Abstract
Machine learning (ML) has revolutionized medical image-based diagnostics. In this review, we cover a rapidly emerging field that can be potentially significantly impacted by ML – eye tracking in medical imaging. The review investigates the clinical, algorithmic, and hardware properties of the existing studies. In particular, it evaluates 1) the type of eye-tracking equipment used and how the equipment aligns with study aims; 2) the software required to record and process eye-tracking data, which often requires user interface development, and controller command and voice recording; 3) the ML methodology utilized depending on the anatomy of interest, gaze data representation, and target clinical application. The review concludes with a summary of recommendations for future studies, and confirms that the inclusion of gaze data broadens the ML applicability in Radiology from computer-aided diagnosis (CAD) to gaze-based image annotation, physicians’ error detection, fatigue recognition, and other areas of potentially high research and clinical impact.
Keywords: Machine learning, eye tracking, medical imaging, radiology, surgery
I. INTRODUCTION
MACHINE learning (ML) has revolutionized computer-aided diagnosis (CAD) with more than 10,000 papers published in recent years. The scientific advances have been followed by commercial successes with hundreds of ML CAD solutions receiving FDA and CE approvals (www.aiforradiology.com).
The recent reviews and reports of the FDA-approved solution indicate that ML CAD is moving in the direction of optimizing tedious and time-consuming tasks [1], [2]. Physicians remain the ultimate judges of the ML decisions, as they can accept/reject and potentially adjust these decisions. In the ML solutions where decision adjustments are possible, the main instruments for the physician-computer interaction are the keyboard and mouse, by which physicians can manually correct ML-based medical image analysis results. However, the keyboard and mouse are not the only input devices available for the user to communicate with a diagnostic workstation. Moreover, manual input via keyboard and mouse is often slower and less convenient in contrast to other means of potential communication such as voice and gaze. There is strong interest in the inclusion of voice and gaze signals into human-computer communication by using ML technologies to convert such signals into computer-interpretable commands. For example, DeepMind has recently demonstrated that neural networks can transform verbal instructions to fill web forms, book plane tickets, etc. [3]. Similarly, gaze-based detection and annotation of objects accelerate database creation by 30 times for plant images [4], 10 times for apple images [4], and 10 times for histopathology images [5], compared to manual annotation with a keyboard and mouse.
Eye tracking of physicians has been a topic of interest for more than 30 years [6], because it gives insights into the perceptual and cognitive processes used in medical decision-making. The research in the field resulted in discovering consistent patterns of medical image reading such as “scanning” (which involves directing high-resolution focus of the visual system, the fovea, throughout each slice) and “drilling” (whereby radiologists keep their gaze steady in one point of the slice and scroll through the stack) when radiologists read volumetric (3D) images [7]. It was also reported that physicians’ gaze patterns depend on their level of expertise and fatigue [8], with less experienced radiologists spending more time examining the images than their more experienced counterparts. At the same time, a comprehensive quantitative analysis of gaze paths remains challenging due to the high complexity of such paths. Gaze paths can be decomposed into fixations, i.e., moments in time when the eyes are fixed on a given area, and saccades, i.e., ballistic movements between fixations. The survey of 124 eye-tracking researchers indicated that the conceptual difference between fixations and saccades is based on three components, namely oculomotor component: whether eye moves slow or fast; functional component: what purpose does the current eye movement have; and coordinate system component [9]. Despite an intuitively clear difference, there are more than a dozen different implementations of fixation and saccade calculations [10]. In addition, the size of the useful visual field of radiologists, i.e., a visual angle sufficient to capture an abnormality using peripheral vision, also remains uncertain [11], varying for different imaging modalities and lesion types [12].
The reason why eye tracking research is so important is that it reflects physicians’ perceptual processes, that is, information capture that they are not cognitively aware of. For example, physicians cannot be aware that the quality of their image reading abilities deteriorates due to fatigue, but an eye-tracking framework can capture the reduction of anatomical coverage with the gaze – a factor that has been associated with fatigue – and alert the reader [13], [14]. As the reading of most medical images is guided by perception, eye tracking is the only way to monitor how observers interact with the image. For example, it has been shown that in the detection of breast cancer in mammograms [15] and the detection of lung nodules in chest radiographs [16], 30% of the misses are never fixated by radiologists. In other words, 70% of errors in these two domains attract visual attention; that is, they are perceptual errors.
The physician’s gaze represents high-dimensional longitudinal data. The modern developments in ML offer instruments for finding consistent patterns in high-dimensional data, which opens an avenue for using ML for gaze analysis in the medical imaging domain. The applicability of ML solutions for gaze analysis is not limited to CAD, as they could go beyond diagnosis and focus on radiologist training, detecting fatigue, performance analysis, etc. The research in such subdomains may face lower regulatory pressure, and the integration of ML solutions for gaze analysis into clinical practice could be simpler than that for CAD, which seeks to aid readers in interpreting a medical image. This review will summarize the published ML studies on physicians’ gaze analysis during medical image reading. The objective is to understand the key properties of this research field, the components needed to conduct similar studies, and the targeted applications.
II. METHOD
A. Eligibility Criteria
The selection process included studies on using ML methods in experiments where eye-tracking equipment is utilized during medical image reading. An eligible study must, therefore, include the visual reading of an image generated by a diagnostic or interventional medical device. The studies that target the reading of electronic health records are not eligible. The eligible use of ML is not limited to the direct analysis of gaze features but extends to image processing and/or gaze feature selection.
B. Search Strategy and Selection Process
We separately searched for eligible papers in the Scopus, PubMed, and Web of Science (WoS) databases. The search pattern was “(artificial intelligence OR deep learning OR machine learning) AND (eye tracking OR gaze tracking) AND (medical imaging OR radiology OR surgery OR diagnosis)”. After removing duplicates, all records that did not match the eligibility criteria were excluded. There were three major reasons for the exclusion. First, a paper was excluded if there was no (or negligible) ML component, e.g., regression used for minor processing. Second, a paper was excluded if it did not work with medical images or was not related to medicine. For example, we excluded papers that worked with surgeon workload analysis through pupil size, emotion prediction, etc. Finally, we excluded papers that did not work with eye tracking, e.g., brain activity recording against different visual stimuli.
For each eligible paper, we extracted its reference list and all papers that cited this paper by using the Google Scholar database. The extracted citations went through the eligibility analysis mentioned above, and the search was repeated until no new papers were added to the final literature list (Fig. 1).
Fig. 1.
Flowchart of the performed review of machine learning studies on the use of eye tracking for medical image analysis.
C. Literature Analysis and Synthesis
The final list of papers was critically analyzed against different research dimensions. These dimensions included: a) the eye tracking equipment, including head-mounted and remote eye-trackers; b) medical imaging modality and dimensionality; c) the software needed to conduct the study if eye tracking was integrated into the existing clinical workflow, and what kind of challenges this integration resulted in; d) the ways ML methods were utilized; e) clinical problems of interest. Apart from these core research dimensions, individual details of the paper were analyzed (Tables I, II, III, and IV).
TABLE I.
SUMMARY OF THE PAPERS THAT USED MACHINE LEARNING IN EYE TRACKING IN X-RAY (EXCLUDING MAMMOGRAPHY) IMAGE ANALYSIS
| Reference | Year | Tracker | Anatomy | # cases | # readers | Gaze data form | ML method | Objective |
|---|---|---|---|---|---|---|---|---|
| Wang et al. [66] | 2024 | Gazepoint GP3 | chest | 1038 | 1 | heatmaps | Graph networks | Classification of chest abnormalities |
| Pershin et al. [41] | 2023 | Tobii 4C | chest | 400 | 4 | heatmaps | SimCLR CNN | Using contrastive learning to predict gaze heatmaps and decision error analysis |
| Luís et al. [70] | 2023 | EyeLink 1000 Plus | chest | 2616 | 5 | heatmaps | Mask R-CNN | Improving automated diagnosis by using gaze heatmaps as additional network output |
| Pershin et al. [14] | 2023 | Tobii 4C | chest | 400 | 4 | fixations | U-Net | Mapping lung X-ray coverage for different chest abnormalties with radiologists’ fatigue |
| Bhattachaiya et al. [69] | 2022 | EyeLink 1000 Plus | chest | 2616 | 5 | heatmaps | Student-teacher CNN | Using a student-teacher network trained on X-rays and gaze heatmaps to improve classification |
| Pershin et al. [13,38] | 2022 | Tobii 4C | chest | 400 | 4 | fixations | U-Net | Mapping lung X-ray coverage with radiologists’ fatigue |
| Ma et al. [95] | 2022 | breast, chest | 1580 | 1 | heatmaps | ViT | Improving automated diagnosis by using gaze heatmaps as network output | |
| Kholiavchenko et al. [39] | 2022 | Tobii 4C | chest | 400 | 1 | heatmaps | U-Net | Improving automated diagnosis by using gaze heatmaps as additional network output |
| Zhu et al. [64, 65] | 2022 | Gazepoint GP3 | chest | 1038 | 1 | heatmaps | ResNet, U-Net, EfficientNet_v2 | Improving automated diagnosis by using gaze heatmaps as additional network output |
| Bhattacharya et al. [63] | 2022 | Gazepoint GP3 | chest | 1038 | 1 | heatmaps | ResNet, DenseNet, ViT | Improving automated diagnosis by using gaze heatmaps as network output |
| Lanfredi et al. [68] | 2022 | Eyelink 1000 Plus | chest | 2616 | 5 | heatmaps | CNN | Comapring ML-generated activation maps with gaze maps |
| Lanfredi et al. [67] | 2022 | Eyelink 1000 Plus | chest | 2616 | 5 | path, fixations | database | Public database for machine learning algorithm development |
| Huang et al. [58] | 2021 | EyeTribe | brain, chest | 494/ 1038 | 1 | heatmaps | U-Net, ResNet, DMFNet, MobileNet | Improving automated diagnosis by using gaze heatmaps as network output |
| Karargyris et al. [62] | 2021 | Gazepoint GP3 | chest | 1038 | 1 | heatmaps, fixations | U-Net | Public database with gaze, image, and diagnosis data while reading chest X-rays |
| Wang et al. [97] | 2021 | Tobii 4C | knee | 2000 | 2 | heatmaps | ResNet | Improving automated diagnosis by using gaze heatmaps as network output |
| Saab et al. [44] | 2021 | Tobii Pro Nano | brain, chest | 2794/ 1083 | 1 | heatmaps, paths | ResNet | Improving automated diagnosis by using gaze heatmaps as network output |
| Castner et al. [51] | 2020 | SMI RED250 | dental | 25 | 79 | fixations | VGG-16 | Extracting features from dental patches of visual attention, and predicting the user’s expertise from these patches |
| Pietrzyk et al. [20,21] | 2012 | Tobii x50 | lung | 50 | 8 | fixations | SVM | Using gaze fixation/mouse clicks as seeds for the classification of false negatives and positives |
| Alzubaidi et al. [18] | 2010 | Tobii 1750 | lung | 20 | 5 | heatmaps | SVM | Discovering image features that summon radiologists’ gaze |
Abbreviations: ML (machine learning), CNN (convolutional neural network), ViT (visual image transformer), MR (magnetic resonance), SVM (support vector machine).
TABLE II.
SUMMARY OF THE PAPERS THAT USED MACHINE LEARNING IN EYE TRACKING IN ULTRASOUND (US) IMAGE ANALYSIS
| Reference | Year | Tracker | # cases | # readers | Gaze data form | ML method | Objective |
|---|---|---|---|---|---|---|---|
| Alsharid et al. [40] | 2022 | Tobii 4C | 341 | 10 | heatmaps | VGG-16, LSTM-RNN | Combining gaze and US videos for US captioning |
| Men et al. [99] | 2022 | Tobii 4C | 551 | 17 | fixations | GCGRU, MNetV2 | Predicting gaze and probe movements during US exams |
| Teng et al. [37] | 2022 | Tobii 4C | 366 | 10 | fixations | CNN | Discovering gaze patterns associated with sonographers’ skills when reading US |
| Teng et al. [36] | 2022 | Tobii 4C | 407 | 10 | paths | kNN | Clustering gaze data to improve fixation recognition |
| Savochkina et al. [106] | 2022 | Tobii 4C | 115 | heatmaps | U-Net, LSTM | Automatic generation of gaze heatmap | |
| Teng et al. [35] | 2021 | Tobii 4C | 366 | 10 | paths | GRU | Recognizing the anatomy user looks at |
| Patra et al. [34] | 2021 | Tobii 4C | 366 | 10 | heatmaps | ResNet50 | Applying incremental learning with gaze input for pre-training CNN for predicting US frame content |
| Savochkina et al. [33] | 2021 | Tobii 4C | 45630 frames | heatmaps | U-Net | Automatically generating gaze heatmap | |
| Drukker et al. [32] | 2021 | Tobii 4C | 3000 | 10 | paths | LSTM | Recognizing the depicted anatomy from gaze, image, and transducer data |
| Sharmaetal. [31] | 2021 | Tobii 4C | 380 | 12 | pupil features | SVM, ResNet | Recognizing the depicted anatomy and user experience from gaze data |
| Sharma et al. [30] | 2021 | Tobii 4C | 370 | 12 | heatmaps, paths | ResNet | Predicting user experience from US images and gaze data |
| Droste et al. [29] | 2020 | Tobii 4C | 33 | 8 | heatmaps | VGG-16, GRU | Automatically generating gaze heatmap |
| Cai et al. [28] | 2020 | Tobii 4C | 280 | 1 | heatmaps | VGG-16, LSTM, GRU | Predicting key anatomical planes from gaze heatmap sequences |
| Droste et al. [27] | 2020 | Tobii 4C | 212 | fixations | SE-ResNet | Predicting points of visual attention from US frames | |
| Droste et al. [23] | 2019 | Tobii 4C | 135 | 1 | gaze paths, heatmaps | SE-ResNext, VGG-16 | Automatically generating gaze heatmap |
| Patra et al. [22] | 2019 | Tobii 4C | 60360 | 1 | heatmaps | VGG-16, MobileNet, SqueezeNet | Applying a student-teacher network trained on both US frames and gaze heatmaps to classify the frames |
| Cai et al. [56] | 2018 | EyeTribe | 30 | 1 | heatmaps | GAN | Improving automated diagnosis by using gaze heatmaps as network output |
| Ahmed et al. [55] | 2016 | eyeTribe | 150 | 10 | fixations | adaBoost, RF | Using gaze fixations to identify core regions of standardized US planes |
Abbreviations: ML (machine learning), CNN (convolutional neural network), kNN (k-nearest neighbor), LSTM (Long short-term memory), SVM (support vector machine), GRU (gated recurrent unit), GAN (generative adversarial network), RF (random forest).
TABLE III.
SUMMARY OF THE PAPERS THAT USED MACHINE LEARNING IN EYE TRACKING IN MAMMOGRAPHIC IMAGE ANALYSIS
| Reference | Year | Tracker | # cases | # readers | Gaze data form | ML method | Objective |
|---|---|---|---|---|---|---|---|
| Ji et al. [45] | 2023 | Tobii Pro Nano | 1364 | 1 | heatmaps | CNN | Contrastive learning on two mammography projections for breast masses analysis |
| Lou et al. [53] | 2023 | SMI RED | 196 | 10 | heatmaps | VGG-16, U-Net ResNet, MobileNetV2 | Comparing different algorithms for the automated generation of gaze heatmaps |
| Malletal. [86] | 2019 | ASL H6 | 120 | 8 | fixations | ResNet, Inception v4, VGG | Using fixations to generate regions of different attention and then applying CNNs to predict the user’s opinion about such regions |
| Mall et al. [85] | 2019 | ASL H6 | 102 | 8 | fixations | SVM, SGD, Gradboost, InseptionsNet, ResNet | Using gaze fixations to extract image patches and predicting radiologist’s decision errors from such patches |
| Malletal. [82,84] | 2018 | ASL H6 | 120 | 8 | fixations | InceptionV2 | Using gaze fixations to extract image patches and predict radiological attention level |
| Yoonetal. [81] | 2018 | ASL H6 | 100 | 10 | gaze paths | HMM, CNN, DBN | Converting gaze paths into 2D arrays and classifying target images using these arrays |
| Gandomkar et al. [77–79] | 2017 | ASL H6 | 120 | 8 | fixations | SVM | Classifying false positive and negative tumor detections using gaze as seeds |
| Voisin et al. [54] | 2013 | Mirametrix S2 | 40 | 6 | fixations | SVM, kNN, decision trees | Predicting radiological errors from image patches and gaze features |
| Tourassi et al. [76] | 2013 | ASLH6 | 20 | 6 | fixations | RF, AdaBoost, BayesNet | Predicting radiologists’ attention, decision, and error from gaze and image data |
| Mello-Thoms et al. [87, 88] | 2003 | ASL 4000SU | 40 | 9 | fixations | PLP | Using gaze fixation as seeds for breast lesion classification |
Abbreviations: ML (machine learning), CNN (convolutional neural network), kNN (k-nearest neighbor), SVM (support vector machine), SGD (stochastic gradient descent), HMM (hidden Markov model), DBN (deep belief network), RF (random forest), PLP (parallel level perceptron).
TABLE IV.
SUMMARY OF THE PAPERS THAT USED MACHINE LEARNING IN EYE TRACKING IN FUNDUS PHOTOGRAPHY, HISTOPATHOLOGY, SURGICAL VIDEO, ENDOSCOPY, COMPUTED TOMOGRAPHY (CT), MAGNETIC RESONANCE (MR) AND OPTICAL COHERENCE TOMOGRAPHY (OCT) ANALYSIS
| Reference | Year | Tracker | Anatomy | Image modality | # cases | # readers | Gaze data form | ML method | Objective |
|---|---|---|---|---|---|---|---|---|---|
| Jiang et al. [48] | 2023 | Tobii Pro Spectrum | eye | fundus photography | 1097 | 1 | paths | CNN | Diagnosing diabetic retinopathy and age-related macular degeneration |
| Akerman et al. [107] | 2023 | Pupil Labs Core | eye | fundus photography | 20 | 13 | paths | ID CNN | Classifying the expertise of clinicians whiel reading OCT using statistical features from eye movements |
| Jiang et al. [48] | 2023 | Tobii Pro Spectrum | eye | fundus photography | 1020 | 1 | heatmaps | Inception-V3, ResNet | Improving automated diagnosis by using gaze heatmaps as additional network input |
| Mariam et al. [5] | 2022 | Gazepoint GP3 | oral cavity | histopathology | 4 | 1 | paths | Fast R-CNN, YOLOv3, YOLOv5 | Annotating histopathological image analysis with gaze assistance |
| Hosp et al. [75] | 2021 | Tobii Glasses 2 | shoulder | videos | 150 | 15 | path statistics | SVM | Recognizing the level of expertise from gaze data |
| Xin et al. [46] | 2021 | Tobii X-60 | colon | endoscopic video | 1 | 10 | paths | GAN, LSTM | Predicting loss of navigation during colonoscopy using gaze paths over endoscopic videos |
| Sharma et al. [52] | 2020 | SMI RED | gallbladder | endoscopic videos | 2 | 29 | paths | HMM | Predicting if experts can recognize surgical error using their gaze data |
| Zimmermann et al. [74] | 2020 | SMI Glass 2 | vasculature | videos | 33 | 5 | fixations | Mask R-CNN | Optimizing fluorography use by capturing the surgeon attention on the fluoroscopic screen |
| Pedrosa et al. [26] | 2020 | Tobii 4C | lungs | 3D CT | 1 | 2 | fixations | YOLO | Annotating 3D lung image with gaze assistance |
| Aresta et al. [25] | 2020 | Tobii 4C | lungs | 3D CT | 20 | 4 | fixations | YOLO | Using gaze fixations for lung nodule detection |
| Dmitriev et al. [96] | 2019 | pancreas | 2D CT | 134 | 4 | heatmaps | CNN | Comparing activation maps of tumor classification CNN with gaze maps | |
| Stember et al. [60] | 2019 | Fovio | brain | 2D MR | 356 | 1 | gaze paths | U-Net | Investigating if tumors segmented with gaze can be used for CNN training |
| Dimas et al. [57] | 2019 | EyeTribe | bowel | endoscopic video | 226 | 1 | heatmaps | VGG-16 | Automatically generating gaze heatmap |
| Khosravan et al. [59] | 2018 | Fovio | lung | 3D CT | 6960 nodules | 3 | gaze paths | CNN | Using 3D gaze paths to find regions of attention and applying CNN to detect nodules in such regions |
| Lejeune et al. [80, 83] | 2018 | eyeTribe | endoscopy, brain, eye, cochlea | video, 2D MR, OCT, 2D CT | 4/4/4/4 | 1 | fixations | U-Net | Separating an image into superpixels and classifying superpixels that received the maximum attention |
| Ahmidi et al. [50] | 2012 | RED | sinuses | videos | 1 | 20 | fixations | HMM | Recognizing the level of experience from surgical tool motion and gaze data |
| Thiemjarus et al. [19] | 2012 | Tobii 1750 | gallbladder | endoscopic video | 1 | 3 | fixations | SVM, RF | Recognizing surgical steps from gaze data over endovideos |
| Ahmidi et al. [49] | 2010 | RED | sinuses | videos | 2 | 11 | fixations | HMM | Recognizing surgical tasks from tool motion and gaze data |
| James et al. [17] | 2007 | Tobii 1750 | gallbladder | endoscopic video | 5 | 3 | fixations | PLP | Recognizing surgical steps from gaze data over endovideos |
| Vilariño et al. [71] | 2007 | EyeLink2 | colon | endoscopic video | 6 | 1 | fixations | SVM, SoM | Using gaze data as seed and then classify the resulting image patches to represent polyps |
Abbreviations: ML (machine learning), CNN (convolutional neural network), SVM (support vector machine), GAN (generative adversarial network), HMM (hidden Markov model), RF (random forest), PLP (parallel level perception), SoM (self-organized maps);
III. RESULTS
The initial search resulted in 150, 276, and 56 papers from Scopus, PubMed, and WoS databases, or 446 unique papers after removing duplicated search results. After performing the paper screening, we discarded 121 papers through the abstract analysis, 69 papers – as they did not have an ML component, 207 papers – for not working with medical images, and 98 papers – for having no eye tracking. Note that the same paper could be discarded for several ineligibility reasons. The remaining 33 papers on the topic went through reference cross-inspection, allowing us to include 40 additional papers on the topic. The cross-inspection and the publication list refinement were conducted in April 2023 and December 2023. The summary of the search process results is given in Fig. 1. In the following subsections, we analyzed different aspects of the existing studies, namely a) eye-tracking equipment used in the different studies and how the equipment selection depends on the study aims and requirements; b) clinical problems of interest; c) eye-tracking experiment software; and d) machine learning algorithms used for imaging and gaze data analysis.
A. Eye Tracking Equipment
There are many commercially available eye trackers with the most prominent providers being Tobii, Argus Science, and Gazepoint. Among the studies analyzed in this review, the researchers utilized remote eye trackers, namely Tobii 1750 [17], [18], [19], Tobii x50 [20], [21], Tobii 4C [13], [14], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], Tobii EyeX [42], [43], Tobii Pro Nano [44], [45], Tobii Pro Fusion [13], [38], [39], Tobii X-60 [46], Tobii Pro Spectrum [47], [48] in addition to SMI RED [49], [50], [51], [52], [53], Mirametrix S2 [54], EyeTribe [55], [56], [57], [58], Fovio [59], [60], [61], Gazepoint GP3 [5], [62], [63], [64], [65], [66], EyeLink 1000 Plus [67], [68], [69], [70]; and head-mounted eye trackers EyeLink 2 [71], Mobileye XG [72], ETG 2.7 [73], SMI Glasses 2 [74], Tobii Glasses 2 [75], ASL H6 [76], [77], [78], [79], [80], [81], [82], [83], [84], [85], [86], ASL 4000SU [87], [88].
1) Remote Eye Trackers:
A remote eye tracker represents a device that is installed at the bottom of a monitor or laptop screen oriented towards the user’s face. The data acquisition speed of such eye trackers is high, and the fastest can reach 2000 Hz [89]. Such eye trackers are usually relatively affordable and, therefore, easy to integrate into practice. At the same time, the stationary position of the tracker limits its field of view. The user’s eyes should be inside the trackable range, usually sitting on a chair in front of the screen, while the user should also avoid significant head movements. Being outside the trackable range results in missed eye-tracking records and may compromise the experimental data. For example, King et al. had to remove 24% of the gaze data samples mainly due to the trackable range problem [42]. Some remote eye-trackers require the use of chin support [90], which ensures gaze capturing but compromises the integration of eye-tracking solutions into the existing radiological workflow. The remote eye trackers require calibration to fit the target user and potential recalibrations during lengthy gaze recording experiments. Chatelain et al. [91] recorded gaze-capturing accuracy degradation of 0.13° per month of use for an eye tracker mounted on a cart-based ultrasound (US) scanner. Finally, a single remote eye tracker can only cover one screen, which is in contrast with the multiscreen environment found in the clinical practice of Radiology.
2) Eye-Tracking Glasses:
Head-mounted eye trackers usually represent some form of glasses with a front-facing camera. Head-mounted eye trackers are often more expensive than simple remote eye trackers and record gaze data with much lower frequency than remote eye trackers [89]. The key advantage of eye-tracking glasses is that they are not affiliated with a specific computer screen and can be used in a broader class of clinical applications. One such application studied by Mall et al. [82], [84], [85], [86] is mammography image analysis with two monitors, where monitors are used for craniocaudal and mediolateral oblique views. The authors used convolutional neural networks (CNNs) to predict the level of visual attention received by different breast regions. Head-mounted eye trackers remain the only option in image-guided surgery studies [73], [74], [75]. Hosp et al. [75] analyzed data from Tobii Pro Glasses 2 to classify surgeons’ skills while navigating to anatomical landmarks during shoulder arthroscopy. Zimmerman et al. [74] used SMI Glasses 2 to capture the time periods when the surgeon’s attention moves from the patient to the intraoperative fluoroscopic screen aiming to optimize the use of interventional imaging. Mello-Thoms et al. [92] contrasted the use of a remote eye tracker and a head-mounted eye tracker for the reading of one-view mammograms. They found that, while no changes in the visual search parameters were observed, radiologists reported neck strain when using the remote eye tracker (due to the need to keep the head stable so as to prevent loss of signal). The flexibility offered by the head-mounted eye trackers comes with disadvantages when the image reading happens from the same screen. There is a need to algorithmically estimate the exact location of the user’s attention on the target screen, while for remote eye trackers, this location is much easier to estimate due to the stationary nature of the tracker.
The selection of eye tracking equipment in research studies is based on three questions: if the user continuously sits in front of the screen or expects to move around; how important is the gaze recording speed and if specific features such as pupil diameter are needed. The comparison of eye trackers confirms that tracking glasses are slightly less accurate and way slower than remote eye trackers [93]. Holmqvist compared the performance of 12 eye trackers in terms of gaze data loss and tracking accuracy against various demographic and anatomic parameters of the readers, such as age, eye dominance, and correction glasses strength [94].
B. Clinical Applications
There are three main subdomains targeted by the eye-tracking studies in medical imaging. The first subdomain uses eye tracking to study reader performance. The aim is to discover certain patterns in how physicians read images and connect these discoveries to clinically-valuable conclusions. Physician error prediction has been of particular interest in the community. The researchers investigated the gaze and image patterns that characterize breast lesions overlooked by mammologists [54], [76], [77], [85], [86], [87], [88], navigation loss in colonoscopy [46], surgical errors that result in injury [52], and overlooked lung nodules [21]. Decision errors are closely related to physician training, as eye movements are shown to be significantly different between experts and novices in the fields of endoscopic sinus surgery [49], [50], ultrasound video reading [30], [31], and shoulder arthroscopy [75]. The third area of interest in this subdomain is the recognition of radiological fatigue from eye movements. The existing studies have demonstrated that anatomy coverage deteriorates for various lung abnormalities with the growth of radiological fatigue [13], [14], [38]. This subdomain has shown significant practical potential as eye tracking offers a unique apparatus for how physicians read images.
The second subdomain uses eye tracking as an additional data source for improving ML algorithms. High concentrations of reader attention can pinpoint the diagnostically important image locations, allowing ML to explicitly study such locations when diagnosing lung diseases [18], [59] and breast lesions [95], and detecting fetus anatomy [35], [36], [55]. A variation of this idea is to train ML algorithms to predict gaze data over target images together with the diagnosis of breast lesions [53], [95], lung diseases [68], [69], [70], brain tumors [44], [58], pancreatic tumors[96], diabetic retinopathy [47], and detecting fetus anatomy [29], [33], [40], which allows the algorithms to explicitly focus on anatomically important locations.
The third subdomain employs eye tracking for medical image annotation with the aim to enrich ML training database. Gaze assistance was tested on the annotation of oral carcinoma cells [5], brain tumors [60], [61], bowel centerline [24], and US landmarking [27]. Alsharid et al. [40] combined gaze and US videos to automatically generate US captions.
C. Software for Image Reading With Eye Tracking
While the hardware for eye-tracking research is commercially available, no eye-tracking software fits all medical image analysis studies. Indeed, the researcher needs to implement medical image viewers that will also record gaze data following a specific experimental protocol. Naturally, the in-house developed viewers have limited functionality and cannot match commercially available solutions.
Most of the existing studies work with two-dimensional (2D) images, which significantly simplifies the viewer development (Fig. 2). The simplest viewer for 2D was used by Stember et al. [60], [61], who converted medical images into a common image format and inserted them into PowerPoint slides. Such a viewer lacks any instruments for image enhancement and, more importantly, integrated time recording that is needed to synchronize gaze and screen data. Other researchers who reported how the gaze data was collected, developed custom graphical user interfaces (GUIs) that allowed image transition, decision recording, and potentially some simple image enhancement. Image transition, i.e., indicating when the current image reading is finished and/or the new image reading starts, is usually controlled by GUI or keyboard button pressing [13], [38], [62], [67], [77], [78], [79], [97]. The keyboard-based control is preferred [13], [38], [62], [67], [81], [97] as GUI buttons require the user to move their gaze towards such buttons, which introduces noise to gaze records. The decision recording highly depends on the exact problem of interest. If the decision pool is limited and all decisions are mutually exclusive, they could be recorded using keyboard buttons or mouse clicks [21], [79], [81], [97]. A more universal approach is to ask physicians to vocalize their decision-making and then parse it manually [13], [14], [38], [39] or use ML-based speech processing, e.g., Google’s speech-to-text [62], [67], [98]. The use of voice instead of explicit controller commands is suitable for diffuse diseases that affect significant parts of the target organs but is less applicable for localized abnormalities. For example, a radiologist’s gaze could continue traveling around the image while vocalizing the presence of a small tumor. Complex image enhancements such as zooming are rarely integrated [81], as they result in changes in the field of view, which is not known to the eye tracker, and therefore require complex gaze data post-processing.
Fig. 2.
Breakdown of the papers included in this review by (a) the anatomy depicted in the analyzed medical images and (b) the problem of interest.
A relatively limited number of studies work with three-dimensional (3D) images as 3D gaze recordings require sophisticated image viewers [24], [25], [26], [59]. Indeed, 3D images are usually visualized as three orthogonal cross-sections, so the gaze over all cross-sections must be recorded. In contrast to 2D image reading, where a decision can be made from a single view without a need for image manipulation, 3D image reading involves zooming and slice scrolling that significantly complicates matching the gaze coordinates in the screen-based coordinate space to the gaze coordinates in the patient’s anatomy-based coordinate space. The 3D eye-tracking studies, therefore, impose a strict experiment protocol, for example, physicians could be asked to use only axial cross-section [25], or visually traverse image landmarks for calibration purposes [24]. The research field of 3D eye-tracking in radiology remains largely under-investigated.
D. Machine Learning Applications
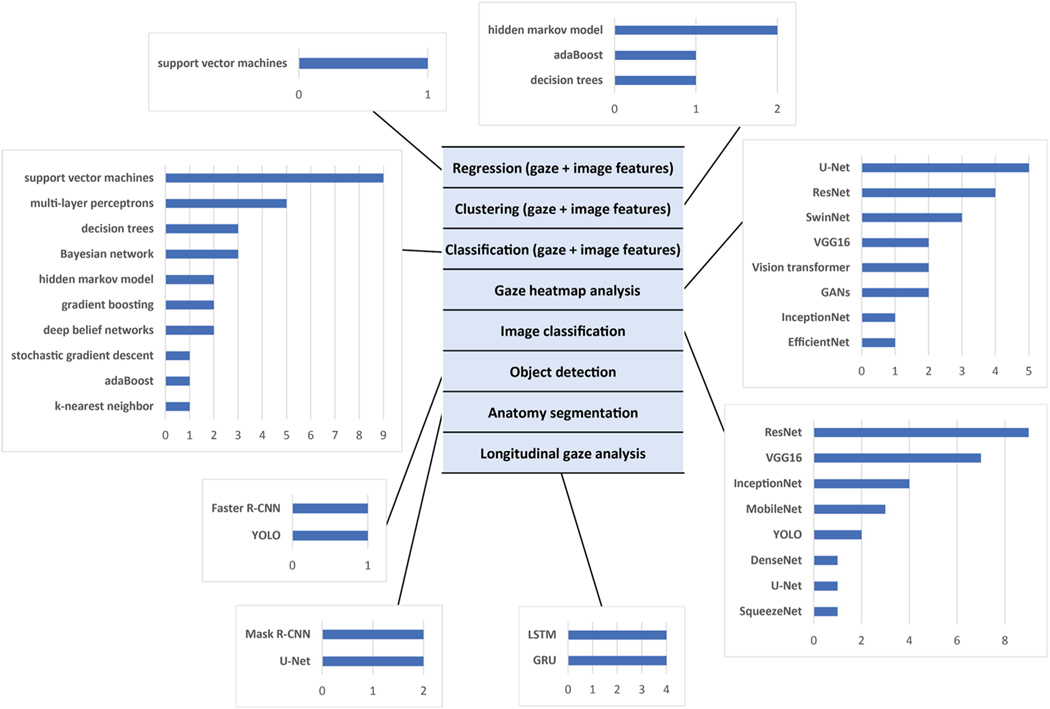
There are two main approaches to how ML is utilized in eye tracking in medical image research. ML can directly analyze gaze data. In such studies, ML methods can, for example, predict physicians’ level of expertise and fatigue and capture the causes of human error. Alternatively, information from physicians’ gaze can improve algorithm training, allowing focus on the key areas of interest in the target images. Finally, ML can analyze medical images as a prerequisite to gaze data processing (Fig. 3). In such studies, ML enables understanding the context of image reading, e.g., whether all areas of importance were sufficiently covered with the gaze.
Fig. 3.
Breakdown of the papers included in this review by problem type and the algorithm used. The selection of the algorithm depends on (a) form of the gaze data, i.e., individual gaze fixation and their features, gaze heatmaps or longitudinal gaze paths; and (b) clinical problem of interest. Machine learning algorithms can be utilized to directly analyze gaze data or to analyze the medical images in eye tracking studies. Note that some papers solve multiple problems using multiple machine learning algorithms.
1) Machine Learning for Image Analysis:
The ML can automatically segment objects of interest from medical images, which then allows for measuring how much visual attention these objects receive [13], [14]. Zimmermann et al. [74] used Mask R-CNN to segment the fluoroscopic screen from SMI Glasses 2 videos and estimated that fluoroscopy use can be reduced by 43% due to surgeon inattention to the screen. Pershin et al. [13], [14] applied U-Net CNN to segment lung fields from chest X-rays and discovered that lung coverage linearly decreases with increasing radiologists’ fatigue. This coverage decrease exhibits the same pattern for different radiologists and different chest abnormalities depicted. The regions of a physician’s attention, defined as gaze fixations or prolonged visual dwells, are used as seed locations for the segmentation of abnormalities [59], [72], [83], and surgical instruments [80]. As attention to abnormalities is not easily separable from attention to surrounding tissues, such studies, therefore, propose complex frameworks based on superpixels [83] and graph analysis [59] for attention refinement.
The physician’s gaze dwells not only over the image regions with pathologies, as in radiological reading the observer has to contrast the “detected” finding with selected areas of the background in order to determine its uniqueness. One can hypothesize that regions of attention have certain image properties, which make them visually distinctive. To discover and quantify image properties that summon attention, researchers compared the numerical image features from regions with high and low visual attention. The comparison was performed with ML assistance. Image features can be predefined, e.g., intensity histograms, Haar-filters, Gabor filters, etc., [18], [71] or automatically extracted from neural network activation maps [51]. The ML mechanisms for the quantitative analysis of image regions of attention opened an avenue for understanding and capturing physicians’ reading errors. Radiologists make false positive (FP) and false negative (FN) diagnostic errors, where FN errors can be due to faulty search when the abnormality is never fixated; perceptual, when an abnormality is not disembedded from the background; or cognitive when the abnormality is seen but not correctly labeled. Mall et al. [82], [84], [85], [86] bridged the diagnostic error nomenclature with eye tracking terminology and defined three types of areas of interest, namely foveal areas of prolonged visual dwelling with multiple fixations, peripheral areas of the short dwelling and never fixated areas. A CNN was able to accurately predict the level of interest mammography regions would receive [76]. In other studies, Mello-Thoms et al. [87], [88] used neural networks to recognize 90% of FN breast cancer lesions, Pietrzyk et al. [20], [21] recognized 90% of FN and FP lung nodules, while Gandomkar et al. [77] showed that support vector machines (SVMs) improved recognition of breast masses for six out of eight radiologists. Despite overall success with FP and FN recognition, CNNs failed to discover image features that lead to perceptual or cognitive FNs [86].
The need for large annotated databases is one of the main bottlenecks of ML-based CAD. The challenges are amplified in ML-based image segmentation, where manual segmentation for algorithm training often requires many minutes and sometimes hours of expert work per image. Can manual segmentation be replaced with gaze-based segmentation? Stember et al. [60] asked a physician to detect tumors in MR images, press the start button, visually traverse the tumor boundary, and press the finish button. The resulting segmentation had an overlap of 0.86 Dice with the manual tumor segmentations. The U-Net CNN for tumor segmentation exhibited similar performance when trained on manual and gaze-based annotations. However, the authors acknowledged that the proposed database generation remains tedious and the participating physician coarsens segmentation masks when getting tired. They simplified the workflow in a follow-up study [61], where segmentation was controlled by voice and not button pressing. The accuracy of such segmentation increased to 92%. Mariam et al. [5] asked a physician to read histopathological images and visually dwell over the keratin pearls they found. The resulting gaze maps were converted to masks and used to train R-CNN and YOLO networks. Gaze-based annotation was more than six times faster than manual annotation, while the performance of networks trained on gaze-based annotations dropped by around 10%.
2) Machine Learning for Gaze Data Analysis:
Gaze data can be analyzed offline or online. They can be in the form of individual gaze points, gaze paths, or accumulated gaze maps, i.e., gaze heatmaps (Fig. 3). The offline analysis usually occurs after the image reading has finished. In contrast, online analysis is performed on a continuous basis during the image reading. The online analysis imposes strict computational requirements as ML algorithms should run in real-time to process gaze changes. Although it is not always specified, most of the studies are designed and executed offline. The online studies mainly belong to the field of image-guided surgery [19], [52], [74], where the decision needs to be made on the fly. Thiemjarus et al. [19] used SVMs and decision trees to recognize laparoscopic cholecystectomy activities from the surgeon’s gaze and instrument movement data. Sharma et al. [52] used hidden Markov models on the surgeon’s gaze over the laparoscopy screen to detect the moment when the surgeon anticipates high risks of bile duct injury. Zimmermann et al. [74] used real-time monitoring of the surgeon’s attention on the fluoroscopic screen during cardiovascular interventions.
While online gaze analysis mainly works with gaze points and gaze paths, offline studies often rely on accumulated gaze maps. A gaze map is a heatmap of the same size as the observed medical image, where colors encode the level of the reader’s attention (Fig. 4(a)). Being 2D image-like arrays, the gaze maps can be used as additional data sources for CNN training and potentially improve the network auto-diagnostic performance. Such a convenient form of gaze heatmaps lead to a straight-forward idea of developing a CNN with two input channels, where the first channel is the target anatomy image, and the second channel is the gaze heatmap from the radiologist who read the target images [47], [70]. Despite being technically simple, such use of gaze heatmaps has relatively limited areas of application. Indeed, the need for a radiologist to first read the target medical image before an ML solution becomes applicable precludes the use of this ML solution in popular research fields such as CAD. The researchers, therefore, investigated several strategies for gaze-assisted CAD where gaze data is only used for CNN training and not required during CNN deployment. The simplest way to achieve this aim is to move gaze heatmaps from CNN input to CNN output [28], [33], [44], [56], [58], [62], [63], [65], [97]. During training, such a CNN both predicts the observed abnormality and generates gaze maps, explicitly learning the areas of high visual attention. When deployed, the CNN predicts diagnosis and generates the expected gaze map, which is usually discarded. An alternative idea is to keep gaze maps for algorithm input and train two different CNNs, where a larger teacher CNN has access to gaze heatmaps while a smaller student CNN does not have access to gaze heatmaps but gets distilled knowledge from the teacher that improves student’s training [22], [63], [69]. Only the student network is then deployed. Following a different approach, Patra et al. [34] first trained a CNN using both gaze maps and anatomy images as input and then fine-turned the network without gaze maps. Ma et al. [95] used gaze maps to mask out regions in the anatomic image that did not receive the reader’s attention so that the network stayed focused on the significant image regions. Finally, Ji et al. [45] employed contrastive learning to match CNN activation maps and the corresponding gaze maps. Even without directly using gaze maps for network training, they can be compared to the CNN activation maps in order to confirm that the network pays attention to the relevant image regions [53], [68], [96].
Fig. 4.
Visualization of gaze data superimposed a chest X-ray read by a radiologist. (a) Gaze heatmaps with parts of the gaze path. (b) Gaze path divided by equal time intervals. (c) Fixation points with the circle size encoding local gaze distribution and color encoding gaze duration.
Several studies applied ML to analyze gaze fixation points during the reading of medical images [17], [54], [76], [99] (Fig. 4(c)). Voisin et al. [54] extracted gaze and image features from mammograms some of which depicted breast masses, and tested 11 ML algorithms on the prediction of radiologists’ diagnostic errors. They found that the errors are much better predicted from the gaze of experienced readers than from novices. Tourassi et al. [76] performed a similar study and confirmed that seven ML algorithms can predict radiological errors in breast mass detection. James et al. [17] applied parallel layer perceptron to recognize surgical steps from gaze data during laparoscopic procedures. Men et al. [99] combined the observed US frame, US probe signal, and sonographer gaze location via graph convolutional Gated Recurrent Units (GRU) to predict the following probe and gaze transformations. Pershin et al. [13] used fixation points to estimate the coverage of lung fields and discovered that the coverage reduction significantly correlated with increased fatigue. In a follow-up study [14], the authors confirmed that the coverage reduction remains invariant to the radiologist’s expertise level as well as to the type of chest abnormality depicted.
While gaze heatmaps and fixation points contain valuable information about image reading, they lack a potentially important longitudinal component that encodes the transition between fixation points that is present in gaze paths [35], [37], [46], [50], [51], [75], [81] (Fig. 4(b)). Yoon et al. [81] observed that neural networks trained only on gaze velocity features are capable of identifying the reading radiologist with 67% accuracy. The gaze paths were also predictive of the reader’s expertise, which was confirmed with 95%-accurate recognition of expert surgeons using tool and gaze movements during sinus surgeries [49], [50], 93%-accurate recognition of expert dentists using panoramic dental radiograph reading [51] and 92%-accurate detection of navigation loss during colonoscopy from trainee gaze movements [46]. In such studies, the gaze paths were sometimes preprocessed by finding clusters of attention [50], consistent path segments that pass through the same anatomical structures [51], or consistent gaze moves such as U-turns and oscillations [37]. As expected, experts exhibit more consistent gaze paths than novices [50], [51].
IV. DISCUSSION
This literature review confirmed a growing interest in the use of ML for physicians’ gaze analysis. It is also clear that the field is in a stage of rapid development, and researchers are probing new technologies and new clinical problems of interest. Indeed, gaze analysis is not limited to the common application of ML-assisted CAD, but extends in radiology to the recognition of physician errors [54], [59], [76], [77], [78], [79], [85], [87], [88], expertise level [30], [49], [50], [51] and confidence level [20], [21], [82], [84], and in surgery to the recognition of expertise level [75], surgical steps [17], [19] and errors [46]. In other words, gaze data provides unique insights into how physicians make their decisions while understating the decision-making process, which may represent a critical link toward explainable and trustworthy ML. The review of the existing papers on the topic confirms an unprecedented set of opportunities for the ML-eye-tracking synergy in radiology and surgery and allows us to identify the core challenges that hamper this research field democratization. We can broadly separate them into hardware, software, and experimental design challenges. To transition eye-tracking research into clinical practice, the benefits of using gaze analysis in a particular task should outweigh the costs of software development, hardware purchase and alterations to the existing workflow. If an eye tracker is challenging to use in clinical conditions, it is unlikely to be adopted irrespective to the potential benefits it could bring. Despite high accuracy and ultrafast recording benefits, remote eye trackers have two significant disadvantages. First, they usually require repeated calibration sessions. Physicians may be reluctant to alter their work routine and complete calibration sessions after every break or distraction. Radiologists spend around 30% of their working day reading the images, so we can expect a relatively high number of reading interruptions and calibration sessions [100]. While calibration may be seen as a minor issue, the limited capture range of remote eye trackers represents a more significant challenge. An eye tracker mounted to a radiological screen can only capture the eyes of the reader in front of the screen. The gaze data may not be recorded if the reader leans back on the chair or looks to a screen from an angle. Such a problem was observed in a number of studies [13], [14], where special sound was installed to alert the participating radiologists if they moved too far away from the screen. A posture restriction cannot be imposed on radiologists in everyday practice. Eye-tracking glasses do not have these issues. Modern glasses, such as Pupil Labs Neon, do not need calibration and are agnostic to the user’s head tilt and position. Moreover, they accommodate snap-on corrective lenses, so even vision-impaired users can use such glasses. The main challenge with eye-tracking glasses is the portable power supply’s limitation, so recording and analysis are usually limited to several hours of use [93]. In general, while the existing studies show the applicability of eye-tracking technology in radiology and surgery, the question of its practical scalability remains under-investigated.
The primary bottleneck of the field is the lack of medical image viewers with incorporated eye-tracking functionality. To the best of our knowledge, all published studies used in-house developed image viewers. Such viewers have limited capabilities and allow no or minimal access to image zooming, enhancement, rotation, etc. While limited functionality may not be as critical in 2D, it represents a significant problem in 3D, where advanced image manipulation tools are essential [24], [25], [59]. The decision-making in 3D relies on non-orthogonal plane or curved planar reconstruction views, where it is difficult to pinpoint which anatomical location the user views at any given moment. Moreover, many studies fail to provide software descriptions, likely due to manuscript space constraints. Without this knowledge, verifying the eye-tracking experiment design and replicating the results is often difficult. Custom image viewers with eye-tracking capabilities cannot be directly translated into clinical practice. Indeed, radiological workstations provide image viewing and manipulation tools, clinical database access, diagnostic recording, network communication, etc. Research groups can hardly develop such solutions in-house, so gaze analysis must eventually be integrated into commercial radiological workstations. Currently, eye-tracking is challenging to incorporate into commercial/free image readers such as Seg3D or 3D Slicer [101], even if such readers provide functionality for in-house developed plug-ins. Indeed, such plug-in functionality is tailored towards image processing and analysis so that a user can add his own segmentation or detection algorithm into a viewer. The eye movement of the user is, however, not limited to the image window and can travel to GUI elements like checkboxes, buttons, sliding bars, and the keyboard. There is, therefore, a need to access both the front and back end of a viewer, which is currently not permitted in commercial solutions. Finally, some eye-tracking applications rely on real-time data analysis, which will impose high requirements on software optimization.
The eye-tracking experiments are very demanding on correct experimental design and execution and software testing. A buggy image analysis algorithm can easily be re-programmed. If an eye-tracking protocol is ill-designed or the gaze recording framework crashes, the whole experiment can be compromised. Re-running a partially completed experiment usually requires a “forgetting” time window for the participants, significantly slowing the research. Although radiologists tend to forget images they interpreted before, especially if they are mixed with previously unseen images, they still may recognize some of the previously seen images [102]. It sometimes may be necessary to conceal the experimental aims from the participants to avoid the Hawthorn effect [61]. In radiological fatigue studies [13], [38], the participants were not informed about the exact experimental aims, to prevent them from involuntarily reading images in a “tired” fashion. To summarize, it is important to carry out dry runs before inviting physicians to check how different participants read the images and communicate with the experiment software.
The field of eye-tracking in radiology and surgery is in a rapid growth stage due to the development of ML methodology, allowing the analysis of unstructured gaze data. Many papers in this review address unique challenges limiting the possibility of comparing the reported results. Recognition of physician’s errors from eye movements over mammograms is one of the exceptions [87], [103], [104], [105]. The existing studies on the topic have demonstrated that the radiological decisions on the patches extracted from the locations fixated by the observers can be predicted with up to 90% accuracy, with the highest accuracy achieved for true positive (97%), false positive (91%) and false negative (90%) patches [87], [104]. These numbers remain consistent for various algorithm types, including SVM, gradient boosting, and shallow and deep neural networks. Classification of lung pathologies using gaze as an additional data source is the second challenge that received considerable attention [62], [64], [69], [95]. The existing studies proposed to train neural networks to predict both abnormalities from chest X-rays and physician gaze maps, or to use gaze maps to train teacher networks in the student-teacher classification framework. Both approaches allow for improvements in classification accuracy by 2–4%. One of the possible explanations for the community’s attention to this particular problem of eye tracking in chest X-ray CAD is the availability of public gaze databases [62].
The literature analysis allows us to outline and assess the developments in this research field. One of the clear developments is the change in the database sizes - a well-known bottleneck of ML in medicine. The problem with database generation is amplified in eye-tracking studies, where, apart from collecting medical images and clinical diagnostic records, physicians need to visually read the images. The early studies on the topic published until 2015 worked with small-size databases with a median number of patients of 21. The median database grew to 127 patients for papers published from 2016 to 2019, and 400 patients for the recent papers published from 2020. Since several physicians can read a single image multiple times, the total number of case-readings varied from 10 to 4400. The recent growth of the database size has been additionally stimulated by the release of two public databases, namely the IBM database of 1083 X-ray readings by one radiologist [62] and the REFLACX database of 3032 X-ray readings by five radiologists from 2616 patients [67] (Table v). Both databases work with chest X-rays with reference diagnoses extracted from public sources. Moreover, both databases follow a similar diagnostic protocol where the radiologists dictate the diagnoses during image reading. These public databases democratized the eye-tracking research for chest X-rays with eight papers published using IBM [58], [63], [64], [65], [66], or REFLACX data [68], [69], [70]. Not only does the number of patients analyzed grow, but the number of physicians who read the images also grows. While there are standards for reading medical images, e.g., ABCD for lung fields, each physician reads differently. Among the reviewed papers, the number of observers ranged from 1 to 79, with 5 being the median. The human performance studies empirically confirmed the need for multiple observers by discovering the differences between experts and novices [50], [51]. The median number of the total image readings per study has increased from 185 for early studies to 1600 for the most recent studies. Considering that collecting eye-tracking data is expensive, researchers apply data augmentation to enrich moderate-size databases. Several unique strategies for data augmentation are applicable in gaze analysis research. The first idea is to modify fixation point calculation by employing any of the dozen alternative algorithms [10]. Fixations can be based on simple gaze velocity thresholding or adaptive analysis that better handles smooth pursuit eye movements [106]. Each algorithm can generate a slightly different set of fixation points and, consequently, slightly different gaze heatmaps. The second strategy is to augment 2D gaze maps using rigid and non-rigid deformations [33]. We can anticipate that the trend of growing database size enriched with augmentations will continue in the future. We also expect the occurrence of guidelines for eye-tracking database collection similar to the protocols used by IBM [62] and REFLACX [67] database creators.
TABLE V.
SUMMARY OF TWO PUBLICLY AVAILABLE DATABASES WITH GAZE RECORDINGS OF RADIOLOGISTS WHILE READING MEDICAL IMAGES. BOTH DATABASES WORK FOCUSED ON DIAGNOSTIC CHEST RADIOGRAPHY
| REFLACX database | |
|---|---|
| Modality | X-ray |
| Anatomy | Chest |
| Task | Diagnose lung diseases |
| # cases | 2616 |
| # readers | 5 |
| # readings (total) | 3032 |
| Gaze paths | Yes |
| Pupil properties | Yes |
| Manual organ annotation | Bounding boxes |
| IBM database | |
|
| |
| Modality | X-ray |
| Anatomy | Chest |
| Task | Diagnose lung diseases |
| # cases | 1083 |
| # readers | 1 |
| # readings (total) | 1083 |
| Gaze paths | Yes |
| Pupil properties | Yes |
| Manual organ annotation | Segmentations |
Another development of this research field goes to the changes in the utilized ML methodology and how gaze data is processed. In the early studies, gaze data was almost always analyzed as individual fixation points and their features. The researchers mainly used SVM, RF, and shallow neural networks. In 2016, the methodology moved to utilize deep neural networks exclusively. From 2016 to 2020, the most common way of using deep neural networks is to classify image patches at the location of fixation points with CNNs. This was mainly justified by the limited database size so that every image was essentially converted into multiple data samples according to the number of fixations. The growth of the database size and the development of CNN methodology allowed researchers to analyze gaze heatmaps. They will become the prime focus for CNNs from 2021. The most recent papers exhibit the increased utilization of longitudinal components in gaze that is captured with recurrent neural networks [29], [35], [46], [98], [107], [108]. The longitudinal component has been augmented with image and heatmap analysis, resulting in a hybrid combination of recurrent and convolutional neural networks [33], [40], [99]. Two recent papers capitalize on the contrastive learning concept, trying to optimize the difference between CNN activation maps and gaze heatmaps [41], [45]. Wang et al. [66] have recently tested the performance of a graph neural network on gaze paths for chest X-ray reading classification. Eye tracking with ML in 3D currently remains a white spot. None of the few papers working with gaze analysis in 3D proposed any ML solution that capitalizes on the 3D nature of gaze paths [24], [25], [26], [59]. The focus is on the technology and protocols for gaze recording in 3D, while ML in the form of YOLO or autoencoder CNNs serves to analyze 3D image patches that receive readers’ attention. We can anticipate that more studies will be published on the topic of 3D image reading. Overall, there is a tendency to adapt to the recent developments in neural networks and perform multimodal analysis of gaze features, paths, heatmaps, and target images.
A. What is the Future of Eye-Tracking Studies
For the longest time, eye trackers were considered laboratory devices due to their cost and complexity, and collecting data from a large pool of subjects was unfeasible due to the slow data collection process, the requirement for subjects to be present in the lab for data collection, and costs. Therefore, data sets generated by eye position recording tended to be significantly smaller than those required for ML tasks. This small dataset size caused issues for training data-intensive algorithms and may have led to overfitting when the algorithm was being evaluated. The recent technology developments and affordability of equipment such as webcam-based eye trackers have contributed to the democratization of this field [109], [110], [111], [112]. Some of the reasons why moving to an online webcam platform for data collection include but are not limited to access to a broader and more diverse population, which can better generalize results, ability to collect data in parallel, independence of established hardware or software, and overall lower costs due to smaller time involvement for both researchers and participants [113]. For example, Darici et al. [114] have used webcam-based eye tracking to measure the visual expertise of medical students during histology training. Sudoke et al. [115] have used it to enhance the visual situational awareness of trainees in anesthesiology. Khosravi et al. [116] have compared eye-tracking glasses and a webcam-based one to monitor self-learning, and they have found that the low-cost webcam solution, combined with ML, can achieve good accuracy relative to the glasses. Meng and Zhao also used CNNs to correct data from a webcam-based eye-tracker [117]. Kaduk et al. [118] compared a webcam-based eye tracker to a remote tracker, EyeLink 1000, with promising results.
Hence, the question becomes: will webcam-based eye trackers replace head-mounted or remote ones? The answer, for the most part, is no. However, webcam-based eye trackers will be the solution when precision and accuracy requirements are not very strict, for example, in applications where it’s important to know the general vicinity of where subjects are looking. For more detailed applications, like determining if a subject is looking at calcifications in a mammogram or digital breast tomosynthesis scan, we still need the precision and accuracy of head-mounted or remote eye trackers.
B. Considerations for Future Eye-Tracking Studies
Reproducibility is a paramount part of research that allows discoveries to stand the test of time [119]. The scientific field studied in this review is rapidly growing, with many papers targeting unique clinical problems of interest. Critical study components are sometimes omitted in the paper descriptions, making them difficult to replicate. One way of increasing reproducibility is introduction of standard guidelines.
Several guidelines exist for conducting eye-tracking studies in the behavioral sciences [120], [121]. These guidelines are universal and applicable to all the studies reported in our review. For example, the guidelines require an eye-tracking study to report the type of equipment used, description of the eye-tracking environment, definition of the events of interest, data collection protocol, study population, potential exclusion of participants due to eye-related diseases, eye-tracker calibration, etc. However, several new recommendations can be specific for medical imaging studies. The first recommendation goes to image reading software development. The existing guidelines [120] also emphasize the importance of comprehensive software documentation, but they mainly focus on the reporting of gaze data processing protocols such as data extraction, conversion, and normalization, i.e., data preprocessing. In radiology, diagnosing is a lengthy, complex process where eye movements related to image reading are mixed with eye movements related to decision making, software navigation, 3D image slice scrolling, etc. Researchers may therefore report the measures that were taken to ensure that diagnostic gaze data is properly refined, and the software is optimized towards the eye-tracking task. The second recommendation goes to the number of participants of the studies. Among the papers analyzed in the review, 19 work with eye movements collected from a single reader, while four papers do not specify the number of readers. Eye-tracking studies have confirmed significant variability in inter-observer visual congruency for images with low scene complexity and a small number of depicted objects of interest [122]. The researchers are advised to include at least two readers and explain how they assess reproducibility. Finally, there is a need to report which algorithm was used for fixation calculation. In summary, future researchers are advised to a) follow the existing guidelines for eye-tracking research [120], [121]; b) report data collection and user training protocols; c) run experiments with multiple observers; and d) publish the code associated with eye-tracking data calculation and potentially a data sample of image reading.
V. CONCLUSION
In this review, we summarized the existing ML frameworks that have been developed for eye-tracking analysis for medical imaging. The need for in-house developed software, sufficiently large databases, and multiple observers are the main obstacles to the democratization of this field of research. With very few exceptions, the proposed solutions are not interactive, i.e., the ML analyzes eye tracking data after the image is read, and it does not communicate with the physician while they read the image. The ML-physician interaction based on gaze data is a likely direction for advancing this field of research.
Acknowledgments
This work was supported in part by the Novo Nordisk Foundation under Grant NNF20OC0062056 and in part by the National Institutes of Health under Grant 1R01CA259048.
Contributor Information
Bulat Ibragimov, Department of Computer Science, University of Copenhagen, 2100 Copenhagen, Denmark.
Claudia Mello-Thoms, Department of Radiology, University of Iowa, Iowa City, IA 52242 USA.
REFERENCES
- [1].Bajwa J, Munir U, Nori A, and Williams B, “Artificial intelligence in healthcare: Transforming the practice of medicine,” Future Healthcare J, vol. 8, no. 2, pp. e188–e194, Jul. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sezgin E, “Artificial intelligence in healthcare: Complementing, not replacing, doctors and healthcare providers,” Digit. Health, vol. 9, Jul. 2023, Art. no. 20552076231186520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Humphreys PC et al. , “A data-driven approach for learning to control computers,” in Proc. Int. Conf. Mach. Learn., 2022, pp. 9466–9482. [Google Scholar]
- [4].Samiei S, Ahmad A, Rasti P, Belin E, and Rousseau D, “Low-cost image annotation for supervised machine learning. Application to the detection of weeds in dense culture,” in Proc. Brit. Mach. Vis. Conf. Comput. Vis. Problems Plant Phenotyping, 2018. [Google Scholar]
- [5].Mariam K. et al. , “On smart gaze based annotation of histopathology images for training of deep convolutional neural networks,” IEEE J. Biomed. Health Inform, vol. 26, no. 7, pp. 3025–3036, Jul. 2022. [DOI] [PubMed] [Google Scholar]
- [6].van der Gijp A. et al. , “How visual search relates to visual diagnostic performance: A narrative systematic review of eye-tracking research in radiology,” Adv. Health Sci. Educ. Theory Pract, vol. 22, no. 3, pp. 765–787, Aug. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Drew T, Vo ML-H, Olwal A, Jacobson F, Seltzer SE, and Wolfe JM, “Scanners and drillers: Characterizing expert visual search through volumetric images,” J. Vis., vol. 13, no. 10, Aug. 2013, Art. no. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Krupinski EA, “Perceptual factors in reading medical images,” in The Handbook of Medical Image Perception and Techniques. Cambridge, U.K.: Cambridge Univ. Press Dec. 2018, pp. 81–90. [Google Scholar]
- [9].Hessels RS, Niehorster DC, Nyström M, Andersson R, and Hooge ITC, “Is the eye-movement field confused about fixations and saccades? A survey among 124 researchers,” Roy. Soc. Open Sci, vol. 5, no. 8, Aug. 2018, Art. no. 180502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Andersson R, Larsson L, Holmqvist K, Stridh M, and Nyström M, “One algorithm to rule them all? An evaluation and discussion of ten eye movement event-detection algorithms,” Behav. Res. Methods, vol. 49, no. 2, pp. 616–637, Apr. 2017. [DOI] [PubMed] [Google Scholar]
- [11].Brunyé TT, Drew T, Weaver DL, and Elmore JG, “A review of eye tracking for understanding and improving diagnostic interpretation,” Cogn. Res.: Princ. Implic., vol. 4, Feb. 2019, Art. no. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lago MA, Sechopoulos I, Bochud FO, and Eckstein MP, “Measurement of the useful field of view for single slices of different imaging modalities and targets,” J. Med. Imag, vol. 7, no. 2, Mar. 2020, Art. no. 022411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pershin I, Kholiavchenko M, Maksudov B, Mustafaev T, Ibragimova D, and Ibragimov B, “Artificial intelligence for the analysis of workload-related changes in radiologists’ gaze patterns,” IEEE J. Biomed. Health Inform, vol. 26, no. 9, pp. 4541–4550, Sep. 2022. [DOI] [PubMed] [Google Scholar]
- [14].Pershin I, Mustafaev T, Ibragimova D, and Ibragimov B, “Changes in radiologists’ gaze patterns against lung X-rays with different abnormalities: A randomized experiment,” J. Digit. Imag, vol. 36, pp. 767–775, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Krupinski EA, “Visual scanning patterns of radiologists searching mammograms,” Acad. Radiol, vol. 3, no. 2, pp. 137–144, Feb. 1996. [DOI] [PubMed] [Google Scholar]
- [16].Kundel HL, Nodine CF, and Krupinski EA, “Computer-displayed eye position as a visual aid to pulmonary nodule interpretation,” Invest. Radiol, vol. 25, no. 8, pp. 890–896, Aug. 1990. [DOI] [PubMed] [Google Scholar]
- [17].James A, Vieira D, Lo B, Darzi A, and Yang GZ, “Eye-gaze driven surgical workflow segmentation,” in Proc. Med. Image Comput. Comput.-Assist. Interv.: Int. Conf. Med. Image Comput. Comput.-Assist. Interv, 2007, pp. 110–117. [DOI] [PubMed] [Google Scholar]
- [18].Alzubaidi M, Balasubramanian V, Patel A, Panchanathan S, and Black JA Jr, “What catches a radiologist’s eye? A comprehensive comparison of feature types for saliency prediction,” Proc. SPIE, vol. 7624, pp. 262–271, Mar. 2010. [Google Scholar]
- [19].Thiemjarus S, James A, and Yang G-Z, “An eye–hand data fusion framework for pervasive sensing of surgical activities,” Pattern Recognit, vol. 45, no. 8, pp. 2855–2867, Aug. 2012. [Google Scholar]
- [20].Pietrzyk MW, Donovan T, Brennan PC, Dix A, and Manning DJ, “Classification of radiological errors in chest radiographs, using support vector machine on the spatial frequency features of false-negative and false-positive regions,” Proc. SPIE, vol. 7966, pp. 89–99, 2011. [Google Scholar]
- [21].Pietrzyk MW, Rannou D, and Brennan PC, “Implementation of combined SVM-algorithm and computer-aided perception feedback for pulmonary nodule detection,” Proc. SPIE, vol. 8318, pp. 2855–335, 2012. [Google Scholar]
- [22].Patra A. et al. , “Efficient ultrasound image analysis models with sonographer gaze assisted distillation,” in Proc. Med. Image Comput. Comput.-Assist. Interv.: Int. Conf. Med. Image Comput. Comput.-Assist. Interv, 2019, pp. 394–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Droste R. et al. , “Ultrasound image representation learning by modeling sonographer visual attention,” in Proc. Inf. Process. Med. Imag. Proc. Conf, Jun. 2019, pp. 592–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lucas A, Wang K, Santillan C, Hsiao A, Sirlin CB, and Murphy PM, “Image annotation by eye tracking: Accuracy and precision of centerlines of obstructed small-bowel segments placed using eye trackers,” J. Digit. Imag, vol. 32, no. 5, pp. 855–864, Oct. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Aresta G. et al. , “Automatic lung nodule detection combined with gaze information improves radiologists’ screening performance,” IEEE J. Biomed. Health Inform, vol. 24, no. 10, pp. 2894–2901, Oct. 2020. [DOI] [PubMed] [Google Scholar]
- [26].Pedrosa J. et al. , “LNDetector: A flexible gaze characterisation collaborative platform for pulmonary nodule screening,” in Proc. XV Mediterranean Conf. Med. Biol. Eng. Comput., 2020, pp. 333–343. [Google Scholar]
- [27].Droste R, Chatelain P, Drukker L, Sharma H, Papageorghiou AT, and Noble JA, “Discovering salient anatomical landmarks by predicting human gaze,” in Proc. IEEE 17th Int. Symp. Biomed. Imag., 2020, pp. 1711–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Cai Y. et al. , “Spatio-temporal visual attention modelling of standard biometry plane-finding navigation,” Med. Image Anal, vol. 65, Oct. 2020, Art. no. 101762. [DOI] [PubMed] [Google Scholar]
- [29].Droste R, Cai Y, Sharma H, Chatelain P, Papageorghiou AT, and Noble JA, “Towards capturing sonographic experience: Cognition-inspired ultrasound video saliency prediction,” in Proc. Annu. Conf. Med. Image Understanding Anal, 2020, pp. 174–186. [Google Scholar]
- [30].Sharma H, Drukker L, Papageorghiou AT, and Noble JA, “Multimodal learning from video, eye tracking, and pupillometry for operator skill characterization in clinical fetal ultrasound,” in Proc. IEEE 18th Int. Symp. Biomed. Imag., 2021, pp. 1646–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sharma H, Drukker L, Papageorghiou AT, and Noble JA, “Machine learning-based analysis of operator pupillary response to assess cognitive workload in clinical ultrasound imaging,” Comput. Biol. Med., vol. 135, Aug. 2021, Art. no. 104589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Drukker L. et al. , “Transforming obstetric ultrasound into data science using eye tracking, voice recording, transducer motion and ultrasound video,” Sci. Rep, vol. 11, Jul. 2021, Art. no. 14109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Savochkina E, Lee LH, Drukker L, Papageorghiou AT, and Noble JA, “First trimester gaze pattern estimation using stochastic augmentation policy search for single frame saliency prediction,” in Proc. Annu. Conf. Med. Image Understanding Anal., 2021, pp. 361–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Patra A. et al. , “Multimodal continual learning with sonographer eye-tracking in fetal ultrasound,” Simplifying Med. Ultrasound, vol. 12967, pp. 14–24, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Teng C, Sharma H, Drukker L, Papageorghiou AT, and Noble JA, “Towards scale and position invariant task classification using normalised visual scanpaths in clinical fetal ultrasound,” Simplifying Med. Ultrasound, vol. 12967, pp. 129–138, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Teng C, Sharma H, Drukker L, Papageorghiou AT, and Noble JA, “Visualising spatio-temporal gaze characteristics for exploratory data analysis in clinical fetal ultrasound scans,” in Proc. Eye Tracking Res. Appl. Symp, 2022, pp. 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Teng C, Lee LH, Lander J, Drukker L, Papageorghiou AT, and Noble JA, “Skill characterisation of sonographer gaze patterns during second trimester clinical fetal ultrasounds using time curves,” in Proc. Symp. Eye Tracking Res. Appl, 2022, pp. 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Pershin I, Kholiavchenko M, Maksudov B, Mustafaev T, and Ibragimov B, “AI-based analysis of radiologist’s eye movements for fatigue estimation: A pilot study on chest X-rays,” Proc. SPIE, vol. 12035, pp. 240–243, Apr. 2022. [Google Scholar]
- [39].Kholiavchenko M, Pershin I, Maksudov B, Mustafaev T, Yuan Y, and Ibragimov B, “Gaze-based attention to improve the classification of lung diseases,” Proc. SPIE, vol. 12032, pp. 77–8, 2022. [Google Scholar]
- [40].Alsharid M, Cai Y, Sharma H, Drukker L, Papageorghiou AT, and Noble JA, “Gaze-assisted automatic captioning of fetal ultrasound videos using three-way multi-modal deep neural networks,” Med. Image Anal, vol. 82, Nov. 2022, Art. no. 102630. [DOI] [PubMed] [Google Scholar]
- [41].Pershin I, Mustafaev T, and Ibragimov B, “Contrastive learning approach to predict radiologist’s error based on gaze data,” in Proc. IEEE Congr. Evol. Comput, 2023, pp. 1–6. [Google Scholar]
- [42].King AJ et al. , “Leveraging eye tracking to prioritize relevant medical record data: Comparative machine learning study,” J. Med. Internet Res, vol. 22, no. 4, Apr. 2020, Art. no. e15876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Visweswaran S. et al. , “Evaluation of eye tracking for a decision support application,” JAMIA Open, vol. 4, no. 3, Jul. 2021, Art. no. ooab059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Saab K. et al. , “Observational supervision for medical image classification using gaze data,” in Proc. Int. Conf. Med. Image Comput. Comput.-Assist. Interv., 2021, pp. 603–614. [Google Scholar]
- [45].Ji C. et al. , “Mammo-net: Integrating gaze supervision and interactive information in multi-view mammogram classification,” in Proc. Int. Conf. Med. Image Comput. Comput.-Assist. Interv., 2023, pp. 68–78. [Google Scholar]
- [46].Xin L. et al. , “Detecting task difficulty of learners in colonoscopy: Evidence from eye-tracking,” J. Eye Movement Res, vol. 14, no. 2, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Jiang H. et al. , “Eye tracking based deep learning analysis for the early detection of diabetic retinopathy: A pilot study,” Biomed. Signal Process. Control, vol. 84, Jul. 2023, Art. no. 104830. [Google Scholar]
- [48].Jiang H, Gao M, Huang J, Tang C, Zhang X, and Liu J, “DCAMIL: Eye-tracking guided dual-cross-attention multi-instance learning for refining fundus disease detection,” Expert Syst. Appl, vol. 243, Jun. 2024, Art. no. 122889. [Google Scholar]
- [49].Ahmidi N, Hager GD, Ishii L, Fichtinger G, Gallia GL, and Ishii M, “Surgical task and skill classification from eye tracking and tool motion in minimally invasive surgery,” in Proc. 13th Int. Conf. Med. Image Comput. Comput.-Assist. Interv., 2010, pp. 295–302. [DOI] [PubMed] [Google Scholar]
- [50].Ahmidi N, Ishii M, Fichtinger G, Gallia GL, and Hager GD, “An objective and automated method for assessing surgical skill in endoscopic sinus surgery using eye-tracking and tool-motion data,” Int. Forum Allergy Rhinol, vol. 2, no. 6, pp. 507–515, Nov. 2012. [DOI] [PubMed] [Google Scholar]
- [51].Castner N. et al. , “Deep semantic gaze embedding and scanpath comparison for expertise classification during OPT viewing,” in Proc. ACM Symp. Eye Tracking Res. Appl, 2020, pp. 1–10. [Google Scholar]
- [52].Sharma C, Singh H, Orihuela-Espina F, Darzi A, and Sodergren MH, “Visual gaze patterns reveal surgeons’ ability to identify risk of bile duct injury during laparoscopic cholecystectomy,” HPB, vol. 23, no. 5, pp. 715–722, May 2021. [DOI] [PubMed] [Google Scholar]
- [53].Lou J. et al. , “Predicting radiologists’ gaze with computational saliency models in mammogram reading,” IEEE Trans. Multimedia, vol. 26, pp. 256–269, 2024. [Google Scholar]
- [54].Voisin S, Pinto F, Morin-Ducote G, Hudson KB, and Tourassi GD, “Predicting diagnostic error in radiology via eye-tracking and image analytics: Preliminary investigation in mammography,” Med. Phys., vol. 40, no. 10, Oct. 2013, Art. no. 101906. [DOI] [PubMed] [Google Scholar]
- [55].Ahmed M. and Noble JA, “An eye-tracking inspired method for standardised plane extraction from fetal abdominal ultrasound volumes,” in Proc. IEEE 13th Int. Symp. Biomed. Imag., 2016, pp. 1084–1087. [Google Scholar]
- [56].Cai Y, Sharma H, Chatelain P, and Noble JA, “Multi-task SonoEyeNet: Detection of fetal standardized planes assisted by generated sonographer attention maps,” in Proc. Int. Conf. Med. Image Comput. Comput.-Assist. Interv., 2018, pp. 871–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Dimas G, Iakovidis D, and Koulaouzidis A, “MedGaze: Gaze estimation on WCE images based on a CNN autoencoder,” in Proc. IEEE 19th Int. Conf. Bioinf. Bioeng., 2019, pp. 363–367. [Google Scholar]
- [58].Huang Y. et al. , “Leveraging human selective attention for medical image analysis with limited training data,” 2021, arXiv:2112.01034. [Google Scholar]
- [59].Khosravan N, Celik H, Turkbey B, Jones EC, Wood B, and Bagci U, “A collaborative computer aided diagnosis (C-CAD) system with eye-tracking, sparse attentional model, and deep learning,” Med. Image Anal, vol. 51, pp. 101–115, Jan. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Stember JN et al. , “Eye tracking for deep learning segmentation using convolutional neural networks,” J. Digit. Imag, vol. 32, no. 4, pp. 597–604, Aug. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Stember JN et al. , “Integrating eye tracking and speech recognition accurately annotates MR brain images for deep learning: Proof of principle,” Radiol.. Artif. Intell., vol. 3, no. 1, Jan. 2021, Art. no. e200047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Karargyris A. et al. , “Creation and validation of a chest X-ray dataset with eye-tracking and report dictation for AI development,” Sci. Data, vol. 8, no. 1, Mar. 2021, Art. no. 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Bhattacharya M, Jain S, and Prasanna P, “RadioTransformer: A cascaded global-focal transformer for visual attention-guided disease classification,” in Proc. Eur. Conf. Comput. Vis, 2022, pp. 679–698. [Google Scholar]
- [64].Zhu H, Rohling R, and Salcudean S, “Multi-task UNet: Jointly boosting saliency prediction and disease classification on chest X-ray images,” 2022, arXiv:2202.07118. [Google Scholar]
- [65].Zhu H, Salcudean S, and Rohling R, “Gaze-guided class activation mapping: Leverage human visual attention for network attention in chest X-rays classification,” in Proc. 15th Int. Symp. Vis. Inf. Commun. Interact., 2022, pp. 1–8. [Google Scholar]
- [66].Wang B. et al. , “GazeGNN: A gaze-guided graph neural network for chest X-ray classification,” in Proc. IEEE/CVF Winter Conf. Appl. Comput. Vis., 2024, pp. 2194–2203. [Google Scholar]
- [67].Bigolin Lanfredi R. et al. , “REFLACX, a dataset of reports and eye-tracking data for localization of abnormalities in chest X-rays,” Sci. Data, vol. 9, no. 1, Jun. 2022, Art. no. 350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Lanfredi RB, Arora A, Drew T, Schroeder JD, and Tasdizen T, “Comparing radiologists’ gaze and saliency maps generated by interpretability methods for chest X-rays,” in Proc. NeurIPS 2022 Gaze Meets ML Workshop, 2022. [Google Scholar]
- [69].Bhattacharya M, Jain S, and Prasanna P, “GazeRadar: A gaze and radiomics-guided disease localization framework,” in Proc. Int. Conf. Med. Image Comput. Comput.-Assist. Interv., 2022, pp. 686–696. [Google Scholar]
- [70].Luís A. et al. , “Integrating eye-gaze data into CXR DL approaches: A preliminary study,” in Proc. IEEE Conf. Virtual Reality 3D User Interfaces Abstr. Workshops, 2023, pp. 196–199. [Google Scholar]
- [71].Vilariño F, Lacey G, Zhou J, Mulcahy H, and Patchett S, “Automatic labeling of colonoscopy video for cancer detection,” in Proc. Iberian Conf. Pattern Recognit. Image Anal, 2007, pp. 290–297. [Google Scholar]
- [72].Khosravan N. et al. , “Gaze2Segment: A pilot study for integrating eye-tracking technology into medical image segmentation,” in Proc. Med. Comput. Vis. Bayesian Graphical Models Biomed. Imag, 2017, pp. 94–104. [Google Scholar]
- [73].Lanzer P, Al-Naser M, Bukhari SS, Dengel A, and Krupinski EA, “Eye tracking in catheter-based cardiovascular interventions: Early results,” J. Med. Imag. Bellingham Wash, vol. 4, no. 3, Jul. 2017, Art. no. 035502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Zimmermann JM et al. , “Quantification of avoidable radiation exposure in interventional fluoroscopy with eye tracking technology,” Invest. Radiol, vol. 55, no. 7, pp. 457–462, Jul. 2020. [DOI] [PubMed] [Google Scholar]
- [75].Hosp B, Yin MS, Haddawy P, Watcharopas R, Sa-ngasoongsong P, and Kasneci E, “Differentiating surgeons’ expertise solely by eye movement features,” in Proc. Companion Pub. Int. Conf. Multimodal Interact., 2021, pp. 371–375. [Google Scholar]
- [76].Tourassi G, Voisin S, Paquit V, and Krupinski E, “Investigating the link between radiologists’ gaze, diagnostic decision, and image content,” J. Amer. Med. Inform. Assoc, vol. 20, no. 6, pp. 1067–1075, Dec. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Gandomkar Z, Tay K, Ryder W, Brennan PC, and Mello-Thoms C, “Predicting radiologists’ true and false positive decisions in reading mammograms by using gaze parameters and image-based features,” Proc. SPIE, vol. 9787, Mar. 2016, pp. 298–307. [Google Scholar]
- [78].Gandomkar Z, Tay K, Brennan PC, and Mello-Thoms C, “A model based on temporal dynamics of fixations for distinguishing expert radiologists’ scanpaths,” Proc. SPIE, vol. 10136, pp. 22–30, 2017. [Google Scholar]
- [79].Gandomkar Z, Tay K, Ryder W, Brennan PC, and Mello-Thoms C, “iCAP: An individualized model combining gaze parameters and image-based features to predict radiologists’ decisions while reading mammograms,” IEEE Trans. Med. Imag, vol. 36, no. 5, pp. 1066–1075, May 2017. [DOI] [PubMed] [Google Scholar]
- [80].Lejeune L, Christoudias M, and Sznitman R, “Expected exponential loss for gaze-based video and volume ground truth annotation,” in Intravascular Imaging and Computer Assisted Stenting, and Large-Scale Annotation of Biomedical Data and Expert Label Synthesis. Berlin, Germany: Springer, 2017, pp. 106–115. [Google Scholar]
- [81].Yoon H-J, Alamudun F, Hudson K, Morin-Ducote G, and Tourassi G, “Deep gaze velocity analysis during mammographic reading for biometric identification of radiologists,” J. Hum. Perform. Extreme Environ, vol. 14, no. 1, Jan. 2018, Art. no. 3. [Google Scholar]
- [82].Mall S, Brennan PC, and Mello-Thoms C, “A deep (learning) dive into visual search behaviour of breast radiologists,” Proc. SPIE, vol. 10577, pp. 33–43, 2018. [Google Scholar]
- [83].Lejeune L, Grossrieder J, and Sznitman R, “Iterative multi-path tracking for video and volume segmentation with sparse point supervision,” Med. Image Anal, vol. 50, pp. 65–81, Dec. 2018. [DOI] [PubMed] [Google Scholar]
- [84].Mall S, Brennan PC, and Mello-Thoms C, “Modeling visual search behavior of breast radiologists using a deep convolution neural network,” J. Med. Imag. Bellingham Wash, vol. 5, no. 3, Jul. 2018, Art. no. 035502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Mall S, Krupinski E, and Mello-Thoms C, “Missed cancer and visual search of mammograms: What feature-based machine-learning can tell us that deep-convolution learning cannot,” Proc. SPIE, vol. 10952, pp. 281–287, 2019. [Google Scholar]
- [86].Mall S, Brennan PC, and Mello-Thoms C, “Can a machine learn from radiologists’ visual search behaviour and their interpretation of mammograms-a deep-learning study,” J. Digit. Imag, vol. 32, no. 5, pp. 746–760, Oct. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Mello-Thoms C, “Perception of breast cancer: Eye-position analysis of mammogram interpretation,” Acad. Radiol, vol. 10, no. 1, pp. 4–12, Jan. 2003. [DOI] [PubMed] [Google Scholar]
- [88].Mello-Thoms C, Dunn SM, Nodine CF, and Kundel HL, “The perception of breast cancers–a spatial frequency analysis of what differentiates missed from reported cancers,” IEEE Trans. Med. Imag, vol. 22, no. 10, pp. 1297–1306, Oct. 2003. [DOI] [PubMed] [Google Scholar]
- [89].Carter BT and Luke SG, “Best practices in eye tracking research,” Int. J. Psychophysiol., vol. 155, pp. 49–62, Sep. 2020. [DOI] [PubMed] [Google Scholar]
- [90].Moreira C, Nobre IB, Sousa SC, Pereira JM, and Jorge J, “Improving X-ray diagnostics through eye-tracking and XR,” in Proc. IEEE Conf. Virtual Reality 3D User Interfaces Abstr. Workshops, 2022, pp. 450–453. [Google Scholar]
- [91].Chatelain P, Sharma H, Drukker L, Papageorghiou AT, and Noble JA, “Evaluation of gaze tracking calibration for longitudinal biomedical imaging studies,” IEEE Trans. Cybern, vol. 50, no. 1, pp. 153–163, Jan. 2020. [DOI] [PubMed] [Google Scholar]
- [92].Mello-Thoms C. et al. , “Head-mounted versus remote eye tracking of radiologists searching for breast cancer: A comparison,” Acad. Radiol, vol. 13, pp. 203–209, Feb. 2006. [DOI] [PubMed] [Google Scholar]
- [93].Dowiasch S, Wolf P, and Bremmer F, “Quantitative comparison of a mobile and a stationary video-based eye-tracker,” Behav. Res. Methods, vol. 52, pp. 667–680, Apr. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Holmqvist K, “Common predictors of accuracy, precision and data loss in 12 eye-trackers,” in Proc. 7th Scandinavian Workshop Eye Tracking, 2017, pp. 1–25. [Google Scholar]
- [95].Ma C. et al. , “Eye-gaze-guided vision transformer for rectifying shortcut learning,” IEEE Trans. Med. Imag, vol. 42, no. 11, pp. 3384–3394, Nov. 2023. [DOI] [PubMed] [Google Scholar]
- [96].Dmitriev K, Marino J, Baker K, and Kaufman AE, “Visual analytics of a computer-aided diagnosis system for pancreatic lesions,” IEEE Trans. Vis. Comput. Graph, vol. 27, no. 3, pp. 2174–2185, Mar. 2021. [DOI] [PubMed] [Google Scholar]
- [97].Wang S, Ouyang X, Liu T, Wang Q, and Shen D, “Follow my eye: Using gaze to supervise computer-aided diagnosis,” IEEE Trans. Med. Imag, vol. 41, no. 7, pp. 1688–1698, Jul. 2022. [DOI] [PubMed] [Google Scholar]
- [98].Drukker L. et al. , “Transforming obstetric ultrasound into data science using eye tracking, voice recording, transducer motion and ultrasound video,” Sci. Rep, vol. 11, no. 1, Jul. 2021, Art. no. 14109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Men Q, Teng C, Drukker L, Papageorghiou AT, and Noble JA, “Multimodal-GuideNet: Gaze-probe bidirectional guidance in obstetric ultrasound scanning,” in Proc. Int. Conf. Med. Image Comput. Comput.-Assist. Interv., 2022, pp. 94–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Dhanoa D, Dhesi TS, Burton KR, Nicolaou S, and Liang T, “The evolving role of the radiologist: The vancouver workload utilization evaluation study,” J. Amer. College Radiol, vol. 10, no. 10, pp. 764–769, Oct. 2013. [DOI] [PubMed] [Google Scholar]
- [101].Pieper S, Halle M, and Kikinis R, “3D slicer,” in Proc. IEEE 2nd Int. Symp. Biomed. Imag.: Nano to Macro, 2004, pp. 632–635. [Google Scholar]
- [102].Hardesty LA, Ganott MA, Hakim CM, Cohen CS, Clearfield RJ, and Gur D, “‘Memory effect’ in observer performance studies of mammograms1,” Acad. Radiol, vol. 12, no. 3, pp. 286–290, Mar. 2005. [DOI] [PubMed] [Google Scholar]
- [103].Voisin S, Pinto F, Morin-Ducote G, Hudson KB, and Tourassi GD, “Predicting diagnostic error in radiology via eye-tracking and image analytics: Preliminary investigation in mammography,” Med. Phys., vol. 40, no. 10, Oct. 2013, Art. no. 101906. [DOI] [PubMed] [Google Scholar]
- [104].Mall S, Brennan PC, and Mello-Thoms C, “Modeling visual search behavior of breast radiologists using a deep convolution neural network,” J. Med. Imag. Bellingham Wash, vol. 5, no. 3, Jul. 2018, Art. no. 035502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Mall S, Brennan PC, and Mello-Thoms C, “Can a machine learn from radiologists’ visual search behaviour and their interpretation of mammograms-a deep-learning study,” J. Digit. Imag, vol. 32, no. 5, pp. 746–760, Oct. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Larsson L, Nyström M, and Stridh M, “Detection of saccades and postsaccadic oscillations in the presence of smooth pursuit,” IEEE Trans. Biomed. Eng, vol. 60, no. 9, pp. 2484–2493, Sep. 2013. [DOI] [PubMed] [Google Scholar]
- [107].Savochkina E, Lee LH, Zhao H, Drukker L, Papageorghiou AT, and Alison Noble J, “First trimester video saliency prediction using clstmunet with stochastic augmentation,” in Proc. IEEE Int. Symp. Biomed. Imag., 2022, pp. 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Akerman M, Choudhary S, Liebmann JM, Cioffi GA, Chen RWS, and Thakoor KA, “Extracting decision-making features from the unstructured eye movements of clinicians on glaucoma OCT reports and developing AI models to classify expertise,” Front. Med., vol. 10, 2023, Art. no. 1251183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Valenti R, Staiano J, Sebe N, and Gevers T, “Webcam-based visual gaze estimation,” in Proc. Int. Conf. Image Anal. Process., 2009, pp. 662–671. [Google Scholar]
- [110].Xu P, Ehinger KA, Zhang Y, Finkelstein A, Kulkarni SR, and Xiao J, “TurkerGaze: Crowdsourcing saliency with webcam based eye tracking,” 2015, arXiv:1504.06755. [Google Scholar]
- [111].Papoutsaki A, Sangkloy P, Laskey J, Daskalova N, Huang J, and Hays J, “Webgazer: Scalable webcam eye tracking using user interactions,” in Proc. 25th Int. Joint Conf. Artif. Intell., 2016, pp. 3839–3845. [Google Scholar]
- [112].Lim S, Walber T, Schaefer C, and Riehl L, “Webcam eye tracking: Study conduction and acceptance of remote tests with gaze analysis,” 2022, arXiv:2207.14380. [Google Scholar]
- [113].Semmelmann K. and Weigelt S, “Online webcam-based eye tracking in cognitive science: A first look,” Behav. Res. Methods, vol. 50, no. 2, pp. 451–465, Apr. 2018, doi: 10.3758/s13428-017-0913-7. [DOI] [PubMed] [Google Scholar]
- [114].Darici D, Reissner C, and Missler M, “Webcam-based eye-tracking to measure visual expertise of medical students during online histology training,” GMS J. Med. Educ, vol. 40, no. 5, 2023, Art. no. Doc60, doi: 10.3205/zma001642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Sodoké K, Nkambou R, Tanoubi I, and Dufresne A, “Toward a webcam based ITS to enhance novice clinician visual situational awareness,” in Proc. Int. Conf. Intell. Tutoring Syst., 2021, pp. 239–243. [Google Scholar]
- [116].Khosravi S, Khan AR, Zoha A, and Ghannam R, “Self-directed learning using eye-tracking: A comparison between wearable head-worn and webcam-based technologies,” in Proc. IEEE Glob. Eng. Educ. Conf, 2022, pp. 640–643. [Google Scholar]
- [117].Meng C. and Zhao X, “Webcam-based eye movement analysis using CNN,” IEEE Access, vol. 5, pp. 19581–19587, 2017. [Google Scholar]
- [118].Kaduk T, Goeke C, Finger H, and König P, “Webcam eye tracking close to laboratory standards: Comparing a new webcam-based system and the EyeLink 1000,” Behav. Res. Methods, pp. 1–21,Oct. 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Open Science Collaboration, “Estimating the reproducibility of psychological science,” Science, vol. 349, no. 6251, Aug. 2015, Art. no. aac4716. [DOI] [PubMed] [Google Scholar]
- [120].Fiedler S, Schulte-Mecklenbeck M, Renkewitz F, and Orquin JL, “Guideline for reporting standards of eye-tracking research in decision sciences,” 2020. Accessed: Jan. 05, 2024. [Online]. Available: https://osf.io/preprints/psyarxiv/f6qcy [Google Scholar]
- [121].Sharafi Z, Sharif B, Guéhéneuc Y-G, Begel A, Bednarik R, and Crosby M, “A practical guide on conducting eye tracking studies in software engineering,” Empirical Softw. Eng, vol. 25, no. 5, pp. 3128–3174, Sep. 2020. [Google Scholar]
- [122].Meur OL, Baccino T, and Roumy A, “Prediction of the inter-observer visual congruency (IOVC) and application to image ranking,” in Proc. 19th ACM Int. Conf. Multimedia, 2011, pp. 373–382. [Google Scholar]