Abstract
Human cytomegalovirus encodes a protein kinase (UL97) that confers sensitivity to ganciclovir by phosphorylating it to the monophosphate. The function of this unusual kinase in viral replication is unknown. We constructed two independent isolates of a recombinant virus, RCΔ97, that contain large deletions in this gene and carry a 4.8-kb insertion containing a selectable genetic marker. These mutant viruses were isolated by using a population of primary cells (HEL97) that express this gene from integrated copies of a defective retroviral vector. The recombinant viruses were severely impaired in their ability to replicate in primary fibroblasts, attaining virus titers that were 2 to 3 orders of magnitude lower than those produced by the parent virus. Despite the severe replication deficit, both of these viruses retained the ability to form small, slowly growing plaques in primary fibroblasts, demonstrating that UL97 is not absolutely essential for replication in cell culture. The replication deficit was relieved when UL97 was provided in trans in the complementing cell line, showing that the phenotype was due to a deficiency in UL97. Thus, the UL97 gene product plays a very important role in viral replication in tissue culture and may be a good target for antiviral chemotherapy.
Human cytomegalovirus (HCMV) causes sight- and life-threatening disease in immunocompromised patients, including individuals infected with human immunodeficiency virus (13, 23, 26). A number of antiviral therapies, including ganciclovir (GCV) (10), cidofovir (41), and foscarnet (40), are presently available to treat HCMV infections. GCV must be phosphorylated to the level of the triphosphate to inhibit the viral DNA polymerase (4, 5). In cells infected with herpes simplex virus (HSV), the viral thymidine kinase phosphorylates this compound (9), but in cells infected with HCMV, the UL97 gene product is responsible for the phosphorylation of the drug and confers sensitivity to the drug (1, 19, 28, 29, 42, 44, 45). UL97 does not have homology with any other known nucleoside kinase; rather, it resembles some protein kinases and bacterial phosphotransferases. GCV-resistant mutants arise quite frequently in patients receiving prolonged therapy (7), and most mutations that confer resistance map to UL97 (1, 6, 8, 19, 38).
Little is known about the function of UL97 with respect to viral replication. Its expression is consistent with β/γ kinetics and is targeted to the nucleus via a nuclear localization signal in the amino terminus of the protein (24, 30). UL97 is also phosphorylated soon after synthesis and is present in virions, possibly as a constituent of the tegument (46). Clear homologs are present in other betaherpesviruses, but only limited homology exists in the alpha- and gammaherpesvirus subfamilies (3). Despite the low degree of sequence homology between UL97 and the HSV homolog UL13, UL97 can partially substitute for the HSV UL13 gene, which suggests that some functions may be conserved (33). Biochemical studies have demonstrated protein kinase activity from UL97 that can result in both autophosphorylation (21, 46) and transphosphorylation (20). However, neither the biological consequences of autophosphorylation nor the natural substrate for this kinase is presently known.
Recombinant viruses with mutations in UL97 have been difficult to isolate (30, 43). Mutant viruses with a limited spectrum of point mutations and small deletions in this gene have been isolated and exhibit resistance to GCV (1, 6, 19, 38, 49). The nature and the location of these mutations suggested that they alter UL97 substrate specificity without ablating enzymatic activity. These and other results raised the possibility that this gene is essential for viral replication. To determine the function of this protein in viral replication, we constructed recombinant viruses in which more than 70% of the UL97 open reading frame (ORF) was deleted. Two recombinant viruses, RCΔ97.08 and RCΔ97.19, were independently isolated with the help of a population of primary cells expressing the UL97 gene product. Our results demonstrate that the deletion mutants exhibit a severe replication deficiency in primary fibroblasts and indicate that this gene plays an important role in viral replication in cell culture.
MATERIALS AND METHODS
Plasmids.
pON2132 was constructed by ligating a 3,498-bp EcoRI-HindIII fragment (coordinates 139499 to 142997 in the AD169 genome [2]) from pCM1007 (14) into the unique EcoRI and HindIII sites in pGEM3Zf(+). pON855 has been described elsewhere (47) and contains the Escherichia coli gpt (guanine phosphoribosyltransferase) selectable marker as well as the E. coli lacZ gene. A 4.8-kb BamHI-PstI fragment containing both lacZ and gpt was ligated into the unique BamHI and PstI sites in pON2132 to yield pON2133. This deletes more than 70% of the UL97 ORF, including crucial subdomains homologous to genes encoding other protein kinases, and replaces it with the selectable genetic markers gpt and lacZ. pON2161 was constructed by inserting a 2,366-bp SalI-XhoI fragment (coordinates 140350 to 142716) into a defective retroviral vector (32). This construct contains the UL97 ORF under the control of the murine leukemia virus (MuLV) long terminal repeat (LTR) and retains a portion of the endogenous UL97 promoter (48).
Cells and virus.
Primary human foreskin fibroblasts (HFF) and human embryonic lung (HEL) cells were grown in monolayer cultures in Dulbecco’s modified Eagle medium (Gibco BRL, Gaithersburg, Md.) supplemented with 100 U of penicillin G/ml, 100 μg of streptomycin sulfate/ml, and 10% fetal bovine serum. Parental virus (AD169) and PA317 cells were obtained from the American Type Culture Collection, and virus stocks were obtained and titered as described previously (39). GDGrK17 was grown and titered as described previously (44).
Construction of HEL97 cells.
pON2161 was transfected into PA317 cells by using Lipofectin (Gibco BRL), and G418-resistant clones were used to produce stocks of a defective retrovirus containing the UL97 gene under the control of both the MuLV LTR and its endogenous promoter. The retrovirus stocks were used to transduce this gene into low-passage-number primary HEL cells (31, 35). Cells infected with the retrovirus were selected with 400 μg of G418/ml, starting at 24 h postinfection (hpi) and continuing for 10 days. Transduced cells were frozen at passage 6 and were grown in the presence of 400 μg of G418/ml every 3rd passage.
Construction of recombinant virus.
HEL cells were seeded into a 6-well tissue culture cluster 24 h prior to transfection. pON2133 (1 μg) was linearized with HindIII and BglII, purified by phenol-chloroform extraction, precipitated with ethanol, and resuspended in 25 μl of Tris-EDTA buffer (10 mM Tris–1 mM EDTA) (TE). Linearized plasmid DNA was then transfected with Lipofectin. DNA-Lipofectin complexes were formed for 10 min at final concentrations of 5 and 6 μg/ml, respectively, in a volume of 200 μl of Opti-MEM (Gibco BRL) without the addition of serum or antibiotics. Monolayers were rinsed with medium without serum, and the Lipofectin complexes were placed on the cell sheet and incubated for 5 h in a humidified incubator with 5% CO2 at 37°C. Medium with 10% fetal bovine serum was then added to a final volume of 4 ml, and the cells were allowed to recover overnight. Monolayers were then inoculated with AD169 at a multiplicity of infection (MOI) of 5 PFU/cell at 24 h posttransfection and incubated for 7 days. Infected monolayers were lysed by freezing at 5 days postinfection, and the supernatant was used to inoculate HEL97 cells. Recombinant viruses containing the gpt gene were subjected to selection with 200 μM mycophenolic acid and 5 μM xanthine (16, 17, 34, 47). Plaques that were resistant to mycophenolic acid were isolated by plaque purification on HEL97 cells three times after RCΔ97 was shown to be free from contaminating AD169 as determined by Southern hybridization. Two independent isolates from separate transfections were isolated and designated RCΔ97.08 and RCΔ97.19.
Infected-cell DNA and DNA blotting.
Procedures for preparing viral DNA have been described previously (34). Briefly, infected monolayers were disrupted with TE containing 1% sodium dodecyl sulfate (SDS) and digested with 20 μl of a 10-mg/ml solution of proteinase K at 65°C for 2 h. The solutions were extracted once with phenol equilibrated with TE, pH 7.8, and approximately 100 μl of Phase-lock gel (5 Prime→3 Prime, Boulder, Colo.). The tubes were gently agitated to form an emulsion and were centrifuged at 12,000 × g for 10 min. The aqueous phase was subsequently extracted with chloroform and precipitated with ethanol. DNA blotting was performed as described previously (34), except that the DNA probes were labeled with [32P]dCTP by using a random priming protocol (Amersham, Little Chalfont, Buckinghamshire, United Kingdom).
Growth curves.
Replication kinetics of the mutant viruses were assayed as described previously (34, 37). Briefly, confluent monolayers of HFF, HEL cells, or HEL97 cells in 96-well plates were infected at an MOI of 0.1 PFU/cell with either AD169, RCΔ97.08, or RCΔ97.19 as cell lysates in tissue culture medium. Infected monolayers were frozen and stored at −80°C until progeny virus was diluted and titered. Frozen dishes were thawed at 37°C, and a 200-μl sample from each well was titered by diluting it in 96-well plates containing monolayers of HEL97 cells or HFF. At 10 days postinfection, the monolayers infected with the wild-type (wt) virus were rinsed with phosphate-buffered saline (PBS), fixed for 5 min in 95% ethanol, and stained with 0.2% methylene blue or crystal violet. Plaques were enumerated on an inverted microscope and were used to calculate virus titers. Monolayers infected with the mutant virus were rinsed with PBS containing 2 mM MgCl2 and fixed for 10 min in 0.5% glutaraldehyde. The monolayers were washed twice in the PBS-MgCl2 buffer and detected with a solution of PBS with the addition of 2 mM MgCl2, 12.5 mM K3FeCN6, 12.5 mM K4FeCN6, and 12.5 mM 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). The monolayers were subsequently counterstained with crystal violet, and plaques were enumerated.
RT-PCR and PCR.
Primers for reverse transcription-PCR (RT-PCR) are as follows: for the 65-bp product corresponding to sequences retained in RCΔ97, the forward primer 5′-CCACTATGTCCTCCGCACTT-3′ and the reverse primer 5′-AGCCCTGAGTCGTCGTTC-3′; for the 153-bp product corresponding to sequences deleted in RCΔ97, the forward primer 5′-GGCGGCGGCGTCACCACTTT-3′ and the reverse primer 5′-CGGGCCGACGCAGGTTCTCC-3′. HEL cells were infected with tissue culture lysates at an MOI of 0.1 PFU/cell, and total RNA from cells was harvested at 48 hpi and purified with Qiagen RNeasy columns by the protocol provided by the manufacturer. RNA samples were treated with RNase-free DNase (Ambion, Austin, Tex.) for 15 min at 37°C, and the DNase was subsequently inactivated by heating to 75°C for 10 min. The RNA was then reverse transcribed with oligo(dT) primers at 42°C for 15 min and then heated to 95°C for 5 min. The reverse-transcribed products were then amplified with the PCR primers listed above by 30 cycles of 95°C for 1 min, 53°C for 1 min, and 72°C for 1 min. For confirmation of the integration site in RCΔ97, a primer upstream of UL97 (5′-ACAGGGAAGACTGTCGCC-3′) and a primer within the gpt gene (5′-CAAACCTGAGCGAAACCC-3′) were used to amplify a specific 250-bp fragment in RCΔ97.
TaqMan RT-PCR.
PCR primers for TaqMan analysis were as follows. Human cyclophilin forward (5′-CCCACCGTGTTCTTCGACAT-3′) and reverse (5′-TCTTTGGGACCTTGTCTGCAA-3′) primers were designed to produce a product of 85 bp. The HCMV UL44 forward (5′-GAATTTTCTCACCGAGGAACCTT-3′) and reverse (5′-CGCTGTTCCCGACGTAATTT-3′) primers produced a 67-bp product. The HCMV UL97 forward (5′-TACAGCCTCAGCGAGCCCTAT-3′) and reverse (5′-GCCGTACCCGTCTCCTGAA-3′) primers were designed to yield a 71-bp product that amplifies a region of the gene that was deleted in the recombinant viruses. The cyclophilin, UL44, and UL97 target probes (5′-CCCTTGGGCCGCGTCTCCTTT-3′, 5′-CCAGCGTGGCGATCCCTTCG-3′, and 5′-ACGGCCACACAGCGCTCGTTG-3′, respectively) were labeled on the 5′ end with 6-FAM and on the nonextendable 3′ end with the quencher fluor TAMRA. The probes and primers were obtained through the Applied Biosystems Division of Perkin-Elmer (Foster City, Calif.).
The TaqMan EZ RT-PCR kit from the Applied Biosystems Division of Perkin-Elmer was used to quantitate HCMV UL44 and UL97 RNA by following the manufacturer’s procedure. This real-time RT-PCR procedure was performed in a single-tube format by using the human cyclophilin gene as an internal housekeeping gene. Each 50-μl RT-PCR mixture was optimized for primer, probe, and manganese acetate concentrations. Forward primer concentrations for cyclophilin, UL44, and UL97 were 200 nM, while reverse primer concentrations were 400, 400, and 200 nM, respectively. The TaqMan probe and manganese acetate were present in each reaction mixture at 150 nM and 4 mM, respectively. In addition to the above components, each reaction mixture contained 5 U of rTth polymerase; 0.5 U of uracil DNA N-glycosylase; 0.3 mM (each) dCTP, dGTP, and dATP; 0.6 mM dUTP; 50 mM bicine, pH 8.2; and 60 nM passive internal reference (ROX fluor). Total RNA was pretreated with RNase-free DNase (Ambion) to remove trace DNA contaminants. The PCR process was initiated with a uracil DNA glycosylase reaction at 50°C for 2 min followed by reverse transcriptase for 30 min at 60°C. Deactivation of reverse transcriptase and activation of the rTth DNA polymerase were carried out at 95°C for 5 min. The amplification process ran 40 cycles of 20 s at 94°C and 1 min at 62°C.
During the extension phase of the PCR cycle, the nucleolytic activity of the DNA polymerase cleaves the hybridization probe and releases the reporter fluor from the quencher moiety of the probe. Physical separation between the reporter and the quencher dyes produces an increase in the fluorescent emission that is monitored in real time during the PCR amplification by using the 7700 sequence detector (15, 22) (Perkin-Elmer Applied Biosystems). A TaqMan computer algorithm analyzes the fluorescent emission for each reaction and calculates a cycle threshold (Ct) value. The Ct value represents the point at which the PCR amplification reaches a significant threshold, which is typically recorded as the point where the cycle fluorescence reaches 10 standard deviations above baseline fluorescence. It has been shown that the Ct value is proportional to the number of target copies present in the sample (22). Standard curves were run for cyclophilin, UL44, and UL97, and the data were extracted from these.
RESULTS
Primary cells containing the UL97 ORF.
Preliminary experiments suggested that a UL97-deficient virus would be unable to replicate efficiently in HEL cells (36). Therefore, a population of primary cells was produced in order to complement a recombinant virus in trans. The UL97 ORF and part of the endogenous promoter (48) was cloned into a defective MuLV retroviral vector containing the neomycin resistance gene (32). This construct was used to generate stocks of a defective retrovirus that efficiently transduced the UL97 gene into low-passage-number primary HEL cells (31, 35). G418-resistant HEL97 cells resembled the parental HEL cells in morphology and remained fully permissive for HCMV infection (36). The ability of these cells to potentially complement the recombinant viruses was tested by examining the GCV sensitivity of the wt virus and a mutant virus, GDGrK17, that phosphorylates GCV inefficiently (44). GDGrK17 was confirmed to be resistant to GCV when the experiments were performed on monolayers of HFF (Table 1). The 50% effective dose (ED50) of GCV on this virus was also the same (P > 0.05) when it was assayed on a population of primary HEL cells containing the HCMV UL45 gene (HEL45 cells). However, when this virus was assayed in HEL97 cells, it was modestly but significantly more sensitive to GCV (P < 0.02), although not as sensitive as the wt virus, AD169. This suggested that the UL97 expressed in this cell population was functional and partially restored GCV sensitivity, presumably by restoring phosphorylation of the drug.
TABLE 1.
GCV sensitivity in HFF, HEL97 cells, and HEL45 cells
| Cell type | ED50
(mean ± SEM)a of:
|
|
|---|---|---|
| AD169 | GDGrK17 | |
| HFF | 5 ± 0.6 | 34 ± 1.9 |
| HEL97 | 5 ± 0.9 (P > 0.5) | 19 ± 3 (P < 0.02) |
| HEL45 | 5 ± 1.3 (P > 0.5) | 35 ± 8.6 (P > 0.05) |
Data are averages from three separate experiments, except for the HEL45 data, which are averages from two experiments. P values were calculated from a two-sample t test (two-tailed) comparing the average ED50 to that seen in HFF.
Isolation of RCΔ97.08 and RCΔ97.19.
Two independently constructed recombinant viruses containing a large deletion within the UL97 ORF were constructed by transfecting pON2133 into HEL cells with Lipofectin, followed by superinfection with wt virus at an MOI of 5 PFU/cell. The high MOI permitted replication of any recombinant viruses that were replication deficient in HEL cells. Recombinant viruses that were resistant to mycophenolic acid were isolated by several rounds of selection on HEL97 cells (16, 17, 34, 47). The genomes of RCΔ97.08 (or RCΔ97.19) and the parental virus are shown schematically in Fig. 1. A 4.8-kb fragment containing the E. coli gpt gene under the control of the tk promoter and the E. coli lacZ gene under the control of the rat β-actin promoter was used to replace 1,519 bp within UL97 (coordinates 140546 to 142065) (47). This insertion interrupted the ORF 22 amino acids (aa) downstream of the translational start site and resulted in the deletion of more than 70% of the gene. Transcripts from this gene are also predicted to be prematurely terminated by the polyadenylation sequences from simian virus 40 (SV40) in the insert. This deletion removed crucial domains conserved among protein kinases, including those required for protein kinase activity (21) as well as the nuclear localization signals at the amino terminus (24, 30). It is possible that a 40-aa peptide containing the first 22 aa of the UL97 and 18 aa encoded by the polylinker could be translated from these RNAs, but this truncated protein would not be expected to be catalytically active.
FIG. 1.
Structure of RCΔ97. At the top is a HindIII map of the AD169 genome, with the region surrounding UL97 (coordinates 139495 to 142993) expanded below. The bottom diagram represents the insertion, with dotted lines showing the portion of the UL97 ORF that was replaced. Open arrow, the E. coli gpt gene under the control of the tk promoter; shaded arrow, the E. coli lacZ gene under the control of the rat β-actin promoter. The DNA probe used for the DNA blots is shown as a heavy solid bar.
Small lacZ-positive plaques on HEL97 cells were identified by using the fluorescent substrate for β-galactosidase, 4-methylumbelliferyl β-d-galactoside, in an agarose overlay. Plaques that fluoresced when illuminated through the polystyrene from a long-wave UV source were picked and subjected to additional rounds of selection. To isolate this slowly growing virus, limiting dilutions were performed under the selective pressure of mycophenolic acid. Both isolates obtained from separate transfections appeared to replicate much more slowly than the parental virus and had an unusual plaque morphology. Cells infected with the mutant virus developed highly refractile bodies near the nucleus that were clearly visible under a phase-contrast microscope (Fig. 2). Because the refractile bodies accumulated in the infected cells, the plaques appeared to glisten at low magnification and were easy to identify. Since both independent isolates of the mutant had the same characteristic, we infer that it is the result of the engineered mutation.
FIG. 2.
Plaques formed by AD169 (left) and RCΔ97.19 (right) on HEL97 cells. AD169 or RCΔ97.19 was used to infect monolayers of HEL97 cells at an MOI of 0.001 PFU/cell, with a 0.4% low-melting-point agarose overlay. The monolayer infected with AD169 was fixed with 0.1% glutaraldehyde at 8 days postinfection and photographed under a phase-contrast microscope. The monolayer infected with RCΔ97 was fixed with glutaraldehyde at 16 days postinfection and photographed. These images and the other continuous-tone images in this article were digitized with a Umax scanner and Adobe Photoshop on a Macintosh computer.
Fragments from HindIII and PstI digests of DNA isolated from cells infected with RCΔ97.08 and AD169 were separated on agarose gels, transferred to nylon membranes, and hybridized with a PCR fragment inside UL97 (coordinates 140479 to 140871) (Fig. 3A). Fragments consistent with the predicted 6,284- and 3,729-bp fragments from HindIII and PstI digests, respectively, were observed in Southern blots from the parent virus. Neither of these fragments were observed in DNA isolated from RCΔ97.08, which indicated that it was not contaminated with the wt virus. Two larger fragments resulting from the insertion in the recombinant virus also hybridized to these sequences and had migration rates consistent with the 9,491- and 6,936-bp fragments predicted from HindIII and PstI digests, respectively (Fig. 3A). These new fragments also hybridized to sequences contained within the insert (pON855), and the same patterns were seen from digests of RCΔ97.19 DNA (36). No other differences in PstI and HindIII restriction patterns of virion DNA from RCΔ97.08 and RCΔ97.19 were observed. PCR was also used to confirm the genomic structure of the recombinant virus. Primers were designed to detect the junction between the 5′ end of UL97 and the gpt gene in the insert. A specific 250-bp fragment was amplified from cells infected with RCΔ97.08 but was not amplified from cells infected with the parent virus (data not shown). These data are consistent with the structure of the engineered mutation and confirm the hybridization analysis. RT-PCR analysis was used to examine UL97 transcripts in HEL cells infected with either RCΔ97.19 or the parent virus. To confirm that this region had been deleted, two primer pairs were designed to detect the presence of the UL97 transcript. One pair amplified a 65-bp region near the transcriptional start site, and the other pair amplified a 153-bp segment of the RNA that was deleted in the recombinant viruses. RNA from infected cells was harvested at 48 hpi, treated with DNase, and reverse transcribed by using random hexamers as primers. The 65-bp PCR product resulting from the amplification of sequences upstream of the insertion was easily detected in both the recombinant and the wt virus (Fig. 3B). The 153-bp PCR product amplified from sequences in the region deleted in RCΔ97.08 was observed only in the wt virus, confirming that this ORF had been disrupted. It also provided additional evidence that the virus stock was not contaminated with the parent virus.
FIG. 3.
(A) DNA blot of viral DNA from AD169 and RCΔ97.08. Viral DNA was cut with PstI (lanes 1 and 3) or HindIII (lanes 2 and 4) and separated on a 0.6% agarose gel prior to transfer to a nylon membrane. The blot was hybridized with a 392-bp PCR fragment (coordinates 140479 to 140871 [Fig. 1]) that was labeled with [α-32P]dCTP. Fragments that hybridized to the probe were visualized by autoradiography. Positions of predicted fragments are indicated by the arrows. (B) Total RNA was harvested from HEL cells infected with RCΔ97.08 or the parent virus at 48 hpi. RNA was treated with DNase and reverse transcribed by using random hexamer primers. Two adjacent amplicons were amplified by PCR, a 65-bp fragment near the translational start site and a 153-bp fragment that lies within the deleted sequences in the UL97 ORF. Amplified fragments were separated on an agarose gel, transferred to a nylon membrane, and hybridized to pON2161 random labeled with [α-32P]dCTP. The autoradiogram is shown. Amplified products from total RNA from uninfected (lanes 1 and 4), AD169-infected (lanes 2 and 5), and RCΔ97.08-infected (lanes 3 and 6) HEL cells are shown.
Replication characteristics of RCΔ97.08 and RCΔ97.19.
Replication kinetics of the recombinants were first examined in HFF, and progeny viruses from all experiments were titered on HEL97 cells. In this experiment, titers of the recombinant virus produced on the complementing cell line were approximately 5 × 104, so cells were infected at an MOI of less than 1 PFU/cell. Both independent isolates of RCΔ97 replicated poorly in HFF; titers were approximately 2 orders of magnitude lower than those seen with the parent virus at 72 hpi and approximately 4 orders of magnitude lower by 120 hpi (Fig. 4A). This level of progeny virus was only slightly higher than those observed during the eclipse phase of replication measured at 24 hpi. Since both independent isolates of this virus have the same phenotype, we assign it to the engineered mutation in UL97.
FIG. 4.
Growth curves for RCΔ97.08 and RCΔ97.19. HFF or HEL cells were infected, and progeny virus from infected monolayers was harvested at the indicated times and frozen at −80°C. Resulting progeny viruses were titered on monolayers of HEL97 cells. (A) Titers from cells infected at an MOI of approximately 0.1 PFU/cell for AD169 (●), RCΔ97.19 (░⃞), and RCΔ97.08 ( ) are shown in log units of PFU per milliliter. Data at 0 hpi represent titers of the input viruses. Error bars, standard deviations of virus titers from three replicate wells. (B) Titers from infections at an MOI of approximately 1 PFU/cell for AD169 grown on HFF (●) or HEL cells ( ) and titers of RCΔ97.08 on HFF (■) or HEL cells (░⃞) are shown in log units of PFU per milliliter.
The experiment was repeated at an MOI of approximately 2 PFU/cell in both HFF and HEL cells, and the supernatant virus was titered on HEL97 cells. At high MOIs, RCΔ97 was clearly able to replicate in both HEL cells and HFF but attained titers that were 2 to 3 orders of magnitude lower than those attained by the parent virus (Fig. 4B). The appearance of progeny virus was also delayed approximately 48 h, indicating that the kinetics of RCΔ97 replication were also defective, consistent with the slow formation of plaques in these cells.
Complementation of RCΔ97 in trans.
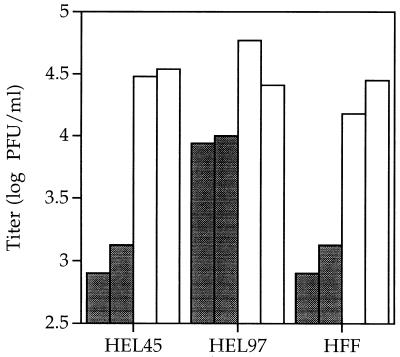
To determine if the replication deficit exhibited by RCΔ97 was due to a deficiency of UL97, HEL97 cells and another cell population containing an unrelated viral gene, UL45, were infected with RCΔ97 at an MOI of 0.1 PFU/cell. The wt virus appeared to replicate with essentially the same kinetics in both HEL and HEL97 cells (Fig. 5). This is consistent with other complementing cell lines constructed in a similar manner (31, 35). Replication of RCΔ97.08 was severely compromised in HEL cells, with titers more than 2 orders of magnitude lower than those of the wt virus at 72 and 96 hpi and 4 orders of magnitude lower by 168 hpi. In contrast, the recombinant virus replicated with the same kinetics as the parent virus in HEL97 cells and achieved titers at 72 hpi that were 100-fold greater than those attained in HEL cells without UL97 (Fig. 5). After this point, replication of the mutant slowed, which may be related to a depletion of permissive cells or to decreased UL97 expression from the integrated copies of the gene in the HEL97 cells. To examine the question more thoroughly, HEL97 cells were infected at an MOI of 2 PFU/cell with virus stocks that were concentrated by centrifugation. In HEL97 cells, RCΔ97 could attain titers ≤3-fold lower than those of the parent virus, which is within the range of variability of this assay. Moreover, this virus attained titers 10-fold higher than those produced in either HFF or HEL45 cells (Fig. 6). These data indicate that a deficiency in the UL97 gene product is responsible for the reduced titers of the recombinant virus.
FIG. 5.
Growth curves for RCΔ97.19 in HEL and HEL97 cells. HEL cells and HEL97 cells were infected with either RCΔ97.19 or AD169 at an MOI of approximately 0.1 PFU/cell, and infected monolayers were harvested at the indicated times and frozen at −80°C. Resulting progeny viruses were titered on monolayers of HEL97 cells, and titers for AD169 grown in HEL cells (●), AD169 grown in HEL97 cells ( ), RCΔ97.19 grown in HEL cells (■), and RCΔ97.19 grown in HEL97 cells (░⃞) are shown in log units of PFU per milliliter. Error bars, standard deviations of virus titers from three replicate wells.
FIG. 6.
HEL97 cells, HEL45 cells (containing the UL45 gene), and HFF were infected at an MOI of 2 PFU/cell with either AD169 (open bars) or RCΔ97 (shaded bars). Progeny virus was harvested at 96 hpi and titered on HEL97 cells. The log of the numbers of infectious progeny harvested from duplicate wells in a 96-well plate is shown.
Expression of UL97 and UL44 transcripts.
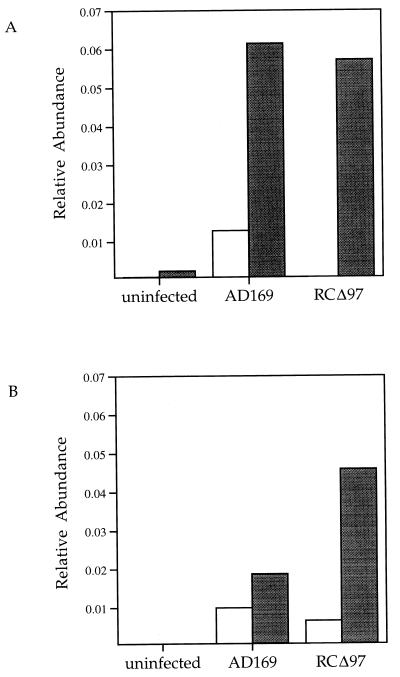
Steady-state levels of UL97 mRNA were measured to characterize expression from the recombinant virus and from the integrated copies of this gene in the complementing cells. RNA from HEL cells and HEL97 cells infected with either the recombinant or the parent virus was harvested at 48 hpi. RNA was treated with DNase I, and mRNA levels were determined by quantitative real-time PCR (TaqMan; see Materials and Methods). Primers for UL97 amplified a product corresponding to sequences within the region that was deleted in RCΔ97. This assay was shown to be linear over 3 orders of magnitude and yielded standard deviations of less than 2% for every sample. All transcript levels were compared to the relative abundance of the cyclophilin gene, a housekeeping gene, and levels of UL44 mRNA were used as a control. In HEL cells, the parent virus and RCΔ97.19 expressed similar levels of UL44 message, yet no UL97 messages were detected in cells infected with RCΔ97.19 (Fig. 7). This confirmed that the UL97 gene in the recombinant was disrupted and that mRNA corresponding to the transcript was undetectable (see also Fig. 2B). Very low levels of UL97 mRNA were expressed in HEL97 cells in the absence of viral infection. Moreover, Western blotting with a polyclonal antibody specific to UL97 (21) was unable to demonstrate significant quantities of the protein (data not shown). However, when these cells were infected with either the recombinant virus or the parent virus, the steady-state levels of UL97 mRNA increased approximately 25-fold. Since UL97 mRNA was not observed in HEL cells infected with the recombinant virus, the increase in UL97 mRNA must be the result of increased expression from integrated copies of this gene in the HEL97 cells. High levels of UL97 mRNA may be the result of either the transcriptional activation of the MuLV LTR or the residual sequences of the endogenous UL97 promoter. The ratio of UL44 mRNA to UL97 mRNA was comparable in HEL cells infected with the wt virus and in HEL97 cells infected with the recombinant virus. UL97 transcripts appeared to be overexpressed when AD169 was grown in the complementing cells. This may be due to altered regulatory elements in the promoter driving expression of UL97 in the construct used to produce the complementing cells. Complementation of the recombinant virus by the HEL97 cells resulted in levels of UL44 mRNA almost sevenfold higher that those seen in HEL cells.
FIG. 7.
Relative abundance of transcripts from UL44 and UL97. Total RNA was harvested at 48 hpi by using Qiagen RNeasy columns and the protocol provided by the manufacturer. The abundance of mRNA was quantitated by TaqMan real-time quantitative PCR as described in Materials and Methods. The relative abundance compared to control cyclophilin mRNA is shown. (A) Levels of UL97 mRNA in HEL (open bars) and HEL97 (shaded bars) cells. (B) Levels of UL44 mRNA in HEL (open bars) and HEL97 (shaded bars) cells. The values shown are the averages of triplicate measurements. Standard deviations were less than 2% for all samples, including the cyclophilin controls.
DISCUSSION
To help define the role that UL97 plays in viral replication, we constructed a recombinant virus with a large deletion in the UL97 gene. This mutation disrupted the gene by replacing it with an insertion containing selectable genetic markers (16, 17, 34, 47). A population of primary cells expressing the UL97 gene was constructed and used for the isolation of the UL97-deficient mutants. This was necessary because preliminary experiments suggested that the recombinants exhibited a severe replication deficiency. A defective retroviral vector was used to transduce the UL97 gene, under the control of both the MuLV LTR and part of the UL97 endogenous promoter, into low-passage-number primary HEL cells (31, 35). Others have reported similar methods for constructing complementing cell lines (11, 31), but we chose not to immortalize cells transduced with the UL97 retrovirus. Our experience with other cell lines of this type suggested that immortalization with the E6 and E7 genes from human papilloma virus type 16 on another retrovirus (LXSN16E6/E7) interfered with subsequent efforts to characterize the recombinant viruses (18). The HEL97 cells grew much more slowly than another primary HEL cell line expressing UL114, suggesting that this gene was poorly tolerated. In an attempt to minimize expression of this potentially toxic gene, the construct used to produce the HEL97 cell lines, pON2161, retained a portion of the endogenous UL97 promoter. It may also retain some elements that further modulate expression following infection. Consistent with this hypothesis, a 25-fold induction of UL97 mRNA was observed when these cells were infected with the mutant virus.
The most striking feature of the recombinant viruses, RCΔ97.08 and RCΔ97.19, was a marked decrease in replication efficiency compared to that of the parent virus. At low MOIs, very low levels of progeny virus were observed in HEL cells and titers were not significantly higher than those at the eclipse phase of replication until 168 hpi. The fact that two independent isolates of this recombinant virus had the identical phenotype permits the inference that the phenotype is the result of the engineered mutation. It is unlikely that two unrelated and deleterious mutations occurred in both independent isolates of the recombinant virus. The data presented here are also consistent with the results from another recently isolated recombinant virus with a smaller deletion in UL97 that was also deficient in replication (25). Although the engineered mutation presented here is not predicted to disrupt the transcription of adjacent genes, it remains possible that their expression is also disrupted and contributes to the observed phenotype. Additional deletions in this region demonstrated that the premature termination of upstream transcripts is not detrimental to the replication of the virus (24). Furthermore, the phenotype of the RCΔ97 viruses was due to a deficiency in UL97, because a cell line expressing UL97 in trans was shown to reverse the replication deficit to a significant degree, and the replication of the RCΔ97 viruses was not statistically different from that of the parent virus in these cells (P > 0.1). An independent line of evidence also suggests that UL97 is crucial to the replication of the virus. A specific inhibitor of UL97 (1263W94) that potently inhibits viral replication both in tissue culture and in humans has recently been described, although the inhibition of UL97 by this drug does not completely abrogate the replication of the virus in cell culture (12). All the available data indicate that UL97 plays an important role in the replication of the virus.
The HEL97 cells used in the isolation of the recombinant virus were able to complement the replication of the recombinants in both high- and low-MOI infections. At high MOIs, the cell line was able to complement the replication of the mutant such that it attained titers that were ≤3-fold lower than those produced by the parent virus. Thus, UL97 supplied in trans can substitute for UL97, and the observed phenotypes were a result of a deficiency in this gene product. In low-MOI infections, complementation was less complete and is likely related to the heterogeneous nature of the HEL97 cells. Perfect complementation was not expected from HEL97 cells and was not observed in this study. This polyclonal cell population was formed from a large number of cells that were independently transduced by the defective retrovirus, so significant variation in expression levels was expected. Although relatively high levels of UL97 mRNA were observed in the HEL97 population as a whole, expression in individual cells probably varies widely, such that UL97 levels are insufficient to support replication of the recombinants in many of these cells. It is also possible that the bicistronic transcripts expressing UL97 and neomycin resistance are not fully functional. Since this gene appears to be poorly tolerated, cells that express low levels of this gene would have a significant growth advantage and would be overrepresented in the population, despite selection with G418.
The recombinant virus appeared to infect HEL cells and expressed comparable levels of the major immediate-early gene products (36). It also appeared to enter the early phase of the viral replication cycle, since it expressed significant quantities of mRNA from the early gene UL44. Steady-state levels of UL44 mRNA were somewhat lower in HEL cells infected with the recombinant virus than in the same cells infected with the parent virus (Fig. 7). This may indicate that the deletion mutant is unable to synthesize DNA as efficiently as the wt virus, since transcription of UL44 is regulated by β/γ kinetics (27). Consistent with this hypothesis, ppUL44 levels expressed by the mutant in HEL cells were always lower than the levels expressed by the parent virus (36). It is also consistent with our observation that low quantities of viral DNA were produced by the recombinants, even in the complementing cell line, and this suggests that a defect in viral replication exists even at early times. We cannot distinguish if these defects are responsible for the low levels of progeny virus. However, it is clear that the disruption of UL97 severely impairs the ability of HCMV to replicate in tissue culture. Replication of the virus also appears to be dependent on the condition of the host cells, and it is possible that a cellular kinase can partially substitute for this gene.
Additional experiments with the UL97 deletion mutants will reveal the precise restriction in the replication cycle and help define the role that this gene plays in viral replication. These recombinants may also be useful in identifying the natural substrate for this unusual kinase. Results presented here also impact the development of chemotherapeutic agents for treating HCMV infections. This protein clearly plays a vital role in viral replication in tissue culture and may be a good target for antiviral chemotherapy. These results also may affect how we view the development of resistance to GCV. The function of this gene appears to be more important than the function of thymidine kinase in cells infected with HSV, and thus it may be less tolerant of mutations. This gene and its function remain the subject of considerable interest because of its clinical significance and its role as a target for antiviral chemotherapy.
ACKNOWLEDGMENTS
We thank Dan Tenney and Edward Mocarski for helpful discussions, and Tom Jones and Michelle Davis for communicating unpublished data.
This work was supported in part by Public Health Service grants AI09008 (to M.N.P.) and U01AI26077 (to D.M.C.) and by a grant from Glaxo Wellcome (to D.M.C.).
REFERENCES
- 1.Baldanti F, Silini E, Sarasini A, Talarico C L, Stanat S C, Biron K K, Furione M, Bono F, Palu G, Gerna G. A three-nucleotide deletion in the UL97 open reading frame is responsible for the ganciclovir resistance of a human cytomegalovirus clinical isolate. J Virol. 1995;69:796–800. doi: 10.1128/jvi.69.2.796-800.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchison C A I, Kouzarides T, Martignetti J A, Preddie E, Satchwell S C, Tomlinson P, Weston K M, Barrell B G. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–170. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 3.Chee M S, Lawrence G L, Barrell B G. Alpha-, beta- and gammaherpesviruses encode a putative phosphotransferase. J Gen Virol. 1989;70:1151–1160. doi: 10.1099/0022-1317-70-5-1151. [DOI] [PubMed] [Google Scholar]
- 4.Cheng Y-C, Grill S P, Dutschman G E, Frank K B, Chiou J-F, Bastow K F, Nakayama K. Effects of 9-(1,3-dihydroxy-2-propoxymethyl)guanine, a new antiherpesvirus compound, on synthesis of macromolecules in herpes simplex virus-infected cells. Antimicrob Agents Chemother. 1984;26:283–288. doi: 10.1128/aac.26.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng Y-C, Huang E-S, Lin J-C, Mar E-C, Pagano J S, Dutschman G E, Grill S P. Unique spectrum of activity of 9-[(1,3-dihydroxy-2-propoxy)methyl]-guanine against herpesviruses in vitroand its mode of action against herpes simplex virus type 1. Proc Natl Acad Sci USA. 1983;80:2767–2770. doi: 10.1073/pnas.80.9.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chou S, Erice A, Jordan M C, Vercellotti G M, Michels K R, Talarico C L, Stanat S C, Biron K K. Analysis of the UL97 phosphotransferase coding sequence in clinical cytomegalovirus isolates and identification of mutations conferring ganciclovir resistance. J Infect Dis. 1995;171:576–583. doi: 10.1093/infdis/171.3.576. [DOI] [PubMed] [Google Scholar]
- 7.Chou S, Guentzel S, Michels K R, Miner R C, Drew W L. Frequency of UL97 phosphotransferase mutations related to ganciclovir resistance in clinical cytomegalovirus isolates. J Infect Dis. 1995;172:239–242. doi: 10.1093/infdis/172.1.239. [DOI] [PubMed] [Google Scholar]
- 8.Chou S, Marousek G, Guentzel S, Follansbee S E, Poscher M E, Lalezari J P, Miner R C, Drew W L. Evolution of mutations conferring multidrug resistance during prophylaxis and therapy for cytomegalovirus disease. J Infect Dis. 1997;176:786–789. doi: 10.1086/517302. [DOI] [PubMed] [Google Scholar]
- 9.Coen D M, Irmiere A F, Jacobson J G, Kerns K M. Low levels of herpes simplex virus thymidine-thymidylate kinase are not limiting for sensitivity to certain antiviral drugs or for latency in a mouse model. Virology. 1989;168:221–231. doi: 10.1016/0042-6822(89)90261-4. [DOI] [PubMed] [Google Scholar]
- 10.Collaborative DHPG Treatment Study Group. Treatment of serious cytomegalovirus infections with 9-(1,3-dihydroxy-2-propoxymethyl)guanine in patients with AIDS and other immunodeficiencies. N Engl J Med. 1986;314:801–805. doi: 10.1056/NEJM198603273141301. [DOI] [PubMed] [Google Scholar]
- 11.Compton T. An immortalized human fibroblast cell line is permissive for human cytomegalovirus infection. J Virol. 1993;67:3644–3648. doi: 10.1128/jvi.67.6.3644-3648.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis, M., and W. Miller. 1998. Personal communication.
- 13.Drew W L, Ives D, Lalezari J P, Crumpacker C, Follansbee S E, Spector S A, Benson C A, Friedberg D N, Hubbard L, Stempien M J, et al. Oral ganciclovir as maintenance treatment for cytomegalovirus retinitis in patients with AIDS. Syntex Cooperative Oral Ganciclovir Study Group. N Engl J Med. 1995;333:615–620. doi: 10.1056/NEJM199509073331002. [DOI] [PubMed] [Google Scholar]
- 14.Fleckenstein B, Muller I, Collins J. Cloning of the complete human cytomegalovirus genome in cosmids. Gene. 1982;18:39–46. doi: 10.1016/0378-1119(82)90054-3. [DOI] [PubMed] [Google Scholar]
- 15.Gibson U, Heid C, Williams P. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- 16.Greaves R F, Brown J M, Vieira J, Mocarski E S. Insertion and deletion mutagenesis of the US3 and IRS1 regulatory genes of human cytomegalovirus using the selectable marker guanosine phosphoribosyl transferase (gpt) J Gen Virol. 1995;76:2151–2160. doi: 10.1099/0022-1317-76-9-2151. [DOI] [PubMed] [Google Scholar]
- 17.Greaves R F, Brown J M, Vieira J, Mocarski E S. Selectable insertion and deletion mutagenesis of the human cytomegalovirus genome using the Escherichia coli guanosine phosphoribosyl transferase (gpt) gene. J Gen Virol. 1995;76:2151–2160. doi: 10.1099/0022-1317-76-9-2151. [DOI] [PubMed] [Google Scholar]
- 18.Halbert C, Demers W, Galloway D. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J Virol. 1991;65:473–478. doi: 10.1128/jvi.65.1.473-478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanson M N, Preheim L C, Chou S, Talarico C L, Biron K K, Erice A. Novel mutation in the UL97 gene of a clinical cytomegalovirus strain conferring resistance to ganciclovir. Antimicrob Agents Chemother. 1995;39:1204–1205. doi: 10.1128/aac.39.5.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He, Z., and D. Coen. 1996. Unpublished data.
- 21.He Z, He Y S, Kim Y, Chu L, Ohmstede C, Biron K K, Coen D M. The human cytomegalovirus UL97 protein is a protein kinase that autophosphorylates on serines and threonines. J Virol. 1997;71:405–411. doi: 10.1128/jvi.71.1.405-411.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heid C, Stevens J, Livak K, Williams P. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 23.Jacobson M A. Cytomegalovirus retinitis: new developments in prophylaxis and therapy. AIDS Clin Rev. 1997;1997:249–269. [PubMed] [Google Scholar]
- 24.Jones, T. 1998. Personal communication.
- 25.Jones, T. 1997. Personal communication.
- 26.Katlama C. Management of CMV retinitis in HIV infected patients. Genitourin Med. 1997;73:169–173. doi: 10.1136/sti.73.3.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leach F S, Mocarski E S. Regulation of cytomegalovirus late-gene expression: differential use of three start sites in the transcriptional activation of ICP36 gene expression. J Virol. 1989;63:1783–1791. doi: 10.1128/jvi.63.4.1783-1791.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Littler E, Stuart A, Chee M. Human cytomegalovirus UL97 open reading frame encodes a protein that phosphorylates the antiviral nucleoside analogue ganciclovir. Nature. 1992;358:160–162. doi: 10.1038/358160a0. [DOI] [PubMed] [Google Scholar]
- 29.Lurain N, Spafford L, Thompson K. Mutation in the UL97 open reading frame of human cytomegalovirus strains resistant to ganciclovir. J Virol. 1994;68:4427–4431. doi: 10.1128/jvi.68.7.4427-4431.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michel D, Pavic I, Zimmermann A, Haupt E, Wunderlich K, Heuschmid M, Mertens T. The UL97 gene product of human cytomegalovirus is an early-late protein with a nuclear localization but is not a nucleoside kinase. J Virol. 1996;70:6340–6346. doi: 10.1128/jvi.70.9.6340-6346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mocarski E S, Kemble G W, Lyle J M, Greaves R F. A deletion mutant in the human cytomegalovirus gene encoding IE1(491aa) is replication defective due to a failure in autoregulation. Proc Natl Acad Sci USA. 1996;93:11321–11326. doi: 10.1073/pnas.93.21.11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morgenstern J, Land H. Advanced mammalian gene transfer: high-titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng T I, Talarico C, Burnette T C, Biron K, Roizman B. Partial substitution of the functions of the herpes simplex virus 1 U(L)13 gene by the human cytomegalovirus U(L)97 gene. Virology. 1996;225:347–358. doi: 10.1006/viro.1996.0609. [DOI] [PubMed] [Google Scholar]
- 34.Prichard M N, Duke G M, Mocarski E S. Human cytomegalovirus uracil DNA glycosylase is required for the normal temporal regulation of both DNA synthesis and viral replication. J Virol. 1996;70:3018–3025. doi: 10.1128/jvi.70.5.3018-3025.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prichard, M. N., N. Gao, G. Duke, E. S. Mocarski, D. Yu, S. Jairath, and G. Pari. Human cytomegalovirus uracil DNA glycosylase participates in the origin specific initiation of viral DNA synthesis and is associated with ppUL44. Submitted for publication.
- 36.Prichard, M. N., N. Gao, S. Jairath, B. Parker, and G. Pari. 1997. Unpublished data.
- 37.Prichard M N, Turk S R, Coleman L A, Engelhardt S L, Shipman C, Jr, Drach J C. A microtiter virus yield reduction assay for the evaluation of antiviral compounds against human cytomegalovirus and herpes simplex virus type 1. J Virol Methods. 1990;28:101–106. doi: 10.1016/0166-0934(90)90091-s. [DOI] [PubMed] [Google Scholar]
- 38.Smith I L, Cherrington J M, Jiles R E, Fuller M D, Freeman W R, Spector S A. High-level resistance of cytomegalovirus to ganciclovir is associated with alterations in both the UL97 and DNA polymerase genes. J Infect Dis. 1997;176:69–77. doi: 10.1086/514041. [DOI] [PubMed] [Google Scholar]
- 39.Spaete R R, Mocarski E S. Regulation of cytomegalovirus gene expression: α and β promoters are transactivated by viral functions in permissive human fibroblasts. J Virol. 1985;56:135–143. doi: 10.1128/jvi.56.1.135-143.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Studies of Ocular Complications of AIDS Research Group and AIDS Clinical Trials Group. Foscarnet-ganciclovir cytomegalovirus retinitis trial. 5. Clinical features of cytomegalovirus retinitis at diagnosis. Am J Ophthalmol. 1997;124:141–157. [PubMed] [Google Scholar]
- 41.Studies of Ocular Complications of AIDS Research Group and AIDS Clinical Trials Group. Parenteral cidofovir for cytomegalovirus retinitis in patients with AIDS: the HPMPC peripheral cytomegalovirus retinitis trial. A randomized, controlled trial. Ann Intern Med. 1997;126:264–274. doi: 10.7326/0003-4819-126-4-199702150-00002. [DOI] [PubMed] [Google Scholar]
- 42.Sullivan V, Biron K K, Talarico C, Stanat S C, Davis M D, Pozzi L M, Coen D M. A point mutation in the human cytomegalovirus DNA polymerase gene confers resistance to ganciclovir and phosphonylmethoxyalkyl derivitives. Antimicrob Agents Chemother. 1993;37:19–25. doi: 10.1128/aac.37.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sullivan, V., and D. Coen. 1992. Unpublished data.
- 44.Sullivan V, Talarico C L, Stanat S C, Davis M, Coen D M, Biron K K. A protein kinase homologue controls phosphorylation of ganciclovir in human cytomegalovirus-infected cells. Nature. 1992;359:85. doi: 10.1038/359085a0. [DOI] [PubMed] [Google Scholar]
- 45.Talarico, C., S. Smith, T. Burnette, W. Miller, M. Davis, S. Stanat, Z. He, T. Ng, D. Coen, B. Roizman, and K. Biron. 1998. Unpublished data. [DOI] [PMC free article] [PubMed]
- 46.van Zeijl M, Fairhurst J, Baum E Z, Sun L, Jones T R. The human cytomegalovirus UL97 protein is phosphorylated and a component of virions. Virology. 1997;231:72–80. doi: 10.1006/viro.1997.8523. [DOI] [PubMed] [Google Scholar]
- 47.Vieira J, Farrell H E, Rawlinson W D, Mocarski E S. Genes in the HindIII J fragment of the murine cytomegalovirus genome are dispensable for growth in cultured cells: insertion mutagenesis with a lacZ/gptcassette. J Virol. 1994;68:4837–4846. doi: 10.1128/jvi.68.8.4837-4846.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wing B A, Huang E S. Analysis and mapping of a family of 3′-coterminal transcripts containing coding sequences for human cytomegalovirus open reading frames UL93 through UL99. J Virol. 1995;69:1521–1531. doi: 10.1128/jvi.69.3.1521-1531.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolf D G, Smith I L, Lee D J, Freeman W R, Flores-Aguilar M, Spector S A. Mutations in human cytomegalovirus UL97 gene confer clinical resistance to ganciclovir and can be detected directly in patient plasma. J Clin Investig. 1995;95:257–263. doi: 10.1172/JCI117648. [DOI] [PMC free article] [PubMed] [Google Scholar]