Abstract
The phosphoprotein, P, of vesicular stomatitis virus (VSV) is a key subunit of the viral RNA-dependent RNA polymerase complex. The protein is phosphorylated at multiple sites in two different domains. We recently showed that specific serine and threonine residues within the amino-terminal acidic domain I of P protein must be phosphorylated for in vivo transcription activity, but not for replication activity, of the polymerase complex. To examine the role of phosphorylation of the carboxy-terminal domain II residues of the P protein in transcription and replication, we have used a panel of mutant P proteins in which the phosphate acceptor sites (Ser-226, Ser-227, and Ser-233) were altered to alanines either individually or in various combinations. Analyses of the mutant proteins for their ability to support replication of a VSV minigenomic RNA suggest that phosphorylation of either Ser-226 or Ser-227 is necessary for optimal replication activity of the protein. The mutant protein (P226/227) in which both of these residues were altered to alanines was only about 8% active in replication compared to the wild-type (wt) protein. Substitution of alanine for Ser-233 did not have any adverse effect on replication activity of the protein. In contrast, all the mutant proteins showed activities similar to that of the wt protein in transcription. These results indicate that phosphorylation of the carboxy-terminal domain II residues of P protein are required for optimal replication activity but not for transcription activity. Furthermore, substitution of glutamic acid residues for Ser-226 and Ser-227 resulted in a protein that was only 14% active in replication but almost fully active in transcription. Taken together, these results, along with our earlier studies, suggest that phosphorylation of residues at two different domains in the P protein regulates its activity in transcription and replication of the VSV genome.
The RNA-dependent RNA polymerase of the prototypic rhabdovirus, vesicular stomatitis virus (VSV), is comprised of two virally encoded proteins: the large protein, L, and the phosphoprotein, P (1, 18, 20). A complex containing both the L and P proteins carries out transcription and replication of the viral genome (see reference 1 for a review). The template recognized by the RNA polymerase complex is the viral nucleocapsid, which contains the ∼11-kb-long negative-sense genomic RNA surrounded by the viral RNA-binding nucleocapsid protein, N. During transcription, the nucleocapsid template is utilized by the RNA polymerase complex to generate subgenomic, capped, and polyadenylated mRNAs, which are translated to produce the viral proteins. During replication, the same RNA polymerase is believed to be switched from transcriptive to replicative mode to synthesize a full-length, positive-sense complementary RNA (or antigenomic RNA) encapsidated by the N protein, which serves as the template for synthesis of progeny nucleocapsids.
Biochemical and mutational studies have suggested that the L protein is multifunctional and carries the catalytic center for polymerization of ribonucleotides during RNA synthesis (50, 51). It presumably also carries other enzymatic activities, including methyltransferase, guanylyltransferase, and poly(A)polymerase activities necessary for generation of mature viral mRNAs (27–29, 32). The multifunctional P protein, on the other hand, is required for the manifestation of the functions of the L protein (1) and plays key roles in both transcription and replication of the viral genome. It interacts specifically with the L protein to form the polymerase complex of the virion (21) as well as stabilizing the L protein against proteolytic degradation (9). It interacts with the terminal sequences of the viral genome (33, 34), presumably for viral RNA synthesis. The P protein also interacts with the N protein to form soluble N-P complexes required for encapsidation of nascent RNA chains during replication (36, 48). Furthermore, P interacts with itself to form oligomers that are required for transcription (15, 23, 24).
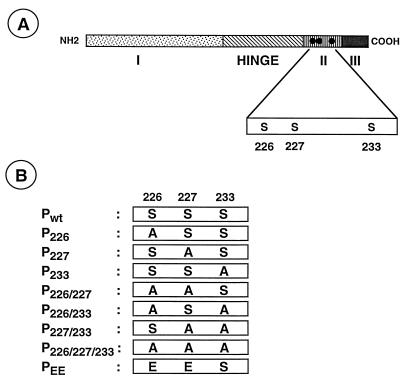
The P proteins of both the Indiana (PI) and New Jersey (PNJ) serotypes of VSV are highly acidic and consist of 265 and 274 amino acids, respectively. Although these two proteins have only 30% similarity at the primary sequence level, three functionally homologous domains (Fig. 1A) have been defined in mutational studies (25, 47). Domain I, spanning residues from 1 to about 150, is the amino-terminal acidic domain and is phosphorylated. Domain II spans from residues 210 to about 244 and is also phosphorylated. Domain III consists of 21 to 25 residues at the extreme carboxy terminus of the protein and is highly basic. Between domains I and II, a hypervariable region (called the hinge) of approximately 50 to 60 amino acids is present, but its function is unknown.
FIG. 1.
(A) Domain structure of the P protein of VSV (Indiana serotype). The P protein with the three functionally defined domains (I, II, and III) and the hinge region is shown. The phosphate acceptor sites (Ser-226, Ser-227, and Ser-233) within domain II are represented by solid circles. (B) Various mutant P proteins with substitutions of alanine (A) or glutamic acid (E) residues in place of serine residues are shown.
In the virion core as well as in infected cells, different phosphorylated forms of the P protein have been shown to exist (6, 13, 30, 31). Furthermore, functional P proteins with different degrees of phosphorylation have been documented (10, 35, 41, 49). It has been shown that the unphosphorylated form of P protein is inactive in transcription in vitro (2, 3). However, it becomes transcriptionally active when first phosphorylated by cellular casein kinase II (CKII) (3, 4, 15). The acceptor sites for CKII-mediated phosphorylation have been mapped to Ser-60, Thr-62, and Ser-64 in domain I of PI (12, 52) and Ser-59 and Ser-61 of PNJ (53). Phosphorylation of these residues was recently shown to mediate the multimerization of P protein that facilitates complex formation with the L protein (15, 23, 24). Using a panel of mutant PI proteins in which the phosphorylation sites in domain I were altered to alanines, we recently showed that phosphorylation of these domain I residues is necessary for the function of the P protein in transcription in vivo (45). The replication functions of the P protein in vivo were unaffected by the phosphorylation status of the domain I residues. Based on these studies, it was proposed that an increase in net negative charge in this region of the protein as a result of phosphorylation is crucial for transcriptional activation of the P protein (45). However, in a separate study, low levels (∼25%) of transcription under in vitro conditions were observed with a phosphorylation-defective P mutant protein (52).
Phosphorylation of serine residues in domain II also seems to control the activity of the P protein in RNA synthesis. It was shown that phosphorylation of two specific residues (Ser-236 and Ser-242) in domain II of PNJ by L-associated kinase (LAK) is necessary for in vitro transcription activity of the P protein (11). For PNJ, domain I must be phosphorylated prior to LAK-mediated phosphorylation of domain II residues (4). PI seems to be different in this respect, since domain II residues can be phosphorylated by LAK without prior phosphorylation of domain I residues (12). However, in a separate study, it was shown that substitution of aspartic acid residues at S-60 and T-62 in domain I of PI results in a protein that is not phosphorylated at other sites (24). Since phosphorylation in domain I is necessary and sufficient for transcription, it appears that phosphorylation of domain II residues may not be involved in transcription (23, 40). In PI, the Ser-226 and Ser-233 residues are homologous to Ser-236 and Ser-242 residues in PNJ and were therefore proposed to be the potential sites of phosphorylation (23). However, the actual sites of phosphorylation have recently been mapped to Ser-226 and Ser-227 residues in PI (12).
To examine the role of phosphorylation of domain II residues in the function of P protein in VSV RNA transcription and replication in vivo, we have used a panel of mutant P proteins with alanine substitutions at the phosphate acceptor sites. Analysis of the mutant P proteins for their ability to support replication of a VSV minigenomic template in vivo showed that phosphorylation of the serine residue at either 226 or 227 is necessary for optimal replication activity of P protein. In vivo-transcription activities of the mutant P proteins were unaffected. Furthermore, substitution of negatively charged glutamic acid residues for these serine residues resulted in a protein that was only 14% as active as the wild-type protein in replication while retaining almost full activity in transcription. These results suggest that phosphorylation of domain II residues is required for optimal replication, but not for transcription, function of the P protein.
MATERIALS AND METHODS
Cells and viruses.
Growth and maintenance of baby hamster kidney (BHK-21) cells (ATCC CCL10) have been described previously (46). VSV (Indiana serotype; San Juan strain) was grown, and infectivity titers were determined in BHK-21 cells. The recombinant vaccinia virus (vTF7-3) containing the bacteriophage T7 RNA polymerase gene (22) was grown in BHK-21 cells, and infectivity titers of stock vTF7-3 were determined as described earlier (45).
VSV protein- and minigenome-expressing plasmids.
The plasmids encoding the wild-type P protein (Pwt) or various mutant P proteins (P226, P227, P233, P226/227, P226/233, P227/233, and P226/227/233) of VSV (Indiana serotype; Mudd-Summers strain) in pET-3a vector have been described previously (12). These mutant P proteins contain serine-to-alanine amino acid substitutions at positions 226, 227, 233, 226-227, 226-233, 227-233, and 226-227-233, respectively, within domain II of the P protein (Fig. 1). The PEE mutant contains glutamic acid residues in place of serine residues at positions 226 and 227 of wild-type P protein. Plasmids pN and pL, carrying the coding regions for VSV (Indiana serotype; San Juan strain) proteins N and L, respectively, under the control of the T7 RNA polymerase promoter, have been reported before (46). Construction of the plasmid p9BN, encoding an antigenomic-sense VSV minigenome, has also been described earlier (39).
Virus infections and DNA transfections.
The details of the virus infection and DNA transfection procedures have been described in our earlier publications (44, 45). Briefly, BHK-21 cells in 60-mm-diameter plates or in 35-mm-diameter six-well plates were grown to approximately 90% confluency. These cells were infected with vTF7-3 at a multiplicity of infection of 10 PFU per cell. Following virus adsorption at 37°C for 45 min, the cells were washed with serum-free Dulbecco’s modified minimal essential medium (DMEM) and transfected with various combinations of plasmid DNAs encoding the VSV proteins and the minireplicon RNA by using Lipofectin reagent (Gibco/BRL, Gaithersburg, Md.) or a transfection reagent prepared in the laboratory as described previously (8). At 4 h posttransfection, the medium was removed from the transfected cells and the cells were washed twice in DMEM supplemented with 2% fetal bovine serum (FBS) and incubated at 37°C with an appropriate volume of the same medium for different lengths of time before metabolic labeling with radioactive precursors. Unless specifically described in the figure legends, in most experiments, 3 μg of pN, 2 μg of pET-3P, 1 μg of pL, and 5 μg of p9BN plasmids were used in transfection of cells in 60-mm-diameter plates. These plasmid amounts were reduced by half for cells in six-well plates.
Metabolic labeling and analysis of proteins.
[35S]methionine labeling of proteins in transfected cells, immunoprecipitation, and subsequent sodium dodecyl sulfate-polyacrylamide gel analysis of the proteins have been described previously (45). P proteins were immunoprecipitated by using a rabbit anti-P (Indiana serotype; Mudd-Summers strain) antibody.
Metabolic labeling and analysis of transcription and replication products.
Following incubation of transfected cells for 15 to 16 h at 37°C, the cells were pretreated with 15 μg of actinomycin D per ml of DMEM plus FBS at 37°C for 45 min. The cells were then labeled with 15 μCi of [3H]uridine and/or 15 μCi of [3H]adenosine per ml of DMEM plus FBS containing actinomycin D. The labeling of RNAs was performed for 6 to 8 h at 37°C. Cytoplasmic extracts from these cells were prepared in lysis buffer as described earlier (46). The labeled RNA replication products in nucleocapsids were immunoprecipitated by polyclonal anti-VSV antibodies and recovered by extraction with phenol and chloroform and precipitation with ethanol. The recovered RNAs were then separated by electrophoresis in agarose-urea gels (38) and detected by fluorography (37) as described in detail previously (46). For transcription analysis, total RNA from the cytoplasmic extracts was purified by extraction with phenol and chloroform and recovered by precipitation with ethanol. To generate VSV mRNAs, the cells were infected with VSV at a multiplicity of infection of 10 and labeled with [3H]uridine at 2 to 6 h postinfection in the presence of actinomycin D. Total RNA was recovered from the cytoplasmic extracts. The RNAs were analyzed by electrophoresis and fluorography as described above. The quantitation of replication and transcription product RNAs was carried out by densitometric scanning of the fluorograms with an IS-1000 digital imaging system version 2.02 (Alpha Innotech Corporation, San Leandro, Calif.). The readout values from the scanner provide integrated intensities of the appropriate bands in the fluorograms. The relative levels of replication and transcription supported by various mutants were calculated, with the values obtained for the wild-type P protein as 100. For normalization of levels of transcription supported by the mutant P proteins, we used the following formula: normalized levels of transcription = relative levels of transcription/relative levels of replication × 100.
RESULTS
The phosphorylation of serine residues at positions 236 and 242 within domain II of PNJ was shown previously to play a major role in the function of P protein in transcription in vitro (11). Based on amino acid sequence homology, corresponding serine residues at positions 227 and 233 of PI were proposed to be the potential sites for phosphorylation (23). Recently, however, biochemical and mutational studies identified serine residues at positions 226 and 227 to be the sites of phosphorylation (12). To examine the role of phosphorylation of these residues in P protein function in transcription and replication, we used a series of mutant P proteins (Fig. 1B) in which serine residues at positions 226, 227, and 233 were altered to alanines either individually (in mutants P226, P227, and P233) or in various combinations (in mutants P226/227, P226/233, P227/233, and P226/227/233). In initial experiments, we found that in transfected cells, the mutant P proteins were expressed at levels similar to that of the wild-type P protein and the amount of P protein expression was roughly proportional to the amount of plasmid transfected into the cells within a range of 1 to 15 μg. Furthermore, in pulse-chase experiments, the mutant proteins were stable over an 8-h chase period (data not shown).
In our earlier studies, we showed that the wild-type P protein of the Mudd-Summers strain of VSV functions as well as the San Juan strain of VSV P protein in transcription and replication with San Juan N and L proteins (45); therefore, the above-mentioned mutant P proteins of the Mudd-Summers strain were used in order to test their abilities to support transcription and replication in the presence of the N and L proteins of San Juan VSV.
Phosphorylation-defective mutant P proteins do not support optimal replication of VSV.
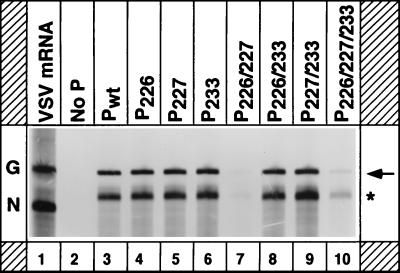
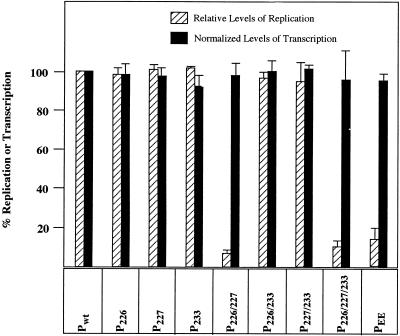
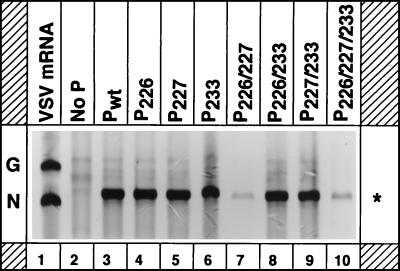
To examine the ability of the mutant P proteins to support replication of a VSV minigenome, we used a plasmid encoding an antigenomic-sense VSV minireplicon (9BN) RNA (39). The minireplicon RNA contains the terminal sequences of the VSV antigenome flanking the sequences of the N gene and part of the L gene (39). In cells transfected with the plasmid along with the plasmids encoding the viral proteins N, P, and L, the antigenomic-sense minireplicon RNA is replicated and amplified to generate the genomic- and antigenomic-sense RNAs (39). However, the genomic-sense RNA replication products can be more readily detected by metabolic labeling of the transfected cells, since the genomic RNAs are synthesized in much greater molar abundance than the antigenomic-sense RNAs. To perform the experiment, BHK-21 cells infected with vTF7-3 were transfected with plasmids encoding the minireplicon and N, L, and the wild-type or mutant P proteins. [3H]uridine- and [3H]adenosine-labeled RNAs synthesized in the presence of actinomycin D were recovered from immunoprecipitated nucleocapsids and analyzed by electrophoresis. The results (Fig. 2) from a typical experiment show that the mutant P proteins P226, P227, P233, P226/233, and P227/233 (Fig. 2, lanes 4 to 6, 8, and 9) were as active as the wild-type P protein (Fig. 2, lane 3) in replication, since the amounts of replication product obtained with the mutant proteins were comparable to that of the wild-type P protein. However, the replication activities of the mutant proteins P226/227 (Fig. 2, lane 7) and P226/227/233 (lane 10) were significantly less than that of the wild-type P protein. In three to five independent experiments, the average replication activity of P226/227 and P226/227/233 mutants was found to be 8 and 13%, respectively (Table 1; also see Fig. 5) of that of the wild-type protein.
FIG. 2.
Replication of a VSV antigenomic minireplicon with P mutants. BHK-21 cells in 60-mm-diameter plates were infected with vTF7-3 and transfected with plasmids encoding the minigenomic RNA and the N, L, and wild-type or mutant P proteins. At 16 h posttransfection, the cells were labeled with [3H]uridine and [3H]adenosine in the presence of actinomycin D for 6 h. The replicated RNAs were recovered by immunoprecipitation and analyzed by electrophoresis. The arrow indicates the negative-sense replication product. The asterisk shows the transcription product which is sometimes nonspecifically immunoprecipitated by anti-VSV antibodies. G and N at left represent the G mRNA and N mRNA of VSV isolated from infected cells labeled with [3H]uridine.
TABLE 1.
Relative levels of replication and transcription by various mutant P proteinsa
| P Protein | Replicationb | Relative replication (%) | Transcriptionb | Relative transcription (%) | Normalized transcription (%)c |
|---|---|---|---|---|---|
| Pwt | 149,535 | 100 | 273,961 | 100 | 100 |
| P226 | 148,183 | 99 | 265,748 | 97 | 98 |
| P227 | 151,218 | 101 | 268,873 | 98 | 97 |
| P233 | 152,562 | 102 | 257,498 | 94 | 92 |
| P226/227 | 12,177 | 8 | 21,449 | 7.8 | 97 |
| P226/233 | 146,783 | 98 | 267,902 | 98 | 100 |
| P227/233 | 142,374 | 95 | 265,835 | 97 | 102 |
| P226/227/233 | 19,132 | 13 | 33,589 | 12.3 | 95 |
Relative levels of replication and transcription were calculated with wild-type levels as 100.
Average values from densitometric scanning of fluorograms of four independent experiments. Note that replication and transcription gels were exposed for different lengths of time.
Normalized levels of transcription were calculated as described in Materials and Methods.
FIG. 5.
Relative levels of replication and normalized levels of transcription supported by various mutant P proteins. The data were obtained by densitometric scanning of fluorograms as described in Materials and Methods. The histograms represent data from four to five separate experiments, including those shown in Fig. 2, 4, and 6. The average and range of each value are shown.
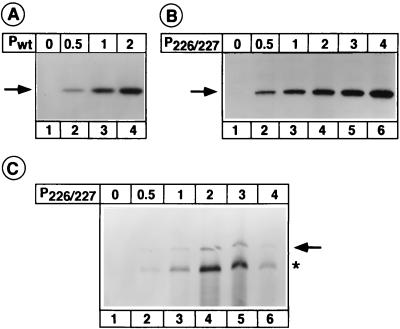
We also examined whether it is possible to rescue higher levels of replication with increasing or decreasing amounts of P226/227 in transfected cells. Initially, we determined the amounts of the Pwt and P226/227 proteins synthesized in cells transfected with various amounts of the corresponding plasmids. We found that the wild-type and mutant proteins were synthesized in equivalent amounts, and a linear relationship was found to exist between the amounts of the expressed proteins and the amounts of the corresponding plasmid DNA used in transfection (Fig. 3A and 3B). When the levels of replication with various amounts of P226/227 were examined, it was found that the levels of replication increased as the amount of P226/227 plasmid was increased and reached a maximum with 2 μg of the plasmid (Fig. 3C, lane 4). However, this level represented only 8% of that obtained with 2 μg of the plasmid encoding the wild-type protein (Fig. 2 and Table 1). The results (Fig. 3C) indicated that higher levels of replication could not be rescued with altered amounts of P226/227 protein. These results suggest that substitution of alanine for serine residues at 226 and 227 renders the protein significantly less active in replication and that serine residue at position 233 may not have a role in the replication activity of the protein. Since the serine residues at both 226 and 227 have been shown to be phosphorylated (12), the results further suggest that phosphorylation of the serine residue at either 226 or 227 is sufficient to achieve optimal replication activity of the protein.
FIG. 3.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of immunoprecipitated wild-type (A) and P226/227 (B) proteins expressed in cells transfected with various amounts (indicated in micrograms above each lane) of the plasmids. The expressed proteins are shown by the arrows in these panels. (C) Replication of VSV minireplicon RNA in the presence of various amounts (indicated in micrograms above each lane) of transfected P226/227 plasmid. Analysis of replication was performed as described in the legend to Fig. 2. The arrow indicates the negative-sense replication product. The asterisk shows the transcription product which is sometimes nonspecifically immunoprecipitated by anti-VSV antibodies.
Taken together, these results suggest that at least one of the serine residues at positions 226 and 227 must be phosphorylated to obtain optimal replication activity of the P protein. In addition, the results indicate that Ser-233 does not play any role in the replication function of the P protein, consistent with the observation that this residue is not phosphorylated (12).
Transcription activity of the mutant P proteins remains unaffected.
We next examined the ability of these mutant proteins to support transcription in vivo. BHK-21 cells infected with vTF7-3 were transfected with plasmids encoding 9BN minireplicon RNA and N, L, and wild-type or mutant P proteins. The transfected cells were then labeled with [3H]uridine and [3H]adenosine in the presence of actinomycin D. Total labeled RNAs isolated from these cells were analyzed by electrophoresis. Synthesis of NΔL mRNA (Fig. 4) was used as a measure of the transcription activity of the mutant P proteins. The results (Fig. 4) show that P226, P227, P233, P226/233, and P227/233 (lanes 4 to 6, 8, and 9) are almost as active as the wild-type P protein (lane 3) in transcription. However, the amount of NΔL mRNA synthesized by the P226/227 (lane 7) and P226/227/233 (lane 10) mutants was significantly reduced. A quantitative determination of the levels of transcription suggests that the P226/227 and P226/227/233 mutants were 8 and 12%, respectively, as active as the wild-type protein (Table 1).
FIG. 4.
Transcription activity of P mutants. BHK-21 cells in 35-mm-diameter six-well plates were infected with vTF7-3 and then transfected with 1.5 μg of pN, 0.5 μg of pL, 2.5 μg of p9BN, and 1.0 μg of pET-P plasmids coding for the wild-type or mutant P proteins. The transfected cells were labeled with [3H]uridine and [3H]adenosine at 16 h posttransfection for 6 h in the presence of actinomycin D. Total RNA from the cytoplasmic extracts was recovered and analyzed by electrophoresis as described in Materials and Methods. G and N represent the corresponding VSV mRNAs isolated from infected cells. The asterisk shows the NΔL RNA transcription product.
Since the genomic-sense transcription template in our experiment is generated through replication of an antigenomic-sense minireplicon synthesized from the transfected plasmid, the low level of transcription by P226/227 and P226/227/233 mutants appears to be due to low levels of transcription template generated by these mutants (Fig. 2, lanes 7 and 10, and Table 1). To obtain a more accurate estimation of the relative levels of transcription activity of these mutants, we normalized the levels of transcription relative to the levels of replication as described in Materials and Methods. In a separate experiment, we first established that the amount of transcription, as measured by the synthesis of NΔL mRNA, directly correlated with the amount of genomic-sense transcription template in transfected cells. In this experiment, vTF7-3-infected cells were transfected with varying amounts of plasmids encoding the minireplicon RNA along with fixed amounts of plasmids encoding the N, L, and wild-type P proteins. The cells were metabolically labeled with [3H]uridine and [3H]adenosine in the presence of actinomycin D. Replication products (RNAs recovered from immunoprecipitated nucleocapsids) and transcription products were analyzed by electrophoresis and fluorography and quantitated by densitometric scanning of the fluorograms as described in Materials and Methods. The results (not shown) demonstrated a linear relationship between the genomic-sense replication product (or transcription template) and the transcription product. When the normalized levels of transcription for various mutant P proteins were calculated, it was found that all the mutant proteins supported levels of transcription comparable to that of the wild-type P protein (Fig. 5). These results were consistently obtained in four to five independent experiments. We conclude from these studies that phosphorylation of serine residues at positions 226 and/or 227 in domain II of the P protein do not play a role in the transcription functions of the protein.
Substitution of glutamic acid at the phosphate acceptor sites does not rescue optimal replication functions of P protein.
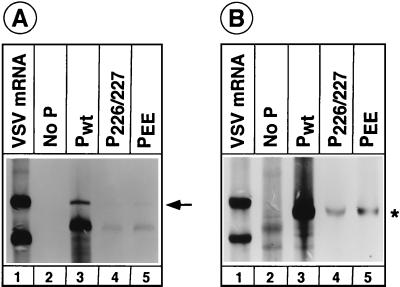
One of the consequences of phosphorylation of amino acid residues in a protein is the introduction of negative charges. We wondered whether phosphorylation per se of the Ser-226 and Ser-227 residues of P protein is essential for optimal replication functions of the protein or whether substitution of negatively charged residues in place of these serine residues can generate replicationally active P protein. To address this possibility, we generated a mutant P protein (PEE) in which serine residues at positions 226 and 227 were replaced with glutamic acid residues. The ability of the mutant protein to support replication of the VSV minigenome was analyzed. The results (Fig. 6A) show that the PEE mutant (lane 5) is slightly more active in replication than the analine-substituted mutant protein (lane 4). However, the level of replication supported by PEE is only 12 to 14% of that of the wild-type protein (lane 3). When the transcription activity of these mutant proteins was measured, PEE consistently produced slightly higher levels of NΔL mRNA (Fig. 6B, lane 5) than P226/227 (lane 4). This is most likely due to higher levels of transcription templates generated by the PEE mutant than the P226/227 mutant. When the level of transcription was normalized to the amount of transcription template, PEE was found to be about 90 to 95% as active as the wild-type P protein (Fig. 5).
FIG. 6.
Replication (A) and transcription (B) supported by PEE. These experiments were performed as described in the legend to Fig. 2 and 4. The arrow in panel A and the asterisk in panel B show the replication and transcription products, respectively.
These results (Fig. 6) suggest that substitution of negatively charged (glutamic acid) residues at the phosphate acceptor sites does not rescue the optimal replication activity of the P protein. This indicates that phosphorylation of these residues rather than negative charges in this region is important for the function of P protein in replication. The observation that PEE was almost as active as P226/227 or wild-type P in transcription further confirms that phosphorylation of these residues does not play any role in P protein function in transcription.
P226/227 mutant protein acts as an inhibitor of wild-type P protein in replication.
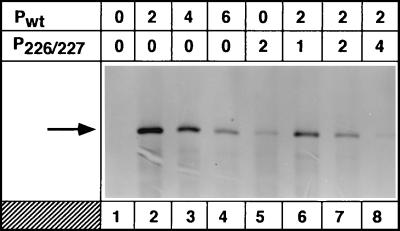
We next wanted to determine whether the P226/227 mutant, which is defective in supporting optimal levels of replication, when coexpressed along with the wild-type protein, can interfere with the normal functioning of the wild-type protein in replication. To examine this, replication of the VSV minigenome in transfected cells coexpressing various amounts of Pwt and P226/227 proteins was analyzed. The results from such an experiment (Fig. 7) show that coexpression of a fixed amount of Pwt and varying amounts of P226/227 (lanes 6 to 8) led to significant inhibition of minigenome replication. It must be noted that increasing the amounts of Pwt or P226/227 led to inhibition of minigenome replication (data not shown), most likely due to imbalance in the molar amounts of the viral proteins required for replication (46). However, the inhibitory activity of P226/227 is clearly evident when one compares levels of replication supported by a given amount of Pwt with that supported by the same amount of both Pwt and P226/227. The amount of replication with 4 μg of total plasmids (2 μg each of Pwt and P226/227) (Fig. 7, lane 7) was approximately one-third of that obtained with 4 μg of Pwt plasmid alone (lane 3). Likewise, the amount of replication with 6 μg of total plasmids (2 μg of Pwt and 4 μg of P226/227) (Fig. 7, lane 8) was approximately one-fourth of that obtained with 6 μg of Pwt plasmid alone (lane 4). In addition, when 2 μg of Pwt plasmid was used along with increasing amounts of P226/227, significant inhibition of replication was observed (compare the levels of replication in Fig. 7, lane 2, with those in lanes 6 to 8). These results, therefore, indicate that P226/227 protein inhibits the function of Pwt protein in replication in vivo.
FIG. 7.
Inhibitory activity of P226/227 in replication when coexpressed with wild-type P. The value above each lane indicates the amount (in micrograms) of each P plasmid used in the transfection. The experiment was performed as described in the legend to Fig. 2. The arrow shows the replication product.
DISCUSSION
The phosphoprotein (P) of VSV plays a central role in the transcription and replication functions of the RNA-dependent RNA polymerase of the virion. Although the P protein does not carry any enzymatic activities of the viral polymerase, its interaction with the large protein, L, generates a specific P-L complex(es) which is absolutely essential for the polymerase activity. The P protein interacts with soluble N protein to form N-P complexes that are required for encapsidation of nascent RNA chains during replication. P protein is known to exist as oligomers, and oligomerization of P protein appears to be required for interaction with the L protein to generate transcription activity of the P-L complex. In addition, the P protein exists in multiple phosphorylated forms within the virion core as well as in infected cells, and phosphorylation of residues within specific domains of the P protein regulates its RNA-synthetic activities. Because of the multifunctional nature of the P protein, a great deal of investigation has been directed towards understanding its structure-function relationships.
We and others have shown recently that phosphorylation of specific residues within domain I of P protein of Indiana serotype VSV is not essential for the in vivo replication function of the protein (45, 52). However, in vivo transcription activity of the protein required phosphorylation of the residues in this domain (45), although low levels of in vitro transcription activity were obtained with domain I phosphorylation-defective P protein (52). In the present work, we have examined the role of phosphorylation of domain II residues in in vivo transcription and replication activities of the protein. Our results suggest that phosphorylation of a specific serine residue(s) (Ser-226 or Ser-227) in this domain is required for optimal replication activity of the protein. Interestingly, transcription activity of the protein was not adversely affected by the phosphorylation status of the domain II residues. The major conclusion that can be drawn from our studies is that phosphorylation at two different domains in the P protein of VSV regulates its activity in transcription and replication. However, it must be pointed out that the requirement for phosphorylation of domain I residues in transcription could be overcome by substitution of negatively charged residues for serine residues (24, 45) whereas appreciable levels of replication could not be rescued by substitution of glutamic acid residues for serine residues in domain II (Fig. 6). These results may suggest a unique role of phosphorylation in the function of the P protein in replication but not in transcription.
Recently, it was demonstrated (23) that serine residues at positions 227 and 233 within domain II of PI are the major targets of phosphorylation by LAK. However, in a separate study, it was shown by mutational and biochemical analyses that serine residues at 226 and 227 are the sites of phosphorylation by LAK (12). The reason(s) for this discrepancy is not clear at this time but the results of our functional analysis of the mutant P proteins, in which each of these three sites was altered to alanine individually or in various combinations, indicate that Ser-233 does not appear to play any role in transcription or replication functions of the protein. Our results are consistent with those reported earlier from in vitro-transcription studies, suggesting that phosphorylation of domain II residues by LAK has no role in the transcription activity of PI (23, 40). These results appear to conflict with those obtained previously, which showed that phosphorylation of Ser-236 and Ser-242 in domain II of PNJ by LAK is necessary for efficient transcription in vitro (11). It is possible that the activity of the mutant proteins differs under in vivo and in vitro conditions, as has been demonstrated recently with domain I phosphorylation-defective P mutant proteins (45, 52). Alternatively, it is possible that the functional requirement for phosphorylation of domain II residues of PNJ may be different from that of PI. This may be borne out by the observation that phosphorylation of domain I residues of PNJ is a prerequisite for phosphorylation of domain II residues (4) whereas domain II of PI can be phosphorylated without prior phosphorylation of domain I residues (12).
The observation that domain II phosphorylation-defective P protein is defective in replication but not in transcription indicates that phosphorylation of domain II residues may be important for formation of replicase complexes. We recently suggested that the replicase complex of VSV may exist as a tripartite complex consisting of the N, P, and L proteins (16) and that other cellular factors may also be present in the replicase complex (26). It is possible that phosphorylation of domain II residues results in a conformation that facilitates specific interaction with the L and/or N protein for assembly of the replicase complex. The P protein, lacking phosphates in domain II residues, may interact poorly with the other viral proteins so that low levels of replication complexes are formed or that the replication complexes are less active. It is obviously important to examine the interaction of the mutant P proteins with the N and L proteins to understand how phosphorylation in domain II of P might influence the assembly and activity of the replicase complex. It must be noted here that the mutant protein (P227/233) multimerized and interacted well with the L protein (23). However, the data should be considered in the context that this mutant protein was phosphorylated at Ser-226 (12).
Several recent studies have provided a correlation between the degree of phosphorylation of the P protein and its activity in transcription and replication. Using okadaic acid, a serine/threonine phosphatase inhibitor, it was shown that the hyperphosphorylated form of the P protein (presumably having phosphates in both domain I and II residues) inhibited replication (10). Conversely, dephosphorylation of P protein by bacterial alkaline phosphatase (BAP) led to increased genome replication, with a concomitant decrease in transcription (49). The amount of BAP needed to stimulate genome replication and inhibit transcription was substantially below that needed to obtain a noticeable increase in reactivity to a monoclonal antibody (6D11) directed against an epitope that maps to residues 177 to 227 of the P protein (49, 55), corresponding to part of domain II. Although these studies did not map the sites from which phosphate moieties were removed by BAP treatment, they indicated that the epitope bound by 6D11 antibody is changed by dephosphorylation (49). Since treatment of infected-cell P protein with BAP increased genome replication and inhibited transcription without a noticeable increase in 6D11 antibody reactivity, these results indicate that BAP may have removed phosphates only from domain I residues. These observations, together with our previous results (45) and the data presented here, suggest that the P1 protein, with phosphorylation of domain I residues, may function as part of a transcription complex whereas the P2 protein, with phosphorylation of residues in domain II, may function exclusively as part of a replication complex. Another subset of P proteins (hyperphosphorylated P2 protein with phosphorylation of both domain I and II residues) may be involved in maturation of nucleocapsids for packaging and thereby indirectly inhibit replication (10). Further studies focusing on isolation of complexes specific for transcription and replication and examination of the phosphorylation status of P protein in these complexes may provide more direct evidence as to the role of phosphorylation of residues in various domains in transcription and replication.
The inhibitory activity of P226/227 in replication by wild-type P protein (Fig. 7) indicates that the domain II phosphorylation-defective P protein perhaps interacts with the other components of the replication machinery to generate inactive complexes or competes with the wild-type P protein for replication machinery. Studies to examine specific interactions of the mutant protein with the N, L, or N-RNA template may provide a better understanding of the inhibitory effect of the mutant protein in replication.
The phosphate acceptor sites (Ser-226 and Ser-227) within domain II are located adjacent to each other, and our results (Fig. 2) show that phosphorylation of either of these two residues is sufficient to obtain optimal replication activity of the protein. Furthermore, previous data indicate that phosphorylations at these sites are independent of each other (12). The question then is why two functionally redundant phosphorylation sites are maintained in the protein. It is possible that in a given P molecule, only one of these two sites is phosphorylated in vivo and the other site may remain unphosphorylated because of steric hindrance. Alternatively, the presence of two functionally redundant phosphorylation sites next to each other may protect the protein from becoming inactivated in replication by phosphatases, which must remove both of the phosphate moieties at the same time.
In summary, the results reported in this article suggest that phosphorylation of either the Ser-226 or Ser-227 residue in domain II is necessary for optimal replication activity, but not for transcription activity, of the P protein. Substitution of negatively charged residues for these residues cannot rescue replication. From these results and those reported previously (10, 23, 24, 45, 49, 52), we conclude that phosphorylation within different domains of the P protein controls its activity in transcription and replication. Studies of infectious molecular clones of VSV containing various phosphorylation-defective mutants of P protein may provide additional information as to the role of this modification in the RNA-synthetic processes of VSV. In several other negative-strand RNA viruses, the P protein has been found to be phosphorylated (5, 7, 14, 17, 19, 42, 43, 54), and the functional significance of phosphorylation in transcription has also been documented (5, 7, 17, 42). Whether transcriptase and replicase activities are regulated by differential phosphorylation of the P proteins of these viruses remains to be examined.
ACKNOWLEDGMENTS
We thank Michelle Perez for preparation of the manuscript and Jin-Lian Chen for generating the P mutants. We also thank Merck and Co., Inc., Rahway, N.J., for a gift of actinomycin D.
This investigation was supported by Public Health Service Grants AI 34956 (to A.K.P.) and AI 26585 (to A.K.B.) from the National Institutes of Health. L.N.H. was supported by a predoctoral fellowship from the training grant T32 EY07129 from the National Eye Institute.
REFERENCES
- 1.Banerjee A K, Barik S. Gene expression of vesicular stomatitis virus genome RNA. Virology. 1992;188:417–428. doi: 10.1016/0042-6822(92)90495-b. [DOI] [PubMed] [Google Scholar]
- 2.Barik S, Banerjee A K. Cloning and expression of the vesicular stomatitis virus phosphoprotein gene in Escherichia coli: analysis of phosphorylation status versus transcriptional activity. J Virol. 1991;65:1719–1726. doi: 10.1128/jvi.65.4.1719-1726.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barik S, Banerjee A K. Phosphorylation by cellular casein kinase II is essential for transcriptional activity of vesicular stomatitis virus phosphoprotein. Proc Natl Acad Sci USA. 1992;89:6570–6574. doi: 10.1073/pnas.89.14.6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barik S, Banerjee A K. Sequential phosphorylation of the phosphoprotein of vesicular stomatitis virus by cellular and viral protein kinases is essential for transcriptional activation. J Virol. 1992;66:1109–1118. doi: 10.1128/jvi.66.2.1109-1118.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barik S, McLean T, Dupuy L. Phosphorylation of Ser232 directly regulates the transcriptional activity of the P protein of human respiratory syncytial virus: phosphorylation of Ser237 may play an accessory role. Virology. 1995;213:405–412. doi: 10.1006/viro.1995.0013. [DOI] [PubMed] [Google Scholar]
- 6.Bell J C, Prevec L. Phosphorylation sites on phosphoprotein NS of vesicular stomatitis virus. J Virol. 1985;54:697–702. doi: 10.1128/jvi.54.3.697-702.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byrappa S, Pan Y-B, Gupta K C. Sendai virus P protein is constitutively phosphorylated at serine 249: high phosphorylation potential of the P protein. Virology. 1996;216:228–234. doi: 10.1006/viro.1996.0052. [DOI] [PubMed] [Google Scholar]
- 8.Campbell M J. Lipofection reagents prepared by a simple ethanol injection technique. BioTechniques. 1995;18:1027–1032. [PubMed] [Google Scholar]
- 9.Canter D M, Perrault J. Stabilization of vesicular stomatitis virus L polymerase protein by P protein binding: a small deletion in the c-terminal domain of L abrogates binding. Virology. 1996;219:376–386. doi: 10.1006/viro.1996.0263. [DOI] [PubMed] [Google Scholar]
- 10.Chang T L, Reiss C S, Huang A S. Inhibition of vesicular stomatitis virus RNA synthesis by protein hyperphosphorylation. J Virol. 1994;68:4980–4987. doi: 10.1128/jvi.68.8.4980-4987.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chattopadhyay D, Banerjee A K. Phosphorylation within a specific domain of the phosphoprotein of vesicular stomatitis virus regulates transcription in vitro. Cell. 1987;49:407–414. doi: 10.1016/0092-8674(87)90293-5. [DOI] [PubMed] [Google Scholar]
- 12.Chen J-L, Das T, Banerjee A K. Phosphorylated states of vesicular stomatitis virus P protein in vitro and in vivo. Virology. 1997;228:200–212. doi: 10.1006/viro.1996.8401. [DOI] [PubMed] [Google Scholar]
- 13.Clinton G M, Huang A S. Distribution of phosphoserine, phosphothreonine, and phosphotyrosine in proteins of vesicular stomatitis virus. Virology. 1981;108:510–514. doi: 10.1016/0042-6822(81)90459-1. [DOI] [PubMed] [Google Scholar]
- 14.Curran J, Pelet T, Kolakofsky D. An acidic activation-like domain of the Sendai virus P protein is required for RNA synthesis and encapsidation. Virology. 1994;202:875–884. doi: 10.1006/viro.1994.1409. [DOI] [PubMed] [Google Scholar]
- 15.Das T, Gupta A K, Sims P W, Gelfand C A, Jentoft J E, Banerjee A K. Role of cellular casein kinase II in the function of the phosphoprotein (P) subunit of RNA polymerase of vesicular stomatitis virus. J Biol Chem. 1995;270:24100–24107. doi: 10.1074/jbc.270.41.24100. [DOI] [PubMed] [Google Scholar]
- 16.Das T, Pattnaik A K, Takacs A M, Li T, Hwang L N, Banerjee A K. Basic amino acid residues at the carboxy-terminal eleven amino acid region of the phosphoprotein (P) are required for transcription but not for replication of vesicular stomatitis virus genome RNA. Virology. 1997;238:103–114. doi: 10.1006/viro.1997.8823. [DOI] [PubMed] [Google Scholar]
- 17.Das T, Schuster A, Schneider-Schaulies S, Banerjee A K. Involvement of cellular casein kinase II in the phosphorylation of measles virus P protein: identification of phosphorylation sites. Virology. 1995;211:218–226. doi: 10.1006/viro.1995.1394. [DOI] [PubMed] [Google Scholar]
- 18.De B P, Banerjee A K. Requirements and functions of vesicular stomatitis virus L and NS proteins in the transcription process in vitro. Biochem Biophys Res Commun. 1985;26:40–49. doi: 10.1016/0006-291x(85)90568-6. [DOI] [PubMed] [Google Scholar]
- 19.De B P, Gupta S, Gupta S, Banerjee A K. Cellular protein kinase C isoform ζ regulates human parainfluenza virus type 3 replication. Proc Natl Acad Sci USA. 1995;92:5204–5208. doi: 10.1073/pnas.92.11.5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emerson S U, Yu Y H. Both NS and L proteins are required for in vitro RNA synthesis by vesicular stomatitis virus. J Virol. 1975;15:1348–1356. doi: 10.1128/jvi.15.6.1348-1356.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emerson S U, Schubert M. Location of the binding domains for the RNA polymerase L and the ribonucleocapsid template within different halves of the NS phosphoprotein of vesicular stomatitis virus. Proc Natl Acad Sci USA. 1987;84:5655–5659. doi: 10.1073/pnas.84.16.5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao Y, Lenard J. Cooperative binding of multimeric phosphoprotein (P) of vesicular stomatitis virus to polymerase (L) and template. J Virol. 1995;69:7718–7723. doi: 10.1128/jvi.69.12.7718-7723.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao Y, Lenard J. Multimerization and transcriptional activation of the phosphoprotein (P) of vesicular stomatitis virus by casein kinase II. EMBO J. 1995;14:1240–1247. doi: 10.1002/j.1460-2075.1995.tb07107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gill D S, Chattopadhyay D, Banerjee A K. Identification of a domain within the phosphoprotein of vesicular stomatitis virus that is essential for transcription in vitro. Proc Natl Acad Sci USA. 1986;83:8873–8877. doi: 10.1073/pnas.83.23.8873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta A K, Banerjee A K. Expression and purification of vesicular stomatitis virus N-P complex from Escherichia coli: role in genome RNA transcription and replication in vitro. J Virol. 1997;71:4264–4271. doi: 10.1128/jvi.71.6.4264-4271.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammond D C, Lesnaw J A. Functional analysis of hypomethylation variants of the New Jersey serotype of vesicular stomatitis virus. Virology. 1987;160:330–335. doi: 10.1016/0042-6822(87)90003-1. [DOI] [PubMed] [Google Scholar]
- 28.Hercyk N, Horikami S M, Moyer S A. The vesicular stomatitis virus L protein possesses the mRNA methyltransferase activities. Virology. 1988;163:222–225. doi: 10.1016/0042-6822(88)90253-x. [DOI] [PubMed] [Google Scholar]
- 29.Horikami S M, Moyer S A. Host range mutants of vesicular stomatitis virus defective in in vitro RNA methylation. Proc Natl Acad Sci USA. 1982;79:7694–7698. doi: 10.1073/pnas.79.24.7694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu C-H, Kingsbury D W. NS phosphoprotein of vesicular stomatitis virus: subspecies separated by electrophoresis and isometric focusing. J Virol. 1982;42:342–345. doi: 10.1128/jvi.42.1.342-345.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsu C H, Kingsbury D W. Constitutively phosphorylated residues in the NS protein of vesicular stomatitis virus. J Biol Chem. 1985;260:8990–8995. [PubMed] [Google Scholar]
- 32.Hunt D M, Smith E G, Buckley D W. Aberrant polyadenylation by a vesicular stomatitis virus mutant is due to an altered L protein. J Virol. 1984;52:515–521. doi: 10.1128/jvi.52.2.515-521.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Isaac C L, Keene J D. RNA polymerase-associated interactions near template promoter sequences of defective interfering particles of vesicular stomatitis virus. J Virol. 1982;43:241–249. doi: 10.1128/jvi.43.1.241-249.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keene J D, Thornton B J, Emerson S U. Sequence-specific contacts between the RNA polymerase of vesicular stomatitis virus and the leader RNA gene. Proc Natl Acad Sci USA. 1981;78:6191–6195. doi: 10.1073/pnas.78.10.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kingsford L, Emerson S U. Transcriptional activities of different phosphorylated species of NS protein purified from vesicular stomatitis virions and cytoplasm of infected cells. J Virol. 1980;33:1097–1105. doi: 10.1128/jvi.33.3.1097-1105.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.La Ferla F M, Peluso R W. The 1:1 N-NS protein complex of vesicular stomatitis virus is essential for efficient genome replication. J Virol. 1989;63:3852–3857. doi: 10.1128/jvi.63.9.3852-3857.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laskey R. The use of intensifying screens or organic scintillators for visualizing radioactive molecules resolved by gel electrophoresis. Methods Enzymol. 1980;65:363–371. doi: 10.1016/s0076-6879(80)65047-2. [DOI] [PubMed] [Google Scholar]
- 38.Lerach H, Diamond D, Wozney J, Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical examination. Biochemistry. 1977;16:4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- 39.Li T, Pattnaik A K. Replication signals in the genome of vesicular stomatitis virus and its defecting interfering particles: identification of a sequence element that enhances DI RNA replication. Virology. 1997;232:248–259. doi: 10.1006/viro.1997.8571. [DOI] [PubMed] [Google Scholar]
- 40.Massey D M, Deans N, Lenard J. Phosphorylation of NS protein by vesicular stomatitis virus nucleocapsids. Lack of effect during RNA synthesis and separation of kinase from L protein. J Virol. 1990;64:3259–3264. doi: 10.1128/jvi.64.7.3259-3264.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masters P S, Banerjee A K. Phosphoprotein of vesicular stomatitis virus: phosphorylated states and transcriptional activities of intracellular and virion forms. Virology. 1986;154:259–270. doi: 10.1016/0042-6822(86)90452-6. [DOI] [PubMed] [Google Scholar]
- 42.Mazumder B, Adhikary G, Barik S. Bacterial expression of human respiratory syncytial virus phosphoprotein P and identification of Ser237 as the site of phosphorylation by cellular casein kinase II. Virology. 1994;205:93–103. doi: 10.1006/viro.1994.1623. [DOI] [PubMed] [Google Scholar]
- 43.Mazumder B, Barik S. Requirement of casein kinase II-mediated phosphorylation for the transcriptional activity of human respiratory syncytial virus phosphoprotein P: transdominant negative phenotype of phosphorylation-defective P mutants. Virology. 1994;205:104–111. doi: 10.1006/viro.1994.1624. [DOI] [PubMed] [Google Scholar]
- 44.Pattnaik A K, Ball L A, LeGrone A, Wertz G. Infectious defective interfering particles of VSV from transcripts of a cDNA clone. Cell. 1992;69:1011–1020. doi: 10.1016/0092-8674(92)90619-n. [DOI] [PubMed] [Google Scholar]
- 45.Pattnaik A K, Hwang L, Li T, Englund N, Mathur M, Das T, Banerjee A K. Phosphorylation within the amino-terminal acidic domain I of the phosphoprotein of vesicular stomatitis virus is required for transcription, but not replication. J Virol. 1997;71:8167–8175. doi: 10.1128/jvi.71.11.8167-8175.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pattnaik A K, Wertz G W. Replication and amplification of defective interfering particle RNAs of vesicular stomatitis virus in cells expressing viral proteins from vectors containing cloned cDNAs. J Virol. 1990;64:2948–2957. doi: 10.1128/jvi.64.6.2948-2957.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paul P R, Chattopadhyay D, Banerjee A K. The functional domains of phosphoprotein (NS) of vesicular stomatitis virus (Indiana serotype) Virology. 1988;166:350–357. doi: 10.1016/0042-6822(88)90505-3. [DOI] [PubMed] [Google Scholar]
- 48.Peluso R W, Moyer S A. Viral proteins required for the in vitro replication of vesicular stomatitis virus defective interfering particle genome RNA. Virology. 1988;162:369–376. doi: 10.1016/0042-6822(88)90477-1. [DOI] [PubMed] [Google Scholar]
- 49.Richardson J C, Peluso R W. Inhibition of VSV genome RNA replication by monoclonal antibodies specific for the viral P protein. Virology. 1996;216:26–34. doi: 10.1006/viro.1996.0031. [DOI] [PubMed] [Google Scholar]
- 50.Schubert M, Harmison G G, Richardson C D, Meier E. Expression of a cDNA encoding a functional 21-kilodalton vesicular stomatitis RNA polymerase. Proc Natl Acad Sci USA. 1985;82:7984–7988. doi: 10.1073/pnas.82.23.7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sleat D, Banerjee A K. Transcriptional activity and mutational analysis of recombinant vesicular stomatitis virus RNA polymerase. J Virol. 1993;67:1334–1339. doi: 10.1128/jvi.67.3.1334-1339.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spadafora D, Canter D M, Jackson R L, Perrault J. Constitutive phosphorylation of vesicular stomatitis virus P protein modulates polymerase complex formation but is not essential for transcription or replication. J Virol. 1996;70:4538–4548. doi: 10.1128/jvi.70.7.4538-4548.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takacs A M, Barik S, Das T, Banerjee A K. Phosphorylation of specific serine residues with the acidic domain of the phosphoprotein of vesicular stomatitis virus regulates transcription in vitro. J Virol. 1992;66:5842–5848. doi: 10.1128/jvi.66.10.5842-5848.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Villanueva N, Navarro J, Mendez E, Garcia-Albert I. Identification of a protein kinase involved in the phosphorylation of the C-terminal region of human respiratory syncytial virus P protein. J Gen Virol. 1994;75:555–562. doi: 10.1099/0022-1317-75-3-555. [DOI] [PubMed] [Google Scholar]
- 55.Williams P M, Williamson K A, Emerson S U, Schubert M. Deletion mapping analyses indicate that epitopes for monoclonal antibodies to the NS phosphoprotein of VSV are linear and clustered. Virology. 1988;164:176–181. doi: 10.1016/0042-6822(88)90634-4. [DOI] [PubMed] [Google Scholar]