G., Alzheimer’s Disease Neuroimaging Initiative∗ Assessing and validating reliable change across ADNI protocols. J. Clin. Exp. Neuropsychol. 2022;44:85–102. doi: 10.1080/13803395.2022.2082386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fowler C., Rainey-Smith S.R., Bird S., Bomke J., Bourgeat P., Brown B.M., Burnham S.C., Bush A.I., Chadunow C., Collins S., et al. Fifteen years of the Australian Imaging, biomarkers and lifestyle (AIBL) study: Progress and observations from 2,359 older adults spanning the spectrum from cognitive normality to Alzheimer’s disease. J. Alzheimers Dis. Rep. 2021;5:443–468. doi: 10.3233/ADR-210005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ellis K.A., Szoeke C., Bush A.I., Darby D., Graham P.L., Lautenschlager N.T., Macaulay S.L., Martins R.N., Maruff P., Masters C.L., et al. Rates of diagnostic transition and cognitive change at 18-month follow-up among 1,112 participants in the Australian Imaging, Biomarkers and Lifestyle Flagship Study of Ageing (AIBL) Int. Psychogeriatr. 2014;26:543–554. doi: 10.1017/S1041610213001956. [DOI] [PubMed] [Google Scholar]
- 44.Brown E.M., Pierce M.E., Clark D.C., Fischl B.R., Iglesias J.E., Milberg W.P., McGlinchey R.E., Salat D.H. Test-retest reliability of FreeSurfer automated hippocampal subfield segmentation within and across scanners. Neuroimage. 2020;210 doi: 10.1016/j.neuroimage.2020.116563. [DOI] [PubMed] [Google Scholar]
- 45.Hedges E.P., Dimitrov M., Zahid U., Brito Vega B., Si S., Dickson H., McGuire P., Williams S., Barker G.J., Kempton M.J. Reliability of structural MRI measurements: The effects of scan session, head tilt, inter-scan interval, acquisition sequence, FreeSurfer version and processing stream. Neuroimage. 2022;246 doi: 10.1016/j.neuroimage.2021.118751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chiappiniello A., Tarducci R., Muscio C., Bruzzone M.G., Bozzali M., Tiraboschi P., Nigri A., Ambrosi C., Chipi E., Ferraro S., et al. Automatic multispectral MRI segmentation of human hippocampal subfields: an evaluation of multicentric test-retest reproducibility. Brain Struct. Funct. 2021;226:137–150. doi: 10.1007/s00429-020-02172-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evans A.C., Janke A.L., Collins D.L., Baillet S. Brain templates and atlases. Neuroimage. 2012;62:911–922. doi: 10.1016/j.neuroimage.2012.01.024. [DOI] [PubMed] [Google Scholar]
- 48.Rao N.P., Jeelani H., Achalia R., Achalia G., Jacob A., Bharath R.D., Varambally S., Venkatasubramanian G., K. Yalavarthy P. Population differences in brain morphology: Need for population specific brain template. Psychiatry Res. Neuroimaging. 2017;265:1–8. doi: 10.1016/j.pscychresns.2017.03.018. [DOI] [PubMed] [Google Scholar]
- 49.Severson K.A., Chahine L.M., Smolensky L.A., Dhuliawala M., Frasier M., Ng K., Ghosh S., Hu J. Discovery of Parkinson’s disease states and disease progression modelling: a longitudinal data study using machine learning. Lancet Digit. Health. 2021;3:e555–e564. doi: 10.1016/S2589-7500(21)00101-1. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y., Li B., Luo X., Wang X. Personalized mobile targeting with user engagement stages: Combining a structural hidden Markov model and field experiment. Inf. Syst. Res. 2019;30:787–804. doi: 10.1287/isre.2018.0831. [DOI] [Google Scholar]
- 51.Moon T.K. The expectation-maximization algorithm. IEEE Signal Process. Mag. 1996;13:47–60. doi: 10.1109/79.543975. [DOI] [Google Scholar]
- 52.Janco N., Bendory T. An accelerated expectation-maximization algorithm for multi-reference alignment. IEEE Trans. Signal Process. 2022;70:3237–3248. doi: 10.1109/TSP.2022.3183344. [DOI] [Google Scholar]
- 53.Shi X. A method of optimizing network topology structure combining Viterbi algorithm and Bayesian algorithm. Wirel. Commun. Mob. Comput. 2021;2021:1–12. doi: 10.1155/2021/5513349. [DOI] [Google Scholar]
- 54.Carrer L., Bruzzone L. Automatic enhancement and detection of layering in radar sounder data based on a local scale hidden Markov model and the Viterbi algorithm. IEEE Trans. Geosci. Remote Sens. 2017;55:962–977. doi: 10.1109/TGRS.2016.2616949. [DOI] [Google Scholar]
- 55.Bigler E.D., Skiles M., Wade B.S.C., Abildskov T.J., Tustison N.J., Scheibel R.S., Newsome M.R., Mayer A.R., Stone J.R., Taylor B.A., et al. FreeSurfer 5.3 versus 6.0: are volumes comparable? A Chronic Effects of Neurotrauma Consortium study. Brain Imaging Behav. 2020;14:1318–1327. doi: 10.1007/s11682-018-9994-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The data supporting the findings of this study were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) and the Australian Imaging Biomarkers and Lifestyle Study of Ageing (AIBL), which are available from the ADNI database (https://adni.loni.usc.edu) and AIBL database (https://aibl.csiro.au/) upon registration and compliance with the data use agreement.
-
•
The source code pertaining to both the personalized hidden Markov model and data analysis in this manuscript has been deposited on GitHub and is publicly available as of the date of publication; URLs are provided at https://github.com/ZJU-BMI/Personalized_HMM_disease_progression.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.