Abstract
We describe, for the first time, the generation of a viral DNA chip for simultaneous expression measurements of nearly all known open reading frames (ORFs) in the largest member of the herpesvirus family, human cytomegalovirus (HCMV). In this study, an HCMV chip was fabricated and used to characterize the temporal class of viral gene expression. The viral chip is composed of microarrays of viral DNA prepared by robotic deposition of oligonucleotides on glass for ORFs in the HCMV genome. Viral gene expression was monitored by hybridization to the oligonucleotide microarrays with fluorescently labelled cDNAs prepared from mock-infected or infected human foreskin fibroblast cells. By using cycloheximide and ganciclovir to block de novo viral protein synthesis and viral DNA replication, respectively, the kinetic classes of array elements were classified. The expression profiles of known ORFs and many previously uncharacterized ORFs provided a temporal map of immediate-early (α), early (β), early-late (γ1), and late (γ2) genes in the entire genome of HCMV. Sequence compositional analysis of the 5′ noncoding DNA sequences of the temporal classes, performed by using algorithms that automatically search for defined and recurring motifs in unaligned sequences, indicated the presence of potential regulatory motifs for β, γ1, and γ2 genes. In summary, these fabricated microarrays of viral DNA allow rapid and parallel analysis of gene expression at the whole viral genome level. The viral chip approach coupled with global biochemical and genetic strategies should greatly speed the functional analysis of established as well as newly discovered large viral genomes.
Human cytomegalovirus (HCMV) has one of the largest known viral genomes, with a complexity that approximates 0.25 Mb of double-stranded DNA. The complete genome sequence of the AD169 laboratory strain of HCMV was made available in 1990 (4). Analysis of this sequence and of related laboratory and clinical strains for potential protein-coding content has revealed at least 226 distinct open reading frames (ORFs) (3, 4, 23). To date, expression analysis of the HCMV genome has resulted in the characterization of approximately 30% of the genome (reviewed in reference 22 and Table 1).
TABLE 1.
Kinetic class of HCMV ORF expression
| ORF (strain) | Class by indicated assay or source
|
ORF (strain) | Class by indicated assay or source
|
|||
|---|---|---|---|---|---|---|
| Chip | Northern (reference) | Chip | Northern (reference) | |||
| JIL | E | UL86 | E-L | L (22) | ||
| TRL2-IRL2 | E | UL89 | E-L | |||
| TRL3-IRL3 | L | UL91 | L | |||
| TRL4-IRL4 | E | E (22) | UL92 | L | ||
| TRL5-IRL5 | E (14) | UL93 | L | |||
| TRL6-IRL6 | L | E-L (14) | UL94 | L | L (38) | |
| TRL7-IRL7 | E | UL95 | E-L (37) | |||
| TRL8-IRL8 | L | L (N) | UL96 | E-L | E-L (21, 32) | |
| TRL9-IRL9 | L | L (N) | UL97 | E-L | E-L (21, 32) | |
| TRL10-IRL10 | E-L | UL98 | E-L | E-L (5, 37) | ||
| TRL11-IRL11 | L | UL99 | L (22) | |||
| TRL12-IRL12 | E-L | UL100 | E-L | L (22) | ||
| TRL13-IRL13 | E-L | UL102 | L | E, I (29) | ||
| TRL14 | L | UL103 | L | |||
| IRL14 | E | UL104 | E | |||
| UL1 | E-L | UL105 | E | E (30) | ||
| UL2 | L | UL106 | E-L | |||
| UL3 | L | UL107 | L | |||
| UL4 | E | E (22) | UL108 | L | ||
| UL5 | E | UL109 | L | |||
| UL7 | L | UL110 | IE, E, L | IE, L(N) (15) | ||
| UL9 | L | UL111 | L | |||
| UL11 | E (11) | UL111A | E-L | L (26) | ||
| UL13 | E | UL112 | E | E (22) | ||
| UL14 | L | UL113 | E-L | E (22) | ||
| UL15 | L | L (N) | UL114 | E | ||
| UL16 | E (17) | UL115 | L (22) | |||
| UL17 | E | UL116 | E-L | L (22) | ||
| UL18 | L | L (22) | UL117 | L (22) | ||
| UL21 | L | UL118 | E | L (22) | ||
| UL25 | L (1) | UL119 | E | L (22) | ||
| UL26 | E | UL120 | L | |||
| UL27 | E | UL121 | L | |||
| UL29 | L | UL122 | IE, L | IE, L (22) | ||
| UL31 | L | L (N) | UL123 | IE | IE (22) | |
| UL32 | L | L (22) | UL124 | E | ||
| UL33 | E | L (22) | UL128 | E | ||
| UL34 | E-L | L (22) | UL129 | L | ||
| UL35 | E | UL130 | E-L | |||
| UL36 | E | IE (22) | UL131 | L | ||
| UL37 | IE (22) | UL132 | E-L | |||
| UL38 | IE (22) | UL147 (Towne) | E-L | |||
| UL40 | E-L | UL152 (Towne) | E-L | |||
| UL41 | L | UL153 (Towne) | E-L | |||
| UL43 | L | UL154 (Towne) | E-L | |||
| UL44 | E-L | E-L (22) | IRS1-TRS1 | IE (22) | ||
| UL46 | E-L | US2 | E (22) | |||
| UL47 | E-L | US3 | IE | IE (22) | ||
| UL48 | L | L (N) | US6 | E-L | E-L (16) | |
| UL49 | E-L | US7 | E-L | E-L (16) | ||
| UL52 | L | US8 | E | E (16) | ||
| UL53 | E | US9 | E | E (16) | ||
| UL54 | E | E (22) | US10 | E | E (16) | |
| UL55 | E | E (22) | US11 | E | E (16) | |
| UL56 | E | E (22) | US12 | E | ||
| UL57 | E | E (22) | US13 | E | ||
| UL58 | E | E | US14 | E | ||
| UL59 | L | US15 | E-L | |||
| UL60 | L | US16 | E | |||
| UL63 | E | US17 | E | |||
| UL66 | E-L (N) | US18 | E | L (9) | ||
| UL67 | L | US19 | E | E (9) | ||
| UL68 | L | L (N) | US20 | E | E (9) | |
| UL69 | E-L | E-L (39) | US22 | E | E (22) | |
| UL70 | E-L (39) | US23 | E | |||
| UL72 | E-L | US24 | E | |||
| UL73 | E-L | L (N) | US25 | E-L | ||
| UL75 | E-L | L (22) | US26 | E | ||
| UL77 | E | US27 | E | L (33) | ||
| UL78 | E | US28 | E | E (33) | ||
| UL80 | L (35) | US29 | E-L | |||
| UL81 | L | US30 | E | |||
| UL82 | L | L (22) | US32 | L | ||
| UL83 | L | E-L (22) | US33 | E | ||
| UL84 | E-L | E-L (10, 22) | US34 | E | ||
| UL85 | E-L | US35 | E (N) | |||
The expression of HCMV genes upon infection is temporally regulated. The first genes expressed (immediate-early [IE or α] genes) are independent of any viral de novo protein synthesis and encode mostly regulatory trans-acting factors. The next set of genes expressed (early [E or β] genes) requires the presence of the viral IE proteins and contributes an essential source of factors, including viral DNA replication, repair enzymes, and other nonstructural proteins, such as those that serve in immune evasion. Late (L or γ) genes are essentially expressed after the onset of viral DNA replication and contribute primarily to assembly and morphogenesis of the virion. Thus, the time of viral gene expression during infection is an important clue to its functional role. Systematic approaches that permit high throughput evaluation of specific ORF expression would greatly assist efforts to elucidate viral gene function of highly complex viruses, such as CMV, on a genome-wide scale. Historically, prominent regions of HCMV IE, E, and L gene expression were initially identified by hybridization to genomic subfragments, and therefore these early studies provided a first analysis of the HCMV transcription program (5, 19, 31, 34, 36).
Recently, DNA chips have been constructed and used to measure genome-wide expression levels of genes in plants, bacteria, yeast, and human cells (6, 25, 27, 28 and references therein). Of these methods, DNA microarrays, consisting of individual ORF sequences printed in a miniaturized format on glass, is a relatively simple but powerful tool for studying gene expression on a large scale (28). Here, we report on the construction of a viral DNA chip for HCMV. These fabricated microarrays of viral DNA allow, in a single hybridization, the analysis of gene expression for many of the predicted HCMV ORFs. In the present study we have applied this technique to examine the temporal transcription program of HCMV gene expression. HCMV array elements that displayed differential patterns of viral gene expression and were thus classified in different kinetic classes were characterized. This study therefore provides a more complete analysis of the transcription program of HCMV.
(Part of these results were presented at the 7th International Cytomegalovirus Workshop in Brighton, United Kingdom, 28 April to 1 May 1999.)
MATERIALS AND METHODS
Selection and synthesis of oligonucleotides for DNA microarrays.
The complete set of ORFs from the HCMV genome was analyzed with a custom sequence analysis program that selected a 75-base sequence to be used as a microarray deposition target. The analysis preferentially selects unique sequences with a 3′ gene bias and a G-C content of 40 to 60% and rejects sequences that contain homopolymeric stretches and potential hairpin structures. The 3′ gene bias is preferred, as fluorescently labelled cDNA prepared for hybridization is generated by using oligo(dT) to prime poly(A) tails of mRNA. The selected target sequences were synthesized by using a PE Perseptive BioSystem (Framingham, Mass.) Expedite MOSS DNA synthesizer with membrane columns. Synthesized gene target oligonucleotides were cleaved, deprotected, and purified by standard procedures. Target oligonucleotides were transferred in triplicate to 96-well master plates at a concentration of 1 μg/μl (in 3× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) for robotic deposition. The sequence of oligonucleotides comprising the deposited HCMV ORF microarray is shown in Fig. 1. The small ORF UL48/49 (8) and the UL74 ORF described by Huber and Compton (13) were not included in the present chip design. Also shown in Fig. 1 is a subset of cellular genes that were included as internal controls for normalization between chips, as follows: elongation factor 1-alpha (accession no. M29548), human acidic ribosomal phosphoprotein (RiboPO; accession no. M17885), alpha tubulin (accession no. K00558), glyceraldehyde-3-phosphate dehydrogenase (GAPDH; accession no. J04038), retinoic acid receptor (RARa1; accession no. X06614), and CAAT box DNA binding protein (NFY; accession no. X59711). Two plant homeobox genes, HAT 1 (accession no. UO9332) and HAT 4 (accession no. Z19602) were deposited as further specificity controls.
FIG. 1.
Oligonucleotides used in the microarray deposition.
Generation of microarrays, hybridization, and scanning.
The preparation of coated glass slides and the subsequent deposition printing of DNA was carried out in a manner similar to that described previously (28). Briefly, a custom-made microarrayer was built by using a Galil (Mountain View, Calif.) DMC1030-18 motion controller. The motion control signals were used to control NEMA 23 sized DC servo motors which drove Parker Dadeal (Harrison City, Pa.) 500,000 ET series linear stage with optical encoder feedback. The array table held up to 40 slides and one titer tray of source DNA targets for spotting. The custom-designed spotting tip was based on the concept of a rod with a closed tweezer tip manufactured to fit into a standard 384-well plate. The spotting tips were washed in a custom-built wash-and-vacuum station before each round of spotting. The entire spotting assembly was placed under a custom-made acrylic enclosure with a class 100 HEPA filter (Envirco, Albuquerque, N. Mex.).
The HCMV chips used in this study were prepared by using a single-tip format. The microarrayer tip delivered approximately 4 nl per spot on prescreened silylated aldehyde-coated glass slides (CEL Associates, Houston, Tex.). Viral microarrays were hybridized for 4 h under coverslips with a Cy3-dCTP (Amersham)-labelled cDNA probe. The entire assembly was enclosed in a custom-made hybridization chamber. After hybridization, the microarray slide assembly was washed and dried. Microarrays were subsequently scanned by using a confocal laser ScanArray 3000 (General Scanning Inc.) system. Data were collected at a maximum resolution of 10 μm/pixel with 16 bits of depth by using ImaGene software (BioDiscovery Inc.).
Virus and cells.
Human foreskin fibroblasts (HFF) were grown in Dulbecco’s modified essential medium supplemented with 2 mM glutamine, 100 U of penicillin per ml, 100 μg of gentamicin, and 10% fetal bovine serum. The Towne strain of HCMV was used for all the experiments.
Viral infections, probe preparation, and labelling.
HFF were mock infected or infected with HCMV at a multiplicity of 5 PFU/cell. To assess IE transcription, cultures were treated with cycloheximide (100 μg/ml) 1 h before infection, and whole-cell RNA was harvested 13 h postinfection. For early transcription, ganciclovir (100 μM) was added at the time of virus infection for 72 h prior to total RNA isolation. Under these conditions, ganciclovir reduces virus yield by greater than 99% (2). For late RNA isolation, whole-cell RNA was harvested from cultures 72 h after infection. Mock-infected cells were treated with cycloheximide or ganciclovir for the same period of time as infected HFF cultures were. Total RNA was isolated from mock-infected and infected cells by using the RNAzol B method (Tel-Test, Inc.; Friendswood, Tex.) according to the manufacturer’s protocol. RNAs were passed through an RNAeasy Qiagen column after DNase I treatment, and cDNA probes were synthesized. Fluorescently labelled cDNA was prepared from RNA by oligo(dT)-primed polymerization by using superscript II reverse transcriptase. The pool of nucleotides in the labelling reaction consisted of 0.5 mM dGTP, dATP, and dTTP and 0.04 mM dCTP and fluorescent nucleotide Cy3-dCTP (Amersham) at 0.04 mM. Probes were purified by using the Qiaquick PCR purification kit (Qiagen) and ethanol precipitation.
Statistical analysis.
Quantitated hybridization levels were normalized so that the 75th percentiles of the cellular gene expression levels were equal across chips. HCMV gene expression was determined by comparing normalized values between mock-infected control chips and infected chips by using a Wilcoxon-Mann-Whitney test (12).
Northern blot analysis.
Total RNA, 14 μg per lane, was separated by electrophoresis on a 1% agarose gel containing 2.2 M formaldehyde, transferred onto a nylon membrane (Hybond-N; Amersham), immobilized by UV cross-linking (Stratagene, La Jolla, Calif.), and hybridized with 32P-labelled probes. The probes for UL110 and US35 were composed of PCR-generated fragments. Primers used to amplify a 451-bp fragment from pCM1050 (7), a cosmid containing the UL110 gene, were 18087 (5′CATCAATCATCGTAGTGACGTC3′) and 18088 (5′GCCTATTGATAATAATCTACCCC3′). Primers used to amplify a 211-bp fragment from pCM1035 (7), a cosmid containing the US35 gene, were 18119 (5′GTACCGTTGTACGCATTACAC3′) and 18120 (5′GACGAAGATGCCGATGTGTGAC3′). The resulting PCR fragments were isolated from agarose gels and then radiolabelled with [α-32P]dATP by the random-primed labelling method (Boehringer, Mannheim, Germany) according to the manufacturer’s protocol. For TRL8-IRL8, TRL9-IRL9, UL15, UL31, UL48, UL66, and UL73, the corresponding oligonucleotides shown in Fig. 1 were used as probes, after being [α-32P]ATP end labelled with polynucleotide kinase (Stratagene). Oligonucleotide probes were hybridized to the filters for 1 h at 45°C by using Quick Hybridization solutions (Stratagene) under conditions recommended by the manufacturer. PCR-generated probes were hybridized with the filters for 12 h at 65°C in 1× Denhardt’s solution, 6× SSC, and 100 μg of denatured salmon sperm DNA/ml. Filters were washed to a stringency of 0.1% sodium dodecyl sulfate (SDS) at 60°C or 1% SDS at 42°C depending whether PCR-generated DNA fragments or oligonucleotides, respectively, were used during the hybridization. Hybridization signals were quantitated by using a Molecular Dynamics PhosphorImager system with ImageQuant software.
MEME analysis of the upstream noncoding DNA sequences.
The computer program Multiple EM for Motif Elicitation (MEME) was used to search for sequence motifs in 500 bp of noncoding sequences upstream of the initiation codon. MEME analysis was performed by using the sequence of strain AD169 of HCMV. The 5′ noncoding regions were categorized according to class of expression as follows: E (TRL4-IRL4, UL104-5, UL11, UL112, UL124, UL13, UL16-7, UL24, UL26-7, UL35, UL4-5, UL45, UL53-7, UL77-9, US8-14, US16-7, US19, US23-4, US26, US28, and US30), early-late (E-L) (TRL-IRL6, TRL-IRL10, TRL-IRL12, TRL-IRL13, UL1, UL106, UL130, UL40, UL44, UL46-7, UL49, UL72, UL83-5, UL95-8, US6-7, and US29), and L (TRL-IRL8, TRL-IRL11, TRL-IRL14, UL100, UL103, UL111A, UL117, UL119, UL131, UL14, UL18, UL2-3, UL7, UL9, UL25, UL29, UL32-3, UL43, UL48, UL52, UL59, UL67, UL73, UL80, UL82, UL91-3, UL99, US18, and US27). By using MEME, 30 motifs (10 of 8 bases in length, 10 of 10 bases in length or longer, and 10 of 12 bases in length or longer) were derived from each gene set. The distribution of the combined 90 patterns was identified, allowing for 10% mismatch. MEME is available on the World Wide Web (20a). The resulting motifs that developed a significant polarized distribution pattern are summarized in Table 2. In addition, the transcription factor database (TFD) was used to search for known regulatory sequences. The TFD was downloaded from the National Center for Biotechnology Information.
TABLE 2.
MEME analysis of E, E-L, and L 5′ noncoding regions
| ORF class (total no.) | ORFs with indicated MEME pattern (respective % of E, E-L, and L ORFs)
|
||||
|---|---|---|---|---|---|
| ACGACGTCGG (25, 11, 0) | AAAACAACGT (25, 19, 3) | TGACGGTG (13, 30, 6) | CGGTCTTCTTTT (0, 0, 14) | CCGCGGGCGCGG (0, 0, 17) | |
| E (n = 40) | UL104 | UL11 | US11 | ||
| UL16 | UL112 | US13 | |||
| UL26 | UL35 | US17 | |||
| UL27 | UL5 | US24 | |||
| UL35 | UL57 | US30 | |||
| UL56 | UL77 | ||||
| UL57 | US11 | ||||
| US19 | US17 | ||||
| US28 | US24 | ||||
| US9 | US30 | ||||
| E-L (n = 27) | UL46 | IRL10 | IRL12 | ||
| UL49 | IRL13 | IRL13 | |||
| UL96 | TRL10 | TRL12 | |||
| TRL13 | TRL13 | ||||
| UL44 | UL49 | ||||
| UL83 | |||||
| UL97 | |||||
| UL98 | |||||
| L (n = 36) | UL52 | UL92 | IRL8 | IRL8 | |
| US18 | TRL8 | TRL8 | |||
| UL25 | UL25 | ||||
| UL32 | UL32 | ||||
| UL33 | UL33 | ||||
| UL48 | |||||
RESULTS
Viral microarray (chip) of the HCMV genome.
Microarray technology provides an excellent method by which nucleic acids can be attached to a solid surface in a highly dense format. Given that the HCMV genome consists of ∼200 ORFs, the entire set of potential genes can be easily arrayed in a small area. With this capability and the availability of the complete sequence of HCMV, our strategy was to use a directed approach for generating the viral genome array. This procedure involved synthesizing a 75-base oligonucleotide corresponding to the sense strand of each ORF. The length of this deposition element provides a more efficient target for specific hybridization than does a 25-base oligonucleotide and therefore affords greater sensitivity. For these experiments, almost all ORFs present in the AD169 laboratory strain of HCMV and four ORFs from the Towne strain (UL147 and UL152-4) were selected for deposition. In addition, a set of cellular genes (see Materials and Methods for details; Fig. 1) and two plant genes were included as controls. The generation of sense-strand oligonucleotides as deposition elements has the advantage of the assignation of polarity of transcription. In the present study, the target oligonucleotide representing the ORF of interest was arrayed in triplicate on glass slides. Three independent experiments were performed for each experimental data point. The viral arrays were less than 1 cm2 and contained approximately 1,000 elements, at a spacing of ∼350 μm. This spacing allows the hybridization volumes to be minimized (15 μl); thus, 2 μg of total RNA (from cells infected at a multiplicity of infection [MOI] of 5) is sufficient for analysis, and subsequent amplification steps are unnecessary.
Gene expression analysis by CMV microarrays.
In this study, the viral microarrays were used to delineate kinetic class by examination of the drug sensitivity of viral gene expression. The infection of permissive cells with HCMV in the presence of an inhibitor of protein synthesis leads to the specific accumulation of viral IE RNA. The IE genes of HCMV are well characterized, and therefore a comparison with the results from the viral microarray hybridization is a good first test of the accuracy of the chip. Previously, it has been shown that viral IE RNA arises from only a few distinct regions of the genome and, on the basis of their levels of expression, can be classified into two groups. The major IE genes, referred to as IE1 (UL123) and IE2 (UL122), are transcribed and expressed at relatively high levels. The other class of genes, including primarily the US3, TRS1-IRS1, and UL36-38 loci, is expressed at lower relative levels (reviewed in reference 22).
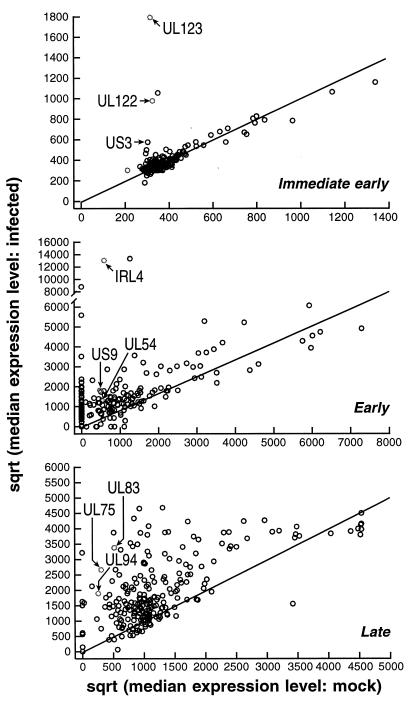
Accordingly, we used cycloheximide as an inhibitor of protein synthesis to investigate IE transcription of HCMV. For these experiments, primary HFF cells were pretreated for 1 h with 100 μg of cycloheximide/ml and subsequently mock infected or infected with HCMV (Towne strain) at an MOI of 5. RNA was isolated in five independent experiments, converted to fluorescently labelled cDNA, and hybridized to the HCMV chips. Fluorescence intensities were normalized by using a set of cellular genes. An absolute fluorescent signal whose intensity was greater than that observed over the mock-infected control microarray element was considered to represent specific viral hybridization. Figure 2 (top panel) shows a scatterplot of these results, with the points above the line of equivalence indicating expression. Conservatively, viral gene-specific hybridization was considered significantly different between mock-infected and infected samples only if the following two criteria were met: (i) the median level of intensity in the infected samples was at least threefold greater than that in the mock-infected samples, and (ii) a nonparametric test for an increase in level of intensity under conditions of virus infection versus mock infection was significant at a P value of <0.05. On the basis of these criteria, four viral ORFs, as follows, were specifically detected at IE: US3, UL122, UL123, and UL110. The UL36-38, TRS1-IRS1 ORFs all showed ratios of approximately 2, with P values of less than 0.05, and thus while they showed a statistically significant increase, were not scored as such under these selection conditions. It is noteworthy that only two other ORFs (UL111A and US8) exhibited twofold ratios. In the case of the UL110 ORF, the results of Northern analysis corroborated the IE gene expression identified by the microarray hybridization (Fig. 4). By the Northern analysis, a 5-kb transcript was detected that corresponds to a previously characterized IE transcript reported from this region (15). As a whole, these analyses show that while the CMV chip does not score positive for all known IE genes, no false-positive signals were detected. The false-negative results are likely due to poor design of the target probe and/or related to level of sensitivity for the detection of low-abundance transcripts. Nevertheless, these experiments clearly demonstrate an efficacious approach for the stringent detection of viral-specific RNA species and thereby validate the microarray hybridization assay.
FIG. 2.
Microarray analysis of HCMV gene expression. (Top panel) Scatterplot of the normalized square root (sqrt) of the median level of expression of mock-infected cells (triplicates, n = 6) on abscissa in contrast to normalized square root of the median level of expression of infected cells (triplicates, n = 6) under IE conditions. (Middle and lower panels) The same comparison shown in the top panel is shown for E expression (n = 3) (in the presence of ganciclovir) and L expression (n = 2) (in the absence of drug at 72 h postinfection). The points above the line of equivalence exhibit the ORFs expressed. Representative ORFs are indicated (see text for details). The point adjacent to IRL-4 represents expression from a triplicate set of TRL-4 (n = 3).
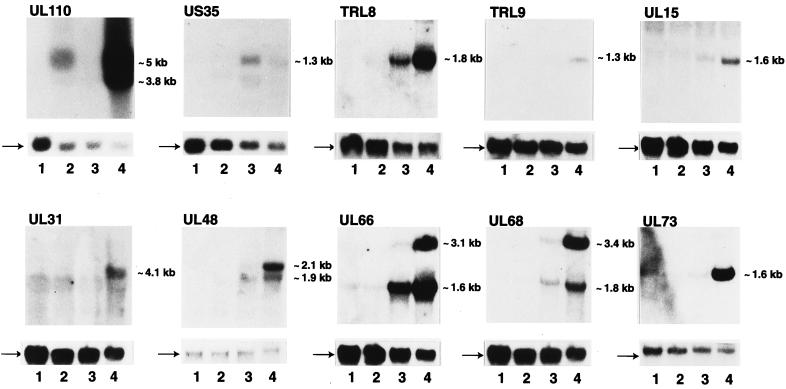
FIG. 4.
Northern blot analysis of selected HCMV ORF transcripts. Whole-cell RNA was harvested either from uninfected cells (lane 1) or HCMV-infected cells at 13 h postinfection in the presence of cycloheximide (lane 2) or 72 h postinfection in the absence (lane 4) or presence (lane 3) of ganciclovir. RNAs were separated on formaldehyde-agarose gels, transferred to a nylon membrane, and hybridized with radiolabelled probes specific for selected viral transcripts (details in Materials and Methods). The probes used are indicated at the top of the lanes. The position and approximate size of the major viral transcript(s) detected in each case are indicated on the right of each panel. The 1.4-kb GAPDH band detectable in all lanes is indicated by an arrow to the left of the lower panels.
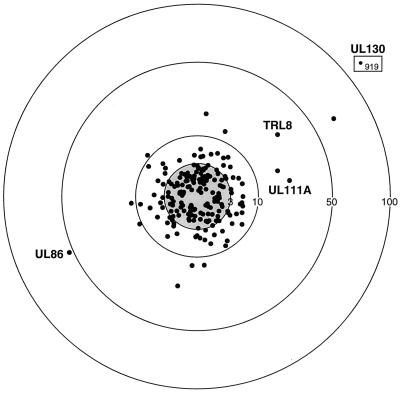
In the next set of experiments, we sought to determine the profiles of the E and L kinetic classes of gene expression. The influence of an inhibitor of viral DNA replication (e.g., ganciclovir) on HCMV gene expression differentiates E and L kinetic classes of viral transcripts. Strictly defined, an L gene is one that is expressed after the onset of DNA replication although, importantly, L genes can differ with respect to the stringency of the requirement for DNA replication. Hence, in the following experiments all L expression classes (E-L and L) designate genes whose expression was reduced or eliminated as shown by results for ganciclovir-treated versus untreated genes. These ORFs are further classified as E-L or L, depending whether expression was detectable in the presence of a replication inhibitor or not, respectively. For the purpose of these experiments, HFF cells were infected with HCMV in the presence of ganciclovir and harvested 72 h after infection for early RNA or 72 h after infection with no drug treatment for L-expression classes. Accordingly, RNAs from the various infected and mock-infected controls were fluorescently labelled and individually hybridized to the viral chip. The results of these experiments are shown in Fig. 2, with the points above the line of equivalence indicating E (middle panel) and L (bottom panel) expression. By the chip, a viral gene was considered to be in the L-expression class if the following two criteria were met: (i) the median level of intensity in the ganciclovir-treated samples was at least threefold lower than that in the untreated samples and (ii) a nonparametric test for a decrease in the level of intensity in the ganciclovir-treated samples versus that in the untreated samples was significant at a P value less than 0.05. Array elements expressed at 72 h postinfection in the presence or absence of ganciclovir were analyzed by using the same criteria described for the IE gene analysis, except that the median level of intensity was at least twofold greater than that in the mock-infected samples. A summary of the results for each ORF is shown in Table 1 and Fig.2. Figure 3 shows a graphic representation of the sensitivity of gene expression to ganciclovir treatment at L times. Note the marked variation in the extent of the inhibition of L gene expression (points distributed outside the shaded circle) to the viral replication inhibitor (Fig. 3). Under these conditions, only a few ORFs showed greater-than-10-fold sensitivity to ganciclovir (TRL-IRL8, TRL-IRL12, UL43, UL49, UL52, UL85, UL86, UL94, UL106, UL111A, UL130, UL152 [Towne], and US6). The aberrantly high ratio for UL130 is due to very low levels of E expression that are likely the result of a suboptimal deposition target. In Table 1, ORFs expressed at 72 h postinfection in the absence of ganciclovir include L designations (E-L and L), while ORFs unaffected by drug treatment are marked as an E class. These data reveal that greater than 75% of the genome is transcriptionally active at 72 h postinfection. Of the genes mapped, 36% were classified as E, 26% were classified as E-L, and 32% were classified as L. Thus, the majority of genes belong to L expression classes. No correlation between members of a gene family (RL11, US6, US12, and US22 families) and temporal class of gene expression was observed, indicating divergence of regulatory control pathways of these family members. In marked contrast to the genes in the UL region, the US genes are predominantly (70%) allocatable to the E kinetic class of expression. It should be noted that approximately 20% of the arrayed ORFs scored negative for expression. In several cases (UL11, UL16, UL25, UL70, UL99, UL117-9, and US2), the ORFs are known to be active genes (Table 1), suggesting that other previously uncharacterized ORFs which did not develop a positive signal in the microarray analysis may well be transcriptionally active. To determine whether this is the case, we performed Northern blot analysis for selected ORFs (US35, UL66, and UL127) that were not scored in the chip analysis or previously studied. In the presence of ganciclovir at 72 h postinfection, RNA levels for UL66 are detectable but not as high as those without drug treatment, indicating that UL66 ORF is transcribed with E-L expression kinetics (Fig. 4). In addition, the US35 ORF is weakly transcribed (Fig. 4). Levels of the US35 RNA are unaltered by ganciclovir treatment, implying that this transcript can be classified as E. We were unable to detect a specific transcript for UL127 by probing Northern blots, consistent with the chip analysis (data not shown). We also note that a number of these ORFs are very small and may not constitute bona fide genes (e.g., UL12, UL90, US4, US5, and US36). Overall, these results indicate that the present chip analysis detects approximately 75% of known or predicted ORFs in the HCMV genome.
FIG. 3.
Sensitivity of HCMV gene expression to ganciclovir treatment. The fold decrease in levels of gene expression in cells 72 h postinfection treated with ganciclovir or left untreated are represented by concentric circles. Each ORF was arbitrarily assigned a radial spacing of approximately 1°. The inner, shaded circle shows values that exhibit threefold-or-less change in normalized median levels of expression (n = 3). The outer boxed point is off scale, and the name of the ORF (UL130) and fold decrease (919-fold) are marked. Representative ORFs are indicated.
The expression profiles of many of the ORFs identified by chip analysis were not previously known, although the transcription of a limited number of E and E-L genes and that of even fewer L genes have been described previously (Table 1). Notably, the expression patterns we observed by chip analysis for previously characterized genes showed almost perfect concordance with previously published results. However, there are a few exceptions, namely, UL102, TRL6, UL33, UL83, UL86, US18, and US27. By the present chip analysis, UL102 is classified as an E-L ORF, while previous work (29) indicates that UL102 is an E gene that is not expressed at L times. However, in a study by Smith and Pari (29), an overlapping transcript is present exclusively at L times and is selectively inhibited by DNA replication inhibitors. Therefore, the chip array cannot distinguish overlapping transcripts, and consequently UL102 would be allocated to both expression classes. Similarly, US18 is a true L gene, but its transcript is expressed at relatively low levels compared with those of the overlapping E transcripts of US19 and US20 (9). Thus, it is not surprising that the chip would designate US18 as an E gene. In the case of the US27 and UL33 L genes, the chip analysis indicated a ratio of approximately 2.6- and 2.4-fold decreases, respectively, with P values of less than 0.05; thus, while these genes show a statistically significant sensitivity to ganciclovir, they are not assessed as such under the selection conditions. These exceptions underscore some limitations of the present chip approach, although the chip results show approximately 90% agreement overall with published studies. Nevertheless, to further corroborate the microarray results, RNA levels of selected ORFs (TRL8-IRL8, TRL9-IRL9, UL15, UL31, UL48, UL68, and UL73) that have not been previously studied were measured by Northern blot analysis. Based on the microarray hybridization, we expected all these selected ORFs to exhibit L expression characteristics. Figure 4 (lanes 2) indicates that none of the messages are transcribed at IE times in the presence of cycloheximide. In the presence of ganciclovir at 72 h postinfection, transcripts for TRL8-IRL8, TRL9-IRL9, UL15, UL31, UL48, UL66, UL68, and UL73 were not well developed (except for the TRL8-IRL8 transcript) compared with those at 72 h postinfection without drug treatment, indicating that these messages have an L classification (Fig. 4, lanes 3 and 4). Altogether, these results are in concordance with the microarray results and further confirm the reliability and accuracy of the viral chip approach.
Examining upstream noncoding DNA sequence of HCMV E, E-L, and L genes.
The above experiments designate the kinetic class of expression of more than 150 ORFs, most of which have not been previously characterized. To date, little is known about how the kinetics of HCMV gene expression are controlled, in part because thus far, relatively few genes have been studied. Relatedly, the regulation of DNA-virus gene expression involves the interaction of host-encoded and viral proteins with discrete elements (DNA or RNA) in the 5′-end region of the gene. Therefore, to gain further insight into the regulation of the kinetics of HCMV gene expression, we examined a set of upstream DNA sequences corresponding to 40 E, 27 E-L, and 36 L ORFs that contain an initiation codon. The rationale for examining this set of upstream regions was that perhaps, as in some viral systems (24), a single regulatory motif may act either as a negative or a positive regulator of gene expression, depending on its kinetic class. The upstream DNA sequence corresponding to each ORF was bounded at the 3′ end by the ORF’s translation start. The 5′ end of the upstream region was designated as being 500 bp from the translation start. This choice of the upstream-region boundaries was justified, as most genes have control elements that lie between 0 and 500 bp upstream of the translation start. In the present study, we used the MEME algorithm (one of several algorithms developed for discovering recurring motifs in unaligned sequences) to search for common motifs within a given kinetic class (see Materials and Methods for details). The results of this analysis, summarized in Table 2, indicate no readily apparent single, dominant element unique to a kinetic class. However, a subset of E, E-L, and L promoter regions have in common conserved sequence motifs of about 8 to 12 bp in length. For example, 25 and 11% of E and E-L promoter regions, respectively, contain the sequence pattern ACGACGTCGG, which harbors a core ATF recognition site (underlined sequence) (Table 2). By contrast, the palindromic sequence pattern CCGCGGGCGCGG is present in 17% of L promoters alone and does not match any known transcription factor binding site (Table 2). The upstream noncoding regions were further analyzed for the presence of binding sites common to known transcription factors by using the TFD. In general, L promoters contained fewer binding sites to known transcription factors than did promoters in the E and E-L expression classes (data not shown). This observation may correlate with a reduction in the complexity of the transcriptional control regions associated with the L expression class. Overall, we conclude that HCMV kinetic classes of promoters may be characterized not by discrete consensus sequence motifs but, instead, by common subsets of related sites, indicating more-elaborate regulatory pathways.
DISCUSSION
This study evaluates a novel approach for profiling the gene expression of large DNA viruses. By applying DNA chip technology, which has been successfully used with a number of microorganisms (references 27 and 28 and references therein) to monitor genome-wide transcription, we were able to specifically detect HCMV-expressed messages in the context of abundant cellular RNAs by using oligonucleotides corresponding to each ORF of the HCMV genome. The obvious advantage this system has over traditional methods is the speed with which global changes in CMV transcription can be simultaneously monitored. These fabricated DNA microarrays greatly extend and complement existing RNA transcript-mapping procedures, such as Northern analysis, slot blot, reverse transcription-PCR, primer extension, and nuclease protection-based methods.
In this study we were able to detect, in parallel, the expression of a total of 151 HCMV ORFs. A comparison of the results we obtained by viral microarray hybridization with previously reported results provided a good test for the sensitivity and accuracy of the chip approach. The expression patterns we observed by chip analysis for previously characterized genes showed almost-perfect concordance with results published earlier. In addition, we further characterized by Northern analysis TRS8-IRS8, TRS9-IRS9, UL15, UL31, UL48, UL68, and UL73 expression patterns. In each case tested, there was complete agreement with the chip analysis. Overall, the HCMV chip proved to be a reliable and robust assay, as the false-negative rate was low (∼10%), providing confidence in the reliability of the analysis determined in this study. The rate of false-negative results can be decreased in the future by selecting different oligonucleotides or by using PCR fragments for deposition. It is important that the microarray approach for defining the kinetic class of gene expression has limitations in distinguishing overlapping viral messages that may be under the control of multiple promoter elements, allowing the assignation of expression to more than one kinetic class (e.g., UL102 and US18). In this case, the false-positive rate (<15%) was primarily due to overlapping transcription units. The present analysis probably underestimates the full expression profile of HCMV, since we measured only predicted ORFs, and some of the viral ORF array DNAs may not be optimal (e.g., UL66, US2, and UL99). More-detailed RNA-mapping experiments will be required to fully characterize the program of HCMV transcription.
Identification and characterization of regulatory sequences are critical to elucidating global mechanisms of transcriptional regulation. The distribution of ORFs to a specific expression class was used to search for regulatory motifs possibly associated with a set of coregulated genes. Automatic alignments obtained by using MEME algorithms of the upstream noncoding DNA sequences of many of the coregulated ORFs did not readily identify a unique class-specific consensus sequence motif. However, a subset of E, E-L, and L genes was found to contain a conserved sequence motif, indicating redundancy in the use of specific elements or perhaps a more complex regulatory hierarchy. Further investigation will be required to assess the role of the regulatory sequences suggested by these experiments.
The position and pairwise polarity of genes may strongly influence their transcription, especially in viral genomes, since limited intergenic sequence necessitates the sharing of upstream regulatory elements. Little evidence was observed for a direct correlation between kinetic class and location or polarity of transcription in infection, as illustrated by the UL112-113 and UL122-123 E and IE regions, respectively. The divergent UL111A ORF is close to the UL112-113 E promoter, yet it displays L expression characteristics. The divergent ORF UL127 is immediately proximal to the major IE enhancer for UL122-123 but is not apparently expressed. Consistent with the chip data, we failed to detect UL127 transcripts by Northern analysis. However, 70% of the US genes exhibit the E kinetic class of expression. Thus, the HCMV genome may be able to accommodate more elaborate control of its transcription program than smaller viral genomes. Evolutionarily, large viral genomes such as HCMV may thus have been provided with a selective advantage.
Viral microarrays have many other potential uses. For instance, viral DNA microarrays may be used to characterize transcripts associated with latency and viral programs of transcription in different tissues and cell types. Viral chips will be particularly useful in analyzing the transcriptional consequences of mutations affecting the activity of host and viral regulatory molecules. This combination of genetics with DNA chip analysis will provide a powerful approach to the dissection and characterization of the infectious program and associated regulatory networks in a variety of biologically important cell types. This strategy also has important practical applications in antiviral drug screening. DNA microarrays can be used to define the signature pattern of known viral inhibitors (e.g., this study) and can also be used to screen for compounds that develop an alternatively desired signature (18). Moreover, mutations in specific genes encoding potential drug targets can serve as surrogates for chemical inhibitors of their activity. Viral chips can also be used to monitor drug resistance by expression profiling and typing genotypic strain variation (40) in clinical samples and thus serve as a valuable diagnostic tool.
In summary, we have developed a viral microarray-based approach to transcriptionally map the genome of HCMV. In the present study, we used viral microarray chips to profile the drug sensitivity of viral gene expression and delineate the kinetic classes of the majority of the predicted ORFs in the HCMV genome. In the future, viral and cellular DNA microarrays (41) will be a rich source of useful insights into viral biology and contribute to a deeper understanding of the gene pathways involved in viral growth and pathogenesis.
ACKNOWLEDGMENTS
J.C. and A.A. contributed equally to this work.
This work was supported by grants from the National Institutes of Health to P.G. (CA-66167 and AI-30627). P.G. is a Scholar of the Leukemia Society of America. A.A. is a Fellow of the Universitywide AIDS Research Program.
We thank Kathy Witmeyer, Jon Fleismann, and Dave Ladmer for cooperation in designing, synthesizing and purifying the gene targets; Mehrdad Khaleghi and John Wittig for help with data collection; Kenny Simmen for help with Northern blot analysis; Fatima Garcia del Rey for cooperation in preparing viral master plates for robotic printing; and Tom Hasse for the CAD layouts and machine work. Last, we apologize to the investigators whose work on the characterization of transcription units in the HCMV genome we unintentionally neglected to cite. We thank Kelly White for assistance in the preparation of the manuscript.
Footnotes
This is publication no. 12111-IMM from The Scripps Research Institute.
REFERENCES
- 1.Baldick C J, Jr, Shenk T. Proteins associated with purified human cytomegalovirus particles. J Virol. 1996;70:6097–6105. doi: 10.1128/jvi.70.9.6097-6105.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnard D L, Huffman J H, Sidwell R W, Reist E J. Selective inhibition of cytomegaloviruses by 9-(3′-ethylphosphono-1′-hydorxymethyl-1′-propyloxy-methyl) guanine. Antiviral Res. 1993;22:77–89. doi: 10.1016/0166-3542(93)90086-x. [DOI] [PubMed] [Google Scholar]
- 3.Cha T-A, Tom E, Kemble G W, Duke G M, Mocarski E S, Spaete R R. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J Virol. 1996;70:78–83. doi: 10.1128/jvi.70.1.78-83.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchison III C A, Kouzarides T, Martignetti J A, Preddie E, Satchwell S C, Tomlinson P, Weston K M, Barrell B G. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–170. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 5.DeMarchi J M, Schmidt C A, Kaplan A S. Patterns of transcription of human cytomegalovirus in permissively infected cells. J Virol. 1980;35:277–286. doi: 10.1128/jvi.35.2.277-286.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeRisi J, Penland L, Borwn P O, Bittner M L, Meltzer P S, Ray M, Chen Y, Su Y A, Trent J M. Use of a cDNA microarray to analyse gene expression. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 7.Fleckenstein B, Mullerand I, Collins J. Cloning of the complete human cytomegalovirus genome in cosmids. Gene. 1982;18:39–46. doi: 10.1016/0378-1119(82)90054-3. [DOI] [PubMed] [Google Scholar]
- 8.Gibson W, Clopper K S, Britt W J, Baxter M K. Human cytomegalovirus (HCMV) smallest capsid protein identified as product of short open reading frame located between HCMV UL48 and UL49. J Virol. 1996;70:5680–5683. doi: 10.1128/jvi.70.8.5680-5683.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo Y-W, Huang E-S. Characterization of a structurally tricistronic gene of human cytomegalovirus composed of US18, US19, and US20. J Virol. 1993;67:2043–2054. doi: 10.1128/jvi.67.4.2043-2054.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He Y S, Xu L, Huang E-S. Characterization of human cytomegalovirus UL84 early gene and identification of its putative protein product. J Virol. 1992;66:1098–1108. doi: 10.1128/jvi.66.2.1098-1108.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hitomi S, Kozuka-Hata H, Chen Z, Sugano S, Yamaguchi N, Watanabe S. Human cytomegalovirus open reading frame UL11 encodes a highly polymorphic protein expressed on the infected cell surface. Arch Virol. 1997;142:1407–1427. doi: 10.1007/s007050050169. [DOI] [PubMed] [Google Scholar]
- 12.Hollander M, Wolfe D A. Nonparametric statistical analysis. New York, N.Y: John Wiley; 1973. [Google Scholar]
- 13.Huber M T, Compton T. The human cytomegalovirus UL74 gene encodes the third component of the glycoprotein H-glycoprotein L-containing envelope complex. J Virol. 1998;72:8191–8197. doi: 10.1128/jvi.72.10.8191-8197.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hutchinson N I, Sondermeyer R T, Tocci M J. Organization and expression of the major genes from the long inverted repeat of the human cytomegalovirus genome. Virology. 1986;155:160–171. doi: 10.1016/0042-6822(86)90176-5. [DOI] [PubMed] [Google Scholar]
- 15.Jahn G, Knust E, Schmolla H, Sarre T, Nelson J A, McDougall J K, Fleckenstein B. Predominant immediate-early transcripts of human cytomegalovirus AD169. J Virol. 1984;49:363–370. doi: 10.1128/jvi.49.2.363-370.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones T R, Muzithras V P. Fine mapping of transcripts expressed from the US6 gene family of human cytomegalovirus strain AD169. J Virol. 1991;65:2024–2036. doi: 10.1128/jvi.65.4.2024-2036.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaye J, Browne H, Stoffel M, Minson T. The UL16 gene of human cytomegalovirus encodes a glycoprotein that is dispensable for growth in vitro. J Virol. 1992;66:6609–6615. doi: 10.1128/jvi.66.11.6609-6615.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marton M J, DeRisi J L, Bennett H A, Iyer V R, Meyer M R, Roberts C J, Stoughton R, Burchard J, Slade D, Dai H, Bassett D E, Jr, Hartwell L H, Brown P O, Friend S H. Drug target validation and identification of secondary drug target effects using DNA microarrays. Nat Med. 1998;4:1293–1301. doi: 10.1038/3282. [DOI] [PubMed] [Google Scholar]
- 19.McDonough S H, Spector D H. Transcription in human fibro-blasts permissively infected by human cytomegalovirus strain AD169. Virology. 1983;125:31–46. doi: 10.1016/0042-6822(83)90061-2. [DOI] [PubMed] [Google Scholar]
- 20.McDonough S H, Staprans S I, Spector D H. Analysis of the major transcripts encoded by the long repeat of human cytomegalovirus strain AD169. J Virol. 1985;53:711–718. doi: 10.1128/jvi.53.3.711-718.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.MEME algorithm. 1994, copyright date. [Online.] Regents of the University of California. www.sdsc.edu/meme/website/intro.html. [25 Feb. 1998, last date accessed.]
- 21.Michel D, Pavic I, Zimmermann A, Haupt E, Wunderlich D, Heuschmid M, Merthens T. The UL97 gene product of human cytomegalovirus is an early-late protein with a nuclear localization but is not a nucleoside kinase. J Virol. 1996;70:6340–6346. doi: 10.1128/jvi.70.9.6340-6346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mocarski E S. Cytomegalovirus and their replication. In: Fields B N, Knipe K M, Howley P M, editors. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. p. 2447. [Google Scholar]
- 23.Mocarski E S, Prichard M N, Tan C S, Brown J M. Reassessing the organization of the UL42-UL43 region of the human cytomegalovirus strain AD169 genome. Virology. 1997;239:169–175. doi: 10.1006/viro.1997.8875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moss B. Regulation of vaccinia virus transcription. Annu Rev Biochem. 1990;59:661–688. doi: 10.1146/annurev.bi.59.070190.003305. [DOI] [PubMed] [Google Scholar]
- 25.Ramsay G. DNA chips: state-of-the-art. Nat Biotechnol. 1997;16:40–44. doi: 10.1038/nbt0198-40. [DOI] [PubMed] [Google Scholar]
- 26.Razzaque A, Jahn N, McWeeney D. Localization and DNA sequence analysis of the transforming domain (mtrII) of human cytomegalovirus. Proc Natl Acad Sci USA. 1988;85:5705–5793. doi: 10.1073/pnas.85.15.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schena M. Genome analysis with gene expression microarrays. Bioessays. 1996;18:427–431. doi: 10.1002/bies.950180513. [DOI] [PubMed] [Google Scholar]
- 28.Schena M, Shalon D, Davis R W, Brown P O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 29.Smith J A, Pari G S. Human cytomegalovirus UL102 gene. J Virol. 1995;69:1734–1740. doi: 10.1128/jvi.69.3.1734-1740.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith J A, Jairath S, Crute J J, Pari G S. Characterization of the human cytomegalovirus UL105 gene and identification of the putative helicase protein. Virology. 1996;220:251–255. doi: 10.1006/viro.1996.0310. [DOI] [PubMed] [Google Scholar]
- 31.Stinski M F, Thomsen D R, Stenberg R M, Goldstein L C. Organization and expression of the immediate early genes of human cytomegalovirus. J Virol. 1983;46:1–14. doi: 10.1128/jvi.46.1.1-14.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Zeijl M, Fairhurst J, Baum E Z, Sun L, Jones T R. The human cytomegalovirus UL97 protein is phosphorylated and is a component of virions. Virology. 1997;231:72–80. doi: 10.1006/viro.1997.8523. [DOI] [PubMed] [Google Scholar]
- 33.Vieira J, Schall T J, Corey L, Geballe A P. Functional analysis of the human cytomegalovirus US28 gene by insertion mutagenesis with the green fluorescent protein gene. J Virol. 1998;72:8158–8165. doi: 10.1128/jvi.72.10.8158-8165.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wathen M W, Stinski M F. Temporal patterns of human cytomegalovirus transcription: mapping the viral RNAs synthesized at immediate early, early, and late times after infection. J Virol. 1982;41:462–477. doi: 10.1128/jvi.41.2.462-477.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Welch A R, McNally L M, Gibson W. Cytomegalovirus assembly protein nested gene family: four 3′-coterminal transcripts encode four in-frame, overlapping proteins. J Virol. 1991;65:4091–4100. doi: 10.1128/jvi.65.8.4091-4100.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilkinson G W G, Akrigg A, Greenaway P J. Transcription of the immediate early genes of human cytomegalovirus strain AD169. Virus Res. 1984;1:101–116. doi: 10.1016/0168-1702(84)90067-4. [DOI] [PubMed] [Google Scholar]
- 37.Wing B A, Huang E-S. Analysis and mapping of a family of 3′-coterminal transcripts containing coding sequences for human cytomegalovirus open reading frames UL93 through UL99. J Virol. 1995;69:1521–1531. doi: 10.1128/jvi.69.3.1521-1531.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wing B A, Lee G C Y, Huang E-S. The human cytomegalovirus UL94 open reading frame encodes a conserved herpesvirus capsid/tegument-associated virion protein that is expressed with true late kinetics. J Virol. 1996;70:3339–3345. doi: 10.1128/jvi.70.6.3339-3345.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winkler M, Rice S A, Stamminger T. UL69 of human cytomegalovirus, an open reading frame with homology to ICP27 of herpes simplex virus, encodes a transactivator of gene expression. J Virol. 1994;68:3943–3954. doi: 10.1128/jvi.68.6.3943-3954.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winzler E A, Richards D R, Conway A R, Goldstein A L, Kalman S, McCullough M J, McCusker J H, Stevens D A, Wodicka L, Lockhart D J, Davis R W. Direct allelic variation scanning of the yeast genome. Science. 1998;281:1194–1197. doi: 10.1126/science.281.5380.1194. [DOI] [PubMed] [Google Scholar]
- 41.Zhu H, Cong J P, Mamtora G, Gingeras T, Shenk T. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;24:14470–14475. doi: 10.1073/pnas.95.24.14470. [DOI] [PMC free article] [PubMed] [Google Scholar]