FIGURE 5.
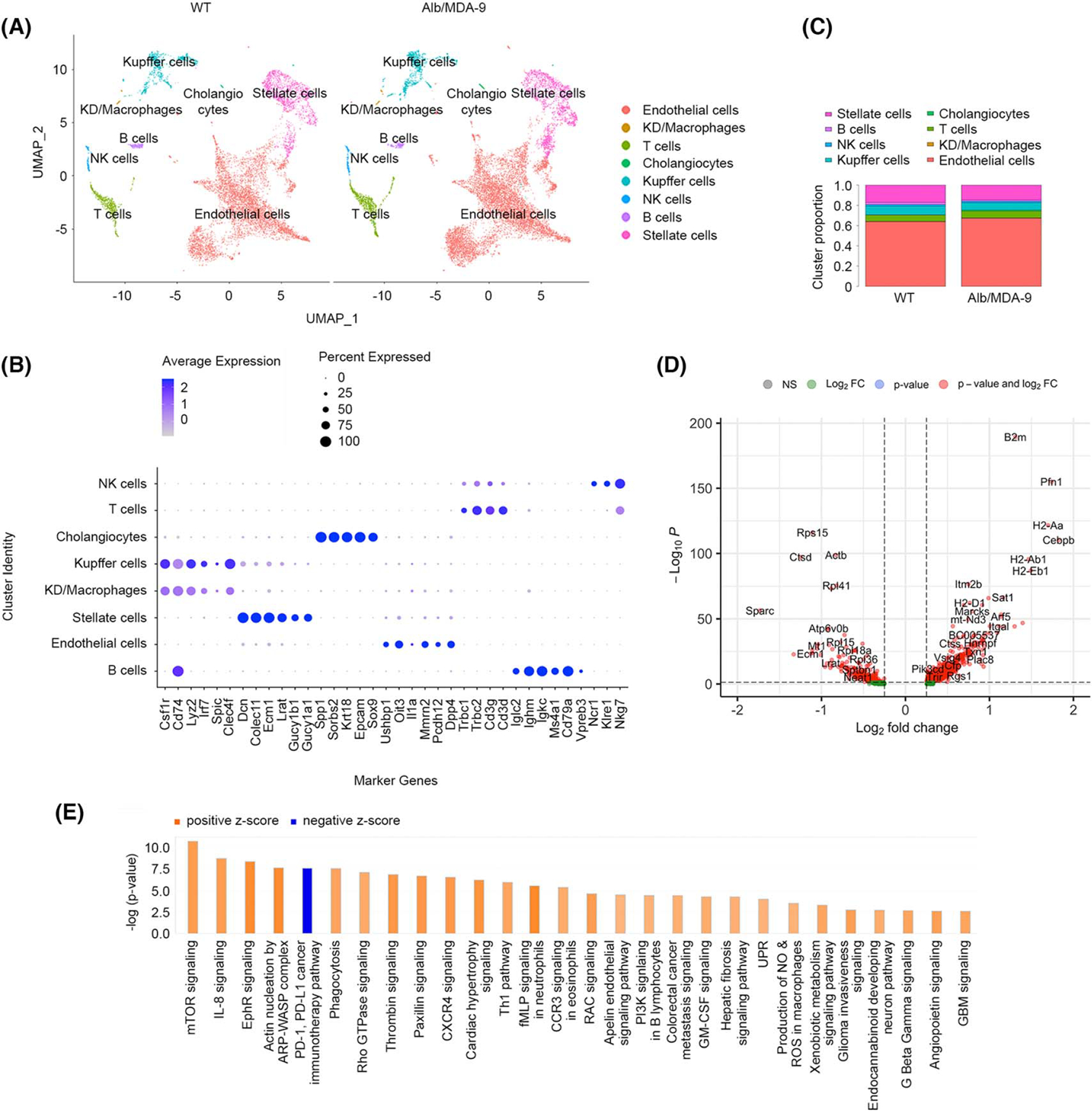
Overexpression of MDA-9 in hepatocytes results in activation of macrophages. Nonparenchymal cells were isolated from naive adult WT and Alb/MDA-9 livers, and scRNA-seq was performed. (A) Uniform Manifold Approximation and Projection (UMAP) plots showing eight different cell clusters. (B) Expression of specific gene markers in the indicated cell clusters. (C) Proportion of each cell cluster in WT and Alb/MDA-9 livers. (D) Volcano plot of DEGs in Kupffer cells. Log2 fold change more than 0: the genes are overexpressed in Alb/MDA-9 versus WT; Log2 fold change less than 0: the genes are downregulated in Alb/MDA-9 versus WT. (E) Canonical pathways activated or inhibited in Alb/MDA-9 Kupffer cells versus WT identified by IPA of scRNA-seq data. Pathways with −log(p value) of 2.5 and higher and z score of 2.0 and higher or −2.0 or lower are shown. Alb, albumin; ARP, actin-related protein; CCR3, C-C motif chemokine receptor 3; CXCR4, C-X-C motif chemokine receptor 4; DEG, differentially expressed gene; EphR, ephrin receptor; GBM, glioblastoma multiforme; GM-CSF, granulocyte-macrophage colony stimulating factor; IPA, ingenuity pathway analysis; KD, Kupffer/dendritic cells; MDA-9, Melanoma differentiation associated gene-9; mTOR, mammalian target of rapamycin; NK, natural killer; NO, nitric oxide; PD-1, programmed cell death 1; PDL-1, PD-1 ligand-1; PI3K, phosphatidylinositol 3-kinase; scRNA-seq, single-cell RNA sequencing; RAC, rac family small GTPase; ROS, reactive oxygen species; WASP, Wiskott-Aldrich syndrome protein; WT, wild-type.
