Abstract
To survive and thrive in a dynamic environment, plants must continuously monitor their surroundings and adjust their development and physiology accordingly. Changes in gene expression underlie these developmental and physiological adjustments, and are traditionally attributed to widespread transcriptional reprogramming. Growing evidence, however, suggests that post-transcriptional mechanisms also play a vital role in tailoring gene expression to a plant’s environment. Untranslated regions (UTRs) act as regulatory hubs for post-transcriptional control, harbouring cis-elements that affect an mRNA’s processing, localization, translation, and stability, and thereby tune the abundance of the encoded protein. Here, we review recent advances made in understanding the critical function UTRs exert in the post-transcriptional control of gene expression in the context of a plant’s abiotic environment. We summarize the molecular mechanisms at play, present examples of UTR-controlled signalling cascades, and discuss the potential that resides within UTRs to render plants more resilient to a changing climate.
Keywords: Abiotic stress, alternative splicing, gene expression, post-transcriptional regulation, RNA-binding protein (RBP), RNA processing, RNA structure, translation, untranslated region (UTR)
Untranslated regions (UTRs) of transcripts act as molecular hubs for the post-transcriptional control of gene expression and thereby help to tailor a plant’s development and physiology to its surroundings.
Introduction
Plants are highly sensitive to the environment in which they grow. Sunlight, temperature, water availability, soil composition, and a plethora of other environmental factors affect plant growth, development, and fitness. Plants have evolved intricate mechanisms to sense these environmental cues and trigger appropriate responses at the developmental and physiological level to maximize chances for survival and reproduction. These responses are driven by major changes in gene expression. Exposure to light, heat, cold, or drought triggers vast transcriptional reprogramming in plants, which relies on shifts in the abundance and activity of transcription factors, epigenetic marks, and chromatin structure (Baulcombe and Dean, 2014; Kim et al., 2015; Lämke and Bäurle, 2017; Strader et al., 2022). Yet, effects of these environmental signals reach far beyond transcription: post-transcriptional processes such as mRNA processing, transport, localization, translation, and turnover (Box 1; Fig. 1) are also tightly regulated in accordance with the plant’s surroundings.
Box 1. Post-transcriptional control of gene function in plants.
Gene expression is tightly controlled at the transcriptional and post-transcriptional level, with the term ‘post-transcriptional’ usually referring to regulatory processes occurring at the level of mRNA (Fig. 1). They are controlled by a plethora of RNA-binding proteins (RBPs) that associate with the nascent RNA as soon as it emerges from RNA polymerase II; in fact, some RNA processing steps occur while the transcript is still associated with RNA polymerase II and can thus be considered co-transcriptional (Marquardt et al., 2023).
5' capping: the 7-methylguanosine (m7G) cap structure, linked to the first RNA nucleoside through a 5'–5' triphosphate bridge, is formed through the subsequent activity of RNA 5' triphosphatase, RNA guanylyltransferase, and RNA (guanine-N7) methyltransferase; it protects the RNA from degradation by 5' exonucleases and facilitates translation initiation (Shuman, 2002).
3' polyadenylation: cis-acting elements (the far and near upstream elements and the cleavage element) in the 3' UTR define the site at which the cleavage and polyadenylation complex cleaves the mRNA and adds a stretch of adenosine nucleotides. This poly(A) tail is critical for nuclear export, translation, and stability of the mRNA (Bernardes and Menossi, 2020; Yang et al., 2021).
Splicing: the removal of introns and ligation of exons is essential to produce a translation-competent mRNA. It is catalysed by a macromolecular ribonucleoprotein complex, the spliceosome, which is assembled anew for each splicing event upon recognition of core cis-acting elements including the 3' splice site with a conserved GU and the 5' splice site with a conserved AG nucleotide pair (Lorković et al., 2000; Reddy et al., 2013).
RNA modification and editing: the term RNA modification denotes chemical modifications of single nucleotides, collectively referred to as the epitranscriptome. Known modifications in Arabidopsis include m7G, N6-methyladenosine (m6A), N1-methyladenosine (m1A), 5-methylcytosine (m5C), 5-hydroxymethylcytosine (hm5C), and uridylation. These modifications can affect most aspects of mRNA metabolism including splicing, nuclear export, translation, and turnover (Shen et al., 2019). The term RNA editing refers to enzymatic changes of the primary nucleotide sequence; in flowering plants, it appears to be restricted to U-to-C editing in mitochondria and plastids (Takenaka et al., 2013).
Nuclear export: mature mRNAs are exported from the nucleus as a tightly packaged mRNA–ribonucleoprotein complex. Translocation through nuclear pore complexes, mediated by transport receptor proteins, represents a major route of nuclear export, but mechanisms involving vesicular transport also exist (Natalizio and Wente, 2013).
RNA localization: once exported from the nucleus, mRNA localization is determined by cis-acting ‘zipcode’ elements; many mRNAs are targeted to the endoplasmatic reticulum for translation, but also to mitochondria, plastids, or plasmodesmata (Tian et al., 2020). Additionally, RBPs can sequester mRNAs into biomolecular condensates such as processing bodies and stress granules, where translationally inactive mRNAs may be stored for rerelease or subjected to degradation (Kearly et al., 2022).
RNA turnover: degradation of mRNAs is required for both mRNA quantity and quality control. Several pathways contribute to mRNA decay: (i) bulk degradation via exoribonucleases (XRNs) or the exosome complex after deadenylation; (ii) co-translational decay by XRN4; (iii) mRNA quality control, which triggers RNA decay upon recognizing transcripts with premature stop codons (nonsense-mediated decay, NMD), without stop codons (no-stop decay), or with stalled ribosomes (no-go decay); and (iv) miRNA-mediated cleavage (Zhang and Guo, 2017).
Translation: the process of translation is divided into steps of initiation, elongation, and termination, with the main regulatory processes thought to occur during initiation. Eukaryotic initiation factor 4 (eIF4) recognizes the 5' cap structure and, aided by poly(A)-binding proteins, circularizes the mRNA molecule. Subsequently, the pre-initiation complex (PIC), consisting of the small ribosomal subunit, initiation factors eIF1, eIF1A, eIF3, and eIF5, and a ternary complex of eIF2, GTP, and the initiator Met-tRNA, is recruited to the mRNA and starts scanning along the 5' UTR to identify the correct translation start site (Dutt et al., 2015). Once the start codon is identified, initiation factors are released and the large ribosomal subunit is recruited to form a translationally active ribosome, which catalyses the formation of the first peptide bond upon accepting an elongator tRNA and thereby enters the elongation phase. Ribosome-catalysed polypeptide formation proceeds as the mRNA is translocated codon by codon until a stop codon is reached, upon which the polypeptide chain is detached from the ribosome. This is followed by ribosome release, and ribosomal subunits can subsequently be recycled for multiple rounds of translation (Browning and Bailey-Serres, 2015; Merchante et al., 2017).
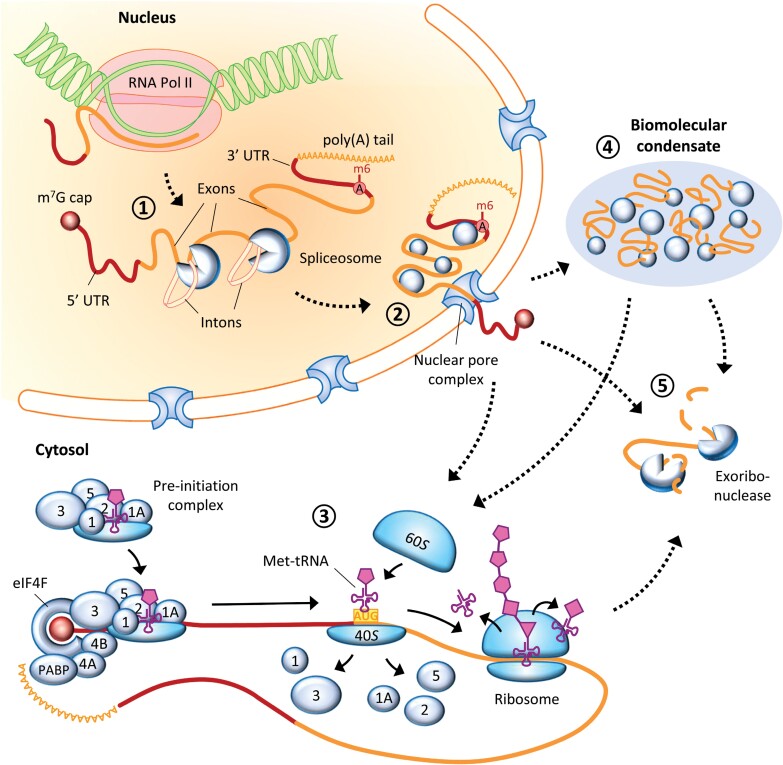
Fig. 1.
Post-transcriptional mechanisms controlling gene expression in plants. After transcription by RNA polymerase II (RNA Pol II), a pre-mRNA undergoes processing, which includes attachment of the m7G cap structure at the 5' end, cleavage and polyadenylation at the 3' end, as well as splicing and nucleotide modifications such as m6A (1). The resulting mature mRNA is then tightly packed into a ribonucleoprotein complex and exported into the cytoplasm through nuclear pore complexes (2). Some cytoplasmic mRNAs will immediately undergo translation (3), which is tightly regulated at the level of initiation (and includes recruitment of the pre-initiation complex, scanning of the 5' UTR, and assembly of the full ribosome at the AUG initiation codon). RNAs can also be sequestered into biomolecular condensates, from which they are later re-released (4). Finally, mRNAs are subjected to degradation (5), which can be a result of mRNA quality control or specific RNA modifications; it can also occur co-translationally or within biomolecular condensates. Solid arrows indicate movement of molecular components during translation; dotted arrows indicate the mRNA’s progression through consecutive stages of gene expression. Blue spheres represent proteins or (ribonucleo)protein complexes; numbers within the spheres indicate specific eukaryotic initiation factors (eIFs).
Post-transcriptional control of mRNAs is mediated by trans-acting factors such as proteins and non-coding RNAs, but also relies on cis-acting elements in the mRNA, many of which reside in its untranslated regions (UTRs). UTRs, defined as the transcribed sequences 5' and 3' of an mRNA’s main coding region, serve vital functions in controlling gene expression at multiple levels. In eukaryotes, pre-initiation complexes (PICs) form at an mRNA’s 5' end and scan along the 5' UTR to identify translation initiation sites; the 5' UTR thus governs the initiation of protein synthesis. In yeast, for instance, 5' UTR polymorphisms and isoforms can result in up to 1000-fold differences in translation activity (Niederer et al., 2022), although such dramatic changes have so far not been observed in other eukaryotic systems (Zhong et al., 2023). 3' UTRs can also affect translation rates, and are main determinants of mRNA localization and stability (Barrett et al., 2012; Mayr, 2017). Together, these regions act as a regulatory interface, influencing the fate of mRNA molecules and ultimately shaping the abundance of the encoded proteins (Fig. 2A).
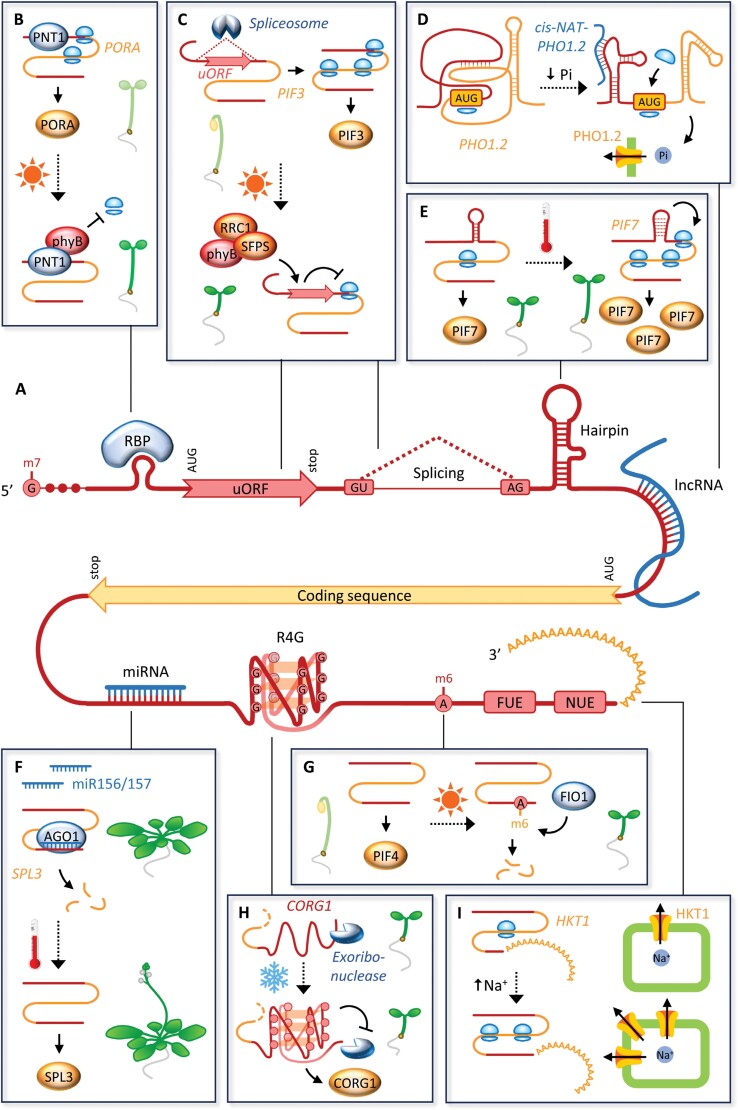
Fig. 2.
Regulatory features of 5' and 3' UTRs and their role in environmental responses. (A) UTRs act as hubs for post-transcriptional regulation of mRNA function and thereby ultimately determine the abundance of the encoded protein. Regulatory features of UTRs include binding sites for RBPs, miRNAs, and lncRNAs, uORFs, RNA secondary structures such as hairpins and RNA G-quadruplexes (RG4s), as well as sites for splicing, nucleotide modification, and polyadenylation, the latter determined with the help of far (FUE) and near upstream elements (NUE). (B–I) Examples of UTR-mediated regulatory processes that operate in response to environmental signals. (B) PNT1 recruits the active phyB photoreceptor under red light, inhibiting translation of PORA mRNA and thereby regulating plant greening. (C) PhyB, together with the splicing factors RRC1 and SFPS, triggers intron retention in the PIF3 5' UTR; the retained intron contains a uORF that down-regulates PIF3 translation in the light and thereby promotes seedling de-etiolation. (D) Binding of the lncRNA cis-NATPHO1.2 triggers structural rearrangements in the 5' UTR and coding region of the rice PHO1.2 mRNA under phosphate starvation and thereby increases access of the large ribosomal subunit to the initiation codon. Translation of PHO1.2 increases, and elevated levels of PHO1.2 transporter allow for efficient redistribution of inorganic phosphate (Pi). (E) A thermosensitive hairpin in the PIF7 5' UTR adopts a more relaxed conformation at elevated temperatures, which acts as a translational enhancer. Increased levels of PIF7 subsequently promote temperature-induced hypocotyl elongation. (F) MiR156/157 bind to the 3' UTR of SPL3 and trigger the transcript’s degradation via ARGONAUTE 1 (AGO1). MiR156/157 levels are reduced at elevated temperature, allowing SPL3 protein to accumulate and induce flowering. (G) An m6A modification in the PIF4 3' UTR under red light destabilizes the transcript, thereby promoting photomorphogenesis. (H) Formation of an RG4 in the CORG1 3' UTR at low temperature prevents degradation of the transcript and thereby attenuates growth in the cold. (I) Selection of a distal polyadenylation site in the HKT1 3' UTR enhances translation under salt stress; increased production of the HKT1 transporter subsequently increases export of sodium ions and thereby promotes salt tolerance in Spartina alterniflora. Arrows indicate positive regulation; blunt arrows indicate negative regulation; dotted arrows denote environmental effects. Ribosomes are depicted in bright blue.
In this review, we present an overview of UTR features that affect gene expression post transcription in the context of a plant’s abiotic environment; we discuss specific examples of how such features regulate development and stress responses in accordance with the plant’s surroundings; and we highlight the largely untapped potential of UTRs to customize these responses for the breeding of climate-resilient crops.
UTR length and nucleotide composition
Average 5' UTR length of 100–200 nucleotides is roughly constant across diverse taxonomic groups, while 3' UTR length is more variable, ranging from an average of 200 nucleotides in many plants and fungi to 800 nucleotides in humans and other vertebrates (Mignone et al., 2002). In the model plant species Arabidopsis thaliana (Arabidopsis) and Oryza sativa (rice), UTRs constitute 18–19% of the entire transcriptome; considerable differences are seen in UTR length between the two species, with Arabidopsis harbouring generally shorter 5' (155 bp versus 259 bp) and 3' UTRs (242 bp versus 469 bp), even when only orthologous genes are considered (Srivastava et al., 2018). In Arabidopsis, Gene Ontology (GO) terms related to abiotic signalling (including responses to salt and cold) are enriched among genes with short 5' and 3' UTR sequences, suggesting that UTR length can provide some level of functional specificity. A similar relationship, however, has not been observed in rice, suggesting that this is not a universal phenomenon (Srivastava et al., 2018).
High GC content of the 5' UTR, and consequently a highly negative normalized free energy of folding, has also been associated with gene function. In Arabidopsis, transcripts with GC-rich 5' UTRs are poorly translated under environmental stresses such as darkness, hypoxia, and dehydration, a process that may be linked to energy conservation (Branco-Price et al., 2005; Kawaguchi and Bailey-Serres, 2005; Juntawong and Bailey-Serres, 2012). Reduced translation of these transcripts is generally attributed to the 5' UTRs’ higher potential for secondary structure formation, which may in turn interfere with 5' UTR scanning by the PIC (Leppek et al., 2018). However, regulatory effects depend on the structures that are formed, and we will discuss specific examples further below.
UTR-binding proteins
RNA-binding proteins (RBPs) represent a large and highly diverse set of proteins, and constitute crucial regulators of mRNA function. In Arabidopsis, >2700 potential RBPs have been identified, with 836 being detected as bona fide RNA binding (Marondedze, 2020). The RNA-binding proteome is tightly regulated in response to environmental factors; multiple RBPs have been found to coordinate gene expression in response to abiotic stimuli (Muthusamy et al., 2021), and several of these RBPs, which we discuss further below, bind preferentially or exclusively to UTRs.
The 5' UTR acts as a hub for the control of translation initiation through eukaryotic initiation factors (eIFs): the eIF3 complex mediates binding of the PIC to the mRNA; eIF4A, eIF4B, and the cap-binding complex eIF4F assist in binding and have pivotal roles in mRNA unwinding during the scanning process; and eIF1, eIF1A, eIF5, and eIF5B are crucial factors for start site selection (Browning and Bailey-Serres, 2015; Merchante et al., 2017). Given that initiation is considered the rate-limiting step of the translation process (Dutt et al., 2015), many of these factors are likely to underpin the dramatic changes in global translation rates observed upon exposure to stimuli such as light (Juntawong and Bailey-Serres, 2012; M.-J. Liu et al., 2012, 2013), heat (Yángüez et al., 2013; Merret et al., 2015; Lukoszek et al., 2016; Chung et al., 2020), hypoxia, and submergence (Branco-Price et al., 2005; Juntawong and Bailey-Serres, 2012; Lee and Bailey-Serres, 2019). However, given their importance for translation in general, few initiation factors have been assigned specific roles during environmental responses. A missense mutation in eIF5B1 results in impaired heat acclimation (Zhang et al., 2017), while a loss-of-function allele of eIFiso4G1 (which encodes a component of the eIF4F complex) displays increased sensitivity to submergence (Cho et al., 2019). The phosphorylation state of several initiation factors is also environmentally controlled: eIFiso4G1 undergoes hypoxia-induced phosphorylation by Snf1-RELATED PROTEIN KINASE 1 (SnRK1), a process thought to promote translation of hypoxia-induced transcripts (Cho et al., 2019), while light and CO2, presumably through their effect on photosynthetic assimilation, have been implicated in regulating the phosphorylation status of eIF3b, eIF4A, eIF4B, and eIF4G (Boex-Fontvieille et al., 2013). In the case of eIF4B and eIF4G, phosphorylation is probably catalysed by the TARGET OF RAPAMYCIN (TOR) kinase, a pivotal sensor of the cell’s energy status (Van Leene et al., 2019). The role of these phosphorylation events in the context of environmental responses, however, remains unclear.
Not all regulation at the 5' UTR is mediated through translation initiation factors. The cytosolic zinc finger protein PENTA 1 (PNT1) binds to the 5' UTR of the mRNA encoding PROTOCHLOROPHYLLIDE REDUCTASE A (PORA), an enzyme in the chlorophyll biosynthesis pathway that operates primarily in darkness. PNT1 recruits the active form of the red/far-red light phytochrome photoreceptor to the PORA mRNA and thereby prevents its translation after the dark-to-light transition (Fig. 2B) (Paik et al., 2012).
3' UTRs are also targeted by multiple sets of RBPs including those mediating 3' processing and modification, discussed further below; here, we focus on direct regulation of mRNA decay or translation. PUF/PUMILIO (PUM) proteins are a family of highly conserved eukaryotic RBPs that bind to specific 3' UTR motifs containing a UGUA core element and negatively regulate gene expression at the post-transcriptional level, affecting mRNA stability or translation (Joshna et al., 2020). Abiotic stresses such as salinity and dehydration induce ARABIDOPSIS PUM 5 (APUM5) expression, and APUM5 itself reduces abiotic stress resistance by down-regulating the expression of stress tolerance genes (Huh and Paek, 2014). APUM9 and APUM23, on the other hand, have been implicated in promoting heat and salt tolerance, respectively (Huang et al., 2018; Nyikó et al., 2019).
Bruno-like proteins constitute another group of RBPs that exert post-transcriptional control via 3' UTRs. Three members have been identified in Arabidopsis: FLOWERING CONTROL LOCUS A (FCA), BRUNO-LIKE 1 (BRN1), and BRN2 (Good et al., 2000). While FCA is involved in polyadenylation of mRNAs (see section below), BRN1 and BRN2 bind to specific response elements in the 3' UTR of the mRNA encoding the floral promoter SUPPRESSOR OF CONSTANS 1 (SOC1). Their binding triggers decay of the SOC1 mRNA and thereby delays flowering (Kim et al., 2013). Expression of BRN1 and BRN2 is induced upon prolonged exposure to cold, and hence contributes to the vernalization-dependent control of flowering in Arabidopsis.
Other 3' UTR-binding RBPs are involved in the formation of stress granules, cytoplasmic biomolecular condensates of RNA and proteins that form when cells experience acute environmental stress. RNAs sequestered in stress granules are translationally repressed; they may later undergo targeted degradation but may also re-enter the translational cycle to ensure coordinated synthesis of selected proteins (Chantarachot and Bailey-Serres, 2018; Yan et al., 2022). OLIGOURIDYLATE BINDING PROTEIN 1 (UBP1) is a core component of stress granules; it is uniformly distributed throughout the cytoplasm and nucleus during control conditions, but rapidly relocates into stress granules upon exposure to heat and hypoxia (Yan et al., 2022). UBP1c specifically appears to act as a molecular switch that interacts with selected target mRNAs depending on the environment: it interacts with U-rich 3' UTRs under non-stress conditions, but under hypoxia it binds preferentially to non-U-rich mRNAs and sequesters them into stress granules. These RNAs are rapidly released upon reoxygenation, allowing for a rapid transition between storage and translation (Sorenson and Bailey-Serres, 2014).
ACETYLATION LOWERS BINDING AFFINITY (ALBA) proteins are another family of RBPs known to phase-separate into stress granules upon heat exposure (Náprstková et al., 2021; Tong et al., 2022). ALBA5 binding in Arabidopsis is enriched in 3' UTRs: it binds almost 3000 transcripts under both control and heat stress conditions, but only two-thirds of these transcripts are shared among the two RNA interactomes. Several mRNAs encoding heat shock factors, and other components of the heat shock response are selectively bound by ALBA5 under heat stress; these mRNAs are destabilized in alba mutants, suggesting that ALBA proteins promote thermotolerance by stabilizing these transcripts (Tong et al., 2022). How this change in ALBA5 binding specificity is controlled remains unknown; no RNA motifs were selectively enriched under stress versus control conditions (Tong et al., 2022).
UTR-binding RNAs
Proteins are not the only molecules acting in trans on UTRs: several non-coding RNAs exert a regulatory function through binding of an mRNA’s 3' or 5' UTR. MiRNAs are small, ~21 nucleotide long, non-coding RNAs that bind target mRNAs to interfere with their translation or to trigger their decay (Fabian et al., 2010). In animals, miRNA target sites are enriched in 3' UTRs, and target sites in the 5' UTR or coding region are generally considered to be less effective (Bartel, 2009). In plants, however, miRNA target sites appear to be equally effective in the 5' UTR, coding region, and 3' UTR when translational repression is considered (Iwakawa and Tomari, 2013).
Cleavage of the PHOSPHATE 2 (PHO2) transcript is induced by binding of miR399 to its 5' UTR (Chiou et al., 2006; Lin et al., 2008). PHO2 encodes an E2 ubiquitin-conjugating enzyme that maintains low levels of Pht1;4 and other phosphate transporters under control conditions (Huang et al., 2013; Park et al., 2014). MiR399 is induced under phosphate starvation, reduces PHO2-dependent degradation of these transporters, and thereby raises intracellular phosphate concentrations (Bari et al., 2006; Chiou et al., 2006). MiR399 expression is also responsive to ambient temperature, and the miR399/PHO2 module was found to regulate flowering time in a temperature-dependent manner (Kim et al., 2011), suggesting that this signalling circuit has been rewired to control both physiological and developmental responses.
MiR156/157 represent another family of miRNAs that control flowering time, mainly through regulation of their direct targets, the mRNAs encoding SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) transcription factors (Wang et al., 2009). In the case of SPL3, SPL4, and SPL5, the miRNA target site is located in the transcript’s 3' UTR, and binding of miR156/157 causes cleavage and translational inhibition of the SPL3 transcript, and probably also those of SPL4 and SPL5 (Fig. 2F) (Wu and Poethig, 2006; Gandikota et al., 2007). While the miR156/157–SPL pathway constitutes foremost an age-dependent pathway (Wu et al., 2009; He et al., 2018), it also integrates environmental factors to regulate flowering time: a warming from 16 °C to 23 °C is sufficient to reduce miR156 and increase SPL3 levels, an effect that accelerates flowering at high temperature (Kim et al., 2012). Similarly, a low red:far-red light ratio, such as observed under a canopy or in dense vegetation, reduces miR156 levels and triggers early flowering (Xie et al., 2017).
While control of translation through miRNAs has been known for decades, recent evidence suggests that long non-coding RNAs (lncRNAs) can also regulate gene expression by interaction with a transcript’s UTR: the rice cis-NATPHO1.2 antisense transcript is induced under phosphate starvation and enhances translation of its target mRNA. This process depends on structural rearrangements in the transcript’s 5' UTR (Reis et al., 2021) and is discussed in detail further below.
Polyadenylation
While the mRNA-binding partners discussed above directly control translation and decay of their target mRNAs, others specifically modify their targets’ UTRs and thereby regulate mRNA function. 3' polyadenylation is a key modification of mRNAs: it is required for proper mRNA localization, nuclear export, stability control, and translation (Box 1). Consequently, alternative polyadenylation (APA) can affect mRNA metabolism and function (Xing and Li, 2011). Environmental stresses such as dehydration (Sun et al., 2017), hypoxia (de Lorenzo et al., 2017), salinity (Chakrabarti et al., 2020; Ma et al., 2022), nitrogen starvation (Conesa et al., 2020), and elevated temperature (Wu et al., 2020; Yan et al., 2021) globally change APA profiles of the plant’s transcriptome. Consistent with these findings, mutations in components of the cleavage and polyadenylation specificity factor (CPSF) complex, a core regulator of APA, result in altered responses to various abiotic stresses (Liu et al., 2014; Téllez-Robledo et al., 2019; Yu et al., 2019). Together, these observations suggest that APA may be a widespread mechanism to tailor gene expression to a plant’s surroundings.
Whereas global APA changes in response to external stimuli have been widely observed, specific examples that establish a causal link between environmentally controlled APA and defined developmental or physiological outputs remain scarce. Arabidopsis transcripts encoding AT3G47610 (a putative transcriptional regulator) and ANKYRIN REPEAT-CONTAINING PROTEIN 2 (AKR2) are alternatively polyadenylated under salt stress, and their APA requires a functional CPSF complex. T-DNA insertions that preclude the use of distal polyadenylation sites in these two genes increase seed germination under salt stress, suggesting that polyadenylation site selection affects salt tolerance (Yu et al., 2019). In the salt-tolerant monocot Spartina alterniflora, exposure to high salinity induces the use of distal polyadenylation sites in several ion transporter transcripts including HIGH-AFFINITY K+TRANSPORTER 1 (SaHKT1), and extension of the 3' UTR of SaHKT1 increases the transcript’s stability and translation (Fig. 2I) (T. Wang et al., 2023). Similarly, several Arabidopsis transcripts that encode core regulators of the heat shock response such as HEAT SHOCK PROTEIN 70 (HSP70) and HSP17.6C display increased 3' UTR length under heat stress, and selection of their distal polyadenylation sites appears to be required for protein synthesis (Wu et al., 2020).
APA also represents a core mechanism in the post-transcriptional control of flowering time through the floral repressor FLOWERING LOCUS C (FLC). FCA and FPA, two RBPs in the autonomous pathway—a flowering time pathway distinct from the well-defined day length-, temperature-, and age-dependent pathways—regulate APA of FLC antisense transcripts collectively termed COOLAIR. FCA and FPA promote proximal polyadenylation of COOLAIR, and these proximally polyadenylated COOLAIR isoforms in turn down-regulate FLC expression (Liu et al., 2007; Swiezewski et al., 2009; Hornyik et al., 2010). While their function was initially thought to be completely independent of the environment, we now know that FCA and FPA signalling intersects with temperature-dependent regulation of flowering time: COOLAIR expression is induced by cold (Swiezewski et al., 2009; Jeon et al., 2023), and a switch from distal to proximal polyadenylation of COOLAIR occurs in response to vernalization (i.e. prolonged cold exposure) (Csorba et al., 2014). Loss of FCA function also disrupts the flowering time response to elevated temperature (Blázquez et al., 2003), but whether this effect relies on RNA processing is as yet unknown.
RNA modifications
RNA undergoes modifications at single nucleotides that alter RNA form and function, and >160 different modifications have been reported (Boccaletto et al., 2018). Several of these modifications are present in plant mRNAs, but specific roles have been assigned to few of them: N7-methylguanosine (m7G) is part of the mRNA’s 5' cap structure and hence a key determinant of mRNA stability and translation (Box 1), while N6-methyladenosine (m6A) and 5-methylcytosine (m5C) are found across the entire mRNA molecule and affect a variety of processes including mRNA transport, processing, translation, and decay (Chmielowska-Bąk et al., 2019; Shen et al., 2019; Liang et al., 2020). In Arabidopsis, most m5C sites have been detected in the coding region (Cui et al., 2017; David et al., 2017), while m6A sites are enriched in 3' UTRs and, arguably, around start and stop codons (Luo et al., 2014; Shen et al., 2016; Anderson et al., 2018; Parker et al., 2020). In plants, m6A interferes with site-specific RNA cleavage and promotes transcript stability (Anderson et al., 2018; Parker et al., 2020). In vitro studies using mammalian translation components revealed that m6A in a transcript’s 5' UTR promotes cap-independent translation through direct recruitment of eIF3 (Meyer et al., 2015), but whether this process occurs in plants is as yet unknown.
Global m6A patterns in plant transcriptomes are highly dynamic, and change in response to light (Artz et al., 2023, Preprint), temperature (Liu et al., 2020; S. Wang et al., 2023), salinity (Anderson et al., 2018; Kramer et al., 2020), and drought (Zhang et al., 2021). In agreement with these findings, mutants in many m6A writers, readers, and erasers—proteins that incorporate, recognize, and remove m6A, respectively—impact the plant’s sensitivity to these environmental signals (Dhingra et al., 2023). FIONA1 (FIO1) is an Arabidopsis m6A methyltransferase that installs m6A in U6 small nuclear RNAs, but also in a small number of polyadenylated RNAs (Wang et al., 2022). A knockout of FIO1 results in increased hypocotyl elongation under red and far-red light and in early flowering (Kim et al., 2008; Wang et al., 2022). These phenotypic changes are underpinned by altered methylation rates in the mRNAs encoding the blue light photoreceptor CRYPTOCHROME 2 (CRY2), the transcription factor PHYTOCHROME INTERACTING FACTOR 4 (PIF4; a core component of phytochrome-dependent light and temperature signalling), the floral promoter CONSTANS (CO), and the floral repressor FLC, and FIO1 was shown to directly methylate these mRNAs. Interestingly, methylation leads to a stabilization of FLC mRNA, but destabilizes the mRNAs of CRY2, PIF4 (Fig. 2G), and CO (Wang et al., 2022), in contrast to the notion that m6A modifications generally promote transcript stability.
In Arabidopsis, the majority of mRNAs are m6A methylated by a writer complex consisting of the mRNA adenosine methyltransferase MTA, its closest homologue MTB, FKBP12 INTERACTING PROTEIN 37KD (FIP37), VIRLIZER (VIR), and HAKAI (Shen et al., 2019). The blue light receptors cry1 and cry2 interact with MTA and MTA, MTB and FIP37, respectively, and ~10% of mRNAs are m6A modified in response to blue light in a cry-dependent manner (X. Wang et al., 2021). While their interaction is light independent, blue light promotes co-condensation of the cry/writer complex into photobodies—biomolecular condensates formed by many components of the light signalling pathway—thereby potentially increasing local concentration and activity of the writer complex (X. Wang et al., 2021).
MTA also contributes to cold acclimation: while m6A modification decreases in the cold, this effect is exacerbated in an MTA RNAi line and alters translation efficiency for approximately a third of cold-regulated transcripts. One of these transcripts encodes ACYL-CoA:DIACYLGLYCEROL ACYLTRANSFERASE 1 (DGAT1), and dgat1 loss-of-function mutants, like the MTA RNAi line, show reduced growth in the cold (S. Wang et al., 2023). Heat, on the other hand, increases overall m6A modification in flowers, but not leaves, of Arabidopsis. This increase negatively impacts variability in gene expression, which has been suggested to underpin reduced fertility rates at elevated temperature (Wang et al., 2022). While changes in m6A modification are clearly relevant for heat and cold sensitivity, it remains unknown how the observed transcriptome-wide changes are brought about.
RNA secondary structures
RNA has the capacity to fold into a variety of secondary and tertiary structures such as hairpins, apical and internal loops, junctions, and pseudoknots (Cruz and Westhof, 2009). Folding depends directly on a structure’s thermodynamic free energy, and only the most energetically favourable conformations will be realized under any given condition. Consequently, RNA structure is highly sensitive to its environment, and changes in temperature and osmolarity directly impart wide structural changes across plant transcriptomes (Su et al., 2018; Tack et al., 2020; Yang et al., 2022). In vivo structure probing revealed that heat shock triggers global transcript unfolding in rice, and unfolding is inversely correlated with transcript abundance (Su et al., 2018). In Arabidopsis, the formation of RNA G-quadruplexes—non-canonical secondary structures formed by G-rich nucleic acid sequences—is globally enhanced in response to cold, and their formation promotes mRNA stability (Yang et al., 2022). Notably, the correlation between structure formation and mRNA abundance found in these studies was strongest in 3' UTRs (Su et al., 2018; Yang et al., 2022), emphasizing that the 3' UTR is a strong determinant of mRNA stability. Yang et al. (2022) were also able to link a G-quadruplex in the transcript encoding the translocase subunit protein COLD RESPONSIVE RNA G-QUADRUPLEX 1 (CORG1) to cold-responsive growth: the presence of the G-quadruplex is essential for CORG1-mediated repression of root growth at 4 °C as well as for the expression of cold-induced genes (Fig. 2H).
RNA structural changes under salt stress appear to be more diverse, with structure probing in Arabidopsis suggesting a slight overall increase in RNA structure formation in the shoot, but a decrease in the root. As with temperature effects, unfolded transcripts exhibit reduced abundance under salt stress, but in this case structures in coding sequences were found to be most relevant for transcript stability, followed by the 5' UTR and the 3' UTR (Tack et al., 2020). Transcript unfolding under salt stress has been linked to increased m6A methylation (Kramer et al., 2020), although this study did not observe a correlation between structure formation and transcript abundance. In any case, a direct link between salinity-induced structural changes and the salt stress response has yet to be established.
Besides their effect on mRNA abundance, structural rearrangements can also affect mRNA translation. This type of regulation is well established in prokaryotes, where dynamic structures such as RNA thermometers and riboswitches have been described to regulate translation initiation (Winkler and Breaker, 2005; Kortmann and Narberhaus, 2012), but similar examples are rare in plants. In the unicellular green alga Chlamydomonas reinhardtii, an RNA thermometer controls translation of the chloroplastic psaA mRNA: this structure forms across, and thereby masks, the transcript’s Shine–Dalgarno sequence (the prokaryotic ribosome-binding site) at standard temperatures, but melts at elevated temperatures of 40 °C, enhancing protein synthesis (Chung et al., 2023). A thermosensitive hairpin has also been identified in the 5' UTR of the Arabidopsis transcript encoding the transcription factor PIF7, but appears to operate differently from prokaryotic RNA thermomoters: this RNA thermoswitch adopts a more relaxed conformation at elevated temperature that enhances PIF7 translation, although the underlying mechanism is not fully understood (Fig. 2E) (Chung et al., 2020). Elevated PIF7 protein levels subsequently trigger cell elongation and a more open growth habit to promote the plant’s cooling capacity (Chung et al., 2020; Fiorucci et al., 2020). Similar hairpin structures enhance the translation of transcription factors WRKY22 and HEAT SHOCK FACTOR 2A (HSFA2) in vitro, but their function in vivo has not been tested (Chung et al., 2020).
RNA structural rearrangements can also be tailored to the plant’s light environment: translation of the plastidic psbA RNA, which encodes the photosystem protein D1, increases under high light due to structural changes in the 5' UTR (Gawroński et al., 2021). It has been proposed that these changes are brought about by an RBP, but experimental evidence for this has yet to be obtained.
Binding of other molecules can trigger structural rearrangements to promote translation. Expression of the non-coding natural antisense transcript (NAT) cis-NATPHO1.2 in rice is induced under phosphate starvation and enhances translation of the OsPHO1.2 sense transcript (Jabnoune et al., 2013). OsPHO1.2 encodes the main root transporter loading phosphate into the xylem for distribution across the plant and is essential for phosphate homeostasis (Secco et al., 2010). In the absence of cis-NATPHO1.2, a GC-rich inhibitory region 350 bp downstream of the initiation codon was found to restrict translation, probably through local interactions with upstream regions in the 5' UTR. Binding of cis-NATPHO1.2 triggers rearrangements that enhance access for the large ribosomal subunit to the initiation codon (Fig. 2D), increasing the formation of 80S ribosomal complexes (Reis et al., 2021). These observations present a first mechanistic insight into the activity of plant antisense transcripts on translation.
Upstream open reading frames
Upstream ORFs (uORFs) are short translated sequences found in a transcript’s 5' UTR (Zhang et al., 2020). Present in eukaryotic organisms ranging from yeast (Lin et al., 2019) to humans (McGillivray et al., 2018), uORFs have been identified in >30% of mRNAs in Arabidopsis, 29% in maize, and 6% in tomato (Zhang et al., 2020). The vast majority of uORF-encoded amino acid sequences are not evolutionarily conserved, suggesting that the peptide sequences of translated uORFs serve little regulatory function (Hayden and Jorgensen, 2007). However, so-called conserved peptide uORFs (CPuORFs), estimated at <1% of all uORFs, are significantly enriched in the UTRs of transcription factor-encoding mRNAs and may thus present important regulators of transcription (Hayden and Jorgensen, 2007).
Acting as cis-regulatory elements, uORFs can alter the translation of main ORFs (mORFs), thereby impacting gene expression. Some uORFs cause ribosomes to stall, while others trigger a complete dissociation of the ribosome once the uORF’s stop codon is reached, both effectively reducing the translation of a transcript’s mORF. Additionally, stalling or early termination may be recognized as a sign of abnormal translation, triggering nonsense-mediated decay (NMD) of the mRNA (von Arnim et al., 2014; Zhang et al., 2020). Though rarer, uORFs can also increase the rate of mORF translation (Lin et al., 2019). In many cases, uORFs do not act as an on/off switch for translation, but represent a fine-tuning mechanism for the control of gene expression (McManus et al., 2014; Moro et al., 2021) and were recently shown to contribute to noise reduction in gene expression (Wu et al., 2022).
Translation of uORF-containing transcripts can be regulated by abiotic stresses: expression of Arabidopsis HEAT SHOCK FACTOR B1 (HSFB1) is suppressed by a uORF under control temperatures, and this suppression is lost under heat stress, boosting HSFB1 protein levels and the heat shock response (Zhu et al., 2012). Additionally, uORFs have been found in 7 of the 21 members of the Arabidopsis HSF family, though their functional relevance is not yet known (Zhu et al., 2012; Guo et al., 2016). CPuORFs have also been implicated in heat and drought responses: when incorporated in luciferase reporter constructs, Arabidopsis CPuORF19 and CPuORF47 strongly increase reporter gene expression under water limitation, while CPuORF46 promotes translation in response to heat (Causier et al., 2022). CPuORF47 is part of an mRNA encoding a putative methyl transferase that promotes drought tolerance when overexpressed; however, whether CPuORF47 or the other two CPuORFs are required for the regulation of drought or heat tolerance in planta remains unknown (Causier et al., 2022).
In addition to the regulation of abiotic stress responses, uORFs impact the uptake and distribution of nutrients. A uORF in NOD26-LIKE INTRINSIC PROTEIN 5;1 (NIP5;1), which codes for a boron transporter, suppresses the transcript’s translation and promotes its degradation at high boron concentrations to minimize toxic effects of boron within the cell (Tanaka et al., 2016). Multiple uORFs also directly or indirectly control the abundance of phosphate transporters and thereby impact phosphate acquisition and distribution in Arabidopsis (Reis et al., 2020), rice (Yang et al., 2020), and soybean (Guo et al., 2022).
Several uORFs influence the expression of developmental regulators in response to light. Genes encoding the photomorphogenesis-promoting transcription factors ELONGATED HYPOCOTYL 5 (HY5) and HY5 HOMOLOGUE (HYH) are transcribed at a higher rate from a downstream transcriptional start site under blue light compared with dark conditions. This leads to the inclusion of uORFs in the respective transcripts, causing an overall reduction of mORF translation (Kurihara et al., 2018). While the importance of HY5 and HYH uORFs has not been investigated at the phenotypic level, CPuORF33 in ARABIDOPSIS THALIANA HOMEOBOX 1 (ATHB1) was shown to control multiple aspects of plant development. ATHB1 is a transcription factor with known roles in light signalling and leaf development (Aoyama et al., 1995), and Arabidopsis plants expressing an ATHB1 transcript with a mutated CPuORF33 displayed a phenotype similar to plants overexpressing ATHB1, including serrated leaves, delayed bolting, and senescence, as well as shorter siliques and reduced seed set (Romani et al., 2016; Ribone et al., 2017). The effect of CPuORF33 is tissue specific: it represses ATHB1 translation selectively in aerial tissues in response to light, and this process involves an as yet unknown signal from the chloroplast (Ribone et al., 2017).
How environmental signals can affect uORF (and thereby mORF) translation remains an open question. Metabolites such as polyamines, sucrose, and ascorbate regulate gene expression via uORFs, and it has been proposed that these molecules are directly sensed by the ribosome and act as cofactors for ribosomal stalling (van der Horst et al., 2020). The rate of reinitiation after uORF translation is also tightly regulated: initiation factor eIF3h is essential for reinitiation (Zhou et al., 2010), and its phosphorylation status is controlled by the TOR pathway (Schepetilnikov et al., 2013), which balances plant growth and stress responses. Stress also promotes phosphorylation of eIF2α, which results in ribosomal read-through of uORFs (Young and Wek, 2016), a mechanism controlling the growth-to-defence transition in Arabidopsis (Pajerowska-Mukhtar et al., 2012). Causal links between any of these phosphorylation events and the perception of abiotic cues, however, have yet to be established.
Alternative splicing
Alternative splicing (AS) due to the selection of alternative splice sites, intron retention, or exon skipping can generate multiple mRNA isoforms from a single gene and thereby contributes to transcriptome and proteome diversity (Reddy et al., 2013). AS in UTRs often has profound consequences for a transcript’s fate within the cell: intron retention in the 5' UTR can insert uORFs and other cis-elements that impact translational efficiency (Jacob and Smith, 2017). In Arabidopsis, intron retention events in the 5' UTR are increased under biotic and abiotic stress and are particularly enriched in genes harbouring uORFs, suggesting this as a common mechanism to fine-tune translation in response to the plant’s environment (Martín et al., 2021). While AS events in the 3' UTR can also affect translation, they have a particularly strong effect on transcript stability: unusually long 3' UTRs (>300–350 bp) and the presence of splice junctions >50 bp downstream of the stop codon act as quality control signals for cytosolic mRNAs, triggering NMD of the respective mRNA (Shaul, 2015).
Changes in the light environment trigger substantial changes in AS: two transcriptome-wide studies detected 700 and 2200 AS events, respectively, when dark-grown Arabidopsis seedlings were exposed to red, blue, or white light (Shikata et al., 2014; Hartmann et al., 2016), while an acute light pulse during the night changed AS for almost 400 transcripts (Mancini et al., 2016). In the moss Physcomitrium patens, exposure of dark-adapted protonemata to red or blue light leads to AS in almost 50% of transcripts in either condition (Wu et al., 2014). In monochromatic light, AS strongly depends on the activity of phytochrome A (phyA) and phyB photoreceptors, while, under white light, control of AS may be more strongly coupled to energy supply (Shikata et al., 2014; Hartmann et al., 2016).
Several light signalling components are regulated by light-dependent AS in their 5' UTRs: under continuous red light, phyB induces retention of an intron in the 5' UTR of the mRNA encoding the Arabidopsis transcription factor PIF3. In turn, uORF sequences within this intron attenuate PIF3 translation and thereby reduce PIF3-mediated repression of light signalling, resulting in decreased hypocotyl elongation (Fig. 2C) (Dong et al., 2020). In the Physcomitrium transcript encoding transcription factor HYH2 (PpHYH2), a positive regulator of light responses, blue and red light reduce intron retention in its 5' UTR; the effect of these retention events is unclear, but may involve production of an N-terminally extended protein as well as uORF-mediated repression of translation (Wu et al., 2014). How AS events are controlled by light is not completely understood, but involves the splicing factors REDUCED RED-LIGHT RESPONSES IN CRY1 CRY2 BACKGROUND 1 (RRC1) and SPLICING FACTOR FOR PHYTOCHROME SIGNALING (SFPS). Both proteins interact with phyB and promote phyB-mediated AS (Shikata et al., 2012; Xin et al., 2017, 2019). Interestingly, RRC1 undergoes light-dependent AS itself: inclusion of the third exon produces a transcript with a premature stop codon and a very long 3' UTR that is subject to NMD. Light reduces inclusion of exon 3 and hence increases the levels of functional RRC1 protein (Shikata et al., 2014).
Temperature changes also induce widespread AS. Cold triggers AS in >2400 Arabidopsis genes when compared with 20 °C levels (Calixto et al., 2018). One of the transcripts undergoing AS in response to cold encodes REGULATOR OF CBF EXPRESSION 1 (RCF1), an RNA helicase required for cold tolerance (Guan et al., 2013). Its transcript isoforms differ in their 3' UTRs, and consequences may involve retention of transcripts in the nucleus as well as NMD, but they have not been investigated experimentally (Calixto et al., 2018). In some cases, a drop in temperature by as little as 2 °C can trigger AS events: intron retention in the 5' UTR of the circadian clock component LATE ELONGATED HYPOCOTYL (LHY) is scalable between 20 °C and 4 °C, and has been suggested to function as a molecular thermostat (James et al., 2018a). Decreasing temperatures result in a gradual increase in intron retention in the LHY 5' UTR, contributing to an overall reduction in LHY transcript levels via NMD (James et al., 2012, 2018a).
Mildly elevated temperature triggers comparatively fewer AS events, which include several key flowering time regulators, the most prominent being FLOWERING LOCUS M (FLM) (Verhage et al., 2017). FLM produces two competing protein splice variants, FLM-β and FLM-δ, both of which bind to the floral repressor SHORT VEGETATIVE PHASE (SVP). Under control temperatures, an SVP–FLM-β complex prevents flowering by repressing the floral integrator gene SOC1. Elevated temperature increases the ratio of FLM-δ to FLM-β; FLM-δ, however, is impaired in DNA binding and was thus thought to accelerate flowering by forming an inactive complex with SVP (Posé et al., 2013). Later observations, however, challenged the role of FLM-δ because plants solely expressing FLM-δ were indistinguishable from an flm knockout (Capovilla et al., 2017). Sureshkumar et al. (2016) showed that high temperature induces formation of multiple splice variants that undergo NMD and hence accelerate flowering simply by reducing levels of FLM-β. Several variants display AS selectively in the 3' UTR, and this agrees with the observation that naturally occurring polymorphisms in the FLM 3' UTR affect the temperature sensitivity of flowering (Sureshkumar et al., 2016).
Other abiotic factors influence AS patterns, but specific examples of functionally relevant AS events in the 5' or 3' UTR are limited. Salt stress as well as sudden illumination cause a shift in PHYTOENE SYNTHASE (PSY) splicing in Arabidopsis, favouring the accumulation of a variant with a shorter 5' UTR. This variant lacks a translation-inhibitory structure, increases PSY protein levels and thus allows for fast production of carotenoids under stress (Álvarez et al., 2016). A reverse mechanism operates in the transcript encoding the Zn transporter ZINC-INDUCED FACILITATOR 2 (ZIF2): here, zinc induces intron retention in the 5' UTR, and the retained sequence forms a stable stem–loop immediately upstream of the start codon, which enhances translation. Consequently, selectively expressing the long ZIF2 splice variant enhances plant zinc tolerance (Remy et al., 2014). These examples highlight that post-transcriptional mechanisms can act cooperatively in the control of gene expression.
UTRs as regulatory hubs for the environmental control of gene expression
In the preceding sections, we highlighted the important role that 5' and 3' UTR sequences play in the control of gene expression, and we discussed multiple post-transcriptional mechanisms operating via UTRs to tune the amount of protein made from the respective transcripts. 5' UTRs are major regulators of translation, providing a platform for the binding of translation initiation factors, but also affecting ribosomal scanning and translation initiation through cis-elements such as uORFs and RNA secondary structures. 3' UTRs, on the other hand, harbour binding sites for many RBPs involved in mRNA sequestration and turnover as well as cis-elements that define the site of polyadenylation, another strong determinant of mRNA stability.
Providing evidence for the functional relevance of UTR regulatory elements remains a challenging task. Recent advancements in next-generation sequencing techniques revealed that changes in the environment can trigger post-transcriptional processes that affect hundreds of transcripts. However, examples for which environmental regulation through UTRs has been causally linked to a developmental or physiological output are few and far between (Table 1) when compared with examples of transcriptional control. Transcriptome-wide studies thus need to be supplemented by detailed gene-focused analyses to prove the functional relevance of post-transcriptional effects, but these require investment of additional time and resources. The issue is further complicated by the fact that some post-transcriptional mechanisms fine-tune or buffer gene expression against external influences and do not act as major on/off switches of protein production (Guerra et al., 2015; Prall et al., 2019; Moro et al., 2021; Zhong et al., 2023); therefore, removing components of these regulatory systems may have minor consequences at the developmental or physiological level. In such cases, computational modelling may be required to fully appreciate the complexity and importance of gene expression regulation (Becker et al., 2018; Furlan et al., 2021).
Table 1.
Transcripts whose function is environmentally controlled via their UTRs
| Transcripta | Regulatory mechanism | Environmental stimulus | Developmental/physiological output | Reference |
|---|---|---|---|---|
| AKR2 | APA | Salinity | Salt tolerance | Yu et al. (2019) |
| AT3G47610 | APA | Salinity | Salt tolerance | Yu et al. (2019) |
| ATHB1 | uORF | Light | Leaf development | Ribone et al. (2017) |
| CO | m6A methylation | Light | Flowering | Wang et al. (2022) |
| COOLAIR | APA | Temperature | Flowering | Csorba et al. (2014) |
| CORG1 | RNA structure | Temperature | Root growth | Yang et al. (2022) |
| CRY2 | m6A methylation | Light | Hypocotyl elongation; flowering | Wang et al. (2022) |
| DGAT1 | m6A methylation | Cold | Growth | S. Wang et al. (2023) |
| FLC | m6A methylation | Light | Flowering | Wang et al. (2022) |
| FLM | AS | Temperature | Flowering | Posé et al. (2013); Sureshkumar et al. (2016) |
| HSFB1 | uORF | Temperature | Heat shock response | Zhu et al. (2012) |
| LHY | AS | Temperature | Circadian clock | James et al. (2018a) |
| NIP5:1 | uORF | Boron | Boron tolerance | Tanaka et al. (2016) |
| OsPHO1.2 | Interaction with cis-NATPHO1.2; RNA structure | Phosphate | Phosphate homeostasis | Reis et al. (2021) |
| PHO2 | Interaction with miR399 | Phosphate; temperature | Phosphate acquisition; flowering | Bari et al. (2006); Chiou et al. (2006); Kim et al. (2011) |
| PIF3 | AS, uORF | Light | Hypocotyl elongation | Dong et al. (2020). |
| PIF4 | m6A methylation | Light | Hypocotyl elongation; flowering | Wang et al. (2022) |
| PIF7 | RNA structure | Temperature | Elongation growth; stomata formation | Chung et al. (2023) |
| PORA | Interaction with PNT1 and phytochrome | Light | Chlorophyll biosynthesis | Paik et al. (2012) |
| PSY | AS | Light; salinity | Carotenoid biosynthesis | Álvarez et al. (2016) |
| SaHKT1 | APA | Salinity | Salt tolerance | T. Wang et al. (2023) |
| SOC1 | Interaction with BRN1/BRN2 | Temperature | Flowering | Kim et al. (2013) |
| SPL3 | Interaction with miR156/157 | Light; temperature | Flowering | Gandikota et al. (2007); Kim et al. (2012); Xie et al. (2017) |
| ZIF2 | AS, RNA structure | Zinc | Zinc tolerance | Remy et al. (2014) |
This table only contains transcripts for which the functional relevance of UTR-mediated regulation in response to specific environmental signals has been shown.
The prefix Os refers to Oryza sativa, the prefix Sa refers to Spartina alterniflora; all other transcripts stem from Arabidopsis thaliana.
Post-transcriptional mechanisms operating via UTRs are highly sensitive to the plant’s environment, but how environmental signals selectively affect processes such as RNA modification, splicing, or translation in a transcript-specific manner often remains enigmatic. In some cases, trans-acting components such as RBPs or miRNAs are regulated by environmental factors and confer this regulation onto their target mRNAs, which are recognized via specific binding sites. In other cases, the UTR itself exhibits environmental responsiveness: this is best exemplified by the effects of RNA secondary structures such as hairpins or G-quadruplexes, whose formation is directly dependent on temperature and osmolarity and can in turn affect accessibility of binding sites or enzymatic processing of the RNA. Understanding the relationship between environmental input and molecular output is essential to exploit these mechanisms for improving plants’ resilience to a changing environment.
UTRs: a means to promote environmental resilience in crops?
Beyond elucidating the regulatory mechanisms through which UTR elements operate, a major goal is to harness these mechanisms for crop improvement. With the progression of climate change, crops are exposed to hostile environments that include cold and heat waves, drought, high salinity, and nutrient scarcity. While changing the expression of key development and stress regulators is at the core of many efforts to render plants more resilient to such conditions, they largely rely on modifying gene transcription. Yet, employing post-transcriptional mechanisms represents a promising expansion of the plant breeding toolbox—these mechanisms act directly on existing transcripts and thus allow for rapid and dynamic responses to the plant’s surroundings, and modification in UTR elements can nowadays be easily accomplished using gene editing techniques (Y. Zhang et al., 2018).
A direct way to exploit UTR function for crop improvement is to take advantage of natural variation present in UTRs. In Arabidopsis, UTR polymorphisms have been linked to temperature adjustments within the circadian clock (James et al., 2018b), thermo-responsiveness of growth (Z. Wang et al., 2021), and root hydraulic conductivity in the context of drought tolerance (Tang et al., 2018). Natural variation in UTRs is also a wide-spread phenomenon in crop species, and several polymorphisms have been linked to stress- and yield-related traits: in maize, a 4 bp deletion in the 3' UTR of Na+CONTENT UNDER SALINE-ALKALINE CONDITION 1 (ZmNSA1) promotes saline–alkaline tolerance (Cao et al., 2020) while deletion of an endoplasmic reticulum (ER) stress response element in the 5' UTR of the protein phosphatase-encoding transcript ZmPP2C-A and a single nucleotide polymorphism in the 5' UTR of DEHYDRATION RESPONSIVE ELEMENT BINDING PROTEIN 27 (ZmDREB27) enhance drought tolerance (S. Liu et al., 2013; Xiang et al., 2017). Polymorphisms in uORFs have also been linked to changes in phosphate acquisition in Arabidopsis (Reis et al., 2020) and soybean (Guo et al., 2022), and globally to whole-plant phenotypic diversity in maize (Gage et al., 2022). Thus, UTR polymorphisms represent a promising genetic resource for conventional breeding, and increasing our understanding of post-transcriptional regulation through UTRs will facilitate selection for specific cis-regulatory UTR elements that convey the desired crop traits.
Gene editing represents an alternative approach to exploit UTRs for crop improvement as it allows for the precise modification of cis-regulatory elements in situ (Y. Zhang et al., 2018). Engineering uORF sequences has already been used to improve vitamin C content in lettuce (H. Zhang et al., 2018) and sugar content in strawberry (Xing et al., 2020), to increase variation in height and tiller number (Xue et al., 2023) and boost broad-spectrum disease resistance in rice (Xu et al., 2017). Other UTR modifications include the editing of splice sites in the WAXY 5' UTR to improve rice grain quality (Zeng et al., 2020) and efforts to implement artificial riboswitches to control gene expression in planta (Bocobza and Aharoni, 2014). The cis-regulatory elements of viruses have also been employed to alter gene expression in plants; the Omega element from tobacco mosaic virus has been widely used to boost translation of transgenes (Gallie, 2002), while recently a targeted knock-in of a translational enhancer from alfalfa mosaic virus has been used to enhance salt tolerance in rice (Shen et al., 2023). The approaches discussed above aimed at constitutive changes in gene expression, and evidence that genetic modification of UTRs can tailor such changes to specific environmental conditions has yet to be obtained. Nevertheless, as our mechanistic insight into the function of cis-regulatory UTR elements increases, we expect many more of these to be targeted by breeding and gene editing efforts in the future, with particular focus on environmental resilience.
Contributor Information
Emma C Hardy, Division of Plant Sciences, University of Dundee at the James Hutton Institute, Dundee DD2 5DA, UK.
Martin Balcerowicz, Division of Plant Sciences, University of Dundee at the James Hutton Institute, Dundee DD2 5DA, UK.
Pablo Manavella, Instituto de Agrobiotecnología del Litoral, Argentina.
Author contributions
MB: conceptualization; ECH and MB: writing; MB: figure design.
Conflict of interest
The authors declare no conflict of interest.
Funding
MB and his lab are supported by the Royal Society (URF\R1\211672) and the BBSRC (BB/Y001672/1). ECH is supported by the BBSRC EASTBIO Doctoral Training Partnership.
Data availability
There are no primary data associated with this manuscript.
References
- Álvarez D, Voß B, Maass D, Wüst F, Schaub P, Beyer P, Welsch R.. 2016. Carotenogenesis is regulated by 5ʹUTR-mediated translation of phytoene synthase splice variants. Plant Physiology 172, 2314–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SJ, Kramer MC, Gosai SJ, et al. 2018. N6-methyladenosine inhibits local ribonucleolytic cleavage to stabilize mRNAs in Arabidopsis. Cell Reports 25, 1146–1157.e3. [DOI] [PubMed] [Google Scholar]
- Aoyama T, Dong CH, Wu Y, Carabelli M, Sessa G, Ruberti I, Morelli G, Chua NH.. 1995. Ectopic expression of the Arabidopsis transcriptional activator Athb-1 alters leaf cell fate in tobacco. The Plant Cell 7, 1773–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artz O, Ackermann A, Taylor L, Koo PK, Pedmale UV.. 2023. Light and temperature regulate m6A-RNA modification to regulate growth in plants. bioRxiv doi: 10.1101/2023.01.17.524395. [Preprint]. [DOI] [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible W-R.. 2006. PHO2, MicroRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiology 141, 988–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett LW, Fletcher S, Wilton SD.. 2012. Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cellular and Molecular Life Sciences 69, 3613–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe DC, Dean C.. 2014. Epigenetic regulation in plant responses to the environment. Cold Spring Harbor Perspectives in Biology 6, a019471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker K, Bluhm A, Casas-Vila N, Dinges N, Dejung M, Sayols S, Kreutz C, Roignant J-Y, Butter F, Legewie S.. 2018. Quantifying post-transcriptional regulation in the development of Drosophila melanogaster. Nature Communications 9, 4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardes WS, Menossi M.. 2020. Plant 3' regulatory regions from mRNA-encoding genes and their uses to modulate expression. Frontiers in Plant Science 11, 1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez MA, Ahn JH, Weigel D.. 2003. A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nature Genetics 33, 168–171. [DOI] [PubMed] [Google Scholar]
- Boccaletto P, Machnicka MA, Purta E, et al. 2018. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Research 46, D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocobza SE, Aharoni A.. 2014. Small molecules that interact with RNA: riboswitch-based gene control and its involvement in metabolic regulation in plants and algae. The Plant Journal 79, 693–703. [DOI] [PubMed] [Google Scholar]
- Boex-Fontvieille E, Daventure M, Jossier M, Zivy M, Hodges M, Tcherkez G.. 2013. Photosynthetic control of Arabidopsis leaf cytoplasmic translation initiation by protein phosphorylation. PLoS One 8, e70692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco-Price C, Kawaguchi R, Ferreira RB, Bailey-Serres J.. 2005. Genome-wide analysis of transcript abundance and translation in Arabidopsis seedlings subjected to oxygen deprivation. Annals of Botany 96, 647–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning KS, Bailey-Serres J.. 2015. Mechanism of cytoplasmic mRNA translation. The Arabidopsis Book 13, e0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calixto CPG, Guo W, James AB, Tzioutziou NA, Entizne JC, Panter PE, Knight H, Nimmo HG, Zhang R, Brown JWS.. 2018. Rapid and dynamic alternative splicing impacts the Arabidopsis cold response transcriptome. The Plant Cell 30, 1424–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Zhang M, Liang X, Li F, Shi Y, Yang X, Jiang C.. 2020. Natural variation of an EF-hand Ca2+-binding-protein coding gene confers saline–alkaline tolerance in maize. Nature Communications 11, 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capovilla G, Symeonidi E, Wu R, Schmid M.. 2017. Contribution of major FLM isoforms to temperature-dependent flowering in Arabidopsis thaliana. Journal of Experimental Botany 68, 5117–5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B, Hopes T, McKay M, Paling Z, Davies B.. 2022. Plants utilise ancient conserved peptide upstream open reading frames in stress-responsive translational regulation. Plant, Cell & Environment 45, 1229–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti M, de Lorenzo L, Abdel-Ghany SE, Reddy ASN, Hunt AG.. 2020. Wide-ranging transcriptome remodelling mediated by alternative polyadenylation in response to abiotic stresses in Sorghum. The Plant Journal 102, 916–930. [DOI] [PubMed] [Google Scholar]
- Chantarachot T, Bailey-Serres J.. 2018. Polysomes, stress granules, and processing bodies: a dynamic triumvirate controlling cytoplasmic mRNA fate and function. Plant Physiology 176, 254–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou T-J, Aung K, Lin S-I, Wu C-C, Chiang S-F, Su C.. 2006. Regulation of phosphate homeostasis by MicroRNA in Arabidopsis. The Plant Cell 18, 412–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmielowska-Bąk J, Arasimowicz-Jelonek M, Deckert J.. 2019. In search of the mRNA modification landscape in plants. BMC Plant Biology 19, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H-Y, Lu M-YJ, Shih M-C.. 2019. The SnRK1–eIFiso4G1 signaling relay regulates the translation of specific mRNAs in Arabidopsis under submergence. New Phytologist 222, 366–381. [DOI] [PubMed] [Google Scholar]
- Chung BYW, Balcerowicz M, Di Antonio M, Jaeger KE, Geng F, Franaszek K, Marriott P, Brierley I, Firth AE, Wigge PA.. 2020. An RNA thermoswitch regulates daytime growth in Arabidopsis. Nature Plants 6, 522–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung KP, Loiacono FV, Neupert J, Wu M, Bock R.. 2023. An RNA thermometer in the chloroplast genome of Chlamydomonas facilitates temperature-controlled gene expression. Nucleic Acids Research 51, gkad816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa CM, Saez A, Navarro-Neila S, et al. 2020. Alternative polyadenylation and salicylic acid modulate root responses to low nitrogen availability. Plants (Basel, Switzerland) 9, 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz JA, Westhof E.. 2009. The dynamic landscapes of RNA architecture. Cell 136, 604–609. [DOI] [PubMed] [Google Scholar]
- Csorba T, Questa JI, Sun Q, Dean C.. 2014. Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proceedings of the National Academy of Sciences, USA 111, 16160–16165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Liang Z, Shen L, et al. 2017. 5-Methylcytosine RNA methylation in Arabidopsis thaliana. Molecular Plant 10, 1387–1399. [DOI] [PubMed] [Google Scholar]
- David R, Burgess A, Parker B, Li J, Pulsford K, Sibbritt T, Preiss T, Searle IR.. 2017. Transcriptome-wide mapping of RNA 5-methylcytosine in Arabidopsis mRNAs and noncoding RNAs. The Plant Cell 29, 445–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo L, Sorenson R, Bailey-Serres J, Hunt AG.. 2017. Noncanonical alternative polyadenylation contributes to gene regulation in response to hypoxia. The Plant Cell 29, 1262–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhingra Y, Gupta S, Gupta V, Agarwal M, Katiyar-Agarwal S.. 2023. The emerging role of epitranscriptome in shaping stress responses in plants. Plant Cell Reports 42, 1531–1555. [DOI] [PubMed] [Google Scholar]
- Dong J, Chen H, Deng XW, Irish VF, Wei N.. 2020. Phytochrome B induces intron retention and translational inhibition of PHYTOCHROME-INTERACTING FACTOR3. Plant Physiology 182, 159–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutt S, Parkash J, Mehra R, Sharma N, Singh B, Raigond P, Joshi A, Chopra S, Singh BP.. 2015. Translation initiation in plants: roles and implications beyond protein synthesis. Biologia Plantarum 59, 401–412. [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W.. 2010. Regulation of mRNA translation and stability by microRNAs. Annual Review of Biochemistry 79, 351–379. [DOI] [PubMed] [Google Scholar]
- Fiorucci A-S, Galvão VC, Ince YC, Boccaccini A, Goyal A, Petrolati LA, Trevisan M, Fankhauser C.. 2020. PHYTOCHROME INTERACTING FACTOR 7 is important for early responses to elevated temperature in Arabidopsis seedlings. New Phytologist 226, 50–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlan M, de Pretis S, Pelizzola M.. 2021. Dynamics of transcriptional and post-transcriptional regulation. Briefings in Bioinformatics 22, bbaa389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage JL, Mali S, McLoughlin F, Khaipho-Burch M, Monier B, Bailey-Serres J, Vierstra RD, Buckler ES.. 2022. Variation in upstream open reading frames contributes to allelic diversity in maize protein abundance. Proceedings of the National Academy of Sciences, USA 119, e2112516119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR. 2002. The 5ʹ-leader of tobacco mosaic virus promotes translation through enhanced recruitment of eIF4F. Nucleic Acids Research 30, 3401–3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandikota M, Birkenbihl RP, Höhmann S, Cardon GH, Saedler H, Huijser P.. 2007. The miRNA156/157 recognition element in the 3ʹ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. The Plant Journal 49, 683–693. [DOI] [PubMed] [Google Scholar]
- Gawroński P, Enroth C, Kindgren P, Marquardt S, Karpiński S, Leister D, Jensen PE, Vinther J, Scharff LB.. 2021. Light-dependent translation change of Arabidopsis psbA correlates with RNA structure alterations at the translation initiation region. Cells 10, 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good PJ, Chen Q, Warner SJ, Herring DC.. 2000. A family of human RNA-binding proteins related to the Drosophila Bruno translational regulator. Journal of Biological Chemistry 275, 28583–28592. [DOI] [PubMed] [Google Scholar]
- Guan Q, Wu J, Zhang Y, Jiang C, Liu R, Chai C, Zhu J.. 2013. A DEAD box RNA helicase is critical for pre-mRNA splicing, cold-responsive gene regulation, and cold tolerance in Arabidopsis. The Plant Cell 25, 342–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra D, Crosatti C, Khoshro HH, Mastrangelo AM, Mica E, Mazzucotelli E.. 2015. Post-transcriptional and post-translational regulations of drought and heat response in plants: a spider’s web of mechanisms. Frontiers in Plant Science 6, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Liu J-H, Ma X, Luo D-X, Gong Z-H, Lu M-H.. 2016. The plant heat stress transcription factors (HSFs): structure, regulation, and function in response to abiotic stresses. Frontiers in Plant Science 7, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Cao H, Zhao J, et al. 2022. A natural uORF variant confers phosphorus acquisition diversity in soybean. Nature Communications 13, 3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann L, Drewe-Boß P, Wießner T, et al. 2016. Alternative splicing substantially diversifies the transcriptome during early photomorphogenesis and correlates with the energy availability in Arabidopsis. The Plant Cell 28, 2715–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden CA, Jorgensen RA.. 2007. Identification of novel conserved peptide uORF homology groups in Arabidopsis and rice reveals ancient eukaryotic origin of select groups and preferential association with transcription factor-encoding genes. BMC Biology 5, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Xu M, Willmann MR, McCormick K, Hu T, Yang L, Starker CG, Voytas DF, Meyers BC, Poethig RS.. 2018. Threshold-dependent repression of SPL gene expression by miR156/miR157 controls vegetative phase change in Arabidopsis thaliana. PLoS Genetics 14, e1007337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornyik C, Terzi LC, Simpson GG.. 2010. The spen family protein FPA controls alternative cleavage and polyadenylation of RNA. Developmental Cell 18, 203–213. [DOI] [PubMed] [Google Scholar]
- Huang K-C, Lin W-C, Cheng W-H.. 2018. Salt hypersensitive mutant 9, a nucleolar APUM23 protein, is essential for salt sensitivity in association with the ABA signaling pathway in Arabidopsis. BMC Plant Biology 18, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T-K, Han C-L, Lin S-I, et al. 2013. Identification of downstream components of ubiquitin-conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. The Plant Cell 25, 4044–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh SU, Paek K-H.. 2014. APUM5, encoding a Pumilio RNA binding protein, negatively regulates abiotic stress responsive gene expression. BMC Plant Biology 14, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa H, Tomari Y.. 2013. Molecular insights into microRNA-mediated translational repression in plants. Molecular Cell 52, 591–601. [DOI] [PubMed] [Google Scholar]
- Jabnoune M, Secco D, Lecampion C, Robaglia C, Shu Q, Poirier Y.. 2013. A rice cis-natural antisense RNA acts as a translational enhancer for its cognate mRNA and contributes to phosphate homeostasis and plant fitness. The Plant Cell 25, 4166–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob AG, Smith CWJ.. 2017. Intron retention as a component of regulated gene expression programs. Human Genetics 136, 1043–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James AB, Calixto CPG, Tzioutziou NA, Guo W, Zhang R, Simpson CG, Jiang W, Nimmo GA, Brown JWS, Nimmo HG.. 2018a. How does temperature affect splicing events? Isoform switching of splicing factors regulates splicing of LATE ELONGATED HYPOCOTYL (LHY). Plant, Cell & Environment 41, 1539–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James AB, Sullivan S, Nimmo HG.. 2018b. Global spatial analysis of Arabidopsis natural variants implicates 5ʹUTR splicing of LATE ELONGATED HYPOCOTYL in responses to temperature. Plant, Cell & Environment 41, 1524–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James AB, Syed NH, Bordage S, Marshall J, Nimmo GA, Jenkins GI, Herzyk P, Brown JWS, Nimmo HG.. 2012. Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. The Plant Cell 24, 961–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon M, Jeong G, Yang Y, Luo X, Jeong D, Kyung J, Hyun Y, He Y, Lee I.. 2023. Vernalization-triggered expression of the antisense transcript COOLAIR is mediated by CBF genes. eLife 12, e84594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshna CR, Saha P, Atugala D, Chua G, Muench DG.. 2020. Plant PUF RNA-binding proteins: a wealth of diversity for post-transcriptional gene regulation. Plant Science 297, 110505. [DOI] [PubMed] [Google Scholar]
- Juntawong P, Bailey-Serres J.. 2012. Dynamic light regulation of translation status in Arabidopsis thaliana. Frontiers in Plant Science 3, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi R, Bailey-Serres J.. 2005. mRNA sequence features that contribute to translational regulation in Arabidopsis. Nucleic Acids Research 33, 955–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearly A, Nelson ADL, Skirycz A, Chodasiewicz M.. 2022. Composition and function of stress granules and P-bodies in plants. Seminars in Cell & Developmental Biology 156, 167–175. [DOI] [PubMed] [Google Scholar]
- Kim H-S, Abbasi N, Choi S-B.. 2013. Bruno-like proteins modulate flowering time via 3ʹ UTR-dependent decay of SOC1 mRNA. New Phytologist 198, 747–756. [DOI] [PubMed] [Google Scholar]
- Kim J, Kim Y, Yeom M, Kim J-H, Nam HG.. 2008. FIONA1 is essential for regulating period length in the Arabidopsis circadian clock. The Plant Cell 20, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JJ, Lee JH, Kim W, Jung HS, Huijser P, Ahn JH.. 2012. The microRNA156–SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 module regulates ambient temperature-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Plant Physiology 159, 461–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J-M, Sasaki T, Ueda M, Sako K, Seki M.. 2015. Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Frontiers in Plant Science 6, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Ahn HJ, Chiou T-J, Ahn JH.. 2011. The role of the miR399–PHO2 module in the regulation of flowering time in response to different ambient temperatures in Arabidopsis thaliana. Molecules and Cells 32, 83–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortmann J, Narberhaus F.. 2012. Bacterial RNA thermometers: molecular zippers and switches. Nature Reviews. Microbiology 10, 255–265. [DOI] [PubMed] [Google Scholar]
- Kramer MC, Janssen KA, Palos K, Nelson ADL, Vandivier LE, Garcia BA, Lyons E, Beilstein MA, Gregory BD.. 2020. N6-methyladenosine and RNA secondary structure affect transcript stability and protein abundance during systemic salt stress in Arabidopsis. Plant Direct 4, e00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Makita Y, Kawashima M, Fujita T, Iwasaki S, Matsui M.. 2018. Transcripts from downstream alternative transcription start sites evade uORF-mediated inhibition of gene expression in Arabidopsis. Proceedings of the National Academy of Sciences, USA 115, 7831–7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lämke J, Bäurle I.. 2017. Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biology 18, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TA, Bailey-Serres J.. 2019. Integrative analysis from the epigenome to translatome uncovers patterns of dominant nuclear regulation during transient stress. The Plant Cell 31, 2573–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppek K, Das R, Barna M.. 2018. Functional 5ʹ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nature Reviews. Molecular Cell Biology 19, 158–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Riaz A, Chachar S, Ding Y, Du H, Gu X.. 2020. Epigenetic modifications of mRNA and DNA in plants. Molecular Plant 13, 14–30. [DOI] [PubMed] [Google Scholar]
- Lin S-I, Chiang S-F, Lin W-Y, Chen J-W, Tseng C-Y, Wu P-C, Chiou T-J.. 2008. Regulatory network of MicroRNA399 and PHO2 by systemic signaling. Plant Physiology 147, 732–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, May GE, Kready H, Nazzaro L, Mao M, Spealman P, Creeger Y, McManus CJ.. 2019. Impacts of uORF codon identity and position on translation regulation. Nucleic Acids Research 47, 9358–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Quesada V, Crevillén P, Bäurle I, Swiezewski S, Dean C.. 2007. The Arabidopsis RNA-binding protein FCA requires a lysine-specific demethylase 1 homolog to downregulate FLC. Molecular Cell 28, 398–407. [DOI] [PubMed] [Google Scholar]
- Liu G, Wang J, Hou X.. 2020. Transcriptome-wide N6-methyladenosine (m6A) methylome profiling of heat stress in Pak-choi (Brassica rapa ssp. chinensis). Plants 9, 1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Xu R, Merrill C, Hong L, Lanken CV, Hunt AG, Li QQ.. 2014. Integration of developmental and environmental signals via a polyadenylation factor in Arabidopsis. PLoS One 9, e115779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M-J, Wu S-H, Chen H-M, Wu S-H.. 2012. Widespread translational control contributes to the regulation of Arabidopsis photomorphogenesis. Molecular Systems Biology 8, 566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M-J, Wu S-H, Wu J-F, Lin W-D, Wu Y-C, Tsai T-Y, Tsai H-L, Wu S-H.. 2013. Translational landscape of photomorphogenic Arabidopsis. The Plant Cell 25, 3699–3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Wang X, Wang H, et al. 2013. Genome-wide analysis of ZmDREB genes and their association with natural variation in drought tolerance at seedling stage of Zea mays L. PLoS Genetics 9, e1003790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorković ZJ, Wieczorek Kirk DA, Lambermon MHL, Filipowicz W.. 2000. Pre-mRNA splicing in higher plants. Trends in Plant Science 5, 160–167. [DOI] [PubMed] [Google Scholar]
- Lukoszek R, Feist P, Ignatova Z.. 2016. Insights into the adaptive response of Arabidopsis thaliana to prolonged thermal stress by ribosomal profiling and RNA-Seq. BMC Plant Biology 16, 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G-Z, MacQueen A, Zheng G, et al. 2014. Unique features of the m6A methylome in Arabidopsis thaliana. Nature Communications 5, 5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Cai L, Lin J, Zhou K, Li QQ.. 2022. Divergence in the regulation of the salt tolerant response between Arabidopsis thaliana and its halophytic relative Eutrema salsugineum by mRNA alternative polyadenylation. Frontiers in Plant Science 13, 866054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini E, Sanchez SE, Romanowski A, Schlaen RG, Sanchez-Lamas M, Cerdán PD, Yanovsky MJ.. 2016. Acute effects of light on alternative splicing in light-grown plants. Photochemistry and Photobiology 92, 126–133. [DOI] [PubMed] [Google Scholar]
- Marondedze C. 2020. The increasing diversity and complexity of the RNA-binding protein repertoire in plants. Proceedings of the Royal Society B: Biological Sciences 287, 20201397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt S, Petrillo E, Manavella PA.. 2023. Cotranscriptional RNA processing and modification in plants. The Plant Cell 35, 1654–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín G, Márquez Y, Mantica F, Duque P, Irimia M.. 2021. Alternative splicing landscapes in Arabidopsis thaliana across tissues and stress conditions highlight major functional differences with animals. Genome Biology 22, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C. 2017. Regulation by 3ʹ-untranslated regions. Annual Review of Genetics 51, 171–194. [DOI] [PubMed] [Google Scholar]
- McGillivray P, Ault R, Pawashe M, Kitchen R, Balasubramanian S, Gerstein M.. 2018. A comprehensive catalog of predicted functional upstream open reading frames in humans. Nucleic Acids Research 46, 3326–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus CJ, May GE, Spealman P, Shteyman A.. 2014. Ribosome profiling reveals post-transcriptional buffering of divergent gene expression in yeast. Genome Research 24, 422–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchante C, Stepanova AN, Alonso JM.. 2017. Translation regulation in plants: an interesting past, an exciting present and a promising future. The Plant Journal 90, 628–653. [DOI] [PubMed] [Google Scholar]
- Merret R, Nagarajan VK, Carpentier M-C, et al. 2015. Heat-induced ribosome pausing triggers mRNA co-translational decay in Arabidopsis thaliana. Nucleic Acids Research 43, 4121–4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian S-B, Jaffrey SR.. 2015. 5ʹ UTR m6A promotes cap-independent translation. Cell 163, 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignone F, Gissi C, Liuni S, Pesole G.. 2002. Untranslated regions of mRNAs. Genome Biology 3, REVIEWS0004.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro SG, Hermans C, Ruiz-Orera J, Albà MM.. 2021. Impact of uORFs in mediating regulation of translation in stress conditions. BMC Molecular and Cell Biology 22, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthusamy M, Kim J-H, Kim JA, Lee S-I.. 2021. Plant RNA binding proteins as critical modulators in drought, high salinity, heat, and cold stress responses: an updated overview. International Journal of Molecular Sciences 22, 6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Náprstková A, Malínská K, Záveská Drábková L, Billey E, Náprstková D, Sýkorová E, Bousquet-Antonelli C, Honys D.. 2021. Characterization of ALBA family expression and localization in Arabidopsis thaliana generative organs. International Journal of Molecular Sciences 22, 1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natalizio BJ, Wente SR.. 2013. Postage for the messenger: designating routes for nuclear mRNA export. Trends in Cell Biology 23, 365–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederer RO, Rojas-Duran MF, Zinshteyn B, Gilbert WV.. 2022. Direct analysis of ribosome targeting illuminates thousand-fold regulation of translation initiation. Cell Systems 13, 256–264.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyikó T, Auber A, Bucher E.. 2019. Functional and molecular characterization of the conserved Arabidopsis PUMILIO protein, APUM9. Plant Molecular Biology 100, 199–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik I, Yang S, Choi G.. 2012. Phytochrome regulates translation of mRNA in the cytosol. Proceedings of the National Academy of Sciences, USA 109, 1335–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajerowska-Mukhtar KM, Wang W, Tada Y, Oka N, Tucker CL, Fonseca JP, Dong X.. 2012. The HSF-like transcription factor TBF1 is a major molecular switch for plant growth-to-defense transition. Current Biology 22, 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BS, Seo JS, Chua N-H.. 2014. NITROGEN LIMITATION ADAPTATION recruits PHOSPHATE2 to target the phosphate transporter PT2 for degradation during the regulation of Arabidopsis phosphate homeostasis. The Plant Cell 26, 454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MT, Knop K, Sherwood AV, Schurch NJ, Mackinnon K, Gould PD, Hall AJ, Barton GJ, Simpson GG.. 2020. Nanopore direct RNA sequencing maps the complexity of Arabidopsis mRNA processing and m6A modification. eLife 9, e49658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posé D, Verhage L, Ott F, Yant L, Mathieu J, Angenent GC, Immink RGH, Schmid M.. 2013. Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 503, 414–417. [DOI] [PubMed] [Google Scholar]
- Prall W, Sharma B, Gregory BD.. 2019. Transcription is just the beginning of gene expression regulation: the functional significance of RNA-binding proteins to post-transcriptional processes in plants. Plant and Cell Physiology 60, 1939–1952. [DOI] [PubMed] [Google Scholar]
- Reddy ASN, Marquez Y, Kalyna M, Barta A.. 2013. Complexity of the alternative splicing landscape in plants. The Plant Cell 25, 3657–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis RS, Deforges J, Schmidt RR, Schippers JHM, Poirier Y.. 2021. An antisense noncoding RNA enhances translation via localized structural rearrangements of its cognate mRNA. The Plant Cell 33, 1381–1397. [DOI] [PubMed] [Google Scholar]
- Reis RS, Deforges J, Sokoloff T, Poirier Y.. 2020. Modulation of shoot phosphate level and growth by PHOSPHATE1 upstream open reading frame. Plant Physiology 183, 1145–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy E, Cabrito TR, Batista RA, Hussein MAM, Teixeira MC, Athanasiadis A, Sá-Correia I, Duque P.. 2014. Intron retention in the 5ʹUTR of the novel ZIF2 transporter enhances translation to promote zinc tolerance in Arabidopsis. PLoS Genetics 10, e1004375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribone PA, Capella M, Arce AL, Chan RL.. 2017. A uORF represses the transcription factor AtHB1 in aerial tissues to avoid a deleterious phenotype. Plant Physiology 175, 1238–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani F, Ribone PA, Capella M, Miguel VN, Chan RL.. 2016. A matter of quantity: common features in the drought response of transgenic plants overexpressing HD-Zip I transcription factors. Plant Science 251, 139–154. [DOI] [PubMed] [Google Scholar]
- Schepetilnikov M, Dimitrova M, Mancera-Martínez E, Geldreich A, Keller M, Ryabova LA.. 2013. TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. The EMBO Journal 32, 1087–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secco D, Baumann A, Poirier Y.. 2010. Characterization of the rice PHO1 gene family reveals a key role for OsPHO1;2 in phosphate homeostasis and the evolution of a distinct clade in dicotyledons. Plant Physiology 152, 1693–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul O. 2015. Unique aspects of plant nonsense-mediated mRNA decay. Trends in Plant Science 20, 767–779. [DOI] [PubMed] [Google Scholar]
- Shen L, Liang Z, Gu X, Chen Y, Teo ZWN, Hou X, Cai WM, Dedon PC, Liu L, Yu H.. 2016. N6-Methyladenosine RNA modification regulates shoot stem cell fate in Arabidopsis. Developmental Cell 38, 186–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Liang Z, Wong CE, Yu H.. 2019. Messenger RNA modifications in plants. Trends in Plant Science 24, 328–341. [DOI] [PubMed] [Google Scholar]
- Shen R, Yao Q, Zhong D, Zhang X, Li X, Cao X, Dong C, Tian Y, Zhu J-K, Lu Y.. 2023. Targeted insertion of regulatory elements enables translational enhancement in rice. Frontiers in Plant Science 14, 1134209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikata H, Hanada K, Ushijima T, Nakashima M, Suzuki Y, Matsushita T.. 2014. Phytochrome controls alternative splicing to mediate light responses in Arabidopsis. Proceedings of the National Academy of Sciences, USA 111, 18781–18786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikata H, Shibata M, Ushijima T, Nakashima M, Kong S-G, Matsuoka K, Lin C, Matsushita T.. 2012. The RS domain of Arabidopsis splicing factor RRC1 is required for phytochrome B signal transduction. The Plant Journal 70, 727–738. [DOI] [PubMed] [Google Scholar]
- Shuman S. 2002. What messenger RNA capping tells us about eukaryotic evolution. Nature Reviews. Molecular Cell Biology 3, 619–625. [DOI] [PubMed] [Google Scholar]
- Sorenson R, Bailey-Serres J.. 2014. Selective mRNA sequestration by OLIGOURIDYLATE-BINDING PROTEIN 1 contributes to translational control during hypoxia in Arabidopsis. Proceedings of the National Academy of Sciences, USA 111, 2373–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava AK, Lu Y, Zinta G, Lang Z, Zhu J-K.. 2018. UTR dependent control of gene expression in plants. Trends in Plant Science 23, 248–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader L, Weijers D, Wagner D.. 2022. Plant transcription factors—being in the right place with the right company. Current Opinion in Plant Biology 65, 102136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Tang Y, Ritchey LE, Tack DC, Zhu M, Bevilacqua PC, Assmann SM.. 2018. Genome-wide RNA structurome reprogramming by acute heat shock globally regulates mRNA abundance. Proceedings of the National Academy of Sciences, USA 115, 12170–12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H-X, Li Y, Niu Q-W, Chua N-H.. 2017. Dehydration stress extends mRNA 3ʹ untranslated regions with noncoding RNA functions in Arabidopsis. Genome Research 27, 1427–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureshkumar S, Dent C, Seleznev A, Tasset C, Balasubramanian S.. 2016. Nonsense-mediated mRNA decay modulates FLM-dependent thermosensory flowering response in Arabidopsis. Nature Plants 2, 16055. [DOI] [PubMed] [Google Scholar]
- Swiezewski S, Liu F, Magusin A, Dean C.. 2009. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462, 799–802. [DOI] [PubMed] [Google Scholar]
- Tack DC, Su Z, Yu Y, Bevilacqua PC, Assmann SM.. 2020. Tissue-specific changes in the RNA structurome mediate salinity response in Arabidopsis. RNA 26, 492–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka M, Zehrmann A, Verbitskiy D, Härtel B, Brennicke A.. 2013. RNA editing in plants and its evolution. Annual Review of Genetics 47, 335–352. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Sotta N, Yamazumi Y, et al. 2016. The minimum open reading frame, AUG-stop, induces boron-dependent ribosome stalling and mRNA degradation. The Plant Cell 28, 2830–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N, Shahzad Z, Lonjon F, Loudet O, Vailleau F, Maurel C.. 2018. Natural variation at XND1 impacts root hydraulics and trade-off for stress responses in Arabidopsis. Nature Communications 9, 3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Téllez-Robledo B, Manzano C, Saez A, et al. 2019. The polyadenylation factor FIP1 is important for plant development and root responses to abiotic stresses. The Plant Journal 99, 1203–1219. [DOI] [PubMed] [Google Scholar]
- Tian L, Chou H-L, Fukuda M, Kumamaru T, Okita TW.. 2020. mRNA localization in plant cells. Plant Physiology 182, 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong J, Ren Z, Sun L, et al. 2022. ALBA proteins confer thermotolerance through stabilizing HSF messenger RNAs in cytoplasmic granules. Nature Plants 8, 778–791. [DOI] [PubMed] [Google Scholar]
- van der Horst S, Filipovska T, Hanson J, Smeekens S.. 2020. Metabolite control of translation by conserved peptide uORFs: the ribosome as a metabolite multisensor. Plant Physiology 182, 110–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J, Han C, Gadeyne A, et al. 2019. Capturing the phosphorylation and protein interaction landscape of the plant TOR kinase. Nature Plants 5, 316–327. [DOI] [PubMed] [Google Scholar]
- Verhage L, Severing EI, Bucher J, Lammers M, Busscher-Lange J, Bonnema G, Rodenburg N, Proveniers MCG, Angenent GC, Immink RGH.. 2017. Splicing-related genes are alternatively spliced upon changes in ambient temperatures in plants. PLoS One 12, e0172950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim AG, Jia Q, Vaughn JN.. 2014. Regulation of plant translation by upstream open reading frames. Plant Science 214, 1–12. [DOI] [PubMed] [Google Scholar]
- Wang C, Yang J, Song P, Zhang W, Lu Q, Yu Q, Jia G.. 2022. FIONA1 is an RNA N6-methyladenosine methyltransferase affecting Arabidopsis photomorphogenesis and flowering. Genome Biology 23, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J-W, Czech B, Weigel D.. 2009. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138, 738–749. [DOI] [PubMed] [Google Scholar]
- Wang S, Wang H, Xu Z, Jiang S, Shi Y, Xie H, Wang S, Hua J, Wu Y.. 2023. m6A mRNA modification promotes chilling tolerance and modulates gene translation efficiency in Arabidopsis. Plant Physiology 192, 1466–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Ye W, Zhang J, Li H, Zeng W, Zhu S, Ji G, Wu X, Ma L.. 2023. Alternative 3ʹ-untranslated regions regulate high-salt tolerance of Spartina alterniflora. Plant Physiology 191, 2570–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Jiang B, Gu L, Chen Y, Mora M, Zhu M, Noory E, Wang Q, Lin C.. 2021. A photoregulatory mechanism of the circadian clock in Arabidopsis. Nature Plants 7, 1397–1408. [DOI] [PubMed] [Google Scholar]
- Wang Z, Yang L, Wu D, Zhang N, Hua J.. 2021. Polymorphisms in cis-elements confer SAUR26 gene expression difference for thermo-response natural variation in Arabidopsis. New Phytologist 229, 2751–2764. [DOI] [PubMed] [Google Scholar]
- Winkler WC, Breaker RR.. 2005. Regulation of bacterial gene expression by riboswitches. Annual Review of Microbiology 59, 487–517. [DOI] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang J-W, Weigel D, Poethig RS.. 2009. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138, 750–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Poethig RS.. 2006. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133, 3539–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H-P, Su Y, Chen H-C, Chen Y-R, Wu C-C, Lin W-D, Tu S-L.. 2014. Genome-wide analysis of light-regulated alternative splicing mediated by photoreceptors in Physcomitrella patens. Genome Biology 15, R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H-W, Fajiculay E, Wu J-F, Yan C-CS, Hsu C-P, Wu S-H.. 2022. Noise reduction by upstream open reading frames. Nature Plants 8, 474–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Wang J, Wu X, Hong Y, Li QQ.. 2020. Heat shock responsive gene expression modulated by mRNA poly(A) tail length. Frontiers in Plant Science 11, 1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Sun X, Gao S, Qin F, Dai M.. 2017. Deletion of an endoplasmic reticulum stress response element in a ZmPP2C-A gene facilitates drought tolerance of maize seedlings. Molecular Plant 10, 456–469. [DOI] [PubMed] [Google Scholar]
- Xie Y, Liu Y, Wang H, Ma X, Wang B, Wu G, Wang H.. 2017. Phytochrome-interacting factors directly suppress MIR156 expression to enhance shade-avoidance syndrome in Arabidopsis. Nature Communications 8, 348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin R, Kathare PK, Huq E.. 2019. Coordinated regulation of Pre-mRNA splicing by the SFPS–RRC1 complex to promote photomorphogenesis. The Plant Cell 31, 2052–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin R, Zhu L, Salomé PA, Mancini E, Marshall CM, Harmon FG, Yanovsky MJ, Weigel D, Huq E.. 2017. SPF45-related splicing factor for phytochrome signaling promotes photomorphogenesis by regulating pre-mRNA splicing in Arabidopsis. Proceedings of the National Academy of Sciences, USA 114, E7018–E7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing D, Li QQ.. 2011. Alternative polyadenylation and gene expression regulation in plants. Wiley Interdisciplinary Reviews. RNA 2, 445–458. [DOI] [PubMed] [Google Scholar]
- Xing S, Chen K, Zhu H, Zhang R, Zhang H, Li B, Gao C.. 2020. Fine-tuning sugar content in strawberry. Genome Biology 21, 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Yuan M, Ai C, Liu L, Zhuang E, Karapetyan S, Wang S, Dong X.. 2017. uORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature 545, 491–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue C, Qiu F, Wang Y, Li B, Zhao KT, Chen K, Gao C.. 2023. Tuning plant phenotypes by precise, graded downregulation of gene expression. Nature Biotechnology 41, 1758–1764. [DOI] [PubMed] [Google Scholar]
- Yan C, Wang Y, Lyu T, Hu Z, Ye N, Liu W, Li J, Yao X, Yin H.. 2021. Alternative polyadenylation in response to temperature stress contributes to gene regulation in Populus trichocarpa. BMC Genomics 22, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Gan J, Tao Y, Okita TW, Tian L.. 2022. RNA-binding proteins: the key modulator in stress granule formation and abiotic stress response. Frontiers in Plant Science 13, 882596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Cao Y, Ma L.. 2021. Co-transcriptional RNA processing in plants: exploring from the perspective of polyadenylation. International Journal of Molecular Sciences 22, 3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S-Y, Lu W-C, Ko S-S, Sun C-M, Hung J-C, Chiou T-J.. 2020. Upstream open reading frame and phosphate-regulated expression of rice OsNLA1 controls phosphate transport and reproduction. Plant Physiology 182, 393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Yu H, Duncan S, et al. 2022. RNA G-quadruplex structure contributes to cold adaptation in plants. Nature Communications 13, 6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yángüez E, Castro-Sanz AB, Fernández-Bautista N, Oliveros JC, Castellano MM.. 2013. Analysis of genome-wide changes in the translatome of Arabidopsis seedlings subjected to heat stress. PLoS One 8, e71425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SK, Wek RC.. 2016. Upstream open reading frames differentially regulate gene-specific translation in the integrated stress response. Journal of Biological Chemistry 291, 16927–16935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Lin J, Li QQ.. 2019. Transcriptome analyses of FY mutants reveal its role in mRNA alternative polyadenylation. The Plant Cell 31, 2332–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng D, Liu T, Ma X, et al. 2020. Quantitative regulation of Waxy expression by CRISPR/Cas9-based promoter and 5'UTR-intron editing improves grain quality in rice. Plant Biotechnology Journal 18, 2385–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Lv Z, Diao S, Liu H, Duan A, He C, Zhang J.. 2021. Unique features of the m6A methylome and its response to drought stress in sea buckthorn (Hippophae rhamnoides Linn.). RNA Biology 18, 794–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Si X, Ji X, Fan R, Liu J, Chen K, Wang D, Gao C.. 2018. Genome editing of upstream open reading frames enables translational control in plants. Nature Biotechnology 36, 894–898. [DOI] [PubMed] [Google Scholar]
- Zhang L, Liu X, Gaikwad K, et al. 2017. Mutations in eIF5B confer thermosensitive and pleiotropic phenotypes via translation defects in Arabidopsis thaliana. The Plant Cell 29, 1952–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Wu A, Yue Y, Zhao Y.. 2020. uORFs: important cis-regulatory elements in plants. International Journal of Molecular Sciences 21, 6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Guo H.. 2017. mRNA decay in plants: both quantity and quality matter. Current Opinion in Plant Biology 35, 138–144. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Massel K, Godwin ID, Gao C.. 2018. Applications and potential of genome editing in crop improvement. Genome Biology 19, 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong V, Archibald BN, Brophy JAN.. 2023. Transcriptional and post-transcriptional controls for tuning gene expression in plants. Current Opinion in Plant Biology 71, 102315. [DOI] [PubMed] [Google Scholar]
- Zhou F, Roy B, von Arnim AG.. 2010. Translation reinitiation and development are compromised in similar ways by mutations in translation initiation factor eIF3h and the ribosomal protein RPL24. BMC Plant Biology 10, 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Thalor SK, Takahashi Y, Berberich T, Kusano T.. 2012. An inhibitory effect of the sequence-conserved upstream open-reading frame on the translation of the main open-reading frame of HsfB1 transcripts in Arabidopsis. Plant, Cell & Environment 35, 2014–2030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
There are no primary data associated with this manuscript.