Abstract
Four bovine viral diarrhea virus type 2 (BVDV-2) pairs consisting of cytopathogenic (cp) and noncp BVDV-2 were isolated during an outbreak of mucosal disease. Comparative sequence analysis showed that the four noncp BVDV-2 isolates were almost identical. For the cp BVDV-2 isolates, viral subgenomic RNAs were shown by Northern blot to have a length of about 8 kb, which is about 4.3 kb shorter than the genome of noncp BVDV. Cytopathogenicity and the expression of NS3 were both strictly correlated to the presence of viral subgenomic RNAs. By reverse transcription-PCR, Southern blot analysis, and nucleotide sequencing, a set of 11 unique subgenomes was identified with up to 5 different subgenomes isolated from one animal. To our knowledge, this is the first report on isolation of a set of pestiviral subgenomes from individual animals. Common features of the BVDV-2 subgenomic RNAs include (i) deletion of most of the genomic region encoding the structural proteins, as well as the nonstructural proteins p7 and NS2, and (ii) insertion of cellular (poly)ubiquitin coding sequences. Three subgenomes also comprised 15 to 75 nucleotides derived from the 5′ part of the NS2 gene. Comparisons of the obtained nucleotide sequences revealed that the different BVDV-2 subgenomes evolved from the respective noncp BVDV-2 by RNA recombination. The presence of short regions of sequence similarity at several crossing-over sites suggests that base pairing between the nascent RNA strand and the acceptor RNA template facilitates template switching of the BVDV RNA-dependent RNA polymerase.
The genera Pestivirus and Flavivirus and the hepatitis C virus group constitute the family Flaviviridae (41). The genus Pestivirus currently consists of three members, namely, bovine viral diarrhea virus (BVDV), classical swine fever virus, and border disease virus. In addition, a fourth pestivirus species comprising isolates from cattle and sheep has recently been described (2, 30, 34). The Flaviviridae study group of the International Committee on Taxonomy of Viruses has proposed to term this additional species BVDV-2.
The pestivirus genome consists of a positive-stranded nonpolyadenylated RNA molecule with a size of approximately 12.3 kb which contains one large open reading frame (7, 12, 23, 35). In the virus-encoded polyprotein, the mature viral proteins are arranged in the following order (from the N terminus to the C terminus): Npro, C, Erns, E1, E2, p7, NS2-3, (NS2), (NS3), NS4A, NS4B, NS5A, and NS5B (see references 26 and 39 for reviews); the abbreviation Npro refers to an N-terminal autoprotease, and Erns refers to a structural glycoprotein with RNase activity. The structural proteins are represented by the capsid protein C, and the three envelope proteins Erns, E1, and E2. The remaining proteins are presumably nonstructural (NS).
Two biotypes of the pestiviruses, cytopathogenic (cp) and noncytopathogenic (noncp) viruses, are distinguished by their ability to cause a cytopathic effect (CPE) in tissue culture. Both cp and noncp BVDV strains are involved in the pathogenesis of mucosal disease (MD), a particularly severe clinical manifestation of BVDV infection (9, 11). A prerequisite for the development of MD is a transplacental infection with noncp BVDV during the first trimester of gestation which results in the birth of a persistently infected animal with acquired immunotolerance to the respective BVDV strain. Such animals can spontaneously come down with MD and then harbor not only the persisting noncp BVDV strain but also a cp BVDV strain. Accordingly, cp viruses are usually isolated as a mixture together with noncp viruses (26). The cp and noncp BVDV strains isolated from one animal are called a virus pair.
One phenotypic difference between cp and noncp BVDV concerns the expression of NS3, which is colinear with the carboxy-terminal part of NS2-3. While NS2-3 is expressed in both cp BVDV- and noncp BVDV-infected cells, NS3 is found exclusively after infection with cp BVDV (26). Accordingly, NS3 is regarded as the marker protein for cp BVDV strains and is supposed to be required for the induction of a CPE.
Molecular analyses of several BVDV pairs isolated from field cases of MD indicated that cp viruses evolved within the affected animal from the respective persisting noncp virus primarily by RNA recombination. The mutations identified in the genomes of cp BVDV strains include insertions of cellular sequences, frequently together with large duplications and genomic rearrangements with duplications and deletions of viral sequences (see reference 26 for a review), as well as point mutations within the NS2 gene (18). In spite of major genomic alterations, most cp BVDV strains are replication competent; thus far only the ones with deletions have been found to be defective (19, 24, 38).
According to the literature, MD occurs sporadically and in general single animals in one herd come down with the disease. Outbreaks affecting several animals would only be expected if a cp virus generated in one animal is transmitted to other persistently infected animals of the same herd (10). Such outbreaks of MD were rarely described, and the genomes of the respective viruses have never been analyzed. In the present study we report on molecular characterization of four cp BVDV isolates obtained during a single outbreak of MD. It was of particular interest to investigate whether the cp viruses isolated from the four diseased animals carry identical genomic alterations. Surprisingly, our analysis led to the identification of a variety of subgenomic RNAs with up to five different subgenomes derived from a single animal. This unexpected finding sheds new light on the generation of cp pestiviruses in persistently infected animals.
MATERIALS AND METHODS
Cells and viruses.
MDBK cells were obtained from the American Type Culture Collection (Rockville, Md.). Cells were grown in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum (FCS). During an outbreak of MD on a single farm in Schleswig-Holstein, Germany, in 1996, BVDV-2 Giessen-1 was isolated from blood samples of four affected calves after inoculation of MDBK cells. The samples were kindly provided by H.-P. Heckert and G. Appel (Freie Universität, Berlin, Germany). The respective virus isolates are termed BVDV-2 Giessen-1 CP-A, CP-B, CP-C, and CP-D, respectively. An accompanying noncp virus was isolated by limiting dilution from the second cell passage for each of the four cp virus isolates. These noncp viruses are termed BVDV-2 Giessen-1 NCP-A, NCP-B, NCP-C, and NCP-D.
Infection of cells.
Cell culture supernatants and lysates of infected cells were combined and used for infection of MDBK cells. Material for infection was prepared by freezing and thawing cultures at 48 h postinfection and was stored at −70°C. A multiplicity of infection of ca. 0.1 was used for infections. The proportion of virus-infected cells was assessed by indirect immunofluorescence with monoclonal antibody 8.12.7 (directed against NS3), kindly provided by E. J. Dubovi (Cornell University, Ithaca, N.Y.). Cells and FCS samples were tested regularly for the absence of pestiviruses by reverse transcription-PCR (RT-PCR) and immunofluorescence. For the FCS samples, the absence of anti-pestivirus antibodies was shown by a lack of virus neutralization.
Immunoblot.
Infected MDBK cells were lysed 48 h postinfection in loading buffer containing 6 M urea, 2% sodium dodecyl sulfate (SDS), 10% glycerol, and 5% β-mercaptoethanol. Samples were subjected to SDS-polyacrylamide gel electrophoresis (8% polyacrylamide) under reducing conditions (36) and transferred to a nitrocellulose filter (Schleicher & Schuell, Dassel, Germany). The filters were blocked with 5% nonfat dry milk–0.05% Tween 20 in phosphate-buffered saline (PBS) for 16 h. After being washed with PBS–0.05% Tween 20, the filters were incubated with monoclonal antibody 8.12.7 (directed against NS3). After several washes the filters were incubated with the substrates of the ECL Kit (Amersham Buchler, Braunschweig, Germany) according to the protocol of the manufacturer. Filters were exposed to Kodak BioMax MR films. The prestained molecular weight standard was obtained from Life Technologies (Eggenstein, Germany).
RNA preparation, gel electrophoresis, and Northern (RNA) hybridization.
RNA from pestivirus-infected cells was prepared by using either the RNeasy Total RNA Kit (Qiagen GmbH, Hilden, Germany) or the RNA Extraction Kit (Pharmacia, Freiburg, Germany) as recommended by the supplier. Five micrograms of glyoxylated RNA (21) was separated in a phosphate-buffered 1.0% agarose gel containing 5.5% formaldehyde and transferred to Duralon-UV membranes (Stratagene, Heidelberg, Germany). An RNA ladder (Life Technologies) served as a size standard. Radioactive labelling of the probe, hybridization, and posthybridization washes were done as described previously (3). A 2.5-kb NotI-NsiI fragment from the cDNA clone pA/BVDV was used as a probe (24).
Oligonucleotides.
Oligonucleotides were purchased from MWG Biotech GmbH (Ebersberg, Germany). The sense primer Ol P3400 (5′-CAATAYTGGTTTGACCTRGA-3′; R = A or G; Y = T or C) corresponds to positions 3428 to 3447 within the genomic sequence of BVDV-2 strain 890 (35). The antisense primers Ol 1400R (4) and Ol NS3R (6) and the sense primer Ol 100 (5) have been described previously. The sequence of Ol 1400R corresponds to nucleotide positions 1430 to 1448 of the published genomic sequence of BVDV-1 strain NADL (12). The positions of Ol NS3R and Ol P100 are indicated in Fig. 2A.
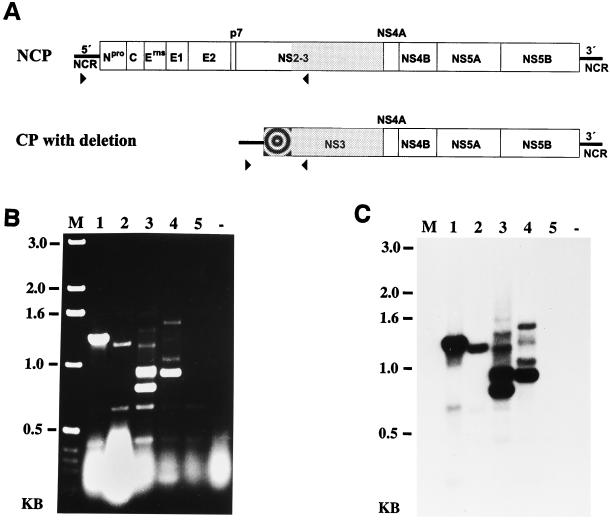
FIG. 2.
(A) RT-PCR strategy for identification of subgenomic viral RNAs with large deletions in the genomic region encoding Npro, the structural proteins, p7, and NS2. The positions and orientations of primers Ol P100 (▸) and Ol NS3R (◂) are indicated. The bull’s-eye icon indicates a putative insertion. NCP, noncp pestivirus. (B and C) RT-PCR products obtained from RNA of cells infected with BVDV-2 CP-A (lane 1), CP-B (lane 2), CP-C (lane 3), CP-D (lane 4), and NCP-A (lane 5). RNA from noninfected cells (−) served as a negative control. (B) The cDNA fragments were separated on a 1.0% agarose gel and stained with ethidium bromide. (C) Southern blot analysis of the agarose gel shown in panel B. After transfer to a nylon membrane, the blot was hybridized with a 2.5-kb NotI-NsiI fragment from the cDNA clone pA/BVDV (24). M, size standard; KB, kilobases.
RT-PCR.
RT of ca. 500 ng of heat-denatured RNA and PCR was done as described previously (4). For amplification of part of the 5′ noncoding region (NCR) and the genomic region encoding Npro, C, and part of Erns, primer Ol 1400R and primer Ol 100 were used. For analysis of the subgenomic RNAs, an RT-PCR with primer Ol NS3R and primer Ol 100 was performed. In addition, the genomic region encoding p7, NS2, and the N-terminal part of NS3 of isolate BVDV-2 NCP-A was amplified by RT-PCR with primer Ol NS3R and primer Ol P3400. After amplification, the PCR products were characterized in agarose-ethidium bromide gels in Tris-acetate buffer.
Southern blot.
Prior to transfer, the DNA samples were separated on a 1.0% agarose gel. After denaturation with 1.5 M NaCl–0.5 M NaOH for 20 min, the gel was neutralized in 1.0 M ammonium acetate–0.02 M NaOH two times each for 15 min. Transfer to Duralon-UV membranes was performed as recommended by the supplier. Hybridization was done as described for the Northern hybridization (see above). Filters were exposed to Kodak BioMax MR films.
Molecular cloning, nucleotide sequencing, and sequence analysis.
The cDNA fragments obtained after RT-PCR were separated by agarose gel electrophoresis and purified by using the Qiaex DNA Purification Kit (Qiagen). The respective cDNA fragments were cloned by using the TA Cloning Kit (Invitrogen, De Schelp, The Netherlands). Nucleotide sequences were determined by cycle sequencing with Thermo Sequenase Kit (Amersham Buchler, Braunschweig, Germany) and the DNA sequencer Li-Cor 4000 (MWG Biotech). All sequences were determined by sequencing both complementary strands of at least two independent cDNA clones. Computer analysis of the sequence data was performed with HUSAR (DKFZ, Heidelberg, Germany), which provides the GCG software package (14).
Nucleotide sequence accession numbers.
Sequence data from this study have been deposited in the EMBL and GenBank data libraries and are assigned accession no. AF104019 to AF104030.
RESULTS
Isolation of BVDV pairs.
In the present study, four cp BVDV isolates, termed CP-A, CP-B, CP-C, and CP-D, and their noncp counterparts were analyzed. The cp BVDV isolates were obtained from four young calves which were housed on the same farm and came down with MD within a period of 3 weeks. During this outbreak of MD, 13 additional animals showed clinical signs of MD and finally died. Cytopathogenicity of the BVDV isolates was observed during the first passages on MDBK cells. For each of the four cp BVDV isolates, an accompanying noncp BVDV strain was isolated by limiting dilution from the second cell passage; these are termed NCP-A, NCP-B, NCP-C, and NCP-D. Both cp and noncp BVDV were apparently present in each of the four animals. In order to purify the four cp BVDV isolates, plaque purification was performed three consecutive times. Limiting dilution of the resulting cp viruses and subsequent immunofluorescence analysis indicated, however, that noncp virus was still present. Additional attempts to obtain cp viruses in the absence of noncp BVDV failed.
Expression of NS3.
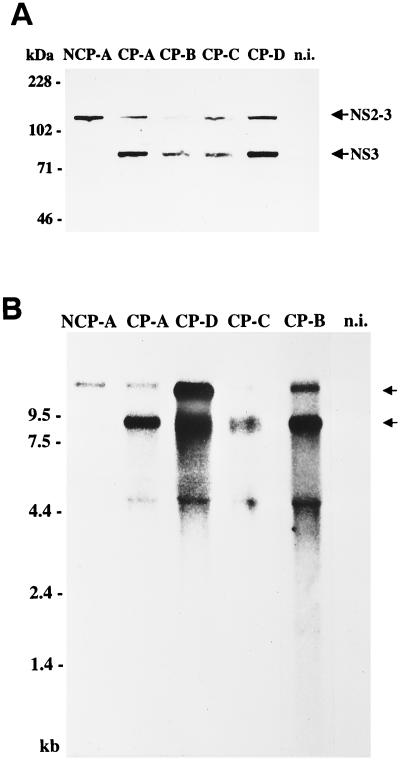
NS3 is regarded as the marker protein for cp BVDV. In order to study the expression of the NS2-3 and NS3 proteins of the BVDV isolates, an immunoblot assay of extracts from MDBK cells infected with CP-A, CP-B, CP-C, CP-D, and NCP-A was performed. Expression of NS2-3 and NS3 was monitored by use of a monoclonal antibody against NS3. After infection of cells with the four cp BVDV isolates, both NS2-3 and NS3 were detected, while in NCP-A-infected cells only NS2-3 was found (Fig. 1A). Accordingly, the cytopathogenicity of the four BVDV isolates correlates with the expression of NS3.
FIG. 1.
Expression of nonstructural proteins NS2-3 and NS3 (A) and detection of viral RNAs (B). (A) Immunoblot analysis. MDBK cells were infected with BVDV-2 Giessen-1 NCP-A, CP-A, CP-B, CP-C, or CP-D. The cells were lysed 48 h postinfection, and the samples were subjected to SDS-PAGE (8% polyacrylamide) under reducing conditions, transferred to a nitrocellulose membrane, and analyzed by using anti-NS3 monoclonal antibody 8.12.7. The sizes (in kilodaltons) of marker proteins are indicated on the left. The positions of NS2-3 and NS3 are marked with arrows. n.i., noninfected MDBK cells. (B) Northern blot analysis of total RNA from infected and noninfected MDBK cells. Prior to transfer and hybridization, RNA was separated on a 1.0% agarose gel under denaturing conditions. The blot was hybridized with a 2.5-kb NotI-NsiI fragment from the cDNA clone pA/BVDV (24). RNA ladder sizes in kilobases are indicated. Migration positions of the viral genomic and subgenomic RNAs are marked with arrows. The additional band with a size of ca. 4.5 kb is presumably due to the presence of large amounts of rRNA.
Nucleotide sequence homology between noncp BVDV isolates.
In order to determine the genetic relatedness among the four noncp BVDV isolates, RT-PCR with primer Ol 1400R and primer Ol 100, molecular cloning, and nucleotide sequencing of the 3′-terminal two-thirds of the 5′ NCR, together with the Npro- and C-coding regions, were performed. The respective consensus sequences were obtained by sequencing the complementary strands of three independent clones. Comparative sequence analysis showed that the four noncp BVDV isolates belong to the species BVDV-2. The nucleotide sequence identities between these four BVDV-2 isolates and the other BVDV-2 strains, including 890 (35), SCP, and 59386 (2), were less than 92%. In contrast, the nucleotide sequences of the noncp viruses were more than 99.5% identical to each other (data not shown). This strongly suggests that the four animals were infected with the same noncp BVDV-2 strain.
Northern blot analysis.
The genomes of several cp pestiviruses contain large duplications or deletions of viral sequences (26). Such mutations result in viral RNAs differing significantly in size from the ones of noncp pestiviruses. To investigate whether similar alterations are present in the genomes of the four cp BVDV-2 isolates, a Northern blot analysis with total RNA from infected MDBK cells was performed. Hybridization of RNA from cells infected with CP-A, CP-B, CP-C, CP-D, and NCP-A with a BVDV-specific probe led to detection of viral genomic RNAs with a size of ca. 12.3 kb. Interestingly, subgenomic RNAs with sizes of ca. 8.0 kb were also identified but only after infection with the four cp viruses (Fig. 1B). Thus, the cytopathogenicity of the four cp BVDV-2 isolates correlates with the presence of viral subgenomic RNAs. It has been previously demonstrated that pestiviral subgenomic RNAs can be responsible for induction of cytopathogenicity (19, 38).
RT-PCR and Southern blot analysis.
For two cp BVDV isolates, subgenomic RNAs were identified which lacked all or almost all of the genomic region encoding the structural proteins, p7 and NS2 (19, 38). In order to analyze whether the BVDV-2 subgenomes detected here contain a similar deletion, an RT-PCR assay with antisense primer Ol NS3R located in the NS3 encoding region and sense primer Ol P100 located in the 5′ NCR was performed (Fig. 2A). This strategy may also lead to amplification of cDNA obtained from a pestivirus genome without a large deletion. Since the time for extension during the amplification cycles was restricted to 50 s, the latter fragment with a calculated length of ca. 5.2 kb was not amplified.
Surprisingly, for the four cp isolates a variety of more than 10 different cDNA fragments was generated; the sizes of these fragments ranged from 0.4 to 1.5 kb (Fig. 2B). RT-PCR analyses with primer pair Ol NS3R and Ol P100 that had longer elongation times (up to 300 s) did not result in detection of additional cDNA fragments. In order to determine whether the amplified fragments were BVDV specific, a Southern blot analysis was performed which revealed that the fragments with sizes between 0.8 and 1.5 kb specifically reacted with the BVDV-specific probe (Fig. 2C). For CP-A and CP-B, one major fragment of ca. 1.3 kb (CP-A) and 1.2 kb (CP-B) could be observed. In addition, a minor fragment of about 0.7 kb was detected for CP-A that was not visible in the ethidium bromide-stained gel. Interestingly, five different cDNA fragments with sizes of ca. 0.8, 0.9, 1.2, 1.4, and 1.6 kb were found after infection with CP-C, while for CP-D four fragments with sizes of about 0.9, 1.0, 1.3, and 1.5 kb reacted with the BVDV-specific probe. It should be noted that the Northern blot analysis (Fig. 1B) did not give a clear indication for the presence of several subgenomes within CP-C and CP-D, a finding which can be explained by the predicted small size differences between the respective subgenomes.
Genome organization of the BVDV-2 subgenomes.
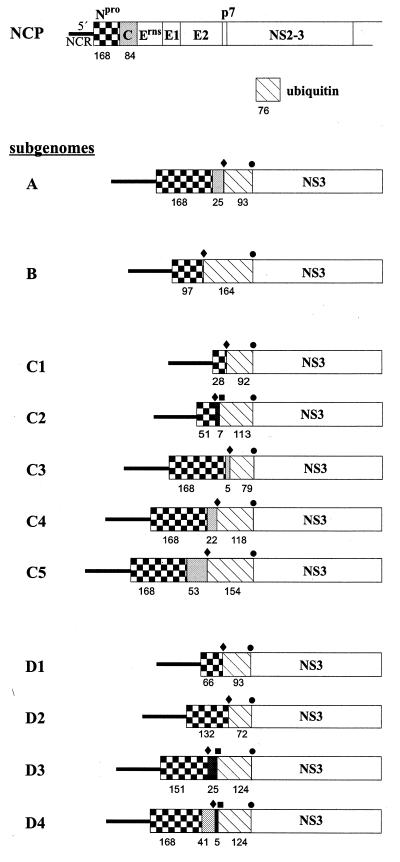
To further characterize the subgenomic RNAs of the four cp BVDV-2 isolates, the respective cDNA fragments were cloned and subjected to nucleotide sequence analysis. Only the 0.7-kb minor fragment detected after infection with CP-A was not accessible to cloning due to low amounts of cDNA. In order to determine the genome organization of the BVDV-2 subgenomes, the obtained nucleotide sequences were compared with the genomic sequence of BVDV-2 reference strain 890 (35). With respect to CP-A, the 5′ part of the sequence obtained for the 1.3-kb fragment corresponds to positions 107 to 964 of the BVDV-2 890 genomic sequence. This region comprises part of the 5′ NCR together with the genomic region encoding Npro and 25 amino acids (aa) from the N terminus of C. This is followed by a nonviral sequence comprising 279 nucleotides. Comparative sequence analyses, together with data bank searches, revealed that the latter sequence is almost identical to a bovine (poly)ubiquitin coding sequence. The sequence located downstream of this cellular sequence corresponds to positions 5375 to 5545 of the BVDV-2 890 sequence, which is colinear with the 5′ region of the NS3 gene (Fig. 3).
FIG. 3.
Schematic representation of the genome organization of a noncp pestivirus (NCP) and the BVDV-2 subgenomic RNAs (only part of the genome/subgenomes is shown). The subgenomes A and B were isolated from animals A and B, respectively. C1, C2, C3, C4, and C5 were derived from animal C, while D1, D2, D3, and D4 were obtained from animal D. All subgenomic RNAs contain either the entire Npro gene or part of it (checkered box), as well as an insertion of cellular (poly)ubiquitin coding sequences (hatched box). The presence of capsid protein coding sequences (A, C3, C4, C5, and D4) is indicated by gray boxes. The presence of NS2 coding sequences (C2, D3, and D4) is indicated by black boxes. The numbers of amino acids corresponding to the respective viral protein(s) or (poly)ubiquitin are shown below the bars. The positions of the 5′ recombination junctions (⧫), the 3′ recombination junctions (●), and the additional internal recombination junctions of subgenomes C2, D3, and D4 (■) are indicated above the bars.
The genome organization of the CP-A subgenome is similar to that of the other 10 identified subgenomes. All subgenomic RNAs described here contain insertions of cellular ubiquitin coding sequences of variable length. Insertions of ubiquitin coding sequences have been identified for several cp BVDV-1 strains (6, 25, 33, 37). However, in contrast to all cp BVDV strains described so far, we have identified the first naturally occurring pestiviral subgenomes with cellular ubiquitin coding insertions. Interestingly, the ubiquitin encoded by CP-D2 (ubiquitin*) lacks the N-terminal 4 aa, while all other subgenomes encode at least one complete ubiquitin monomer consisting of 76 aa.
Like the genome structures of previously described BVDV subgenomes, either the entire or most of the genomic region encoding the structural proteins, p7, and NS2 is deleted in the BVDV-2 subgenomes (Fig. 3). Subgenomes A, C3, C4, C5, and D4 encode the complete Npro, followed by 5 to 53 N-terminal aa of C. In contrast, the Npro genes of the other BVDV-2 subgenomes are truncated at their 3′ ends, and the entire region encoding the structural proteins is missing. Furthermore, subgenomes C2, D3, and D4 contain 21, 75, and 15 nucleotides derived from the 5′ part of the NS2 gene, respectively; within these subgenomes the NS2-specific sequences are located directly upstream of the ubiquitin coding sequences (Fig. 3). Accordingly, these subgenomes comprise two separate deletions, together with an insertion of ubiquitin coding sequences. Remarkably, all BVDV-2 subgenomes characterized here maintain one large open reading frame.
When the pestivirus-specific nucleotide sequences determined for the individual subgenomes were compared with the corresponding sequence of BVDV-2 NCP-A, in each case more than 99% identity was found. It can be concluded that the BVDV-2 subgenomes were derived from the same noncp BVDV-2 strain by RNA recombination, including the deletion(s) of viral sequences as well as the insertion of cellular ubiquitin coding sequences. Interestingly, the genome structure of each of the 11 BVDV-2 subgenomes is unique. For animals A and B one subgenomic RNA was obtained, while for animals C and D five and four different subgenomic RNAs were found, respectively (Fig. 3; also, see above).
Analysis of recombination sites.
The most widely accepted model of RNA recombination postulates the dissociation of the polymerase-nascent strand complex from one region of the template RNA molecule followed by its reassociation either with another region of the same template or with a different template (16, 17). With regard to nonhomologous RNA recombination, short regions of sequence identity have been observed near the crossing-over sites, suggesting that base pairing between the nascent strand and the acceptor template may facilitate RNA recombination (29, 31). While pestiviruses appear to undergo nonhomologous recombination quite frequently, such nucleotide sequence similarities at the recombination junctions have not been observed for pestiviruses analyzed so far. To study the BVDV-2 subgenomes in this respect, the nucleotide sequences at the recombination junctions were compared with the corresponding dissociation and reassociation regions of the parent RNA molecules. For all subgenomes the 3′ recombination junction (between ubiquitin coding and NS3 coding sequences) is conserved, while the 5′ recombination junction is unique for each subgenome. Subgenomes C2, D3, and D4 each comprise an additional internal recombination junction (between NS2 coding and ubiquitin coding sequences); this junction is identical for subgenomes D3 and D4 (Fig. 3). Taken together, 14 different recombination regions were analyzed. Surprisingly, the sequences at six recombination junctions showed regions of similarity (Fig. 4). The respective sequence motifs comprised 6 to 16 nucleotides exhibiting 67 to 86% identity and were unique for each recombination site. For 8 of the 14 recombination junctions it was not possible to define the recombination site precisely, since short regions of sequence identity comprising one to three nucleotides were found at the junction site of the subgenomic RNA and in the corresponding regions of both parent RNAs (Fig. 4 and data not shown). The identification of sequence similarity at the recombination junctions suggests that sequence complementarity between the nascent strand and the acceptor template can contribute to template switching of the BVDV RNA-dependent RNA polymerase (RdRp).
FIG. 4.
Alignment of sequences in the regions of the recombination junctions of BVDV-2 subgenomes. Nucleotide sequences of the parental template RNA molecules (upper and lower lines) are shown together with the sequences of the progeny subgenomic RNAs (middle lines). The sequence identity between the subgenomic RNAs and the respective template RNAs is indicated by the vertical lines between the sequences. The sites of recombination are indicated by either boxes (5′ sites of subgenomes C1, C3, D2, and D4 and the conserved 3′ site) or a vertical line (subgenome C4). The regions of sequence similarity between the nascent and the assumed acceptor RNA strand are highlighted by a gray background. The 3′ recombination site is conserved among all BVDV-2 subgenomes analyzed here. Recombination sites without regions of sequence similarity are not included.
DISCUSSION
The genomes of cp pestiviruses exhibit interesting characteristics such as insertions of cellular sequences, duplications of viral sequences, or deletions of viral sequences, while the genomes of noncp pestiviruses generally do not show such changes (26). Remarkably, the occurrence of cp BVDV is directly linked to MD, a lethal disease in cattle. In the present study we have characterized four BVDV-2 pairs isolated from a single outbreak of MD. It has been speculated that such outbreaks are caused by one cp BVDV strain which evolved in a single animal and is then transmitted to other persistently infected animals (10). Accordingly, the cp viruses obtained from different animals during one outbreak of MD in a single herd are expected to be almost identical. Surprisingly, the four cp BVDV-2 isolates analyzed in our study are clearly different from each other. We were able to identify 11 unique BVDV-2 subgenomes with up to 5 different subgenomes derived from one animal. This is in contrast to all previous reports about pestiviruses where either one cp virus or one subgenome together with a noncp virus was identified. It should be noted here that isolates of cp pestiviruses are usually plaque purified prior to molecular characterization and that this may result in the loss of additional cp pestiviruses and/or subgenomes. It will be interesting to analyze additional samples from animals with MD where plaque purification is avoided and in vitro passages are kept to a minimum. Such an approach will show whether isolates of cp pestiviruses generally contain more than one cp virus or subgenome.
All BVDV-2 subgenomes described here carry cellular (poly)ubiquitin coding sequences and maintain one large open reading frame, thereby allowing the expression of both viral proteins and ubiquitin. While insertions of ubiquitin coding sequences have been found for several cp BVDV strains (6, 25, 33, 37), this is the first report on the identification of naturally occurring pestiviral subgenomic RNAs carrying this cellular insertion. For all cp BVDV strains with ubiquitin coding sequences, including the subgenomes analyzed here, the 3′ recombination site is conserved, resulting in the fusion of the N terminus of NS3 to the C terminus of ubiquitin. Previous studies have shown that ubiquitin serves in this context as a processing signal to yield NS3, the marker protein of cp BVDV. Furthermore, it has been reported that at least one entire ubiquitin monomer is required for processing at the C terminus of ubiquitin (37). Interestingly, subgenome D2 encodes a truncated ubiquitin (ubiquitin*) which lacks the four N terminal amino acids, while the other subgenomes encode at least one complete ubiquitin monomer. In a recent study we have demonstrated for BVDV strain CP Rit that a fusion protein composed of ubiquitin and NS3 was not cleaved when the N terminus of ubiquitin was truncated by 3 aa (6). Addition of ribosomal protein S27a-derived amino acids to the N terminus of this truncated ubiquitin restored processing, whereas NS3 was not generated after replacement of the S27a coding sequence by an unrelated sequence (6). It was therefore interesting to determine whether the Npro*-ubiquitin*-NS3 hybrid protein encoded by subgenome D2 was cleaved to yield NS3. After expression of the fusion protein of subgenome D2, NS3 was clearly detectable (data not shown). Taken together, it can be assumed that all identified subgenomes express NS3 with Gly1590 as N-terminal amino acid (the number corresponds to the sequence of BVDV-1 strain SD-1 [GenBank accession number M96751]).
The occurrence of subgenomic RNAs has already been described for two cp BVDV isolates. BVDV CP9 and BVDV CP13 were both shown to consist of a defective interfering (DI) RNA of ca. 8 kb and a noncp BVDV strain (19, 38). Additional studies revealed that DI9 and DI13 induce a CPE in cells infected with a noncp BVDV strain. An engineered subgenome that mirrors the genome structure of DI9 has recently been demonstrated to be responsible for the induction of cytopathogenicity (24) and to be replication competent (8). In the subgenome of DI9, the genes encoding the structural proteins, p7, and NS2 are completely deleted. Another engineered subgenome where the C terminal part of Npro was replaced by an ubiquitin monomer (termed DI9cΔNproubi) was also capable of replicating autonomously (8). The BVDV-2 subgenomes analyzed here encode at least 28 aa from the N terminus of Npro, the NS proteins 3, 4A, 4B, 5A, and 5B, as well as ubiquitin fused to the N terminus of NS3 (Fig. 3). Accordingly, the genome structures of the BVDV-2 subgenomes are very similar to the one of DI9cΔNproubi. It is suggested that the BVDV-2 subgenomes analyzed here also replicate autonomously and induce a CPE.
The BVDV-2 subgenomes were generated by RNA recombination that included integration of ubiquitin coding sequences and large deletion(s) of viral sequences. With respect to generation of the BVDV-2 subgenomes analyzed here, their overall similar genome structures argue against several independent recombination processes. It is considered more likely that the various subgenomes developed in the course of two separate recombination processes. In a first step, integration of ubiquitin coding sequences into the genome of the noncp BVDV-2 strain resulted in a replication-competent precursor virus. Subsequently, several different deletions may have occurred which led to the generation of a set of unique viral subgenomes. Transmission of the hypothetical precursor virus among animals would also explain the development of a variety of BVDV-2 subgenomes with ubiquitin coding sequences. However, our analysis failed to identify such a precursor virus.
It has been proposed that homologous and nonhomologous RNA recombination involve template switching of the viral RdRp (1, 17, 27, 29). This model postulates the dissociation of the nascent RNA from the template RNA and its reassociation to a different region of the same template or to another template. For several RNA viruses, base pairing between the nascent RNA and the acceptor RNA has been suggested to facilitate the reassociation step (20, 28, 29, 32, 42, 43). Furthermore, the formation of heteroduplex secondary structures between the two recombination partners, as well as signals for the pausing of transcription (donor template) and reinitiation of RNA synthesis (acceptor template), have been proposed (1, 29). Comparison of the recombination junctions in the BVDV-2 subgenomes with the corresponding regions of the recombination partners revealed short regions of sequence similarity at several crossing-over sites, suggesting that base pairing between the nascent RNA and the acceptor template guided the polymerase as it switched templates (Fig. 4). In the pestivirus system, there is only one other report on sequence complementarity between the nascent RNA and the acceptor template molecule (13). In that study a PCR-based strategy was used to examine total RNA from tissue samples of cattle persistently infected with noncp BVDV. In contrast to our study, the majority of the recombinant sequences in that study contained a frameshift at the recombination site which would cause premature termination of the viral polyprotein and thus not result in the production of a viable virus.
When the nascent RNA undergoes recombination, one of its functions is to serve as a primer for elongation. Our analysis revealed that in 6 of 14 cases the 3′ nucleotide of the nascent strand was obviously not base paired with the acceptor template. While binding of the 3′ end of a primer is a prerequisite for DNA synthesis performed by DNA polymerases, some viral RdRps, including the pestiviral RNA polymerase, appear to be able to extend primers with or without a base-paired 3′ end (29).
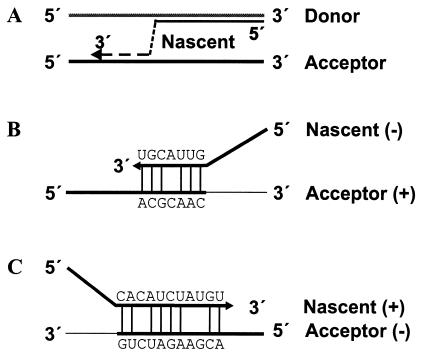
Assuming that base pairing between the nascent strand and the acceptor RNA has guided the reassociation process, the complement of the region of sequence similarity is incorporated into the nascent strand directly before its dissociation from the template. Accordingly, the recombination site should lie upstream of the region of sequence similarity shared by the donor and acceptor RNAs. Furthermore, a comparison of the sequences in the region of the recombination junctions may help to determine whether template switching occurred during synthesis of either negative- or positive-strand RNA. With respect to the positive strands of the template RNAs, the conserved 3′ recombination site as well as the 5′ recombination sites of subgenomes C3 and D4 lie upstream of the region of sequence similarity, and it is therefore assumed that template switching in these cases occurred during minus-strand synthesis (Fig. 4 and 5B). In contrast, for the 5′ recombination sites of subgenomes C1, C4, and D2 it is assumed that the regions of sequence identity have influenced template switching during positive-strand RNA synthesis, since the respective recombination sites are found downstream of the regions with sequence similarity (Fig. 4 and 5C).
FIG. 5.
(A) Replicase-mediated template switching model of RNA recombination. The model includes dissociation of the nascent RNA strand (3′ part as dashed line) from the donor template (gray line) followed by reassociation to an acceptor template (black line). (B and C) Sequence similarity-guided reassociation of the nascent RNA to the acceptor template during the synthesis of either negative (B)- or positive (C)-sense RNA strands. As examples, the sequences of the 5′ recombination junctions of subgenomes D4 (B) and D2 (C) and the reassociation regions of the respective acceptor templates are shown. Sequence complementarity between the nascent RNA and the acceptor template is indicated by the vertical lines between the sequences.
For several positive-strand RNA viruses it has been suggested that RNA recombination predominantly occurs during the synthesis of RNA negative strands (1, 15, 17, 20). With respect to pestiviral genomes with integrated cellular protein coding sequences, RNA recombination must have occurred during negative-strand synthesis, since the cellular recombination partners exist only as positive strands (26). This argument, however, would not apply to the development of the BVDV-2 subgenomes when a common viral precursor with ubiquitin coding sequences is assumed. Our analysis of the recombination sites actually suggests that recombination occurred not only during the synthesis of negative-strand RNAs but also during positive-strand RNA synthesis (Fig. 5). This assumption is in agreement with the existence of a viral precursor genome with ubiquitin coding sequences.
Previous studies of cp pestiviruses focused on the identification of genomic alterations in connection with MD and the generation of NS3 but contributed little to aspects of RNA recombination. The results of our analysis of the junction sequences indicate that (i) base pairing between the nascent strand and the acceptor template may facilitate the reassociation step and thereby guide the RdRp as it switches templates, (ii) the pestiviral RdRp appears to be able to extend primers with or without a base-paired 3′ end, (iii) template switching may occur during both negative- and positive-sense strand synthesis. The availability of infectious full-length and subgenomic BVDV cDNA clones (22, 24, 40) will now allow the study of RNA recombination of pestiviruses in more detail.
ACKNOWLEDGMENTS
This study was supported by Intervet International BV (project 75/73,1808.720) and SFB 535 “Invasionsmechanismen und Replikationsstrategien von Krankheitserregern” from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Agol V I. Recombination and other genomic rearrangements in picornaviruses. Semin Virol. 1997;8:77–84. [Google Scholar]
- 2.Becher P, König M, Paton D, Thiel H-J. Further characterization of border disease virus isolates: evidence for the presence of more than three species within the genus pestivirus. Virology. 1995;209:200–206. doi: 10.1006/viro.1995.1243. [DOI] [PubMed] [Google Scholar]
- 3.Becher P, Meyers G, Shannon A D, Thiel H-J. Cytopathogenicity of border disease virus is correlated with integration of cellular sequences into the viral genome. J Virol. 1996;70:2992–2998. doi: 10.1128/jvi.70.5.2992-2998.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becher P, Orlich M, Shannon A D, Horner G, König M, Thiel H-J. Phylogenetic analysis of pestiviruses from domestic and wild ruminants. J Gen Virol. 1997;78:1357–1366. doi: 10.1099/0022-1317-78-6-1357. [DOI] [PubMed] [Google Scholar]
- 5.Becher P, Orlich M, Thiel H-J. Complete genomic sequence of border disease virus, a pestivirus from sheep. J Virol. 1998;72:5165–5173. doi: 10.1128/jvi.72.6.5165-5173.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becher P, Orlich M, Thiel H-J. Ribosomal S27a-coding sequences upstream of ubiquitin-coding sequences in the genome of a pestivirus. J Virol. 1998;72:8697–8704. doi: 10.1128/jvi.72.11.8697-8704.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becher P, Shannon A D, Tautz N, Thiel H-J. Molecular characterization of border disease virus, a pestivirus from sheep. Virology. 1994;198:542–551. doi: 10.1006/viro.1994.1065. [DOI] [PubMed] [Google Scholar]
- 8.Behrens S E, Grassmann C W, Thiel H-J, Meyers G, Tautz N. Characterization of an autonomous subgenomic pestivirus RNA replicon. J Virol. 1998;72:2364–2372. doi: 10.1128/jvi.72.3.2364-2372.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolin S R, McClurkin A W, Cutlip R C, Coria M F. Severe clinical disease induced in cattle persistently infected with noncytopathogenic bovine viral diarrhea virus by superinfection with cytopathogenic bovine viral diarrhea virus. Am J Vet Res. 1985;46:573–576. [PubMed] [Google Scholar]
- 10.Brownlie J, Clarke M C. Experimental and spontaneous mucosal disease of cattle: a validation of Koch’s postulates in the definition of pathogenesis. Intervirology. 1993;35:51–59. doi: 10.1159/000150295. [DOI] [PubMed] [Google Scholar]
- 11.Brownlie J, Clarke M C, Howard C J. Experimental production of fatal mucosal disease in cattle. Vet Rec. 1984;114:535–536. doi: 10.1136/vr.114.22.535. [DOI] [PubMed] [Google Scholar]
- 12.Collett M S, Larson R, Gold C, Strick D, Anderson D K, Purchio A F. Molecular cloning and nucleotide sequence of the pestivirus bovine viral diarrhea virus. Virology. 1988;165:191–199. doi: 10.1016/0042-6822(88)90672-1. [DOI] [PubMed] [Google Scholar]
- 13.Desport M, Collins M E, Brownlie J. Genomic instability in BVDV: an examination of the sequence and structural influences on RNA recombination. Virology. 1998;246:352–361. doi: 10.1006/viro.1998.9219. [DOI] [PubMed] [Google Scholar]
- 14.Devereux J, Haeberli P, Smithies O A. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarvis T C, Kirkegaard K. Poliovirus RNA recombination: mechanistic studies in the absence of selection. EMBO J. 1992;11:3135–3145. doi: 10.1002/j.1460-2075.1992.tb05386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King A M Q, Ortlepp S A, Newman J W I, McCahon D, editors. Genetic recombination in RNA viruses. London, United Kingdom: Academic Press; 1987. [Google Scholar]
- 17.Kirkegaard K, Baltimore D. The mechanism of RNA recombination in poliovirus. Cell. 1986;47:433–443. doi: 10.1016/0092-8674(86)90600-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kümmerer B, Stoll D, Meyers G. Bovine viral diarrhea virus strain Oregon: a novel mechanism for processing of NS2-3 based on point mutations. J Virol. 1998;72:4127–4138. doi: 10.1128/jvi.72.5.4127-4138.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kupfermann H, Thiel H-J, Dubovi E J, Meyers G. Bovine viral diarrhea virus: characterization of a cytopathogenic defective interfering particle with two internal deletions. J Virol. 1996;70:8175–8181. doi: 10.1128/jvi.70.11.8175-8181.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Ball L A. Nonhomologous RNA recombination during negative-strand synthesis of flock house virus RNA. J Virol. 1993;67:3854–3860. doi: 10.1128/jvi.67.7.3854-3860.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 22.Mendez E, Ruggli N, Collett M S, Rice C M. Infectious bovine viral diarrhea virus (strain NADL) RNA from stable cDNA clones: a cellular insert determines NS3 production and viral cytopathogenicity. J Virol. 1998;72:4737–4745. doi: 10.1128/jvi.72.6.4737-4745.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyers G, Rümenapf T, Thiel H-J. Molecular cloning and nucleotide sequence of the genome of hog cholera virus. Virology. 1989;171:555–567. doi: 10.1016/0042-6822(89)90625-9. [DOI] [PubMed] [Google Scholar]
- 24.Meyers G, Tautz N, Becher P, Thiel H-J, Kümmerer B. Recovery of cytopathogenic and noncytopathogenic bovine viral diarrhea viruses from cDNA constructs. J Virol. 1996;70:8606–8613. doi: 10.1128/jvi.70.12.8606-8613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyers G, Tautz N, Dubovi E J, Thiel H-J. Viral cytopathogenicity correlated with integration of ubiquitin-coding sequences. Virology. 1991;180:602–616. doi: 10.1016/0042-6822(91)90074-L. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyers G, Thiel H-J. Molecular characterization of pestiviruses. Adv Virus Res. 1996;47:53–118. doi: 10.1016/s0065-3527(08)60734-4. [DOI] [PubMed] [Google Scholar]
- 27.Monroe S S, Schlesinger S. Common and distinct regions of defective-interfering RNAs of Sindbis virus. J Virol. 1984;49:865–872. doi: 10.1128/jvi.49.3.865-872.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagy P D, Bujarski J J. Efficient system for homologous RNA recombination in brome mosaic virus: sequence and structure requirements and accuracy of crossovers. J Virol. 1995;69:131–140. doi: 10.1128/jvi.69.1.131-140.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagy P D, Simon A E. New insights into the mechanisms of RNA recombination. Virology. 1997;235:1–9. doi: 10.1006/viro.1997.8681. [DOI] [PubMed] [Google Scholar]
- 30.Pellerin C, Van Den Hurk J, Lecomte J, Tijssen P. Identification of a new group of bovine viral diarrhea virus strains associated with severe outbreaks and high mortalities. Virology. 1994;203:260–268. doi: 10.1006/viro.1994.1483. [DOI] [PubMed] [Google Scholar]
- 31.Pilipenko E V, Gmyl A P, Agol V I. A model for rearrangements in RNA genomes. Nucleic Acids Res. 1995;23:1870–1875. doi: 10.1093/nar/23.11.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pogany J, Romero J, Huang Q, Sgro J-Y, Shang H, Bujarski J J. De novo generation of defective interfering-like RNAs in broad bean mottle bromovirus. Virology. 1995;212:574–586. doi: 10.1006/viro.1995.1515. [DOI] [PubMed] [Google Scholar]
- 33.Qi F, Ridpath J F, Lewis T, Bolin S R, Berry E S. Analysis of the bovine viral diarrhea virus genome for possible cellular insertions. Virology. 1992;189:285–292. doi: 10.1016/0042-6822(92)90704-s. [DOI] [PubMed] [Google Scholar]
- 34.Ridpath F F, Bolin S R, Dubovi E J. Segregation of bovine viral diarrhea virus into genotypes. Virology. 1994;205:66–74. doi: 10.1006/viro.1994.1620. [DOI] [PubMed] [Google Scholar]
- 35.Ridpath J F, Bolin S R. The genomic sequence of a virulent bovine viral diarrhea virus (BVDV) from the type 2 genotype: detection of a large genomic insertion in a noncytopathic BVDV. Virology. 1995;212:39–46. doi: 10.1006/viro.1995.1451. [DOI] [PubMed] [Google Scholar]
- 36.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 37.Tautz N, Meyers G, Thiel H-J. Processing of poly-ubiquitin in the polyprotein of an RNA virus. Virology. 1993;197:74–85. doi: 10.1006/viro.1993.1568. [DOI] [PubMed] [Google Scholar]
- 38.Tautz N, Thiel H-J, Dubovi E J, Meyers G. Pathogenesis of mucosal disease: a cytopathogenic pestivirus generated by internal deletion. J Virol. 1994;68:3289–3297. doi: 10.1128/jvi.68.5.3289-3297.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thiel H-J, Plagemann P G W, Moennig V. Pestiviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1059–1073. [Google Scholar]
- 40.Vassilev V B, Collett M S, Donis R O. Authentic and chimeric full-length genomic cDNA clones of bovine viral diarrhea virus that yield infectious transcripts. J Virol. 1997;71:471–478. doi: 10.1128/jvi.71.1.471-478.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wengler G, Bradley D W, Collett M S, Heinz F X, Schlesinger R W, Strauss J H. Flaviviridae. In: Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D, editors. Virus taxonomy. Sixth Report of the International Committee on Taxonomy of Viruses. Vienna, Austria: Springer-Verlag; 1995. pp. 415–427. [Google Scholar]
- 42.White K A, Morris T J. Nonhomologous RNA recombination in tombusvirus: generation and evolution of defective interfering RNAs by stepwise deletions. J Virol. 1994;68:14–24. doi: 10.1128/jvi.68.1.14-24.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.White K A, Morris T J. RNA determinants of junction site selection in RNA virus determinants and defective interfering RNAs. RNA. 1995;1:1029–1040. [PMC free article] [PubMed] [Google Scholar]