Abstract
In the immediate-early phase of reactivation or primary infection, herpesviruses express a small number of genes without requiring prior viral protein synthesis. Immediate-early genes usually encode regulatory proteins critical for the viral life cycle. Kaposi’s sarcoma-associated herpesvirus (KSHV) gene transcription in the immediate-early stage of viral reactivation was examined by using a chemical induction combined with a gene expression screening method. RNA transcripts from at least four KSHV genomic loci accumulate when latently infected B-lymphoma cells are induced for reactivation in the presence of an inhibitor of protein synthesis (cycloheximide) and thus represent immediate-early class transcripts. Among them, a 3.6-kb mRNA encodes three putative open reading frames (ORFs), namely, ORF50, K8, and K8.2. ORF50 is a homologue of Rta, a transcription activator encoded by Epstein-Barr virus (EBV). The K8 gene codes for a 237-amino-acid protein with a basic-leucine zipper domain near its C terminus and an acidic domain near its N terminus and which closely resembles the ZEBRA protein of EBV and Jun/Fos family proteins. Other immediate-early mRNAs of KSHV include a 1.7-kb mRNA encoding ORF45, a 2.0-kb mRNA encoding ORF K4.2, and a 4.5-kb mRNA. Functional roles of products of these KSHV immediate-early transcripts remain to be studied.
Kaposi’s sarcoma-associated herpesvirus (KSHV) is a newly identified human herpesvirus (6). It is also designated human herpesvirus 8. Epidemiological studies of KSHV suggest that this virus is an etiologic agent of Kaposi’s sarcoma (KS), the most common AIDS-related malignancy. For instance, DNA sequences of this virus were consistently found in KS lesions of all epidemiological forms (i.e., classic, AIDS-associated, African endemic, and posttransplant KS). Furthermore, serological assays suggest that KSHV is not ubiquitous but is closely associated with those at risk for developing KS (reviewed in references 9 and 27). In addition, the KSHV genome is present in several other proliferative lesions including those of primary effusion lymphoma (also known as body cavity-based lymphoma) and multicentric Castleman’s disease (4, 28).
The complete nucleotide sequence of one KSHV isolate (BC-1) has been published (24). Based on sequence analysis, KSHV has been classified as a member of the gammaherpesvirus 2 subfamily, Rhodinovirus genus. Another member of this viral group found in primates is herpesvirus saimiri. KSHV is also closely related to Epstein-Barr virus (EBV). Gammaherpesviruses characteristically establish latent infections in lymphoid cells. In latently infected cells that contain a limited number of herpesvirus genomes, there is no infectious virus. Latent viral DNA expresses a limited number of genes, which are referred to as latent genes. In KSHV, four genes that code for v-cyclin, latency-associated nuclear antigen (LANA), v-FLIP, and kaposin have been identified as latent genes (11, 17, 20, 26). When latency is disrupted, the virus can switch to a lytic life cycle. The switch of KSHV in primary effusion lymphoma cells from latency to lytic replication can be induced by various chemicals, such as tetradecanoyl phorbol acetate (TPA) and n-butyrate (14, 21).
The switch between latency and lytic life cycle is a crucial event that allows for the propagation of viruses. Our interest in the switch mechanism of KSHV also derives from the notion that the diseases caused by KSHV may be associated with the reactivation of latent virus. First, unique among all human herpesviruses, KSHV encodes several cytokines and a cytokine receptor. These include genes homologous to interleukin-6 (IL-6), three beta-chemokines, and a CXC cytokine receptor (reviewed in reference 18). The presence of cytokine signaling genes in the KSHV genome is intriguing because the virus is closely associated with a disease (KS) which has long been known as a cytokine disorder (7, 8). Unlike tumor-associated genes in other DNA tumor viruses, such as those for EBV-encoded EBNA-2, EBNA-3, and LMPs, which are latent genes (12), the KSHV cytokine signaling genes are expressed after the virus is induced to enter a lytic cycle in primary effusion lymphoma cell lines (16, 34). This raises the possibility that the KSHV lytic life cycle may be responsible for KS pathogenesis. Second, because KS is an endothelial neoplasm and latent KSHV infection is established in lymphocytes well before the onset of KS, it has been proposed that reactivation of lytic KSHV infection from the latently infected lymphoid reservoir is a necessary antecedent step in KS development (12a). Thus, the switch of the virus from latent to lytic replication appears to be important not only for viral propagation but also for viral pathogenicity. However, very little is known about the nature of the switch.
Herpesvirus genes can be classified into four categories: latent genes, immediate-early (IE) genes, early genes, and late genes. IE genes are the first class of the viral genes expressed after primary infection or reactivation. As transcription of IE genes does not require prior viral protein synthesis, this class of genes is experimentally defined by their transcription following primary infection or reactivation in the presence of inhibitors of protein synthesis. IE genes usually encode regulatory proteins that alter the expression of viral and cellular genes during the course of infection or reactivation. Therefore, herpesvirus IE genes play crucial roles in the switch from latency to lytic life cycle. The purpose of the work described in this paper was to identify KSHV IE genes so that their roles in the regulation of KSHV switch from latency to lytic life cycle could be studied. Through the use of a gene expression screening method which was developed based on cDNA subtractive selection, several KSHV IE transcripts were identified and characterized. Identification of KSHV IE transcripts was the first step in efforts toward understanding mechanisms of the viral reactivation.
MATERIALS AND METHODS
Cell cultures and cosmid DNAs.
BC-1 (5) cells were purchased from the American Type Culture Collection and grown in RPMI 1640 medium (Gibco-BRL, Gaithersburg, Md.) supplemented with 15% fetal bovine serum (Gibco-BRL). BCBL-1 (21) cells were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program and grown in RPMI 1640 medium supplemented with 10% fetal bovine serum. All cultures contained penicillin-streptomycin (50 U/ml) and amphotericin B (Fungizone) (1.25 μg of amphotericin B per ml and 1.25 μg of sodium desoxycholate per ml).
Six KSHV cosmid clones, namely, GB11, GA29, GB22, GA1, GA2, and GB1, prepared from BC-1 cells, were kindly provided by Ren Sun and George Miller at Yale University.
Chemical induction.
BC-1 and BCBL-1 cells were induced with 3 mM sodium butyrate (Sigma, St. Louis, Mo.). When induction was accompanied by inhibition of protein synthesis, cycloheximide (Sigma) was added in the culture to 100 μg/ml 4 h prior to induction.
Subtractive cDNA cloning.
Total RNAs were purified with Trizol reagent (Gibco-BRL) from 108 BC-1 cells that had been treated with 3 mM sodium butyrate for 4 h in the presence of 100 μg of cycloheximide per ml, as well as from the same number of cells that had not been treated with sodium butyrate but had been incubated with cycloheximide for 8 h. Poly(A)+ RNAs were purified with the PolyAtract mRNA isolation system (Promega, Madison, Wis.). The double-stranded cDNAs were synthesized with the Universal riboclone cDNA synthesis system (Promega).
The cDNA subtraction procedure was established based on the work of Wang and Brown (31) and Patel and Sive (1) with modifications. The double-stranded cDNAs were digested completely with AluI and then ligated with 0.5 μg of double-stranded linker which was prepared by annealing oligodeoxyribonucleotide Ad-1A (5′ GATCCCAGTCACGACGAATTCC 3′) and phosphorylated Ad-1B (5′ pGGAATTCGTCGTGACTGG 3′). The ligated samples were loaded on a 1.4% low-melting-point agarose gel, and cDNA fragments in the size range of 0.2 to 1 kb were collected. Linker-ligated cDNA fragments were amplified by PCR with oligonucleotide Ad-1A as primer.
cDNA fragments prepared from the induced and uninduced cells were amplified by PCR and served as tracer and driver DNAs, respectively. The driver DNA was biotinylated during PCR by incorporating bio-11-dUTP (Enzo Diagnostics, Farmingdale, N.Y.), followed by a complete digestion with EcoRI to cleave the linker. Twenty micrograms of biotinylated driver cDNA(−) (cDNA prepared from noninduced BC-1 cells) was mixed with 1 to 1.5 μg of nonbiotinylated tracer cDNA(+) (cDNA prepared from butyrate-induced BC-1 cells) in 10 μl of H2O. The DNA mixture was heated at 100°C for 3 min, and then 10 μl of 2× hybridization buffer (1× buffer is 50 mM HEPES [pH 7.5], 0.2% sodium dodecyl sulfate, 2 mM EDTA, 500 mM NaCl) was added. The DNA solution was overlaid with mineral oil and incubated in a 68°C water bath for 20 h. The hybridization mixture was diluted with 80 μl of HE buffer (10 mM HEPES, 1 mM EDTA, pH 7.5) to bring the final NaCl concentration to 100 mM. Twenty microliters of streptavidin (2 mg/ml in 0.15 M NaCl, 10 mM HEPES, 1 mM EDTA, pH 7.5) was mixed with the hybridized DNA solution and incubated at room temperature for 10 min. The streptavidin and associated DNAs were then removed from the solution by extraction with an equal volume of phenol. Streptavidin-phenol extraction was repeated three times. The subtracted tracer cDNA(+) was mixed with 10 μg of biotinylated driver cDNA(−) and coprecipitated with ethanol. The DNAs were denatured and hybridized as described above, except that the hybridization was carried out for 2 h. Biotinylated DNAs were removed by using streptavidin-phenol extraction as described above. The enriched DNAs were amplified by PCR and used for the next cycle of subtractive hybridization.
After the seventh cycle of subtraction, enriched DNA was used to prepare 32P-labeled probe for Southern analysis. The DNA was also digested with EcoRI and cloned into pBluescript at the EcoRI site. Colonies were screened by hybridization with 32P-labeled KSHV genomic DNA fragments excised from six cosmids, and the clones that were hybridized detectably were picked up for miniassay and sequencing analysis.
Southern analysis.
Six KSHV cosmid DNAs were digested with EcoRI, BamHI, or both and separated in 0.8% agarose gels. DNAs were transferred onto a Nytran membrane (Schleicher & Schuell, Keene, N.H.) and probed with 32P-labeled probes. The probes were PCR-amplified total cDNA fragments prepared from sodium butyrate-induced and uninduced BC-1 cells in the presence of cycloheximide and the enriched cDNA fragments derived from the cDNA subtractive selection. These cDNAs were labeled by the random priming method with [α-32P]dCTP (Amersham, Arlington Heights, Ill.).
Northern analysis.
Total RNA was isolated from cells with Trizol reagent, and poly(A)+ mRNA was purified with the PolyAtract mRNA isolation system. The mRNA was separated by electrophoresis in a 1% agarose–6% formaldehyde gel in 20 mM morpholinepropanesulfonic acid (MOPS) buffer, pH 7.0. Each lane was loaded with mRNA from 2 × 107 cells. The RNA was transferred to a Nytran membrane and hybridized with a single-stranded 32P-labeled probe. Single-stranded DNA probes were prepared by asymmetric PCR with linearized plasmid templates and specific oligonucleotide primers, which were either a plasmid vector primer (i.e., KS or SK primers in pBluescript) or a primer specific to an insert sequence. The labeling reactions were performed in 15 μl of reaction solution (1× Taq polymerase buffer; 16.67 μM (each) dATP, dGTP, and dTTP; 1.67 μM dCTP; 5 μl of [α-32P]dCTP [800 Ci/mmol; 10 μCi/μl; Amersham]; 100 ng of DNA; 20 pmol of primer; 2.5 U of Taq polymerase). The PCR was initiated with a denaturing step of 2 min at 94°C, followed by 15 cycles of sequential steps of 1 min at 94°C, 1 min at 50°C, and 3 min at 74°C. Finally, the reaction was extended for 10 min at 74°C. RNA loading equivalence was controlled by probing with β-actin cDNA. An RNA ladder (0.24 to 9.5 kb; Gibco-BRL) was included in each agarose-formaldehyde gel and detected in Northern blots by hybridization with labeled λ DNA.
Cloning of IE full-length cDNAs.
The full-length cDNAs of IE mRNAs were generated by a PCR-based cDNA amplification strategy. Poly(A)+ RNA was isolated from BC-1 cells that had been treated with sodium butyrate for 4 h. Double-stranded cDNA was synthesized with avian myeloblastosis virus reverse transcriptase and cDNA synthesis primer [a modified lock-docking oligo(dT) primer; Clontech, Palo Alto, Calif.]. After ligation of the cDNAs with an adapter, the 5′ portion and 3′ portion of each cDNA were obtained by using the Marathon cDNA amplification kit (Clontech). Nested primers designed to clone open reading frame 50 (ORF50) transcript were ORF50-RACE1 (5′ CGACCACGACACCTGGTACCTCTTTGGG 3′, nucleotides 74178 to 74205), ORF50-RACE2 (5′ CATGTTTCAGGGCCCGCTTCGTCTAACA 3′, nucleotides 74358 to 74331), ORF50-RACE3 (5′ ATGCGCAGAGGCATCCCAAGGCATTATT 3′, nucleotides 72859 to 72886), and ORF50-RACE4 (5′ CAGCCCGGCGGTATCGTACGTGTTGTAG 3′, nucleotides 73117 to 73090). To obtain the 5′ rapid-amplification-of-cDNA-end fragment of ORF50 transcript, the cDNA pool was amplified first with ORF50-RACE2 and AP1 from the Marathon cDNA kit. The PCR products were then amplified with ORF50-RACE4 and AP2 from the kit. Similarly, the 3′ portion was obtained through two PCRs, ORF50-RACE3 and AP1 being used in the first reaction and ORF50-RACE1 and AP2 being used in the second. DNA fragments of 0.6 and 2.0 kb were obtained in the 5′ and 3′ rapid-amplification-of-cDNA-end reactions, respectively. The central portion was generated by PCR with ORF50-RACE2 and ORF50-RACE3. These three PCR products were cloned into T/A-type PCR cloning vectors (pCR2.1; Invitrogen, Carlsbad, Calif.) and sequenced.
The full-length cDNAs for ORF45 and ORF K4.2 mRNAs were obtained by the same strategy. Nested primers designed to clone these two transcripts were ORF45-RACE1 (5′ GGCGTCCATGGGATGGGTTAGTCAGGAT 3′, nucleotides 68097 to 68070), ORF45-RACE2 (5′ ACGTCCGGAGAGTTGGAACTGTCATCGC 3′, nucleotides 67813 to 67840), ORFK4.2-RACE1 (5′ CGACCTTTTCTGGGACCGCAAGTGGATT 3′, nucleotides 23029 to 23002), and ORFK4.2-RACE2 (5′ CAACTTGACACAGGGGAAACACCAGGGG 3′, nucleotides 22806 to 22833).
Reverse transcription-PCR (RT-PCR).
Poly(A)+ RNA was extracted from BC-1 cells 4 h postinduction with sodium butyrate. RT was performed at 42°C for 60 min with avian myeloblastosis virus reverse transcriptase (Boehringer-Mannheim, Mannheim, Germany) in a 20-μl reaction mixture containing 0.1 μg of poly(A)+ RNA and primed with oligo(dT). A 50-μl PCR mixture containing 20 μmol of primer ORF50-SEQ7 (5′ GCACTAAGGCCAAACAGGGCGCAGG 3′, nucleotides 75271 to 75295) and primer ORF50-SEQ5 (5′ TCGCCGCTAGGAAACATAGTTGTGC 3′, nucleotides 75687 to 75663) and 2.5 U of Taq DNA polymerase was added, and PCRs were carried out for 25 cycles (94°C for 30 s, 60°C for 30 s, and 72°C for 1 min). Amplified DNAs were separated on a 2% agarose gel.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this article have been submitted to GenBank. The cDNA sequences of ORF45, ORF K4.2, ORF50 type I, ORF50 type II, and ORF50 type III were assigned the accession no. AF091346, AF091347, AF091348, AF091349, and AF091350, respectively.
RESULTS
Experimental design.
The aim of this study was to identify IE transcripts of KSHV, which typically code for regulatory proteins. We initiated this study with two strains of KSHV that are harbored in BC-1 and BCBL-1 cell lines. The BC-1 cell line, established from an AIDS-related primary effusion lymphoma, carries two gammaherpesviruses, KSHV and EBV (5). In more than 98% of the cells, both viruses are latent. It was reported by Miller et al. (14) that the two viruses could be differentially induced to switch to their lytic cycles by chemicals. EBV can be induced by phorbol ester (TPA), and KSHV can be activated by sodium butyrate. BCBL-1 was also established from a primary effusion lymphoma but carries only KSHV. The latent virus in BCBL-1 cells can be induced to enter lytic replication with TPA and sodium butyrate (12a, 21). To detect KSHV IE transcripts, BC-1 and BCBL-1 cells were treated with TPA (20 ng/ml) or sodium butyrate (3 mM), respectively, in the presence of a protein synthesis inhibitor (cycloheximide) at a concentration of 50 or 100 μg/ml. The expression of a putative KSHV IE gene, ORF50, in BC-1 and BCBL-1 cells under the conditions mentioned above was measured by Northern analysis. KSHV ORF50 is the homologue of an EBV IE gene, BRLF1, which encodes a transcription activator. In agreement with the findings of Miller et al. (14) and Renne et al. (21), ORF50 mRNA was detected in sodium butyrate-induced BC-1 cells and TPA- or sodium butyrate-induced BCBL-1 cells in the absence of cycloheximide starting at 4 h postinduction. In the presence of cycloheximide at 50 or 100 μg/ml, ORF50 mRNA could be detected in the induced BC-1 cells; however, the RNA level was at 30 to 40% of its amount in the BC-1 cells induced in the absence of cycloheximide. In contrast, no mRNA could be detected in BCBL-1 cells in the presence of cycloheximide as judged by Northern analyses of KSHV mRNAs of different classes (LANA, ORF50, and ORF59) as well as cellular β-actin mRNA, indicating that BCBL-1 cells are very sensitive to the toxicity of cycloheximide. Northern analysis also revealed that a considerable polyadenylated level of KSHV lytic RNAs (such as ORF50, ORF59, and 1.1-kb nuclear RNA) can be detected in noninduced BCBL-1 cells, suggesting a relatively high degree of spontaneous reactivation of the virus in BCBL-1 cells. These two problems make it difficult to identify IE transcripts of KSHV in BCBL-1 cells.
The BC-1 cell line is ideal for studies of KSHV IE transcripts for the following reasons. (i) BC-1 cells exhibit less sensitivity to cycloheximide treatment. After 8 h of incubation in 100 μg of cycloheximide per ml, the BC-1 cell viability rate was 74% in comparison to the cell viability rate of 82% in the absence of cycloheximide. (ii) KSHV latency is under tight control in standard tissue culture conditions. (iii) The complete nucleotide sequence of the KSHV genome of this strain is available (24). However, BC-1 cells are dually infected by EBV and KSHV, and it was unclear if the presence of EBV affected KSHV gene expression. To clarify this issue, we compared the expression patterns of some KSHV genes of known classes in BC-1 and BCBL-1 cells by Northern analysis. The result showed that the transcription patterns of genes for latent LANA, IE ORF50, and delayed-early ORF59 were similar between BC-1 and BCBL-1 cells. The expression of these genes in both cell lines was consistent with their classification (data not shown, but similar data are shown in Fig. 3 and 4). In addition, EBV and KSHV coinfection occurs naturally in most primary effusion lymphomas and hence reflects the biological status of KSHV in vivo (25). Therefore, we used BC-1 cells to search for IE transcripts in reactivation of KSHV.
FIG. 3.
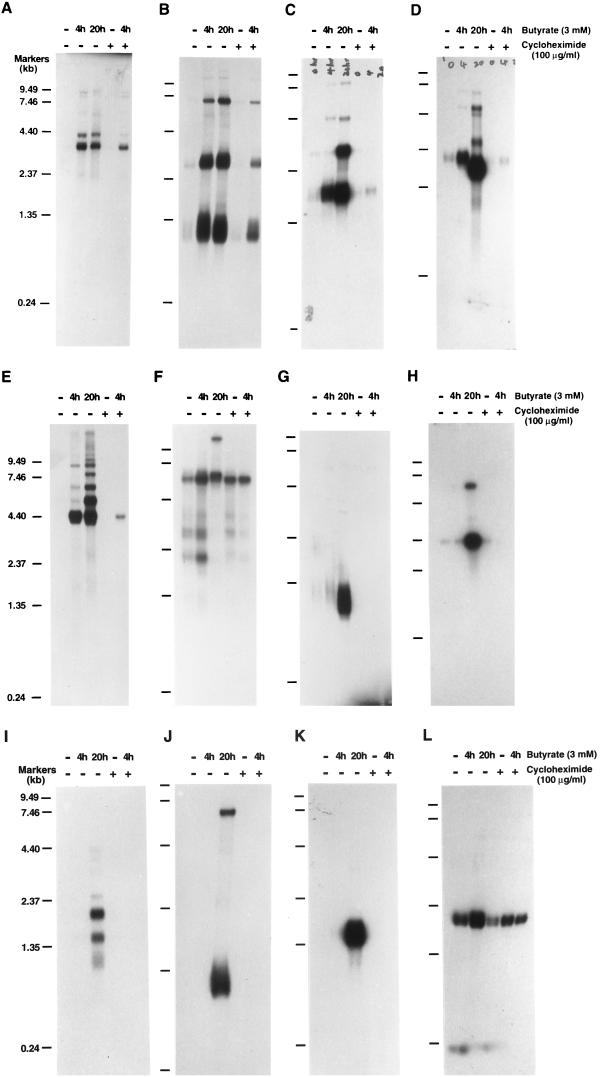
Northern analysis of KSHV IE mRNAs. Poly(A)+ RNA was isolated from nonstimulated BC-1 cells and BC-1 cells that had been treated with sodium butyrate for 4 or 20 h in the absence or presence of cycloheximide as indicated above each lane. RNA from BC-1 cells that were treated with cycloheximide for 8 h but not induced was also included. These RNA samples were separated on a 1.0% agarose-formaldehyde gel and transferred onto Nytran membranes. The membranes were probed with different 32P-labeled single-stranded DNA probes that were prepared by asymmetric PCR as described in Materials and Methods. The probes are as follows: single-stranded DNA complementary to nucleotides 73682 to 73956 in the KSHV genome within ORF50 (A); single-stranded DNA complementary to nucleotides 73956 to 73682, the antisense sequence of ORF50 (B); single-stranded DNA complementary to nucleotides 68502 to 68228 within ORF45 (C); single-stranded DNA complementary to nucleotides 22751 to 23073 within ORF K4.2 (D); single-stranded DNA complementary to nucleotides 50016 to 50261 (E); random priming-labeled ORF73 (LANA) cDNA (nucleotides 123809 to 127297) (F); random priming-labeled ORF K2 (vIL-6) cDNA (nucleotides 17261 to 17875) (G); random priming-labeled ORF59 cDNA (nucleotides 95549 to 96739) (H); random priming-labeled K3 cDNA (nucleotides 18608 to 19609) (I); random priming-labeled K5 cDNA (nucleotides 25713 to 26483) (J); random priming-labeled ORF57 cDNA (nucleotides 82717 to 83544) (K); and β-actin cDNA as a control for RNA integrity and loading (L). Molecular markers were an 0.24- to 9.5-kb RNA ladder.
FIG. 4.
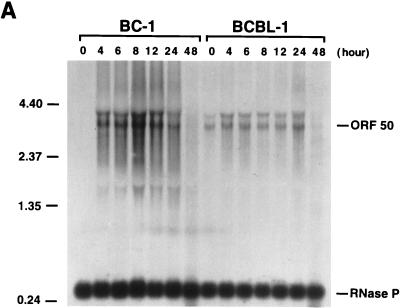
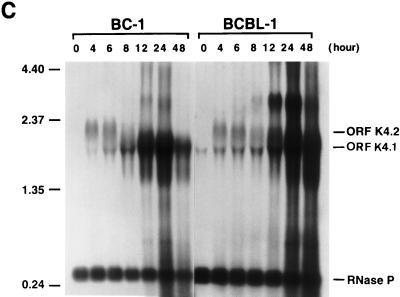
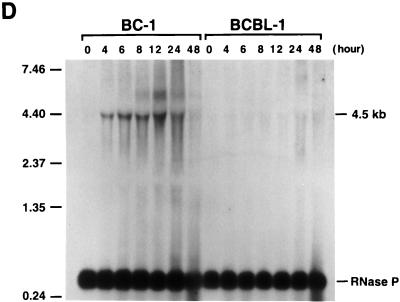
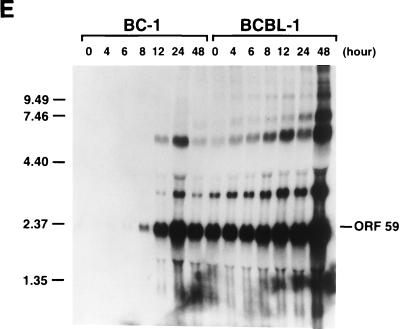
Kinetics of the expression of KSHV IE mRNAs following induction of viral reactivation in BC-1 and BCBL-1 cells. Cells were treated with sodium butyrate for 0, 4, 6, 8, 12, 24, and 48 h. Total RNA was isolated at each time point and analyzed by Northern blotting. RNA blots were hybridized with single-stranded DNA complementary to ORF50 mRNAs (A), single-stranded DNA complementary to ORF45 mRNA (B), single-stranded DNA complementary to ORF K4.2 mRNA (C), single-stranded DNA complementary to nucleotides 50016 to 50261 (the 4.5-kb mRNA) (D), and random priming-labeled KSHV ORF59 cDNA (E). In each hybridization, random priming-labeled cDNA of RNase P RNA was included to serve as a control for loading. Molecular markers were an 0.24- to 9.5-kb RNA ladder (sizes are given in kilobases to the left of each panel).
Induction of the expression of KSHV IE genes.
BC-1 cells were induced with sodium butyrate (3 mM) for 4 h in the presence of cycloheximide at the concentration of 100 μg/ml. Total poly(A)+ RNAs were isolated from the induced and noninduced BC-1 cells and converted to cDNAs. cDNAs made from both induced and uninduced cells were used to scan the KSHV genome for transcription as follows. Six overlapping cosmid clones, which represent the whole genome of KSHV (Fig. 1A), were digested with EcoRI, BamHI, or both; electrophoresed through agarose gels; and stained with ethidium bromide (Fig. 1B). These DNAs were transferred onto Nytran membranes and probed respectively with these two pools of radiolabeled cDNAs. The probes represent all viral as well as cellular transcripts in the induced and noninduced BC-1 cells. At first glance, the pattern of major hybridized bands looked simple and similar between the blots probed with two cDNA probes (Fig. 1C and D). The major sequences hybridizing with cosmids 1 and 2 were the polyadenylated nuclear transcript nut-1 (also designated PAN RNA) (29, 32, 33), and the major transcripts originating from KSHV DNA in cosmids 4 and 5 were the mRNAs for the latent protein LANA (the 3-kb EcoRI/BamHI fragments in cosmid 4 or 5), ORF K12 (kaposin), v-FLIP, and v-cyclin (the 8-kb EcoRI/BamHI fragment in cosmid 4). Closely inspecting the blots, we observed a number of minor bands that were present only in the blot hybridized with the cDNA pool from the induced cells (Fig. 1D). Examples included the 2.2-kb EcoRI/BamHI fragment in cosmid 1 (nucleotides 22409 to 24637), the 2.6-kb EcoRI/BamHI fragments in cosmids 2 and 3 (nucleotides 66444 to 69094), the 3.1-kb EcoRI/BamHI fragment in cosmid 2 (nucleotides 47518 to 50637), and the 3.8-kb EcoRI/BamHI fragment in cosmid 3 (nucleotides 69095 to 72888) (Fig. 1D). The transcripts originating from these DNA fragments may correspond to KSHV genes induced by sodium butyrate in the presence of cycloheximide and thus are candidates for the IE mRNAs of KSHV. Some induced transcripts may not be detected by the Southern hybridization if they originated from the same DNA fragments (especially large restriction fragments) to which abundant latent transcripts also hybridized. Therefore, we attempted to identify the IE mRNAs of KSHV by isolating their cDNA from the induced BC-1 cells.
FIG. 1.
Identification of regions that are actively transcribed in the KSHV genome before and after induction with sodium butyrate in the presence of cycloheximide (100 μg/ml). (A) Summary of overlapping cosmid clones of KSHV DNA. Cosmids GB11, GA29, GB22, GA1, GA2, and GB1 are simply referred to as cosmids 1, 2, 3, 4, 5, and 6, respectively. (B) Ethidium bromide-stained agarose gel (0.8%) of KSHV cosmid DNAs digested with EcoRI, BamHI, or both as indicated. (C) Southern hybridization of restricted KSHV cosmid DNAs with cDNA probe prepared from uninduced BC-1 cells. (D) Southern hybridization of the restricted DNAs with cDNA probe prepared from sodium butyrate-induced BC-1 cells. (E) Southern hybridization of the restricted DNAs with cDNA probe which was prepared from induced BC-1 cells and had been subjected to a subtractive selection, representing the enrichment of induced KSHV cDNAs after the selection. For panels B to E, lanes 1 to 6 correspond to cosmids 1 to 6, respectively. Lines to the left of panels B to E indicate molecular sizes in kilobases (size markers are in lane M of panel B).
Identification of IE transcripts of KSHV by cDNA subtractive selection.
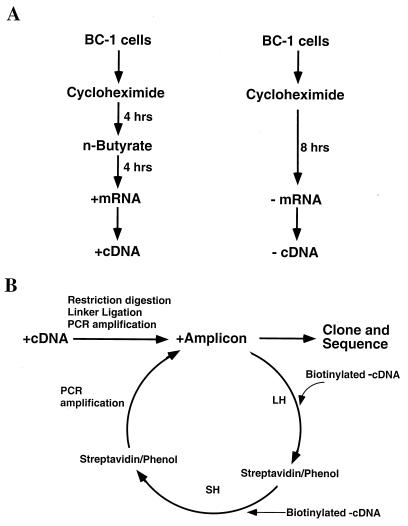
To isolate the cDNAs of KSHV IE mRNAs from a complex cDNA pool that contains cellular mRNA sequences and numerous KSHV sequences that can be from latent viral gene expression or low-level lytic gene expression (25), we employed a gene expression screening method which was developed based on cDNA subtractive hybridization (1, 31). This method was designed to isolate mRNAs that differ in abundance between two RNA populations, so that the KSHV transcripts whose amounts were dramatically increased after induction in the presence of cycloheximide could be obtained. Figure 2 depicts the strategy used in this study. Basically, cDNA fragment amplicons were prepared with poly(A)+ RNAs from cells, both induced and uninduced, as described in Materials and Methods. The cDNAs from sodium butyrate-induced cells were designated cDNA(+), and the cDNAs from uninduced cells were designated cDNA(−). The cDNA(−) were biotinylated and hybridized to the cDNA(+). After hybridization, the cDNA(−) and some of the cDNA(+) that hybridized to the cDNA(−) were removed by addition of streptavidin, followed by phenol extraction. As a result, the common sequences present in both cDNA populations were eliminated and theoretically only unique sequences (sodium butyrate-induced sequences) in the cDNA(+) pool were retained. The subtractive selections were repeated several times until a small number of unique sequences had been greatly enriched.
FIG. 2.
Scheme for chemical induction of KSHV reactivation in latently infected BC-1 cells (A) and subtractive cDNA cloning of differentially expressed KSHV mRNAs in the IE stage of reactivation (B). A plus sign refers to the mRNA or cDNA prepared from the BC-1 cells induced with sodium butyrate. A minus sign refers to mRNA or cDNA from uninduced BC-1 cells. LH, long hybridization (20 h); SH, short hybridization (2 h).
After the seventh cycle of subtractive selection, the extent of enrichment of the unique sequences in the cDNA(+) pool was measured by hybridization of the subtracted cDNA(+) pool to KSHV cosmid DNAs restricted with EcoRI, BamHI, or both (Fig. 1E). The Southern blots showed that the cDNAs of the latent mRNAs had apparently been eliminated from the pool, as judged by the disappearance of hybridization signals in the 3- and 8-kb EcoRI/BamHI fragments in cosmid 4 as well as the 3- and 7-kb EcoRI/BamHI fragments in cosmid 5 (compare Fig. 1E with 1D). It was also shown that the abundance of nut-1 (PAN RNA) cDNA sequence in the cDNA pool had been dramatically reduced (6-kb EcoRI/BamHI fragment in cosmid 1 and 5-kb EcoRI/BamHI fragment in cosmid 2). In contrast, most of the unique cDNA species which were induced by sodium butyrate were greatly enriched, as measured by the hybridization intensity of the induced hybridization bands, such as the 2.6-kb EcoRI/BamHI fragments in cosmids 2 and 3 (nucleotides 66444 to 69094), the 3.1-kb EcoRI/BamHI fragment in cosmid 2 (nucleotides 47518 to 50637), and the 3.8-kb EcoRI/BamHI fragment in cosmid 3 (nucleotides 69095 to 72888) (compare Fig. 1E with 1D). These results indicated that the cDNA subtractive hybridization was successful.
The enriched cDNAs were then digested with EcoRI and cloned into pBluescript at the EcoRI site. Clones that contain KSHV cDNA inserts were screened by colony hybridization with 32P-labeled KSHV genomic DNA fragments excised from the six cosmids. The plasmid DNAs from these clones were sequenced. Fifty clones were analyzed by sequencing. The majority of the sequences originated from six regions in the KSHV genome. Five of these regions were found within the KSHV fragments which corresponded to sodium butyrate-induced hybridization bands shown in Fig. 1D and E. The other region was within the terminal repeat sequence and was located in the 9-kb EcoRI/BamHI fragment in cosmid 6 (Fig. 1B). This fragment hybridized with the subtraction-enriched cDNA(+) (Fig. 1E) but was not detected with unenriched sodium butyrate-induced cDNAs (Fig. 1D). Nevertheless, a conclusive assignment of these cDNA fragments requires knowing the transcription orientation of their mRNAs and obtaining their full-length cDNAs.
Northern analyses of putative IE mRNAs.
To characterize the mRNAs corresponding to the isolated cDNAs and determine whether these mRNAs are indeed IE transcripts of KSHV, a series of Northern analyses were performed. Poly(A)+ RNA was prepared from BC-1 cells that had been induced with sodium butyrate for 4 and 20 h in the absence or presence of 100 μg of cycloheximide per ml. Cycloheximide was added 4 h prior to induction. These mRNAs were electrophoresed through denaturing agarose gels and transferred onto Nytran membranes. Radiolabeled single-stranded DNA probes were prepared from cloned plasmids by asymmetric PCR (see Materials and Methods) and used to probe the RNA blots. Using single-stranded probes allows the determination of transcription orientation of relevant genes. The results showed that five cDNA probes detected mRNAs that were induced either in the absence or in the presence of cycloheximide. These mRNAs included (i) two rightward transcripts of 3.6 and 4.3 kb that encompass ORF50 (Fig. 3A), (ii) three leftward transcripts of 1.2, 3.0, and 7.0 kb that are complementary to ORF50 mRNA (Fig. 3B), (iii) a rightward mRNA of 1.7 kb which contains ORF45 (Fig. 3C), (iv) a leftward transcript of 2.0 kb which carries ORF K4.2 (Fig. 3D), and (v) a rightward transcript of 4.5 kb which is partially complementary to ORF29 mRNA (Fig. 3E). These mRNAs have similar transcription patterns. First, their transcriptions were induced to a high level or had reached a plateau at 4 h postinduction in BC-1 cells. Second, their transcription could be induced in the presence of a high concentration of cycloheximide (100 μg/ml) in BC-1 cells. The cycloheximide-resistant mRNAs were detected at 4 h after the induction. They were not detected after 20 h of induction in the presence of cycloheximide due to severe toxicity to the cells after a long incubation time (24 h). According to the extent of their transcription in the presence of cycloheximide, these KSHV transcripts could be classified into two categories. The first category included the 3.6-kb ORF50 mRNA. The transcription of this mRNA in the presence of 100 μg of cycloheximide per ml was at the level of 30 to 40% of that in the absence of cycloheximide. The second category consisted of mRNAs for 4.3-kb ORF50, ORF45, and ORF K4.2 and the 4.5-kb mRNA. The transcription levels of these RNAs in the presence of cycloheximide (100 μg/ml) were between 12 and 20% of their amounts in the cells induced in the absence of cycloheximide.
To ensure that the transcription patterns of these IE mRNA candidates were distinct from those of transcripts from other classes, such as latent and delayed-early, the RNA blots were probed with cDNAs from KSHV mRNAs of known classes. The probes included cDNAs for a latent gene (LANA) and two delayed-early lytic genes (vIL-6 and ORF59). Northern blots of LANA, ORF59, and vIL-6 mRNAs are shown in Fig. 3F to H. As anticipated, the LANA mRNA was expressed constitutively regardless of sodium butyrate induction and cycloheximide treatment. Expression of vIL-6 and ORF59 was not detected at 4 h, only at 20 h postinduction, and their transcription was completely blocked in the presence of cycloheximide (Fig. 3G and H). A β-actin probe detected a 1.9-kb mRNA (Fig. 3L), which was used as a control for the integrity of the various RNA samples and as a marker for host gene transcription.
The extreme sensitivity of BCBL-1 cells to the toxicity of cycloheximide made it impossible to examine the transcription of these mRNAs in the absence of de novo protein synthesis in this cell line. To determine whether these IE mRNAs were expressed in BCBL-1 cells in the same stage as they were in BC-1 cells, kinetics of the transcription of these mRNAs were compared between these two cell lines by Northern analysis. Total RNAs were isolated from BC-1 and BCBL-1 cells after 4, 6, 8, 12, 24, and 48 h of treatment with sodium butyrate and from untreated cells. The Northern blots of these RNAs were hybridized with cDNA probes complementary to mRNAs for ORF50, ORF45, and ORF K4.2 and the 4.5-kb mRNA. Identical blots were also hybridized with a cDNA for ORF59, which is known to be a delayed-early gene and to be activated by ORF50 (12a). The results are shown in Fig. 4. ORF50 mRNAs of 3.6 and 4.3 kb were induced by sodium butyrate and reached a plateau at 4 h postinduction in both BC-1 and BCBL-1 cells. The transcription patterns of ORF50 mRNAs in BC-1 and BCBL-1 cells were similar except that the mRNAs could be detected at a high level in untreated BCBL-1 cells but not in untreated BC-1 cells (Fig. 4A). The induction of an ORF45 mRNA of 1.7 kb could be observed at 4 h postinduction in BC-1 and BCBL-1 cells. The mRNA level appeared to increase with time in both cell lines (Fig. 4B). The transcription of a K4.2 mRNA of 2.0 kb was induced and reached a peak at 4 h postinduction in both BC-1 and BCBL-1 cells. After 8 h, the amount of K4.2 mRNA appeared to decline with time (Fig. 4C). With the decrease in the transcription of K4.2 mRNA, a 1.7-kb mRNA that carries ORFs K4.1 and K4 began to accumulate in BC-1 and BCBL-1 cells (Fig. 4C). The 1.7-kb K4.1 mRNA is a delayed-early transcript and can be activated by ORF50 (data not shown). The transcription of the ORF K4.1 gene initiates at or near nucleotide 22908 on the KSHV genome in BC-1 cells. The 4.5-kb mRNA was induced by sodium butyrate and reached a plateau at 4 h postinduction in both BC-1 and BCBL-1 cells. However, the transcription level of this mRNA species appeared lower in BCBL-1 cells (Fig. 4D). Overall, the transcription patterns of these mRNAs are similar between BC-1 and BCBL-1 cells. In both cell lines, the time of their first appearance is earlier than that of delayed-early mRNAs such as ORF59 (Fig. 4E). The kinetics data, together with induction of their transcription in the presence of cycloheximide in BC-1 cells, suggest that the mRNAs for ORF50, ORF45, and ORF K4.2 and the 4.5-kb mRNA are IE transcripts of KSHV.
cDNA fragments corresponding to two other loci in the KSHV genome were also isolated in the subtractive selection. One corresponds to a leftward transcript of approximately 9.5 kb in BC-1 cells which is partially complementary to glycoprotein B. The transcription of the 9.5-kb mRNA was detectable in the presence of cycloheximide in BC-1 cells. However, this mRNA was not detected in BCBL-1 cells induced with either sodium butyrate or TPA (data not shown). Thus, more study on this transcript is needed before we are able to draw any conclusion. The other isolated cDNA fragment was from the terminal repeat sequence. Northern analysis with single-stranded probes corresponding to each strand of the sequence failed to detect any cycloheximide-resistant transcript, suggesting that these cDNAs may not correspond to any IE mRNA. They may be selected in the cDNA subtraction due to their unusual GC-rich sequence. This notion was supported by the observation that, in the Southern analysis (Fig. 1), the hybridization signals corresponding to the terminal repeat sequence (the 9-kb EcoRI/BamHI fragment in cosmid 6) were detected only in the hybridization with the subtraction-enriched cDNA(+) (Fig. 1E) and not detected with unenriched sodium butyrate-induced cDNAs (Fig. 1D). In the absence of cycloheximide, Northern analysis with a cDNA probe of terminal repeats detected two transcripts (9.5 and 12 kb) that were induced with sodium butyrate. In addition, in uninduced cells, the probe detected a cluster of latent transcripts as an 800-nucleotide repeat ladder, ranging from 2.4 to over 20 kb (data not shown).
Structure of the IE mRNA of ORF50.
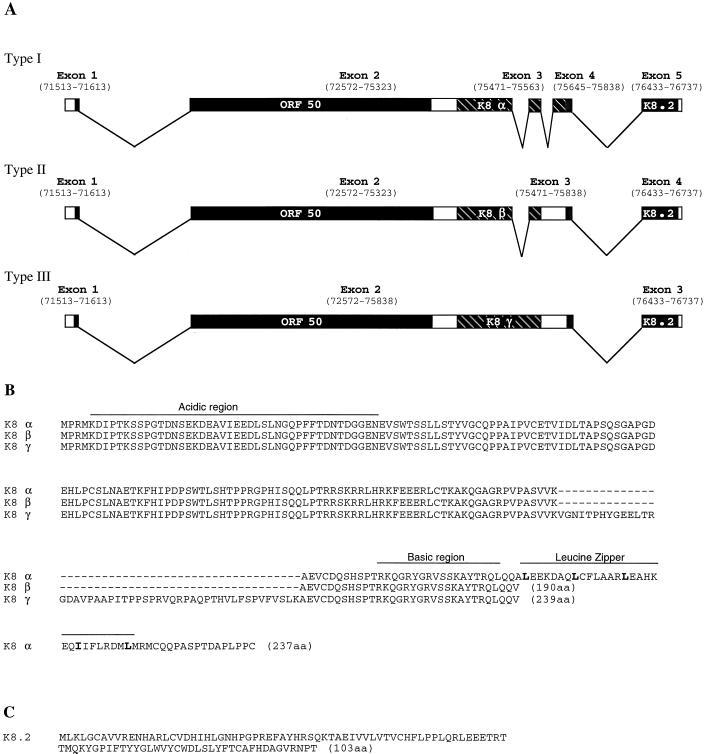
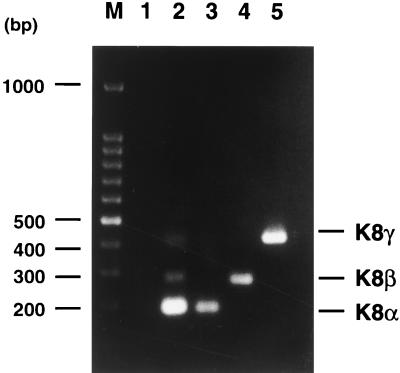
Hybridization of 32P-labeled single-stranded ORF50 cDNA to Northern blots of the induced mRNA displayed two major transcripts of 3.6 and 4.3 kb (Fig. 3A and 4A). Although both of them could be detected in the cells which were induced in the presence of cycloheximide, the 3.6-kb transcript was expressed to a higher level than was the 4.3-kb mRNA and was more resistant to cycloheximide treatment. The full-length cDNAs for the 3.6-kb IE mRNA of ORF50 were obtained by a PCR-based cDNA amplification procedure and then cloned and sequenced. Sequencing analysis revealed that the 5′ end of the mRNA is at or near nucleotide 71513 (numbering as in reference 24), and the 3′ poly(A) tail begins at nucleotide 76737 (Fig. 5A). The sequences of different cDNA clones revealed that the ORF50 IE mRNAs (∼3.6 kb) actually consist of three splice variants ranging from 3.6 to 3.8 kb. As illustrated in Fig. 5A, type I mRNA is composed of five exons, and in type II mRNA, intron 3 was not removed, while the type III mRNA contained both intron 2 and intron 3. To confirm the existence of three splice variants, semiquantitative RT-PCR was performed with two primers adjacent to introns 2 and 3 and mRNA isolated from BC-1 cells induced with sodium butyrate for 4 h. The results showed that the majority of 3.6- to 3.8-kb ORF50 mRNA is type I and that types II and III are minor species (Fig. 6). Based on the frequency of isolation of the cDNA clones for each type of ORF50 mRNA as well as the result of the semiquantitative RT-PCR assay, the type I mRNA accounts for approximately 70% of the 3.6- to 3.8-kb ORF50 mRNAs and types II and III together compose 30%.
FIG. 5.
Structure of ORF50 IE mRNAs and the ORFs they encode. (A) Schematic representation of the intron-exon structures of ORF50 mRNAs. Numbers indicate nucleotide positions in the KSHV genome (24). All three types of mRNA initiate at or near position 71513 and end at position 76737. Exons are presented with boxes, and introns are presented with lines. Each type of mRNA has three putative ORFs, namely, ORF50, K8, and K8.2 as indicated. ORF K8 shows three distinct forms (designated α, β, and γ) as a result of alternative splicing. (B) Amino acid sequences of three forms of KSHV ORF K8. The acidic domain and the basic-leucine zipper domain are indicated. (C) Amino acid sequence of putative ORF K8.2.
FIG. 6.
RT-PCR of ORF50-K8-K8.2 mRNA. Poly(A)+ RNAs were extracted from sodium butyrate-induced BC-1 cells and used to perform RT-PCR with two primers adjacent to introns 2 and 3 of ORF50-K8-K8.2 mRNA (lane 2). A PCR without reverse transcription was also performed with the same RNA and primers as a control (lane 1). The size of three RT-PCR fragments in lane 2 is in agreement with the size of fragments amplified with cloned cDNAs for type I (lane 3), type II (lane 4), and type III (lane 5) ORF50-K8-K8.2 mRNAs. Lane M is the molecular markers, a 100-bp DNA ladder (Promega).
All three types of mRNAs share the ORF50 of 691 amino acid residues and the ORF K8.2 of 103 amino acid residues. They differ in the middle portion that encodes different forms of the putative ORF K8 (designated α, β, and γ) as a result of alternative splicing (Fig. 5A). The ORF50 is located in the first two exons. The encoded protein shows significant sequence homology with Rta of EBV. The overall homology between KSHV ORF50 and EBV Rta is 20% identity in amino acid sequence. It has been shown that ORF50 activates some viral early and late genes (12a, 30).
Three forms of ORF K8 result from alternative splicing and usage of different stop codons. First, two intron sequences in K8 were doubly removed in K8α, singly spliced out in K8β, and not spliced in K8γ. Second, the stop codon used for K8β and K8γ is located in intron 3. This stop codon has been spliced in type I mRNA so that K8α is predicted to be translated through most of exon 4 (Fig. 5A). Interestingly, the unique sequence near the carboxy terminus of K8α contains a perfect basic-leucine zipper (bZip) structure (residues 169 and 218), which is absent in the β and γ forms (Fig. 5B). In addition, an acidic domain (12 of 42 residues [28.6%]) could be discerned between residues 6 and 47 (Fig. 5B). This suggests that K8α is a transcription factor of the bZip family. Amino acid sequence analysis revealed significant similarities between ORF K8α and the ZEBRA protein of EBV, a transactivator responsible for the switch of EBV from latency to lytic life cycle. The overall sequence homology between K8α and ZEBRA is 21.6% identity at the amino acid level with gaps. Similarities were also seen between K8α and the c-Jun proto-oncogene product (20% identity).
The putative ORF K8.2 was not identified before, and the predicted amino acid sequence is shown in Fig. 5C.
Northern analysis with a single-stranded DNA probe corresponding to ORF50 mRNA strand detected three transcripts in the absence and presence of cycloheximide. These RNAs are transcribed from the right to the left in the KSHV genome and can serve as antisense RNAs to ORF50 messenger (Fig. 3B).
Structures of the IE mRNAs of ORF45 and K4.2.
The cDNAs for ORF45 and K4.2 IE mRNAs were also obtained. Both of these mRNAs are unspliced. The 1.7-kb ORF45 mRNA encodes a putative peptide of 407 amino acid residues (the sequence is available in GenBank under accession no. AF091346). The predicted amino acid sequence of ORF45 has unique features. It has sequence similarity to nuclear proteins and transcription factors. Between amino acids 90 and 115, there is an extended acidic domain, which is glutamic acid and aspartic acid rich, typical for transcription activators. This type of acidic region is often found in nuclear proteins and transcription factors, including YY-1, yeast origin recognition complex protein subunit 1, nucleolin, and UBF-1; the latter two are responsible for rRNA synthesis. In addition, ORF45 is predicted to be a nuclear protein based on analysis with PSORT, a program designed to predict the subcellular localization sites of proteins.
The 2.0-kb IE mRNA for ORF K4.2 was synthesized in the leftward direction. The cDNA sequences showed that this mRNA encodes three putative ORFs, namely, K4.2, K4.1, and K4 (the cDNA sequence is available in GenBank under accession no. AF091347). ORFs K4 and K4.1 are predicted to encode two cytokine-like proteins, both closely resembling cellular MIP-1. The peptide encoded by K4, designated v-MIP-II, has been shown to preserve the functions predicted by its homology (16). For instance, v-MIP-II can bind to cytokine receptors CCR-3 and CCR-5 and block human immunodeficiency virus type 1 from using these receptors to enter CD4+ cells. v-MIP-II also demonstrated angiogenic activity (3). However, sequence analysis with GCG Blast could not identify any similarity of ORF K4.2 to any known protein. Kyte-Doolittle hydrophilicity prediction revealed hydrophobic domains at the N terminus and near the C terminus of ORF K4.2, suggesting that it is a membrane protein. In addition to the tricistronic transcript, K4 and K4.1 are also transcribed individually with two downstream promoters in the delayed-early phase of the lytic life cycle (16). A delayed-early ORF K4.1 mRNA of 1.7 kb can be detected on Northern blots hybridized with K4.2 cDNA probe because the 5′ end of ORF K4.1 mRNA is within the K4.2 ORF (Fig. 3D and 4C). The ORF K4.1 mRNA of 1.7 kb cannot be detected in the presence of cycloheximide (Fig. 3D).
The structures of the 4.5-kb IE mRNAs and three ORF50 antisense transcripts are under investigation.
mRNAs for ORF57, K3, and K5 of KSHV are delayed-early transcripts.
Three other KSHV ORFs were previously predicted to encode IE proteins based on their homology to IE proteins of other herpesviruses. ORFs K3 and K5 were found to have sequence similarity near the amino terminus with the major IE gene encoded by bovine herpesvirus 4. The region of similarity includes a newly recognized PHD/LAP class of zinc finger motif (19). KSHV ORF57 was referred to as an IE protein homologue because it has homology with EBV BMLF1 and herpes simplex virus type 1 (HSV-1) ICP27 (24). However, no cDNA for ORF57, K3, or K5 was detected in our cDNA subtractive selection. To clarify whether these three genes are IE genes in the KSHV genome, Northern analysis was performed with labeled cDNAs of these three mRNAs as probes. The results showed that a 1.3-kb K5 mRNA and 1.5-kb ORF57 mRNA were detected at 20 h after induction with sodium butyrate (Fig. 3J and 3K). A K3 cDNA probe detected two major transcripts of 1.5 and 2.0 kb at 20 h postinduction (Fig. 3I). The transcriptions of these three genes were completely blocked in the presence of cycloheximide (100 μg/ml) (Fig. 3I to K). It is suggested that ORF57, K3, and K5 are not IE genes in the KSHV genome in BC-1 cells.
DISCUSSION
IE genes of herpesviruses.
Reactivation of latent herpesviruses or infection of permissive cells by the viruses in the presence of protein synthesis inhibitors leads to restricted transcription of the viral genome. The genes that are expressed under these conditions are usually the first ones expressed after reactivation or after primary infection and are referred to as the IE viral genes. In many herpesviruses, such as HSVs and cytomegalovirus, IE genes are usually defined by their transcription after primary infection of permissive cells in the presence of complete inhibition of de novo protein synthesis. However, this criterion cannot be applied to KSHV, at least at the present time, because establishment of a permissive cell system for KSHV lytic infection has been problematic (22). KSHV infection in vitro so far can be studied only by induction of permissivity in B cells that are latently infected. In this study, IE genes of KSHV are defined as those that are activated and expressed during chemical-induced viral reactivation in the presence of complete inhibition of de novo protein synthesis. By this definition, KSHV IE mRNAs were identified in BC-1 cells that were induced with sodium butyrate in the presence of cycloheximide through the use of a cDNA subtraction-based gene expression screening method. These IE messengers originated from four regions in the KSHV genome, which we refer to as KIE-1 to KIE-4 (Table 1).
TABLE 1.
KSHV cDNAs isolated from BC-1 cells induced for viral reactivation in the presence of cycloheximide
| Locus | Transcription direction | mRNA size (kb) | ORF | No. of cDNA clones isolated and genomic locations (nucleotide) |
|---|---|---|---|---|
| KIE-1 | Rightward | 3.6–3.8 | ORF50, K8, and K8.2 | 10 (72570–75569) |
| KIE-2 | Leftward | 1.7 | ORF45 | 10 (68840–67485) |
| KIE-3 | Leftward | 2.0 | K4.2, K4.1, and K4 | 3 (23077–22745) |
| KIE-4 | Rightward | 4.5 | Unknown | 9 (49419–54688) |
Several lines of evidence support the classification of these mRNAs, namely, ORF50, ORF45, ORF K4.2, and the 4.5-kb mRNA, as IE transcripts of KSHV. First, these transcripts are induced by a stimulation of viral reactivation and accumulate in the absence of de novo protein synthesis. Second, transcription of the genes for these mRNAs obviously precedes the activation of delayed-early genes such as ORF59 and vIL-6. After stimulation of KSHV activation, these IE mRNAs begin to accumulate within 4 h, while delayed-early mRNAs are usually detected at or after 8 h. In addition, these four IE mRNAs were also detected in BCBL-1 cells, which carry only KSHV. The kinetics of the transcription of these IE genes are similar in BC-1 and BCBL-1 cells, except that the level of the 4.5-kb mRNA was found to be lower in BCBL-1 cells. Therefore, ORF50, ORF45, ORF K4.2, and 4.5-kb mRNAs are defined as IE transcripts in KSHV.
In general, most viral IE gene products are regulatory proteins, which either regulate subsequent viral gene expression or modulate host cells ready for supporting lytic viral replication. Five major IE genes (α genes) of HSV-1 were identified. Most HSV-1 IE genes encode transcription activators (ICP0, ICP4, ICP22, and ICP27) regulating viral β and γ gene expression and viral DNA replication (23). The product of the other α gene, ICP47, was shown to modulate the host defense mechanism by blocking presentation of viral peptides to major histocompatibility complex class I-restricted cells (10). Similarly, human cytomegalovirus IE proteins originating from five genomic loci are involved in the regulation of viral and cellular gene activation, viral DNA replication (i.e., IE1, IE2, UL36 to -38, and TRS1/IRS1), and modulation of the host immune system (i.e., US3, which impairs maturation and transport of major histocompatibility complex class I heavy chains) (15). In EBV, which is a member of the gammaherpesvirus family like KSHV, two IE genes, namely, BZLF1 and BRLF1, have been identified and studied intensively (2). These two genes encode two transcription activators, ZEBRA and Rta, which activate viral lytic genes in a synergistic manner (13). Identification of KSHV IE genes opens avenues for studying the mechanism of viral reactivation and infection and the accompanying expression of viral cytokine signaling genes that have been thought of as crucial components in the pathogenesis of KSHV-associated diseases.
ORF50 and K8 IE mRNAs.
Among the IE mRNAs of KSHV identified in this study, ORF50 is the only one that has been previously predicted to encode an IE regulatory protein based on its homology to EBV Rta. A previous study showed that KSHV ORF50 can activate early lytic genes including virus-encoded IL-6 and polyadenylated nuclear RNA (nut-1) and a late gene coding for a small viral capsid antigen (30).
In EBV, the product of the BZLF1 gene, known as ZEBRA, is capable of driving the entire lytic cycle in B cells and in epithelial cells, while Rta synergizes with ZEBRA to activate early genes in B cells and the lytic cascade in epithelial cells (12, 13). Homologues of EBV BZLF1 have not been found in other members of the gammaherpesvirus family. In this study, we found that the 3.6-kb IE mRNA, which encodes ORF50, also carries two additional ORFs, K8 and K8.2. A highly spliced form of ORF K8 (K8α) shows significant similarity to the ZEBRA protein of EBV. In addition to significant homology of amino acid sequence between K8α and ZEBRA (21.6% identity), the predicted K8α protein has a typical bZip domain near the carboxyl terminus and an acidic domain near the amino terminus, closely resembling ZEBRA and c-Jun proteins in structure (Fig. 5B). However, the unspliced and singly spliced forms of K8 (β and γ) lack the bZip domain. Furthermore, aside from the 3.6- to 3.8-kb tricistronic mRNAs, the ORF K8 sequence was found to be transcribed as a 0.9-kb delayed-early mRNA by Northern analysis with a K8 probe (data not shown). The role(s) played by the K8 proteins in viral infection, reactivation, and pathogenesis as well as the functional relationship among these three forms of K8 protein is very intriguing and is under investigation.
Recently, Lukac et al. reported the structure of a 3.4-kb ORF50 mRNA which carries ORF50, K8, and K8.1 (12a). That structure seems different from those of the ORF50 mRNAs described above. In all three types of mRNAs that we characterized, the ORF K8.1 has been spliced out.
ORF45 IE mRNA.
The 1.7-kb mRNA coding for ORF45 is a major IE mRNA induced in BC-1 cells. Twenty percent of the KSHV cDNA clones in our subtracted cDNA library contained sequences originating from this mRNA. This observation, together with the results of a Northern analysis (Fig. 3C), reflects the abundance of the mRNA in BC-1 cells in the IE time. ORF45 has unique features, including an extended acidic domain, which is mainly composed of glutamic acid and aspartic acid. The glutamic acid-aspartic acid-rich domain is often found in nuclear proteins and transcription factors. In the KSHV genome, such a glutamic acid-aspartic acid-rich domain is also seen in LANA, a latent nuclear protein of KSHV. Although the overall ORF45 is conserved among the gammaherpesvirus family members, the glutamic acid-aspartic acid-rich domain is not found in the herpesvirus saimiri and EBV homologues, suggesting that it may be a unique feature for KSHV ORF45. Due to its nuclear protein features, ORF45 is likely to play a role in regulating gene expression or lytic DNA replication.
K4.2 IE mRNA.
Although the 2.0-kb K4.2 IE transcript encodes three ORFs, we do not know if the second and third frames, i.e., K4.1 and K4, are translatable in this tricistronic messenger. The transcription pattern of the gene for ORF K4.2 is distinct from that of the other KSHV IE genes because of its transient expression period. Like other KSHV IE mRNAs, the ORF K4.2 mRNA accumulates at a considerable level at 4 h postinduction in BC-1 and BCBL-1 cells. However, the amount of ORF K4.2 mRNA begins to decline at 8 h postinduction. Kyte-Doolittle hydrophilicity prediction revealed a small hydrophobic domain at the N terminus and a large hydrophobic domain near the C terminus of ORF K4.2. These regions may serve as a signal peptide and a transmembrane domain, respectively, predicting that K4.2 may be a type I cell receptor.
ACKNOWLEDGMENTS
We are grateful to Ren Sun and George Miller (Yale University) for providing the cosmid clones of KSHV. We thank Gary Cohen, Robert Ricciardi (University of Pennsylvania), Tonia Symensma, and Ren Sun (UCLA) for critical reading of the manuscript and helpful discussion.
The work presented in this paper was partially supported by a University of Pennsylvania Cancer Center pilot project program grant.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley Interscience; 1996. pp. 5.9.1–5.9.20. [Google Scholar]
- 2.Biggin M, Bodescot M, Perricaudet M, Farrell P. Epstein-Barr virus gene expression in P3HR1-superinfected Raji cells. J Virol. 1987;61:3120–3132. doi: 10.1128/jvi.61.10.3120-3132.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boshoff C, Endo Y, Collins P D, Takeuchi Y, Reeves J D, Schweickart V L, Siani M A, Sasaki T, Williams T J, Gray P W, Moore P S, Chang Y, Weiss R A. Angiogenic and HIV-inhibitory functions of KSHV-encoded chemokines. Science. 1997;278:290–294. doi: 10.1126/science.278.5336.290. [DOI] [PubMed] [Google Scholar]
- 4.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 5.Cesarman E, Moore P S, Rao P H, Inghirami G, Knowles D M, Chang Y. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi’s sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood. 1995;86:2708–2714. [PubMed] [Google Scholar]
- 6.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 7.Ensoli B, Gendelman R, Markham P, Fiorelli V, Colombini S, Raffeld M, Cafaro A, Chang H K, Brady J N, Gallo R C. Synergy between basic fibroblast growth factor and HIV-1 Tat protein in induction of Kaposi’s sarcoma. Nature. 1994;371:674–680. doi: 10.1038/371674a0. [DOI] [PubMed] [Google Scholar]
- 8.Ensoli B, Nakamura S, Salahuddin S Z, Biberfeld P, Larsson L, Beaver B, Wong-Staal F, Gallo R C. AIDS-Kaposi’s sarcoma-derived cells express cytokines with autocrine and paracrine growth effects. Science. 1989;243:223–226. doi: 10.1126/science.2643161. [DOI] [PubMed] [Google Scholar]
- 9.Ganem D. Human herpesvirus 8 and the biology of Kaposi’s sarcoma. Semin Virol. 1996;7:325–332. [Google Scholar]
- 10.Hill A, Jugovic P, York I, Russ G, Bennink J, Yewdell J, Ploegh H, Johnson D. Herpes simplex virus turns off the TAP to evade host immunity. Nature. 1995;374:411–415. doi: 10.1038/375411a0. [DOI] [PubMed] [Google Scholar]
- 11.Kedes D H, Lagunoff M, Renne R, Ganem D. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi’s sarcoma-associated herpesvirus. J Clin Investig. 1997;100:2606–2610. doi: 10.1172/JCI119804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Howley P M, Knipe D M, Channock R M, Melnick J L, Monath T P, Roizman B, editors. Fields’ virology. Vol. 3. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2343–2396. [Google Scholar]
- 12a.Lukac D M, Renne R, Kirshner J R, Ganem D. Reactivation of Kaposi’s sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology. 1998;252:304–312. doi: 10.1006/viro.1998.9486. [DOI] [PubMed] [Google Scholar]
- 13.Miller G. The switch between latency and replication of Epstein-Barr virus. J Infect Dis. 1990;161:833–844. doi: 10.1093/infdis/161.5.833. [DOI] [PubMed] [Google Scholar]
- 14.Miller G, Heston L, Grogan E, Gradoville L, Rigsby M, Sun R, Shedd D, Kushnaryov V M, Grossberg S, Chang Y. Selective switch between latency and lytic replication of Kaposi’s sarcoma-associated herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J Virol. 1997;71:314–324. doi: 10.1128/jvi.71.1.314-324.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mocarski E. Cytomegaloviruses and their replication. In: Fields B N, Howley P M, Knipe D M, Channock R M, Melnick J L, Monath T P, Roizman B, editors. Fields’ virology. Vol. 3. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2447–2492. [Google Scholar]
- 16.Moore P S, Boshoff C, Weiss R A, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 17.Muralidhar S, Pumfery A M, Hassani M, Sadaie M R, Azumi N, Kishishita M, Brady J N, Doniger J, Medveczky P, Rosenthal L J. Identification of kaposin (open reading frame K12) as a human herpesvirus 8 (Kaposi’s sarcoma-associated herpesvirus) transforming gene. J Virol. 1998;72:4980–4988. doi: 10.1128/jvi.72.6.4980-4988.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neipel F, Albrecht J-C, Fleckenstein B. Cell-homologous genes in the Kaposi’s sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicholas J, Ruvolo V, Zong J, Ciufo D, Guo H G, Reitz M S, Hayward G S. A single 13-kilobase divergent locus in the Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genome contains nine open reading frames that are homologous to or related to cellular proteins. J Virol. 1997;71:1963–1974. doi: 10.1128/jvi.71.3.1963-1974.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rainbow L, Platt G M, Simpson G R, Sarid R, Gao S-J, Stoiber H, Herrington C S, Moore P S, Schulz T F. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J Virol. 1997;71:5915–5921. doi: 10.1128/jvi.71.8.5915-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 22.Renne R, Blackbourn D, Whitby D, Levy J, Ganem D. Limited transmission of Kaposi’s sarcoma-associated herpesvirus in cultured cells. J Virol. 1998;72:5182–5188. doi: 10.1128/jvi.72.6.5182-5188.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Howley P M, Knipe D M, Channock R M, Melnick J L, Monath T P, Roizman B, editors. Fields’ virology. Vol. 3. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2231–2295. [Google Scholar]
- 24.Russo J J, Bohenzky R A, Chien M-C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;91:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarid R, Flore O, Bohenzky R A, Chang Y, Moore P S. Transcription mapping of the Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1) J Virol. 1998;72:1005–1012. doi: 10.1128/jvi.72.2.1005-1012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarid R, Wiezorek J S, Moore P S, Chang Y. Characterization and cell cycle regulation of the major Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J Virol. 1999;73:1438–1446. doi: 10.1128/jvi.73.2.1438-1446.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schulz T F. Kaposi’s sarcoma-associated herpesvirus (human herpesvirus-8) J Gen Virol. 1998;79:1573–1591. doi: 10.1099/0022-1317-79-7-1573. [DOI] [PubMed] [Google Scholar]
- 28.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d’Agay M F, Clauvel J P, Raphael M, Degos L, Sigaux F. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 29.Sun R, Lin S-F, Gradoville L, Miller G. Polyadenylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc Natl Acad Sci USA. 1996;93:11883–11888. doi: 10.1073/pnas.93.21.11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun R, Lin S-F, Gradoville L, Zhu F X, Yuan Y, Miller G. A viral gene that activates lytic cycle expression of Kaposi’s sarcoma associated herpesvirus. Proc Natl Acad Sci USA. 1998;95:10866–10871. doi: 10.1073/pnas.95.18.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z, Brown D D. A gene expression screen. Proc Natl Acad Sci USA. 1991;88:11505–11509. doi: 10.1073/pnas.88.24.11505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong W, Wang H, Herndier B, Ganem D. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc Natl Acad Sci USA. 1996;93:6641–6646. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong W, Ganem D. Characterization of ribonucleoprotein complexes containing an abundant polyadenylated nuclear RNA encoded by Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) J Virol. 1997;71:1207–1212. doi: 10.1128/jvi.71.2.1207-1212.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu, F. X., and Y. Yuan. 1998. Unpublished data.