Abstract
It is thought that complete cleavage of retroviral envelope protein into mature surface protein (SU) and transmembrane protein (TM) is critical for its assembly into virions and the formation of infectious virus particles. Here we report the identification of highly infectious, cleavage-deficient envelope mutant proteins. Substitution of aspartate for lysine 104, arginines 124 and 126, or arginines 223 and 225 strongly suppressed cleavage of the envelope precursor and yet allowed efficient incorporation of precursor molecules as the predominant species in virions that were almost as infectious as the wild-type virus. These results indicate that cleavage of the envelope precursor into mature SU and TM is not necessary for assembly into virions. Moreover, they call into question how many mature envelope protein subunits are required to complete virus entry, suggesting that a very few molecules suffice. The failure of host cell proteases to cleave these mutant proteins, whose substitutions are distal to the actual site of cleavage, suggests that the envelope precursor is misfolded, sequestering the cleavage site. In agreement with this, all cleavage mutant proteins exhibited significant losses of receptor binding, suggesting that these residues play roles in proper envelope protein folding. We also identified a charged residue, arginine 102, whose substitution suppressed envelope cleavage and allowed precursor incorporation but resulted in virions that were virtually noninfectious and that exhibited the greatest reduction in receptor binding. Placement of these cleavage mutations into envelope proteins of targeted retroviral vectors for human gene therapy may prevent loss of the modified surface proteins from virions, improving their infectivity and storage hardiness.
Ecotropic murine leukemia viruses (MLV) are simple type C retroviruses carrying three genes, gag, pol, and env. The first and last genes encode the structural proteins for the assembly of infectious progeny virus. The protein products of the gag gene make up the core of the virus particle, while the env gene encodes the envelope proteins found in the virus membrane. Synthesized as a polyprotein, the envelope proteins are targeted for translation in the endoplasmic reticulum via a cleaved amino-terminal signal peptide. Following glycosylation in the endoplasmic reticulum, the precursor molecules form trimers. A second internal cleavage by an unidentified host cell protease processes each molecule of the trimer into mature surface glycoprotein (SU) and transmembrane anchor protein (TM), which remain covalently associated via a disulfide bridge. Heterotrimers of mature SU and TM are presented on the plasma membranes of host cells, where they incorporate into virus particles budding from the cell surface. Virions that do not contain heterotrimeric envelope proteins in their membranes are not infectious (6). When ecotropic virus particles bud from the producer cell surface, there is a third cleavage of envelope protein in which carboxy-terminal residues, called the R peptide, are removed to yield a 12-kDa TM that is more fusogenic than the 15-kDa TM (17, 35, 36).
Cleavage of retroviral envelope precursors into SU and TM is thought to be essential for assembly of infectious retroviral particles because mutations that abolish cleavage result in noninfectious particles. Frequently, the failure to cleave the envelope precursor prevented its transport to the cell surface, presumably because of gross misfolding, precluding its incorporation into virions (2, 3, 40). Deletion of a 20-amino-acid segment that included the cleavage site of ecotropic Moloney MLV (MoMLV) envelope protein resulted in the synthesis of uncleaved protein and the production of “bald” particles lacking envelope protein (16). Subtle changes in the cleavage site of ecotropic AKV MLV reduced the amount of mature SU present in virions, resulting in incorporation of very small amounts of an extraordinarily large envelope species (100 kDa) and reducing the infectivity of virions on NIH 3T3 cells by 100-fold but rendering them noninfectious on XC cells (13). Mutations that altered the cleavage sites of human immunodeficiency virus, mouse mammary tumor virus, and Rous sarcoma virus envelope proteins did not prevent cell surface presentation of the uncleaved precursors but resulted in complete loss of infection, even though virions of one of the Rous sarcoma virus mutants contained appreciable amounts of precursor protein (8, 9, 15, 18). One exception has been reported. Uncleaved envelope proteins were reported on virions from a spontaneous, infectious mutant of ecotropic Rauscher MLV, although the exact mutation(s) responsible for this phenomenon was not identified (27).
In addition, cleavage of the precursor protein is thought to be essential for assembly of infectious particles because it frees a stretch of hydrophobic residues at the amino terminus of TM (6, 32). Mutations in this sequence result in the loss of infection and a loss of the ability of a retrovirus to induce fusion of two adjacent cells bearing virus receptors, suggesting that these residues comprise a fusion peptide (4, 12, 22, 43). Cleavage just upstream of such an internal fusion peptide in the envelope proteins of other viruses, such as influenza virus, is a prerequisite for their role in penetration of the host cell (42). By analogy to the model for influenza virus entry, binding of the envelope protein to the virus receptor triggers a change in the conformation of the fusion peptide, swinging it approximately 180 degrees to intrude into the host cell membrane for formation of a pore through which the core of the virus is passed (5). Such a dramatic change in the structure of the envelope protein would be difficult, if at all possible, if the fusion peptide remained covalently secured to the carboxy-terminal portion of the surface protein as it is in the envelope precursor.
The association of SU and TM after cleavage is tenuous. The disulfide bridge connecting them is labile and is readily disrupted by the mechanical stress, such as freezing and thawing (21, 26), that occurs during storage of retrovirus stocks. Disruption of this covalent linkage causes SU to dissociate or shed from the surfaces of virus particles, leaving them noninfectious. The association of SU and TM is also destabilized by insertion of foreign sequences into the envelope protein, such as by the fusion of a binding domain from a heterologous ligand for the production of a hybrid envelope protein that directs virus attachment and infection to specific cell types for targeted gene delivery (7).
Infection begins with binding of SU protein to the virus receptor on the plasma membrane of a host cell. At least one region consisting of approximately the first 200 residues in SU is essential for recognition of the ecotropic retrovirus receptor (20). Only a few critical residues within this domain have been identified. Aspartate 84 and arginines at positions 83 and 95 appear to be required for receptor binding (2, 28). In addition, three pairs of positively charged arginine and lysine residues at positions 102 and 104, 124 and 126, and 223 and 225 appear to play an as yet undetermined role in virus infection (39).
The virus attachment site lies within the third extracellular loop of the receptor, a polytopic membrane protein that normally functions as the principal transporter of cationic amino acids in the host cell (1a, 24). We previously showed that within this binding site tyrosine 235 provides a critical hydrophobic side chain and that glutamate 237 provides a critical side chain carboxyl group (29). Moreover, we noted that a similar motif of a hydrophobic amino acid and a nearby charged residue has been found in the virus binding sites of other retrovirus receptors (29).
Here we report the use of site-directed mutagenesis to identify important functional residues on SU, in an attempt to locate those that bind the critical glutamate residue on the receptor. Unexpectedly, we identified a number of novel envelope cleavage mutant proteins. Remarkably, suppression of cleavage in four of the mutant proteins did not prevent efficient incorporation of the envelope precursor into virus particles. Moreover, virions containing the precursor molecules were highly infectious. We also identified a residue whose replacement results in a cleavage mutant protein that is virtually noninfectious. These virions contained the envelope precursor but no detectable mature SU. Particles containing each of the envelope cleavage mutant proteins showed marked losses of receptor binding. These results indicate that the residues altered in the cleavage mutant proteins are involved in correct folding of the cleavage recognition and receptor binding sites. They also indicate that cleavage of the envelope precursor into mature SU and TM is not necessary for assembly into virus particles. Furthermore, virions coated with precursor protein can be highly infectious, suggesting that only a few molecules of free fusion peptide are required to complete virus fusion. Inclusion of these cleavage mutations into the retroviral envelope proteins on targeted retroviral vectors should prevent loss of the modified surface proteins from virions, improving their infectivity and storage hardiness.
MATERIALS AND METHODS
Cell lines and viruses.
All cell lines were maintained at 37°C in 5% CO2. Mouse NIH 3T3 fibroblasts and nonpermissive human 293 fetal kidney cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 8% donor calf serum. The 293 cell-derived stable transfectant expressing the receptor cDNA has been described elsewhere (29). These cells and virus-producer H1-BAG cells were maintained in Dulbecco’s modified Eagle’s medium with 8% fetal bovine serum and 250 μg of G418 (Sigma) per ml.
Plasmids bearing virus genomes.
Initially, we constructed a master plasmid, pcDNA-MoMLV, bearing a virus genome derived from ecotropic MoMLV that provides wild-type gag and pol genes, from which proteins for the virion core were made, and an env gene, from which the envelope proteins for assembly into the membrane of the virions were synthesized. The 8-kbp fragment from the BssHII restriction site to the env termination codon containing the MoMLV proviral genome that lacks the packaging signal was derived from plasmid pEM-5 (gift of V. Garcia). The env gene termination codon is followed by an artificially engineered BamHI site. The genome was inserted between the HindIII and BamHI sites of the eukaryotic expression vector pcDNA3 (Invitrogen), which resulted in transcription of the viral genome being placed under the control of the cytomegalovirus promoter and a downstream polyadenylation site being provided by the bovine growth hormone poly(A). This genome is not incorporated into virions because it lacks the encapsidation sequence, Ψ. It also lacks the U3 region of the 5′ long terminal repeat and the entire 3′ long terminal repeat. Envelope gene fragments containing specific mutations were subcloned into the HpaI sites in pcDNA-MoMLV for production of virions.
Virus production.
The amphotropic packaging cell line PA317 (33) was transiently transfected by calcium phosphate precipitation with the pBAG plasmid (gift of C. Cepko) (41), which bears a replication-defective but packageable MoMLV genome in which the structural genes were replaced by the Escherichia coli lacZ gene and the neomycin resistance gene (neo). The virus-containing supernatant was harvested and used to infect human 293 cells, into which the virus transduced the lacZ and neo genes. Infected 293 cells were selected in the medium containing 1 mg of G418 per ml. Twenty-four drug-resistant colonies were propagated and analyzed for β-galactosidase expression by staining with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). The five clones developing the most intense staining in the shortest period, indicative of high levels of transcription of the virus genome, were selected as virus producers. Expression of packageable viral RNA was confirmed by Northern blot analysis (data not shown). One of these cell clones, H1-BAG, was used in all experiments reported here.
To produce virus particles, we transiently transfected the H1-BAG cells with pcDNA-MoMLV DNA containing wild-type or mutated env genes using calcium phosphate precipitation (38). Virus-containing supernatant was freed of producer cells by low-speed centrifugation followed by filtration through a 0.45-μm-pore-size filter. An aliquot of 3 ml was removed, stored at −80°C, and then used for virus titration. Viruses from the remaining 7 ml were immediately pelleted as described below for immunoblotting. For each virus binding experiment, the entire virus preparations were frozen and concentrated as described below. Virions produced in this manner transduce β-galactosidase activity upon infection of cells.
Site-directed mutagenesis of env genes.
Nucleotide substitutions in the env gene were generated by the method described by Kunkel (25). The envelope residues whose codons were mutagenized are indicated by their positions in the MoMLV envelope protein; the alanine residue at the N terminus of mature SU was considered residue 1. For this purpose we subcloned the 1,300-bp HpaI-HpaI restriction fragment from the MoMLV env gene on plasmid pMOV3 (gift of H. Stuhlmann) (19) into the bacteriophage vector M13mp18 with an engineered HpaI site. Site-directed mutagenesis was performed, and the fragment was transferred to pcDNA3-MoMLV. The entire 1,300-bp fragment was sequenced with an fmol sequencing kit (Promega), with the resulting plasmid being used as a template to ensure the absence of unscheduled substitutions and to confirm the presence of desired mutations.
Virus titers.
Endpoint dilution titration of all virus stocks was performed essentially as previously described for ecotropic MLV (29) with modifications as follows. Briefly, 2 × 104 cells were seeded in each well of 24-well culture plates, and quadruplicate wells were exposed to 10-fold serial dilutions of virus stock in medium containing Polybrene (20 μg/ml; Sigma). Forty-eight hours after exposure, cells were fixed and stained with X-Gal for β-galactosidase activity. Titers were calculated for the endpoint dilution. Every titration included exposure of straight virus stocks and a 10−1 dilution into parental human 293 cells lacking an ecotropic receptor. We did not observe entry by any mutant or wild-type virus into the parental 293 cells in any of the titrations, indicating that the entry reported for the human 293 cell line stably expressing the ecotropic receptor did not occur by a non-receptor-mediated pathway.
Western blot analysis.
Virus particles were pelleted from 7 ml of cell-free virus supernatant through 3 ml of 25% sucrose in TEN (10 mM Tris [pH 8.0], 1 mM EDTA, 100 mM NaCl) in a Beckman SW41 rotor (30,000 rpm, 2 h, 4°C). Pellets were taken up in 40 μl of phosphate-buffered saline. Virus producer cells were lysed immediately after virus harvest in 300 μl of RIPA buffer (20 mM Tris [pH 7.0], 1% Triton X-100, 0.05% sodium dodecyl sulfate [SDS], 0.5% Na deoxycholate, 150 mM NaCl, 2.5 mM phenylmethylsulfonyl fluoride) by incubation for 30 min on ice. Cell lysates were obtained after pelleting nuclei by centrifugation in an Eppendorf model 5415C centrifuge for 10 min at 10,000 rpm. Total protein concentrations in cell lysates were determined by the Bradford assay (Bio-Rad). Ten microliters of virus pellets or 100 μg of total protein from cell lysates was diluted 1:1 in 2× gel loading buffer (38), boiled, and subjected to SDS-polyacrylamide gel electrophoresis, and the separated proteins were transferred onto a nitrocellulose membrane (Protran; Schleicher & Schuell) or Immobilon (Millipore). Envelope proteins (SU and precursor) were detected with goat anti-Rauscher-gp70 antiserum (1:100, identification no. 80S000018; Quality Biotech Inc.), structural capsid protein (CA) was detected with goat anti-Rauscher-p30 antiserum (1:10,000, identification no. 81S000263; Quality Biotech Inc.), and envelope TM protein was detected with rabbit anti-p15E antiserum (1:250; gift of Alan Rein). Membranes from subsequent incubation with mouse anti-goat or mouse anti-rabbit antiserum conjugated to horseradish peroxidase (1:10,000; Sigma) were developed with a chemiluminescent substrate from a Renaissance kit (NEN).
Virus binding assays.
Binding assays were performed essentially as described previously (7, 23) with the following modifications. Virus-containing supernatants were concentrated 10- to 15-fold on Centricon-100 concentrators (Amicon). Concentration promotes binding of multiple virions to a single cell, increasing the mean fluorescence per cell (45), and eliminates most of the free SU protein. To ensure that equal numbers of particles from each of the virus stocks were incubated with cells during the assay, the concentration of virus stocks was adjusted to achieve comparable particle concentrations based on the reverse transcriptase activity and Western blot quantitation of capsid protein (data not shown). Human 293 cells (106) expressing the wild-type virus receptor or parental 293 cells were detached from culture plates with phosphate-buffered saline containing 0.02% EDTA and then incubated with 1 ml of concentrated virus stocks containing equal amounts of virions in the presence of Polybrene (5 μg/ml) for 1 h at 4°C. Cells and bound virus were incubated with goat anti-gp70 antiserum (1:100) for 30 min at 4°C and then with donkey anti-goat antiserum conjugated to fluorescein isothiocyanate (1:200; Jackson Laboratories) for 30 min at 4°C. Propidium iodide (Sigma) was added to the binding reaction mixture for 5 min at a final concentration of 20 μg/ml. The fluorescence of 5,000 live cells (negative for propidium iodide) was analyzed by flow cytometry (Epics Profile Analyzer; Coulter Cytometry). Experiments were repeated two times.
RESULTS
In these studies, we attempted to identify residues on the virus envelope proteins that interact with critical residue glutamate 237 in the putative virus-binding site on the receptor. We previously proposed that the negatively charged carboxyl group on the side chain of glutamate 237 participates in a salt bridge with a positively charged side chain of a lysine or arginine residue on the virus SU (29). To test this hypothesis, we examined the effect of replacing positively charged residues in SU with amino acids having negatively charged side chains or with alanine. We focused on lysine 111, arginine 149, and lysine 153 (envelope residues are numbered beginning with the alanine residue at the N terminus of mature SU [residue 1]). In addition, we examined the three pairs of arginines and lysines previously identified by Skov and Andersen as critical for virus infection, i.e., arginine 102 and lysine 104, arginines 124 and 126, and arginines 223 and 225 (39). We also examined the effect of replacing a negatively charged amino acid, aspartate 135, because it is conserved in SUs from a number of retroviruses that use receptors other than the ecotropic receptor.
Stocks of virus assembled from wild-type gag and pol gene products and wild-type or mutant env gene products were produced by transient transfection of pcDNA-MoMLV into human 293 cells stably expressing the pBAG retroviral genome. Mutant virus stocks that contained levels of CA comparable to that in the wild-type virus stocks (indicative of equivalent transfection efficiencies and levels of virus particle production) were analyzed by endpoint dilution titration for their ability to infect host cells and by Western blot analysis for envelope protein incorporation. Envelope protein processing was assessed by Western blot analysis of the lysates from the transfected human cells. Selected virus stocks were also analyzed for receptor binding by flow cytometry. To determine if the high-molecular-weight protein species found in virions was the envelope precursor or hyperglycosylated mature SU, selected stocks were also analyzed for the presence of TM sequences in the putative precursor species, as well as for the size of the envelope protein after treatment with glycosidase F.
Replacement of arginine 102 resulted in incorporation of a putative envelope precursor protein into virus particles.
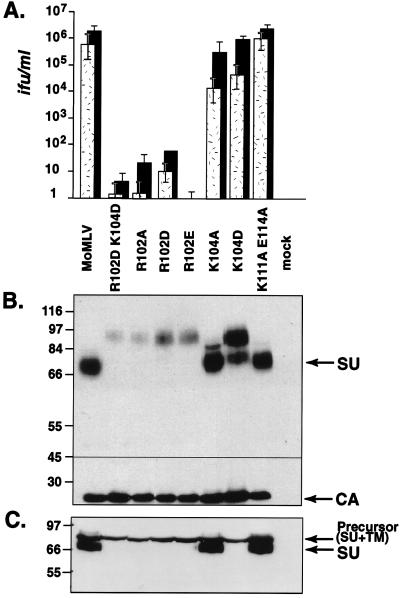
Replacement of arginine 102 with alanine (R102A), aspartate (R102D), or glutamate (R102E) almost completely abolished virus infection of mouse NIH 3T3 fibroblasts and of human 293 cells stably expressing the exogeneous ecotropic virus receptor (Fig. 1A). Surprisingly, these virions contained a protein species the size of the envelope precursor (85 kDa) that reacted with anti-SU antiserum; no species the size of mature SU (70 kDa) was detected in virions (Fig. 1B). The producer cells from which the viruses had been harvested did not contain detectable amounts of mature SU either (Fig. 1C). Since the anti-SU antiserum was specific for envelope proteins (data not shown), these results suggested that cleavage of the precursor into SU and TM was suppressed by these substitutions. More importantly, if the 85-kDa species represented uncleaved envelope protein, then these results indicate that the envelope precursor can be assembled into virions, albeit poorly in the mutant virions, particularly in that containing the alanine substitution.
FIG. 1.
Substitution of amino acids at positions 102 and 104 appear to suppress envelope precursor protein cleavage but do not preclude putative envelope precursor incorporation into virions, and in the case of residue 104, do not preclude a productive virus-receptor interaction. Replacement of R102 abolishes virus infection, whereas viruses remain highly infectious upon replacement of K104. Substitutions at positions 111 and 114 do not influence envelope protein processing or virus infectivity. (A) Naive NIH 3T3 cells (stippled bars) and human 293 cells stably expressing the wild-type ecotropic receptor (black bars) were exposed to 10-fold serial dilutions of stocks of virions pseudotyped with envelope proteins containing the indicated substitutions. Each value is the average of results of five independent experiments. MoMLV, wild-type ecotropic MoMLV. Titers were calculated from the endpoint dilution (n = 4). (B) Western blot analysis of virions containing mutant env genes. Proteins were separated on 8% acrylamide gels. The membrane was cut into two parts at the position indicated by the black line (approximately 45 kDa). The top portion was incubated with anti-SU antiserum, and the bottom part was incubated with anti-CA antiserum. Production of viruses containing mutant envelope proteins was comparable to that of the wild-type virus, as upon short exposure of immunoblots, the intensities of the bands corresponding to CA were within twofold of each other in all pellets, except those from mock-transfected cells (data not shown). The immunoblot shown is representative of five made from independent virus preparations. (C) Western blot analysis of virus producer cell lysates. Proteins were separated on an 8% polyacrylamide gel, and the membrane was probed with anti-SU antiserum.
Replacement of lysine 104 with aspartate yielded putative cleavage mutant virions that are highly infectious.
Replacement of lysine 104 on SU with alanine or with aspartate did not alter infection of human 293 cells stably expressing the virus receptor. These cells were as susceptible to virions coated with a mutant protein containing the K104D or K104A mutation as they were to virions coated with wild-type envelope protein (Fig. 1A). NIH 3T3 cells were slightly less susceptible to these viruses (Fig. 1A). Western blot analysis showed that anti-SU antiserum reacted with two protein species in K104D virions. One species was the size of mature SU, and the second species was the size of the envelope precursor (Fig. 1B), suggesting that these substitutions also suppress envelope protein cleavage. If the 85-kDa species represents uncleaved envelope protein, then these results not only provide additional evidence that the precursor can be assembled into virions but, more importantly, also indicate that virions carrying the precursor can be highly infectious.
We also analyzed the envelope protein forms in lysates of the producer cells from which the K104D virus had been harvested. Mature SU was not detectable in cells producing K104D viruses, although the precursor was present (Fig. 1C), suggesting that the K104D mutation is a potent suppressor of envelope cleavage. Interestingly, K104D virions contained appreciable amounts of mature SU even though steady-state levels of mature SU were too low to be detected in the K104D producer cells.
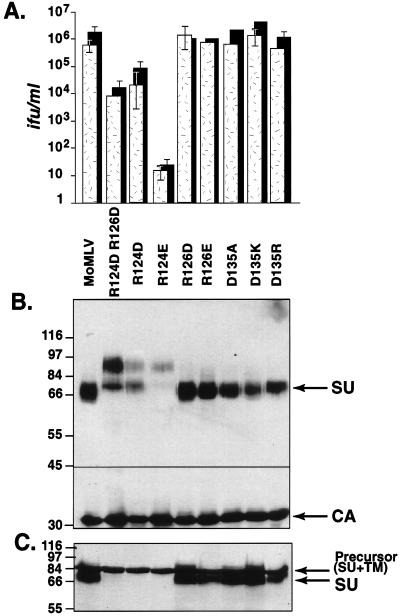
Replacement of arginines 124 and 126 or arginines 223 and 225 also results in highly infectious putative cleavage mutant virions.
Replacement of arginine 124 with glutamate (R124E) gave a phenotype similar to that of the R102A mutant virion. Infection was dramatically decreased and precursor cleavage was almost completely abolished by an R124E substitution (Fig. 2). Envelope precursor was incorporated into virions, albeit poorly. No mature SU was detectable in virions or in producer cells. Surprisingly, virions containing an arginine 124-to-aspartate (R124D) substitution were only slightly less infectious than were wild-type viruses, a 10,000-fold improvement over the infectivity observed for the glutamate change (Fig. 2). Both precursor and mature SU were incorporated into these highly infectious R124D virions. As with the K104D mutant virions contained a surprising amount of mature SU, considering that none was detectable in producer cells. Moreover, double mutant virions with an R124D and an R126D substitution consistently contained more precursor molecules than mature SU (Fig. 2), suggesting that the addition of the seemingly innocuous R126D substitution somehow enhances precursor incorporation into virions. Furthermore, replacement of arginines 223 and 225 with aspartate (R223D R225D) also resulted in incorporation of an 85-kDa envelope protein species, suggesting that precursor cleavage is also suppressed by these changes (Fig. 3). Here too, the envelope precursor was incorporated into highly infectious virions.
FIG. 2.
Substitution of arginine 124 also appears to suppress protein cleavage. Glutamate at this position produces noninfectious particles, whereas aspartate produces highly infectious virions coated with uncleaved envelope protein. Mutations at positions 126 and 135 do not affect protein maturation and virus entry. (A) Virus titers on NIH 3T3 cells (stippled bars) and human 293 cells expressing the exogenous wild-type receptor (black bars). Titers were calculated from the endpoint dilution (n = 4) after exposure to virions pseudotyped with envelope proteins containing the indicated substitutions. Each value is the average of results from five independent experiments. MoMLV indicates the wild-type virus. (B) Western blot analysis of virions containing mutant env genes. The membrane was cut at the position indicated by the black line, and then the top portion was incubated with anti-SU antiserum and the bottom portion was incubated with anti-CA antiserum. (C) Western blot analysis of virus producer cell lysates with anti-SU antiserum.
FIG. 3.
Substitutions of residues at positions 223 and 225 also appear to suppress cleavage, resulting in highly infectious virions coated with the putative envelope precursor. (A) Virus titers on NIH 3T3 cells (stippled bars) and human 293 cells expressing the exogenous wild-type receptor (black bars) calculated from the endpoint dilution (n = 4). Each value is the average of results from five independent experiments. MoMLV, wild-type virus; mock, supernatant or lysate of cells transfected with pcDNA3. (B) Western blot analysis of virions with anti-SU antiserum and anti-CA antiserum. (C) Western blot analysis of virus producer cell lysates with anti-SU antiserum.
The phenotype of a number of substitutions was indistinguishable from that of the wild-type envelope protein. Replacement of lysine 111 with alanine (K111A), arginine 126 with aspartate (R126D) or glutamate (R126E), aspartate 135 with alanine (D135A) or lysine (D135K), or arginine 149 with aspartate and lysine 153 with aspartate (R149D K153D) did not alter infection or envelope precursor cleavage (Fig. 1 to 3). These results suggest that those residues are not essential to productive virus-receptor interaction and that they are not involved in envelope protein folding.
The 85-kDa protein species consisted of uncleaved envelope precursor protein.
It was possible that the 85-kDa species present in virions coated with mutant envelope proteins was not uncleaved envelope precursor protein but rather a hyperglycosylated form of mature SU. Misfolding of the envelope precursor might lead to greater than normal carbohydrate addition at any or all of the glycosylation sites. Cleavage of such a hyperglycosylated precursor would then yield mature SU that by coincidence was the size of the precursor with normal glycosylation. If this was the case, then removal of carbohydrates would yield a single 52-kDa protein species representing unglycosylated SU. If the size difference was not due to hyperglycosylation, then removal of carbohydrates would yield 68- and 52-kDa species representing the unglycosylated forms of the precursor and SU, respectively.
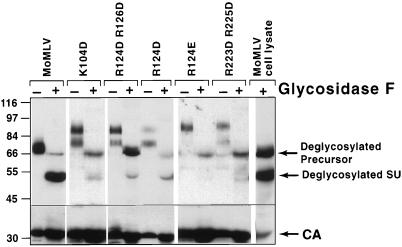
We used wild-type producer cell lysate to assess glycosylation of envelope proteins because it would provide precise size standards for the deglycosylated precursor and mature SU. We also used the cell lysate to determine if the glycosidase F digestion proceeded to completion in order to be reasonably certain that any species detected after glycosidase treatment was the 68-kDa deglycosylated precursor rather than 70-kDa molecules of mature SU that escaped deglycosylation. Lysate from human cells expressing the wild-type envelope protein was treated with excess glycosidase F overnight, the same conditions used to treat virions. The expected 68- and 52-kDa envelope protein species resulted (last lane of Fig. 4). Most importantly, there was no detectable 85-kDa protein, indicating that the glycosidase reaction had proceeded to completion. Treatment of virions coated with the mutant envelope proteins also yielded two species of 68 and 52 kDa corresponding, respectively, to the unglycosylated precursor and SU species seen in the producer cells (Fig. 4). Moreover, the ratio of putative precursor species (85 kDa) to mature SU (70 kDa) in untreated aliquots of virions was comparable to the ratio of the 68-kDa species to the 52-kDa species observed upon deglycosylation for all mutants. The predominant species in K104D, R124D R126D, R124E, and R223D R225D virions was 85 kDa prior to digestion and 68 kDa (representing the deglycosylated envelope precursor) after glycosidase F digestion, whereas the predominant species in R124D virions were 70 kDa prior to digestion and 52 kDa after glycosidase F digestion. These results demonstrated that hyperglycosylation of mature SU was not responsible for the 85-kDa protein species.
FIG. 4.
The 85-kDa envelope protein species does not represent hyperglycosylated SU. Wild-type MoMLV producer cell lysate (last lane) or purified wild-type and mutant virions were incubated with glycosidase F overnight, separated by SDS-polyacrylamide gel electrophoresis on 8% gels, and then submitted to Western blot analysis with anti-SU antiserum (top portion) and anti-CA antiserum (bottom portion).
In addition, we determined if TM sequences were present in the 85-kDa envelope protein species. As a positive control, we again used wild-type producer cell lysate. Initially, we used monoclonal anti-TM antibody 42-114 (34) to detect TM sequences. Although 42-114 readily recognized its epitope in mature TM, it failed to recognize the epitope when it was present in the precursor species of wild-type producer cell lysate under a variety of conditions and with nitrocellulose and nylon (Immobilon; Millipore) membranes (last lane of Fig. 5A). We then tried polyclonal anti-TM antiserum (gift of A. Rein) to detect TM sequences. No reaction of this anti-TM antiserum with precursor species was observed when producer cell lysate was transferred to nitrocellulose (data not shown). However, when lysate was transferred to Immobilon, it weakly recognized TM sequences in the precursor (second lane of Fig. 5A). Importantly, this antiserum did not react with mature SU (70 kDa). It reacted strongly with the 15-kDa species of mature TM and cross-reacted with CA present in its precursor forms, particularly the 65-kDa capsid precursor, gag65 (middle lane of Fig. 5A). Because the 85-kDa species does not contain Gag- or CA-specific sequences (data not shown), we could still use this antiserum for immunoblot analysis of the mutant virion proteins. It weakly recognized the 85-kDa species in virions coated with the R124D R126D, R124D, R124E, R223D R225D, and K104D putative envelope cleavage mutant proteins, in addition to reacting with mature TM (15 and 12 kDa), the 65-kDa capsid precursor, and mature CA (left membrane of Fig. 5B). Notably, the antiserum consistently reacted more strongly with the sparsely incorporated precursor species of the R124D, R124E, and R223E R225D mutant proteins than with the abundantly incorporated precursor species of the R124D R126D and K104D mutant proteins. We do not know the cause of this variation but suspect that the principal epitopes that this antiserum recognizes tend to bind to the membrane when TM remains covalently bonded to SU, so that they are unavailable for antibody binding. In R124D, R124E, and R223D R225D mutant proteins, these epitopes appear to be more accessible for unknown reasons. Only mature TM and the CA forms were recognized in wild-type virions. These results demonstrate that the 85-kDa species that was incorporated into these mutant virions were envelope precursor protein, establishing that these mutations suppress envelope protein cleavage.
FIG. 5.
Immunoblot analysis of virions with anti-TM antiserum confirmed that mutant virions contained the uncleaved envelope precursor. (A) Replicate samples of wild-type producer cell lysate were separated on a 6 to 15% polyacrylamide gradient SDS gel and transferred to an Immobilon membrane; lanes were then cut apart and reacted with monoclonal anti-TM 42/114 (last lane), anti-TM (middle lane), or anti-SU (first lane) antiserum. (B) Wild-type and mutant virion proteins were separated on 6 to 15% polyacrylamide gradient SDS gels and transferred to Immobilon membranes. Membranes were analyzed first by incubation with anti-TM antiserum (left membrane) and then deprobed and reacted with anti-SU antiserum (right membrane). gag 65, the 65-kDa capsid precursor.
Virus particles coated with mutant envelope precursor proteins were deficient in receptor binding.
To determine if any of the mutations affected receptor binding, we performed equilibrium virus binding assays. Under conditions where 107 infectious units corresponding to approximately 109 physical particles (44) were incubated with 106 host cells, we observed a 50-fold increase in mean fluorescence intensity upon incubation of susceptible host cells expressing the receptor with MoMLV over the level of fluorescence intensity upon incubation with nonsusceptible cells lacking the receptor (Fig. 6). As an additional positive control, we measured receptor binding of the D135A virions that did not exhibit any detectable differences in levels of envelope processing or incorporation and that were as infectious as the wild-type virus. We observed a 60-fold increase in mean fluorescence for these viruses. Remarkably, host cells incubated with R223D R225D particles exhibited only a 2-fold increase in mean fluorescence—25-fold less than that of the wild-type virus—even though they were only 20-fold less infectious than the wild-type virus. Highly infectious particles coated with the K104D, R124D R126D, and R124D mutant proteins exhibited only 9- to 10-fold increases (Fig. 6), indicating that receptor binding of these mutant virions was reduced by >5-fold compared with that of the wild-type or D135A virus. The R124E mutant virions exhibited a 6.6-fold-higher level of mean fluorescence and a 7.6-fold-lower level of equilibrium binding to the virus receptor than the levels of the wild-type virus and 1.5-fold-lower levels than those of the highly infectious R124D virions. These results suggested that the longer side chain of glutamate results in a more marked change in the structure of the receptor binding domain than does the side chain of aspartate. In agreement with this, the R124E substitution produced a more profound suppression of cleavage than did the R124D substitution. It does not appear that the 1.5-fold reduction in binding of R124E virions over that of R124D virions accounts for their dramatic difference in infection since the R223D R225D virions exhibited an even greater loss of receptor binding without a concomitant loss of infection. It is more likely that the difference in infectivity is due to greater incorporation of mature SU and TM in R124D virions.
FIG. 6.
Virions coated with envelope cleavage mutants bind the ecotropic virus receptor inefficiently. Purified virions were incubated with human 293 cells stably expressing the exogenous ecotropic virus receptor and then with goat anti-SU antiserum and mouse anti-goat antiserum conjugated to fluorescein isothiocyanate. Last, virus cell-antibody complexes were incubated with propidium iodide. Bound virus was quantified by measuring the mean fluorescence intensity of live cells (those cells lacking propidium iodide staining) by flow cytometry. “No receptor” indicates parental human 293 cells not expressing the exogenous receptor incubated with wild-type MoMLV and antibodies; “No virus” indicates 293 cells stably expressing the exogenous receptor incubated with antibodies alone. The mean fluorescence intensity of the no receptor peak was 0.5, and that of the no virus peak was 0.6. Infectivity was summarized from the data in Fig. 1 to 3. +, infectious titer within 100-fold of that of wild-type MoMLV; −, infectious titer >10,000-fold less than that of wild-type MoMLV.
It was possible that the cleavage mutations caused dissociation of SU from TM. If this was the case, virions might have lacked mature SU because it was shed during pelleting through the sucrose cushion prior to immunoblotting or during concentration prior to binding assays. This is particularly important because we performed the virus titration studies using virions directly from the producer cell supernatant. To determine if this was the case, we performed immunoblot analyses on virions that were pelleted by direct centrifugation from the cell supernatant (no sucrose cushion) and so contained virion-associated, plus shedded, SU from the cell supernatant. We then evaluated shedded SU by comparing the amounts of mature SU in these samples to those found in virions after they were pelleted through sucrose; any difference would indicate that a mutation caused shedding that might account for the pausity of SU in mutant virions. After being pelleted through sucrose, mutant and wild-type virions contained amounts of mature SU comparable to those found in cell supernatants pelleted directly (data not shown), indicating that none of the cleavage mutations led to detectable SU shedding. The cell supernatant pellets also showed that mature SU was undetectable or greatly reduced and that the 85-kDa precursor was the predominant envelope species in the cleavage mutants.
DISCUSSION
Incorporation of envelope precursor protein does not preclude productive virus-receptor interaction.
It was previously thought that cleavage of the precursor into mature SU and TM was required for assembly of envelope proteins into retroviral particles. However, we identified two types of novel envelope cleavage mutant proteins, each exhibiting some degree of failure to cleave precursor protein into mature SU and TM accompanied by assembly of precursor into virions. The first type resulted in a slight reduction of infectivity (10- to 100-fold), while the second type contained changes that resulted in a >10,000-fold loss of infectivity. These data provide evidence that cleavage is not a prerequisite for envelope protein transport to the cell surface and assembly into virus particles. Moreover, virions containing predominantly precursor protein can support productive virus-receptor interactions.
We cannot make definitive conclusions as to whether the altered residues interact directly with the receptor. The failure of host cell proteases to cleave precursors in mutant proteins containing substitutions that are distal to the actual site of cleavage suggests that these envelope precursors are misfolded. Hence, the defect in virus-receptor binding is most likely attributable to incorrect conformation of envelope proteins.
Mature SU and TM appear to be the preferred substrates for virion assembly.
K104D virions consistently contained the envelope precursor and less of the mature SU, whereas, cells producing these viruses contained detectable levels of precursor but not mature SU. Interestingly, K104D, R124D, and R124D R126D virions also contained appreciable amounts of mature SU even though steady-state levels of mature SU were too low to be detected in their respective producer cells. These results suggest that although the precursor can be incorporated, the cleaved envelope proteins are the preferred substrate for assembly into virions. Alternatively, the suppression of precursor cleavage may be somewhat relieved during or after virion assembly by changes in envelope protein conformation so that secreted cellular proteases can perform the cleavage that normally occurs in the Golgi apparatus.
Only a few molecules of mature TM appear to be required to mediate virus entry.
In all but two cases, the infectivities of the cleavage mutant viruses correlated with the amounts of mature SU present on virions regardless of the amount of precursor protein present. Mutants that were highly infectious contained appreciable amounts of mature SU and TM in addition to the precursor, whereas mutants that were poorly infectious contained undetectable amounts of mature SU and TM. It is likely that the observed infection was accomplished by free fusion peptide on a relatively small number of cleaved envelope molecules incorporated into virions acting in trans after being bound by the uncleaved precursor molecules. Recent studies have shown that monomers within the envelope trimer can cooperate in trans to accomplish virus entry (37, 46). However, we cannot rule out the possibility that infection by some mutant virions was mediated by a constrained fusion peptide that was able to function in the context of the appropriate changes in SU. For example, the envelope protein of vesicular stomatitis virus contains an internal fusion peptide that remains constrained during virus entry (11).
We consistently observed 10- to 50-fold-lower infectious titers of cleavage mutant stocks on NIH 3T3 cells than on human 293 cells stably expressing the ecotropic receptor. This difference may be the result of the greater number of virus receptors available on 293-ecotropic receptor cells. These cells express three to four times as many virus binding sites as do NIH 3T3 cells (1b). However, NIH 3T3 cells also appear to be less fusogenic in that they are naturally resistant to cell-cell fusion in the presence of high-titer wild-type MoMLV stocks (31), whereas the human 293-ecotropic receptor cells readily fuse (1). Thus, the difference in susceptibilities to cleavage mutant viruses might be the result of the less fusogenic NIH 3T3 cells requiring more molecules of free fusion peptide to efficiently complete virus entry than 293-receptor cells require.
Interestingly, the weaker band intensity observed with the anti-TM antiserum revealed that the 85-kDa species is actually a doublet, a fact that was likely obscured by the intensity of these species when they were reacted with anti-SU. This result brings up the possibilities that the higher-molecular-mass species represents the intact precursor and that the slightly faster-migrating species represents the precursor from which R peptide has been removed. Indeed, the small amounts of mature TM detected in the virions coated with mutant proteins was the 12-kDa, R-less species, indicating that these mutant proteins receive this cleavage as particles mature. Alternatively, it is possible that the two species represent differences in levels of glycosylation of the precursors found in virions.
Receptor binding might not be the rate-limiting step in MoMLV entry.
Structural changes resulting from a number of substitutions affected receptor binding. Remarkably, for entry, most of the cleavage mutant virions could tolerate as much as 5- to 25-fold reductions in the levels of equilibrium binding to the receptors on host cells without experiencing more than a 100-fold reduction in infection. These results suggest that virus binding to the receptor might not be the rate-limiting step in retroviral entry. They also suggest that efficient virus binding is not required for entry of at least these mutant virions and raise the question of whether the same is true for the wild-type virus. No infection data or measurements of the relative levels of binding efficiency of particles containing various amounts of wild-type envelope protein are available to address this question. Determining its answer should provide important insights as to the numbers of envelope molecules and receptors required for productive infection of retroviruses.
A possible mechanism for suppression of SU-TM cleavage by substitutions at residues 102, 104, 124, 126, 223, and 225.
Our results suggest that the residues altered in the cleavage mutant proteins can influence the folding of the cleavage recognition site. In the crystal structure of amino acids 9 through 236 of SU from ecotropic Friend MLV (10), the residues that correspond to all but two of the cleavage mutations are at the “top” along the surface that has been proposed to be the receptor binding site since it contains the residue corresponding to MoMLV aspartate 84 (10). If this is the surface that makes the initial contact with the receptor, then it would be expected to act like a sensor, transducing changes occurring upon receptor binding through the β-sandwich of the binding domain into the carboxy-terminal region of SU and then into TM to activate fusion peptide function. Mutations that produce structural changes mimicking those occurring during interaction with the receptor might be expected to translate through the protein to change folding of these same domains, including that of the cleavage recognition site between arginine 436 and glutamate 437 of the precursor. Arginines 223 and 225, the two sites we altered that were not on the receptor binding surface, are near the base of the β-sandwich. Their replacement by aspartate residues might create similar conformational changes, sequestering the cleavage site. It is striking that as great as a 25-fold decrease in binding of R223D R225D virions over that of wild-type virions was sufficient to support almost as efficient an entry as that of the wild-type virus. It might be that repulsion of the side chain carboxyl groups of the aspartates at positions 223 and 225 disturbs the orientation of the β-sandwich, creating a rearrangement in the envelope oligomers similar to that induced by receptor binding. The resulting “conformational intermediate” might be a more entry-competent molecule in that a single receptor binding event would be sufficient to induce it to undergo all conformational changes required for virus entry.
Retroviral vectors designed for use in human gene therapy employ chimeric or hybrid envelope proteins to redirect virus binding to a protein or to a chemical moiety found uniquely on the type of cell that is the target for gene delivery (14). Virions containing these modified envelope proteins are poorly infectious or completely noninfectious. It has been proposed that conformational changes in modified envelope proteins cause excessive loss of the hybrid surface protein (7, 30). Furthermore, use of any retroviral vector for gene therapy will require storage at low temperature, which disrupts the tenuous association of SU with TM, resulting in SU shedding upon thawing. Inclusion of these cleavage mutations into the retroviral envelope proteins on targeted retroviral vectors may prevent loss of the modified surface proteins from virions due to shedding, improving their infectivity and storage hardiness. In these virus particles, the loss of receptor binding resulting from the cleavage mutations might well be innocuous since only rodent cells express a functional ecotropic virus receptor.
ACKNOWLEDGMENTS
We thank Alan Rein for providing the anti-TM antiserum and Krish Kizhatil, Zhaohui Qian, and Byoung Ryu for critical readings of the manuscript.
This work was supported by Public Health Service grant AI33410 from the National Institutes of Health (to L.M.A.).
REFERENCES
- 1.Albritton, L. M. Unpublished observations.
- 1a.Albritton L M, Kim J W, Tseng L, Cunningham J M. Envelope-binding domain in the cationic amino acid transporter determines the host range of ecotropic murine retroviruses. J Virol. 1993;67:2091–2096. doi: 10.1128/jvi.67.4.2091-2096.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1b.Albritton, L. M., and K. Kizhatil. Unpublished data.
- 2.Bae Y, Kingsman S M, Kingsman A J. Functional dissection of the Moloney murine leukemia virus envelope protein gp70. J Virol. 1997;71:2092–2099. doi: 10.1128/jvi.71.3.2092-2099.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkowitz R D, Goff S P. Point mutations in Moloney murine leukemia virus envelope protein: effects on infectivity, virion association, and superinfection resistance. Virology. 1993;196:748–757. doi: 10.1006/viro.1993.1532. [DOI] [PubMed] [Google Scholar]
- 4.Bosch M L, Earl P L, Fargnoli K, Picciafuoco S, Giombini F, Wong-Staal F, Franchini G. Identification of the fusion peptide of primate immunodeficiency viruses. Science. 1989;244:694–697. doi: 10.1126/science.2541505. [DOI] [PubMed] [Google Scholar]
- 5.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 6.Coffin J M, Hughes S H, Varmus H E. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 7.Cosset F L, Morling F J, Takeuchi Y, Weiss R A, Collins M K L, Russell S J. Retroviral retargeting by envelopes expressing an N-terminal binding domain. J Virol. 1995;69:6314–6322. doi: 10.1128/jvi.69.10.6314-6322.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong J Y, Dubay J W, Perez L G, Hunter E. Mutations within the proteolytic cleavage site of the Rous sarcoma virus glycoprotein define a requirement for dibasic residues for intracellular cleavage. J Virol. 1992;66:865–874. doi: 10.1128/jvi.66.2.865-874.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubay J W, Dubay S R, Shin H J, Hunter E. Analysis of the cleavage site of the human immunodeficiency virus type 1 glycoprotein: requirement of precursor cleavage for glycoprotein incorporation. J Virol. 1995;69:4675–4682. doi: 10.1128/jvi.69.8.4675-4682.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fass D, Davey R A, Hamson C A, Kim P S, Cunningham J M, Berger J M. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 angstrom resolution. Science. 1997;277:1662–1666. doi: 10.1126/science.277.5332.1662. [DOI] [PubMed] [Google Scholar]
- 11.Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E. Fields virology. Vol. 1. Philadelphia, Pa: Lippincott-Raven; 1996. [Google Scholar]
- 12.Freed E O, Myers D J, Risser R. Characterization of the fusion domain of the human immunodeficiency virus type 1 envelope glycoprotein gp41. Proc Natl Acad Sci USA. 1990;87:4650–4654. doi: 10.1073/pnas.87.12.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freed E O, Risser R. The role of envelope glycoprotein processing in murine leukemia virus infection. J Virol. 1987;61:2852–2856. doi: 10.1128/jvi.61.9.2852-2856.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedmann T. Overcoming the obstacles to gene therapy. Sci Am. 1997;276:96–101. [PubMed] [Google Scholar]
- 15.Goodman L J, Kain S R, Firestone G L. Trafficking of wild-type and an endoproteolytic-site mutant of the mouse mammary tumor virus glycoprotein. J Biol Chem. 1993;268:2329–2336. [PubMed] [Google Scholar]
- 16.Granowitz C, Colicelli J, Goff S P. Analysis of mutations in the envelope gene of Moloney murine leukemia virus: separation of infectivity from superinfection resistance. Virology. 1991;183:545–554. doi: 10.1016/0042-6822(91)90983-i. [DOI] [PubMed] [Google Scholar]
- 17.Green N, Shinnick T M, Witte O, Ponticelli A, Sutcliffe J G, Lerner R A. Sequence-specific antibodies show that maturation of Moloney leukemia virus envelope polyprotein involves removal of a COOH-terminal peptide. Proc Natl Acad Sci USA. 1981;78:6023–6027. doi: 10.1073/pnas.78.10.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo H G, Veronese F M, Tschachler E, Pal R, Kalyanaraman V S, Gallo R C, Reitz M S., Jr Characterization of an HIV-1 point mutant blocked in envelope glycoprotein cleavage. Virology. 1990;174:217–224. doi: 10.1016/0042-6822(90)90070-8. [DOI] [PubMed] [Google Scholar]
- 19.Harbers K, Schnieke A, Stuhlman H, Jahner D, Jaenish R. DNA methylation and gene expression: endogenous retroviral genome becomes infectious after molecular cloning. Proc Natl Acad Sci USA. 1981;78:7609–7613. doi: 10.1073/pnas.78.12.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heard J, Danos O. An amino-terminal fragment of the Friend murine leukemia virus binds the ecotropic receptor. J Virol. 1991;65:4026–4032. doi: 10.1128/jvi.65.8.4026-4032.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson P, Rosner M. Characterization of murine-specific leukemia virus receptor from L cells. J Virol. 1986;58:900–908. doi: 10.1128/jvi.58.3.900-908.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones J S, Risser R. Cell fusion induced by the murine leukemia virus envelope glycoprotein. J Virol. 1993;67:67–74. doi: 10.1128/jvi.67.1.67-74.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kadan M J, Sturm S, Anderson W F, Eglitis M A. Detection of receptor-specific murine leukemia virus binding to cells by immunofluorescence analysis. J Virol. 1992;66:2281–2287. doi: 10.1128/jvi.66.4.2281-2287.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J W, Closs E I, Albritton L M, Cunningham J M. Transport of cationic amino acids by the mouse ecotropic retrovirus receptor. Nature. 1991;352:725–728. doi: 10.1038/352725a0. [DOI] [PubMed] [Google Scholar]
- 25.Kunkel T A. Rapid and efficient site specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:477–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Linder M, Wenzel V, Linder D, Stirm S. Structural elements in glycoprotein 70 from polytropic Friend mink cell focus-inducing virus and glycoprotein 71 from ecotropic Friend murine leukemia virus, as defined by disulfide-bonding pattern and limited proteolysis. J Virol. 1994;68:5133–5141. doi: 10.1128/jvi.68.8.5133-5141.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Machida C A, Kabat D. Role of partial proteolysis in processing murine leukemia virus membrane envelope glycoproteins to the cell surface. A viral mutant with uncleaved glycoprotein. J Biol Chem. 1982;257:14018–14022. [PubMed] [Google Scholar]
- 28.MacKrell A J, Soong N W, Curtis C M, Anderson W F. Identification of a subdomain in the Moloney murine leukemia virus envelope protein involved in receptor binding. J Virol. 1996;70:1768–1774. doi: 10.1128/jvi.70.3.1768-1774.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malhotra S, Scott A G, Zavorotinskaya T, Albritton L M. Analysis of the murine ecotropic leukemia virus receptor reveals a common biochemical determinant on diverse cell surface receptors that is essential to retrovirus entry. J Virol. 1996;70:321–326. doi: 10.1128/jvi.70.1.321-326.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marin M, Noel D, Valsesia-Wittman S, Brockly F, Etienne-Julan M, Russell S, Cosset F L, Piechaczyk M. Targeted infection of human cells via major histocompatibility complex class I molecules by Moloney murine leukemia virus-derived viruses displaying single-chain antibody fragment-envelope fusion proteins. J Virol. 1996;70:2957–2962. doi: 10.1128/jvi.70.5.2957-2962.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McClure M O, Sommerfelt M A, Marsh M, Weiss R A. The pH independence of mammalian retrovirus infection. J Gen Virol. 1990;71:767–773. doi: 10.1099/0022-1317-71-4-767. [DOI] [PubMed] [Google Scholar]
- 32.McCune J M, Rabin L B, Feinberg M B, Lieberman M, Kosek J C, Reyes G R, Weissman I L. Endoproteolytic cleavage of gp160 is required for the activation of human immunodeficiency virus. Cell. 1988;53:55–67. doi: 10.1016/0092-8674(88)90487-4. [DOI] [PubMed] [Google Scholar]
- 33.Miller A D, Buttimore C. Redesign of retrovirus packaging cell lines to avoid recombination leading to helper virus production. Mol Cell Biol. 1986;6:2895–2902. doi: 10.1128/mcb.6.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pinter A, Honnen W J, Tung J S, O’Donnell P V, Hammerling U. Structural domains of endogenous murine leukemia virus gp70s containing specific antigenic determinants defined by monoclonal antibodies. Virology. 1982;116:499–516. doi: 10.1016/0042-6822(82)90143-x. [DOI] [PubMed] [Google Scholar]
- 35.Ragheb J A, Anderson W F. pH-independent murine leukemia virus ecotropic envelope-mediated cell fusion: implications for the role of the R peptide and p12E TM in viral entry. J Virol. 1994;68:3220–3231. doi: 10.1128/jvi.68.5.3220-3231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rein A, Mirro J, Haynes J G, Ernst S M, Nagashima K. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J Virol. 1994;68:1773–1781. doi: 10.1128/jvi.68.3.1773-1781.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rein A, Yang C, Haynes J A, Mirro J, Compans R W. Evidence for cooperation between murine leukemia virus Env molecules in mixed oligomers. J Virol. 1998;72:3432–3435. doi: 10.1128/jvi.72.4.3432-3435.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Skov H, Andersen K B. Mutational analysis of Moloney murine leukemia virus surface protein gp70. J Gen Virol. 1993;74:707–714. doi: 10.1099/0022-1317-74-4-707. [DOI] [PubMed] [Google Scholar]
- 40.Thomas A, Roth M J. Analysis of cysteine mutations on the transmembrane protein of Moloney murine leukemia virus. Virology. 1995;211:285–289. doi: 10.1006/viro.1995.1402. [DOI] [PubMed] [Google Scholar]
- 41.Turner D L, Cepko C L. A common progenitor for neuron and glia persists in rat retina late in development. Nature. 1987;328:131–136. doi: 10.1038/328131a0. [DOI] [PubMed] [Google Scholar]
- 42.White J M. Membrane fusion. Science. 1992;258:917–924. doi: 10.1126/science.1439803. [DOI] [PubMed] [Google Scholar]
- 43.White J M. Viral and cellular membrane fusion proteins. Annu Rev Physiol. 1990;52:675–697. doi: 10.1146/annurev.ph.52.030190.003331. [DOI] [PubMed] [Google Scholar]
- 44.Yu H, Soong N, Anderson W F. Binding kinetics of ecotropic (Moloney) murine leukemia retrovirus with NIH 3T3 cells. J Virol. 1995;69:6557–6562. doi: 10.1128/jvi.69.10.6557-6562.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zavorotinskaya, T. Unpublished results.
- 46.Zhao Y, Lee S, Anderson W F. Functional interactions between monomers of the retroviral envelope protein complex. J Virol. 1997;71:6967–6972. doi: 10.1128/jvi.71.9.6967-6972.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]