Abstract
Paramyxoviruses cotranscriptionally edit their P gene mRNAs by expanding the number of Gs of a conserved AnGn run. Different viruses insert different distributions of guanylates, e.g., Sendai virus inserts a single G, whereas parainfluenza virus type 3 inserts one to six Gs. The sequences conserved at the editing site, as well as the experimental evidence, suggest that the insertions occur by a stuttering process, i.e., by pseudotemplated transcription. The number of times the polymerase “stutters” at the editing site before continuing strictly templated elongation is directed by a cis-acting sequence found upstream of the insertions. We have examined the stuttering process during natural virus infections by constructing recombinant Sendai viruses with mutations in their cis-acting sequences. We found that the template stutter site is precisely determined (C1052) and that a relatively short region (∼6 nucleotides) just upstream of the AnGn run can modulate the overall frequency of mRNA editing as well as the distribution of the nucleotide insertions. The positions more proximal to the 5′ AnGn run are the most important in this respect. We also provide evidence that the stability of the mRNA/template hybrid plays a determining role in the overall frequency and range of mRNA editing. When the template U run is extended all the way to the stutter site, adenylates rather than guanylates are added at the editing site and their distribution begins to resemble the polyadenylation associated with mRNA 3′ end formation by the viral polymerase. Our data suggest how paramyxovirus mRNA editing and polyadenylation are related mechanistically and how editing sites may have evolved from poly(A)-termination sites or vice versa.
The concept of RNA editing was introduced in 1986 to describe the posttranscriptional insertion of nongenomically encoded uridylates within the coding region of trypanosome mitochondrial mRNAs; this insertion restores the coding capacity of these genes (3). Many other examples of RNA editing by a variety of mechanisms, including nucleotide insertion, deletion, and base substitution, were subsequently found. The term RNA editing now encompasses a large number of separate processes that produce RNA transcripts whose sequence (and informational capacity) differs from that encoded by the corresponding gene, other than by splicing or 5′- and 3′-end formation (2, 18, 50). Although “editing” implies that the process occurs posttranscriptionally, there are two examples, both by nucleotide insertion, where it has not been possible to separate the modification of the RNA from its synthesis, namely, the paramyxovirus P genes and those of the mitochondria of Physarum polycephalum. The editing of the Physarum mRNAs is relatively complex. Single cytidylates or uridylates, and certain dinucleotides, are added to ca. 1,000 different sites of the mRNAs from this ∼60-kb genome, and the information that determines the specificity of the insertions at each site is unclear. The mechanism(s) operating here is tightly coupled to the synthesis of the mRNA, but this editing does not appear to be due to the reiterative copying of a template base(s) (i.e., pseudotemplated transcription [56, 57]). In contrast, the editing of paramyxovirus P gene mRNAs is relatively simple, since only a single site per 15-kb viral genome is modified and only guanylates are added within a short run of guanylates. Moreover, different paramyxoviruses insert different distributions of guanylates at their editing sites, and the sequences conserved here as well as experimental evidence suggests that the insertions occur by a stuttering process, i.e., by pseudotemplated transcription (30, 32).
The Paramyxovirinae are organized into three genera: Respiroviruses, including Sendai virus (SeV) and human and bovine parainfluenza virus type 3 (h- and bPIV3); Morbilliviruses (e.g., measles virus and the distemper viruses); and Rubulaviruses (e.g., mumps virus and simian virus 5 [SV5]). They contain nonsegmented 15- to 16-kb negative-strand RNA genomes found as helical nucleocapsids, in which each nucleocapsid protein (N) subunit is associated with precisely six nucleotides, and only viral genomes which are multiples of six nucleotides in length are found in nature (the “rule of six” [5, 12, 31]). Ca. 300 copies of the P (phosphoprotein) and ca. 50 copies of the L (large) protein are also bound to each nucleocapsid. Six mRNAs (in the order N, P, M, F, HN, and L) are transcribed from the N:RNA genome by the P-L polymerase. All of these viral mRNAs, except the P gene mRNA, express a single primary translation product from a single open reading frame (ORF). The P gene mRNAs, in contrast, generally contain one or two alternate ORFs that overlap the middle region of the P-protein ORF and that are expressed as fusion proteins with the N-terminal half of P. In contrast to retroviruses or coronaviruses, where alternate downstream ORFs are accessed by ribosomal frameshifting (4, 23, 60), the trans-frame P-fusion proteins result from transcriptional frameshifting due to the programmed insertion of different numbers of guanylates.
Most paramyxoviral P genes contain a 5′ AnGn purine run (Fig. 1A) at the start of the internal, overlapping V ORF (by convention, plus strands are written 5′ to 3′ and minus strands are written 3′ to 5′). mRNAs with expanded G runs are transcribed from these genes in addition to those which are faithful copies of their templates, and the number of G insertions which occur for each virus group mirrors their requirements to switch between the in-frame and trans-frame downstream ORFs (30). For the morbilliviruses and Sendai virus (SeV), which require a single nucleotide insertion to frameshift to the V ORF from the genome-encoded P ORF, a single G is added as the predominant editing event (Fig. 1A). For the rubulaviruses, which require the insertion of two nucleotides to access the remainder of the P ORF from the genome-encoded V ORF, two Gs are added at high frequency when insertions occur. For bPIV3, where both V and another ORF (called D) overlap the middle of the genome-encoded P ORF, one to six Gs are added at roughly equal frequency so that mRNAs encoding all three overlapping ORFs are expressed.
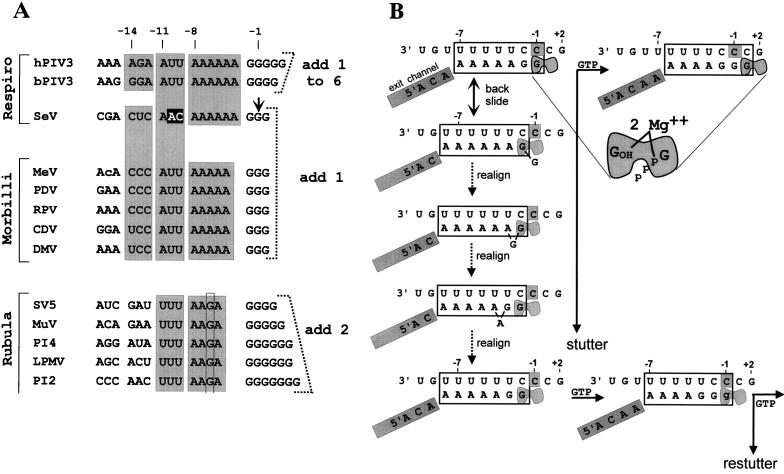
FIG. 1.
(A) Sequence homologies at the paramyxovirus editing sites. The sequences are written as [+] RNA, 5′ to 3′, and are grouped into the three genera of the Paramyxovirinae. Spaces have been introduced to emphasize the different elements of the sequence, and shaded boxes indicate sequence conservations. The short G run which is expanded on mRNA editing is shown on the right, together with the pattern of G insertions which occurs for each group. Note that the A run preceding the G run is the only part of this cis-acting sequence that is strictly conserved according to genera. Also note that the second A residue upstream of the rubulavirus G run is replaced by a G (highlighted with a rectangle), which presumably accounts for why rubulaviruses insert a minimum of two G residues when stuttering begins. The precise SeV editing site determined in this study (arrow) is listed as position −1, and positions upstream are numbered according to their distance from this mRNA 3′ end when the polymerase active site is at the editing site. Virus abbreviations: MeV, measles virus; PDV, phocine distemper virus; RPV, rinderpest virus; CDV, canine distemper virus; DMV, dolphin morbillivirus; MuV, mumps virus; PI4, human parainfluenza virus type 4; LPMV, La Piedad, Michoacan virus; PI2, human parainfluenza virus type 2. (b) Competitive kinetic model for SeV RNAP stuttering-elongation decision. The template and mRNA chains of the transcription elongation complex at the editing site are shown schematically. The putative 7-bp hybrid between the polypyrimidine tract of the [−] genome (top strand) and the polypurine run of the nascent mRNA chain (bottom strand) when the transcription elongation complex is at the editing site is boxed. The mRNA upstream of the hybrid is proposed to enter an exit channel (gray-shaded box) before it reaches the surface of the RNAP, which maintains the length of the hybrid as transcription elongation proceeds. The exit channel, analogous to other RNAPs, would contain ∼10 nt (see text). The RNAP bipartite active site, in which the nascent mRNA 3′ end (position −1) and the NTP α-phosphate (position +1) are coordinated via two Mg2+ ions, is highlighted in gray. The transcription complex at the top left is at the editing site (the middle template C1052, boxed in gray) and has just incorporated a strictly templated G1052 (top left). The transcription complex at the editing site presumably pauses due to backsliding of RNAP by one position along both the template and the mRNA chains, undoing the last base pair of the hybrid (and removing the mRNA 3′ end from the active site) and reforming 1 bp on the upstream side. RNAP at pause sites is envisaged as oscillating between the inactive backtracked alignment (second line) and the active alignment (top line). If a strictly templated GMP is the next nucleotide incorporated, RNAP moves past the stutter site and resumes normal elongation (top line). Alternatively, realignment of the hybrid also correctly repositions the mRNA 3′ end in the active site. Hybrid realignment when RNAP is in the backtracked state is initiated when the unpaired 3′ G re-pairs with C−2 (third line), causing the penultimate G to bulge out. Realignment is completed upon translocation of the single nucleotide bulge to the upstream side of the hybrid, reforming a 7-bp hybrid which is nearly as stable as its predecessor. The mRNA 3′ G is now correctly repositioned in the active site, and nucleotide addition at this point leads to a single pseudotemplated G insertion, or stutter (lower case g, bottom line). Having stuttered once, the transcription complex is back to where it started from and has the same choices (second branchpoint, bottom right). Escape from stuttering occurs when the transcription complex moves to a template position where hybrid realignment (stuttering) is no longer favored. Numbers above the genome sequence always indicate the positions relative to mRNA 3′ end at the start of the stutter (top left).
Paramyxovirus mRNAs are made in the cytoplasm, and these viruses must consequently fend for themselves in all aspects of mRNA synthesis. Negative-strand virus RNA polymerases (RNAPs) polyadenylate their mRNAs by stuttering on a short run of template U residues (4 to 7 nucleotides [nt] long) at the end of each gene. It was this characteristic that first suggested that the G insertions would similarly occur by pseudotemplated transcription (6, 24, 51, 54), and there is now strong experimental evidence that the insertions occur cotranscriptionally, by a stuttering mechanism (21, 55). By analogy to the elongation-termination decision of Escherichia coli RNAP (58, 59), paramyxovirus mRNA editing can be described by a competitive kinetic model (Fig. 1B). The paramyxovirus RNAP elongation complex has two choices at any template position; it can extend the nascent chain by 1 nt, or it can be induced by features of the template or nascent chain sequence to pause (Fig. 1B). At this point (the first branchpoint, see below) there is a significant probability that the active site together with its nascent chain are repositioned one (respiroviruses and morbilliviruses) or two (rubulaviruses) residues upstream of the template C which has just been copied (shown as C −1 in Fig. 1B, top left). In the realignment of the nascent mRNA/template hybrid, U:G (but not A:C) pairs are permitted, and in analogy to ribosomal frameshifting, the region where alternate base pairing occurs after realignment is called the slippery sequence. The template cytidylate(s) is then copied a second time when nucleotide addition recurs, resulting in pseudotemplated G insertions. If a strictly templated rather than a pseudotemplated nucleotide monophosphate (NMP) is nevertheless added; this allows RNAP to move past the potential site of editing (the stutter site) and to escape downstream (Fig. 1B, top line). This first branchpoint thus determines the overall frequency of edited mRNAs. However, if a stutter occurs, the frequency with which RNAP now escapes to strictly templated elongation or restutters represents a second branchpoint, because h- and bPIV3 behave differently than SeV at this juncture. The second branchpoint thus determines the range of G insertions once stuttering has commenced.
The replacement of the SeV editing region with that of bPIV3 in an SeV minigenome leads to mRNAs with G insertions whose distribution resembles those found in bPIV3 infection (21, 25). The counting mechanism that determines this difference in distribution of G insertions is thus apparently controlled in large part by a cis-acting sequence. This study reports that the editing site is precisely determined (C1052) and examines how the controlling sequences which lie immediately upstream of the 5′ A6G3 slippery sequence affect the counting mechanism. Our results suggest how mRNA editing and polyadenylation might be related mechanistically.
MATERIALS AND METHODS
Generation of pFL-3Δ2.
To generate pFL-3Δ2, pFL-3Δ1 (16) was digested with SmaI, religated, and transformed into XL1-Blue bacteria. This resulted in a Sendai full-length clone with a deletion between the NcoI site (nt 358, in the N gene) and the SmaI site (nt 3553, in the P gene), 5 nt downstream of the P ORF.
Generation of mutations in the shuttle vector pN/PxhoM and in PHA.
Construction of the pN/PxhoM shuttle vector is described in Garcin et al. (16). The A3G6, A4G5, A5G4, A7G2, A8G1, and A9 mutants were constructed in pGEM-PHA by inserting their respective P cassettes in place of the corresponding 1028XbaI-EcoRI1130 fragment of wild-type PHA. The A3G6, A4G5, A5G4, A7G2, and A8G1 cassettes were obtained by PCP amplification from pGEM-PHA with primer A3G6 (5′-GACTCTAGAGAGCGACTCTAACAAAGGGGGGCATAGGAGAG), primers A4G5, A5G4, A7G2, A8G1, and A9 (which are identical to A3G6 except for the polypurine run [underlined]), and primer PEcoP (5′-GGGCACGTCTTGCAAACAC). The PCR products were then digested with XbaI and EcoRI and introduced into pGEM-PHA. These series of mutations were then transferred to the shuttle vector pN/PxhoM by inserting the SmaI fragment of PHA derivatives.
Other mutations were constructed as described above with primers AAt (5′-GACTCTAGAGAGCGACTCTAATAAAAAAGGGCATAGGAGAG) and Att, AtC, and ttt (which are identical to AAt except at the upstream sequences underlined); primers Comp(−20 to −12) (5′-GACTCTAGActcgctgagAACAAAAAAGGGCATAGGAGAG), Comp(−20 to −15) (5′-GACTCTAGActcgctCTCAACAAAAAAGGGCATAGGAGAG), and Comp(−20 to −18) (5′-GACTCTAGActcCGACTCAACAACAAAAAAGGGCATAGGAGAG); primer SV5 (5′-GACTCTAGAccccatCgattttAAgAggggCATAGGAGAGAAC); primers Swap(−16 to −9) (5′-GACTCTAGAGAGCagggaAttAAAAAAGGGCATAGGAGAG) and Swap(−16 to −12) (5′-GACTCTAGAGAGCagggaAACAAAAAAGGGCATAGGAGAG); primer Co.Sw(−16 to −9) (5′-GACTCTAGAGAGCtcccttggAAAAAAGGGCATAGGAGAG); and finally primer SeV(Z) (5′-GACTCTAGAGAcCGACTCTAACAAAAAAGGGCATAGGAGAG).
Generation of recombinant SeV (rSeV).
Briefly, one 9-cm-diameter dish of A549 cells was infected with 3 PFU of vaccinia virus TF7-3 (14) per cell and transfected 1 h later with 1.5 μg of pGEM-L, 5 μg of pGEM-N, 5 μg of pGEM-PHA (which do not express C proteins), 15 μg of pFL3-Δ2, and 5 μg of the various pN/PXho/M shuttle vectors (15, 16). All mutations were introduced into both pN/PXho/M and pGEM-PHA to prevent possible loss of the mutation by recombination. Twenty-four hours later, 1-β-d-arabinofuranosylcytosine (100 μg/ml) was added to inhibit vaccinia virus replication, and after a further 24 h the cells were scraped into their medium and directly injected into the allantoic cavity of 10-day-old embryonated chicken eggs. Three days later, the allantoic fluids were harvested and reinjected undiluted into eggs. For further passages, the viruses were diluted 1/500 before injection. The presence of viruses was determined by pelleting the allantoic fluids through a TNE–25% glycerol cushion at 14,000 rpm. Virus pellets were then lysed in sample buffer, and the proteins were separated by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue.
Analysis of mRNAs and genomes and antigenomes by limited primer extension.
A549 cell monolayers in 9-cm-diameter dishes were infected with 10 to 30 PFU of the various rSeV per cell. At 24 hours postinfection (hpi), the cells were solubilized by scraping into 150 mM NaCl–50 mM Tris (pH 7.4)–10 mM EDTA–0.6% Nonidet P-40. Nuclei were removed by pelleting at 12,000 × g for 5 min. To separate mRNA and the viral nucleocapsids, cytoplasmic extracts were centrifuged in a step gradient composed of a 5.7 M CsCl cushion, 40% CsCl, and 20% CsCl at 35,000 rpm overnight in an SW55 rotor. RNAs from either viral nucleocapsids (the 20 to 40% interface) or pelleted mRNAs were analyzed by limited primer extension after reverse transcription-PCR amplification. The reverse transcription reaction used the oligonucleotide PEcoP (5′-GGGCACGTCTTGCAAACAC) and a 1/10 aliquot was used for PCR with 50 pM of PEcoP and PEag primer (5′-CCAGCCAACGGCCGCCC) in 10 mM Tris-Cl (pH 8.3)–50 mM KCl–2 mM MgCl2–100 μM deoxynucleoside triphosphates (dNTP). PCR was carried out in 50 μl with 1 U of Taq polymerase in a GeneAmp PCR system 9600, with denaturation at 94°C for 20 s, elongation at 72°C for 30 s, and annealing at 45°C for 18 cycles. The PCR products were purified on a 2% agarose gel and annealed to 32P-labeled primers (SeV-edit; 5′-GATGTGTTCTCTCCTATG) complementary to the sequence immediately downstream of the editing site. Primer extension was performed in 10 μl at 37°C for 6 min with 1 U of T7 DNA polymerase (Pharmacia) in the presence of 40 μM concentrations (each) of dGTP, dTTP, and dCTP and 4 μM ddATP. Then, 300 μM dNTP was added, and the mix was incubated an additional 2 min to chase stalled complexes. The reaction was stopped by adding 4 μl of STOP solution (95% formamide, 20 mM EDTA, 0.1% bromophenol blue, and xylene cyanol FF). The products were boiled 1 min and electrophoresed on a 12.5% sequencing gel (1).
RESULTS
5′ A run.
The three respiroviruses that edit their P mRNAs contain a 5′ A6G3–5 run at the editing site (Fig. 1A). Consistent with this conservation, a minimum of three Gs are required for SeV editing activity, and minigenomes containing G4 or G5 runs continued to edit their mRNAs at slightly reduced frequencies (21). The importance of the length of the A run, in contrast, is unclear. An SeV minigenome in which the 5′ A6 run is replaced with 5′ AAAAGA or a recombinant measles virus in which the 5′ A5 run is replaced with 5′ AAAGA (both in attempts to induce the insertion of 2 Gs in a single stutter as postulated for rubulaviruses) are editing inactive (25, 47). This inactivity is presumably because unstable A:C pairs would be formed during the realignment of the mRNA/template hybrid required for editing, and these pairs must be avoided in designing our mutants.
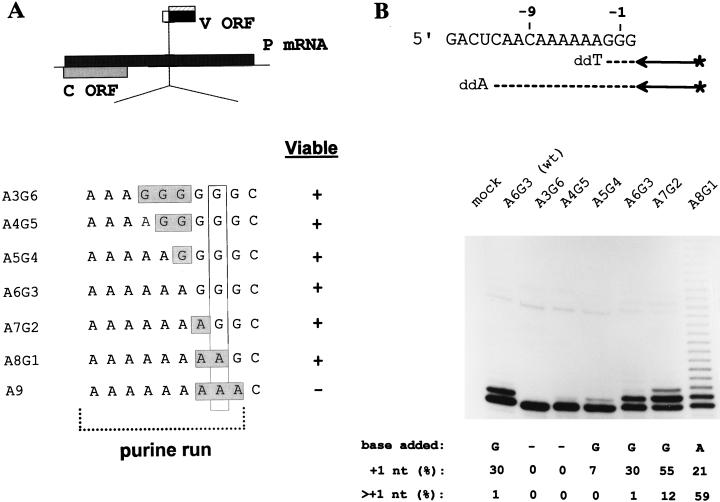
It has recently become possible to recover infectious mononegaviruses from DNA (15, 44, 48) and therefore to study SeV mRNA editing within the context of a natural virus infection by reverse genetics. This approach is possible because the products of the edited mRNAs (the V and W proteins) are not required for the infection of cultured cells or hen’s eggs but rather play a crucial role during infections of mammals (10, 11, 28, 29). We have examined the importance of the length of the A run for mRNA editing by systematically mutating the adenylates abutting the G run to guanylates and vice versa (from 5′ A3G6 to A9G0, without altering other positions [Fig. 2A]) in rSeV. As the A and G runs in these rSeV remain intact, unstable A:C pairs are not encountered during hybrid realignment. Except for rSeV-A9G0, the other viruses were recovered from DNA as efficiently as rSeV-wt and were amplified in hen’s eggs similarly to rSeV-wt. In contrast, we were unable to prepare rSeV-A9G0 despite repeated attempts.
FIG. 2.
mRNA editing in rSeV-A3G6 to -A8G1-infected cells. (A) The ORFs expressed from the P gene mRNA (shaded boxes) are shown above. The sequences of the various rSeVs in which the lengths of the A and G runs were altered (as described in the text) are shown. (B) Parallel cultures of A549 cells were infected with 20 PFU of the various rSeVs per cell as indicated. CsCl pellet RNA was prepared at 24 hpi, and the distribution of lengths of their P gene mRNA purine runs was determined by primer extension analysis limited with ddATP, as schematized in the upper panel. RNA from uninfected cells (mock) served as a negative control. The relative intensities of the various bands was determined, and the fraction of the mRNA population with a single (+1G) or multiple (>+1G) purine insertion is listed below. The lengths of their G runs alone was determined by primer extension analysis limited with ddTTP (not shown), as schematized in the upper panel, and the nature of the insertion deduced (see the text) is also listed below.
Parallel cultures of A549 cells were infected with 20 PFU of the various rSeV per cell. The editing regions of the P gene mRNAs (at 24 hpi) were then examined by limited primer extension terminated with ddATP to measure the combined length of the A and G runs (Fig. 2). rSeV-A3G6 and -A4G5 infections were found to contain little or no edited mRNAs. rSeV-A5G4 displayed some editing activity (7% of the mRNAs contained a single insertion), but clearly less than the wild-type control (30%). rSeV-A7G2, on the other hand, was found to edit 67% of its mRNAs, and mRNAs with two additional purines now represented 12% of the total. Most remarkably, rSeV-A8G1 infections contained mRNAs with a very broad range of insertions (up to at least 20 nt) which slowly decreased in frequency, such that mRNAs with multiple insertions now represented 59% of the total. Thus, systematically mutating the upstream adenylates to guanylates progressively decreases the frequency of mRNA editing, whereas mutating the downstream guanylates to adenylates has the opposite effect. The nature of the bases inserted was determined by also carrying out primer extension limited with ddTTP, which measures the length of the G run alone. The pattern of bands obtained with ddTTP for the A5G4 to A7G2 viruses was identical to that obtained with ddATP, indicating that only guanylates were added to the purine run in these cases. The pattern of bands obtained with ddTTP for rSeV-A8G1, in contrast, showed that very few guanylates had been inserted into these mRNAs (only a very weak band at position +1 was visible with ddTTP, which could account for only ca. 1% of the total insertions [not shown]). Predominantly adenylates were thus added to the purine run for rSeV-A8G1. Limited primer extension carried out with antigenomes showed that no insertions had occurred during genome replication (data not shown), and thus the extra nucleotides must have been added during mRNA synthesis.
G insertion into the SeV P gene mRNA occurs by a pseudotemplated process where one (or more) of the three cytidylates of the template 3′ 1045UUUUUUCCCG1054 slippery sequence are copied more than once (numbers refer to distance from the 5′ end of the P mRNA). In virion polymerase reactions in which the various NTPs were severely limited to increase the step time for nucleotide addition, low concentrations of CTP did not affect the frequency of editing, indicating that the downstream C1053 was not copied twice. Low GTP concentrations, on the other hand, strongly enhanced editing, suggesting that either C1051 or C1052 (or both) acted as the insertion or stutter site (55). The results shown in Fig. 2 indicate that C1052 is used as a stutter site, since the nucleotide insertions change from G to A when U replaces C1052 (rSeV-A8G1). By the same criteria C1051 is not being used, because guanylates continue to be inserted into the mRNA when U replaces C1051 (rSeV-A7G2). The SeV stutter site thus appears to be precisely determined (C1052 [or C−1 in Fig. 1B]), in a way similar to the pause and termination sites in Escherichia coli (8).
Importance of the upstream trinucleotide.
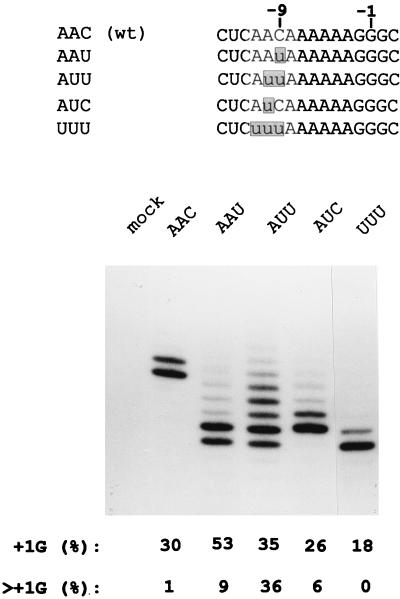
Except for SeV which contains the trinucleotide 5′ AAC immediately upstream of the 5′ AnGn run, the other respiroviruses and morbilliviruses have 5′ AUU (Fig. 1A). All RNAPs are thought to add nucleotides to the nascent RNA in a bipartite active site which coordinates the RNA 3′ end (position −1) and the NTP α-phosphate (position +1) via two Mg2+ ions (24). If the SeV stutter site is C1052 (3′ 1045UUUUUUCCC1053) and the mRNA/template hybrid is 7 bp long, as shown in Fig. 1B (see below), then the AAC in question is at positions −11 to −9 relative to the mRNA 3′ end (−11AAC−9), i.e., just upstream of the hybrid. When −11AAC−9 is changed to AUU in rSeV, this virus edits its P gene mRNA similarly to PIV3 in that mRNAs with two to four additional guanylates are now as numerous as mRNAs without insertions (lane AUU; Fig. 3). To investigate whether both uridylates are important for this phenotype switch, rSeV in which −11AAC−9 was changed to AAU or AUC were prepared. The adenylate that begins this trinucleotide (position −11) is invariant for respiroviruses and morbilliviruses, whereas 5′ UUU precedes the purine run of all rubulaviruses (Fig. 1A). An rSeV in which −11AAC−9 was changed to UUU was also prepared to examine whether the conservation of A−11 was important for editing.
FIG. 3.
mRNA editing in rSeV-infected cells with mutations in positions −11 to −9. The various rSeVs with mutations at positions −11 to −9 (relative to the middle G of the G run at position −1) are shown above. The distribution of lengths of their P gene mRNA purine runs determined by primer extension analysis limited with ddATP (as in Fig. 2) are shown below. The lengths of the various extended primers representing the uninserted mRNA (zero bands) vary here due to differences in the position of the limiting ddATP incorporated. The fastest band in each lane represents the zero or uninserted mRNA.
When compared to wild-type rSeV, rSeV-AAU clearly increased the fraction of mRNAs with a single G insertion (from 30 to 53%). However, in contrast to rSeV-AUU where mRNAs with more than one G insertions were predominantly affected (from 1 to 36%), mRNAs with more than one G were increased only marginally for rSeV-AAU (from 1 to 9%). According to the competitive kinetic model of Fig. 1B, the mutation from C to U at position −9 would predominantly affect the first branchpoint. rSeV-AUC, in contrast, edited its mRNA very similarly to the wild-type virus (rSeV-AAC), although there was a small increase in the “>1G” population (to 6%). The presence of uridylates at both positions −9 and −10 are thus required for the strong increase in >+1G mRNAs seen in rSeV-AUU infections (i.e., the switch to a PIV3-like phenotype). According to the competitive kinetic model, the enhancing effect of a uridylate at position −10 (in rSeV-AUU) would predominantly affect the second branchpoint. Lastly, rSeV-UUU was found to edit a smaller fraction of its mRNAs than rSeV-wt (AAC), and only the +1G population could be detected. The presence of the conserved A−11 is thus critical for the enhancing effects of the uridylates at positions −9 and −10. The sequence of the 3 nt just upstream of the 5′ AnGn run can clearly affect the distribution of G insertions during SeV mRNA editing.
Positions −12 to −14.
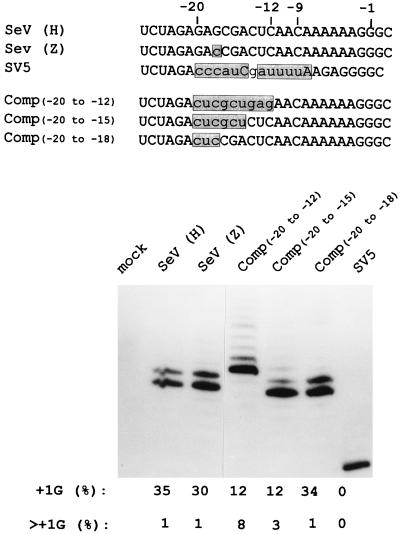
SeV and the morbilliviruses, which predominantly add a single G to their edited mRNAs, contain only pyrimidines at positions −12 to −14 of the plus strand, whereas h- and bPIV3 contain purines at these positions (Fig. 1A). Farther upstream, however, there is no obvious sequence that is conserved according to editing phenotype. To examine the extent of the upstream sequences which can modulate mRNA editing, rSeV were prepared in which the bases at positions −20 to −12, −20 to −15, and −20 to −18 were radically mutated to their Watson-Crick complements (Fig. 4). When P mRNAs from these various rSeV-infected cells were compared with wild-type virus infections for their distribution of G insertions, the distribution in the Comp(−20 to −18) mRNAs was found to be very similar to that of the wild-type virus, and the overall fraction of edited mRNAs was unchanged. Sequences beyond position −17 thus appear to have little or no effect on the pattern of editing. rSeV-Comp(−20 to −15) and rSeV-Comp(−20 to −12) edited a smaller fraction of their mRNAs; hence, the nature of the bases at positions −17 to −12 can affect the overall frequency of editing (presumably at the first branchpoint). The bases at positions −17 to −12 also appear to affect the second branchpoint, since rSeV-Comp(−20 to −12) infections contained a significant fraction of edited mRNAs with a broad distribution of insertions (from 2 to >6 Gs, 8% of the total) (Fig. 4). As rSeV-Comp(−20 to −12) has replaced the 5′ −14CUC−12 pyrimidine run of SeV-wt with purines (5′ −14GAG−12), the difference in editing patterns between rSeV-Comp(−20 to −12) and rSeV-Comp(−20 to −15) is presumably due to differences at positions −14 to −12, i.e., the presence of a purine as opposed to a pyrimidine run here may contribute to enhancing multiple G insertions independent of the presence of UU at positions −9 and −10.
FIG. 4.
mRNA editing in rSeV-infected cells with complementary sequences at positions −20 to −12 and an rSeV with a SV5 editing site. The various rSeVs in which the sequences at positions −20 to −18, −20 to −15, or −20 to −12 were mutated to their complements, as well as one containing positions −20 to −1 of the rubulavirus SV5 sequence (SV5), are shown above. The sequences of SeV strains H and Z, which vary at position −18, are also shown. The distribution of lengths of their P gene mRNA purine runs, as determined by primer extension analysis limited with ddATP (as in Fig. 2 and 3), are shown below.
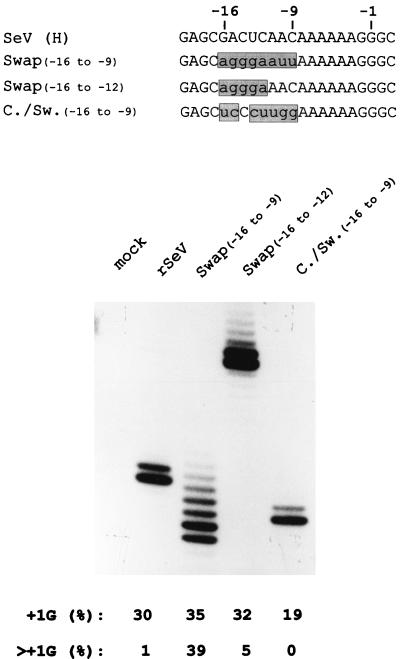
We have previously described rSeV-Swap8 [or Swap(−16 to −9)], so named because the 8 nt upstream of the SeV 5′ −8AAAAAAGGG+1 purine run (5′ −16GACUCAAC−9) were replaced with those of bPIV3 (5′ −16AGGGAAUU−9), and which led to a significant fraction of the mRNAs (∼40%) with >+1G (21). The distribution of >+1G mRNAs in rSeV-Comp(−20 to −12)-infected cells, however, was much weaker than those of rSeV-Swap(−16 to −9) infections (only 8%), possibly because rSeV-Swap(−16 to −9) contains both the −14GGA−12 purine run and −11AUU−9. The relative importance of these two elements in contributing to multiple G insertions was estimated by preparing and examining rSeV-Swap(−16 to −12) (5′ −16AGGGAAAC−9), which contains a −14GGA−12 purine run, but the wild-type 5′ −11AAC−9. When Swap(−16 to −12) and Swap(−16 to −9) were compared (Fig. 5), the extent of multiple G insertions was much reduced when −11AAC−9 rather than −11AUU−9 is present (from 39 to 5%), but it was still clearly detectable. A 5′ −14RRR−12 run thus also contributes to enhancing multiple G insertions, but this element is clearly less important than the presence of 5′ −11AUU−9. Finally, we also prepared rSeV-Comp/Swap(−16 to −9) in which the 8 nt upstream present in bPIV3 (5′ −16AGGGAAUU−9) were changed to the complementary sequence (5′ −16UCCCUUGG−9) except for positions −10 and −9, which were 5′ GG rather than 5′ AA, so the length of the 5′ A6 run would remain unaltered. rSeV-Comp/Swap(−16 to −9), like rSeV-UUU (Fig. 3), continued to edit its mRNA with single G insertions at a somewhat reduced frequency (15 to 20%). Since neither of these latter two rSeV contain either a 5′ −14RRR−12 run or −11AAC−9 or −11AUU−9, the 5′ A6G3 slippery sequence by itself appears to be sufficient to direct a moderate level of single G insertions during mRNA editing. The 5′ A6G3 slippery sequence by itself has previously been shown to allow for insertions (and deletions) in the slippery sequence during antigenome synthesis from non-hexamer-length minigenomes (20).
FIG. 5.
mRNA editing in rSeV-Swap(−16 to −9)-, Swap(−16 to −12)-, and Comp/Swap(−16 to −9)-infected cells. The sequences of the various rSeVs in which positions −16 to −9 were exchanged for those of bPIV3 [Swap(−16 to −9)] or the complement of the bPIV3 sequence [comp/swap(−16 to −9)] or in which the sequences at positions −16 to −12 were exchanged for those of bPIV3 [swap(−16 to −12)] are shown at the top of the figure. The distributions of lengths of their P gene mRNA purine runs, as determined by primer extension analysis limited with ddATP (as in Fig. 2 to 4), are shown below.
DISCUSSION
The catalytic subunits of all RNAPs, both cellular and viral, are thought to have evolved from a common ancestor (9). Although detailed structural information for most RNAPs is not available, it is generally assumed that all RNAPs have retained the basic structural features of this enzyme (e.g., the aspartate triad that coordinates two Mg2+ ions [26, 27]).
SeV RNAP, like other highly processive RNAPs, must grip the nascent RNA and template tightly enough to prevent even a low frequency of dissociation during elongation, yet loosely enough to translocate along the chains quickly (ca. 6 nt/s for VSV or SeV in vitro [8a, 22]). To explain these paradoxical properties, the concept of the sliding clamp has been proposed, analogous to the DNA replication apparatus (34, 35). In current models of E. coli RNAP, the sliding clamp is proposed to consist of three elements which bind the template, the nascent RNA/template hybrid (∼8 to 9 bp in length [17, 40]), and the nascent RNA as it leaves the RNAP. This latter site is also referred to as the tight product binding site (7, 39). Together, these latter two elements should cover 16 to 18 3′-proximal nucleotides of the mRNA, in accord with the RNase protection data (references 40 and 53 and references therein) and the availability of the nascent RNA to anneal to short oligonucleotides (33, 45). The observation that a strong RNA secondary structure (hairpins) just upstream of the hybrid (at 7 to 9 nt from the RNA 3′ end) destabilizes the elongation ternary complex at termination sites indicates that the nascent RNA binding site may be an area of important protein-RNA interactions (8, 26, 46).
SeV RNAP, like other RNAPs which respond to signals during elongation, must also be able to reverse one or both of the above properties (high ternary complex stability and smooth translocation along the chains) at high efficiency at the single template positions where pausing and mRNA editing (and presumably also polyadenylation and/or termination) are programmed. Since only sequences upstream of the paramyxovirus 5′ AnGn run are conserved according to virus group and editing pattern (Fig. 1A), the cis-acting sequences (other than the AnGn run) that help determine this precise position are presumably found here. This work has provided evidence that ca. six positions upstream of the AnGn run (positions −14 to −9 relative to the stutter site at position −1) can modulate the overall frequency of mRNA editing as well as the distribution of the nucleotide insertions. Moreover, the positions more proximal to the 5′ AnGn run are the most important in this respect. Converting −11AAC−9 to the conserved −11AUU−9 in rSeV strongly increases the overall fraction of mRNAs with insertions and particularly those with two to six extra guanylates. Positions −14 to −12 appear to exert a small but noticeable effect, whereas sequences upstream of position −14 appear not to play a role. The model of Fig. 1B, based on current models of the bacterial and eucaryotic RNAP elongation complex (53), assumes a constant 7 bp between the nascent mRNA and the SeV genome (the maximum number of base pairs consistent with this slippage mechanism). This number is based on the position of the stutter site (C−1/C1052), the conserved length of the respirovirus A6 run, and the need to leave U−7 of the template U6 run initially unpaired to accommodate realignment of the mRNA/template hybrid. The length of the hybrid could be maintained by the entry of the mRNA chain into an exit channel (or tight product-binding site) at this point, which prevents further hybridization of the nascent RNA to the template (41; see also Fig. 1B). If it is assumed that the basic structural features of all RNAPs have been conserved throughout evolution (9), the 5′ −14CUCAAC−9 region of the SeV cis-acting sequence would be located in the exit channel when the SeV RNAP catalytic center is at the stutter site. Base-specific interactions within the exit channel then provide a possible explanation of how the upstream cis-acting sequences can modulate the insertion process by SeV RNAP. We note, however, that there is as yet no detailed structural information for any mononegavirus RNAP.
RNAPs are thought to normally advance continuously along the template during elongation as each nucleotide is added. At pause sites, however, RNAP is thought to backtrack along the template and nascent mRNA chains, unwinding the hybrid at the mRNA 3′ end and reforming an equal number of base pairs upstream (40). The number of positions that RNAP backtracks varies according to the particular pause site (19, 37, 42). The temporary loss of catalytic activity at pause sites has been ascribed to the loss of contact between the mRNA 3′ end and the enzyme’s catalytic center. This process is reversible, and RNAP at pause sites is envisaged as oscillating between the inactive backtracked state and its return to the original active location, from where RNAP can escape downstream (33). The defining feature of paramyxovirus mRNA editing is that the pause site coincides with a slippery sequence, i.e., one where mRNA 3′ end realignment can occur because the resulting hybrid is almost as stable as the original one. Within this model for editing, the SeV RNAP would backtrack by only a single position, leaving a single 3′ nucleotide unpaired. Realignment can then be envisaged as the translocation of the unpaired nucleotide from the mRNA 3′ end to the upstream side of the hybrid by a series of looping-out transitions (Fig. 1B), which is energetically more favorable than simultaneously breaking and reforming all of the base pairs. Realignment correctly repositions the mRNA 3′ end in the catalytic center, and nucleotide addition at this point leads to a single pseudotemplated insertion. For SeV RNAP, backtracking at the stutter site may be limited to a single position, since this RNAP cannot edit its mRNA like rubulavirus RNAPs (i.e., by initiating the process by the simultaneous addition of two guanylates) even when they contain the appropriate cis-acting sequences (Fig. 4 and data not shown). If rubulavirus RNAPs, in contrast to those of respiroviruses and morbilliviruses, can backtrack by two positions at the stutter site, this will allow the translocation of a 2-nt bulge which bypasses the unstable A:C pair (55). These differences between the various paramyxovirus RNAPs could be due to different specific base contacts in their respective exit channels.
The model as outlined in Fig. 1B predicts that varying the stability of the mRNA/template hybrid should affect the editing process. Consistent with this, partial replacement of GTP with ITP in the virion RNAP reaction (which could act to weaken the hybrid by replacing G:C with I:C base pairs) led to a strong enhancement of mRNA editing (55). Since then, however, it was found that yeast RNAP III and calf thymus RNAP II pause after incorporation of IMP (38, 52), and the enhancing effects of IMP incorporation on SeV mRNA editing may have been due as well to extending the pause at the stutter site. The experiment of Fig. 2, however, provides more direct evidence that the stability of the hybrid per se plays a determining role in the overall frequency and range of mRNA editing. Strengthening the hybrid by converting the contiguous adenylates of the 5′ A6G3 site to guanylates progressively reduces the editing process, whereas weakening the hybrid by converting the contiguous guanylates to adenylates progressively increases the editing. There is thus an inverse relationship in rSeV-infected cells between estimated hybrid stability (assuming a constant number of base pairs) and the overall fraction of edited mRNAs, as well as the distribution of the insertions. Furthermore, when the template U run extends all the way to the stutter site (rSeV-A8G1), the distribution of A insertions is not limited to 1 to 6 nt, but mRNAs with an additional 1 to >20 nt are found at roughly equal frequency. This dramatic extension of the distribution of the insertions is presumably due not only to the further weakening of the hybrid such that only A:U pairs are present. There is now no loss of stability at all in forming the realigned hybrid, because the hybrid is composed of homopolymers. The presence of poly(A) in the exit channel may also stabilize realignment and further extend the distribution of the insertions.
Although the editing of the A8G1 mRNAs begins to resemble 3′ end formation of these viral mRNAs, it remains well removed from the 100 or so adenylates found at the 3′ ends of the majority of paramyxovirus and rhabdovirus mRNAs. It is also not associated with chain termination. For vesicular stomatitis virus (VSV), a model mononegavirus with an exceptionally vigorous virion transcriptase reaction, a similarly sized poly(A) tail is formed as well under these in vitro conditions, indicating that this distribution is due to virion proteins (13, 22). VSV mRNA polyadenylation takes place at a similar conserved cis-acting sequence (3′-AUACUUUUUUUG versus 3′-AUUCUUUUUG for SeV), and Barr et al. (1) have recently investigated the effect of mutating the four bases upstream of the U run on VSV RNAP response at a gene junction. They found that the upstream C (in boldface) was particularly important, as any other base here prevented poly(A) termination and led to read-through of the junction. Remarkably, when this C was mutated to another base, VSV RNAP nevertheless inserted a relatively even distribution of ca. 6 to 15 adenylates before continuing on to transcribe the downstream gene, as did SeV RNAP at the A8G1 editing site. Alteration of this critical base-specific interaction for VSV RNAP at a poly(A)-termination site eliminates termination and reduces stuttering to a maximum of ∼20 cycles before resuming strictly templated synthesis. These VSV mRNAs can be considered as having been edited.
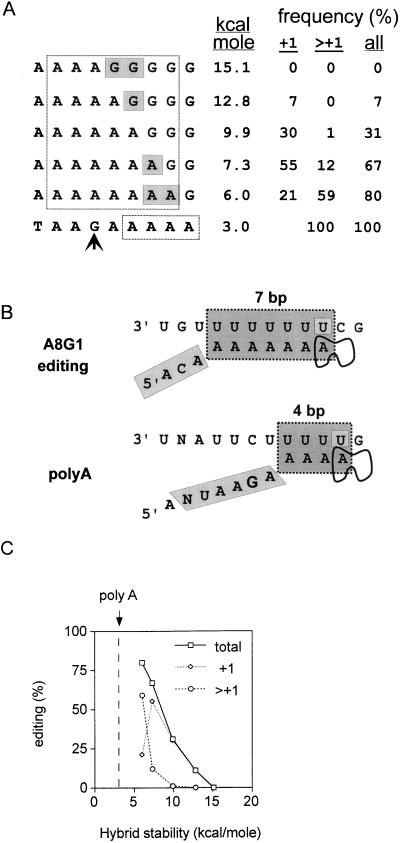
How, then, can editing sites have evolved from poly(A)-termination sites or vice versa? The model of Fig. 1B predicts that one way of extending the number of insertions is to further weaken the stability of the hybrid. Converting all the base pairs in the hybrid to A:U pairs at the editing site, however, is insufficient for extensive polyadenylation. Another way to weaken the hybrid is to reduce the number of base pairs it contains. Inspection of the conserved SeV cis-acting poly(A)-termination sequence (3′-UNAUUCUUUUUG) in relation to the proposed editing mechanism shows that there would be a maximum hybrid length of only 4 bp (even if the stutter site is displaced to the most downstream template uridylate (indicated in boldface) (Fig. 6). This leads to a relatively unstable hybrid (ΔG of only −3 kcal/mol; dotted vertical line in Fig. 6). Nascent mRNA chain realignment after NMP incorporation could then become so favored over further NMP incorporation that stuttering is prolonged, as presumably occurs during SeV mRNA 3′-end formation. The manner in which mononegavirus RNAPs eventually cease stuttering and release their mRNAs, however, remains unclear. The key difference between mRNA editing and polyadenylation according to the scheme of Fig. 6 is the shortening of the nascent chain-template hybrid in the latter process to help ensure the repetitive stuttering required for poly(A) tail formation. This could occur by decreasing the distance between the exit channel and the catalytic center, since there is considerable internal flexibility in E. coli RNAP, at least during chain initiation (7). Experiments to directly test this notion for SeV cannot be carried out by modifying the resident P gene, since extensive polyadenylation and/or termination at the editing site will prevent virus viability. These experiments can best be carried out by modifying an editing site within a supplemental (e.g., luciferase) trans gene.
FIG. 6.
Importance of hybrid stability in paramyxovirus RNAP stuttering. The 5′ nonapurine runs of the rSeV-AnGn of Fig. 2 are aligned above the conserved SeV poly(A)-termination signal (bottom line of panel A). The seven purines thought to be hybridized to the [−] genome when the transcription elongation complex is at the editing site are boxed, as are the proposed four adenylates hybridized to the [−] genome when the elongation complex is at the polyadenylation site (A). A comparison of the proposed stuttering structures of the rSeV-A8G1 transcription elongation complex at the editing and poly(A)-termination sites is shown in panel B. Note that in the poly(A) structure the stutter site has been displaced downstream by one position relative to the alignment shown in panel A, which would allow for a hybrid with a maximum of only 3 bp. The stabilities of the various hybrids (Serra et al. [49]), along with the distributions of insertions which result, are listed in panel A and are plotted in panel C. The vertical dashed line in panel C indicates the stability of the proposed 4-bp hybrid during polyadenylation as shown in panel B.
REFERENCES
- 1.Barr J N, Whelan S P, Wertz G W. cis-Acting signals involved in termination of vesicular stomatitis virus mRNA synthesis include the conserved AUAC and the U7 signal for polyadenylation. J Virol. 1997;71:8718–8725. doi: 10.1128/jvi.71.11.8718-8725.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benne R. RNA editing: an overview. In: Benne R, editor. RNA editing. Chichester, England: Ellis Horwood; 1993. pp. 13–24. [Google Scholar]
- 3.Benne R, Van den Burg J, Brakenhoff J P, Sloof P, Van Boom J H, Tromp M C, Major M C. Transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986;46:819–826. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- 4.Brierley I, Digard P, Inglis S C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calain P, Roux L. The rule of six: a basic feature for efficient replication of Sendai virus defective interfering RNA. J Virol. 1993;67:4822–4830. doi: 10.1128/jvi.67.8.4822-4830.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cattaneo R, Kaelin K, Baczko K, Billeter M A. Measles virus editing provides an additional cysteine-rich protein. Cell. 1989;56:759–764. doi: 10.1016/0092-8674(89)90679-x. [DOI] [PubMed] [Google Scholar]
- 7.Chamberlin M J. New models of transcription elongation and termination. Harvey Lect. 1995;88:1–21. [PubMed] [Google Scholar]
- 8.Chan C L, Wang D, Landick R. Multiple interactions stabilize a single paused transcription intermediate in which hairpin to 3′ end spacing distinguishes pause and termination pathways. J Mol Biol. 1997;268:54–68. doi: 10.1006/jmbi.1997.0935. [DOI] [PubMed] [Google Scholar]
- 8a.Curran, J. Unpublished data.
- 9.Delarue M, Poch O, Tordo N, Moras D, Argos P. An attempt to unify the structure of polymerases. Protein Eng. 1990;3:461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- 10.Delenda C, Hausmann S, Garcin D, Kolakofsky D. Normal replication of Sendai virus in cultured cells and embryonated chicken eggs without the trans-frame nonstructural V protein. Virology. 1997;228:55–62. doi: 10.1006/viro.1996.8354. [DOI] [PubMed] [Google Scholar]
- 11.Delenda C, Taylor G, Hausmann S, Garcin D, Kolakofsky D. Sendai viruses with altered P, V, and W protein expression. Virology. 1998;242:327–337. doi: 10.1006/viro.1998.9027. [DOI] [PubMed] [Google Scholar]
- 12.Egelman E H, Wu S S, Amrein M, Portner A, Murti G. The Sendai virus nucleocapsid exists in at least four different helical states. J Virol. 1989;63:2233–2243. doi: 10.1128/jvi.63.5.2233-2243.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehrenfeld E, Summers D F. Adenylate-rich sequences in vesicular stomatitis virus messenger ribonucleic acid. J Virol. 1972;10:683–688. doi: 10.1128/jvi.10.4.683-688.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcin D, Pelet T, Calain P, Roux L, Curran J, Kolakofsky D. A highly recombinogenic system for the recovery of infectious Sendai paramyxovirus from cDNA: generation of a novel copy-back nondefective interfering virus. EMBO J. 1995;14:6087–6094. doi: 10.1002/j.1460-2075.1995.tb00299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcin D, Itoh M, Kolakofsky D. A point mutation in the Sendai virus accessory C proteins attenuates virulence for mice, but not virus growth in cell culture. Virology. 1997;238:424–431. doi: 10.1006/viro.1997.8836. [DOI] [PubMed] [Google Scholar]
- 17.Gelles J, Landick R. RNA polymerase as a molecular motor. Cell. 1998;93:13–16. doi: 10.1016/s0092-8674(00)81140-x. [DOI] [PubMed] [Google Scholar]
- 18.Grosjean H, Benne R, editors. Modification and editing of RNA. Washington, D.C: ASM Press; 1998. [Google Scholar]
- 19.Gu W, Reines D. Identification of a decay in transcription potential that results in elongation factor dependence of RNA polymerase II. J Biol Chem. 1995;270:11238–11244. doi: 10.1074/jbc.270.19.11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hausmann S, Jacques J-P, Kolakofsky D. Paramyxovirus RNA editing and the requirement for hexamer genome length. RNA. 1996;2:1033–1045. [PMC free article] [PubMed] [Google Scholar]
- 21.Hausmann S, Garcin D, Morel A-S, Kolakofsky D. Two nucleotides immediately upstream of the essential A6G3 slippery sequence modulate the pattern of G insertions during Sendai virus mRNA editing. J Virol. 1999;73:343–351. doi: 10.1128/jvi.73.1.343-351.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iverson L E, Rose J K. Localized attenuation and discontinuous synthesis during vesicular stomatitis virus transcription. Cell. 1981;23:477–484. doi: 10.1016/0092-8674(81)90143-4. [DOI] [PubMed] [Google Scholar]
- 23.Jacks T, Varmus H E. Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science. 1985;230:1237–1242. doi: 10.1126/science.2416054. [DOI] [PubMed] [Google Scholar]
- 24.Jacques J-P, Kolakofsky D. Pseudo-templated transcription in prokaryotic and eukaryotic organisms. Genes Dev. 1991;5:707–713. doi: 10.1101/gad.5.5.707. [DOI] [PubMed] [Google Scholar]
- 25.Jacques J-P, Hausmann S, Kolakofsky D. Paramyxovirus mRNA editing leads to G deletions as well as insertions. EMBO J. 1994;13:5496–5503. doi: 10.1002/j.1460-2075.1994.tb06884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeruzalmi D, Steitz T A. Structure of T7 RNA polymerase complexed to the transcriptional inhibitor T7 lysozyme. EMBO J. 1998;17:4101–4113. doi: 10.1093/emboj/17.14.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joyce C M, Steitz T A. Function and structure relationships in DNA polymerases. Annu Rev Biochem. 1994;63:777–822. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]
- 28.Kato A, Kiyotani K, Sakai Y, Yoshida T, Nagai Y. The paramyxovirus Sendai virus V protein encodes a luxury function required for viral pathogenesis. EMBO J. 1997;16:678–587. doi: 10.1093/emboj/16.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato A, Kiyotani K, Sakai Y, Yoshida T, Shioda T, Nagai Y. Importance of the cysteine-rich carboxyl-terminal half of V protein for Sendai virus pathogenesis. J Virol. 1997;71:7266–7272. doi: 10.1128/jvi.71.10.7266-7272.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolakofsky D, Curran J, Pelet T, Jacques J-P. Paramyxovirus P gene mRNA editing. In: Benne R, editor. RNA editing. Chichester, England: Ellis Horwood; 1993. pp. 105–123. [Google Scholar]
- 31.Kolakofsky D, Pelet T, Garcin D, Hausmann S, Curran J, Roux L. Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited. J Virol. 1998;72:891–899. doi: 10.1128/jvi.72.2.891-899.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolakofsky D, Hausmann S. Cotranscriptional paramyxovirus mRNA editing: a contradiction in terms? In: Grosjean H, Benne R, editors. Modification and editing of RNA. Washington, D.C: ASM Press; 1998. pp. 413–420. [Google Scholar]
- 33.Komisarrova N, Kashlev M. Arrest of transcription: E. coli RNA polymerase translocates backward leaving the 3′ end of the RNA intact and extruded. Proc Natl Acad Sci USA. 1997;94:1755–1760. doi: 10.1073/pnas.94.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kong X P, Onrust R, O’Donnell M, Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 35.Kuriyan J, O’Donnell M. Sliding clamps of DNA polymerases. J Mol Biol. 1993;234:915–925. doi: 10.1006/jmbi.1993.1644. [DOI] [PubMed] [Google Scholar]
- 36.Lamb R A, Kolakofsky D. Paramyxoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. New York, N.Y: Raven Press; 1996. pp. 1177–1204. [Google Scholar]
- 37.Landick R. RNA polymerase slides home: pause and termination site recognition. Cell. 1997;88:741–744. doi: 10.1016/s0092-8674(00)81919-4. [DOI] [PubMed] [Google Scholar]
- 38.Matsuzaki H, Kassavetis G A, Geiduschek E P. Analysis of RNA chain elongation and termination by Saccharomyces cerevisiae RNA polymerase III. J Mol Biol. 1994;235:1173–1192. doi: 10.1006/jmbi.1994.1072. [DOI] [PubMed] [Google Scholar]
- 39.Nudler E, Avetissova E, Markovtsov V, Goldfarb A. Transcription processivity: RNA polymerase-DNA interactions holding together the elongation complex. Science. 1996;273:211–217. doi: 10.1126/science.273.5272.211. [DOI] [PubMed] [Google Scholar]
- 40.Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 41.Nudler E, Gusarov I, Avetissova E, Kozlov M, Goldfarb A. Spatial organization of transcription elongation complex in Escherichia coli. Science. 1998;281:424–428. doi: 10.1126/science.281.5375.424. [DOI] [PubMed] [Google Scholar]
- 42.Palangat M, Meier T I, Keene R G, Landick R. Transcriptional pausing at +62 of the HIV-1 nascent RNA modulates formation of the TAR RNA structure. Mol Cell. 1998;1:1033–1042. doi: 10.1016/s1097-2765(00)80103-3. [DOI] [PubMed] [Google Scholar]
- 43.Pelet T, Curran J, Kolakofsky D. The P gene of bovine parainfluenza virus 3 expresses all three reading frames from a single mRNA editing site. EMBO J. 1991;10:443–448. doi: 10.1002/j.1460-2075.1991.tb07966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Radecke F, Spielhofer P, Schneider H, Kaelin K, Huber M, Dötsch C, Christiansen G, Billeter M A. Rescue of measles viruses from cloned DNA. EMBO J. 1995;14:5773–5784. doi: 10.1002/j.1460-2075.1995.tb00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reeder T C, Hawley D K. Promoter proximal sequences modulate RNA polymerase II elongation by a novel mechanism. Cell. 1996;87:767–777. doi: 10.1016/s0092-8674(00)81395-1. [DOI] [PubMed] [Google Scholar]
- 46.Sastry S, Ross B M. RNA-binding site in T7 RNA polymerase. Proc Natl Acad Sci USA. 1998;95:9111–9116. doi: 10.1073/pnas.95.16.9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneider H, Kaelin K, Billeter M A. Recombinant measles viruses defective for RNA editing and V protein synthesis are viable in cultured cells. Virology. 1997;227:314–322. doi: 10.1006/viro.1996.8339. [DOI] [PubMed] [Google Scholar]
- 48.Schnell M J, Mebatsion T, Conzelmann K K. Infectious rabies viruses from cloned cDNA. EMBO J. 1994;13:4195–4203. doi: 10.1002/j.1460-2075.1994.tb06739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Serra M J, Turner D H, Freier S M. Predicting thermodynamic properties of RNA. Methods Enzymol. 1995;259:243–261. doi: 10.1016/0076-6879(95)59047-1. [DOI] [PubMed] [Google Scholar]
- 50.Smith H C, Gott J M, Hanson M R. A guide to RNA editing. RNA. 1997;3:1105–1123. [PMC free article] [PubMed] [Google Scholar]
- 51.Thomas S M, Lamb R A, Paterson R G. Two mRNAs that differ by two nontemplated nucleotides encode the amino coterminal proteins P and V of the paramyxovirus SV5. Cell. 1988;54:891–892. doi: 10.1016/S0092-8674(88)91285-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thomas M J, Platas A A, Hawley D K. Transcriptional fidelity and proofreading by RNA polymerase II. Cell. 1998;15:627–637. doi: 10.1016/s0092-8674(00)81191-5. [DOI] [PubMed] [Google Scholar]
- 53.Uptain S M, Kane C M, Chamberlin M J. Basic mechanisms of transcript elongation and its regulation. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 54.Vidal S, Curran J, Kolakofsky D. Editing of the Sendai virus P/C mRNA by G insertion occurs during mRNA synthesis via a virus-encoded activity. J Virol. 1990;64:239–246. doi: 10.1128/jvi.64.1.239-246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vidal S, Curran J, Kolakofsky D. A stuttering model for paramyxovirus P mRNA editing. EMBO J. 1990;9:2017–2022. doi: 10.1002/j.1460-2075.1990.tb08330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Visomirski-Robic L M, Gott J M. Insertional editing in isolated Physarum mitochondria is linked to RNA synthesis. RNA. 1997;3:821–837. [PMC free article] [PubMed] [Google Scholar]
- 57.Visomirski-Robic L M, Gott J M. Insertional editing of nascent mitochondrial RNAs in Physarum. Proc Natl Acad Sci USA. 1997;94:4324–4329. doi: 10.1073/pnas.94.9.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.von Hippel P H, Yager T D. The elongation-termination decision in transcription. Science. 1992;255:809–812. doi: 10.1126/science.1536005. [DOI] [PubMed] [Google Scholar]
- 59.von Hippel P H. An integrated model of the transcription complex in elongation, termination, and editing. Science. 1998;281:660–665. doi: 10.1126/science.281.5377.660. [DOI] [PubMed] [Google Scholar]
- 60.Weiss R B, Dunn D M, Atkins J F, Gesteland R F. Ribosomal frameshifting from −2 to +50 nucleotides. Prog Nucleic Acid Res Mol Biol. 1990;30:159–183. doi: 10.1016/s0079-6603(08)60626-1. [DOI] [PubMed] [Google Scholar]