Abstract
Newcomb and coworkers (W. W. Newcomb, F. L. Homa, D. R. Thomsen, F. P. Booy, B. L. Trus, A. C. Steven, J. V. Spencer, and J. C. Brown, J. Mol. Biol. 263:432–446, 1996; W. W. Newcomb, F. L. Homa, D. R. Thomsen, Z. Ye, and J. C. Brown, J. Virol. 68:6059–6063, 1994) have recently described an in vitro herpes simplex virus (HSV) capsid assembly product which, because of certain parallels between its properties and those of bacteriophage proheads, they have designated the procapsid. As in their bacteriophage counterparts, there are marked differences between the structures of the two types of particle, and conversion from the procapsid to the capsid form requires extensive reconfiguration of the subunits. This reconfiguration occurs spontaneously upon extended in vitro incubation. One of the distinctive features of the HSV procapsids is that, unlike mature capsids, they are unstable and disassemble upon storage at 2°C. Using a mutant of HSV type 1 (ts1201), which has a lesion in the protease responsible for maturational cleavage of the scaffolding protein, we have demonstrated that capsids present within cells infected at nonpermissive temperatures are also cryosensitive and disappear if the cells are incubated at 0°C. This suggests that ts1201 capsids may resemble procapsids in structure. However, ts1201 capsids remain cryosensitive following extended incubation at an elevated temperature and, therefore, do not appear to undergo the spontaneous reconfiguration seen with in vitro-assembled procapsids. The lesion in ts1201 is reversible, and capsids formed at the nonpermissive temperature can undergo maturational cleavage and go on to form infectious virions following downshift to permissive temperatures. The sensitivity of ts1201 capsids to low temperatures is closely correlated with the cleavage status of the scaffolding protein, suggesting that proteolysis may act to trigger their conversion to the stable form. The experiments described here provide the firmest evidence yet that the procapsid has a biologically relevant role in the virus life cycle.
Recently, our understanding of the process of capsid assembly has been considerably advanced by the development of an in vitro capsid assembly model (17, 18, 26). The early events suggested by this model are coassembly of scaffolding protein and capsid shell protein subunits to form segments of shell-scaffold which increase in extent until they become complete spherical shells. These spherical shells have a T=16 icosahedral organization similar to that of typical herpes simplex virus (HSV) capsids, but they differ markedly in appearance and properties and have been designated procapsids. The procapsid has a less well defined structure (26) than do typical capsids isolated from cells infected with HSV (31) or with recombinant baculoviruses expressing HSV capsid proteins (27, 29, 30). In the procapsid, connections between subunits are tenuous, giving rise to a very open structure. This is reflected in the fact that they are unstable and disassemble if incubated at low temperatures (17). Following assembly, the procapsid undergoes a spontaneous reconfiguration to adopt the stable polyhedral form of typical capsids (26). This reconfiguration involves changes that have the effect of bringing the capsid subunits into closer contact and result in the formation of a continuous capsid floor largely made up from the lower portions of the hexon subunits (26, 30). The process has been likened to the extensive structural changes that many bacteriophage capsids undergo and which are functionally linked to capsid maturation and DNA packaging. Based on this apparent similarity, Newcomb et al. (17) suggested that the two processes are functionally equivalent and proposed that the HSV procapsid is the form into which the virus genome is packaged. One implication of this model is that the polyhedral B capsids, which are found in large numbers in infected cells and are the best-understood capsids in structural terms, are not intermediates in virion assembly but are probably dead-end products, incapable of progressing any further. In the absence of an in vitro packaging system which could test the functionality of procapsids and given the lack of direct evidence for procapsid formation during the course of normal HSV infection, there has been no way of testing this hypothesis.
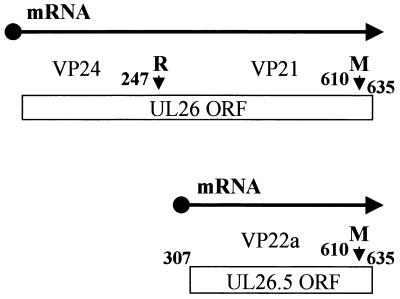
Newcomb et al. (17) also suggested that procapsids, which have not been detected in normal HSV infections, might accumulate in certain temperature-sensitive mutants that are defective for DNA packaging. One such mutant, ts1201, has a defect in the virus-encoded protease, which is responsible for cleavage of the capsid scaffolding protein (21). The scaffold is formed by the products of the overlapping genes UL26 and UL26.5 (Fig. 1) (12, 13, 22). The UL26 protein contains a protease activity in its N-terminal unique portion and cleaves itself in two places to generate the B-capsid proteins VP24 and VP21 (11, 16). The primary product of the UL26.5 open reading frame (ORF) is the major scaffolding protein, pre-VP22a, which is cleaved by the UL26 protease to generate the B-capsid protein VP22a. Cleavage of both proteins near their C-terminal ends breaks the connection between the scaffolding proteins and the capsid shell (9, 15) and is required for capsid maturation and DNA packaging (7, 21).
FIG. 1.
Organization of UL26 and UL26.5. The numbers refer to the amino acid residues in UL26, and the small vertical arrows indicate the proteolytic cleavage sites. The primary product of the UL26 ORF is the full-length (635-amino-acid) protease. This undergoes self-cleavage at the release (R) and maturation (M) sites to generate the BSC capsid proteins VP24 (247 amino acids) and VP21 (363 amino acids), respectively. The primary product of the UL26.5 ORF is the major scaffolding protein, pre-VP22a (328 amino acids), which is cleaved at the M site by the UL26 protease to generate the BSC capsid protein VP22a (303 amino acids). The mutation in ts1201 maps to the VP24 portion of the protease (21).
In ts1201 capsids at the nonpermissive temperature, the scaffolding proteins remain uncleaved and their cores appear larger than normal in thin-section electron micrographs (1). Here we refer to them as large-core B (BLC) capsids to distinguish them from the typical small-core B (BSC) capsids which are formed in wild-type (wt) virus as a result of proteolysis. An interesting feature of ts1201 is the reversibility of the lesion. Thus, downshift to a permissive temperature allows large-core capsids to proceed through the remainder of their maturation pathway, giving rise to A, BSC, and C capsids and ultimately to mature virions (2, 21). To investigate the nature of the relationship between ts1201 capsids and procapsids, we examined one of the distinctive properties of the latter, namely, their instability at low temperatures.
MATERIALS AND METHODS
Cells and virus.
All experiments were carried out in BHK-21 C13 cells cultured in Glasgow modified Eagle’s medium (Life Technologies) supplemented with 10% tryptose phosphate and 10% newborn calf serum. wt HSV type 1 (HSV-1) strain 17 (3) was used. The temperature-sensitive mutants ts1201 and ts1203 were isolated from this strain. ts1201 has a defect in the amino-terminal region of the UL26 protease (21), and ts1203 has a defect in a DNA-packaging-related gene, UL28 (2).
Infections.
Confluent 35-mm-diameter culture dishes of BHK cells were infected with 5 PFU of wt or mutant viruses per cell. After incubation at 39°C for 10 h, the plates were either harvested immediately or overlaid with fresh medium, containing 200 μg of cycloheximide per ml, which had been equilibrated at the appropriate temperature. Cells were further incubated at 39, 31, or 0°C. Incubations at 0°C were carried out by placing the culture dishes on ice. At the time of harvest, a third of each sample was taken for Western blot analysis and the remainder was prepared for electron microscopy.
Electron microscopy.
Cells were washed and scraped into phosphate-buffered saline, pelleted by low-speed centrifugation (2,000 × g for 2 min) in BEEM capsules (Taab Laboratories), and fixed with 2.5% glutaraldehyde in phosphate-buffered saline. Following secondary fixation with 1% osmium tetroxide, the pellets were dehydrated through a graded alcohol series and embedded in Epon 100 (Agar Aids) resin as described previously (21).
Western blotting.
Samples were prepared for electrophoresis, and proteins were separated on 10% polyacrylamide gels cross-linked with 2.5% (wt/wt) N,N′-methylene bisacrylamide (14). Separated proteins were transferred to a nitrocellulose membrane (25). The nitrocellulose membrane was incubated for 1 h in 1028 antibody at a dilution of 1:1,000, washed, and then incubated in goat anti-mouse immunoglobulin G-peroxidase conjugate (Sigma Immunochemicals). Bound antibody was detected by enhanced chemiluminescence (Amersham, Little Chalfont, United Kingdom) according to the manufacturer’s instructions.
RESULTS
Stability of ts1201 capsids.
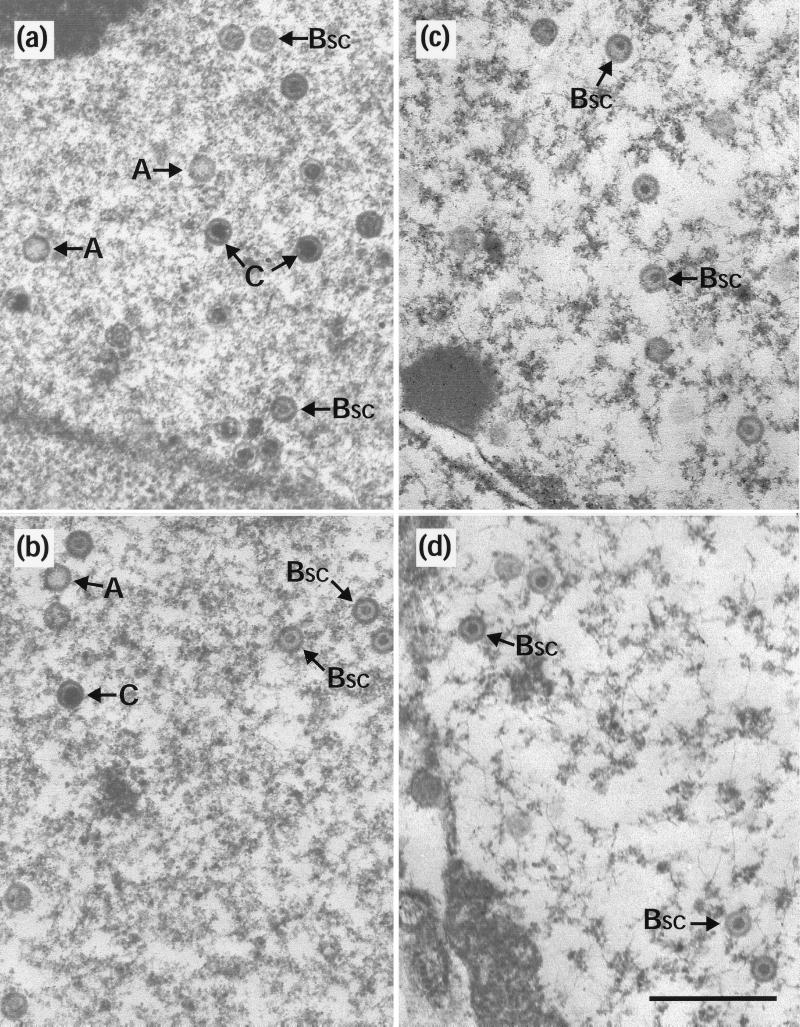
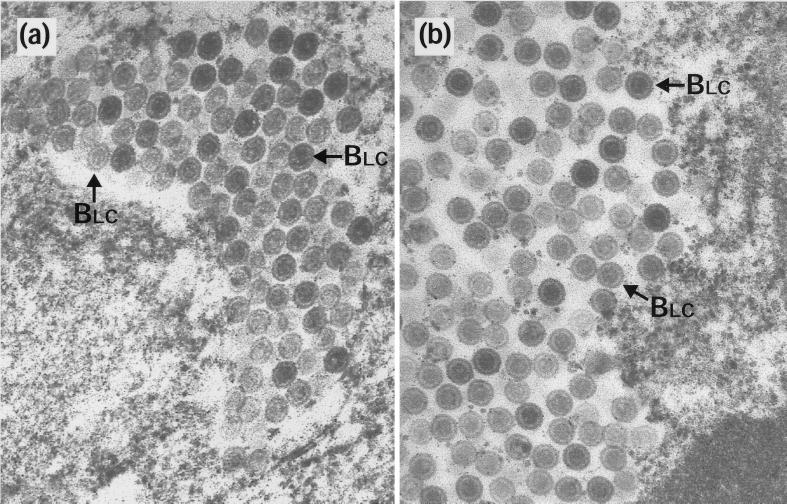
Newcomb et al. (17) observed that incubation of newly assembled procapsids at 2°C caused them to disintegrate. However, if they were first allowed to reconfigure into the polyhedral form, they were stable under these conditions. To investigate whether capsids made within cells had similar properties, duplicate 35-mm-diameter plates of BHK C13 cells were infected with HSV-1 wt virus or with two temperature-sensitive mutants, ts1201 and ts1203. ts1203 has an irreversible defect in DNA packaging but has normal protease activity and assembles large numbers of small-core (BSC) capsids (2). After incubation at 39°C for 10 h, plates were either harvested immediately for electron microscopy or overlaid with fresh medium that had been equilibrated at the desired temperature. In order to prevent the formation of new capsids during the subsequent incubations, the fresh overlay medium contained 200 μg of cycloheximide per ml. After 10 h at 39°C, each of the viruses gave a characteristic pattern of capsid forms. wt had a mixture of A, BSC, and C capsids (Fig. 2a);ts 1203 had exclusively BSC capsids (Fig. 2c); and ts1201 had exclusively large-core (BLC) capsids (Fig. 2e), many of which were in large aggregations. Upon incubation at 0°C, the following responses were seen. In wt and ts1203, the patterns had not changed. Thus, wt still contained similar numbers of A, BSC, and C capsids (Fig. 2b) while in ts1203 only BSC capsids were still present (Fig. 2d). This established that none of the three types of capsid made during normal infection is sensitive to incubation at 0°C and demonstrated that BSC capsids in both wt and a packaging-incompetent mutant have similar properties. However, following incubation at 0°C very few capsids could be seen in the ts1201-infected cells (Table 1). Figure 2f shows a field containing a few of the capsids that did survive the exposure to low temperature. In addition, the area shown contains a number of partial capsids and what appear to be free cores. Regions containing concentrations of these putative cores were readily observed, suggesting that they may have been the locations of capsid concentrations. The intact capsids in Fig. 2f still have large cores, indicating that exposure to low temperature has not resulted in loss or contraction of the scaffold. ts1201 cells that had been shifted to 31°C showed the expected conversion to a wt pattern, with the disappearance of the BLC capsid aggregates and the appearance of A, BSC, and C capsids throughout the nucleus (Fig. 2g) and of extranuclear virions (data not shown and references 2 and 21). This confirms the reversibility of the ts1201 lesion and the ability of the capsids made at 39°C to convert to mature forms. Furthermore, these capsids now appeared resistant to disassociation on subsequent incubation at 0°C (Fig. 2h).
FIG. 2.
Capsid stability at 0°C. Replica 35-mm-diameter plates of BHK C13 cells were infected with 5 PFU of HSV-1 wt virus (a and b), ts1203 (c and d), or ts1201 (e to h) per cell. After incubation at 39°C for 10 h, the plates were either harvested immediately for electron microscopy (a, c, and e) or overlaid with fresh prewarmed or precooled medium containing 200 μg of cycloheximide per ml and incubated further at the desired temperature. For both wt (b) and ts1203 (d), cells were overlaid with medium that had been precooled to 0°C and were then incubated on ice for a further 4 h. For ts1201, one plate was treated the same as for wt and ts1203 by being incubated at 0°C for 4 h (f), while two further plates were incubated at 31°C for 4 h. One of these was then harvested immediately for electron microscopy (g), and the other was overlaid with medium at 0°C and incubated for a further 10 h on ice (h). A, A capsids; C, C capsids; pc, partial capsids; fc, free cores. Scale bar = 500 nm.
TABLE 1.
Capsid stability at 0°Ca
| Virus | No. of capsids after incubation at temp (°C):
|
||
|---|---|---|---|
| 39 | 31 | 0 | |
| wt | 32.0 | 34.2 | |
| ts1203 | 28.5 | 34.9 | |
| ts1201 | 50.2 | 0.7 | |
| ts1201 | 50.2 | 21.2 | 11.9 |
Values represent the numbers of capsids seen in 25 nuclear sections from the samples shown in Fig. 2. For wt, ts1203, and ts1201, the numbers given are values after incubation at 39°C for 10 h and after a further 4-h incubation at 0°C in the presence of cycloheximide. In addition, the numbers present for ts1201 after downshift to 31°C for 4 h and subsequently to 0°C for a further 10 h (see Fig. 2) are shown. The same 39°C starting value for ts1201 is given twice for clarity.
Quantitative estimates of capsid stability.
The stability of different capsid types at 0°C (Table 1) was determined by counting the capsids present in 25 nuclear sections from the samples shown in Fig. 2. The numbers of capsids in wt- and ts1203-infected cells were comparable before and after incubation at 0°C, confirming that the capsids made by these viruses are stable. For ts1201, there was an approximately 50% decrease in the numbers of capsids following downshift to 31°C. This probably reflects the maturation and loss of capsids as virions, which were not counted in these experiments. There was a further decrease following a subsequent 10-h incubation on ice, but at the end of this period, capsids were still readily detectable. However, when ts1201 samples were shifted directly from 39 to 0°C, the number of capsids observed declined by more than 50-fold to <1/nucleus. This demonstrates that the ts1201 BLC capsids are sensitive to incubation at 0°C, while incubation at 31°C allowed them to adopt a stable configuration.
Cleavage status of scaffolding proteins.
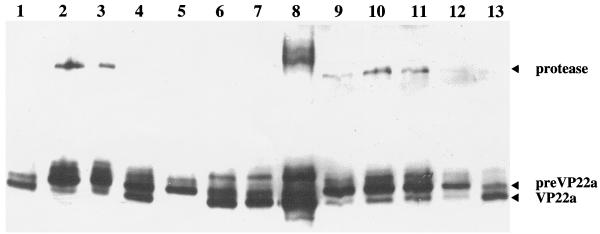
In order to determine the cleavage status of the scaffolding proteins, part of each sample used for electron microscopy was also analyzed by Western blotting with the monoclonal antibody 1028, which recognizes both processed and unprocessed forms of the scaffolding protein. In wt-infected cells (Fig. 3, lanes 1 and 13), the fully processed form of the scaffolding protein, VP22a, was the most abundant, while the uncleaved form, pre-VP22a, was present in smaller amounts and the full-length protease was not detected. Similar patterns were seen for ts1203-infected cells, both at 39°C and following downshift to 0°C (Fig. 3, lanes 6 and 7). In ts1201-infected cells at 39°C, this pattern was reversed, with VP22a present in reduced amounts while both pre-VP22a and the protease were prominent (Fig. 3, lane 2). Thus, proteolysis had been inhibited as suggested by the large-core phenotype (Fig. 2e). When the ts1201-infected cells were maintained at 39°C for a further 4 h following the addition of cycloheximide, the pattern of bands was unchanged (Fig. 3, compare lanes 2 and 3). However, downshift to 31°C allowed proteolysis to take place as indicated by the appearance of VP22a and the reduction in pre-VP22a and full-length protease (Fig. 3, lane 4). Downshift from 39 to 0°C (Fig. 3, lane 5) did not induce the characteristic cleavage as shown by the absence of VP22a. However, disassociation of the capsid shell at 0°C exposes the scaffold to the nuclear environment, and the reduced amounts of scaffolding protein and protease in lanes 5 and 12 (see below) may be due to breakdown caused by cellular factors.
FIG. 3.
Cleavage status of scaffolding proteins. Extracts of BHK C13 cells infected with 5 PFU of HSV-1 wt virus, ts1203, or ts1201 per cell were separated on a 10% polyacrylamide gel, transferred to nitrocellulose, and probed with monoclonal antibody 1028. All samples were infected for 10 h at 39°C. Some were then harvested, while the remainder were overlaid with fresh medium containing 200 μg of cycloheximide per ml and incubated further as described for Fig. 2. Cells were infected and incubated with wt virus for 10 h at 39°C (lanes 1 and 13); ts1203 for 10 h at 39°C (lane 6) followed by 4 h at 0°C (lane 7); or ts1201 for 10 h at 39°C (lane 2) followed by 4 h at 39°C (lane 3), 31°C (lane 4), or 0°C (lane 5). In lanes 9 to 12, samples were infected with ts1201 for 10 h at 39°C and then overlaid with medium containing 200 μg of cycloheximide per ml and incubated for a further 14 h at 39°C. They were then harvested immediately (lane 9) or incubated in the continued presence of cycloheximide for a further 4 h at 39°C (lane 10), 31°C (lane 11), or 0°C (lane 12). BSC capsids were run in lane 8 as a control. The UL26 protease and the uncleaved (pre-VP22a) and cleaved (VP22a) forms of the UL26.5 scaffolding protein are indicated.
ts1201 capsids do not undergo spontaneous reconfiguration.
In vitro-assembled procapsids spontaneously reconfigure following incubation at room temperature, or 37°C, into polyhedral capsids that are stable at low temperatures (17). To determine whether ts1201 BLC capsids behaved similarly, replicate 35-mm-diameter plates of BHK C13 cells were infected with ts1201 and incubated at 39°C for 10 h as before. The plates were then overlaid with medium containing 200 μg of cycloheximide per ml and incubated for a further 14 h at 39°C. One plate was then harvested, while the others were incubated for a further 4 h at 39, 31, or 0°C, respectively. From Western blotting of their protein profiles, it was clear that incubation at 39°C for 24 h had not affected the cleavage status of the scaffolding proteins (Fig. 3, compare lanes 9 and 2). Thus, both pre-VP22a and full-length protease were prominent while VP22a was found in only reduced amounts compared to wt. As before, incubation at 39°C (Fig. 3, lane 10) or 0°C (Fig. 3, lane 12) for a further 4 h did not greatly affect this pattern. Surprisingly, however, the pattern was also unchanged following downshift to 31°C (Fig. 3, lane 11), indicating that the ts1201 defect had been rendered irreversible by extended incubation at the nonpermissive temperature.
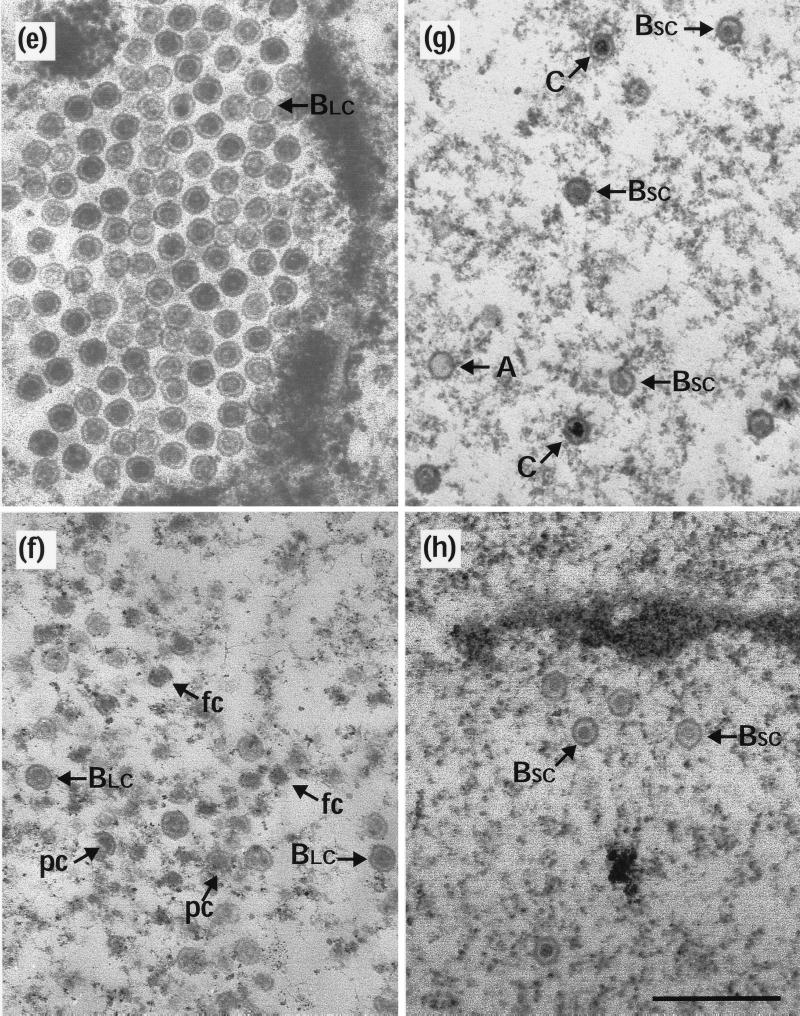
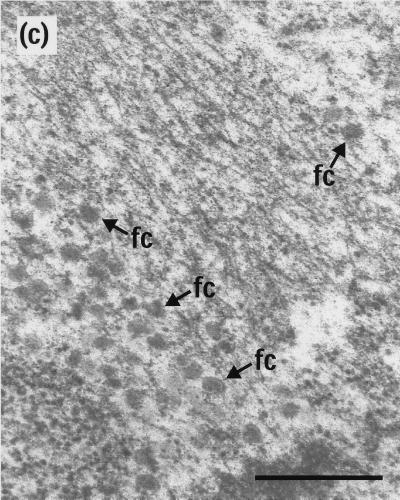
Electron microscopy revealed that BLC capsids, indistinguishable from those present at 10 h postinfection, were still present after a further 14-h incubation at 39°C (Fig. 4a). However, downshift to 31°C did not result in their conversion to A, BSC, or C capsids (Fig. 4b). Thus, it appears that the reversibility of the ts1201 defect had been lost, thereby confirming the findings from Western blotting (Fig. 3). As before, incubation of the cells at 0°C caused the disappearance of virtually all the capsids, although there were regions which appeared to contain residual free cores (Fig. 4c). The absence of capsids at 0°C suggests that the BLC capsids present after the extended incubation at an elevated temperature were still in an unstable conformation.
FIG. 4.
Test for spontaneous angularization of ts1201 capsids. Thirty-five-millimeter-diameter plates of BHK C13 cells were infected with 5 PFU of ts1201 per cell for 10 h at 39°C. They were then overlaid with fresh, prewarmed medium containing 200 μg of cycloheximide per ml and incubated for a further 14 h at 39°C as described for Fig. 3. The cells were then either harvested for electron microscopy (a) or incubated in the presence of cycloheximide for a further 4 h at 31°C (b) or 0°C (c). fc, free cores. Scale bar = 500 nm.
DISCUSSION
The recent development of an in vitro model promises to alter radically our understanding of the mechanism of HSV capsid assembly and maturation. In particular, the identification of the procapsid as a precursor to typical capsids offers a picture of how the proteins first come together in a loosely connected icosahedral arrangement which then undergoes tightening of interactions and strengthening of contacts to form the robust and stable capsid shell. Newcomb et al. (17) have proposed that the procapsid is a normal intermediate in capsid morphogenesis and is the progenitor of all other capsid types and of virions. Although procapsid-like structures have not been identified in virus infections, Newcomb et al. suggested that some virus mutants, such as ts1201, might accumulate procapsids at nonpermissive temperatures.
ts1201 is unusual among HSV mutants defective for DNA packaging in that the mutation is reversible (2, 21). The capsids formed at nonpermissive temperatures therefore genuinely do represent functional progenitors of other capsid types. A second HSV-1 mutant, tsProt.A, which was engineered to contain amino acid substitutions identical to those of ts1201, was shown to have a similar phenotype (7). This mutant has been used to study the progress of DNA packaging and virion maturation following release from the high-temperature block (4, 5). Prior to this study, the only evidence for a structural relationship between in vitro procapsids and ts1201 capsids was their similarity in possessing distinctive large cores in thin section. However, possession of a large core is not diagnostic of a procapsid structure since capsids of normal morphology with large cores can be readily isolated with a baculovirus model system (23, 24, 30). The other features of procapsids, their rounded appearance and open structure, cannot be seen in thin-section images. One characteristic property of procapsids is that they are unstable and fall apart at low temperatures. This behavior is very different from that of typical capsids, which are extremely robust. The fact that ts1201 BLC capsids are also cryosensitive and disassemble at 0°C therefore provides further evidence that they have a similar structure. Therefore, we interpret these results as suggesting that ts1201 capsids are procapsids. After this paper was submitted, Dasgupta and Wilson (6) reported that they were unable to purify capsids from cells infected with tsProt.A at the nonpermissive temperature, after cell extracts had been incubated on ice overnight. From this, they also conclude that the cold-sensitive capsids are procapsids.
Although the great majority of the ts1201 BLC capsids are cryosensitive, a small percentage does survive the low-temperature incubation (Table 1). If the thermolability of ts1201 BLC capsids is an accurate reflection of their procapsid nature, then a logical conclusion would be that the few stable capsids have a polyhedral conformation, presumably as a result of a reconfiguration similar to the spontaneous angularization reported for in vitro procapsids. In support of this suggestion is the observation that small numbers of polyhedral BLC capsids can be purified from ts1201-infected cells (22a).
Newcomb et al. (17) proposed that DNA packaging and scaffold loss take place at the procapsid stage. In an attempt to investigate this, we kept ts1201-infected cells at 39°C for extended periods to allow time for more of the capsids to undergo reconfiguration into the polyhedral form, before downshifting and looking for DNA packaging. However, this experiment had two unexpected outcomes. Firstly, the ts1201 lesion was no longer reversible, presumably because maintaining the protease at 39°C for this length of time caused it to adopt an irreversibly inactivated form. Secondly, the capsids remained cryosensitive, implying that they were still in the procapsid conformation. That the capsids were still unstable after at least 14 h of incubation at 39°C was surprising in view of the observation that in vitro-assembled procapsids reconfigure into stable polyhedral capsids within 8 h at 28°C or within 4 h at 37°C (17). The differing behaviors of ts1201 and in vitro-assembled procapsids could be related to differences between the intracellular and in vitro environments. Alternatively, it might reflect variations in the compositions of the two sets of capsids, since in vitro-assembled procapsids contain none of the other proteins (including packaging proteins and possibly some tegumet proteins) normally associated with capsids in infected cells (8, 28). In ts1201, where spontaneous angularization appears to be inhibited, the trigger for reconfiguration may be the proteolytic cleavage of the scaffolding proteins. It may be significant that angularization occurs efficiently in the presence of uncleaved scaffolding proteins in the baculovirus model for capsid assembly (30). Here, as in the in vitro situation, other capsid-associated proteins are absent, suggesting that their presence may help stabilize the capsid in a procapsid conformation, thereby extending the period during which it can participate in DNA packaging. Recently, it has been reported that epitope-specific conformational changes and packaging of viral DNA in tsProt.A capsids are both ATP dependent, although it was not clear whether these two processes are functionally linked. However, conversion of the capsids to a cold-resistant form does not require ATP (6).
Since ts1201 capsids can give rise to DNA containing C capsids and virions (2, 21), the experiments described here support the proposal that DNA enters capsids while they are in a procapsid-like state. However, the possibility that formation of stable capsids precedes packaging remains, as suggested by pulse-chase labelling studies on equine herpesvirus type 1 (19, 20) and pseudorabies virus (10). One sequence of events that is consistent with available data would be (i) assembly of the procapsid, (ii) cleavage of scaffold and reconfiguration to polyhedral capsids, and (iii) loss of scaffold and DNA packaging. Therefore, the question of which capsid type participates in DNA packaging remains unresolved.
Analysis of purified BSC capsids has shown that effectively all their scaffolding proteins are cleaved. However, it is not known whether cleavage of the many target sites in the scaffold is random or takes place in a coordinated manner (16). The presence of partial capsids and arcs following incubation at 0°C is intriguing, as these structures closely resemble the earliest forms observed during in vitro assembly (17). Since procapsid assembly seems to occur by sequential growth of the capsid shell, it is interesting to speculate whether conversion to a stable form occurs in a similar fashion. A scenario in which the change from procapsid to polyhedral form occurs progressively in a wave travelling around the capsid shell can be envisaged. A reconfiguring capsid at intermediate stages in this process would represent a chimera of stable and unstable conformations and upon disassembly could give rise to the type of structure seen in Fig. 2f. Scaffold cleavage and exit, capsid shell reconfiguration, and DNA packaging could even be linked in a process which begins at a specific location and proceeds through a coordinated, cooperative series of steps until its completion. However, elucidating the precise sequence of events leading to the formation of mature capsids and the relationship between the various capsid types will probably have to await the development of an in vitro packaging model to complement the in vitro assembly model.
ACKNOWLEDGMENTS
We thank Valerie Preston for providing ts1203, Anne Cross for monoclonal antibody 1028, and Joyce Mitchell for her excellent technical assistance.
REFERENCES
- 1.Addison C, Rixon F J, Palfreyman J W, Ohara M, Preston V G. Characterization of a herpes simplex virus type-1 mutant which has a temperature-sensitive defect in penetration of cells and assembly of capsids. Virology. 1984;138:246–259. doi: 10.1016/0042-6822(84)90349-0. [DOI] [PubMed] [Google Scholar]
- 2.Addison C, Rixon F J, Preston V G. Herpes simplex virus type-1 UL28 gene product is important for the formation of mature capsids. J Gen Virol. 1990;71:2377–2384. doi: 10.1099/0022-1317-71-10-2377. [DOI] [PubMed] [Google Scholar]
- 3.Brown S M, Ritchie D A, Subak-Sharpe J H. Genetic studies with herpes simplex virus type 1. The isolation of temperature-sensitive mutants, their arrangement into complementation groups and recombination analysis leading to a linkage map. J Gen Virol. 1973;18:329–346. doi: 10.1099/0022-1317-18-3-329. [DOI] [PubMed] [Google Scholar]
- 4.Church G A, Dasgupta A, Wilson D W. Herpes simplex virus DNA packaging without measurable DNA synthesis. J Virol. 1998;72:2745–2751. doi: 10.1128/jvi.72.4.2745-2751.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Church G A, Wilson D W. Study of herpes simplex virus maturation during a synchronous wave of assembly. J Virol. 1997;71:3603–3612. doi: 10.1128/jvi.71.5.3603-3612.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dasgupta A, Wilson D W. ATP depletion blocks herpes simplex virus DNA packaging and capsid maturation. J Virol. 1999;73:2006–2015. doi: 10.1128/jvi.73.3.2006-2015.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao M, Matusick-Kumar L, Hurlburt W, DiTusa S F, Newcomb W W, Brown J C, McCann III P J, Deckman I, Colonno R J. The protease of herpes simplex virus type 1 is essential for functional capsid formation and viral growth. J Virol. 1994;68:3703–3712. doi: 10.1128/jvi.68.6.3702-3712.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Homa F L, Brown J C. Capsid assembly and DNA packaging in herpes simplex virus. Med Virol. 1997;7:107–122. doi: 10.1002/(sici)1099-1654(199707)7:2<107::aid-rmv191>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 9.Kennard J, Rixon F J, McDougall I M, Tatman J D, Preston V G. The 25 amino acid residues at the carboxy terminus of the herpes simplex virus type 1 UL26.5 protein are required for the formation of the capsid shell around the scaffold. J Gen Virol. 1995;76:1611–1621. doi: 10.1099/0022-1317-76-7-1611. [DOI] [PubMed] [Google Scholar]
- 10.Ladin B F, Blankenship M L, Ben-Porat T. Replication of herpesvirus DNA. V. Maturation of concatemeric DNA of pseudorabies virus to genome length is related to capsid formation. J Virol. 1980;33:1151–1164. doi: 10.1128/jvi.33.3.1151-1164.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu F, Roizman B. Differentiation of multiple domains in the herpes simplex virus 1 protease encoded by the UL26 gene. Proc Natl Acad Sci USA. 1992;89:2076–2080. doi: 10.1073/pnas.89.6.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu F, Roizman B. The herpes simplex virus 1 gene encoding a protease also contains within its coding domain the gene encoding the more abundant substrate. J Virol. 1991;65:5149–5156. doi: 10.1128/jvi.65.10.5149-5156.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu F, Roizman B. The promoter, transcriptional unit, and coding sequences of herpes simplex virus 1 family 35 proteins are contained within and in frame with the UL26 open reading frame. J Virol. 1991;65:206–212. doi: 10.1128/jvi.65.1.206-212.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marsden H S, Stow N D, Preston V G, Timbury M C, Wilkie N M. Physical mapping of herpes simplex virus-induced polypeptides. J Virol. 1978;28:624–642. doi: 10.1128/jvi.28.2.624-642.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matusick-Kumar L, Newcomb W W, Brown J C, McCann III P J, Hurlburt W, Weinheimer S P, Gao M. The C-terminal 25 amino acids of the protease and its substrate ICP35 of herpes simplex virus type 1 are involved in the formation of sealed capsids. J Virol. 1995;69:4347–4356. doi: 10.1128/jvi.69.7.4347-4356.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCann P J, III, O’Boyle II D R, Deckman I C. Investigation of the specificity of the herpes simplex virus type 1 protease by point mutagenesis of the autoproteolysis sites. J Virol. 1994;68:526–529. doi: 10.1128/jvi.68.1.526-529.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newcomb W W, Homa F L, Thomsen D R, Booy F P, Trus B L, Steven A C, Spencer J V, Brown J C. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J Mol Biol. 1996;263:432–446. doi: 10.1006/jmbi.1996.0587. [DOI] [PubMed] [Google Scholar]
- 18.Newcomb W W, Homa F L, Thomsen D R, Ye Z, Brown J C. Cell-free assembly of the herpes simplex virus capsid. J Virol. 1994;68:6059–6063. doi: 10.1128/jvi.68.9.6059-6063.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Callaghan D J, Randall C C. Molecular anatomy of herpesviruses: recent studies. Prog Med Virol. 1976;22:152–210. [PubMed] [Google Scholar]
- 20.Perdue M, Cohen J, Randall C, O’Callaghan D. Biochemical studies on the maturation of herpesvirus nucleocapsid species. Virology. 1976;74:194–208. [PubMed] [Google Scholar]
- 21.Preston V G, Coates J A V, Rixon F J. Identification and characterization of a herpes simplex virus gene product required for encapsidation of virus DNA. J Virol. 1983;45:1056–1064. doi: 10.1128/jvi.45.3.1056-1064.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Preston V G, Rixon F J, McDougall I M, McGregor M, Al-Kobaisi M F. Processing of the herpes simplex virus assembly protein ICP35 near its carboxy terminal end requires the product of the whole of the UL26 reading frame. Virology. 1992;186:87–98. doi: 10.1016/0042-6822(92)90063-u. [DOI] [PubMed] [Google Scholar]
- 22a.Rixon, F. J., and Z. H. Zhou. Unpublished data.
- 23.Tatman J D, Preston V G, Nicholson P, Elliott R M, Rixon F J. Assembly of herpes simplex virus type 1 capsids using a panel of recombinant baculoviruses. J Gen Virol. 1994;75:1101–1113. doi: 10.1099/0022-1317-75-5-1101. [DOI] [PubMed] [Google Scholar]
- 24.Thomsen D R, Roof L L, Homa F L. Assembly of herpes simplex virus (HSV) intermediate capsids in insect cells infected with recombinant baculoviruses expressing HSV capsid proteins. J Virol. 1994;68:2442–2457. doi: 10.1128/jvi.68.4.2442-2457.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trus B L, Booy F P, Newcomb W W, Brown J C, Homa F L, Thomsen D R, Steven A C. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J Mol Biol. 1996;263:447–462. doi: 10.1016/s0022-2836(96)80018-0. [DOI] [PubMed] [Google Scholar]
- 27.Trus B L, Homa F L, Booy F P, Newcomb W W, Thomsen D R, Cheng N, Brown J C, Steven A C. Herpes simplex virus capsids assembled in insect cells infected with recombinant baculoviruses: structural authenticity and localization of VP26. J Virol. 1995;69:7362–7366. doi: 10.1128/jvi.69.11.7362-7366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu D, Weller S K. Herpes simplex virus type 1 cleavage and packaging proteins UL15 and UL28 are associated with B but not C capsids during packaging. J Virol. 1998;72:7428–7439. doi: 10.1128/jvi.72.9.7428-7439.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Z H, He J, Jakana J, Tatman J D, Rixon F J, Chiu W. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat Struct Biol. 1995;2:1026–1030. doi: 10.1038/nsb1195-1026. [DOI] [PubMed] [Google Scholar]
- 30.Zhou Z H, Macnab S J, Jakana J, Scott L R, Chiu W, Rixon F J. Identification of the sites of interaction between the scaffold and the outer shell in HSV-1 capsids by difference electron imaging. Proc Natl Acad Sci USA. 1998;95:2778–2783. doi: 10.1073/pnas.95.6.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Z H, Prasad B V V, Jakana J, Rixon F J, Chiu W. Protein subunit structures in the herpes simplex virus A-capsid determined from 400 kV spot-scan electron cryomicroscopy. J Mol Biol. 1994;242:456–469. doi: 10.1006/jmbi.1994.1594. [DOI] [PubMed] [Google Scholar]