Abstract
The most abundantly expressed latent transcripts encoded by the Kaposi’s sarcoma (KS)-associated herpesvirus derive from the genomic region surrounding open reading frame (ORF) K12 (kaposin A). Here we show that these transcripts, initially described as limited to ORF K12 itself, more frequently encompass upstream sequences spanning two sets of 23-nucleotide GC-rich direct repeats (DRs) (DR1 and DR2). Although the DRs lack AUG codons and were previously presumed to be noncoding, a monoclonal antibody raised to infected cells detected multiple polypeptides encoded by this region. These proteins are expressed during latency and upon induction of lytic viral replication in both primary effusion lymphoma (PEL) cell lines and KS tumors. Biochemical and genetic analyses reveal that these proteins are derived from variant translational initiation at CUG codons. The predominant translation product in the PEL cell line BCBL-1 derives from the 5′-most CUG codon in the transcript, resulting in a protein (termed kaposin B) which is encoded largely by the repeats themselves and which does not include K12 sequences. Other non-AUG codons in alternate reading frames are also used at lower efficiency, including one that initiates translation of a DR-K12 fusion protein (kaposin C) that is predicted to sort to a different subcellular locale than kaposin B. Thus, the products of the K12 region, which is the most abundantly transcribed region in latency, are surprisingly complex and may encompass multiple biological functions.
Kaposi’s sarcoma (KS) is a complex multicentric neoplasm originally described as a rare tumor of elderly men of Mediterranean origin (17). More recently, KS has been recognized as a frequent malignancy among human immunodeficiency virus-infected individuals (4). Established KS tumors consist predominantly of spindle-shaped cells of apparent endothelial origin which are regarded as the primary proliferative component of the lesion. In addition, the tumors display abundant, abnormal neovascular elements and infiltration by plasma cells and mononuclear cells (see reference 6 for review). The recently discovered KS-associated herpesvirus (KSHV) (10), also known as human herpesvirus 8, is strongly linked to KS epidemiologically (see references 11, 18, and 45 for review) and is consistently detected in the spindle cells of KS tissues (5, 48, 49). In keeping with its classification as a lymphotropic herpesvirus (gamma-2-herpesvirus) (32), the virus is also consistently present in at least two lymphoproliferative diseases, primary effusion lymphoma (PEL; formerly called body cavity-associated lymphoma) and multicentric Castleman’s disease (7, 47). In KS and PEL, the majority of the tumor cells display latent KSHV infection, while lytic replication is limited to a small subset of cells in both cases, presumably reflecting spontaneous reactivation from the latent state (5, 48, 49).
KSHV, like other gamma-2-herpesviruses, encodes a large number of proteins that have homology to cellular genes that influence or regulate proliferation of the cell (35, 41). These include homologs of chemokines (31, 36), a G protein-coupled chemokine receptor (9), the cytokine interleukin-6 (31, 36) and other signaling molecules (K1 protein [27, 28]), antiapoptotic proteins (vBcl-2 [13, 43]), Flice inhibitor proteins (vFLIPs [50]), and a D-type cyclin (Orf 72) (9, 12). Of these genes, however, only the cyclin D homolog and vFLIP are known to be transcribed during viral latency (16, 25, 44). Given the prominence of latent infection in KS tumors and the known role of latent gene products in tumorigenesis by other herpesviruses (30), we (24, 40, 48, 51) and others (15, 25, 39, 42) have sought to define additional viral genes expressed during KSHV latency.
We have previously identified in a pulmonary KS tumor a highly abundant transcript, T0.7, which spanned the K12 open reading frame (ORF K12), the predicted translation product of which is an extremely hydrophobic 60-amino-acid (aa) protein, kaposin (41, 51). RNAs bearing ORF K12 sequences are consistently detectable by in situ hybridization in the majority of spindle cells and in all stages of KS progression, as well as in all PEL cell lines analyzed (48, 49). In both situations, the RNAs are expressed at variable levels in latently infected cells and at much higher levels in cells undergoing viral reactivation (48). Recently, it has been reported that expression of the K12 coding region in the Rat-3 cell line is capable of inducing focus formation in vitro (33).
In this study, we have further examined the transcription and translation of the K12 region. These experiments establish that in PELs and in most KS tumors the predominant mRNA containing K12 is much larger than the T0.7 transcript previously described. Detailed transcript mapping reveals that the 5′ end of the larger mRNA is positioned well upstream of the K12 AUG codon, immediately 5′ of two large sets of GC-rich direct repeats (DRs) (DR1 and DR2). Surprisingly, although these upstream sequences are without AUG codons, they are nonetheless translated in vivo, as a result of variant initiation at non-AUG codons, into a family of novel proteins. In the PEL cell line BCBL-1, the predominant translation product, named kaposin B, does not contain ORF K12 sequences. However, K12 sequences are translated in this cell line by low-level initiation at an alternate start codon, resulting in expression of ORF K12 sequences as a DR-K12 fusion protein.
MATERIALS AND METHODS
Cell lines and transfection.
The following cell lines were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum and penicillin-streptomycin: BCBL-1 (40), BC1 (8), BC3 (1), Raji Burkitt’s lymphoma, BJAB Burkitt’s lymphoma, and SLK KS (46) (a gift of S. Leventon-Kriss). Rodent Rat-3 (a gift of L. Rosenthal) and NIH 3T3 cells and monkey CV-1 fibroblasts were maintained in Dulbecco modified Eagle medium H-21 supplemented as described above. For transfections, cell lines were plated at 5 × 104 cells per 35-mm-diameter dish overnight prior to addition of Superfect transfection reagent (Qiagen) with up to 2 μg of DNA used according to the manufacturer’s instructions.
cDNA libraries and plasmid expression constructs.
KS tissue and BCBL-1 oligo(dT)-primed cDNA libraries from poly(A)+ RNA were prepared with the ZAP cDNA synthesis kit (Stratagene) according to the manufacturer’s instructions and screened with randomly primed labeled KSHV genomic fragments. Subgenomic plasmid constructs were prepared from BCBL-1 cells and a pulmonary KS sample as described elsewhere (51). XhoII-NheI subfragments of the DR-K12 region were subcloned into pcDNA3.0 (Invitrogen), creating pBCBL-1 XhoII-NheI. Xpress epitope tags were added at the NsiI restriction site immediately 5′ of the K12 ORF by PCR with the pcDNA3.1 (Invitrogen) anti-Xpress epitope region as template and three different 5′ PCR primers (differing by 1 nucleotide [nt] to generate tags in all three reading frames) with a common 3′ primer, creating pBCBL-1 XhoII-NsiI-XprFr1, -XprFr2, and -XprFr3. The CUG codon at position 118679 (all numbered KSHV genomic positions are according to the BC1 sequence [41]) of pBCBL-1-XhoII-NheI was mutated to UUG by PCR with pBCBL-1-XhoII-NheI as template and the primers 5′-GGGGTACT GCAGGGTTCGCAGGGTTCGGGGGTACTACCTGGTTTCCTGGGGTGT GCCAAGACGGGTTCCT and 5′-GATCCAAGCTTGGGATCTCTTGGATGGACACGTATCG. The PCR product was ligated into the HindIII site in the polylinker and the PstI site adjacent to DR2 within pBCBL-1-XhoII-NheI. Plasmids were purified with Qiagen plasmid purification kits (Qiagen) and verified by DNA sequencing.
Northern and Southern blot analysis.
RNA was harvested from tetradecanoyl phorbol acetate (TPA)-induced (20 ng/ml, 24 h) or uninduced cell lines with RNAzol B RNA isolation solvent (Tel-Test). To prepare RNA from frozen KS tissue, samples were pulverized in liquid N2 with a Microdismembrator U (Braun) pulverizer for 1 min and solubilized in RNAzol B. RNA gel electrophoresis was performed with approximately 10 μg of total RNA per sample by standard methods, blotted onto Hybond-N nylon membranes (Amersham Life Sciences), and hybridized as previously described (27), at 65°C with single-stranded [32P]UTP-labeled riboprobes at 106 cpm/ml. For Southern blots, 10 μg of genomic DNA, isolated by standard methods as described elsewhere (21), was digested with PstI prior to blotting and hybridization at 65°C with 106 cpm of a random-hexamer primed [32P]dCTP-labeled probe per ml made with the Redi-Prime II random prime labeling kit (Amersham).
RNase protection and primer extension assays.
RNase protection assays were performed with total cellular RNA hybridized to gel-purified single-stranded 32P-labeled riboprobes at 55°C. Hybrids were digested with RNase T2 (Gibco BRL) prior to phenol chloroform extraction and ethanol precipitation and resolved by 6% polyacrylamide gel electrophoresis (2). Primer extension assays were performed with Superscript II reverse transcriptase (Gibco BRL) as described previously (51) with 5 μg of total BCBL-1 RNA and a 32P-end-labeled primer, 5′-TTCGCAGGGTTCGGGGGTACTACCTG, with a 5′ end at position 118635.
Western blot analysis.
Western blotting was performed with total cell extracts solubilized in 2× sodium dodecyl sulfate (SDS) sample buffer (4% SDS, 3% dithiothreitol) followed by SDS-polyacrylamide gel electrophoresis on 12.5 or 10% polyacrylamide or Tricine separation gels (2). Proteins were transferred to polyvinylidene difluoride Immobilon-P transfer membranes (Millipore) which were processed according to the manufacturer’s instructions. Incubations were performed with primary and secondary antibodies diluted as follows: monoclonal antibody (MAb) 5B2, 1:5,000; Anti-Xpress MAb (Invitrogen), 1:5,000; horseradish peroxidase-conjugated anti-mouse immunoglobulin (Babco), 1:5,000. Horseradish peroxidase-conjugated antibodies were visualized with ECL Western blotting detection reagents (Amersham Life Sciences) and Biomax MS scientific imaging film (Kodak).
Immunohistochemistry.
Standard three-step diaminobenzidine colorimetric immunoperoxidase staining was performed on acetone-fixed tissue sections as described previously (23) with MAb 5B2 at a dilution of 1:100.
RESULTS
The K12 region is transcribed in BCBL-1 cells as a 2.3-kb mRNA.
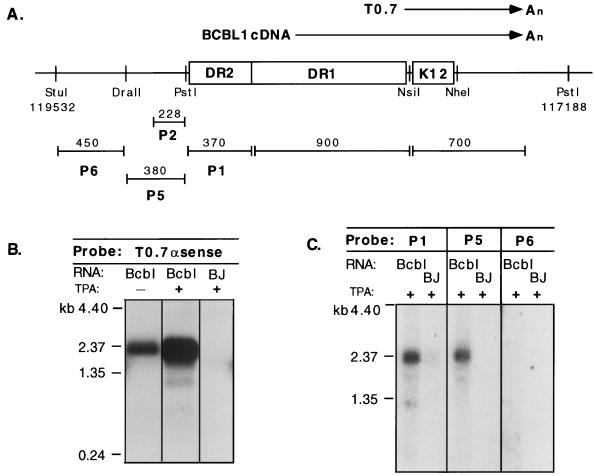
Although our previous study identified a 700-nt mRNA (T0.7) spanning the K12 ORF in a KS tumor (51), examination of RNA from the PEL cell line BCBL-1 by Northern blotting with a K12 region probe did not detect a transcript of this size. Rather, an mRNA of 2.3 kb was identified in uninduced cells, and this transcript increased in abundance upon induction of viral replication with the phorbol ester TPA (Fig. 1B). To define the structure of this transcript, we isolated cDNA clones from an oligo(dT)-primed BCBL-1 cDNA library with a K12 region-specific probe. Sequence analysis of these cDNAs indicated that the 2.3-kb transcript is 3′ coterminal with the previously identified T0.7 mRNA and extends without splicing into a region that contains two sets of 23-bp, GC-rich DRs (DR1 and DR2) located immediately 5′ to the K12 ORF (Fig. 1A). The size of the cDNA clones (1.0 to 1.6 kb) indicated that none of them was full length; all terminated within DR1, presumably due to the inability of reverse transcriptase to extend through these long GC-rich elements. For initial mapping of the 2.3-kb RNA, we carried out Northern blotting with probes from the adjacent genomic regions. A probe (P1) within DR2 annealed strongly to the transcript, as did an additional probe (P5) adjacent to DR2, while a probe further 5′ (P6) was negative (Fig. 1C). These data, plus the size of the RNA, suggest that if the transcript were unspliced it would initiate immediately 5′ of DR2—an inference that was validated by 5′-end mapping of the transcript (see below).
FIG. 1.
The K12 region is transcribed in BCBL-1 cells as a 2.3-kb mRNA. (A) Diagram of the ORF K12 region of BCBL-1 cells, including DRs DR1 and DR2, shown with relevant restriction sites and genomic coordinates derived from the sequenced BC1 isolate (41). The locations of the T0.7 mRNA (51) and BCBL-1 cDNA sequences are shown above the line; the locations of probes used for RNA mapping by Northern blotting are shown below. Note that the transcripts are depicted in a rightward orientation but are leftward relative to the numbered genome of the sequenced BC1 isolate. (B) Northern blotting of uninduced (−) and TPA-induced (+) BCBL-1 (Bcbl) and BJAB (BJ; KSHV negative) RNA with a T0.7 antisense riboprobe which detects the 2.3-kb mRNA species. (C) RNA mapping of the 2.3-kb mRNA in BCBL-1 cells with probes P1, P5, and P6. Abbreviations are the same as those for panel B.
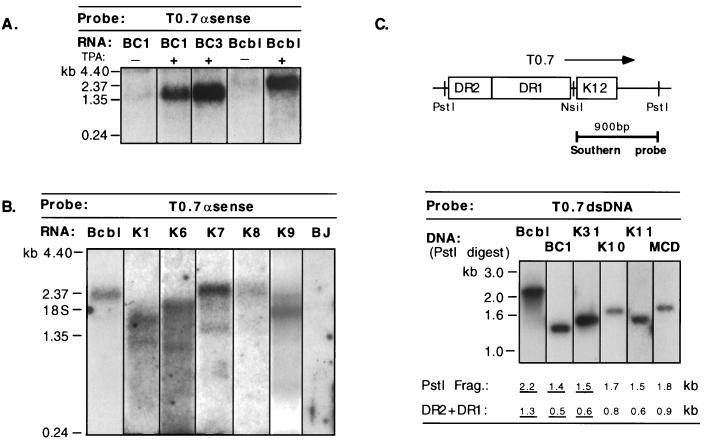
We examined additional PEL cell lines and KS tumors to determine whether the DR-K12 transcripts seen in BCBL-1 cells were representative of those produced in the majority of KSHV-infected cells. RNA from these samples was analyzed by Northern blotting with an antisense riboprobe encompassing the K12 ORF. In the PEL cell lines BC1 and BC3, TPA-induced transcripts of ca. 1.5 and 1.6 kb, respectively, were detected (Fig. 2A), while in five KS tumor samples K12 mRNAs ranging in size from 1.7 to 2.5 kb were observed (Fig. 2B). The T0.7 species was not detected by this assay in any of these samples.
FIG. 2.
The DR-K12 mRNA is transcribed in PEL cell lines and KS tumors. (A and B) Northern blot of uninduced (−) and TPA-induced (+) PEL cell line RNA from BC1, BC3, and BCBL-1 (Bcbl) (A) and KS tumor RNA (K1, K6, K7, K8, and K9) (B) with a T0.7 antisense riboprobe. BJ, BJAB (KSHV negative). (C) Southern blot with a double-stranded DNA (dsDNA) probe from the K12 region of PstI-digested genomic DNA isolated from KSHV-infected cell lines and tumors. The size of the PstI fragment encompassing the DRs and the corresponding size of the DR region itself (DR2 + DR1) are indicated. Isolates that have been sequenced through this region are underlined. K3, K10, and K11 are KS tumors. MCD, multicentric Castleman’s disease. Bcbl, BCBL-1 cells.
The variability in the size of the DR-K12 transcript in these samples can be understood in terms of the genomic organization of KSHV. The length of the GC-rich repeats, DR1 and DR2, is polymorphic among KSHV isolates, as demonstrated by Southern blot analysis of numerous KS and PEL DNA samples (Fig. 2C). DNA sequencing reveals that this size polymorphism is due to variation in the number of repeats present in both DR1 and DR2. In the BCBL-1 isolate, for example, the DR1 region is ca. 900 bp (39 repeat units of 23 bp) and the DR2 region is ca. 400 bp (17.5 repeat units) (50a), resulting in a total repeat length of 1.3 kb. (The corresponding PstI restriction fragment is 2.2 kb [Fig. 2C].) In the BC1 isolate, fewer copies of both repeats are present (41), resulting in DR1 and DR2 regions of ca. 300 and 200 bp, respectively. (The corresponding PstI restriction fragment is 1.4 kb.) These differences in viral genome size correspond to the variation in mRNA size in the PELs and are likely to account for the variation in the size of the mRNAs in the KS samples as well. Interestingly, in the KSHV isolate from the pulmonary KS tumor in which we initially observed T0.7, the DR-K12 mRNA (1.6 kb) was also evident at low abundance (see Fig. 2B of reference 51).
5′-end mapping of the 2.3-kb RNA from BCBL-1 cells.
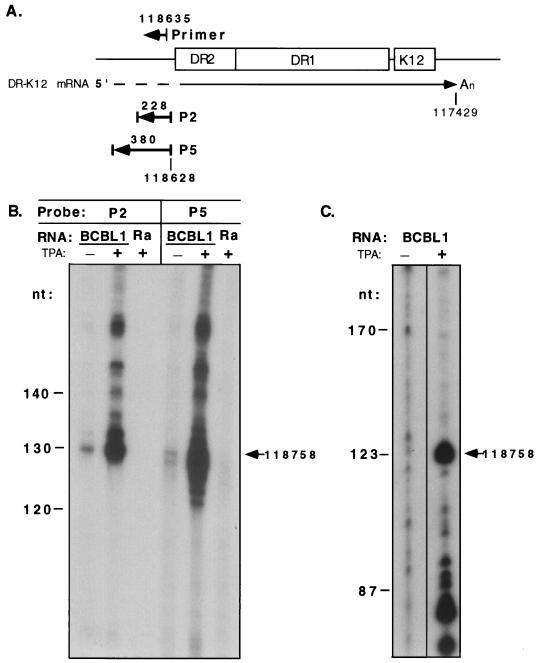
The 5′ end of the 2.3-kb BCBL-1 DR-K12 mRNA was mapped by RNase protection assay with riboprobes P2 and P5, which were shown to hybridize to the 2.3-kb mRNA by Northern blotting (Fig. 1C). These probes, which are 5′ coterminal and 228 and 380 nt in length, respectively (Fig. 3A), were hybridized with BCBL-1 RNA; nonannealed, single-stranded regions were digested with T2 RNase. Both probes demonstrated protection of 130-nt hybridized fragments (Fig. 3B), which corresponds to a discontinuity (5′ end or splice site) in the RNA at position 118758, 158 bp 5′ of the end of DR2 and 34 bp 3′ of a consensus TATA box sequence. The protected signal was increased in TPA-induced BCBL-1 RNA, consistent with the Northern blot data, and was not present with RNA from the KSHV-negative Burkitt’s lymphoma cell line Raji.
FIG. 3.
5′-end mapping of the 2.3-kb DR-K12 mRNA in BCBL-1 cells. (A) Diagram showing locations of probes and primer used for RNase protection and primer extension assays, respectively, with genomic coordinates according to the BC1 isolate. (B) RNase protection assays with uninduced (−) and TPA-induced (+) BCBL-1 and Raji (Ra; KSHV negative) RNA with single-stranded riboprobes P2 and P5. The protected fragment of 130 nt corresponding to a start site at position 118758 is indicated. The larger fragments in the overexposed samples from the TPA-treated cells represent incomplete digestion of the probe. (C) Primer extension assay with uninduced (−) and TPA-induced (+) BCBL-1 RNA and an end-labeled primer with the 5′ end at position 118635. The 123-nt extended product corresponding to a start site at position 118758 is indicated. Fragment sizes were determined by comparison with a DNA sequencing ladder.
To confirm that this discontinuity in the RNA was indeed a start site and not a splice site, we carried out a primer extension assay with an end-labeled, antisense oligonucleotide with a 5′ end at position 118635. Upon annealing to TPA-induced BCBL-1 mRNA, the oligonucleotide was extended in the presence of reverse transcriptase to a length of 123 nt, as determined by comigration with a sequencing ladder (Fig. 3C). This length predicts a start site at position 118758, in excellent agreement with the RNase protection data. The signal of the extended product was also greatly increased compared to RNA from uninduced cells. This start site would result in an unspliced transcript of approximately 2,130 nt in BCBL-1 cells, which, upon polyadenylation at nt 117429 (as indicated by BCBL-1 cDNAs), corresponds well to the size of the 2.3-kb mRNA estimated by Northern blotting. Owing to the high GC content of the repeats, we were unable to perform RNase mapping across DR1 and DR2 to confirm the absence of splicing within the repeats. However, the excellent agreement between the observed and predicted transcript sizes and the absence of canonical splice site sequences within this region make this exceedingly unlikely.
Coding potential of the 2.3-kb DR-K12 mRNA.
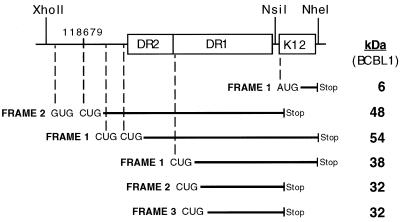
Analysis of the coding potential of the DR-K12 transcript reveals continuous coding regions extending through the entire length of DR1 and DR2 in all three reading frames. The absence of AUG codons within and immediately 5′ of the DRs, however, precluded recognition of the coding potential of this region in previous studies (41). There is considerable evidence, however, that in certain situations eukaryotic ribosomes can initiate at codons other than AUG, most notably, CUG (e.g., in c-myc [20] and basic fibroblast growth factor [38]) and GUG (reference 19 and references therein) as well as ACG (3, 14). In the DR-K12 transcript, a single GUG codon and three CUG codons (in two different reading frames) are present immediately 5′ of the DRs, and numerous CUG codons are present in each reading frame of every repeat unit of DR1. The potential translation products produced by initiations at these codons are depicted schematically in Fig. 4. In addition, the DR-K12 transcript also includes the AUG codon of ORF K12, which could direct translational initiation of a 60-aa (6-kDa) product (originally named kaposin [41]), although without an internal ribosome entry site element within DR1 such initiation would be expected to be very inefficient in a transcript of this structure (22, 26).
FIG. 4.
Coding potential of the DR-K12 mRNA. (A) CUG and GUG potential translation initiation codons within the DR-K12 mRNA are indicated with the reading frame and size of resultant translation products for the BCBL-1 isolate. Note that the K12 ORF is assigned to reading frame 1. Additional CUG codons are present in all three reading frames within each repeat of DR1.
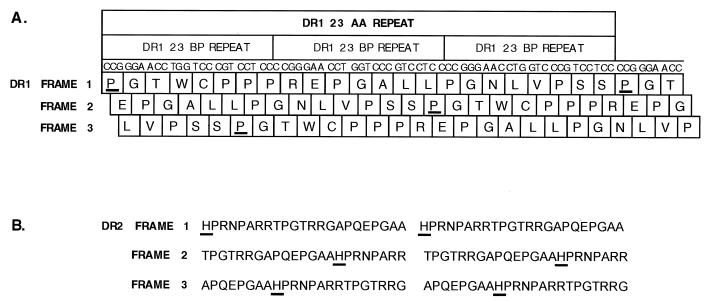
The presence of 23-nt DRs that lack stop codons in any reading frame gives rise to another translational curiosity in this region—namely, that translation of DR1 or DR2 results in a repeating 23-aa peptide of common sequence in all three reading frames (Fig. 5). This is because the number of nucleotides in each repeat (23 nt) is not a multiple of 3; therefore, ribosomes entering a repeat in one reading frame will translate seven codons (21 nt) and utilize the first nucleotide of the next repeat to translate the eighth codon, thereby shifting the reading frame by 1 nt. After translating three repeats, ribosomes are once again in the original frame, and thus, the resulting protein sequence consists of repeats of 23 codons. In BCBL-1 cells, there are ca. 13 reiterations of the DR1 peptide repeat and 6 reiterations of DR2, while in the shorter BC1 isolate, the DR1 and DR2 peptides are repeated ca. four and three times, respectively.
FIG. 5.
Translation of DR1 or DR2 results in a repeating 23-aa peptide in all three reading frames. (A) The single-letter amino acid code of DR1 is shown below the appropriate reading frame of the DNA sequence. The proline residue (P) that is arbitrarily assigned as the start of each repeat unit is underlined in each reading frame. (B) The amino acid code of DR2 is shown in all three reading frames; the histidine residue (H) at the start of the repeat is underlined. The sequences of the 23-bp repeat elements are identical in the BC1 and BCBL-1 isolates.
Translated products from the DR region are expressed in vivo.
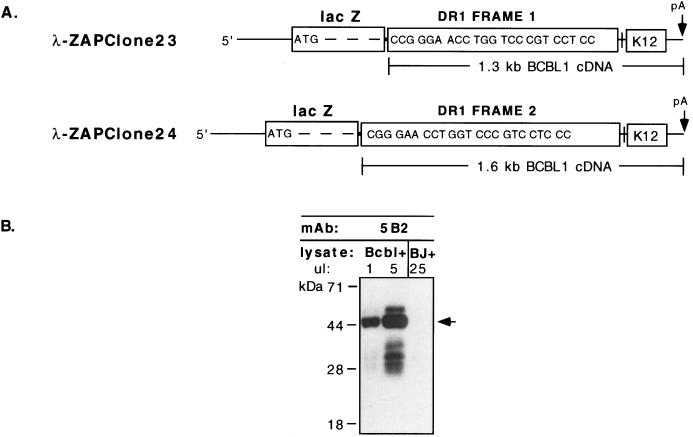
Evidence that the DR region of the 2.3-kb DR-K12 mRNA is in fact translated in infected cells emerged from parallel studies aimed at generating MAbs to KSHV proteins in BCBL-1 lysates. TPA-induced BCBL-1 whole-cell lysate was used to immunize mice in order to generate a library of MAbs. Hybridoma culture supernatants were screened by immunofluorescence analysis on fixed and permeabilized BCBL-1 cell preparations with appropriate controls (KSHV-negative BJAB cells) to exclude recognition of nonviral proteins. Positively staining supernatants were subsequently used to screen a λ-ZAP cDNA expression library, in which oligo(dT)-primed BCBL-1 cDNAs were fused to lacZ sequences that direct their expression in Escherichia coli. Supernatant from MAb 5B2 identified two lambda clones that contained partial BCBL-1 cDNA sequences from the DR-K12 mRNA. Interestingly, in one clone lacZ was fused to reading frame 1 of the DR1 region, while in the other clone lacZ was fused to reading frame 2 of DR1 (Fig. 6A). This indicated that MAb 5B2 recognized an epitope within the repeating 23-aa peptide of DR1 that is common to all three reading frames, as noted above. Of the 46 hybridoma supernatants that were screened, 21 displayed the same specificity as MAb 5B2, indicating that this is an immunodominant epitope in mice.
FIG. 6.
Translation products from the DR region are expressed in vivo. (A) Structure of λ-ZAP BCBL-1 cDNA expression clones 23 and 24 showing fusion of lacZ coding sequence to DR1 sequences in reading frames 1 and 2, respectively. Both clones were identified with MAb 5B2. (B) Western blot with MAb 5B2 of TPA-induced BCBL-1 (Bcbl) and BJAB (BJ) cell lysates. Microliter amounts of loaded lysates are indicated; two loads of BCBL-1 lysate are shown to allow an estimation of relative abundance of translated products. The arrow indicates the predominant 48-kDa protein in BCBL-1 cells.
We performed Western blot analyses with MAb 5B2 to identify its viral protein target in PEL. A strongly predominant band of approximately 48 kDa was detected in TPA-induced and in uninduced (not shown) BCBL-1 cell lysates (Fig. 6B). This band corresponds in size to that of a product initiating at the first CUG codon of the mRNA, in reading frame 2 (Fig. 4). If CUG initiations behave similarly to AUG initiations with regard to ribosomal scanning, this would be predicted to be the predominant site of translational initiation on the message (26). (Further evidence that this CUG codon is in fact the principal initiation codon is presented below.) The next most abundant species observed in the BCBL-1 lysate is a minor band of ca. 32 kDa. This size is consistent with initiations at CUGs within DR1 in reading frame 2 or 3, although the possibility that these bands are derived from the larger species by proteolysis is not excluded. Additional species of 54 and 38 kDa were also reproducibly detected by MAb 5B2, but at relatively low abundance. The sizes of these products are consistent with (but not diagnostic of) translation initiations in reading frame 1, from a CUG codon 5′ of DR2 and from a CUG codon within DR1, respectively. As products of reading frame 1, these 54- and 38-kDa proteins would contain the K12 ORF at their C termini (Fig. 4). Although these reading frame assignments are based on apparent molecular weights and are therefore provisional, they are also supported by genetic evidence, presented below.
Western blotting with MAb 5B2 also detected multiple proteins in TPA-induced BC1 and BC3 PEL cell lysates (data not shown). The apparent levels of 5B2-reactive proteins in these cell lines were lower overall than those in BCBL-1 cells, which can be attributed at least in part to (i) the ca. threefold-lower number of 5B2 epitopes within their shorter DR1 regions and (ii) a lower percentage of lytic induction achieved by TPA treatment of the cells.
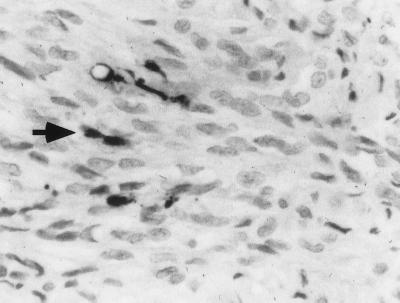
To determine if these MAb 5B2-reactive products could also be detected in KS lesions, we examined KS tissues by immunohistochemistry. These samples displayed strongly positive staining in a small number of cells, some of which had spindle-like morphology (Fig. 7). These cells are likely to represent sites of lytic viral reactivation within the KS lesion. The presence of the 5B2 epitope in KS cells provides further evidence that the DR-K12 transcript of the type described herein is common in KS tumors. Although we have previously shown that the majority of spindle cells express detectable levels of K12 region mRNA (48), we were unable to detect 5B2-immunoreactive protein in the bulk of the tumor. This result is not surprising in view of (i) the relative insensitivity of the immunohistochemical detection method compared to in situ hybridization and (ii) the relative inefficiency of CUG versus AUG initiation (29, 37). (We considered the alternate possibility that the DR-K12 mRNA might require viral lytic cycle products for its translation; however, this model is excluded by the transfection studies presented in the following section.)
FIG. 7.
Colorimetric immunohistochemistry of KS tissue with MAb 5B2. A typical strongly positive-staining cell with MAb 5B2 at a 1:100 dilution is indicated by the arrow. Magnification, ca. ×300.
Analysis of the translational strategy of DR-K12 mRNAs.
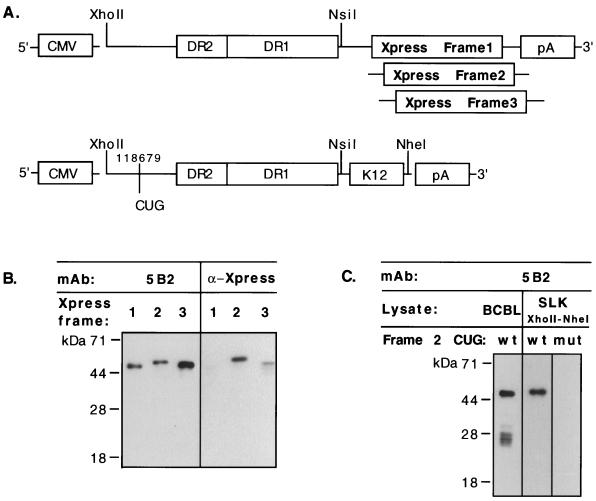
To more explicitly determine which of the many potential reading frames and alternate start codons in the DR-K12 mRNA are actually translated in vivo, we examined protein expression from transfected plasmid constructs in which the products of translation from each frame were individually marked with epitope tags. In these vectors, the cytomegalovirus immediate-early promoter was used to drive expression of an mRNA whose structure mimics that of the 2.3-kb transcript, including the transcription start site, DR2, and DR1 sequences; all CUG and GUG codons were left in their normal context. In place of the K12 ORF, however, an Xpress epitope tag was fused to the 3′ end of DR1 in each of the three reading frames (Fig. 8A, top). These three constructs were separately transfected into several KSHV-negative cell lines, including SLK (a spindle cell line derived from a KS tumor [46]), Rat-3 and NIH 3T3 rodent fibroblasts, and CV-1 monkey fibroblasts. Cell extracts were examined at 48 h posttransfection by immunoblotting with either MAb 5B2 or an anti-Xpress epitope antibody. Identical results were obtained in all cell lines, indicating that no endothelium-specific factors were necessary for translation. Western blotting of transfected SLK cells with MAb 5B2 demonstrated that all three constructs expressed the DR region to equivalent levels (Fig. 8B, left). Blotting with the anti-Xpress antibody, however, detected a translation product primarily from the construct that contained the Xpress epitope in reading frame 2 (Fig. 8B, right). This result indicates that the predominant protein expressed from the DR-K12 mRNA is a frame 2 translation product, as predicted from the size of the dominant protein observed by Western blotting of BCBL-1 cells (Fig. 4 and 6B). Lower-abundance products generated by the frame 3 construct and, upon long exposure (data not shown), by the frame 1 construct were also evident, faithfully reflecting the complexity of translation that is observed in BCBL-1 cells in vivo. Because the frame 3 product migrates more slowly than expected given the available CUG codons (Fig. 4), this protein either may use a different initiator or migrates aberrantly.
FIG. 8.
The predominantly expressed protein in BCBL-1 cells is the 48-kDa product of reading frame 2. (A) (Top) Diagram indicating the structure of expression constructs (pBCBL-1 XhoII-NsiI-XprFr1, -XprFr2, and -XprFr3) which contain the Xpress epitope (Invitrogen) in different reading frames at the 3′ end. (B) (Left) Equivalent expression of the three constructs in transfected SLK cells was verified by Western blotting with MAb 5B2, the epitope for which is present in all three reading frames (see text). (Right) Western blotting of equivalent lysate loads was also performed with the anti-Xpress epitope MAb, demonstrating that translation of the Xpress epitope occurred predominantly in frame 2. Note that the frame 3 construct is overloaded to allow detection of the epitope tag. The tag is also detectable in frame 1 upon longer exposure (data not shown). (C) Western blotting of SLK cells transfected with pBCBL-1 XhoII-NheI (wild type [wt] [A, bottom]) demonstrates authentic translation of the predominant 48-kDa product as detected in BCBL-1 lysate (BCBL). Mutation of CUG (to UUG) in reading frame 2 at position 118679 in an equivalent transfected construct (mut) eliminates detection of the protein.
Examination of the sequence shows two potential alternate (non-AUG) start codons in reading frame 2 that could generate the observed, predominant translation product: the 5′-most CUG codon in the mRNA (at position 118679) and a nearby GUG codon (Fig. 4). To test which codon might be used in vivo, we transfected SLK cells with untagged constructs containing the entire DR-K12 region under the control of the cytomegalovirus immediate-early promoter; in these constructs, the CUG codon at position 118679 was either wild type (CUG) or mutant (UUG). The wild-type construct demonstrated predominant expression of a 48-kDa protein that comigrated with the authentic protein from BCBL-1 lysate (Fig. 8C). Rat-3 mouse fibroblasts and all other cell lines tested also demonstrated the same capacity to translate the 48-kDa protein from this construct (data not shown). Transfection of the construct containing the mutated CUG codon, however, resulted in virtual ablation of translation from this region, although very low abundance products were visible upon long exposure (Fig. 8C and data not shown). These results indicate that this CUG codon is the principal initiator used in vivo and gives rise to the 48-kDa protein observed in BCBL-1 cells.
DISCUSSION
These results demonstrate that the expression of the kaposin-K12 locus of KSHV displays a remarkable and previously unanticipated complexity. The principal transcript from this region spans a series of 23-nt GC-rich repeats whose number is highly variable among KSHV isolates. These repeats, previously assumed to be noncoding due to an absence of AUG codons, in fact direct the synthesis of multiple polypeptides by initiation at alternate start codons in several reading frames. In BCBL-1 cells, the predominant product of the locus appears to initiate at the most 5′ CUG codon, resulting in a product of reading frame 2 derived primarily from translation of DR sequences. We propose the term kaposin B for this polypeptide. In addition, the data indicate that initiations also occur at other non-AUG codons, including the second CUG codon in the message that generates a DR-K12 fusion protein from reading frame 1—a protein for which we propose the name kaposin C. By virtue of the extremely hydrophobic C terminus provided by the K12 ORF, this protein, unlike kaposin B, is predicted to be membrane associated.
Our current estimate of the translational complexity of the locus is a minimal one, and more products may well be produced, especially given the multiplicity of CUGs within DR1. Given the sequence variation and variation in repeat lengths among KSHV isolates, other isolates may express certain reading frames at levels or ratios differing from that which is observed in BCBL-1 cells. Even with invariant use of the different CUG codons, the variability in repeat number among KSHV isolates will result in kaposin B and kaposin C species of strikingly different sizes, which potentially could produce differences in activity. For example, if the repeats encode motifs involved in protein-protein interactions, then differences in repeat number could create large differences in affinity for their protein target(s). It is also possible that differences in repeat length might affect the stability of the translated products.
Our findings raise the question of whether the 60-aa product of ORF K12 is ever produced as such (as a 6-kDa protein). In cells in which the T0.7 mRNA is not present—which appears from the present work to be the majority case—the only way such a protein could be generated would be by internal initiation from the K12 AUG codon. As yet, we have not detected expression of such a polypeptide from RNAs initiating upstream of the DRs. However, we cannot exclude the possibility that, at very low efficiency, scanning ribosomes could reach the K12 AUG codon. It is also formally possible that the DR region might also be able to function as an internal ribosome entry site to allow direct internal access to the K12 AUG codon.
The fact that the AUG codon for ORF K12 has been conserved in all sequenced isolates of KSHV also suggests that circumstances exist under which this ORF may be separately translated (34, 41, 50a). Of course, the most obvious way for such an ORF to be expressed would be by translation of an RNA like T0.7, which lacks DR sequences. Clearly, such an RNA was present in one KS tumor previously studied (51), although it is noteworthy that even this tumor also had evidence for an RNA initiating from the upstream start site. Given its apparent rarity in other specimens, we think it possible that the 0.7-kb transcript in that tumor might have resulted from aberrant RNA processing from the larger transcript rather than from de novo initiation. Further work will be required to determine if there are certain cell types or physiologic conditions in which K12-specific mRNAs might be preferentially produced. In the meantime, it seems prudent to remain open to the possibility that ORF K12 may be translated as such; accordingly, we propose the name kaposin A for its putative 60-aa product.
Lastly, is there any biological rationale for the seemingly baroque coding arrangement described here? While we lack definitive evidence, certain experimental observations may be pertinent. Recently, we have attempted to explore the phenotype mediated by expression of the DR-K12 locus in transfected cells, by using constructs designed to overexpress kaposins B and C. Although we have had little difficulty with transient expression of these proteins, stable transformants bearing these constructs often display little or no 5B2 immunoreactivity. This raises the possibility that one or more of the kaposins may be powerful regulatory molecules whose expression at high levels is not compatible with cell survival or growth. If so, then translation schemes employing inefficient, noncanonical initiators and/or ribosomal scanning may have evolved to titrate levels of expression down to those consistent with host cell survival.
ACKNOWLEDGMENT
We gratefully acknowledge N. Abbey for expert technical assistance.
REFERENCES
- 1.Arvanitakis L, Mesri E A, Nador R G, Said J W, Asch A S, Knowles D M, Cesarman E. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi’s sarcoma-associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood. 1996;88:2648–2654. [PubMed] [Google Scholar]
- 2.Ausubel F M. Current protocols in molecular biology. Vol. 2. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 3.Becerra S P, Rose J A, Hardy M, Baroudy B M, Anderson C W. Direct mapping of adeno-associated virus capsid proteins B and C: a possible ACG initiation codon. Proc Natl Acad Sci USA. 1985;82:7919–7923. doi: 10.1073/pnas.82.23.7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beral V. Epidemiology of Kaposi’s sarcoma. Cancer Surv. 1991;10:5–22. . (Erratum, 12:226, 1992.) [PubMed] [Google Scholar]
- 5.Boshoff C, Schulz T F, Kennedy M M, Graham A K, Fisher C, Thomas A, McGee J O, Weiss R A, O’Leary J J. Kaposi’s sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat Med. 1995;1:1274–1278. doi: 10.1038/nm1295-1274. [DOI] [PubMed] [Google Scholar]
- 6.Boshoff C, Weiss R A. Aetiology of Kaposi’s sarcoma: current understanding and implications for therapy. Mol Med Today. 1997;3:488–494. doi: 10.1016/S1357-4310(97)01116-7. [DOI] [PubMed] [Google Scholar]
- 7.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 8.Cesarman E, Moore P S, Rao P H, Inghirami G, Knowles D M, Chang Y. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi’s sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood. 1995;86:2708–2714. [PubMed] [Google Scholar]
- 9.Cesarman E, Nador R G, Bai F, Bohenzky R A, Russo J J, Moore P S, Chang Y, Knowles D M. Kaposi’s sarcoma-associated herpesvirus contains G protein-coupled receptor and cyclin D homologs which are expressed in Kaposi’s sarcoma and malignant lymphoma. J Virol. 1996;70:8218–8223. doi: 10.1128/jvi.70.11.8218-8223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 11.Chang Y, Moore P S. Kaposi’s sarcoma (KS)-associated herpesvirus and its role in KS. Infect Agents Dis. 1996;5:215–222. [PubMed] [Google Scholar]
- 12.Chang Y, Moore P S, Talbot S J, Boshoff C H, Zarkowska T, Godden K, Paterson H, Weiss R A, Mittnacht S. Cyclin encoded by KS herpesvirus. Nature. 1996;382:410. doi: 10.1038/382410a0. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 13.Cheng E H, Nicholas J, Bellows D S, Hayward G S, Guo H G, Reitz M S, Hardwick J M. A Bcl-2 homolog encoded by Kaposi sarcoma-associated virus, human herpesvirus 8, inhibits apoptosis but does not heterodimerize with Bax or Bak. Proc Natl Acad Sci USA. 1997;94:690–694. doi: 10.1073/pnas.94.2.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Curran J, Kolakofsky D. Ribosomal initiation from an ACG codon in the Sendai virus P/C mRNA. EMBO J. 1988;7:245–251. doi: 10.1002/j.1460-2075.1988.tb02806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis M A, Sturzl M A, Blasig C, Schreier A, Guo H G, Reitz M, Opalenik S R, Browning P J. Expression of human herpesvirus 8-encoded cyclin D in Kaposi’s sarcoma spindle cells. J Natl Cancer Inst. 1997;89:1868–1874. doi: 10.1093/jnci/89.24.1868. [DOI] [PubMed] [Google Scholar]
- 16.Dittmer D, Lagunoff M, Renne R, Staskus K, Haase A, Ganem D. A cluster of latently expressed genes in Kaposi’s sarcoma-associated herpesvirus. J Virol. 1998;72:8309–8315. doi: 10.1128/jvi.72.10.8309-8315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franceschi S, Geddes M. Epidemiology of classic Kaposi’s sarcoma, with special reference to Mediterranean population. Tumori. 1995;81:308–314. doi: 10.1177/030089169508100502. [DOI] [PubMed] [Google Scholar]
- 18.Ganem D. KSHV and Kaposi’s sarcoma: the end of the beginning? Cell. 1997;91:157–160. doi: 10.1016/s0092-8674(00)80398-0. [DOI] [PubMed] [Google Scholar]
- 19.Gupta K C, Ono E, Ariztia E V, Inaba M. Translation initiation from non-AUG codons in COS1 cells is mRNA species dependent. Biochem Biophys Res Commun. 1994;201:567–573. doi: 10.1006/bbrc.1994.1739. [DOI] [PubMed] [Google Scholar]
- 20.Hann S R, King M W, Bentley D L, Anderson C W, Eisenman R N. A non-AUG translational initiation in c-myc exon 1 generates an N-terminally distinct protein whose synthesis is disrupted in Burkitt’s lymphomas. Cell. 1988;52:185–195. doi: 10.1016/0092-8674(88)90507-7. [DOI] [PubMed] [Google Scholar]
- 21.Hansen L J, Tennant B C, Seeger C, Ganem D. Differential activation of myc gene family members in hepatic carcinogenesis by closely related hepatitis B viruses. Mol Cell Biol. 1993;13:659–667. doi: 10.1128/mcb.13.1.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herman R C. Alternatives for the initiation of translation. Trends Biochem Sci. 1989;14:219–222. doi: 10.1016/0968-0004(89)90030-3. [DOI] [PubMed] [Google Scholar]
- 23.Herndier B G, Werner A, Arnstein P, Abbey N W, Demartis F, Cohen R L, Shuman M A, Levy J A. Characterization of a human Kaposi’s sarcoma cell line that induces angiogenic tumors in animals. AIDS. 1994;8:575–581. doi: 10.1097/00002030-199405000-00002. [DOI] [PubMed] [Google Scholar]
- 24.Kedes D H, Ganem D, Ameli N, Bacchetti P, Greenblatt R. The prevalence of serum antibody to human herpesvirus 8 (Kaposi sarcoma-associated herpesvirus) among HIV-seropositive and high-risk HIV-seronegative women. JAMA. 1997;277:478–481. [PubMed] [Google Scholar]
- 25.Kellam P, Boshoff C, Whitby D, Matthews S, Weiss R A, Talbot S J. Identification of a major latent antigen (LNA-1) in the human herpesvirus-8 genome. J Hum Virol. 1997;1:19–29. [PubMed] [Google Scholar]
- 26.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lagunoff M, Ganem D. The structure and coding organization of the genomic termini of Kaposi’s sarcoma-associated herpesvirus. Virology. 1997;236:147–154. doi: 10.1006/viro.1997.8713. [DOI] [PubMed] [Google Scholar]
- 28.Lee H, Veazey R, Williams K, Li M, Guo J, Neipel F, Fleckenstein B, Lackner A, Desrosiers R C, Jung J U. Deregulation of cell growth by the K1 gene of Kaposi’s sarcoma-associated herpesvirus. Nat Med. 1998;4:435–440. doi: 10.1038/nm0498-435. [DOI] [PubMed] [Google Scholar]
- 29.Mehdi H, Ono E, Gupta K C. Initiation of translation at CUG, GUG, and ACG codons in mammalian cells. Gene. 1990;91:173–178. doi: 10.1016/0378-1119(90)90085-6. [DOI] [PubMed] [Google Scholar]
- 30.Miller G. Epstein-Barr virus biology, pathogenesis and medical aspects. In: Fields B, Knipe D, editors. Fields’ virology. Vol. 2. New York, N.Y: Raven Press; 1990. pp. 1921–1958. [Google Scholar]
- 31.Moore P S, Boshoff C, Weiss R A, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 32.Moore P S, Gao S J, Dominguez G, Cesarman E, Lungu O, Knowles D M, Garber R, Pellett P E, McGeoch D J, Chang Y. Primary characterization of a herpesvirus agent associated with Kaposi’s sarcoma. J Virol. 1996;70:549–558. doi: 10.1128/jvi.70.1.549-558.1996. . (Erratum, 70:9083.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muralidhar S, Pumfery A M, Hassani M, Sadaie M R, Azumi N, Kishishita M, Brady J N, Doniger J, Medveczky P, Rosenthal L J. Identification of kaposin (open reading frame K12) as a human herpesvirus 8 (Kaposi’s sarcoma-associated herpesvirus) transforming gene. J Virol. 1998;72:4980–4988. doi: 10.1128/jvi.72.6.4980-4988.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neipel F, Albrecht J-C, Enser A, Huang Y-Q, Freidman-Kien A E, Fleckenstein B. Primary structure of the Kaposi’s sarcoma-associated human herpesvirus 8. GenBank accession no. U93872. 1997. [Google Scholar]
- 35.Neipel F, Albrecht J C, Fleckenstein B. Cell-homologous genes in the Kaposi’s sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicholas J, Ruvolo V R, Burns W H, Sandford G, Wan X, Ciufo D, Hendrickson S B, Guo H G, Hayward G S, Reitz M S. Kaposi’s sarcoma-associated human herpesvirus-8 encodes homologues of macrophage inflammatory protein-1 and interleukin-6. Nat Med. 1997;3:287–292. doi: 10.1038/nm0397-287. [DOI] [PubMed] [Google Scholar]
- 37.Peabody D S. Translation initiation at non-AUG triplets in mammalian cells. J Biol Chem. 1989;264:5031–5035. [PubMed] [Google Scholar]
- 38.Prats H, Kaghad M, Prats A C, Klagsbrun M, Lelias J M, Liauzun P, Chalon P, Tauber J P, Amalric F, Smith J A, et al. High molecular mass forms of basic fibroblast growth factor are initiated by alternative CUG codons. Proc Natl Acad Sci USA. 1989;86:1836–1840. doi: 10.1073/pnas.86.6.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rainbow L, Platt G M, Simpson G R, Sarid R, Gao S J, Stoiber H, Herrington C S, Moore P S, Schulz T F. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J Virol. 1997;71:5915–5921. doi: 10.1128/jvi.71.8.5915-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 41.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarid R, Flore O, Bohenzky R A, Chang Y, Moore P S. Transcription mapping of the Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1) J Virol. 1998;72:1005–1012. doi: 10.1128/jvi.72.2.1005-1012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarid R, Sato T, Bohenzky R A, Russo J J, Chang Y. Kaposi’s sarcoma-associated herpesvirus encodes a functional bcl-2 homologue. Nat Med. 1997;3:293–298. doi: 10.1038/nm0397-293. [DOI] [PubMed] [Google Scholar]
- 44.Sarid R, Wiezorek J S, Moore P S, Chang Y. Characterization and cell cycle regulation of the major Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J Virol. 1999;73:1438–1446. doi: 10.1128/jvi.73.2.1438-1446.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schulz T F. Kaposi’s sarcoma-associated herpesvirus (human herpesvirus-8) J Gen Virol. 1998;79:1573–1591. doi: 10.1099/0022-1317-79-7-1573. [DOI] [PubMed] [Google Scholar]
- 46.Siegal B, Levinton-Kriss S, Schiffer A, Sayar J, Engelberg I, Vonsover A, Ramon Y, Rubinstein E. Kaposi’s sarcoma in immunosuppression. Possibly the result of a dual viral infection. Cancer. 1990;65:492–498. doi: 10.1002/1097-0142(19900201)65:3<492::aid-cncr2820650320>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 47.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d’Agay M F, Clauvel J P, Raphael M, Degos L, et al. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 48.Staskus K A, Zhong W, Gebhard K, Herndier B, Wang H, Renne R, Beneke J, Pudney J, Anderson D J, Ganem D, Haase A T. Kaposi’s sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J Virol. 1997;71:715–719. doi: 10.1128/jvi.71.1.715-719.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sturzl M, Blasig C, Schreier A, Neipel F, Hohenadl C, Cornali E, Ascherl G, Esser S, Brockmeyer N H, Ekman M, Kaaya E E, Tschachler E, Biberfeld P. Expression of HHV-8 latency-associated T0.7 RNA in spindle cells and endothelial cells of AIDS-associated, classical and African Kaposi’s sarcoma. Int J Cancer. 1997;72:68–71. doi: 10.1002/(sici)1097-0215(19970703)72:1<68::aid-ijc10>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 50.Thome M, Schneider P, Hofmann K, Fickenscher H, Meinl E, Neipel F, Mattmann C, Burns K, Bodmer J L, Schroter M, Scaffidi C, Krammer P H, Peter M E, Tschopp J. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature. 1997;386:517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- 50a.Zhong, W., and D. Ganem. Unpublished data.
- 51.Zhong W, Wang H, Herndier B, Ganem D. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc Natl Acad Sci USA. 1996;93:6641–6646. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]