Abstract
The microbiome encompasses the genomes of the microorganisms that inhabit specific environments. One Health is an emerging concept, recognised as a cohesive, harmonising approach aimed at sustainably improving the well-being of humans, animals, and the environment. The microbiome plays a crucial role in the One Health domain, facilitating interactions among humans, animals, and the environment, along with co-evolution, co-development, co-metabolism, and co-regulation with their associated humans and animals. In addition, the microbiome regulates environmental health through interactions with plant microbiota, which actively participate in substance cycling (particularly the carbon and nitrogen cycles) and influence the overall energy flow in the biosphere. Moreover, antibiotic resistance genes present in microbiota can lead to widespread drug resistance in both humans and animals. This review explores the impact of the microbiome on humans, animals, and the environment, highlighting the significance of focusing on this field in One Health research.
Keywords: One Health, Microbiome, Co-evolution, Substance cycling
1. Background
One Health is defined as an integrated, unifying approach that aims to sustainably balance and optimise the health of people, animals, and ecosystems according to a statement newly launched by the One Health High Level Expert Panel in 2021 [1]. The One Health concept recognises close links and interdependent relationships in the health of humans, domestic and wild animals, plants, and the wider environment (including ecosystems). To address the challenges at the intersection of human, animal, and environmental health, the One Health approach relies on collaboration across multiple sectors, disciplines, and communities [2,3].
The origins of the concept of a close relationship between humans and nature can be linked to ancient China based on publication of the Yellow Emperor's Canon of Internal Medicine, written between the 21st century BC (the China pre-Qin period) and the Han Dynasty [4]. In modern times, the One Health concept has evolved from One Medicine and EcoHealth approach to gradually encompass concerns related to animal and environmental health [5].
The microbiota and microbiome are two significant topics in the field of microbiology. The microbiota refers to all microorganisms present in a specific habitat at a given time, whereas the microbiome represents the collective set of gene sequences (including homologous sequences) within a microbial community in a specific habitat and timeframe [6,7]. Microorganisms are widespread in humans, animals, and the environment. Therefore, studying the microbiota and microbiome offers potential solutions to address One Health issues, and conversely adopting a One Health perspective can also address outstanding questions related to the microbiota and microbiome. Some microorganisms lead to similar diseases in humans, livestock, and pets, suggesting that microbiota act as a connection between clinical human and veterinary medicine [[8], [9], [10]].
As the concept of the microbiome encompasses a broader scope than that of the microbiota, this review mainly focuses on the former to address the connection between microorganisms and One Health at the genetic level. Specifically, we summarise the impact of the microbiome on human, animal, and environmental health, highlighting how the microbiome serves as a unifying factor for One Health.
2. The microbiome and human health
The connection between microorganisms and human health can be traced back to Koch's postulates, introduced in the late 19th century by Robert Koch, the founder of the field of Medical Microbiology [11]. Koch's postulates consist of four key points: (I) a pathogen of certain diseases can be isolated from patients; (II) this pathogen will not be detected in patients with other diseases; (III) the pathogen can cause similar diseases in experimental animals; and (IV) the pathogen can also be isolated from infected experimental animals [12]. Koch's postulates serve as a specific set of criteria for verifying the relationship between pathogens and diseases, representing an advanced scientific approach that guides the exploration of epidemic aetiologies [11]. However, from a modern scientific perspective, Koch's postulates not only ignored the pathogen community (i.e. different pathogens may work together to cause certain diseases) and its relationship to humans, but also overlooked the existence of probiotics and the beneficial role of microorganisms in human health [13].
Some probiotics have functions in the intestines, oral cavity, and skin and have a promoting effect on human, animal, and environmental health. Commonly used probiotics include Lactobacillus spp., Bifidobacterium spp., and Saccharomyces cerevisiae. Additionally, bacteria with beneficial effects on the human body, such as Roseburia spp., Akkermansia spp., Propionibacterium spp., and faecal Bacillus spp., are continuously being developed as probiotics. Bacillus spp. with probiotic effects secrete various antibacterial substances such as organic acids, bacteriocins, and antimicrobial peptides to directly kill harmful bacteria in the surroundings [14].
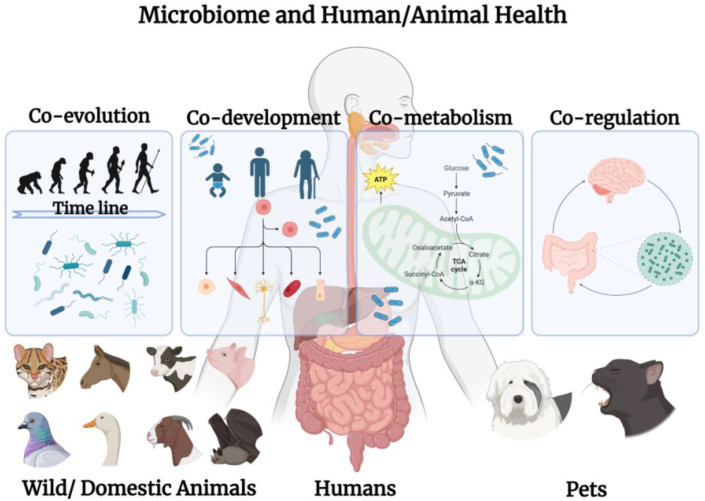
The significance of the microbiome has often been ignored in the development of human medicine, despite recognition of the active roles of microbiota in human and animal activities, resulting in co-evolution, co-development, co-metabolism, and co-regulation (Fig. 1) [7].
Fig. 1.
Interactive models between the microbiome and human health. The microbiome experiences co-evolution, co-development, co-metabolism, and co-regulation with humans and animals (including wild/domestic animals and pets). Co-evolution depicts the evolution timeline of both human/animals and bacteria. Co-development, co-metabolism, and co-regulation represent the interaction of bacteria and individual humans/animals. Bacteria intervene with the development of the immune, nervous, and endocrine systems, as well as host metabolism and regulation among these systems.
Co-evolution between the microbiome and its host has been observed from the perspectives of human and animal evolution. Bacteria have existed for approximately 3.8 billion years, whereas eukaryotes such as humans appeared between 2.2 billion and 2.4 billion years ago. Microorganisms insert genetic fragments into their hosts by interacting with living organisms. Co-evolution of microorganisms with their hosts has resulted in the formation of specific phenotypes within the lineage of eukaryotes [15]. In particular, the intestinal microbiota, which has evolved over millions of years, is a prominent area of research in the field of microbiology. Analysis of the intestinal microbiota of certain primates can provide insights into the origins of human evolution [16].
From an individual standpoint, microbiomes also exert an influence throughout the lifespan of the host. Microorganisms accompany the development of physical functions, and infants are exposed to external microorganisms from birth. The composition of an infant's intestinal microbiota is influenced by vertical transmission from the mother, dietary habits, antibiotic use, and health status. Bifidobacteria, potentially acquired through vertical transmission, play a beneficial role in the growth and development of infants by exerting saccharolytic activity towards glycans abundant in the infant intestine [17].
In addition, the microbiome influences development of the host immune system. It is widely accepted that the initial encounter with microbes occurs at birth, which assists in the differentiation and maturation of T and B cells as well as in the establishment of immune tolerance. The diversity of the gut microbiome, characterised by different antigen types and metabolic characteristics, affects the maturation of CD4+ T cells, leading to variations in the immune system and susceptibility to various diseases among individuals [18].
Furthermore, the microbiome affects the development of the nervous and endocrine systems. The gut microbiome plays a fundamental role in neurogenerative processes such as blood–brain barrier formation, myelination, neurogenesis, microglial maturation, and the regulation of animal behaviours. Therefore, the gut bacteria are considered integral contributors to the development and function of the nervous system [19,20]. Additionally, the gut microbiome regulates ovarian dysfunction and insulin resistance in polycystic ovary syndrome [21], as well as participates in the neuroendocrine regulation associated with depression and obesity [22].
Microbiomes play crucial roles in the metabolism and functional regulation of the body, influencing the nutritional state and lifestyle of the host. The gut microbiota actively participate in the regulation of multiple host metabolic pathways, influence signal transduction and inflammatory response mechanisms, and serve as a vital link between important tissues and organs such as the intestine, liver, muscle, and brain [23]. Microbes are involved in the onset, progression, and metastasis of tumours in the epithelial tissues. Certain tumour therapies, including chemotherapy, radiotherapy, and immunotherapy, are performed with the assistance of microbiomes [24]. For instance, the rate of absorption and bioavailability of many cancer treatments depend on their exposure to host and bacterial enzymes in the gut before entering the circulation. A diet-driven microbiome community plays a role in the response to immune checkpoint inhibitor treatment for melanoma [24]. These findings inspired the application of the diet-microbiome-immune system interaction axis to maximise clinical benefits. Furthermore, evidence from human and animal studies suggests that the composition of the intestinal microbiota may affect the severity of radiation-induced mucosal toxicity [24]. Various studies have demonstrated that regulation of the microbiome on the brain–gut and brain–lung axes plays a role in conditions such as depression, obesity, autism, and other diseases [25].
Therefore, the interactive models between the microbiome and human health can be summarised as co-evolution, co-development, co-metabolism, and co-regulation, all of which have significant impacts on human health. It is important to note that the microbiome not only affects vertebrates, including humans, but also has similar effects on other eukaryotes, including livestock and domestic pets, which are closely associated with humans, as well as various wild animals that come into contact with human society [7]. Owing to the universal correlations between different organisms, microbiome regulation exists in both humans and animals, meaning that changes in human health can have repercussions on animal health.
3. The microbiome and animal health
Microorganisms serve as sources of infection in many zoonotic diseases. Microorganisms that cause infectious diseases typically originate from wild animals. As cities and farms expand, humans encroach on the territories of wild animals, resulting in increased encounters among humans, animals in human societies (livestock and pets), and wild animals, leading to the emergence of infectious diseases. Research conducted on severe acute respiratory syndrome coronavirus (SARS-CoV) supports this view. In 2003, SARS-CoV originated in bats was transmitted to humans through civets and other intermediate hosts [26]. Some studies have also suggested that severe acute respiratory coronavirus-2 (SARS-CoV-2) may originate from bats [27,28]. Middle East respiratory syndrome coronavirus (MERS-CoV) can be transmitted between humans and camels [29]. It is estimated that approximately 61% of human pathogens are zoonotic; therefore, research on microorganisms is valuable for addressing zoonotic issues [7,30].
In the comprehensive study of animal diseases, researchers have gradually recognised the significance of animal medicine, and have shifted their focus towards exploring the prevention and control of infectious diseases from the perspective of animal health rather than merely using animals as tools for clinical medicine. Rudolf Virchow's support for veterinary pathology in the 19th century fostered the connection between animal and human medicine, which introduced the concept of ‘One Medicine’ [31]. Karl Friedrich Meyer further acknowledged that human, animal, and ecosystem health are interdependent, emphasising the need for collaboration among medical professionals to promote the health and well-being of all species; accordingly, he is regarded as a pioneer of the concept of ‘One Health and One Medicine’ [32].
The development of animal vaccines is an exemplary application of microorganisms to animal health. In the 18th century, Edward Jenner, the so-called ‘father of immunology’, successfully prevented and ultimately eradicated smallpox through vaccinia vaccination [33], employing an approach that considered animal health. Louis Pasteur began studying rabies in 1880. After successfully developing an effective human rabies vaccine, a preventive vaccination for rabies in dogs was subsequently established [34]. Pasteurella was innovatively added to the cholera vaccine to effectively suppress the spread of cholera in poultry [34]. Koch utilised quinine as a preventive measure to inhibit protein synthesis in Plasmodium and introduced bed nets to prevent mosquito bites, which had an inhibitory effect on malaria transmission [35].
4. The microbiome and environmental health
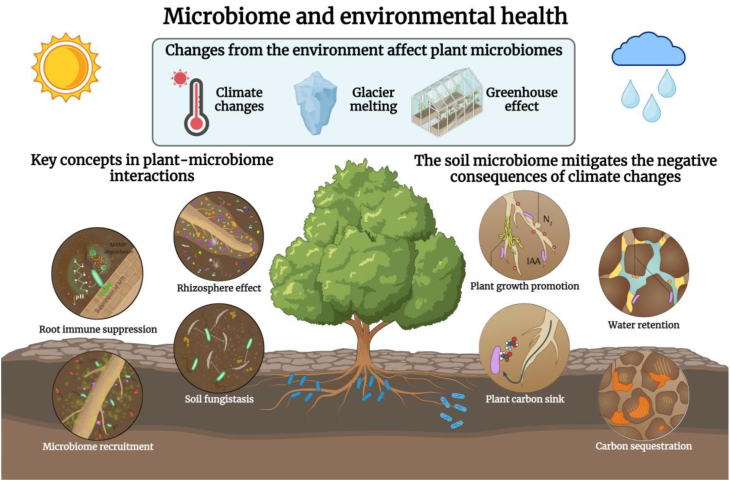
The role of environmental microbiomes in the context of the human-animal-environment interface in the field of health should be emphasised (Fig. 2). Traditionally, little was known about the correlation between microorganisms and environmental changes. However, natural environmental changes such as climate change, sea level rise, and greenhouse effects have undeniable impacts on biodiversity and human health [36,37]. In nature, the microbiome is influenced and adapted to environmental changes, and an altered microbiome, in turn, has adverse effects on environmental health [38].
Fig. 2.
The microbiome and environmental health. Changes from the environment such as climate changes, melting glaciers, and greenhouse gases have impacts on the plant microbiome. These impacts include four key concepts in the plant–microbiome interaction (root immune suppression, rhizosphere effect, microbiome recruitment, and soil fungi stasis). The soil microbiome mitigates the negative consequences of climate changes in four aspects (plant growth promotion, water retention, plant carbon sink, and carbon sequestration) [40,43].
Large-scale environmental changes have a widespread impact on the microbiome of a region, or even the entire world. For example, consider glacial melting. The carbon stored in frozen permafrost is largely preserved because of its low microbial activity. However, when the permafrost thaws, the microbial activity increases, leading to the decomposition of organic carbon. This process releases greenhouse gases such as carbon dioxide and methane into the atmosphere. Additionally, the unique marine environment exerts a distinct influence on the microbial community through nitrification and deoxidation (details are discussed in the following sections) [36,39].
Previous research introduced the concept of the ‘disease triangle’ [37]. This highlights the significance of genetic sensitivity and toxicity to plant health, in which the microbiome plays a critical role. Environmental conditions are also of great importance to the disease triangle. Pathogenesis and symbiosis occur between microorganisms and plants. Research on the rhizosphere microbial community has revealed that the microbial population directly surrounding plant roots is far more abundant than that in the non-rhizosphere soil. This phenomenon is known as the ‘rhizosphere effect’. Plants recruit beneficial bacteria from the soil to assist in growth and self-protection by inhibiting fungi [40]. Moreover, the microbiome influences the plant immune system. For instance, high temperatures can affect the pathogen-related molecular mechanisms of plant immune systems. The immune categories include pathogen-associated molecular pattern-triggered immunity (PTI) and effector-triggered immunity (ETI). PTI and ETI respond to pathogens by activating the production of defence metabolites that orchestrate immune responses [41]. The temperature and humidity of the natural environment also affect the circadian rhythms of plants. For example, humidity changes enhance ETI, as higher humidity levels cluster more effectors in bacteria. High atmospheric humidity significantly influences host–pathogen interactions in plants by creating an aqueous living space that benefits pathogens [40,42].
The soil microbiome can be used to mitigate the negative impacts of climate change. For instance, the soil microbiome helps to alleviate the negative impacts of drought by improving water retention in the soil. As a plant carbon sink, the soil microbiome absorbs carbon output from plant roots through microorganisms, which is stored as cell biomass or transformed into stable metabolites. When a microorganism dies, the carbon within is sealed during soil carbon sequestration. Plant growth-promoting microorganisms can be employed to enhance plant growth in the soil that is negatively affected by climate change [43]. For example, Rhizobium spp. inoculants have been used for biological nitrogen fixation in association with legumes. Some soil bacteria produce extracellular polymeric substances, leading to the formation of hydrophobic biofilms that protect plants from desiccation. Beneficial soil microorganisms can also be harnessed to increase crop tolerance to drought stress by producing phytohormones that stimulate plant growth [43].
5. The microbiome and One Health
Research shows that the microbiome not only plays a role in human health but also affects animal and environmental health. As a result, microbiomes have been included in human, animal, and environmental research, particularly at the intersection of these three areas. Understanding the microbiome has promoted the development of the One Health concept, which integrates human, animal, and environmental health [7].
It is now recognised that microbial genes can be integrated into the genomes of humans, animals, and plants, which has motivated further exploration of the relationship between the microbiome and health from the perspective of material, energy, and information cycles in nature.
5.1. The microbiome participates in the circulation of materials
Carbon and nitrogen cycles are the cornerstones of the One Health framework. In addition, the microbiome is the main promoter of carbon and nitrogen transformation [44], which is associated with many One Health issues such as soil and air pollution. Studies on biological and microbiome processes within the carbon and nitrogen cycles are crucial for addressing climate change, maintaining ecological balance, and promoting soil health.
5.1.1. The microbiome participates in the carbon cycle
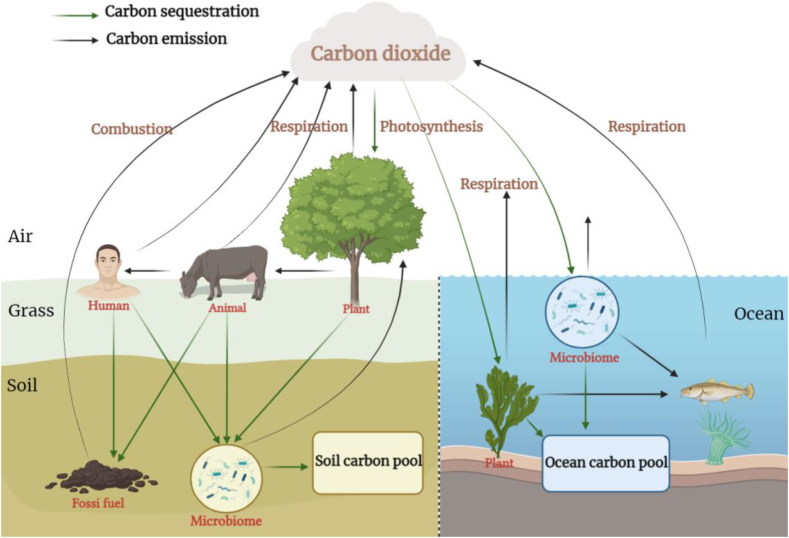
The carbon cycle is one of the most important ecological processes. The microbiome participates in several important metabolic steps of this cycle, such as carbon fixation (converting CO2 into organic matter) and degradation (decomposing organic matter) [7,45] (Fig. 3).
Fig. 3.
The carbon cycle and the microbiome. Carbon dioxide from the atmosphere is sequestered as organic carbon by plants in the oceans and on land. The organic carbon is returned to the atmosphere through biological respiration. Some organic carbon is taken up by the soil microbiome, which is then decomposed to form carbon dioxide and inorganic carbon, providing a source of the soil carbon pool. The carbon cycle in marine ecosystems is similar to that in terrestrial ecosystems.
Microbial carbon fixation is of great significance for advocates of halting the increase in carbon dioxide emissions and subsequently reaching carbon neutrality. Therefore, the mechanism of carbon sequestration into marine and terrestrial carbon sinks has been an area of substantial research focus in recent years [46,47], which includes microbial carbon sequestration as a crucial pathway.
The total carbon storage capacity of the oceans is nearly 4 trillion tons [46], making the oceans the largest active carbon sink on Earth's surface, with marine biological carbon sequestration a critical factor. In addition to the well-known photosynthesising surface microbiome including prokaryotic cyanobacteria and some eukaryotes such as algae, the carbon sequestration process extends deeper into the ocean. Carbon storage mechanisms in the ocean include biological, carbonate, dissolution, and miniature biological carbon pumps [46]. Microbiological pumps related to the microbiome refer to the physiological and ecological processes that transform organic carbon from biodegradable active carbon to inert dissolved organic carbon that is not easily degraded, thus constituting a form of marine carbon storage [48]. The carbon storage mechanism includes active and passive processes, in which the former includes cell metabolism to produce inert compounds and the latter includes inert dissolved organic carbon produced by viral cleavage products and zooplankton feeding metabolism [49].
In the terrestrial carbon sequestration system, the total global carbon storage of soil at a depth of 2 meters is 206 billion tons, which is far greater than that of the combined atmospheric and vegetation carbon sinks. A slight fluctuation in the soil carbon sink can cause significant changes in the atmospheric carbon dioxide concentration [50]. The microbiome plays an important role in carbon sequestration. Stable organic carbon in the soil is composed of keratin, soft wood, waxy lipids, lignin from plants [51], and cytosolic acids and glucosamine from microorganisms [47]. In addition, with the development of soil analysis technology, accumulating evidence has shown that the remains of microorganisms after death are important sources of organic carbon in the soil. For example, researchers found that the average contributions of residual microbial carbon to organic carbon in the surface soils of grasslands, farmlands, and forests were 47%, 51%, and 35%, respectively [52]. Moreover, the carbon sequestration of microbial biomass and metabolites has been described as a microbial carbon pump [53]. Liang et al. [54] proposed this framework in 2017 with the core theory that the soil microbial carbon pump mediates and participates in the formation of organic carbon in the soil, emphasising that microbial decomposers play a role in the decomposition of plant-derived organic carbon. Soil minerals also play a protective role in the accumulation of organic carbon from metabolites and dead residues.
The microbiome plays a pivotal role in the carbon cycle by curtailing carbon dioxide emissions and attaining carbon neutrality. Owing to continued developments in microbiology, our understanding of microorganisms is no longer limited to isolated systems, but instead to a broader concept of the microbiome and its impact on the wider environment [55]. When applied to One Health, climate change affects the soil microbiota, supply of soil carbon, and physical and chemical properties of the soil, subsequently affecting the activity and structure of the microbiome [31], while engendering environmental threats to humans and animals through global warming and the accompanying extreme weather events and rising sea levels. With the deepening of research on carbon sequestration in the environment, microbial carbon sequestration can be used as a means to achieve the goals of carbon dioxide emission peaks and carbon neutrality in the future to further guarantee the survival of humans and animals. This enables considering the impact of the microbiome on the carbon cycle from a macro perspective.
5.1.2. The microbiome participates in the nitrogen cycle
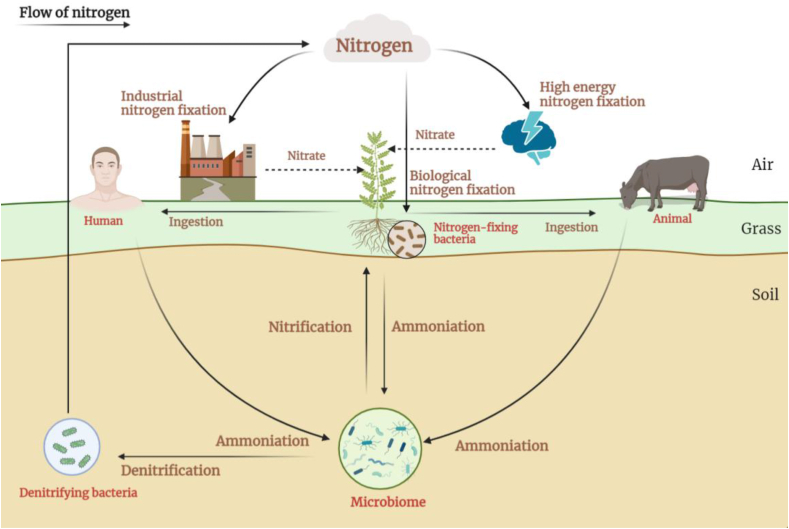
Nitrogen-containing organic matter such as proteins and nucleic acids constitutes an important component of biological organisms. The conversion of organic and inorganic environmental nitrogen is inseparable from the role of the microbiome. The nitrogen cycle refers to the indefinite natural conversion of gaseous, inorganic, and organic nitrogen compounds [56] (Fig. 4).
Fig. 4.
The nitrogen cycle and the microbiome. Nitrogen in the air can be transformed into nitrogenous compounds by biological fixation, high-energy fixation, and industrial fixation; these nitrogenous compounds are then utilized by plants and the microbiome. Nitrogenous compounds flow between humans and animals through the food chain, and they can return to the atmosphere as nitrogen through ammonification, nitrification, and denitrification to complete the nitrogen cycle.
The microbiome is involved in the majority of key nitrogen conversion processes [57]. At present, there are six known types of nitrogen transformations: nitrogen fixation, nitrification, denitrification, assimilation, alienation, and ammonification. Among them, the soil microbiome participates in fixation, ammonification, nitrification, and denitrification [58].
Nitrogen-fixing organisms are categorised into autogenic, symbiotic, and combined nitrogen-fixing bacteria, according to the relationship between the microbiome and higher plants [58]. Most autogenous nitrogen-fixing bacteria synthesise proteins from molecular nitrogen using the ferromolybdenum nitrogenase system. However, symbiotic nitrogen-fixing bacteria can only effectively fix nitrogen if they live in symbiosis with plants. Nitrogen molecules in the atmosphere are fixed by rhizobia and supplied to the legumes for use. Nitrogen molecules are fixed by nitrogen-fixing bacteria into ammonia, which is then oxidised by nitrifying bacteria into nitrate. Plants can absorb these substances, and then inorganic nitrogen is transformed into proteins and other nitrogen-containing organic matter. Finally, the combined nitrogen-fixing bacteria are authigenic and symbiotic in nature. The recently discovered species Azotobacter paspali belongs to this category [58].
Ammonisation is a process in which organic nitrogen compounds are deaminated to form ammonia by ammoniating bacteria such as Clostridium. Nitrification is a process in which nitrifying bacteria and nitrosomes deammoniate amino acids into nitric acid under aerobic conditions. Denitrification is a process in which facultative anaerobic nitrate-reducing bacteria reduce nitrate to nitrogen.
Nitrogen plays contrasting roles in environmental health. Although nitrogenous compounds are essential for plant life, they can cause pollution. Agricultural fertilisers elevate soil nitrogen, which leeches into other areas as pollution and changes the soil biome, leading to many global health problems. In addition, the microbiome and nitrogen cycle have other deleterious effects. For example, nitrous oxide produced by the microbiome accounts for 70% of the total nitrous oxide emission flux [59], and atmospheric nitrous oxide causes global warming. Although the nitrous oxide concentration is relatively low compared to the concentration of other greenhouse gases, its warming effect is 310 times that of carbon dioxide and nitrous oxide is a potent ozone-depleting molecule [60].
The nitrogen cycle is involved in all aspects of One Health. As an important part of the nitrogen cycle, the microbiome plays key roles in several processes. For some environmental issues such as the greenhouse effect, the relationship between the microbiome and nitrogen cycle requires further exploration, especially regarding how environmental factors such as temperature and humidity affect the different metabolic processes of the nitrogen cycle.
5.2. The microbiome and energy flow
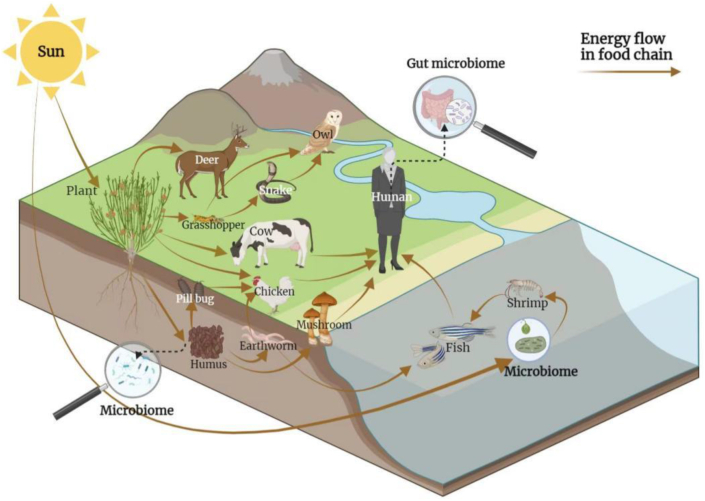
The microbiome is involved in the energy cycle. Solar energy is mainly fixed by plants on land and by algae in water before being passed on to consuming organisms for food. The microbiome is involved in these processes within both the energy fixers and consumers along the energy flow. For example, Huang et al. [61] and Hu [62] proposed that the microbiome decomposes lignin to form humus. The humus is then ingested by subterranean organisms, which are then ingested by other animals to form an energy flow (Fig. 5).
Fig. 5.
The microbiome and energy flow. The microbiome participates in the formation of humus, which is then ingested by earthworms and other soil-dwelling organisms. These animals lower on the trophic chain are then ingested by higher animals to form an energy flow. The microbiome is also involved in energy flow when animals and humans digest food. The intestinal microbiome is involved in animal metabolism, regulating the transformation, absorption, and excretion of nutrients, and affecting the overall nutrient distribution of animals.
The microbiome is also involved in the digestion of food and absorption of energy by animals and humans. The intestinal microbiome regulates the transformation, absorption, and excretion of nutrients in the digestive tract, and affects nutrient distribution [63]. Microbial metabolism of both nutrients and non-nutrients can produce a variety of metabolites with bioregulatory and nutritional effects in the host through metabolic, immune, endocrine, and nerve signalling pathways [64]. Additionally, studies have demonstrated the co-evolution of mammalian digestive flora with their hosts [65].
An example of symbiotic metabolism is Escherichia coli, which, among other factors, uses undigested proteins and carbohydrates to produce biogenic amines, short-chain fatty acids, and ammonia. These metabolites constitute an energy source for host intestinal epithelial cells [66]. After amino acids are utilised by the microbiome, bioamines such as histamine, tryptamine, and tyramine are produced through decarboxylation, and ammonia is produced through deamination [67], which regulates colon peristalsis and intestinal barrier function.
In a non-nutrient metabolic process, some plant components ingested by humans and animals, such as soybean isoflavones, can be converted into equol by the intestinal microbiome and therefore play a regulatory role in the body [68].
Thus, the microbiome plays an important role in overall energy pathways. In addition, in the environment, the microbiome can help to advance research on the development of new energy sources aimed at tackling atmospheric problems caused by the excessive burning of fossil fuels [[69], [70], [71], [72]].
Fossil fuel depletion and air and water pollution are some of the main contemporary issues facing humanity. Due to the continued reliance on fossil fuels, polluting gases are discharged into the atmosphere, causing acid rain, ocean acidification, and global warming. Unsustainable industrialisation pollutes water resources, resulting in severe ecological problems. At present, the proportion of energy consumption in sewage treatment approaches 3% [69]; therefore, microbial electrochemical systems, as a new form of energy-conversion equipment that can be used in both energy generation and wastewater treatment, have attracted widespread attention [69].
A classical microbial electrochemical system consists of an anode, a cathode, and an ion exchange membrane [70]. At the anode, the microorganism oxidises and degrades organic matter in the sewage. At the cathode, the microorganism catalyses the transfer of electrons to ions through a reduction reaction to produce water, biofuels (H2 and CH4), and other chemicals with high economic value. The intermediate ion-exchange membrane separates and prevents interactions between the two stages. In the past 20 years, microbial fuel cells have developed rapidly, and substantial research has been carried out in this field in terms of optimising the electrode materials, electrode structure, reactor structure, and system amplification [71,72].
Although there is still a long way to go before the practical application of microbial electrochemical systems, humans have already taken the steps towards realising biological power generation. In general, with regards to environmental protection, microbial productivity has achieved gratifying results; however, there are still some key technical problems to be solved by scientists on how to improve productivity. For example, the available electricity generation power is too low and appropriate power collection technology is lacking.
As part of the current goal of a shared future for humankind, it is essential solve ecological and environmental problems caused by excessive fossil fuel burning and emissions as soon as possible. Based on the research summarised above, it is clear that microbiota promote energy conversion between organisms; thus, microbial fuel cells have great potential in the development of new energy sources and the utilisation of waste resources.
5.3. The microbiome and genetic material
Antimicrobial resistance is a growing concern in the health domain involving humans, animals, and the environment. Although antibiotics are crucial tools in treating bacterial infections and in agriculture, their overuse has led to the development of antibiotic-resistant bacteria (ARB) and antibiotic resistance genes (ARGs) [73]. The increasing emergence of ARB has created a grim picture of bacterial resistance. Particularly after the ‘superbug’ with the blaNDM-1 gene was reported in The Lancet in 2010 [74], researchers and professionals have been preparing for the post-antibiotic era [75]. By 2050, an estimated 10 million people will lose their lives due to ARB infections every year worldwide, resulting in an economic loss of approximately 2%–3.5% [76]. Understanding and controlling the spread of ARB to preserve the effectiveness of antibiotics is one of the most urgent tasks in human health management of the 21st century [77].
ARGs are mainly transmitted among ARB via horizontal gene transfer by means of conjugation, transformation, and transduction through mobile genetic elements [78]. Many studies have shown that ARGs can circulate in hospitals, animal farms, sewage treatment plants, and other environments, and these genes can even transfer from symbiotic bacteria to pathogenic bacteria or opportunistic pathogens [79]. Thus, ARGs pose an urgent threat to be addressed from a One Health perspective.
When analysing the emergence of ARGs, the first factor is the method of antibiotic production. In this process, the active intermediate products or drugs can be discharged into the environment through wastewater [80]. There have also been cases of antibiotics accidentally leaking into the environment during transportation and sale [81]. However, the primary cause of antibiotic resistance is the use of antibiotics in hospitals [82]. Untransformed antibiotics and their metabolites that form in vivo can be excreted from the body. As sewage enters the natural environment, it promotes the transfer of ARGs from humans into the environment [83]. Indeed, wastewater discharged from hospitals was found to contain high levels of ARGs [84]. Additionally, medical waste is sent to landfills, which increases the diversity and abundance of ARGs in the soil [85].
Domestic animals also contribute to antibiotic resistance, as many countries add large varieties of antibiotics to their fodder [86]. Previous studies have shown that the Chinese live poultry market is a major reservoir of ARGs [87], which have also been detected in pigs [88]. Furthermore, antibiotics are excreted and transferred to the environment through soil, surface runoff, and groundwater [89]. In China, the production of livestock and poultry manure was estimated to reach 3.8 billion tons in 2021, and animal faeces have been shown to be an important pathway for ARGs in the environment [90].
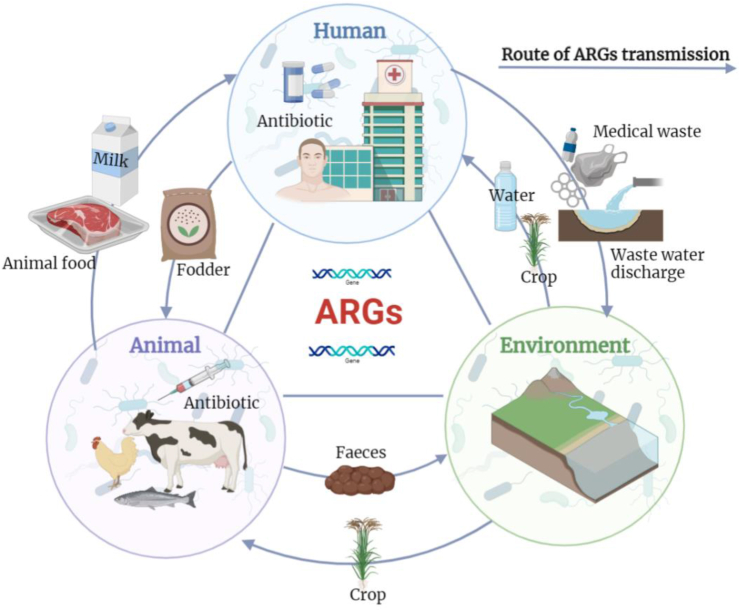
Antibiotics used in the treatment of humans and animals accumulate in the environment and then return to humans and animals through various pathways such as drinking water and the food chain, further expanding the distribution of ARGs. Indeed, multiple ARGs have been detected in drinking water [91]. In addition, ARGs can be adsorbed onto urban dust and spread as aerosols [92]. Studies have found that lactic acid bacteria in fermented milk contain ARGs [93]. In addition, seafood products contain antibiotics, and the human intestinal flora can acquire resistance through the ingestion of these foods [94] (Fig. 6).
Fig. 6.
Antibiotic resistance genes (ARGs) in the microbiome. The medical waste from humans can increase the diversity of ARGs in the soil. Some antibiotic-resistant bacteria that contains ARGs diffuse in the environment through surface runoff and groundwater, which can also return to humans and animals through drinking water, plants, and other food chain pathways. In addition, humans can also be exposed to ARGs through the foods of animals, such as fermented milk and meat products.
6. Discussion
Our understanding of the microbiome has broadened over time, from recognising the role of a single organism in acute infectious diseases to the impacts of various microbiomes in polyinfections, and more recently understanding the broad roles of the microbiome in the health of humans, animals, and the environment.
In the past, the understanding of pathogens in diseases was based on Koch's postulates and the identification of a single microbe. However, the microbiome is the part of the human body that interacts with other microbiomes. Microbiomes in the human gut play many roles such as in immunity and neural regulation. In humans, the characteristics and diversity of the microbiome can be used as indicators of disease. Similarly, the environmental microbiome can be used as an indicator of pollution. Finally, in the wider ecosystem, a large number of microbiomes and their genes play regulatory roles together as a collective medium, affecting environmental material and energy cycles and linking the health of humans, animals, and the environment.
Historically, the microbiome has rarely been mentioned in the concept of One Health despite increasing recognition of its important role. However, research on the microbiome is ongoing, and many of its functions, especially the interactions between different microbiomes in soil, marine, and other environments, remain unclear. In addition, although microbiome modulation strategies such as probiotics, faecal microbiota transplantation, and phage interventions have been proven to improve the environment and treat human and animal diseases effectively, much research remains in this field, especially with respect to their efficacy and safety.
The microbiome has become a key research priority in the One Health system and provides an important intermediary link in the management of One Health issues. With the development of science and technology, humans have increased their impact on animals and plants, and have also extended this impact to previously unaffected environments such as the deep sea and polar permafrost. Many steady states in the environment gradually break down, and the impact of some unknown viruses is immeasurable. In addition, with increased exploration of the Universe, humans may face the risks of physical, chemical, and biological research outside Earth. These aspects must be considered in the context of One Health and are significant for standardising research and promoting social development.
Funding statement
This research received no specific grants from any funding agency in the public, commercial, or not-for-profit sectors.
Data availability statement
The statements in the paper have been properly cited in the manuscript and no additional data were generated for this review.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors are grateful for the support of Prof. Xiao-kui Guo, Prof. Xiao-nong Zhou, and all other faculty members at the School of Global Health, Chinese Centre for Tropical Diseases Research, Shanghai Jiao Tong University School of Medicine, Shanghai, China.
Contributor Information
Ling-chao Ma, Email: malingchao@sjtu.edu.cn.
Han-qing Zhao, Email: zhaohanqing98@sjtu.edu.cn.
Logan Blair Wu, Email: wu.l@wehi.edu.au.
Zi-le Cheng, Email: chengzile@sjtu.edu.cn.
Chang Liu, Email: tiantianlc@sjtu.edu.cn.
References
- 1.World Health Organization Tripartite and UNEP support OHHLEP's definition of "One Health". 2021. https://www.who.int/news/item/01-12-2021-tripartite-and-unep-support-ohhlep-s-definition-of-one-health
- 2.Chen G.Q. To develop theory and practice of One Health is imperative in China. Sci. Technol. Rev. 2020;38(5):1. (in Chinese) [Google Scholar]
- 3.One Health High-Level Expert Panel (OHHLEP) Adisasmito W.B., Almuhairi S., Behravesh C.B., Bilivogui P., Bukachi S.A., et al. One Health: a new definition for a sustainable and healthy future. PLoS Pathog. 2022;18(6) doi: 10.1371/journal.ppat.1010537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H.C., Han Q. One Health--"Quan jian Kang". Chin. J. Veter. Med. 2022;58:5. (in Chinese) [Google Scholar]
- 5.Liu J.S., Zhang X.X., Guo X.K. The origin, connotation and prospect of One Health. Chin. J. Parasitol. Parasit. Dis. 2022;40:1. (in Chinese) [Google Scholar]
- 6.Sorbara M.T., Pamer E.G. Microbiome-based therapeutics. Nat. Rev. Microbiol. 2022;20(6):365–380. doi: 10.1038/s41579-021-00667-9. [DOI] [PubMed] [Google Scholar]
- 7.Guo X.K., Microbiome and One Health . Changchun; China: 2022. China Annual Conference of Medical Microbiology and Immunology. [Google Scholar]
- 8.Gilbert J.A., Blaser M.J., Caporaso J.G., Jansson J.K., Lynch S.V., Knight R. Current understanding of the human microbiome. Nat. Med. 2018;24(4):392–400. doi: 10.1038/nm.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bahram M., Hildebrand F., Forslund S.K., Anderson J.L., Soudzilovskaia A.A., Bodegom P.M., et al. Structure and function of the global topsoil microbiome. Nature. 2018;560(7717):233–237. doi: 10.1038/s41586-018-0386-6. [DOI] [PubMed] [Google Scholar]
- 10.Uberoi A., McKenney C.B., Zheng Q., Flowers L., Campbell A., Knight S.A., et al. Commensal microbiota regulates skin barrier function and repair via signaling through the aryl hydrocarbon receptor. Cell Host Microbe. 2021;29(8):1235–1248. doi: 10.1016/j.chom.2021.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Q., Wu J., Ye D.Q., Fan Y.G. The expeditioner of epidemiological causal inference: Robert Koch. Chin. J. Dis. Control Prev. 2020;24:10. (in Chinese) [Google Scholar]
- 12.Zhao H.Z., You L.P., Yang L., Yu J.Y., Huang Y.N. Discussion on dysbiosis in inflammatory bowel disease and microbial treatment strategy based on modified Koch's postulates. Med. Recapitulate. 2022;28:4. (in Chinese) [Google Scholar]
- 13.Cao M.K., Wang M.X. New thinking of koch rule in modern medicine. Med. Phil. 1999:12. (in Chinese) [Google Scholar]
- 14.Zhou C.B., Fang J.Y. Cross communication of diet-microbiome-immune interactions in cancer immunotherapy. Cell Rep. Med. 2022;3(11):100806. doi: 10.1016/j.xcrm.2022.100806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bello M.G.D., Godoy F.V., Knight R., Blaser M.J. Role of the microbiome in human development. Gut. 2019;68(6):1108–1114. doi: 10.1136/gutjnl-2018-317503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moeller A.H., Sanders J.G. Roles of the gut microbiota in the adaptive evolution of mammalian species. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020;375(1808) doi: 10.1098/rstb.2019.0597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turroni F., Milani C., Duranti S., Ferrario C., Lugli G.A., Mancabelli L., et al. Bifidobacteria and the infant gut: an example of co-evolution and natural selection. Cell. Mol. Life Sci. 2018;75(1):103–118. doi: 10.1007/s00018-017-2672-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Q., Elson C.O. Adaptive immune education by gut microbiota antigens. Immunology. 2018;154(1):28–37. doi: 10.1111/imm.12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharon G., Sampson T.R., Geschwind D.H., Mazmanian S.K. The central nervous system and the gut microbiome. Cell. 2016;167(4):915–932. doi: 10.1016/j.cell.2016.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cryan J.F., O'Riordan K.J., Cowan C.S.M., Sandhu K.V., Bastiaanssen T.F.S., Boehme M., et al. The microbiota-gut-brain Axis. Physiol. Rev. 2019;99(4):1877–2013. doi: 10.1152/physrev.00018.2018. [DOI] [PubMed] [Google Scholar]
- 21.Qi X., Yun C., Sun L., Xia J., Wu Q., Wang Y., et al. Gut microbiota-bile acid-interleukin-22 axis orchestrates polycystic ovary syndrome. Nat. Med. 2019;25(8):1225–1233. doi: 10.1038/s41591-019-0509-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milaneschi Y., Simmons W.K., Rossum E.F.C., Penninx B.W. Depression and obesity: evidence of shared biological mechanisms. Mol. Psychiatr. 2019;24(1):18–33. doi: 10.1038/s41380-018-0017-5. [DOI] [PubMed] [Google Scholar]
- 23.Nicholson J.K., Holmes E., Kinross J., Burcelin R., Gibson G., Jia W., Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336(6086):1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 24.Roy S., Trinchieri G. Microbiota: a key orchestrator of cancer therapy. Nat. Rev. Cancer. 2017;17(5):271–285. doi: 10.1038/nrc.2017.13. [DOI] [PubMed] [Google Scholar]
- 25.Dinan T.G., Cryan J.F. Gut-brain axis in 2016: brain-gut-microbiota axis - mood, metabolism and behaviour. Nat. Rev. Gastroenterol. Hepatol. 2017;14(2):69–70. doi: 10.1038/nrgastro.2016.200. [DOI] [PubMed] [Google Scholar]
- 26.Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C C.L., et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302(5643):276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 27.Zhou J., Li C., Liu X., Chiu M.C., Zhao X., Wang D., et al. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med. 2020;26(7):1077–1083. doi: 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]
- 28.Zhou H., Ji J., Chen X., Bi Y., Li J., Wang Q., et al. Identification of novel bat coronaviruses sheds light on the evolutionary origins of SARS-CoV-2 and related viruses. Cell. 2021;184(17):4380–4391.e14. doi: 10.1016/j.cell.2021.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Azhar E.I., Kafrawy S.A.E., Farraj S.A., Hassan A.M., Saeed M.S.A., Hashem A.M., Madani T.A. Evidence for camel-to-human transmission of MERS coronavirus. N. Engl. J. Med. 2014;370(26):2499–2505. doi: 10.1056/NEJMoa1401505. [DOI] [PubMed] [Google Scholar]
- 30.Rahman M.T., Sobur M.A., Islam M.S., Ievy S., Hossain M.J., Zowalaty M.E., et al. Zoonotic diseases: etiology, impact, and control. Microorganisms. 2020;8(9):1405. doi: 10.3390/microorganisms8091405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bardgett R.D., Freeman C., Ostle N.J. Microbial contributions to climate change through carbon cycle feedbacks. ISME J. 2008;2(8):805–814. doi: 10.1038/ismej.2008.58. [DOI] [PubMed] [Google Scholar]
- 32.Pospischil A. Human and animal health on three continents--a biography of the early life of Karl Friedrich Meyer (1884-1974) Pathog. Dis. 2015;73(6):ftv039. doi: 10.1093/femspd/ftv039. [DOI] [PubMed] [Google Scholar]
- 33.Saied A.A., Metwally A.A., Mohamed H.M.A., Haridy M.A.M. The contribution of bovines to human health against viral infections. Environ. Sci. Pollut. Res. Int. 2021;28(34):46999–47023. doi: 10.1007/s11356-021-14941-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Atlas R.M. One Health: its origins and future. Curr. Top. Microbiol. Immunol. 2013;365:1–13. doi: 10.1007/82_2012_223. [DOI] [PubMed] [Google Scholar]
- 35.Vonaesch P., Anderson M., Sansonetti P.J. Pathogens, microbiome and the host: emergence of the ecological Koch's postulates. FEMS Microbiol. Rev. 2018;42(3):273–292. doi: 10.1093/femsre/fuy003. [DOI] [PubMed] [Google Scholar]
- 36.Hutchins D.A., Jansson J.K., Remais J.V., Rich V.I., Singh B.K., Trivedi P. Climate change microbiology - problems and perspectives. Nat. Rev. Microbiol. 2019;17(6):391–396. doi: 10.1038/s41579-019-0178-5. [DOI] [PubMed] [Google Scholar]
- 37.Cheng Y.T., Zhang L., S.Y. Plant-microbe interactions facing environmental challenge. Cell Host Microbe. 2019;26(2):183–192. doi: 10.1016/j.chom.2019.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Y.G., Zhu D., Rillig C.M., Yang Y.F., Chu H.Y., Chen Q.L. Ecosystem microbiome science. mLife. 2023;1(4):1–9. doi: 10.1002/mlf2.12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oppen M.J.H., Blackall L.L. Coral microbiome dynamics, functions and design in a changing world. Nat. Rev. Microbiol. 2019;17(9):557–567. doi: 10.1038/s41579-019-0223-4. [DOI] [PubMed] [Google Scholar]
- 40.Bakker P., Berendsen R.L., Pelt J.A.V., Vismans G., Yu K., Li E., et al. The soil-borne identity and microbiome-assisted agriculture: looking back to the future. Mol. Plant. 2020;13(10):1394–1401. doi: 10.1016/j.molp.2020.09.017. [DOI] [PubMed] [Google Scholar]
- 41.Zhai K., Liang D., Li H., Jiao, Yan F.B., Liu J., et al. NLRs guard metabolism to coordinate pattern- and effector-triggered immunity. Nature. 2022;601:245–251. doi: 10.1038/s41586-021-04219-2. [DOI] [PubMed] [Google Scholar]
- 42.Roussin-Léveillée C., Lajeunesse G., St-Amand M., Veerapen V.P., Silva-Martins G., Nomura K. Evolutionarily conserved bacterial effectors hijack abscisic acid signaling to induce an aqueous environment in the apoplast. Cell Host Microbe. 2022;30(4):489–501. doi: 10.1016/j.chom.2022.02.006. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jansson J.K., Hofmockel K.S. Soil microbiomes and climate change. Nat. Rev. Microbiol. 2020;18(1):35–46. doi: 10.1038/s41579-019-0265-7. [DOI] [PubMed] [Google Scholar]
- 44.Shen J.P., He J.Z. Responses of microbes-mediated carbon and nitrogen cycles to global climate change. Acta Ecol. Sin. 2011;31:11. (in Chinese) [Google Scholar]
- 45.Liu Y.Y., Wang S., Li S.Z., Deng Y. Advances in molecular ecology on microbial functional genes of carbon cycle. Microb. China. 2017;44:7. (in Chinese) [Google Scholar]
- 46.Jiao N.Z., Dai M.H., Jian Z.M., Wang X.X., Zhang R. Research strategy of marine carbon storage mechanism and related biogeochemical processes. Sci. Bull. 2022;67:15. (in Chinese) [Google Scholar]
- 47.Zhou Z.H., Liu L., Hou L. Soil organic carbon stabilization and formation: mechanism and model. J. Beijing For. Univ. 2022;44:10. (in Chinese) [Google Scholar]
- 48.Jiao N., Herndl G.J., Hansell D.A., Benner R., Kattner G., Wilhelm S.W., et al. Microbial production of recalcitrant dissolved organic matter: long-term carbon storage in the global ocean. Nat. Rev. Microbiol. 2010;8(8):593–599. doi: 10.1038/nrmicro2386. [DOI] [PubMed] [Google Scholar]
- 49.Jiao C.R.N., Azam F., Thomas H., Baltar F., Dang H., Hardman-Mountford N.J., et al. Mechanisms of microbial carbon sequestration in the ocean-future research directions. Biogeosciences. 2014;11:5285–5306. [Google Scholar]
- 50.Batjes N.H. Geoderma; 2016. Harmonized Soil Property Values for Broad-Scale Modelling (WISE30sec) with Estimates of Global Soil Carbon Stocks. [Google Scholar]
- 51.Otto A., Simpson M.J. Degradation and preservation of vascular plant-derived biomarkers in grassland and forest soils from Western Canada. Biogeochemistry. 2005;74:377–409. [Google Scholar]
- 52.Wang B., An S.S., Liang C., Liu Y., Kuzyakov Y. Microbial necromass as the source of soil organic carbon in global ecosystems. Soil Biol. Biochem. 2021;162 [Google Scholar]
- 53.Joshua P., Schimel M.N.W. The implications of exoenzyme activity on microbial carbon and nitrogen limitation in soil: a theoretical model. Soil Biol. Biochem. 2003;35(4):549–563. [Google Scholar]
- 54.Liang C., Schimel J.P., Jastrow J.D. The importance of anabolism in microbial control over soil carbon storage. Nat. Microbiol. 2017;2:17105. doi: 10.1038/nmicrobiol.2017.105. [DOI] [PubMed] [Google Scholar]
- 55.Proctor L.M. The national institutes of health human microbiome project. Semin. Fetal Neonatal Med. 2016;21(6):368–372. doi: 10.1016/j.siny.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 56.Zhang J., Lin X.G., Yin R. Advances in functional gene diversity of microorganism in relation to soil nitrogen cycling. Chin. J. Eco-Agric. 2009;17:5. (in Chinese) [Google Scholar]
- 57.Lin W., Li Y.Z., Li Y.J., Zhou W.L., Zhang D.D., Qi Z.Y. Advances in the mechanism of microbe-driven nitrogen cycling. J. Plant Nutr. Fert. 2020;26:6. (in Chinese) [Google Scholar]
- 58.Hou H.J., Qin H.L., Chen C.L., Wei W.X. Research progress of the molecular ecology on microbiological processes in soil nitrogen cycling. Res. Agr. Moder. 2014;35:5. (in Chinese) [Google Scholar]
- 59.Conrad R. Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO) Microbiol. Rev. 1996;60(4):609–640. doi: 10.1128/mr.60.4.609-640.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uraguchi D., Ueki Y., Ooi T. Chiral organic ion pair catalysts assembled through a hydrogen-bonding network. Science. 2009;326(5949):120–123. doi: 10.1126/science.1176758. [DOI] [PubMed] [Google Scholar]
- 61.Huang H.L., Zeng G.M., Huang G.H., Yu H.Y. Effect of lignin degrading microorganisms on humus formation in compost. China Biotechnol. 2004;24:8. (in Chinese) [Google Scholar]
- 62.B.T. Hou, Effect of Aerobic Microorganisms on the Soil Humus Formation, Jilin Agricultural University. (in Chinese).
- 63.Mu C., Zhu W. Antibiotic effects on gut microbiota, metabolism, and beyond. Appl. Microbiol. Biotechnol. 2019;103(23–24):9277–9285. doi: 10.1007/s00253-019-10165-x. [DOI] [PubMed] [Google Scholar]
- 64.Dumas M.E. The microbial-mammalian metabolic axis: beyond simple metabolism. Cell Metabol. 2011;13(5):489–490. doi: 10.1016/j.cmet.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 65.Sandberg R., Neilson J.R., Sarma A., Sharp P.A., Burge C.B. Proliferating cells express mRNAs with shortened 3' untranslated regions and fewer microRNA target sites. Science. 2008;320(5883):1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mu C.L., Li X., Wu H.Q., Liu S.Q., Yu K.F., Zhu W.Y. Gut microbiome and gastrointestinal nutrition in animals. Sci. Sin. Vitae. 2022;52 (in Chinese) [Google Scholar]
- 67.Davila A.M., Blachier F., Gotteland M., Andriamihaja M., Benetti P.H., Sanz Y., et al. Re-print of "Intestinal luminal nitrogen metabolism: role of the gut microbiota and consequences for the host". Pharmacol. Res. 2013;69(1):114–126. doi: 10.1016/j.phrs.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 68.Yu Z.T., Yao W., Zhu W.Y. In vitro culture reveals the conversion of daidzein to equol by fecal microflora of Erhualian pigs. J. Nanjing Agric. Univ. 2009;32:1. (in Chinese) [Google Scholar]
- 69.Xu P.P. Where will the high energy consumption sewage treatment industry go? Gener. Mach. 2015;8:16–18. (in Chinese) [Google Scholar]
- 70.Zhou L. Chongqing University; 2020. A Dissertation Submitted to Chongqing University in Partial Fulfillment of the Requirement for the Doctor's Degree of Engineering. (in Chinese) [Google Scholar]
- 71.Foley J.M., Rozendal R.A., Hertle C.K., Lant P.A., Rabaey K. Life cycle assessment of high-rate anaerobic treatment, microbial fuel cells, and microbial electrolysis cells. Environ. Sci. Technol. 2010;44(9):3629–3637. doi: 10.1021/es100125h. [DOI] [PubMed] [Google Scholar]
- 72.Logan B., Cheng S., Watson V., Estadt G. Graphite fiber brush anodes for increased power production in air-cathode microbial fuel cells. Environ. Sci. Technol. 2007;41(9):3341–3346. doi: 10.1021/es062644y. [DOI] [PubMed] [Google Scholar]
- 73.Zhang Y.S., Ye J., Su J.Q. Antibiotic resistance in agroecosystem: progress and challenges. J. Zhejiang Univ. 2017;43:6. (in Chinese) [Google Scholar]
- 74.Kumarasamy K.K., Toleman M.A., Walsh T.R., Bagaria J., Butt F., Balakrishnan R., et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 2010;10(9):597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Boucher H.W., Talbot G.H., Bradley J.S., Edwards J.E., Gilbert D., Rice L.B., et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009;48(1):1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 76.Wang Y.N. Henan Agricultural University; 2021. The Diversity and Dissemination of Antibiotic Resistance Genes in Chicken, Pig and Human Gut Microbiomes. (in Chinese) [Google Scholar]
- 77.Seale A.C., Blencowe H., Manu A.A., Nair H., Bahl R., Qazi S.A., et al. Estimates of possible severe bacterial infection in neonates in sub-Saharan Africa, south Asia, and Latin America for 2012: a systematic review and meta-analysis. Lancet Infect. Dis. 2014;14(8):731–741. doi: 10.1016/S1473-3099(14)70804-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu S., Wu Y., Cao B., et al. An invisible workforce in soil: the neglected role of soil biofilms in conjugative transfer of antibiotic resistance genes. Crit. Rev. Environ. Sci. Technol. 2022;52:15. [Google Scholar]
- 79.Rolain J.M. Food and human gut as reservoirs of transferable antibiotic resistance encoding genes. Front. Microbiol. 2013;4:173. doi: 10.3389/fmicb.2013.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thai P.K., Ky L.X., Binh V.N., Nhung P.H., Nhan P.T., Hieu N.Q., et al. Occurrence of antibiotic residues and antibiotic-resistant bacteria in effluents of pharmaceutical manufacturers and other sources around Hanoi, Vietnam. Sci. Total Environ. 2018;645:393–400. doi: 10.1016/j.scitotenv.2018.07.126. [DOI] [PubMed] [Google Scholar]
- 81.Kookana R.S., Williams M., Boxall A.B., Larsson D.G., Gaw S., Choi K., et al. Potential ecological footprints of active pharmaceutical ingredients: an examination of risk factors in low-, middle- and high-income countries. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369(1656):20130586. doi: 10.1098/rstb.2013.0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Qian J., Wu Z.Y., Guo X.K., Liu C. Antibiotic-resistant microbes, antibiotic resistance genes and One Health. Microb. China. 2022;49:10. (in Chinese) [Google Scholar]
- 83.Michael C.A., Dominey-Howes D., Labbate M. The antimicrobial resistance crisis: causes, consequences, and management. Front. Public Health. 2014;2:145. doi: 10.3389/fpubh.2014.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lamba M., Graham D.W., Ahammad S.Z. Hospital wastewater releases of carbapenem-resistance pathogens and genes in urban India. Environ. Sci. Technol. 2017;51(23):13906–13912. doi: 10.1021/acs.est.7b03380. [DOI] [PubMed] [Google Scholar]
- 85.Chi T., Zhang A., Zhang X., Li A.D., Zhang H., Zhao Z. Characteristics of the antibiotic resistance genes in the soil of medical waste disposal sites. Sci. Total Environ. 2020;730:139042. doi: 10.1016/j.scitotenv.2020.139042. [DOI] [PubMed] [Google Scholar]
- 86.Cantas L., Shah S.Q., Cavaco L.M., Manaia C.M., Walsh F., Popowska M., et al. A brief multi-disciplinary review on antimicrobial resistance in medicine and its linkage to the global environmental microbiota. Front. Microbiol. 2013;4:96. doi: 10.3389/fmicb.2013.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nordmann P., Perler J., Kieffer N., Poirel L. In-vitro evaluation of a dual carbapenem combination against carbapenemase-producing Acinetobacter baumannii. J. Infect. 2020;80(1):121–142. doi: 10.1016/j.jinf.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 88.Li B., Yang Y., Ma L., Ju F., Guo F., Tiedje J.M., et al. Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes. ISME J. 2015;9(11):2490–2502. doi: 10.1038/ismej.2015.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xie W.Y., Shen Q., Zhao F.J. Antibiotics and antibiotic resistance from animal manures to soil: a review. Eur. J. Soil Sci. 2018;69:1. (in Chinese) [Google Scholar]
- 90.Wang F., Han W., Chen S., Dong W., Qiao M M., Hu C. Fifteen-year application of manure and chemical fertilizers differently impacts soil ARGs and microbial community structure. Front. Microbiol. 2020;11:62. doi: 10.3389/fmicb.2020.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ma L., Li B., Jiang X.T., Wang Y.L., Xia Y., Li A.D., et al. Catalogue of antibiotic resistome and host-tracking in drinking water deciphered by a large scale survey. Microbiome. 2017;5(1):154. doi: 10.1186/s40168-017-0369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kellogg C.A., Griffin D.W. Aerobiology and the global transport of desert dust. Trends Ecol. Evol. 2006;21(11):638–644. doi: 10.1016/j.tree.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 93.Liu C., Zhang Z.Y., Dong K., Yuan J.P., Guo X.K. Antibiotic resistance of probiotic strains of lactic acid bacteria isolated from marketed foods and drugs. Biomed. Environ. Sci. 2009;22(5):401–412. doi: 10.1016/S0895-3988(10)60018-9. [DOI] [PubMed] [Google Scholar]
- 94.Heuer O.E., Kruse H., Grave K., Collignon P., Karunasagar I., Angulo F.J. Human health consequences of use of antimicrobial agents in aquaculture. Clin. Infect. Dis. 2009;49(8):1248–1253. doi: 10.1086/605667. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The statements in the paper have been properly cited in the manuscript and no additional data were generated for this review.