Abstract
Complement receptor 1 (CR1) is a membrane glycoprotein with a highly duplicated domain structure able to bind multiple ligands such as C3b and C4b, the activated fragments of complement components C3 and C4, respectively. We have previously used our knowledge of this domain structure to identify CSL040, a soluble extracellular fragment of CR1 containing the long homologous repeat (LHR) domains A, B, and C. CSL040 retains the ability to bind both C3b and C4b but is also a more potent complement inhibitor than other recombinant CR1-based therapeutics. To generate soluble CR1 variants with increased inhibitory potential across all three complement pathways, or variants with activity skewed to specific pathways, we exploited the domain structure of CR1 further by generating LHR domain duplications. We identified LHR-ABCC, a soluble CR1 variant containing a duplicated C3b-binding C-terminal LHR-C domain that exhibited significantly enhanced alternative pathway inhibitory activity in vitro compared to CSL040. Another variant, LHR-BBCC, containing duplications of both LHR-B and LHR-C with four C3b binding sites, was shown to have reduced classical/lectin pathway inhibitory activity compared to CSL040, but comparable alternative pathway activity. Interestingly, multiplication of the C4b-binding LHR-A domain resulted in only minor increases in classical/lectin pathway inhibitory activity. The CR1 duplication variants characterized in these in vitro potency assays, as well as in affinity in solution C3b and C4b binding assays, not only provides an opportunity to identify new therapeutic molecules but also additional mechanistic insights to the multiple interactions between CR1 and C3b/C4b.
Keywords: CSL040, complement, receptor, recombinant protein expression, long homologous repeat, potency
Complement receptor 1 (CR1) is a type I membrane protein that acts as a central regulator of complement. Its extracellular domain is comprised of 30 short consensus repeat (SCR) domains that are grouped into four long homologous repeat (LHR) domains responsible for ligand binding and biological activity (1, 2, 3). These ligands have been identified as the activated complement fragments and opsonins C3b and C4b (4, 5), which bind to LHR-A, LHR-B, and LHR-C; and C1q, mannose-binding lectin, and L-ficolin which bind to LHR-D (6, 7, 8). LHR-A binds to C4b and not to C3b, although some studies have suggested a weak interaction with the latter (9, 10, 11). LHR-B and LHR-C are functionally identical and are the main binding domains for C3b (2, 9), but they also bind C4b weakly in experiments where C3b is absent (12, 13), raising the question of whether C4b binds these domains when both ligands are present. The receptor–ligand interactions between CR1 and C3b, as well as C4b, are further complicated in that both molecules are present in plasma as monomers, dimers, and even heterodimers to mediate decay acceleration activity of C3 and C5 convertases and cofactor activity (CFA) mediated by factor I for the classical, lectin, and alternative complement pathways (1). The significantly stronger affinity of CR1 for dimeric C3b and C4b than monomeric ligands suggests a bivalent interaction between multiple LHR domains of a single CR1 molecule and each dimeric ligand (12, 14, 15).
Although there is some understanding of CR1 LHR domain contribution to ligand binding, CFA, and decay acceleration activity (reviewed by Hardy et al. 2023) (1), relatively few studies have been performed to understand their contribution to overall potency for the classical, lectin, and alternative pathways, as measured by either red blood cell hemolytic assays or soluble membrane attack complex formation (sC5b-9; containing the terminal complement components C5b, C6, C7, C8, and C9) using the Wieslab and similar assays. One of the earliest studies (14) demonstrated that LHR-A to LHR-C were necessary for inhibition of CHO cell lysis, with a subsequent study showing that removing the LHR-A domain from the soluble CR1 extracellular domain (desLHR-A) resulted in no difference in alternative pathway mediated erythrocyte lysis but did show a significant reduction in classical pathway lytic activity (16). Two later studies (13, 17) took a more systematic approach with the use of single and combined LHR domains as soluble molecules to assess lytic activity, but these experiments used only a single concentration of soluble receptor. Recently, an approach was taken to generate LHR domain truncations as recombinant soluble CR1 proteins and to comparatively assess them for inhibitory activity against the full extracellular domain of CR1 in both hemolytic and Wieslab assays for all complement pathways (18). These experiments not only led to a greater understanding of the contribution of the CR1 LHR domains to in vitro potency but also to the identification of CSL040, a molecule comprised of LHR-ABC that is being assessed clinically as a potential therapeutic candidate (18). CSL040 has been shown to be more potent in vitro and in vivo (18, 19, 20) than the full extracellular domain of CR1, itself a former clinical candidate known as TP10/CDX-1135 (21).
One of the most important conclusions derived from the Wymann et al. (2021) study (18) was that single LHR domains, when compared to CSL040/LHR-ABC, have dramatically reduced inhibitory activity for all three complement pathways. The transition from single LHR domain to two LHR domain variants, in the form of LHR-AB or LHR-BC, led to synergistic increases in inhibitory activity for all three complement pathways, suggesting that both LHR domains bind with high affinity and bivalently to (homo- or hetero-) dimeric ligands. The transition from constructs encoding two LHR domains (LHR-AB, LHR-BC) to CSL040/LHR-ABC led to further synergistic increases in complement inhibitory activity for all three pathways (18). This implies that three ligand binding sites are required to provide the high potency of CSL040/LHR-ABC and raised the question of whether this activity could be further improved by the generation of soluble CR1 variants with additional C3b/C4b binding domains. There was also the possibility that manipulating the domain composition of soluble CR1 variants by duplication and/or replacement could skew their biological activity toward specific pathways and allow for the generation of novel therapeutic candidates.
In this study, therefore, we have generated variants of soluble CR1 containing duplications of either LHR-A or LHR-B/LHR-C and conducted comparative in vitro potency assays to provide mechanistic insights into how multiple LHR domains contribute to overall potency. To support this, affinity in solution assays have been performed to determine the affinity of specific CR1 variants to both C3b and C4b. These experiments also aim to identify new therapeutic candidate molecules with increased or differential complement pathway inhibitory activity.
Results
Duplication of the CR1 LHR-A domain and its effect on complement activity in vitro
To explore the possibility of generating a variant of CR1 with enhanced or specific classical/lectin pathway inhibitory activity compared to CSL040 and given the contribution of the C4b-binding LHR-A domain to this activity, we set out to determine the effect of duplicating the LHR-A domain on in vitro potency, without LHR-B or LHR-C present. Soluble constructs containing single and multiple CR1 LHR-A domains tandemly arranged (LHR-A, LHR-AA, LHR-AAA, LHR-AAAA; Fig. 1) were expressed, purified (Table S1), and assessed for their ability to inhibit complement pathway activity using both Wieslab and hemolytic complement inhibition assays, with CSL040 (LHR-ABC) as a positive control (Fig. 2, Table 1).
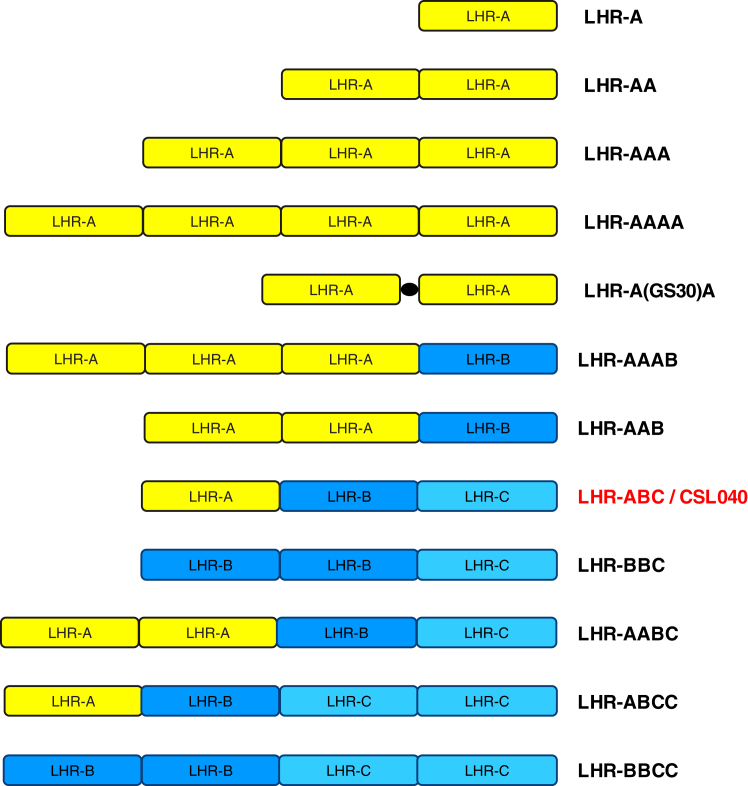
Figure 1.
Expressed variants of CR1-containing duplications of LHR domains. All soluble CR1 variants used in this study that containing LHR domain duplications are shown here schematically, with amino acids 42 to 489, 490 to 939, and 940 to 1392 used to define the boundaries of LHR-A, LHR-B, and LHR-C, respectively, based on Met+1. The solid black circle between LHR-A domains in construct LHR-A(GS30)A indicates the location of an in-frame 30 amino acid Gly-Ser linker. CSL040, containing LHR-A, LHR-B, and LHR-C, was used as the primary comparator for all experiments. CR1, complement receptor 1; LHR, long homologous repeat.
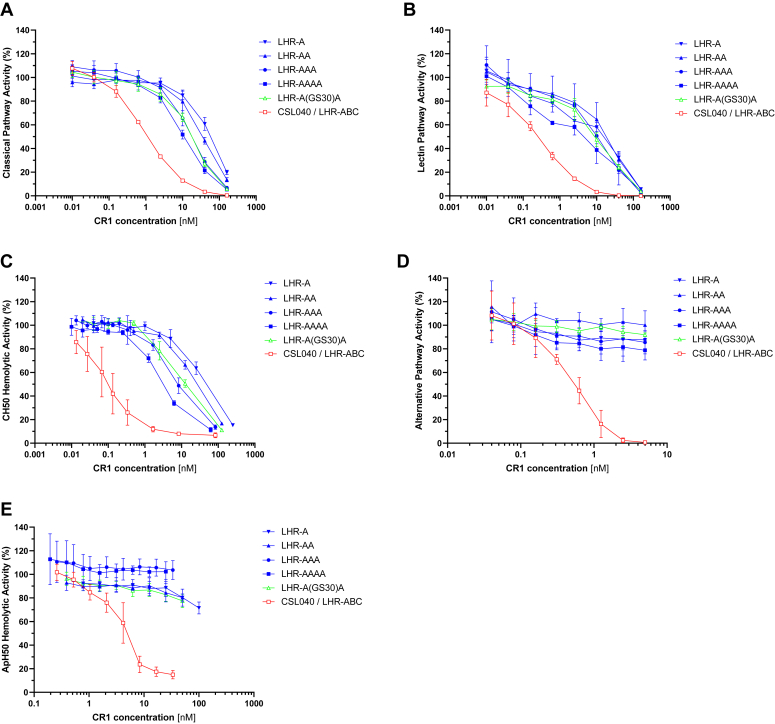
Figure 2.
Dose-dependent activity of human soluble CR1 truncation variants containing tandem multiplications of the LHR-A domain in vitro. The indicated CR1 LHR-A domain variants (Fig. 1) were tested in the following complement assays: the Wieslab classical (A), lectin (B), and alternative (D) pathways assays; and the red blood cell hemolytic assays specific for the classical (C) and alternative (E) pathways (CH50 and ApH50, respectively). LHR-A(GS30)A refers to an LHR-AA construct containing a 30-amino acid Gly-Ser linker between each LHR-A domain. CSL040 containing LHR-ABC was used as a control/comparator. Data points show the mean (±SD) activity (%) for each CR1 variant concentration (nM) tested. IC50 values calculated from these data are shown in Table 1. CR1, complement receptor 1; LHR, long homologous repeat.
Table 1.
Potency of human soluble CR1 truncation variants containing tandem multiplications of the LHR-A domain in complement pathway specific Wieslab and red blood cell hemolytic assays
| Human CR1 truncation |
Wieslab IC50 (nM) ± SD |
Hemolytic IC50 (nM) ± SD |
|||
|---|---|---|---|---|---|
| Domain | Classical | Lectin | Alternative | Classical | Alternative |
| LHR-A | 53.1 ± 9.7 | 8.9 ± 1.9 | No activity | 43.5 ± 9.0 | No activity |
| LHR-AA | 33.7 ± 7.4 | 15.1 ± 10.8 | No activity | 24.9 ± 2.7∗ | No activity |
| LHR-AAA | 18.0 ± 4.3∗∗ | 9.0 ± 2.5 | No activity | 8.8 ± 2.1∗∗ | No activity |
| LHR-AAAA | 11.0 ± 2.0∗∗ | 3.4 ± 2.4∗ | No activity | 3.4 ± 0.3∗∗ | No activity |
| LHR-A(GS30)A | 16.5 ± 3.1∗∗ | 7.5 ± 2.7 | No activity | 12.5 ± 1.5∗∗ | No activity |
| LHR-ABC/CSL040 | 1.2 ± 0.1∗∗ | 0.2 ± 0.1∗∗ | 0.5 ± 0.04 | 0.1 ± 0.1∗∗ | 4.9 ± 1.6 |
LHR-A(GS30)A refers to an LHR-AA construct containing a 30 amino acid Gly-Ser linker between each LHR-A domain. CSL040 containing LHR-ABC was used as a control/comparator. The IC50 values listed are the mean ± SD of three independent experiments, shown graphically in Figure 2. Statistically significant differences for individual HuCR1 fragment IC50 values compared to LHR-A for each pathway assay were calculated by an unpaired t test; ∗ p < 0.05; ∗∗ p < 0.005.
CR1, complement receptor 1; LHR, long homologous repeat.
As shown in Figure 2A, increasing the number of LHR-A domains from one to four in a single molecule conferred modest increases in classical pathway inhibitory activity in vitro, with the effects appearing to be additive rather than synergistic, based on the calculated IC50 values (Table 1). The increased classical pathway inhibitory activities of both LHR-AAA and LHR-AAAA were statistically significant compared to LHR-A alone (p < 0.005). However, the activity of LHR-AAAA remained 10-fold weaker than CSL040 (Fig. 2A; Table 1). The effect of increasing the number of LHR-A domains on lectin pathway activity was not as clear as for the classical pathway (Fig. 2B) but LHR-AAAA was also found to be the most inhibitory of the duplication variants tested, with an approximately 2.6-fold increase in potency compared to LHR-A (p < 0.05; Table 1). To confirm the above findings, a hemolytic assay specific for the classical pathway (CH50) was used to comparatively assess the CR1 LHR-A domain variants. As shown in Figure 2, C and A domain-dependent increase in CH50 activity was also observed, with the IC50 values of LHR-AA, LHR-AAA, and LHR-AAAA increasingly and significantly more potent than LHR-A alone (up to 13-fold; Table 1). Similar to the data observed for the Wieslab assays, LHR-AAAA was at least 10-fold less potent than CSL040 in the CH50 assay (Table 1). In contrast to CSL040, however, none of the LHR-A domain multiplication variants inhibited alternative pathway activity in vitro, using either the alternative pathway-specific Wieslab assay (Fig. 2D) or the hemolytic alternative pathway (ApH50) assay (Fig. 2E); calculated IC50 values confirmed this (Table 1). This data shows that multiplying the LHR-A domain of soluble CR1 confers only small, additive improvements in classical and lectin pathway activity and demonstrates the importance of including LHR-B/-C domains within soluble CR1 molecules for maximal inhibitory activity toward all complement pathways.
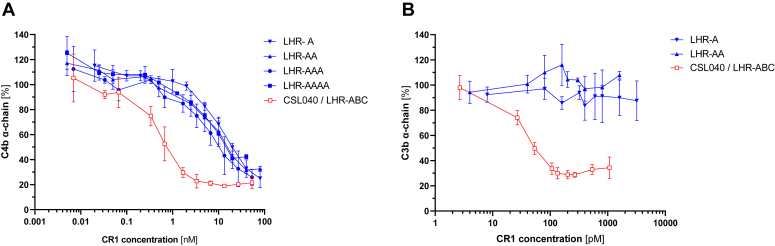
We extended our analysis of the LHR-A domain variants by conducting CFA assays for both C4b and C3b where the ability of CR1 to act as a cofactor for complement factor I–mediated cleavage of either ligand is assessed. As shown in Figure 3A and in Table 2, LHR-A showed dose-dependent C4b CFA. However, this activity was significantly weaker (>20-fold; p < 0.0005; Table 2) than that measured for CSL040, suggesting that the LHR-A domain is not sufficient for full C4b CFA and that LHR-B and/or LHR-C are also required. LHR-AA, LHR-AAA, and LHR-AAAA exhibited no statistically significant increase in C4b CFA (Fig. 3A, Table 2) compared to LHR-A alone. In contrast, when LHR-A and LHR-AA were assessed for C3b CFA, no activity at all was detectable (Fig. 3B, Table 2). CSL040 in comparison showed strong C3b CFA (Fig. 3B), with an IC50 of 78.94 ± 20.20 PM calculated, approximately 10-fold stronger than its C4b CFA IC50 (860 ± 290). The C3b CFA data suggests that the LHR-B and/or LHR-C domains are responsible for its activity.
Figure 3.
C3b and C4b cofactor activity of LHR-A domain variants. The indicated CR1 LHR-A domain variants (Fig. 1) were tested in C4b (A) and C3b (B) cofactor activity assays. CSL040 containing LHR-ABC was used as a control/comparator cofactor activity is expressed as the % of intact C4b or C3b alpha-chain relative to un-cleaved C4b or C3b, respectively. Data points show the mean (±SD) C3b/C4b alpha chain (%) for each CR1 variant concentration (nM or pM) tested in combination with a constant Factor I concentration. IC50 values calculated from these data are shown in Table 2. CR1, complement receptor 1; LHR, long homologous repeat.
Table 2.
Potency of human soluble CR1 LHR-A domain variants in C3b and C4b cofactor activity assays
| Human CR1 truncation |
Cofactor activity (pM ± SD) |
|
|---|---|---|
| Domain | C4b | C3b |
| LHR-ABC/CSL040 | 860 ± 290 | 78.9 ± 20.2 |
| LHR-A | 20,460 ± 3280∗∗∗ | No activity |
| LHR-AA | 16,650 ± 2700∗∗∗ | No activity |
| LHR-AAA | 11,800 ± 4700∗ | Not tested |
| LHR-AAAA | 14,690 ± 4220∗∗ | Not tested |
The IC50 values listed are the mean ± SD co-factor activity (in pM) of three independent experiments, shown graphically in Figure 3. Statistically significant differences for LHR-A domain variant IC50 values compared to LHR-ABC/CSL040 were calculated by an unpaired t test; ∗ p < 0.05; ∗∗ p < 0.005; and ∗∗∗ p < 0.0005.
CR1, complement receptor 1; LHR, long homologous repeat.
Steric hindrance at the N-terminal end of soluble CR1
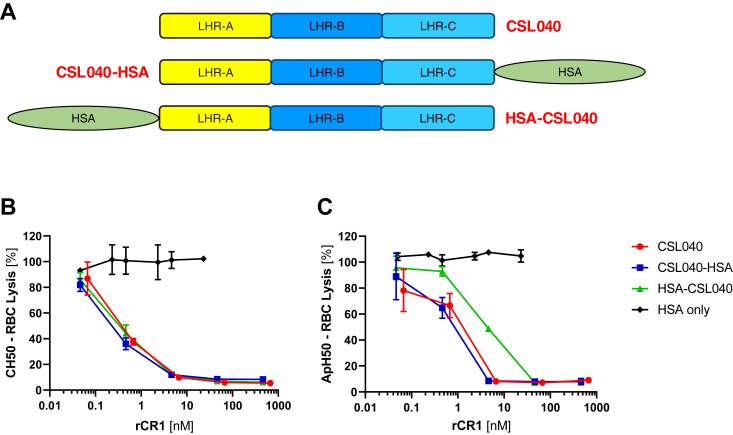
In order to understand whether steric hindrance of C4b binding might be playing a role in reducing potency gains when multiple LHR-A domains are present in tandem, an LHR-AA construct was generated with a 30-amino acid Gly-Ser linker introduced in-frame between each LHR-A domain (LHR-A(GS30)A; Fig. 1) and its potency compared to LHR-AA in vitro. As shown in Figure 2, A–C and in Table 1, a statistically significant (p < 0.005) 2-fold improvement in classical/lectin pathway activity was observed for LHR-A(GS30)A compared to LHR-AA. This data indicates that although additional spacing between LHR-A domains does confer increased inhibitory activity, the gains are relatively modest. We also examined the effect of adding large globular proteins such as human serum albumin (HSA) to the N-terminal end of LHR-A would have to complement pathway activity, since these types of proteins can have a positive impact on plasma clearance (22). Having previously shown that a C-terminal HSA-fusion of CSL040/LHR-ABC did not affect complement pathway activity (20), we constructed an N terminally fused version, HSA-CSL040, and compared its ability to inhibit the activity of all three complement pathways in vitro, using CSL040-HSA, CSL040, and HSA alone as controls (Fig. 4A). The results of CH50 and ApH50 assays are shown in Figure 4, B and C, respectively, and in Table 3 as calculated IC50 values. As expected, HSA alone was noninhibitory, and CSL040-HSA did not show any differences in inhibitory activity compared to CSL040 alone. HSA-CSL040 also showed no significant differences in inhibiting CH50 activity compared to CSL040 (Fig. 4B, Table 3), suggesting that HSA does not sterically block C4b from binding the adjacent LHR-A domain. In contrast, however, a statistically significant (p < 0.05) decrease in ApH50 activity of approximately 5-fold was measured for HSA-CSL040 compared to CSL040 alone (Fig. 4C, Table 3), indicating that the HSA component of the fusion protein may sterically (and selectively) inhibit C3b binding to LHR-B and/or LHR-C within CSL040.
Figure 4.
Comparative in vitro efficacy of N- and C-terminal HSA fusion of CSL040.A, schematic representation of CSL040 and N-terminal and C-terminal fusions of CSL040 with human serum albumin (HSA). These constructs, as well as HSA alone, were comparatively assessed for in vitro inhibitory activity using (B) CH50 classical pathway hemolytic and (C) ApH50 alternative pathway hemolytic activity. Data points show the mean (±SD) activity (%) for each CR1 variant concentration (nM) tested. IC50 values calculated from these data are shown in Table 3. CR1, complement receptor 1.
Table 3.
Potency of CSL040 fused to CSL040/LHR-ABC in red blood cell hemolytic assays
| Human CR1 protein | Hemolytic IC50 (nM) ± SD |
|
|---|---|---|
| Classical | Alternative | |
| CSL040 | 0.4 ± 0.1 | 0.9 ± 0.5 |
| CSL040-HSA | 0.3 ± 0.1 | 0.7 ± 0.3 |
| HSA-CSL040 | 0.4 ± 0.1 | 4.5 ± 0.2∗ |
| HSA | No Activity | No Activity |
The IC50 values listed are the mean ± SD of three independent experiments, shown graphically in Figure 4. Statistically significant differences for each HSA fusion compared to CSL040 were calculated by an unpaired t test; ∗ p < 0.05.
CR1, complement receptor 1; HSA, human serum albumin; LHR, long homologous repeat.
LHR domain contributions to the complement inhibitory activity of CSL040
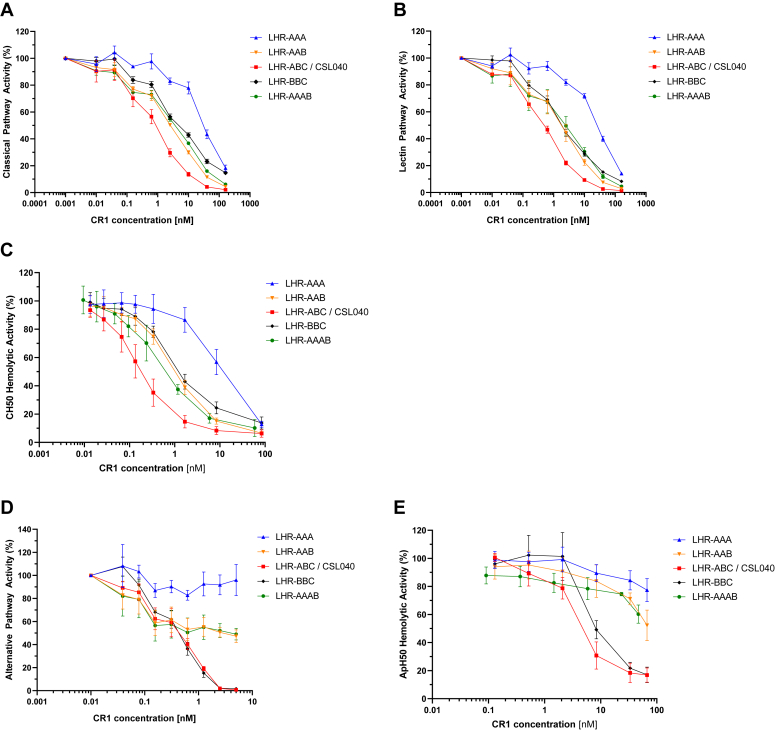
To gain a clearer understanding of the role of each LHR domain involved in both C3b and C4b binding (LHR-A, LHR-B, LHR-C) and its contribution to complement pathway activity, a series of CSL040 variants were constructed where, singly, LHR-A (C4b binding) was replaced with LHR-B or LHR-C (main C3b-binding domains), and vice versa. The variants, LHR-AAA, LHR-AAB, and LHR-BBC (Fig. 1), were then expressed recombinantly, purified, and comparatively assessed against CSL040/LHR-ABC in vitro using both Wieslab and hemolytic assays as above (Fig. 5; Table 4).
Figure 5.
Dose-dependent activity of LHR domain combination variants in vitro. The indicated CR1 LHR domain variants (Fig. 1) were tested in the following complement assays: the Wieslab classical (A), lectin (B), and alternative (C) pathways assays; and the red blood cell hemolytic assays specific for the classical (D) and alternative (E) pathways (CH50 and ApH50 assays, respectively). CSL040 containing LHR-ABC was used as a control/comparator. IC50 values calculated from these data are shown in Table 4. CR1, complement receptor 1; LHR, long homologous repeat.
Table 4.
Potency of LHR domain combination variants in complement pathway-specific Wieslab and red blood cell hemolytic assays
| Human CR1 truncation |
Wieslab IC50 (nM) ± SD |
Hemolytic IC50 (nM) ± SD |
|||
|---|---|---|---|---|---|
| Domain | Classical | Lectin | Alternative | Classical | Alternative |
| LHR-AAA | 30.9 ± 3.8∗∗∗ | 23.9 ± 1.9∗∗∗ | No activity | 11.3 ± 4.4∗ | No activity |
| LHR-AAB | 2.2 ± 0.3∗∗ | 1.5 ± 0.9 | Weak activitya | 1.1 ± 0.3∗ | Weak activitya |
| LHR-AAAB | 2.8 ± 0.6∗∗ | 2.0 ± 1.3 | Weak activitya | 0.8 ± 0.4 | Weak activitya |
| LHR-ABC/CSL040 | 0.8 ± 0.3 | 0.4 ± 0.04 | 0.4 ± 0.1 | 0.2 ± 0.1 | 6.0 ± 2.7 |
| LHR-BBC | 5.3 ± 0.9∗∗ | 2.1 ± 0.4∗∗ | 0.4 ± 0.1 | 1.7 ± 0.5∗ | 10.8 ± 3.6 |
See Figure 4 for graphical data. CSL040 containing LHR-ABC was used as a control/comparator. The IC50 values listed are the mean ± SD of three independent experiments, shown graphically in Figure 5. Statistically significant differences for individual HuCR1 fragment IC50 values compared to CSL040 for each pathway assay were calculated by an unpaired t test; ∗ p < 0.05; ∗∗ p < 0.005; and ∗∗∗ p < 0.0005.
CR1, complement receptor 1; LHR, long homologous repeat.
IC50 values could not be determined.
As shown in Figure 5A, the dose-responsive inhibition of Wieslab classical pathway activity by LHR-AAA was weaker than for CSL040/LHR-ABC (see also Fig. 2). The differences in IC50 values were approximately 40-fold and statistically significant (p < 0.0005; Table 4). Replacing the C-terminal LHR-A domain of LHR-AAA with LHR-B to produce LHR-AAB conferred an increase in potency of more than 10-fold compared to LHR-AAA, but which was still 3-fold lower than CSL040/LHR-ABC (Fig. 5A; Table 4). Similar results were obtained using the Wieslab lectin pathway assay and with the CH50 hemolytic assay (Fig. 5, B and C, Table 4). When the LHR-A domain of CSL040/LHR-ABC was replaced with LHR-B to form LHR-BBC and tested in the above assays, we observed a decrease in classical/lectin pathway inhibitory activity of LHR-BBC compared to CSL040/LHR-ABC (Fig. 5, A–C). The difference in measured IC50 values were between 5- and 8-fold and were statistically significant (p < 0.005; Table 4). This data suggests that for optimal classical/lectin pathway inhibitory activity, soluble complement receptor 1 molecules such as CSL040 require the C4b-binding LHR-A domain as well as at least two C3b-binding LHR-B/LHR-C domains.
A comparison of the CR1 variants LHR-AAA, LHR-AAB, LHR-ABC (CSL040), and LHR-BBC in alternative pathway-specific in vitro Wieslab and ApH50 assays was performed next, with the results shown in Figure 3, D and E, respectively. Here, we observed differences in comparative activity compared to the above results from the classical/lectin pathways assays. As shown previously (Fig. 2, D and E), LHR-AAA did not inhibit the alternative pathway (Fig. 5, D and E), while LHR-AAB was observed to have only weak inhibitory activity, with no IC50 value able to be calculated (Table 4). This suggests that unlike CSL040 which has two LHR-B/LHR-C domains (Fig. 5, D and E), a single LHR-B/LHR-C domain is not sufficient to confer full alternative pathway inhibitory activity. Interestingly, LHR-BBC did not provide a significant alternative pathway potency improvement compared to CSL040/LHR-ABC (Fig. 5, D and E, Table 4), suggesting that in this instance the additional LHR-B domain is able to compensate for the loss of LHR-A.
An additional construct, LHR-AAAB, was also tested to determine whether duplication of the LHR-A domain in the presence of LHR-B could provide any potency improvement in any of the pathway-specific assays. As shown in Figure 5, A–E and in Table 4, no significant differences in inhibitory activity were found when LHR-AAB was compared to LHR-AAAB. Taken together, this data suggests that maximal inhibition of the classical and lectin pathways with CR1 variants containing three LHR domains requires sites with both a single LHR-A domain and two LHR-B/LHR-C domains. For alternative pathway inhibition, LHR-A appears dispensable only when replaced by an LHR-B (or LHR-C) domain.
Comparative analysis of soluble CR1 variants containing four C3b/C4b-binding LHR domains
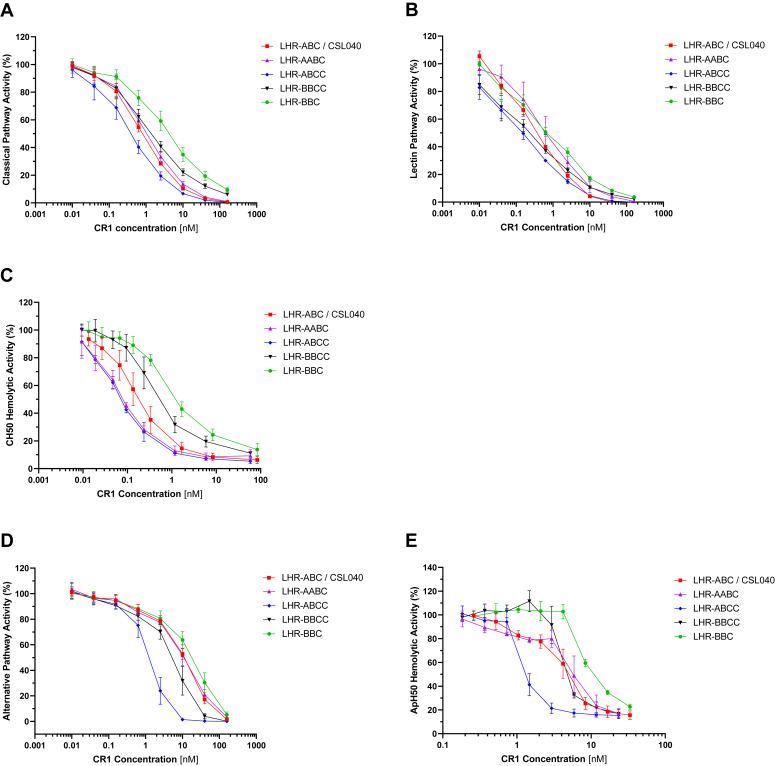
Having determined the importance of having three CR1 LHR domains with at least two of them as C3b-binding domains (LHR-B/LHR-C) plus a single C4b-binding LHR-A domain to ensure robust complement inhibitory activity, we hypothesized that soluble CR1 constructs containing four LHR-A, LHR-B, and/or LHR-C domains might exhibit greater inhibitory activity than CSL040/LHR-ABC. In addition, the combination of LHR domains might skew responses toward one or more complement pathways. To this end, LHR-ABCC and LHR-BBCC were generated (Fig. 1) and tested these domain duplication variants in vitro using both Wieslab and hemolytic assays against with CSL040/LHR-ABC and LHR-BBC as comparators. An additional variant, LHR-AABC (Fig. 1), was constructed to confirm that LHR-A domain duplication in the context of CSL040/LHR-ABC did not confer any potency improvements over CSL040/LHR-ABC alone.
As shown in Figure 6, A–E and in Table 5, LHR-BBCC showed a statistically significant (p < 0.05) improvement (2- to 4-fold) in inhibitory activity for all three complement pathways compared to LHR-BBC, suggesting that an additional ligand binding domain was contributing to its improved activity. Compared to CSL040/LHR-ABC, however, LHR-BBCC showed no improvement in classical pathway activity in Wieslab assays and a significant reduction (p < 0.05) in activity in the CH50 hemolytic assay. Alternative pathway Wieslab assay results showed a small but statistically significant (p < 0.05) improvement in LHR-BBCC activity compared to CSL040/LHR-ABC, while no differences were observed in the ApH50 assay (Table 5). Taking CSL040/LHR-ABC and duplicating the LHR-A domain to form LHR-AABC resulted in no statistically significant differences in complement activity for any of the pathways tested (Fig. 6, A–E, Table 5), confirming earlier results on the lack of improved potency brought about by duplicating this domain. A further construct, LHR-ABCC, was produced which retained the C4b binding site in LHR-A but was also comprised of three C3b binding sites in its LHR-B and -C domains. When tested in Wieslab and hemolytic assays specific for the classical and lectin pathways, an improvement in inhibitory activity was observed compared to CSL040/LHR-ABC (Fig. 6, A–C). The calculated differences in IC50 values were found to be approximately 2-fold and statistically significant for the Wieslab assay results (p < 0.05). Statistical significance could not be demonstrated for the IC50 values determined from the CH50 assay due to the large SD in the CSL040/LHR-ABC results (Table 5). In alternative pathway assays, LHR-ABCC showed a marked improvement in dose-dependent inhibitory activity compared to CSL040/LHR-ABC (Fig. 6, D and E). This improvement was approximately 10-fold based and statistically significant (p < 0.0005), based on calculated Wieslab IC50 values, and 3-fold for the ApH50 assay (Table 5). In summary, soluble CR1 constructs containing four C3b/C4b-binding LHR domains exhibited potency benefits compared to CSL040/LHR-ABC with one, LHR-ABCC, demonstrating 2- to 3-fold and 3- to 10-fold improvements in classical/lectin, and alternative pathway inhibitory activity, respectively. This is likely attributed to the presence of an additional C3b-binding domain.
Figure 6.
Dose-dependent activity of CR1 variants containing duplicated LHR-B/C domains in vitro. The indicated CR1 LHR domain variants (Fig. 1) were tested in the following complement assays: the Wieslab classical (A), lectin (B), and alternative (C) pathways assays; and the red blood cell hemolytic assays specific for the classical (D) and alternative (E) pathways (CH50 and ApH50 assays, respectively). CSL040 containing LHR-ABC was used as a control/comparator. IC50 values calculated from these data are shown in Table 5. CR1, complement receptor 1; LHR, long homologous repeat.
Table 5.
Potency of human soluble CR1 truncation variants containing duplicated LHR-B/C domains in complement pathway-specific Wieslab and red blood cell hemolytic assays
| Human CR1 truncation |
Wieslab IC50 (nM) ± SD |
Hemolytic IC50 (nM) ± SD |
|||
|---|---|---|---|---|---|
| Domain | Classical | Lectin | Alternative | Classical | Alternative |
| LHR-ABC/CSL040 | 0.8 ± 0.2 | 0.4 ± 0.04 | 9.4 ± 1.2 | 0.2 ± 0.1a | 5.0 ± 1.1 |
| LHR-AABC | 1.1 ± 0.3 | 0.7 ± 0.5 | 9.7 ± 3.1 | 0.1 ± 0.03 | 5.9 ± 2.2 |
| LHR-ABCC | 0.4 ± 0.2∗ | 0.1 ± 0.05∗∗ | 0.9 ± 0.4∗∗∗ | 0.1 ± 0.03 | 1.7 ± 0.5∗ |
| LHR-BBCC | 1.5 ± 0.5 | 0.2 ± 0.1 | 4.7 ± 1.7∗ | 0.7 ± 0.3∗ | 5.4 ± 0.3 |
| LHR-BBC | 4.4 ± 1.7∗ | 0.8 ± 0.3 | 15.5 ± 6.0 | 1.7 ± 0.5a∗ | 12.6 ± 0.5∗∗∗ |
CSL040 containing LHR-ABC was used as a control/comparator. The IC50 values listed are the mean ± SD of three independent experiments. Statistically significant differences for individual HuCR1 fragment IC50 values compared to CSL040 for each pathway assay were calculated by an unpaired t test; ∗ p < 0.05; ∗∗ p < 0.005; ∗∗∗ p < 0.0005.
CR1, complement receptor 1; LHR, long homologous repeat.
Affinity in solution of plasma derived C3b and C4b to truncation and duplication variants of sCR1
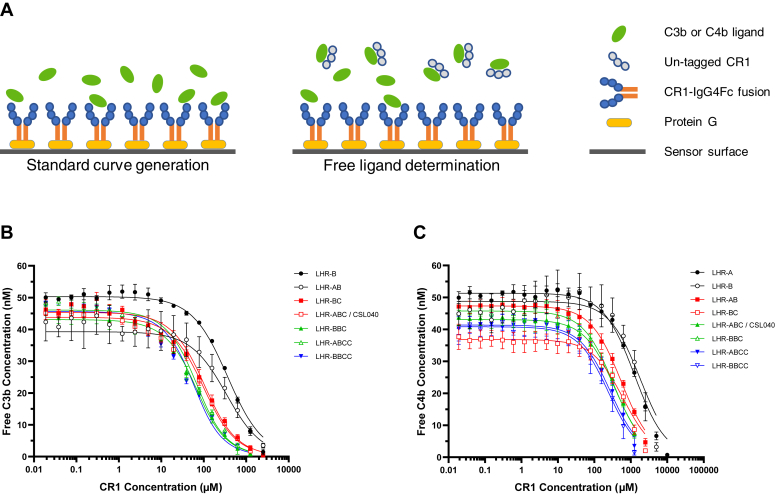
To gain mechanistic insights into the potency differences of the various soluble CR1 constructs tested in vitro, we employed an affinity in solution method (Fig. 7A) to determine and compare their respective affinities to plasma-derived C3b (pdC3b) and plasma-derived C4b (pdC4b). Each pdC3b and pdC4b preparation contained a mixture of monomeric and dimeric species that were consistent with preparations used previously (18). The results of these experiments are summarized in Figure 7, B and C and in Table 6. No binding to pdC3b was observed for LHR-A alone and comparatively weak affinities were determined for LHR-B (336.6 ± 28.8 nM) and LHR-AB (305.3 ± 22.6 nM). LHR-BC and CSL040/LHR-ABC showed an approximately 4-fold stronger affinity to pdC3b than LHR-B or LHR-AB (78.2 ± 10.4 nM for LHR-BC and 76.4 ± 4.3 nM for CSL040/LHR-ABC: Table 6) but not to each other. This data suggests a switch from a monovalent interaction of LHR-B (or LHR-AB) with C3b monomer to a bivalent interaction with dimeric C3b for the LHR-B and LHR-C domains of LHR-BC and LHR-ABC/CSL040. A significant improvement in the apparent affinity to pdC3b of approximately 1.7-fold was determined for LHR-BBC (44.4 ± 4.2 nM) and LHR-ABCC (44.6 ± 2.1 nM) compared to CSL040/LHR-ABC (p < 0.005; Table 6). The LHR-BBCC variant was found to have further improvements in affinity to pdC3b (34.8 ± 1.8 nM; Table 6), likely due to increased avidity brought about by pdC3b binding to CR1 variants with increasing numbers of C3b binding sites. In summary, the in-solution-affinity data suggest that the apparent affinity of CR1 variants to pdC3b increases with the number of C3b binding sites present in each molecule.
Figure 7.
Affinity in-solution of CR1 and duplication variants.A, the method for generating affinity in solution data. On the left-hand side, CR1-IgG4Fc fusions of each CR1 variant/duplication to be tested were captured onto a protein G–coupled sensor surface, and then increasing concentrations of pdC3b or pdC4b added to generate standard response curves. As shown in the right-hand panel, affinity in-solution data was then generated by premixing increasing concentrations of untagged versions of each CR1 variant with 50 nM pdC3b or pdC4b and running each over their respective CR1-IgG4Fc captured protein G–coupled sensor surface to determine free C3b, using the previously generated standard curves as a reference. CR1 concentration was then plotted against free C3b (B) or free C4b (C). In both graphs, ligand concentration (y-axis), measured as mean ± SD from N = 3 experiments each is plotted against total concentration of the respective CR1 truncation (x-axis). See Table 6 for calculated dissociation constants derived from these graphs. CR1, complement receptor 1.
Table 6.
Affinity in-solution dissociation constants of plasma-derived C3b and C4b to CR1 variants
| Human CR1 truncation | Affinity in solution to pdC3b KD (nM) | Affinity in solution to pdC4b KD (nM) |
|---|---|---|
| LHR-A | No binding | 1231.0 ± 116.9∗∗ |
| LHR-AA | ND | 1545.0 ± 54.7∗∗ |
| LHR-B | 336.6 ± 28.8∗∗ | 1913.7 ± 175.2∗∗ |
| LHR-AB | 305.3 ± 22.6∗∗ | 530.2 ± 32.1∗ |
| LHR-BC | 78.2 ± 10.4 | 581.2 ± 43.0∗ |
| LHR-ABC/CSL040 | 76.4 ± 4.3 | 352.8 ± 15.5 |
| LHR-BBC | 44.4 ± 4.2∗ | 317.0 ± 23.2 |
| LHR-ABCC | 44.6 ± 2.1∗∗ | 249.1 ± 20.1∗ |
| LHR-BBCC | 34.8 ± 1.8∗∗ | 218.6 ± 4.8∗∗ |
Shown in this table are the mean ± SD in-solution affinities (nM) for each CR1 variant binding to plasma-derived C3b (pdC3b) and C4b (pdC4b), based on N = 3 experiments. KD is the equilibrium dissociation constant of the interaction, calculated from sensorgram data fitted to a 1:1 affinity in-solution binding model. See Figure 7, B and C for the graphical data used to generate these values. Statistically significant differences for individual CR1 fragment affinities compared to CSL040 were calculated by an unpaired t test; ∗ p < 0.005; ∗∗ p < 0.0005.
CR1, complement receptor 1; LHR, long homologous repeat; ND, not done.
A similar result was observed for the affinities in solution of the same CR1 variants to pdC4b (also shown in Table 6), where the apparent affinity was shown to be dependent on the number of C4b binding sites found in each molecule tested. In contrast to pdC3b binding, LHR-A did bind pdC4b alone although the measured affinity was weak (1231.0 ± 116.9 nM). Binding of pdC4b to LHR-B was slightly weaker of with an affinity of 1913.7 ± 175.2 nM than LHR-A (Table 6). Both LHR-AB (530.2 ± 32.1 nM) and LHR-BC (581.2 ± 43.0 nM) showed a significant improvement in binding affinity to pdC4b of 2- and 4-fold compared to either LHR-A or LHR-B, respectively (Table 6; p < 0.005), suggesting that either each LHR domain binds monomeric C4b separately or bivalently to dimeric C4b. CSL040/LHR-ABC and LHR-BBC showed similar binding affinities to pdC4b (352.8 ± 15.5 nM and 317.0 ± 23.2 nM, respectively) but approximately 1.5-fold stronger than LHR-AB or LHR-BC (530.2 ± 32.1 nM and 581.2 ± 43.0 nM, respectively; p < 0.05; Table 6). The CR1 variants LHR-ABCC and LHR-BBCC displayed further improvements in pdC4b binding, with measured affinities of 249.1 ± 20.1 nM and 218.6 ± 4.8 nM, respectively (Table 6). These data suggest that all four domains within these two CR1 variants interact with multiple C4b molecules, either as monomers and/or dimers, similar to what was observed for pdC3b (Table 6). Apart from the observed binding of pdC4b to LHR-A, the overall affinity to pdC4b of each CR1 variant tested was significantly weaker than pdC3b (Table 6), suggesting a possible preference for binding of LHR-B and LHR-C for the latter when both ligands are present.
Discussion
Understanding the interaction between receptor and ligand in the case of CR1 is important from both a biological and a drug development perspective, but is complicated by the presence of multiple ligands as well as multiple binding sites delineated by the LHR domains, and the SCR domains contained within them (1). We set out to analyze the in vitro inhibitory activity of domain variants of soluble CR1 based on CSL040 (LHR-ABC), since this protein lacks the LHR-D domain responsible for C1q, mannose binding lectin, and ficolin binding (6, 7, 8) and confined this work to understanding ligand binding domain utilisation in the context of the main CR1 ligands, C3b, and C4b. Previous potency data (18) showed significant improvements in the ability of soluble CR1 truncation variants to inhibit complement pathway activity when LHR-A was compared to LHR-AB and then to LHR-ABC, or alternatively from LHR-B (or LHR-C) to LHR-BC to LHR-ABC. This clearly showed the importance of increasing the number of C3b/C4b-binding domains for optimal activity but also raised the question of the effect of domain usage on individual pathway activity and whether this could be exploited to generate a more potent molecule/therapeutic and/or variant with skewed activity toward either the classical/lectin or to the alternative complement pathway.
We therefore commenced our study with an examination of the comparative activity of CR1 LHR-A domain multiplications (A, AA, AAA, AAAA) on complement pathway inhibitory activity. LHR-A is considered the primary C4b binding site within CR1 (9, 12, 23). The studies were limited to constructs with up to four LHR domains, as it was hypothesized that larger constructs may provide limited additional potency benefits and/or have deleterious effects on protein clearance if tested in vivo as potential therapeutic candidates (18). Whole LHR domains were used, rather than the minimal binding regions contained therein, because previous studies conclusively demonstrated that reducing the spacing between ligand binding sites by deleting the intervening SCR domains attenuated activity in vitro (24, 25). Constructs containing duplicated LHR-A domains have been generated previously and showed increased activity compared to LHR-A alone (24). However, these studies were limited to decay acceleration experiments for the classical pathway C3 and C5 convertases only, and no comparison was made to full-length soluble CR1. Our studies using LHR-A domain multiplication variants (Fig. 2, Table 1) also showed improvements in classical pathway inhibitory activity as the number of LHR-A domains within each molecule increased, although all were weakly inhibitory compared to CSL040/LHR-ABC. These activity improvements appear disconnected to C4b CFA, since no significant activity differences were determined between any of the LHR-A duplication variants tested (Fig. 3). Compared to a 2-fold improvement in classical pathway inhibitory activity for LHR-AA relative to LHR-A, previous experiments showed a 10-fold improvement in activity for LHR-AB relative to LHR-A for the same pathway (18). This suggests that LHR-AB interacts with a C3b/C4b heterodimer to promote synergistic improvements in classical pathway potency compared to monomeric ligand for each single LHR-containing domain. In the case of LHR-AA (and LHR-AAA and LHR-AAAA), the interactions of these sCR1 variants may only be directed toward monomeric C4b rather than to C4b homodimer since the increases in potency are additive rather than synergistic and so do not suggest a bivalent interaction with dimeric ligand.
In contrast to the results with LHR-A and LHR-AA, previous data showed significant improvements in classical/lectin pathway inhibitory activity for LHR-BC compared to either LHR-B or LHR-C alone (18), again demonstrating that the likely mechanism underlying this observation is a bivalent interaction with dimeric ligand for LHR-BC compared to monomeric ligand for LHR-B or LHR-C alone. It is important to reiterate that LHR-B and LHR-C are almost identical in both amino acid sequence and function (1, 2, 18) and so are essentially interchangeable. Constructs containing only LHR-A, regardless of the number of these moieties present, were unable to inhibit the alternative pathway in any of the assays used (Fig. 2). This makes sense mechanistically, given that we did not observe any binding for this domain to C3b, the ligand primarily responsible for inhibition of alternative pathway activity (Table 6).
Since the effect of duplicating the LHR-A domain contrasted with that of duplicating the LHR-B/LHR-C domains, a mechanistic explanation was desired, and so we investigated whether making an N-terminal HSA fusion to CSL040/LHR-ABC might sterically inhibit receptor–ligand interaction and result in reduced complement inhibitory activity. Previous experiments showed that C-terminal HSA fusions of CSL040 do not deleteriously affect potency (20). However, our data indicated that although an N-terminal HSA-CSL040 fusion was similar to both CSL040-HSA and CSL040 alone in terms of classical pathway activity, it displayed a 5-fold decrease in alternative pathway inhibitory activity (Fig. 4, Table 3). This suggested that the high degree of flexibility of CR1 previously observed (26) may be causing HSA to sterically hinder interactions with C3b but not C4b. That this does not occur with the LHR-AABC molecule (Fig. 6) may be explained by the most N-terminal LHR-A domain within LHR-AABC being able to bind C4b with no steric hindrance. Modification of the C4b binding site on LHR-A to increase its affinity of interaction may increase classical/lectin pathway inhibitory activity. To support this hypothesis, CR1 variants containing amino acid substitutions within LHR-A have previously been identified that display increases in both ligand binding and decay acceleration activity (25).
The next experiments were designed to elucidate the contribution of the LHR domains to the pathway-specific inhibitory activity of CSL040/LHR-ABC using a domain swapping approach (Fig. 5, Table 4). In terms of inhibitory activity toward the classical and lectin pathways, soluble constructs encoding LHR-AAA were the least potent, while CSL040/LHR-ABC was the most potent with LHR-AAB and LHR-BBC showing intermediate effects. This result showed that a single C4b and two C3b binding sites were required together for optimal potency. For alternative pathway inhibitory activity, LHR-ABC and LHR-BBC were equipotent, while LHR-AAB and LHR-AAA showed weak or no activity (Fig. 5, Table 4). Given that molecules containing two LHR domains such as LHR-AB and LHR-BC are also 10-fold less active than LHR-ABC (18), this data demonstrates that a minimum of three LHR domains are essential for maximal alternative pathway activity; two of them must be C3b-binding domains (LHR-B or LHR-C) with the third either a C3b or a C4b binding site (LHR-A, LHR-B, or LHR-C). Moreover, LHR-BBC is identified as a soluble molecule whose in vitro activity is skewed toward the alternative pathway due to a reduction in classical/lectin pathway inhibitory activity.
Having investigated variants of soluble CR1 containing three C3b/C4b LHR-binding domains, we asked whether molecules containing four domains might exhibit additional and/or differential potency improvements over CSL040/LHR-ABC and provide further insights into receptor–ligand interactions. To that end, the variants LHR-AAAB, LHR-AABC, LHR-ABCC, and LHR-BBCC (Fig. 1) were constructed and characterized in vitro. No differences in potency were observed between LHR-AAAB and LHR-AAB (Fig. 5) or between LHR-AABC and CSL040/LHR-ABC (Fig. 6). LHR-AAB and LHR-ABC are more potent compared to the LHR-A multiplication variants; additional LHR-A domains would therefore add negligible increases in potency. LHR-BBCC was found to have significantly improved complement inhibitory activity compared to LHR-BBC (Fig. 6, Table 5; p < 0.05), suggesting that the additional C3b binding site present in the duplicated LHR-C domain was the contributing factor. However, LHR-BBCC remained approximately 2- to 3-fold less potent than CSL040/LHR-ABCC as an inhibitor of the classical pathway and did not show improvements in alternative pathway activity, indicating the importance of retaining the C4b-binding LHR-A domain. In contrast, LHR-ABCC was shown to confer significant improvements in inhibitory activity for all three complement pathways compared the CSL040/LHR-ABC (Fig. 6, Table 5), with IC50 values approximately 2-fold greater for the classical/lectin pathways, and up to 10-fold greater for the alternative pathway. The additional LHR-C domain present in LHR-ABCC, as a C3b binding module, is likely responsible for the disproportionally greater improvements in alternative pathway inhibitory activity relative to the classical/lectin pathways (1). It is also possible that LHR-ABCC allows for a second bivalent interaction with ligand, with a C3b/C4b heterodimer-binding LHR-A and LHR-B, and the duplicated LHR-C domains binding dimeric C3b.
To correlate the in vitro potency findings described above with information on ligand binding affinity, we turned to an affinity in solution method (27) to provide a more reliable measure of total affinity of receptor to ligand compared to standard surface plasmon resonance (SPR) kinetics techniques where the interaction occurs at the surface of the biosensor chip which could introduce confounding effects. Using this method, affinities are determined based on the amount of free ligand following the generation of standard curves (Fig. 7). We used commercially available pdC3b and pdC4b, which both have an approximately 80:20 distribution of monomer to dimer and so the affinities generated would represent binding to both species and would take avidity into account.
Despite being able to separate and purify monomeric and dimeric C3b and C4b species from unfractionated ligand by size-exclusion chromatography (Fig. S1), generation of dimeric C3b and C4b of sufficient purity and amount precluded us from conducting comparative assessments of the multiple CR1 variants used in this study. Sufficient pdC3b dimer was purified to perform a single kinetic binding experiment which demonstrated high affinity binding (3.8 ± 0.3 nM) to CSL040/LHR-ABC compared to unfractionated pdC3b (134.3 ± 13.6 nM; Table S1; Fig. S2, A and B). This finding is consistent with previous observations (1). No meaningful affinity could be ascertained for binding of monomeric C3b to CSL040/LHR-ABC, even at high concentrations (Table S1; Fig. S2, C and D), with a biphasic response possibly attributed to either dimerization at higher concentrations or dimerization happening on the CR1-bound surface. No affinity data for either monomeric or dimeric C4b to CSL040/LHR-ABC was able to be generated. Given our aim was to provide a comparative assessment of both ligands binding to multiple CR1 variants, the affinity-in-solution approach was determined to be suitable, despite some limitations in overall data interpretation.
The determined affinities of selected soluble CR1 truncation and duplication variants to pdC3b aligned with the potency data, above. No binding of LHR-A was measured to pdC3b with binding observed for LHR-B (336.6 ± 28.8). Adding LHR-A to LHR-B did not significantly affect this affinity (305.3 ± 22.6) but adding a second C3b binding site in the form of LHR-BC or CSL040/LHR-ABC resulted in a 4-fold improvement in affinity, suggesting that the receptor–ligand interaction had converted from a monovalent to a bivalent one. This result is consistent with an early study where binding assays on transfected cells using iodinated C3b showed dissociation constants for chimeric LHR-BD and LHR-CD constructs 2- to 3-fold weaker than for the CR1 extracellular domain (28). Another study indirectly showed no contribution of LHR-A to C3b binding by demonstrating similar C3b binding to LHR-ABCD and to a similar construct lacking LHR-A (16). More recently, Schramm et al. (29) measured an affinity of 69 nM for plasma-derived C3b to the full extracellular domain of CR1, which is in line with the value of 76.4 nM that obtained here for CSL040/LHR-ABC (Table 6). LHR-BBC and LHR-ABCC, with three C3b binding sites each, showed a further 1.7-fold increase in affinity compared to CSL040/LHR-ABC, while LHR-BBCC showed an approximately 2-fold increase in affinity which correlated with the doubling of C3b binding sites present in the molecule. Based on these results, further increases in the number of C3b binding sites in the form of constructs containing five or more LHR-B or LHR-C domains might provide only limited affinity improvements.
The affinity data generated for pdC3b and pdC4b toward the CR1 variants tested correlated with the number of ligand binding sites present (Table 6). However, the pdC4b data also differed from pdC3b in two key aspects. Firstly, pdC4b bound both LHR-A and LHR-B (and by inference LHR-C) with weak affinity, which suggests an interaction with monomeric C4b ligand (Table 6). The affinity of C4b to LHR-B is similar to that previously reported for the SCR15 to 18 fragment of LHR-C (12). Secondly, the affinity of each CR1 variant was significantly weaker for pdC4b than it was for pdC3b (Table 6). This phenomenon has also been observed previously for soluble CR1 (1, 12, 30, 31) and indicates that C3b may outcompete C4b for binding to LHR-B and LHR-C depending on the local concentrations of each ligand. The use of constructs containing two, three, and four LHR domains in affinity in-solution experiments also suggested situations where C4b dimer might bind bivalently to two LHR domains, leaving a third LHR domain to bind monomer, or a third and fourth domain to bind bivalently to a second dimer. One limitation of this affinity in-solution experiments was that the affinity of each CR1 variant to C3b/C4b was performed on each ligand separately, and so we lack an understanding of the interplay of receptor and ligand when both ligands are present. Secondly, since preparations of pdC3b and pdC4b containing the natural mixture of monomeric and dimeric ligand were used for these studies, the separate contribution of monomeric and dimeric ligand to CR1 binding is unclear and was not able to be experimentally determined.
In conclusion, we have exploited the role of individual CR1 LHR domains as C3b and/or C4b binding sites to better understand the relative contribution of individual domains to ligand binding and complement inhibitory activity. This also allowed us to generate new molecules with greater inhibitory and therapeutic potential than CSL040 and/or with their inhibitory activity skewed to the alternative pathway. Although LHR-A domain duplications/multiplications did not significantly improve classical/lectin pathway activity in vitro via increased avidity, it is possible that the affinity of LHR-A to C4b could be increased by introducing point mutation(s). A previously described D150N point mutation within the LHR-A domain of soluble CR1 is an example of this (11). The LHR-ABCC variant is a potential new therapeutic candidate with significantly enhanced alternative pathway inhibitory activity. However, in vivo pharmacokinetic/pharmacodynamic analyses and comparative assessments in relevant preclinical animal models of disease are required in order to further characterize and validate this molecule.
Experimental procedures
Generation of copy DNA expression plasmids
The generation of a copy DNA (cDNA) encoding untagged CSL040 which contains the LHR-ABC domains of human CR1 has been previously described (18). This was used as a template to design constructs encoding truncations and/or duplications of the LHR-A, LHR-B, and LHR-C domains as listed in Figure 1, as well as LHR-B, LHR-AB, and LHR-BC, based on amino acids 42 to 489 for LHR-A, amino acids 490 to 939 for LHR-B, and amino acids 940 to 1392 for LHR-C. All constructs encoding LHR-A at the N-terminal end utilized the endogenous CR1 signal peptide; those not encoding LHR-A at the N-terminal end used amino acids 1 to 19 of Ceruloplasmin (GenBank Accession number NP_000087). A final cDNA encoding duplicated LHR-AA but with a 30 amino acid Gly-Ser linker—(SGG)7SGS—between each LHR-A domain was also designed. The generation of constructs encoding HSA, CSL040-HSA, and CSL040-IgG4Fc have also been previously described (20). A cDNA was designed encoding a fusion protein with HSA switched to the N-terminal end of CSL040. The CSL040-IgG4Fc construct was used as a template to design cDNAs encoding C-terminal IgG4Fc fusions of LHR-A, LHR-B, LHR-BC, LHR-BBC, LHR-ABCC, and LHR-BBCC. All cDNA constructs described above were codon-optimized for human expression and synthesized by GeneArt (Thermo Fisher Scientific). These cDNAs were cloned into pcDNA3.1 (Thermo Fisher Scientific) as previously described (18).
Cell culture, recombinant protein expression, and purification
Cell culture and transient transfection of Expi293F cells were carried out as previously described (18). The purification of untagged CSL040 was carried out as previously described (18). This method was also used to purify all other untagged CR1 duplication/truncation protein variants. Purification of HSA and IgG1Fc fusions of CR1 variants also used methods previously described (20). The yield, purity, and aggregation content of each purified sample is shown in Table S1.
Wieslab complement inhibition assays
Wieslab complement assays (Svar Life Sciences) specific for the classical, lectin, and alternative pathway were performed according to the manufacturer's recommendations and as previously described (18). Briefly, diluted study samples were mixed with prediluted pooled human complement serum (Innovative Research) and incubated for 1 h at 37 °C on immunoglobulin M-coated microtiter plates, mannan-coated microtiter plates, or lipopolysaccharide-coated microtiter plates (for the classical, lectin, and alternative pathways, respectively). After washing, an alkaline phosphatase-labeled antibody against a neoantigen expressed during C5b-9 formation was added to each well and incubated for 30 min at room temperature (RT). Another washing step was followed by the incubation of the p-Nitrophenyl phosphate solution during 30 min at RT. Absorbance at 405 nm was read using an EnVision plate reader (PerkinElmer) or the Synergy HT plate reader (Agilent BioTek). Raw values were expressed as a percentage of C5b-9 formation by the serum and control (normal human serum [NHS]). Results were analyzed where appropriate in GraphPad Prism (https://www.graphpad.com) for IC50 values using a log(inhibitor) versus response; Variable slope (four parameters) fit.
Hemolytic assays
The complement classical pathway-specific and alternative pathway-specific assays were performed similar to that previously described (18). For the classical pathway-specific assay, sheep erythrocytes (Acila Schweiz AG) were sensitized with rabbit anti-sheep antibodies (Hemolytic Ambozeptor; Virion/Serion) and diluted to 4 × 108 cells/ml GVB++ (GVB, 0.15 mM CaCl2, 0.5 mM MgCl2; CompTech). To assess the inhibition of the classical pathway, CSL040 and variants thereof were preincubated in 1/40 diluted NHS (Swiss Blood Donation Center) for 30 min at RT, added to the erythrocytes at a 1/1 (v/v) ratio, and incubated during 1 h at 37 °C in a microtiter-plate shaking device. For the alternative pathway-specific assay, rabbit erythrocytes (Jackson Laboratories, Boston USA) were washed and diluted to 2 × 108 cells/ml GVB/MgEGTA (GVB, 5 mM MgEGTA; CompTech). Prediluted CSL040 and variants thereof were preincubated in one-sixth diluted NHS for 30 min at RT), added to the erythrocytes at a 2/1 (v/v) ratio, and incubated during 1 h at 37 °C in a microtiter-plate shaking device. For both assays, ice-cold gelatin veronal buffer with EDTA (0.1% gelatin, 5 mM veronal, 145 mM NaCl, 0.025 % NaN3, 10 mM EDTA, pH 7.3) was added to each sample followed by centrifugation (10 min at 1250g). Hemolysis was determined by measuring the absorbance of released hemoglobin in the supernatant at 412 nm using an EnVision plate reader (PerkinElmer), or a Synergy HT plate reader (Agilent BioTek). Cells incubated with NHS and the corresponding buffers served as 100% lysis controls. The inhibition of lysis of each soluble CR1 variant was calculated relative to this control. Results were analyzed in GraphPad Prism as above.
CFA assays
Four micrograms C4b and 100 ng factor I (CompTech) were incubated for 60 min at 37 °C with 0 to 8000 ng CR1-protein. After incubation, 2x prediluted NuPAGE lithium dodecyl sulfate sample buffer and prediluted NuPAGE lithium dodecyl sulfate sample reducing agent (both Thermo Fisher Scientific) were mixed and added at a 1/1 (v/v) ratio and heated at 60 °C during 15 min. Reduced samples were then run on 8% BisTris Plus SDS gels (Thermo Fisher Scientific) to separate C4b α-chain from the β- and γ-chains. A Coomassie stain was then performed according to manufacturer`s instructions, using the GelCode blue stain reagent (Thermo Fisher Scientific). For the quantification of the cofactor activity of the analyzed CR1 variants, stained gels were imaged using a Fusion FX Imager (VILBER) and the intact C4 α-chain quantified via densitometry using EvolutionCapt edge Software (VILBER, http://www.vilber.de/en/products/chemiluminescence/fusion-fx/). For calculation of uncleaved C4b α-chain, a control sample (C4b incubated with factor I only) was run in parallel and set as 100% C4b α-chain. The C3b CFA assay was performed as previously described (18).
Affinity in solution
In-solution binding affinities were measured using SPR at 37 °C using a Biacore 8K+ (Cytiva) docked with protein G (carboxymethylated dextran matrix preimmobilised with protein G) sensor chip (Cytiva). The sensor surface was equilibrated with running buffer (10 mM Hepes; 150 mM NaCl; 0.1% bovine serum albumin pH 7.3) and preconditioned with three times 1-min pulse of 10 mM glycine pH 1.5. CR1-IgG4Fc fusions were captured on the protein G surface to approximately 1000 to 2000 response units. For affinity in solution assays, a standard curve was generated using a 2-fold dilution series of plasma derived C3b or C4b (0.4875–125 nM; CompTech) prepared in running buffer and injected over the CR1-IgG4Fc captured surface. Untagged CR1 variants at a concentration range of 0.15 to 10,000 nM prepared in running buffer were premixed for 60 min with a constant concentration of plasma derived C3b or C4b (50 nM) and separately injected over their respective CR1-IgG4Fc captured surface to measure free C3b or C4b concentrations. Responses from the reference surface (in which no CR1-IgG4Fc was captured) were subtracted from the CR1-IgG4Fc captured surface, to produce reference-subtracted data. Report points used for the analysis were the binding level (5 s before injection end) plotted against relative response derived from the calibration curves. Responses on the y-axis (calculated free C3b or C4b concentrations) were plotted against CSL040 concentration (Log scale) and fitted to an in-solution binding affinity model (evaluation software, Cytiva) to determine the affinity of the interaction.
Size-exclusion chromatography
Commercially purchased human pdC3b and pdC4b (CompTech) was loaded to a prepacked HiLoad 16/600 Superdex 200 pg column (Cytiva) equilibrated with 10 mM Hepes, 150 mM NaCl, pH7.4 with flow rate of 1.5 ml/min and 0.5 ml sample per fraction collected. Individual fractions were applied to a Superdex 200 Increase 5/150 Gl column in an Agilent 1260 Infinity LC system (Agilent) to determine sizing profile against a set of molecular size standard loaded as 1 μl at a flow rate of 0.4 ml/min.
SPR analyses; kinetics
SPR binding kinetics measurements were carried out using a Biacore T200 biosensor system (Cytiva). A series S-NTA chip (Cytiva) was used for the capture of His-tagged CSL040/LHR-ABC. The sensor chip surface was preconditioned with a 1-min pulse of 350 mM EDTA and charged for 1 min with 0.5 mM NiCl2 after equilibration with running buffer (10 mM Hepes, 150 mM NaCl, 0.1% bovine serum albumin pH 7.3) at the beginning of each cycle. CSL040 was captured on the NTA surface to levels ranging from 75 to 100 resonance units. Human pdC3b or pdC4b samples were diluted in running buffer in concentrations ranging from 15.6 nM to 1000 nM and injected over the CR1 for 150 s, and the dissociation was monitored for 180 s. For kinetic analysis of the dimeric fractions, dilutions in the running buffer were made at concentrations ranging from 1.56 to 50 nM. At the end of each cycle, the surface was regenerated with 60 s injection of 350 mM EDTA, followed by a buffer wash injection. All experiments were carried out at 37 °C with data analysis performed using the BIAevaluation software v4.1 (Cytiva, https://biaevaluation.software.informer.com/4.1). Responses from the reference surface (no CSL040 capture) were subtracted from the active surface, to produce reference-subtracted data. Reference subtracted responses from buffer blank injections were subtracted from the resultant sensorgrams to produce double-referenced data. Data was fitted to a 1:1 Langmuir binding model.
Data availability
All data are available on request from the authors. Authors confirm that all data are included in the manuscript.
Supporting information
This article contains supporting information.
Conflict of interest
S. W., A. B. M., T. R., and M. P. H. are listed as inventors on International Patent Publication number WO2019/218009. All authors are CSL shareholders.
Acknowledgments
Author contributions
S. W. and M. P. H. writing-review & editing; S. W., T. R., and M. P. H. writing-original draft; S. W., A. G. N., and M. P. H. visualization; S. W., A. G. N., and M. P. H. validation; S. W., A. G. N., S. E., G. A. P., and S. S. W. investigation; S. W., M. P., A. B. M., T. R., M. P. H. conceptualization; S. W., A. G. N., S. E., G. A. P., S. S. W., and M. P. H. formal analysis; T. R. and M. P. H. project administration; M. P., A. B. M., and M. P. H. supervision; S. W., A. G. N., M. P., A. B. M., T. R., and M. P. H. methodology.
Funding and additional information
This work was wholly supported by CSL Ltd. No external sources of funding were used.
Reviewed by members of the JBC Editorial Board. Edited by George DeMartino
Supporting information
References
- 1.Hardy M.P., Mansour M., Rowe T., Wymann S. The molecular mechanisms of complement receptor 1-it is complicated. Biomolecules. 2023;13:1522. doi: 10.3390/biom13101522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krych-Goldberg M., Atkinson J.P. Structure-function relationships of complement receptor type 1. Immunol. Rev. 2001;180:112–122. doi: 10.1034/j.1600-065x.2001.1800110.x. [DOI] [PubMed] [Google Scholar]
- 3.Liu D., Niu Z.X. The structure, genetic polymorphisms, expression and biological functions of complement receptor type 1 (CR1/CD35) Immunopharmacol. Immunotoxicol. 2009;31:524–535. doi: 10.3109/08923970902845768. [DOI] [PubMed] [Google Scholar]
- 4.Fearon D.T. Identification of the membrane glycoprotein that is the C3b receptor of the human erythrocyte, polymorphonuclear leukocyte, B lymphocyte, and monocyte. J. Exp. Med. 1980;152:20–30. doi: 10.1084/jem.152.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ross G.D. Analysis of the different types of leukocyte membrane complement receptors and their interaction with the complement system. J. Immunol. Methods. 1980;37:197–211. doi: 10.1016/0022-1759(80)90307-5. [DOI] [PubMed] [Google Scholar]
- 6.Ghiran I., Barbashov S.F., Klickstein L.B., Tas S.W., Jensenius J.C., Nicholson-Weller A. Complement receptor 1/CD35 is a receptor for mannan-binding lectin. J. Exp. Med. 2000;192:1797–1808. doi: 10.1084/jem.192.12.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacquet M., Lacroix M., Ancelet S., Gout E., Gaboriaud C., Thielens N.M., et al. Deciphering complement receptor type 1 interactions with recognition proteins of the lectin complement pathway. J. Immunol. 2013;190:3721–3731. doi: 10.4049/jimmunol.1202451. [DOI] [PubMed] [Google Scholar]
- 8.Klickstein L.B., Barbashov S.F., Liu T., Jack R.M., Nicholson-Weller A. Complement receptor type 1 (CR1, CD35) is a receptor for C1q. Immunity. 1997;7:345–355. doi: 10.1016/s1074-7613(00)80356-8. [DOI] [PubMed] [Google Scholar]
- 9.Klickstein L.B., Bartow T.J., Miletic V., Rabson L.D., Smith J.A., Fearon D.T. Identification of distinct C3b and C4b recognition sites in the human C3b/C4b receptor (CR1, CD35) by deletion mutagenesis. J. Exp. Med. 1988;168:1699–1717. doi: 10.1084/jem.168.5.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krych M., Clemenza L., Howdeshell D., Hauhart R., Hourcade D., Atkinson J.P. Analysis of the functional domains of complement receptor type 1 (C3b/C4b receptor; CD35) by substitution mutagenesis. J. Biol. Chem. 1994;269:13273–13278. [PubMed] [Google Scholar]
- 11.Krych M., Hauhart R., Atkinson J.P. Structure-function analysis of the active sites of complement receptor type 1. J. Biol. Chem. 1998;273:8623–8629. doi: 10.1074/jbc.273.15.8623. [DOI] [PubMed] [Google Scholar]
- 12.Reilly B.D., Makrides S.C., Ford P.J., Marsh H.C., Jr., Mold C. Quantitative analysis of C4b dimer binding to distinct sites on the C3b/C4b receptor (CR1) J. Biol. Chem. 1994;269:7696–7701. [PubMed] [Google Scholar]
- 13.Yazdanbakhsh K., Kang S., Tamasauskas D., Sung D., Scaradavou A. Complement receptor 1 inhibitors for prevention of immune-mediated red cell destruction: potential use in transfusion therapy. Blood. 2003;101:5046–5052. doi: 10.1182/blood-2002-10-3068. [DOI] [PubMed] [Google Scholar]
- 14.Makrides S.C., Scesney S.M., Ford P.J., Evans K.S., Carson G.R., Marsh H.C., Jr. Cell surface expression of the C3b/C4b receptor (CR1) protects Chinese hamster ovary cells from lysis by human complement. J. Biol. Chem. 1992;267:24754–24761. [PubMed] [Google Scholar]
- 15.Weisman H.F., Bartow T., Leppo M.K., Marsh H.C., Jr., Carson G.R., Concino M.F., et al. Soluble human complement receptor type 1: in vivo inhibitor of complement suppressing post-ischemic myocardial inflammation and necrosis. Science. 1990;249:146–151. doi: 10.1126/science.2371562. [DOI] [PubMed] [Google Scholar]
- 16.Scesney S.M., Makrides S.C., Gosselin M.L., Ford P.J., Andrews B.M., Hayman E.G., et al. A soluble deletion mutant of the human complement receptor type 1, which lacks the C4b binding site, is a selective inhibitor of the alternative complement pathway. Eur. J. Immunol. 1996;26:1729–1735. doi: 10.1002/eji.1830260810. [DOI] [PubMed] [Google Scholar]
- 17.Mqadmi A., Abdullah Y., Yazdanbakhsh K. Characterization of complement receptor 1 domains for prevention of complement-mediated red cell destruction. Transfusion. 2005;45:234–244. doi: 10.1111/j.1537-2995.2004.04163.x. [DOI] [PubMed] [Google Scholar]
- 18.Wymann S., Dai Y., Nair A.G., Cao H., Powers G.A., Schnell A., et al. A novel soluble complement receptor 1 fragment with enhanced therapeutic potential. J. Biol. Chem. 2021;296 doi: 10.1074/jbc.RA120.016127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bongoni A.K., Vikstrom I.B., McRae J.L., Salvaris E.J., Fisicaro N., Pearse M.J., et al. A potent truncated form of human soluble CR1 is protective in a mouse model of renal ischemia-reperfusion injury. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-01423-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wymann S., Mischnik M., Leong D., Ghosh S., Tan X., Cao H., et al. Sialylation-dependent pharmacokinetics and differential complement pathway inhibition are hallmarks of CR1 activity in vivo. Biochem. J. 2022;479:1007–1030. doi: 10.1042/BCJ20220054. [DOI] [PubMed] [Google Scholar]
- 21.Hardy M.P., Rowe T., Wymann S. Soluble complement receptor 1 therapeutics. J. Immunological Sci. 2022;6:1–17. [Google Scholar]
- 22.Rogers B., Dong D., Li Z., Li Z. Recombinant human serum albumin fusion proteins and novel applications in drug delivery and therapy. Curr. Pharm. Des. 2015;21:1899–1907. doi: 10.2174/1381612821666150302120047. [DOI] [PubMed] [Google Scholar]
- 23.Krych M., Hourcade D., Atkinson J.P. Sites within the complement C3b/C4b receptor important for the specificity of ligand binding. Proc. Natl. Acad. Sci. U. S. A. 1991;88:4353–4357. doi: 10.1073/pnas.88.10.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krych-Goldberg M., Hauhart R.E., Porzukowiak T., Atkinson J.P. Synergy between two active sites of human complement receptor type 1 (CD35) in complement regulation: implications for the structure of the classical pathway C3 convertase and generation of more potent inhibitors. J. Immunol. 2005;175:4528–4535. doi: 10.4049/jimmunol.175.7.4528. [DOI] [PubMed] [Google Scholar]
- 25.Krych-Goldberg M., Hauhart R.E., Subramanian V.B., Yurcisin B.M., 2nd, Crimmins D.L., Hourcade D.E., et al. Decay accelerating activity of complement receptor type 1 (CD35). Two active sites are required for dissociating C5 convertases. J. Biol. Chem. 1999;274:31160–31168. doi: 10.1074/jbc.274.44.31160. [DOI] [PubMed] [Google Scholar]
- 26.Furtado P.B., Huang C.Y., Ihyembe D., Hammond R.A., Marsh H.C., Perkins S.J. The partly folded back solution structure arrangement of the 30 SCR domains in human complement receptor type 1 (CR1) permits access to its C3b and C4b ligands. J. Mol. Biol. 2008;375:102–118. doi: 10.1016/j.jmb.2007.09.085. [DOI] [PubMed] [Google Scholar]
- 27.Day E.S., Capili A.D., Borysenko C.W., Zafari M., Whitty A. Determining the affinity and stoichiometry of interactions between unmodified proteins in solution using Biacore. Anal. Biochem. 2013;440:96–107. doi: 10.1016/j.ab.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalli K.R., Hsu P.H., Bartow T.J., Ahearn J.M., Matsumoto A.K., Klickstein L.B., et al. Mapping of the C3b-binding site of CR1 and construction of a (CR1)2-F(ab')2 chimeric complement inhibitor. J. Exp. Med. 1991;174:1451–1460. doi: 10.1084/jem.174.6.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schramm E.C., Roumenina L.T., Rybkine T., Chauvet S., Vieira-Martins P., Hue C., et al. Mapping interactions between complement C3 and regulators using mutations in atypical hemolytic uremic syndrome. Blood. 2015;125:2359–2369. doi: 10.1182/blood-2014-10-609073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clemenza L., Isenman D.E. The C4A and C4B isotypic forms of human complement fragment C4b have the same intrinsic affinity for complement receptor 1 (CR1/CD35) J. Immunol. 2004;172:1670–1680. doi: 10.4049/jimmunol.172.3.1670. [DOI] [PubMed] [Google Scholar]
- 31.Reilly B.D., Mold C. Quantitative analysis of C4Ab and C4Bb binding to the C3b/C4b receptor (CR1, CD35) Clin. Exp. Immunol. 1997;110:310–316. doi: 10.1111/j.1365-2249.1997.tb08333.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available on request from the authors. Authors confirm that all data are included in the manuscript.