Dear Editors,
The detection of specialist pro‐resolving mediators has recently been the subject of discussion in the literature. In our study in 2015, 1 we published the detection of lipoxin A2 (LXA4) in mouse lung homogenate following house dust mite (HDM) challenge in mice (Figure 4H). Recently, a commenter on PubPeer asked if we would post the raw mass spectrometry data to evidence detection of the lipid.
The samples were analysed in 2010, on a 4000QTrap using an analytical RP‐HPLC column. An example chromatogram shows the presence of a relatively low‐intensity peak in a “HDM challenge” sample (blue), which is absent in the “PBS challenge” control (red) (Figure 1). However, on reviewing the dataset, it became apparent that an error had been made during integration in 2010. Specifically, the retention time of the standard LXA4 (in our standard curve) is not identical to the lipid we detected in the mouse lung homogenate, although the peaks somewhat co‐elute (Figure 2). From our re‐analysis of this old dataset, we conclude that the lipid detected in mouse lungs cannot reliably be identified as LXA4.
FIGURE 1.
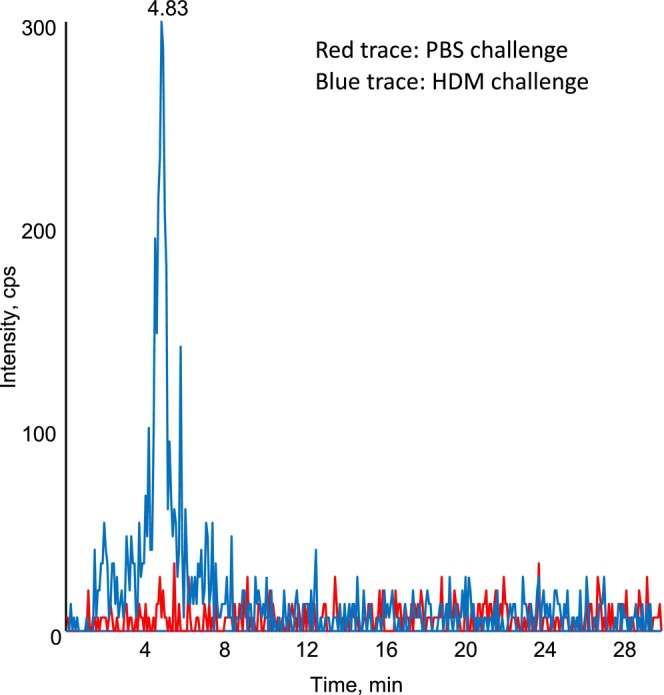
Chromatogram of m/z 351–115 showing the presence of a detectable peak in mouse lung homogenate following HDM challenge. Lipid extracts from mouse lung homogenate from mice challenged with HDM (blue) or PBS control (red) were analysed using LC/MS/MS as outlined in the methods.
FIGURE 2.
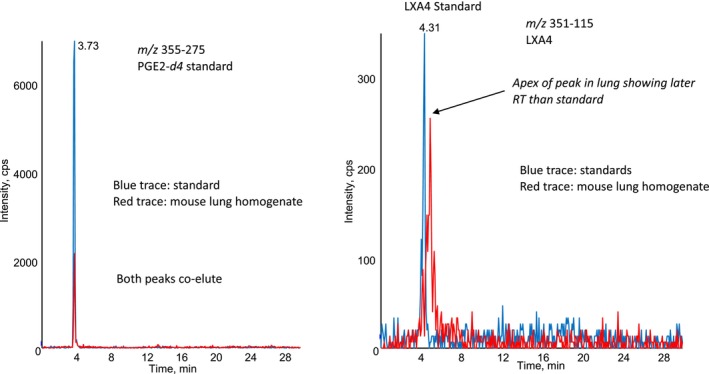
Monitoring PGE2‐d4 standard and LXA4 in the same samples shows that the lipid with m/z 351–115 in lung homogenate elutes later than authentic LXA4. The left panel shows co‐elution of PGE2‐d4, and the right panel shows later elution of lipids in mouse lung homogenate, than the LXA4 standard. Red trace: mouse lung, blue trace: LXA4 standard.
We apologise sincerely for this error, which was an entirely genuine mistake made during the integration of the peaks, and was not detected at that time. The appropriate course of action is to correct the paper, to remove this panel and the statement relating to the detection of LXA4 in the Results section. The lipid was not referred to in the Discussion.
The question remains as to what the structure of the lipid might be. A potential candidate is 15R‐LXA4 (aspirin‐triggered lipoxin) which elutes shortly after LXA4 on reverse phase separation. 2 However, we did not have a standard for 15‐epi‐LXA4 in 2010 so this cannot be verified. Also, the mice were not on aspirin. Another potential candidate for the lipid is an isoprostane. Martin Giera and colleagues previously reported a 5‐D2‐isoprostane that co‐elutes with LXA4 using the same MRM transition. 3 While he suggested this as a likely oxidation product of PGD2, it is possible a related E‐series isomer could be formed from PGE2 autoxidation since this is generated in the HDM model. It is notable that difficulties in verifying LX isomers in complex tissues using reverse phase LC/MS/MS were reported in 2014, where Ferreiros and Geisslinger showed that a lipid co‐eluting with LXA4 on reverse phase LC from cell culture supernatant was found to not co‐elute with known LX isomers including LXA4 on chiral phase separation. 4 Further work is needed to determine the structure of the lipid we detected in the HDM model.
Our original study aimed to investigate the role of airway macrophages in regulating type2 lung inflammation. While this data on LXA4 was included, we did not infer that the lipid was playing a defined biological role in controlling the inflammatory response. No experiments tested this idea or made any claims relating to a role. Removing the panel does not impact the conclusions relating to the role of airway macrophages and IL27 in regulating the immune response to HDM in the murine lung.
On behalf of the authors:
Valerie O'Donnell, Clare Lloyd, Victoria Tyrrell.
ACKNOWLEDGEMENTS
V O'D wrote the first draft. All authors made substantial contributions to the conception and interpretation of data, reviewed the article critically for important intellectual content and gave the final approval of the version.
REFERENCES
- 1. Mathie SA, Dixon KL, Walker SA, et al. Alveolar macrophages are sentinels of murine pulmonary homeostasis following inhaled antigen challenge. Allergy. 2015;70:80‐89. doi: 10.1111/all.12536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kutzner L, Rund KM, Ostermann AI, et al. Development of an optimized LC‐MS method for the detection of specialized pro‐resolving mediators in biological samples. Front Pharmacol. 2019;10:10. doi: 10.3389/fphar.2019.00169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chakraborty T, Thuer E, Heijink M, et al. Eicosanoid biosynthesis influences the virulence of Candida parapsilosis. Virulence. 2018;9:1019‐1035. doi: 10.1080/21505594.2018.1475797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ferreirós N, Homann J, Labocha S, et al. Lipoxin A4: problems with its determination using reversed phase chromatography–tandem mass spectrometry and confirmation with chiral chromatography. Talanta. 2014;127:82‐87. doi: 10.1016/j.talanta.2014.03.051 [DOI] [PubMed] [Google Scholar]


