Abstract
Sequence comparison of 16S rRNA PCR amplicons is an established approach to taxonomically identify bacterial isolates and profile complex microbial communities. One potential application of recent advances in long-read sequencing technologies is to sequence entire rRNA operons and capture significantly more phylogenetic information compared to sequencing of the 16S rRNA (or regions thereof) alone, with the potential to increase the proportion of amplicons that can be reliably classified to lower taxonomic ranks. Here we describe GROND (Genome-derived Ribosomal Operon Database), a publicly available database of quality-checked 16S-ITS-23S rRNA operons, accompanied by multiple taxonomic classifications. GROND will aid researchers in analysis of their data and act as a standardised database to allow comparison of results between studies.
Keywords: amplicons, database, long read sequencing, microbiome, nanopore, rRNA
Significance as a BioResource to the community
DNA sequencing of ribosomal RNA (rRNA) genes, particularly the widely used 16S rRNA gene, plays a pivotal role in bacterial identification and phylogenetic analysis. However, the limitations of traditional Sanger sequencing and short-read sequencing, i.e. low throughput and limited phylogenetic resolution, respectively, have hindered accurate profiling of microbial communities, especially in cases of highly related species. High throughput long-read sequencing technologies from, for example, PacBio and Oxford Nanopore Technologies have revolutionized genome examination capabilities, enabling the sequencing of entire 16S rRNA genes and overcoming these constraints. Here we describe an open-access, quality-checked database containing full-length 16S-ITS-23S rRNA sequences and their associated taxonomy. Extending amplicon-based metagenomic studies to include the 23S gene and ITS region enhances species- and strain-level resolution compared to conventional 16S rRNA sequencing, making it a versatile and valuable resource for researchers wanting to explore microbial diversity at subspecies resolution. This database represents an important resource for the scientific community, facilitating standardized and reliable analysis of microbial communities using long-read sequencing technologies. Its open-access nature promotes general availability and collaboration, allowing researchers to explore and compare results across studies, ultimately advancing our understanding of microbial diversity and evolution.
Data Summary
The GROND databases, pretrained Naïve-Bayes machine learning classifiers, and statistics describing genome length and rrn copy number for each taxon in the databases, are available to download from Zenodo (https://zenodo.org/records/10889037)https://zenodo.org/records/10889037) The code used to generate the databases is available at https://github.com/cazzlewazzle89/GROND.
Introduction
DNA sequencing of ribosomal RNA (rRNA) genes is an important technique for the identification and phylogenetic analysis of bacteria. Sanger sequencing of the entire ~1.5 kb-encompassing 16S rRNA gene is commonly used to identify cultured isolates and will provide species-level resolution in most cases. However, the 16S rRNA genes of some highly related species, such as members of the Streptococcus mitis group or Escherichia coli and Shigella spp., share >99 % sequence identity and can therefore not be reliably distinguished. Sequencing the longer 23S rRNA gene instead of, or in tandem with, the 16S rRNA gene provides enhanced phylogenetic resolution, thereby allowing reliable separation of these very closely related species. Sanger sequencing is well-established for the purpose of generating long, high fidelity reads from the PCR amplicons that span these rRNA genes. However, Sanger sequencing is inefficient for profiling complex microbial communities and so high-throughput short read sequencing, such as that provided by Illumina, of single hypervariable regions of the 16SrRNA gene is used instead. This approach can generate massive quantities of highly accurate reads but the increased quantity comes with reduced phylogenetic resolution due to substantially shorter read lengths. The 16S rRNA gene has also been the target of choice for amplicon-based metagenomic (a.k.a. metabarcoding) studies due to its mix of alternating highly conserved and hypervariable regions [1,2].
The introduction of long-read sequencing in 16S rRNA gene-targeted metagenomic studies has revolutionised the ability to sequence the entire 16S rRNA gene, overcoming the constraints of short-read technologies, such as Illumina, that were limited to hypervariable regions [3,9]. Pioneered by PacBio and Oxford Nanopore Technologies (ONT), long-read technologies offer the benefits of sequencing long stretches of DNA in a relatively high-throughput, culture-independent, manner. Despite initial challenges with high error rates, both ONT and PacBio have significantly improved their accuracy. ONT’s Q20 +chemistry has notably increased its reliability for 16S rRNA gene studies [10,11], while PacBio has benefited from higher accuracy for a longer period, partly through the application of consensus or HiFi sequencing [12,13]. Additionally, PacBio has made strides in reducing the costs of 16S rRNA sequencing with the introduction of the Kinnex 16S rRNA kit, which utilises the MAS-seq method [14] and their latest sequencing platforms, enhancing the accessibility of full length 16S rRNA sequencing. As the benefits of 16S rRNA gene sequencing become increasingly recognized, the scope of long-read platforms is expanding. They are now being used to sequence both the 16S and 23S genes on a single stretch of DNA while also capturing the internal transcribed spacer (ITS) region. This allows a greater proportion of amplicon sequences to be assigned to species and even strain-level compared to 16S rRNA sequencing [15]. Recent studies have applied this approach to profiling microbial communities [16,17], though they had to rely on custom-built or commercial databases such as Intus Biosciences (formerly Shoreline Biome), which do not allow the direct comparison of results between studies. Recently, others have highlighted the requirement for a regularly updated and publicly available database to format the rrn structure for amplicon-mediated metagenomic analyses [18].
Here we present GROND (Genome-derived Ribosomal Operon Database), an open access quality-checked database of full-length 16S-ITS-23S rRNA sequences, accompanied by taxonomic and contextual data, to act as a database to standardise analysis between studies in the same way silva [19], RDP [20], and Greengenes [21] function for single rRNA genes.
Database construction
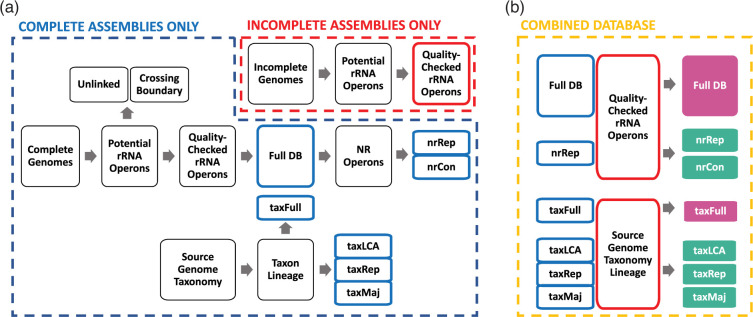
Two datasets were used as the basis for the GROND database, differing primarily by the taxonomy systems employed. The first of these, referred to as RefSeq throughout this manuscript, was constructed from all 253 840 RefSeq genome assemblies marked ‘Latest’ on 14 July 2022. The second, referred to as GTDB, was constructed from all 317 541 genomes included in the most recent release (07-RS207) of the GTDB database [22]. The pipeline described below, and illustrated in Fig. 1, was used to first construct databases of quality-checked 16S-ITS-23S rRNA operon (rrn) sequences from complete genomes and then expand this with sequences from incomplete assemblies, thereby capturing as much diversity as possible while placing a premium on genome assembly quality. The associated annotation information of RefSeq and GTDB source genomes was downloaded in GFF3 format and rRNA gene features were extracted. If annotations were not available, rRNA genes were identified using barrnap (v 0.9) with default parameters except genes with a coverage of <80 % were marked ‘partial’. The combined annotation information from complete assemblies was imported into R [23] using the read.gff function from the ape package [24], partial genes were discarded, and rrn sequences were identified by iteratively associating 16S genes with their neighbouring 23S gene if it was:
Fig. 1. GROND database construction pipeline. Panel (a) depicts intermediary steps and files which are combined and/or dereplicated to generate the final database depicted in Panel (b) (more details in DATABASE CONSTRUCTION section). Colour-outlined rectangles represent intermediary files used to build the colour-filled sequence and taxonomy files available for download.
Located on the same assembled sequence (contig or scaffold),
Encoded on the same strand,
Encoded in the order 16S-ITS-23S rRNA.
The potential rrn operons identified based on these criteria were further filtered to remove ‘unlinked’ operons with ITS regions longer than 1.5kbp [25] and operons that crossed the start/end boundary of the contig or scaffold, to leave a final dataset of quality-checked rrn sequences. As previously reported [25], unlinked rrn operons were highly prevalent in the Deinococcus-Thermus and Planctomycetes phyla (Table S1, available in the online version of this article). Each sequence in the final dataset was assigned a unique operon identifier and their nucleotide sequences were written to a single multifasta file using BEDTools [26]. To reduce computation time and aid analysis, a non-redundant (NR 99.9 %) database was created for each dataset. First, a multifasta file of high-quality rrn operon sequences recovered from complete genomes was sorted by sequence length using BBTools sortbyname.sh [27] and clustered based on 99.9 % nucleotide identity using vsearch --cluster_smallmem [28]. First sorting by length ensured that the longest sequence in each cluster was retained as the representative sequence. The high-quality rrn operon sequences from incomplete assemblies were then appended to the representative sequence multifasta file and re-clustered, meaning that operon sequences from incomplete assemblies were only retained if they expanded on the sequence diversity of sequences from complete genomes. Consensus sequences for each NR cluster were generated using the --consout option. We believe that most users would benefit from using the NR 99.9 % databases built from both complete and incomplete assemblies to maximise phylogenetic range while minimising computation burden. These are available for download as either representative (nrRep) or consensus (nrCon) sequences.
Database description
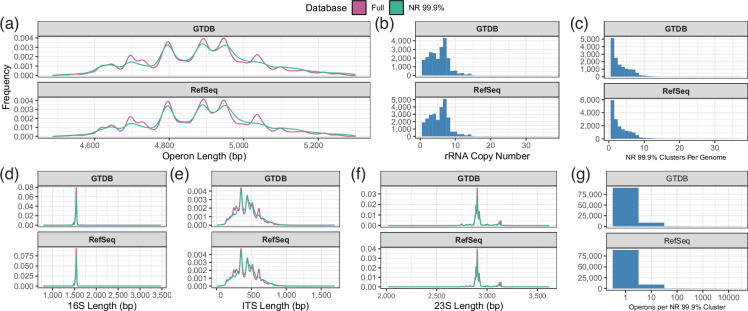
The median length of rrn sequences ranged between 4892 and 4899 bp depending on database version (Fig. 2a, Table 1). Of the three constituent regions, the ITS exhibited the greatest length variability across all database versions, followed by the 23S and 16S genes (Fig. 2d–f). When considering complete genomes only, the corresponding source genomes have a mean per-genome rRNA copy number of 5.29 (s=2.76) and 5.41 (s=2.77) for the GTDB and RefSeq datasets, respectively (Fig. 2b), while 63.62 % or 63.80 % of these genomes were represented in more than one NR cluster (Fig. 2c), supporting previous reports of intragenomic diversity in rRNA genes and ITS regions [29,31]. This is explored further below. For both datasets, approximately 81 % of NR clusters contained just a single rrn sequence (Fig. 2g). Dereplication of sequences at 99.9 % identity reduced the GTDB dataset from 291 365 sequences to 103 991, a 64.3 % reduction, and reduced the RefSeq dataset from 317 986 sequences to 103 293, a 67.5 % reduction. Comparatively, this same dereplication was applied to the 16S gene from these rrn operon sequences, yielding 58 531 and 55 193 sequences from the GTDB and RefSeq datasets respectively – an approximately two-fold greater reduction.
Fig. 2. Summary statistics of the full and non-redundant (NR 99.9 %) GROND databases. (a) Length distribution of 16S-ITS-23S rRNA operons using the GTDB and RefSeq derived databases. (b) Number of distinct rRNA operons in genomes represented in the database. (c) Number of NR clusters per genome. (d–f) Length distribution of individual 16S-ITS-23S rRNA operon regions. (g) Number of rRNA operons represented by each NR cluster. Operons identified as outliers based on length (quartile 1/3±1.5 × interquartile range [IQR]) are not included in this plot to increase readability.
Table 1. Summary statistics describing length (bp) of rrn operons and constituent regions for each dataset and database. SD=Standard Deviation, CV=Coefficient of Variation.
| Dataset | Database | Region | Mean | Median | SD | CV |
| GTDB | Full | 16S | 1534.91 | 1539 | 30.22 | 1.97 |
| 23S | 2924.31 | 2903 | 97.91 | 3.35 | ||
| ITS | 439.85 | 427 | 182.56 | 41.51 | ||
| rrn | 4899.05 | 4893 | 186.64 | 3.81 | ||
| NR 99.9 % | 16S | 1532.32 | 1539 | 37.21 | 2.43 | |
| 23S | 2924.54 | 2903 | 117.71 | 4.02 | ||
| ITS | 452.85 | 435 | 186.57 | 41.2 | ||
| rrn | 4909.69 | 4895 | 205.98 | 4.2 | ||
| RefSeq | Full | 16S | 1536.3 | 1540 | 27.16 | 1.77 |
| 23S | 2924.58 | 2903 | 90.61 | 3.1 | ||
| ITS | 437.56 | 428 | 179.54 | 41.03 | ||
| rrn | 4898.43 | 4892 | 179.92 | 3.67 | ||
| NR 99.9 % | 16S | 1534.44 | 1539 | 32.9 | 2.14 | |
| 23S | 2927.21 | 2903 | 109.82 | 3.75 | ||
| ITS | 450.6 | 436 | 186.45 | 41.38 | ||
| rrn | 4912.25 | 4899 | 197.58 | 4.02 |
Taxonomy
Taxonomy was assigned to each operon based on the source genome in seven-level format (Kingdom>Phylum>Class >Order>Family>Genus>Species). GTDB taxonomy was readily available to download in this format for archaea and bacteria. RefSeq TaxIDs were converted to this format by TaxonKit [32].
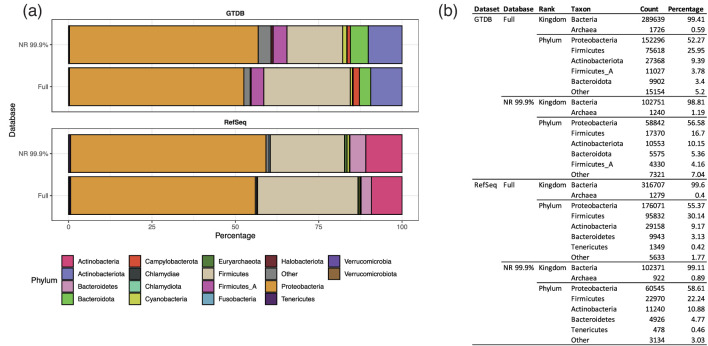
We found that 99.66 % of GTDB NR clusters (and 97.18 % of RefSeq clusters) exhibit 1 % taxonomic agreement in the species-level taxonomy of their constituent operon sequences. In an effort to account for the absence of taxonomic agreement among the remaining clusters, three methods are used to assign taxonomy.
taxRep: source genome taxonomy of the cluster representative sequence,
taxLCA: lowest common ancestor of all sequences in the cluster,
taxMaj: lowest taxonomic rank at which there is a simple majority agreement of all sequences in the cluster.
Files describing these taxonomy systems are available for download with the NR database and phylum-level descriptions are provided in Fig. 3. For most analyses, we recommend the taxRep system. The taxLCA and taxMaj systems are provided to compensate for clusters representing species with unclear or ‘disputed’ taxonomy that may have arisen from incorrect taxonomic assignment of the genome when uploaded to the RefSeq database or an imperfect understanding of what defines a species. For example, a cluster which contains 97 sequences from E. coli and three sequences from the Shigella genus, two genera which are historically separated but should not be distinct based on genome-level comparative analysis [33], would be classified as s__Escherichia coli by taxMaj, but as f_Enterobacteriaceae by taxLCA.
Fig. 3. Taxonomic profile of GROND database. (a) Phylum-level composition of full and NR 99.9 % databases using GTDB and RefSeq databases and taxonomy systems. (b) Kingdom and Phylum-level composition expressed as number of operons and percentage of total.
Intragenomic diversity
Intragenomic diversity of the rrn operon, and the individual constituent 16S, ITS, and 23S regions, was assessed to better understand the diversity of sequences expected for genome identification and microbiome profiling. This was performed on complete GTDB genomes based on the within-cluster taxonomic consistency observed above. From each genome containing more than one rrn operon: the regions being compared (rrn, 16S, ITS, or 23S) were written to a single multifasta file by BEDtools, a multiple sequence alignment was constructed by MAFFT [34], before being converted to a pairwise distance matrix by EMBOSS distmat [35] with option -nucmethod 0. These pairwise distance matrices were then imported into R using the read_phylip_distmat function in the tidygenomes library [36], where summary statistics and plots were generated using the tidyverse [37] and ggcorrplot [38] libraries. Pairwise distances are reported by distmat as SNVs per 100 nucleotides, so pairwise identities were calculated by subtracting the distances from 100.
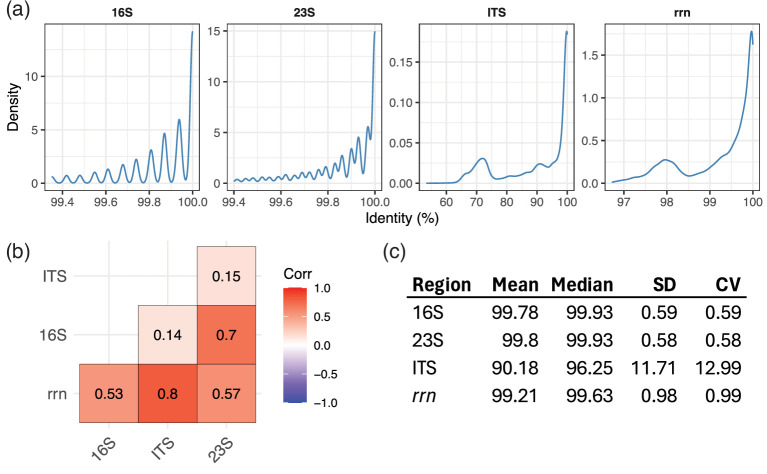
Of the 21 242 complete genomes analysed, 16 680 (78.5 %) exhibited intragenomic diversity of the rrn operon, compared to 14 611 (68.8 %), 13 943 (65.6 %), and 13 276 (62.5 %) for the ITS, 23S, and 16S regions respectively. Pairwise intragenomic distances of the entire rrn operon followed a roughly bimodal distribution (Fig. 4a), with the major peak at ~99.92 % sequence identity and the minor peak at ~98 % sequence identity, the latter representing ~98 SNVs based on the mean operon sequence length of 4899 bp. This pattern was mirrored by the ITS region which also followed a roughly bimodal distribution, with the major peak at 100 % sequence identity and the minor peak at ~72 % sequence identity, the latter representing ~123 SNVs based on the mean ITS length of 440 bp. This suggests that the main factor driving rrn operon intragenomic diversity are variations in the ITS regions, supported by correlation analysis showing a strong (ρ=0.8) relationship between intragenomic diversity of the rrn and ITS (Fig. 4b). The 16S and 23S genes exhibited similar intragenomic similarity distributions and summary statistics (Fig. 4c). Their diversities were also highly correlated, meaning genomes with higher 16S diversity tended to have higher 23S diversity, and vice versa. Information on the intragenomic diversity of all taxa with complete GTDB genomes is included in GROND.
Fig. 4. Intragenomic diversity of rrn operons and constituent regions from complete GTDB genomes. Plots are based on nucleotide identity between all pairwise intragenomic combinations of rrn operons and constituent regions. (a) Distribution of pairwise identities. Regions identified as outliers based on length (quartile 1/3±1.5 × interquartile range [IQR]) are not included in this plot to increase readability. (b) Pearson correlation of pairwise identities between regions from the same pairwise operon comparisons. (c) Summary statistics of intragenomic pairwise identities for the rrn operon and the constituent regions. SD=standard deviation, CV=coefficient of variation.
Similar to previous reports examining intragenomic diversity of the 16S gene [31], we observed that the highest degree of intragenomic rrn diversity tended to be present in extremophiles. For example, the halophilic family Haloarculaceae, the psychrophilic genus Acerihabitans, and the thermophilic genus Thermoanaerobacter all possessed mean rrn nucleotide diversities>3 %.
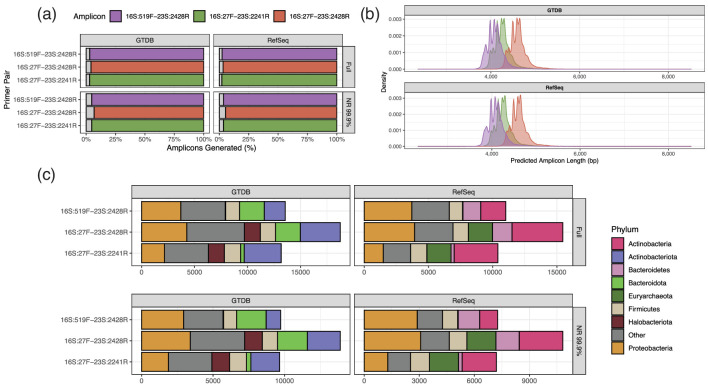
PCR primer evaluation for amplicon generation
To assess the impact of primer choice on the functionality of the database, we tested three different primer combinations in silico (Table 2) using the perl script in_silico_pcr.pl (https://github.com/egonozer/in_silico_pcr) to evaluate their amplicon generation efficiency, phylogenetic bias, and amplicon length distribution. Two primer pairs from previous studies focused on the full-length rRNA operon were evaluated, in addition to a new pair that combines the forward and reverse primers from each pair to potentially generate a longer amplicon.
Table 2. Primer sequences used for in silico PCR analysis.
Based on the predicted primer binding characteristics there is a strong inverse relationship between predicted amplicon length and database coverage meaning there is a trade-off to be considered between phylogenetic range and resolution when selecting primers (Fig. 5a, b). The primer pair 16S:27 F-23S:2428R, which generates the largest amplicons, is predicted to be 1–2 % less sensitive than the other two pairs (Fig. 5a and Table S2).
Fig. 5. Predicted amplicon generation statistics. (a) Percentage of database sequences which were predicted to generate an amplicon by each primer pair. (b) Predicted length distribution of amplicons generated from the NR 99.9 % GROND database by each primer pair. (c) Phylum-level composition of GROND database sequences where an amplicon was not predicted to be generated by each primer pair.
Primer binding biases were predicted to be relatively consistent between primer pairs at phylum level (Fig. 5c).
Outlook
We predict that the future of rRNA-based phylogenetic analysis will become increasingly dependent on long-read sequencing technologies due to their superior discriminatory ability. Advancements in sequencing accuracy achieved through ONT R10.4 flow cells [10,11, 39] and PacBio HiFi reads [12,13] have played a pivotal role in this shift from short-read to long-read sequencing. This progression has notably facilitated the rise of full-length 16S rRNA sequencing in recent times. Consequently, phylogenetic analysis based on full-length 16S rRNA currently benefits from well-established laboratory and computational workflows. Meanwhile, rrn sequencing is gaining attention for its enhanced resolution in distinguishing closely related species compared to full-length 16S rRNA sequencing [40,44]. However, as rrn sequencing is still developing, more advancements and validation are needed before it becomes broadly adopted.
One such advance, essential for the success of any microbiome study, is the construction of reliable databases. The current state of rrn sequencing relies on custom-built [16,45,47] or commercial databases [48,49], which results in a lack of standardisation across the field. Moreover, these databases require ongoing updates and maintenance to remain relevant [18]. With these considerations in mind, GROND has been developed to address these shortcomings in rrn sequencing. As the most comprehensive rrn database to date, and incorporating a GTDB-based version that includes numerous metagenome-assembled genomes (MAGs), this database should prove to be a valuable resource for the rrn sequencing community. Indeed, the preprint version of this database has already been used to study bacterial transmission in low-biomass human milk samples [50].
Another key advancement in the adoption of rrn sequencing is the standardisation of PCR primers. Variations in microbial profiles have previously been reported depending on the primer pairs used to generate rrn amplicons. Kinoshita et al. [16] and Martijn et al. [17] have demonstrated that the 519 F-2428R primer pair reveals a broader diversity of bacteria and archaea. As a result, while most existing rrn databases feature sequences extracted using the 27 F-2241R primer pair [18,46], GROND is intentionally untrimmed so as to be useable by all studies regardless of primer choice, enabling more levelled comparisons in future studies of primer biases in rrn sequencing.
The choice of classifier for phylogenetic analysis is significantly influenced by the type of long-read platform employed. The high accuracy of PacBio sequencing data makes it compatible with denoising or clustering pipelines, such as DADA2 [50,51] and vsearch [17,28], respectively. Meanwhile, the lower read quality from ONT demands specialised pipelines to account for its higher error rates. Such pipelines include, alignment using Minimap2 [18,45, 46, 52] or EMU [39,40, 53], or clustering through NanoCLUST [54] or Natrix2 [55]. The application of Unique Molecular Identifiers (UMIs) has shown promise in achieving more accurate ASVs or 97 % OTUs than NanoCLUST-based methods [11,15]. However, this strategy needs further evaluation before it can be applied in rrn sequencing for a range of complex communities. The single-nucleotide resolution offered by PacBio HiFi sequencing and UMI-supported ONT sequencing is important as the scope of rrn sequencing is widening to include strain-level profiling of microbial communities [48,56, 57], where PacBio’s higher sequencing quality has currently made it the more suitable choice for strain-level detection [12,48, 58].
As we move towards achieving strain-level distinction through rrn sequencing, acknowledging the influence of intragenomic variation becomes essential. Bacterial and archaeal genomes typically feature multiple copies of the rrn operon and the sequence variation among these copies within a single genome may lead to strain-level misassignments and inflated species-level abundance figures [12,58,60]. It is therefore critical to factor in these intragenomic variations. Sequencing the rrn operon, rather than full-length 16S rRNA gene, has proven to more precisely capture the heterogeneity among these copies [12,60]. Techniques based on OTU clustering, rather than exact sequence variants (ESVs), offer a superior way to accommodate intragenomic variation, resulting in more accurate taxonomic profiling when high resolution is necessary [58]. Appropriate clustering thresholds have been established for human microbiome samples specifically for the 16S rRNA gene. However, research on these thresholds for environmental samples and rrn sequencing methods is still lacking, presenting an opportunity for further exploration and advancement. Here we have analysed intragenomic diversity of the rrn operon and its constituent regions using only complete genome assemblies and operon sequences which passed strict quality control thresholds. However, there are still some factors that must be considered when interpreting these results. First, estimating true copy number can be problematic as these are essentially repetitive regions, which may be collapsed into a smaller number of copies by short-read assemblers. Second, since these repeats may be non-exact, the sequences may be fragmented across multiple contigs. Third, minority variants may not be represented because the assembly algorithm thinks they are errors and ‘corrects’ them, for example if a given genome contains seven copies of the rRNA operon, and at a particular position six have G and one has C, the resulting assembly might lose the C variant and only return the G variants. Using complete genomes for this analysis likely negates the first and second issues but the third may still be present.
As the field of rrn sequencing progresses, we can anticipate improved resolution, wider application, and early adoption of standardised methods such as those currently recommended by the Earth Microbiome Project [61]. The GROND database will additionally support this advancement through continual updates with each major RefSeq and GTDB release to keep pace with the ever-growing collection of genome sequences and constantly evolving taxonomy systems.
supplementary material
Acknowledgements
We would like to thank Ryan Wick for his valuable insights into the relationship between genome assembly, rrn copy number, and intragenomic diversity.
Abbreviations
- CoV
coefficient of variation
- ESV
exact sequence variants
- GROND
genome-derived ribosomal operon database
- GTDB
genome taxonomy database
- IQR
interquartile range
- ITS
internal transcribed spacer
- NR
non-redundant
- ONT
Oxford Nanopore Technologies
- rrn
16S-ITS-23S rRNA operon
- rRNA
ribosomal RNA
- SD
standard deviation
- taxLCA
lowest common ancestor of all sequences in the cluster
- taxMaj
lowest taxonomic rank at which there is a simple majority agreement of all sequences in the cluster
- taxRep
source genome taxonomy of the cluster representative sequence,
- UMIs
unique molecular identifiers
Footnotes
Funding: C.J.W. and T.P.S. are funded by National Health and Medical Research Council (GNT1145631). M.S. is the recipient of a Teagasc Walsh Scholarship award (2020018) funded by Teagasc Agriculture and Food Development Authority and received support from Science Foundation Ireland (SFI) under [grant number SFI/12/RC/2273_P2] (APC Microbiome Ireland).
Contributor Information
Calum J. Walsh, Email: Calum.walsh@unimelb.edu.au.
Meghana Srinivas, Email: meghana.srinivas@teagasc.ie.
Timothy P. Stinear, Email: tstinear@unimelb.edu.au.
Douwe van Sinderen, Email: d.vansinderen@ucc.ie.
Paul D. Cotter, Email: paul.cotter@teagasc.ie.
John G. Kenny, Email: john.kenny@teagasc.ie.
References
- 1.Chakravorty S, Helb D, Burday M, Connell N, Alland D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods. 2007;69:330–339. doi: 10.1016/j.mimet.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van de Peer Y, Chapelle S, De Wachter R. A quantitative map of nucleotide substitution rates in bacterial rRNA. Nucleic Acids Res. 1996;24:3381–3391. doi: 10.1093/nar/24.17.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stevens BM, Creed TB, Reardon CL, Manter DK. Comparison of Oxford Nanopore Technologies and illumina MiSeq sequencing with mock communities and agricultural soil. Sci Rep. 2023;13:9323. doi: 10.1038/s41598-023-36101-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Catozzi C, Ceciliani F, Lecchi C, Talenti A, Vecchio D, et al. Short communication: milk microbiota profiling on water buffalo with full-length 16S rRNA using nanopore sequencing. J Dairy Sci. 2020;103:2693–2700. doi: 10.3168/jds.2019-17359. [DOI] [PubMed] [Google Scholar]
- 5.Lemoinne A, Dirberg G, Georges M, Robinet T. Fine-scale congruence in bacterial community structure from marine sediments sequenced by short-reads on illumina and long-reads on nanopore. Mol Biol. 2023 doi: 10.1101/2023.06.06.541006. [DOI] [Google Scholar]
- 6.Waechter C, Fehse L, Welzel M, Heider D, Babalija L, et al. Comparative analysis of full-length 16s ribosomal RNA genome sequencing in human fecal samples using primer sets with different degrees of degeneracy. Front Genet. 2023;14:1213829. doi: 10.3389/fgene.2023.1213829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huggins LG, Colella V, Atapattu U, Koehler AV, Traub RJ. Nanopore sequencing using the full-length 16S rRNA gene for detection of blood-borne bacteria in dogs reveals a novel species of hemotropic mycoplasma. Microbiol Spectr. 2022;10:e0308822. doi: 10.1128/spectrum.03088-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsuo Y, Komiya S, Yasumizu Y, Yasuoka Y, Mizushima K, et al. Full-length 16S rRNA gene amplicon analysis of human gut microbiota using MinIONTM nanopore sequencing confers species-level resolution. BMC Microbiol. 2021;21:35. doi: 10.1186/s12866-021-02094-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Handy MY, Sbardellati DL, Yu M, Saleh NW, Ostwald MM, et al. Incipiently social carpenter bees (Xylocopa) host distinctive gut bacterial communities and display geographical structure as revealed by full-length PacBio 16S rRNA sequencing. Mol Ecol. 2023;32:1530–1543. doi: 10.1111/mec.16736. [DOI] [PubMed] [Google Scholar]
- 10.Zhang T, Li H, Ma S, Cao J, Liao H, et al. The newest Oxford Nanopore R10.4.1 full-length 16S rRNA sequencing enables the accurate resolution of species-level microbial community profiling. Appl Environ Microbiol. 2023;89:e00605-23. doi: 10.1128/aem.00605-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin X, Waring K, Tyson J, Ziels RM. High-accuracy meets high-throughput for microbiome profiling with near full-length 16S rRNA amplicon sequencing on the Nanopore platform. Microbiology. 2023 doi: 10.1101/2023.06.19.544637. [DOI] [Google Scholar]
- 12.Callahan BJ, Wong J, Heiner C, Oh S, Theriot CM, et al. High-throughput amplicon sequencing of the full-length 16S rRNA gene with single-nucleotide resolution. Nucleic Acids Res. 2019;47:e103. doi: 10.1093/nar/gkz569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Earl JP, Adappa ND, Krol J, Bhat AS, Balashov S, et al. Species-level bacterial community profiling of the healthy sinonasal microbiome using Pacific Biosciences sequencing of full-length 16S rRNA genes. Microbiome. 2018;6:190. doi: 10.1186/s40168-018-0569-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al’Khafaji AM, Smith JT, Garimella KV, Babadi M, Popic V, et al. High-throughput RNA isoform sequencing using programmed cDNA concatenation. Nat Biotechnol. 2024;42:582–586. doi: 10.1038/s41587-023-01815-7. [DOI] [PubMed] [Google Scholar]
- 15.Karst SM, Ziels RM, Kirkegaard RH, Sørensen EA, McDonald D, et al. High-accuracy long-read amplicon sequences using unique molecular identifiers with nanopore or PacBio sequencing. Nat Methods. 2021;18:165–169. doi: 10.1038/s41592-020-01041-y. [DOI] [PubMed] [Google Scholar]
- 16.Kinoshita Y, Niwa H, Uchida-Fujii E, Nukada T. Establishment and assessment of an amplicon sequencing method targeting the 16S-ITS-23S rRNA operon for analysis of the equine gut microbiome. Sci Rep. 2021;11:11884. doi: 10.1038/s41598-021-91425-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martijn J, Lind AE, Schön ME, Spiertz I, Juzokaite L, et al. Confident phylogenetic identification of uncultured prokaryotes through long read amplicon sequencing of the 16S-ITS-23S rRNA operon. Environ Microbiol. 2019;21:2485–2498. doi: 10.1111/1462-2920.14636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seol D, Lim JS, Sung S, Lee YH, Jeong M, et al. Microbial identification using rRNA operon region: database and tool for metataxonomics with long-read sequence. Microbiol Spectr. 2022;10:e0201721. doi: 10.1128/spectrum.02017-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, et al. Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDonald D, Jiang Y, Balaban M, Cantrell K, Zhu Q, et al. Greengenes2 unifies microbial data in a single reference tree. Nat Biotechnol. 2023 doi: 10.1038/s41587-023-01845-1. https://www.nature.com/articles/s41587-023-01845-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parks DH, Chuvochina M, Rinke C, Mussig AJ, Chaumeil P-A, et al. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 2022;50:D785–D794. doi: 10.1093/nar/gkab776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.R Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2021. [Google Scholar]
- 24.Paradis E, Schliep K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019;35:526–528. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- 25.Brewer TE, Albertsen M, Edwards A, Kirkegaard RH, Rocha EPC, et al. Unlinked rRNA genes are widespread among bacteria and archaea. ISME J. 2020;14:597–608. doi: 10.1038/s41396-019-0552-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brian Bushnell Bbmap. 2021.
- 28.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stewart FJ, Cavanaugh CM. Intragenomic variation and evolution of the internal transcribed spacer of the rRNA operon in bacteria. J Mol Evol. 2007;65:44–67. doi: 10.1007/s00239-006-0235-3. [DOI] [PubMed] [Google Scholar]
- 30.Pereira TJ, De Santiago A, Schuelke T, Hardy SM, Bik HM. The impact of intragenomic rRNA variation on metabarcoding‐derived diversity estimates: a case study from marine nematodes. Environ DNA. 2020;2:519–534. doi: 10.1002/edn3.77. [DOI] [Google Scholar]
- 31.Sun D-L, Jiang X, Wu QL, Zhou N-Y. Intragenomic heterogeneity of 16S rRNA genes causes overestimation of prokaryotic diversity. Appl Environ Microbiol. 2013;79:5962–5969. doi: 10.1128/AEM.01282-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen W, Ren H. TaxonKit: a practical and efficient NCBI taxonomy toolkit. J Genet Genom. 2021;48:844–850. doi: 10.1016/j.jgg.2021.03.006. [DOI] [PubMed] [Google Scholar]
- 33.Pettengill EA, Pettengill JB, Binet R. Phylogenetic analyses of Shigella and Enteroinvasive Escherichia coli for the identification of molecular epidemiological markers: whole-genome comparative analysis does not support distinct genera designation. Front Microbiol. 2015;6:1573. doi: 10.3389/fmicb.2015.01573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 36.Wittouck S. tidygenomes: a grammar of genome Data manipulation. 2024. https://github.com/SWittouck/tidygenomes
- 37.Wickham H, Averick M, Bryan J, Chang W, McGowan L, et al. Welcome to the Tidyverse. J Open Source Softw. 2019;4:1686. doi: 10.21105/joss.01686. [DOI] [Google Scholar]
- 38.Kassambara A. ggcorrplot: visualization of a correlation matrix using ‘ggplot2. 2023. https://CRAN.R-project.org/package=ggcorrplot
- 39.Petrone JR, Rios Glusberger P, George CD, Milletich PL, Ahrens AP, et al. RESCUE: a validated Nanopore pipeline to classify bacteria through long-read, 16S-ITS-23S rRNA sequencing. Front Microbiol. 2023;14:1201064. doi: 10.3389/fmicb.2023.1201064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olivier SA, Bull MK, Strube ML, Murphy R, Ross T. Long-read MinIONTM sequencing of 16S and 16S-ITS-23S rRNA genes provides species-level resolution of Lactobacillaceae in mixed communities. Front Microbiol. 2023;14:1290756. doi: 10.3389/fmicb.2023.1290756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kalia VC, Kumar R, Kumar P, Koul S. A genome-wide profiling strategy as an aid for searching unique identification biomarkers for Streptococcus. Indian J Microbiol. 2016;56:46–58. doi: 10.1007/s12088-015-0561-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lal D, Verma M, Lal R. Exploring internal features of 16S rRNA gene for identification of clinically relevant species of the genus Streptococcus. Ann Clin Microbiol Antimicrob. 2011;10:28. doi: 10.1186/1476-0711-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Devanga Ragupathi NK, Muthuirulandi Sethuvel DP, Inbanathan FY, Veeraraghavan B. Accurate differentiation of Escherichia coli and Shigella serogroups: challenges and strategies. New Microbes New Infect. 2018;21:58–62. doi: 10.1016/j.nmni.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deurenberg RH, Bathoorn E, Chlebowicz MA, Couto N, Ferdous M, et al. Application of next generation sequencing in clinical microbiology and infection prevention. J Biotechnol. 2017;243:16–24. doi: 10.1016/j.jbiotec.2016.12.022. [DOI] [PubMed] [Google Scholar]
- 45.Cuscó A, Catozzi C, Viñes J, Sanchez A, Francino O. Microbiota profiling with long amplicons using nanopore sequencing: full-length 16S rRNA gene and the 16S-ITS-23S of the rrn operon. F1000Res. 2018;7:1755. doi: 10.12688/f1000research.16817.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Planý M, Sitarčík J, Pavlović J, Budiš J, Koreňová J, et al. Evaluation of bacterial consortia associated with dairy fermentation by ribosomal RNA (rrn) operon metabarcoding strategy using MinION device. Food Biosci. 2023;51:102308. doi: 10.1016/j.fbio.2022.102308. [DOI] [Google Scholar]
- 47.Benítez-Páez A, Sanz Y. Multi-locus and long amplicon sequencing approach to study microbial diversity at species level using the MinIONTM portable nanopore sequencer. Gigascience. 2017;6:1–12. doi: 10.1093/gigascience/gix043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gehrig JL, Portik DM, Driscoll MD, Jackson E, Chakraborty S, et al. Finding the right fit: evaluation of short-read and long-read sequencing approaches to maximize the utility of clinical microbiome data. Microb Genom. 2022;8:000794. doi: 10.1099/mgen.0.000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Graf J, Ledala N, Caimano MJ, Jackson E, Gratalo D, et al. High-resolution differentiation of enteric bacteria in premature infant fecal microbiomes using a novel rRNA amplicon. mBio. 2021;12:e03656-20. doi: 10.1128/mBio.03656-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spreckels JE, Fernández-Pato A, Kruk M, Kurilshikov A, Garmaeva S, et al. Analysis of microbial composition and sharing in low-biomass human milk samples: a comparison of DNA isolation and sequencing techniques. ISME Commun. 2023;3:116. doi: 10.1038/s43705-023-00325-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Curry KD, Wang Q, Nute MG, Tyshaieva A, Reeves E, et al. Emu: species-level microbial community profiling of full-length 16S rRNA Oxford nanopore sequencing data. Nat Methods. 2022;19:845–853. doi: 10.1038/s41592-022-01520-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodríguez-Pérez H, Ciuffreda L, Flores C. NanoCLUST: a species-level analysis of 16S rRNA nanopore sequencing data. Bioinform Oxf Engl. 2021;37:1600–1601. doi: 10.1093/bioinformatics/btaa900. [DOI] [PubMed] [Google Scholar]
- 55.Deep A, Bludau D, Welzel M, Clemens S, Heider D, et al. Natrix2 – improved amplicon workflow with novel Oxford Nanopore Technologies support and enhancements in clustering, classification and taxonomic databases. Metabarcoding Metagenom. 2023;7:e109389. doi: 10.3897/mbmg.7.109389. [DOI] [Google Scholar]
- 56.Kerkhof LJ, Roth PA, Deshpande SV, Bernhards RC, Liem AT, et al. A ribosomal operon database and MegaBLAST settings for strain-level resolution of microbiomes. FEMS Microbes. 2022;3:xtac002. doi: 10.1093/femsmc/xtac002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dowden RA, McGuinness LR, Wisniewski PJ, Campbell SC, Guers JJ, et al. Host genotype and exercise exhibit species-level selection for members of the gut bacterial communities in the mouse digestive system. Sci Rep. 2020;10:8984. doi: 10.1038/s41598-020-65740-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson JS, Spakowicz DJ, Hong B-Y, Petersen LM, Demkowicz P, et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat Commun. 2019;10:5029. doi: 10.1038/s41467-019-13036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun DL, Jiang X, Wu QL, Zhou NY. Intragenomic heterogeneity of 16S rRNA genes causes overestimation of prokaryotic diversity. Appl Environ Microbiol. 2013;79:5962–5969. doi: 10.1128/AEM.01282-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Oliveira Martins L, Page AJ, Mather AE, Charles IG. Taxonomic resolution of the ribosomal RNA operon in bacteria: implications for its use with long-read sequencing. NAR Genom Bioinform. 2020;2:lqz016. doi: 10.1093/nargab/lqz016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thompson LR, Sanders JG, McDonald D, Amir A, Ladau J, et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature. 2017;551:457–463. doi: 10.1038/nature24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.