Abstract
Background and Aims:
The oncogene Melanoma differentiation associated gene-9/syndecan binding protein (MDA-9/SDCBP) is overexpressed in many cancers, promoting aggressive, metastatic disease. However, the role of MDA-9 in regulating hepatocellular carcinoma (HCC) has not been well studied.
Approach and Results:
To unravel the function of MDA-9 in HCC, we generated and characterized a transgenic mouse with hepatocyte-specific overexpression of MDA-9 (Alb/MDA-9). Compared with wild-type (WT) littermates, Alb/MDA-9 mice demonstrated significantly higher incidence of N-nitrosodiethylamine/phenobarbital-induced HCC, with marked activation and infiltration of macrophages. RNA sequencing (RNA-seq) in naive WT and Alb/MDA-9 hepatocytes identified activation of signaling pathways associated with invasion, angiogenesis, and inflammation, especially NF-κB and integrin-linked kinase signaling pathways. In nonparenchymal cells purified from naive livers, single-cell RNA-seq showed activation of Kupffer cells and macrophages in Alb/MDA-9 mice versus WT mice. A robust increase in the expression of Secreted phosphoprotein 1 (Spp1/osteopontin) was observed upon overexpression of MDA-9. Inhibition of NF-κB pathway blocked MDA-9–induced Spp1 induction, and knock down of Spp1 resulted in inhibition of MDA-9–induced macrophage migration, as well as angiogenesis.
Conclusions:
Alb/MDA-9 is a mouse model with MDA-9 overexpression in any tissue type. Our findings unravel an HCC-promoting role of MDA-9 mediated by NF-κB and Spp1 and support the rationale of using MDA-9 inhibitors as a potential treatment for aggressive HCC.
INTRODUCTION
Melanoma differentiation associated gene-9/syndecan binding protein (MDA-9/SDCBP) is a postsynaptic density-95 (PSD-95), discs-large, zona occludens1 (ZO-1) (PDZ) domain containing adapter protein that plays a central role in regulating cell–cell and cell–matrix adhesion. MDA-9 transduces signals from the cell surface to the interior by protein–protein interaction, thereby regulating intracellular trafficking and cell-surface targeting.[1–4] MDA-9 plays a seminal role in cancer metastasis. Overexpression of MDA-9 has been identified in a variety of metastatic and invasive cancers compared with the primary tumor or less invasive cancers.[5–12] MDA-9 does not augment proliferation of cancer cells, but forced overexpression of MDA-9 results in increased migration of nonmetastatic cancer cells, which correlated with a more polarized distribution of actin filaments and increased pseudopodia formation.[6,10] MDA-9 also stimulates tumor angiogenesis.[13]
MDA-9 augments invasion and metastasis through interactions with c-Src and promotes the formation of an active focal adhesion kinase/c-Src signaling complex, leading to NF-κB and matrix metalloproteinase activation.[5–8] In human melanoma cells, this signaling axis is also important in mediating MDA-9–induced angiogenesis.[13] Crossing MDA-9 knockout (MDA-9−/−) mice with a spontaneous melanoma model significantly delayed tumor initiation and suppressed metastasis to the lymph nodes and lungs.[14] Additionally, tumor growth and lung metastasis by B16 melanoma cells were significantly inhibited in MDA-9−/− mice compared with wild-type (WT) mice.[14] These findings indicate that MDA-9 is important in both tumor cells and tumor microenvironment cells to promote metastasis.
Although MDA-9 has been studied in many cancers, the role of MDA-9 in hepatocellular carcinoma (HCC) has not been studied yet. The human MDA-9 gene is located in chromosome 8q12.1, and amplification of 8q is observed in many cancers, including HCC.[15,16] In this study we interrogated the role of MDA-9 in HCC using a transgenic mouse with hepatocyte-specific overexpression of MDA-9 (Alb/MDA-9). Our studies unravel an important role of MDA-9 in the tumor cells in modulating the tumor microenvironment and identify Secreted phosphoprotein 1 (Spp1/osteopontin) as a mediator of MDA-9-induced inflammation and angiogenesis.
MATERIALS AND METHODS
Generation of Alb/MDA-9 mice
All animal studies were approved by the Institutional Animal Care and Use Committee at Virginia Commonwealth University (VCU) and performed in accordance with the principles and procedures outlined in the National Research Council “Guide for the Care and Use of Laboratory Animals.” To generate the Alb/MDA-9 transgene construct (Figure 1A), the human MDA-9 coding region (fused to a C-terminal hemagglutinin (HA) tag) was inserted into the EcoRI site in β-globin exon 3 of the pBSpKCR3 vector. pBSpKCR3 contains part of exon 2, intron 2, and exon 3 of the rabbit β-globin gene for efficient transgene expression and has been described previously.[17] A 2.3-kb region of the mouse albumin gene (kindly provided by Dr. Snorri Thorgeirsson) was then inserted into the BamHI site in β-globin exon 2. This region contains an upstream enhancer region (−10,400 to −8500) fused to the 335-bp core promoter.[18] The transgene sequence was confirmed by automated DNA sequencing. Generation of the Alb/MDA-9 transgenic mice was performed by the VCU Transgenic/Knockout Mouse Core using standard procedures. A 4.5-kb XhoI fragment was excised from the construct and microinjected at a concentration of 3 ng/μl into the male pronucleus of single-cell mouse embryos obtained from mating B6CBAF1/J (C57BL/6 × CBA) male and female mice. The injected embryos were then reimplanted into the oviducts of pseudopregnant CD1 female mice. Genomic tail DNA from the resulting offspring was screened by a polymerase chain reaction (PCR) protocol using a β-globin intron 2 forward primer (5’-GATATACACTGTTTGAGATGAGG-3’) in combination with a human MDA-9 reverse primer (5’-AGAATTAGCCTGGACTAGCTGAAC-3’) that generated a 592-bp product in mice carrying the Alb/MDA-9 transgene. Three founders were obtained and bred to B6CBAF1/J mates to establish independent lines that were then analyzed for liver-specific MDA-9 expression.
FIGURE 1.
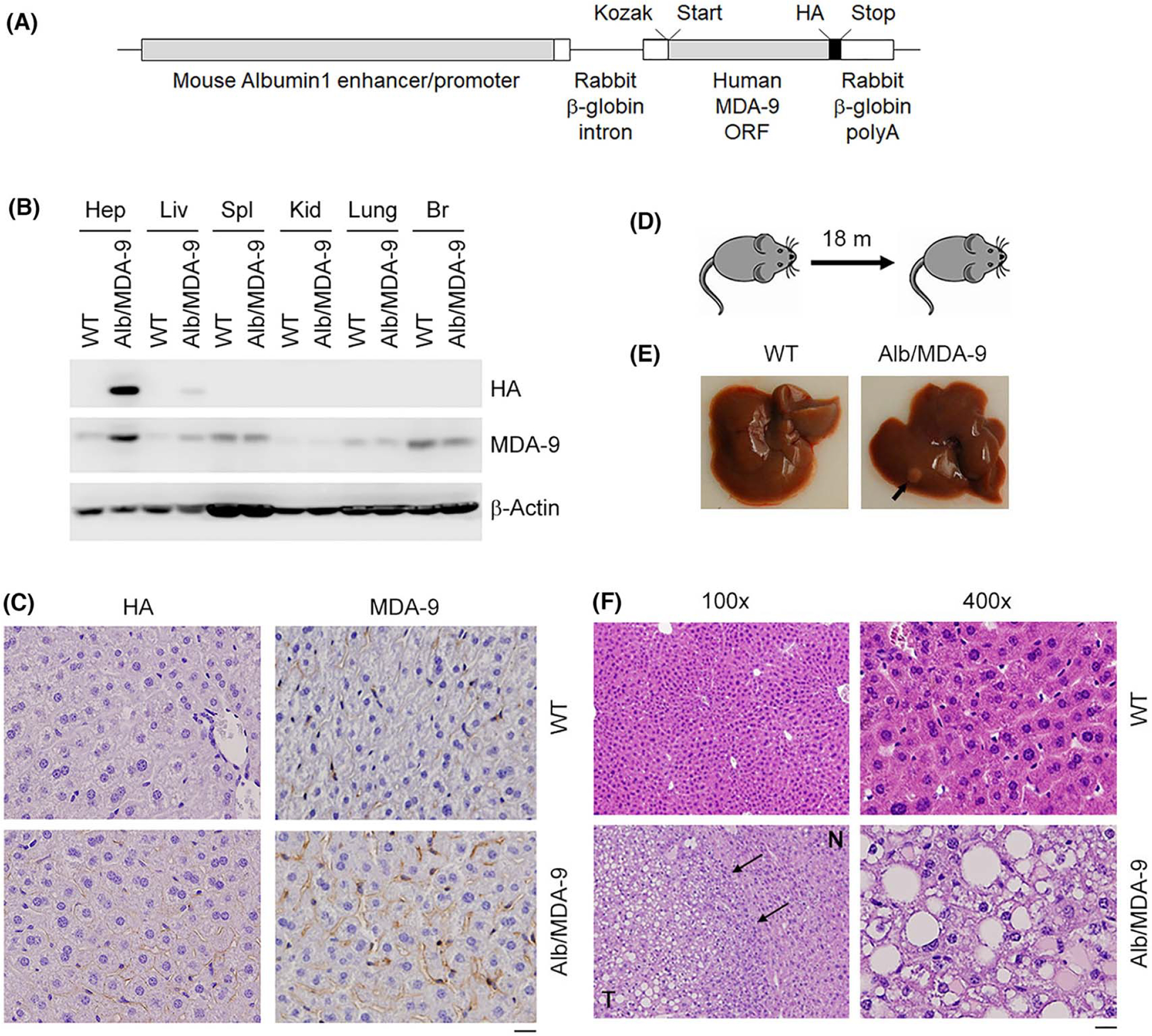
Generation and initial characterization of Alb/MDA-9 mouse. (A) The Alb/MDA-9 transgene contains a mouse Albumin1 enhancer/promoter, a rabbit β-globin intron to enhance expression, the human MDA-9 open reading frame (ORF), a C-terminal hemagglutinin (HA) tag, and a rabbit β-globin polyadenylation (polyA) region. The Kozak consensus sequence and translational start and stop codons are indicated. (B) Human MDA-9 was detected by western blot in mouse hepatocytes (Hep) and mouse tissues (liver [Liv], spleen [Spl], kidney [Kid], brain [Br]) using anti-HA and anti–MDA-9 antibodies. β-Actin was used as loading control. (C) Immunohistochemical analysis of WT and Alb/MDA-9 liver sections using anti-HA and anti–MDA-9 antibodies. Magnification: 400×. Scale bar: 20 μm. (D) Schematic of experimental protocol to detect spontaneous tumorigenesis in WT and Alb/MDA-9 mice. (E) Representative photograph of WT and Alb/MDA-9 livers at 18 months of age. Arrow indicates solitary tumor in Alb/MDA-9 liver. (F) Hematoxylin and eosin (H&E) staining of liver sections of WT and Alb/MDA-9 livers at 18 months of age. N: normal liver; T: tumor. Arrows indicate tumor margin. Scale bar: 20 μm. Alb, albumin; MDA-9, Melanoma differentiation associated gene-9; WT, wild-type.
Induction of chemical carcinogenesis
A single intraperitoneal injection of 10 μg/gm body weight of N-nitrosodiethylamine (DEN; Sigma, Catalog: N0756) was given at 14 days of age to male WT and Alb/MDA-9 littermates followed by addition of (PB, 0.05%; Sigma, Catalog: P1636) to the drinking water from 6 weeks of age.[19] The animals were regularly monitored and euthanized at 32 weeks of age. Liver, internal organs, and blood were collected for further analysis. Serum liver enzymes were measured in the Molecular Diagnostic Laboratory, Department of Pathology, VCU, using standard procedures. All experiments were performed using sibling littermates and fed ad libitum regular chow diet (Teklad global 2919 for breeder chow, Teklad irradiated 7912 for maintenance chow; Envigo) and water during light cycle.
Primary cells isolation, cell lines, and culture conditions
All primary cells were used immediately after isolation in-house and were mycoplasma-free as detected by Mycoplasma Detection Kit (ThermoFisher Scientific). Primary mouse hepatocytes and peritoneal macrophages were isolated and cultured as described.[20,21] Human umbilical vein endothelial cells (HUVECs) were obtained from Lonza (Catalog: CC-2517) and were cultured according to the provided protocol. Normal immortalized human fetal hepatocyte (Hc3716-hTERT) is a kind gift from Dr. Waki from Hiroshima University.[22] QGY-7703 cells were obtained from Fudan University, and HuH-7 cells were obtained from the Japanese Collection of Research Bioresources Cell Bank. All other human HCC cells and human embryonic kidney cells HEK-293T were obtained from the American Type Culture Collection and cultured according to the supplier’s protocols. Mouse HCC cell line RIL-175, developed from C57BL/6 mice, is a kind gift from Dr. Zender from University Hospital Tübingen.[23] The cells come with an authentication certificate from the original source.
Total RNA extraction, complementary deoxyribonucleic acid (cDNA) preparation and quantitative real-time PCR and RNA sequencing
Total RNA was extracted from hepatocytes or mouse tissues using the QIAGEN miRNAeasy Mini Kit (QIAGEN). cDNA preparation was done using ABI cDNA synthesis kit (Applied Biosystems). Real-time polymerase chain reaction (RT-PCR) was performed using an ABI ViiA7 fast real-time PCR system and Taqman gene expression assays according to the manufacturer’s protocol (Applied Biosystems). The probes are listed in Table S1. Total RNA from naive hepatocytes, isolated from WT and Alb/MDA-9 litter-mates, was used for RNA sequencing (RNA-seq). The RNA-Seq library was prepared using the Illumina TruSeq stranded messenger RNA (mRNA) sample preparation kit and sequenced on the Illumina HiSeq3000 platform. RNA-Seq libraries were pooled together to aim about 25 to 40 million read-passed filtered reads per sample. All sequencing reads were quality controlled using FastQC, version 0.11.2. Illumina adapters were trimmed using TrimGalore!, version 0.6.6; replicates were merged and aligned with their reference genome (University of California, Santa Cruz mouse genome build mm10) using STAR, version 2.7.9a. The BAM files from alignment were processed using STAR, version 2.7.9a, to obtain the counts per gene in all samples. Mus_musculus. GRCm38.100.gtf gene definition file was used. The differential expression analysis was performed using edgeR, version 3.36.0. Genes having less than 2 counts per million in all samples were excluded. Differentially expressed genes (DEGs) were defined using p value less than 0.01 and false discovery rate (FDR)-corrected p value less than 0.1 cutoffs. All bioinformatics analyses were conducted in R/Bioconductor computing environment, version 4.0.1. The Gene Expression Omnibus (GEO) Series accession number of this dataset is GSE199175.
Single-cell RNA-seq
Total liver cell population was isolated from 9-week-old WT and Alb/Mda-9 littermates using a liver perfusion technique.[20] Hepatocytes were pelleted and removed from the population by 70 × g centrifugation (3 min, 4° C). The cells in the supernatant were pelleted at 600 × g centrifugation (10 min, 4°C) and further purified using OptiPrep density gradient centrifugation.[20,24] The nonparenchymal cell (NPC) fraction collected at the gradient interface was centrifuged at 780 × g (10 min, 4°C) and resuspended in Dulbecco’s phosphate-buffered saline (DPBS) containing 0.04% bovine serum albumin (BSA).[24] Cell viability was determined to be more than 90% using a Corning CytoSMART cell counter, and concentration was adjusted to 1000 cells/μl. Samples were prepared using the 10x Genomics Single Cell 3’ Chromium system (PN-1000128, 1000127, 1000213) according to the manufacturer’s protocol. Gel beads in emulsion (GEMs) were generated with a targeted cell recovery of approximately 5000 cells. The GEMs were broken and barcoded; cDNA was purified with Dynabeads MyOne SILANE (PN-2000048), amplified, and purified with SPRIselect reagent (Beckman Coulter, B23317); and yield was quantified on an Agilent Bioanalyzer high sensitivity chip in preparation for 3′ gene expression library construction. After fragmentation, size selection with SPRIselect reagent, adaptor ligation, and sample index PCR, average fragment size was determined using the Agilent Bioanalyzer high sensitivity chip. The resulting library was sequenced using the Illumina NextSeq2000 on a P2 flow cell, producing approximately 200 million reads per sample. The GEO Series accession number of this dataset is GSE206096.
Western blotting, histopathology, and immunohistochemistry
Cell lysates and tissue extracts were prepared and Western blotting was performed as described.[25] The primary antibodies are listed in Table S2. β-Actin and GAPDH were used for housekeeping protein normalization and Ponceau S (Sigma, Catalog#: P7170) staining was used for total protein normalization. Images were taken using C-Digit (Li-Cor) with Image Studio software (Version 5.2). For histology, tissues were immediately placed in 10% Zn-Formalin (10:1 ratio of formalin to tissue for routine histology) or 10% Neutral Buffered Formalin (20:1 ratio of formalin to tissue for Opal staining) after collection. Formalin-fixed tissues were processed for paraffin embedding, sectioning, slide preparation and hematoxylin and eosin (H & E) staining by the VCU Tissue and Data Acquisition and Analysis Core Laboratory. Images were taken using Olympus BX41. Immunohistochemistry (IHC) was performed on formalin-fixed paraffin-embedded (FFPE) sections as described,[25] and the primary antibodies are listed in Table S2. Images were taken using Olympus BX41. H-scoring (percentage positive cells × intensity) was used to quantify individual markers within each region of interest.[26]
Multiplex immunofluorescent staining
Multiplex immunofluorescent staining using Opal (Akoya Biosciences) on FFPE sections was performed using a 7-plex comprising CD4, CD8, forkhead box P3 (FOXP3), CD11b, lymphocyte antigen 6 complex, locus G (Ly6g/Gr-1), pan cytokeratin (CK), and 4’,6-diamidino-2-phenylindole. This panel was designed to detect CD8+ and CD4+ cells as an indicator of T-cell infiltration, T regulatory cells (Treg; CD4+ and FOXP3+), and myeloid-derived suppressor cells (MDSCs; CD11b+, and Gr-1+). The pan CK antibody was used to distinguish the tumor cells. The primary and secondary antibodies are listed in Table S3. All slides were digitally scanned using PhenoImager HT (Akoya Biosciences) at a resolution of 20× magnification. Slide images were viewed, and regions of interest were selected using PhenoChart software (Akoya Biosciences). Color separation, adaptive cell segmentation, and positivity scoring were performed using inForm software (Akoya Biosciences, version 2.6) for quantitative analysis. Slides were examined for the infiltration of specific cells in the tumor region of WT and Alb/MDA-9 tissue sections. All settings applied to the training images were saved within an algorithm to allow batch analysis of at least three different tumor regions within the same experimental group.
Immune cell analysis by flow cytometry
Livers were perfused with phosphate-buffered saline (PBS) and harvested. The gallbladder was removed; the liver tissue was finely minced and immersed in digestion buffer (collagenase D, Roche; deoxyribonuclease I (DNase I), Sigma) for 40 min at 37°C.[27] After digestion, livers were filtered, washed, and subjected to a slow (30 g for 5 min) spin to settle debris and dead cells. The liquid component was transferred to a new tube and subjected to a normal spin (350 g for 5 min). Pelleted cells were then washed in PBS and stained for flow cytometry. Live/dead exclusion stain was performed per manufacturer’s directions (Zombie NIR; Biolegend). Cells were washed; Fc receptors were blocked (2.4G2; in-house) and stained in Brilliant Violet Stain Buffer (BD Biosciences) for 30 min on ice in the dark. The list of the antibodies is shown in Table S4. Cells were washed, fixed with Fixation Buffer (Biolegend) for 15 min at room temperature, washed again, and resuspended in PBS for analysis. All acquisition was performed on the Cytek Aurora 5 laser instrument (Cytek) in the Massey Cancer Center Flow Cytometry Shared Resource at Virginia Commonwealth University, and data were analyzed in FlowJo, version 10.8.0 (BD Biosciences).
Statistical analysis
Data were represented as the mean ± SEM and analyzed for statistical significance using a t-test.
RESULTS
MDA-9 is overexpressed in human HCC
In cBioPortal, analysis of The Cancer Genome Atlas (TCGA) data of liver HCC from 366 patients identified amplification of MDA-9 (SDCBP) in 8% patients (Figure S1A). Analysis of TCGA data in Oncomine comparing 115 normal liver samples versus 97 HCC samples showed significant upregulation of MDA-9 in HCC samples, MDA-9 being in the top 1% overex-pressed genes (Figure S1B). We checked MDA-9 mRNA and protein expression levels in normal immortalized human fetal hepatocytes (Hc3716-hTERT) and a variety of human HCC cells. Compared with Hc3716-hTERT cells, human HCC cells showed variable levels of MDA-9 overexpression, and a concordance between MDA-9 mRNA and protein levels was observed (Figure S1C,D).
Overexpression of MDA-9 promotes HCC in mice
To analyze how MDA-9 regulates HCC development and progression in vivo, we generated a hepatocyte-specific human MDA-9 transgenic (TG) mouse using mouse albumin promoter/enhancer (Alb/MDA-9) (Figure 1A). The human MDA-9 gene has a C-terminal hemagglutinin (HA) tag for monitoring of the transgene expression. In Alb/MDA-9 mice but not in WT mice, Western blot detected HA-tagged human MDA-9 only in the liver and hepatocytes and not in any other tissues, establishing the authenticity of the mouse line (Figure 1B). IHC staining using anti-HA and anti–MDA-9 antibodies also detected overexpression of MDA-9 in Alb/MDA-9 livers with MDA-9 staining observed along the cell margins, the known localization of MDA-9 (Figure 1C). We checked how MDA-9 overexpression affects spontaneous tumorigenesis in the liver. Initially, we generated three independent Alb/MDA-9 lines (Alb/MDA-9-03, Alb/MDA-9-06, and Alb/MDA-9-09), and we allowed mice from WT mice and all three TG lines of both sexes to age up to 18 months (Figure 1D). At this age, there was no difference in body and liver weights of WT and Alb/MDA-9 mice (Figure S2A,B). High-dimensional flow cytometry analysis of the liver did not show any major significant difference in immune cell population upon MDA-9 overexpression (S2C-I and Figure S3). Examination of the liver revealed that none of the 11 WT mice developed any tumor, whereas one male and one female Alb/MDA-9 mouse developed one isolated tumor each (Figure 1E). These tumors showed loss of architecture and hepatocyte ballooning in hematoxylin and eosin (H&E) staining, indicating HCC (Figure 1F). These findings suggest that MDA-9 may not be a strong driver oncogene; rather, it might promote tumor progression once the tumor is initiated.
To confirm this hypothesis, male WT and Alb/MDA-9 littermates (from line Alb/MDA-9–09, which was selected because of the highest expression of MDA-9) were subjected to DEN/PB-induced hepatocarcinogenesis (Figure 2A). Male mice were used because female mice are protected from DEN/PB-induced HCC because estrogen inhibits IL-6, a key driver of HCC.[28] Tumor burden in the livers of Alb/MDA-9 mice was significantly higher than that of WT mice (Figure 2B,C and Table S5). These tumors showed features of HCC, such as loss of hepatic architecture and hepatocyte ballooning (Figure 2D). Significantly increased levels of serum liver enzymes, aspartate aminotransferase (AST) and alanine aminotransferase (ALT), and significantly increased staining for the proliferation marker proliferating cell nuclear antigen (PCNA), angiogenesis marker CD31, and HCC marker alpha-feto protein (AFP) in tumor sections were observed in Alb/MDA-9 mice versus WT mice (Figures 2E,F and S4).
FIGURE 2.
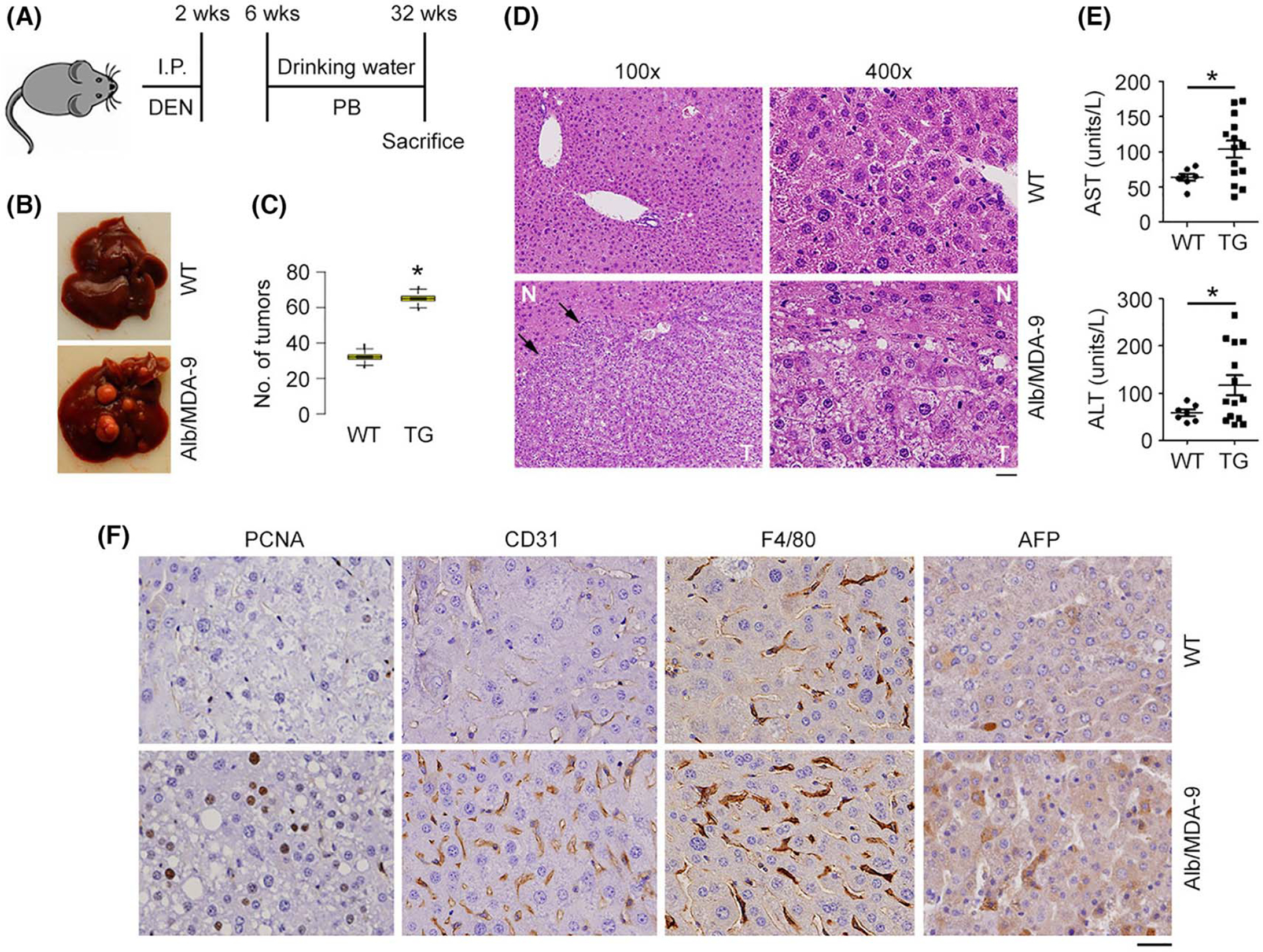
HCC is augmented in Alb/MDA-9 mouse. (A) Schematic of experimental protocol. WT and Alb/MDA-9 littermates were injected with the hepatocarcinogen DEN (10 μg/g bw) at 2 weeks of age, and starting at 6 weeks of age, they were given PB (0.05%) in drinking water, which serves as a mitogen. The mice were sacrificed at 32 weeks. (B) Representative photograph of DEN/PB-treated WT and Alb/MDA-9 livers at 32 weeks of age. (C) Number of tumors in WT and Alb/MDA-9 (TG) livers. Data represent mean ± SEM. *p < 0.01. (D) H&E staining of DEN/PB-treated WT and Alb/MDA-9 liver sections at 32 weeks of age. Arrows indicate tumor margin. Scale bar: 20 μm. (E) AST and ALT levels in the sera of DEN/PB-treated WT and Alb/MDA-9 (TG) mice at 32 weeks of age. Data represent mean ± SEM. *p < 0.01. (F) Representative IHC staining of the indicated markers in WT and Alb/MDA-9 liver tumors. Magnification: 400×. Scale bar: 20 μm. AFP, alpha-feto protein; Alb, albumin; ALT, alanine aminotransferase; AST, aspartate aminotransferase; DEN, N-nitrosodiethylamine; H&E, hematoxylin and eosin; HCC, hepatocellular carcinoma; IHC, immunohistochemistry; I.P., intraperitoneal; MDA-9, Melanoma differentiation associated gene-9; PB, phenobarbital; TG, transgenic; WT, wild-type.
MDA-9 overexpression in hepatocytes activates macrophages and myeloid cells in tumor microenvironment
To check the immune profile of the tumors, Opal multiplex stain was performed in tumor sections from WT and Alb/MDA-9 livers. Alb/MDA-9 tumors stained strongly for CK, indicating that it is HCC, whereas the CK staining was weak in WT tumors (Figure 3A,B). Alb/MDA-9 tumors showed marked infiltration of CD11b+ myeloid cells, FoxP3+ Tregs, and GR-1+ MDSCs, indicating an immune-suppressive environment (Figure 3A,B). Macrophage-mediated inflammation plays a central role in hepatocarcinogenesis.[29] Indeed, a significant increase in F4/80 positive macrophages were observed in Alb/MDA-9 livers compared with WT mice (Figures 2F and S4), indicating that MDA-9 creates an inflammatory milieu favoring recruitment of macrophages.
FIGURE 3.
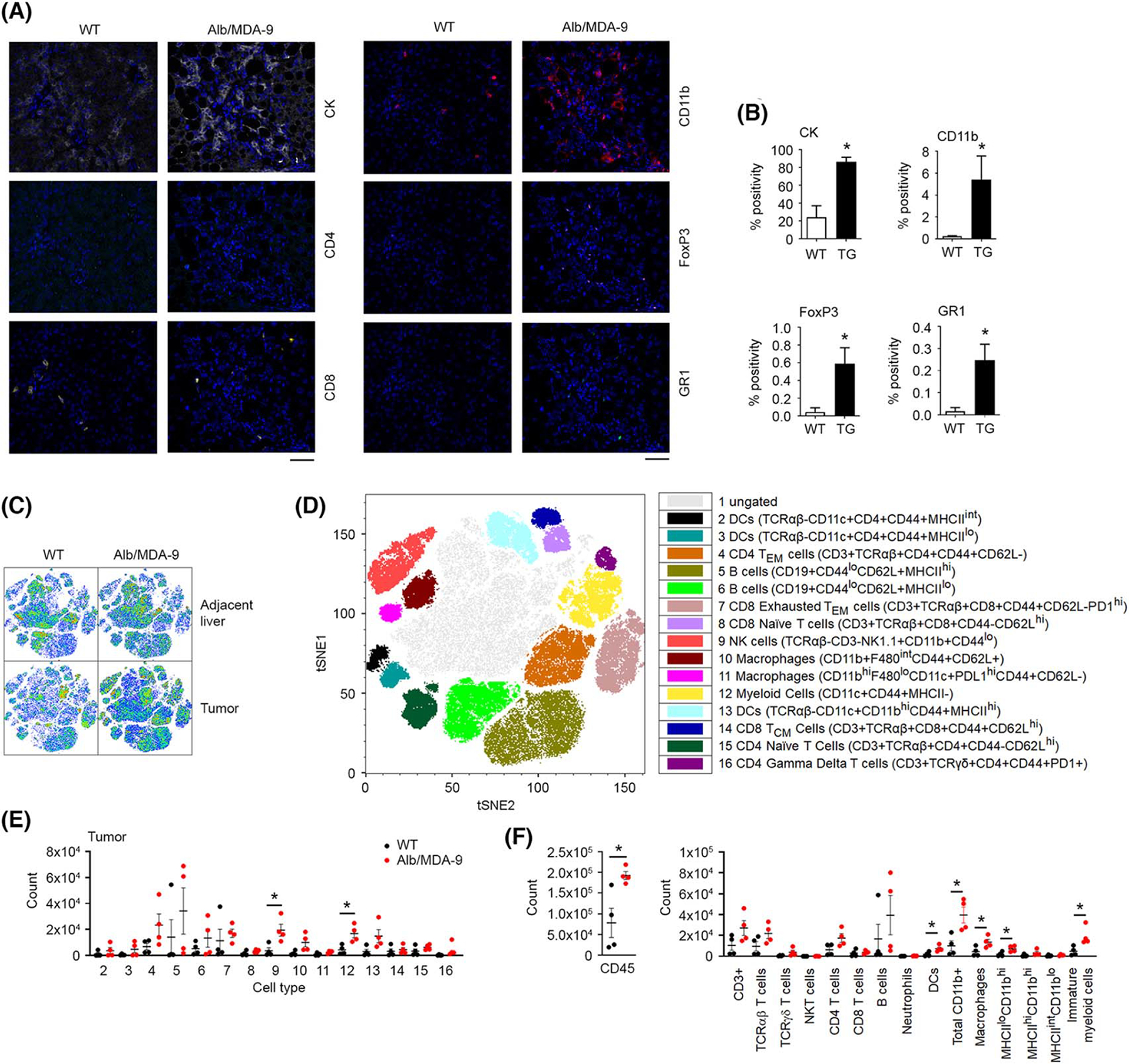
Myeloid cell infiltration is augmented in Alb/MDA-9 liver tumors. (A) Representative photomicrograph of multiplex immunofluoroscent staining of the indicated cell markers in DEN/PB-induced WT and Alb/MDA-9 liver tumors. Magnification: 200×. Scale bar: 10 μm. (B) Quantification of (A). Data represent mean ± SEM. *p < 0.01. (C) High-dimensional flow cytometry was performed on liver tumors and adjacent normal liver. Shown are t-distributed stochastic neighbor embedding (tSNE) plots of cell populations from tumor and adjacent liver concatenated by groups. (D) Concatenated tSNE plot with 16 identified cell populations. (E) Cell count from WT and Alb/MDA-9 tumors identified in (D). (F) Cell count from tumors for cell populations gated in Figure S5. Black dots: WT mice, red dots: Alb/MDA-9 mice. For (E) and (F), data represent mean ± SEM. *p < 0.05 versus WT. N = 4 mice/group. Alb, albumin; CK, cytokeratin; DC, dendritic cells; DEN, N-nitrosodiethylamine; FoxP3, forhead box P3; GR1, lymphocyte antigen 6 complex, locus G (Ly6g/Gr-1); MDA-9, Melanoma differentiation associated gene-9; NK, natural killer; PB, phenobarbital; TG, transgenic; WT, wild-type.
To corroborate these findings, we analyzed immune/inflammatory cell profile in the tumors and adjacent liver tissue of WT and Alb/MDA-9 littermates by high-dimensional flow cytometry (Figures S5 and S6). We identified 16 cell populations (Figure 3C,D), none of which showed any difference in adjacent normal liver between WT and Alb/MDA-9 mice (Figure S7A–C). In the tumor tissue, a significant increase was observed predominantly for CD11b positive cells, including macrophages and immature myeloid cells (Figure 3E, F and S7D), confirming the findings of the Opal staining (Figure 3A,B).
RNA-seq identifies activation of NF-κB and integrin-linked kinase signaling by MDA-9 in hepatocytes
We performed RNA-seq in naive hepatocytes isolated from WT and Alb/MDA-9 littermates to unravel the global gene expression changes induced by overexpressed MDA-9. Using an FDR value less than 0.01, 669 genes were upregulated and 745 genes were downregulated upon MDA-9 overexpression (Figure 4A). These DEGs were subjected to ingenuity pathway analysis (IPA) to identify which signaling pathways are activated by MDA-9 overexpression (Figure 4B). A z score of more than 2 indicates pathway activation. MDA-9 functions as a scaffold protein mediating signaling from extracellular matrix (ECM) to the interior of the cells. Indeed, integrin-linked kinase (ILK) signaling was significantly activated by MDA-9 (Figure 4B). Activation of ILK signaling leads to activation of serine/threonine kinase AKT, and AKT activation was observed in Alb/MDA-9 hepatocytes versus WT hepatocytes (Figure 4C). Activation of extracellular regulated kinase (ERK) and Src signaling pathways was also detected in Alb/MDA-9 hepatocytes compared with WT hepatocytes (Figure 4C). MDA-9 is known to stimulate epithelial-mesenchymal transition (EMT), cell migration, invasion, and angiogenesis. Rac signaling mediating cell motility, extravasation signaling, and IL-8 and inducible nitric oxide synthase (iNOS) signaling, known to induce angiogenesis, were significantly activated by MDA-9 (Figure 4B). E-cadherin levels were downregulated in Alb/MDA-9 hepatocytes versus WT hepatocytes, indicating EMT (Figure 4C). MDA-9 is known to activate NF-κB, a transcription factor regulating proinflammatory cytokines (PICs), and thus stimulate macrophage recruitment and inflammation. NF-κB activation is also observed in Alb/MDA-9 hepatocytes (Figure 4B). In luciferase reporter activity assay, NF-κB activity was significantly higher in Alb/MDA-9 hepatocytes than in WT hepatocytes (Figure 4D). Expression levels of PICs, such as Tnfa, Il1b, Il8, and Il6, were significantly higher in Alb/MDA-9 hepatocytes than in WT hepatocytes (Figure 4E).
FIGURE 4.
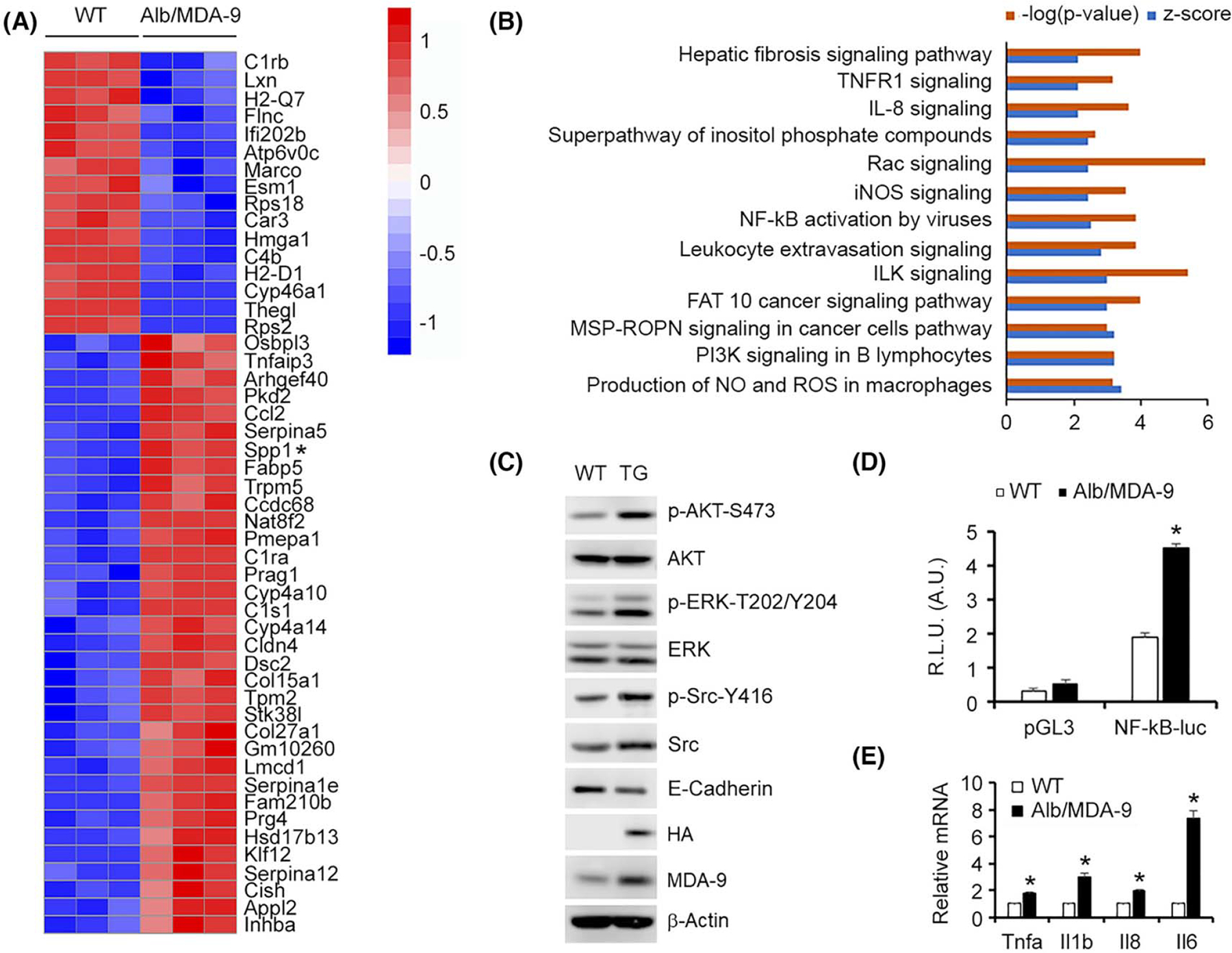
Overexpression of MDA-9 activates ILK and NF-κB pathways. (A) Heatmap of top upregulated and downregulated genes in naive Alb/MDA-9 hepatocytes versus WT identified by RNA-seq. Spp1 is indicated by an asterix. (B) Signaling pathways significantly activated in naïve Alb/MDA-9 hepatocytes versus WT identified by IPA of RNA-seq data. Signaling pathways with −log(p value) of 2.5 and higher and z score of 2.0 and higher are shown. (C) Representative Western blot of the indicated proteins in WT and Alb/MDA-9 hepatocytes. β-Actin was used as loading control. (D) NF-κB luciferase reporter activity measured in WT and Alb/MDA-9 hepatocytes. Firefly luciferase activity was normalized by Renilla luciferase activity. Data represent mean ± SEM. *p < 0.01. E. Expression analysis of the indicated mRNAs by Q-RT-PCR in WT and Alb/MDA-9 hepatocytes. Data represent mean ± SEM. *p < 0.01. Alb, Albumin; A.U., arbitrary units; AKT, serine threonine kinase AKT; ERK, extracellular regulated kinase; FAT10, ubiquitin D; HA, hemagglutinin; ILK, integrin-linked kinase; iNOS, inducible nitric oxide synthase; IPA, ingenuity pathway analysis; MDA-9, Melanoma differentiation associated gene-9; MSP-RON, macrophage-stimulating protein-recepteur d’origine nantais; NF-κB, nuclear factor kappa-light-chain-enhancer of activated B cells; NO, nitric oxide; pGL3, empty pGL3-basic vector; PI3K, phosphatidylinositol 3-kinase; R.L.U., relative luciferase units; Q-RT-PCR, quantitative real time polymerase chain reaction; Rac, Rac family small GTPase; RNA-seq, RNA sequencing; ROS, reactive oxygen species; Spp1, Secreted phosphoprotein 1; TG, transgenic; TNFR1, TNF receptor 1; WT, wild-type.
Single-cell RNA-seq in NPCs unravels robust activation of macrophages/Kupffer cells upon MDA-9 overexpression in hepatocytes
We interrogated how these changes in hepatocytes, brought forth by MDA-9 overexpression, affect the NPCs by purifying NPCs from naive WT and Alb/MDA-9 littermates and performing single-cell RNA-seq (Figure 5A–C). We used the following markers to classify clusters: Kupffer cells Csf1r, Cd74, Lyz2, Irf7, Spic, and Clec4f; stellate cells Dcn, Colec11, Ecm1, Lrat, Gucy1b1, and Gucy1a1; cholangiocytes Spp1, Sorbs2, Krt18, Epcam, Sox9; endothelial cells Ushbp1, Oit3, Il1a, Mmrn2, Pcdh12, and Dpp4; T cells Trbc1, Trbc2, Cd3g, and Cd3d; B cells Iglc2, Ighm, Igkc, Ms4a1, Cd79a, and Vpreb3; natural killer cells Ncr1, Klre1, Klre1, and Nkg7.[30–32] Based on the gene expression signature, we identified eight different clusters (Figure 5A,B) that did not show any significant difference in proportion between WT and Alb/MDA-9 mice (Figure 5C), which correlates with findings of flow cytometry analysis of the aged livers (Figure S2). However, when we performed IPA on DEGs of each cluster, a distinct pattern emerged. Using a p value less than 0.01, 438 genes were upregulated and 211 genes were downregulated in Kupffer cells isolated from Alb/MDA-9 livers compared with WT livers (Figure 5D). In canonical pathway analysis by IPA, using a −log(p value) cutoff of 2.5 and z score of more than 2 for activation and less than −2 for inhibition, 28 pathways were identified to be significantly changed (Figure 5E), many of which indicative of an activated state of macrophages, for example, interleukin-8 (IL-8), C-X-C motif chemokine receptor 4 (CXCR4), and granulocyte-macrophage colony stimulating factor (GM-CSF) signaling, production of nitric oxide (NO) and reactive oxygen species (ROS), and phagocytosis, as well as pathways regulating fibrosis, invasiveness, and metastasis. Very interestingly, the only pathway showing marked inhibition was programmed cell death 1 (PD-1), PD-1 ligand-1 (PDL-1) cancer immunotherapy pathway (Figure 5E), indicating a potential role of MDA-9 overexpression in modulating immune checkpoint inhibitor therapy. Using the same parameters of IPA, only five pathways showed activation in T cells and only four pathways in stellate cells (data not shown) indicating that hepatocyte MDA-9 has a major effect on Kupffer cells/macrophages.
FIGURE 5.
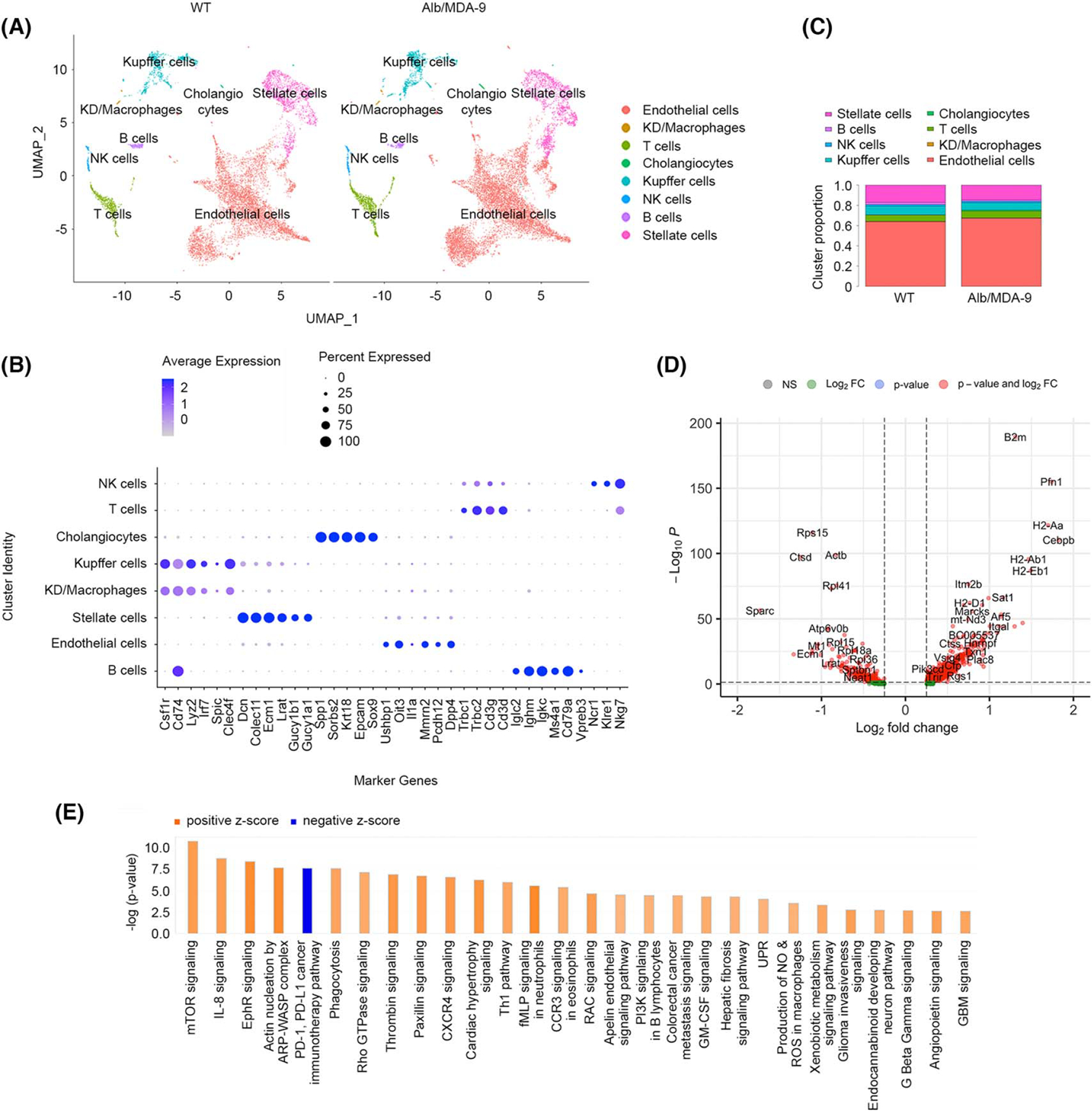
Overexpression of MDA-9 in hepatocytes results in activation of macrophages. Nonparenchymal cells were isolated from naive adult WT and Alb/MDA-9 livers, and scRNA-seq was performed. (A) Uniform Manifold Approximation and Projection (UMAP) plots showing eight different cell clusters. (B) Expression of specific gene markers in the indicated cell clusters. (C) Proportion of each cell cluster in WT and Alb/MDA-9 livers. (D) Volcano plot of DEGs in Kupffer cells. Log2 fold change more than 0: the genes are overexpressed in Alb/MDA-9 versus WT; Log2 fold change less than 0: the genes are downregulated in Alb/MDA-9 versus WT. (E) Canonical pathways activated or inhibited in Alb/MDA-9 Kupffer cells versus WT identified by IPA of scRNA-seq data. Pathways with −log(p value) of 2.5 and higher and z score of 2.0 and higher or −2.0 or lower are shown. Alb, albumin; ARP, actin-related protein; CCR3, C-C motif chemokine receptor 3; CXCR4, C-X-C motif chemokine receptor 4; DEG, differentially expressed gene; EphR, ephrin receptor; GBM, glioblastoma multiforme; GM-CSF, granulocyte-macrophage colony stimulating factor; IPA, ingenuity pathway analysis; KD, Kupffer/dendritic cells; MDA-9, Melanoma differentiation associated gene-9; mTOR, mammalian target of rapamycin; NK, natural killer; NO, nitric oxide; PD-1, programmed cell death 1; PDL-1, PD-1 ligand-1; PI3K, phosphatidylinositol 3-kinase; scRNA-seq, single-cell RNA sequencing; RAC, rac family small GTPase; ROS, reactive oxygen species; WASP, Wiskott-Aldrich syndrome protein; WT, wild-type.
MDA-9-induced Spp1 mediates macrophage activation and angiogenesis
In RNA-seq analysis, Spp1, also known as osteopontin, showed one of the highest levels of induction by MDA-9 (Figure 4A). Spp1 is a secreted protein known to play a seminal role in regulating chronic liver diseases, including hepatic inflammation and fibrosis, as well as angiogenesis, tumor progression, and metastasis.[33] We, therefore, focused our further studies on Spp1. Validation by Q-RT-PCR confirmed marked overexpression of Spp1 mRNA in Alb/MDA-9 hepatocytes versus WT hepatocytes (Figure 6A). Western blot using conditioned medium (CM) also showed increased levels of SPP1 protein in Alb/MDA-9 hepatocytes compared with WT hepatocytes (Figure 6B). MDA-9/SDCBP and SPP1 mRNA levels showed significant correlation in TCGA liver cancer database (Figure 6C). We checked which signaling pathway, activated by MDA-9, induces Spp1. We treated Alb/MDA-9 hepatocytes with IκB kinase inhibitor BMS-345541, which inhibits NF-κB pathway, resulting in ~90% downregulation of Spp1 mRNA levels (Figure 6D, left panel). Inhibition of the ILK pathway using either a chemical inhibitor Cpd 22 or Ilk small interfering RNA (siRNA) caused ~30% to 50% reduction, respectively, in Spp1 mRNA levels (Figure 6D, middle and right panels), indicating that MDA-9 induces Spp1 predominantly by activating the NF-κB pathway. Inhibition of the phosphatidylinositol 3-kinase (PI3K) pathway by LY294002 did not affect Spp1 mRNA level (Figure S8), indicating that even though both ILK and PI3K pathways activate AKT, the PI3K pathway does not play a role in MDA-9–induced Spp1. Next, we checked whether MDA-9–induced NF-κB and ILK pathways interacted with each other. Treatment of Alb/MDA-9 hepatocytes with BMS-345541 resulted in a decrease in p-AKT levels but no change in p-ILK levels (Figure S9, left panel). This finding indicates that regulation of AKT by NF-κB is independent of ILK. Similarly, treatment of Alb/MDA-9 hepatocytes with Cpd 22 induced no change in p-p65 levels (Figure S9, right panel). Collectively, these data suggest that MDA-9 activates NF-κB and ILK pathways independently and that these two pathways do not interact with each other.
FIGURE 6.
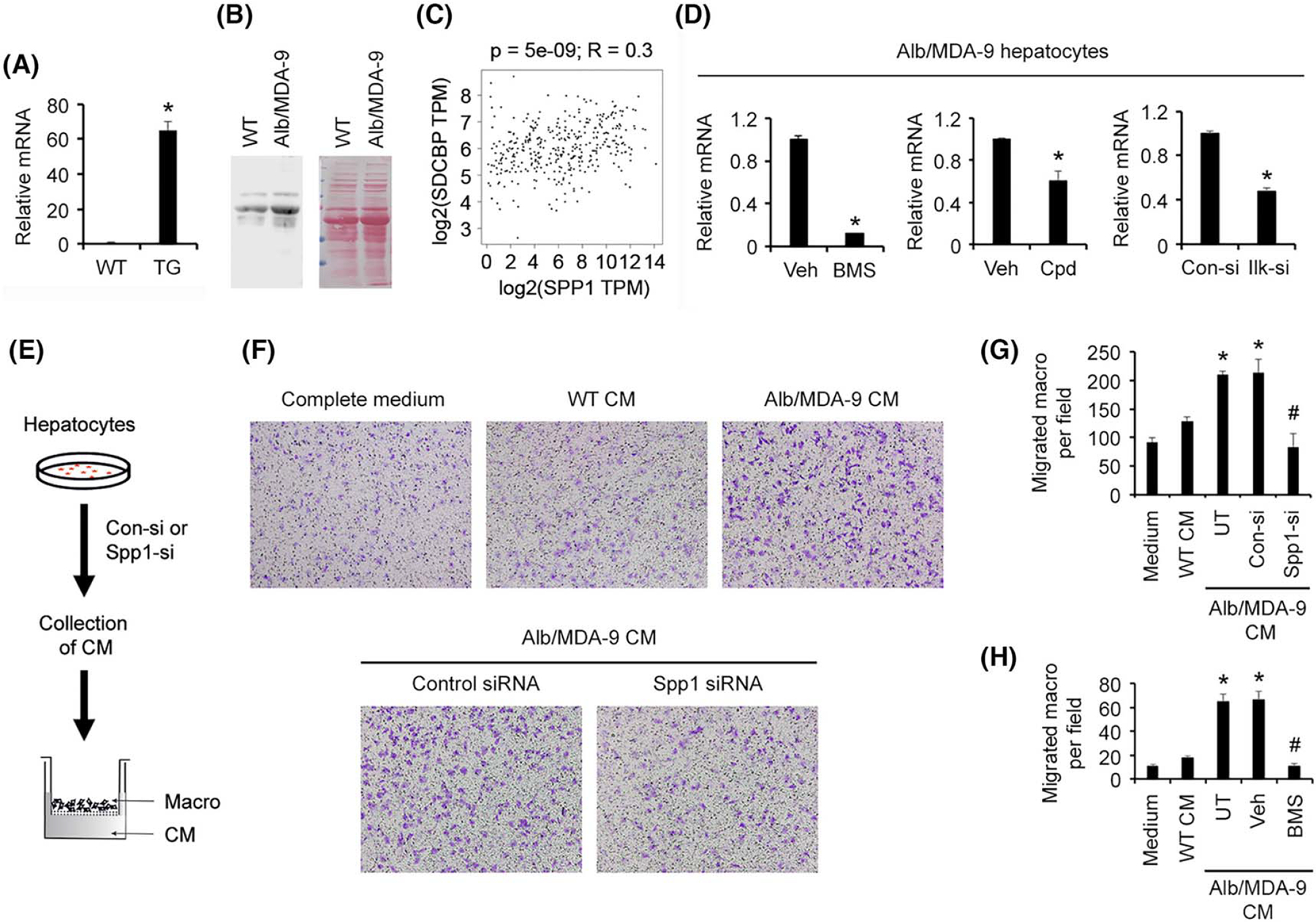
Spp1 mediates MDA-9–induced macrophage migration. (A) Expression analysis of Spp1 mRNA by Q-RT-PCR in WT and Alb/MDA-9 hepatocytes. Data represent mean ± SEM. *p < 0.01. (B) Analysis of SPP1 protein in the conditioned medium (CM) of WT and Alb/MDA-9 hepatocytes (left panel). Equal protein loading is shown by ponceau S staining (right panel). (C) Gene Expression Profiling Interactive Analysis (GEPIA) showing positive correlation between MDA-9/SDCBP and SPP1 mRNA levels in TCGA liver cancer database. (D) Alb/MDA-9 hepatocytes were treated for 48 h with either BMS-345541 (5 μm; left panel), Cpd 22 (2.5 μm; middle panel), or control and Ilk siRNAs (right panel), and Spp1 mRNA levels were analyzed by Q-RT-PCR. Data represent mean ± SEM. *p < 0.01. (E) Schematic representation of the experimental protocol of macrophage transwell migration assay. WT macrophages (macro) were plated in the top chamber. The bottom chamber contained either CM, CM from WT hepatocytes or CM from Alb/MDA-9 hepatocytes, untreated (UT) or treated with control, or Spp1 siRNA. (F) Representative photomicrographs of macrophage migration assay. (G) Quantification of macrophage migration assay. Data represent mean ± SEM. *p < 0.01 between WT CM and Alb/MDA-9 CM, either UT or treated with control siRNA; #p < 0.01 between Alb/MDA-9 CM, either UT or treated with control siRNA, and Alb/MDA-9 CM treated with Spp1 siRNA. (H) Quantification of macrophage migration assay upon treatment with BMS-345541 (5 μm). Data represent mean ± SEM. *p < 0.01 between WT CM and Alb/MDA-9 CM, either UT or treated with DMSO; #p < 0.01 between Alb/MDA-9 CM, either UT or treated with DMSO, and Alb/MDA-9 CM treated with BMS-345541. Alb, albumin; Con-si, control siRNA; DMSO, dimethyl sulfoxide; Ilk-si, Ilk siRNA; mRNA, messenger RNA; MDA-9, Melanoma differentiation associated gene-9; Q-RT-PCR, quantitative real time polymerase chain reaction; SDCBP, syndecan binding protein; siRNA, small interfering RNA; Spp1, Secreted phosphoprotein 1; TCGA, The Cancer Genome Atlas; TG, transgenic; TPM, transcript count per million; Veh, vehicle; WT, wild-type.
We checked macrophage migration toward CM from WT and Alb/MDA-9 hepatocytes (Figure 6E). A significantly increased number of WT macrophages migrated toward Alb/MDA-9 CM compared with WT CM, and knocking down Spp1 in Alb/MDA-9 hepatocytes markedly inhibited macrophage migration (Figure 6F,G). As a corollary, treatment with BMS-345541 also profoundly abrogated MDA-9–induced macrophage migration (Figure 6H), indicating that MDA-9–induced NF-κB activation with subsequent Spp1 induction contributes to increased macrophage migration. We checked angiogenesis by human umbilical vein endothelial cells (HUVECs) tube formation assay by treating HUVECs with CM from WT and Alb/MDA-9 hepatocytes (Figure 7A). Although WT hepatocyte CM failed to induce tube formation, Alb/MDA-9 hepatocyte CM significantly induced tube formation, which was profoundly inhibited by both Spp1 siRNA and Ilk siRNA (Figure 7B,C). Altogether, these studies indicate that MDA-9 activation of NF-κB and ILK signaling induces Spp1, which mediates augmentation of macrophage recruitment and angiogenesis (Figure 7D). Analysis of tumor samples showed increased PCNA staining, indicating increased proliferation, in Alb/MDA-9 livers versus WT livers (Figure 2F). MDA-9 has been shown not to stimulate proliferation in other cancers. We, therefore, analyzed in vitro proliferation of WT and Alb/MDA-9 hepatocytes. It should be noted that hepatocytes do not proliferate in vitro and enter into senescence after 4 days. Indeed, over a period of 1 week, both WT and Alb/MDA-9 hepatocytes showed decreased proliferation without any statistically significant difference (Figure S10A), indicating that MDA-9 overexpression did not augment proliferation. To check whether there is a differential effect of MDA-9 on hepatocytes versus HCC cells, we established stable MDA-9 overexpressing clones in HuH-7 and HepG2 human HCC cell lines (HuH-7-MDA-9 and HepG2-MDA-9, respectively) using a lentivirus expressing human MDA-9-HA (Figure S10B). In both 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and colony formation assays, HuH-7-MDA-9 and HepG2-MDA-9 cells did not show any difference in proliferation compared with the parental cells (Figure S10C–F). We observed significantly increased expression of IL-6 and C-C motif chemokine ligand 2 (CCL2) in Alb/MDA-9 tumors compared with WT tumors (Figure S10G,H), suggesting that these cytokines might contribute to increased proliferation in the tumors, as evidenced by increased PCNA staining (Figure 2F).
FIGURE 7.
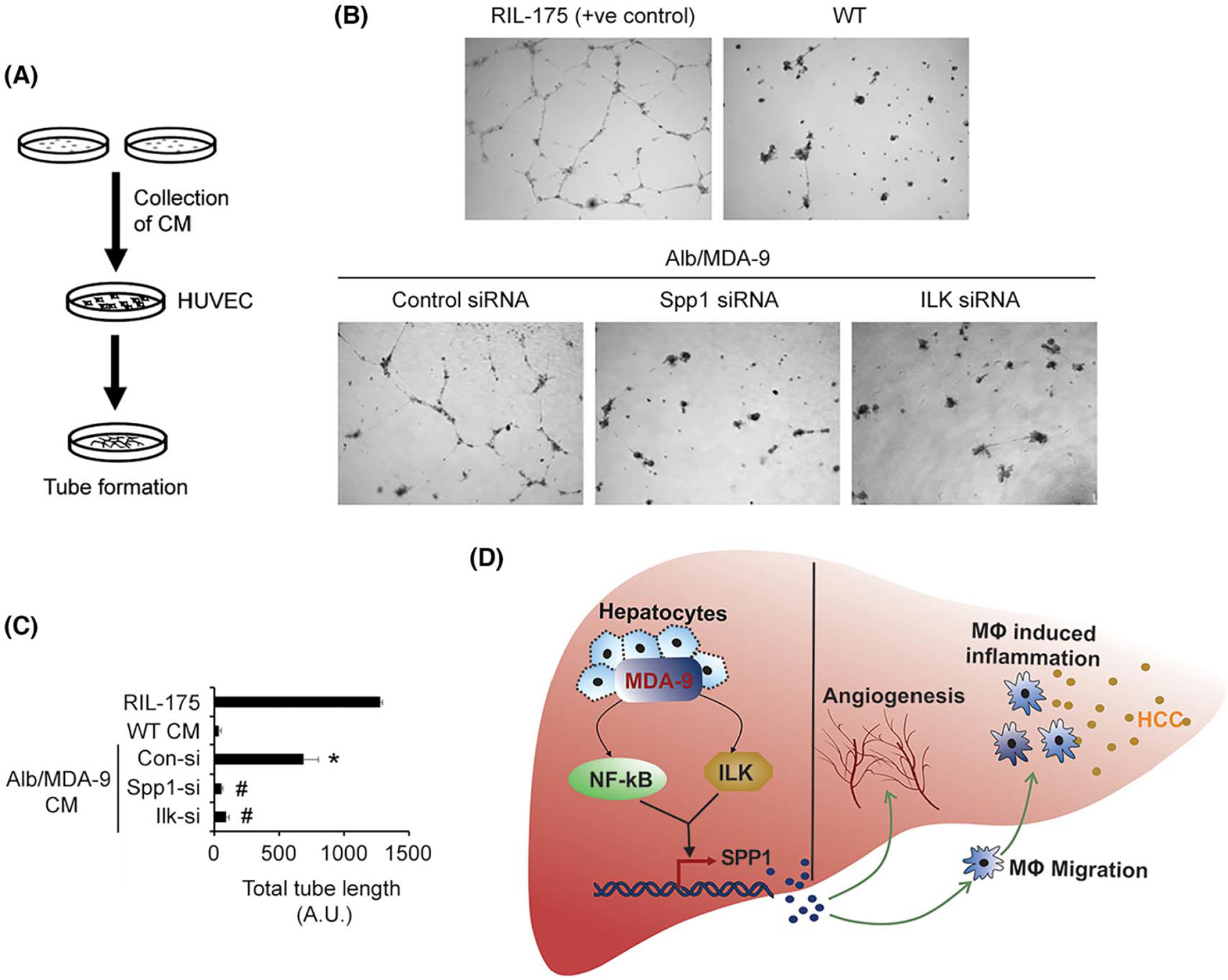
Spp1 mediates MDA-9–induced angiogenesis. (A) Schematic representation of the experimental protocol. (B) Representative photomicrographs of tube formation assay in the indicated treatment conditions. (C) Quantification of tube formation assay in the indicated treatment conditions. Data represent mean ± SEM. *p < 0.01 between WT CM and Alb/MDA-9 CM treated with control siRNA; #p < 0.01 between Alb/MDA-9 CM, treated with control siRNA, and Alb/MDA-9 CM treated with Spp1 or Ilk siRNA. (D) Cartoon showing mechanism by which MDA-9 promotes HCC. MDA-9 overexpression activates NF-κB and ILK signaling pathways, which leads to induction of Spp1 predominantly via the NF-κB pathway (thick arrow). Spp1 stimulates angiogenesis and macrophage recruitment and hence inflammation, leading to promotion of HCC. Alb, albumin; CM, conditioned medium; Con-si, control siRNA; HCC, hepatocellular carcinoma; HUVEC, human umbilical vein endothelial cells; ILK, integrin-linked kinase; Ilk-si, Ilk siRNA; MDA-9, Melanoma differentiation associated gene-9; siRNA, small interfering RNA; Spp1, Secreted phosphoprotein 1; WT, wild-type.
DISCUSSION
HCC is a disease of chronic inflammation.[34] Liver-resident macrophages (Kupffer cells) and infiltration of monocyte-derived macrophages play a central role in generating a proinflammatory and protumorigenic milieu as a consequence of HCC-inducing risk factors, such as viral hepatitis, alcoholism, and NAFLD.[29,35] These underlying conditions cause hepatocyte damage with release of cytokines, such as IL-1β, that activates NF-κB in macrophages, resulting in the production of proinflammatory cytokines like IL-6. IL-6 in turn activates signal transducer and activator of transcription 3 (STAT3) in hepatocytes, leading to tumorigenesis.[28,36–42] In this context, our studies identify that MDA-9 plays a key role by activating NF-κB, which induces Spp1, causing macrophage migration. HCC, generated in Spp1 knockout mice, showed significant decrease in the numbers of tumor-associated macrophages (TAMs), documenting a role of Spp1 in macrophage recruitment.[43] Our previous study using a total MDA-9 knockout mouse and B16 mouse melanoma metastasis model documented that MDA-9 in a tumor microenvironment is required for tumor-promoting inflammation.[14] We now show that MDA-9 overexpression in hepatocytes induces PICs and Spp1, thereby stimulating macrophage recruitment and inflammation. Collectively, these studies establish MDA-9 as a key inflammation activator.
We did not observe increase in hepatocyte proliferation upon MDA-9 overexpression, which correlates with previous findings that MDA-9 does not stimulate proliferation in other cancers. This observation is not a limitation of the assay because we previously demonstrated that a strong driver oncogene, such as myelocytomatosis oncogene (MYC), could increase in vitro proliferation of mouse hepatocytes.[44] We also show that overexpression of MDA-9 does not augment proliferation in human HCC cells. The increase in PCNA staining, hence proliferation, in Alb/MDA-9 tumors may not be because of MDA-9 overexpression in hepatocytes but rather because of TAM-released cytokines and chemokines, including IL1-β, TNFα, IL-6, CCL2, and C-X-C motif chemokine ligand 10 (CXCL10), as well as angiogenic and growth factors, which stimulate growth and proliferation of tumor cells.[29,35] In support of this argument, we observed significantly increased staining for IL-6 and CCL2 in Alb/MDA-9 tumors compared with WT tumors.
One signaling pathway that showed strong activation in Alb/MDA-9 hepatocytes is ILK. ILK has an amino acid sequence suggesting that it functions as a kinase. However, it has been identified that ILK lacks specific amino acid residues required for phosphotransferase activity, and it is now considered an adapter protein that mediates integrin-induced signaling, leading to activation of AKT.[45] A previous report showed that, in human breast cancer cells, cultured on type I collagen, MDA-9 interacts with ILK and facilitates AKT activation.[46] We document that ILK contributes to MDA-9–induced increase in Spp1 mRNA by using both Ilk siRNA and an ILK inhibitor Cpd 22. We also show that knocking down Ilk inhibits MDA-9–induced angiogenesis. This is an observation highlighting the importance of ILK in mediating MDA-9–induced hepatocarcinogenesis.
In this study, we confirm that MDA-9 plays an important role in aggressive progression of HCC. These studies establish the rationale that MDA-9 inhibition, either alone or in combination, might be an effective treatment approach for HCC. The next step in our studies is to address this hypothesis by using PDZ1i, a MDA-9 inhibitor that we developed.[47,48]
Supplementary Material
FUNDING INFORMATION
The present study was supported in part by the National Cancer Institute (NCI) grants 1R01CA230561-01A1 (D.S.), 1R01CA240004-01 (D.S.), and 1R01CA244993-01 (D.S. and P.B.F.) and by the National Institute of Diabetes and Digestive and Kidney Diseases grant 2R01DK107451-05 (D.S.). Services in support of this project were provided by the VCU Massey Cancer Center Transgenic/Knockout Mouse Facility, Tissue and Data Acquisition and Analysis Core, Bioinformatics Core, and Flow Cytometry Shared Resource, supported in part with funding from NIH-NCI Cancer Center support grant P30 CA016059. Data were generated at the Genomics Core Facility at Virginia Commonwealth University and in the Genome Sequencing Facility at University of Texas Health San Antonio, which is supported by National Cancer Institute (NCI) grant P30 CA054174, NIH Shared Instrument grant 1S10OD021805-01, and Cancer Prevention and Research Institute of Texas Core Facility Award (RP160732).
Abbreviations:
- Alb
albumin
- CK
cytokeratin
- CM
conditioned medium
- DEG
differentially expressed gene
- DEN
N-nitrosodiethylamine
- FOXP3
forkhead box P3
- HA
hemagglutinin
- HCC
hepatocellular carcinoma
- ILK
integrin-linked kinase
- IPA
ingenuity pathway analysis
- MDA-9
Melanoma differentiation associated gene-9
- mRNA
messenger RNA
- NPC
nonparenchymal cell
- PI3K
phosphatidylinositol 3-kinase
- PIC
proinflammatory cytokine
- RNA-seq
RNA sequencing
- RT-PCR
real-time polymerase chain reaction
- SDCBP
syndecan binding protein
- Spp1
Secreted phosphoprotein 1
- TCGA
The Cancer Genome Atlas
- WT
wild-type
Footnotes
CONFLICTS OF INTEREST
Nothing to report.
Supplemental Digital Content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s website, www.hepjournal.com.
REFERENCES
- 1.Sarkar D, Boukerche H, Su ZZ, Fisher PB. mda-9/syntenin: recent insights into a novel cell signaling and metastasis-associated gene. Pharmacol Ther. 2004;104:101–5. [DOI] [PubMed] [Google Scholar]
- 2.Sarkar D, Boukerche H, Su ZZ, Fisher PB. mda-9/Syntenin: more than just a simple adapter protein when it comes to cancer metastasis. Cancer Res. 2008;68:3087–93. [DOI] [PubMed] [Google Scholar]
- 3.Das SK, Bhutia SK, Kegelman TP, Peachy L, Oyesanya RA, Dasgupta S, et al. MDA-9/syntenin: a positive gatekeeper of melanoma metastasis. Front Biosci. 2012;17:1–15. [DOI] [PubMed] [Google Scholar]
- 4.Das SK, Maji S, Wechman SL, Bhoopathi P, Pradhan AK, Talukdar S, et al. MDA-9/Syntenin (SDCBP): Novel gene and therapeutic target for cancer metastasis. Pharmacol Res. 2020; 155:104695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boukerche H, Aissaoui H, Prevost C, Hirbec H, Das SK, Su ZZ, et al. Src kinase activation is mandatory for MDA-9/syntenin-mediated activation of nuclear factor-kappaB. Oncogene. 2010; 29:3054–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boukerche H, Su ZZ, Emdad L, Baril P, Balme B, Thomas L, et al. mda-9/Syntenin: a positive regulator of melanoma meta-stasis. Cancer Res. 2005;65:10901–1. [DOI] [PubMed] [Google Scholar]
- 7.Boukerche H, Su ZZ, Emdad L, Sarkar D, Fisher PB. mda-9/Syntenin regulates the metastatic phenotype in human melanoma cells by activating nuclear factor-kappaB. Cancer Res. 2007;67:1812–22. [DOI] [PubMed] [Google Scholar]
- 8.Boukerche H, Su ZZ, Prevot C, Sarkar D, Fisher PB. mda-9/Syntenin promotes metastasis in human melanoma cells by activating c-Src. Proc Natl Acad Sci U S A. 2008;105: 15914–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koo TH, Lee JJ, Kim EM, Kim KW, Kim HD, Lee JH. Syntenin is overexpressed and promotes cell migration in metastatic human breast and gastric cancer cell lines. Oncogene. 2002;21:4080–8. [DOI] [PubMed] [Google Scholar]
- 10.Grootjans JJ, Zimmermann P, Reekmans G, Smets A, Degeest G, Durr J, et al. Syntenin, a PDZ protein that binds syndecan cytoplasmic domains. Proc Natl Acad Sci U S A. 1997;94: 13683–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kegelman TP, Das SK, Hu B, Menezes M, Emdad L, Dasgupta S, et al. MDA-9/syntenin is a key regulator of glioma pathogenesis. Neuro Oncol. 2014;16:50–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dasgupta S, Menezes ME, Das SK, Emdad L, Janjic A, Bhatia S, et al. Novel role of MDA-9/syntenin in regulating urothelial cell proliferation by modulating EGFR signaling. Clin Cancer Res. 2013;19:4621–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das SK, Bhutia SK, Azab B, Kegelman TP, Peachy L, Santhekadur PK, et al. MDA-9/syntenin and IGFBP-2 promote angiogenesis in human melanoma. Cancer Res. 2013;73: 844–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das SK, Guo C, Pradhan AK, Bhoopathi P, Talukdar S, Shen XN, et al. Knockout of MDA-9/Syntenin (SDCBP) expression in the microenvironment dampens tumor-supporting inflammation and inhibits melanoma metastasis. Oncotarget. 2016;7: 46848–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knuutila S, Bjorkqvist AM, Autio K, Tarkkanen M, Wolf M, Monni O, et al. DNA copy number amplifications in human neoplasms: review of comparative genomic hybridization studies. Am J Pathol. 1998;152:1107–23. [PMC free article] [PubMed] [Google Scholar]
- 16.Zimonjic DB, Keck CL, Thorgeirsson SS, Popescu NC. Novel recurrent genetic imbalances in human hepatocellular carcinoma cell lines identified by comparative genomic hybridization. Hepatology. 1999;29:1208–4. [DOI] [PubMed] [Google Scholar]
- 17.Howes KA, Ransom N, Papermaster DS, Lasudry JG, Albert DM, Windle JJ. Apoptosis or retinoblastoma: alternative fates of photoreceptors expressing the HPV-16 E7 gene in the presence or absence of p53. Genes Dev. 1994;8:1300–10. [DOI] [PubMed] [Google Scholar]
- 18.Pinkert CA, Ornitz DM, Brinster RL, Palmiter RD. An albumin enhancer located 10 kb upstream functions along with its promoter to direct efficient, liver-specific expression in transgenic mice. Genes Dev. 1987;1:268–76. [DOI] [PubMed] [Google Scholar]
- 19.Srivastava J, Siddiq A, Emdad L, Santhekadur PK, Chen D, Gredler R, et al. Astrocyte elevated gene-1 promotes hepatocarcinogenesis: novel insights from a mouse model. Hepatology. 2012;56:1782–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mendoza R, Banerjee I, Reghupaty SC, Yetirajam R, Manna D, Sarkar D. Isolation and culture of mouse hepatocytes and kupffer cells (KCs). Methods Mol Biol. 2022;2455:73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robertson CL, Mendoza RG, Jariwala N, Dozmorov M, Mukhopadhyay ND, Subler MA, et al. Astrocyte elevated gene-1 regulates macrophage activation in hepatocellular carcinogenesis. Cancer Res. 2018;78:6436–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waki K, Anno K, Ono T, Ide T, Chayama K, Tahara H. Establishment of functional telomerase immortalized human hepatocytes and a hepatic stellate cell line for telomere-targeting anticancer drug development. Cancer Sci. 2010;101:1678–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zender L, Xue W, Cordon-Cardo C, Hannon GJ, Lucito R, Powers S, et al. Generation and analysis of genetically defined liver carcinomas derived from bipotential liver progenitors. Cold Spring Harb Symp Quant Biol. 2005;70:251–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer J, Lacotte S, Morel P, Gonelle-Gispert C, Buhler L. An optimized method for mouse liver sinusoidal endothelial cell isolation. Exp Cell Res. 2016;349:291–301. [DOI] [PubMed] [Google Scholar]
- 25.Robertson CL, Srivastava J, Siddiq A, Gredler R, Emdad L, Rajasekaran D, et al. Genetic deletion of AEG-1 prevents hepatocarcinogenesis. Cancer Res. 2014;74:6184–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fedchenko N, Reifenrath J. Different approaches for interpretation and reporting of immunohistochemistry analysis results in the bone tissue—a review. Diagn Pathol. 2014;9:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rajesh Y, Reghupaty SC, Mendoza RG, Manna D, Banerjee I, Subler MA, et al. Dissecting the balance between metabolic and oncogenic functions of astrocyte-elevated gene-1/metadherin. Hepatol Commun. 2022;6:561–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naugler WE, Sakurai T, Kim S, Maeda S, Kim K, Elsharkawy AM, et al. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science. 2007;317:121–4. [DOI] [PubMed] [Google Scholar]
- 29.Krenkel O, Tacke F. Liver macrophages in tissue homeostasis and disease. Nat Rev Immunol. 2017;17:306–21. [DOI] [PubMed] [Google Scholar]
- 30.Halpern KB, Shenhav R, Matcovitch-Natan O, Toth B, Lemze D, Golan M, et al. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature. 2017;542: 352–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang W, He H, Wang T, Su N, Zhang F, Jiang K, et al. Single-cell transcriptomic analysis reveals a hepatic stellate cell-activation roadmap and myofibroblast origin during liver fibrosis in mice. Hepatology. 2021;74:2774–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nault R, Fader KA, Bhattacharya S, Zacharewski TR. Single-nuclei RNA sequencing assessment of the hepatic effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin. Cell Mol Gastroenterol Hepatol. 2021;11:147–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song Z, Chen W, Athavale D, Ge X, Desert R, Das S, et al. Osteopontin takes center stage in chronic liver disease. Hepatology. 2021;73:1594–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011; 365:1118–27. [DOI] [PubMed] [Google Scholar]
- 35.Wan S, Kuo N, Kryczek I, Zou W, Welling TH. Myeloid cells in hepatocellular carcinoma. Hepatology. 2015;62:1304–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakurai T, He G, Matsuzawa A, Yu GY, Maeda S, Hardiman G, et al. Hepatocyte necrosis induced by oxidative stress and IL-1 alpha release mediate carcinogen-induced compensatory proliferation and liver tumorigenesis. Cancer Cell. 2008;14:156–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maeda S, Kamata H, Luo JL, Leffert H, Karin M. IKKbeta couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell. 2005;121: 977–90. [DOI] [PubMed] [Google Scholar]
- 38.He G, Dhar D, Nakagawa H, Font-Burgada J, Ogata H, Jiang Y, et al. Identification of liver cancer progenitors whose malignant progression depends on autocrine IL-6 signaling. Cell. 2013;155: 384–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He G, Yu GY, Temkin V, Ogata H, Kuntzen C, Sakurai T, et al. Hepatocyte IKKbeta/NF-kappaB inhibits tumor promotion and progression by preventing oxidative stress-driven STAT3 activation. Cancer Cell. 2010;17:286–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park EJ, Lee JH, Yu GY, He G, Ali SR, Holzer RG, et al. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell. 2010;140: 197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lanaya H, Natarajan A, Komposch K, Li L, Amberg N, Chen L, et al. EGFR has a tumour-promoting role in liver macrophages during hepatocellular carcinoma formation. Nat Cell Biol. 2014; 16(972–981):971–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pikarsky E, Porat RM, Stein I, Abramovitch R, Amit S, Kasem S, et al. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature. 2004;431:461–6. [DOI] [PubMed] [Google Scholar]
- 43.Zhu Y, Yang J, Xu D, Gao XM, Zhang Z, Hsu JL, et al. Disruption of tumour-associated macrophage trafficking by the osteopontin-induced colony-stimulating factor-1 signalling sensitises hepatocellular carcinoma to anti-PD-L1 blockade. Gut. 2019;68: 1653–66. [DOI] [PubMed] [Google Scholar]
- 44.Srivastava J, Siddiq A, Gredler R, Shen XN, Rajasekaran D, Robertson CL, et al. Astrocyte elevated gene-1 and c-Myc cooperate to promote hepatocarcinogenesis in mice. Hepatology. 2015;61:915–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fukuda K, Gupta S, Chen K, Wu C, Qin J. The pseudoactive site of ILK is essential for its binding to alpha-Parvin and localization to focal adhesions. Mol Cell. 2009;36:819–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hwangbo C, Park J, Lee JH. mda-9/Syntenin protein positively regulates the activation of Akt protein by facilitating integrin-linked kinase adaptor function during adhesion to type I collagen. J Biol Chem. 2011;286:33601–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kegelman TP, Wu B, Das SK, Talukdar S, Beckta JM, Hu B, et al. Inhibition of radiation-induced glioblastoma invasion by genetic and pharmacological targeting of MDA-9/Syntenin. Proc Natl Acad Sci U S A. 2017;114:370–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pradhan AK, Maji S, Bhoopathi P, Talukdar S, Mannangatti P, Guo C, et al. Pharmacological inhibition of MDA-9/Syntenin blocks breast cancer metastasis through suppression of IL-1beta. Proc Natl Acad Sci U S A. 2021;118:e2103180118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


