Abstract
The herpesvirus entry mediator A (HveA) is a recently characterized member of the tumor necrosis factor receptor family that mediates the entry of most herpes simplex virus type 1 (HSV-1) strains into mammalian cells. Studies on the interaction of HSV-1 with HveA have shown that of all the viral proteins involved in uptake, only gD has been shown to bind directly to HveA, and this binding mediates viral entry into cells. In addition to gD binding to HveA, the latter has been shown to interact with proteins of tumor necrosis factor receptor-associated factor family, lymphotoxin-α (LT-α), and a membrane-associated protein referred to as LIGHT. To study the relationship between HveA, its natural ligands, and the viral proteins involved in HSV entry into cells, we have screened two phage-displayed combinatorial peptide libraries for peptide ligands of a recombinant form of HveA. Affinity selection experiments yielded two peptide ligands, BP-1 and BP-2, which could block the interaction between gD and HveA. Of the two peptides, only BP-2 inhibited HSV entry into CHO cells transfected with an HveA-expressing plasmid. When we analyzed these peptides for the ability to interfere with HveA binding to its natural ligand LT-α, we found that BP-1 inhibited the interaction of cellular LT-α with HveA. Thus, we have dissected the sites of interaction between the cell receptor, its natural ligand LT-α and gD, the virus-specific protein involved in HSV entry into cells.
The herpesvirus entry mediator A (HveA; formerly named HVEM) is a member of the tumor necrosis factor receptor (TNFR) superfamily and has been shown to act as a receptor for herpes simplex virus (HSV) (16). Expressed in otherwise nonpermissive CHO cells, it rendered these cells susceptible to entry by several HSV strains. This binding was inhibited by recombinant soluble HveA and antibodies to HveA. In addition to the involvement of HveA in the entry of extracellular virus, it was found that it participates in cell-to-cell transmission of the virus (22, 30). The HSV protein mediating its binding with HveA has been shown to be the glycoprotein D (gD), as it binds directly to a soluble form of HveA [HveA(200t)] (34) in a specific and saturable manner and inhibits the binding of HSV to HveA-expressing cells (20, 21, 27–29, 34).
In addition to its involvement in HSV entry, several findings suggest that HveA plays a role in the activation of the host immune response. For example, HveA, predominantly expressed in lymphocyte-rich tissues, has been shown to interact with several members of the TNFR-associated factor (TRAF) family of proteins. This interaction leads to the activation of transcriptional regulators such as NF-κB, Jun N-terminal kinase, and AP-1 (8, 14). There are two known ligands for the extracellular domain of HveA, lymphotoxin-α (LT-α) and the membrane-associated protein referred to as LIGHT. LIGHT is a newly identified lymphotoxin homolog which is expressed by T cells upon induction with phorbol myristate acetate and Ca2+ ionophore and competes with a soluble form of HSV gD (gDt) for binding to HveA. Thus, either LT-α or LIGHT may modulate HSV infection by competing for HveA binding and vice versa, which has led to the hypothesis that gD may modify HveA-signaling activities during entry or egress of HSV, thus modulating the immune response of the host (15).
The mode of HveA interaction with its ligands, as well as whether HveA interacts with them via multiple sites or whether these ligands share binding sites, is not known. The rich but uncharted molecular diversity that is offered by the surface of the HveA molecule calls for an equally diverse approach to searching for ligands that are complementary and specifically interactive with particular sites. Within the last 10 years, random peptide libraries have provided a rich source of structural diversity (10). They have proved to be a useful tool in identifying the peptide epitopes recognized by particular monoclonal antibodies as well as mimetics of ligands for various proteins.
In this study, our goal was to study the interaction between HveA, its natural ligands, and HSV gD. To this end, we have used recombinant HveA to screen two phage-displayed combinatorial peptide libraries and have selected two peptide ligands that differentially inhibit binding of gDt and LT-α to the receptor. Furthermore, one of these peptides was able to block HSV entry into HveA-expressing CHO cells.
MATERIALS AND METHODS
Chemicals and buffers.
All chemicals and reagents used for peptide synthesis were purchased from Applied Biosystems (Foster City, Calif.) with the exception of the F-moc (9-fluorenylmethoxycarbonyl) amino acids, which were obtained from Nova Biochem (San Diego, Calif.).
Protein expression.
The production and purification of HveA(200t), gD-1(306t), gD-1(Δ290-299t), and LT-α from recombinant baculovirus-infected cells have been described elsewhere (5, 19, 21, 25, 31, 34, 35). Briefly, the cDNA of interest was amplified by PCR for each protein with five or six C-terminal His codons, and a stop codon was added to the downstream primer. The run of His codons was added to provide a binding site for a nickel-nitrilotriacetic acid-agarose resin used in the purification of the expressed protein. The PCR-amplified products were cloned into the pVT-Bac vector (31), with the honeybee mellitin signal sequence replacing their own signal sequences.
The resulting constructs were recombined into baculovirus (Autographa californica nuclear polyhedrosis virus) by cotransfection with Baculogold DNA (Pharmingen, San Diego, Calif.).
Spodoptera frugiperda Sf9 (GIBCO BRL) cells grown in suspension cultures were infected with one of the recombinant baculoviruses at a multiplicity of infection of 4. At 48 to 72 h after infection, the supernatants were cleared by centrifugation, concentrated, and then dialyzed against phosphate-buffered saline (PBS). The proteins were affinity purified over a column of nickel-nitrilotriacetic acid-agar resin and eluted with increasing concentrations of imidazole (0.01 to 0.25 M) in 0.02 M phosphate buffer (pH 7.5)–0.5 M NaCl. The eluates were dialyzed against PBS and concentrated.
Antibodies.
A polyclonal antibody (R140) against HveA was generated by immunizing a rabbit with baculovirus-produced HveA (200t) as previously described (30, 34). A polyclonal antibody specific for gD (R7) was raised against gD-2 isolated from virus-infected cells as previously described (9). The production of a monoclonal antibody to LT-α (AG9) has been described elsewhere (4).
Cells and viruses.
CHO cells were grown in Ham’s F-12 medium supplemented with 10% fetal calf serum (FCS). CHO-K1 cells expressing HveA (CHO-HveA cells) and CHO-K1 cells expressing HveC (CHO-HveC cells) were grown in Ham’s F-12 medium supplemented with 10% FCS and 200 μg of G418 per ml. KOS-hrR3 virus is a mutant KOS virus in which the Escherichia coli lacZ gene is positioned in the ribonucleotide reductase large subunit locus (ICP6) and is under the transcriptional control of the ICP6 promoter. This virus was propagated in African green monkey kidney (Vero) cells expressing the large subunit of HSV type 1 ribonucleotide reductase (7). Vero cells were grown in Dulbecco’s minimal essential medium, supplemented with 5% FCS, at 37°C.
Construction and screening of the phage libraries.
The 27-mer library consisted of 2 × 108 recombinants, each expressing the peptide sequence SRX12(S/P/T/A)A(V/A/D/E/G)X12SR at the N terminus of mature pIII of bacteriophage M13. The library was constructed by annealing and extending two long degenerate oligonucleotides, complementary to each other at their 3′ termini (11, 23). The six nucleotides of complementarity form a SacII restriction site and encode the tripeptide (S/P/T/A)A(V/A/D/E/G) (1). The 12-mer library consisted of 108 recombinants, each expressing the peptide sequence X12 at the N terminus of mature pIII. The library was constructed by annealing and extending two oligonucleotides, one of which was degenerate.
The coding scheme of the random amino acids in both libraries was NNK, where N represents equimolar ratios of A, C, G, or T and K represents G or T. In this coding strategy, the 20 amino acids are encoded by 32 codons, each with certain amino acids occurring once (C, D, E, F, H, I, K, M, N, Q, W, Y), twice (A, G, P, V, T), or three times (L, R, S).
HveA-binding phage were isolated by screening the 27-mer and 12-mer libraries as previously described (2, 11). Microtiter wells (Nunc, Inc., Naperville, Ill.) were coated overnight at 4°C or for 2 h at 22°C with 500 ng of HveA(200t) and blocked with PBS containing 1% bovine serum albumin (BSA) for 1 h at 22°C. After washing, 6 × 1011 PFU of each library was added to each well and incubated for 1 h at 22°C. The wells were washed two times with PBS containing 0.05% Tween 20. Bound phage particles were eluted with 100 mM glycine-HCl (pH 2.3), and the samples were immediately neutralized with 200 mM sodium phosphate (pH 7.4). The eluted phage particles were amplified in E. coli DH5α F′, and the affinity selection procedure repeated twice more. The amplified phage mixture obtained after the third round of amplification was plated, and positive phages were identified by confirming their ability to bind to HveA(200t) in an enzyme-linked immunosorbent assay (ELISA) in which bound phages were detected by peroxidase-labeled anti-M13 antibody (Amersham Pharmacia Biotech, Piscataway, N.J.). DNA was prepared from positive phage stocks and subjected to dideoxy sequencing as previously described (24).
Synthesis and purification of peptides.
All peptides were synthesized on an Applied Biosystems peptide synthesizer (model 431A), using F-moc amide resin. The side chain protecting groups were Cys(Trt), Cys(Acm), Arg(Pmc), Ser(tBu), and Tyr(tBu).
The peptides cyclic BP-2 and scrambled BP-2 were cyclized on resin by treatment with 1.5 equivalents of thallium III trifluoroacetate in dimethylformamide for 3 h at 22°C. The peptide-resin beads were then washed with dimethylformamide, methanol, and methanol-dichloromethane (60:40) and dried under vacuum. The peptides were cleaved from the peptide-resin by treatment with 87.5% trifluoroacetic acid (TFA), 5% phenol, 5% water, and 2.5% triisopropylsilane for 3 h at 22°C, harvested from the reaction mixture by filtration, and precipitated in cold ether. The peptide precipitates were extracted three times with cold ether, the pellets were dissolved in 50% acetonitrile containing 0.1% TFA, and the samples were lyophilized.
Disulfide oxidation of BP-1 was performed after cleavage from the resin by stirring a 0.15 mM solution of peptide in 0.1 M ammonium bicarbonate (pH 8.0, and bubbling with oxygen at 22°C for 48 h. Linear BP-1 was obtained by maintaining the side chain protecting groups on the Cys residues Cys(Acm). All of the crude peptides were dissolved in 10% acetonitrile containing 0.1% TFA and purified by reversed-phase high-performance liquid chromatography on an automated system (Prep-LC 4000; Waters, Milford, Mass.) with a C18 column (Vydac, Western Analytical Products, Inc., Murrieta, Calif.). The column was initially equilibrated with 5% buffer B (0.1% TFA in water) for 15 min at a flow rate of 20 ml/min, and peptide fractions were eluted with a 50-min linear gradient of 1 liter of 5 to 90% buffer B (0.1% TFA in acetonitrile) at a flow rate of 20 ml/min. The elution profile of the peptide fractions was monitored by UV detection at 230 nm, and the major peak containing the desired peptide was collected and lyophilized. The purity of the final products was assessed by analytical high-performance liquid chromatography and matrix-assisted laser desorption mass spectrometry, using a time-of-flight mass spectrometer (MicroMass TofSpec; Micromass Inc., Beverly, Mass.) (3, 17, 18).
ELISAs.
Several ELISAs were performed to analyze the interaction between HveA, the isolated phage peptides, gD, and LT-α. In these assays, microtiter wells were coated for 2 h at 22°C with 40 ng of HveA(200t), human factor H, trout complement C3, BSA, milk, or ovalbumin. Nonspecific binding in the wells was blocked with PBS containing 1% BSA for 1 h at 22°C. For competition assays, serial dilutions of gD-1(306t), gD-1(Δ290-299), BSA, peptide BP-1 or BP-2, or an unrelated cyclic peptide (ICVVQDWGHHRCT) was added to each well and incubated for 30 min at 22°C. Recombinant protein [gD-1(306t) at 0.4 μg/ml, gD-1(Δ290-299t) at 0.1 μg/ml, or LT-α at 20 nM] or phage supernatant was added to each well, and the samples were incubated for 1 h. The wells were washed twice with PBS containing 0.05% Tween 20 and incubated with either (i) a 1:1,000 dilution of a peroxidase-labeled anti-M13 antibody, (ii) a 1:400 dilution of an anti-gD polyclonal antibody (R7), or (iii) an anti-LT-α monoclonal antibody (AG9; 1 μg/ml) for 1 h at 22°C. The wells were washed with PBS containing 0.05% Tween 20 and then incubated with a 1:1,000 dilution of peroxidase-conjugated goat anti-rabbit immunoglobulin G (for polyclonal antibody detection) or goat anti-mouse immunoglobulin G (for monoclonal antibody detection) (Bio-Rad Laboratories, Richmond, Calif.). The plates were incubated for an additional 30 min at 22°C. Color was developed by adding 2.2′-azino-di-[3-ethylbenzthiazolinesulfonate (6)] (ABTS; Boehringer Mannheim) and 0.05% hydrogen peroxide, and the optical density was read at 405 nm. Net gD-1 or LT-α binding was calculated by subtracting the readings of secondary antibody to HveA binding from the readings of ligand binding to HveA.
HSV entry assays.
CHO-HveA, CHO-HveC, or Vero cells, were plated in 96-well dishes and incubated overnight. The cells were chilled at 4°C for 10 min, and the medium was replaced with Ham’s F-12 (CHO-HveA and CHO-HveC cells) or Dulbecco’s modified Eagle medium DMEM (Vero cells) with 10% fetal bovine serum and 10 mM HEPES, containing various concentrations of peptide, previously filtered through a 0.2-μm-pore-size filter (Corning Glass Works, Corning, N.Y.). The plates were rocked at 4°C for 90 min, at which time KOS-hrR3 virus (5 × 105 PFU/well) was added. The plates were rocked at 4°C for an additional 90 min and then incubated at 37°C for 5 h. Cells were lysed with 20% Nonidet P-40, and substrate (o-nitrophenyl-β-d-glucopyranoside) was added to each well. β-Galactosidase activity (milli-optical density units per minute) was measured at various time points with a Spectromax 250 ELISA reader at 560 nm.
RESULTS
Isolation and characterization of HveA-binding phages.
To isolate peptide ligands to the extracellular domain of HveA, we screened two phage libraries displaying combinatorial peptides 27 and 12 amino acids in length. Phage particles expressing HveA-binding peptides were affinity selected against HveA(200t) immobilized on microtiter plate wells. After three rounds of selection, individual phage clones were isolated and tested for binding to HveA.
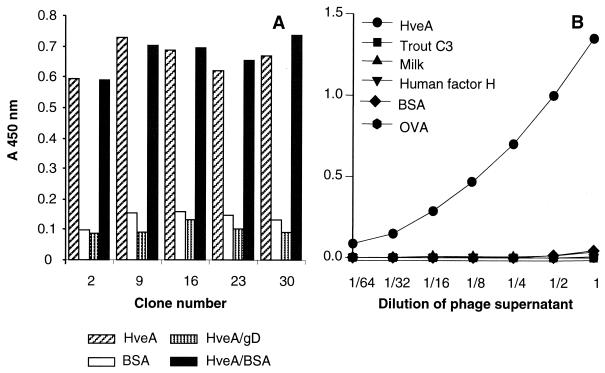
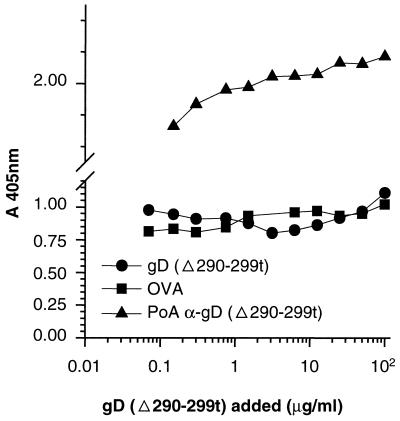
The binding of the phages was deemed to be specific, as they did not bind to trout complement C3, milk, ovalbumin, BSA, or human factor H (Fig. 1B). In addition, soluble gD-1(306t) could compete the binding of the phage to immobilized HveA(200t) (Fig. 1A).
FIG. 1.
Specific binding to HveA of positive clones isolated from two phage-displayed random peptide library. (A) Microtiter wells were coated with HveA(200t) and incubated with dilutions of gD-1(306t) or BSA. Phage supernatant from five positive clones isolated from the 26-mer library was added. Similar results were obtained with positive clones from the 12-mer library. (B) Microtiter wells were coated with HveA(200t), BSA, milk, trout complement C3, human complement factor H, or ovalbumin (OVA). Phage supernatant from a positive clone isolated from a 12-mer library was added. Bound phage was detected with peroxidase-conjugated anti-M13 antibody and ABTS peroxidase substrate.
DNA obtained from several positive clones was purified and sequenced. All positive clones from each of the libraries had the same sequence, indicating that the clones had been amplified during the second and third rounds of biopanning. The deduced amino acid sequences of the clones were SISCSRGLVCLLPRLTNESGNDRFDS (BP-1) from the 27-mer library and GSCDGFRVCYMH (BP-2) from the 12-mer library. The amino acid sequence of the positive clones from the 27-mer library does not agree with the original design of the library, X12(S/P/T/A)A(V/A/D/E/G)X12. The central sequence of the peptide in this region was PR instead of the expected (S/P/T/A)A(V/A/D/E/G). Thus, BP-1 is only 26 amino acids in length. We have searched the GenBank (DNA) and SwissProt (protein) databases for sequences showing similarity to BP-1 and BP-2, using BlastP, BlastN, and BlastX, and found none.
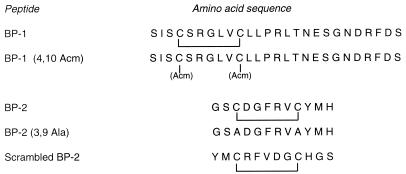
To further characterize the binding properties of the isolated clones, we synthesized two peptides corresponding to the deduced amino acid sequences of the phage-displayed peptides (Fig. 2). It is interesting that although the two isolated clones are not similar in sequence, binding of phage displaying BP-1 to HveA was inhibited in an ELISA by peptide BP-2, and vice versa (Fig. 3).
FIG. 2.
Amino acid sequences of HveA-binding peptides and their variants.
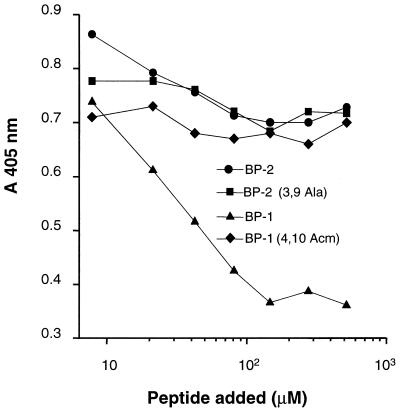
FIG. 3.
Inhibition of phage binding to HveA by BP-1 and BP-2. Microtiter wells were coated with HveA(200t) and incubated with decreasing concentrations of peptide and phage displaying BP-2 (A) or BP-1 (B). Bound phage particles were detected with peroxidase-conjugated anti-M13 antibody and ABTS peroxidase substrate.
Inhibition of gD-1 binding to HveA.
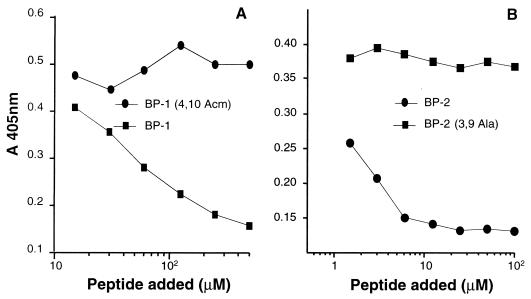
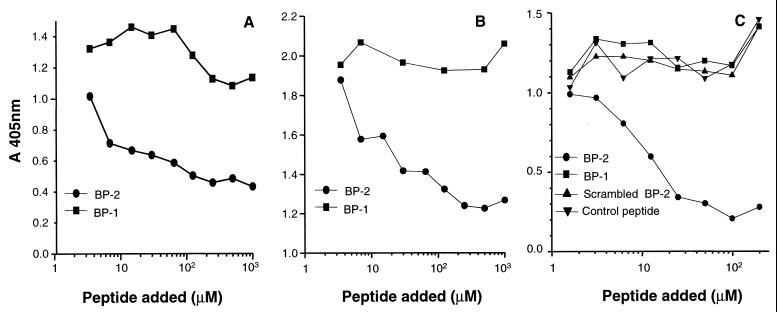
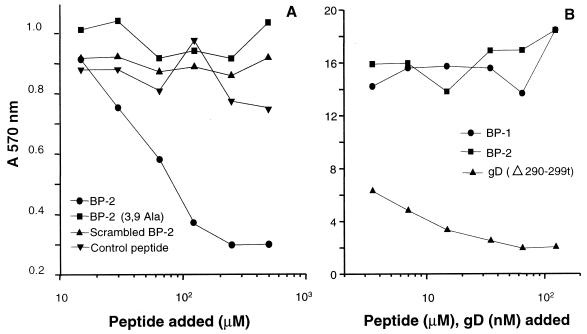
The two synthetic peptides, BP-1 and BP-2, were next tested for the ability to compete the binding of gD-1 to the HveA receptor (Fig. 4). Binding of soluble wild-type gD-1, gD-1(306t), to immobilized HveA(200t) was reduced by BP-2 to a level of 80% and by BP-1 to a level of 20% at a concentration of 0.5 mM (Fig. 4A). Binding of the mutant form gD-1(Δ290-299t), which has a 100-fold-higher affinity for HveA, was inhibited to a level of 40% by BP-2 (Fig. 4B). Conversely, no inhibitory effect was observed with a blocked cysteine [BP-1 (4,10 Acm)], alanine replacement [BP-2 (3,9 Ala)], a peptide with a scrambled sequence of BP-2, or a cyclic peptide with a completely different sequence (Fig. 4C). Since these negative results included peptides BP-1 (4,10 Acm) and BP-2 (3,9 Ala), in which no disulfide bond is present, it appears that disulfide bond formation is important for maintaining the inhibitory activity of both BP-1 and BP-2.
FIG. 4.
Inhibition of gD-1(306t) binding to HveA by BP-2 and BP-1. Microtiter wells were coated with HveA(200t) and incubated with decreasing concentrations of BP-2 or BP-1. gD-1(306t) (A) or gD-1(Δ290-299t) (B) was added. (C) In this ELISA, HveA-coated plates were incubated with decreasing concentrations of BP-2 and control peptides before gD-1(306t) was added. Bound gD-1 was detected with a polyclonal antibody anti-gD (R7) followed by incubation with peroxidase-conjugated secondary antibody and ABTS. The data were obtained by subtracting nonspecific binding of anti-gD antibody to peptide bound to HveA.
Inhibition of LT-α binding to HveA.
The effect of synthetic peptides BP-1 and BP-2 on LT-α binding to HveAt was analyzed by ELISA. HveAt was coated onto the wells of a microtiter plate and incubated with various concentrations of peptides along with 20 nM LT-α, and the amount of LT-α bound was detected with a monoclonal antibody. As seen in Fig. 5, binding of soluble LT-α to HveAt was inhibited by BP-1, whereas neither BP-2 nor the control peptides had an inhibitory effect. This assay indicates again that peptide BP-1 requires cyclization to bind to HveAt. On the other hand, BP-2 did not inhibit LT-α binding to the receptor, suggesting that BP-1 and BP-2 bind to different sites on HveAt.
FIG. 5.
Inhibition of LT-α binding to HveA by BP-1, BP-2, and control peptides. HveA-coated microtiter plate wells were incubated with concentrations of peptides, and LT-α was added. Bound LT-α was detected with an anti-LT-α monoclonal antibody (AG9), followed by incubation with peroxidase-conjugated secondary antibody and ABTS peroxidase substrate.
Inability of gD to inhibit LT-α binding to HveA.
Given that we had discovered that the peptides appeared to bind two different sites on HveA, we investigated whether gD and LT-α bind HveA in a competitive manner (Fig. 6). An ELISA was designed to compare the bindings of gDt and LT-α to HveAt. In this assay, HveAt was first immobilized on the plate and incubated with a constant amount of LT-α in the presence of various amounts of gDt. The amount of LT-α bound to HveAt was detected with a monoclonal antibody to LT-α (AG9). To confirm that gDt remained bound to HveAt during the wash steps, bound gDt was detected with a polyclonal anti-gD antibody, R7, in separate wells of the same ELISA plate. We found that the same amount of LT-α bound to HveAt regardless of how much gDt was added to the plate wells. Thus, gDt had no inhibitory effect on LT-α binding to the receptor, suggesting that these proteins do not compete for the same binding sites on HveAt.
FIG. 6.
Lack of competition by gD on LT-α binding to HveA. HveA-coated microtiter plate wells were incubated with various concentrations of gD, and LT-α was added. Bound LT-α and gD were detected with an anti-LT-α monoclonal antibody (AG9) and a polyclonal antibody (R7), respectively, followed by incubation with a peroxidase-conjugated secondary antibody and ABTS peroxidase substrate.
Inhibition of HSV-1 entry into cells by BP-1 and BP-2.
The inhibition of gD-1 binding to HveAt by BP-2 raised the question of whether this peptide has any effect on viral entry into mammalian cells. To test this possibility, we examined the effect of the 12-mer peptide on HSV-1 entry into CHO-HveA, CHO-HveC, and Vero cells. Cell monolayers were incubated with peptide at 4°C for 90 min and then infected with the β-galactosidase reporter virus KOS-hrR3. BP-2 blocked HSV entry into CHO-HveA cells; in contrast, neither BP-1 nor any of the control peptides blocked virus entry (Fig. 7A). Neither peptide had any effect on HSV entry into Vero (Fig. 7B) and CHO-HveC cells (data not shown) cells. Thus, although both BP-1 and BP-2 bind to HveA, only BP-2 is able to inhibit gD-1 binding to HveA effectively enough to block viral entry into CHO-HveA cells. This result is consistent with the data in Fig. 4 showing that only BP-2 effectively blocked the binding of gDt to HveAt.
FIG. 7.
BP-2 can block HSV entry into CHO-HveA cells (A) but not into Vero cells (B). Cell monolayers were incubated with dilutions of peptides at 4°C for 90 min. KOS-hrR3 was then added and incubated for an additional 90 min at 4°C for adsorption. Five hours later (at 37°C), the cells were lysed and β-galactosidase was measured. The data are the averages of duplicate wells; background β-galactosidase activity (cells alone) has been subtracted.
DISCUSSION
Several receptors of HSV that mediate postattachment entry into the cell have recently been described. The first to be characterized was HveA, a member of the TNFR family, which allows entry of most HSV strains into nonpermissive CHO cells (16). This process was shown to be mediated by direct binding of the viral glycoprotein gD to HveA (19, 34). In addition to binding to gD, the same cell surface protein was found to interact with several host proteins including, LT-α, LIGHT, and TRAF proteins (8, 14, 15).
In this study, we used two different phage-displayed combinatorial peptide libraries to identify HveA-binding peptides that affect the interaction of the receptor with its ligands. We have dissected the interaction between the proteins gD, LT-α, and HveA in the context of viral entry.
After three rounds of biopanning, several positive clones from each library that bound specifically to HveA(200t) were isolated. These clones did not bind to any of several other proteins, including trout complement C3, human complement factor H, ovalbumin, and milk. This specificity was corroborated by the fact that soluble gD-1(306t) inhibited phage binding to HveAt. To our surprise, all of the positive clones from each library had the same sequence. This result suggests that the unique HveA-binding clones were amplified during the second and third rounds of library biopanning. It is interesting that the two identified peptide ligands, while differing in primary structure, each had two cysteine residues. Experiments with Ala-substituted Cys or side chain-blocked Cys demonstrated that the intramolecular disulfide bond is necessary for peptide binding to HveA. It was found that both synthetic peptides BP-1 and BP-2 were unable to inhibit gDt binding to HveAt when their two Cys residues were not disulfide linked. The same effect was observed for BP-1 in relation to inhibition of LT-α binding to HveAt; the linear form of this peptide was unable to inhibit the LT-α–HveA interaction. These results suggest that the intramolecular disulfide bond is essential to maintain the binding and inhibitory activities of these peptides. The binding of phage clones to HveA suggests that the disulfide bond in the peptides is also formed when they are expressed on the surface of the phages; the oxidizing environment of the bacterial periplasm has been shown to allow disulfide bond formation between Cys residues in phage-displayed peptides (12).
The marginal effect observed on the inhibition of fluid-phase gDt binding to HveAt by BP-1 contrasts that seen for BP-2, which inhibited not only the binding of soluble gD-1(306t) but also the binding of soluble gD-1(Δ290-299t), a mutant form of gD having a 100-fold-higher affinity for HveAt (21, 34, 36). Furthermore, BP-2 blocked viral entry into CHO-HveA cells, where gD-1(306t) had failed to do so in previous studies. It is possible that BP-2 has a higher affinity for HveA than does the wild-type gD-1(306t), and therefore large amounts of gD-1(306t) are needed for inhibition. The failure of BP-2 to block viral entry into CHO-HveC cells suggests that this peptide binds specifically to HveA. Since BP-2 also fails to block HSV entry into Vero cells, it appears likely that entry mediators other than or in addition to HveA are expressed. Alternatively, it is possible that the site in HveA to which the peptides bind is not conserved in the primate Vero cell homolog of HveA.
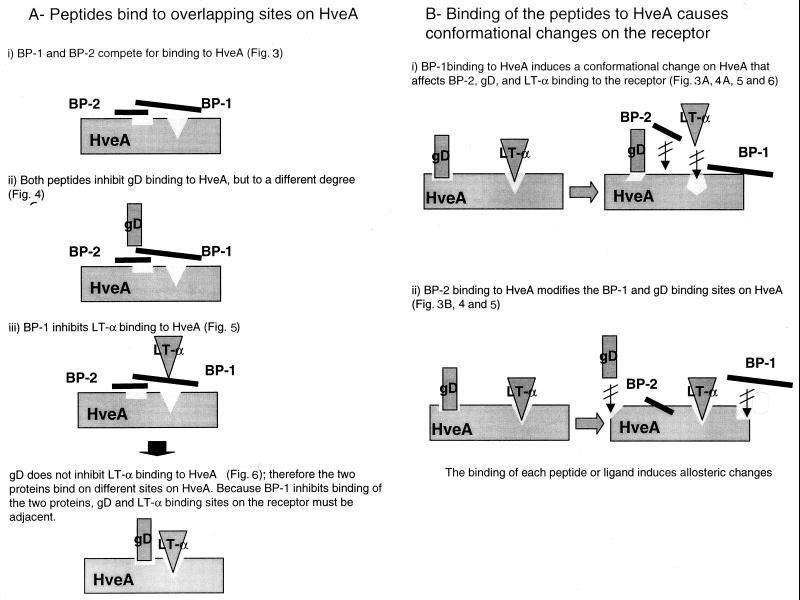
The differential inhibitory effects of the two peptides on LT-α and gDt binding to HveAt suggest that BP-1 and BP-2 bind to different sites on HveA. Because binding of BP-1 to HveAt is inhibited by BP-2, and vice versa, we suggest that either the peptides bind to overlapping sites on HveAt or the binding of one peptide to HveAt induces conformational changes in the receptor that influence the binding of the other peptide (26) (Fig. 8). In the first model, BP-1 and BP-2 may partially share binding sites on HveAt and thus compete for binding to those sites. Accordingly, although LT-α binding to HveAt was inhibited only by BP-1, the HveA site to which gD binds could include structural elements involved in the binding of both peptides. In this way, both peptides could affect the interaction between gDt and HveAt. However, BP-1 inhibits only partially the binding of gD to HveA, and thus it appears that the gD-binding site on HveA is nearer the BP-2 site than the BP-1 site. The remarkable inhibitory effect of BP-2 on gDt binding to HveAt, which led to inhibition of HSV entry into CHO-HveA cells, suggests that BP-2 may have a high affinity for the receptor. In addition, BP-2 may compete with gDt for more than one binding site on HveAt. This possibility is consistent with a recent study in which monoclonal antibodies recognizing two different regions on gD, Ib (amino acids 222 to 252) and VII (amino acids 11 to 19), were found to block HSV binding to HveA (19). If the peptides bound to overlapping sites on HveA, then gD and LT-α would bind to adjacent sites on the receptor.
FIG. 8.
Two models of the proposed interactions between HveA and BP-1, BP-2, gD, and LT-α.
A second interpretation of our data is that binding of the peptides to the receptor causes conformational changes in HveA. Thus, BP-1 binding to HveAt could cause a conformational change in the receptor that interferes with BP-2, LT-α, and gDt binding to HveA. On the other hand, BP-2 binding to HveAt could alter BP-1 and gDt binding sites on HveA.
HveA is not the only mediator of HSV entry into the cell. Other cell surface receptors have recently been isolated and are currently being studied (6, 33); the role and importance of these new receptors in HSV entry have yet to be determined. Viral entry is a key step in infection, and we have now succeeded in isolating a peptide that blocks the interaction between HveA and HSV in vitro. Furthermore, we have identified one site on HveA, where BP-2 binds, which when it is occupied prevents viral uptake but not the binding of a cellular ligand, LT-α. Therefore, screening chemical compound libraries for those that displace BP-2 but not BP-1 could lead to the development of a useful drug to prevent HSV infection. We conclude that although further studies are needed to assess the potential therapeutic usefulness of BP-2, we believe that peptides BP-1 and BP-2 are promising tools for analyzing the mechanisms of viral entry into cells and HveA-host ligand interactions and functions.
ACKNOWLEDGMENTS
We thank A. Sahu, W. T. Moore, and A. Soulika for helpful suggestions, D. McClellan for editorial assistance, and Yvonne Harrison-Shahan for excellent technical assistance.
This work was supported by National Institutes of Health grants NS-36731 and NS-30606, National Institute of Allergy and Infectious Diseases grants AI-30040 and AI-18289, and Cancer and Diabetes Centers Core Support grants CA-16520 and DK-19525.
REFERENCES
- 1.Adey N, Kay B. Identification of calmodulin-binding peptide consensus sequences from a phage-displayed random peptide library. Gene. 1996;169:133–134. doi: 10.1016/0378-1119(95)00804-7. [DOI] [PubMed] [Google Scholar]
- 2.Adey N B, Guo R, Hanson H L, Rider J E, Sparks A B, Kay B K. Construction and screening of M13 phage-displayed random peptide libraries. Methods Mol Cell Biol. 1996;6:3–14. [Google Scholar]
- 3.Angeletti R H, Bibbs L, Bonewald L F, Fields G B, McMurray J S, Moore W T, Stults J T. Formation of disulfide bond in an octreotide-like peptide: a multicenter study. In: Marshak D R, editor. Techniques in protein chemistry VII. San Diego, Calif: Academic Press; 1996. pp. 261–274. [Google Scholar]
- 4.Browning J L, Dougas I, Ngamek A, Bourdon P R, Ehrenfels B N, Miatkowski K, Zafari M, Yampaglia A M, Lawton P, Meier W, Benjamin C P, Hession C. Characterization of surface lymphotoxin forms—use of specific monoclonal-antibodies and soluble receptors. J Immunol. 1995;154:33–46. [PubMed] [Google Scholar]
- 5.Crowe P D, Vanarsdale T L, Walter B N, Dahms K M, Ware C F. Production of lymphotoxin (LT-alpha) and a soluble dimeric form of its receptor using the baculovirus expression system. J Immunol Methods. 1994;168:79–89. doi: 10.1016/0022-1759(94)90212-7. [DOI] [PubMed] [Google Scholar]
- 6.Geraghty R J, Krummenacher C, Cohen G H, Eisenberg R J, Spear P G. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 7.Golstein D J, Weller S K. Herpes simplex virus type 1-induced ribonucleotide reductase activity is dispensable for virus growth and DNA synthesis: isolation and characterization of an ICP6 lacZ insertion mutant. J Virol. 1988;62:196–205. doi: 10.1128/jvi.62.1.196-205.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu H L, Solovyev I, Colombero A, Elliott R, Kelley M, Boyle W J. ATAR, a novel tumor necrosis factor receptor family member, signals through TRAF2 and TRAF5. J Biol Chem. 1997;272:13471–13474. doi: 10.1074/jbc.272.21.13471. [DOI] [PubMed] [Google Scholar]
- 9.Isola V J, Eisenberg R J, Siebert G R, Heilman C J, Wilcox W C, Cohen G H. Fine mapping of antigenic site II of herpes simplex virus glycoprotein D. J Virol. 1989;63:2325–2334. doi: 10.1128/jvi.63.5.2325-2334.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kay B K. Mapping protein-protein interactions with biologically expressed random peptide libraries. Persp Drug Discov Des. 1995;2:251–268. [Google Scholar]
- 11.Kay B K, Adey N B, He Y-S, Manfredi J P, Mataragnon A H, Fowlkes D M. An M13 phage library displaying random 38-amino acid peptides as a source of novel sequences with affinity to select targets. Gene. 1993;128:59–65. doi: 10.1016/0378-1119(93)90153-t. [DOI] [PubMed] [Google Scholar]
- 12.Kremser A, Rasched I. The adsorption of filamentous phage fd: assignment of its disulfide bridges and identification of the domain incorporated in the coat. Biochemistry. 1994;33:13954–13958. doi: 10.1021/bi00250a051. [DOI] [PubMed] [Google Scholar]
- 13.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;277:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 14.Marsters S A, Ayres T M, Skubatch M, Gray C L, Rothe M, Ashkenazi A. Herpesvirus entry mediator, a member of the tumor necrosis factor receptor (TNFR) family, interacts with members of the TNFR-associated factor family and activates the transcription factors NF-kappa B and AP-1. J Biol Chem. 1997;272:14029–14032. doi: 10.1074/jbc.272.22.14029. [DOI] [PubMed] [Google Scholar]
- 15.Mauri D N, Ebner R, Montgomery R I, Kochel K D, Cheung T C, Yu G L, Ruben S, Murphy M, Eisenberg R J, Cohen G H, Spear P G, Ware C F. LIGHT, a new member of the TNF superfamily, and lymphotoxin alpha are ligands for herpesvirus entry mediator. Immunity. 1998;8:21–30. doi: 10.1016/s1074-7613(00)80455-0. [DOI] [PubMed] [Google Scholar]
- 16.Montgomery R I, Warner M S, Lum B J, Spear P G. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 17.Moore W T. Integration of mass spectrometry into strategies for peptide synthesis. Biol Mass Spectrom. 1993;22:149–162. doi: 10.1002/bms.1200220303. [DOI] [PubMed] [Google Scholar]
- 18.Moore W T. Laser desorption mass spectrometry. Methods Enzymol. 1997;289:520–542. doi: 10.1016/s0076-6879(97)89062-3. [DOI] [PubMed] [Google Scholar]
- 19.Nicola A V, de Leon M P, Xu R L, Hou W F, Whitbeck J C, Krummenacher C, Montgomery R I, Spear P G, Eisenberg R J, Cohen G H. Monoclonal antibodies to distinct sites on herpes simplex virus (HSV) glycoprotein D block HSV binding to HVEM. J Virol. 1998;72:3595–3601. doi: 10.1128/jvi.72.5.3595-3601.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicola A V, Peng C R, Lou H, Cohen G H, Eisenberg R J. Antigenic structure of soluble herpes simplex virus (HSV) glycoprotein D correlates with inhibition of HSV infection. J Virol. 1997;71:2940–2946. doi: 10.1128/jvi.71.4.2940-2946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicola A V, Willis S H, Naidoo N N, Eisenberg R J, Cohen G H. Structure-function analysis of soluble forms of herpes simplex virus glycoprotein D. J Virol. 1996;70:3815–3822. doi: 10.1128/jvi.70.6.3815-3822.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roller R J, Rauch D. Herpesvirus entry mediator HVEM mediates cell-cell spread in BHK(TK−) cell clones. J Virol. 1998;72:1411–1417. doi: 10.1128/jvi.72.2.1411-1417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sahu A, Kay B K, Lambris J D. Inhibition of human complement by a C3-binding peptide isolated from a phage displayed random peptide library. J Immunol. 1996;157:884–891. [PubMed] [Google Scholar]
- 24.Sanger F S, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sisk W P, Bradley J D, Leopold R J, Stoltzfus A M, DeLeon M P, Hilf M, Peng C, Cohen G H, Eisenberg R J. High-level expression and purification of secreted forms of herpes simplex virus type 1 glycoprotein gD synthesized by baculovirus-infected insect cells. J Virol. 1994;68:766–775. doi: 10.1128/jvi.68.2.766-775.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slepenkov S V, Witt S N. Peptide-induced conformational changes in the molecular chaperone DnaK. Biochemistry. 1998;37:16749–16756. doi: 10.1021/bi981738k. [DOI] [PubMed] [Google Scholar]
- 27.Sodora D L, Cohen G H, Eisenberg R J. Influence of asparagine-linked oligosaccharides on antigenicity, processing, and cell surface expression of herpes simplex virus type 1 glycoprotein D. J Virol. 1989;63:5184–5193. doi: 10.1128/jvi.63.12.5184-5193.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sodora D L, Cohen G H, Muggeridge M I, Eisenberg R J. Absence of asparagine-linked oligosaccharides from glycoprotein D of herpes simplex virus type 1 results in a structurally altered but biologically active protein. J Virol. 1991;65:4424–4431. doi: 10.1128/jvi.65.8.4424-4431.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tal-Singer R, Eisenberg R J, Valyinagy T, Fraser N W, Cohen G H. N-linked oligosaccharides on herpes-simplex virus glycoprotein GD are not essential for establishment of viral latency or reactivation in the mouse eye model. Virology. 1994;202:1050–1053. doi: 10.1006/viro.1994.1437. [DOI] [PubMed] [Google Scholar]
- 30.Terry-Allison T, Montgomery R I, Whitbeck J C, Xu R L, Cohen G H, Eisenberg R J, Spear P G. HveA (herpesvirus entry mediator A), a coreceptor for herpes simplex virus entry, also participates in virus-induced cell fusion. J Virol. 1998;72:5802–5810. doi: 10.1128/jvi.72.7.5802-5810.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tessier D C, Thomas D Y, Khouri H E, Lalibertie F, Vernet T. Enhanced secretion from insect cells of a foreign protein fused to the honeybee melittin signal peptide. Gene. 1991;98:177–183. doi: 10.1016/0378-1119(91)90171-7. [DOI] [PubMed] [Google Scholar]
- 32.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Warner M S, Geraghty R J, Martinez W M, Montgomery R I, Whitbeck J C, Xu R L, Eisenberg R J, Cohen G H, Spear P G. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by mutants of herpes simplex virus type 1, herpes simplex virus type 2, and pseudorabies virus. Virology. 1998;246:179–189. doi: 10.1006/viro.1998.9218. [DOI] [PubMed] [Google Scholar]
- 34.Whitbeck J C P, Peng C, Lou H, Xu R, Willis S H, Ponce de Leon M, Peng T, Nicola A V, Montgomery R I, Warner M S, Sooulika A M, Spruce L A, Moore W T, Lambris J D, Spear P G, Cohen G H, Eisenberg R J. Glycoprotein D of herpes simplex virus (HSV) binds directly to HVEM, a member of the tumor necrosis factor receptor superfamily and a mediator of HSV entry. J Virol. 1997;71:6083–6093. doi: 10.1128/jvi.71.8.6083-6093.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams-Abbott L, Walter B N, Cheung T C, Goh C R, Porter A G, Ware C F. The lymphotoxin-alpha (LT alpha) subunit is essential for the assembly, but not for the receptor specificity, of the membrane-anchored LT alpha 1 beta 2 heterotrimeric ligand. J Biol Chem. 1997;272:19451–19456. doi: 10.1074/jbc.272.31.19451. [DOI] [PubMed] [Google Scholar]
- 36.Willis S H, Rux A H, Peng C, Whitbeck J C, Nicola A V, Lou H, Hou W F, Salvador L, Eisenberg R J, Cohen G H. Examination of the kinetics of herpes simplex virus glycoprotein D binding to the herpesvirus entry mediator, using surface plasmon resonance. J Virol. 1998;72:5937–5947. doi: 10.1128/jvi.72.7.5937-5947.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]