Abstract
Recently, mRNA has gained a lot of attention in the field of vaccines, gene therapy, and protein replacement therapies. Herein, we are demonstrating a comprehensive approach to designing, cloning, and characterizing an antigenic cassette for the development of mRNA vaccine for COVID-19. The gene encoding the antigenic spike protein of the SARS-CoV-2 Omicron variant (B.1.1.529) was designed using the databases, characterized by in-silico tools, and assembled using overlapping oligonucleotide-based assembly by PCR. Next, the gene was cloned, mRNA was synthesized, and characterized using orthogonal approaches (Capillary electrophoresis, Sanger DNA sequencing, Next-generation sequencing, HPLC, qPCR, etc.). Furthermore, the antigen expression was monitored in-vitro using an animal cell model by western blot, flow cytometer, and surface plasmon resonance. The demonstrated approach has also been followed for developing the mRNA vaccines for various other indications such as Malaria, Herpes, Dengue, HPV, etc.
Keywords: mRNA, Vaccine, SOE-PCR, SARS-CoV-2, Omicron
Highlights
-
•
Self-amplifying mRNA-based vaccines are the emerging field that has received considerable attention during COVID-19.
-
•
A comprehensive workflow to support mRNA vaccine development has been demonstrated.
-
•
The workflow includes designing, cloning, and characterizing the final product as mRNA.
-
•
The workflow uses in-silico tools, molecular biology techniques, and in-vitro approaches to confirm the correctness of the gene and conformationally active form of the encoded protein.
-
•
The scope of the demonstrated workflow is not only limited to the vaccines but other mRNA modalities including cell/gene therapy and protein replacement therapies.
1. Introduction
The recent success of the COVID-19 mRNA-guided vaccines has established the platform potential and become the foremost choice for the biotechnology industry. mRNA platform is a disease agonist platform where its potential can be harnessed for most of the modalities like vaccines, cell/gene therapy, and protein replacement therapy across various disease indications. mRNA vaccines launched like mRNA-1273, BNT 162b2, Gemcovac®-19, Gemcovac®-OM, and ARCT-154 against COVID-19 have extensively explored the non-amplifying mRNA (NAM) and self-amplifying mRNA (SAM) platforms [1]. COVID-19, a disease caused by the SARS-CoV-2 virus spread in late 2019 and arose as an emergency that halted the entire world. Today, the disease has been weakened but long-term COVID effects are still percolating. The virus is continuously changing its genotypic content as a result of the immune escape mechanism [2]. Most of the authorized vaccines are showing compromised efficacy on the emerging variants, thus the vaccine developers shall be cognizant of the rapidly evolving virus framework to provide a concrete solution.
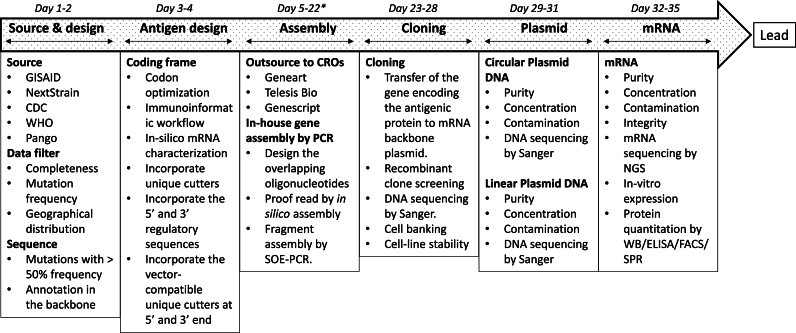
Herein, we demonstrate a comprehensive process flow/approach to assist the mRNA vaccine development program using COVID-19 as a model (Fig. 1). The study starts with the extraction and analysis of the metadata of SARS-CoV-2 infected patients from publicly available databases such as the GISAID, NextStrain, CDC, WHO, and Pango. Next, a potent antigenic sequence was designed from the metadata by an in-silico structure-guided approach that resulted in a few coding cassettes varying in their codon usage and mRNA secondary structure content. Subsequently, the overlapping oligonucleotides were designed for the gene assembly by PCR [3], cloned into a self-amplifying mRNA backbone plasmid, and confirmed through plasmid DNA sequencing. Further, the plasmid DNA was extracted and mRNA was synthesized, purified, and characterized. Each subsequent step of our comprehensive approach has various quality checks to minimize errors and deliver a potent antigen. The developed clones were characterized and stored following a 3-tier banking system (RCB, MCB, and WCB). The plasmid and mRNA were checked for various quality parameters by using orthogonal approaches to verify their identity, purity, and functionality. Later, mRNA-encoded antigenic protein expression was evaluated by an in-vitro cell-based assay using western blot, flow cytometry, and surface plasmon resonance. In the totality of the evidence or data, the demonstrated comprehensive approach will generate quantifiable datasets for antigenic cassette identification for vaccine development.
Fig. 1.
An integrated antigenic cassette identification workflow. The flow diagram shows the steps originating from extracting sequences from various databases, analyzing the sequences, and designing the codon-optimized sequence of the antigen. The critical attributes and the checkpoints at each step of the workflow like the assembly, cloning, plasmid, and mRNA preparation are shown. * denotes the 1.5 to 2 weeks of the oligonucleotide/gene synthesis, purification, and shipping from commercial suppliers.
2. Materials and methods
2.1. Database source and designing
The metadata of 109 SARS-CoV-2 (B.1.1.529) infected patients from September to December 2021 was downloaded from the GISAID database. Later, mutations occurring only in the spike region with a frequency of >50 % were filtered and annotated in the Wuhan-1 spike sequence of the virus (IVDC-HB-01/2019). The final mutated protein sequence was reverse translated and codon-optimized as per the human's codon usage by MacVector [4] [18.2.5(43)]. During the design, the ApaI and SacII restriction sites were ensured to be the non-cutters for the cloning purpose. The GC content and codon adaptation index (CAI) was calculated using the VectorBuilder tool (https://en.vectorbuilder.com/tool/codon-optimization.html). The target range of GC was kept at 30–70 % and CAI >0.8. Further, by employing an energy-minimization-based RNAfold web service, the stability of the mRNA was predicted through the analysis of the secondary structures of the self-amplifying backbone and antigenic spike encoding mRNA [5].
2.2. Antigen design and gene assembly
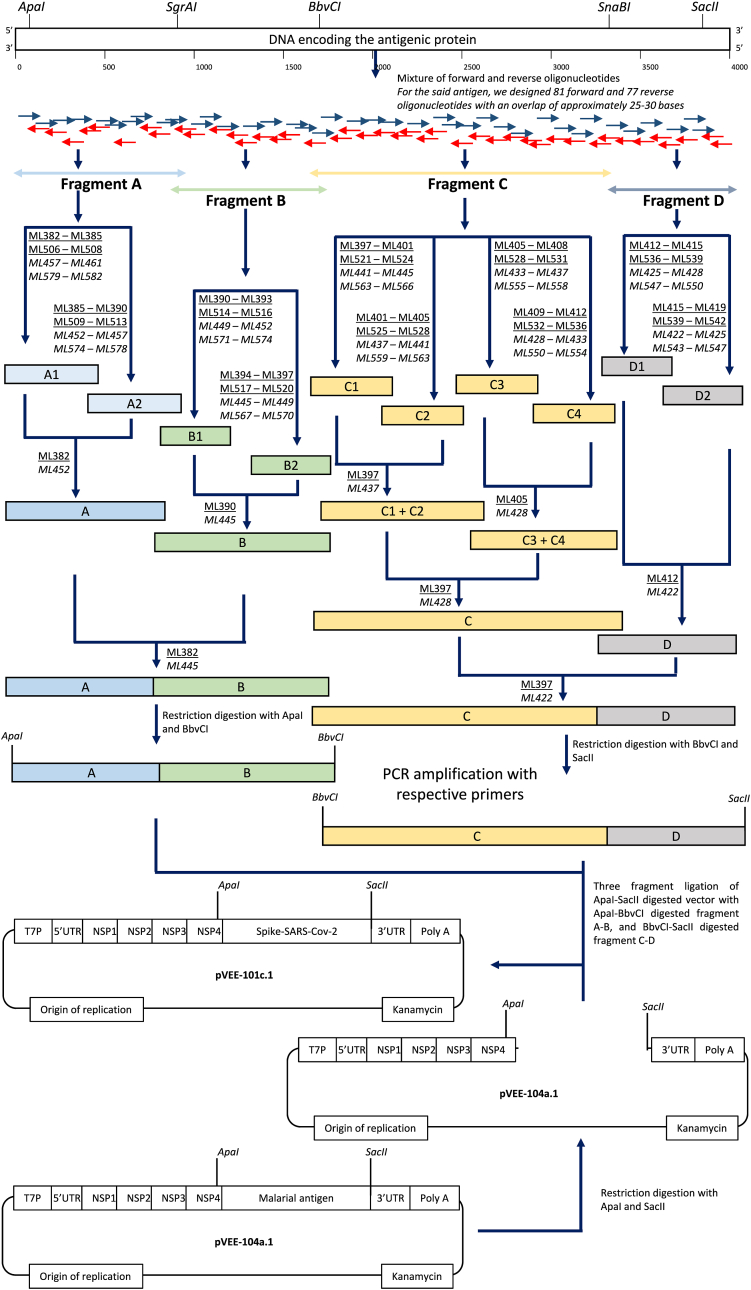
For the gene assembly, 158 overlapping oligonucleotides (Supplementary Table 1) were designed ranging from 60 to 120 bases in length with approximately 20–30 bases overlapping between each forward and reverse oligonucleotide. The repeat sequences were adjusted in the middle or towards the 3’ end of the oligonucleotides with overall GC content for each oligonucleotide between 45 and 60 %. All these oligonucleotides were synthesized from Sigma Aldrich at a 1 μM scale with PAGE purification. The detailed strategy for gene assembly and cloning is shown in (Fig. 2). For the assembly of the coding frame of approximately 4000 bp from ApaI to SacII, we marked fragment A from ApaI to SgrAI, fragment B from SgrAI to BbvCI, fragment C from BbvCI to SnaBI and fragment D from SnaBI to SacII. All the fragments were further split into sub-fragments to ensure error-free gene assembly. Fragment A was subdivided into A1 and A2; B into B1 and B2; C into C1, C2, C3, and C4; and D into D1 and D2. The boundaries of each of these fragments and sub-fragments were designed to have 25–30 bp overlap with their next-in-line considered sub-fragments and fragments. First, all the aforementioned sub-fragments were amplified and later assembled to generate final fragments (Supplementary Table 3). The final assembly resulted in two fragments AB and CD flanked by ApaI – BbvCI and BbvCI – SacII, respectively (Fig. 2). For PCR-based assembly, the oligonucleotide concentrations (Inner-20 nM with Outer-100 nM; Inner-50 nM with Outer-100 nM; and Inner-200 nM with Outer-400 nM) and annealing temperature (59 °C, 62 °C, 64 °C, 66 °C, 68 °C, 70 °C, and 72 °C) for the synthesis of sub-fragments of A and fragment A were optimized. We used Advantage® 2 DNA polymerase (Takara, Cat. 639206) for all kinds of PCR amplification that possess substantial proofreading activity. The PCR conditions were, initial denaturation at 95 °C for 4 min; followed by 16 cycles at 95 °C for 45 s; annealing at 59 °C, 62 °C, 64 °C, 66 °C, 68 °C, 70 °C, and 72 °C for 1 min; extension at 68 °C for 1 min and final extension at 68 °C for 2 min. The higher annealing temperature was chosen based on the properties of long oligonucleotides and 2-step PCR amplification, if required. For the assembly of A1 and A2 to generate an A fragment, equal volumes of both the PCR products were used in the presence of ML382 and ML452 oligonucleotides at 200 nM concentration. The PCR cycle conditions were similar as detailed above. Similarly, all the remaining sub-fragments and fragments were synthesized and assembled to generate fragments B (ML390 and ML445 oligonucleotides), C (ML397 and ML428 oligonucleotides), and D (ML412 and ML422 oligonucleotides). For the assembly of the A and B fragments, the equimolar concentration of A and B was assembled by PCR using ML382 and ML445 oligonucleotides. Similarly, fragments C and D were assembled using ML397 and ML422 oligonucleotides. Oligonucleotide concentrations and PCR conditions were similar as detailed previously. The extension time for the assembly of AB and CD was increased to 3 min for 28 cycles. Next, PCR-purified fragment AB was digested using ApaI (NEB, Cat. R0114S) and BbvCI (NEB, Cat. R0601S), CD fragment was digested using, BbvCI (NEB, Cat. R0601S) and SacII (NEB, Cat. R01578), and pVEE 104a.1 plasmid DNA was digested using ApaI-SacII. The pVEE 104a.1 plasmid is 11720 base pairs in length and encodes the malarial antigenic protein, non-structural proteins (nsp1-4) from Venezuelan Encephalitis alpha-virus, nptII gene, and pMB1 plasmid origin. These three fragments were ligated by T4 DNA ligase (Roche, Cat. 10481220001) and electroporated into E. coli DH5α cells (ThermoFisher Scientific, Cat. 11319019). The recombinants were selected on the LB agar plate containing 50 μg/mL kanamycin. Recombinant clone screening was performed by colony PCR using plasmid backbone-specific primers in the presence of AmpliTaq Gold DNA polymerase (Applied Biosystem™, Cat. 4311806). Colony PCR-positive clones were used for the plasmid DNA isolation (PureYield™, Promega miniprep system, Cat. A1223), and sequenced (Applied Bio-systems 3500xL) using BigDye™ Terminator v3.1 Cycle Sequencing Kit (ThermoFisher Scientific, Cat. 4337455). The sequencing data was analyzed on the MacVector software with an assembler [18.2.5(43)]. The sequencing primers have been listed in Supplementary Table 2.
Fig. 2.
Strategy for the assembly and cloning of the antigenic gene into mRNA backbone plasmid. The DNA encoding the antigenic protein was designed based on the metadata of B.1.1.529 infected humans. The unique restriction sites at the 5′ and 3′ end of the coding gene and a few unique sites were incorporated in-between regions to facilitate the cloning. A total of 158 oligonucleotides that included 81 forward and 77 reverse oligonucleotides were designed and used for the assembly of the entire DNA. For the PCR-based assembly of the final DNA, fragments A, B, C, and D were annotated, and they were flanked by unique restriction sites. Fragment A was assembled using sub-fragments A1, and A2. Fragment B was assembled using sub-fragments B1, and B2. Fragment C1–C2 was assembled using sub-fragments C1, and C2. Fragment C3–C4 was assembled using sub-fragments C3, and C4. Fragment C was finally assembled using sub-fragments C1–C2, and C3–C4. Fragment D was assembled using sub-fragments D1, and D2. Finally, AB was assembled using A and B, and similarly, CD using C and D fragments. For the cloning of the synthesized fragment AB and CD into the pVEE-104a.1 plasmid backbone, each was digested with their respective restriction enzyme cutters and finally ligated by three-fragment ligation to create pVEE-101c.1. Oligonucleotides used here are given the “ML” prefix followed by the numerical numbers. The oligonucleotides mentioned in the underlined text are the forward and those which are in italics are the reverse oligonucleotides. The single pair of primers that are used for the final assembly is mentioned for each of the sub-fragments and fragments. The unique restriction sites are also mentioned flanking the fragments.
2.3. Plasmid DNA
The large-scale circular plasmid DNA was isolated using the QIAGEN Plasmid Giga Kit (Qiagen, Cat. 12191). The circular plasmid was linearized and purified by a QIAquick PCR Purification Kit (Qiagen, Cat. 28104). The plasmid DNA was visualized by agarose gel electrophoresis and quantified using Quant-iT™ PicoGreen™ dsDNA Assay Kit (ThermoFisher Scientific Inc., Cat. P7589). Further, quantitative PCR (qPCR) was performed to detect the presence of any residual E. coli genomic DNA. qPCR was performed using SsoFast™ EvaGreen® Supermix (Biorad, Cat. 1725201) using forward (5′-AAGCTGCCTGCACTAATGTTCC-3′) and reverse (5′-TCGCGTACCGTCTTCATGG-3′) primers. Genomic DNA of E. coli DH5α was used as standard in the concentration range from 50.0 ng to 5.0 fg. The PCR was performed at 98 °C for 2 min followed by 39 cycles at 98 °C for 5 s, 61.5 °C for 5 s, one cycle at 95 °C for 10 s, and a melting curve at 65 °C to 95 °C (BioRad, CFX96). The plasmid DNA preparation was also sequence verified as detailed previously and it was named pVEE 101c.1.
2.4. mRNA drug product
For mRNA preparation, in-vitro transcription was carried out using HiScribe™ T7 High Yield RNA Synthesis Kit (NEB, Cat. E2040S), DNA was degraded with DNase I enzyme (ThermoFisher Scientific, Cat. EN0525), and mRNA was capped using the Vaccinia Capping System (NEB, Cat. M2080S). The mRNA was finally purified using LiCl precipitation and quantified by Quant-it™ RiboGreen RNA Assay Kit (ThermoFisher Scientific, Cat. R11490). mRNA integrity was checked by denaturing agarose gel electrophoresis and capillary electrophoresis on an Agilent 5200 Fragment analyzer (Agilent, Cat. DNF-471 RNA kit_15 nt). The plasmid DNA contamination was checked by qPCR. For this, we designed forward (5′-AACGGCTCGTAACATAGG-3′) and reverse (5′-TGGTCGAGCCAACAGAG-3′) primers from the nsp 1 region of the pVEE101c.1 plasmid. qPCR was performed using SsoFast™ EvaGreen® Supermix as detailed previously. The nucleotide sequence and poly-A tail length were checked on the Illumia Miniseq platform. For the sequencing process, mRNA was quantified using Qubit™ RNA Broad Range (BR), assay Kits (ThermoFisher Scientific, Cat. Q10210) on Qubit 4 Fluorometer (ThermoFisher Scientific). TruSeq® Stranded mRNA Library Prep (48 Samples) was used for the mRNA library preparation (Illumina, Cat. 20020594) along with the TruSeq RNA CD Index Plate (96 Indexes, 96 Samples, Illumina, Cat. 20019792). This library was run using MiniSeq High Output Reagent Kit (300-cycles) (Illumina, Cat. FC-420-1003). The quality analysis of the raw sequencing reads was performed using the FASTQc toolkit. Next, the low-quality reads were trimmed using Trimmomatic-0.39 [6]. The parameters selected for trimming were LEADING:5, TRAILING:5, SLIDINGWINDOW:4:15, and MINLEN:36 [7]. The PCR duplicates present in the reads were removed by employing the Picard tool. The reads were mapped using Bowtie2 aligner (v.2.4.5) (Langmead & Salzberg, 2012), and the generated SAM file was converted to a BAM file [8]. The Bam files were visualized on the Integrated Genomics Viewer (IGV) software (v.2.8.10) [9,10] and sequence alignment was performed using the Nucleotide Basic Local Alignment Search Tool (BLASTn).
2.5. In-vitro mRNA expression
HEK 293T cells (ATCC, Cat. CRL-3216) were cultured in a humidified incubator at 37 °C with 5 % CO2 and seeded at the density of 2.5 × 105 cells/ml. The next day, mRNA (10 μg) was diluted in Opti-MEM (Gibco™, Cat. 31985070) and Lipofectamine 3000 transfection reagent (Invitrogen™, Cat. L3000075) in a ratio of (1:5) and incubated for 15 min and added to the cells. For the western blot, the lysates were prepared using SDS lysis buffer (SDS 4 %, 0.004 Bromophenol blue, 0.125 M Tris HCL, pH 6.8). The expressed protein was detected by anti-SARS-CoV-2 spike mouse monoclonal IgG (R&D Systems, Cat. MAB10557) followed by HRP-conjugated goat anti-mouse IgG (Millipore, Cat. 31430). For FACS, the cells were detached using a cell dissociation buffer (Gibco™, Cat. 13151–014). Next, the harvested cells were washed thrice with ice-chilled PBS with 5 % MACS BSA (Miltenyl Biotech, Cat. 130-091-376, FACS buffer) and blocked with Fc-blocker (BD Pharmingen, Cat. 564219). The expressed protein on the cell surface was detected using fluorophore–conjugated anti-SARS-CoV-2 spike IgG (R&D Biosystem, Cat. FAB105403 N) on BD FACS Lyric™. The assay was performed thrice and data analysis was done using the FlowJo (v10) single-cell analysis software. For SPR analysis, the lysates were prepared using RIPA buffer (Sigma Aldrich, Cat. R0278) with 2 mM PMSF. Receptor protein ACE-2 (Sigma Aldrich, Cat. SAE0064) was immobilized on the CM5 chip surface and the lysate was passed at a flow rate of 30 μl/min for 300 s followed by dissociation for 300 s on Biacore 8K+ (Cytiva). The assay was performed in triplicates.
3. Results and discussion
The COVID-19 pandemic wave reversal by efficacious vaccination across the globe has brought day-to-day life back to normal. Throughout the COVID-19 journey, various healthcare modalities have been explored in detail and many of them have provided insight and potential for their future inclination. mRNA platform has come up as one such modality that has gained tremendous attention in the field of vaccinology. The development path, manufacturing process, quality testing, and regulatory guidance are evolving frequently. We have been working on the mRNA platform from last many years and developed a comprehensive workflow starting from the antigen database to candidate identification. This workflow has helped us in identifying the mRNA lead candidate for various infectious diseases. Our two EUA-granted mRNA vaccines: Gemcovac®-19, an mRNA vaccine for COVID-19, and Gemcovac®-OM, Omicron specific mRNA vaccine for COVID-19; have been developed following a similar approach. Herein, we demonstrated a comprehensive workflow of antigenic cassette design, cloning, and characterization for the development of the mRNA vaccine candidate (Fig. 1).
3.1. Antigen design and cloning
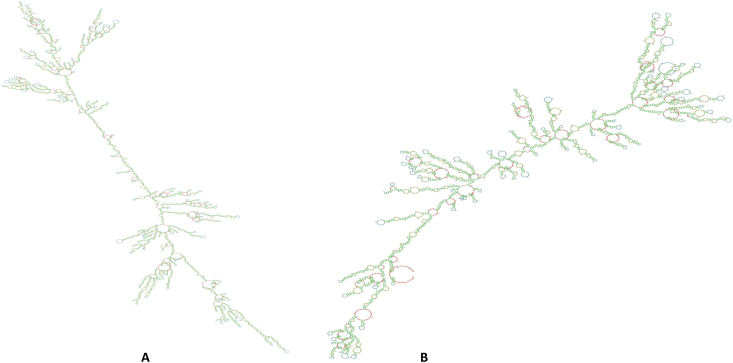
The antigenic cassette screening starts with the SARS-CoV-2 metadata analysis from the publicly available database, GISAID. The analysis identified 30 point mutations, 6 deletions, and 3 insertions in the spike region of the Omicron variant (B.1.1529) of SARS-CoV-2 in comparison to the Wuhan-1 spike protein [7]. These mutations were used to design the antigenic mRNA vaccine cassette encoding the spike protein of Omicron – (B.1.1529). The designed sequence has been submitted to the DDBJ database, accession number - LC731729. Codon optimization was performed for the antigenic protein, wherein GC% was kept at 45.85 % and CAI at 0.54. CAI is used for evaluating highly expressed genes, closer the CAI value to 1, the probability of gene expression will be higher [11]. However, we took a conservative approach to codon optimization and harmonization by keeping the CAI closer to the native sequences. The stability of mRNA was assessed for the self-amplifying backbone sequence and antigenic spike encoding mRNA by calculating free energy minimization using the RNAfold web server. After constructing secondary structures for mRNA (Fig. 3), the minimum free energy of their thermodynamic ensembles was found to be −2394.80 kcal/mol and −1044.04 for the self-amplifying backbone sequence and antigenic spike encoding mRNA, respectively. The higher stability of the predicted structure was indicated by a larger negative energy value.
Fig. 3.
Secondary structure prediction.
(A) self-amplifying backbone.
(B) antigenic spike encoding.
Green: Stems (canonical helices), Red: Multiloops (junctions), Yellow: Interior Loops, Blue: Hairpin loops, Orange: 5′ and 3′ unpaired region.
The designed gene cassette can be outsourced for its synthesis or assembled in-house using oligonucleotide-based gene assembly [8]. Here, we have shown the in-house gene assembly by PCR. We designed 158 overlapping oligonucleotides covering both strands of the gene cassette. The full-length gene was assembled by SOE-PCR as detailed in (Fig. 2). Briefly, we segmented the entire cassette of approximately 4000 base pairs into four fragments (A-D). We optimized various oligonucleotide concentrations along with the annealing temperature gradient for the synthesis of sub-fragments of A and fragment A. Finally, 200 nM inner with 400 nM outer oligonucleotide concentration at 70 °C annealing temperature was selected for the assembly of remaining sub-fragments and fragments. The desired size of each of the sub-fragments and fragments is tabulated in Supplementary Table 3. Next, the two insert fragments (AB and CD) and vector backbone of pVEE 104a.1 were ligated and electroporated into E. coli. DH5α cells. Recombinant clones were confirmed by colony PCR, restriction digestion, and plasmid DNA sequencing.
3.2. Plasmid DNA and mRNA drug substance
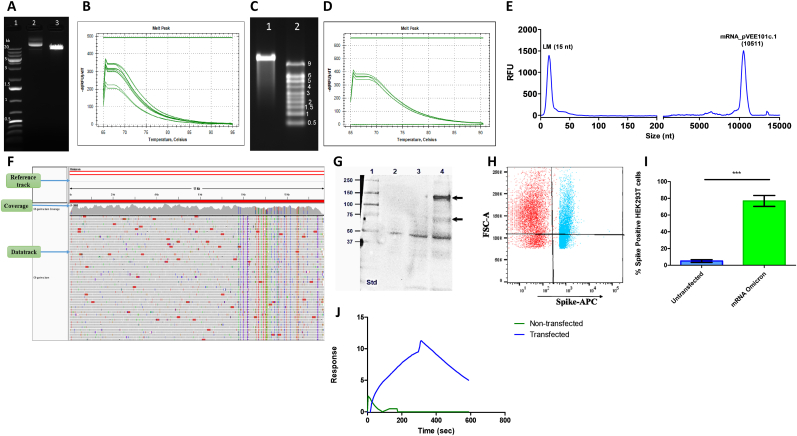
mRNA synthesis is an enzymatic cell-free process that requires linearized plasmid DNA and DNA-dependent RNA polymerase. The identity and purity of the plasmid DNA are important determinants of the process's success. Plasmid DNA is mostly isolated from the E. coli host and is linearized by enzymatic methods. Alternatively, DNA templates are also generated by PCR-based approaches [9] and enzymatic methods such as doggy bone DNA [10]. Herein, we have isolated the circular plasmid DNA from the E. coli host followed by its linearization. The plasmid DNAs were isolated from the cell banks that have been previously confirmed and characterized. We checked the circular and linearized plasmid DNA quality by agarose gel electrophoresis, qPCR for genomic DNA contamination, and DNA sequencing. The agarose gel showed the band of desired quality (Fig. 4A), and qPCR confirmed less than 50 pg of genomic DNA contamination in 1 mg of purified pVEE 101c.1 plasmid (Fig. 4B), and complete identity with the reference sequence by plasmid DNA sequencing. Later, mRNA was synthesized by in-vitro transcription in the presence of T7-RNA polymerase followed by enzymatic capping and purification. The mRNA sample when loaded on the agarose gel showed a clear band at the desired position of approximately 11700 bases (Fig. 4C). Plasmid DNA presence in the mRNA was checked by qPCR and our results met the specification of less than 10 pg of plasmid DNA in 1 mg of purified mRNA (Fig. 4D). The mRNA integrity was checked by capillary electrophoresis on a fragment analyzer wherein a major peak was observed for the desired size confirming the mRNA integrity (Fig. 4E). Next Generation Sequencing (NGS) on the Illumina Miniseq platform determined the mRNA sequence of the entire cassette, including the poly-A tail. We obtained a total data output of 10.5 Gb, 282 K/mm2 cluster density, and a cluster passing filter of 79.70 %. The Q30 passed data (88.75 %) were processed with Trimmomatic and PICARD tools to remove the low-quality reads and PCR duplicates, respectively. The processed data were mapped using Bowtie2 and visualized on the IGV. The consensus sequence was found to contain a complete match with the reference sequence file (Fig. 4F). Furthermore, we checked the mRNA expression using an in-vitro HEK 293T cell-based assay, wherein the expressed protein was detected by western blot (Fig. 4G). We also confirmed the surface expression of protein by FACS (Fig. 4H and I) and functional or receptor binding capability by SPR assay (Fig. 4J).
Fig. 4.
Plasmid DNA and mRNA characterization.
(A) Ethidium bromide-stained agarose gel image of plasmid DNA pVEE 101c.1, Lane:1 1 kb plus DNA ladder. Lane 2: pVEE 101c.1 uncut, Lane 3: pVEE 101c.1 linearized plasmid.
(B) Melt peaks of the pVEE 101c.1 plasmid DNA sample from qPCR reaction.
(C) Ethidium bromide stained MOPS gel image of mRNA. Lane 1: mRNA encoded by pVEE 101c.1, Lane 2: RNA Millennium 9 kb ladder.
(D) Melt peaks of mRNA sample on qPCR for the detection of pVEE101c.1 plasmid DNA contamination.
(E) Capillary electrophoresis profile of mRNA on 5200 fragment analyzer system.
(F) Snapshot of NGS data alignment of mRNA with the reference sequence file.
(G) The whole cell extract of mRNA transfected and non-transfected HEK293T was probed using anti-SARS-CoV-2 spike mouse monoclonal IgG and detected using HRP-conjugated goat anti-mouse IgG, lane 1: marker, lane 2. non-transfected, lane 3: mRNA from empty vector (without antigenic gene), and lane 4: mRNA encoding Omicron spike.
(H) Shows the scatter plot of FACS for non-transfected and mRNA-transfected HEK293T cells.
(I) The % of spike-positive HEK 293T cells is plotted as an average of three independent sets of experiments for both transfected and un-transfected cells. The error bar represents the standard deviation. ***p < 0.05
(J) The whole cell extract of transfected and non-transfected cells was analyzed on Biacore 8K + by immobilizing ACE-2 on the CM5 chip. Shown is one of the representative sensorgrams of the transfected and non-transfected cell lysate.
The blot detected using anti-spike IgG confirmed the presence of full-length spike protein at approximately 140 kDa in the samples from transfected cells, however, there was no band observed in the un-transfected cells (Fig. 4G). The surface expression was confirmed using FACS and the analysis showed that 77 ± 6.50 % of total HEK 293T cells were positive for anti-spike IgG staining as compared to the non-transfected cells (Fig. 4H and I). Next, the binding kinetics of transfected whole-cell extract with the surface-immobilized ACE-2 receptor showed significant binding of 10 ± 2.75 RU in comparison to cell extract of un-transfected samples using SPR (Fig. 4J). These assays have also established the fact that mRNA-encoded protein was full-length and functionally active. In the totality of the evidence or quantifiable datasets, our comprehensive approach as described in our workflow (Fig. 1) is capable enough of finding the best cassette of the mRNA to be used for the downstream applications. We have also developed an in-silico structure-guided immune-informatics workflow for designing a potent antigen sequence for robust humoral and cellular immunological responses [7].
4. Conclusion
We demonstrated a comprehensive integrated workflow for the identification of antigenic cassettes starting from database search to antigen expression. The workflow has checkpoints at each of the subsequent steps to ensure the timely, error-free selection of the most potent antigenic cassette. The dataset obtained from the approach can also be used to rank the tested candidates for their downstream suitability. The scope of the workflow is not just limited to vaccine development but to other modalities of mRNA-guided therapies as well.
Funding information
We thank the Department of Biotechnology of India (BT/COVID0041/01/20) for partial financial support.
Competing interests
The authors declare no competing interests.
Data availability
All data are included in the manuscript as figures, tables, and supplementary material.
CRediT authorship contribution statement
Renuka Khanzode: Conceptualization, Methodology, Writing – original draft. Garima Soni: Conceptualization, Methodology, Writing – original draft. Shalini Srivastava: Conceptualization, Methodology, Writing – original draft, Writing – review & editing. Sharad Pawar: Methodology. Rucha Wadapurkar: Data curation, Methodology, Software, Writing – original draft. Ajay Singh: Conceptualization, Investigation, Methodology, Writing – original draft, Writing – review & editing.
Declaration of competing interest
All authors, Garima Soni, Renuka Khanzode, Shalini Srivastava, Sharad Pawar, Rucha Wadapurkar, and Ajay Singh have declared that the experimental work and data in this manuscript have not been previously published elsewhere. All authors declare no conflict of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2024.101766.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Deo S., Desai K., Patare A., Wadapurkar R., Rade S., Mahudkar S., Sathe M., Srivastava S., Prasanna P., Singh A. Evaluation of self-amplifying mRNA platform for protein expression and genetic stability: implication for mRNA therapies. Biochem. Biophys. Res. Commun. 2023;680:108–118. doi: 10.1016/j.bbrc.2023.09.016. [DOI] [PubMed] [Google Scholar]
- 2.de Souza G.A.P., Le Bideau M., Boschi C., Ferreira L., Wurtz N., Devaux C., Colson P., La Scola B. Emerging sars-cov-2 genotypes show different replication patterns in human pulmonary and intestinal epithelial cells. Viruses. 2022;14 doi: 10.3390/v14010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rouillard J.-M., Lee W., Truan G., Gao X., Zhou X., Gulari E. Gene2Oligo: oligonucleotide design for in vitro gene synthesis. Nucleic Acids Res. 2004;32:W176–W180. doi: 10.1093/nar/gkh401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Athey J., Alexaki A., Osipova E., Rostovtsev A., Santana-Quintero L.V., Katneni U., Simonyan V., Kimchi-Sarfaty C. A new and updated resource for codon usage tables. BMC Bioinf. 2017;18 doi: 10.1186/s12859-017-1793-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samavarchi Tehrani S., Gharibi S., Movahedpour A., Goodarzi G., Jamali Z., Khatami S.H., Maniati M., Ranjbar M., Shabaninejad Z., Savardashtaki A., Taheri-Anganeh M. Design and evaluation of scFv-RTX-A as a novel immunotoxin for breast cancer treatment: an in silico approach. J. Immunoassay Immunochem. 2021;42:19–33. doi: 10.1080/15321819.2020.1812640. [DOI] [PubMed] [Google Scholar]
- 6.Wu Z.-W., Mou Q., Fang T., Wang Y., Liang H., Wang C., Du Z.-Q., Yang C.-X. Global 3’-untranslated region landscape mediated by alternative polyadenylation during meiotic maturation of pig oocytes. Reprod. Domest. Anim. 2022;57:33–44. doi: 10.1111/rda.14026. [DOI] [PubMed] [Google Scholar]
- 7.Wadapurkar R., Singh S., Singh A. Leveraging the immunoinformatics approach for designing the SARS-CoV-2 omicron-specific antigenic cassette of mRNA vaccine. Vaccine. 2024 doi: 10.1016/j.vaccine.2024.01.087. [DOI] [PubMed] [Google Scholar]
- 8.Hoose A., Vellacott R., Storch M., Freemont P.S., Ryadnov M.G. DNA synthesis technologies to close the gene writing gap. Nat. Rev. Chem. 2023;7:144–161. doi: 10.1038/s41570-022-00456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henderson J.M., Ujita A., Hill E., Yousif-Rosales S., Smith C., Ko N., McReynolds T., Cabral C.R., Escamilla-Powers J.R., Houston M.E. Cap 1 messenger RNA synthesis with Co-transcriptional CleanCap® analog by in vitro transcription. Curr. Protoc. 2021;1 doi: 10.1002/cpz1.39. [DOI] [PubMed] [Google Scholar]
- 10.Walters A.A., Kinnear E., Shattock R.J., Mcdonald J.U., Caproni L.J., Porter N., Tregoning J.S. Comparative analysis of enzymatically produced novel linear DNA constructs with plasmids for use as DNA vaccines. Gene Ther. 2014;21:645–652. doi: 10.1038/gt.2014.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Puigbò P., Bravo I.G., Garcia-Vallve S. CAIcal: a combined set of tools to assess codon usage adaptation. Biol. Direct. 2008;3 doi: 10.1186/1745-6150-3-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the manuscript as figures, tables, and supplementary material.