Abstract
EBNA3C can specifically repress the expression of reporter plasmids containing EBV Cp latency-associated promoter elements. Cp is normally the main promoter for EBNA mRNA initiation, so it appears that EBNA3C contributes to a negative autoregulatory control loop. By mutational analysis it was previously established that this repression is consistent with EBNA3C being targeted to Cp by binding the cellular sequence-specific DNA-binding protein CBF1 (also known as recombination signal-binding protein [RBP]-Jκ. Further analysis suggested that in vivo a corepressor interacts with EBNA3C in this DNA binding complex. Results presented here are all consistent with a component of such a corepressor exhibiting histone deacetylase activity. The drug trichostatin A, which specifically inhibits histone deacetylases, relieved two- to threefold the repression of Cp induced by EBNA3C in two different cell types. Moreover, repression of pTK-CAT-Cp4× by EBNA3C was specifically enhanced by cotransfection of an expression plasmid for human histone deacetylase-1 (HDAC1). Consistent with these functional assays, in vitro-translated HDAC1 bound to a glutathione S-transferase (GST) fusion protein including full-length EBNA3C, and in the reciprocal experiment EBNA3C bound to a GST fusion with the N terminus of HDAC1. Coimmunoprecipitations also revealed an EBNA3C-HDAC1 interaction in vivo, and GST-EBNA3C bound functional histone deacetylase enzyme activity from HeLa cell nuclear extracts. The region of EBNA3C involved in the interaction with HDAC1 appears to correspond to the region which is necessary for binding to CBF1/RBP-Jκ. A direct physical interaction between EBNA3C and HDAC1 was demonstrated with recombinant proteins purified from bacterial cells, and we therefore conclude that HDAC1 and CBF1/RBP-Jκ bind to the same or adjacent regions of EBNA3C. These data suggest that recruitment of histone deacetylase activity makes a significant contribution to the repression of transcription from Cp because EBNA3C bridges an interaction between CBF1/RBP-Jκ and HDAC1.
Infection in vitro by Epstein-Barr virus (EBV) induces the activation and continuous proliferation of a subset of resting human B cells. The resulting lymphoblastoid cell lines (LCLs), which have a phenotype resembling that of activated B blasts, express only nine latent EBV proteins. There are six nuclear proteins (EBNA1, -2, -3A, -3B, -3C, and -LP) and three membrane-associated proteins (LMP1, -2A, and -2B). Together they activate the quiescent B cells from G0 into the cell cycle, maintain continuous proliferation in the absence of specific growth factors or T-cell help, maintain the viral genome in a latent episomal form, and probably prevent the cells from undergoing terminal differentiation or apoptosis (reviewed in reference 22). In vivo this ability of the virus to drive cell proliferation is important for the amplification and spread of clones of cells capable of entering the memory B-cell population and sustaining a long-term persistent infection (6). Expression of the “proliferation program” in vivo is thought to produce dividing cells equivalent to the in vitro LCLs, and as a result, EBV can be the causative agent in infectious mononucleosis. Uncontrolled proliferation of such cells in the immunocompromised can result occasionally in the development of immunoblastic or large-cell lymphoma. In addition, EBV is associated with at least three types of human tumors in the immunocompetent: Burkitt’s lymphoma, nasopharyngeal carcinoma, and Hodgkin’s disease (reviewed in reference 39).
Three of the nuclear antigens, EBNA3A, -3B, and -3C, are considered to constitute a family which probably arose by gene duplication, since they have limited but significant amino acid sequence homology, have the same gene structure (a short 5′ exon and a long 3′ exon), and are arranged tandemly in the EBV genome (3, 18, 36, 40, 43). Genetic studies using recombinant EBV have shown that EBNA3A and EBNA3C are essential for in vitro immortalization of B cells, whereas EBNA3B is dispensable (22, 44, 45). EBNA3C is a large protein (992 amino acids [aa]) which is likely to be multifunctional. It is first expressed in EBV-infected resting human B cells during activation into their first cell cycle; the steady-state level of EBNA3C in the LCLs produced is then low and remarkably constant (1, 4). There is some evidence that EBNA3C may contribute to the immortalization process by disrupting cell cycle regulation, since it can substitute for papillomavirus E7 and adenovirus E1A in cooperation with activated Ras to immortalize and transform primary rodent fibroblasts (34).
In transient transfection assays, EBNA3C can modulate the EBNA2-mediated activation of the LMP1 and LMP2A promoters (24, 29, 41). The N-terminal 250 aa of EBNA3C contain a binding site for the cellular sequence-specific DNA-binding protein CBF1 (also known as recombination signal-binding protein [RBP]-Jκ) (19, 42, 57). EBNA2 also binds to CBF1/RBP-Jκ and is targeted to DNA by the interaction (14, 17, 25, 26, 47, 58). This directs a strong activation domain in EBNA2 to promoters and also interferes with the interaction between CBF1/RBP-Jκ and a corepressor; the net result is strong activation of transcription (46, 58). It has been suggested that EBNA3C binding to CBF1/RBP-Jκ prevents the association of EBNA2–CBF1/RBP-Jκ complexes with DNA and that in this way EBNA3C represses the EBNA2-mediated activation of various natural and synthetic promoters, which include CBF1/RBP-Jκ sites (19, 41, 48, 57). However, fusions of EBNA3C with the DNA binding domain of the Saccharomyces cerevisiae transactivator GAL4 have shown that, when tethered to DNA, EBNA3C is a very potent repressor of reporter gene transcription in its own right (7, 48). Subsequently, we also showed that EBNA3C specifically represses Cp, the major promoter for EBNA expression in LCLs. Repression occurred even when Cp was not activated by EBNA2, and the data were consistent with EBNA3C being targeted to Cp by CBF1/RBP-Jκ (37). However, the precise nature of the interaction between EBNA3C and Cp was not determined, and although all the data were consistent with repression involving additional proteins, the nature of the corepressor(s) in this regulatory complex was not investigated.
Recent studies have shown that the regulation of chromatin structure is an important mechanism in controlling gene transcription (reviewed in references 15, 20, and 35). Specifically, nucleosomes—which organize the structure of chromosomal DNA—have been shown to inhibit transcription. Nucleosomes are composed of histones H2A, H2B, H3, and H4, and their formation is regulated by posttranslational modifications of histone amino termini; the best understood of these modifications is acetylation and deacetylation. Acetylation of histones neutralizes the positive charge on lysines, and this disrupts nucleosome structure and allows DNA access to transcription factors; consequently, acetylase activity stimulates transcription. Several transcriptional coactivators, such as p300/CBP and P/CAF, have been shown to exhibit histone acetyltransferase activity (8, 33, 38, 53).
Conversely, various transcriptional repressors have been shown to associate with histone deacetylases. This class of repressor protein includes Mad (which forms heterodimers with Max) (2, 5, 16, 23), unliganded nuclear hormone receptors (12, 32), and the complex of E2F with the retinoblastoma protein pRb (9, 27, 28). These multiprotein complexes, consisting of sequence-specific DNA-binding proteins, accessory proteins, and corepressors, are thought to deacetylate histones on the promoter and thereby promote nucleosome formation, leading to transcriptional repression.
The focus of this study has been to determine whether EBNA3C resembles repressors like Mad-Max, nuclear hormone receptors, and pRb-E2F and associates in a complex with proteins which repress transcription by modifying chromatin. Specifically we investigate whether EBNA3C interacts with histone deacetylase to repress transcription.
MATERIALS AND METHODS
Reporter plasmids.
p-1425-Luc contains the EBV Cp latency promoter upstream of a luciferase reporter gene (37).
pTK-CAT-Cp4× includes multiple CBF1/RBP-Jκ binding sites from Cp cloned upstream of the herpesvirus thymidine kinase (TK) promoter and has been described previously (46, 48).
Expression vectors.
pSG5-EBNA3C (37), pBKCMV-EBNA3CΔ346-543 (37), pING14A-HDAC1 (9), and pcDNA3–HDAC1-F (9) have been described previously. Plasmid pcDNA3–HDAC1-F contains the entire open reading frame of histone deacetylase 1 (HDAC1) (aa 1 to 482) and was used for transient transfection in mammalian cells. The HDAC1 protein is “flag” tagged for immunodetection.
The pGEX–RBP-Jκ fusion protein contains the entire 500-aa CBF1/RBP-Jκ open reading frame cloned in-frame with the glutathione S-transferase (GST) gene in the pGEX vector (Pharmacia Biotech).
pGEX-2TKP-HDAC1 aa 1-382 and pGEX-2TKP-HDAC1 aa 1-432 contain the coding sequences indicated cloned in-frame with the GST moiety in pGEX-2TK.
pET30a-HDAC1 contains the sequence coding for HDAC1 protein, fused in-frame to six histidine residues, cloned in the vector pET30a.
GST-EBNA3C fusion protein was obtained from a recombinant baculovirus (30). A full-length EBNA3C cDNA was derived by PCR amplification from the EBNA3C-pZip-neoSV plasmid (36) with primers designed to restore the missing 18 nucleotides at the 5′ end and to introduce SmaI sites immediately 5′ of the AUG codon and immediately 3′ of the TTA stop codon (B95.8 coordinates 98371 to 101423). The EBNA3C cDNA was cloned into the SmaI site of the pAcG2T transfer vector (13) to produce a chimeric gene, encoding the N terminus of the GST protein fused to full-length EBNA3C, upstream of the polyhedrin promoter. The pAcG2T-E3C vector was then used to replace the polyhedrin gene in a wild-type baculovirus by recombination in Sf9 cells cotransfected with 25 μg of pAcG2T-E3C and 50 μg of linearized wild-type baculovirus DNA.
pGEX-EBNA3C aa 146-565 encodes the amino acids of EBNA3C indicated as an in-frame fusion with GST in pGEX.
Antibodies.
For immunoprecipitations, 2.5 μg of purified mouse anti-EBNA3C monoclonal antibody (MAb) A10 (31), 2.5 μg of polyclonal rabbit anti-flag antibody (D-8; Santa Cruz Biotechnology), or 2.5 μg of mouse anti-p107 MAb (SD9; Santa Cruz Biotechnology) was used. The anti-EBNA3C MAb A10 recognizes the EBNA3C epitope WAPSV (aa 682 to 686) (31). The polyclonal rabbit antiserum to HDAC1 was raised against a peptide epitope from the C terminus of HDAC1.
Cell culture.
B cells (DG75, a Burkitt’s lymphoma-derived cell line) and T cells (Jurkat, a T-cell leukemia-derived cell line) were grown in suspension and maintained in RPMI 1640 (Gibco BRL) supplemented with 10% (vol/vol) heat-inactivated fetal calf serum (FCS), 2 mM l-glutamine (Gibco BRL), and 100 U of penicillin and streptomycin (Gibco BRL)/ml. Cells were cultured at 37°C under a 10% CO2 humidified atmosphere.
Insect cells, recombinant baculovirus, and purification of GST-EBNA3C.
Insect Sf9 cells were cultured at 27°C in TC100 (Gibco BRL) growth medium, supplemented with 10% (vol/vol) heat-inactivated FCS and 100 U of penicillin and streptomycin (Gibco BRL)/ml. For infection, Sf9 cells were seeded in 3 ml of supplemented prewarmed growth medium at a cell density of 5 × 106 to 10 × 106 and were allowed to settle for 30 min before infection with recombinant virus. For maximum efficiency of infection, approximately 3 to 10 PFU/cell was used. One milliliter of high-titer recombinant virus (1 × 107 to 2 × 107) was added to each dish and incubated for 60 min with gentle rocking at room temperature (RT). The virus was then diluted with 6 to 7 ml of growth medium, and the extracts were prepared 24 to 36 h postinfection. Infected Sf9 cells were collected by centrifugation at 900 rpm for 5 min at 4°C in an IEC-centra-892, with an IEC-216 rotor, and were washed twice in ice-cold phosphate-buffered saline (PBS); after the second wash, cells were transferred to 1.5-ml Eppendorf tubes and pelleted in an MSE benchtop microcentrifuge for 1 min at 6,500 rpm. The supernatant was aspirated, and the cell pellet was resuspended in ∼4 volumes of lysis buffer (0.5 M NaCl, 20 mM HEPES [pH 7.9], 10 mM NaF, 1 mM Na3VO4, 20% glycerol, and 1 mM phenylmethylsulfonyl fluoride [PMSF], freshly made for each purification procedure) and freeze-thawed twice with vigorous vortexing between each freeze-thawing step. Debris was sedimented by centrifugation at 1,600 × g for 15 min at 4°C, and the supernatant was incubated with 0.5 ml of GST-Sepharose beads for 1 h at 4°C with end-to-end mixing. When binding was completed, recombinant protein bound to the beads was washed twice in 0.1% Triton X-100–PBS and twice in PBS. The quality of the recovered protein was estimated by loading a 10-μl aliquot on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel, and the purified protein was quantified by including a known amount of bovine serum albumin on the same gel.
Transient transfection assays and TSA.
B and T cells were split 1:3 with freshly supplemented prewarmed medium 8 to 12 h prior to transfection. Cycling cells were pelleted (at 500 × g at 4°C for 5 min), and growth medium was retained as “conditioned medium.” The cell pellet was resuspended in ice-cold unsupplemented RPMI 1640 (150 μl for DG75 and Jurkat cells), and the suspension was added to a chilled cuvette (0.4-cm gap; Bio-Rad) which contained the appropriate DNA suspended in 50 μl of ice-cold unsupplemented RPMI 1640. The cell-DNA mix was incubated on ice for 10 min and then resuspended by gentle shaking before electroporation with a Bio-Rad Gene Pulser set at a capacitance of 960 μF and 250 V. Transfected cells were placed at 37°C for 15 to 20 min before they were resuspended in 10 ml of prewarmed conditioned medium. Trichostatin A (TSA; Wako Chemical Industries, Tokyo, Japan) was added immediately after cells were resuspended in conditioned medium. Cell were routinely harvested, and extracts were prepared, 24 h after transfections. Transfected B and T lymphocytes were harvested by centrifugation (at 500 × g for 5 min at 4°C), washed in 10 ml of ice-cold PBS, pelleted under the same conditions, and transferred to 1.5-ml Eppendorf tubes in 1 ml of ice-cold PBS. For chloramphenicol acetyltransferase (CAT) and luciferase assays, cell pellets were resuspended in 65 μl of 1× reporter lysis buffer (Promega, Southampton, United Kingdom) and incubated for 15 min at RT. Cell debris was removed by centrifugation (at 11,600 × g for 20 min at RT), and the supernatant was transferred to a fresh Eppendorf tube for a CAT or luciferase assay. Luciferase and CAT assays have been described previously (7, 37). All values were normalized to β-galactosidase activity expressed from 2 μg of pSV-β-Gal plasmid, which was included as an internal control for transfection efficiency and any cytotoxic effects of the tested DNA.
Immunoprecipitations.
Coimmunoprecipitation assays were carried out essentially as described previously (16). Briefly, 5 μg each of the EBNA3C and the HDAC1-F expression vector was cotransfected in DG75 cells. Samples were harvested 48 h after transfection in DG75 lysis buffer (DGLB) (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 10% glycerol, 0.5% Triton X-100) containing 2 mM PMSF and 1 μg of a mix of protease inhibitors (complete protease inhibitor cocktail; Boehringer Mannheim)/ml. Lysates were incubated at 4°C for 15 min on an orbital rotor for end-to-end inversion and then centrifuged at 11,600 × g for 15 min at 4°C to isolate supernatants. Whole-cell supernatants were split into three aliquots, and the EBNA3C and HDAC1-F proteins were coimmunoprecipitated with the anti-EBNA3C antibody A10, the anti-flag polyclonal antibody D-8, or the control anti-p107 MAb. Lysates and antibodies were incubated at 4°C for 90 min on an orbital rotor. The immune complexes were then cross-linked with 50 μl of protein G-agarose beads (Pharmacia Biotech) at 4°C for 60 min with gentle mixing. All immunoprecipitates were washed three times in DGLB, and the specifically bound proteins were eluted from the affinity matrix in 100 μl of SDS protein sample buffer, boiled for 3 min loaded on an SDS-PAGE gel, and visualized by Western immunoblot analysis.
Expression of GST fusion proteins in Escherichia coli.
The expression of fusion proteins was performed basically as described previously (7). Briefly, 50 ml of an overnight culture was inoculated into 500 ml of 2× TY medium containing 50 μg of ampicillin/ml. The culture was placed on a shaking incubator at 37°C until an absorbance of 1 at 600 nm was reached. Production of recombinant protein was induced with 1 mM (final concentration) isopropylthiol-β-d-galactoside (IPTG; Gibco BRL), and the culture was then incubated for a further 3 to 4 h at RT. Cells were harvested by centrifugation and resuspended in 5 ml of 1% Triton X-100–PBS (T-PBS) and 1 mM PMSF. The suspension was incubated on ice for 30 min before cells were lysed by sonication. The bacterial lysate was centrifuged at 11,600 × g for 20 min at 4°C to remove the insoluble fraction.
Purification and elution of the fusion proteins.
All the supernatant prepared as described above was mixed with 600 μl of 50% (vol/vol) glutathione-Sepharose beads (Pharmacia Biotech) and incubated for 1 h at 4°C on an orbital rotor for end-to-end inversion. The beads were washed three times in T-PBS and three times in PBS and were then resuspended in 50% (vol/vol) PBS. In this form, the purified protein was used for GST pulldown experiments. The quality and the quantity of the recovered protein were estimated as described for insect cell GST fusion purification.
GST pulldown experiments.
GST pulldown experiments were performed essentially as described previously (7, 34, 37).
GST pulldown of deacetylase activity.
GST pulldown of deacetylase activity was performed essentially as described previously (9). GST fusion proteins (1 to 3 μg) prebound to glutathione-Sepharose beads (Pharmacia Biotech) were added to 50 μl of HeLa nuclear extract (Computer Cell Culture Centre, Mons, Belgium) and 200 μl of IPH buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 5 mM EDTA, 0.5% Nonidet P-40, 1 mM PMSF) and incubated on a rotating wheel for 1 to 2 h at 4°C. Beads were recovered by centrifugation and washed three times in 1 ml of IPH buffer. Beads were then resuspended in 100 μl of IPH buffer containing tritium-labelled acetylated peptides corresponding to bovine histone H4 (aa 1 to 24). The reaction mixture was then incubated at 37°C for 1 to 2 h with occasional agitation. The deacetylase reaction was stopped by the addition of 150 μl of IPH buffer with 65 μl of 1 M HCl–0.16 M acetic acid. To extract free acetate, 700 μl of ethyl acetate was added, the sample was vortexed, and the organic and aqueous phases were separated by centrifugation. A 500-μl portion of the upper phase was added to 2 ml of scintillation cocktail (Optiphase Hisafe 3; Fisher Chemicals). After vortexing, the amount of tritium released was determined by liquid scintillation counting.
Purification of His-tagged fusion protein.
E. coli HB101 transformed with the pET30a-HDAC1 plasmid (encoding full-length HDAC1 with a six-His-tag at the C terminus) was incubated on a shaker overnight (at 37°C) in 50 ml of 2× TY medium containing 50 μg of kanamycin/ml. The culture was then diluted 1/10 in 2× TY medium and incubated with shaking for a further 3 to 4 h (at 37°C). Bacteria were then induced to produce the fusion protein by incubation for 4 to 5 h at RT with 1 mM (final concentration) IPTG. Cells were collected by centrifugation and resuspended in 10 ml of T-PBS with 1 mM PMSF. Cells were then sonicated on ice for 30 s. Cell debris was removed by centrifugation, and the cleared supernatant was incubated with 300 ml of Ni beads (Qiagen) for 45 min in the presence of 10 mM imidazole. After binding, the beads were washed five times in T-PBS with 10 mM imidazole and five times in T-PBS with 40 mM imidazole. After the washes, the fusion protein was eluted with T-PBS plus 160 mM imidazole for 15 min at room temperature. The beads were removed by centrifugation, and the eluted fusion protein was stored at −70°C in small aliquots containing 10 to 20% glycerol.
RESULTS
TSA interferes with EBNA3C-mediated repression.
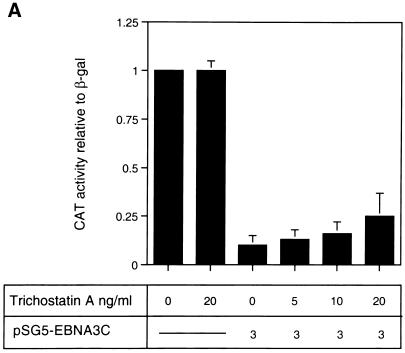
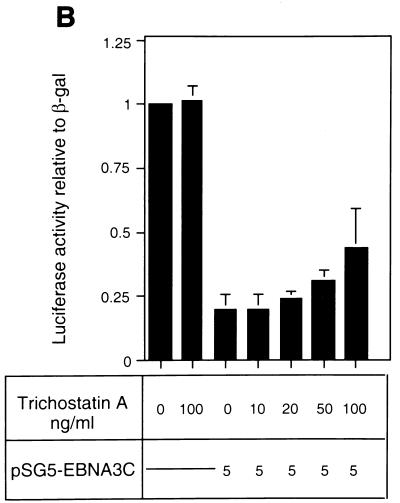
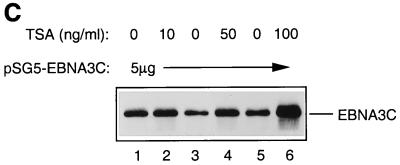
The analysis of repression by both GAL4-EBNA3C and wild-type unfused EBNA3C led us to the hypothesis that EBNA3C would repress transcription as part of a multiprotein complex including CBF1/RBP-Jκ and one (or more) corepressor protein(s). Recent studies with both yeast and mammalian cells have implicated multiprotein complexes with histone deacetylase activity in transcriptional repression (15, 20, 35). Such repression results from the modulation of chromatin architecture (see the introduction). We therefore investigated whether the repression by EBNA3C is associated with histone deacetylase activity. Deacetylases are sensitive to nanomolar concentrations of the specific inhibitor TSA (9, 54); therefore, in order to investigate whether EBNA3C utilizes deacetylase activity in the repression of Cp, TSA was added to transient transfection assays. DNA from pTK-CAT-Cp4× (a reporter plasmid which includes multiple CBF1/RBP-Jκ binding sites from Cp cloned upstream of the herpes simplex virus TK promoter) was cotransfected with a concentration of the pSG5-EBNA3C expression plasmid which produced about 10-fold repression of CAT activity in DG75 B-lymphoma cells. Increasing amounts of TSA were added to parallel transfections (Fig. 1A). Although the effects of TSA were very modest, they consistently increased the CAT activity two- to threefold. TSA is very toxic, and above 20 ng/ml it starts to induce nonspecific transcriptional effects and cell death in DG75 cells (data not shown). However, at the concentrations used in these experiments, although it stimulated or derepressed Cp, it had no effect on the pSV-β-Gal reporter plasmid included as a control in each transfection or on the basal activity of pTK-CAT-Cp4×. Similar experiments were repeated with a second reporter plasmid, p-1425-Luc, in which the Cp promoter (bp −1425 to +1) is upstream of a luciferase reporter gene. These were performed with the Jurkat T-cell line, which is less sensitive to TSA. This analysis produced results similar to those in DG75 cells; that is, despite its toxicity, TSA relieved repression by EBNA3C. However, in Jurkat cells, higher concentrations of TSA were necessary (Fig. 1B). In both series of experiments, the levels of EBNA3C expressed from pSG5-EBNA3C were monitored by Western immunoblotting (see, for example, Fig. 1C). TSA did not suppress the level of EBNA3C expressed; in fact, it generally resulted in slightly increased accumulation of the protein. Therefore, since repression by EBNA3C is dose dependent, the apparent relief of repression may be slightly underrepresented. The results of these transfection experiments are all consistent with EBNA3C (at least in part) using deacetylase to repress Cp.
FIG. 1.
(A) Effect of TSA on EBNA3C-mediated repression in DG75 B cells. DG75 cells were transfected with 2 μg of the pTK-CAT-Cp4× reporter plasmid and with the pSG5-EBNA3C effector plasmid as indicated. Transfected cells were treated for 24 h with the amounts of TSA shown. CAT assays were performed and standardized to β-galactosidase activity expressed from the cotransfected plasmid pSV-β-Gal (2 μg). The data shown are relative to the activity from untreated pTK-CAT-Cp4×, which was given an arbitrary value of 1. Each result is the mean and standard deviation from three independent experiments. (B) Effect of TSA on EBNA3C-mediated repression in Jurkat T cells. Jurkat cells were transfected with 2 μg of the p1425-Cp-Luc reporter plasmid and with the pSG5-EBNA3C effector plasmid as indicated. Transfected cells were treated with the indicated amounts of TSA for 24 h. Luciferase assays were performed and standardized to β-galactosidase activity expressed from the cotransfected plasmid pSV-β-Gal (2 μg). Data shown are relative to the activity from untreated p1425-Cp-Luc, which was given an arbitrary value of 1. Each result is the mean and standard deviation from three independent experiments. (C) Western blot analysis of EBNA3C in a representative experiment performed with p1425-Cp-Luc in Jurkat cells. The protein extract from transfected cells was resolved by SDS-PAGE (7.5% polyacrylamide). After transfer to a nitrocellulose membrane, the proteins were probed with the anti-EBNA3C MAb A10.
Expression of HDAC1 enhances repression by EBNA3C.
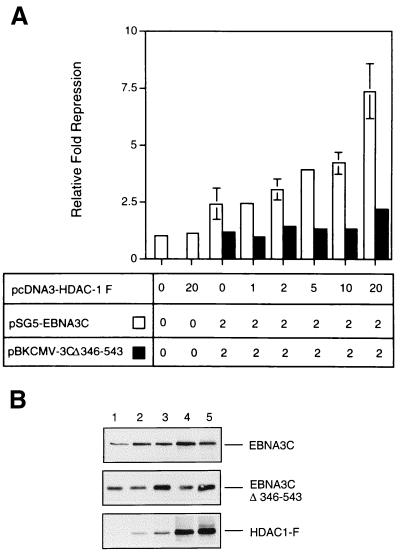
In order to investigate further whether deacetylase activity is involved in EBNA3C-mediated repression, an expression vector encoding HDAC1—one of the three characterized human histone deacetylases—was used in transient transfection assays in DG75 cells. The reporter plasmid including multiple CBF1/RBP-Jκ binding sites from Cp (pTK-CAT-Cp4×) was cotransfected with an amount of pSG5-EBNA3C DNA judged to produce suboptimal repression of CAT activity. When increasing amounts of pcDNA3–HDAC1-F DNA (which encodes a flag epitope-tagged HDAC1) were included in the cotransfections, repression of CAT activity was specifically enhanced (Fig. 2A). In a parallel series of similar experiments, HDAC1-F expression had no effect when a nonrepressing mutant of EBNA3C (Δ346-543) (37) was used. Also, the maximum amount of pcDNA3–HDAC1-F DNA used (20 μg/transfection) consistently had a negligible effect on the 4× Cp reporter plasmid alone or on the β-galactosidase control. The level of protein expressed in these experiments was monitored by Western blotting (see, for example, Fig. 2B). These results are also consistent with EBNA3C specifically recruiting HDAC1 to the CBF1/RBP-Jκ elements of Cp.
FIG. 2.
(A) Repression by EBNA3C is enhanced by HDAC1 expression. DG75 cells were transiently transfected with 2 μg of the pTK-CAT-Cp4× reporter plasmid, 2 μg of pSV-β-Gal, and 2 μg of either the pSG5-EBNA3C or the pBKCMV-EBNA3CΔ346-543 effector plasmid. The indicated amount (in micrograms) of pcDNA3–HDAC-1F was added to each transfection. Cell extracts were prepared 48 h after transfection, and CAT activity was determined. After normalization to β-galactosidase activity, the data were expressed as fold repression relative to the activity from pTK-CAT-Cp4× with empty control vectors, which was given an arbitrary value of 1. Means and standard deviations from three independent experiments are shown. (B) Western blot analysis of the expression of EBNA3C, EBNA3CΔ346-543, and flag-tagged HDAC1 in a representative experiment. Protein extract was resolved by SDS-PAGE (10% polyacrylamide). After transfer to a nitrocellulose membrane, the proteins were probed with the anti-EBNA3C MAb A10 and the anti-flag antibody D-8. Lane 1, 0 μg of pcDNA3–HDAC1-F DNA; lanes 2 through 5, 1, 2, 10, and 20 μg, respectively.
EBNA3C and HDAC1 interact in vitro and in vivo.
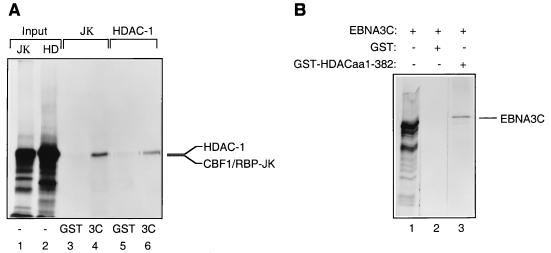
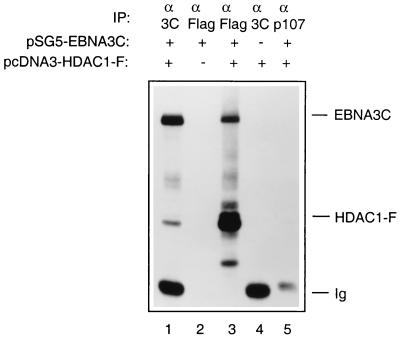
The functional analyses described above suggested that HDAC1 might physically interact with EBNA3C and therefore might be targeted to promoter elements containing CBF1/RBP-Jκ binding sites. Various assays were performed in order to demonstrate this. Figure 3A shows that a fusion of EBNA3C with GST and HDAC1 translated in vitro can form a complex in a manner similar to that of GST-EBNA3C and RBP-Jκ incubated and washed under the same conditions (Fig. 3A; compare lanes 4 and 6). When the reciprocal experiment was performed, GST-HDAC1 (aa 1 to 382) bound to in vitro-translated EBNA3C (Fig. 3B, lane 3). In binding reactions using the same conditions, wild-type GST failed to bind in vitro-translated EBNA3C, HDAC1, or CBF1/RBP-Jκ. Also, control radiolabelled luciferase was not precipitated by either GST-EBNA3C or GST-HDAC1 (aa 1 to 382) (data not shown). In order to establish whether EBNA3C and HDAC1 could interact under more physiologically relevant conditions, DG75 cells were cotransfected with pSG5-EBNA3C and pcDNA3–HDAC1-F, and protein extracts were made 48 h later. These were then subjected to immunoprecipitation followed by Western blot analysis. Figure 4 clearly shows that the anti-EBNA3C MAb A10 precipitates both EBNA3C and HDAC1-F (lane 1) and that the antiflag MAb precipitates a similar complex containing both proteins (lane 3). The specificity of each MAb was established by control transfections with each plasmid alone (lanes 2 and 4), and a control antibody (anti-p107) precipitated neither EBNA3C nor HDAC1-F (lane 5). The proteins apparently interacted with greater efficiency in this in vivo interaction, perhaps suggesting that other cellular proteins participate and stabilize the complex.
FIG. 3.
(A) HDAC1 interacts with GST-EBNA3C. GST-EBNA3C expressed from a baculovirus was incubated with equal amounts of [35S]methionine-labelled RBP-Jκ (as a positive control) (lane 4) or HDAC1 (lane 6). Bound proteins were resolved by SDS-PAGE (7.5% polyacrylamide gel). Lanes 1 and 2 contain 10% of the input protein in each binding reaction, and lanes 3 and 5 contain the protein which bound to GST. (B) EBNA3C interacts with the N terminus of HDAC1 fused to GST. Bacterially expressed GST-HDAC1 (aa 1 to 382) was incubated with equal amounts of [35S]methionine-labelled EBNA3C (lane 3). Bound proteins were resolved by SDS-PAGE (7.5% polyacrylamide gel). Lane 1 contains 10% of the input protein in each binding reaction, and lane 2 contains the protein which bound to GST.
FIG. 4.
EBNA3C and HDAC1 can interact in vivo. DG75 cells were transfected with a mixture of 5 μg of pSG5-EBNA3C and 5 μg of pcDNA–HDAC1-F (lanes 1, 3, and 5) or with pSG5-EBNA3C alone (lane 2) or pcDNA3–HDAC1-F alone (lane 4). Cells were harvested 48 h after transfection and immunoprecipitated (IP) with the appropriate MAbs as indicated. After separation on an SDS–7.5% polyacrylamide gel, the proteins were Western blotted onto a nitrocellulose membrane and probed sequentially with anti-EBNA3C (MAb A10) and anti-flag (MAb D-8).
GST-EBNA3C binds histone deacetylase activity from HeLa cell nuclear extracts.
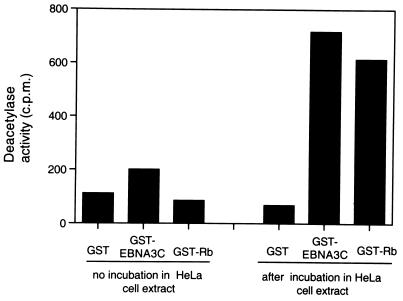
All the above data support a model in which CBF1/RBP-Jκ targets EBNA3C to the Cp promoter and EBNA3C in turn recruits HDAC1, which converts the surrounding chromatin from a hyperacetylated, transcriptionally active state to a hypoacetylated and transcriptionally repressive state. A prediction from this model is that EBNA3C should bind and facilitate the partial purification of histone deacetylase enzymatic activity from nuclear extracts. To test this, GST-EBNA3C prebound to glutathione-Sepharose beads was incubated with HeLa cell nuclear extracts. Bound histone deacetylase activity was assayed by using an acetylated histone H4 peptide (corresponding to amino acids 1 to 24 of the bovine protein) as described previously (9). Unfused GST and GST-pRb were used as negative and positive controls for binding, respectively. The experiment was repeated several times, and GST-EBNA3C consistently purified an amount of deacetylase activity similar to that obtained with GST-pRb; the interaction between pRb and HDAC1 has been widely reported (9, 27, 28). Results of a representative experiment are shown in Fig. 5. Control assays to which no HeLa extract was added are shown on the left. It was noted that GST-EBNA3C generally showed a slightly higher background deacetylase activity than GST or GST-pRb. We assume that this is due to contaminating deacetylase enzyme copurified from the Sf9 insect cells used for the expression of the GST-EBNA3C from a recombinant baculovirus. Wild-type GST and GST-pRb, on the other hand, were purified from E. coli.
FIG. 5.
EBNA3C can precipitate histone deacetylase enzyme activity from HeLa nuclear extracts. Nuclear extracts from HeLa cells were subjected to a GST pulldown analysis with beads coated with GST-EBNA3C, GST-Rb, or wild-type GST alone. In a parallel control experiment, the coated beads were incubated without nuclear extract in the reaction mixtures. Bound proteins were assayed for histone deacetylase activity as described in Materials and Methods. All samples were assayed in duplicate.
Both HDAC1 and CBF1/RBP-Jκ interact with the N terminus of EBNA3C.
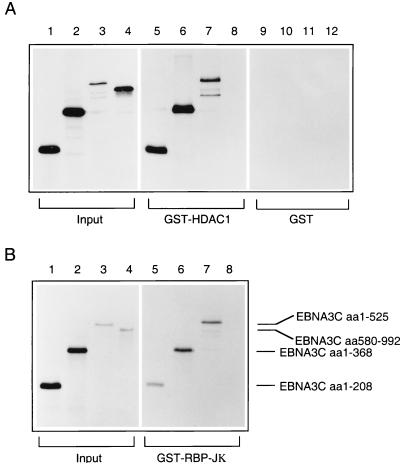
In order to determine which region of EBNA3C was responsible for the interaction with HDAC1, GST pulldown experiments using bacterially expressed GST-HDAC1 (aa 1 to 432) and a series of EBNA3C deletion mutants transcribed and translated in vitro were performed. Fragments comprising aa 1 to 208, aa 1 to 368, and aa 1 to 525 all showed efficient binding to the GST-HDAC1 fusion protein (Fig. 6A, lanes 5 through 7). In contrast, the C-terminal fragment (aa 580 to 992) exhibited no detectable binding activity in similar assays (Fig. 6A, lane 8). This spectrum of binding to EBNA3C corresponds exactly to the binding pattern of CBF1/RBP-Jκ with the same in vitro-translated polypeptides (Fig. 6B, lanes 5 through 8). None of the EBNA3C fragments showed any significant binding to GST (Fig. 6A, lanes 9 through 12).
FIG. 6.
Both HDAC1 and RBP-Jκ interact with the N terminus of EBNA3C. (A) In a GST pulldown experiment, bacterially expressed GST-HDAC1 (aa 1 to 432) was incubated with equal amounts of [35S]methionine-labelled EBNA3C deletion mutants: aa 1 to 208 (lane 5), aa 1 to 368 (lane 6), aa 1 to 525 (lane 7), and aa 580 to 992 (lane 8). The protein pulled down by wild-type GST is shown in lanes 9 through 12. (B) In a parallel pulldown experiment, the same EBNA3C deletion mutants were incubated with bacterially expressed GST-RBP-Jκ. In both experiments, bound proteins were resolved by SDS-PAGE (7.5% polyacrylamide), and comparable amounts of GST-HDAC1 and GST-RBP-Jκ were used in the binding reactions (data not shown). In both experiments, lanes 1 through 4 contain 10% of the input protein included in each binding reaction.
Do HDAC1 and CBF1/RBP-Jκ have adjacent binding sites in EBNA3C or does CBF1/RBP-Jκ bridge between HDAC1 and EBNA3C?
It has been reported that the binding of CBF1/RBP-Jκ to EBNA3C is mediated through amino acids located in the N terminus of the latter; this domain (aa 182 to 231) is included in one of the regions conserved among EBNA3A, -B, and -C (see the introduction). Although the precise contact residues have not been defined (42, 57), the mutation to alanine of four conserved amino acids in this region (aa 208 to 211) was shown to abolish the interaction between EBNA3C and CBF1/RBP-Jκ (37, 57). When a polypeptide with this 4-amino-acid substitution was translated in vitro and subjected to GST pulldown analysis, as expected, it failed to bind to GST-CBF1/RBP-Jκ (references 37 and 57 and data not shown). Interestingly, this protein also failed to bind to GST-HDAC1 (aa 1 to 432) (Fig. 7, lane 8).
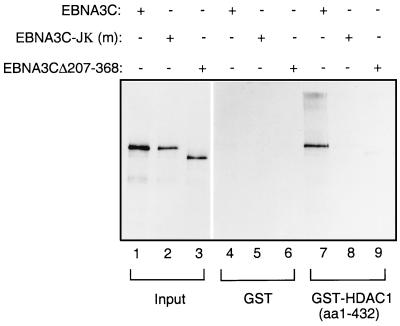
FIG. 7.
HDAC1 binds to the region of EBNA3C which interacts with CBF1/RBP-Jκ. In a GST pulldown experiment, bacterially expressed GST-HDAC1 (aa 1 to 432) was incubated with equal amounts of [35S]methionine-labelled EBNA3C (lane 7); EBNA3C-Jκ (m), a mutant which no longer binds CBF1/RBP-Jκ (lane 8); and EBNA3CΔ207-368, which also fails to bind CBF1/RBP-Jκ (lane 9). Lanes 4 through 6 contain protein bound by wild-type GST, and lanes 1 through 3 show 10% of the input protein in each binding reaction.
In similar assays a protein with amino acids 207 to 368 deleted also failed to bind to the HDAC1 fusion protein. This was not surprising. However, since both HDAC1 and CBF1/RBP-Jκ bind efficiently to aa 1 to 208 (Fig. 6, lanes 5), we conclude that although retention of amino acids 208 to 211 is clearly necessary for these interactions, they cannot include the specific binding site (see Discussion). The contact residues probably reside to the N-terminal side of aa 207. These data suggest that HDAC1 and CBF1/RBP-Jκ bind either to the same region or to adjacent regions of EBNA3C. Alternatively, it is possible that HDAC1 is unable to bind directly to EBNA3C and requires CBF1/RBP-Jκ to act as a bridge between the deacetylase complex and viral antigen. If the binding is indirect, this would mean that in GST pulldown experiments the reticulocyte lysate used to translate one or another of the proteins provided the CBF1/RBP-Jκ.
In order to distinguish between these alternatives (that is, direct or indirect binding between HDAC1 and EBNA3C), in vitro binding assays using fusion proteins purified from E. coli were performed. A histidine-tagged HDAC1 protein was purified by virtue of its affinity for nickel-agarose, and a GST fusion with EBNA3C (aa 146 to 565) was purified by using glutathione-Sepharose. It should be noted that it was not possible to use GST–full-length EBNA3C in these assays, since this fusion is not stably expressed in E. coli (our unpublished data).
Each binding reaction mixture using these purified proteins was incubated either with or without reticulocyte lysate added, in order to determine whether this provided a bridging protein(s) or had any detectable effect on the interaction. The results of a Western blot probed with a rabbit serum raised against the C terminus of HDAC1 showed unambiguously that His-HDAC1 binds directly to GST-EBNA3C (aa 146 to 565) (Fig. 8A, lanes 3 and 4). In similar assays performed in parallel, HDAC1 bound to GST-Rb (10) but not to much larger amounts of GST protein (Fig. 8A, lanes 1 and 2) or GST-E7 (10). Although GST-EBNA3C (aa 146 to 565) binds with high affinity to His-HDAC1, it does not bind to various other proteins in similar assays (our unpublished data). The addition of reticulocyte lysate had no significant effect in any of the experiments performed.
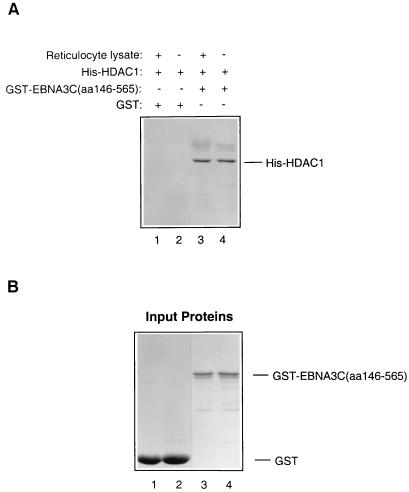
FIG. 8.
EBNA3C binds directly to HDAC1. (A) Equal amounts of His-HDAC1, purified from a bacterial lysate, were incubated with Sepharose beads loaded with GST-EBNA3C (aa 146 to 565) or GST. Similar binding reactions were set up with a supplement of TNT reticulocyte lysate (Promega). After washing, the bound proteins were resolved by SDS-PAGE (12% polyacrylamide), transferred to a nitrocellulose membrane, and probed with a polyclonal antiserum which recognizes the C terminus of HDAC1. His-HDAC1 bound to GST-EBNA3C (aa 146 to 565) (lanes 3 and 4) irrespective of whether reticulocyte lysate was present. No binding to GST was detected (lanes 1 and 2). (B) Brilliant blue-stained polyacrylamide gel showing the amounts of purified protein used in each binding reaction.
DISCUSSION
We previously reported that EBNA3C could repress—in transient transfection assays—transcription from Cp, the major promoter for EBNA expression (37). All the evidence in that study suggested that repression was absolutely dependent on the binding of EBNA3C to the cellular factor CBF1/RBP-Jκ and also on the binding of CBF1/RBP-Jκ to the EBNA2-response element (E2RE; a CBF/RBP-Jκ binding site) in Cp. Mutations in EBNA3C which prevented binding to CBF1/RBP-Jκ abrogated repression; similarly, a mutation in the E2RE (which prevented CBF1/RBP-Jκ binding to Cp DNA) also abolished EBNA3C-mediated repression. Moreover, transferring multiple E2REs to a heterologous promoter also transferred repressibility to that promoter (reference 48, our unpublished data, and this study). In none of these assays—which were performed in various B and T cells—was it necessary to transactivate Cp with EBNA2.
These data led us to the hypothesis that CBF1/RBP-Jκ targets EBNA3C to Cp and that EBNA3C interacts with one or more cellular cofactors to repress transcription. In the present study the nature of the hypothetical cofactor(s) has been explored. We provide compelling evidence that the repression complex includes human HDAC1, and we therefore conclude that at least one component of the ability of EBNA3C to repress transcription results from modulation of chromatin architecture. This is, to our knowledge, the first demonstration of a viral transcription factor which functionally and physically associates with a deacetylase-mediated chromatin-modifying complex through a direct interaction with a cellular histone deacetylase.
The participation of a histone deacetylation complex in the repression mediated by EBNA3C was first suggested by experiments in which a specific inhibitor of deacetylases, TSA, relieved repression induced by EBNA3C in transient transfection assays using two different cell lines and two different reporter plasmids (Fig. 1). Although the effects of TSA were modest, they were specific to EBNA3C repression. It has been demonstrated in many cases that the relief of repression by TSA is only partial and is of a magnitude similar to that of the effects we have described here (49, 51, 56). The generally held view is that many repressors use multiple mechanisms to repress and that not all of these mechanisms operate via histone deacetylation. This is probably the case for EBNA3C. Despite the rather crude nature of these experiments, the results were reproducible and consistent with a deacetylase interacting with EBNA3C in the repression of Cp or a synthetic promoter which includes CBF1/RBP-Jκ binding sites.
Since drugs such as TSA are highly toxic and may have nonspecific effects on transcription, further experiments were performed to establish whether a known histone deacetylase, HDAC1, could participate in EBNA3C-mediated repression. A series of cotransfections showed that HDAC1 can cooperate with EBNA3C to repress a reporter containing multiple Cp-derived E2REs (Fig. 2). These results suggested a mechanism for repression of CBF1/RBP-Jκ-responsive promoters in which HDAC1 associates with EBNA3C on the promoter and deacetylase activity converts the surrounding chromatin from a transcriptionally active to a transcriptionally repressed state. Multiple in vitro and in vivo assays were then performed in order to determine whether EBNA3C binds to the histone deacetylase enzyme HDAC1. The data from all of these experiments (Fig. 3 to 8) were consistent with HDAC1 being recruited to DNA through an association with EBNA3C, which is itself targeted to DNA by CBF1/RBP-Jκ. It should be noted, however, that TSA only partially relieved the repression of Cp induced by EBNA3C (Fig. 1). This suggests that other interactions and another mechanism(s) of repression are involved in achieving optimal repression by EBNA3C.
In the course of mapping the region of EBNA3C responsible for the interaction with HDAC1, it became apparent that a 4-aa substitution (aa 208 to 211) in EBNA3C, which disrupts the binding to CBF1/RBP-Jκ, also prevents the association of EBNA3C with HDAC1. A possible explanation for this result is that EBNA3C does not actually contact HDAC1, but that CBF1/RBP-Jκ simultaneously binds to both HDAC1 and EBNA3C and acts as a bridge between them. However, experiments using proteins purified from bacterial cells showed that no eukaryotic factor is necessary and that a direct physical interaction can occur between HDAC1 and EBNA3C (Fig. 8). It has been suggested previously that aa 208 to 211 of EBNA3C might lie within the binding site for CBF1/RBP-Jκ (37, 57). However, it now seems likely that replacement of these residues with alanine alters the conformation of this region and disrupts the interaction with CBF1/RBP-Jκ, HDAC1, and perhaps other factors. Since the precise binding site in EBNA3C for these proteins is unknown, we can only speculate that they bind to adjacent regions of the N terminus or perhaps even to the same region but on opposite sides of an alpha-helix.
It has recently been reported that CBF1/RBP-Jκ can participate in a repression complex which includes HDAC1 and SMRT—although it is unclear whether there is a direct interaction between CBF1/RBP-Jκ and HDAC1 (21). We therefore suggest that EBNA3C converts CBF1/RBP-Jκ from a neutral DNA binding factor to a repressor by bridging an interaction between CBF1/RBP-Jκ and a repression complex which includes HDAC1 (see the model proposed in Fig. 9). It is now becoming clear that several HDAC complexes with different subunit compositions exist. These complexes are likely to differ in their precise functions; for example, some have remodelling activity in addition to deacetylase activity (the Mi2 complex), while others have no remodelling activity (the Sin3 complex) (2, 10, 16, 20, 32, 35, 55). We suggest that it is possible that EBNA3C could recruit a different complex to CBF1/RBP-Jκ (possibly replacing one that is bound to CBF1/RBP-Jκ) and that this represses more efficiently.
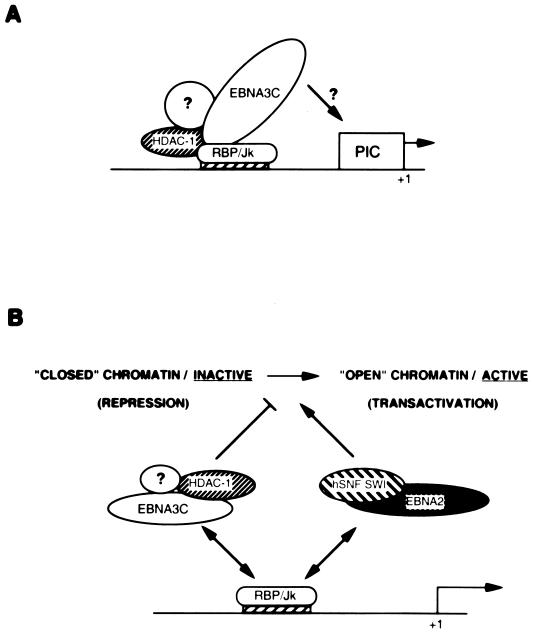
FIG. 9.
A possible role for EBNA3C in modulating transcription. (A) All the data reported previously (21, 46) and in this study are consistent with a model in which CBF1/RBP-Jκ can reversibly associate with HDAC1 and target it to DNA. Since EBNA3C binds to both proteins, we propose that EBNA3C can bridge this interaction and so mediate repression. It is unclear whether the other members of the deacetylase repression machinery (such as SMRT or Sin3) are also associated with HDAC1 in these complexes or whether EBNA3C establishes a novel complex with deacetylase activity. It is also unclear whether EBNA3C additionally contributes to repression by interacting with the basal transcription machinery (represented by the preinitiation complex [PIC]). (B) In cells latently infected with EBV, there will be a dynamic equilibrium between EBNA3C–HDAC1–RBP-Jκ complexes and EBNA2–hSNF-SWI–RBP-Jκ complexes (50) associated with Cp, so the surrounding chromatin configuration will be in a state of flux. The net result will be finely regulated EBNA expression. It should be noted that EBNA3A and EBNA3B (which can also bind RBP-Jκ) may also influence the protein-protein and protein-DNA interactions on Cp. For the sake of simplicity, the other factors which associate with EBNA2 have not been included, and neither have the endogenous cell factors TAN-1 and activated Notch receptor, which may also influence chromatin conformation in a manner analogous to that of EBNA2 (21).
In addition to engaging multiple members of the RNA polymerase II transcription complex, EBNA2 can also associate with a chromatin remodelling complex which includes the human equivalents of the yeast SWI/SNF complex (50). Such complexes are thought to function by destabilizing the interactions between DNA and histones in the nucleosome in an ATP-dependent reaction; this activity leads to an enhanced affinity of transcription factors for their binding sites and so activates transcription (reviewed in reference 11). Since EBNA2 can also bind to CBF1/RBP-Jκ and is thereby targeted to responsive promoters, we propose that in cells latently infected with EBV there will be a dynamic equilibrium between CBF1/RBP-Jκ–EBNA3C–HDAC1 and CBF1/RBP-Jκ–EBNA2–hSNF-SWI complexes shaping chromatin in the region of responsive promoters. With the EBNA2 complexes activating and the EBNA3C complexes repressing transcription, gene expression from promoters such as Cp can be finely regulated (Fig. 9B).
Finally, it has been shown that the tumor suppressor pRb associates with HDAC1 and that this complex is targeted to responsive promoters by E2F transcription factors; this interaction of pRb with a deacetylase is therefore likely to be important in the regulation of the cell cycle (9, 27, 28). The pRb-HDAC1 repression complex can be disrupted by the papillomavirus E7 oncoprotein, and E7 also associates with Mi2 and HDAC1 and -2 to promote cell growth (9, 10). Since EBNA3C can substitute for E7 in the immortalization and transformation of primary rat embryo fibroblasts transfected with cooperating oncogenes (34), this suggests that EBNA3C, like E7, might modulate the cyclin–cyclin-dependent kinase–pRb–E2F pathway which converges on the restriction point in G1 of the cell cycle. EBNA3C can bind pRb in vitro (34), and here we have shown that it can bind directly to HDAC1. It will be very interesting, therefore, to determine the effect of EBNA3C on repression complexes which include HDAC1, pRb, and E2F and also to determine the role of the EBNA3C-HDAC1 complex in cell proliferation.
ACKNOWLEDGMENTS
We thank E. Manet (Lyon, France) for the pTK-CAT-Cp4× plasmid, D. Hayward (Baltimore, Md.) for the GEX-CBF1/RBP-Jκ plasmid, and Eric Verdin (Cambridge, United Kingdom) for the anti-HDAC1 serum. We are grateful to Roger Watson for helpful comments on the manuscript.
This work was supported in part by the Wellcome Trust, which provided a project grant to M.J.A.
REFERENCES
- 1.Alfieri C, Birckenbach M, Kieff E. Early events in Epstein-Barr virus infection of human lymphocytes. Virology. 1991;181:595–608. doi: 10.1016/0042-6822(91)90893-g. [DOI] [PubMed] [Google Scholar]
- 2.Alland L, Muhle R, Hou H, Potes J, Chin L, Schreiber-Agus N, DePinho R. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 3.Allday M J, Crawford D H, Griffin B E. Prediction and demonstration of a novel Epstein-Barr virus nuclear antigen. Nucleic Acids Res. 1988;16:4353–4367. doi: 10.1093/nar/16.10.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allday M J, Crawford D H, Griffin B E. Epstein-Barr virus latent gene expression during the initiation of B cell immortalisation. J Gen Virol. 1989;70:1755–1764. doi: 10.1099/0022-1317-70-7-1755. [DOI] [PubMed] [Google Scholar]
- 5.Ayer D E, Lawrence Q A, Eisenman R N. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- 6.Babcock G J, Decker L L, Volk M, Thorley-Lawson D A. EBV persistence in memory B cells in vivo. Immunity. 1998;9:395–404. doi: 10.1016/s1074-7613(00)80622-6. [DOI] [PubMed] [Google Scholar]
- 7.Bain M, Watson R J, Farrell P J, Allday M J. Epstein-Barr virus nuclear antigen 3C is a powerful repressor of transcription when tethered to DNA. J Virol. 1996;70:2481–2489. doi: 10.1128/jvi.70.4.2481-2489.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 9.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:601–605. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 10.Brehm A, Nielsen S J, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. The E7 oncoprotein associates with Mi2 and histone deacetylase activity to promote cell growth. EMBO J. 1999;18:2449–2458. doi: 10.1093/emboj/18.9.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlson M, Laurent B C. The SNF/SWI family of global transcriptional activators. Curr Opin Cell Biol. 1994;6:396–402. doi: 10.1016/0955-0674(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 12.Chen H, Lin R J, Schlitz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 13.Davies A H, Jowett J B M, Jones I M. Recombinant baculovirus vectors expressing glutathione-S-transferase fusion proteins. Bio/Technology. 1993;11:933–936. doi: 10.1038/nbt0893-933. [DOI] [PubMed] [Google Scholar]
- 14.Grossman S R, Johannsen E, Tong X, Yalamanchili R, Kieff E. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the J kappa recombination signal binding protein. Proc Natl Acad Sci USA. 1994;91:7568–7572. doi: 10.1073/pnas.91.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;839:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 16.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 17.Henkel T, Ling P D, Hayward S D, Peterson M G. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science. 1994;265:92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- 18.Joab I, Rowe D T, Bodescot M, Nicolas J-C, Farrell P J, Perricaudet M. Mapping of the gene coding for Epstein-Barr virus-determined nuclear antigen EBNA3 and its transient overexpression in a human cell line by using an adenovirus expression vector. J Virol. 1987;61:3340–3344. doi: 10.1128/jvi.61.10.3340-3344.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johannsen E, Miller C L, Grossman S R, Kieff E. EBNA-2 and EBNA-3C extensively and mutually exclusively associate with RBPJκ in Epstein-Barr virus-transformed B lymphocytes. J Virol. 1996;70:4179–4183. doi: 10.1128/jvi.70.6.4179-4183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadonga J T. Eukaryotic transcription: an interlaced network of transcription factors and chromatin modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 21.Kao H-Y, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner C R, Evans R M, Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2343–2396. [Google Scholar]
- 23.Laherty C D, Yang W M, Sun J M, Davie J R, Seto E, Eisenman R N. Histone deacetylases associated with the mSin3 corepressor mediate Mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 24.Le Roux A, Kerdiles B, Walls D, Dedieu J F, Perricaudet M. The Epstein-Barr virus determined nuclear antigens EBNA-3A, -3B, and -3C repress EBNA-2-mediated transactivation of the viral terminal protein 1 gene promoter. Virology. 1994;205:596–602. doi: 10.1006/viro.1994.1687. [DOI] [PubMed] [Google Scholar]
- 25.Ling P D, Hsieh J J-D, Ruf I K, Rawlins D R, Hayward S D. EBNA-2 upregulation of Epstein-Barr virus latency promoters and the cellular CD23 promoter utilizes a common targeting intermediate, CBF1. J Virol. 1994;68:5375–5383. doi: 10.1128/jvi.68.9.5375-5383.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ling P D, Rawlins D R, Hayward S D. The Epstein-Barr virus immortalizing protein EBNA-2 is targeted to DNA by a cellular enhancer-binding protein. Proc Natl Acad Sci USA. 1993;90:9237–9241. doi: 10.1073/pnas.90.20.9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo R X, Postigo A A, Dean D C. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 28.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain J P, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 29.Marshall D, Sample C. Epstein-Barr virus nuclear antigen 3C is a transcriptional regulator. J Virol. 1995;69:3624–3630. doi: 10.1128/jvi.69.6.3624-3630.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maunders M J. Ph.D. thesis. Birmingham, United Kingdom: University of Birmingham; 1993. [Google Scholar]
- 31.Maunders M J, Petti L J, Rowe M. Precipitation of the Epstein-Barr virus protein EBNA2 by an EBNA3C-specific monoclonal antibody. J Gen Virol. 1994;75:769–778. doi: 10.1099/0022-1317-75-4-769. [DOI] [PubMed] [Google Scholar]
- 32.Nagy L, Kao H-Y, Chakravati D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 33.Ogryzko V V, Shiltz R L, Russanova V, Howard B H, Nakatini Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 34.Parker G A, Crook T, Bain M, Sara E A, Farrell P J, Allday M J. Epstein-Barr virus nuclear antigen (EBNA) 3C is an immortalizing oncoprotein with similar properties to adenovirus E1A and papillomavirus E7. Oncogene. 1996;13:2541–2549. [PubMed] [Google Scholar]
- 35.Pazin M J, Kadonga J T. What’s up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 36.Petti L, Sample J, Wang F, Kieff E. A fifth Epstein-Barr virus nuclear protein (EBNA3C) is expressed in latently infected growth-transformed lymphocytes. J Virol. 1988;62:1330–1338. doi: 10.1128/jvi.62.4.1330-1338.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Radkov S A, Bain M, Farrell P J, West M, Rowe M, Allday M J. Epstein-Barr virus EBNA3C represses Cp, the major promoter for EBNA expression, but has no effect on the promoter of the cell gene CD21. J Virol. 1997;71:8552–8562. doi: 10.1128/jvi.71.11.8552-8562.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reid J L, Bannister A J, Zegerman P, Martinez-Balbas M A, Kouzarides T. E1A directly binds and regulates the P/CAF acetyltransferase. EMBO J. 1998;17:4469–4477. doi: 10.1093/emboj/17.15.4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2397–2446. [Google Scholar]
- 40.Ricksten A, Kallin B, Alexander H, Dillner J, Fahraeus R, Klein G, Lerner R, Rymo L. BamHI E region of the Epstein-Barr virus genome encodes three transformation-associated nuclear proteins. Proc Natl Acad Sci USA. 1988;85:995–999. doi: 10.1073/pnas.85.4.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robertson E S, Grossman S, Johannsen E, Miller C, Lin J, Tomkinson B, Kieff E. Epstein-Barr virus nuclear protein 3C modulates transcription through interaction with the sequence-specific DNA-binding protein Jκ. J Virol. 1995;69:3108–3116. doi: 10.1128/jvi.69.5.3108-3116.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robertson E S, Lin J, Kieff E. The amino-terminal domains of Epstein-Barr virus nuclear proteins 3A, 3B, and 3C interact with RBPJκ. J Virol. 1996;70:3068–3074. doi: 10.1128/jvi.70.5.3068-3074.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sample J, Young L, Martin B, Chatman T, Kieff E, Rickinson A, Kieff E. Epstein-Barr virus types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J Virol. 1990;64:4084–4092. doi: 10.1128/jvi.64.9.4084-4092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomkinson B, Kieff E. Use of second-site homologous recombination to demonstrate that Epstein-Barr virus nuclear protein 3B is not important for lymphocyte infection or growth transformation in vitro. J Virol. 1992;66:2893–2903. doi: 10.1128/jvi.66.5.2893-2903.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomkinson B, Robertson E, Kieff E. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J Virol. 1993;67:2014–2025. doi: 10.1128/jvi.67.4.2014-2025.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waltzer L, Bourillot P Y, Sergeant A, Manet E. RBP-J kappa repression activity is mediated by a co-repressor and antagonized by the Epstein-Barr virus transcription factor EBNA2. Nucleic Acids Res. 1995;23:4939–4945. doi: 10.1093/nar/23.24.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waltzer L, Logeat F, Brou C, Israel A, Sergeant A, Manet E. The human J kappa recombination signal sequence binding protein (RBP-J kappa) targets the Epstein-Barr virus EBNA2 protein to its DNA responsive elements. EMBO J. 1994;13:5633–5638. doi: 10.1002/j.1460-2075.1994.tb06901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waltzer L, Perricaudet M, Sergeant A, Manet E. Epstein-Barr virus EBNA3A and EBNA3C proteins both repress RBP-Jκ–EBNA2-activated transcription by inhibiting the binding of RBP-Jκ to DNA. J Virol. 1996;70:5909–5915. doi: 10.1128/jvi.70.9.5909-5915.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wotton D, Lo R S, Lee S, Massague J. A Smad transcriptional corepressor. Cell. 1999;97:29–39. doi: 10.1016/s0092-8674(00)80712-6. [DOI] [PubMed] [Google Scholar]
- 50.Wu D Y, Kalpana G V, Goff S P, Schubach W H. Epstein-Barr virus nuclear protein 2 (EBNA2) binds to a component of the human SNF-SWI complex, hSNF5/Ini1. J Virol. 1996;70:6020–6028. doi: 10.1128/jvi.70.9.6020-6028.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xue Y, Wong J, Moreno G T, Young M K, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin remodelling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 52.Yang W M, Yao Y L, Sun J M, Davie J R, Seto E. Isolation and characterisation of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J Biol Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- 53.Yang X-J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 54.Yoshida M, Horinouchi S, Beppu T. Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. Bioessays. 1995;17:423–430. doi: 10.1002/bies.950170510. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Y, Leroy G, Seelig H P, Lane W S, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodelling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Y, Sun Z W, Iratni R, Erdjument-Bromage H, Tempst P, Hampsey M, Reinberg D. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol Cell. 1998;1:1021–1031. doi: 10.1016/s1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]
- 57.Zhao B, Marshall D R, Sample C E. A conserved domain of the Epstein-Barr virus nuclear antigens 3A and 3C binds to a discrete domain of Jκ. J Virol. 1996;70:4228–4236. doi: 10.1128/jvi.70.7.4228-4236.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zimber-Strobl U, Strobl L J, Meitinger C, Hinrichs R, Sakai T, Furukawa T, Honjo T, Bornkamm G W. Epstein-Barr virus nuclear antigen 2 exerts its transactivating function through interaction with recombination signal binding protein RBP-J kappa, the homologue of Drosophila Suppressor of Hairless. EMBO J. 1994;13:4973–4982. doi: 10.1002/j.1460-2075.1994.tb06824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]